SUMMARY
As a result of developmental synapse formation, the presynaptic neurotransmitter release machinery becomes accurately matched with postsynaptic neurotransmitter receptors. Trans-synaptic signaling is executed through cell adhesion proteins such as Neurexin::Neuroligin pairs but also through diffusible and cytoplasmic signals. How exactly pre/post coordination is ensured in vivo remains largely enigmatic. We here identified a “molecular choreography” coordinating pre- with postsynaptic assembly during the developmental formation of Drosophila neuromuscular synapses. Two presynaptic Neurexin-binding scaffold proteins, Syd-1 and Spinophilin (Spn), spatio-temporally coordinated pre/post assembly in conjunction with two postsynaptically operating, antagonistic Neuroligin species: Nlg1 and Nlg2. The Spn/Nlg2 module promoted active zone (AZ) maturation by driving the accumulation of AZ scaffold proteins critical for synaptic vesicle release. Simultaneously, these regulators restricted postsynaptic glutamate receptor incorporation. Both functions of the Spn/Nlg2 module were directly antagonized by Syd-1/Nlg1. Nlg1 and Nlg2 also had divergent effects on Nrx-1 in vivo motility. Concerning diffusible signals, Spn and Syd-1 antagonistically controlled the levels of Munc13-family protein Unc13B at nascent AZs, whose release function facilitated glutamate receptor incorporation at assembling postsynaptic specializations. In result, we here provide direct in vivo evidence illustrating how a highly regulative and interleaved communication between cell adhesion protein signaling complexes and diffusible signals allows for a precise coordination of pre- with postsynaptic assembly. It will be interesting to analyze whether this logic also transfers to plasticity processes.
eTOC Blurb
Ramesh et al. identify transsynaptic and cytoplasmic proteins which coordinate pre- with postsynaptic assembly. Syd-1 & Spn cooperate with Nlg1 & Nlg2, and likely execute their functions by changing the biochemical milieu of Nrx-1. Their antagonism also controls Unc13B, whose glutamate release function facilitates glutamate receptor incorporation.
INTRODUCTION
Synaptic vesicle (SV) release at chemical synapses depends on the formation of active zone (AZ) scaffolds composed of a canonical apparatus of proteins including (M)Unc13, RIM-binding protein (RIM-BP), Liprin-α and CAST/ELKS (called Bruchpilot (BRP) in Drosophila) [1]. The size of individual AZ scaffolds scales with SV release probability [2, 3]. Once matured, each AZ apparently forms an integer number of release sites apposed by postsynaptic glutamate receptors (GluRs), likely spatially coordinated through a trans-synaptic micropattern (“nanocolumns” [4]). Importantly, in the course of maturation, AZ size becomes closely matched to the size of the postsynaptic density (PSD) scaffold clustering neurotransmitter (NT) receptors [5].
How the in vivo synapse assembly process and associated regulatory steps achieve this precise pre-post matching during developmental assembly is not fully understood. Notably, trans-synaptic cell-adhesion molecules (CAMs) have the capacity to bidirectionally tune synapse assembly [1], with Neurexin (Nrx) and Neuroligin (Nlg) interactions representing a regulatory principle conserved across vertebrate and invertebrate synapses [6, 7]. Although many synaptic CAMs and cytoplasmic proteins have been studied in isolation, how different CAMs selectively engage with each other and their cytoplasmic partners to ensure pre-post matching during synapse assembly has remained enigmatic, partly due to the high genetic redundancy among mammalian CAMs. Besides CAM signaling, diffusible signals including NT release at nascent AZs might play a regulatory role in postsynaptic assembly [8].
Here we characterize mechanisms that ensure pre-post matching during the assembly of individual glutamatergic synapses at the Drosophila neuromuscular junction (NMJ). In developing larvae, synapse maturation ultimately establishes a precisely defined pre-post stoichiometry over the course of several hours. To interrogate these mechanisms, we utilize the unique advantages of the larval NMJ system which allows for a synergy of reduced genetic redundancy, super-resolution and dynamic intravital microscopy and electrophysiology. We target Nlg1 and Nlg2, two Nlg species previously shown to functionally interact with the only Nrx family protein in Drosophila, Nrx-1 [7, 9–12]. Our results reveal that these two Nlgs serve antagonistic roles and operate in conjunction with two antagonistic presynaptic proteins that bind Nrx-1: Syd-1 cooperating with Nlg1, and Spinophilin (Spn) with Nlg2. While the Spn/Nlg2 functional module promoted AZ maturation (BRP/RIM-BP/Unc13A incorporation) but restrained GluRIIA-containing receptor incorporation, Syd-1/Nlg1 initiated AZ assembly and promoted GluRIIA receptor incorporation through Unc13B recruitment and its glutamate release function. Genetic interaction experiments identified a remarkable degree of crosstalk between these modules, exemplifying a regulatory principle obviously evolved to ensure precise pre-post matching, and integrating Unc13B-dependent glutamate release acting as a diffusible signal. Together, our data indicate that synaptic matching is not established via a trans-synaptic “stoichiometric building principle” to continuously accumulate synaptic components, but via a regulatory crosstalk between antagonistic assembly modules.
RESULTS:
Drosophila glutamatergic terminals grow throughout the larval stages while continuously adding new synaptic sites involving a presynaptic AZ and postsynaptic GluRs. Individual AZs are precisely apposed by a corresponding GluR cluster [13, 14]. We here analyze how this pre- and postsynaptic matching is achieved in vivo.
Reciprocal antagonism between Spinophilin and Syd-1 in presynaptic assembly
Syd-1, a regulator of presynaptic signaling, was shown to arrive early at nascent AZ sites and seed new AZ scaffolds at NMJ terminals [15]. BRP, member of the generic ELKS/Cast and a major component of mature AZ scaffolds, in contrast only incorporates into AZs hours after Syd-1 [16, 17]. Confocal analysis of BRP showed that Syd-1 mutants formed fewer AZs, with each overgrown AZ having elevated BRP content as expected ([15], Figure S1A–C). Stimulated emission depletion (STED) microscopy was then used to measure AZ sizes in Syd-1 mutants where individual BRP scaffolds were enlarged and extended ([15], Figure S1E–F). Since in Syd-1, AZs are more sparse than normal, we previously interpreted this phenotype as axonally transported BRP building a “cargo pressure” and unloading excessive BRP per AZ.
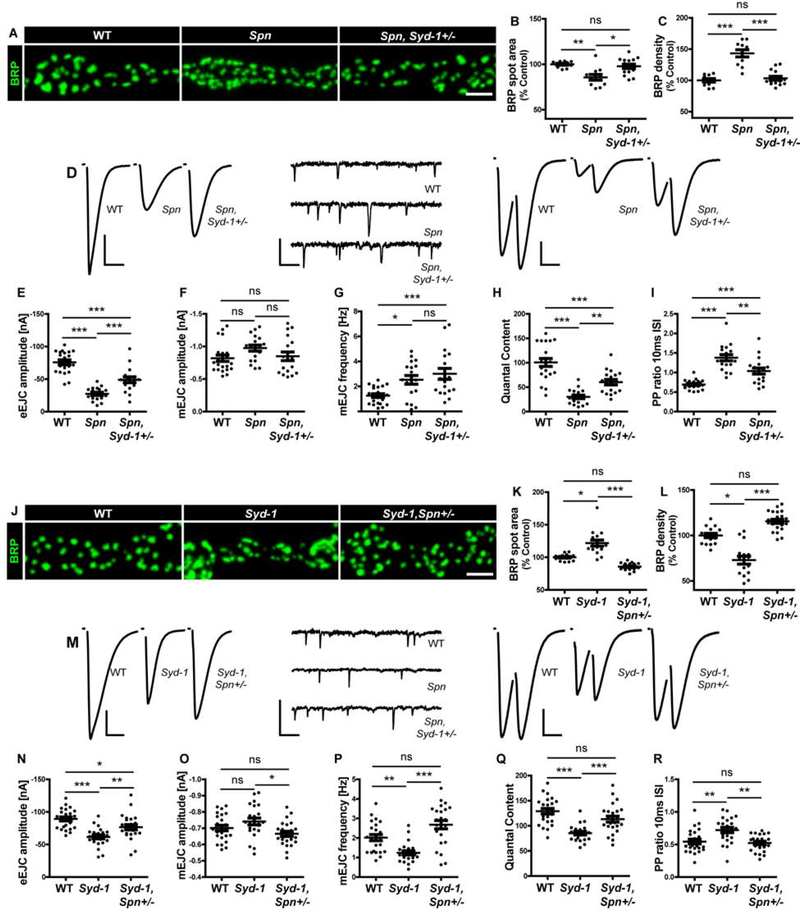
We previously identified another presynaptic signaling protein Spn, whose absence provokes an inverse phenotype: supernumerary AZs but reduced BRP amounts per AZ ([18], Figure S1S–U). STED analysis showed smaller individual AZ sizes (Figure S1W–Y). Previously, the Spn phenotype of smaller, supernumerary AZs (BRP) could be suppressed through genetic interaction experiments where Syd-1 function was attenuated through a single loss-of-function (LoF) allele (Syd-1Ex3.4) in Spn mutants ([18], Figure 1A–C). Visualizing another AZ marker, RIM-BP [19] confirmed this suppression (Figure S2C–E). Two electrode voltage-clamp (TEVC) analysis (Figure 1D) showed that the Spn phenotypes of reduced evoked excitatory junctional current (eEJC) amplitudes (Figure 1E), reduced quantal contents (Figure 1H) and increased paired-pulse facilitation (Figure 1I, Figure S2A) were indeed rescued upon removal of one copy of Syd-1. Increased spontaneous release frequency remained unaltered (Figure 1G).
Figure 1. Reciprocal antagonism between Spinophilin and Syd-1 in presynaptic assembly.
(A,J) Representative images of third-instar larval muscle 4 NMJs immunostained with an antibody against BRP. Scale bars: 2μm. (D,M) Representative traces of eEJC, mEJC and paired-pulse (inter stimulus interval (ISI) of 10ms) measurements at third-instar larval muscle 6/7 NMJs. Scale bars: eEJC, paired-pulse traces 10ms, 20mV; mEJC traces 250ms, 2mV. (A,D) Spn mutant phenotype is suppressed by Syd-1 heterozygosity as shown through quantification of (B) BRP spot area (C) BRP density (E) eEJC amplitudes (H) Quantal content and (I) Paired pulse ratio (10ms ISI). (F) mEJC amplitudes are unaltered and (G) mEJC frequency phenotype is not suppressed. (J,M) Syd-1 mutant phenotype is suppressed by Spn heterozygosity as shown through quantification of (K) BRP spot area (L) BRP density (N) eEJC amplitudes (O) mEJC amplitudes (P) mEJC frequency (Q) Quantal content (R) Paired pulse ratio (10ms ISI). See also figures S1 and S2, and table S1.
We here intended to develop our understanding of the interaction between Syd-1 and Spn by executing reciprocal suppression experiments by attenuating Spn function through a single LoF allele (SpnEx3.1) in Syd-1 mutants. Surprisingly, the “seeder” Syd-1 phenotype of enlarged, sparse AZs (BRP) could also be efficiently suppressed by Spn heterozygosity (Figure 1J–L). The same pattern was observed staining for AZ protein RIM-BP (Figure S2F–H). Thus, increased Spn signaling at the remaining Syd-1 AZs, induced by Syd-1 absence, apparently promotes excessive BRP incorporation. TEVC analysis (Figure 1M) showed that the physiological Syd-1 phenotypes characterized by reduced eEJC amplitudes (Figure 1N), decreased mini frequencies (Figure 1P), reduced quantal contents (Figure 1Q) and increased paired-pulse facilitation (Figure 1R) were rescued upon one Spn copy removal. Thus, while both single mutants (Spn and Syd-1) individually reduced quantal contents, they increased quantal contents in conjunction with LoF heterozygosity of the respective other protein (Figure 1H,Q). Thus, both Spn and Syd-1 operate as truly antagonistic presynaptic signaling platforms, which tune a setpoint ideal for structural and functional assembly. Both factors seemingly continuously antagonize each other in the presynaptic assembly sequence of AZs, rather than Syd-1 executing the early assembly independent of Spn signaling.
Loss of postsynaptic Nlg2 mimics the loss of presynaptic Spinophilin
Trans-synaptic Nrx/Nlg signaling is thought to be a major regulatory process for tuning synapse assembly and maturation [7, 10, 12, 20–23]. We previously showed that presynaptic Syd-1 and Spn, through their PDZ domains, bind to the same Nrx-1 binding motif. Their PDZ domain integrity was found to be essential for effective signaling [18, 24].
Notably, besides Nlg1, Nlg family-member Nlg2, has also been shown to operate at NMJ synapses, with both Nlgs executing their functions through Nrx-1 binding [9, 11]. Nlg2 mutants were shown to form supernumerary AZ scaffolds (“T-bars”) in electron microscopy analysis and have elevated number of AZs in confocal microscopy analysis [9, 11], reminiscent of Spn mutant NMJs. We could directly reproduce this finding (Figure 2A–C). STED microscopy indeed showed that AZ organization of Nlg2 mutants appeared similar to the smaller AZs of Spn mutants (Figure 2D–E).
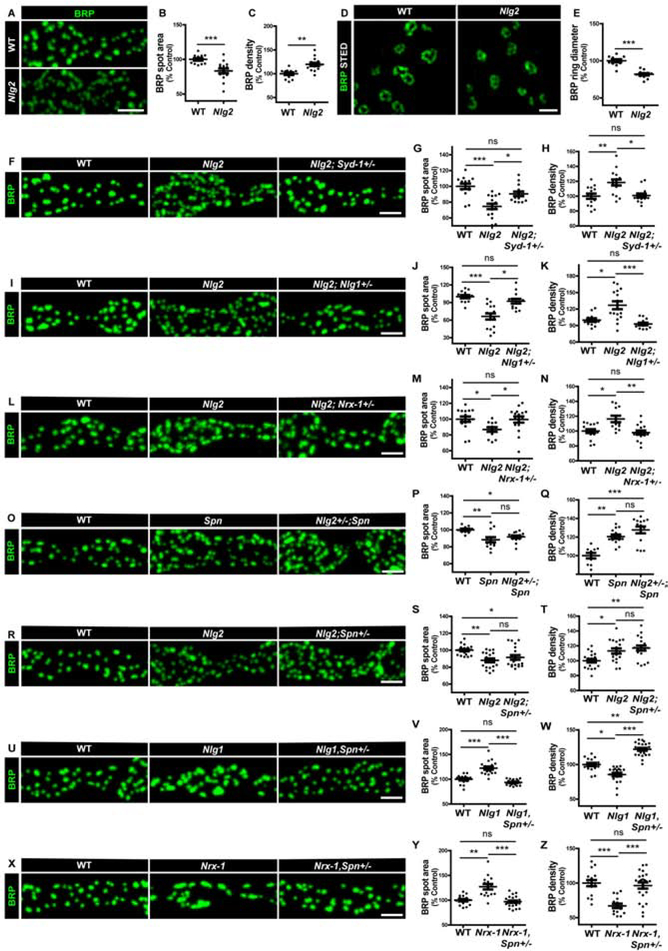
Figure 2. Syd-1/Nlg1 and Spn/Nlg2 take on antagonistic trans-synaptic signalling roles in presynapse development.
(A-X) Representative images of third-instar larval muscle 4 NMJs immunostained with an antibody against BRP. Scale bars: 2μm (A) Nlg2 mutants show (B) smaller, (C) supernumerary AZs compared to controls. (D) BRP rings resolved through STED microscopy show (E) smaller ring diameters in Nlg2 mutants. Scale bar: 500nm. Quantification of BRP spot areas and BRP ring diameters show that Nlg2 mutant phenotype of smaller, supernumerary AZs is suppressed by (F-H) Syd-1, (I-K) Nlg1 and (L-N) Nrx-1 heterozygosity. Spn and Nlg2 mutant phenotypes of smaller, supernumerary AZs are not suppressed by (O-Q) Nlg2 and (R-T) Spn heterozygosity, respectively. (U-W) Nlg1 and (X-Z) Nrx-1 mutant phenotypes of larger and fewer AZs are suppressed by Spn heterozygosity. See also figures S1, S2 and S3, and table S1.
We then tested whether Nlg2 functions in motoneurons or muscles. Both the presynaptic and postsynaptic phenotypes of Nlg2 were rescued by transgenic reexpression of Nlg2 in the postsynaptic muscle cell (Figure S3F–J). In our hands, reexpression of Nlg2 in motoneurons did not significantly influence the Nlg2 phenotype (Figure S3A–E), indicating that Nlg2 functions postsynaptically for the phenotypes addressed here.
Given that Nlg2 and Spn display similar phenotypes, we asked whether Nlg2 and Spn functioned cooperatively during synapse assembly. Thus, we analyzed whether Nlg2 would behave like Spn in genetic interaction experiments. Notably, the Nlg2 phenotype could also be effectively suppressed by single copy loss of Syd-1 (Figure 2F–H), suggesting that Spn and Nlg2 function together to antagonize Syd-1 function.
Antagonistic trans-synaptic signaling roles of Nlg1 and Nlg2
Like Syd-1 mutants, absence of postsynaptic Nlg1 provokes a phenotype of fewer but enlarged AZs ([10], Figure S1G–L). As Nlg1 and Nlg2 appear to have antagonistic phenotypes that mimic Syd-1 and Spn, we went on to explore the functional relationship between these Nlgs in tuning synapse assembly.
The Spn phenotype could be suppressed by reducing Nlg1 levels using the LoF allele Nlg1ex2.3 ([18]; Figure S2I–K). Thus, we tested the functional relationships between Nlg1/Syd-1 and Nlg2/Spn. Indeed, Nlg2 mutants were also rescued by Nlg1 heterozygosity (Figure 2I–K). Thus, the two Nlgs execute at least partially antagonistic functions during synapse assembly.
Absence of Nrx-1 mimics Syd-1 and Nlg1 mutant phenotypes ([23, 24], Figure S1M–R). Previously, Nrx-1 heterozygosity (using LoF allele Nrx-1241) was also found to suppress Spn ([18], Figure S2L–N). Here we found that Nrx-1 heterozygosity also suppressed the Nlg2 phenotype (Figure 2L–N), suggesting that Spn/Nlg2 may antagonize Syd-1/Nlg1.
To further scrutinize whether Spn/Nlg2 functioned in a common signaling module, we tested genetic interactions between them. Consistent with a cooperative function, we found that neither heterozygosity of Nlg2 nor Spn suppressed Spn or Nlg2 mutants, respectively (Figure 2O–T). Furthermore, double mutants of Spn and Nlg2 showed a non-additive phenotype (Figure S3K–M). Notably, Spn levels were reduced at Nlg2 mutant terminals (Figure S3P–Q), suggesting a molecular link between them. Thus, the presynaptic Nlg2 phenotype could, at least in part, be explained as a consequence of reduced Spn levels, and a trans-synaptic Spn/Nlg2 cooperation seemingly orchestrates pre-post matching.
As Syd-1/Nrx-1/Nlg1 seem to function antagonistically to Spn, and Spn antagonizes Syd-1 function (Figure 1) in developmental AZ assembly, we considered whether Spn heterozygosity might also rescue Nlg1 and Nrx-1 mutant phenotypes. Indeed, both Nlg1 and Nrx-1 mutant phenotypes were suppressed by Spn heterozygosity (Figure 2U–Z). Spn therefore seems to function via a dynamic antagonism to the Syd-1/Nrx-1/Nlg1 seeder module.
Interestingly, Nlg2 heterozygosity could not suppress the Nrx-1 and Nlg1 mutant phenotypes (Figure S2O–T). This might imply that Nlg2 normally functions to extract Nrx-1 from the Syd-1/Nrx-1/Nlg1 seeder complex and/or inhibits the seeder complex function by binding Nlg1, potentially through cis-heteromerization ([25], also see discussion).
Spinophilin/Nlg2 restrict the Syd-1-promoted incorporation of GluRIIA
As a result of their temporally extended assembly process [16, 24, 26], NMJ synapses ultimately adopt a robust trans-synaptic architecture with precise stoichiometry, as is evident in super-resolution images picturing BRP “in scale” with GluRs (Figure S4F). We went on asking how presynaptic Syd-1 and Spn would engage with trans-synaptic signals to tune GluR incorporation.
At mature NMJ synapses, two major GluR complexes are present, containing either GluRIIA or GluRIIB subunits [27]. Previously, intravital imaging demonstrated that both Nlg1 and Syd-1 mutants suffered from delayed incorporation of GluRIIA-containing complexes during early AZ assembly [10, 24], with GluRIIA incorporation unconventionally following GluRIIB incorporation. In contrast, Spn and Nlg2 mutants were shown to have increased GluR levels in immunostainings [9, 11]. When directly compared, Spn and Nlg2 mutants both showed elevated levels of GluRIIA (Figure S4A–E). The relative NMJ area covered by GluRIIA was also elevated in Spn mutants (Figure S4C). We analyzed the ratios of GluRIIA spot area versus BRP spot area across individual AZs for Nlg2 and Spn and found GluRIIA levels to be clearly skewed over BRP levels (Figure S4G–H). To determine whether these excessive amounts of GluRIIA in Spn mutants was in fact part of the PSD, we performed STED analysis with another PSD marker, p21-activated kinase (PAK1). PAK1, which promotes GluRIIA accumulation [28], but does not form a molecular complex with it, strictly colocalized with GluRIIA (Figure S4O). PAK patches appeared enlarged in Spn mutants (Figure S4M), indicating that the PSD was indeed physically enlarged here. GluRIID, which forms a complex with GluRIIA (but also GluRIIB), largely colocalizes with GluRIIA (Figure S4N). Thus, the excessive GluRIIA in Spn mutants is seemingly associated with a physically extended PSD organization (Figure S4).
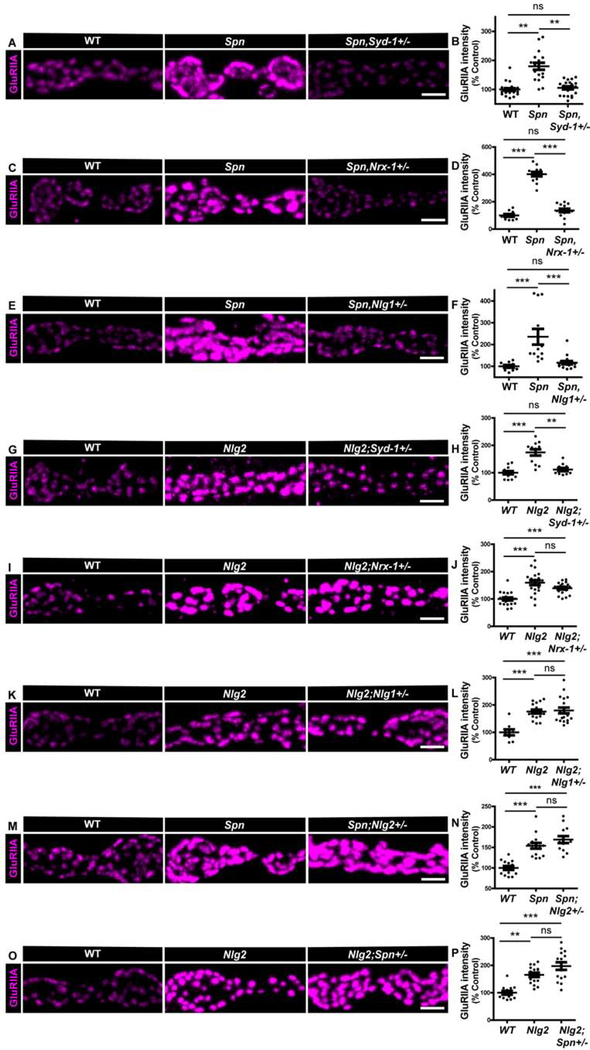
GluRIIA incorporates during the seeding phase of the developmental synapse assembly, peaking hours before BRP incorporation as measured by intravital imaging [16, 29]. We again used genetic interaction analysis to test whether Spn and Nlg2 would indeed antagonize Syd-1-dependent GluRIIA incorporation. For both Spn (Figure 3A–B) and Nlg2 (Figure 3G–H), heterozygosity of Syd-1 fully suppressed their excessive GluRIIA accumulation. Staining for another PSD marker, PAK, showed no increase of average PAK intensities at Spn mutant terminals, although the relative area covered by PAK staining was increased (Figure S4K–M). PAK levels were also unchanged in Spn mutants with one copy of Syd-1 removed, but the increased PAK area in Spn mutants was suppressed (Figure S4K–M). Thus, the signaling roles of Spn and Nlg2, crucial to specifically restrict GluRIIA incorporation, seemingly work in an antagonistic relationship to Syd-1-driven GluRIIA incorporation. Syd-1, Nrx-1 and Nlg1 mutants all share a specific deficit in GluRIIA incorporation [24]. Consistently, half dose of both Nrx-1 and Nlg1 (Figure 3C–F) also efficiently rescued the Spn GluRIIA phenotype. In contrast, the Nlg2 homozygous phenotype was not rescued by Nrx-1 or Nlg1 heterozygosity (Figure 3I–L).
Figure 3. Syd-1/Nlg1 and Spn/Nlg2 take on antagonistic trans-synaptic signalling roles in postsynapse development.
(A-O) Representative images of third-instar larval muscle 4 NMJs immunostained with an antibody against GluRIIA. Scale bars: 2μm. Quantification of GluRIIA intensity shows Spn mutant phenotype of overgrown GluRIIA fields is suppressed by (A,B) Syd-1, (C-D) Nrx-1 and (E-F) Nlg1 heterozygosity. Nlg2 mutant phenotype of overgrown GluRIIA fields is suppressed by (G-F) Syd-1 heterozygosity, but not (I-J) Nrx-1 or (K-L) Nlg1 heterozygosity. Spn and Nlg2 mutant phenotypes are not suppressed by (M-N) Nlg2 and (O-P) Spn heterozygosity, respectively. See also figure S4 and table S1.
We once again analyzed genetic interaction between Spn and Nlg2, finding that half dose of Nlg2 or Spn did not suppress the GluRIIA phenotype of Spn and Nlg2 mutants, respectively (Figure 3M–P). Once again, double mutants of Spn and Nlg2 showed a non-additive phenotype (Figure S3N,O). Thus, the two regulators seemingly function cooperatively in tuning GluRIIA incorporation, and likely both counteract the principal Syd-1/Nrx-1/Nlg1 module activity which attracts and immobilizes GluRs.
In short, postsynaptic GluRIIA accumulation, centered around the seeding phase of NMJ synapses [29, 30], is controlled by the antagonism between Syd-1/Nlg1-mediated accumulation and Spn/Nlg2-mediated suppression.
Antagonistic roles of Nlg1 and Nlg2 for Nrx-1 in vivo motility
As mentioned above, Nlg1 and Nlg2 function have been connected to the only Nrx family member of Drosophila, Nrx-1. Notably, we previously found through in vivo fluorescence recovery after photobleaching (FRAP) analyses that Nrx-1GFP shows elevated motility in Syd-1 [24] but decreased motility in Spn [18] mutants. Thus, the biochemical milieu of Nrx-1 changes in the absence of either Syd-1 or Spn. If the distinct assembly roles of the postsynaptic binding partners of Nrx-1, Nlg1 or Nlg2, were indeed mediated via altering the Nrx-1 status, differences of Nrx-1 motility might also be expected in Nlg1 and Nlg2 mutants. Intravital FRAP analysis showed that Nlg1 mutants (Figure 4B,C) showed significantly decreased but Nlg2 mutants (Figure 4D,E) significantly increased Nrx-1GFP motility. Thus, these data suggest that Nlg1 and Nlg2 differentially change the biochemical milieu of Nrx-1, and that Nlg2 might extract Nrx-1from its AZ seeding function to allow a switch to the AZ maturation phase.
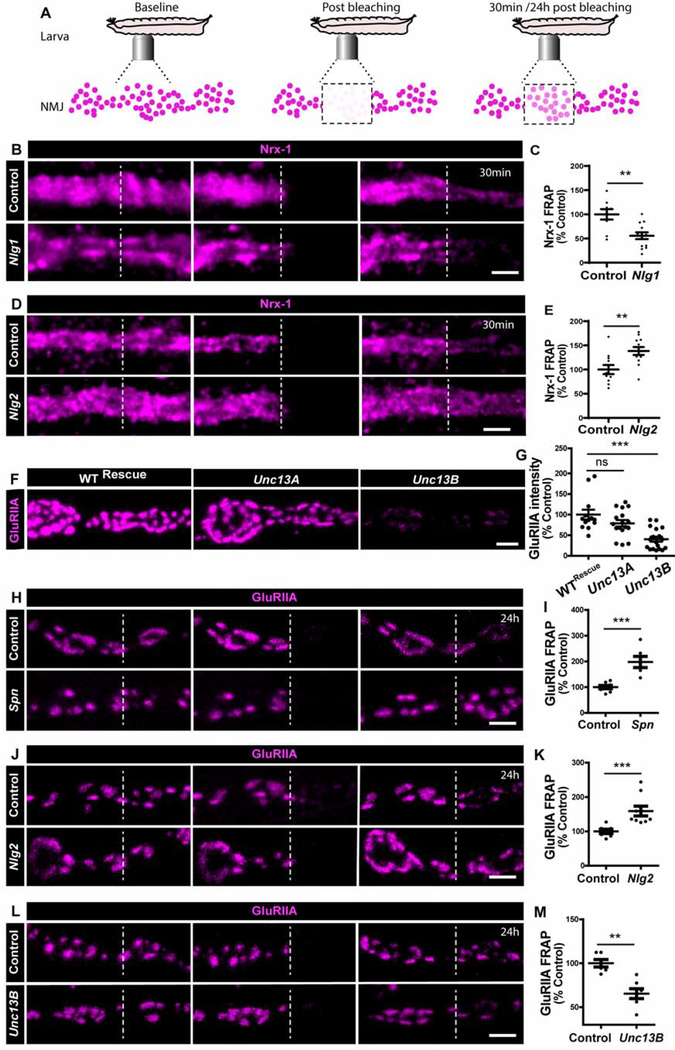
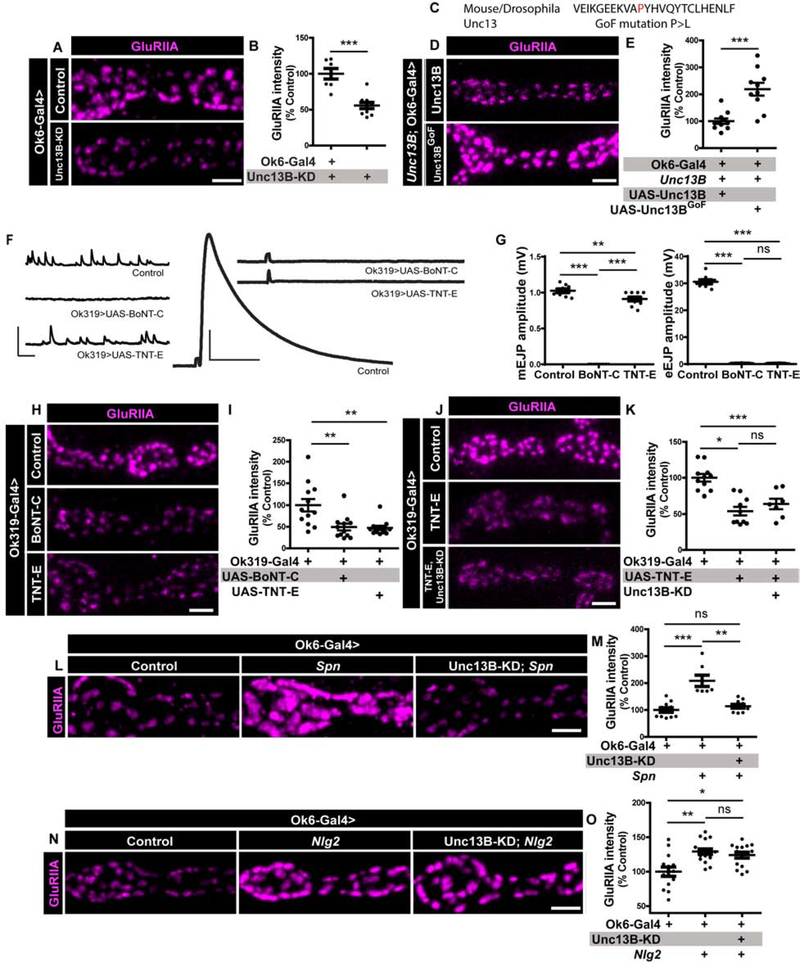
Figure 4. Unc13B facilitates GluRIIA accumulation.
(A) Scheme depicting in vivo fluorescence recovery after photobleaching (FRAP) imaging procedure at developing Drosophila larval NMJs at baseline (before photobleaching), immediately post bleaching and then again after 30min/24h to track recovery of fluorescently labelled protein. Representative images of muscle 26/27 NMJs labelled with (B,D) Nrx-1GFP or (H,J,L) GluRIIAGFP. Quantification of FRAP shows (C) lower Nrx-1GFP FRAP in Nlg1 mutants and (E) higher Nrx-1GFP FRAP in Nlg2 mutants. (F) Representative images of third-instar larval muscle 4 NMJs immunostained with an antibody against GluRIIA. Quantification of GluRIIA intensity shows (G) lower GluRIIA accumulation Unc13B mutants. (H-I) Spn and (J-K) Nlg2 mutants show higher GluRIIA FRAP while (L-M) Unc13B mutants show lower FRAP. Scale bars: 2μm. See also figure S5 and table S1.
Unc13B promotes developmental GluRIIA incorporation
As said above, intravital imaging showed that GluRIIA incorporation is delayed along the AZ assembly timeline in Syd-1 and Nlg1 [24]. At matured PSDs, however, Syd-1, Nrx-1 and Nlg1 finally establish roughly normal GluRIIA levels (Figure S4I,J). Notably, Unc13B, one of two main Unc13 isoforms expressed at NMJ AZs, still accumulates in Syd-1 mutants, though at lower levels [26]. Unc13B is largely dispensable for SV release at matured AZs [26]. We therefore speculated that Unc13B might execute an assembly function at NMJ synapses in conjunction with Syd-1/Nlg1 during the seeding phase. Indeed, genetic elimination of Unc13B resulted in clearly reduced GluRIIA levels (Figure 4F–G). In contrast, Unc13A mutants, while suffering a drastic reduction of evoked release at mature NMJs [26], did not show significant changes in GluRIIA incorporation (Figure 4F–G).
Previous work had shown that GluRIIA incorporates at assembling sites from diffuse receptor pools likely positioned at the muscle plasma membrane [30]. We thus applied intravital FRAP to dynamically measure GluRIIA incorporation in Spn, Nlg2 and Unc13B mutants. Absence of both Spn (Figure 4H–I) and Nlg2 (Figure 4J–K) increased intravital FRAP of GluRIIA in a 24hr period compared to controls. Notably, we measured a reduced GluRIIA incorporation in Unc13B mutants (Figure 4L–M). Thus, presence of Unc13B is a precondition to effectively incorporate GluRIIA but was not critical for assembly of the presynaptic AZ scaffold [26].
The role of Unc13B might per se be explained via an assembly or scaffolding function during AZ development or involve its glutamate release function. We went on to ask whether the release function of Unc13B would be rate-limiting in controlling developmental GluRIIA accumulation.
Boosting Unc13B-mediated evoked release facilitates GluRIIA incorporation
At NMJ synapses, mutations changing the gating behavior of GluRIIA receptors alter their distribution and trafficking [31], indicating that presynaptic glutamate release might tune GluRIIA incorporation during the early-to-mid assembly phase of individual PSDs. As in the case of Unc13B mutants, knock-down (KD) of Unc13B in motoneurons using RNA-interference (RNAi) resulted in reduced accumulation of GluRIIA (Figure 5A–B). Directly comparing Unc13A with Unc13B KD, we found that only Unc13B KD, and not Unc13A KD, significantly suppressed GluRIIA levels (Figure S5A). These data suggest that Unc13B, which localizes to nascent presynaptic sites at the NMJ terminals [26], is required for efficient GluRIIA incorporation at apposing postsynaptic specializations.
Figure 5. Unc13B-mediated evoked release facilitates GluRIIA incorporation, antagonized by Spn.
(A-N) Representative images of third-instar larval muscle 4 NMJs immunostained with an antibody against GluRIIA. Scale bars: 2μm. (F) Representative traces of eEJP and mEJP measurements at muscle 6/7 NMJs. Scale bars: eEJP 50ms, 10mV; mEJC traces 50ms, 2mV. (A-B) Quantification of GluRIIA intensity shows that motoneuronal knock-down (KD) of Unc13B impairs GluRIIA accumulation. (C) Partial sequence of Unc13 showing a single amino acid change (P>L) resulting in a gain-of-function (GoF) construct. (D-E) Motoneuron reexpression of an Unc13BGoF construct results in higher accumulation of GluRIIA. (F-G) Blocking evoked and spontaneous release with motoneuron expression of BoNT-C or specifically blocking only evoked release with TNT-E resulted in (H-I) reduced GluRIIA incorporation. (J-K) Concomitant expression of TNT-E and Unc13B KD in motoneurons shows that Unc13B-mediated evoked release function might facilitate GluRIIA accumulation during development. (L-M) Motoneuron KD of Unc13B in Spn mutants results in a suppression of the Spn mutant phenotype. (N-O) Motoneuron KD of Unc13B in Nlg2 mutants does not suppress Nlg2 mutant phenotype. See also figures S5 and S6, and table S1.
Once matured, all NMJ PSDs contain both Unc13A and Unc13B. We tested whether GluRIIA incorporation would, as predicted from our findings, persist in the near absence of Unc13A. We used Rab3 mutants, where BRP/Unc13A co-cluster in only about half of the AZs [26, 32] (Figure S5L). We observed that GluRIIA clusters were equally present at sites unapposed by BRP/Unc13A (Figure S5M). Unc13B accumulated at BRP/Unc13A-negative spots (Figure S5N) and every GluRIIA cluster showed an apposed Unc13B spot (Figure S5O). Thus, apart from the fact that Unc13A mutants are not defective in GluRIIA accumulation, these results also suggests that Unc13B can facilitate GluRIIA accumulation, independent of Unc13A.
To further probe the role of Unc13B in GluRIIA incorporation, we created gain-of-function (GoF) constructs of both Unc13A and Unc13B through a single amino acid exchange of P to L at a conserved position within the MUN domain of Unc13 (Figure 5C). A homologous mutation was shown to boost the SV release function of (m)unc13 family members [33]. Electrophysiological analysis of the Unc13AGoF (Figure S6A–E) showed a significant increase in evoked release amplitudes and an increase in spontaneous release events (mini frequency). The same analysis of the Unc13BGoF mutation, however, did not recover any significant changes in electrophysiological parameters (Figure S6F–J), consistent with our previous finding that Unc13A absolutely dominates release function at third instar larval NMJ terminals where the major fraction of synapses is already mature [26]. Electrophysiological recordings, however, point towards a role of Unc13B in ensuring normal release probability in early developmental stages, as we observed atypical paired-pulse facilitation in Unc13B mutants specifically in L2 developmental stage (Figure S6K,N,O). This does not rule out a concomitant role of Unc13A in defining normal release probabilities.
Despite the lack of gross effects on NMJ transmission, expressing the Unc13BGoF construct specifically in motoneurons strongly promoted GluRIIA incorporation over the normal level (Figure 5D–E), suggesting that developmental GluRIIA incorporation is indeed coupled to the degree of Unc13B-mediated release. We suspect that Unc13B, whose appearance as mentioned above clearly precedes Unc13A along the assembly trajectory of NMJ synapses, triggers glutamate release at immature synapses to consequently immobilize GluRIIA (see discussion).
To further explore the role of release for GluRIIA incorporation, we used botulinum toxin-C (BoNT-C), which targets Syntaxin to efficiently block both evoked and spontaneous release [34], or tetanus toxin (TNT-E) [35], which targets n-synaptobrevin and blocks evoked release. To allow the larvae to develop, we expressed these toxins in only a subset of motoneurons using a specific mosaic motoneuron driver line, Ok319-Gal4 (drives expression in motoneurons innervating muscles 4, 6 and 7, [36]). Electrophysiological measurements showed that BoNT-C expression fully suppressed both spontaneous and evoked release, whereas TNT-E fully suppressed evoked release, but only slightly impaired spontaneous release (Figure 5F–G). Both BoNT-C and TNT-E expression (Figure 5H–I) resulted in a decrease in GluRIIA incorporation, while no changes were seen at NMJs of muscle 1/2 (Figure S5C–D). The mean levels of GluRIIA were significantly reduced upon Unc13B-KD (Figures 5A–B, S5A–B) and also upon TNT-E expression (Figure 5H–I) in the motoneuron. Expressing TNT-E together with a Unc13B-RNAi KD construct did not further decrease the GluRIIA immunoreactivity (Figure 5J–K). Thus, our data suggest that evoked release at nascent synapses, mediated at least in part by Unc13B, is critical to allow efficient GluRIIA incorporation during developmental synapse assembly.
Spinophilin controls GluRIIA incorporation via restricting Unc13B levels
Lastly, we addressed whether Unc13B function (as part of Syd-1 module) would be subject to Spn-mediated antagonism. We found that Spn mutants had elevated Unc13B levels at AZs (Figure S5E–H), while in contrast, Unc13A levels were unchanged (Figure S5I–J). Thus, we asked whether increased Unc13B levels would contribute to the excessive GluRIIA incorporation in Spn mutants. Indeed, the GluRIIA increase observed in Spn mutants could be efficiently suppressed by concomitant reduction of Unc13B by RNAi KD (Figure 5L–M). This finding is consistent with the idea that Unc13B, as a critical effector for developmental GluRIIA incorporation, is part of the antagonistic control executed by Syd-1 and Spn. In contrast, we found that reducing Unc13B levels in Nlg2 mutants could not suppress the excessive GluRIIA incorporation in Nlg2 mutants (Figure 5N–O), suggesting that Nlg2 has an additional function in attenuating GluRIIA incorporation in an Unc13B-independent mode (see discussion).
Taken together, we identified a regulatory signaling system that tunes the trans-synaptic assembly process at a model glutamatergic synapse, which also could provide a template to search for similar regulatory schemes at mammalian central synapses.
DISCUSSION
Synapses form out of three interdependent molecular assemblies, each precisely crafted to execute fast and precise information transfer between two cells [4]: the presynaptic AZ where SVs fuse at defined release sites, the synaptic cleft through which NT diffuses, and the postsynaptic compartment where the NT binds its receptors. Importantly, these compartments do not form in isolation, but the size of the AZ (and thus the number of presynaptic release sites per AZ) must closely scale with the number of postsynaptic NT receptors. Super-resolution microscopy identified presynaptic AZ protein nanoclusters to align with concentrated postsynaptic receptors and scaffolding proteins, suggesting the existence of trans-synaptic molecular “nanocolumns” [5, 37]. Indeed, the exact nanometer location of vesicular release relative to receptors may be a critical determinant of synaptic strength, which may also contribute to synaptic plasticity [38].
A central question now pertains to how trans-synaptic signalling is precisely executed in molecular terms to coordinate pre- with post-synaptic assembly. Candidate molecular scenarios include interactions that directly bridge pre- and postsynaptic membranes like trans-synaptic CAMs, which bidirectionally control synapse formation, remodeling, and elimination [1]. We here exploit the unique features of the Drosophila NMJ system: unique accessibility to intravital imaging to accurately analyze the assembly path, a cytoarchitecture ideal for super-resolution analysis, high resolution electrophysiological measurements and a low level of genetic redundancy, to address how presynaptic AZs are matched to postsynaptic GluRs. Moreover, the amount of ELKS protein BRP, easily accessible for STED microscopy, directly scales with presynaptic release at AZs [2], making it an ideal readout to assess both structural and functional assembly.
Trans-synaptic assembly modules achieve pre-post matching
In principle, a strategy of continuously accumulating stoichiometric amounts of pre- and postsynaptic material along the assembly trajectory, potentially via a single transcellular bridge connecting to nucleation processes on both sides, might appear the easiest way to establish pre-post matching. Indeed, such an idea has recently been proposed, where the age of AZs determines their size and strength at the Drosophila NMJ [39]. However, such a solution might lack regulatory flexibility and is also not what we find in this work. Instead, our analysis identifies antagonistic regulatory inputs to be executed by two postsynaptically active Nlg species operating synergistically with their respective “cognate” presynaptic scaffold proteins, Syd-1 and Spn, previously shown to steer synapse assembly via their Nrx-1-binding function [9–11, 18, 24]. It here appears likely that autonomy over the presynaptic versus the postsynaptic compartment might be particularly relevant during plasticity processes, shown to involve the specific incorporation of BRP at NMJ synapses [38, 40, 41]. This antagonistic operation may serve to embed contextual information while steering the assembly process and could be particularly robust when utilized in such a highly regulative scheme.
Presynaptic AZ assembly tuned via Syd-1/Nlg1 versus Spn/Nlg2 signalling
In figure 6 we provide a model for the functional relations analyzed in this study. Syd-1 and Nlg1 form new AZs in the seeding phase (Figure 6A), whereas Spn and Nlg2 promote incorporation of BRP to appropriate levels (Figure 6C) in the maturation phase. Notably, BRP is the rate-limiting building block of the AZ scaffold determining the size and functional strength of the AZ specialization [42]. Overactivity of Syd-1/Nrx-1/Nlg1 signaling likely is directly responsible for the Spn AZ phenotype, as it could be suppressed by lowering the dose of any of these molecules. The same is true for the Nlg2 phenotype as well, suggesting that the Nlg2 AZ phenotype similarly reflects Syd-1/Nlg1 axis overactivity. Furthermore, reduction of Spn efficiently suppressed the normally excessive BRP incorporation at the AZs remaining in Syd-1, Nrx-1 and Nlg-1 mutants. Mechanistically, future analysis will have to clarify whether direct physical interactions of Spn with BRP complexes, co-clustering RIM-BP and Unc13A [38], are of relevance here. Alternatively, the Syd-1 and Spn modules might antagonistically control a downstream process such as the status of F-actin [43].
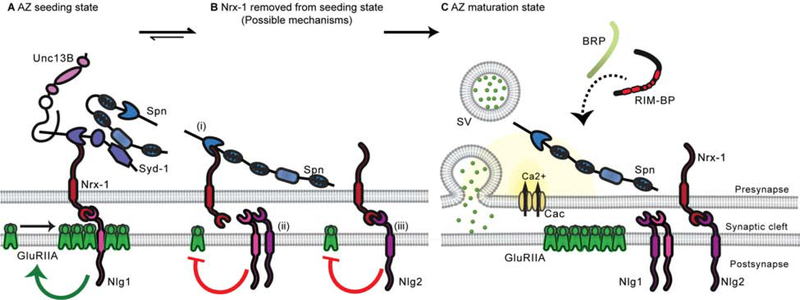
Figure 6. Model.
(A) Syd-1, Nrx-1 and Nlg1 cooperate during the seeding phase to initiate AZ formation (B) Nrx-1/Nlg1 seeding function is stalled by (i) Spn (in competition with Syd-1) binding Nrx-1 C-terminus (ii) Cis-heteromerization between Nlg1 and Nlg2 (iii) Nrx-1 and Nlg2 interaction (in competition with Nlg1) (C) Spn and Nlg2 cooperate to change the biochemical milieu of Nrx-1 to extract it from its AZ seeding function to allow a switch to the AZ maturation phase.
While in the past we interpreted that Spn might function after Syd-1 during the AZ development process, our data now suggest that Syd-1 and Spn in fact continuously antagonize each other throughout assembly to tune final AZ size and function. Still, our intravital imaging of nascent AZs showed that the peak of Syd-1 accumulation precedes the peak of BRP accumulation by hours [24]. The fact that the Syd-1 scaffold is favored over the Spn scaffold during the seeding phase might be explained via a “quasi-epistatic” relation between these regulators: Syd-1 mutants show lower levels of Spn (Figure S5P–Q), while Spn mutants show elevated levels of Syd-1 [18], suggesting that Syd-1 is required for Spn accumulation at the AZ, potentially allowing Syd-1-mediated AZ seeding to precede Spn-mediated BRP accumulation. Spn and Syd-1 were shown to interact with each other in Drosophila [18] and In C. elegans [43]. It would be interesting to investigate whether the Spn/Syd-1 interaction plays a role in regulating access to Nrx-1 (Figure 6A–B), thereby contributing to define the actual “assembly mode”: seeding or maturation (Figure 6A,C). Obviously, the assembly modules must communicate to ultimately ensure a well-defined assembly product, e.g., via associated kinase/phosphatase activities. For example, the phosphorylation status of BRP can control transport [44]. Furthermore, while Spn attenuation did efficiently suppress the Syd-1, Nrx-1 and Nlg1 AZ phenotypes, Nlg2 attenuation did not suppress the Nrx-1 and Nlg1 phenotypes. This suggests that the trans-synaptic signaling through Nrx/Nlgs might ensure that assembly proceeds from seeding towards maturation during development. This also opens up the possibility that Nlg2 attenuates Syd-1/Nrx-1/Nlg1 function by removing Nrx-1 from the seeding module (Figure 6Biii) and/or suppressing Nlg1 activity through cis-heteromerization [25] (Figure 6Bii). Our FRAP data also indicate that the postsynaptic binding partner identity (Nlg1 or Nlg2) has differential effects on Nrx-1 mobility (Figure 4B–E). Lack of Nlg2 likely boosts the Nrx-1::Nlg1 seeding activity, directly explaining the supernumerary AZs typical for Nlg2 mutant.
Postsynaptic receptor assembly through Syd-1/Nlg1 and Spn/Nlg2 signalling
Nlg1 promoted but Nlg2 blocked GluRIIA incorporation, which precedes BRP accumulation. Previous analysis showed that Syd-1 seemingly instructs Nrx-1 to interact with Nlg1 and promotes GluRIIA incorporation [10, 24] before BRP incorporation [29]. Our genetic interaction analysis showed that Syd-1/Nlg1 and Spn/Nlg2 execute a mutual regulatory counterplay here. Our study now extends the understanding of GluRIIA incorporation to involve the release function of Unc13B, enriched at nascent AZs by Syd-1, a process antagonized by Spn [26]. Spn and Nlg2 functionally cooperate to limit the amount of GluRIIA incorporation in the nascent postsynaptic specialization and match receptor amounts to the AZ size. However, while the Spn mutant phenotype was rescued by Syd-1, Nrx-1 and Nlg1 heterozygosity, the Nlg2 mutant phenotype was only rescued by Syd-1 heterozygosity, suggesting that Nlg1 and Nlg2 have an additional function in mediating GluRIIA incorporation independent of Unc13B. Mechanistically, it might well be that Nlg2 at the nascent postsynaptic compartment directly competes with Nlg1 for the binding of a critical effector, e.g. the ectodomain of the GluR complex or other membrane proteins such as Neto [45].
Unc13B-mediated neurotransmitter release and GluRIIA incorporation
In mice, most synapses formed normally in the absence of NT release during development, but the synapses did not persist as they matured [46, 47]. Experiments in mice have shown in the past that massive local glutamate release could induce spine formation at the postsynapse [48]. However, whether vesicular transmitter release tunes the incorporation dynamics of GluRs during developmental synapse assembly remains inconclusive [49, 50].
Unc13B arrives early at nascent NMJ AZs [26]. This recruitment of Unc13B is antagonistically controlled by the two complexes, as Syd-1 mutants showed reduced [26] but Spn mutants strongly increased synaptic Unc13B levels (Figure S5E–H). Importantly, the excessive GluRIIA incorporation in Spn mutants critically depended on Unc13B (Figure 5L–M). Notably, treatment of cell cultures with BoNT-C and TNT-E previously was shown to prevent effective postsynaptic insertion of glutamatergic receptors in cultivated hippocampal neurons [51]. However, we cannot exclude that once Unc13A accumulates at the AZ, Unc13B might continue to mediate GluRIIA incorporation into later stages of synapse assembly.
Concerning the mode of Unc13B action, our data suggest that evoked Unc13B-mediated glutamate release at nascent sites attracts GluRIIA receptors, which are recruited from diffuse pools at the plasma membrane [30]. Notably, proper gating behavior of GluRIIA in response to presynaptic glutamate release previously was shown to be essential for matching pre- with post-assembly [31]. Unc13B-mediated release is coupled more loosely to Ca2+ channel activity compared with release mediated by the functionally dominant isoform, Unc13A [52]. Likely, sensing glutamate at nascent sites renders GluRIIA into an active state which allows for postsynaptic incorporation, previously shown to be nearly irreversible [30]. Whether the GluRIIA incorporation subsequent to the glutamate sensing is truly stage dependent, e.g., via specific scaffold or cleft proteins, or whether differences in the spatio-temporal detail of glutamate release between Unc13B and Unc13A are more important here remains to be addressed.
Nrx-1, Nlg1 and Syd-1 mutants all show reduced NMJ area, while Spn and Nlg2 mutants showed normal NMJ sizes (Figure S1), with all of them showing reduced evoked potentials [9, 11, 15, 18, 23]. Previous studies have shown that synaptic terminals can compensate for a change in size by adjusting NT output [53, 54]. A recent study showed that spontaneous neurotransmission is required for the normal structural maturation of Drosophila NMJ synapses exclusive from the role of evoked neurotransmission [55]. Increasing miniature events was sufficient to induce synaptic terminal growth, and this synapse maturation was locally regulated via a Trio guanine nucleotide exchange factor (GEF) and Rac1 GTPase molecular signaling pathway. Interestingly, Syd-1 was found to interact with Trio signaling [56]. Together with the Rac guanine exchange factor (RacGEF) Trio, Syd-1 GAP activity promotes BRP clustering and independent of its GAP activity, Syd-1 recruits Nrx-1 to boutons. Additionally, mammalian Spn forms a complex with Rac1-GEF Kalirin-7 and along with Rho-GEF Lfc, control dendritic spine morphology and function [57, 58]. Therefore, it will be interesting to study how Syd-1 and Spn antagonism translates into GAP/GEF signaling which in turn might control the synapse assembly at Drosophila NMJs.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stephan J. Sigrist stephan.sigrist@fu-berlin.de.
Materials Availability
The new lines described in this study are available on request from the lead contact.
Data and Code Availability
The datasets used for analyses are available on request from the lead contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Strains
Drosophila melanogaster strains were maintained as stocks at room temperature. Fly strains for experiments were reared at 25°C under standard conditions [59] on semi-defined medium (Bloomington recipe). For RNA interference experiments, parents laid eggs under standard conditions and larvae were raised at 29°C throughout development. No selection was done based on sex. For fly strain details, please refer the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BRPNc82 | Developmental Studies Hybridoma Bank | RRID: AB_2314866 |
| GluRIIA | Developmental Studies Hybridoma Bank | RRID: AB_528269 |
| GluRIID | [27] | N/A |
| Spinophilin | [18] | N/A |
| Unc13A | [26] | N/A |
| Unc13B | [26] | N/A |
| RIM-BP | [26] | N/A |
| PAK | [30] | N/A |
| Goat anti-mouse Alexa-Fluor-488 | Invitrogen | Cat# A-11001; RRID: AB_2534069 |
| Goat anti-rabbit-Cy3 | Jackson ImmunoResearch | Cat# 111–165-144; RRID: AB_2338006 |
| Goat anti-guineapig-Cy3 | Jackson ImmunoResearch | Cat# 106–165-003; RRID: AB_2337423 |
| Anti-Horseradish Peroxidase Alexa-Fluor-647 | Jackson ImmunoResearch | Cat# 123–605-021; RRID: AB_2338967 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Vectashield | VectorLabs | Cat# H-1900–10 |
| ProLong™ Gold Antifade Mountant | Thermo Fisher Scientific | Cat# P36930 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: w1118 | Bloomington Drosophila Stock Center | BDSC: 5905; FlybaseID: FBst0005905 |
| D. melanogaster: spnΔ3.1 or spex3.1 | [18] | N/A |
| D. melanogaster: Df(3L)BSC116 | Bloomington Drosophila Stock Center | BDSC: 8973; FlybaseID: FBst0008973 |
| D. melanogaster: syd-1ex1.2 | [15] | N/A |
| D. melanogaster: syd-1ex3.4 | [15] | N/A |
| D. melanogaster: nrx-1241 | [23] | N/A |
| D. melanogaster: Df(3R)5C1 | Bloomington Drosophila Stock Center | BDSC: 1605; FlybaseID: FBst0001605 |
| D. melanogaster: nlg1ex1.9 | [10] | N/A |
| D. melanogaster: nlg1ex2.3 | [10] | FlybaseID: FBal0246577 |
| D. melanogaster: nlg2CL5 | [11] | FlybaseID: FBal0285173 |
| D. melanogaster: GluRIIAGFP | [30] | N/A |
| D. melanogaster: Unc13Pacman | [26] | N/A |
| D. melanogaster: Del100BPacman | [26] | N/A |
| D. melanogaster: unc-13EMS7.5 | [26] | FlybaseID: FBal0338122 |
| D. melanogaster: unc-13 P84200 | KYOTO stock center | DGRC# 101911 |
| D. melanogaster: unc-13AGoF | This paper | N/A |
| D. melanogaster: unc13BGoF | This paper | N/A |
| D. melanogaster: Ok6-Gal4 | [64] | N/A |
| D. melanogaster: Ok319-Gal4 | [65] | N/A |
| D. melanogaster: UAS-TNT-E | [35] | N/A |
| D. melanogaster: UAS-BONT-C | This paper | N/A |
| D. melanogaster: UAS-Unc13B-RNAi | [66] | N/A |
| D. melanogaster: unc13BCRISPR | This paper | N/A |
| Oligonucleotides: | ||
| GGGCGAGGAAAAAGTAGCTCTTTACCACGTACAATATACCTGTTTACACG | This paper | N/A |
| CGTGTAAACAGGTATATTGTACGTGGTAAAGAGCTACTTTTTCCTCGCCC | This paper | N/A |
| AATTGAGCTCCCACCATGCCAATAACAATTAACAAC | This paper | N/A |
| TTAAGGTACCTTATTTATTATATAATGATCTACCATC | This paper | N/A |
| AGAGCTCCGCTCTTAAGCCA[GGG] | This paper | N/A |
| Software and Algorithms | ||
| Fiji | [61] | http://fiji.sc/ |
| GraphPad Prism 5 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Imspector Image Acquisition & Analysis Software v16.1. | Abberior Instruments | http://www.imspector.de |
| pClamp 10 | Molecular Devices | https://www.moleculardevices.com/ |
Genotypes used for experiments were wild type WT: (+/+ (w1118)). Spn: (SpnΔ3.1/dfBSc116). Nlg2: (Nlg2CL5/ Nlg2CL5). Syd-1: (Syd-1ex3.4/ Syd-1ex1.2). Nrx-1: (Nrx-1241/ Nrx-1df). Nlg1: (Nlg1ex2.3 /Nlg1ex1.9). Spn, Syd-1+/−: (SpnΔ3.1, Syd-1ex3.4/dfBSc116). Spn, Syd-1+/−: (SpnΔ3.1, Syd-1ex3.4/dfBSc116). Spn, Nrx-1+/−: (SpnΔ3.1, Nrx-1241/dfBSc116). Spn, Nlg1+/−: (SpnΔ3.1, Nlg1ex2.3/dfBSc116). Syd-1, Spn+/−: (SpnΔ3.1, Syd-1ex3.4/ Syd-1ex1.2). Nrx-1, Spn+/−: (SpnΔ3.1, Nrx-1241/ Nrx-1df). Nlg1, Spn+/−: (SpnΔ3.1, Nlg1ex2.3 /Nlg1ex1.9). Nlg2; Syd-1+/−: (Nlg2CL5/ Nlg2CL5. Syd-1ex3.4). Nlg2; Nrx-1+/−: (Nlg2CL5/ Nlg2CL5; Nrx-1241). Nlg2; Nlg1+/−: (Nlg2CL5/ Nlg2CL5; Nlg1ex2.3). Nlg2; Spn+/−: (Nlg2CL5/ Nlg2CL5; SpnΔ3.1). Nlg2+/−; Spn: (Nlg2CL5; SpnΔ3.1/dfBSc116). Nlg2+/−; Nrx-1: (Nlg2CL5/ +; Nrx-1241/ Nrx-1df). Nlg2+/−; Nlg1: (Nlg2CL5/ +; Nlg1ex2.3 /Nlg1ex1.9). Ok6-Gal4>UAS-Nlg2, Nlg2: (Ok6-Gal4, Nlg2CL5/ Nlg2CL5; UAS-Nlg2). Mef2-Gal4>UAS-Nlg2, Nlg2: (Mef2-Gal4, Nlg2CL5/ Nlg2CL5; UAS-Nlg2). WTRescue: (Unc13Pacman/+; P84200/P84200). Unc13A: (EMS7.5/P84200). Unc13B: (Del100BPacman/+; P84200/P84200). Unc13B (Figure S6): Unc13BCRISPR/ Unc13BCRISPR). Nlg1 (Figure 4): (Ok6>Nrx-1GFP;;Nlg1ex2.3 /Nlg1ex1.9). Nlg2 (Figure 4): (Ok6>Nrx-1GFP; Nlg2CL5/ Nlg2CL5). Spn (Figure 4): (GluRIIAGFP; SpnΔ3.1/dfBSc116). Nlg2 (Figure 4): (GluRIIAGFP; Nlg2CL5/ Nlg2CL5). Unc13B (Figure 4L): (GluRIIAGFP; Del100BPacman/+; P84200/P84200.) Ok6-Gal4>Control: (Ok6-Gal4/+). Ok6-Gal4>Unc13B-KD: (Ok6-Gal4/+; UAS-Unc13B-RNAi/+). Ok319-Gal4>Control: (Ok319-Gal4/+). Ok319-Gal4>Unc13A-KD: (Ok319-Gal4/+; UAS-Unc13A-RNAi/+). Ok319-Gal4>Unc13B-KD: (Ok319-Gal4/+; UAS-Unc13BRNAi/+). Unc13B; Ok6-Gal4>Unc13B: (Ok6-Gal4, UAS-Unc13B; Del100BPacman/+; P84200/P84200). Unc13B; Ok6-Gal4>Unc13BGoF: (Ok6-Gal4, UAS-Unc13BGoF; Del100BPacman/+; P84200/P84200). Unc13A; Ok6-Gal4>UAS-Unc13A: (Ok6-Gal4, UAS-Unc13A; EMS7.5/P84200). Unc13A; Ok6-Gal4>Unc13AGoF: (Ok6-Gal4, UAS-Unc13AGoF; EMS7.5/P84200). Ok319-Gal4>UAS-BoNT-C: (Ok319-Gal4/+; UAS-BoNT-C/+). Ok319-Gal4>UAS-TNT-E: (Ok319-Gal4/+; UAS-TNT-E/+). Ok6-Gal4>Spn: (Ok6-Gal4/+; SpnΔ3.1/dfBSc116). Ok6-Gal4>UAS-Unc13B-KD; Spn: (Ok6-Gal4/+; Unc13B-RNAi, SpnΔ3.1/dfBSc116). Ok6-Gal4>Nlg2: (Ok6-Gal4, Nlg2CL5/ Nlg2CL5). Ok6-Gal4>UAS-Unc13B-KD; Nlg2: (Ok6-Gal4, Nlg2CL5/ Nlg2CL5; Unc13B-RNAi).
METHOD DETAILS
Generation of Unc13 gain-of-function constructs (GoF/ P>L)
A GoF point mutation based on Lipstein et al., 2017 was introduced into Unc13 entry clones [26] in pENTR/D-Topo (Invitrogen). A single nucleotide exchange was induced in Drosophila Unc13A and Unc13B C-terminus by the following primers:
Unc13AB P>L fw: 5′-GGGCGAGGAAAAAGTAGCTCTTTACCACGTACAA TATACCTGTTTACACG-3′,
Unc13AB P>L rev: 5′-CGTGTAAACAGGTATATTGTACGTGGTAAAGAGC TACTTTTTCCTCGCCC-3′.
The point-mutated constructs were shuttled into pUAST vectors containing a C-terminal GFP tag by Gateway reaction. For the generation of transgenic flies both constructs were sent for DNA micro-injection into embryos to Rainbow Transgenic Flies, Inc, CA, USA (injected fly strain: Strain 24862; y[1] M{vas-int.Dm}vZH-2A w[*]; PBac{y[+]-attP-9A}VK00005)
Generation of UAS-BoNT-C
A plasmid containing the entire light chains of BoNT-C (BoNT/C-LC) was a gift from Dr. Thomas Binz (Hannover, Germany). This plasmid was used as a template to amplify the N- and C-terminal fragment of BoNT-C and cloned into pENTR/D-Topo (Invitrogen) using the following primers:
BoNT-C fw: 5′-AATTGAGCTCCCACCATGCCAATAACAATTAACAAC-3′,
BoNT-C rev: 5′-TTAAGGTACCTTATTTATTATATAATGATCTACCATC-3′.
The BoNT-C light chain was then shuttled into the pUASt vector using a Gateway reaction. Transgenic flies were sent for DNA micro-injection to BestGene, Inc (CA, USA). w1118 was injected and an insertion on the third chromosome was isolated and used for further analysis.
Generation of Unc13BCRISPR
The Unc13B null mutant was generated by WellGenetics (Taiwan) using CRISPR/Cas9-mediated genome editing by homology-dependent repair (HDR) using 1 guide RNA(s) and a dsDNA plasmid donor[60]. 3-frame STOPs were introduced immediately following the start codon of Unc13B to create a specific Unc13B null mutant. In brief, gRNA sequence AGAGCTCCGCTCTTAAGCCA[GGG] was cloned into U6 promoter plasmid(s). Cassette Stop-RFP containing two loxP sites, 3xP3-RFP, 3-frame stop codon and two homology arms were cloned into pUC57-Kan as donor template for repair. unc-13/CG2999-targeting gRNAs and hs-Cas9 were supplied in DNA plasmids, together with donor plasmid for microinjection into embryos of control strain w[1118]. F1 flies carrying selection marker of 3xP3-RFP were further validated by genomic PCR and sequencing. CRISPR generates a 61-bp deletion allele of unc-13/CG2999, deleting partial coding exon of unc-13/CG2999 and is replaced by cassette Stop-RFP.
Immunostaining
Larval dissections and immunostaining were performed as previously described[27]. Briefly, the larvae were immobilized with insect pins on a rubber dissection pad and cut open dorsally between the dorsal tracheal trunks. The muscle wall flattened using insect pins and the tissue was fixed. Except for staining against GluRIIA where larvae were fixed in ice-cold methanol for 5 min, all staining was performed after fixation in ice-cold 4%- paraformaldehyde (PFA) in 0.1 mM phosphate-buffered saline (PBS) for 10 minutes. Larvae were then processed for immunohistochemistry and mounted in Vectashield (Vector Labs, CA, USA). For STED imaging, larvae were mounted in ProLong™ Gold Antifade Mountant (Thermo Fisher Scientific).
Image acquisition, processing and analysis.
Confocal microscopy was performed with a Leica TCS SP8 inverted confocal microscope (Leica DMI 6000, Leica Microsystems, Germany) and STED microscopy with a Leica TCS SP5 microscope. All images were acquired at room temperature using LCS AF software. (Leica Microsystems, Germany). Confocal imaging was performed using a 63×1.4 NA oil immersion objective. Images of muscle 4 Type-1b NMJs were obtained from abdominal segments A3-A4 of the fixed larval preparations for all experiments. Images were acquired in line scanning mode with a pixel size 75.16nm*75.16nm and with a z step of 0.25μm. Stacks were processed with Fiji [61] software. Images were quantified for the average size of an AZ, density of AZs in an NMJ (using BRP/RIM-BP as markers), average intensity of receptor fields over an NMJ (using GluRIIA, PAK), relative NMJ area covered by receptor fields (using GluRIIA, PAK) and the average intensity of other AZ proteins: Spn, Unc13A and Unc13B. The stacks of images for each NMJ were converted to a 2D image using maximum intensity Z-projection. The area stained by HRP was taken as the area of contact between a motoneuron and muscle (NMJ area). The intensity of the protein in question was obtained by measuring average pixel intensity inside the NMJ area marked by the HRP signal using ‘measure’ command. The size and number of BRP spots was determined as follows: lower intensity pixels were removed by thresholding, restricting the signal to being relevant to the protein in question. The spots were segmented by using an inbuilt ‘Find Maxima’ command which determines the local maxima in an image and creates a binary image of the same size with one segmented particle per maximum. This binary mask was then projected onto the original unmodified image using the ‘min’ operation from the Fiji image calculator and any particles left unsegmented were segmented by hand using the pencil tool with a line thickness of 1 pixel. The inbuilt ‘analyse particle’ command was used to obtain the average size and total number of BRP spots. All data was copied into Microsoft Excel and spot density was calculated by dividing the total number of spots by the NMJ area.
Single-color STED imaging was performed with a 100× 1.4 NA oil immersion objective. Images were obtained from muscle 4 Type 1b NMJs of the abdominal segments. Images were obtained with a pixel size of 25.3 nm*25.3 nm. The diameters were measured on deconvolved images by drawing a line passing through the centre of a planar ring. The intensity profile of the line was obtained using ‘plot profile’ available in Fiji which Displays a graph of the intensities of pixels along the line. The x-axis represents distance along the line and the y-axis is the pixel intensity. The distance between the two peak intensities is taken as the diameter of the BRP ring. Two lines were drawn through each ring and the average of the two diameters was taken as the diameter of each BRP ring. BRP ring diameters obtained from one NMJ were averaged to obtain a value of average ring diameter per NMJ. The average size of BRP spots was obtained by using the inbuilt ‘analyse particle’ command above a set threshold.
Two-colour STED microscopy was performed using an Abberior Instruments Expert Line STED setup equipped with two STED lasers for depletion. The dyes STAR RED and Alexa Fluor 594 were depleted with a pulsed STED laser at 775 nm. Images were acquired with a 100×, 1.4 NA oil immersion objective. According to the field of view (10 × 10 μm resolution) two-dimensional STED images were scanned with a pixel dwell time of 2 μs using 10× line accumulation. The lateral pixel size was set to values of 20 nm. STED images were processed using the Imspector deconvolution software (16.1.6477, Germany) using a theoretical PSF automatically computed with a full width half maximum of 40 nm, based on 40 nm Crimson-beads measurements. Default deconvolution settings were applied.
In vivo live imaging was performed using a Leica SP8 microscope and a 63×1.4 NA oil immersion objective. Images of muscle 26 and 27 Type-1b NMJs were obtained from larval abdominal segments A2-A4. Confocal images were acquired in line scanning mode with a pixel size of 75.16nm*75.16nm and with a z step of 0.25μm. Images were obtained from third instar larvae by immobilizing them in an airtight imaging chamber with a small amount of Voltalef H10S oil (Arkema, Inc., France) which has the same refractive index as the larval cuticle. The larvae were anaesthetized with short pulses of a mixture of air and desflurane (Baxter,IL, UAS) until the heartbeat completely stopped. The number of pulses differed between animals, but the animals were checked for viability after each imaging session was completed. For experiments looking at larval NMJ development over a 24-hour period, the larvae were kept separately for about 24 hours on normal food at 25°C. The very same NMJs were then reimaged through the same technique to capture changes that took place over the 30min/24-hour period. For fluorescence recovery after photobleaching (FRAP) experiments, a z-stack of confocal images of an NMJ was acquired before bleaching. The images were set to approximately the same average intensity value at the beginning of the experiment. An area of about 2 boutons was then selected and bleached by increasing the laser power to 100% using the ROI function in the LCS AF software and bleaching through the z-stack. The same ROI area was used across NMJs to bleach a consistent area. Bleaching a confined area of 2 boutons allowed for an internal control of both bleached and unbleached regions within the same NMJ to aid analysis. For analysis, the ratios of intensities were calculated between the bleached and unbleached areas at three time points: before bleaching, after bleaching and after recovery. The ratio before bleaching was used as a control to check that the intensity over the NMJ was constant. Recovery was calculated as the ratio of signal that returned to the NMJ in the time period after bleaching to the signal before bleaching.
Electrophysiology
Electrophysiological recordings were performed at room temperature on muscle 6 of 2nd and 3rd instar larval NMJs in the abdominal segments A2 and A3. The larvae were dissected in Ca2+-free HL3 solution and recordings were obtained with a bath solution of HL3 with added 1.5mM CaCl2. Glass electrodes were pulled using a Flaming Brown Model P-97 micropipette puller (Sutter Instrument, CA, USA). Recordings were made using an Axoclamp 2 B amplifier with HS-2A x0.1 headstage (Molecular Devices, CA, USA) and on a BX51WI Olympus microscope with a 40X LUMPlanFL/IR water immersion objective (Olympus Corporation, Shinjuku, Tokyo, Japan). mEPSPs/mEPSCs were recorded for 90 seconds. eEPSPs/eEPSCs were recorded after stimulating the appropriate motoneuron bundle with 5 V, 300 μs at 0.2 Hz using an S48 Stimulator (Grass Instruments, Astro-Med, Inc., RI, USA). Signals were digitized at 10 kHz using an Axon Digidata 1322 A digitizer (Molecular Devices, CA, USA) and low pass filtered at 1 kHz using an LPBF-48DG output filter (NPI Electronic, Tamm, Germany). The recordings were analysed with pClamp 10 (Molecular Devices, Sunnyvale, CA, USA), Graphpad Prism 6 (Graphpad Software, Inc., San Diego, CA, USA) and MATLAB R2010b (Mathworks, Natick, MA, USA). Stimulation artifacts of eEPSPs were removed for clarity.
Two-Electrode Voltage Clamp (TEVC) recordings were performed as previously described[62]. Recordings were made from cells with an initial Vm between −50 and −70 mV, and input resistances of ≥ 4 MΩ, using intracellular electrodes with resistances of 8–20 MΩ, filled with 3 M KCl. eEJCs which reflect the compound EJC of both motoneurons innervating muscle 6 were recorded at a voltage clamp of −60 mV. Current clamp recordings were performed as previously described [63]. Recordings were made from cells with an initial Vm between −40 and −80 mV, and input resistances of ≥ 4 MΩ, using intracellular electrodes with resistances of 30–70 MΩ, filled with 3 M KCl. mEPSPs were further filtered with a 500 Hz Gaussian low-pass filter. Using a single template for all cells, mEPSPs were identified and analysed, noting the mean mEPSP amplitude per cell. An average trace was generated from 20 eEPSP traces per cell. The amplitude of the average eEPSP trace was divided by the mean mEPSP amplitude, for each respective cell, to determine the quantal content. Electrophysiological recordings in figure 5 were performed in 0.5 mM calcium modified HL-3 as described previously[41].
QUANTIFICATION AND STATISTICAL ANALYSIS
Data was analysed using Prism (GraphPad Software, CA, USA). In all data sets with two groups, nonparametric t-test or Mann-Whitney U test was used. For all data sets with three or more groups, nonparametric Kruskal-Wallis test followed by Dunn’s multiple comparison test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was used. For all data failing normality test and all immunostaining data, Mann-Whitney U test or nonparametric Kruskal-Wallis test followed by Dunn’s multiple comparison test was used. Statistical parameters are stated in the figure legends. Data is represented as mean ± SEM. Statistical significance is denoted in the graphs as asterisks: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns. (not significant), p > 0.05.
Supplementary Material
Highlights.
Neurexin-interactors Spinophilin (Spn) & Syd-1 cooperate with Neuroligin (Nlg)-1& 2
Syd-1/Nlg1 & Spn/Nlg2 antagonism tunes initial assembly and maturation of synapses
Spn and Syd-1 modulate Unc13B and glutamate receptors via Unc13B release function
Functional Nlg1 & 2 antagonism might be a novel principle to tune synapse assembly
ACKNOWLEDGEMENTS
We acknowledge the assistance of the core facility BioSupramol supported by the DFG. This work was supported by grants to S.J.S (SFB958 A03 and A06, FOR2705; SFB1315 A08).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sudhof TC (2018). Towards an Understanding of Synapse Formation. Neuron 100, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holderith N, Lorincz A, Katona G, Rozsa B, Kulik A, Watanabe M, and Nusser Z (2012). Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat Neurosci 15, 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peled ES, Newman ZL, and Isacoff EY (2014). Evoked and spontaneous transmission favored by distinct sets of synapses. Current biology : CB 24, 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biederer T, Kaeser PS, and Blanpied TA (2017). Transcellular Nanoalignment of Synaptic Function. Neuron 96, 680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang AH, Chen H, Li TP, Metzbower SR, MacGillavry HD, and Blanpied TA (2016). A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han KA, Um JW, and Ko J (2019). Intracellular protein complexes involved in synapse assembly in presynaptic neurons. Adv Protein Chem Struct Biol 116, 347–373. [DOI] [PubMed] [Google Scholar]
- 7.Knight D, Xie W, and Boulianne GL (2011). Neurexins and neuroligins: recent insights from invertebrates. Mol Neurobiol 44, 426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petzoldt AG, Lutzkendorf J, and Sigrist SJ (2016). Mechanisms controlling assembly and plasticity of presynaptic active zone scaffolds. Curr Opin Neurobiol 39, 69–76. [DOI] [PubMed] [Google Scholar]
- 9.Sun M, Xing G, Yuan L, Gan G, Knight D, With SI, He C, Han J, Zeng X, Fang M, et al. (2011). Neuroligin 2 is required for synapse development and function at the Drosophila neuromuscular junction. J Neurosci 31, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banovic D, Khorramshahi O, Owald D, Wichmann C, Riedt T, Fouquet W, Tian R, Sigrist SJ, and Aberle H (2010). Drosophila neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron 66, 724–738. [DOI] [PubMed] [Google Scholar]
- 11.Chen YC, Lin YQ, Banerjee S, Venken K, Li J, Ismat A, Chen K, Duraine L, Bellen HJ, and Bhat MA (2012). Drosophila neuroligin 2 is required presynaptically and postsynaptically for proper synaptic differentiation and synaptic transmission. J Neurosci 32, 16018–16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng X, Sun M, Liu L, Chen F, Wei L, and Xie W (2007). Neurexin-1 is required for synapse formation and larvae associative learning in Drosophila. FEBS Lett 581, 2509–2516. [DOI] [PubMed] [Google Scholar]
- 13.Collins CA, and DiAntonio A (2007). Synaptic development: insights from Drosophila. Curr Opin Neurobiol 17, 35–42. [DOI] [PubMed] [Google Scholar]
- 14.Owald D, and Sigrist SJ (2009). Assembling the presynaptic active zone. Curr Opin Neurobiol 19, 311–318. [DOI] [PubMed] [Google Scholar]
- 15.Owald D, Fouquet W, Schmidt M, Wichmann C, Mertel S, Depner H, Christiansen F, Zube C, Quentin C, Korner J, et al. (2010). A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J Cell Biol 188, 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, and Sigrist SJ (2009). Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol 186, 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. (2006). Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054. [DOI] [PubMed] [Google Scholar]
- 18.Muhammad K, Reddy-Alla S, Driller JH, Schreiner D, Rey U, Bohme MA, Hollmann C, Ramesh N, Depner H, Lutzkendorf J, et al. (2015). Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function. Nat Commun 6, 8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu KS, Siebert M, Mertel S, Knoche E, Wegener S, Wichmann C, Matkovic T, Muhammad K, Depner H, Mettke C, et al. (2011). RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science 334, 1565–1569. [DOI] [PubMed] [Google Scholar]
- 20.Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, and Scheiffele P (2003). Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci 6, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z, Hom S, Kudze T, Tong XJ, Choi S, Aramuni G, Zhang W, and Kaplan JM (2012). Neurexin and neuroligin mediate retrograde synaptic inhibition in C. elegans. Science 337, 980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheiffele P, Fan J, Choih J, Fetter R, and Serafini T (2000). Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Ashley J, Budnik V, and Bhat MA (2007). Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron 55, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owald D, Khorramshahi O, Gupta VK, Banovic D, Depner H, Fouquet W, Wichmann C, Mertel S, Eimer S, Reynolds E, et al. (2012). Cooperation of Syd-1 with Neurexin synchronizes pre- with postsynaptic assembly. Nat Neurosci 15, 1219–1226. [DOI] [PubMed] [Google Scholar]
- 25.Poulopoulos A, Soykan T, Tuffy LP, Hammer M, Varoqueaux F, and Brose N (2012). Homodimerization and isoform-specific heterodimerization of neuroligins. Biochem J 446, 321–330. [DOI] [PubMed] [Google Scholar]
- 26.Bohme MA, Beis C, Reddy-Alla S, Reynolds E, Mampell MM, Grasskamp AT, Lutzkendorf J, Bergeron DD, Driller JH, Babikir H, et al. (2016). Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca(2+) channel-vesicle coupling. Nat Neurosci 19, 1311–1320. [DOI] [PubMed] [Google Scholar]
- 27.Qin G, Schwarz T, Kittel RJ, Schmid A, Rasse TM, Kappei D, Ponimaskin E, Heckmann M, and Sigrist SJ (2005). Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J Neurosci 25, 3209–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos CI, Igiesuorobo O, Wang Q, and Serpe M (2015). Neto-mediated intracellular interactions shape postsynaptic composition at the Drosophila neuromuscular junction. PLoS Genet 11, e1005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid A, Hallermann S, Kittel RJ, Khorramshahi O, Frolich AM, Quentin C, Rasse TM, Mertel S, Heckmann M, and Sigrist SJ (2008). Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat Neurosci 11, 659–666. [DOI] [PubMed] [Google Scholar]
- 30.Rasse TM, Fouquet W, Schmid A, Kittel RJ, Mertel S, Sigrist CB, Schmidt M, Guzman A, Merino C, Qin G, et al. (2005). Glutamate receptor dynamics organizing synapse formation in vivo. Nature neuroscience 8, 898–905. [DOI] [PubMed] [Google Scholar]
- 31.Petzoldt AG, Lee YH, Khorramshahi O, Reynolds E, Plested AJ, Herzel H, and Sigrist SJ (2014). Gating characteristics control glutamate receptor distribution and trafficking in vivo. Current biology : CB 24, 2059–2065. [DOI] [PubMed] [Google Scholar]
- 32.Graf ER, Daniels RW, Burgess RW, Schwarz TL, and DiAntonio A (2009). Rab3 dynamically controls protein composition at active zones. Neuron 64, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipstein N, Verhoeven-Duif NM, Michelassi FE, Calloway N, van Hasselt PM, Pienkowska K, van Haaften G, van Haelst MM, van Empelen R, Cuppen I, et al. (2017). Synaptic UNC13A protein variant causes increased neurotransmission and dyskinetic movement disorder. J Clin Invest 127, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Backhaus P, Langenhan T, and Neuser K (2016). Effects of transgenic expression of botulinum toxins in Drosophila. J Neurogenet 30, 22–31. [DOI] [PubMed] [Google Scholar]
- 35.Sweeney ST, Broadie K, Keane J, Niemann H, and O’Kane CJ (1995). Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14, 341–351. [DOI] [PubMed] [Google Scholar]
- 36.Goel P, and Dickman D (2018). Distinct homeostatic modulations stabilize reduced postsynaptic receptivity in response to presynaptic DLK signaling. Nat Commun 9, 1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H, Tang AH, and Blanpied TA (2018). Subsynaptic spatial organization as a regulator of synaptic strength and plasticity. Curr Opin Neurobiol 51, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohme MA, McCarthy AW, Grasskamp AT, Beuschel CB, Goel P, Jusyte M, Laber D, Huang S, Rey U, Petzoldt AG, et al. (2019). Rapid active zone remodeling consolidates presynaptic potentiation. Nature communications 10, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akbergenova Y, Cunningham KL, Zhang YV, Weiss S, and Littleton JT (2018). Characterization of developmental and molecular factors underlying release heterogeneity at Drosophila synapses. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weyhersmuller A, Hallermann S, Wagner N, and Eilers J (2011). Rapid active zone remodeling during synaptic plasticity. J Neurosci 31, 6041–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goel P, Dufour Bergeron D, Bohme MA, Nunnelly L, Lehmann M, Buser C, Walter AM, Sigrist SJ, and Dickman D (2019). Homeostatic scaling of active zone scaffolds maintains global synaptic strength. J Cell Biol 218, 1706–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Piao C, Beuschel CB, Gotz T, and Sigrist SJ (2020). Presynaptic Active Zone Plasticity Encodes Sleep Need in Drosophila. Curr Biol. [DOI] [PubMed] [Google Scholar]
- 43.Chia PH, Patel MR, and Shen K (2012). NAB-1 instructs synapse assembly by linking adhesion molecules and F-actin to active zone proteins. Nat Neurosci 15, 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Driller JH, Lutzkendorf J, Depner H, Siebert M, Kuropka B, Weise C, Piao C, Petzoldt AG, Lehmann M, Stelzl U, et al. (2019). Phosphorylation of the Bruchpilot N-terminus in Drosophila unlocks axonal transport of active zone building blocks. J Cell Sci 132. [DOI] [PubMed] [Google Scholar]
- 45.Kim YJ, Bao H, Bonanno L, Zhang B, and Serpe M (2012). Drosophila Neto is essential for clustering glutamate receptors at the neuromuscular junction. Genes Dev 26, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, et al. (2000). Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287, 864–869. [DOI] [PubMed] [Google Scholar]
- 47.Sigler A, Oh WC, Imig C, Altas B, Kawabe H, Cooper BH, Kwon HB, Rhee JS, and Brose N (2017). Formation and Maintenance of Functional Spines in the Absence of Presynaptic Glutamate Release. Neuron 94, 304–311 e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon HB, and Sabatini BL (2011). Glutamate induces de novo growth of functional spines in developing cortex. Nature 474, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Featherstone DE, Rushton E, and Broadie K (2002). Developmental regulation of glutamate receptor field size by nonvesicular glutamate release. Nat Neurosci 5, 141–146. [DOI] [PubMed] [Google Scholar]
- 50.Saitoe M, Schwarz TL, Umbach JA, Gundersen CB, and Kidokoro Y (2001). Absence of junctional glutamate receptor clusters in Drosophila mutants lacking spontaneous transmitter release. Science 293, 514–517. [DOI] [PubMed] [Google Scholar]
- 51.Schenk U, Verderio C, Benfenati F, and Matteoli M (2003). Regulated delivery of AMPA receptor subunits to the presynaptic membrane. In EMBO J, Volume 22. pp. 558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reddy-Alla S, Bohme MA, Reynolds E, Beis C, Grasskamp AT, Mampell MM, Maglione M, Jusyte M, Rey U, Babikir H, et al. (2017). Stable Positioning of Unc13 Restricts Synaptic Vesicle Fusion to Defined Release Sites to Promote Synchronous Neurotransmission. Neuron 95, 1350–1364 e1312. [DOI] [PubMed] [Google Scholar]
- 53.Herrera AA, and Grinnell AD (1985). Effects of changes in motor unit size on transmitter release at the frog neuromuscular junction. J Neurosci 5, 1896–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickman DK, Lu Z, Meinertzhagen IA, and Schwarz TL (2006). Altered synaptic development and active zone spacing in endocytosis mutants. Current biology : CB 16, 591–598. [DOI] [PubMed] [Google Scholar]
- 55.Choi BJ, Imlach WL, Jiao W, Wolfram V, Wu Y, Grbic M, Cela C, Baines RA, Nitabach MN, and McCabe BD (2014). Miniature neurotransmission regulates Drosophila synaptic structural maturation. Neuron 82, 618–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spinner MA, Walla DA, and Herman TG (2018). Drosophila Syd-1 Has RhoGAP Activity That Is Required for Presynaptic Clustering of Bruchpilot/ELKS but Not Neurexin-1. Genetics 208, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, and Eipper BA (2001). The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229–242. [DOI] [PubMed] [Google Scholar]
- 58.Ryan XP, Alldritt J, Svenningsson P, Allen PB, Wu GY, Nairn AC, and Greengard P (2005). The Rho-specific GEF Lfc interacts with neurabin and spinophilin to regulate dendritic spine morphology. Neuron 47, 85–100. [DOI] [PubMed] [Google Scholar]
- 59.Sigrist SJ, Reiff DF, Thiel PR, Steinert JR, and Schuster CM (2003). Experience-dependent strengthening of Drosophila neuromuscular junctions. J Neurosci 23, 6546–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondo S, and Ueda R (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kittel RJ, Hallermann S, Thomsen S, Wichmann C, Sigrist SJ, and Heckmann M (2006). Active zone assembly and synaptic release. Biochem Soc Trans 34, 939–941. [DOI] [PubMed] [Google Scholar]
- 63.Zhang B, and Stewart B (2010). Electrophysiological recording from Drosophila larval body-wall muscles. Cold Spring Harb Protoc 2010, pdb prot5487. [DOI] [PubMed] [Google Scholar]
- 64.Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, and Goodman CS (2002). wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron 33, 545–558. [DOI] [PubMed] [Google Scholar]
- 65.Schmid A, Qin G, Wichmann C, Kittel RJ, Mertel S, Fouquet W, Schmidt M, Heckmann M, and Sigrist SJ (2006). Non-NMDA-type glutamate receptors are essential for maturation but not for initial assembly of synapses at Drosophila neuromuscular junctions. J Neurosci 26, 11267–11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fulterer A, Andlauer TFM, Ender A, Maglione M, Eyring K, Woitkuhn J, Lehmann M, Matkovic-Rachid T, Geiger JRP, Walter AM, et al. (2018). Active Zone Scaffold Protein Ratios Tune Functional Diversity across Brain Synapses. Cell Rep 23, 1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used for analyses are available on request from the lead contact without restriction.