Abstract
Large-scale efforts to identify breast cancer (BC) risk alleles have historically taken place among women of European ancestry. Recently, there are new efforts to verify if these alleles increase risk in African American (AA) women as well. We investigated the effect of previously reported AA breast cancer and triple-negative breast cancer (TNBC) risk alleles in our African-enriched International Center for the Study of Breast Cancer Subtypes (ICSBCS) cohort. Using case–control, case-series and race-nested approaches, we report that the Duffy-null allele (rs2814778) is associated with TNBC risk (OR = 3.814, p = 0.001), specifically among AA individuals, after adjusting for self-indicated race and west African ancestry (OR = 3.368, p = 0.007). We have also validated the protective effect of the minor allele of the ANKLE1 missense variant rs2363956 among AA for TNBC (OR = 0.420, p = 0.005). Our results suggest that an ancestry-specific Duffy-null allele and differential prevalence of a polymorphic gene variant of ANKLE1 may play a role in TNBC breast cancer outcomes. These findings present opportunities for therapeutic potential and future studies to address race-specific differences in TNBC risk and disease outcome.
Subject terms: Genetics, Cancer, Breast cancer, Cancer epidemiology, Cancer genetics, Risk factors, Genetics research
Introduction
Breast cancer (BC) is caused by a combination of dynamic influences, which are typically unique for each individual, but frequently may include underlying heritable genetic risks. Particularly, breast cancer patients who have early onset, or pre-menopausal incidence, typically are carriers of germline mutations in key cancer genes1,2. However, population studies have shown disparities in BC incidence and mortality among ethnic and racial groups persistently over the past five decades. In the US, White/European Americans (EA) have historically demonstrated the highest incidence of breast cancer, while Black or African Americans (AA) have the highest mortality rates reported in any race/ethnic group3,4. Interestingly, this mortality gap only emerged in the late 1970s, coinciding with implementation of targeted hormone therapies. The consequential decrease of mortality in EA was not matched in AA, which aside from unequal access to these new therapies, unmasked a race-group bias in breast tumor biology and incidence rates of tumor subtypes. Population studies of hormone receptor (HR) status in breast cancer diagnoses indicates a two-fold increased risk of Triple Negative Breast Cancer (TNBC) in AA compared to EA patients, which persists after adjusting for stage and age at diagnosis5–8. This trend also extends beyond certain social determinants, with AA having the highest rate of TNBC at every poverty level as well9. This finding translates to disproportionate survival benefits in EA patients from the standard-of-care targeted therapies that are primarily designed to target HRs10, which AA diagnosed with TNBC are not eligible to receive. Clinically, TNBC is a confirmed adverse prognostic feature in patients overall11, and in AA patients specifically12, and it underscores a need to identify any unique risk of certain breast cancer subtypes. An investigation of genetic risk across self-identified AA groups becomes more informative with the inclusion of an individual’s genetic ancestry composition, as levels of African versus European or other ancestry may be found at varying levels among this admixed population. For example, genetic risk in particular ancestral groups could be unmasked by investigating risk alleles within the predominant ancestral group, as opposed to the traditional risk studies that were devoid of ancestry data13. However, there is a severe shortage of genetic and GWAS data in non-white populations13,14, where less than 10–15% of individuals in population studies are Black, Indigenous, and People of Color (BIPOC), if race or ethnicity groups are reported at all13. This tragic limitation stifles our efforts to identify population-specific risk alleles outside of European descendant groups. However, recent studies have investigated race-specific risk; including, the Multi-Ethnic Cohort (MEC)15, the African American Breast Cancer Epidemiology and Risk (AMBER) Consortium16–19 (which includes the MEC), and our International Center for the Study of Breast Cancer Subtypes (ICSBCS), along with others14,20–22, are paving the way to more inclusion of AA and African participants in genomic research.
Previous studies inferred that AA-specific risk alleles held race-group specificity due to shared African genetic ancestry among AAs15,23. Through our Oncologic Anthropology epidemiological studies of breast cancer incidence and prevalence across the African Diaspora24,25, we have revealed a common trend of lower incidence but higher mortality among women of African descent26. Globally, there is also higher frequency of TNBC among women of western sub-Saharan African descent within every country that has a substantial population of individuals of African descent, and where we could investigate HR status, coupled with higher distribution of poor prognosis in these groups as well7,27–30. This strikingly correlates with the social history and unparalleled numbers of Africans dispersed during forced migrations of the Trans-Atlantic Slave trade, where over hundreds of years and a dozen generations, enslaved Africans were scattered across Europe, the Americas and the Caribbean.
We previously reported our independent analysis of AA race-group specific risk and our previous findings were able to replicate some, but not all, BC and TNBC-specific risk alleles in our African-enriched ICSBCS cohort31. Distinctions in risk associations from hazard models between cohorts could be confounded by bias in shared ancestry, due to differences in composition of genetic admixture among AAs. In this report, we reconsidered our previous risk findings to determine their relevance from a more global perspective, by (i) including additional ancestral populations from contemporary African women, and (ii) adjusting risk models for bias in ancestry background within admixed AAs. These efforts will provide further evidence and methodological insight in the role of shared African ancestry in the shared racial disparity of TNBC incidence across the African diaspora.
Results
Multi-ethnic cohort analysis of population-specific BC risk alleles reaffirms race group specific effects
Our overall BC risk assessment model was an all-inclusive analysis, including all breast cancer subtypes and self-indicated race (SIR)/ancestral groups, where we have expanded the number of BC cases from Eastern and Western African nations, investigating previously published BC risk alleles that have been validated among African American women in the AMBER consortium32 (Tables 1, 2, Fig. 1A (left)). No strong linkage disequilibrium was observed among these alleles (maximum r2 of 0.44). Three alleles replicated previous associations of increased overall BC risk in our unadjusted models. These include rs2981578 (FGFR2), rs4849887 (GLI2), and rs3745185 (BABAM1). Interestingly, we found that the T allele of rs2981578 in the FGFR2 gene was associated with increased risk (OR = 1.508, p = 0.008491), which contrasts with previous reports of the C allele as the risk allele. The C allele of rs4849887 in the GLI2 gene was associated with increased risk (OR = 1.654, p = 0.006122), replicating previous findings. We also replicated the protective A allele of rs3745185 in the BABAM1 gene (OR = 0.67, p = 0.008402).
Table 1.
Population frequencies of candidate alleles for BC and TNBC-specific risk analyses.
| SNP | Associated/neighboring gene | Chr:position | Variant consequence | Minor allele | ICSBCS Cohort* | 1000 Genomes** | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA MAF | EA MAF | G MAF | E MAF | Global MAF | AFR MAF | EUR MAF | |||||
| rs13000023 | TNP1, DIRC3 | 2:217,924,394 | Intron | A | 0.219 | 0.268 | 0.244 | 0.155 | 0.150 | 0.130 | 0.210 |
| rs2363956 | ANKLE1 | 19:17,394,124 | Missense | G | 0.482 | 0.510 | 0.490 | 0.683 | 0.460 | 0.500 | 0.570 |
| rs2981578 | FGFR2 | 10:123,340,311 | Intron | T | 0.271 | 0.516 | 0.364 | 0.250 | 0.370 | 0.080 | 0.480 |
| rs2981579 | FGFR2 | 10:123,337,335 | Intron | G | 0.398 | 0.551 | 0.449 | 0.440 | 0.510 | 0.340 | 0.550 |
| rs3112572 | CASC16, LOC643714 | 16:52,600,447 | Intron | A | 0.216 | 0.011 | 0.246 | 0.207 | 0.120 | 0.270 | 0.030 |
| rs3745185 | BABAM1 | 19:17,384,267 | Intron | A | 0.191 | 0.400 | 0.261 | 0.369 | 0.310 | 0.210 | 0.480 |
| rs4245739 | MDM4 | 1:204,518,842 | 3′ UTR | C | 0.271 | 0.321 | 0.252 | 0.145 | 0.210 | 0.230 | 0.260 |
| rs4849887 | LOC84934, GLI2 | 2:121,245,122 | Intergenic | C | 0.349 | 0.495 | 0.329 | 0.440 | 0.790 | 0.700 | 0.900 |
| rs609275 | MYEOV, CCND1 | 11:69,402,915 | Regulatory | T | 0.491 | 0.037 | 0.552 | 0.085 | 0.210 | 0.490 | 0.000 |
| rs6676002 | DARC/ACKR1 | 1:159,173,144 | Upstream | T | 0.027 | 0.212 | 0.000 | 0.195 | 0.090 | 0.010 | 0.180 |
| rs3027008 | DARC/ACKR1 | 1:159,173,539 | Upstream | T | 0.027 | 0.212 | 0.012 | 0.195 | 0.090 | 0.010 | 0.180 |
| rs3027013 | DARC/ACKR1 | 1:159,174,209 | 5′ UTR | T | 0.022 | 0.087 | 0.000 | 0.110 | 0.030 | 0.000 | 0.080 |
| rs71782098 | DARC/ACKR1 | 1:159,174,347 | 5′ UTR | DEL | 0.080 | 0.021 | 0.103 | 0.060 | 0.030 | 0.100 | 0.000 |
| rs2814778 | DARC/ACKR1 | 1:159,174,683 | 5′ UTR | C | 0.797 | 0.005 | 0.971 | 0.533 | 0.270 | 0.960 | 0.010 |
| rs17838198 | DARC/ACKR1 | 1:159,175,005 | Intron | T | 0.084 | 0.263 | 0.005 | 0.131 | 0.220 | 0.010 | 0.230 |
| rs3027016 | DARC/ACKR1 | 1:159,175,193 | Splice/Intron | G | 0.045 | 0.163 | 0.000 | 0.037 | 0.060 | 0.000 | 0.160 |
| rs12075 | DARC/ACKR1 | 1:159,175,354 | Missense | G | 0.128 | 0.398 | 0.012 | 0.207 | 0.460 | 0.020 | 0.400 |
*African Americans (AA), European Americans (EA), Ghanaians (G), Ethiopians (E).
*MAF = Minor allele frequency.
**1000 Genomes Allele frequencies obtained from ensemble.
Table 2.
Breast cancer risk assessment (case–control) of previously identified variant alleles.
| SNV ID | Associated/neighboring genes | Minor allele | Overall BC risk (all samples) | SIR African Americans | SIR Ghanaians | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Models without covariates | Models with covariates* | Models with covariates** | Models with covariates** | |||||||||||
| N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | |||
| rs13000023 | TNP1, DIRC3 | A | 420 | 1.048 (0.756, 1.449) | 0.775 | 271 | 1.061 (0.601, 1.874) | 0.838 | 104 | – | – | 75 | 0.909 (0.466, 1.770) | 0.780 |
| rs2363956 | ANKLE1 | G | 415 | 0.847 (0.629, 1.140) | 0.273 | 274 | 1.272 (0.718, 2.254) | 0.409 | 103 | 0.916 (0.135, 6.197) | 0.928 | 76 | 1.409 (0.688, 2.890) | 0.349 |
| rs2981578 | FGFR2 | T | 416 | 1.508 (1.111, 2.047) | 0.008 | 267 | 1.036 (0.611, 1.759) | 0.895 | 99 | – | – | 78 | 0.891 (0.481, 1.650) | 0.713 |
| rs2981579 | FGFR2 | G | 418 | 1.246 (0.928, 1.675) | 0.144 | 269 | 1.899 (1.063, 3.393) | 0.030 | 99 | – | – | 77 | 1.792 (0.905, 3.545) | 0.095 |
| rs3112572 | CASC16, LOC643714 | A | 385 | 1.007 (0.691, 1.467) | 0.972 | 246 | 2.410 (1.086, 5.347) | 0.031 | 96 | – | – | 65 | 2.088 (0.948, 4.597) | 0.068 |
| rs3745185 | BABAM1 | A | 409 | 0.666 (0.492, 0.901) | 0.008 | 265 | 0.618 (0.341, 1.119) | 0.112 | 99 | 0.787 (0.104, 5.947) | 0.816 | 77 | 0.590 (0.272, 1.280) | 0.182 |
| rs4245739 | MDM4 | C | 409 | 1.193 (0.860, 1.654) | 0.291 | 264 | 0.995 (0.569, 1.740) | 0.986 | 97 | 0.008 (0.000, 20.16) | 0.228 | 75 | 0.838 (0.427, 1.644) | 0.607 |
| rs4849887 | LOC84934, GLI2 | C | 403 | 1.654 (1.154, 2.371) | 0.006 | 257 | 0.790 (0.351, 1.776) | 0.568 | 100 | 0.399 (0.013, 12.43) | 0.601 | 69 | 0.754 (0.274, 2.073) | 0.584 |
| rs609275 | MYEOV, CCND1 | T | 396 | 1.129 (0.822, 1.552) | 0.453 | 253 | 1.121 (0.532, 2.361) | 0.764 | 98 | 0.224 (0.014, 3.646) | 0.293 | 67 | 0.920 (0.389, 2.176) | 0.849 |
Significant alleles (p value < 0.05) are given in bold and italic
*Overall analysis models with covariates adjusts for age and SIR.
**SIR models with covariates adjusts for age.
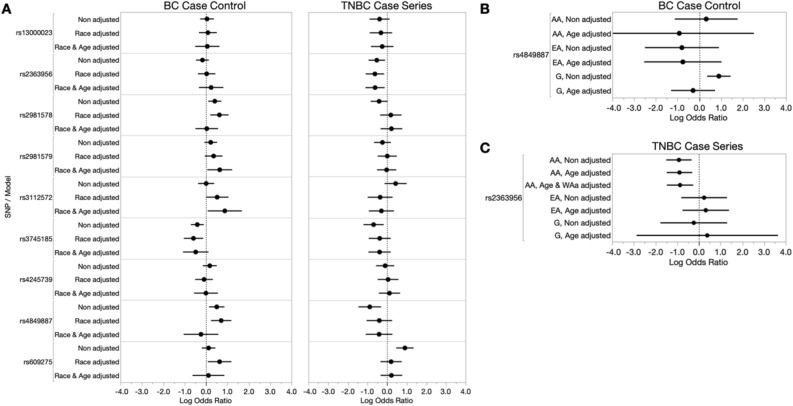
Figure 1.
Breast cancer case–control and TNBC case-series risk analysis of previously identified BC risk alleles among our ICSBCS cohort. (A) The log odds ratio (x-axis) depicting SNV association with BC- or TNBC-risk among all samples is shown in non-adjusted models, and models adjusted for covariates (race and age) in our BC case–control analysis (left) and TNBC case-series analysis (right). (B) Within SIR BC case–control risk analysis for rs4849887. (C) Within SIR TNBC case-series analysis for rs2363956. For both (B) and (C), non-adjusted and age-adjusted models within SIR groups are shown for African Americans (AA), European Americans (EA), and Ghanaians (labelled as G). In our TNBC case-series analysis among SIR AA, we additionally adjusted for West African ancestry (WAa).
To determine whether these all-inclusive association models may be confounded by race-specific bias in age or allele frequency, we adjusted the risk model to correct for race and age. Interestingly, each unadjusted risk association loses significance in the combined race group model after adjusting for race and age, indicating that the risk alleles may have higher frequency in one of the SIR groups (See Table 1). Specifically, in the case of the risk (C) allele of rs4849887, we find it is 10–15% lower in populations of West African descent (AA = 34.9%, Ghanaians = 32.9%), compared to European Americans (49.5%) and East Africans (44.0%) in our cohort. Two additional alleles gained significance in overall BC risk associations after race and age adjustments in our all-inclusive model, rs2981579 in the FGFR2 gene (OR = 1.899, p = 0.03038) and rs3112572 in the LOC643714 gene (OR = 2.410, p = 0.03055).
Next, we tested whether the associated BC risk of our candidate alleles was different among SIR groups by performing a nested BC risk assessment within each of the SIR groups (Table 2 and Supplemental Table 1). While we observed rs4849887 was associated with overall BC risk prior to adjusting for age and race, this allele is associated with higher overall BC risk only in Ghanaians prior to adjusting for age (OR = 2.472, p = 0.001032) (Fig. 1B, Supplemental Table 1). While we did not observe a significant association between rs609275 and overall BC risk for the whole cohort assessment, a very high overall BC risk was observed specifically for AA prior to adjusting for age (OR = 5.383, p = 0.048). There were no significant associations found between the previously identified variants and breast cancer risk among SIR EA in both unadjusted and age-adjusted models (Supplemental Table 1).
TNBC-specific case-series analysis of population-specific BC risk alleles shows associations within ancestral groups
The higher rate of TNBC among women of African descent worldwide begs the question of whether there is a shared genetic risk among the African diaspora, and we have previously shown that quantified West African ancestry was strongly associated with TNBC disease31. Using a case-series analysis in our African-enriched cohort, we tested whether previously reported AA-specific risk alleles were associated specifically with TNBC disease risk (Table 3, Supplemental Table 2, Fig. 1A (right)). Prior to adjusted covariate modeling, five of the nine AA-risk variants showed significant association with TNBC disease risk. Four of these variants were not previously reported as having ER-negative disease specific risk, and four were predicted to have a protective effect; including, rs2981578 in FGFR2 (OR = 0.667, p = 0.0627), rs3745185 in BABAM1 (OR = 0.503, p = 0.009), rs4849887 in GLI2 (OR = 0.414, p = 0.003), and rs2363956 in ANKLE1 (OR = 0.593, p = 0.0149). Only the SNV rs609275 in MYEOV/CCND1 showed higher hazard/risk for TNBC in the unadjusted model (OR = 2.479, p = 5.68E-05). The ANKLE1 variant rs2363956 replicated in the TNBC/ER-negative specific protective effect that was previously reported and was the only variant to retain significance after adjusting for race and age (OR = 0.542, p = 0.014).
Table 3.
TNBC-specific risk assessment (case-series) of previously identified variant alleles.
| SNV ID | Associated/neighboring genes | Minor allele | Overall TNBC risk (all samples) | SIR African Americans | SIR Ghanaians | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Models without covariates | Models with covariates* | Models with covariates** | Models with covariates*** | |||||||||||
| N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | |||
| rs13000023 | TNP1, DIRC3 | A | 197 | 0.682 (0.410, 1.133) | 0.139 | 190 | 0.781 (0.439, 1.387) | 0.399 | 96 | 1.109 (0.538, 2.286) | 0.780 | 6 | – | – |
| rs2363956 | ANKLE1 | G | 201 | 0.593 (0.389, 0.903) | 0.015 | 194 | 0.542 (0.332, 0.883) | 0.014 | 95 | 0.420 (0.230, 0.769) | 0.005 | 8 | 1.471 (0.057, 38.26) | 0.816 |
| rs2981578 | FGFR2 | T | 190 | 0.667 (0.435, 1.022) | 0.063 | 183 | 1.248 (0.718, 2.169) | 0.432 | 92 | 1.304 (0.656, 2.590) | 0.449 | 6 | 17.55 (0.010, 29,760) | 0.450 |
| rs2981579 | FGFR2 | G | 193 | 0.790 (0.515, 1.212) | 0.280 | 186 | 0.978 (0.599, 1.597) | 0.930 | 92 | 0.942 (0.530, 1.672) | 0.838 | 6 | 6.261E−08 (0, –) | 0.993 |
| rs3112572 | CASC16, LOC643714 | A | 181 | 1.546 (0.880, 2.716) | 0.130 | 174 | 0.748 (0.394, 1.421) | 0.375 | 89 | 0.714 (0.367, 1.390) | 0.322 | 5 | 6.976E−49 (0, –) | 0.976 |
| rs3745185 | BABAM1 | A | 189 | 0.503 (0.306, 0.843) | 0.009 | 182 | 0.682 (0.386, 1.203) | 0.180 | 92 | 0.584 (0.288, 1.182) | 0.135 | 6 | 3.304 (0.090, 120.90) | 0.515 |
| rs4245739 | MDM4 | C | 190 | 0.906 (0.571, 1.438) | 0.675 | 183 | 1.130 (0.658, 1.941) | 0.657 | 90 | 0.861 (0.440, 1.685) | 0.663 | 6 | 0.356 (0.004, 34.530) | 0.658 |
| rs4849887 | LOC84934, GLI2 | C | 189 | 0.414 (0.232, 0.738) | 0.003 | 182 | 0.666 (0.338, 1.313) | 0.240 | 93 | 0.542 (0.241, 1.221) | 0.140 | 6 | 0.676 (0.0470, 9.710) | 0.773 |
| rs609275 | MYEOV, CCND1 | T | 187 | 2.479 (1.593, 3.857) | < 0.001 | 180 | 1.245 (0.717, 2.163) | 0.437 | 91 | 1.121 (0.610, 2.061) | 0.714 | 6 | 14.23 (0.0261, 7750) | 0.409 |
Significant alleles (p value < 0.05) are given in bold and italic
*Overall analysis models with covariates adjusts for age and SIR.
**SIR AA models with covariates adjusts for age and West African ancestry.
***SIR Ghanaian models with covariates adjusts for age.
Similar to our BC case–control analysis, we used a nested risk analysis within SIR groups to test for SIR-specific risk. For the admixed AA population, we included quantified West African ancestry (WAa) in the adjusted covariate modeling. The rs2363956 variant in the ANKLE1 gene retained a protective effect for TNBC in AAs, even after covariate adjustments, (age and WAa adjusted OR = 0.4204, p = 0.005), indicating this is not a mere artifact of disequilibrium, or biased distribution of the allele in African populations (Fig. 1C and Table 3).
DARC/ACKR1 alleles in BC and TNBC risk
In addition to the previously implicated AA-risk alleles, we have also included DARC/ACKR1 alleles, including the TNBC risk associated Duffy-null allele31, to investigate whether alternative variants may capture risk due to unique biological contributions of either isoforms or distinct gene regulation (Table 1). Our new analysis found that four DARC/ACKR1 SNVs also had significant potential to confer overall BC risk in our all-inclusive analysis models (rs2814778 OR = 1.512, p < 0.001, rs17838198, OR = 4.798, p < 0.001, rs3027016 OR = 4.586, p = 0.005 and rs12075 OR = 2.534, p < 0.001, respectively), however, after adjusting for age and race, this is mostly lost (Table 4, Fig. 2A (left)). In our SIR nested analysis model, the DARC/ACKR1 variant rs3027013 showed a significant protective effect in EA patients, even after age-adjusted modeling (age-adjusted OR = 0.131, p = 0.03897) (Fig. 2B and Supplemental Table 3).
Table 4.
Breast cancer risk assessment (case–control) of DARC/ACKR1 alleles.
| SNV ID | Associated/neighboring genes | Minor allele | Overall BC risk (all samples) | SIR African Americans | SIR Ghanaians | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Models without covariates | Models with covariates* | Models with covariates** | Models with covariates** | |||||||||||
| N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | |||
| rs6676002 | DARC/ACKR1 | T | 282 | 1.290 (0.702, 2.368) | 0.412 | 231 | 0.634 (0.164, 2.454) | 0.509 | 96 | – | – | 47 | – | – |
| rs3027008 | DARC/ACKR1 | T | 282 | 1.187 (0.661, 2.131) | 0.566 | 231 | 0.737 (0.230, 2.369) | 0.609 | 96 | – | – | 49 | 2.270 (0.277, 18.60) | 0.445 |
| rs3027013 | DARC/ACKR1 | T | 283 | 0.869 (0.372, 2.033) | 0.747 | 231 | 0.250 (0.050, 1.241) | 0.090 | 96 | – | – | 49 | – | – |
| rs71782098 | DARC/ACKR1 | DEL | 291 | 0.575 (0.307, 1.077) | 0.084 | 238 | 0.753 (0.280, 2.028) | 0.575 | 96 | – | – | 52 | 0.882 (0.298, 2.608) | 0.820 |
| rs2814778 | DARC/ACKR1 | C | 712 | 1.512 (1.263, 1.809) | < 0.001 | 492 | 0.772 (0.392, 1.520) | 0.454 | 153 | 0.696 (0.330, 1.467) | 0.340 | 54 | 3.657 (0.231, 57.81) | 0.357 |
| rs17838198 | DARC/ACKR1 | T | 299 | 4.798 (2.125, 10.83) | < 0.001 | 244 | 3.413 (0.678, 17.20) | 0.137 | 97 | 1.052 (0.044, 25.41) | 0.975 | 58 | – | – |
| rs3027016 | DARC/ACKR1 | G | 281 | 4.586 (1.587, 13.26) | 0.005 | 229 | 2.311 (0.269, 19.88) | 0.446 | 96 | – | – | 47 | – | – |
| rs12075 | DARC/ACKR1 | G | 292 | 2.534 (1.498, 4.287) | < 0.001 | 238 | 1.131 (0.382, 3.351) | 0.824 | 97 | 1.108 (0.048, 25.80) | 0.949 | 53 | – | – |
Significant alleles (p value < 0.05) are given in bold and italic
*Overall analysis models with covariates adjusts for age and SIR.
**SIR models with covariates adjusts for age.
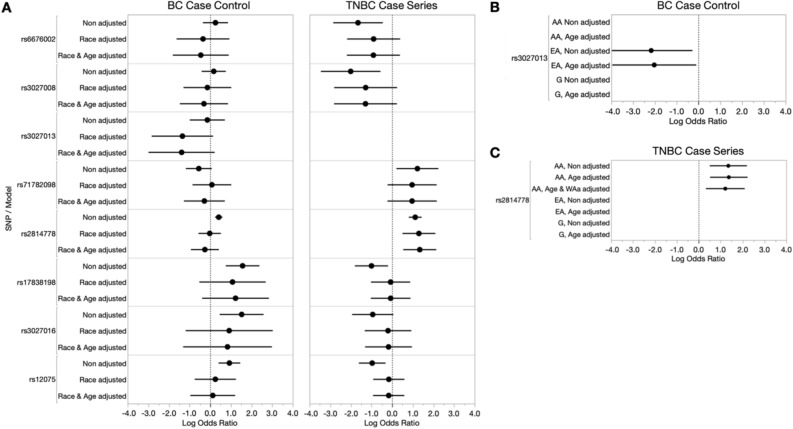
Figure 2.
Breast cancer case–control and TNBC case-series risk analysis of DARC/ACKR1 alleles among our ICSBCS cohort. (A) The log odds ratio (x-axis) depicting SNV association with BC- or TNBC-risk among all samples is shown in non-adjusted models, and models adjusted for covariates (race and age) in our BC case–control analysis (left) and TNBC case-series analysis (right). (B) Within SIR BC case–control analysis for rs3027013. (C) Within SIR TNBC case-series analysis for rs2814778. For both (B) and (C), non-adjusted and age-adjusted models within SIR groups are shown for African Americans (AA), European Americans (EA), and Ghanaians (labelled as G). In our TNBC case-series analysis among SIR AA, we additionally adjusted for West African ancestry (WAa).
For DARC/ACKR1 variant associations in TNBC-specific risk, we similarly observed that seven out of eight variants were associated with TNBC disease, in which five of the minor alleles presented a protective effect and two showed increased risk, prior to race/age adjustments (rs6676002, OR = 0.191, p = 0.007; rs3027008, OR = 0.134, p = 0.006; rs17838198, OR = 0.367, p = 0.015; rs3027016, OR = 0.390, p = 0.065; rs12075, OR = 0.380, p = 0.003, rs71782098, OR = 3.403, p = 0.018; and rs2814778, OR = 3.062, p < 0.001) (Table 5, Fig. 2A (right)). Interestingly, as we previously reported with only AA and EA, the Duffy-Null allele, rs2814778, retained significant TNBC-risk association with the addition of West African samples, even after age and SIR adjustments (OR = 3.814, p = 0.001). The Duffy-Null (rs2814778) TNBC-risk association was also retained in our nested SIR analysis among AA, following both age and quantified West African ancestry adjustment (OR = 3.368, p = 0.007) (Fig. 2C and Table 5). This indicates that the TNBC-specific risk conferred by the Duffy-null allele in the DARC/ACKR1 gene is not an artifact of shared ancestry bias, but rather an ancestry-specific risk allele.
Table 5.
TNBC-specific risk assessment (case-series) of DARC/ACKR1 alleles.
| SNV ID | Associated/neighboring genes | Minor Allele | Overall TNBC risk (all samples) | SIR African Americans | SIR Ghanaians | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Models without covariates | Models with covariates* | Models with covariates** Models with covariates** | Models with covariates*** | |||||||||||
| N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | N | OR (95% CI) | P value | |||
| rs6676002 | DARC/ACKR1 | T | 176 | 0.191 (0.058, 0.635) | 0.007 | 175 | 0.403 (0.111, 1.460) | 0.167 | 90 | – | – | 2 | – | – |
| rs3027008 | DARC/ACKR1 | T | 174 | 0.134 (0.032, 0.568) | 0.006 | 173 | 0.275 (0.060, 1.261) | 0.097 | 90 | – | – | 2 | – | – |
| rs3027013 | DARC/ACKR1 | T | 174 | – | – | 173 | – | – | 90 | – | – | 2 | – | – |
| rs71782098 | DARC/ACKR1 | DEL | 178 | 3.403 (1.231, 9.412) | 0.018 | 177 | 2.629 (0.796, 8.682) | 0.112 | 90 | 2.547 (0.668, 9.716) | 0.171 | 2 | – | – |
| rs2814778 | DARC/ACKR1 | C | 339 | 3.062 (2.249, 4.168) | < 0.001 | 304 | 3.814 (1.710, 8.493) | 0.001 | 95 | 3.368 (1.390, 8.165) | 0.007 | 16 | – | – |
| rs17838198 | DARC/ACKR1 | T | 178 | 0.367 (0.164, 0.821) | 0.015 | 177 | 0.929 (0.355, 2.430) | 0.881 | 91 | 0.722 (0.190, 2.754) | 0.634 | 2 | – | – |
| rs3027016 | DARC/ACKR1 | G | 174 | 0.390 (0.144, 1.058) | 0.065 | 173 | 0.839 (0.270, 2.609) | 0.762 | 90 | 1.204 (0.226, 6.422) | 0.828 | 2 | – | – |
| rs12075 | DARC/ACKR1 | G | 177 | 0.380 (0.199, 0.726) | 0.003 | 176 | 0.846 (0.396, 1.807) | 0.666 | 91 | 0.922 (0.319, 2.669) | 0.881 | 2 | – | – |
Significant alleles (p value < 0.05) are given in bold and italic
*Overall analysis models with covariates adjusts for age and SIR.
**SIR AA models with covariates adjusts for age and West African ancestry.
***SIR Ghanaian models with covariates adjusts for age.
Functional consequences of the TNBC-protective rs2362956 variant in ANKLE1
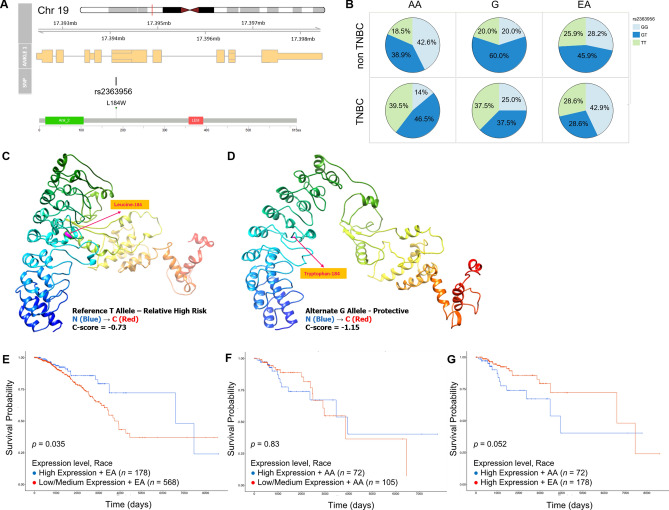
In our TNBC risk analysis, we found that the minor G allele of the rs2363956 ANKLE1 variant was protective against TNBC disease, which has previously been shown for ER-negative disease among AA32. Given its SIR-specific effect, we investigated the frequency of the allele across global 1000 genomes (1 KG) populations33. Population minor allele frequency (MAF) of the protective G allele is relatively equal among European and African groups (57% vs 50%, respectively, Table 1). However, among TNBC cases in our ICSBCS cohort, the frequency of the GG genotype is much lower in AA patients, compared to EA patients (14% and 43%, respectively) (Fig. 3B). This 20% drop in the minor allele frequency in TNBC cases among AA is what explains the interpreted potentially protective effect of the minor allele, inferring the major allele may somehow drive TNBC frequency higher in AAs (MAFEA = 57.1%, MAFAA = 37.2%).
Figure 3.
Functional implications of the ANKLE1 variant rs2363956. (A) rs2363956 is a coding region variant of the ANKLE1 gene, located at 19p13.11. This missense variant encodes a leucine to tryptophan change at amino acid position 184 (ANKLE1 protein domain model shown from cBioPortal61). (B) Genotype frequency pie charts of the rs2363956 allele among SIR African Americans (AA), SIR Ghanaian (G) and SIR European American (EA) individuals. Non-TNBC cases are shown in the top row, and TNBC cases are in the bottom row. Those individuals homozygous for the protective/minor G allele are shown in light blue, heterozygotes are dark blue, and individuals homozygous for the major T allele are in light green. (C) Illustration of the predicted 3D ANKLE1 protein structure from I-TASSER using Chimera with leucine at position 184 (representing the reference allele), and (D) with tryptophan at position 184 (representing the missense rs2363956 G allele). For both C and D, confidence score (C-score) > − 1.5 indicates a model of correct global topology. The 3D structure follows rainbow coloring, where blue coloring represents the N-terminus, and red indicates the C-terminus. Kaplan Meier curves comparing ANKLE1 gene expression and overall survival outcomes between low/medium and high ANKLE1 expressing (E) EA and (F) AA, where high expression is shown in blue, and low/medium expression is shown in red. (G) KM curves comparing of overall survival between high expressing AA (blue) and high expressing EA (red). For (E–G), N values are reported for each comparison group, and the p value is reported on the plot.
To date, despite being repeatedly reported as a risk allele in both breast and ovarian cancer32,34,35, no investigation has linked a functional impact of this variant to risk or survival in this population. Given that the variant causes a dramatic amino acid change of leucine to tryptophan (L184W, Fig. 3A), there is a high probability that the protein structure is impacted, and subsequently have altered the function. We conducted a 3D rendering of the variant, comparing the structure of the protein with leucine at position 184 (Fig. 3C) to the minor allele change to tryptophan, and found a predicted destabilization of the gene product (Fig. 3D).
The allele’s protective effect through destabilization of ANKLE1 structure, together with its significant loss in AAs who suffer from higher rates of TNBC, suggests the major allele ANKLE1 protein could be a genetic driver of TNBC. We hypothesize that wildtype ANKLE1 expression suppresses TNBC progression, which is most frequently found in EA patients when caused by the rs2363956 variant. To further investigate this theory, we determined whether the expression of ANKLE1 had any impact on survival36. We found that survival trends in TCGA breast cancer cases are significantly impacted by ANKLE1 expression, but that the advantage of ANKLE1 expression only benefits EA patients (Fig. 3E–G). Specifically, we found that when comparing high vs low/medium ANKLE1 expression within SIR groups, EA have a significant survival improvement associated with higher expression (p = 0.035), but AA did not (p = 0.83) (Fig. 3E–F). In fact, when only including patients who had high expression of ANKLE1, EA had a longer survival advantage associated with ANKLE1, compared to AA (Fig. 3G, p = 0.052). This suggests that the benefit of ANKLE1, only found in EA, could be due to the 41–53% chance that EA are expressing the polymorphic version of ANKLE1, which harbors the rs2363956 allele.
Discussion
While recent findings have delineated breast cancer risk alleles that pose increased or even decreased risk in African Americans specifically, many of these findings do not always replicate in other independent multi-ethnic cohorts. This is likely because of unmeasured individual admixture among the non-white individuals, who through social history are of mixed ancestry (i.e. Caribbean, Latin American and AAs) resulting from recent genetic admixture originating from multiple ancestor lineages37–39. This complexity of AA ancestry includes heterogeneity of African origins, spanning multiple African parental lineages through dozens of generations. This undoubtedly creates confounding genetic backgrounds that still pose a significant obstacle in identifying causal risk alleles among “African” Americans. However, measuring this genetic and ancestral diversity, and accounting for ancestry substructure would be a key first step toward clarifying the alleles that may be shared among individuals of common ancestry within SIR groups who display common disease/tumor types. Our latest race and West African ancestry adjustments in risk models demonstrate the power of combining diverse ancestral groups and utilizing ancestry estimates to clarify either false-positive or false-negative results if models do not properly consider the underlying ancestry/genetic background of the cohorts.
Our work represents a uniquely powered cohort that is enriched with a diverse cohort of patients and controls of African ancestry to directly investigate the impact of shared African ancestry in genetic risk for TNBC. We anticipate that our observations account for increased prevalence in women of African descent, at least in part. However, our analysis is still limited by the paucity of hormone receptor status in African cases and so the limited number of patients we can include in this analysis, thus far. Despite this limitation, we have robust findings that are compelling to expound upon in follow-up molecular and clinical studies.
First, our intention to replicate and verify the findings of AA-specific risk alleles is somewhat tenuous with associations fluctuating after adjustments for age and/or race. These covariate adjustments altering significance reflect the varying frequency of these alleles across these strata in our cohort and possibly more broadly in the population. Specifically, rs2981578, rs3745195 and rs4849887 were found to be significant prior to and after race adjustment, and lost significance with age adjustment, while rs2981579 and rs3112572 were found to be significant after race and age adjustment. For alleles that are in significantly different frequency across age categories, their distribution may reflect a difference in early vs. late onset cancers. For alleles that have significantly different frequency across race categories, their distribution may reflect ancestry-specific risk or population-private variants. Either scenario warrants a larger and more inclusive dataset to uncover genetic risk, robustly. This is an unmet need that could be essential to cancer prevention and much needed improvement for cancer risk prediction models.
We have validated our previous finding31 of the Duffy-null allele (rs2814778) as a TNBC-risk allele in our SIR all-inclusive analysis (OR = 3.814, p = 0.001). The Duffy-null allele is an ancestry-specific allele restricted to descendants of Sub-Saharan Africans. The allele arose among Sub-Saharan Africans and removed expression of DARC from erythrocytes, lending immunity from Plasmodium vivax malaria, as this malaria parasite utilized DARC as a portal of entry into erythrocytes40,41. The allele quickly swept to fixation across this population and is found at nearly ~ 100% among West Africans, and ~ 80% among AAs42,43. With the associations between WAa and TNBC that we and others have reported31,44, the potential association of the Duffy-null allele and TNBC is of great interest. With our expanded cohort analysis, we were able to perform the TNBC case-series risk assessment among SIR AAs only, and found that the risk was significantly retained among AA women after adjusting for both age and WAa (OR = 3.368, p = 0.007). This highlights that the Duffy null allele represents an ancestry-specific TNBC risk allele, and that the findings in our SIR all-inclusive analysis were not driven by ancestry-bias in our cohort. This is an important finding among our cohort, as the Duffy-null allele would not have been identified among previous GWAS studies underpowered with individuals of African ancestry.
Second, we have investigated the consequences of the protective rs2363956 variant on the ANKLE1 gene coding region and uncovered a potential functional reason for race-group risk distinction. The allele has repeatedly been associated with breast and ovarian cancer risk and survival34,35, and this association has been replicated among AA women32. In the present analysis, we are the first to report that the ‘protective’ polymorphic ANKLE1 would be the more likely version expressed in EA patients, compared to AA or Ghanaian patients (GG genotype, 43%, 14% and 25%, respectively) (Fig. 3B). This suggests that the major T allele corresponds to a TNBC-specific oncogenic version of the ANKLE1 gene. The potential mechanism of action for increased survival would appear to be DNA damage response, as ANKLE1 has repeatedly been shown to be involved in DNA repair pathways in pre-clinical and ex vivo screening, including endonuclease activity45,46, proliferation, and drug response hits in CRISPR screens in cancer cell lines47–50. Most intriguingly, one study in non-small-cell lung cancer indicated the combination of ANKLE1 RNAi with paclitaxel increased the efficacy of the drug response51. Altogether, this is a very promising avenue for further investigation of targeted/combinatorial therapy, with potential to be transformative in treatment of TNBC, and with specific impact in AA who have higher expression of ANKLE1.
If validated through additional clinical studies, finding a novel oncogene specific to TNBC could be transformative in two ways: (i) to improve genetic risk models or create AA-tailored risk models, and (ii) to develop prognostic tests to inform survival prediction models, which currently do not include information about ANKLE1. Specifically, if we find that the patients who have longer survival carry the minor protective allele, correlated with higher expression of this polymorphic ANKLE1, we can quickly investigate if this is ultimately related to treatment response. Our preliminary data on survival trends certainly suggests this could be true.
The reported, albeit controversial, findings of TNBC mortality differences between women of African descent compared to women of European descent may be an important indicator of unknown differences in tumor biology. Here, we show that ANKLE1 expression is linked to distinct survival outcomes, and this could potentially be linked to this polymorphic version of the ANKLE1 gene. Intriguingly, this corresponds with differential impact of the gene’s expression on survival when comparing race groups among patients with high expression of the gene. While the functional consequence on mechanistic change is yet unknown, it is a clear indicator of survival and therefore a prognostic indicator. Excitingly, this also reveals a potential opportunity to develop immune-based inhibition of the oncogenic (major allele) version that is more likely expressed in AA. As the frequency of the oncogenic ANKLE1 allele is higher in AA populations, this could present an opportunity for additional research to address its potential in precision therapies to bridge the survival gap in TNBC among race groups. Inclusion of diverse cohorts have powered this discovery and will drive clinical applications in the future.
Methods
International center for the study of breast cancer subtypes
The mission of the International Center for the Study of Breast Cancer Subtypes (ICSBCS) is to reduce the global breast cancer burden through advances in research and delivery of care to diverse populations worldwide. The ICSBCS brings together an international consortium of breast cancer clinicians and researchers, all of whom share the goal of addressing genetic and phenotypic variation in breast cancer risk and survival outcomes. We accrued prospective breast cancer patients from 2013 to 2017 as previously described31, extracting germline DNA from saliva samples collected at the time of consent at Komfo Anokye Teaching Hospital (KATH) in Kumasi, Ghana (N = 120), and St. Paul’s Millennium Hospital Medical College in Addis Ababa, Ethiopia. Additional cancer patient samples were collected at the Henry Ford Health System Hospital in Detroit, Michigan, and the University Cancer and Blood Center in Athens, GA (NAA = 192 and NEA = 184). The mean age is 47 ± 15.4 (mean ± sd) for Ghanaian patients, 59 ± 12.8 for AA and 60 ± 12.1 for EA. Healthy controls (N = 271) were recruited to the ICSBCS biospecimen registry through various sources of community engagement efforts throughout the US52 and the breast cancer screening clinic at KATH22. Informed consent was obtained from all individuals participating in the study, which was approved and under the regulation of the Weill Cornell Medical College (WCM) Institutional Review Board (IRB; protocol number 1807019405). All experiments were performed in accordance with the approved IRB protocol.
Immunohistochemistry for BC tumor subtyping
For our TNBC case-series risk analysis, we determined hormone receptor status in our ICSBCS biospecimen registry via immunohistochemistry (IHC) methods that were described in detail in our previous study31. Expression of biomarkers was interpreted in accordance with the American Society of Clinical Oncology/College of American Pathologists guidelines53,54. Briefly, for estrogen and progesterone receptor IHC, staining of at least 1% was determined as positive. HER2/neu staining score of 0 or 1 + was determined as negative, and 3 + was determined as positive. HER2/neu staining score of 2 + was deemed equivocal and was further evaluated by fluorescent in situ hybridization. ICSBCS cases accrued in the USA were reviewed by the treating facility. IHC and pathology review of Ghanaian and Ethiopian cases was completed in Michigan (University of Michigan and Henry Ford Health System Hospital) and New York (Weill Cornell Medicine).
Allele selection for BC case–control and TNBC case-series analyses
In our previous publication, we investigated nine reported AA BC risk variants in our African-enriched ICSBCS cohort, to determine BC or TNBC-specific risk within self-identified race (SIR) groups in our cohort. We additionally included the Duffy-Null allele (rs2814778), a promoter region variant of the DARC/ACKR1 gene in our panel and demonstrated this allele to be a TNBC-specific risk allele among AA. Building upon our previous findings, we have both increased our number of samples across our SIR groups with genotypes available, and included an additional eight DARC/ACKR1 gene variants in our panel that are implicated as ancestry-specific alleles, or sit in regions that are potentially involved in DARC/ACKR1 gene regulation. These eight DARC/ACKR1 gene variants represent upstream variants, 5′ UTR variants, and variants in the coding region of the gene. All alleles that were assessed in subsequent analyses are described in Table 1. Additionally, our African-enriched ICSBCS cohort allows us to also incorporate African ancestry measurements into the association model (below). PLINK (version 2.0)55 was used to assess linkage disequilibrium among these alleles, and no strong linkage disequilibrium was observed (maximum r2 of 0.44).
Global ancestry estimation and genotyping of candidate alleles
Methods to determine global genetic ancestry have been previously reported in detail31,56. Briefly, DNA extracted from saliva samples were genotyped on the Sequenom MassARRAY iPLEX platform using an AIMs panel containing 100 markers specifically selected and validated for estimating continental ancestry among admixed populations57,58. The Sequenom TYPER software (version 4.0) was used for genotype calls, and STRUCTURE (version 2.3) was used to calculate admixture estimates for each individual59.
Similar to our global ancestry estimations, to obtain genotypes for our candidate variants for risk analyses (Table 1), DNA from saliva samples were genotyped for each of the variants using the Sequenom platform. For the Duffy-Null allele (rs2814778), we have obtained additional genotypes using single-target allele amplification reactions, as previously described31.
Risk assessment
From our genotyping data, we used PLINK (version 2.0)55 to determine associations between the candidate variants and breast cancer risk in case–control analysis model, and TNBC-specific risk in case-series analysis model as previously described31. In both our BC and TNBC-specific risk analyses, we performed associations without covariates (non-adjusted), with SIR adjustment, and with SIR and age adjustments. We additionally investigated variant and risk associations within each SIR race group, where we performed analyses for non-adjusted and age-adjustments. For our analysis within SIR AA, using the genetic ancestry estimates, we were additionally able to adjust for West African ancestry in our models. For the candidate variants, we conducted the risk association using both a dominant and dosage statistical model31. In the dominance model where the genotypes are AA, Aa, aa (where a is minor allele), the resulting genotypes would be coded as 0, 1, 1 in the analysis model, where risk is weighted based on having at least one minor dominant allele. In the dosage model using the same genotypes, the resulting genotypes would be coded as 0, 1, 2, where the risk is weighted by the number of minor alleles present. In the main figures and tables, we show and discuss risk assessment output from the dosage models, where the full range of genotypes is considered in the analysis. In addition, the Benjamini–Hochberg method was used to adjust for multiple comparisons while controlling false discovery rate (FDR) at 0.05. FDR adjusted p values for Tables 2, 3, 4, and 5 are shown in Supplemental Tables 5–8, respectively.
For both the BC and TNBC-specific analyses, odds ratio output from the dosage risk assessment analyses were log transformed and plotted using the Forest Plot add-in (v8) within JMP Pro 15.0.0 statistical software (SAS Institute Inc., Cary, NC, 1989–2019).
3D modeling of ANKLE1 protein
We used the cBioPortal MutationMapper online program to visualize the ANKLE1 protective variant rs2363956 in the context of the protein domain structure60,61. For 3D modeling of the wild type and rs2363956 missense variant, the ANKLE1 amino acid sequence in FASTA format was obtained from NCBI using the GrCh37.p13 reference and was submitted to I-TASSER62–64. The amino acid sequence is 615 residues long, and we performed 3D modeling to obtain the structure with and without the ANKLE1 missense mutation included in our candidate variant analysis (rs2363956, L184W). The estimate of the accuracy of the predictions using I-TASSER is provided based on the confidence score (C-score) of the modeling. The C-score range is between [− 5, 2], where a C-score of a higher value suggests a model with higher confidence and vice-versa. Furthermore, Chimera program65 (version 1.14) was used for visualization and analysis of the predicted 3D ANKLE1 protein structure from I-TASSER.
ANKLE1 survival analysis
The UALCAN online database was accessed to determine potential associations between gene expression and patient survival outcomes in the TCGA BC cohort36. ANKLE1 gene expression was assessed across the patient cohort, and the upper quartile of expression was used to dichotomize expression into high and low/medium ANKLE1 expressing individuals. The log rank p value obtained between comparison groups is reported on the plots.
Supplementary Information
Acknowledgements
Funding—R21 5R21CA210237-03 NIH/NCI (to MD), Susan G Komen (to LN), Fashion Footwear Association of New York Shoes on Sale (to LN), U54-MD007585-26 NIH/NIMHD (to CY) and U54 CA118623 (NIH/NCI) (to CY). We would like to acknowledge all of our ICSBCS team members who assisted with consent, biospecimen aggregation and logistics, and of course all breast cancer patients and healthy control volunteers for consenting and contributing to this important work.
Author contributions
Y.C., L.N. and M.D. designed the study. R.M., Y.C., B.J., I.E., J.H., R.Z., C.Y., R.K. and M.D. were involved in the methodology. R.M., Y.C. and M.D. analyzed and interpreted the data. R.M., L.N. and M.D. wrote and edited the manuscript. E.C., S.H., P.G., J.O., E.A., A.J., D.C., J.B., B.A., M.B., E.A., I.K., F.A., M.A., K.A., E.O., S.N., L.J., E.J., L.P., E.P., P.N., K.G. are all affiliated with the International Center for the Study of Breast Cancer Subtypes (ICSBCS) Consortium. ICSBCS consortium authors contributed targeted enrollment, biospecimen and data collection, and review of the manuscript. All authors have read and approved the final submitted manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88613-w.
References
- 1.Rummel SK, Lovejoy L, Shriver CD, Ellsworth RE. Contribution of germline mutations in cancer predisposition genes to tumor etiology in young women diagnosed with invasive breast cancer. Breast Cancer Res. Treat. 2017;164:593–601. doi: 10.1007/s10549-017-4291-8. [DOI] [PubMed] [Google Scholar]
- 2.Kudela E, et al. Breast cancer in young women: Status quo and advanced disease management by a predictive, preventive, and personalized approach. Cancers. 2019;11:1791. doi: 10.3390/cancers11111791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeSantis CE, Miller KD, GodingSauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J. Clin. 2019;69:211–233. doi: 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 4.Hunt BR, Silva A, Lock D, Hurlbert M. Predictors of breast cancer mortality among white and black women in large United States cities: An ecologic study. Cancer Causes Control. 2019;30:149–164. doi: 10.1007/s10552-018-1125-x. [DOI] [PubMed] [Google Scholar]
- 5.Amirikia KC, Mills P, Bush J, Newman LA. Higher population-based incidence rates of triple-negative breast cancer among young African-American women: Implications for breast cancer screening recommendations. Cancer. 2011;117:2747–2753. doi: 10.1002/cncr.25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Li CI. Racial disparities in breast cancer diagnosis and treatment by hormone receptor and HER2 status. Cancer Epidemiol. Biomarkers Prev. 2015;24:1666–1672. doi: 10.1158/1055-9965.EPI-15-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohler BA, et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J. Natl. Cancer Inst. 2015;107:048. doi: 10.1093/jnci/djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garlapati C, Joshi S, Sahoo B, Kapoor S, Aneja R. The persisting puzzle of racial disparity in triple negative breast cancer: Looking through a new lens. Front. Biosci. 2019;11:75–88. doi: 10.2741/s527. [DOI] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 10.Newman LA. Parsing the etiology of breast cancer disparities. J Clin Oncol. 2016;34:1013–1014. doi: 10.1200/JCO.2015.65.1877. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res. Treat. 2017;161(2):279–287. doi: 10.1007/s10549-016-4059-6. [DOI] [PubMed] [Google Scholar]
- 12.Akinyemiju T, Moore JX, Altekruse SF. Breast cancer survival in African-American women by hormone receptor subtypes. Breast Cancer Res Treat. 2015;153:211–218. doi: 10.1007/s10549-015-3528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Palmer JR, et al. Genetic susceptibility loci for subtypes of breast cancer in an African American population. Cancer Epidemiol. Biomarkers Prev. 2013;22:127–134. doi: 10.1158/1055-9965.EPI-12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanda R, et al. Genetic testing in an ethnically diverse cohort of high-risk women: A comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294:1925–1933. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz-Narvaez EA, et al. Gene-based analysis of the fibroblast growth factor receptor signaling pathway in relation to breast cancer in African American women: The AMBER consortium. Breast Cancer Res Treat. 2016;155:355–363. doi: 10.1007/s10549-015-3672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Narvaez EA, et al. Genetic variation in the insulin, insulin-like growth factor, growth hormone, and leptin pathways in relation to breast cancer in African-American women: the AMBER consortium. NPJ Breast Cancer. 2016 doi: 10.1038/npjbcancer.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Narvaez EA, et al. Admixture mapping of African-American women in the AMBER consortium identifies new loci for breast cancer and estrogen-receptor subtypes. Front. Genet. 2016;7:170. doi: 10.3389/fgene.2016.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biunno I, et al. BRCA1 point mutations in premenopausal breast cancer patients from Central Sudan. Fam. Cancer. 2014;13:437–444. doi: 10.1007/s10689-014-9717-4. [DOI] [PubMed] [Google Scholar]
- 21.Campbell MC, Tishkoff SA. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genom. Hum. Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiagge E, et al. Breast cancer and African ancestry: Lessons learned at the 10-year anniversary of the Ghana-Michigan research partnership and international breast registry. J. Glob. Oncol. 2016;2:302–310. doi: 10.1200/JGO.2015.002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, et al. A genome-wide association study of breast cancer in women of African ancestry. Hum. Genet. 2013;132:39–48. doi: 10.1007/s00439-012-1214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman LA, Kaljee LM. Health disparities and triple-negative breast cancer in African American women: A review. JAMA Surg. 2017;152:485–493. doi: 10.1001/jamasurg.2017.0005. [DOI] [PubMed] [Google Scholar]
- 25.Rotimi CN, Tekola-Ayele F, Baker JL, Shriner D. The African diaspora: History, adaptation and health. Curr. Opin. Genet. Dev. 2016;41:77–84. doi: 10.1016/j.gde.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindquist KJ, et al. Mutational landscape of aggressive prostate tumors in African American men. Cancer Res. 2016;76:1860–1868. doi: 10.1158/0008-5472.CAN-15-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman LA, Reis-Filho JS, Morrow M, Carey LA, King TA. The 2014 Society of Surgical Oncology Susan G. Komen for the cure symposium: triple-negative breast cancer. Ann. Surg. Oncol. 2015;22:874–882. doi: 10.1245/s10434-014-4279-0. [DOI] [PubMed] [Google Scholar]
- 28.Jiagge E, et al. Comparative analysis of breast cancer phenotypes in African American, White American, and West Versus East African patients: correlation between African ancestry and triple-negative breast cancer. Ann. Surg. Oncol. 2016;23:3843–3849. doi: 10.1245/s10434-016-5420-z. [DOI] [PubMed] [Google Scholar]
- 29.Brewster AM, Chavez-MacGregor M, Brown P. Epidemiology, biology, and treatment of triple-negative breast cancer in women of African ancestry. Lancet Oncol. 2014;15:e625–634. doi: 10.1016/S1470-2045(14)70364-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis, M. B. & Newman, L. A. Oncologic anthropology: An interdisciplinary approach to understanding the association between genetically-defined African ancestry and susceptibility for triple negative breast cancer. Curr. Breast Cancer Rep. In Press (2020).
- 31.Newman LA, et al. hereditary susceptibility for triple negative breast cancer associated with Western Sub-Saharan African Ancestry: Results from an international surgical breast cancer collaborative. Ann. Surg. 2019;270:484–492. doi: 10.1097/SLA.0000000000003459. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Q, et al. Trans-ethnic follow-up of breast cancer GWAS hits using the preferential linkage disequilibrium approach. Oncotarget. 2016;7:83160–83176. doi: 10.18632/oncotarget.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolton KL, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandrashekar DS, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith MW, et al. Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am. J. Hum. Genet. 2001;69:1080–1094. doi: 10.1086/323922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruz-Correa M, et al. Clinical Cancer Genetics Disparities among Latinos. J Genet Couns. 2017;26:379–386. doi: 10.1007/s10897-016-0051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hines LM, et al. The interaction between genetic ancestry and breast cancer risk factors among Hispanic women: The breast cancer health disparities study. Cancer Epidemiol. Biomarkers Prev. 2017;26:692–701. doi: 10.1158/1055-9965.EPI-16-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livingstone FB. The Duffy blood groups, vivax malaria, and malaria selection in human populations: A review. Hum. Biol. 1984;56:413–425. [PubMed] [Google Scholar]
- 41.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 42.Davis MB, et al. Distinct transcript isoforms of the atypical chemokine receptor 1 (ACKR1)/Duffy antigen receptor for chemokines (DARC) gene are expressed in lymphoblasts and altered isoform levels are associated with genetic ancestry and the Duffy-Null Allele. PLoS ONE. 2015;10:e0140098. doi: 10.1371/journal.pone.0140098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howes RE, et al. The global distribution of the Duffy blood group. Nat. Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiagge E, Chitale D, Newman LA. Triple-negative breast cancer, stem cells, and African Ancestry. Am. J. Pathol. 2018;188:271–279. doi: 10.1016/j.ajpath.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 45.Brachner A, et al. The endonuclease Ankle1 requires its LEM and GIY-YIG motifs for DNA cleavage in vivo. J. Cell. Sci. 2012;125:1048–1057. doi: 10.1242/jcs.098392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zlopasa L, Brachner A, Foisner R. Nucleo-cytoplasmic shuttling of the endonuclease ankyrin repeats and LEM domain-containing protein 1 (Ankle1) is mediated by canonical nuclear export- and nuclear import signals. BMC Cell. Biol. 2016;17:23. doi: 10.1186/s12860-016-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toledo CM, et al. Genome-wide CRISPR-cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell. Rep. 2015;13:2425–2439. doi: 10.1016/j.celrep.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang T, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell. 2017;168:890–903. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacLeod G, et al. Genome-wide CRISPR-Cas9 screens expose genetic vulnerabilities and mechanisms of temozolomide sensitivity in glioblastoma stem cells. Cell. Rep. 2019;27:971–986. doi: 10.1016/j.celrep.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 50.Kabir S, et al. The CUL5 ubiquitin ligase complex mediates resistance to CDK9 and MCL1 inhibitors in lung cancer cells. Elife. 2019 doi: 10.7554/eLife.44288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitehurst AW, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 52.Newman LA, Jackson KE. The sisters network: A National African American breast cancer survivor advocacy organization. J. Oncol. Pract. 2009;5:313–314. doi: 10.1200/JOP.091037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammond ME, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch. Pathol. Lab. Med. 2010;134:e48–72. doi: 10.1043/1543-2165-134.7.e48. [DOI] [PubMed] [Google Scholar]
- 55.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Alem U, et al. Association of genetic ancestry with breast cancer in ethnically diverse women from Chicago. PLoS ONE. 2014;9:e112916. doi: 10.1371/journal.pone.0112916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosoy R, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum. Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nassir R, et al. An ancestry informative marker set for determining continental origin: Validation and extension using human genome diversity panels. BMC Genet. 2009;10:39. doi: 10.1186/1471-2156-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J, Zhang Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy A, Yang J, Zhang Y. COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012;40:W471–477. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.