Abstract
The oomycete Aphanomyces astaci is an emerging infectious pathogen affecting freshwater crayfish worldwide and is responsible for one of the most severe wildlife pandemics ever reported. The pathogen has caused mass mortalities of freshwater crayfish species in Europe and Asia, and threatens other susceptible species in Madagascar, Oceania and South America. The pathogen naturally coexists with some North American crayfish species that are its chronic carriers. Presumptions that A. astaci originated in North America are based on disease outbreaks that followed translocations of North American crayfish and on the identification of the pathogen mainly in Europe. We studied A. astaci in the southeastern US, a center of freshwater crayfish diversity. In order to decipher the origin of the pathogen, we investigated (1) the distribution and haplotype diversity of A. astaci, and (2) whether there are crayfish species-specificities and/or geographical restrictions for A. astaci haplotypes. A total of 132 individuals, corresponding to 19 crayfish species and one shrimp species from 23 locations, tested positive for A. astaci. Mitochondrial rnnS and rnnL sequences indicated that A. astaci from the southeastern US exhibited the highest genetic diversity so far described for the pathogen (eight haplotypes, six of which we newly describe). Our findings that A. astaci is widely distributed and genetically diverse in the region supports the hypothesis that the pathogen originated in the southeastern US. In contrast to previous assumptions, however, the pathogen exhibited no clear species-specificity or geographical patterns.
Subject terms: Pathogens, Fungal genetics
Introduction
During the past few decades, fungal and fungal-like pathogens have caused several worldwide pandemics responsible for declines in wildlife populations—even causing extinctions1–5. Globalization facilitates these pandemics—usually consequences of the transport and introduction of exotic and invasive species6–8. Moreover, habitat alterations due to anthropogenic activity break down natural dispersal barriers, allowing invasive species (frequently carrying pathogens)9–15 to further expand their ranges. Climate change alters environmental conditions, further benefitting some invasive species and favoring the development and spread of disease2,4.
Fungal and fungal-like pathogenic species have impacted freshwater ecosystems particularly strongly, causing a global decline in freshwater biodiversity that is far greater than that seen in terrestrial ecosystems16,17. For example, the panzootic chytrid fungus Batrachochytrium dendrobatidis originated in Asia and spread globally due to amphibian trade, causing declines in more than 500 amphibian species over the past half-century18,19. Furthermore, fungal-like pathogens, such as Saprolegnia diclina and Saprolegnia ferax (Oomycetes), are also responsible for mass extinctions in amphibians20,21 and may be spread by the fish trade22. Another pathogenic oomycete, Aphanomyces invadans, causes epizootic ulcerative syndrome (EUS), affecting more than 100 fish species in Asia, Australia, North America and Africa23,24.
Similarly, Aphanomyces astaci causes the crayfish plague in native European, Asian and Australian crayfish species25–28 and has decimated crayfish populations in those continents5,6. This oomycete is a specialized pathogen in freshwater crayfish27,29,30, one third of which are threatened with extinction globally31. The pathogen coexists naturally with North American crayfish but can efficiently colonize non-North American crayfish, almost without resistance26. In addition, A. astaci has spread rapidly throughout the world through translocations of North American chronic carriers25,28–40. In non-North American crayfish, crayfish plague infections typically cause death within a few days41.
Presumptions about the origin of the crayfish plague were based on disease outbreaks that followed historical translocations of North American crayfish species to many countries for aquaculture, sport fishing, or aquarium pet trade42. The first known introduction of a North American crayfish and subsequent crayfish plague outbreak was recorded in Europe in the nineteenth century43. Later, additional large-scale introductions of North American crayfish species were made in European and non-European countries34–37,39,40,44–49. Moreover, illegal translocations resulted in new crayfish plague outbreaks that decimated native crayfish populations in many countries33,46,50 (Fig. 1). Thus, A. astaci was listed among the 100 of the World’s Worst Invasive Alien Species51.
Figure 1.
Distribution of Aphanomyces astaci haplotypes detected from locations in Europe and Japan33,52,53. Each point and color represent the presence of an A. astaci haplotype based on concatenated mitochondrial rnnS and rnnL regions. Colors indicate the haplotype code at each site as follows: green—a-haplotype, blue—b-haplotype, orange—d1-haplotype, red—d2-haplotype and pink—d3-haplotype [Maps were prepared using QGIS 2.14 (https://www.qgis.org/en/site/)].
Knowledge about the virulence and genetic variability of A. astaci has come primarily from studies of crayfish plague outbreaks in Europe and Asia50,52,54–60. Specifically, mitochondrial DNA regions of A. astaci have been informative in assessing genetic diversity in both pure cultures and clinical samples of this clonally reproducing pathogen. To date, the mitochondrial DNA variability found in crayfish plague outbreaks in Europe and Japan has been allocated to six haplotypes (a, b, d1, d2, d3 and e-haplotypes) (Fig. 1) within two lineages33,38,52,53,61. However, only three studies have confirmed the presence of the pathogen in North America, revealing only two of the previously described haplotypes: a and b-haplotypes62–64.
Although evidence strongly supports a North American origin of A. astaci, our understanding of the crayfish plague pathogen in North America is still insufficient. A clearer understanding of the diversity and distribution of A. astaci within its native range is needed, not only to improve our comprehension of the evolution and epidemiology of pandemic pathogens, but also to determine future management and research directions. Similar questions have been faced when studying other emerging pathogens. For example, despite the occurrence of Batrachochytrium dendrobatidis in Asia, the lack of lethal outbreaks evidenced an endemic host–pathogen interaction in that region. Several studies have confirmed that the geographic origin of chytridiomycosis was in Asia, explaining the survival of Asian amphibian populations and stable host–pathogen dynamics19.
Although more than 428 crayfish species are native to North America65, the diversity, distribution and prevalence of A. astaci there is still largely unknown. Within North America, the southeastern US harbors the highest number of endemic crayfish species. The region represents not only a center of diversity, but also one of the two distinct origins of freshwater crayfish6,52,66. The presence of A. astaci has not been investigated in this crayfish-rich region even though such knowledge would improve our understanding of the origin and diversity of the pathogen. Thus, the main aim of this study was to evaluate the southeastern US as the possible center of origin of the crayfish plague pathogen A. astaci. For this purpose, we tested key questions including: (1) what is the distribution and haplotype diversity of A. astaci in the southeastern US, and (2) are A. astaci haplotypes crayfish species-specific and/or geographically restricted. In order to perform this study, we isolated and analyzed the pathogen from 30 distinct crayfish populations comprising a total of 21 crayfish species and one shrimp from five states in the southeastern US.
Results
Aphanomyces astaci detection
We obtained a total of 391 crayfish from 30 locations in five states (Kansas, Kentucky, Louisiana, Mississippi and South Carolina) (Fig. 2). The crayfish represented six genera and 21 species: Cambarellus shufeldtii, Cambarus latimanus, Cambarus striatus, Cambarus tenebrosus, Creaserinus fodiens, Creaserinus oryktes, Faxonius etnieri species complex, Faxonius sp., Faxonius tricuspis, Faxonius wrighti, Lacunicambarus ludovicianus, Procambarus ablusus, Procambarus acutus, Procambarus clarkii, Procambarus hayi, Procambarus hybus, Procambarus pubescens, Procambarus raneyi, Procambarus troglodytes, Procambarus viaeviridis and Procambarus vioscai (Supplementary Table 1). Additionally, one species of freshwater shrimp (Palaemon kadiakensis) was sampled and analyzed for the presence of A. astaci.
Figure 2.
Aphanomyces astaci presence from the study locations. Aphanomyces astaci presence/absence is represented by green (positive) or grey (negative) circles. Numbers indicate the location code at each site (as in Supplementary Table 1). Two-letter state code is provided with the following abbreviation: KS (Kansas), KY (Kentucky), LA (Louisiana), MS (Mississippi) and SC (South Carolina). [Maps were prepared using QGIS 2.14 (https://www.qgis.org/en/site/)].
From 392 individuals, 132 crayfish and one shrimp tested positive for the A. astaci ITS region, 102 tested negative and 158 were not analyzed (i.e., the crayfish did not molt). Aphanomyces astaci-positive samples came from 23 locations and included 19 crayfish species: C. shufeldtii, C. latimanus, C. striatus, C. fodiens, C. oryktes, F. etnieri species complex, Faxonius sp., F. tricuspis, F. wrighti, P. ablusus, P. acutus, P. clarkii, P. hayi, P. hybus, P. pubescens, P. raneyi, P. troglodytes, P. viaeviridis and P. vioscai) and one species of shrimp (Palaemon kadiakensis) (Fig. 2 and Supplementary Table 1). The ITS sequences (specific primers 42 and 640) for the 132 clinical samples were 99.82% identical to sequences of A. astaci available in GenBank (e.g., sequence FM999249-isolate SAP302) and identical to each other.
Sequence analyses and haplotyping of A. astaci
Twenty crayfish clinical samples (taken directly from crayfish) and 12 pure cultures from nine locations and three states (Kentucky, Mississippi and South Carolina) (Fig. 3) contained enough of the pathogen DNA for amplifying both mitochondrial regions (i.e., clinical samples often harbor low pathogen DNA concentration).
Figure 3.
Aphanomyces astaci haplotypes detected from locations (numbers on pie graphs indicate locations; see Table 1) in North America. Haplotype frequencies indicated by relative proportion of the pie graph. Each color of the legend represents a different haplotype based on concatenated mitochondrial rnnS and rnnL regions. Crayfish species hosting different haplotypes are indicated at each location (also in Table 1). Location code LP corresponds to Lake Pitt50, LT to Lake Tahoe63, BL to Big Lake62, TL to Trout Lake62, CC to Chickies Creek64, BR to Brubaker Run64 and SP to Sunfish Pond64. Two-letter State code is provided with the following abbreviation: CA (California) KY (Kentucky), MS (Mississipi), PA (Pennsylvania), NV (Nevada), SC (South Carolina) and WI (Wisconsin). [Maps were prepared using QGIS 2.14 (https://www.qgis.org/en/site/)].
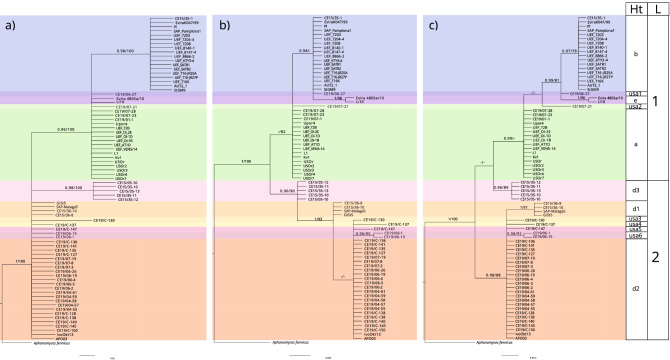
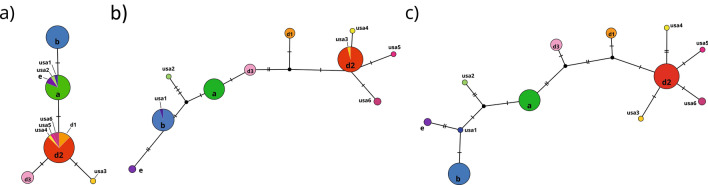
For the phylogenetic approximations [Bayesian Inference (BI) and Maximum likelihood (ML)] and diversity estimations, we included a total of 78 sequences [32 sequences from the present study, 43 obtained from GenBank as reported from previous studies33,52,62 and two sequences from new A. astaci and A. fennicus isolates (CCRJB-75 and CCRJB-76, respectively)67 with 476 and 355 bp fragments of rnnS and rnnL amplicons, respectively. The phylogenetic approximations (BI and ML) supported the differentiation of the two lineages previously described52 (Fig. 4). The genetic diversity analysis confirmed and supported the phylogenetic analysis. Although both mitochondrial ribosomal rnnS and rnnL regions were informative, there were differences between them (Fig. 5). We obtained five haplotypes for the rnnS subunit (Fig. 5a), represented by four segregating sites (S), with a haplotype diversity (Hd) of 0.703, a nucleotide diversity (π) of 0.0022 and 1.018 average nucleotide differences (k). On the other hand, we obtained ten haplotypes for the rnnL subunit (Fig. 5b), represented by13 segregating sites (S), with a haplotype diversity (Hd) of 0.786, a nucleotide diversity (π) of 0.009 and 3.16 average nucleotide differences (k). However, concatenating rnnS and rnnL regions we confirmed a total of 12 haplotypes represented by 17 segregating sites (S), where 13 of them were parsimony informative (Fig. 5c). The concatenated sequences presented a haplotype diversity (Hd) of 0.801 with a nucleotide diversity (π) of 0.005 and 4.178 average nucleotide differences (k).
Figure 4.
Phylogenetic analyses of Aphanomyces astaci mitochondrial regions. Bayesian phylogenetic analyses of A. astaci mitochondrial rnnS, rnnL and concatenated rnnS + rnnL sequences obtained from infected crayfish specimens and previous studies33,52,53,62. (a) Bayesian phylogenetic tree based on the rnnS sequences, (b) Bayesian phylogenetic tree based on the rnnL sequences, (c) Bayesian phylogenetic tree based on the concatenated rnnS + rnnL sequences. Values above the branches represent the Bayesian posterior probabilities (> 0.95) and ML bootstrap support values (> 75), respectively. Scale bar for phylogenetic analysis indicates substitutions per site. Abbreviations: Ht, haplotypes; L, lineages.
Figure 5.
Haplotype network based on rnnS, rnnL and concatenated rnnS + rnnL mitochondrial DNA sequences, generated by statistical parsimony. The area of the circles is proportional to the number of sequences. (a) Haplotype network based on rnnS mtDNA sequences, (b) Haplotype network based on rnnL mtDNA sequences, (c) Haplotype network based on concatenated rnnS + rnnL mtDNA sequences. Mutation steps between haplotypes are shown as hatch marks.
The phylogenetic approximations and the haplotype network confirmed the presence of eight haplotypes for the rrnS and rrnL concatenated regions among the analyzed samples: a, d2, usa1, usa2, usa3, usa4, usa5 and usa6-haplotypes (Figs. 4, 5) (Table 1). Six of these eight haplotypes (usa1, usa2, usa3, usa4, usa5 and usa6-haplotypes) are described and reported here for the first time, bringing the total number of known A. astaci haplotypes to 12. Moreover, these results were confirmed by an independent secondary molecular test (i.e., by repeating the amplification and sequencing of each of the haplotyped samples).
Table 1.
Location and samples ID of the of the crayfish species analyzed for Aphanomyces astaci haplotypes.
| Location code | Collection code | Host species | State | Sample type | DNA extraction | Species ID (rRNA) | Haplotype (rnnS/rnnL) | GenBank rnnS | GenBank rnnL |
|---|---|---|---|---|---|---|---|---|---|
| 1 | SA908-DEAD1 | Procambarus hybus | Mississippi | Clinical sample | CE19/01-1 | Aphanomyces astaci | a | MW346522 | MW346542 |
| 3 | SA927-1 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/06-12 | Aphanomyces astaci | usa6 | MW346516 | MW346536 |
| SA927-5 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/04-58 | Aphanomyces astaci | d2 | MW346519 | MW346539 | |
| SA927-8 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/04-57 | Aphanomyces astaci | d2 | MW346520 | MW346540 | |
| SA927-16 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/06-2 | Aphanomyces astaci | d2 | MW346513 | MW346533 | |
| SA927-18* | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/06-26 | Aphanomyces astaci | d2 | MW346512 | MW346532 | |
| SA927-23 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/06-3 | Aphanomyces astaci | d2 | MW346510 | MW346530 | |
| SA927-25 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/04-55 | Aphanomyces astaci | d2 | MW346521 | MW346541 | |
| SA927-27 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/04-59 | Aphanomyces astaci | d2 | MW346518 | MW346538 | |
| SA927-18* | Cambarellus shufeldtii | Mississippi | Culture | CE19/C-136 | Aphanomyces astaci | d2 | MW346494 | MW346481 | |
| SA927-18* | Cambarellus shufeldtii | Mississippi | Culture | CE19/C-135 | Aphanomyces astaci | d2 | MW346495 | MW346482 | |
| SA927-18* | Cambarellus shufeldtii | Mississippi | Culture | CE19/C-127 | Aphanomyces astaci | d2 | MW346498 | MW346485 | |
| SA927-18* | Cambarellus shufeldtii | Mississippi | Culture | CE19/C-138 | Aphanomyces astaci | d2 | MW346492 | MW346479 | |
| SA927-14 | Cambarellus shufeldtii | Mississippi | Culture | CE19/C-140 | Aphanomyces astaci | d2 | MW346491 | MW346478 | |
| SA927-18* | Cambarellus shufeldtii | Mississippi | Culture | CE19/C-150 | Aphanomyces astaci | d2 | MW346487 | MW346474 | |
| 4 | SA928-7 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/06-4 | Aphanomyces astaci | d2 | MW346509 | MW346529 |
| 6 | SA930-2 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/04-61 | Aphanomyces astaci | d2 | MW346517 | MW346537 |
| SA930-4 | Cambarellus shufeldtii | Mississippi | Clinical sample | CE19/07-03 | Aphanomyces astaci | d2 | MW346504 | MW346524 | |
| SA930-6 | Procambarus clarkii | Mississippi | Clinical sample | CE19/06-15 | Aphanomyces astaci | usa6 | MW346515 | MW346535 | |
| SA930-8* | Procambarus clarkii | Mississippi | Clinical sample | CE19/07-08 | Aphanomyces astaci | d2 | MW346503 | MW346523 | |
| SA930-8* | Procambarus clarkii | Mississippi | Culture | CE19/C-141 | Aphanomyces astaci | d2 | MW346490 | MW346477 | |
| SA930-8* | Procambarus clarkii | Mississippi | Culture | CE19/C-128 | Aphanomyces astaci | d2 | MW346497 | MW346484 | |
| SA930-8 * | Procambarus clarkii | Mississippi | Culture | CE19/C-145 | Aphanomyces astaci | d2 | MW346489 | MW346476 | |
| 7 | M306-3 | Procambarus clarkii | Mississippi | Clinical sample | CE19/06-19 | Aphanomyces astaci | d2 | MW346514 | MW346534 |
| 9 | SA937-5 | Faxonius tricuspis | Kentucky | Clinical sample | CE19/06-27 | Aphanomyces astaci | usa1 | MW346511 | MW346531 |
| 11 | SA946-9 | Procambarus ablusus | Mississippi | Clinical sample | CE19/07-19 | Aphanomyces astaci | d2 | MW346508 | MW346528 |
| SA946-23 | Cambarus striatus | Mississippi | Clinical sample | CE19/07-21 | Aphanomyces astaci | usa2 | MW346507 | MW346527 | |
| SA946-FAX SP JUV | Faxonius sp. | Mississippi | Culture | CE19/C-137 | Aphanomyces astaci | usa4 | MW346493 | MW346480 | |
| 13 | SA948-1 | Procambarus acutus | Mississippi | Clinical sample | CE19/07-23 | Aphanomyces astaci | a | MW346506 | MW346526 |
| SA948-5 | Faxonius etnieri | Mississippi | Clinical sample | CE19/07-28 | Aphanomyces astaci | a | MW346505 | MW346525 | |
| 23 | SC32 MOLT 4* | Procambarus raneyi | South Carolina | Culture | CE19/C-130 | Aphanomyces astaci | usa3 | MW346496 | MW346483 |
| SC32 MOLT 5* | Procambarus raneyi | South Carolina | Culture | CE19/C-147 | Aphanomyces astaci | usa5 | MW346488 | MW346475 |
North American crayfish species from each of the analyzed locations (Location code:1–30), including the corresponding collection codes, state, sample type (clinical sample/culture), DNA isolation code (CE19/), BLAST species ID, result of the mitochondrial rnnS/rnnL haplotype and GenBank references for the rnnS and rnnL regions. NA: not applicable. Each unique collection code refers to an individual crayfish. *Different pieces from the same molt.
Crayfish species, A. astaci haplotype diversity and distribution
The haplotyping results for the 20 crayfish clinical samples included five haplotypes: usa1-haplotype (one Faxonius tricuspis), usa2-haplotype (one Cambarus striatus), a-haplotype (one Procambarus hybus, one Procambarus acutus and one Faxonius etnieri), d2-haplotype (ten Cambarellus shufeldtii, two Procambarus clarkii and one Procambarus ablusus) and usa6-haplotype (one Cambarellus shufeldtii and one Procambarus clarkii) (GenBank accession numbers MW346503-MW346522 for rnnS and MW346523-MW346542 for rnnL) (Table 1). The amount of infection in the remaining samples that tested positive for A. astaci (ITS region) was too low to obtain conclusive results for both rrnS and rrnL.
Additionally, the haplotyping results for the 12 pure cultures included four haplotypes: usa3-haplotype (one Procambarus raneyi culture), usa4-haplotype (one Faxonius sp. culture), usa5-haplotype (one Procambarus raneyi culture) and d2-haplotype (six Cambarellus shufeldtii and three Procambarus clarkii cultures) (GenBank accession number MW346487-MW346498 for rnnS and MW346474-MW346485 for rnnL).
We found five scenarios relative to the distribution of A. astaci haplotypes among crayfish species at various locations: (1) one haplotype from one crayfish species (Locations 1, 4, 7 and 9), (2) two haplotypes from one crayfish species (Locations 3 and 23) (see below), (3) one haplotype from two crayfish species (Location 13), (4) two haplotypes from two crayfish species (Location 6) (see below), and (5) three haplotypes, one from each of three crayfish species (Location 11). At Location 23, the two haplotypes were recovered from one P. raneyi molt (i.e., we isolated the pathogen in two different pure cultures) and at Location 6, one haplotype was recovered from both species, and the second haplotype from only one of the species (Fig. 3) (Table 1).
Discussion
We report and describe for the first time the presence, distribution and genetic diversity of the crayfish plague pathogen, A. astaci, in its potential center of origin, the southeastern US68. Previous studies regarding the origin, diversity, and distribution of A. astaci have addressed different questions. In the current study, we have explored several of them, including (1) the distribution and diversity of A. astaci in the southeastern US, and (2) whether A. astaci haplotypes are crayfish species-specific50,54,55 and/or restricted to a narrow geographic region of the pathogen’s native range52,62,63. Our results indicated that A. astaci is present and widely distributed in the southeastern US (e.g., it was present in 21 out of 23 sampling sites across the nine river basins we investigated) and possesses the highest genetic diversity of A. astaci described from any region to date. Previously, only two haplotypes (a and b-haplotypes) had been found in the five North American crayfish species examined from California, Michigan and Pennsylvania (Pacifastacus leniusculus, Cambarus bartonii, Faxonius obscurus, Faxonius rusticus and Faxonius virilis)62–64. However, in the southeastern US, we examined 21 North American crayfish species and found a total of eight haplotypes, six of which are previously unreported. This represents almost 70% of the A. astaci haplotype diversity known globally. The genetic diversity we describe is comparable to that of other pathogenic oomycetes. For example, in the US, 13 Phytopthtora infestans haplotypes were recently described using five mitochondrial loci69. Moreover, we showed that A. astaci can chronically colonize 19 additional North American crayfish species and also one species of freshwater shrimp (Palaemon kadiakensis). Previously, the only known report of a wild shrimp carrying A. astaci was of a Macrobrachium lanchesteri population from Indonesia that co-occurred with P. clarkii70. Only two (Cambarus tenebrosus and Lacunicambarus ludovicianus) of the 21 crayfish species that we sampled did not test positive for A. astaci. Thus, we confirm the presence of A. astaci in a wider distribution (Fig. 2) than previously described in North America62–64, confirming the origin of this pathogenic disease in North America.
Although the presence of the crayfish plague in North America was previously reported62–64, those studies examined a limited number of crayfish species and found haplotypes that had been previously isolated in Europe and Japan (i.e., a or b-haplotypes)33,62–64. Likewise, we confirmed the presence in North America of two haplotypes (a- and d2-haplotypes) first described in Europe. In the case of the a-haplotype, it was described as the first haplotype introduced to Europe in the nineteenth century43,52, yet no North American carrier has been described in the literature for this introduction. Two recent studies described F. rusticus and F. obscurus as the only North American crayfish known to host the a-haplotype62,64. We expanded the known range of this haplotype to include the southeastern US and added three additional native taxa as hosts: the F. etnieri species complex, P. acutus and P. hybus. In the case of the d2-haplotype, the only two North American crayfish described as carriers so far in Europe are P. clarkii and Procambarus fallax virginalis. As we expected, our study shows for the first time the presence of A. astaci in native populations of P. clarkii54, which carried the d2-haplotype and the newly discovered usa6-haplotype.
We confirmed that at least some A. astaci haplotypes are neither host species-specific nor narrowly distributed. Broadening and deepening our knowledge of A. astaci haplotype diversity and distribution, we recovered the d2-haplotype from two species in different genera (Cambarellus and Procambarus) in one river basin (Locations 3, 4, 6 and 7) and from a third genus (Cambarus) in a different river basin (Location 11) (Table 1). Although the a-haplotype was previously thought to be restricted to F. rusticus and F. obscurus62,64, we found it not only in another congener, but also in another genus (Procambarus). In addition, we documented the a-haplotype in two river basins in the southeastern US, even though it was previously thought to be restricted to the northern US. We also documented the d2-haplotype from two river basins. Our results indicated that A. astaci haplotypes tend to be neither host species-specific nor restricted to small geographic areas.
We also found instances of multiple haplotypes of A. astaci occurring in one species, in one individual, and in one location. We isolated two pure cultures of two different, but closely related, A. astaci haplotypes (usa3 and usa5) from a single individual (SC32 MOLT in Location 23). Also, multiple A. astaci haplotypes often co-occurred in a location. Within Location 11, we found three haplotypes from two lineages: usa2-haplotype from Lineage 1 and d2-haplotype and usa4-haplotype from Lineage 2. We also recovered two closely related haplotypes (d2 and usa6) from Locations 3 and 6 and two others (usa3 and usa5) from Location 23. None of the phenomena described have been documented previously. Our analysis showed no clear haplotype distributional patterns with respect to crayfish host species or geography. Thus, the biogeographic distribution of A. astaci genetic diversity within North America needs further investigation.
The pathogen’s genetic diversity and observed lack of species-specificity may have implications within North America as well as on continents where A. astaci is introduced. Crayfish have been frequently translocated within North America. For example, although P. clarkii and F. virilis are native in parts of North America, both are invasive beyond their native range, including west of the Great Divide where all native crayfish belong to the family Astacidae71,72. The genetic diversity of A. astaci suggests potential for intracontinental impacts from translocations of haplotypes. Presumably all North American crayfish are resistant to all haplotypes of A. astaci, considering that no mass mortalities have been observed after crayfish translocations. However, more subtle effects of translocated haplotypes with differing virulence from native haplotypes would not likely have been detected. Further understanding of the geographical distribution of A. astaci genetic diversity, its virulence, and the immune responses of crayfish to novel haplotypes would be beneficial for managing crayfish translocations and conserving native species in North America.
Our new approach of obtaining A. astaci from crayfish molts produced larger amounts of the pathogen for identification and isolation in pure cultures. This resulted in detection of A. astaci in 56.17% of the analyzed clinical samples. Instead of analyzing parts of the crayfish most susceptible to infection (i.e., soft abdominal cuticle, telson or walking legs)73, we waited until the molting period. Thus, we avoided the unnecessary killing of crayfish and maximized the amount of pathogen grown within the original host. Moreover, by incubating crayfish molts in distilled water for three days, we allowed the pathogen to continue growing both within and outside of the cuticle. By controlling the incubation temperature at 4 °C, we reduced bacterial blooms during the first stages of the pathogen isolation. Bacteria commonly surround the A. astaci hyphae, and antibiotics are often added to the PGA medium to inhibit bacterial growth. However, because bacteria rapidly develop resistance to these antibiotics, their addition becomes less effective over time. By reducing the incubation temperature and introducing a physical barrier74, we controlled bacterial blooms75 and obtained many clean isolates of the pathogen.
By using this new approach, we more readily detected and isolated A. astaci. Moreover, we strongly recommend isolating the pathogen in an axenic culture to assure an optimal concentration of the pathogen for analysis. In this study, we combined the sequencing of the ITS region (for identification of the Aphanomyces species)76 and the mtDNA (for identification of the A. astaci haplotype)52 in both clinical samples and pure cultures. Several studies have examined the genetic diversity of A. astaci using diverse methodologies (i.e., RAPD-PCR, the chitinase gene, AFLP, microsatellites)50,52,56,58–60,77. Although the mtDNA approximation52 requires a large concentration of the pathogen, it provides reliable results67 and enables detection of new diversity within A. astaci that might go undetected with other approximations59,60. Other approaches, such as eDNA monitoring78, could also be used to detect the presence of A. astaci in water samples and potentially to detect additional genetic diversity.
The uniquely high diversity of A. astaci haplotypes found in this study are an important step toward confirming the host–pathogen co-evolution between A. astaci and North American crayfish species. Our results suggest that further sampling in North America will reveal additional undiscovered A. astaci haplotype diversity vital to answering new host–pathogen co-evolutionary questions. Further, long-term monitoring might reveal emerging A. astaci diversity, depending on the rate of pathogen evolution.
Methods
Sample collection
We sampled 25 locations in five states between February and May 2019. Additionally, we included nine crayfish samples previously collected from five more locations and preserved in 95% ethanol. The specimens from Kansas were from an introduced population of Procambarus clarkii (Supplementary Table 1). Crayfish were captured by kick-seining and trapping and then held, separated by species and collection location, in the laboratory (US Forest Service, Southern Research Station, Center for Bottomland Hardwoods Research, in Oxford [Mississippi, USA]) until processed. Crayfish were kept individually in labeled, individual, plastic containers with chlorine-free water, aerators, gravel and medium size rocks. Each crayfish was kept alive until it molted and was subsequently preserved in 95% ethanol.
Microscopic examination and Aphanomyces astaci isolation
For the microscopic examination, the molts were carefully removed and handled individually due to the fragility of the samples. Molts were kept in individual petri dishes with distilled water at 4 °C in order to reduce bacterial growth and optimize the growth of potential mycelium75. After three days, each molt was examined with an inverted microscope to check for the presence of growing hyphae. Each sample was divided into two parts: one for molecular identification and one for the pathogen isolation.
Pieces of the molt intended for molecular identification were transferred into 1.5-ml tubes and were frozen at − 80 °C until the DNA extraction was carried out in the laboratory at the Department of Biology at the University of Mississippi (UM). Samples were subsequently homogenized by manual mechanical disruption. A DNeasy Blood & Tissue Kit (Qiagen, Valencia, California, USA) was used to isolate genomic DNA.
Pieces of the molt intended for culture isolation were grown in Peptone Glucose Agar (PGA) at 4 °C. A selected agar plug was cut out from the resulting mycelia and inserted within an aluminum ring placed on a new PGA plate to protect the growing isolate from bacterial growth74. Plates were incubated at 4 °C for seven days, and each isolate was transferred into new PGA media once the hyphae spread under the metal ring. This process was repeated until no bacterial growth was observed using an inverted microscope. Additionally, a selected agar plug containing mycelia was placed in a 9 mm Petri dish containing 10 mL of liquid Peptone Glucose (PG-1) and incubated at room temperature for 48 h in order to obtain material for molecular identification. The obtained mycelium was transferred into 1.5-ml tubes and frozen at − 20 °C until the DNA extraction was carried out in the laboratory at UM. Samples were subsequently homogenized by manual mechanical disruption, and DNA extractions were carried out using an E.Z.N.A. Fungal DNA Mini Kit (Omega Biotek, Norcross, Atlanta, USA).
Aphanomyces astaci detection and haplotyping
To test for the presence of the A. astaci pathogen, a fragment of the internal transcribed spacer (ITS) region was amplified using the diagnostic primers 42 (5′-GCTTGTGCTGAGGATGTTCT-3′)73 and 640 (5′-CTATCCGACTCCGCATTCTG-3′)79 (which amplify ITS1, the 5.8S rDNA and ITS2) in a single round of amplification according to the assay described by73. DNA extracted from a pure culture of A. astaci was used as the positive control; sterile Milli-Q water was used as the negative control. Amplified products (3 μL of each reaction) were analyzed by electrophoresis in 2% agarose SB gels stained with ethidium bromide and then purified using magnetic beads. Sequencing of both strands of positive products was performed using an automated sequencer (Applied Biosystems 3730xl DNA Analyzer, DNA Analysis Facility at Yale, USA and Applied Biosystems 3730xl DNA, Macrogen, The Netherlands). Sequences were aligned and edited using the program Geneious 10.0.280. A BLAST search (NCBI database) was performed to verify the identity of each sequence.
Genomic DNA samples that tested positive for the presence of A. astaci with diagnostic primers 4273 and 64079 were used to characterize the phylogenetic relationships and haplotypes. The mitochondrial ribosomal small (rnnS) and large (rnnL) subunits were amplified using the primer pairs AphSSUF/AphSSUR (5′-AGCACTCCGCCTGAAGAGTA-3′ and 5′-GGGCGGTGTGTACAAAGTCT-3′) and AphLSUF/AphLSUR (5-AGGCGAAAGCTTACTATGATGG-3′ and 5′-CCAATTCTGTGCCACCTTCT-3′), respectively, as described by52. Positive and negative controls were included. Amplified products were analyzed by electrophoresis, purified, sequenced and aligned as described above.
Phylogenetic approximations based on Bayesian inference (BI) and maximum likelihood (ML) were used to reconstruct relationships. The BI analysis was performed in MrBayes v.3.2.681 using the MCMC method with 10 million generations, three runs (8 chains per run) with a burn-in of 25% and a standard deviation of split frequencies < 0.01. Nodes with posterior probability (pp) values ≥ 0.95 were considered supported. The ML analysis was performed using RAxML v.882, as implemented in raxmlGUI v1.5b183, with 100 independent replicates and 1000 rapid bootstraps. Nodes with bootstrap values ≥ 75 were considered supported. The resulting trees from the BI and ML analyses were visualized with FigTree v1.4.284. Sequences (57) corresponding to the mtDNA regions rnnS and rnnL of isolates analyzed in previous studies33,52,62,67, available in GenBank, were also included in our analyses. Aphanomyces fennicus was used as the outgroup in both phylogenetic approximations67,76. Analyses were performed with rnnS and rnnL individually, as well as with a concatenated rnnS and rnnL dataset, using the same parameters described above.
Genetic diversity was estimated by calculating the number of polymorphic (segregating) sites (S), the number of haplotypes, the haplotype diversity (Hd), the average number of nucleotide differences (k), and the nucleotide diversity (π) utilizing the program DNAsp v.5.10.0185. Mutational changes between sequences in the most parsimonious haplotype network were estimated using TCS v.1.2186, and the genealogical relationships were visualized with PopArt v1.7.287.
Ethics declarations
All experimental procedures and animal manipulations, as well as field sampling, were performed according to the US legislation.
Supplementary Information
Acknowledgements
We thank those who assisted with field work and provided sample identifications, or contributed specimens: Zanethia C. Barnett, Mickey Bland, William Hammond, Carl Smith, and Kenneth Sterling (US Forest Service [FS]), Earl Choice (FS volunteer), Gianna Richardson (FS volunteer and American Fisheries Society Hutton Scholar), Bronwyn Williams (North Carolina Museum of Natural Sciences), Gregory Meyers (West Liberty University), Becky Rosamond (US Fish and Wildlife Service), Dan Jones and Chris Steffen (Kansas Department of Wildlife, Parks and Tourism). LMT and SBA were supported by funding from the US Forest Service. MMR and GCH were supported by projects IND2018/AMB-10056 and IND2019/AMB-17177, Dirección General de Investigación e Innovación Tecnológica, Consejería de Ciencia y Universidades de Innovación, Comunidad de Madrid, Spain. JDU was supported from Project CGL2016-80526-R of the Ministerio de Economía y Competitividad, Spain.
Author contributions
L.M.T. contributed to the design, with the laboratory work and writing of this manuscript. M.M.R. and G.C.H. contributed with the laboratory work and writing of this manuscript. S.B.A., C.R.J. and J.D.U. contributed to the supervision and writing of this manuscript.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Susan B. Adams, Colin R. Jackson and Javier Diéguez-Uribeondo.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88704-8.
References
- 1.Fisher MC, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt ME, Park BJ. Think fungus-prevention and control of fungal infections. Emerg. Infect. Dis. 2013;19:1688–1689. doi: 10.3201/eid1910.131092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallabhaneni S, Mody RK, Walker T, Chiller T. The global burden of fungal diseases. Infect. Dis. Clin. N. Am. 2016;30:1–11. doi: 10.1016/j.idc.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Benedict K, Richardson M, Vallabhaneni S, Jackson BR, Chiller T. Emerging issues, challenges, and changing epidemiology of fungal disease outbreaks. Lancet Infect. Dis. 2017;17:e403–e411. doi: 10.1016/S1473-3099(17)30443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh PN, Fisher MC, Bates KA. Diagnosing emerging fungal threats: A one health perspective. Front. Genet. 2018;9:1–8. doi: 10.3389/fgene.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogden NH, et al. Emerging infectious diseases and biological invasions: A call for a One Health collaboration in science and management. R. Soc. Open Sci. 2019;6:181577. doi: 10.1098/rsos.181577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lwande OW, et al. Globe-trotting Aedes aegypti and Aedes albopictus: Risk factors for arbovirus pandemics. Vector Borne Zoonotic Dis. 2020;20:71–81. doi: 10.1089/vbz.2019.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santini A, Battisti A. Complex insect-pathogen interactions in tree pandemics. Front. Physiol. 2019;10:1–7. doi: 10.3389/fphys.2019.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowl TA, Crist TO, Parmenter RR, Belovsky G, Lugo AE. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 2008;6:238–246. doi: 10.1890/070151. [DOI] [Google Scholar]
- 10.Stout JC, Morales CL. Ecological impacts of invasive alien species on bees. Apidologie. 2009;40:388–409. doi: 10.1051/apido/2009023. [DOI] [Google Scholar]
- 11.D’hondt B, et al. Harmonia+ and Pandora+: Risk screening tools for potentially invasive plants, animals and their pathogens. Biol. Invasions. 2015;17:1869–1883. doi: 10.1007/s10530-015-0843-1. [DOI] [Google Scholar]
- 12.Robinson CV, Uren Webster TM, Cable J, James J, Consuegra S. Simultaneous detection of invasive signal crayfish, endangered white-clawed crayfish and the crayfish plague pathogen using environmental DNA. Biol. Conserv. 2018;222:241–252. doi: 10.1016/j.biocon.2018.04.009. [DOI] [Google Scholar]
- 13.Blaustein AR, et al. Effects of invasive larval bullfrogs (Rana catesbeiana) on disease transmission, growth and survival in the larvae of native amphibians. Biol. Invasions. 2020;22:1771–1784. doi: 10.1007/s10530-020-02218-4. [DOI] [Google Scholar]
- 14.Van Wilgen BW, Measey J, Richardson DM, Wilson JR, Zengeya TA. Biological invasions in South Africa: An overview. In: van Wilgen BW, Measey J, Richardson DM, Wilson JR, Zengeya TA, editors. Biological Invasions in South Africa. SpringerOpen; 2020. pp. 3–32. [Google Scholar]
- 15.Murray AG, Munro LA, Matejusova I. The network of farmed Pacific oyster movements in Scotland and routes for introduction and spread of invasive species and pathogens. Aquaculture. 2020;520:734747. doi: 10.1016/j.aquaculture.2019.734747. [DOI] [Google Scholar]
- 16.Dudgeon D, et al. Freshwater biodiversity: importance, threats, status and conservation challenges. Biol. Rev. Camb. Philos. Soc. 2006;81:163–182. doi: 10.1017/S1464793105006950. [DOI] [PubMed] [Google Scholar]
- 17.Strayer DL, Dudgeon D. Freshwater biodiversity conservation: recent progress and future challenges. J. N. Am. Benthol. Soc. 2010;29:344–358. doi: 10.1899/08-171.1. [DOI] [Google Scholar]
- 18.Scheele BC, et al. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science. 2019;363:1459–1463. doi: 10.1126/science.aav0379. [DOI] [PubMed] [Google Scholar]
- 19.Fisher MC, Garner TWJ. Chytrid fungi and global amphibian declines. Nat. Rev. Microbiol. 2020;18:332–343. doi: 10.1038/s41579-020-0335-x. [DOI] [PubMed] [Google Scholar]
- 20.Kiesecker JM, Blaustein AR. Synergism between UV-B radiation and a pathogen magnifies amphibian embryo mortality in nature. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11049–11052. doi: 10.1073/pnas.92.24.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Beneitez MJ, Ortiz-Santaliestra ME, Lizana M, Dieguez-Uribeondo J. Saprolegnia diclina: another species responsible for the emergent disease ‘Saprolegnia infections’ in amphibians. FEMS Microbiol. Lett. 2008;279:23–29. doi: 10.1111/j.1574-6968.2007.01002.x. [DOI] [PubMed] [Google Scholar]
- 22.Kiesecker JM, Blaustein AR, Miller CL. Transfer of a pathogen from fish to amphibians. Conserv. Biol. 2001;15:1064–1070. doi: 10.1046/j.1523-1739.2001.0150041064.x. [DOI] [Google Scholar]
- 23.Oidtmann B. Review of biological factors relevant to import risk assessments for epizootic ulcerative syndrome (Aphanomyces invadans) Transbound. Emerg. Dis. 2012;59:26–39. doi: 10.1111/j.1865-1682.2011.01241.x. [DOI] [PubMed] [Google Scholar]
- 24.Kamilya D, Baruah A. Epizootic ulcerative syndrome (EUS) in fish: History and current status of understanding. Rev. Fish Biol. Fish. 2014;24:369–380. doi: 10.1007/s11160-013-9335-5. [DOI] [Google Scholar]
- 25.Unestam T. Resistance to the crayfish plague in some American, Japanese and European crayfishes. Rep. Inst. Freshw. Res. 1969;49:202–209. [Google Scholar]
- 26.Unestam T, Weiss DW. The host-parasite relationship between freshwater crayfish and the crayfish disease fungus Aphanomyces astaci: Responses to infection by a susceptible and a resistant species. Microbiology. 1970;60:77–90. doi: 10.1099/00221287-60-1-77. [DOI] [PubMed] [Google Scholar]
- 27.Unestam T. On the host range and origin of the crayfish plague fungus. Rep. Inst. Freshw. Res. 1972;52:192–198. [Google Scholar]
- 28.Nyhlén L, Unestam T. Ultrastructure of the penetration of the crayfish integument by the fungal parasite, Aphanomyces astaci Oomycetes. J. Invertebr. Pathol. 1975;26:353–366. doi: 10.1016/0022-2011(75)90233-5. [DOI] [Google Scholar]
- 29.Dieguez-Uribeondo J, et al. Phylogenetic relationships among plant and animal parasites, and saprotrophs in Aphanomyces (Oomycetes) Fungal Genet. Biol. 2009;46:365–376. doi: 10.1016/j.fgb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Rezinciuc S, Sandoval-Sierra JV, Oidtmann B, Diéguez-Uribeondo J. The biology of crayfish plague pathogen Aphanomyces astaci. In: Kawai T, Faulkes Z, Scholtz G, editors. Current Answers to Most Frequent Questions in Freshwater Crayfish: A Global Overview. CRC Press; 2015. pp. 182–204. [Google Scholar]
- 31.Richman NI, et al. Multiple drivers of decline in the global status of freshwater crayfish (Decapoda: Astacidea) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20140060. doi: 10.1098/rstb.2014.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holdich DM, Reynolds JD, Souty-Grosset C, Sibley PJ. A review of the ever increasing threat to European crayfish from non-indigenous crayfish species. Knowl. Manag. Aquat. Ecosyst. 2010;11:394–395. [Google Scholar]
- 33.Martín-Torrijos, L. et al. Crayfish plague in Japan: A real threat to the endemic Cambaroides japonicus. PLoS ONE13 (2018). [DOI] [PMC free article] [PubMed]
- 34.Baran I, Soylu E. Crayfish plague in Turkey. J. Fish Dis. 1989;12:193–197. doi: 10.1111/j.1365-2761.1989.tb00292.x. [DOI] [Google Scholar]
- 35.Loureiro TG, Anastácio PMSG, Araujo PB, Souty-Grosset C, Almerão MP. Red swamp crayfish: Biology, ecology and invasion—An overview. Nauplius. 2015;23:1–19. doi: 10.1590/S0104-64972014002214. [DOI] [Google Scholar]
- 36.Hsieh CY, Huang CW, Pan YC. Crayfish plague Aphanomyces astaci detected in redclaw crayfish, Cherax quadricarinatus in Taiwan. J. Invertebr. Pathol. 2016;136:117–123. doi: 10.1016/j.jip.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 37.Peiró DF, et al. First detection of the crayfish plague pathogen Aphanomyces astaci in South America: A high potential risk to native crayfish. Hydrobiologia. 2016;781:181–190. doi: 10.1007/s10750-016-2841-4. [DOI] [Google Scholar]
- 38.Martín-Torrijos, L., Kokko, H., Makkonen, J., Jussila, J. & Diéguez-Uribeondo, J. Mapping 15 years of crayfish plague in the Iberian Peninsula: The impact of two invasive species on the endangered native crayfish. PLoS ONE14, (2019). [DOI] [PMC free article] [PubMed]
- 39.Matthews M, Reynolds JD. Ecological impact of crayfish plague in Ireland. Hydrobiologia. 1992;234:1–6. doi: 10.1007/BF00010773. [DOI] [Google Scholar]
- 40.Alderman DJ. Crayfish plague in Britain, the first twelve years. Freshw. Crayfish. 1993;9:266–272. [Google Scholar]
- 41.Cerenius L, Bangyeekhun E, Keyser P, Soderhall I, Soderhall K. Host prophenoloxidase expression in freshwater crayfish is linked to increased resistance to the crayfish plague fungus Aphanomyces astaci. Cell. Microbiol. 2003;5:353–357. doi: 10.1046/j.1462-5822.2003.00282.x. [DOI] [PubMed] [Google Scholar]
- 42.Patoka J, et al. Aquarium hitchhikers: attached commensals imported with freshwater shrimps via the pet trade. Biol. Invasions. 2015;18:457–461. doi: 10.1007/s10530-015-1018-9. [DOI] [Google Scholar]
- 43.Cornalia E. Sulla malattia dei gamberi. Atti della Soc. Ital. Sci. Nat. 1860;2:334–336. [Google Scholar]
- 44.Taugbøl T, Skurdal J, Håstein T. Crayfish plague and management strategies in Norway. Biol. Conserv. 1993;63:75–82. doi: 10.1016/0006-3207(93)90076-D. [DOI] [Google Scholar]
- 45.Alderman DJ. History of the spread of crayfish plague in Europe, in Crustaceans: bacterial and fungal diseases. OIE Sci. Tech. Rev. 1997;15:15–23. doi: 10.20506/rst.15.2.943. [DOI] [PubMed] [Google Scholar]
- 46.Diéguez-Uribeondo, J., Temiño, C. & Múzquiz, J. L. The crayfish plague fungus (Aphanomyces astaci) in Spain. Bull. Fr. Pêche Piscic. 753–763 (1997).
- 47.Kawai T, Mitamura T, Ohtaka A. The taxonomic status of the introduced North American signal crayfish, Pacifastacus leniusculus (Dana, 1852) in Japan, and the source of specimens in the newly reported population in Fukushima prefecture. Crustaceana. 2004;77:861–870. doi: 10.1163/156854004774248735. [DOI] [Google Scholar]
- 48.Kawai T, et al. Parthenogenetic alien crayfish (Decapoda: Cambaridae) spreading in Madagascar. J. Crustac. Biol. 2009;29:562–567. doi: 10.1651/08-3125.1. [DOI] [Google Scholar]
- 49.Kawai, T. & Kobayashi, Y. Origin of the red swamp crayfish Procambarus clarkii in Kamakura, Kanagawa prefecture, Japan. Nat. Hist. Rep. Kanagawa32 (2011).
- 50.Huang T, Cerenius L, Söderhäll K. Analysis of genetic diversity in the crayfish plague fungus, Aphanomyces astaci, by random amplification of polymorphic DNA. Aquaculture. 1994;126:1–9. doi: 10.1016/0044-8486(94)90243-7. [DOI] [Google Scholar]
- 51.Lowe, S., Browne, M., Boudjelas, S. & De Poorter, M. 100 of the World’s Worst Invasive Alien Species. A selection from the Global Invasive Species Database. ISSG.12 (2000).
- 52.Makkonen J, et al. MtDNA allows the sensitive detection and haplotyping of the crayfish plague disease agent Aphanomyces astaci showing clues about its origin and migration. Parasitology. 2018;145:1210–1218. doi: 10.1017/S0031182018000227. [DOI] [PubMed] [Google Scholar]
- 53.Panteleit J, et al. Hidden sites in the distribution of the crayfish plague pathogen Aphanomyces astaci in Eastern Europe: Relicts of genetic groups from older outbreaks? J. Invertebr. Pathol. 2011;157:117–124. doi: 10.1016/j.jip.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Diéguez-Uribeondo J, Huang T-S, Cerenius L, Söderhäll K. Physiological adaptation of an Aphanomyces astaci strain isolated from the freshwater crayfish Procambarus clarkii. Mycol. Res. 1995;99:574–578. doi: 10.1016/S0953-7562(09)80716-8. [DOI] [Google Scholar]
- 55.Kozubikova E, Viljamaa-Dirks S, Heinikainen S, Petrusek A. Spiny-cheek crayfish Orconectes limosus carry a novel genotype of the crayfish plague pathogen Aphanomyces astaci. J. Invertebr. Pathol. 2011;108:214–216. doi: 10.1016/j.jip.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Grandjean F, et al. Microsatellite markers for direct genotyping of the crayfish plague pathogen Aphanomyces astaci (Oomycetes) from infected host tissues. Vet. Microbiol. 2014;170:317–324. doi: 10.1016/j.vetmic.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Makkonen J, Jussila J, Kortet R, Vainikka A, Kokko H. Differing virulence of Aphanomyces astaci isolates and elevated resistance of noble crayfish Astacus astacus against crayfish plague. Dis. Aquat. Organ. 2012;102:129–136. doi: 10.3354/dao02547. [DOI] [PubMed] [Google Scholar]
- 58.Makkonen J, Jussila J, Kokko H. The diversity of the pathogenic Oomycete (Aphanomyces astaci) chitinase genes within the genotypes indicate adaptation to its hosts. Fungal Genet. Biol. 2012;49:635–642. doi: 10.1016/j.fgb.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 59.Minardi D, Studholme DJ, van der Giezen M, Pretto T, Oidtmann B. New genotyping method for the causative agent of crayfish plague (Aphanomyces astaci) based on whole genome data. J. Invertebr. Pathol. 2018;156:6–13. doi: 10.1016/j.jip.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 60.Minardi D, Studholme DJ, Oidtmann B, Pretto T, Van Der Giezen M. Improved method for genotyping the causative agent of crayfish plague (Aphanomyces astaci) based on mitochondrial DNA. Parasitology. 2019;146:1022–1029. doi: 10.1017/S0031182019000283. [DOI] [PubMed] [Google Scholar]
- 61.Makkonen J, et al. Mitochondrial genomes and comparative genomics of Aphanomyces astaci and Aphanomyces invadans. Sci. Rep. 2016;6:36089. doi: 10.1038/srep36089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panteleit J, et al. Invasive rusty crayfish (Faxonius rusticus) populations in North America are infected with the crayfish plague disease agent (Aphanomyces astaci) Freshw. Sci. 2019;38:425–433. doi: 10.1086/703417. [DOI] [Google Scholar]
- 63.Makkonen J, et al. The signal crayfish (Pacifastacus leniusculus) in Lake Tahoe (USA) hosts multiple Aphanomyces species. J. Invertebr. Pathol. 2019;166:107218. doi: 10.1016/j.jip.2019.107218. [DOI] [PubMed] [Google Scholar]
- 64.Butler E, et al. Preliminary survey of Aphanomyces sp. associated with native and invasive crayfish in the Lower Susquehanna watershed of South Central Pennsylvania native and invasive crayfish in the Lower Susquehanna watershed of South Central Pennsylvania. J. Freshw. Ecol. 2020;35:223–233. doi: 10.1080/02705060.2020.1779141. [DOI] [Google Scholar]
- 65.Kawai T, Crandall KA. Global diversity and conservation of freshwater crayfish (Crustacea: Decapoda: Astacoidea) In: Kawai T, Cumberlidge N, editors. A Global Overview of the Conservation of Freshwater Decapod Crustaceans. Berlin: Springer; 2016. pp. 65–114. [Google Scholar]
- 66.Thoma, R. The crayfish fauna of Canada and the United States in North America in Freshwater Crayfish: A Global Overview (eds. Kawai, T., Zen Faulkes & Scholtz, G.) 369–403 (2015).
- 67.Casabella-Herrero G, Martínez-Ríos M, Viljamaa-Dirks S, Martín-Torrijos L, Diéguez-Uribeondo J. Aphanomyces astaci mtDNA: insights into the pathogen´s differentiation and its genetic diversity from other closely related oomycetes. Fungal Biol. 2021;125:316–325. doi: 10.1016/j.funbio.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 68.Crandall KA, Buhay JE. Global diversity of crayfish (Astacidae, Cambaridae, and Parastacidae––Decapoda) in freshwater. Hydrobiologia. 2007;595:295–301. doi: 10.1007/s10750-007-9120-3. [DOI] [Google Scholar]
- 69.Martin FN, et al. Insights into evolving global populations of Phytophthora infestans via new complementary mtDNA haplotype markers and nuclear SSRs. PLoS ONE. 2019;14:1–24. doi: 10.1371/journal.pone.0208606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Putra MD, et al. Procambarus clarkii (Girard, 1852) and crayfish plague as new threats for biodiversity in Indonesia. Aquat. Conserv. Mar. Freshw. Ecosyst. 2018;28:1434–1440. doi: 10.1002/aqc.2970. [DOI] [Google Scholar]
- 71.Larson ER, Olden JD. Do schools and golf courses represent emerging pathways for crayfish invasions? Aquat. Invasions. 2008;3:465–468. doi: 10.3391/ai.2008.3.4.18. [DOI] [Google Scholar]
- 72.Larson ER, Olden JD. The state of crayfish in the Pacific Northwest. Fisheries. 2011;36:60–73. doi: 10.1577/03632415.2011.10389069. [DOI] [Google Scholar]
- 73.Oidtmann B, Geiger S, Steinbauer P, Culas A, Hoffmann RW. Detection of Aphanomyces astaci in North American crayfish by polymerase chain reaction. Dis. Aquat. Organ. 2006;72:53–64. doi: 10.3354/dao072053. [DOI] [PubMed] [Google Scholar]
- 74.Cerenius, L., Fuller, M. S., Söderhäll, K. Aphanomyces astaci and Aphanomyces spp. in Zoosporic Fungi in Teaching and Research (ed. Fuller, M. S., Jaworoski, A.) 303 (South Eastern Publishing Corp., 1987).
- 75.Ratkowsky DA, Olley J, McMeekin TA, Ball A. Relationship between temperature and growth rate of bacterial cultures. J. Bacteriol. 1982;149:1–5. doi: 10.1128/JB.149.1.1-5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viljamaa-Dirks S, Heinikainen S. A tentative new species Aphanomyces fennicus sp. nov. interferes with molecular diagnostic methods for crayfish plague. J. Fish Dis. 2019;42:413–422. doi: 10.1111/jfd.12955. [DOI] [PubMed] [Google Scholar]
- 77.Rezinciuc S, Galindo J, Montserrat J, Diéguez-Uribeondo J. AFLP-PCR and RAPD-PCR evidences of the transmission of the pathogen Aphanomyces astaci (Oomycetes) to wild populations of European crayfish from the invasive crayfish species Procambarus clarkii. Fungal Biol. 2014;118:612–620. doi: 10.1016/j.funbio.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 78.Wittwer C, Stoll S, Strand D, Vrålstad T, Nowak C, Thines M. eDNA-based crayfish plague monitoring is superior to conventional trap-based assessments in year-round detection probability. Hydrobiologia. 2018;807:87–97. doi: 10.1007/s10750-017-3408-8. [DOI] [Google Scholar]
- 79.Oidtmann B, Schaefers N, Cerenius L, Soderhall K, Hoffmann RW. Detection of genomic DNA of the crayfish plague fungus Aphanomyces astaci (Oomycete) in clinical samples by PCR. Vet. Microbiol. 2004;100:269–282. doi: 10.1016/j.vetmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 80.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Silvestro D, Michalak I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 84.Rambaut, A. FigTree_v1.4.0 (2012). http://tree.bio.ed.ac.uk/software/figtree/.
- 85.Librado P, Rozas J. DnaSP v5: A software for comprenhensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 86.Clement M, Snell Q, Walker P, Posada D, Crandall K. TCS: Estimating gene genealogies. Parallel Distrib. Process. Symp. Int. Proc. 2002;2:184. [Google Scholar]
- 87.Leigh JW, Bryant D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).