Abstract
Emerging and re-emerging viral diseases can create devastating effects on human lives and may also lead to economic crises. The ongoing COVID-19 pandemic due to the novel coronavirus (nCoV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which originated in Wuhan, China, has caused a global public health emergency. To date, the molecular mechanism of transmission of SARS-CoV-2, its clinical manifestations and pathogenesis is not completely understood. The global scientific community has intensified its efforts in understanding the biology of SARS-CoV-2 for development of vaccines and therapeutic interventions to prevent the rapid spread of the virus and to control mortality and morbidity associated with COVID-19. To understand the pathophysiology of SARS-CoV-2, appropriate animal models that mimic the biology of human SARS-CoV-2 infection are urgently needed. In this review, we outline animal models that have been used to study previous human coronaviruses (HCoVs), including severe acute respiratory syndrome coronavirus (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-CoV). Importantly, we discuss models that are appropriate for SARS-CoV-2 as well as the advantages and disadvantages of various available methods.
Keywords: animal model, ferret, hamster, macaque, mice, pathophysiology, SARS-CoV-2
1 |. INTRODUCTION
In late December 2019, a viral outbreak of unknown aetiology in Wuhan, China, was reported and several patients were admitted to hospitals with a diagnoses of pneumonia and respiratory distress (Bogoch et al., 2020). This ‘viral pneumonia’ was linked to the Huanan Seafood Market in Wuhan, China (Huang et al., 2020). This emerging novel coronavirus (nCoV) was named SARS-CoV-2, and on 11 February 2020, the WHO named the disease associated with SARS-CoV-2 as coronavirus disease 19 (COVID-19) (Li et al., 2020; Rothan & Byrareddy, 2020). Similar to previously detected HCoVs, such as SARS-CoV and MERS-CoV, this nCoV infects human and spread rapidly from person-to-person through droplets (Lewis, 2020). The COVID-19 outbreak was recognized as a pandemic on 11 March 2020 (Cucinotta & Vanelli, 2020). As of 8 October 2020, the virus has surpassed 36 million infections and more than a million deaths worldwide. At the same time, in the United States of America there are more than 7 million confirmed cases and deaths. (https://coronavirus.jhu.edu/us-map).
CoVs are enveloped and positive-stranded RNA viruses that have large viral genomes ranging from 26–32 kb in length (Denison et al., (2011). Common HCoVs, such as 229E and NL63, belong to alpha coronavirus genus, comparatively OC43 and HKU1 belong to the genus betacoronavirus, that usually cause mild and self-limiting respiratory tract infections (Corman et al., 2018). In the last two decades, the pandemics associated with SARS-CoV and MERS-CoV (betacoronaviruses) caused severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS), respectively (Peiris et al., 2003; Raj et al., 2014). The antigenicity of the virus depends on the enveloped spike protein, which is the major determinant of attachment and entry of the virus into host cells (Sui et al., 2004). The viruses belonging to the family coronaviridae have a wide variety of hosts including birds, pigs, cats, dogs, bats, pangolins and humans (Woo et al., 2012). These beta coronaviruses manifest symptoms of varying severity ranging from symptoms comparable to the common cold affecting the upper respiratory tract (URT), to more severe symptoms such as bronchitis, pneumonia and associated fatalities affecting the lower respiratory tract (LRT) (Peiris et al., 2003; Raj et al., 2014). The outbreak of SARS pandemic in 2002 resulted in a 9.6% fatality rate from 8,096 cases and almost 774 deaths worldwide (World Health Organization, 2003). MERS, which emerged in 2012, and was monitored for new cases until October 2018, had 2,229 confirmed cases and 791 deaths resulting in a higher rate fatality rate of around 35.5% (World Health Organization, 2019). These viruses are known for being cross-species (zoonotic) and human-to-human communicable where transmission can be coupled with severe pathological effects (Corman et al., 2018). SARS-CoV and MERS-CoV use angiotensin-converting enzyme 2 (ACE2) (Li et al., 2003) and dipeptidyl peptidase 4 (DPP-4) (Raj et al., 2013) as receptors to enter host cells, respectively.
SARS-CoV-2 belongs to the same clade of beta coronaviruses as SARS-CoV and MERS-CoV, with more than 80% and 50% sequence similarity, respectively (Cui et al., 2019). The genome of SARS-CoV-2, which was isolated from a cluster of patients with pneumonia in Wuhan, had more than 90% nucleotide identity with the bat CoV, RaTG13 (Zhou et al., 2020). The exact route of animal-to-human transmission of SARS-CoV-2 remains unclear but genomic data suggest their evolution is from bats to humans (Zhou et al., 2020) (Figure 1). A recent study suggested that there is 99% sequence similarity between SARS-CoV-2 structural proteins and coronavirus isolated from pangolins, suggesting pangolins may be intermediate hosts (Xiao et al., 2020) (Figure 1). Another study suggested that snakes could be an intermediate host for SARS-CoV-2 based on codon usage (Ji et al., 2020) (Figure 1). In contrast, the bamboo rat has also been proposed to be the intermediate host (Nanshan, 2020;) (Figure 1). Despite active investigation, the definitive animal reservoir responsible for cross-species transmission of SARS-CoV-2 from animals to humans remains elusive (Bassetti et al., 2020). Like SARS and MERS, clinical symptoms of COVID-19 range from acute to severe. Characteristic mild symptoms include fever, sore throat with a dry cough, respiratory stress and myalgia (Huang et al., 2020). In severe conditions, bilateral lung ground-glass opacity is observed under chest-computed tomography (Shi et al., 2020). Neurological manifestations like dizziness, headache, ataxia, seizure with taste, smell and visual impairment were reported (Acharya et al., 2020; Mao et al., 2020) Histopathological reports showed diffuse alveolar damage and pulmonary oedema, indicating acute respiratory distress syndrome (Rothan & Byrareddy, 2020; Xu et al., 2020), which was also seen in SARS-CoV infection (Nicholls et al., 2003).
FIGURE 1.
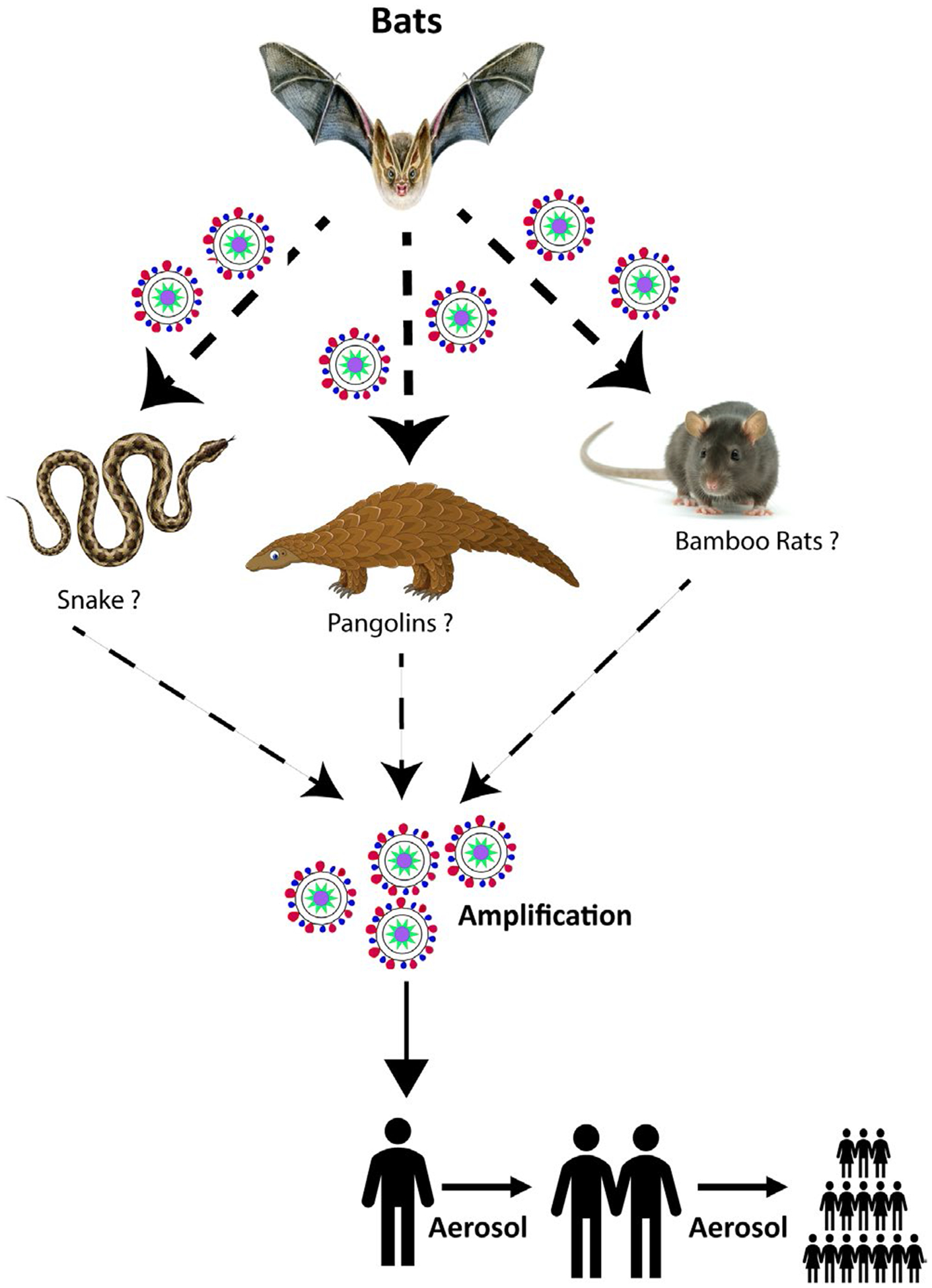
Transmission of SARS-CoV-2 from main reservoir to humans. Bats are the main reservoirs for many other coronaviruses. SARS-CoV-2 may have originated from bats and with the help of unknown intermediate hosts have contributed to human-to-human transmission
SARS-CoV-2 uses ACE2 as a receptor for binding to and entering host cells; this is facilitated by the host protease transmembrane serine protease 2 (TMPRSS2) (Hoffmann et al., 2020). ACE2 is expressed in airway epithelial cells, lung parenchyma and vascular endothelial cells in the kidney and small intestine (Hamming et al., 2004). Therapeutic interventions for COVID-19 such as vaccines, anti-virals and antibodies are an active area of investigation (Mukherjee, 2020), and several have already reached the stage of clinical trials (Scavone et al., 2020). In vitro cell culture models and organoids are also helpful in understanding virus entry and life cycle and for testing the efficacy of therapeutics (Takayama, 2020); however, they are unable to exhibit the exact pathological changes and lesions that are specific to particular organs. Therefore, suitable biological and relevant animal models are necessary for testing the safety and efficacy of the vaccines, anti-virals and other potential therapeutics that are currently in the pipeline.
2 |. REQUIREMENTS OF A SARS-COV-2 ANIMAL MODEL
An ideal animal model should be permissive to infection and be able to mimic the clinical pathology of the disease observed in humans (Sutton & Subbarao, 2015). These models play a crucial role in studying host-virus interactions that contribute to disease pathogenesis and transmission. A protocol for the development of animal models is mentioned by the ‘FDA animal efficacy rule’ (Product Development Under the Animal Rule, 2015). Accordingly, these models should have the same receptors as those present in humans, which help viruses during the attachment and entry process, and the outcome of infection should match the severity observed in humans. In many emerging diseases, in vitro studies cannot completely simulate human pathophysiology. Also, the immunological components are very complex in humans, which cannot be proven in the in vitro experiments. However, despite differences between animal models and humans, critical information related to the pathogenesis, prevention and treatment of newly emerging infectious diseases can still be discovered.
Animal models can be categorized as large or small. Small animal models include the use of mice, rats, ferrets, hamsters and rabbits, which are relatively smaller in size and require limited space. Small animal models offer several advantages because they, and the animal-specific reagents, are readily available, the animals can be handled with less effort and cost, and they may be used in large numbers for stronger statistical power during data analysis (Sutton & Subbarao, 2015). The limitation with using a small animal model is the significant intrinsic biological differences between humans and rodents or small mammals. This divergence led to requirements declaring that either virus must be adapted to the animals or the animals must be genetically manipulated to recapitulate the human system (Subbarao et al., 2004). In addition, the animals’ short life span hampers the ability to monitor the long-term prognosis of the disease (Gretebeck & Subbarao, 2015). Large animal models such as non-human primates (NHP) are more reliable models to replicate human disease pathogenesis as they are physiologically, immunologically and genetically more closely related to humans (Fujiyama et al., 2002; Lu et al., 2008). The major limitations of using NHP models are the high cost and resources involved, which limit the number of animals that may be included in a study and thus adversely affect the statistical power of the outcome. In addition, most NHPs are outbred animals and have a wide variability in genetic backgrounds, which sometimes make it difficult to interpret the outcome of a study due to variability in results among individual animals (Lu et al., 2008).
For the ongoing COVID-19 pandemic, direct human clinical trials are complicated by the mild to severe forms of the disease due to genetic diversities, age of the host, comorbidities and multiple infections along with other pre-existing diseases (Chan & Chan, 2013; Gold et al., 2020; Begley, 2020). The virus uses the ACE-2 receptor that is expressed in cells found in the heart, lungs, gastrointestinal tract and urinary tract (Hoffmann et al., 2020). After entry, viral replication kinetics in most of the target cells remains unknown. Air droplets and aerosols are critical aspects of the global spread of SARS-CoV-2 (Hamid et al., 2020; Meselson, 2020). Regardless, there is an urgent need to understand the risks factors of transmission for SARS-CoV-2 (Figure 2). The symptoms of COVID-19 with other specific and non-specific upper respiratory tract symptoms are similar in nature, which make it difficult to distinguish it from other diseases (Bhatraju et al., 2020). A case report from Eiju General Hospital Tokyo, Japan, reported the existence of the influenza virus along with SARS-CoV-2 showing similar types of symptoms that posed a difficulty in the differential diagnosis of the two diseases (Azekawa et al., 2020). Another case in Rhode Island showed the co-infection of SARS-CoV-2 with human metapneumovirus, where a patient tested positive for human metapneumovirus but failed to test for SARS-CoV-2. The symptoms did not subside even after treatment for metapneumovirus. Subsequently, the patient tested for SARS-CoV-2 by PCR (Touzard-Romo et al., 2020). A travel-related case of an 80-year-old man from Japan, with a history of diabetes, showed co-infection of SARS-CoV-2 and Legionella with respiratory distress and gastrointestinal symptoms. The patient passed away after 13 days and later was confirmed to be infected with COVID-19 (Arashiro et al., 2020). These individual case reports reveal the need for suitable animal studies of SARS-CoV-2 co-infection with other pathogens that have similar disease manifestations for better management of clinical outcomes.
FIGURE 2.
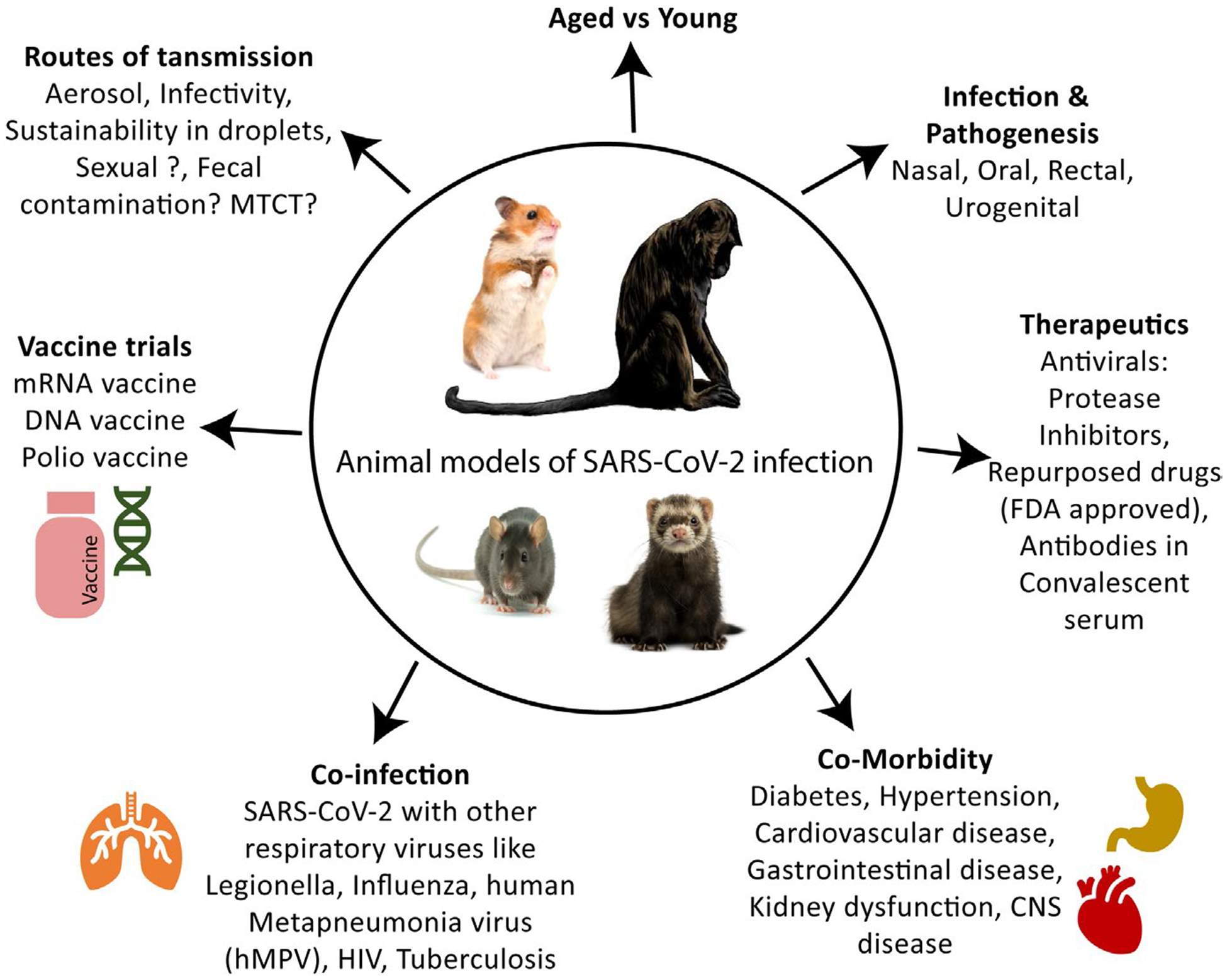
Utility of small and large animal models of SARS-CoV-2 infection ad pathogenesis. The possible use of animal models in SARS-CoV-2 infection has been described
COVID-19 has a variety of clinical outcomes (Chan et al., 2020; Huang et al., 2020). Most of the patients who are admitted to hospitals with severe clinical manifestations have other comorbid conditions such as diabetes, cardiovascular disease, gastrointestinal disease or hypertension (Gold et al., 2020). In the case of Influenza, one publication showed that the risk of the acute respiratory distress increases 3.4-fold in the H7N9-infected person with similar comorbidity (Gao et al., 2013). Age-related comorbidity has mostly affected the transmission cycle of the disease (Sun et al., 2020). The exact mechanism of how these comorbidities deteriorate SARS-CoV-2 patient conditions remains unknown. These comorbid conditions that lead to severe to fatal outcomes are another aspect of COVID-19 pathogenesis that require a suitable animal model for study.
There is a concerted effort in the research community to develop therapeutics and vaccines against COVID-19. For example, Remdesivir is an anti-viral drug that failed the clinical trial for Ebola a decade ago (Mulangu et al., 2019) and now has been proposed for COVID-19 treatment (Beigel et al., 2020) (ClinicalTrials.gov: NCT04257656, NCT04252664, NCT04280705). It was repurposed for COVID-19 treatment because it directly blocks RNA synthesis (Agostini et al., 2018). Preliminary reports for Remdesivir in a clinical trial of 1,063 adult patients with lower respiratory tract infection showed a shortened recovery time in 538 patients given Remdesivir intravenously (median 11 days), compared to the 521 patients given a placebo (median 15 days) (Beigel et al., 2020). There were still serious adverse effects reported in COVID-19 patients given Remdesivir, but Remdesivir worked better in comparison with the placebo. A study using Lopinavir (a protease inhibitor) and Ritonavir (primes the action of Lopinavir which is widely used to treat HIV infection) is in clinical trials for COVID-19 (Bhatnagar et al., 2020). ACE-2 inhibitors as well as some fusion inhibitors such as Arbidol are also in clinical trials (Chinese Government Clinical Trials: ChiCTR2000029573) (Wang et al., 2020). Clinical trials using convalescent serum as treatment for COVID-19 have already begun at the Icahn School of Medicine at Mount Sinai, New York, USA (Liu et al., 2020). Similarly, polyclonal antibodies have been employed in clinical trials because polyclonal human immunoglobulin G (IgG) showed effectiveness against MERS-CoVs(Luke et al., 2016). Vaccine development is important for achieving control of the disease and protection against the transmission of the virus to immunized individuals. Many groups are working to develop potential vaccines for SARS-CoV-2, out of which some candidates are already in clinical trials (Mukherjee, 2020). Moderna, a vaccine manufacturer, has started phase II clinical trials of their vaccine candidate mRNA-1273, which passed phase I trial recently by generating desired immune responses by natural infection (ClinicalTrials.gov Identifier: NCT04283461) (Moderna, Inc., 2020, Press release).
Recombinant protein-based vaccines by University of Queensland and viral vector-based vaccines by University of Oxford, England, are also being actively tested for vaccine efficacy (NIAID, 2020 Press release). Another study has suggested that polio vaccine could be used to prevent SARS-CoV-2 (Yeh et al., 2020).
In order to study various unanswered questions about disease pathogenesis, suitable animal models are essential. Several studies are in progress to find suitable animal models to study the transmission, disease pathogenesis and pre-clinical trials of potential therapeutics for the management of COVID-19. However, to successfully end the COVID-19 pandemic, efforts to develop vaccines to prevent the virus spread should synchronize with studies to uncover disease pathogenic mechanisms in comorbid and high-risk co-infection conditions. Although difficult, identifying a model that adequately mimics the human disease of SARS-CoV-2 is bolstered by several studies of SARS-CoV and MERS-CoV, which has provided some preliminary insights on the models. All laboratory animal models, from mice and hamsters to ferrets and NHPs, are valuable in addressing various questions to better understand disease mechanisms.
3 |. ANIMAL MODELS FEASIBLE FOR SARS-COV-2 STUDIES
3.1 |. Mouse model
Mice are the most widely used animal models in infectious disease research. Mice have a well-characterized immune system, high reproduction capability and short gestation period. Many inbred mouse strains have been tested for SARS-CoV infections. BALB/c mice, C57BL/6 (B6) and 129SvEv mice were challenged intranasally with SARS-CoV Urbani strain (Glass et al., 2004; Roberts et al., 2005). This resulted in a productive infection with peak titres on day 3, but there was an early clearance of the virus. Furthermore, mice did not lose weight, display signs of clinical disease nor develop pulmonary pathology observed in humans (Yin and Wunderink, (2018). Mutant mice such as B6, Beige and CD1−/− (lack NK cell function and NK-T cells) and RAG1−/− mice (lack T and B lymphocytes) did not develop clinical disease (Glass et al., 2004). STAT1−/− mice showed some clinical signs of weight loss and bronchiolitis, but were unable to show the pathological signs and mortality observed in human cases (Frieman et al., 2010). Examination of the ACE-2 amino acid sequence of mouse and rat revealed differences in the amino acids at positions 353 and 82 compared to human. Mice have histidine, and humans have lysine at position 353 (Li et al., 2005), which may explain why SARS-CoV replicated less efficiently in murine cells (Li et al., 2004). At position 82, mice ACE2 has serine, whereas human ACE2 has methionine; this difference did not prevent the S-protein of SARS-CoV from binding to mice cells and entering, but there was reduced permissiveness of these cells to infection, thus making it semi-permissive to the SARS-CoV.
Various transgenic mouse models like mice expressing the human ACE2 (hACE2) receptor and transgenic hACE2 under the control of an epithelial cell-specific promoter K18 (K18-hACE2) showed infectivity with disease pathogenesis similar to that in humans (Bao et al., 2020; McCray et al., 2007), thus making them suitable for pathogenesis studies and evaluation of vaccines and other therapeutics against SARS-CoVs. Genetically engineered mice expressing human ACE-2 (hACE2) are suitable for SARS-CoV infection and produced mortality (Dediego et al., 2008). Different contract research companies have produced hACE-2 expressing mice models and are already supplying them for SARS-CoV-2 studies. Taconic Biosciences, Rensselaer, New York, is supporting CoV research by providing ACE2-expressing mice, which express human ACE2 cDNA under the control of the CAG promoter. The Jackson Laboratory (JAX), Maine, Florida, is working to provide a transgenic mouse expressing hACE-2 that was originally developed by Dr. Stanley Perlman at the University of Iowa. A study by Bao et al., used the mice model for SARS-CoV-2 and reported that transgenic hACE2 mice, when inoculated with BetaCoV/Wuhan/IVDC-HB-01/2020|EPI_ISL_402119 strain, developed clinical signs of COVID-19, including weight loss and interstitial pneumonia with histopathological signs in lungs (Table 1) (Bao et al., 2020) (Table 1). Jiang et. al. used a SARS-CoV-2 hACE2 transgenic mouse (HFH4-hACE2 in C3B6 mice) infection model, where infected mice demonstrated clinical pathology like that of COVID-19 patients (Jiang et al., 2020). Mice were infected with 3 × 10^4 TCID50 virus, each intranasally, and samples were collected after necropsy; lung samples were deemed a major site for viral replication; additionally, some viral RNA was found in the heart, brain and eye. They were rechallenged with the same dose of the virus via the same route, which revealed generation of antibodies against SARS-CoV-2, which protected them from pneumonia on re-exposure. Viral vector-mediated hACE2 delivery systems have been developed and used in SARS-CoV-2 studies. A study done by Doremalan et. al. showed the immunogenicity of the adenovirus-vectored vaccine ChAdOx1 nCoV-19, encoding the spike protein of SARS-CoV-2 in mice (Doremalen et al., 2020). They used two mouse strains- BALB/c and outbred CD1 which were vaccinated with ChAdOx1 nCoV-19 or ChAdOx1 GFP, a control vaccine expressing green fluorescent protein, intramuscularly. There was robust generation of humoral and cellular immunity after 9–14 days of vaccination, which was detected by examination of total IgG titres. Similarly, researchers at Icahn School of Medicine at Mount Sinai compared K18-hACE2 transgenic model and adenovirus-mediated delivery of hACE2 to the mouse lung. They showed that while adenovirus-mediated delivery produced lower viral replication in the lungs of mice and showed no profound clinical signs of infection even though challenged with 104 plaque forming units (PFU) virus dose, K18-hACE2 mice, when infected, showed high viral replication in both the lung and brain which led to fatal conditions in mice. This study shows the potency of K18-hACE2 mice models in studying anti-viral and vaccine efficacy (Rathnasinghe et al., 2020). Researchers at the University of Pittsburgh School of Medicine have developed a vaccine candidate ‘PittCoVacc’ (Pittsburgh CoV Vaccine) using in vitro-generated fragments of the viral proteins. Administration of the vaccine to transgenic BABL/c mice resulted in the successful production of neutralizing antibodies against SARS-CoV-2 (Kim et al., 2020).
TABLE 1.
Animal models permissive for SARS-CoV-2 infections
| Models | Virus strain | Route of Inoculation | Dose of virus | Clinical signs | Viral RNA | Histopathology | References | |
|---|---|---|---|---|---|---|---|---|
| Mouse | Transgenic hACE2 mice | BetaCoV/Wuhan/IVDC-HB-01/2020|EPI_ISL_402119 | Intranasal | 10^5 TCID50 | Weight loss and interstitial pneumonia | Lungs tissue | Focal to multifocal dark red discoloration in certain lung lobes | Denison et al., (2011) |
| Transgenic hACE2 mice | SARS-CoV-2 | Intranasal | 3 × 10^4 TCID50/ml | No clinical signs until day-3 post-infection | Lungs tissue | Slight changes were observed in the lung tissues, multifocal lesions, alveolar walls thickening and monocyte and lymphocyte infiltration |
Doremalen et al., (2020) | |
| BALB/c and outbred CDI. | SARS-CoV-2 strain nCoV-WA1–2020 (MN985325.1) | Intranasal | 2.6 × 10^6 TCID50/ml | No clinical signs in immunized animals | N/A | Rathnasinghe et al., (2020) | ||
| K18-hACE2 transgenic model | SARS-CoV-2, isolate USA-WA1/2020 | Intranasal | No clinical signs | Kim et al., (2020) | ||||
| BALB/c Mice | Mouse-adapted 2019-nCoV/USA-WA1/2020 | Intranasal | 1 × 10^4 PFU | Nasal congestion and difficulty in breathing | Lungs tissue | N/A | Gu et al., (2020) | |
| BALB/c Mice | Mouse-adapted-BetaCov/human/CHN/Beijing_IME-BJ05/2020 | Intranasal | 1.6 × 10^4 PFU | N/A | Lungs, trachea, heart, liver, spleen, brain and faeces. | Denatured and collapsed epithelial cells, thickened alveolar septa, alveolar damage, focal exudation and haemorrhage and activated inflammatory cell infiltration | Dinnon et al., (2020) | |
| BALB/c Mice | Mouse-adapted infectious clone of SARS-CoV-2 | Intranasal | 10^5 PFU | No clinical signs | Lungs | Inflammation of airways | Wang et al., (2020) | |
| BALB/c Mice and C57BL/6J | Mouse-adapted SARS-CoV-2 HRB26M | Intranasal | 10^4.4 pFU | N/A | Nasal turbinates and lungs | Loss of cilia in the nasal tissues, infiltrations of monocytes, congestion of intra-alveolar septa. | Gao et al., (2020) | |
| Hamster | Syrian/Golden Hamster | Isolated from the nasopharyngeal aspirate specimen of a laboratory-confirmed | Intranasal | 1 × 10^5 PFU | Weight loss, ruffled furs, stressed breathing | Lungs tissue and intestines | Patches of focal inflammation and pleural invaginations in lungs | Chan et al., (2020) |
| COVID-19 patient in Hong Kong | ||||||||
| Syrian/Golden Hamster | BetaCoV/Hong Kong/VM20001061/2020 virus | Intranasal | 8 × 10^4 TCID50 | N/A | Nasal washes, lungs, faecal samples | Presence of inflammatory cells and consolidation in the lungs | Sia et al., (2020) | |
| Syrian/Golden Hamster | SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo (UT-NCGM02) | Intranasal and ocular | 10^5.6 PFU or with 10^3 PFU | Mild to no weight loss | Nasal turbinates, trachea, lungs and brain | Ground-glass opacity and consolidation of lungs under microcomputed tomographic imaging | Imai et al., (2020) | |
| Ferret | Normal Ferrets | SARS-CoV-2/F13/environment/2020/Wuhan (F13-E) or SARS-CoV-2/CTan/human/2020/Wuhan (CTan-H) | Intranasal | 10^5 PFU | Weight loss was not reported. | Nasal turbinate, tonsils and soft palate | N/A | Shi et al., (2020) |
| Normal Ferrets | NMC-nCoV02 | Intranasal | 10^5.5TCID50 | Elevated body temperatures, body weight loss, cough | Blood, nasal washes, saliva, urine and faeces | Infiltration as well as cell debris in the alveolar wall, bronchial epithelium and bronchial lumen | Kim et al., (2020) | |
| Normal Ferrets | SARS-CoV-2 strain from a German traveller returning from China | Intranasal | 6×10^5 TCID50 | N/A | Throat, nasal and rectal swab | N/A | Mathilde et al., (2020) | |
| Normal Ferrets | Intranasal | Schlottau et al., 2020 | ||||||
| Normal Ferrets | Victoria/1/2020 SARS-CoV-2 | Three different titres; high (5 × 10^6 PFU/ml), medium (5 × 10^4 PFU/ml) and low dose (5 × 102 PFU/ml | Reduced activity, elevated body temperature, | Nasal wash, throat swab, rectal swabs, bronchoalveolar lavage | Multifocal bronchopneumonia, infiltration of inflammatory cells in lungs, thickening of alveolar septa. | Kathryn et al., 2020) | ||
| Non-Human Primate | Cynomolgus macaques | SARS-CoV-2 strain from a German traveller returning from China | Combined intratracheal and intranasal | N/A | Little or no symptoms with serous nasal discharge only | Nasal and throat swabs | Consolidation of pulmonary tissues with oedema, fibrin | Rockx et al., (2020) |
| Rhesus macaques | Purified inactivated SARS-CoV-2 strains from the bronchoalveolar lavage fluid samples | PiCoVacc, vaccine candidate, through IM route SARS-CoV-2 CN1 challenged by Intratracheal route | 10^6 TCID50 | N/A | Controlled animals showed viral genomic RNA in the pharynx, crissum and lung | Very mild in lungs | Moderna Inc., (2020) | |
| Rhesus macaques | BetaCoV/Wuhan/IVDC-HB-01/2020|EPI_ISL_402119 | Intratracheal | 10^6 TCID50 | Weight loss, fever | Nose, pharynx, and crissum, anal swabs | Oedema filled alveolar cavities, filled with inflammatory cells | Yu et al., (2020) | |
| Rhesus macaques | SARS-CoV-2 strain nCoV-WA1–2020 | Intranasal, oral, ocular and intratracheal routes | 2.6×10^6 TCID50 | Weight loss, ARDS | Nasal and broncholavage | Mild-to-moderate interstitial pneumonia | Williamson et al., (2020) | |
| Rhesus macaques | SARS-CoV-2/WH-09/human/2020/CHN | Intratracheal | 10^6 TCID50 | Weight loss, reduced appetite, increased respiration rate and hunched posture | Nasal, pharyngeal and anal swabs. tissues of nose, gut, spinal cord, heart, bladder | Thickened alveolar septa, accumulation of alveolar macrophages in the alveoli, degeneration of the alveolar epithelia, and infiltration of inflammatory cells | Linlin et al., (2020) | |
| Rhesus macaques | SARS-CoV-2 isolate nCoV-WA1–2020 | Intranasal, oral, ocular and intratracheal routes | 2.6 × 10^6 TCID50 | reduced appetite, hunched posture, pale appearance and dehydration | nose, throat and rectal swabs | Multifocal interstitial pneumonia, oedematous alveoli, pneumocyte hyperplasia | Vincent et al., (2020) | |
| Rhesus macaques | SARS-CoV-2 USA-WA1/2020 | Intranasal and intratracheal | 1.1 × 10^6 PFU (n = 3), 1.1 × 10^5 PFU (n = 3) PFU or 1.1 × 10^4 PFU (n = 3) | Loss of appetite | Bronchoalveolar lavage (BAL) and nasal swabs | Mononuclear infiltrates and multifocal regions of inflammation in alveoli | Chandrashekar et al., (2020) | |
| Rhesus macaques | SARS-CoV-2 | Intratracheal and intranasal | 1.2×10^8 VP (1.1×10^4PFU) SARS-CoV-2 | N/A | Nasal swabs and bronchoalveolar lavage (BAL) | N/A | Yu et al., (2020) | |
| Chinese-origin rhesus macaques | SARS-CoV-2/WH-09/human/2020/CHN | Intratracheal | 1 × 10^6 TCID50 | Weight loss but not elevated rectal temperature., reduced appetite and increased in respiration | Nasal swabs and pharyngeal swabs | Interstitial pneumonia, widened alveolar septa, increased alveolar macrophages and degenerated alveolar epithelium. | Deng et al., (2020) | |
| Chinese-origin rhesus macaques | SARS-CoV-2 (strain 2019-nCoV-WIV04) | Intratracheal | 2 × 10^4 TCID50 | N/A | Pharyngeal swabs, bronchus and lungs | Severe interstitial pneumonia, expansion of alveolar septum, infiltration of monocytes and lymphocytes. | Feng et al., (2020) |
Many researchers have been trying to adapt the virus in a mouse model that can produce comparably severe clinical signs as seen in humans. A study by Sheahan et. al. showed that an orally bioavailable broad-spectrum, ribonucleoside analog β-D-N4-hydroxycytidine (NHC, EIDD-1931) is able to inhibit SARS-CoV-2 replication. Following intranasal challenge with a mouse-adapted 2019-nCoV/USA-WA1/2020 strain of SARS-CoV-2 virus, there was nasal congestion and difficulty in breathing, both of which are symptoms that have been reported in COVID-19 patients (Sheahan et al., 2020). Researchers at the NIH tested an mRNA vaccine, developed by Moderna in Phase I clinical trials, and confirmed immune responses by natural infection where BALB/c mice were challenged with mouse-adapted SARS-CoV-2, which is now in its phase III trial (Moderna Inc., 2020, Press release). Gu et. al. also generated a mouse-adapted SARS-CoV-2 from serial passage of human clinical isolate BetaCov/human/CHN/Beijing_IME-BJ05/2020 in aged mice (Gu et al., 2020). Nine-month-old (aged) and six-week-old (young) BALB/c mice were intranasally inoculated with mouse-adapted strain of SARS-CoV-2, and after 3, 5 and 7 days post-infection, lungs and tracheal samples were measured for viral loads. A high virus titre was detected in the trachea and lungs, and in heart, liver, spleen and brain, as well as in faeces from both young and old-aged mice. Mild-to-moderate pneumonia was seen in both aged mice, whereas more severe pathological signs like interstitial pneumonia characterized by thickened alveolar walls due to inflammatory cell infiltration were seen in aged mice. Similar to this study, Dinnon et. al. showed the pathogenicity of mouse-adapted SARS-CoV-2 on aged mice to prove the comorbidities and its effect in aged patients(Dinnon et al., 2020). They used mouse-adapted SARS-CoV-2 in young and 12-month old mice where they found virus replication in upper and lower respiratory tracts of both aged mice, but severe pathological conditions were only observed in old-aged mice. Wang et. al. also showed mouse-adapted SARS-CoV-2 replication in upper and lower respiratory tract of aged and young BALB/c and C57BL/6J mice with high pathogenicity in aged mice(Wang, et al., 2020). These studies demonstrate that severity of pathogenicity in COVID-19 in patients correlates with age.
A purified, inactivated SARS-CoV-2 virus candidate (PiCoVacc) tested in mice, rats and non-human primates has also shown a protective immune response suggesting neutralizing activity against SARS-CoV-2 (Gao et al., 2020). Administration of an inactivated SARS-CoV-2 vaccine, PiCoVacc, to ten BALB/c mice at multiple doses on day-0 and day-7 produced a good amount of SARS-CoV-2 spike and receptor binding domain-specific immunoglobulin G (IgG) responses (Gao et al., 2020). All these studies signify the importance of mouse models in the testing of vaccines against SARS-CoV-2.
Transgenic mice model, mouse-adapted SARS-CoV-2 virus, immunodeficient mice and delivery of viral proteins through vector in mice represent promising mice models that are beneficial in the study of SARS-CoV-2. The major limitation with transgenic mice is that they are susceptible to lethal encephalitis. However, they are suitable models for therapeutics and pathogenesis studies. Similarly, mouse-adapted SARS-CoV-2 is produced by serial passaging of virus in mouse lungs, which reduces virus potency but on the other hand also increases chances of mutations. They could be helpful in vaccine studies and comparison of COVID-19 pathogenicity (Table 2).
TABLE 2.
Types of studies in which different animal models can be used
| SN | Animal models | Types/species of animal | Types of study | References( | Limitation of the model |
|---|---|---|---|---|---|
| 1 | Mouse | Transgenic hACE2 mice, normal mouse can be infected with mouse-adapted mice | Infection and pathogenesis, vaccine trials, therapeutics | (Bao et al., 2020; Doremalen et al., 2020; Rathnasinghe et al., 2020; Sheahan et al., 2020; Gu et al., 2020; Dinnon et al., 2020; Wang, et al., 2020; Gao et al., 2020) | Mice cannot be used directly, needs to be made transgenic or mouse-adapted virus should be used for infection |
| 2 | Hamster | Syrian/Golden hamsters are permissive to SARS-CoV-2 | Infection and pathogenesis, vaccines, therapeutics, transmission study | (Chan et al., 2020; Sia et al., 2020; Imai et al., 2020) | Hamsters do not mimic all the clinical signs; mortality is not observed. Viral clearance was soon, cannot be used for long-term pathogenesis |
| 3 | Ferret | Ferrets are permissive to SARS-CoV-2 | Vaccines, therapeutics, transmission study | (Kim et al., 2020; Shi et al., 2020; Mathilde et al., 2020; Schlottau et al., 2020; Kathryn et al., 2020) | Ferrets do not mimic all the clinical signs; mortality is not observed |
| 4 | Non-Human Primate | Rhesus macaques and cynomolgus macaques are permissive for infection | Comorbidity studies, infection and pathogenesis, co-infection studies, vaccine trials, therapeutics | (Moderna Inc., 2020; Bao et al., 2020; Deng et al., 2020; Yu et al., 2020; Chandrashekar et al., 2020; Feng et al., 2020; Vincent et al., 2020; Williamson et al., 2020; Rockx et al., 2020) | Not all the macaque’s species could be infected, common clinical signs were mild/moderate |
3.2 |. Hamster model
Golden Syrian hamsters are rodents that belong to the hamster subfamily and are widely used in research related to human diseases, accounting for 19% of research in the United States (Valentine et al., 2012). These hamsters showed productive viral replication as well as pathological signs in lungs and disease pathogenesis when challenged with SARS-CoV. However, despite the fact that lung pathology looked similar to that in humans, hamsters did not recapitulate clinical disease nor mortality in SARS-CoV infection (Roberts et al., 2006). Many other studies used different strains such as Urbani, HKU-39849, Frankfurt 1 and a recombinant clone GD03T0013, but failed to recapitulate the SARS-CoV-mediated fatality in hamsters (Roberts et al., 2008). Collectively, these studies demonstrate that the hamster represents a suitable model for SARS-CoV infection, but it is not suitable to model severe disease that is at times fatal in the human.
Hamsters support SARS-CoV-2 replication, (Chan et al., 2020) which was recently demonstrated. In a study from Hong Kong, 8 hamsters were infected with a viral isolate purified from the nasopharyngeal aspirate specimen of a laboratory-confirmed COVID-19 patient. Clinical signs included weight loss, ruffled furs and laboured breathing with high viral loads in the hamster’s lungs and intestines. Histopathological examination revealed multifocal areas of inflammatory cell infiltration with pleural invaginations in lung tissues (Chan et al., 2020) (Table 1). Another study used hamsters as a model to understand the mechanism of intraspecies transmission (Sia et al., 2020). In this study, nine hamsters were infected intranasally with BetaCoV/Hong Kong/VM20001061/2020 virus where peak viral load was attained at day 2 post-infection in nasal washes, lungs and faecal samples. Histopathological changes like consolidation and infiltration in lungs, kidneys and intestines were detectable 7 days post-infection. At 24 hr post-inoculation, 3 of the experimentally infected animals were group housed with 3 naïve hamsters, and after remaining in full contact for one day, they successfully transmitted the infection to naïve hamsters with substantial viral loads in nasal swabs, and clinical signs including weight loss and respiratory distress (Table 1). At day-13 post-contact, both donors and contact hamsters had developed neutralizing antibodies against SARS-CoV-2 (Sia et al., 2020). These studies clearly suggest that hamsters can be used to better understand the transmissibility of the virus.
Syrian hamsters have also been used to investigate age-related effect in COVID-19 patients and the prophylactic models against SARS-CoV-2. Imai et al., used two different groups of Syrian hamsters—Juvenile (a month old) and Mature adults (7–8 months old)— and infected them with SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo (UT-NCGM02) strain at 10^5.6 (PFU) or with 10^3.0 PFU by intranasal and ocular routes (Imai et al., 2020). There was mild to no weight loss in both groups, but high virus titres were observed in the nasal turbinates, trachea, lungs and brain. Severe pathological lung lesions were seen with the higher infectious doses used. Microcomputed tomographic imaging showed severe lung lesions such as consolidation of lungs and ground-glass opacity. When rechallenged with the same dose of virus, they were shown to develop protective immunity against the virus. Next, the authors transferred convalescent serum from the infected animals to the naïve animals, which suppressed virus replication 2 days post-infection suggesting the advantages of the hamster model in testing vaccine efficacy and anti-virals.
The sequence of hamster ACE2 revealed the presence of asparagine at position 82, which contrasts with human ACE2 that has lysine at this position. Although hamsters were permissible for SARS-CoV and SARS-CoV-2 infections, they did not show severe fatal disease as seen in the human, which may be due to the amino acid change at position 82. Also, there is a lack of sequence data and other tools needed to carry out experiments in hamsters. Despite these shortcomings, hamsters may be used as models for SARS-CoV-2 because they do not need any adapted virus or transgenic models. Taken together, a hamster model would be a good model to study transmission and consequently immunoprophylaxis (Table 2).
3.3 |. Ferret model
Ferrets have been used mainly in respiratory disease-related studies because their lungs share many similarities with that of humans (Vinegar et al., 1985). Ferrets mimic clinical symptoms of the SARS-CoVs such as coughing and fever. They were previously used in human (Lee et al., 2020) and Swine influenza virus studies (Brand et al., 2010). Ferrets are permissive to SARS-CoV infection (Martina et al., 2003; Weingartl et al., 2004) and may be a potential model for the study of the SARS-CoV-2. Like mice models, ferrets were shown to support SARS-CoV replication with varying degrees of clinical signs, but without significant mortality. When ferrets were infected with SARS-CoV at a high dose, they showed productive infection in the lungs, trachea and nasal turbinates (Martina et al., 2003). Viral replication peaked in the lungs on days 5 or 6 as seen in humans, but another study using the same conditions failed to produce infection and mortality (Weingartl et al., 2004).
Ferrets may be a suitable model to study SARS-CoV-2 pathogenesis and human-human transmission (Kim, et al., 2020; Shi, et al., 2020). Shi et al., tested the susceptibility of ferrets to SARS-CoV-2 by infecting pairs of ferrets with two viral strains: SARS-CoV-2/F13/environment/2020/Wuhan (F13-E) or SARS-CoV-2/CTan/human/2020/Wuhan (CTan-H) intranasally with 10^50 PFU. After four days of inoculation, the animals were euthanized and tissues from the nasal turbinate, soft palate, tonsils, trachea, lung, heart, liver, spleen, kidneys, pancreas, small intestine and brain were collected. Viral RNA was detected in areas of the upper respiratory tract-nasal turbinate, soft palate and tonsils (Shi, et al., 2020). Another study from Korea showed that when infected intranasally with NMC-nCoV02, a strain isolated from a COVID-19 patient in South Korea, clinical signs developed two days after infection (Kim, et al., 2020). Although they showed an increment in body temperature, lethargy and occasional coughs, all animals recovered day 8 post-infection (Table 1). In follow-up studies, naïve ferrets were exposed to the infected ones to study the transmission dynamics. Interestingly, nasal swabs and saliva samples of the exposed ferrets showed high viral loads. In support of this, two other studies from the Netherlands and Germany demonstrated aerosol mediated transmission of SARS-CoV-2 from infected to naïve exposed ferrets (Schlottau et al., 2020; Richard et al., 2020) (Table 1). These studies also showed that the pathogenicity of the virus was the same in the inoculated and subsequently exposed naïve ferrets. These studies showed the rapidity of virus transmission and the requirement of the appropriate distancing to avoid getting infected. A study by Ryan et al., used a ferret model to understand the suitable dose of virus necessary to conduct experiments (Kathryn et al., 2020). Ferrets were challenged intranasally with 1ml of Victoria/1/2020 SARS-CoV-2 at three different titres: high (5 × 10^6 PFU/ml), medium (5 × 104 PFU/ml) and low dose (5 × 10^2 PFU/ml). The high and medium dose induced virus replication in upper respiratory tract consistently in all the infected animals that developed pathological bronchopneumonia, whereas in low dose group only one animal out of six got infected. In the high dose virus-infected group, viral shedding was observed from days 14 to 21 which represents the mild clinical symptoms that are mostly seen in the patients.
Ferrets are permissive to SARS-CoV-2 infection, but they are unable to produce clinical signs and symptoms with the same degree of severity as seen in humans. Other difficulties in working with ferrets are their larger size compared to mice and hamsters, difficulty in handling, expenses and uniquely ferret-specific reagents. Nonetheless, ferrets can reproduce the virus dynamics of infection and transmission pattern (Table 2). The respiratory illness seen in ferrets resembles that in humans because their lungs are proportionately larger compared to other organs in their body. Moreover, their lungs abundantly express ACE2 protein similar to the human, which makes them excellent models to study the early events in SARS-CoV-2 attachment and entry into host cells (Peters, 2020, Press release). Further, ferrets can cough and sneeze and in doing so transmit the disease to nearby ferrets via aerosol/droplets, which makes them useful in transmission studies as well. Furthermore, their immune systems share a lot of similarities with the human, thereby making them potential animal models for testing vaccines, therapeutics and anti-virals against SARS-CoV-2. Finally, the fact that ferrets are long-lived animals makes them well suited for studying the impact of ageing on COVID-19 pathogenesis.
3.4 |. Non-human primate (NHP) models
Several species of NHP’s have been used in SARS-CoV and MERS-CoV studies. This includes old world monkeys such as rhesus macaques, cynomolgus macaques and african green monkeys, as well as new world monkeys that include the common marmosets, squirrel monkeys and moustached tamarins (Lawler et al., 2006; McAuliffe et al., 2004; Qin et al., 2005). Squirrel monkeys and moustached tamarins are not permissive to SARS-CoV (Roberts & Subbarao, 2006). MERS-CoV can only replicate in rhesus macaques and the common marmosets (Falzarano et al., 2014; Wit et al., 2013; Yao et al., 2014). In a study that utilized cynomolgus macaques for SARS-CoV studies, the virus was retrieved from nasal secretions and lung samples via RT-PCR including the detection of pulmonary pneumonia, which resembled the human disease (Lawler et al., 2006). African green monkeys, cynomolgus macaques and rhesus macaques infected with SARS-CoV Urbani strain did not develop clinical signs but viral replication was detected in nasal swabs and tracheal lavage samples (McAuliffe et al., 2004). The virus replicated to the highest titre in african green macaques followed by cynomolgus macaques and finally rhesus macaques. Infection of the common marmoset with SARS-CoV Urbani resulted in mild clinical disease accentuated by the development of fever and diarrhoea (Greenough et al., 2005). High levels of viral RNA were detected in lung samples on days 4 and 7 after infection, and there was evidence of both pulmonary (interstitial pneumonia) and hepatic pathology. These macaques developed significant hepatic lesions on days 4 and 7 post-infection. The marmoset was the only NHP that showed liver pathology resembling that described in humans (Greenough et al., 2005).
The rhesus macaque and common marmoset are susceptible to MERS-CoV infection and showed a wide spectrum of disease manifestations (Falzarano et al., 2014; Wit et al., 2013). When infected intratracheally with a high dose of the EMC-2012 virus strain, rhesus macaques developed mild clinical signs such as anorexia, respiratory distress and elevated WBCs on days 1–2 post-infection. Viral RNA could not be detected in the kidney or bladder (Yao et al., 2014). Another study by Falzarano et al., utilized marmosets and inoculated via intranasal, oral and ocular routes resulting in high virus titres (Falzarano et al., 2014). The animals showed respiratory distress, fever, nausea and lethargy at 4–6 days after infection. Viral RNA was detected in the throat and nasal swabs. Out of 9 marmosets used in the study, two of them developed multiple organ failure (kidney, liver and heart). Common marmosets were also used to study the therapeutic potential of combined anti-virals Lopinavir and Ritonavir. Chan et al., used Lopinavir and Ritonavir with IFN-beta and showed improvement in the pulmonary functions of the infected animals (Chan et al., 2015). Thus, from these studies, it was suggested that NHP models could provide deeper insights into the virus-induced pathology associated with severe human coronavirus infections. In summary, african green monkeys and rhesus macaques were identified as good models for SARS-CoV replication studies. Between the two, rhesus macaques showed viral replication kinetics that was consistent with the MERS-CoV replication in humans. However, the common marmoset remains the best model to study disease severity and multiple organ failure in both SARS-CoV and MERS-CoVs.
Many studies have been done to better understand the disease pathogenesis of SARS-CoV-2 in NHPs. A recent study by Bao et. al. used the non-human primate models with SARS-CoV-2 infection followed by a repeat challenge with the same virus to ascertain the possibility of re-infection (Bao et al., 2020). In this study, four adult Chinese rhesus macaques were intratracheally challenged with SARS-CoV-2/WH-09/human/2020/CHN at 1 × 10^6 TCID50 via intratracheal route. Weight loss, reduced appetite, increased respiration rate and hunched posture were observed, and the viral loads from nasal and anal swabs showed peak viremia (RNA) at 3 days post-infection (Table 1). After 28 days, two infected monkeys were intratracheally challenged with the same dose (1 × 10^6 TCID50) of SARS-CoV-2 to verify the possibility of re-infection. Viral loads in nasopharyngeal and anal swabs tested negative after re-exposure to SARS-CoV-2. The presence of the high levels of neutralizing antibodies in infected animals revealed that they had protective antibodies. Deng et. al also performed a similar type of challenge and rechallenge study, where six out of seven Chinese rhesus macaques were intratracheally challenged with SARS-CoV-2 at the dose of 1 × 10^6 TCID50. About 28 days post-primary challenge, four out of six were rechallenged with the same dose and strain of SARS-CoV-2 (Deng et al., 2020). Clinical diagnosis, viral loads, haematological analysis and binding and neutralizing antibodies were examined following some time point’s post-secondary challenge, which confirmed negative infection in the rechallenged animals.
Numerous research groups as well as pharmaceutical companies are using NHP models for testing suitable vaccine candidates. PiCoVacc, a purified inactivated SARS-CoV-2 vaccine candidate, was tested in rhesus macaques and showed a good immunogenic response (Gao et al., 2020) (Table 1). The vaccine candidate neutralized ten representative SARS-CoV-2 strains and offered complete protection in rhesus macaques. A second set of studies was performed in the rhesus macaque model to test the efficacy of a DNA vaccine candidate against SARS-CoV-2 (Yu et al., 2020). A total of 35 adult rhesus macaques were injected with the DNA vaccine candidates, with various constructs of SARS-CoV-2 proteins such as S (n = 4), S.dCT (n = 4), S.dTM (n = 4), S1 (n = 4), RBD (n = 4), S.dTM.PP (n = 5) and Control (n = 10) intramuscularly, and after 6 weeks, they were challenged with 1.1 × 10^4 PFU SARS-CoV-2 intratracheally and intranasally. Nasal swabs and bronchoalveolar lavage (BAL) taken from both control and vaccinated animals found lower viral RNA in the vaccinated groups compared to the control animals. Furthermore, using the same control animals Chandrashekar et al., showed development of protective immunity in SARS-CoV-2-infected macaques when the same animals were challenged a second time with the same virus (Chandrashekar et al., 2020). In this study, nine adult rhesus macaques, divided into three groups, were inoculated with 1.1 × 10^6 PFU (n = 3), 1.1 × 10^5 PFU (n = 3) or 1.1 × 10^4 PFU (n = 3) SARS-CoV-2 USA-WA1/2020 intranasally and intratracheally. Two days post-infection, viral RNA was detected in bronchoalveolar lavage (BAL) and nasal swabs with animals experiencing loss of appetite and transient lymphopenia and neutropenia. Around day 35 post-infection, these animals were re-inoculated with the same dose of SARS-CoV-2 as in the initial challenge. Interestingly, 2 days after the second challenge, the viral RNA in BAL was more than five logs lower than that detected after the primary challenge, suggesting that recovery from the primary exposure helps the development of protective immunity against secondary exposure (Table 1). A group from Guangzhou, China, reported the generation of replication-incompetent recombinant serotype 5 adenovirus (Ad5-S-nb2) that has a codon-optimized gene encoding Spike protein (S), as well as its effectiveness against SARS-CoV-2 challenge when tested in rhesus macaques (Feng et al., 2020). They showed protective immunity in macaques at 30 days after a single dose of the vaccine when inoculated intramuscularly or intranasally. They further demonstrated that a 10-fold lower dose of vaccine when tested in rhesus macaque also provides protective immunity against SARS-CoV-2 evidenced by the cell-mediated immune responses and S-protein-specific responses generated against the structural proteins of SARS-CoV-2. Likewise, another adenovirus-vectored vaccine (ChAdOx1 nCoV-19) encoding the spike protein of SARS-CoV-2 produced both cellular and humoral response in rhesus macaques after a single administration. There was reduction in viral loads in bronchoalveolar fluids and respiratory tract tissues with no pneumonia in the animals challenged with the vaccine (Doremalen et al., 2020).
Anti-viral testing is also being done using NHP models. A research group from Montana, USA, showed infection in rhesus macaque in the upper and lower respiratory tract (Vincent et al., 2020). In their study, rhesus macaques were infected with the SARS-CoV-2 isolate nCoV-WA1–2020 through various routes (intranasal, oral, ocular and intratracheal routes) showed reduced appetite, hunched posture, pale appearance and dehydration (Table 1). The viral loads were highest in the nasal swabs followed by throat and rectal swabs. Histopathology revealed multifocal interstitial pneumonia, oedematous alveoli and type II pneumocyte hyperplasia. Chest radiographs revealed the presence of pulmonary infiltrates and consolidation, which is the main clinical sign of COVID-19 in infected rhesus macaques. Near-identical clinical signs and histopathology were observed when rhesus macaques were challenged with a similar virus strain to show the therapeutic efficacy of Remdesivir (Williamson et al., 2020). To test the efficacy of Remdesivir in disease outcome, a rhesus macaque model of SARS-CoV-2 infection was developed, where infected macaques developed mild-to-moderate clinical symptoms (Williamson et al., 2020). The macaques treated with Remdesivir did not show any respiratory distress, and viral titres in bronchoalveolar lavage were reduced after 12 hr of treatment, suggesting that Remdesivir may have some beneficial effect in the treatment of COVID-19 (Williamson et al., 2020).
There are some studies showing age-related correlations in rhesus (Qi, et al., 2020) and cynomolgus macaques (Rockx et al., 2020), which demonstrated the impact of age-related comorbidities on SARS-CoV-2 disease severity (Table 1). In the study by Yu et. al, an aged group (~15 years) and younger group (3–5 years), of rhesus macaques, were compared after intratracheal infection with SARS-CoV-2. The results from this study support that age increases the infection rate, which was demonstrated by the viral loads after 7 days of inoculation. The histopathological changes in the older group were alveolar oedema, and inflammatory cells infiltrate which were severe compared to younger macaques that showed mild interstitial pulmonary infiltrates (Qi, et al., 2020). Rockx et al., also challenged SARS-CoV-2 in young adult (4–5 years) and aged (15–20 years) cynomolgus macaques via intranasal and intratracheal routes. After 4 days post-infection, two animals from each group were necropsied and showed limited focal lesions. There was minimal or no clinical sign observed in both groups with serous nasal discharge (Rockx et al., 2020). There is considerable interest within the research community to determine the immune mechanism/s associated with severity of responses in the human population when infected with SARS-CoV-2. Rosa et al., characterized transcriptional parameters induced in the lungs of juvenile and old rhesus macaques following SARS-CoV-2 infection. They showed, following SARS-CoV-2 infections, type I interferon signalling and the Notch signalling pathways were upregulated in juvenile when compared to old rhesus macaques. Neutrophil degranulation pathways were upregulated in the infected lungs which might be a major cause of severe inflammation and damage (Rosa et al., 2020).
More recently, african green monkeys (AGMs) (Woolsey et al., 2020) and baboons (Singh et al., 2020) were demonstrated to show more pronounced respiratory disease than rhesus macaques, suggesting that both AGMs and baboons may be the models of choice to test novel immunomodulatory therapies for reducing disease severity. Interestingly, african green monkeys were successfully infected with a lower SARS-CoV-2 inoculum compared to rhesus macaques. Researchers from Texas Biomedical Research Institute (Texas Biomed) and Southwest National Primate Research Center showed that baboons and macaques are suitable animal model for the SARS-CoV-2 studies, and recommended macaques for the study of vaccines, and baboons for drugs and therapeutics studies as well as studies of pathogenicity of COVID-19 with comorbidities (Texas Biomed, 2020, Press release). A major limitation of NHP model is that their larger size and greater space requirements compared to small animal models make NHPs more costly compared to other animal models. However, macaques develop respiratory disease that is comparable to the human disease and therefore represent a better translational animal model to study pathogenesis, vaccine efficacy, therapeutics and co-infections, including the impact of age and other pre-existing comorbidities on SARS-CoV-2 disease course.
4 |. CONCLUSIONS AND FUTURE PERSPECTIVES
Intensive efforts have been invested in the short timespan after the outbreak of COVID-19 to characterize and develop anti-SARS-CoV-2 therapeutics. Many drugs are being developed or repurposed with varying degrees of success in in vitro studies and clinical trials. As the speed for discovering drugs and vaccines is increasing, understanding their safety and effectiveness is urgently needed to control the spread of the SARS-CoV-2 infection. However, many questions still need to be answered regarding the route of transmission of SARS-CoV-2. Are other routes like faecal, sexual or mother-to-child transmission possible? The disease has dynamic outcomes, which has made it challenging to understand the pathogenesis. Why does the disease have higher infectivity than MERS and SARS? Why are elderly people and people with underlying conditions more susceptible? (Rothan et al., 2020) Does sex (Rothan et al., 2020) or host genetics (Olwenyi et al., 2020) also contribute to the pathogenicity and need to be addressed using biologically relevant animal models and warrants further research. There are many unknowns regarding the comorbidities and co-infections of SARS-CoV-2. Finally, there are questions as to why some repurposed drugs are effective in some individuals in the treatment of COVID-19. In order to answer these questions, in vivo studies using the best animal models are needed because it is impossible to address all these questions in human clinical studies. Because of the large variability in the genetic makeup of humans, it is impossible to understand mechanisms. Animal models such as ferrets, mice and hamsters can aid in answering many questions pertaining to the mechanism of action of anti-virals, efficacy and safety of vaccines and the impact of comorbidities on COVID-19 pathogenesis. It is possible that no single animal model will be able to answer all the human translational questions due to inherent differences in the development and physiology of the organism. Furthermore, there is a considerable amount of variability among the animals such as differences in their biology, genetics and level of ACE2 receptor expression, all of which can influence the rate of infection. In vitro models, primary cell lines and organoids represent an important aspect of study for SARS-CoV-2 pathogenesis (Leist et al., 2020). Generation of humanized mouse model, reminiscent of human tissues, may also provide an important avenue for the rapid testing of anti-virals and vaccines.
ACKNOWLEDGEMENTS
We thank Michellie Thurman for editorial help. This work is partially supported by National Institute of Allergy and Infectious Diseases Grant R01 AI129745, 5P30 CA036727-33 and Frances E. Lageschulte and Evelyn B. Weese New Frontiers in Medical Research Fund to SNB.
Footnotes
CONFLICTS OF INTERESTS
Authors declared no conflict of interest exists.
ETHICAL APPROVAL
We declare ethical statement is not applicable.
DATA AVAILABILITY STATEMENT
All the data to support the findings are within the manuscript.
REFERENCES
- Acharya A, Kevadiya BD, Gendelman HE, & Byrareddy SN (2020). SARS-CoV-2 infection leads to neurological dysfunction. Journal of Neuroimmune Pharmacology: the Official Journal of the Society on NeuroImmune Pharmacology, 15(2), 167–173. 10.1007/s11481-020-09924-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, Ray AS, Cihlar T, Siegel D, Mackman RL, Clarke MO, Baric RS, & Denison MR (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio, 9(2). 10.1128/mbio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arashiro T, Nakamura S, Asami T, Mikuni H, Fujiwara E, Sakamoto S, Miura R, Shionoya Y, Honda R, Furukawa K, Nakamura A, & Saito H (2020). SARS-CoV-2 and legionella co-infection in a person returning from a nile cruise. Journal of Travel Medicine, 27(3), 10.1093/jtm/taaa053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azekawa S, Namkoong H, Mitamura K, Kawaoka Y, & Saito F (2020). Co-infection with SARS-CoV-2 and influenza A virus. Idcases., 20, e00775. 10.1016/j.idcr.2020.e00775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Qin C (2020). The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature, 583, (7818), 830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bassetti M, Vena A, & Giacobbe DR (2020). The novel Chinese coronavirus (2019-nCoV) infections: Challenges for fighting the storm. European Journal of Clinical Investigation, 50(3), e13209. 10.1111/eci.13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley S What explains Covid-19’s lethality for the elderly? Scientists look to ‘twilight’ of the immune system [press release] (2020). https://www.boston.com/news/health/2020/03/30/coronavirus-covid-19-elderly
- Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, & ACTT-1 Study Group Members (2020). Remdesivir for the treatment of covid-19 - preliminary report. New England Journal of Medicine. 383, 992–994. https://www.nejm.org/doi/full/10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- Bhatnagar T, Murhekar MV, Soneja M, Gupta N, Giri S, Wig N, & Gangakhedkar R (2020). Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: Protocol for restricted public health emergency use. Indian Journal of Medical Research, 151(2 & 3), 184–189. 10.4103/ijmr.IJMR_502_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK, Greninger AL, Pipavath S, Wurfel MM, Evans L, Kritek PA, West TE, Luks A, Gerbino A, Dale CR, Goldman JD, O’Mahony S, & Mikacenic C. (2020). Covid-19 in critically Ill patients in the seattle region - case series. New England Journal of Medicine, 382(21), 2012–2022. 10.1056/NEJMoa2004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, & Khan K (2020). Pneumonia of unknown aetiology in Wuhan, China: Potential for international spread via commercial air travel. Journal of Travel Medicine, 27(2). 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J-F-W, Kok K-H, Zhu Z, Chu H, To K-K-W, Yuan S, & Yuen K-Y (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan W, Fan Z, Tsoi H, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai J, Kok K, Chu H, Chan K, & Yuen K (2020). Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clinical Infectious Diseases, 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PK, & Chan MC (2013). Tracing the SARS-coronavirus. Journal of Thoracic Disease, 5(Suppl 2), S118–S121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, Tostanoski LH, Yu J, Maliga Z, Nekorchuk M, Busman-Sahay K, Terry M, Wrijil LM, Ducat S, Martinez DR, Atyeo C, Fischinger S, Burke JS, Slein MD, & Barouch DH (2020). SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science, 369, (6505), 812–817. 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Muth D, Niemeyer D, & Drosten C (2018). Hosts and sources of endemic human coronaviruses. Advances in Virus Research, 100, 163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D, & Vanelli M (2020). WHO declares COVID-19 a pandemic. Acta BioMedica, 91(1), 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, & Shi ZL (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, & Munster VJ (2013). Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proceedings of the National Academy of Sciences, 110(41), 16598–16603. 10.1073/pnas.1310744110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego ML, Pewe L, Alvarez E, Rejas MT, Perlman S, & Enjuanes L (2008). Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology, 376(2), 379–389. 10.1016/j.virol.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Bao L, Liu J, Xiao C, Liu J, Xue J, Lv Q, Qi F, Gao H, Yu P, Xu Y, Qu Y, Li F, Xiang Z, Yu H, Gong S, Liu M, Wang G,Wang S, … Qin C (2020). Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science, 369(6505), 818–823. 10.1126/science.abc5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MR, Graham RL, Donaldson EF, Eckerle LD, & Baric RS (2011). Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biology, 8(2), 270–279. 10.4161/rna.8.2.15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiraj Kumar Singh SRG, Singh B, Cole J, Alfson KJ, Clemmons E, Gazi M, Gonzalez O, Escobedo R, Lee T-H, Chatterjee A, Goez-Gazi Y, Sharan R, Thippeshappa R, Gough M, Alvarez C, Blakley A, Ferdin J, Bartley C, Staples H, … Carrion R Jr. (2020) View ORCID ProfileDeepak Kaushal. SARS-CoV-2 infection leads to acute infection with dynamic cellular and inflammatory flux in the lung that varies across nonhuman primate species. https://www.biorxiv.org/content/10.1101/2020.06.05.136481v1 [Google Scholar]
- Dinnon KH, Leist SR, Schafer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, & Baric RS, (2020). A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. Nature, 586(7830), 560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D, de Wit E, Feldmann F, Rasmussen AL, Okumura A, Peng X, Thomas MJ, van Doremalen N, Haddock E, Nagy L, LaCasse R, Liu T, Zhu J, McLellan JS, Scott DP, Katze MG, Feldmann H, & Munster VJ (2014). Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Path, 10(8), e1004250. 10.1371/journal.ppat.1004250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Wang Q, Shan C, Yang C, Feng Y, Wu J, Liu X, Zhou Y, Jiang R, Hu P, Liu X, Zhang F, Li P, Niu X, Liu Y, Zheng X, Luo J, Sun J, Gu Y, Chen L (2020). An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nature Communications, 11, (1), 4027. 10.1038/s41467-020-18077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman MB, Chen J, Morrison TE, Whitmore A, Funkhouser W, Ward JM, Lamirande EW, Roberts A, Heise M, Subbarao K, Baric RS (2010). SARS-CoV Pathogenesis Is Regulated by a STAT1 Dependent but a Type I, II and III Interferon Receptor Independent Mechanism. PLoS Pathogens, 6(4), e1000849. 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama A, Watanabe H, Toyoda A, Taylor TD, Itoh T, Tsai SF, Park S, Yaspo M, Lehrach H, Chen Z, Fu G, Saitou N, Osoegawa K, Jong PJ, Suto Y, Hattori M, & Sakaki Y et al. (2002). Construction and analysis of a human-chimpanzee comparative clone map. Science, 295(5552), 131–134. 10.1126/science.1065199 [DOI] [PubMed] [Google Scholar]
- Gao H, Lu H, Cao B, Du B, Shang H, Gan J, Lu S, Yang Y, Fang Q, Shen Y, Xi X, Gu Q, Zhou X, Qu H, Yan Z, Li F, Zhao W, Gao Z, Wang G, Li L (2013). Clinical Findings in 111 Cases of Influenza A (H7N9) Virus Infection. New England Journal of Medicine, 368(24), 2277–2285. 10.1056/nejmoa1305584. [DOI] [PubMed] [Google Scholar]
- Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, Li Y, Zhu L, Wang N, Lv Z, Gao H, Ge X, Kan B, Hu Y, Liu J, Cai F, Jiang D, Yin Y, Qin C, (2020). Development of an inactivated vaccine candidate for SARS-CoV-2. Science, 369(6499), 77–81. 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass WG, Subbarao K, Murphy B, & Murphy PM (2004). Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. The Journal of Immunology, 173(6), 4030–4039. 10.4049/jimmunol.173.6.4030 [DOI] [PubMed] [Google Scholar]
- Gold JAW, Wong KK, Szablewski CM, Patel PR, Rossow J, da Silva J, Natarajan P, Morris SB, Fanfair RN, Rogers-Brown J, Bruce BB, Browning SD, Hernandez-Romieu AC, Furukawa NW, Kang M, Evans ME, Oosmanally N, Tobin-D’Angelo M, Drenzek C, & Jackson BR et al. (2020). Characteristics and clinical outcomes of adult patients hospitalized with COVID-19 - Georgia, March 2020. MMWR. Morbidity and Mortality Weekly Report, 69(18), 545–550. 10.15585/mmwr.mm6918e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough TC, Carville A, Coderre J, Somasundaran M, Sullivan JL, Luzuriaga K, Luzuriaga K, & Mansfield K (2005). Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. American Journal of Pathology, 167(2), 455–463. 10.1016/S0002-9440(10)62989-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretebeck LM, & Subbarao K (2015). Animal models for SARS and MERS coronaviruses. Current Opinion in Virology, 13, 123–129. 10.1016/j.coviro.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li XF, Li J, Zhang NN, Yang X, Chen S, Guo Y, Zhao G, Wang X, & Zhou Y et al. (2020). Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 369(6511), 1603–1607. 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid S, Mir MY, & Rohela GK (2020). Novel coronavirus disease (COVID-19): A pandemic (epidemiology, pathogenesis and potential therapeutics). New Microbes and New Infections, 35, 100679. 10.1016/j.nmni.2020.100679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, & van Goor H (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology, 203(2), 631–637. 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, & Pöhlmann S (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–80 e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, … Cao B (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, & Kawaoka Y (2020). Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proceedings of the National Academy of Sciences of the United States of America, 117(28), 16587–16595. 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Wang W, Zhao X, Zai J, & Li X (2020). Cross-species transmission of the newly identified coronavirus 2019-nCoV. Journal of Medical Virology, 92(4), 433–440. 10.1002/jmv.25682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si H-R, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang H-J, Baric RS, … Shi Z-L(2020). Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell, 182(1), 50–8 e8. 10.1016/j.cell.2020.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathryn A, Ryan KRB, Fotheringham SA, Brown P, Hall Y, Marriott AC, Tree JA, Allen L, Aram MJ, Brunt E, Buttigieg KR, Cavell BE, Carter DP, Cobb R, Coombes NS, Godwin KJ, Gooch KE, Gouriet J, Halkerston R, …Humphries HE (2020) Dose-dependent response to infection with SARS-CoV-2 in the ferret model: evidence of protection to re-challenge. 10.1101/2020.05.29.123810 [DOI] [PMC free article] [PubMed]
- Kim E, Erdos G, Huang S, Kenniston TW, Balmert SC, Carey CD, Raj VS, Epperly MW, Klimstra WB, Haagmans BL, Korkmaz E, Falo LD, & Gambotto A (2020). Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development. EBioMedicine., 102743. 10.1016/j.ebiom.2020.102743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang J-H, Kim EJ, Lee S, Casel MAB, Um J, Song M-S, Jeong HW, Lai VD, Kim Y, Chin BS, Park J-S, Chung K-H, … Choi YK (2020). Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host & Microbe, 27(5), 704–709.e2. 10.1016/j.chom.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A, & Beer M (2020). SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. The Lancet. Microbe, 1(5), e218–e225. 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler JV, Endy TP, Hensley LE, Garrison A, Fritz EA, Lesar M, Baric RS, Kulesh DA, Norwood DA, Wasieloski LP, Ulrich MP, Slezak TR, Vitalis E, Huggins JW, Jahrling PB, & Paragas J (2006). Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med, 3(5), e149. 10.1371/journal.pmed.0030149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LYY, Zhou J, Frise R, Goldhill DH, Koszalka P, Mifsud EJ, Mifsud EJ, Baba K, Noda T, Ando Y, Sato K, Yuki A-I, Shishido T, Uehara T, Wildum S, Zwanziger E, Collinson N, Kuhlbusch K, Clinch B, … Barclay WS (2020). Baloxavir treatment of ferrets infected with influenza A(H1N1)pdm09 virus reduces onward transmission. PLoS Path, 16(4), e1008395. 10.1371/journal.ppat.1008395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist SR, Schafer A, & Martinez DR (2020). Cell and animal models of SARS-CoV-2 pathogenesis and immunity. Disease Models & Mechanisms, 13(9), 1–6. 10.1242/dmm.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D (2020). Is the coronavirus airborne? Experts can’t agree. Nature, 580(7802), 175. 10.1038/d41586-020-00974-w [DOI] [PubMed] [Google Scholar]
- Li F, Li W, Farzan M, & Harrison SC (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science, 309(5742), 18 64–1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, … Feng Z (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. New England Journal of Medicine, 382(13), 1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Greenough TC, Moore MJ, Vasilieva N, Somasundaran M, Sullivan JL, Farzan M, & Choe H (2004). Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. Journal of Virology, 78(20), 11429–11433. 10.1128/JVI.78.20.11429-11433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, & Farzan M et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linlin Bao WD, Gao H, Xiao C, Liu J, Xue J, Lv Q, Liu J, Pin Y, Yanfeng X, Qi F, Yajin QU, Li F, Xiang Z, Haisheng YU, Gong S, Liu M, Wang G, Wang S, Song Z, …Qin C. (2020) Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv doi: 10.1101/2020.03.13.990226 [DOI] [Google Scholar]
- Liu S, Lin HM, Baine I, Wajnberg A, Gumprecht JP, Rahman F, Rodriguez D, Tandon P, Bassily-Marcus A, Bander J, Sanky C, Dupper A, Zheng A, Nguyen FT, Amanat F, Stadlbauer D, Altman DR, Chen BK, Krammer F, Mendu DR, … Bouvier NM (2020) Convalescent plasma treatment of severe COVID-19: a propensity score-matched control study. Nature medicine, Advance online publication. 10.1038/s41591-020-1088-9. [DOI] [PubMed] [Google Scholar]
- Lu YR, Wang LN, Jin X, Chen YN, Cong C, Yuan Y, Li YC, Tang WD, Li HX, Wu XT, Li YP, Wang L, & Cheng JQ (2008). A preliminary study on the feasibility of gene expression profile of rhesus monkey detected with human microarray. Transplantation Proceedings, 40(2), 598–602. 10.1016/j.transproceed.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Luke T, Wu H, Zhao J, Channappanavar R, Coleman CM, Jiao JA, Matsushita H, Liu Y, Postnikova EN, Ork BL, Glenn G, Flyer D, Defang G, Raviprakash K, Kochel T, Wang J, Nie W, Smith G, Hensley LE, & Frieman MB (2016). Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS-CoV in vivo. Science Translational Medicine. 8(326), 326ra21. 10.1126/scitranslmed.aaf1061 [DOI] [PubMed] [Google Scholar]
- Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Homg C, Zhou Y, Wang D, Miao X, Li Y, & Hu B (2020). Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurology, 77(6), 683–690. 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina BE, Haagmans BL, Kuiken T, Fouchier RA, Rimmelzwaan GF, Van Amerongen G, Peiris JS, Lim W, & Osterhaus AD (2003). Virology: SARS virus infection of cats and ferrets. Nature, 425(6961), 915. 10.1038/425915a [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe J, Vogel L, Roberts A, Fahle G, Fischer S, Shieh WJ, Butler E, Zaki S, St Clarie M, Murphy B, & Subbarao K (2004). Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology, 330(1), 8–15. 10.1016/j.virol.2004.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray PB Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, & Perlman S (2007). Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. Journal of Virology, 81(2), 813–821. 10.1128/JVI.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M (2020). Droplets and aerosols in the transmission of SARS-CoV-2. New England Journal of Medicine, 382(21), 2063. 10.1056/NEJMc2009324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modema, Inc. (2020) Moderna Moderna announces funding award from CEPI to accelerate development of messenger RNA (mRNA) vaccine against novel coronavirus; [accessed 2020 February15] [press release]. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-funding-award-cepi-accelerate-development
- Mukherjee R (2020). Global efforts on vaccines for COVID-19: Since, sooner or later, we all will catch the coronavirus. Journal of Biosciences, 45(1), 68. 10.1007/s12038-020-00040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S, Dodd LE, Davey RT Jr, Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, Ali R, Coulibaly S, Levine AC, Grais R, Diaz J, Lane HC, Muyembe-Tamfum JJ, PALM Writing Group, Sivahera, B., & PALM Consortium Study Team (2019). A randomized, controlled trial of ebola virus disease therapeutics. New England Journal of Medicine, 381(24), 2293–2303. 10.1056/NEJMoa1910993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E (2020). Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature, 585, (7824), 268–272. 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanshan Z (2020) The new coronavirus is likely to come from game products such as bamboo rats and tadpoles. Netease News. January 20, 2020.https://news.163.com/20/0120/22/F3C9KSI50001899O.html [Google Scholar]
- NIAID.NIH.GOV. Developing Therapeutics and Vaccines for Coronaviruses. [press release]. https://www.niaid.nih.gov/diseases-conditions/coronaviruses-therapeutics-vaccines.
- Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, Yan KW, Chan KH, Tsang NC, Guan Y, Yuen KY, & Peiris JS (2003). Lung pathology of fatal severe acute respiratory syndrome. Lancet, 361(9371), 1773–1778. 10.1016/S0140-6736(03)13413-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olwenyi OA, Dyavar SR, Acharya A, Podany AT, Fletcher CV, Ng CL, Reid SP, & Byrareddy SN (2020). Immuno-epidemiology and pathophysiology of coronavirus disease 2019 (COVID-19). Journal of Molecular Medicine (Berlin), 98(10), 1369–1383. 10.1007/s00109-020-01961-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY, & SARS study group (2003). Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 361(9366), 1319–1325. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters D. [4.25.2020];How Ferrets Are Helping Researchers Battle Covid-19. https://undark.org/2020/04/25/ferrets-covid-19/ It came up on.
- Product Development Under the Animal Rule (2015) Food and Drug Administration. [Google Scholar]
- Qin C, Wang J, Wei Q, She M, Marasco WA, Jiang H, Tu X, Zhu H, Ren L, Gao H, Gao L, Huang L, Yang R, Cong Z, Guo L, Wang Y, Liu Y, Sun Y, Duan S, & He W (2005). An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. The Journal of Pathology, 206(3), 251–259. 10.1002/path.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, & Haagmans BL (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature, 495(7440), 251–254. 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Osterhaus AD, Fouchier RA, & Haagmans BL (2014). MERS: Emergence of a novel human coronavirus. Current Opinion in Virology, 5, 58–62. 10.1016/j.coviro.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R, Strohmeier S, Amanat F, Gillespie VL, Krammer F, Garcia-Sastre A, Coughlan L, Schotsaert M, & Uccellini M (2020). Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerging microbes & infections, 1, 2433–2445. 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Lamirande EW, Vogel L, Jackson JP, Paddock CD, Guarner J, Zaki SR, Sheahan T, Baric R, & Subbarao K (2008). Animal models and vaccines for SARS-CoV infection. Virus Research, 133(1), 20–32. 10.1016/j.virusres.2007.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Paddock C, Vogel L, Butler E, Zaki S, & Subbarao K (2005). Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. Journal of Virology, 79(9), 5833–5838. 10.1128/JVI.79.9.5833-5838.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, & Subbarao K (2006). Animal models for SARS. Advances in Experimental Medicine and Biology, 581, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Thomas WD, Guarner J, Lamirande EW, Babcock GJ, Greenough TC, Vogel L, Hayes N, Sullivan JL, Zaki S, Subbarao K, & Ambrosino DM (2006). Therapy with a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody reduces disease severity and viral burden in golden Syrian hamsters. Journal of Infectious Diseases, 193(5), 685–692. 10.1086/500143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba N, Schipper D, van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Drosten C, & Haagmans BL (2020). Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 368(6494), 1012–1015. 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa BA, Ahmed M, Singh DK, Choreno-Parra JA, Cole J, Jimenez-Alvarez LA, Rodriguez-Reyna TS, Singh B, Golzalez O, Carrion R, Schlesinger LS, Martin J, Zuniga J, Mitreva M, Khader SA, & Kaushal D (2020). IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection. bioRxiv: the preprint server for biology. 10.1101/2020.08.06.239798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, Acharya A, Reid SP, Kumar M, & Byrareddy SN (2020). Molecular aspects of COVID-19 differential pathogenesis. Pathogens, 9(7), 538. 10.3390/pathogens9070538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, & Byrareddy SN (2020). The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. Journal of Autoimmunity, 109, 102433. 10.1016/j.jaut.2020.102433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, Fentener van Vlissingen M, Rockx B, Haagmans BL, Koopmans MPG, Fouchier RAM, Herfst S (2020). SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nature Communications, 11(1), 3496. 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavone C, Brusco S, Bertini M, Sportiello L, Rafaniello C, Zoccoli A, Berrino L, Racagni G, Rossi F, & Capuano A (2020). Current pharmacological treatments for COVID-19: What’s next? British Journal of Pharmacology, 177(21), 4813–4824. 10.1111/bph.15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, & Baric RS (2020). An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Science Translational Medicine, 12(541). 10.1126/scitranslmed.abb5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, & Zheng C (2020). Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. The Lancet Infectious Diseases, 20(4), 425–434. 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, & Bu Z (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science, 368(6494), 1016–1020. 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia SF, Yan LM, Chin A, Fung K, Choy KT, Wong A, Kaewpreedee P, Perera R, Poon L, Nicholls JM, Peiris M, & Yen HL (2020) Pathogenesis and transmission of SARS-CoV-2 virus in golden Syrian hamsters. Nature, 583(7818), 834–838. 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, McAuliffe J, Vogel L, Fahle G, Fischer S, Tatti K, Packard M, Shieh WJ, Zaki S, & Murphy B (2004). Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. Journal of Virology, 78(7), 3572–3577. 10.1128/JVI.78.7.3572-3577.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, Anderson LJ, Bellini WJ, Farzan M, & Marasco WA (2004). Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proceedings of the National Academy of Sciences, 101(8), 2536–2541. 10.1073/pnas.0307140101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ning R, Tao Y, Yu C, Deng X, Zhao C, Meng S, Tang F, & Xu D (2020). Risk factors for mortality in 244 older adults with COVID-19 in Wuhan, China: A retrospective study. Journal of the American Geriatrics Society. 68(6), E19–E23. 10.1111/jgs.16533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton TC, & Subbarao K (2015). Development of animal models against emerging coronaviruses: From SARS to MERS coronavirus. Virology, 479–480, 247–258. 10.1016/j.virol.2015.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K (2020). In vitro and animal models for SARS-CoV-2 research. Trends in Pharmacological Sciences, 41(8), 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texas Biomedical Research Institute (Texas Biomed) (2020) Researchers release results of comprehensive SARS-CoV-2 animal model study [press release]. 06.02.2020. https://www.txbiomed.org/news-press/news-releases/covid19-animal-model-study/
- Touzard-Romo F, Tape C, & Lonks JR (2020). Co-infection with SARS-CoV-2 and human metapneumovirus. R I Med J(2013), 103(2), 75–76. [PubMed] [Google Scholar]
- Valentine H, Daugherity EK, Singh B, Maurer KJ (2012) The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents Chapter 34 - The Experimental Use of Syrian Hamsters.
- van den Brand JM, Stittelaar KJ, van Amerongen G, Rimmelzwaan GF, Simon J, de Wit E, Munster V, Bestebroer T, Fouchier RA, Kuiken T, & Osterhaus AD (2010). Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. Journal of Infectious Diseases, 201(7), 993–999. 10.1086/651132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato V, Bushmaker T, Flaxman A, Ulaszewska M, Feldmann F, Allen ER, Sharpe H, Schulz J, Holbrook M, Okumura A, Meade-White K, Pérez-Pérez L, Bissett C, & Munster VJ (2020). ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv: the preprint server for biology. 10.1101/2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinegar A, Sinnett EE, Kosch PC, & Miller ML (1985). Pulmonary physiology of the ferret and its potential as a model for inhalation toxicology. Laboratory Animal Science, 35(3), 246–250. [PubMed] [Google Scholar]
- Wang J, Shuai L, Wang C, Liu R, He X, Zhang X, Sun Z, Shan D, Ge J, Wang X, Hua R, Zhong G, Wen Z, & Bu Z (2020). Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein & Cell, 11(10), 776–782. 10.1007/s13238-020-00767-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, Li Y, Zhao L, Li W, Sun X, Yang X, Shi Z, Deng F, Hu Z, Zhong W, & Wang M (2020). The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery, 6, 28. 10.1038/s41421-020-0169-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, Smith G, Jones S, Proulx R, Deschambault Y, Grudeski E, Andonov A, He R, Li Y, Copps J, Grolla A, Dick D, Berry J, Ganske S, & Cao J (2004). Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. Journal of Virology, 78(22), 12672–12676. 10.1128/JVI.78.22.12672-12676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson BN, Feldmann F, Schwarz B, Meade-White K, Porter DP, Schulz J, van Doremalen N, Leighton I, Yinda CK, Pérez-Pérez L, Okumura A, Lovaglio J, Hanley PW, Saturday G, Bosio CM, Anzick S, Barbian K, Cihlar T, Martens C, … de Wit E (2020). Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature, 585(7824), 273–276. 10.1038/s41586-020-2423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, & Yuen KY (2012). Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. Journal of Virology, 86(7), 3995–4008. 10.1128/JVI.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C, Borisevich V, Prasad AN, Agans KN, Dobias NS, Heymann JC, Foster SL, Levine CB, Medina L, Melody K, Geisbert JB, Fenton KA, Geisbert TW, & Cross RW (2020). Establishment of an African green monkey model for COVID-19. bioRxiv: the preprint server for biology. 10.1101/2020.05.17.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2003) Geneva (Switzerland): World Health Organization. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. [Google Scholar]
- World Health Organization (2019) Geneva (Switzerland): World Health Organization; MERS situation update, December 2019. [Google Scholar]
- Xiao C, Gao H, Yu P, Cai JP, Chu H, Zhou J, Chen H, Qin C, & Yuen KY (2015). Treatment With Lopinavir/Ritonavir or Interferon-β1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. Journal of Infectious Diseases, 212(12), 1904–1913. 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, Zhang J, Shu F, Huang W, Li Y, Zhang Z, Chen RA, Wu YJ, Peng SM, Huang M, & Shen Y, (2020). Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature, 583(7815), 286–289. 10.1038/s41586-020-2313-xy. [DOI] [PubMed] [Google Scholar]
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, & Wang FS (2020). Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet Respiratory Medicine, 8(4), 420–422. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Bao L, Deng W, Xu L, Li F, Lv Q, Yu P, Chen T, Xu Y, Zhu H, Yuan J, Gu S, Wei Q, Chen H, Yuen KY, & Qin C (2014). An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. Journal of Infectious Diseases, 209(2), 236–242. 10.1093/infdis/jit590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh MT, Bujaki E, Dolan PT, Smith M, Wahid R, Konz J, Weiner AJ, Bandyopadhyay AS, Van Damme P, De Coster I, Revets H, Macadam A, & Andino R (2020). Engineering the live-attenuated polio vaccine to prevent reversion to virulence. Cell Host & Microbe, 27(5), 736–751.e8. 10.1016/j.chom.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, & Wunderink RG (2018). MERS, SARS and other coronaviruses as causes of pneumonia. Respirology, 23(2), 130–137. 10.1111/resp.13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis F, & Barouch DH (2020). DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 309(6505), 806–811. 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, Xiao C, Lv Q, Gong S, Liu J, Song Z, Qu Y, Xue J, Wei Q, Liu M, & Qin C (2020). Age-related rhesus macaque models of COVID-19. Animal Models and Experimental Medicine, 3(1), 93–97. 10.1002/ame2.12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, & Shi ZL (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]


