Summary
The growth of plant organs is driven by cell division and subsequent cell expansion. The transition from proliferation to expansion is critical for the final organ size and plant yield. Exit from proliferation and onset of expansion is accompanied by major metabolic reprogramming, and in leaves with the establishment of photosynthesis. To learn more about the molecular mechanisms underlying the developmental and metabolic transitions important for plant growth, we used untargeted proteomics and metabolomics analyses to profile young leaves of a model plant Arabidopsis thaliana representing proliferation, transition, and expansion stages. The dataset presented represents a unique resource comprising approximately 4,000 proteins and 300 annotated small-molecular compounds measured across 6 consecutive days of leaf growth. These can now be mined for novel developmental and metabolic regulators of plant growth and can act as a blueprint for studies aimed at better defining the interface of development and metabolism in other species.
Keywords: leaf development, mitotic arrest, 12-OPDA, metabolomics, proteomics
Graphical abstract
Highlights
-
•
Untargeted metabolomics and proteomics characterization of early leaf growth
-
•
Translation is the primary determiner of protein abundance during early leaf growth
-
•
12-OPDA accumulation coincides with meristem arrest
The bigger picture
Developmental and metabolic transitions occurring during plant growth are critical for crop yield. The multi-omics dataset presented here was generated to enable the identification of novel molecular players involved in the regulation of plant growth. It comprised approximately 4,000 proteins and 300 annotated small-molecular compounds, measured across early leaf development spanning major developmental transitions. As such, the work provides a blueprint for studies aimed at better defining the interface between metabolism and development, an appreciated yet understudied research frontier across all kingdoms of life.
Plants are the primary source of food and feed. Given limited land and water resources to feed the world but with increasing human population, we need to increase plant yields. To achieve this, we need to understand how plant growth and plant yield are regulated. In this study, we measured proteins and metabolites involved in early leaf growth to be explored by us and others for novel plant-growth regulators.
Introduction
Plant growth is an important component of plant yield and thus has been extensively studied, revealing evolutionary conservation of cellular and molecular pathways driving growth in model and crop plant species.1 Growth of plant organs depends on the integration of cell division and cell expansion; the transition from proliferation to expansion (referred to as cellular differentiation) being a critical point which affects cell number and, thus, organ size.2 Importantly, cellular differentiation is accompanied by major metabolic reprogramming.3,4 In fact extensive cross-regulation between development and metabolism is not unique to plants and has been widely reported across eukaryotes.5 Despite this fact, multi-omic datasets encompassing broad measurements at multiple layers of the molecular hierarchy at important developmental transitions remain rare, even though they are often critical in determining such important applications as crop yield, biotechnological efficiency, and the relative efficacy of medicines. As such we are certain that the data described herein will prove an invaluable resource providing deep kinetic resolution of a key biological process. We anticipate it will not only be invaluable for advancing progress in this field but also as a case study, which can be used for the development of sophisticated modeling tools that can interrogate metabolic and signaling aspects of cellular development.
Here, to learn more about the molecular mechanisms underlying developmental and metabolic transitions important for plant growth, we used untargeted proteomics and metabolomics analyses to profile young leaves of a model plant Arabidopsis thaliana at the proliferation, transition, and expansion stages. Arabidopsis leaves initiate at the flank of the shoot apical meristem. Initially, the growth of a new leaf is driven by rapid cell proliferation with cells doubling in mass before a mitotic division. In approximately 5–6 days following primordium initiation, and starting from the leaf tip, cells exit mitosis and start expanding concomitantly with an increase of the central vacuole and onset of endoreduplication.2 The timing of the transition from proliferation to expansion is critical for the final leaf size,6,7 with premature and delayed differentiation resulting in smaller7 and larger6 leaves, respectively. Importantly, the transition from proliferation to expansion is accompanied by significant rewiring of metabolism. While cell division is characterized by rapid metabolic rates to sustain membrane, nucleic acid, and protein synthesis, the transition from proliferation to expansion is accompanied by chloroplast maturation and establishment of photosynthesis.3 Notably, it was previously demonstrated that mature and photosynthetically active chloroplasts are important for the exit from proliferation.3 However, the exact molecular mechanisms driving differentiation of the leaf meristem, and particularly how these relate to the differentiation of the photosynthetic metabolism, are currently not well understood.8
Forward and reverse genetics remain presiding strategies by which to learn about mechanisms underlying leaf growth.9 However, despite being very powerful, genetic approaches are limited to genes which when affected significantly alter leaf size. Omics profiling constitutes an alternative strategy to identify important regulators based on their differential behavior across the dataset of interest and may thereby reveal finer levels of regulation. In the past, detailed growth and cellular analysis of developing Arabidopsis leaves were combined with transcriptomics.3 Here, we performed similar leaf-development time-course experiments, but rather than at transcripts we focused on proteins and metabolites, which change during early leaf growth. The presented multi-omics dataset can now be mined for novel protein and small-molecule growth regulators.
Results
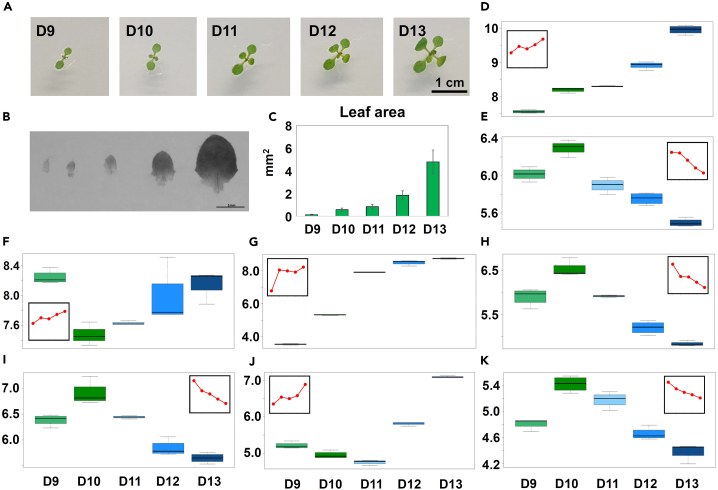
The third true leaves of the in vitro grown Arabidopsis seedlings were harvested daily from day 8 to day 13 after stratification (8–13 DAS) (Figure 1). In the past, cellular and transcriptomics analysis of the analogous leaf-development experiments demonstrated that the “8 to 13 time course” is well suited to study the transition from proliferation to expansion-driven growth.3 While leaves at days 8 and 9 are mainly proliferating, leaves at days 10 and 11 correspond to the transition phase, and leaves at days 12 and 13 are mainly expanding. To investigate how well our experiment matches the developmental trajectory reported by the previous study,3 we measured leaf area (Figures 1B and 1C) and expression of the selected transcripts (Figures 1D–1K) in the leaves harvested from day 9 to day 13. Comparison of growth and expression data revealed good correspondence between the previously published3 and the current experiment.
Figure 1.
Experimental information
(A) Seedlings used for the dissection of third leaf from 9 to 13 DAS. DAS, days after stratification. Scale bar, 1 cm.
(B) Photo of a representative third leaf dissected from the seedlings from 9 to 13 DAS. Scale bar, 1 mm.
(C) Leaf area measured for third leaf from 9 to 13 DAS. Data are mean ± SD; n = 9–20.
(D–K) Boxplots showing the gene expression (qRT-PCR) after normalization (10 − ΔCt) for GUN5 (D), CDKB2 (E), TIP1 (F), LOX2 (G), PCNA1 (H), PCNA2 (I), ABA1 (J), and PRL (K) Boxplots represent data from at least three replicates per developmental stage. Small, linear chart plots represent expression changes measured by Andriankaja et al.3 from 9 to 13 DAS.
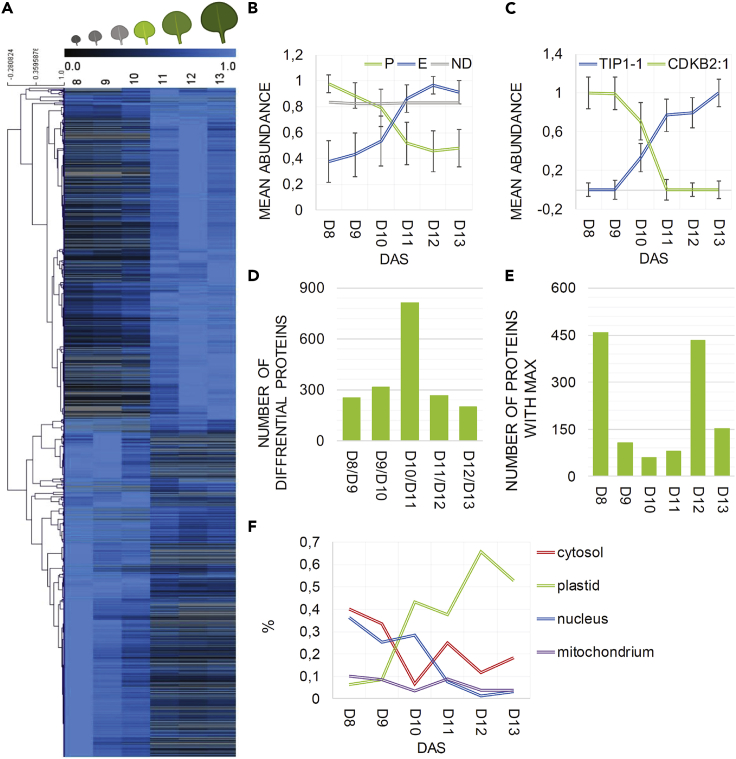
Proteomics analysis of the early leaf development reveals major reprogramming at the transition from cell proliferation to cell expansion
Untargeted proteomics analysis of developing Arabidopsis leaves resulted in a dataset comprising 4,189 proteins (Data S1). Statistical analysis using one-way ANOVA retrieved a list of 797 proteins characterized by a differential accumulation pattern across the six leaf developmental time points (Bonferroni corrected p < 0.05) (Data S1). Differential proteins were also delineated using absence and presence criteria in the young, mainly proliferating (day 8 and day 9) versus older, mainly expanding (day 12 and day 13) leaves. In this manner we identified a further 418 proteins, specific for either proliferation or expansion (Data S1). Clustering analysis of the 1,215 differential proteins distinguished two major groups (Figure 2A). The first group of 583 proteins was associated with cell proliferation while the second group of 632 proteins was related to cell expansion. The average accumulation pattern of both groups crossed over between day 10 and day 11, reflected by the behavior of mitotic cell-cycle marker protein cyclin-dependent kinase B (CDKB2;1), and a vacuolar growth-associated protein aquaporin TIP1;1 (Figures 2B and 2C). Consistently, comparison of the protein abundance between each pair of consecutive days yielded the highest number of differential proteins (unpaired t test, p < 0.05, or absence/presence criteria) between day 10 and day 11 (Figure 2D). In comparison with the previous benchmark study of Andriankaja et al.,3 the majority of the transcriptional changes occurred earlier between day 9 and day 10. The discrepancy can be explained by either the difference in growth, which would not be unexpected considering that the experiments were performed independently, and/or an expected time delay between transcription and translation.
Figure 2.
Protein changes across early leaf development
(A) Heatmap of differential proteins identified using one-way ANOVA and absence/presence criteria (see results). Data were normalized to the maximal intensity. DAS, days after stratification. ANOVA analysis and heatmap were prepared using MeV software.10 Leaf panel was created using BioRender.com.
(B) Mean ± SD of all proteins classified as proliferation or expansion specific (see heatmap), in comparison with the mean of all non-differential (ND) proteins across leaf developmental time series (day 8 to day 13).
(C) Maximum normalized abundance of a selected proliferation and expansion marker proteins.
(D) Number of differential proteins calculated between two consecutive time points (e.g., between day 8 and day 9) using t test and absence/presence criteria (see results).
(E) Number of proteins with the highest abundance measured at a given day (x axis).
(F) SUBA database was used to assign subcellular localization.11 Percentage of proteins (where 100% is equal to 1) characterized by a given subcellular localization in comparison to all proteins peaking at a given time point (see Figure 1E).
In (A–F), data are from four replicas. In (D) and (E), analysis was restricted to the 1,215 proteins assigned as either proliferation or expansion specific using one-way ANOVA and absence/presence criteria. Complete data can be found in Data S1.
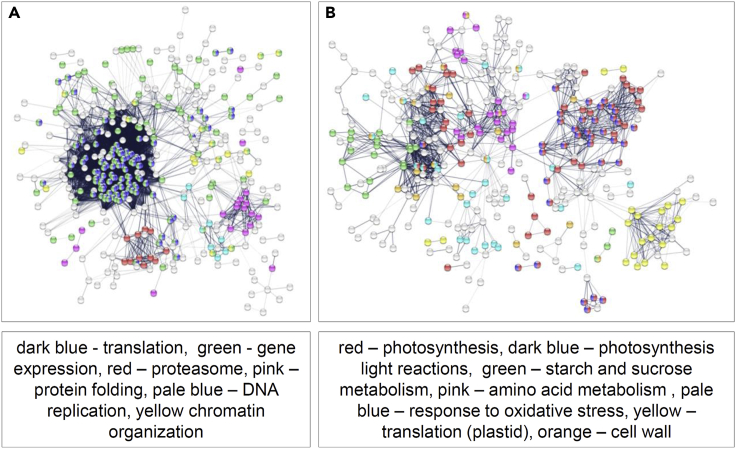
Seventy-two percent of the proliferation-specific proteins peaked at day 8, their levels remaining high at day 9 and 10 and dropping sharply at day 11 (Figure 2D). Functional analysis revealed over-representation of the cytosolic and nuclear proteins (Figure 2E), associated with cell division and cytoplasmic growth. These included subunits of the minichromosome maintenance (MCM) complex involved in replication, RNA polymerase complex and mRNA-processing enzymes, ribosomal proteins (60S and 40S subunits) elongation initiation factors, chaperones, proteasome subunits, proteins associated with chromatin organization, mitotic markers such as CDKB2;1, and PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1 and 2 (PCNA1 and PCNA2) (Figure 3). By contrast, expansion-specific proteins were characterized by a sharp increase at day 11 and an overall highest abundance at day 12. Functional analysis revealed an over-representation of plastidial proteins involved in protein translation, photosynthesis (both light and dark reactions), starch and sucrose metabolism, and the oxidative stress response (Figures 2E and 3).
Figure 3.
STRING visualization of the protein networks
STRING12 visualization of proteins assigned as proliferation (A) and expansion specific (B). Nodes represent proteins and edges represent interactions based on the experimental and database evidence with combined confidence score >0.4 imported from STRING. Unconnected nodes were removed. Colors indicate functional categories. Note changes in the protein interactome associated with the transition from proliferation to expansion.
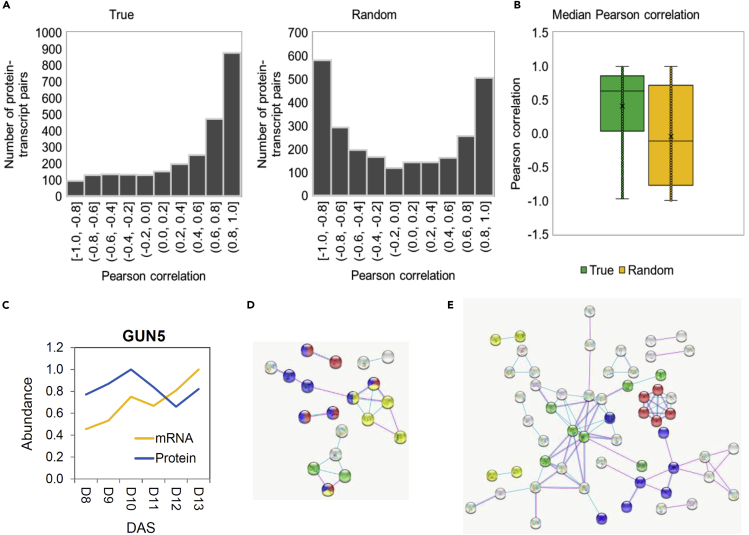
Next, and in order to look for the correlation between previously published transcript and protein expression profiles across early leaf development reported here, we filtered our proteomics dataset for proteins reproducibly detected in all six developmental time points (from day 8 to day 13). We then queried the list against the transcriptomics dataset of Andriankaja et al.3 The resulting list contained 2,562 transcript-protein pairs (referred to as the true dataset) used to calculate Pearson’s correlation between transcript and protein expression profiles (Data S2). Randomly shuffled protein and transcript profiles (referred to as the random dataset) were used to test whether the degree of correlation calculated for the true dataset differs from the random dataset. Indeed, this was the case. The Pearson correlation distribution shifted toward higher values when using the true in comparison with the random dataset; the median Pearson correlation of the true dataset was 0.62 while that of the random dataset was −0.1 (Figures 4A and 4B). Such good correlation points to protein translation being the primary determinant of protein abundance and composition during early leaf development. Consistent with this, 999 of the 1,192 differential proteins identified in our study and present in the transcriptome dataset3 were previously classified as differentially expressed (Data S2). As expected, however, considering that the data come from two independent studies, and considering a time delay between transcription and translation, transcript and protein levels, though highly correlated, were not necessarily identical. For instance, transcripts encoding photosynthetic proteins first started to accumulate between day 9 and day 10,3 while corresponding proteins accumulated between day 10 and day 11. Moreover, analysis of the 999 protein-transcript pairs identified 54 for which protein and transcript levels were behaving in a dissimilar manner (Data S2). A good example is a magnesium-chelatase subunit, GUN5, involved in chlorophyll synthesis, but also plastid-to-nucleus retrograde signaling.13 While the GUN5 transcript level increased gradually during leaf development, the GUN5 protein was characterized by maximum abundance at day 10 (Figure 4C) just before the onset of expansion. Considering the GUN5 protein accumulation pattern and its known involvement in the coordination of plastid-to-nucleus communication, GUN5 constitutes an interesting candidate to be characterized with respect to its role in linking differentiation with photosynthetic metabolism. Finally we looked into the differential proteins, whose transcripts did not change across the leaf developmental time series (69 proliferation- and 124 expansion-specific proteins) (Data S2). While proteins associated with gene expression such as eukaryotic translation initiation factors and those involved in mRNA maturation and DNA-directed RNA polymerases were enriched among proliferation-specific proteins (Figure 4D), the expansion-specific list contained enzymes involved in starch metabolism, photosystem subunits, and multiple components of the ATPase synthase complex (Figure 4E).
Figure 4.
Correlation between protein and transcript data
(A) Histogram distribution of the Pearson correlation across 2,562 protein-transcript pairs in the true versus a random dataset.
(B) Median Pearson correlation calculated across 2,562 protein-transcript pairs in the true versus random dataset.
(C) Maximum normalized abundance of GUN5 mRNA3 and protein abundance across leaf development.
(D and E) STRING12 visualization of proliferation (D) and expansion (E) specific proteins, characterized by non-differential transcripts.3 Nodes represent proteins and edges represent interactions based on the experimental and database evidence with combined confidence score >0.4 imported from STRING. Unconnected nodes were removed. Colors indicate functional categories. In (D) red, mRNA processing; blue, RNA binding; yellow, translation; green, RNA polymerase. In (E) yellow, glutathione metabolism; green, starch metabolism; red, ATP synthase complex; blue, photosynthesis. For complete data see Data S2.
Metabolomics analysis of early leaf development reveals major reprogramming at the transition from cell proliferation to cell expansion
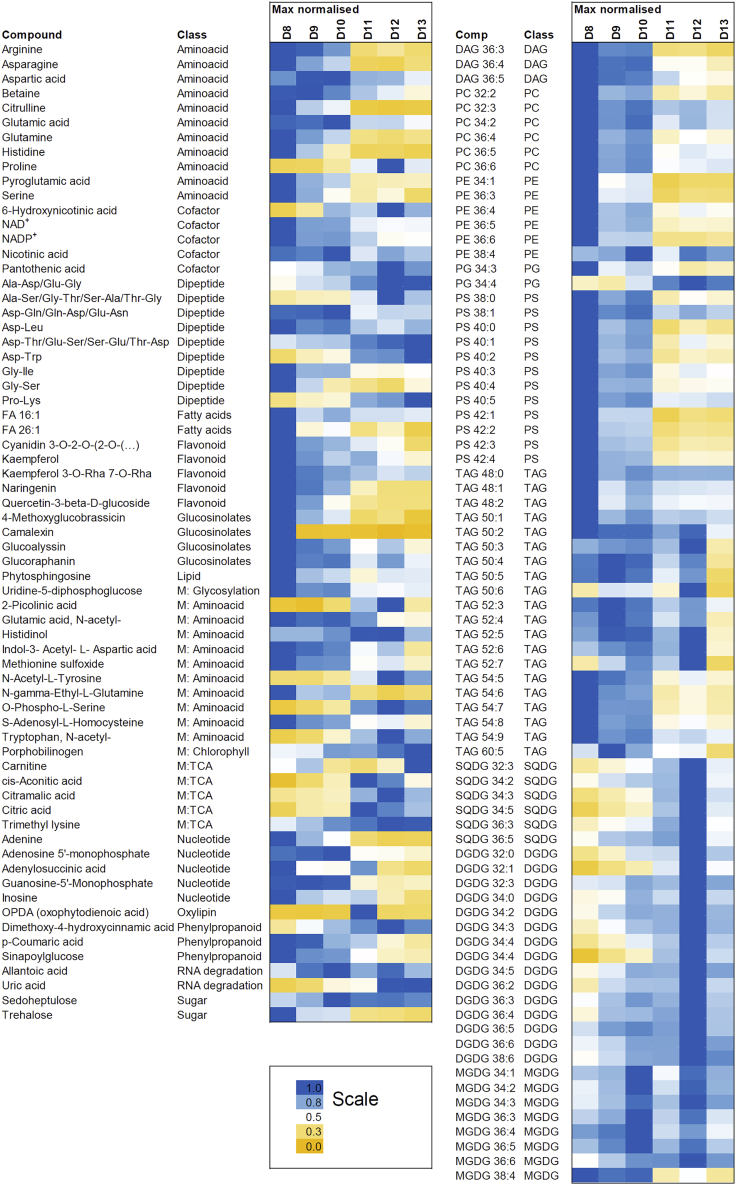
Untargeted liquid chromatography-mass spectrometry (LC-MS)-based metabolomics analysis of developing Arabidopsis leaves resulted in a dataset comprising 285 putatively annotated small molecules covering major lipid classes, primary and specialized metabolites, and dipeptides (Data S3). Statistical analysis using one-way ANOVA retrieved a list of 145 compounds characterized by a differential accumulation pattern across the six leaf developmental time points (Bonferroni corrected p < 0.05) (Data S3). One more compound, camalexin, was assigned as differential based on presence/absence criteria. Analogously to the protein data, the majority of the differential compounds displayed either proliferation- or expansion-specific accumulation patterns with a marked shift notable between day 10 and day 11 (Figures 5 and S1A). Similarity between metabolite and protein accumulation patterns was also visualized using correlation matrix calculated between all differential proteins and metabolites (Data S4 and Figure S2).
Figure 5.
Accumulation of differential (ANOVA, Bonferroni corrected p < 0.05) metabolites across leaf developmental time series
Data were normalized to the maximal intensity across the developmental time series. Data for metabolites, dipeptides, and lipids are from seven to eight replicates. Data for abscisic acid and indole-3-carboxylic acid are from three replicates. All metabolomics data can be found in Data S4.
The majority of the proliferation-specific compounds peaked at day 8 (Figure S1B), their levels remaining high at day 9 and 10 and dropping sharply at day 11. These included phosphatidylcholines, phosphatidylserines, phosphatidylethanolamines, triacylglycerols, diacylglycerols, unsaturated fatty acids, amino acids and their derivatives, nicotinic acid, nucleotides, glucosinolates, flavonoids, sinapol glucose, and trehalose (Figure 5). By contrast, expansion-specific metabolites increased at day 11 and peaked at day 12 (Figure S1B). These were thylakoid membrane lipids (monogalactosyldiacylglycerols, digalactosyldiacylglycerols, sulfoquinovosyldiacylglycerols, and phosphatidylglycerols), intermediates of the tricarboxylic acid cycle, numerous amino acid derivatives, proline, porphobilinogen, and pantothenic acid (Figure 5). Of the 45 proteinogenic dipeptides present in the dataset, four were proliferation specific while five were expansion specific (Figure 5).
A handful of compounds displayed a different accumulation pattern. Camalexin, for instance, was found almost exclusively at day 8, while 12-oxophytodienoic acid (12-OPDA), unlike any other compound, accumulated specifically at day 11. In contrast to 12-OPDA, N-acetylglutamic acid levels, high in the proliferating leaves, decreased sharply at day 12 and stayed low in the expanding leaves, while sedoheptulose peaked at day 10 just before the onset of expansion and remained high in the expanding leaves (Figure 5).
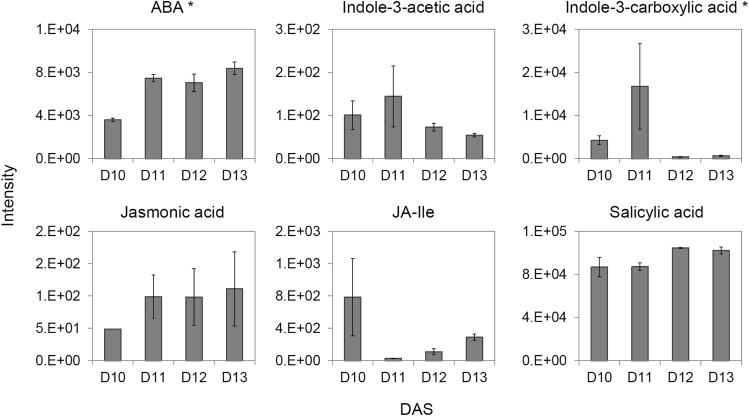
To complement the metabolite analysis, we measured selected plant hormones in four of the six leaf developmental time points (day 10 to day 13) (Data S5). While the levels of abscisic acid (ABA) increased significantly at the transition from proliferation to expansion and stayed high in expanding leaves (day 11 to day 13) (Figure 6), the levels of indole-3-acetic acid (IAA), salicyclic acid, jasmonate, and jasmonate-Ile were unchanged at the time points measured (Figure 6). Interestingly, indole-3-carboxylic acid (ICA), similarly to N-acetylglutamic acid, decreased sharply at the transition from day 11 to day 12 and remained low in the expanding leaves (Figure 6).
Figure 6.
Accumulation of plant hormones across leaf developmental time series
Data are mean ± SE from three replicas. Asterisk indicates significance (ANOVA, Bonferroni corrected p < 0.05). Note that measured jasmonic acid and jasmonate-Ile levels were very low and at the limit of detection. Data can be found in Data S5.
Discussion
While extensively studied, the molecular mechanisms underlying the transition from proliferation to expansion and how these are linked to metabolism are not well understood.8,9 To address the existing knowledge gap, we extended previous transcriptomics analysis of the early Arabidopsis leaf growth3 with proteomics and metabolomics analyses. The below examples are provided to demonstrate how a presented dataset can be mined for novel biological insight into molecular networks important for plant growth, with particular emphasis on the regulatory interplay between development and metabolism.
While the majority of the metabolites reported in our study were either proliferation or expansion specific, a handful of compounds displayed a different accumulation pattern. Most conspicuously, we report a major peak of 12-OPDA accumulation. 12-OPDA is an evolutionary conserved plant signaling small molecule and a precursor of a plant hormone, jasmonate.14 Markedly, accumulation of 12-OPDA was not accompanied by changes in either jasmonate or jasmonate-Ile. Our results are not without precedent. Accumulation of 12-OPDA, but not of jasmonate, was for instance reported during tendril coiling15 and in response to drought.16 Documented 12-OPDA-dependent, but jasmonate-independent, responses include seed germination,17 stomatal closure,16 expression of wounding-responsive genes,18 thermotolerance,19 and embryo development.20 12-OPDA treatment was also shown to inhibit plant growth and promote mitotic arrest.21,22 Specifically, exogenous application of 12-OPDA reduced leaf area21 and root length22 of wild-type Arabidopsis plants. An important characteristic of the peak of 12-OPDA, reported here, is its short-lived nature. The increase in the 12-OPDA accumulation measured between day 10 and day 11 can be attributed to the concomitant maturation of plastids whereby 12-OPDA is synthesized.23 What the biological significance of the 12-OPDA accumulation would be remains to be investigated, but we speculate that it may be involved in the previously reported coordination of plastidial maturation with the leaf meristem arrest.3,4 It is worth mentioning that expression of the LOX2 gene encoding lipoxygenase that catalyzes the first committed step in OPDA and jasmonate synthesis is under the control of TCP transcription factors24 involved in the regulation of the early leaf development and cell-cycle arrest. Finally, similarly to 12-OPDA, ABA synthesis is also associated with plastids, which may explain ABA accumulation in the expanding versus proliferating leaves and shedding new light onto ABA regulation of plant growth.25
12-OPDA and ABA accumulation coincides with the meristem disappearance. In comparison, N-acetylglutamate (NAG) and ICA stand out as unlike other proliferation-specific metabolites, whose levels dropped at day 11. NAG and ICA abundance decreases at day 12, which makes them interesting candidates in the context of leaf meristem maintenance. NAG is the first intermediate in the biosynthesis of arginine. In fact arginine was also high in the proliferating leaves, but unlike NAG arginine levels dropped at day 11. Indeed, a role of NAG in this scenario is not without precedence, since the extracellular NAG produced by Rhizobium was shown to stimulate mitosis in undifferentiated cells found on the outermost cell layer of clover seedling roots.26 ICA is an indole derivative known in for its role in the pathogen response.27
Another interesting group of compounds found in our analysis were proteinogenic dipeptides. In the past proteogenic dipeptides have been considered exclusively as products of protein turnover on the way to further proteolysis. However, this view has been challenged by a number of recent studies reporting dipeptides emerging as novel small-molecule regulators at the interface of protein degradation and metabolism.28, 29, 30, 31, 32, 33 Consistent with such a role, we report a number of dipeptides whose accumulation changes across early leaf development and are either proliferation or expansion specific, making them intriguing targets for detailed functional characterization.
Analogously to dipeptides, gluconeogenesis is also evolutionarily conserved across living organisms. The principal role of gluconeogenesis is to produce sugars from non-carbohydrate stores such as lipids and proteins. The majority of the eukaryotes rely on a single gluconeogenic route operated by phosphoenolpyruvate carboxykinase (PCK). However, a recent study of Arabidopsis demonstrated the existence of an alternative route depending on the activity of the orthophosphate dikinase (PPDK).34 Originally described during seed germination, both enzymes operate together; PCK is involved in lipid while PPDK is involved in protein mobilization. By contrast, our analysis suggests that unlike during seed germination, the two gluconeogenic routes are separated during leaf development. Specifically, we found that PCK is expressed in the proliferating leaves while PPDK is expressed in the expanding leaves, indicating that the proliferation-to-expansion transition is accompanied by a metabolic switch from cells using lipids to using proteins to fuel gluconeogenesis.
The above examples demonstrate how the datasets presented here can be mined to uncover novel biological insight into how development and metabolism are integrated during plant growth. Over the long term such understanding is in our opinion essential for developing strategies to enhance plant yield. The approach is, however, by no means restricted to (model) plants, being equally applicable in crop species. Indeed, it is not restricted to plants at all and could readily be adopted to study other systems in which metabolism is thought to play an important role in developmental progression, such as the dauer phase of nematodes35 or the proliferation of cancer cells in mammals.36 That said, given that these cases are already at least partially understood, the greatest utility of the approach taken will most likely be in studying the interface of development and metabolism under less well characterized circumstances.
Experimental procedures
Resource availability
Lead contact
Aleksandra Skirycz, Max-Planck-Institute of Molecular Plant Physiology, 14476 Potsdam-Golm, Germany. skirycz@mpimp-golm.mpg.de.
Materials availability
This study did not generate new unique reagents.
Data and code availability
Metabolomics and proteomics data used in this study can be found in the supplementary datasets. Moreover, chromatograms (proteomics) were submitted to ProteomeXchange: PXD019545.
Plant growth
Seeds of the Arabidopsis thaliana (L.) Heynh., accession Columbia-0 (Col-0), were sterilized and sown as described previously.37 In brief, Col-0 seeds were surface sterilized for 5 min in 70% (v/v) ethanol and then in sterilization solution (6% [w/v] sodium hypochlorite) for 5 min. After discarding the solution, seeds were washed three times with autoclaved double-distilled water (ddH2O). Seeds were then sown on half-strength Murashige and Skoog medium38 supplemented with 1% (w/v) sucrose, and solidified with 0.7% (w/v) phytoagar. Homogeneous germination was achieved by keeping the plates in darkness at 4°C for 3 days. Seedlings were grown in controlled conditions, in a growth chamber (Percival) with 16-h day length provided by fluorescent light at 120 μmol m−2 s−1, at constant temperature of 21°C.
Leaf sample collection
Third leaves were harvested from almost uniformly developed seedlings (from 8 days after stratification [DAS] until 13 DAS) grown on multiple plates, from independent experiments (approximately 5,000 leaves for 8- and 9-DAS seedlings and 1,500 leaves for 12- and 13-DAS seedlings). The selection of developmental timeline was guided by Andriankaja et al.3,37 and Omidbakhshfard et al.3,37 Dissection of small leaves was done under a stereomicroscope. Each day the harvest started 2 h into the day and lasted for 6–7 h. Leaves were harvested directly into eppies prefilled with liquid nitrogen, and never more than one plate was taken out of the growth chamber. For older leaves, a replica represents leaves harvested from one independent experiment, “experiment” being defined as plants sown and grown at the same time. For the very young leaf primordia, a replica represents leaves harvested from one or two independent experiments to ensure sufficient leaf material for multi-omics analysis. Frozen leaves from each replica were pulverized separately using 4-mm glass beads (Merck, Germany) in a mixer mill MM 400 (Retsch, Germany).
RNA extraction and qRT-PCR
Leaves were collected across five developmental stages (i.e., 9, 10, 11, 12, and 13 DAS), snap frozen in liquid nitrogen, and stored at −80°C. Leaves were ground using a mixer mill MM 400 (Retsch), and RNA extraction was performed using an RNA purification kit (Macherey-Nagel) with DNase treatment. RNA quality and concentration were checked on 1% agarose gel and using a spectrophotometer (NanoDrop OneC; Thermo Scientific). cDNA was synthesized using the PrimerScript RT reagent kit with gDNA Eraser (Takara), diluted (1:10) by adding ddH2O, and stored at −80°C. Primers for qRT-PCR were designed using the Primer3 (v0.4.0) tool, based on the gene coding sequences. GADPH (housekeeping gene)39 primers were used for gene-expression normalization. When possible, primers spanning two consecutive exons were designed. Only those primers that specifically blasted (Primer-Blast tool; NCBI) on the corresponding target gene were retained for the qRT-PCR. The amplification efficiency of the primers was checked by amplification of serial dilution (1:10) of the same cDNA. qRT-PCR was performed on four replicates per developmental stage. The Ct values (threshold cycle) of the candidate genes were normalized against the Ct values of the housekeeping gene in order to obtain the ΔCt normalized expression values (i.e., Ctcandidate − Cthousekeeping). Here, the housekeepers showed higher expression than the candidates, and as the lower the Ct value the higher the gene expression, bigger and positive ΔCt values were obtained when candidate genes had lower expression: for this reason, for the graphical representation of the gene expression (see Figure 1) the “10 − ΔCt” transformed values were used.40
Primer sequences for qRT-PCR
| Gene name | Gene ID | Forward sequence | Reverse sequence |
|---|---|---|---|
| TIP1;1 | AT2G36830 | GGC TCT GGC ATG GCT TTC AAC AAG | TGA AAG CAC CGA AAG TGA CGG CAG |
| GAPDH | AT1G13440 | TTG GTG ACA ACA GGT CAA GCA | AAA CTT GTC GCT CAA TGC AAT C |
| LOX2 | AT3G45140 | TGA CAT TGC TGA TAT CCG CGG CAG | TCA GGC ATC TCA AAC TCG CAC TCG |
| CDKB2;1 | AT1G76540 | TGA AGA AAT GTG GCC AGG AGT GAG C | CTT TGC TGA GAT TCG TTT CGC TGG C |
| GUN5 | AT5G13630 | AGT GCC TGA AGT TGG TGG TCA TGG | TGC TGC TGT TGT GGG AAT AGC CTG |
| PCNA1 | AT1G07370 | TGG ATT CGA GTC ACG TTG CTC TGG | TTC CGG CGC ATT TGA GCA TCT TCG |
| PCNA2 | AT2G29570 | TGT GCT CAG GCA GAA TAC TAC CGT CG | CCA CCA CGA CCG GTA ACT CAG AGG |
| ABA1 | AT5G67030 | GGG TTT AGG AGC CGT CGA GCT TTG C | TCT CCG TCA CCG CCT CTC TCT TCT C |
| PRL | AT4G02060 | AGC ACT GGT GCT GGC TGA CAT GG | TGG ATT GGC TGC AGC AAG AAC AGC |
Leaf area measurements
Plants were grown under controlled conditions as described above. Third leaves were harvested from seedlings between 9 and 14 DAS. Harvested leaves were blanched in 70% ethanol overnight, after which the leaves were washed twice in deionized water and mounted on a microscope slide in 85% lactic acid. Images were acquired by a Leica MZ 12.5 stereomicroscope with trinocular tubes and integrated digital Leica camera. Leaf area was calculated using ImageJ software.41
MTBE extraction
A three-in-one methyl tert-butyl ether (MTBE) extraction protocol42 was used to extract leaf samples (13 ± 1 mg per sample). While lipids separated into upper organic phase, polar and semipolar metabolites remained in the lower polar phase, and proteins could be found in the pellet. Metabolite data are from seven to eight replicates (see above). Protein data are from four replicates (see above).
Proteomics
Protein pellets obtained after MTBE extraction were resuspended in 50 μL of denaturation buffer (6 M urea, 2 M thiourea in 40 mM ammonium bicarbonate). Reduction of cysteines, alkylation, and enzymatic digestion using LysC/Trypsin Mix (Promega, Fitchburg, WI) followed by desalting of a digested peptide was performed according to the protocol described in Sokolowska et al.43 Dried peptides were resuspended in Murashige-Skoog loading buffer (3% acetonitrile [can], 0.1% formic acid) and measured with Q Exactive HF (Thermo Fisher Scientific, Hennigsdorf, Germany) coupled to a reverse-phase nano liquid chromatography ACQUITY ultra-performance liquid chromatography (UPLC) M-Class system (Waters). The equivalent of 1 μg of proteins was injected per run and the gradient ramped from 3.2% ACN to 7.2% ACN over 20 min, then to 24.8% ACN over the next 70 min and to 35.2% ACN over the next 30 min, followed by a 5-min washout with 76% ACN. The MS was run using a data-dependent acquisition method. Full scans were acquired at a 120,000 resolution, m/z ranging from 300.0 to 1,600.0, a maximum fill time of 50 ms, and an automatic gain control (AGC) target value of 3 × 106 ions. Each dd-MS2 scan was recorded at the resolution of 15,000 with an AGC target of 1 × 105, maximum injection time 100 ms, isolation window 1.2 m/z, normalized collision energy 27, and dynamic exclusion of 30 s. Raw proteomics files were analyzed using MaxQuant software (version 1.6.0.16)44 with Andromeda, an integrated peptide search engine using default orbitrap settings. Peptides were identified using A. thaliana Uniprot protein sequences (UP000006548). Moreover, a maximum of two missed cleavages were allowed and the threshold for peptide validation was set to 0.01 using a decoy database. In addition, methionine oxidation and N-terminal acetylation was considered as variable modification while cysteine carbamidomethylation was considered a fixed modification. In the further analysis, MS1-based label-free quantification was used and only proteins with two or more unique peptides were considered. Known contaminants such as keratins were removed. Data are available via ProteomeXchange45 with the identifier PXD019545.
Lipid profiling
Vacuum-dried organic phases (550 μL) were processed using UPLC (on a C8 reverse-phase column) coupled with Fourier transform mass spectrometry (Exactive mass spectrometer; Thermo Fisher, http://www.thermofisher.com) in positive and negative ionization modes as described in Giavalisco et al.42 Processing of chromatograms, peak detection, and integration were performed using RefinerMS (version 5.3; GeneData). Mass features were annotated using an in-house lipid database42 allowing for 10 ppm mass error, and a dynamic retention-time shift window of 0.1. For more details, see Data S6. Log2 normalized raw intensities were used for further analysis.
Metabolite profiling
Vacuum-dried polar phases (400 μL) resulted from MTBE extraction resuspended in 100 μL of LC grade water. After sonication of samples for 10 min in an ice-cooled sonicator bath, tubes were centrifuged for 15 min at full speed (>12,000 rpm). Supernatant (100 μL)—without any sediment from the bottom of the tube—was transferred to LC tubes for analysis. The samples were run on a UPLC-LC-MS machine as described previously.46 In brief, the UPLC system was equipped with an HSS T3 C18 reverse-phase column (100 × 2.1 mm internal diameter, 1.8 μm particle size; Waters) that was operated at a temperature of 40°C. The mobile phases consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The flow rate of the mobile phase was 400 μL/min, and 2 μL of sample was loaded per injection. The UPLC instrument was connected to an Exactive Orbitrap-focus (Thermo Fisher Scientific) via a heated electrospray source (Thermo Fisher Scientific). The spectra were recorded using full-scan positive and negative ion-detection mode, covering a mass range from m/z 100 to 1,500. The resolution was set to 70,000, and the maximum scan time was set to 250 ms. The sheath gas was set to a value of 60 while the auxiliary gas was set to 35. The transfer capillary temperature was set to 150°C while the heater temperature was adjusted to 300°C. The spray voltage was fixed at 3 kV, with a capillary voltage and a skimmer voltage of 25 V and 15 V, respectively. MS spectra were recorded from minutes 0 to 19 of the UPLC gradient. Processing of chromatograms, peak detection, and integration were performed using RefinerMS (version 5.3; GeneData). Metabolite identification and annotation were performed using standard compounds, tandem MS (MS/MS) fragmentation, and metabolomics databases.47 When using the in-house reference compound library we allowed for 10 ppm mass error, and a dynamic retention-time shift of 0.1. For more details, see Data S6. Log2 normalized raw intensities were used for further analysis.
Hormone profiling
The levels of selected plant hormones (see Data S4) were quantified as follows. Fifty milligrams of plant sample was extracted with 1 mL of precooled (−20°C) extraction MTBE/MeOH solvent. After mixing by vortexing, samples were kept on an orbital shaker for 30 min at 4°C followed by a 15-min sonication step in an ice-cooled bath. Thereafter, the samples were kept on an orbital shaker for an additional 30 min at 4°C. The samples were centrifuged at a speed of 10,000 × g for 10 min at 4°C. A fixed volume (0.5 mL) of upper supernatant (MTBE phase) was collected into a 1.5-mL microcentrifuge tube and dried down using a SpeedVac concentrator at room temperature (samples take up to 1 h at 30°C). The dried pellets were resuspended in 100 μL of water/methanol (50:50) solution, and the resuspended samples were subjected to UPLC-electrospray ionization (ESI)-MS/MS hormonal analysis. Separation was achieved using a UPLC system (e.g., Waters Acquity UPLC system; Waters, Manchester, UK) consisting of autoinjector, column oven, two binary pumps, degasser, and low-volume mixer. Analytical UPLC separation was achieved on a reverse-phase C18-column. A high-strength silica (HSS) T3 column (100 × 2.1 mm containing 1.8-μm diameter particles; Waters) fitted with a 2.1 × 5-mm guard column (Acquity UPLC HSS T3 VanGuard Pre-column, 100 Å, 1.8 μm; Waters) was used for this method. A UPLC separation method performed using a binary solvent system consists of water containing 0.1% (v/v) formic acid (solvent A) and methanol containing 0.1% (v/v) formic acid (solvent B). The gradient parameters for reverse-phase UPLC separation in negative ionization mode were as follows: 62% eluent A for 6.5 min; 45% B from 6.5 to 7 min; 10% A from 7 to 7.1 min; held at 0% A from 7.1 to 8.1 min; returned to initial conditions by 8.1 min. From 8.1 to 10 min, the column was re-equilibrated and conditioned to 62% A. The analysis in the positive mode was performed using the same gradient but starting with 80% A instead of 62% A. The column temperature was set at 40°C. The flow rate was 0.4 mL/min. The autosampler temperature was set at 10°C. Injection volume was 7.5 μL. The detection was determined by MS/MS analysis using a 4000 Triple Quad mass analyzer (e.g., 4000 QTRAP; AB Sciex Germany, Darmstadt, Germany) in multiple reaction monitoring scan type equipped with an ESI source (e.g., Turbo V Ion Source; AB Sciex) and attached to the UPLC system. Analyst software (version 1.6.2, AB Sciex) was used for instrument control and data acquisition, processing, and analysis. The operating parameters for MS/MS analysis were as follows: ion polarity, positive or negative; ion source, Turbo spray; capillary ion spray voltage, 5.5 kV or −4.5 kV for positive or negative ion polarity, respectively; probe tip position, 3 mm; source temperature, 500°C; gas, nitrogen; curtain gas, 25 psi; nebulizing gas and focusing gas, 40 psi; interface heater, on; collision activated dissociation gas pressure, medium.
Acknowledgments
We would like to thank Änne Michaelis for technical support. We are grateful to Prof. Ivo Feussner and Prof. Claus Wasternack for the critical discussion of our work, and to Dr. Otto Miersch and Dr. Joerg Ziegler for the oxylipin standards.
Author contributions
M.A.O. developed the idea, designed and planned the experiments, proved the initial concept, harvested the leaf material, performed MTBE extraction, provided the organic and polar fractions for lipid and metabolite analyses as well as the protein pellet for proteomics analysis, and helped in writing the paper. E.M.S. prepared, ran, and analyzed protein samples, measured leaf areas, and assisted in writing the paper. V.D.V. performed qRT-PCR measurements. A.K. analyzed lipid data under the supervision of Y.B. S.A. measured and analyzed metabolite data. L.P.d.S. measured and analyzed plant hormones. A.R.F. conceptualized the project and co-wrote the manuscript. A.S. analyzed and biologically interpreted the data and co-wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2021.100235.
Contributor Information
Mohammad Amin Omidbakhshfard, Email: omidbakhshfard@yahoo.com.
Alisdair R. Fernie, Email: fernie@mpimp-golm.mpg.de.
Aleksandra Skirycz, Email: skirycz@mpimp-golm.mpg.de.
Supplemental information
Log2 transformed labeled free quantification (LFQ) intensities were used to calculate mean, one-way ANOVA, and t test (unpaired, two-tailed distribution assuming equal variance). Data were also normalized to the maximum intensity measured across developmental time series (day 8 to day 13). GO categories were downloaded from TAIR and subcellular localization from the SUBA database. Expansion- and proliferation-specific proteins were assigned based on the results of ANOVA (Bonferroni corrected p value <0.05) or absence/presence criteria.
Transcript data were retrieved from Skirycz et al.3 Pearson correlation was calculated only for proteins reproducibly detected (in at least 3 of the 4 replicas) in all six developmental time points (day 8 to day 13). Column AD, with asterisks, indicates protein-transcript pairs characterized by opposite behavior. Column AE, with asterisks, indicates differential proteins whose transcript level was classified as non-differential.
Log2 transformed raw intensities were used to calculate mean, one-way ANOVA, and t test (unpaired, two-tailed distribution assuming equal variance). Data were also normalized to the maximum intensity measured across developmental time series (day 8 to day 13). For camalexin, p was assigned a value of 0.05 based on absence/presence criteria.
Log2 transformed raw intensities were used to calculate mean and one-way ANOVA. Data were also normalized to the maximum intensity measured across developmental time series (day 10 to day 13).
References
- 1.Nelissen H., Gonzalez N., Inze D. Leaf growth in dicots and monocots: so different yet so alike. Curr. Opin. Plant Biol. 2016;33:72–76. doi: 10.1016/j.pbi.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez N., Vanhaeren H., Inze D. Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 2012;17:332–340. doi: 10.1016/j.tplants.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Andriankaja M., Dhondt S., De Bodt S., Vanhaeren H., Coppens F., De Milde L., Muhlenbock P., Skirycz A., Gonzalez N., Beemster G.T., Inze D. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell. 2012;22:64–78. doi: 10.1016/j.devcel.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Beauvoit B.P., Colombie S., Monier A., Andrieu M.H., Biais B., Benard C., Cheniclet C., Dieuaide-Noubhani M., Nazaret C., Mazat J.P., Gibon Y. Model-assisted analysis of sugar metabolism throughout tomato fruit development reveals enzyme and carrier properties in relation to vacuole expansion. Plant Cell. 2014;26:3224–3242. doi: 10.1105/tpc.114.127761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazawa H., Aulehla A. Revisiting the role of metabolism during development. Development. 2018;145 doi: 10.1242/dev.131110. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez N., De Bodt S., Sulpice R., Jikumaru Y., Chae E., Dhondt S., Van Daele T., De Milde L., Weigel D., Kamiya Y. Increased leaf size: different means to an end. Plant Physiol. 2010;153:1261–1279. doi: 10.1104/pp.110.156018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skirycz A., Claeys H., De Bodt S., Oikawa A., Shinoda S., Andriankaja M., Maleux K., Eloy N.B., Coppens F., Yoo S.D. Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell. 2011;23:1876–1888. doi: 10.1105/tpc.111.084160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalve S., De Vos D., Beemster G.T. Leaf development: a cellular perspective. Front. Plant Sci. 2014;5:362. doi: 10.3389/fpls.2014.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez N., Inze D. Molecular systems governing leaf growth: from genes to networks. J. Exp. Bot. 2015;66:1045–1054. doi: 10.1093/jxb/eru541. [DOI] [PubMed] [Google Scholar]
- 10.Howe E.A., Sinha R., Schlauch D., Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27:3209–3210. doi: 10.1093/bioinformatics/btr490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper C.M., Tanz S.K., Castleden I.R., Vacher M.A., Small I.D., Millar A.H. SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics. 2014;30:3356–3364. doi: 10.1093/bioinformatics/btu550. [DOI] [PubMed] [Google Scholar]
- 12.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mochizuki N., Brusslan J.A., Larkin R., Nagatani A., Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U S A. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasternack C., Feussner I. The oxylipin pathways: biochemistry and function. Annu. Rev. Plant Biol. 2018;69:363–386. doi: 10.1146/annurev-arplant-042817-040440. [DOI] [PubMed] [Google Scholar]
- 15.Stelmach B.A., Muller A., Hennig P., Laudert D., Andert L., Weiler E.W. Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry. 1998;47:539–546. doi: 10.1016/s0031-9422(97)00547-5. [DOI] [PubMed] [Google Scholar]
- 16.Savchenko T., Kolla V.A., Wang C.Q., Nasafi Z., Hicks D.R., Phadungchob B., Chehab W.E., Brandizzi F., Froehlich J., Dehesh K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 2014;164:1151–1160. doi: 10.1104/pp.113.234310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dave A., Hernandez M.L., He Z., Andriotis V.M., Vaistij F.E., Larson T.R., Graham I.A. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell. 2011;23:583–599. doi: 10.1105/tpc.110.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taki N., Sasaki-Sekimoto Y., Obayashi T., Kikuta A., Kobayashi K., Ainai T., Yagi K., Sakurai N., Suzuki H., Masuda T. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005;139:1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monte I., Kneeshaw S., Franco-Zorrilla J.M., Chini A., Zamarreno A.M., Garcia-Mina J.M., Solano R. An ancient COI1-independent function for reactive electrophilic oxylipins in thermotolerance. Curr. Biol. 2020;30:962–971 e3. doi: 10.1016/j.cub.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Goetz S., Hellwege A., Stenzel I., Kutter C., Hauptmann V., Forner S., McCaig B., Hause G., Miersch O., Wasternack C., Hause B. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 2012;158:1715–1727. doi: 10.1104/pp.111.192658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Turner J.G. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller S., Hilbert B., Dueckershoff K., Roitsch T., Krischke M., Mueller M.J., Berger S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell. 2008;20:768–785. doi: 10.1105/tpc.107.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasternack C., Song S.S. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017;68:1303–1321. doi: 10.1093/jxb/erw443. [DOI] [PubMed] [Google Scholar]
- 24.Schommer C., Palatnik J.F., Aggarwal P., Chetelat A., Cubas P., Farmer E.E., Nath U., Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6:1991–2001. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida T., Christmann A., Yamaguchi-Shinozaki K., Grill E., Fernie A.R. Revisiting the basal role of ABA—roles outside of stress. Trends Plant Sci. 2019;24:625–635. doi: 10.1016/j.tplants.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Philip-Hollingsworth S., Hollingsworth R.I., Dazzo F.B. N-Acetylglutamic acid: an extracellular nod signal of Rhizobium trifolii ANU843 that induces root hair branching and nodule-like primordia in white clover roots. J. Biol. Chem. 1991;266:16854–16858. [PubMed] [Google Scholar]
- 27.Pastorczyk M., Kosaka A., Pislewska-Bednarek M., Lopez G., Frerigmann H., Kulak K., Glawischnig E., Molina A., Takano Y., Bednarek P. The role of CYP71A12 monooxygenase in pathogen-triggered tryptophan metabolism and Arabidopsis immunity. New Phytol. 2020;225:400–412. doi: 10.1111/nph.16118. [DOI] [PubMed] [Google Scholar]
- 28.Thirumalaikumar V.P., Wagner M., Balazadeh S., Skirycz A. Autophagy is responsible for the accumulation of proteogenic dipeptides in response to heat stress in Arabidopsis thaliana. FEBS J. 2020;288:281–292. doi: 10.1111/febs.15336. [DOI] [PubMed] [Google Scholar]
- 29.Veyel D., Sokolowska E.M., Moreno J.C., Kierszniowska S., Cichon J., Wojciechowska I., Luzarowski M., Kosmacz M., Szlachetko J., Gorka M. PROMIS, global analysis of PROtein-metabolite interactions using size separation in Arabidopsis thaliana. J. Biol. Chem. 2018;293:12440–12453. doi: 10.1074/jbc.RA118.003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strehmel N., Hoehenwarter W., Monchgesang S., Majovsky P., Kruger S., Scheel D., Lee J. Stress-related mitogen-activated protein kinases stimulate the accumulation of small molecules and proteins in Arabidopsis thaliana root exudates. Front. Plant Sci. 2017;8:1292. doi: 10.3389/fpls.2017.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Zhao Y., Wang X., Lin R., Zhang Y., Ma H., Guo Y., Xu L., Zhao B. The novel dipeptide Tyr-Ala (TA) significantly enhances the lifespan and healthspan of Caenorhabditis elegans. Food Funct. 2016;7:1975–1984. doi: 10.1039/c5fo01302j. [DOI] [PubMed] [Google Scholar]
- 32.Kanegawa N., Suzuki C., Ohinata K. Dipeptide Tyr-Leu (YL) exhibits anxiolytic-like activity after oral administration via activating serotonin 5-HT1A, dopamine D1 and GABAA receptors in mice. FEBS Lett. 2010;584:599–604. doi: 10.1016/j.febslet.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Naka K., Jomen Y., Ishihara K., Kim J., Ishimoto T., Bae E.J., Mohney R.P., Stirdivant S.M., Oshima H., Oshima M. Dipeptide species regulate p38MAPK-Smad3 signalling to maintain chronic myelogenous leukaemia stem cells. Nat. Commun. 2015;6:8039. doi: 10.1038/ncomms9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eastmond P.J., Astley H.M., Parsley K., Aubry S., Williams B.P., Menard G.N., Craddock C.P., Nunes-Nesi A., Fernie A.R., Hibberd J.M. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 2015;6:6659. doi: 10.1038/ncomms7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnell A.M., Houthoofd K., O'Hanlon K., Vanfleteren J.R. Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans. Exp. Gerontol. 2005;40:850–856. doi: 10.1016/j.exger.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 37.Omidbakhshfard M.A., Fujikura U., Olas J.J., Xue G.P., Balazadeh S., Mueller-Roeber B. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018;14:e1007484. doi: 10.1371/journal.pgen.1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murashige T., Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 39.Skirycz A., De Bodt S., Obata T., De Clercq I., Claeys H., De Rycke R., Andriankaja M., Van Aken O., Van Breusegem F., Fernie A.R., Inzé D. Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 2010;152:226–244. doi: 10.1104/pp.109.148965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giavalisco P., Li Y., Matthes A., Eckhardt A., Hubberten H.M., Hesse H., Segu S., Hummel J., Kohl K., Willmitzer L. Elemental formula annotation of polar and lipophilic metabolites using (13)C, (15)N and (34)S isotope labelling, in combination with high-resolution mass spectrometry. Plant J. Cell Mol. Biol. 2011;68:364–376. doi: 10.1111/j.1365-313X.2011.04682.x. [DOI] [PubMed] [Google Scholar]
- 43.Sokolowska E.M., Schlossarek D., Luzarowski M., Skirycz A. PROMIS: global analysis of PROtein-metabolite interactions. Curr. Protoc. Plant Biol. 2019;4:e20101. doi: 10.1002/cppb.20101. [DOI] [PubMed] [Google Scholar]
- 44.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alseekh S., Tohge T., Wendenberg R., Scossa F., Omranian N., Li J., Kleessen S., Giavalisco P., Pleban T., Mueller-Roeber B. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell. 2015;27:485–512. doi: 10.1105/tpc.114.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tohge T., Fernie A.R. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 2010;5:1210–1227. doi: 10.1038/nprot.2010.82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Log2 transformed labeled free quantification (LFQ) intensities were used to calculate mean, one-way ANOVA, and t test (unpaired, two-tailed distribution assuming equal variance). Data were also normalized to the maximum intensity measured across developmental time series (day 8 to day 13). GO categories were downloaded from TAIR and subcellular localization from the SUBA database. Expansion- and proliferation-specific proteins were assigned based on the results of ANOVA (Bonferroni corrected p value <0.05) or absence/presence criteria.
Transcript data were retrieved from Skirycz et al.3 Pearson correlation was calculated only for proteins reproducibly detected (in at least 3 of the 4 replicas) in all six developmental time points (day 8 to day 13). Column AD, with asterisks, indicates protein-transcript pairs characterized by opposite behavior. Column AE, with asterisks, indicates differential proteins whose transcript level was classified as non-differential.
Log2 transformed raw intensities were used to calculate mean, one-way ANOVA, and t test (unpaired, two-tailed distribution assuming equal variance). Data were also normalized to the maximum intensity measured across developmental time series (day 8 to day 13). For camalexin, p was assigned a value of 0.05 based on absence/presence criteria.
Log2 transformed raw intensities were used to calculate mean and one-way ANOVA. Data were also normalized to the maximum intensity measured across developmental time series (day 10 to day 13).
Data Availability Statement
Metabolomics and proteomics data used in this study can be found in the supplementary datasets. Moreover, chromatograms (proteomics) were submitted to ProteomeXchange: PXD019545.