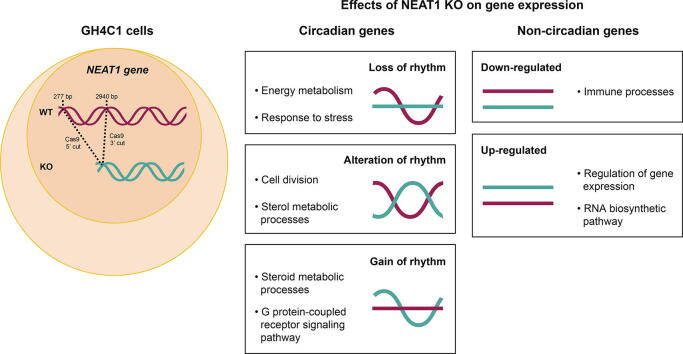
Graphical abstract
Keywords: Long non-coding RNA Neat1, CRISPR/Cas9 edition, Paraspeckles, Circadian gene expression, RNA-sequencing
Highlights
-
•
Neat1 deletion affects numerous circadian and non-circadian genes.
-
•
Neat1 deletion causes loss, modification or acquisition of gene circadian pattern.
-
•
Paraspeckles contribute significantly to the circadian transcriptome.
Abstract
The functions of the long non-coding RNA, Nuclear enriched abundant transcript 1 (Neat1), are poorly understood. Neat1 is required for the formation of paraspeckles, but its respective paraspeckle-dependent or independent functions are unknown. Several studies including ours reported that Neat1 is involved in the regulation of circadian rhythms. We characterized the impact of Neat1 genetic deletion in a rat pituitary cell line. The mRNAs whose circadian expression pattern or expression level is regulated by Neat1 were identified after high-throughput RNA sequencing of the circadian transcriptome of wild-type cells compared to cells in which Neat1 was deleted by CRISPR/Cas9. The numerous RNAs affected by Neat1 deletion were found to be circadian or non-circadian, targets or non-targets of paraspeckles, and to be associated with many key biological processes showing that Neat1, in interaction with the circadian system or independently, could play crucial roles in key physiological functions through diverse mechanisms.
1. Introduction
Whereas Nuclear enriched abundant transcript 1 (Neat1) is one of the most studied long non-coding RNA (lncRNA), its roles in physiology and pathology remain still poorly understood. Initially, its physiological role was considered non-essential due to the observation that Neat1 knock-out mice have adequate health and fertility [1]. It was later found that Neat1 was required for corpus luteum formation affecting fertility in certain sub-populations, and for mammary gland development and lactation in mice [2], [3]. Furthermore, multiple roles for Neat1 in pathology have been since also described, from cancers [4] to neurodegenerative [5], cardiac [6], immune and viral [7] diseases.
Neat1 is required for the formation of paraspeckles, which are nuclear substructures found in most cultured cells [8]. The Neat1 gene is expressed as two variant isoforms: a short one, Neat1_1 (3.8 kb in length in humans) and a long one, Neat1_2 (22.7 kb). Both of these are transcribed from the same promoter, but have different sites of transcriptional termination [8]. The longer Neat1 isoform, Neat1_2, is essential for the assembly of paraspeckles, whereas Neat1_1, albeit also a paraspeckle component, is dispensable for their formation and likely plays paraspeckle-independent roles [9]. At the cellular level, Neat1 has been implicated in various processes, including regulation of transcription and chromatin active state and miRNA biogenesis (for review see [10]) and has been shown to regulate epigenetic marks on histones [11], [12]. The expression of Neat1 has been shown to increase when cells are submitted to a stress [13], [14], [15], [16]. Indeed stress conditions such as hypoxia [17], viral infection [18], heat shock [19], mitochondrial stress [20] or proteasome inhibition [21] leads to an increase in Neat1.
Although the respective functions of both isoforms as well as the functions that are paraspeckle-dependent or independent remain unclear, it was recently reported in several studies including ours that Neat1 is involved in the regulation of circadian rhythms [22], [23], [24], [25], [26].
The mammalian circadian system is a timing system that allows living organisms to anticipate the daily environmental changes, so that behavior and tissue physiology can be adjusted according to the day/night cycle [27]. The circadian system controls the rhythmicity of a wide range of processes, from molecular (e.g., transcriptional mechanisms), cellular (e.g., the cell cycle [28]) physiological (hormonal secretion [29], metabolic [30]), up to the behavioral processes (sleep/wake cycle, cognitive functions, etc.). This circadian clock system is hierarchically organized and composed of tissue and cellular clocks. Within the cells, a set of clock genes and their protein products, which are highly conserved among animals, generate cell-autonomous rhythms by forming transcription – translation feedback loops through circadian variation in abundance of the positive (Clock and Bmal1) and the negative (Per1, Per2 and Cry1, Cry2) components of the loops. This molecular clock, in turn, drives the rhythmic expression of numerous genes, which ultimately exert control over every biological, physiological, and behavioral process.
Most cellular circadian rhythms are therefore underpinned by daily rhythms in gene expression. While the core circadian system concentrates on transcriptional control, it has been apparent that substantial regulation of clock-controlled genes (ccg) is achieved after transcription so that post-transcriptional controls are emerging as crucial modulators of circadian genes [31], [32], [33], [34], [35]. Indeed in eukaryotes, approximately 1%-10% of genes are subjected to circadian control directly or indirectly but only ~ 1/5 of the mRNAs that display rhythmic expression are driven directly by transcription, which suggests that post-transcriptional mechanisms including RNA splicing, polyadenylation, mRNA stability, mRNA cytoplasmic export and RNAs nuclear retention are essential layers for generation of gene expression rhythmicity [33], [34], [35], [36], [37], [38].
We have shown that paraspeckles play a role in the circadian expression of genes through a post-transcriptional mechanism [25]. Indeed, paraspeckles are well known to control gene expression at the post-transcriptional level through the nuclear retention of mRNAs [39], [40]. We have shown in a rat pituitary cell line, the GH4C1 cells, that the expression of numerous paraspeckle components including Neat1 itself, follows a circadian pattern leading to rhythmic variations in paraspeckle number within the cells and that, thanks to their circadian expression pattern and their functions in mRNA nuclear retention, paraspeckles rhythmically retain RNAs in the nucleus. This rhythmic nuclear retention leads to the rhythmic expression of these paraspeckle-target genes [25], [26].
A genome-wide screening of the impact of Neat1 deletion was undertaken to evaluate the role of Neat1 on the circadian and non-circadian gene expression in GH4C1 cells. To this end the mRNAs whose circadian expression pattern or expression level is regulated by Neat1 were identified after high-throughput RNA sequencing of the circadian transcriptome of wild type GH4C1 cells compared to that of GH4C1 cells in which Neat1 was deleted by the CRISPR/Cas9 technology. Using Panther analysis [41], the biological processes with which the numerous circadian and non-circadian genes affected by Neat1 deletion were associated, were characterized. Finally, whether these genes were or weren’t included in the list of paraspeckle RNA targets previously established [25] was investigated.
2. Material and methods
2.1. Cell line culture
GH4C1 cells, a rat pituitary somatolactotroph line, were obtained in 2012 from ATCC® (CCL-82.2™, lot number: 58945448, Molsheim, France) with certificate analysis and were confirmed to be free of mycoplasma (MycoAlert, Lonza, Levallois-Perret, France). They were grown in HamF10 medium supplemented with 15% horse serum and 2% fetal calf serum.
2.2. CRISPR genomic engineering
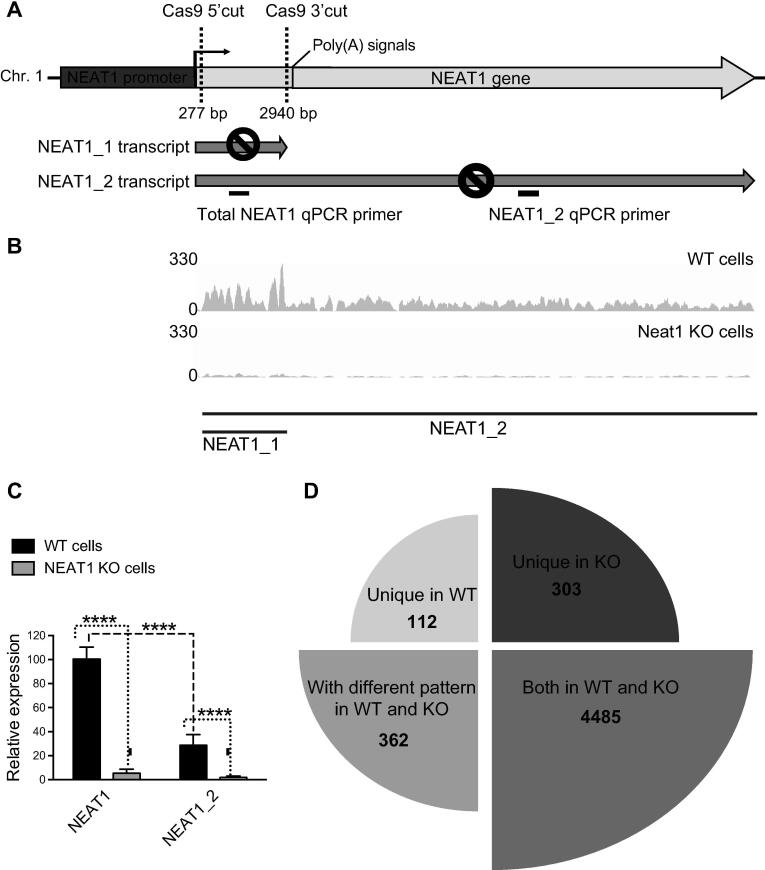
For Neat1 knockout, a fragment of 2663nt was edited using a CRISPR/Cas9-based strategy in the 5′ region common to Neat1_1 and Neat1_2 isoforms (Fig. 1A). Two sgRNAs target sites were designed using a CRISPR design tool available on http://crispr.mit.edu and the pairs of oligonucleotides (Supplementary Table 1) were synthesized by Integrated DNA Technologies (Coralville, Iowa, USA). These oligonucleotides were phosphorylated using T4 PNK, annealed and ligated into the BbsI (New England Biolabs, Ipswich, MA, USA) restriction site in plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (gift from Feng Zhang; Addgene #42230) [42]. The two sgRNA expression plasmids were amplified using Subcloning EfficiencyTM DH5αTM Competent Cells (Thermo Fisher Scientific, Carlsbad, CA, USA). Midi-preparations of plasmids were made with the NucleoBond Xtra Midi kit (Macherey-Nagel, Hoerdt, France) and sequencing was performed (Genewiz, Leipzig, Germany) to confirm the accuracy of the inserted sequence.
Fig. 1.
A-C: Schematic representation of Neat1 genetic edition by CRISPR/Cas9 and characterization of Neat1 KO cell line. A. The positions of guide RNAs used to delete the Neat1 gene are indicated as well as the positions of qPCR primers used to verify deletion of Neat1 gene (primer sequences are given in Supplementary Table 1). B. RNA-Seq peaks for the Neat1 locus, that is not officially annotated in the rat genome, demonstrate preferential expression of Neat1_1 in Neat1WT cells and residual Neat1_1 and Neat1_2 expression in Neat1 KO cells. C. Significant down-regulation of total Neat1 and Neat1_2 in Neat1 KO GH4C1 lines as revealed by RT-qPCR analysis. qPCR primers (Supplemental Table 1) for total Neat1 are in shared sequences between Neat1_1 and Neat1_2 and detect both isoform RNAs. D. Impact of Neat1 genetic edition on circadian expressed genes. Neat genetic deletion leads to a loss of rhythmic pattern in 112 genes (Unique in WT), in a different rhythmic pattern in 362 genes (With different pattern in WT and KO) and in a de novo genesis of rhythmic pattern in 303 genes (Unique in KO). 4485 circadian genes were not affected (Both in WT and KO).
GH4C1 cells were transfected with both plasmids containing sgRNAs, using Lipofectamine 3000 (Thermo Fisher Scientific). Forty-eight hours after the seeding, genomic DNA was extracted using the Nucleospin Tissue kit (Macherey-Nagel) and tested by PCR to screen for genomic editing events, using Hot Start Taq polymerase (New England Biolabs). Subcloning was performed by limit dilution in a 96-well plate. Total RNA of each clonal population was extracted (Macherey-Nagel) and the level of total Neat1 and Neat1_2 expression was monitored by qPCR using specific primers (Supplementary Table 1) whose position was indicated in Fig. 1A. One clone line invalidated for Neat1 (hereinafter referred to as Neat1 KO cells) was selected for the following experiments.
2.3. RNA sequencing experiments
To synchronize cells between themselves, cells were transferred to fresh medium and were harvested after this fresh medium replacement (T0) every 4 h from T10 to T34. For WT and Neat1KO cells, three wells (3.5 cm2) were pooled at each time and three replicates were collected from each pool. Total RNA was extracted using the Nucleospin RNA kit (Macherey Nagel). The construction of Illumina DNA libraries and the sequencing from RNA pools were performed by Genewiz (Leipzig, Germany). Libraries were prepared with Illumina Sample Preparation kit with rRNA depletion. Strand-specific RNA-seq was done on Illumina HiSeq 2500, with a read length of 2x150 bp, strand specific (30 million reads per sample on average were obtained).
2.4. RNA-sequencing data processing
Analyses were performed on a local instance of Galaxy. After quality control checks by FastQC and check for adapter content with Trimmomatic, paired-end reads were aligned to the Rat reference genome (Rnor_6.0.80, Ensembl) using STAR [43]. The length of the genomic sequence around annotated junctions used was 149 and the other parameters were set to default values. FeatureCounts [44] was used to quantify RNA expression at the gene level, using the default values for parameters.
The RNA sequencing data are available at Gene Expression Omnibus (GEO) (accession number n° GSE162751).
2.5. RNA-sequencing analysis
Several tools were used to visually explore sample relationships (Supplemental Fig. 1). First, the distribution for logarithm-transformed counts of triplicate of each sample showed a slightly shift for one of the replicates of the T34 time-point in the WT group (Supplemental Fig. 1A). However, scatterplot of transformed counts from the two groups at T34 showed that while T34 r1 replicate was less correlated to r2 and r3 in WT than in KO, the correlation was still good enough to keep T34 r1 in the analysis (Supplemental Fig. 1B).
2.6. Identification of differentially cycling mRNAs
Rhythmic expression of RNAs was analyzed using ECHO (Extended Circadian Harmonic Oscillators). ECHO is an R powered application to find and visualize circadian rhythms using extended harmonic [45]. We specified a period of 24 h. Differences in rhythmicity between WT and Neat1 KO cells were analyzed using the LimoRhyde (Linear models for Rhythmicity design) workflow [46]. We built a global list of rhythmic genes, applying a 0.1 q-value cutoff and we then used Limma [47] to characterize the changes of rhythmicity between groups. The q-value cutoff for differentially rhythmic genes was 0.1.
2.7. Identification of differentially expressed mRNAs
The non-rhythmic genes where further analyzed for differential expression using Limma with a 0.01 q-value cutoff. The comparison of expressions for non-circadian genes between genotypes was made for all time points (3 replicates X 7 time points for each genotype).
2.8. Real-Time qPCR
To synchronize cells between themselves, cells were transferred to fresh medium and were harvested after this fresh medium replacement (T0) every 4 h from T2 to T30.
Real time reverse transcriptase PCR (RT-qPCR) was performed using gene-specific primers (Supplementary Table 1). The PCR was performed on a CFX96TM Real-Time PCR system using iTaq™ universal SYBR® Green supermix (Bio-Rad, Hercules, CA, USA). Rplp0 and Gapdh were used as endogenous controls. The non-circadian expression for each of those two genes was checked and Rplp0 being the most stable was used for normalizing the expression data of the target genes.
2.9. Cosinor analysis
Cosinor analyses were performed using Prism4 software (GraphPad Software, Inc.). Mean experimental values (±SEM) were fitted using Prism4 by a non-linear sine wave equation: Y = Baseline + Amplitude * sin (Frequency*X + Phase-shift), where Frequency = 2pi/period and period = 24 h. Goodness-of-fit was quantified using R squared, experimental values being considered well fitted by cosinor regression when the R squared was higher than 0.50. A statistically significant circadian oscillation was considered if the 95% confidence interval for the amplitude did not include the zero value (zero-amplitude test) [48], [49].
3. Results
3.1. Loss of Neat1 affects circadian genes in three different ways
Part of the genome corresponding to the Neat1_1 isoform was edited using CRISPR/Cas9 technology (Fig. 1A). RNA-Seq peaks for the Neat1 locus which is not officially annotated in the rat genome, exhibited in WT cells more counts for Neat1_1 compared to Neat1_2 and a residual number of counts for both Neat1_1 and Neat1_2 in Neat1 KO cells (Fig. 1B). Results obtained by RT-qPCR analysis were consistent with RNA-Seq data (Fig. 1C). Indeed, the efficacy of CRISPR/Cas9 edition was evaluated by RT-qPCR with primers that detected total Neat1 (Neat1), namely both Neat1_1 and Neat1_2 or only Neat1_2 (Fig. 1A). In Neat1 KO cells, Neat1 as well as Neat1_2 levels were shown to be less than 8% compared to the levels in WT cells (Fig. 1C). Furthermore, in WT cells, Neat1_2 was shown to represent around 30% of Neat1 (Fig. 1C). Then, the deletion of the part of the genome of Neat1 corresponding to the Neat1_1 isoform (Fig. 1A) leads also to the decrease in the Neat1_2 expression. This was in agreement with results obtained previously by Yamazaki et al. [50].
A global analysis by ECHO, Lymorhyde and Limma applications of the RNA-seq data obtained from both cellular genotypes showed that 5262 genes followed a circadian expression pattern which represented 38% of the 13,776 genes expressed in GH4C1 cells (Supplementary Table 2). Since Neat1 is not officially annotated in the rat genome, it was not included in the list of genes that displayed a rhythmic pattern, but we showed by an additional qPCR analysis that Neat1 as well as Neat1_2 exhibited a circadian expression pattern in WT cells, as we previously reported (Supplemental Fig. 2). This was not only consistent with our previous report [25], but this corroborated another study that has also reported since there, the circadian expression of Neat1 in skin fibroblasts [51].
Among the 5262 genes with a circadian expression pattern, 777 (14.8%) exhibited circadian differences between the two cellular genotypes (Fig. 1D; Supplementary Table 2). It was observed that 112 genes (2%) became arrhythmic when Neat1 was deleted, 362 genes (6.9%) displayed a modified circadian pattern and 303 genes (5.7%) acquired a circadian pattern after Neat1 editing (Fig. 1D and Fig. 2; Supplementary Table 3). It then appeared that, while around 15% of rhythmic genes in GH4C1 cells displayed a circadian pattern dependent on Neat1, these Neat1-dependent circadian genes could be classified in three categories depending on whether they lost their circadian pattern, they displayed a modified one or they acquired one (Fig. 1D and Fig. 2).
Fig. 2.
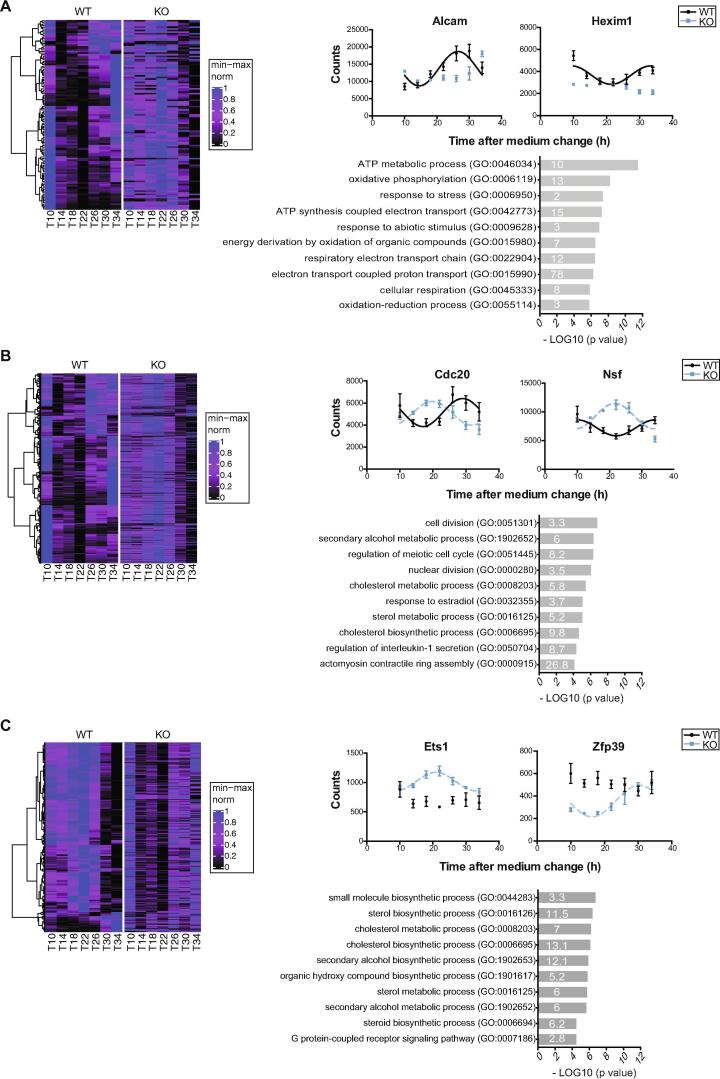
Analysis of circadian genes affected by Neat1 deletion. A. Analysis of genes from the category “Unique in WT”. Left Panel Heat map representation of genes found to belong to the category “Unique in WT”. Heat maps have been built using the euclidean distance as a measurement of how similar gene expressions in WT group are to each other and the ward.D2 method to group them based on that distance. Corresponding genes are displayed in the same order in KO group. The dendrogram obtained from hierarchical clustering is shown on the left side of the heatmap. A min–max normalization was applied for each gene independently and is represented by colors on the heatmap: higher, blue; lower, black. The hour of cell collection is indicated below (T). Right Upper Panel Cosinor analysis of RNA-Seq counts of two genes, Alcam and Hexim1, shows that counts from WT cells could be adequately fitted (R2 > 0.50) with a non-linear cosinor equation in which the period value was set to 24 h (cosinor fit values given in Supplemental Table 4), while counts from Neat1 KO cells can’t. At each time point, data are means ± SEM of samples from the three biological replicates of RNA-Seq. Right Lower Panel Functional characterization by Panther Analysis showing the Gene Ontology biological processes with the highest p value. The numbers inside the columns correspond to the fold enrichment. B. Analysis of genes from the category “With different pattern in WT and KO”. Left Panel Heat map representation of genes found to belong to the category “With different pattern in WT and KO”. Heat maps have been built using the euclidean distance as a measurement of how similar gene expressions in WT group are to each other and the ward.D2 method to group them based on that distance. Corresponding genes are displayed in the same order in KO group. The dendrogram obtained from hierarchical clustering is shown on the left side of the heatmap. A min–max normalization was applied for each gene independently and is represented by colors on the heatmap: higher, blue; lower, black. The hour of cell collection is indicated below (T). Right Upper Panel Cosinor analysis of RNA-Seq counts of two genes, Cdc20 and Nsf, shows that counts from WT and KO cells could be adequately fitted (R2 > 0.50) with a non-linear cosinor equation in which the period value was set to 24 h (cosinor fit values given in Supplemental Table 4). At each time point, data are means ± SEM of samples from the three biological replicates of RNA-Seq. Right Lower Panel Functional characterization by Panther Analysis showing the Gene Ontology biological processes with the highest p value. The numbers inside the columns correspond to the fold enrichment. C. Analysis of genes from the category “Unique in KO”. Left Panel Heat map representation of genes found to belong to the category “Unique in KO”. Heat maps have been built using the euclidean distance as a measurement of how similar gene expressions in KO group are to each other and the ward.D2 method to group them based on that distance. Corresponding genes are displayed in the same order in WT group. The dendrogram obtained from hierarchical clustering is shown on the left side of the heatmap. A min–max normalization was applied for each gene independently and is represented by colors on the heatmap: higher, blue; lower, black. The hour of cell collection is indicated below (T). Right Upper Panel Cosinor analysis of RNA-Seq counts of two genes, Ets1 and Zfp39, shows that counts from KO cells could be adequately fitted (R2 > 0.50) with a non-linear cosinor equation in which the period value was set to 24 h (cosinor fit values given in Supplemental Table 4), while counts from WT cells can’t. At each time point, data are means ± SEM of samples from the three biological replicates of RNA-Seq. Right Lower Panel Functional characterization by Panther Analysis showing the Gene Ontology biological processes with the highest p value. The numbers inside the columns correspond to the fold enrichment. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We previously showed that another lncRNA, Malat1, which is not officially annotated in the rat genome is closely associated with Neat1 [52]. Malat1 was shown here in an additional qPCR analysis to belong to the first category of Neat1 regulated genes since Malat1 exhibited a circadian expression pattern in WT cells that was lost after Neat1 deletion (Supplemental Fig. 2). This altered circadian pattern of Malat1 was further associated with a decrease in its expression level (Supplemental Fig. 2). Since it has been shown that the expression of Malat1 is not altered in different tissues of mice lacking the Neat1 poly-adenylation signal necessary for the formation of the short isoform of Neat1 [53], it may be assumed that the lack of Neat1_2 was specifically responsible for the effects on Malat1 we reported here.
Within two of the three categories of Neat1 gene regulation mentioned above, we found some core-clock genes. This was the case for Per2 whose circadian expression pattern was lost in Neat1 KO cells (Supplemental Fig. 3) and for Per1, Per3 and Cry1 whose circadian expression pattern differed between the two cell lines (Supplemental Fig. 3). Moreover, direct clock-controlled genes considered as transcriptional relays between the molecular core-oscillator and target genes such as Tef or Nfil3 were also shown to display a different circadian pattern in Neat1 KO cells as compared to WT cells (Supplemental Fig. 3).
Within each of the three categories of genes, we isolated 3 to 4 clusters at the third node (Fig. 2A-C Left Panels). Whereas oscillating patterns were consistent within each cluster, they were distinct between clusters, with transcripts peaking at different times during the 24 h cycle (Fig. 2A-C Left Panels). Several genes from each of these three categories were selected and their circadian pattern fitted by cosinor analysis using either RNA-seq counts (Fig. 2A-C Right Upper Panels) or qPCR measurements (Supplemental Fig. 4). Accordingly, two genes (Alcam and Hexim1) were shown here to lose their circadian expression pattern in Neat1 KO cells. Indeed, while counts from WT cells could be adequately fitted (R2 > 0.50) with a non-linear cosinor equation in which the period value was set to 24 h, counts from Neat1 KO cells couldn’t (Fig. 2A Right Upper Panel; cosinor fit values given in Supplementary Table 4). The same hold true for two others genes selected from the same category (Nos1 and Mtnd4) whose expression pattern was determined after RT-qPCR analysis (Supplemental Fig. 4). While being adequately fitted (R2 > 0.50) with a non-linear cosinor equation with a period value of 24 h in both WT and Neat1 KO cells, the genes Cdc20 and Nsf (Fig. 2B Right Upper Panel; cosinor fit values given in Supplementary Table 4) as well as the genes Aura and Ccnb1 (Supplemental Fig. 4) displayed a phase inverted pattern between the two genotypes. As examples of genes belonging to the third category, we showed that Ets1 and Zfp39 were rhythmic in Neat1 KO and not in WT cells (Fig. 2C Right Upper Panel; cosinor fit values given in Supplementary Table 4). This was also the case for Mvk and Fdft1 as shown in Supplemental Fig. 4. Whether these three categories of circadian genes that were differently affected by Neat1 deletion were involved in different functions, was further assessed by Panther analysis [41].
3.2. Circadian genes impacted by Neat1 deletion are associated with different functions
The circadian genes becoming arrhythmic after Neat1 deletion were shown to be mainly involved in energy metabolism and response to stress after Panther analysis (Fig. 2A Right Lower Panel, Supplementary Table 5). We indeed found that in the list of genes from the first category, namely genes that displayed a circadian expression pattern only in WT, the terms “energy metabolism” and “response to stress” were among the most prominent enriched annotation clusters in Gene Ontology biological processes (Fig. 2A Right Lower Panel). These functional annotations were consistent with the recently reported cross-regulation between paraspeckles and mitochondria [20] and with the numerous studies showing that Neat1 acts as an important sensor and effector during stress [19], [15], [16].
The circadian genes with a Neat1 dependent cycling profile were further mainly involved in cell cycle and sterol metabolism. Functional classification of genes from the second category, namely genes with a different circadian pattern in Neat1 KO cells showed indeed the prominence of annotations such as “cell division” and “sterol metabolic processes” (Fig. 2B Right Lower Panel, Supplementary Table 5). The genes from this category could then constitute the substratum of the implication of Neat1 in proliferative processes and in lipid metabolism [54], [55]. Indeed Neat1 has been not only involved in the formation of physiological tissue such as the mammary gland or the corpus luteum [3], [2] but its deregulation has been extensively described in tumor processes (for a review [56]). Finally, circadian genes acquiring a circadian pattern after Neat1 deletion were also mainly involved in “sterol metabolic processes” and in “G protein-coupled receptor signaling pathway” (Fig. 2C Right Lower Panel, Supplementary Table 5).
3.3. Down-regulated and up-regulated genes after Neat1 deletion are functionally distinct
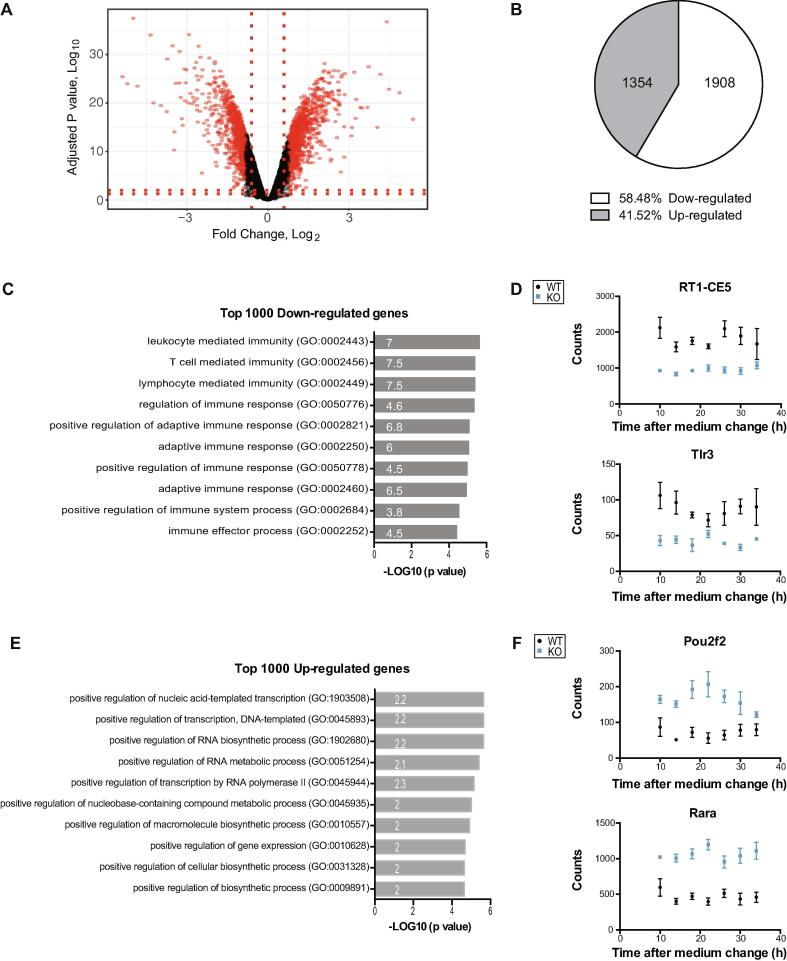
Apart from circadian expressed genes, 3262 genes were found differentially expressed following Neat1 deletion representing near 25% of genes expressed in GH4C1 cells (13776 genes). Among these 3262 genes, 1354 were up- and 1908 down-regulated (Fig. 3A, B; Supplementary Table 2). Panther analysis of the top 1,000 up- or down-regulated genes was carried out. This analysis revealed distinct gene classes whose expression was altered by Neat1 deletion. Down-regulated genes were shown to be mainly involved in “immune processes” (Fig. 3C, Supplementary Table 6) consistent with the well-recognized view that Neat1 is an immunity-associated lncRNA that regulates important cytokine production and the immune response [57], [58]. Up-regulated genes were themselves merely associated with “regulation of gene expression” and “RNA biosynthetic process” (Fig. 3E, Supplementary Table 6). A few genes from both lists of up- and down-regulated genes were selected and their RNA-seq counts reported during the circadian period to illustrate down-regulated genes associated with “immune processes” and up-regulated genes associated with “regulation of gene expression”, (Fig. 3D, F).
Fig. 3.
Analysis of down-regulated and up-regulated genes after Neat1 deletion. A. Volcano plot for Neat1 KO cells versus WT cells is shown; the top 1000 down-regulated and the top 1000 up-regulated genes are highlighted in red. B. The proportion of down-regulated and up-regulated genes is also shown (gene numbers are given in pie charts). C. Functional characterization by Panther Analysis of the top 1000 down-regulated genes showing the Gene Ontology biological processes with the highest p value. The numbers inside the columns correspond to the fold enrichment. D. RNA-Seq counts of two genes, RT1-CE5 and Tlr3, from the top 1000 down-regulated genes are given as an example. At each time point, data are means ± SEM of samples from the three biological replicates of RNA-Seq. E. Functional characterization by Panther Analysis of the top 1000 up-regulated genes showing the Gene Ontology biological processes with the highest p value. The numbers inside the columns correspond to the fold enrichment. F. RNA-Seq counts of two genes, Pou2f2 and Rara, from the top 1000 up-regulated genes are given as an example. At each time point, data are means ± SEM of samples from the three biological replicates of RNA-Seq. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. A quarter of circadian expressed genes are paraspeckle RNA targets
We previously determined the list of RNAs that are paraspeckle targets [25]. Since the circadian variation of the number of paraspeckles within the nucleus of GH4C1 cells, we reported [25], is believed to account for the circadian expression of paraspeckle RNA targets, we compared the list of paraspeckle RNA targets, we previously established, with that of genes that displayed a circadian expression pattern presented here. We found that among the 5262 genes that displayed a circadian expression pattern, 1380 (26%) were paraspeckle targets (Fig. 4A). Since a quarter of circadian expressed genes were associated with paraspeckles, the contribution of the paraspeckles to the circadian transcriptome was believed to be very significant.
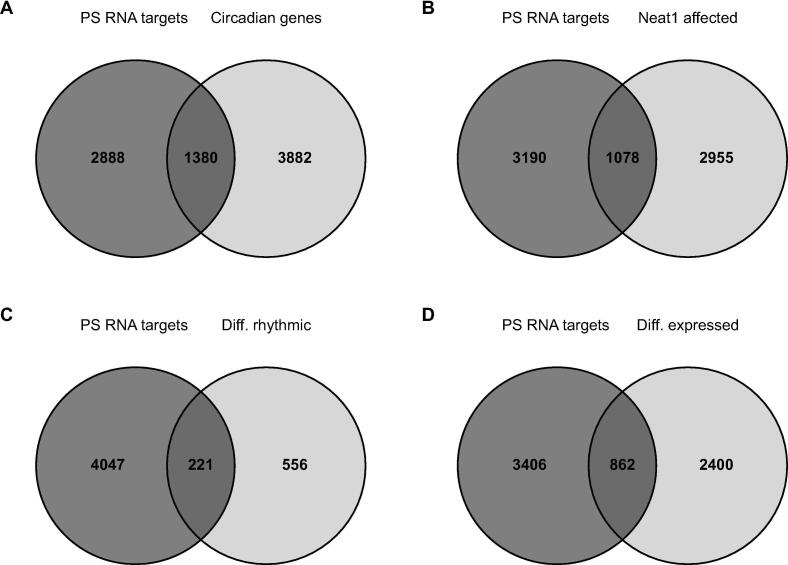
Fig. 4.
A. Contribution of Paraspeckle RNA targets to the circadian transcriptome. Venn diagram representing overlaps between circadian transcripts and Paraspeckle (PS) RNA targets. B-D. Contribution of Paraspeckle RNA targets to Neat1 dependent transcriptome. Venn diagrams representing overlaps between Paraspeckle (PS) RNA targets and total genes affected by Neat1 deletion (B), circadian genes affected by Neat1 deletion (C) and genes whose expression is affected by Neat1 deletion (D).
3.5. A quarter of genes affected by Neat1 deletion are paraspeckle RNA targets
When taking into account both the genes whose expression level was modified by Neat1 deletion (3262 genes) and those whose circadian expression was affected (777 genes), it was concluded that Neat1 was an important regulator since near 30% of genes expressed by GH4C1 cells (13776 genes) were affected by Neat1 deletion.
Since Neat1 deletion is believed to disrupt paraspeckle bodies, we evaluated the number of genes affected by Neat1 deletion (i.e. whose circadian expression or expression level was modified) with that of genes associated with paraspeckles, by overlapping this list with that of paraspeckle RNA targets, we previously reported [25]. Neat1 deletion affected 1078 genes (26%) that were paraspeckle RNA targets (Fig. 4B). We further discriminated among these 1078 genes those that displayed a modified circadian pattern from those that exhibited an altered expression level. We found that among the 777 genes that displayed an altered circadian pattern after Neat1 deletion, 221 (28%) were paraspeckle RNA targets (Fig. 4C) and among the 3262 genes with an altered expression level after Neat1 deletion, 862 genes (26%) were paraspeckle RNA targets (Fig. 4D). It then appeared that the proportion of genes affected by Neat1 deletion that were associated with paraspeckles, was equivalent whether Neat1 regulated their level of expression or their circadian expression.
4. Discussion
The present study shows that in GH4C1 cells, the circadian transcriptome turns out to be really significant representing no less than 38% of expressed genes. Of course the use of ECHO to identify rhythmic genes, an application that has been shown to out-perform existing approaches to identify oscillations in genome-scale circadian datasets [45], may explain the great proportion of rhythmically expressed genes found here. Indeed, the development of high-throughput sequencing techniques [59] coupled to always more efficient and robust methods to detect rhythmic expression profiles has led to the re-evaluation of the extent of the circadian transcriptome in a given tissue or cell line. Depending on the tissue or cell type studied, the proportion of rhythmically expressed genes is estimated to span from 6 to 20%. Moreover, when considering the whole organism, it was first estimated that approximately half of the genome is oscillating somewhere in the body [60], but nowadays substantial improvements in performance have amplified the notion that circadian oscillations are pervasiveness by showing that 93.5% of all possible protein coding transcripts exhibit circadian oscillations in at least one tissue [61]. In any case, the significant extent of the circadian transcriptome in GH4C1 cells is not only consistent with our previous reports showing that these cells harbor a functional circadian oscillator [62], [25] but also supports the relevance to use this cell line for circadian issues.
Although there are various computational methods to detect rhythmicity in genome-scale data, methods to detect changes in rhythmicity or changes in average expression between different conditions are scarcer. We used LimoRhyde, a method that allows detecting changes in rhythmicity across different conditions and therefore allows unraveling the effects of genetic perturbations on biological rhythms [46]. In keeping with the view that Neat1 is deeply involved in the circadian system functioning [22], [23], [24], [25], [26], this computational method allows us to show that the CRISPR/Cas9 editing of Neat1 in GH4C1 cells affects no less than 15% of circadian expressed genes. While the number of genes that became arrhythmic following Neat1 deletion is relatively weak, a more significant number of genes display a modified circadian profile in Neat1 KO cells. The same hold also true for core-clock genes themselves. Whereas only Per2 loses its circadian expression pattern, several other core-clock genes as well as their direct targets such as transcription factors known to relay circadian oscillations to circadian-controlled genes, display a modified circadian profile after Neat1 removal. Of course, these core-clock genes and their direct targets that are altered after Neat1 removal may trigger part of the observed modifications in circadian-clock-controlled genes. Strikingly, in addition to disrupting the circadian expression of numerous genes that become either arrhythmic or display a different profile, Neat1 deletion causes an unexpected genesis of de novo oscillating transcripts. Whether these de novo oscillating transcripts are genes whose circadian expression in WT cells is masked by an anti-phasic circadian rhythm generated by Neat1 or whether they reflect a real reprogramming of the circadian transcriptome after Neat1 deletion remains to be determined. Whatever the case, when analyzed for singular enrichment in biological processes, the circadian genes affected by Neat1 deletion exhibited different functional association depending on whether they lost or gain rhythmicity or whether they display a modified profile.
Interestingly, genes that lose their circadian expression after Neat1 deletion were found associated with both “energy metabolism” and “response to stress”. This is reminiscent of the well-recognized involvement of Neat1 in the cellular response to stress [13], [14], [15] as well as in the cross-regulation that has been reported between paraspeckles and mitochondria [20]. Indeed, it has been shown that Neat1 depletion has important effects on mitochondrial dynamics by altering the sequestration of mitochondrial mRNAs in paraspeckles [20]. It should be noted that the interaction between the circadian clock and the response to stress is well established and that the interest in regulation of mitochondria by the circadian timekeeping system has gained interest as more and more evidence indicates that the biological clock orchestrates the functioning of mitochondria and suggests that the circadian clock coordinates energy metabolism and cellular anti-oxidant mechanisms that prevent oxidative damage [63], [64]. In view of the present findings, it is then tempting to speculate that Neat1 could be one of the links through which the circadian clock is involved in both the response to stress and the regulation of energy metabolism.
The numerous genes that exhibit a modified circadian pattern after Neat1 deletion were rather shown associated with “cell cycle” and “sterol metabolism”. Interestingly, the genetic ablation of Neat1 in mice results in disturbances of cell growth such as aberrant mammary gland morphogenesis [3] or impaired corpus luteum formation associated with a lack of progesterone synthesis [2]. In addition, Neat1 is now well recognized as a critical component in the progress and development of cancer [65]. Of note, it has now become obvious that cholesterol pathways are intertwined with circadian clock and that the circadian clock affects the carcinogenesis by regulating the lipid metabolism [66]. The finding we report here, that Neat1 is involved in the regulation of clock-controlled-genes associated both with “cell cycle” and “sterol metabolism”, suggests that Neat1 plays a role in the interconnection of the circadian system with cell growth.
While 15% of circadian-expressed-genes are shown here altered in Neat1 KO cells, 25% of non-circadian genes exhibit a modified level of expression. This strengthens the regulatory role exerted by Neat1 on GH4C1 transcriptome. It may be suggested that the endocrine origin of these cells may account for the important impact of Neat1 since in mice “in vivo”, Neat1 knockdown mostly results in endocrine perturbations, such as mammary gland [3] or corpus luteum [2] development or the inappropriate behavioral response to stress [24]. In keeping with this view, the number of differentially expressed genes was found greatly smaller in the cortex of Neat1 KO mice; however, this number strongly varied according to the different studies [67], [24]. While in one study [67], the number of genes found to be differentially expressed is 1359 genes, in the other one, the number is very low (289 genes) [24]. These discrepancies may rely on the variability between samples which can be reduced by increasing the sampling as it is the case in our study, where we compared 21 samples (3 replicates X 7 time points) from each genotype. Moreover, it is also probable that samples from cell lines are more homogenous than samples from animals. Consistent with this view, Neat1 loss in mouse embryonic fibroblasts lines was shown to result in significant expression level changes in near 1300 genes [68]. In addition, when the raw data from the latter study [68] were re-analyzed with the 1% FDR used in our own study, 2905 genes were found to be differentially expressed in Neat1-/-MEFs, a number very close to ours.
Of note, in our study, the top down-regulated genes are mostly associated with “immune processes” in keeping with several reports highlighting the emergence of Neat1 as an important regulatory layer in controlling gene expression in innate immunity and adaptive immune response [57], [69]. In particular, Neat1 is considered to be critical for the immune response against virus [7]. It is therefore not surprising to find here that Neat1 positively regulates numerous genes closely associated with immune functions, such as Tlr3 which is one of the Toll‐like receptors (TLRs) that are central players in the early host immune response to acute viral infection and have been shown to play a crucial role in defense against microorganisms [70]. Consistent with the positive regulation of Tlr3 by Neat1 we report here, Neat1 has been recently found inversely correlated to the level of Tlr3 in patients with Hepatitis B virus infection [70].
Neat1 through its binding to an RNA-binding protein, Hexamethylene bis-acetamide-inducible protein 1 (Hexim1) has been shown to play an important role in regulating DNA-mediated induction of the innate immune response [71]. It is then of note that in GH4C1 cells, Hexim1 follows a circadian expression pattern which is under Neat1 control since this pattern is lost after Neat1 removal. Importantly a number of immune functions follow diurnal variations including for instance, counts of lymphocytes, T lymphocytes, and B lymphocytes in human blood [72], cytokine and chemokine expression [73], response to antigen presentation [74] or leukocyte tissue recruitment [75]. Whether the crosstalk between Neat1 and the immune functions implies the rhythmicity of the innate immune response remains to be explored.
The comparison of the paraspeckle RNA targets we previously established in GH4C1 cells [25] with circadian-expressed genes identified here allows us to show that the circadian nuclear retention by paraspeckles of mRNAs contributes significantly to the circadian transcriptome in this cell line. Surprisingly however, the proportion of genes that in addition to be affected by Neat1 deletion, are also paraspeckle RNA targets, is the same in the non-circadian gene group compared to the circadian one and represents only a quarter of Neat1 regulated genes. Therefore, three inferences may be drawn from these data. First, the paraspeckle RNA targets may be arrhythmic, whether they are or they aren’t affected by Neat1 deletion. For those RNAs, the rhythmic pattern generated by paraspeckle rhythm may be counterbalanced by an opposite rhythm in their transcription. As an alternative, the quantity of these RNAs associated to paraspeckles may not be sufficient to generate their rhythmic pattern. The second inference drawn from our data is that some paraspeckle RNA targets are not affected by Neat1 deletion suggesting that the association of these RNAs with paraspeckles is not dispensable for their expression. Finally, since there are both rhythmic and non-rhythmic genes affected by Neat1 that aren’t paraspeckle targets, it may be suggested that some Neat1 regulation exerted on the rhythmic pattern or on the expression level of RNAs may be independent of paraspeckles. In this view, there are several ways by which Neat1 can regulate the expression of genes that aren’t associated with paraspeckles. First, Neat1 can mediate gene regulation through paraspeckle independent mechanisms by acting as a miRNA sponge. This mechanism is utilized in various cancers such as hepatocellular carcinoma [76]. Another way through which Neat1 has been shown to regulate the transcription of some genes, is via the retention of proteins such as transcription factors in paraspeckles. For instance the retention of the transcriptional and splicing regulator SFPQ in paraspeckles impedes the ability of SFPQ to bind the chromatin of target genes, leading to changes in the transcriptional output, such as the repression of Adarb2 [21] and the induction of Il8 [18]. p53 is another example of a transcription factor whose nuclear retention by paraspeckles can allow Neat1 to regulate the expression of many genes. Indeed, p53 not only promotes Neat1 transcription [14] but it has also been shown to be a paraspeckle RNA target [25] which is up-regulated after Neat1 knock-down as reported here and in other studies ([77], [78]). It is therefore probable that thanks to the up-regulation of p53, Neat1 deletion can affect the expression of many rhythmic and non-rhythmic genes; indeed, KEGG pathways classification shows a significant enrichment in the p53 pathway in rhythmic genes affected by Neat1 deletion after a DAVID analysis (data not shown); furthermore, a bioinformatics analysis of the enrichment in transcription factor by the software ChIP-X Enrichment Analysis 3 (ChEA3) [79] shows that among the 777 and 3262 genes whose rhythmic pattern or expression level was affected by Neat1 deletion, 196 genes and 708 genes are potential p53 targets, respectively (data not shown).
Neat1 can also direct gene repression through DNA methylation of gene promoters [80] and has been shown to alter chromatin marks such as histone H3K4 trimethylation and histone H3K9 acetylation in specific genes [11]. Neat1 could play a role in the control of chromatin remodeling. Proteins from the SWI/SNF chromatin remodeling complex were shown to be major components of the paraspeckles [81], leading to the assumption that Neat1 could alter chromatin organization by bringing these proteins to the chromatin of active genes or by retaining them in the paraspeckles. Finally, as both isoforms of Neat1 have been deleted in our KO cells, the Neat1 regulation of genes that are not associated with paraspeckles may be attributed to the deletion of the short isoform that is known to have paraspeckle independent roles [9], [82].
In summary, the genetic deletion of Neat1 in GH4C1 cells affects many circadian- and non-circadian genes that, depending on the type of control exerted by Neat1, are associated with different specific biological processes, including “energy metabolism”, “response to stress”, “cell division”, “sterol metabolism”, “immune processes” and “gene regulation”. Furthermore, by overlapping the list of genes affected by Neat1 deletion with that of paraspeckle RNA targets previously established in this cell line [25], the existence of different mechanisms through which Neat1 can exert its regulatory control is proposed. Overall, the present data shed light on the crucial roles that Neat1 can play through multiple diverse mechanisms in cellular biological processes.
Funding
This work was supported by Aix-Marseille University and Centre National Recherche Scientifique and funded by a grant from Pfizer Laboratories.
6. Availability of data and material
The RNA sequencing data have been deposited at Gene Expression Omnibus (GEO) (accession number n° GSE162751). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162751.
Credit authorship contribution statement
Audrey Jacq: Investigation, Methodology, Data curation, Formal analysis, Visualization, Writing - review & editing. Denis Becquet: Conceptualization, Data curation, Formal analysis, Software, Validation, Visualization, Writing - review & editing. Maria-Montserrat Bello-Goutierrez: Data curation, Formal analysis, Software, Validation, Visualization. Bénédicte Boyer: Investigation, Methodology, Formal analysis. Séverine Guillen: Investigation, Methodology, Formal analysis. Jean-Louis Franc: Conceptualization, Formal analysis, Validation, Visualization, Supervision, Writing - review & editing. Anne-Marie François-Bellan: Conceptualization, Formal analysis, Validation, Visualization, Supervision, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.022.
Contributor Information
Jean-Louis Franc, Email: jean-louis.franc@univ-amu.fr.
Anne-Marie François-Bellan, Email: anne-marie.francois@univ-amu.fr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Nakagawa S., Naganuma T., Shioi G., Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Standaert L., Adriaens C., Radaelli E., Van Keymeulen A., Blanpain C., Hirose T. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong P., Xiong Y., Yue J., Hanley S.J.B., Kobayashi N., Todo Y. Long Non-coding RNA NEAT1: A novel target for diagnosis and therapy in human tumors. Front Genet. 2018;9 doi: 10.3389/fgene.2018.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riva P., Ratti A., Venturin M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res. 2016;13:1219–1231. doi: 10.2174/1567205013666160622112234. [DOI] [PubMed] [Google Scholar]
- 6.Zhou H., Wang B., Yang Y.-x., Jia Q.-J., Zhang A.o., Qi Z.-W. Long Noncoding RNAs in pathological cardiac remodeling: A review of the update literature. Biomed Res Int. 2019:1–11. doi: 10.1155/2019/7159592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prinz F., Kapeller A., Pichler M., Klec C. The implications of the long non-coding RNA NEAT1 in non-cancerous diseases. Int J Mol Sci. 2019;20.;20:627. doi: 10.3390/ijms20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirose T., Yamazaki T., Nakagawa S. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip Rev RNA. 2019;10 doi: 10.1002/wrna.v10.610.1002/wrna.1545. [DOI] [PubMed] [Google Scholar]
- 9.Li R., Harvey A.R., Hodgetts S.I., Fox A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA. 2017;23:872–881. doi: 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox A.H., Nakagawa S., Hirose T., Bond C.S. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarty D., Sboner A., Nair S.S., Giannopoulou E., Li R., Hennig S. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5 doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler A.A., Johnston D.R., Kaur S., Lubin F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci Signal. 2019;12:eaaw9277. doi: 10.1126/scisignal.aaw9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An H, Tan JT, Shelkovnikova TA. Stress granules regulate stress-induced paraspeckle assembly. J Cell Biol 2019. https://doi.org/10.1083/jcb.201904098. [DOI] [PMC free article] [PubMed]
- 14.Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 15.Adriaens C., Marine J.-C. NEAT1-containing paraspeckles: Central hubs in stress response and tumor formation. Cell Cycle. 2017;16:137–138. doi: 10.1080/15384101.2016.1235847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCluggage F, Fox AH. Paraspeckle nuclear condensates: Global sensors of cell stress? Bioessays 2021:e2000245. https://doi.org/10.1002/bies.202000245. [DOI] [PubMed]
- 17.Choudhry H., Albukhari A., Morotti M., Haider S., Moralli D., Smythies J. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Lellahi S.M., Rosenlund I.A., Hedberg A., Kiær L.T., Mikkola I., Knutsen E. The long non-coding RNA NEAT1 and nuclear paraspeckles are upregulated by the transcription factor HSF1 in the heat shock response. J Biol Chem. 2018;293:18965–18976. doi: 10.1074/jbc.RA118.004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Hu S.-B., Wang M.-R., Yao R.-W., Wu D.i., Yang L.i. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol. 2018;20:1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- 21.Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. MBoC. 2014;25:169–183. doi: 10.1091/mbc.e13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X., Chen L., Zeb F., Huang Y., An J., Ren J. Regulation of circadian rhythms by NEAT1 mediated TMAO-induced endothelial proliferation: A protective role of asparagus extract. Exp Cell Res. 2019;382:111451. doi: 10.1016/j.yexcr.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Wu X., Zeb F., Huang Y., An J., Jiang P. Acrolein-induced apoptosis of smooth muscle cells through NEAT1-Bmal1/Clock pathway and a protection from asparagus extract. Environ Pollut. 2020;258:113735. doi: 10.1016/j.envpol.2019.113735. [DOI] [PubMed] [Google Scholar]
- 24.Kukharsky M.S., Ninkina N.N., An H., Telezhkin V., Wei W., Meritens C.R.d. Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice. Transl Psychiatry. 2020;10 doi: 10.1038/s41398-020-0854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres M, Becquet D, Blanchard M-P, Guillen S, Boyer B, Moreno M, et al. Circadian RNA expression elicited by 3’-UTR IRAlu-paraspeckle associated elements. ELife 2016;5:e14837. https://doi.org/10.7554/eLife.14837. [DOI] [PMC free article] [PubMed]
- 26.Torres M., Becquet D., Blanchard M.-P., Guillen S., Boyer B., Moreno M. Paraspeckles as rhythmic nuclear mRNA anchorages responsible for circadian gene expression. Nucleus. 2017;8:249–254. doi: 10.1080/19491034.2016.1277304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asher G., Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Hong C.I., Zamborszky J., Baek M., Labiscsak L., Ju K., Lee H. Circadian rhythms synchronize mitosis in Neurospora crassa. Proc Natl Acad Sci USA. 2014;111:1397–1402. doi: 10.1073/pnas.1319399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonsfeldt K.J., Chappell P.E. Clocks on top: The role of the circadian clock in the hypothalamic and pituitary regulation of endocrine physiology. Mol Cell Endocrinol. 2012;349:3–12. doi: 10.1016/j.mce:2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey S.M., Udoh U.S., Young M.E. Circadian regulation of metabolism. J Endocrinol. 2014;222:R75–R96. doi: 10.1530/JOE-14-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim C., Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 32.Wang D., Liang X., Chen X., Guo J. Ribonucleoprotein complexes that control circadian clocks. Int J Mol Sci. 2013;14:9018–9036. doi: 10.3390/ijms14059018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife 2012;1:e00011. https://doi.org/10.7554/eLife.00011. [DOI] [PMC free article] [PubMed]
- 34.Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science:1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurley J.M., Dasgupta A., Emerson J.M., Zhou X., Ringelberg C.S., Knabe N. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc Natl Acad Sci. 2014;111:16995–17002. doi: 10.1073/pnas.1418963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres M., Becquet D., Franc J.-L., François-Bellan A.-M. Circadian processes in the RNA life cycle. Wiley Interdiscip Rev RNA. 2018;9:e1467. doi: 10.1002/wrna.2018.9.issue-310.1002/wrna.1467. [DOI] [PubMed] [Google Scholar]
- 37.Partch C.L., Green C.B., Takahashi J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauvoisin D. Circadian rhythms and proteomics: It’s all about posttranslational modifications! Wiley Interdiscip Rev Syst Biol Med. 2019;11 doi: 10.1002/wsbm.v11.510.1002/wsbm.1450. [DOI] [PubMed] [Google Scholar]
- 39.Chen L.-L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L-L, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J 2008;27:1694–705. https://doi.org/10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed]
- 41.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 2019;14:703–21. https://doi.org/10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed]
- 42.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science:1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. https://doi.org/10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed]
- 44.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 45.De Los Santos H, Collins EJ, Mann C, Sagan AW, Jankowski MS, Bennett KP, et al. ECHO: an application for detection and analysis of oscillators identifies metabolic regulation on genome-wide Circadian output. Bioinformatics 2019. https://doi.org/10.1093/bioinformatics/btz617. [DOI] [PMC free article] [PubMed]
- 46.Singer J.M., LimoRhyde H.JJ. A flexible approach for differential analysis of rhythmic transcriptome data. J Biol Rhythm. 2018 doi: 10.1177/0748730418813785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47–e47. https://doi.org/10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed]
- 48.Yang X., Zhang Y.-K.J., Esterly N., Klaassen C.D., Wan Y.-J.Y. Gender disparity of hepatic lipid homoeostasis regulated by the Circadian clock. J Biochem. 2009;145:609–623. doi: 10.1093/jb/mvp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kavčič P., Rojc B., Dolenc-Grošelj L., Claustrat B., Fujs K., Poljak M. The impact of sleep deprivation and nighttime light exposure on clock gene expression in humans. Croat Med J. 2011;52:594–603. doi: 10.3325/cmj.2011.52.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70:1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 51.Ray S., Valekunja U.K., Stangherlin A., Howell S.A., Snijders A.P., Damodaran G. Circadian rhythms in the absence of the clock gene Bmal1. Science. 2020;367:800–806. doi: 10.1126/science:aaw7365. [DOI] [PubMed] [Google Scholar]
- 52.Torres M, Becquet D, Guillen S, Boyer, Bénédicte B, Moreno M, Blanchard M-P, et al. RNA pull-down procedure to identify RNA targets of a long non-coding RNA. JoVE 2018:e57379. https://doi.org/10.3791/57379. [DOI] [PMC free article] [PubMed]
- 53.Isobe M., Toya H., Mito M., Chiba T., Asahara H., Hirose T. Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. RNA. 2020;15 doi: 10.1261/rna.072587.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J Cell Biochem. 2018;119:1567–1574. doi: 10.1002/jcb.v119.210.1002/jcb.26317. [DOI] [PubMed] [Google Scholar]
- 55.Sun Y., Song Y., Liu C., Geng J. LncRNA NEAT1-MicroRNA-140 axis exacerbates nonalcoholic fatty liver through interrupting AMPK/SREBP-1 signaling. Biochem Biophys Res Commun. 2019;516:584–590. doi: 10.1016/j.bbrc.2019.06.104. [DOI] [PubMed] [Google Scholar]
- 56.Yu X., Li Z., Zheng H., Chan M.T.V., Wu W.K.K. NEAT1: A novel cancer-related long non-coding RNA. Cell Proliferat. 2017;50:e12329. doi: 10.1111/cpr.2017.50.issue-210.1111/cpr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang F., Wu L., Qian J., Qu B., Xia S., La T. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 58.Ye L., Shi H., Yu C., Fu J., Chen C., Wu S. LncRNA Neat1 positively regulates MAPK signaling and is involved in the T pathogenesis of Sjögren’s syndrome. Int Immunopharmacol. 2020;11 doi: 10.1016/j.intimp.2020.106992. [DOI] [PubMed] [Google Scholar]
- 59.Hughes M.E., Grant G.R., Paquin C., Qian J., Nitabach M.N. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22:1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ceglia N, Liu Y, Chen S, Agostinelli F, Eckel-Mahan K, Sassone-Corsi P, et al. CircadiOmics: circadian omic web portal. Nucleic Acids Res 2018. https://doi.org/10.1093/nar/gky441. [DOI] [PMC free article] [PubMed]
- 62.Guillaumond F., Boyer B., Becquet D., Guillen S., Kuhn L., Garin J. Chromatin remodeling as a mechanism for circadian prolactin transcription: rhythmic NONO and SFPQ recruitment to HLTF. FASEB J. 2011;25:2740–2756. doi: 10.1096/fsb2.v25.810.1096/fj.10-178616. [DOI] [PubMed] [Google Scholar]
- 63.de Goede P., Wefers J., Brombacher E.C., Schrauwen P., Kalsbeek A. Circadian rhythms in mitochondrial respiration. J Mol Endocrinol. 2018;60:R115–R130. doi: 10.1530/JME-17-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sardon Puig L., Valera-Alberni M., Cantó C., Pillon N.J. Circadian rhythms and mitochondria: connecting the dots. Front Genet. 2018;9:452. doi: 10.3389/fgene.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pisani G., Baron B. NEAT1 and paraspeckles in cancer development and chemoresistance. Noncoding RNA. 2020;6:43. doi: 10.3390/ncrna6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kovač U., Skubic C., Bohinc L., Rozman D., Režen T. Oxysterols and gastrointestinal cancers around the clock. Front Endocrinol (Lausanne) 2019;10:483. doi: 10.3389/fendo.2019.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katsel P., Roussos P., Fam P., Khan S., Tan W., Hirose T. The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophr. 2019;5 doi: 10.1038/s41537-019-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mello S.S., Sinow C., Raj N., Mazur P.K., Bieging-Rolett K., Broz D.K. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang M., Zheng Y., Sun Y., Li S., Chen L., Jin X. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics. 2019;9:3425–3442. doi: 10.7150/thno.33178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng Y., Wu W., Fu Y., Chen S., Chen T., Yang B. Toll-like receptors, long non-coding RNA NEAT1, and RIG-I expression are associated with HBeAg-positive chronic hepatitis B patients in the active phase. J Clin Lab Anal. 2019;33 doi: 10.1002/jcla.2019.33.issue-510.1002/jcla.22886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morchikh M., Cribier A., Raffel R., Amraoui S., Cau J., Severac D. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. 2017;67:387–399.e5. doi: 10.1016/j.molcel.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 72.Cermakian N., Lange T., Golombek D., Sarkar D., Nakao A., Shibata S. Crosstalk between the circadian clock circuitry and the immune system. Chronobiol Int. 2013;30:870–888. doi: 10.3109/07420528.2013.782315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rahman S.A., Castanon-Cervantes O., Scheer F.A.J.L., Shea S.A., Czeisler C.A., Davidson A.J. Endogenous circadian regulation of pro-inflammatory cytokines and chemokines in the presence of bacterial lipopolysaccharide in humans. Brain Behav Immun. 2015;47:4–13. doi: 10.1016/j.bbi.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fortier E.E., Rooney J., Dardente H., Hardy M.-P., Labrecque N., Cermakian N. Circadian variation of the response of T cells to antigen. J Immunol. 2011;187:6291–6300. doi: 10.4049/jimmunol.1004030. [DOI] [PubMed] [Google Scholar]
- 75.Scheiermann C., Kunisaki Y., Lucas D., Chow A., Jang J.-E., Zhang D. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37:290–301. doi: 10.1016/j.immuni.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X., Zhou Y., Yang L., Ma Y., Peng X., Yang S. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. 2020;235:3402–3413. doi: 10.1002/jcp.v235.410.1002/jcp.29230. [DOI] [PubMed] [Google Scholar]
- 77.Ma F., Lei Y.-Y., Ding M.-G., Luo L.-H., Xie Y.-C., Liu X.-L. LncRNA NEAT1 Interacted With DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of p53, cGAS, and STING in Lung Cancer. Front Genet. 2020;11:250. doi: 10.3389/fgene.2020.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li M., Peng J., Zeng Z. Overexpression of long non-coding RNA nuclear enriched abundant transcript 1 inhibits the expression of p53 and improves premature ovarian failure. Exp Ther Med. 2020;20:1. doi: 10.3892/etm10.3892/etm.2020.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Research 2019;47:W212–24. https://doi.org/10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed]
- 80.Zhang X.-N., Zhou J., Lu X.-J. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J Cell Physiol. 2018;233:6733–6741. doi: 10.1002/jcp.26371. [DOI] [PubMed] [Google Scholar]
- 81.Kawaguchi T., Tanigawa A., Naganuma T., Ohkawa Y., Souquere S., Pierron G. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. P Natl Acad Sci USA. 2015;112:4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Naveed A., Cooper J.A., Li R., Hubbard A., Chen J., Liu T. NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma. Cell Mol Life Sci. 2021;78:2213–2230. doi: 10.1007/s00018-020-03632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data have been deposited at Gene Expression Omnibus (GEO) (accession number n° GSE162751). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162751.