Graphical abstract

Keywords: Salinity, Metabolomics, Transcriptomics, Raffinose pathway
Highlight
-
•
Τhe response of different Medicago truncatula genotypes to salt stress was examined.
-
•
Systems biology revealed tissue-specific metabolic and transcriptional signatures.
-
•
RFO metabolites linked with tolerance were identified.
-
•
Several genes belonging to the TIR-NBS-LRR class were linked with hyper-sensitivity.
Abstract
Salt stress is an important factor limiting plant productivity by affecting plant physiology and metabolism. To explore salt tolerance adaptive mechanisms in the model legume Medicago truncatula, we used three genotypes with differential salt-sensitivity: TN6.18 (highly sensitive), Jemalong A17 (moderately sensitive), and TN1.11 (tolerant). Cellular damage was monitored in roots and leaves 48 h after 200 mM NaCl treatment by measuring lipid peroxidation, nitric oxide, and hydrogen peroxide contents, further supported by leaf stomatal conductance and chlorophyll readings. The salt-tolerant genotype TN1.11 displayed the lowest level of oxidative damage, in contrast to the salt sensitive TN6.18, which showed the highest responses. Metabolite profiling was employed to explore the differential genotype-related responses to stress at the molecular level. The metabolic data in the salt tolerant TN1.11 roots revealed an accumulation of metabolites related to the raffinose pathway. To further investigate the sensitivity to salinity, global transcriptomic profiling using microarray analysis was carried out on the salt-stressed sensitive genotypes. In TN6.18, the transcriptomic analysis identified a lower expression of many genes related to stress signalling, not previously linked to salinity, and corresponding to the TIR-NBS-LRR gene class. Overall, this global approach contributes to gaining significant new insights into the complexity of stress adaptive mechanisms and to the identification of potential targets for crop improvement.
1. Introduction
Salinity stress is one of the most important limiting factors for plant growth and agricultural productivity worldwide [1]. The deleterious effects of salinity on plant growth are associated with various factors, including ion toxicity, changes in water relations, impairment of mineral nutrition, and inactivation of photosynthetic machinery [2], [3]. Nevertheless, although a huge volume of research is dedicated to the study of salinity effects in plants, a large number of questions regarding how plants sense and respond to salinity remain unanswered [4].
Salt-induced osmotic and ionic stress disturbs the cellular redox balance causing over-reduction of the photosynthetic electron transport chain and thus amplifying the production of reactive oxygen species (ROS) [5], [6]. The highly reactive ROS, when overproduced, can, in turn, damage lipids, proteins, and nucleic acids. Moreover, the free radical-induced peroxidation of polyunsaturated fatty acids in the plasma membrane reflects the stress-induced damage at the cellular level, as an indicator of oxidative stress [7]. In addition to ROS, reactive nitrogen species (RNS; for example, the NO molecule) have been implicated in salt stress responses [8], [9]. Although the enzymatic source of NO production in plants is still controversial [10], [11], [12], nitrate reductase (NR) is established as a critical enzyme responsible for nitrate assimilation and NO generation in plants [13].
Plants respond to salinity by switching on a coordinated set of physiological and molecular responses resulting in acclimation [14], [15]. Salt stress is perceived as ionic (Na+) stress signal, and the SOS-mediated pathway limits Na+ levels in the cytosol under salt stress and helps to maintain a high cytosolic K+/Na+ ratio [16], necessary for salt tolerance. Moreover, to overcome salt-mediated oxidative stress, ROS detoxification is achieved by regulating the antioxidant defence apparatus (superoxide dismutase [SOD], ascorbate peroxidase [APX], glutathione cycle enzymes, etc.; [17]) and producing low molecular mass antioxidants (flavonoids, anthocyanins, α-tocopherol, ascorbate, glutathione, and polyphenolic compounds; [18]).
In response to osmotic imbalance by salt stress, plant cells accumulate compatible osmolytes such as sugars [19], amino acids, amides, organic acids and polyols to combat water loss [20]. Due to the importance of salinity stress in agriculture, there are many metabolomic studies to assess the metabolic effect of salinity in a variety of related plant species and legumes [21], [22], [23]. Rearrangement of the metabolic network would also be anticipated to result in changes of metabolites which are related to the regulated pathways. Hence, metabolites responding to various stresses may be fundamental constituents of the stress responses [24]. In a more global analysis using metabolomics approaches, the difference between salt-sensitive and salt-tolerant species has revealed an interesting diversity pattern (for a review, see Sanchez et al. [25]).
Another aspect of the response to stress occurs at the transcriptional level as implicated by alteration in gene expression [26]. Due to the large number of genes involved in response to various abiotic stresses, microarray-based analyses are commonly used to monitor global gene expression changes in a range of species including Arabidopsis [27], rice [28], maize [29], tomato [30], tobacco [31] and grapevine [32].
Medicago truncatula is considered an important model legume species, due to its small genome size, short life cycle, and autogamous reproduction [33], [34]. Moreover, because of these favourable characteristics, it has been internationally adopted as a model legume for genome sequencing and functional genomic-research programs [35]. Previous studies revealed that acclimation could enhance NaCl tolerance in calli of M. truncatula [36], conferring tolerance to the M. truncatula-Sinorhizobium symbiosis under salinity conditions [37]. Thus, it is of fundamental interest to further understand the salt stress responses and acclimation mechanisms in M. truncatula plants, as this will assist in the identification of potential targets for crop improvement.
The response to salinity may widely vary not only among species but even cultivars or lines of the same species. Knowledge of the reasons underlying the differential responses between different genotypes can be critical to enhance salinity tolerance. For this reason, the physiological and molecular responses to salt stress were assessed in three M. truncatula lines showing contrasting tolerance to salt stress, namely TN6.18 (highly sensitive), Jemalong A17 (moderately sensitive; reference line), and TN1.11 (tolerant, previously described [38], [39]).
In the present study, a concerted effort to understand soil salinization problems is demonstrated. We discuss differences in the adaptation mechanisms of three M. truncatula genotypes in response to salt stress at a local (roots) and systemic (leaves) level. Comparison of the antioxidant defence systems, lipid peroxidation, reactive oxygen and nitrogen species (RONS) content, and global regulatory analysis of transcriptomic and metabolomic profiles will help develop a better understanding of salt stress tolerance mechanisms.
2. Materials and methods
2.1. Plant material and salinity stress conditions
Mature (40 days-old) Medicago truncatula genotype TN1.11 (tolerant), Jemalong A17 (moderate), and TN6.18 (highly sensitive) plants were used in this study. Seeds were sown in sterile potting soil:perlite (3:1) pots and placed at 4 °C for four days for stratification. Plants were grown in a growth chamber at 22/16 °C day/night temperatures, at 60–70% RH, with a photosynthetic photon flux density of 100 μmol m2 s−1 and long day (16/8 h) photoperiod. Plants were watered three times per week until maturity. Water was added by weighting the pots to reach 80 % of soil moisture, to ensure a uniform water status throughout plant development. Salinity was imposed by watering plants once with 80 mL of 200 mM NaCl solution (till leaching), and results were analyzed at 0 h, 24 h and 48 h after the treatment started [38]. Experiments were performed in triplicates using pooled samples. Each sample consisted of separated whole root (without nodules) and leaf tissues from a minimum of three independent plants.
2.2. Physiological measurements
Stomatal conductance was measured using a ΔΤ-Porometer AP4 (Delta-T Devices-Cambridge) according to the manufacturer’s instructions. Chlorophyll fluorescence parameters of leaves representing the maximum photochemical efficiency of photosystem II (PSII) (Fv/Fm) were measured with an OptiSci OS-30p Chlorophyll Fluorometer (Opti-Sciences, U.S.A.). Leaves were incubated in the dark for 30 min before measurements.
2.3. Lipid peroxidation assay
Lipid peroxidation was determined from the measurement of malondialdehyde (MDA) content resulting from the thiobarbituric acid reaction [40] using an extinction coefficient of 155 mM−1 cm−1.
2.4. Quantification of H2O2 and NO
Hydrogen peroxide was quantified using the KI method, as described by Velikova et al. [41]. NO content was measured indirectly (nitrite-derived NO) using the Griess reagent in homogenates prepared in an ice-cold Na-acetate buffer (pH 3.6) as described by Zhou et al. [42].
2.5. Proline content
The levels of free proline were measured using the ninhydrin reaction following the method proposed by Bates et al. [43]. Briefly, frozen plant material was homogenized in 3 % aqueous sulphosalicylic acid (0.01 g/ 0.5 mL) and the residue was removed by centrifugation at 12 000 g for 10 min. Supernatant was reacted with acid-ninhydrin and glacial acetic for 1 h at 100 °C and the reaction terminated in an ice bath. The reaction mixture was extracted with toluene, mixed vigorously and leave at room temperature for 30 min until separation of the two phases. Then, chromophore-containing toluene was measured at 520 nm (TECAN, Infinite 200® PRO) and proline concentration was estimated based on a standard curve of known concentrations of proline and expressed as μmol proline g-1 FW. In addition, proline levels were also obtained with GC-TOF-MS [44].
2.6. Enzyme activity assays
2.6.1. Nitrate reductase (NR)
The assay was performed as described by Liu et al. [45], with some modifications. The buffer used for preparation of crude extracts contained 100 mM potassium phosphate (pH 7.5), 5 mM (CH3COO)2Mg, 10 % (v/v) glycerol, 10 % (w/v) polyvinylpyrrolidone, 0.1 % (v/v) Triton X-100, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM benzamidine (prepared fresh) and 1 mM 6-aminocaproic acid. Leaf tissue was extracted in the buffer using a mortar and pestle, and the mixture was thoroughly homogenized. The leaf extract was centrifuged at 14,000 × g for 15 min, and the clear supernatant was used immediately for measurement [46]. Total protein content was determined according to Bradford’s method [47]. NR activity was expressed as specific enzymatic activity (units/mg protein).
2.6.2. Pyrroline-5-carboxylate synthase (p5CS)
Extraction and p5CS activity measurements were performed according to Filippou et al. [48]. Leaves were homogenized in an extraction buffer (100 mM Tris-Cl, pH 7.5, 10 mM β-mercaptoethanol, 10 mM MgCl2, 1 mM PMSF) in pre-chilled Eppendorf tubes on ice. Extracts were centrifuged at 4 °C for 20 min at 10,000 × g. Supernatants were further clarified by centrifugation at 10,000 × g for 20 min at 4 °C. p5CS enzymatic assay was carried out in 100 mM Tris-Cl (pH 7.2), 25 mM MgCl2, 75 mM Na-glutamate, 5 mM ATP, 0.4 mM NADPH, and the appropriate crude protein extract. The reaction velocity was measured as the rate of consumption of NADPH, monitored as the decrease in absorption at 340 nm as a function of time. Total protein content was determined according to Bradford’s method [47]. p5CS specific enzymatic activity was expressed as units/mg protein.
2.7. RT-qPCR analysis
Total RNA was extracted from leaves using TRIzol (TRI reagent), followed by DNase digestion (RNase-free DNase Set; Qiagen). One microgram of total RNA was transcribed into cDNA using Primescript 1st Strand Synthesis Kit according to the manufacturer’s protocol (Takara, Japan). Real-time qPCR was performed using Biorad IQ5 (Biorad, USA). Relative quantification of gene expression and statistical analysis of the RT-qPCR data (pairwise fixed reallocation randomization test) were performed using the REST software, according to Pfaffl et al. [49]. Actin11 was selected as a housekeeping reference gene due to its common use in Medicago and its proven stability in salt conditions in a variety of plant species [50], [51], [52]. Primer sequences used are listed in Table S1.
2.8. RNA labeling and Affymetrix expression array processing
Three independent samples per line and condition were used for the microarray analysis. RNA integrity screening, probe synthesis, hybridization, and scanning were conducted by the BSRC Alexander Fleming's Expression Profiling Unit. Three hundred nanogram of total RNA was used to generate biotinylated complementary RNA (cRNA) for each treatment group using the GeneChip® 3′ IVT Express Protocol (Affymetrix, Santa Clara, CA) from the GeneChip® 3′ IVT Express Kit User Manual (Rev.8). In short, isolated total RNA was checked for integrity using the RNA 6000 Nano LabChip kit on the Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Palo Alto, CA) and concentration using the ND-1000Nanodrop (Thermo Fisher Scientific, Wilmington, Delaware USA). Poly-A RNA controls were added in each total RNA sample and were reverse transcribed using the included buffer and enzyme mixes. Double-stranded cDNA was synthesized, labeled by in vitro transcription and purified with the appropriate protocol using beads (Affymetrix, Santa Clara, CA). Prior to hybridization, the cRNA was fragmented, and 12.5 µg from each experimental sample was hybridized for 16 h to Medicago Genome arrays in an Affymetrix GeneChip® Hybridization Oven 640. Affymetrix GeneChip® Fluidics Station 450 was used to wash and stain the arrays with streptavidin–phycoerythrin (Molecular Probes, Eugene, OR), biotinylated anti-streptavidin (Vector Laboratories, Burlingame, CA) according to the standard antibody amplification protocol. Arrays were scanned with an Affymetrix GeneChip® Scanner 3000 at 570 nm. All cDNA was synthesized at the same time. Images and data were acquired using the Affymetrix® GeneChip® Command Console® Software (AGCC), where an initial quality check of the experiment was performed. The quality of the hybridizations was checked, and one of the salt-treated samples was removed from subsequent analyses. The raw data was processed using the RMA algorithm ([53]; affypackage of Bioconductor). The absolute expression values were median centred across each gene, and log2 transformed. A t-test for comparison between salt-stressed genotypes was performed using the limma R package [54]. The P values of the t-test statistics were corrected for multiple testing to assess the false-discovery rate with the publicly available software QVALUE (http://genomine.org/qvalue; [55]). Probes with Q value < 0.05 and an absolute fold-change greater or equal to 1.5 in any of the comparisons, were kept for cluster analysis. The expression values of 26,910 filtered probes were centred and scaled across samples and subjected to complete-linkage hierarchical clustering using the ComplexHeatmap R package [56]. Eight clusters were obtained by splitting the heatmap using k-means clustering (row_km = 8). Gene ontology enrichment was performed for probes occurring on each cluster using the R package topGO [57] and the latest Medicago array annotation provided by Affymetrix (NetAffx).
2.9. Metabolite profiling
GC-TOF-MS based metabolite profiling on six independent samples per line and condition was performed as described by Lisec et al. [44]. Polar metabolites were extracted from 50 mg of frozen leaf material, and 150 µl of each extract was used for the analysis. TagFinder [58] was used for peak annotation and quantification with the Golm Metabolome Database (http://gmd.mpimp-golm.mpg.de; [59]) as a reference library. The parameters used for the peak annotation are listed in Table S2, according to Fernie et al. [60]. The intensity of each fragment was normalized by that of the ribitol, which was added into the extraction solution as an internal standard. Principal component analysis (PCA), hierarchical cluster analysis (HCA), and heatmap representation were performed using the software ClustVis [61]. For the heatmap, columns and rows were clustered using Pearson’s correlation and average linkage. For the RFO pathway analysis, the metabolite levels were normalized respect to A17 t = 0 h. The statistical differences were evaluated by a paired t-test analysis (P < 0.05) in SPSS statistical software.
2.10. Statistics
Statistical analyses of the biochemical and physiological data were performed using ANOVA and Tukey’s pairwise comparison test at the 5 % confidence level. SPSS was used as statistical software.
3. Results
3.1. Salt application highlights early differential stress phenotypes in Medicago genotypes
Previous observations have shown significant differences in salt-stress responses within the first 48 h of NaCl stress between tolerant and sensitive Medicago genotypes [62]. Thus, we subjected 40 days-old Medicago plants to salt stress by watering the pots with 200 mM NaCl. After 48 h, macroscopic observation revealed an early phenotypic response consisting of chlorotic leaves in TN6.18 plants (Fig. 1A), which turned into general necrosis after 30 days (Fig. 1B), demonstrating its highly sensitive behavior to salinity and confirming previous observations [38]. Not surprisingly, TN1.11 showed the most tolerant phenotype, whereas A17 exhibited lower damage levels compared with TN6.18 yet still displayed some wilted, chlorotic leaves, clearly visible after 30 days (Fig. 1B). In addition to the macroscopic phenotype, we analyzed in more detail the early-stress response by monitoring stress-related physiological parameters. Thus, mean values of photochemical efficiency of photosystem II (Fv/Fm) and stomatal conductance, both well-known correlated parameters and stress indicators, were determined in leaves of the three genotypes (Fig. 1C, D). Following the application of salt stress for 48 h, leaves of the highly sensitive TN6.18 genotype showed significantly reduced photochemical efficiency (Fv/Fm, i.e., the ratio of variable and maximal chlorophyll fluorescence of PSII), whereas, these parameters remained constant in the moderate (A17) or salt-tolerant (TN1.11) genotypes (Fig. 1C). The decrease in Fv/Fm indicates a reduced PSII efficiency in response to stress in the sensitive genotype. Additionally, stomatal conductance was found to decrease in salinity in all three genotypes. The decrease, however, was more dramatic in TN6.18 plants (Fig. 1D), in agreement with the photosynthetic efficiency data.
Fig. 1.
Effect of salinity stress (200 mM ΝaCl) on 40 days-old Medicago truncatula genotypes TN6.18 (highly sensitive), Jemalong A17 (moderately sensitive) and TN1.11 (tolerant) after 200 mM NaCl treatment for 48 h (A) and 30 days (B). Chlorophyll fluorescence (C) and stomatal conductance (D) were measured in leaves at 0 h and 48 h upon salt application. Values are means of three replicates with SE bars. Different letters denote significant differences at P < 0.05.
3.2. Salt stress phenotypes correlate with cellular oxidative levels
Salinity stress is often correlated to accumulation of reactive oxygen species (ROS). In low amounts, ROS plays an important role as a secondary messenger in the stress signaling pathway, but increasing levels can lead to deleterious effects such as oxidative damage, especially detrimental to the plant when it occurs in the photosynthetic tissue [6]. The analysis of H2O2 content, the primary ROS formed in salt stress, revealed a substantial accumulation (5 fold) in leaves of the sensitive TN6.18 genotype after 48 h treatment, while A17 and the tolerant TN1.11 didn’t show significant changes respect to control conditions (Fig. 2A). During such salinity conditions, the accumulation of ROS in TN6.18 plants caused apparent cellular damage, as demonstrated by the levels of the lipid peroxidation indicator MDA. Consistently, MDA content was significantly lower in A17 and TN1.11 leaves (Fig. 2C). Similar results, albeit lesser in magnitude, were observed in roots (Fig. 2B, D), where higher MDA content was found in TN6.18, whereas the TN1.11 plants showed lower levels (Fig. 2D) in accordance with the lower H2O2 accumulation (Fig. 2B). In general, these results confirm previous observations in the highly sensitive TN6.18 genotype [62].
Fig. 2.
Hydrogen peroxide (H2O2) content in leaves (A) and roots (B) in the different Medicago genotypes at 0 h, 24 h and 48 h 200 mM after NaCl treatment commenced. Lipid peroxidation measured as malondialdehyde (MDA) in leaves (C) and roots (D). Values are means of three replicates with SE bars. Different letters denote significant differences at P < 0.05.
To further characterize the effects of salinity stress in the different genotypes, we analyzed the reactive nitrogen species (RNS) levels. Nitric oxide (NO), a signaling molecule and the primary RNS free radical accumulated in salt stress, was quantified in leaves and roots of Medicago plants. Similar to ROS observations, maximum nitrite-derived NO content was recorded in leaves and roots of TN6.18 plants 48 h after the application of salt stress (3-fold increase), while no significant changes were observed in the tolerant TN1.11 genotype. A17 plants showed an intermediate response in both tissues (Fig. 3A, B). Unlike ROS, we observed a similar magnitude of response in both leaf and root. To support the genotype-dependent NO variations in M. truncatula plants, the specific activity of the NO biosynthetic enzyme nitrate reductase (NR) was monitored during the stress treatment. NR activity measurements correlated with the NO levels recorded in both tissues of the different genotypes (Fig. 3C, D); higher NR activation was observed in leaves and roots of TN6.18 M. truncatula plants after 48 h compared with the other genotypes, where A17 plants displayed an intermediate response.
Fig. 3.
Effect of salinity stress on NO production in A17, TN1.11, and TN6.18, was evaluated by measuring NO content (A, B) and NR activity (C, D) in leaves and roots at the indicated times after salt application. Values are means of three replicates with SE bars. Different letters denote significant differences at P < 0.05.
3.3. Medicago salt-sensitive genotypes show increased p5CS activity and proline levels
It is known that ROS can induce proline accumulation in plants by increasing the activity of its biosynthetic route [63]. Proline, in turn, acts as a radical scavenger protecting cells from oxidative damage [64]. To further investigate proline metabolism and its role as a cellular stress indicator, the activity of p5CS, a rate-limiting proline biosynthetic enzyme, was determined in both root and leaf. Under salt stress, p5CS activity increased to a greater extent in TN6.18, followed by the A17 genotype, in agreement with the more significant oxidative damage observed in these plants. In the tolerant TN1.11 line, the p5CS activity remained unaltered in both tissues upon salt application (Fig. 4A, B). Consistently, free proline levels followed a similar trend with the measured p5CS enzymatic activity throughout the treatment (Fig. 4C, D). Overall, the results obtained support the notion of an increased oxidative response in the salt-sensitive genotypes A17 and TN6.18.
Fig. 4.
Effect of salinity stress on proline biosynthesis was evaluated by measuring p5CS activity (A, B) and proline content (C, D) in leaves and roots of the three Medicago truncatula genotypes at 0 h, 24 h and 48 h after 200 mM ΝaCl treatment commenced. Values are means of three replicates with SE bars. Different letters denote significant differences at P < 0.05.
3.4. Global metabolic profiling in leaves and roots of Medicago genotypes reveal differential stress-response patterns
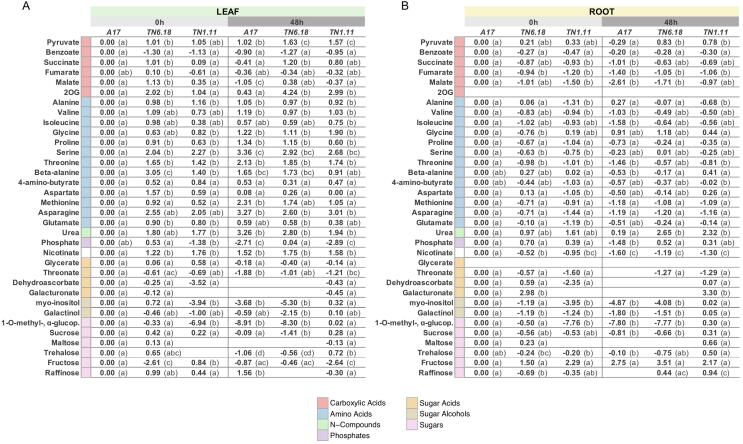
To get further insight into additional mechanisms underlying Medicago truncatula salt stress tolerance from a metabolic view, we performed global metabolite profiling of the root and leaf in the three different genotypes during salt stress. The GC–MS analyses identified 34 metabolites, including sugars, sugar acids, sugar alcohols, amino acids, and carboxylic acids. The data obtained was used for PCA analysis. PC1, which explains 49 % of the total variance, revealed the effect of salt stress in the different tissues, separating root from leaf metabolic responses (Fig. 5A). PC2, with 13.8 % of the total variance, reflected the impact of salt stress in the metabolomic profiles and the difference between genotypes. In leaves, differences observed between genotypes at control conditions substantially decreased after exposure to stress. Interestingly, in roots, the salt tolerant TN1.11 showed a differentiated stress response not correlated with A17 and TN6.18. PC1 loadings with the highest values (and thus contributing to the differentiated root response), corresponded to the sugar forms sucrose, trehalose, and galactinol (Table S3). HCA using Pearson’s correlation and average linkage of metabolites and samples, revealed the occurrence of four major metabolite clusters (Fig. 5B). Sample groupings confirmed PCA data when evidenced apparent differences between leaves and roots, and between the metabolite type in response to salt stress. Amino acids and carboxylic acids were found to be more responsive in leaves, whereas sugar forms changed to a greater extent in roots. Heat map representation showed increased levels of carboxylic acids belonging to the TCA cycle in control conditions in TN6.18 leaves (Fig. 5B, Fig. S1). The metabolic differences between genotypes before applying the stress, however, disappeared when NaCl was added, in accordance with PCA data. In stress conditions, the heat map data clearly indicated that some metabolic responses were more intense in TN1.11 root than the responses observed in A17 or TN6.18, also following previous PCA observations (Fig. 5). This distinct TN1.11 response involved the accumulation of several forms of soluble sugars such as galactinol, myo-inositol, sucrose, methyl-ɑ-glucopyranoside, trehalose, or raffinose, all of them related directly or indirectly to the raffinose family of oligosaccharides (RFOs) metabolism. RFO biosynthetic pathway, which is known to be related in acquiring salt stress tolerance [2], [65], [66], was indeed found to be significantly induced in TN1.11 roots after 48 h salt exposure (Fig. 6). This observation is consistent with its tolerant phenotype and suggests a greater capacity in regulating osmotic stress. Remarkably, in A17 leaves, we found a significant accumulation of amino acids in response to salt, including alanine, valine, glycine, proline, serine, threonine, β-alanine, methionine, asparagine, and the amino acid derivative urea (Fig. 5B, Fig. S1). Since free amino acids are known to have an important osmotic regulatory role in stress [67], the increase in amino acid synthesis observed in A17 but not in TN6.18 may contribute to Medicago's response to salinity, as evidenced by their distinct early-stress phenotypes.
Fig. 5.
(A) Principal component analysis (PCA) of metabolites and samples A17, TN6.18 and TN1.11 exposed to salt stress for 0 h and 48 h. Prediction ellipses indicate 95 % confidence level. (B) HCA Pearson’s correlation and average linkage with heat map representation. Levels of the different metabolites are indicated as row z-score. N = 4–6.
Fig. 6.
Schematic representation of the RFO metabolism and associated pathways in A17, TN6.18 and TN1.11 root samples at 0 h and 48 h after 200 mM NaCl treatment. Biosynthesis of RFO is initiated by the formation of galactinol from myo-inositol and UDP-galactose. Sequential addition of galactose units, provided by galactinol, to sucrose leads to the formation of raffinose and higher order RFO. Values shown are relative to A17 at 0 h and are expressed as log2 ratio ± SD (n = 4–6). Significant differences were determined using a paired t-test and indicated with different letters (P < 0.05).
3.5. Global transcriptomic analysis indicates substantial differences in signaling-related genes between low and highly salt-sensitive genotypes
Having observed important metabolic responses linked to a certain degree of tolerance in the less sensitive genotypes A17 and TN1.11, we next decided to carry out a transcript profiling comparing highly salt-sensitive (TN6.18) with moderately sensitive (A17) M. truncatula plants. The main focus of this assay was to identify key transcripts involved in the ‘switch’ from salt sensitivity to hyper-sensitivity in a spatiotemporal manner, examining both leaves and roots. For that purpose, we used the Affymetrix Medicago array on plants exposed for 48 h to 200 mM NaCl. Four statistical comparisons were performed for the different combination of genotype and tissue. A total of 26,910 probes were found to be differentially expressed in stress conditions (Q value < 0.05 and absolute fold change > 1.5) in any of the comparisons. Particularly, examining the comparison of gene expression differences between A17 and TN6.18 genotypes, we found 3049 and 2911 probes highly expressed in TN16.18 in leaf and root respectively, while 1146 probes were highly expressed in both tissues (Fig. 7). Conversely, 2096 and 2441 probes were highly expressed in A17, respectively in leaf and root, while 1275 were highly expressed in both tissues. Furthermore, we performed a cluster analysis of the 26,910 differentially expressed probes and identified eight clusters showing distinct patterns of gene expression (Fig. 8A). The two largest clusters contained genes highly expressed in leaves (Cluster 7, 6627 probes) or roots (Cluster 2, 7175 number of probes) but no difference between A17 and TN6.18 was observed (Fig. 8B). The remaining clusters contain those genes that showed a difference between genotypes which were summarized in Fig. 8A, B. To elucidate which biological processes were overrepresented in any of the clusters, we performed GO enrichment analysis for the set of genes contained in each cluster (Fig. 8C). The analysis in cluster 6, which showed differences between genotypes at the leaf level, contained a large number of genes (>100) involved in protein phosphorylation processes. Indeed, a more closely look revealed 86 genes significantly higher expressed in TN6.18 in salt stress compared with A17 (Table S4), the top 10 of which showed > 4-fold increased expression levels for receptor-like kinases and protein kinases with direct or indirect phosphorylation roles in signalling cascades (Fig. S2). Additionally, in roots, cluster 5 highlighted potential differential responses associated to nodulation between genotypes as shown by the > 100 differentially more expressed genes in A17 (Table S4). In a much lesser extent and magnitude, in cluster 1, two genes related to inositol metabolism were found to be significantly down-regulated less expressed (>2-fold) in TN6.18 roots (Fig. S2). Interestingly, cluster 4, the only cluster showing a substantial number of differentially regulated genes (DEG) in both roots and leaves, was highly enriched in genes related to stimulus perception (>70). Indeed, 32 genes were significantly highly expressed in A17 with respect to TN6.18 in both tissues (Table S4). A closer analysis revealed that 66 % of those DEG belonged to the TIR-domain containing subfamily of the nucleotide-binding site leucine-rich repeat (NBS-LRR) gene class (Fig. 9), which is known to play an important role in stress signalling [68]. These findings were further supported with results from cluster 8, which was highly enriched in signalling-related genes (>100) and, similarly, showed a strongly increased expression (>4-fold) of TIR-NBS-LRR genes in A17 leaves (Fig. S2). Altogether, this data indicates substantial differences between the moderately-sensitive and highly-sensitive genotypes in signalling-related processes in response to salt.
Fig. 7.
Differential gene expression summary. The upset plot shows the intersection between the sets of probes differentially expressed (Q < 0.05, absolute log2 fold change > 1.5) when comparing A17 versus TN6.18 gene expression in leaf or root tissue.
Fig. 8.
Hierarchical clustering of the microarray data. A) A total of 26,910 differentially expressed genes in the comparison of genotypes A17 and TN6.18, and tissues root and leaf, were clustered using a hierarchical clustering method. The similarity in expression patterns among probes was measured as Euclidean distance. Eight main clusters were obtained using k-means clustering. The section of the dendrogram colored in yellow highlights cluster 4. B) A graphical summary depicting the statistical difference (Q < 0.05) and direction of gene expression change of the four 2-way comparisons between genotypes and tissues; red indicates significant upregulation, blue indicates significant downregulation. C) The eight main clusters were further inspected for gene ontology enrichment. Bars colored yellow highlights GO terms enriched in cluster 4. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Expression differences between genotypes in genes associated with cellular response to stimulus. The bars indicate the log2 fold change of the genes annotated with GO:0051716 (cellular response to stimulus) that were contained in cluster 4. Bars show differential expression in leaf (blue bars) and root (yellow bars) samples. Positive values indicate higher expression in A17 in comparison to TN6.18. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
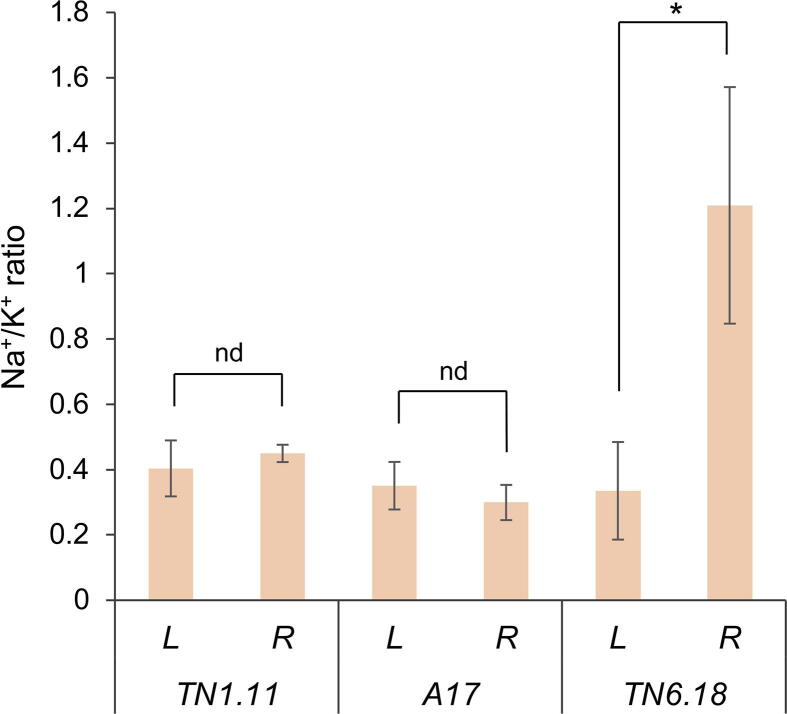
In this work, natural variations in salt tolerance between TN6.18, A17, and TN1.11 Medicago truncatula genotypes were analysed, based on their responses to salt treatment in soil. It is known that salinity impairs plant growth and development via water stress and ion toxicity due to the uptake of ions such as sodium (Na+) and chloride (Cl-). In the first phase of the stress, which takes place after minutes or days, it is appreciated an ion-independent growth reduction due to the osmotic component of salinity. If the stress is prolonged, cytotoxic ion levels build-up in tissues affecting metabolic processes, causing premature senescence, and eventually cell death [1]. In our conditions, 48 h salt treatment triggered an apparent stress phenotype in the highly sensitive TN6.18, consisting of chlorotic and wilting leaves and ultimately leading to cell death (Fig. 1). While no apparent stress symptoms were seen in the moderately-sensitive A17 and the tolerant TN1.11, a detailed physiological analysis revealed a significant reduction in stomatal conductance (Fig. 1). This effect, reflecting the partial stomatal closure to preserve water and thus leading to cell growth arrest [69], was more pronounced in TN6.18 plants and indicated a stress response present in all three genotypes. The reduction in PSII efficiency in the sensitive TN6.18 but not in the other genotypes, is in agreement with its chlorotic phenotype and reflects a potential photoinhibition in the photosynthetic tissue likely due to a cytotoxic ion accumulation. Plant tolerance to salinity involves a multitude of molecular and physiological mechanisms, i.e. osmotic tolerance, ion tolerance, and tissue tolerance, with the prevention of Na+ translocation to photosynthetic tissues also being a key element for plant survival [1]. In that regard, in the early stages of the salt treatment, we observed a significant increase in the Na+/K+ ratio in roots of the highly sensitive TN6.18 (Fig. S3), indicating a lowered capacity to prevent Na+ uptake and maintain ion homeostasis.
Abiotic stress in general and salinity in particular, cause oxidative stress by increasing the cellular levels of ROS, being its accumulation especially detrimental at the leaf level. Thus, the capacity to maintain the cellular oxidative balance, the membrane integrity, and the functionality of the photosynthetic apparatus is crucial for salinity tolerance [70]. In our study, lipid peroxidation, an indicator of membrane damage, was significantly increased in TN6.18 leaves in agreement with the increased H2O2 levels in this tissue (Fig. 2). The oxidative burst observed in the most sensitive genotype in stress conditions was supported with gene expression data from the microarray assay and validated by RT-qPCR. This data shows key antioxidant and redox enzymes such as superoxide dismutase, ascorbate peroxidase, glutathione-S-transferase, and cationic peroxidase being significantly down-regulated less expressed in TN6.18 compared with A17 (Fig. S4), demonstrating the inability of the sensitive genotype to contain an increasing oxidative cellular damage. Accordingly, NO and NR activity correlated in both root and leaf with the oxidative status in the different genotypes, in agreement with the oxidative levels and the phenotype (Fig. 2, Fig. 3). The interplay between NO and H2O2 is well known in salt stress responses [71], while salt stress is well known to stimulate H2O2 and NO production in both leaf and root tissues, indicating that direct salt application is transduced to nitro-oxidative stress in salt-sensitive species in particular [8]. In the current experimental setup where stress was imposed locally in roots, increases in NO and H2O2 content in leaves and in those of the salt-hypersensitive TN6.18 genotype in particular, may be attributed to the notion of the systemic ‘auto-propagation’ of the ROS/RNS waves throughout the plant [8]. Furthermore, increasing NO levels are often associated with cellular responses meant to reduce or contain increased levels of Na+ in plant tissue [72]. Unlike H2O2, NO accumulation in TN6.18 was especially evident in roots, highlighting the signalling role of this molecule in response to salinity and in agreement with the increased Na+/K+ ratio in the highly sensitive genotype.
Proline has a dual role in plant stress, acting as both an osmotic agent and a radical scavenger to protect cells from oxidative damage [63]. Proline’s key biosynthetic enzyme p5CS, is known to be activated by H2O2 in stress conditions [64] and thus, free proline levels tent to follow the accumulation of ROS in plant cells [63]. Interestingly, in the photosynthetic tissue, A17 and TN6.18 plants showed a similar trend in both p5CS activity and proline levels although ROS levels in the sensitive genotype were substantially higher than in A17 (Fig. 2, Fig. 4). This result, again, supports the idea of a reduced capacity in TN6.18 to respond to increasing oxidative levels. A recent study in our group, has also shown a differential metabolic regulation of polyamines, which are stress-related antioxidant compounds, in TN6.18 plants. In particular, salt-treated TN6.18 plants show a remarkable increase in diaminoxidase activity, which has been previously shown to contribute to the accumulation of proline to extend the plant response to an increased oxidative environment [73].
The metabolite profiling analysis provided insights into metabolic responses associated with salt-tolerance mechanisms in M. truncatula. Our results showed that, although some differences in several sugar alcohol and carboxylic acid levels were observed between genotypes before the application of NaCl, those lessened to a great extent in presence of stress and consistently highlighted differential responses between genotypes and tissues in such conditions (Fig. 5). HCA showed a trend that the TCA cycle intermediates and amino acids accumulate in leaves, while sugars tend to accumulate in roots. A similar response pattern, with increased accumulation of soluble sugars in roots and active synthesis metabolism in leaves, has been observed in other plant species and is associated with osmotic adjustments and provision of the energy required for survival at the expense of slow growth [74]. Among genotypes, we observed a clear differentiated response after 48 h in A17 leaves and TN1.11 roots, indicating the activation of different metabolic pathways. In the moderately sensitive genotype, most amino acids showed a significant accumulation in leaves following the salt treatment, which is a common plant response to abiotic stresses [24]. This was also observed in TN6.18 and TN1.11 leaves but in a lesser magnitude. Interestingly, in the salt tolerant TN1.11, an additional metabolic pattern was revealed at the root level. In this line, a significant accumulation of myo-inositol, sucrose, and o-methyl-alpha-glucopyranoside was observed. O-methyl-alpha-glucopyranoside is a compound which can be artificially generated by the reaction between glucose and methanol, which was used for the metabolite extraction in this study [75]. The level of glucose was saturated for quantitative detection and could not be determined. Thus, assuming that O-methyl-alpha-glucopyranoside levels reflect that of glucose, we observe a strong trend with the levels of its derivative myo-inositol (Fig. 5). Numerous studies indicate the involvement of myo-inositol in abiotic stress resilience in plants [76], [77], [78], while a recent report by Hu et al. [79] showed that over-expression of the myo-inositol biosynthetic gene MdMIPS1 resulted in increased tolerance to salt stress in transgenic apple lines. Myo-inositol is a precursor for the biosynthesis of various compounds including raffinose family oligosaccharides, auxin conjugates, phosphatidylinositol, phytic acid, ascorbic acid, and O-methyl inositols [80], [81]. As some of these metabolites include anti-oxidative compounds such as ascorbic acid, the genotypic difference in myo-inositol levels may be related with that in oxidative damages (Fig. 2). Interestingly, RFOs were found to be significantly increased in response to salt in TN1.11 roots (Fig. 6). Galactinol and its sucrose derivative raffinose are well-known stress-related compounds, the accumulation of which has been shown to be correlated with salt- and osmotic-stress tolerance in several plant species, playing a role in membrane protection and radical scavenging [82]. Similarly, trehalose was also found to accumulate in TN1.11 roots (Fig. 6). The levels of this sugar follow a similar trend of that of its precursor sucrose, and it is frequently known to act as an osmolyte, stabilize proteins and membranes, and mediate sugar signalling in stress together with trehalose-6-phosphate [83], [84], having shown to confer enhanced tolerance to salinity [85]. Additionally, the levels of sucrose and trehalose significantly decreased in the salt sensitive TN6.18 leaves (Fig. 5) under salt stress. This event probably reflects lower photosynthetic activity in the salt-treated plants, as observed previously (Fig. 2), resulting in the reduced production of photo-assimilates, including sucrose.
A17 and TN6.18 plants exhibited marked differences in response to a 48 h exposure to salinity (Fig. 1). TN6.18 and A17 have been previously characterized as highly-salt-sensitive and moderately salt-sensitive, respectively [62], although the fine-tuning of the molecular responses of these two genotypes in salt tolerance has not been previously reported. The global transcriptional analysis showed 26,910 DEG at the root and leaf level between the two genotypes in stress conditions. A more detailed analysis using hierarchical clustering and GO categorization revealed substantial differences (>100 DEG) in biological functions related to protein phosphorylation and cell signalling (Fig. 8). Particularly, TN6.18 leaves showed a strong increased expression of genes related to kinase activity (cluster 6, Fig. S2), which are known to be involved in signalling cascades often negatively regulating the cellular responses. In particular, the top 10 mostly expressed genes included members of the lectine and serine/threonine kinase family (Fig. S2), some of which have been shown in plants to respond to abiotic stress in general and salt and osmotic stress in particular [86], [87]. Interestingly, a member of the with- no-lysine kinase (WNK) family was found to be highly expressed in the highly-salt-sensitive genotype. Recent evidences have shown that members of this family negatively influence the plant’s ability to cope with both osmotic stress and ion imbalance in vivo, by affecting proline content and redox activities [88]. This observation is consistent with the salt phenotype seen in TN6.18 leaves, and indicates a potential stress mechanism that will require further analysis.
The differential responses at the root level are particularly interesting since roots are known to perceive the early salinity signals and trigger a systemic response. In that regard, a cluster enriched in genes related to nodulation and nodule morphogenesis was identified to be down-regulated less expressed in the more sensitive genotype. Although the starting material did not contain rhizobial inoculum, this result goes in agreement with previous reports indicating an increased slowdown in TN6.18 nodule-related activity in response to salt stress, and associated with diminished root antioxidant responses [38]. More interestingly, the transcriptomic data also revealed a large amount of TIR-NBS-LRR genes significantly down-regulated less expressed in TN6.18 in comparison with A17 (Fig. 8, Fig. S2, Table S4). TIR-NBS-LRR are well-known plant sensors involved in biotic stress perception and signalling [68]. Its involvement in abiotic stress is less known, although some recent evidences indicate an overlap between disease resistance and abiotic stress pathways [89]. In this context, interestingly, the overexpression of TIR-NBS-LRR receptors in Arabidopsis has been shown to lead to an increase in salt stress tolerance [85], with their depleted expression being consistent with the more salt-sensitive phenotype observed in our work and thus indicating a potential role in salt sensing and signalling. The relevance and the precise mechanism of this interesting player, will require further studies.
Altogether, analysis of the mechanisms underlying salt sensitivity in the Medicago cultivars examined identified several key defence components, potentially linking tolerance with an accumulation of metabolites related to the raffinose pathway, while hyper-sensitivity was linked, among others, with the suppression of a large number of genes not previously linked to salinity and corresponding to the TIR-NBS-LRR gene class. Current main findings, summarized in Fig. 10, show different adaptive strategies adopted by plants in co-evolution with stress environments, allowing an efficient integration of environmental signals into complex transcriptional, biochemical, and metabolic responses.
Fig. 10.
Schematic overview of the main results obtained in this study associated to increased salt tolerance and hyper-sensitivity in Medicago truncatula plants. Plant drawing modified from Gholami et al. [90].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We would like to thank Prof. Laurent Gentzbittel (ENSAT-Toulouse) for the kind provision of seeds of the Medicago genotypes. This work was supported by CUT Internal Start-up grant EX-32 to V.F.
Author Contributions
P.F. and V.F. planned and designed the research; P.F., X.Z., C.A., T.O., A.C. and V.H. performed experiments; X.Z., T.O., C.A.V., G.G. and I.G. analyzed and interpreted the data; V.A., P.M., A.R.F. and A.F.T. were involved in discussions throughout the project; X.Z., P.F. and V.F. wrote the manuscript with contributions by all authors; V.F. supervised the project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
Quantification of the different metabolites in the different genotypes and tissues. Results are expressed as log2 fold increase. Different letters indicate significant differences according to a paired t-student test (P < 0.05).
Supplementary Fig. S2.
Expression differences between genotypes in genes that belong to cluster 1, 6, and 8. The bars show the log2 fold change of gene expression in the comparison between A17 and TN6.18 genotypes. Positive values (yellow bars) indicate higher expression in A17 genotype. Negative values (blue bars) indicate higher expression in TN6.18 genotype. The genes selected belong to the enriched GO terms GO:0046488 (phosphatidylinositol metabolic process, cluster 1, root), GO:0048017 (inositol lipid-mediated signaling, cluster 1, root), GO:0006468 (protein phosphorylation, cluster 6, leaf), and GO:0023052 (signaling, cluster 8, leaf). Up to 10 genes were plotted for each group based on the magnitude of their log2 fold change.
Supplementary Fig. S3.
Na+/K+ quantification in leaves (L) and roots (R) of the different genotypes at 0 and 24h after 200 mM NaCl treatment. Results are expressed as mean ± sd. Asterisk indicates significant differences according to a paired t-student test (P < 0.05; nd, no differences). Ion content was quantified using a commercial service provider (CP. FoodLab, Nicosia, Cyprus) by flame photometry following the AOAC 969.23 protocol.
Supplementary Fig. S4.
Microarray and RT-qPCR relative expression of the indicated antioxidant enzymes for TN6.18 vs A17 plants. Measurements were performed in roots and leaves after 48 h 200 mM NaCl treatment started. Statistical analysis was performed using REST software. Results are expressed as fold change. N = 3.
List of primers used in RT-qPCR analysis.
Parameters for metabolite peak annotation.
Loading values of the different metabolites for PC1 and PC2.
Expression data (fold change), statistical tests results (P-value), and annotation of genes occurring in each of the eight main clusters obtained with hierarchical clustering.
References
- 1.Munns R., Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 2.Krasensky J., Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63:1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang W., Ma X., Wan P., Liu L. Plant salt-tolerance mechanism: a review. Biochem Biophys Res Commun. 2018;495:286–291. doi: 10.1016/j.bbrc.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 4.Isayenkov S.V., Maathuis F.J.M. Plant salinity stress: many unanswered questions remain. Front Plant Sci. 2019;10 doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller G., Shulaev V., Mittler R. Reactive oxygen signaling and abiotic stress. Physiol Plant. 2008;133:481–489. doi: 10.1111/j.1399-3054.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- 6.Hasanuzzaman M., Bhuyan M.H.M.B., Zulfiqar F., Raza A., Mohsin S.M., Al Mahmud J. Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants. 2020;9:1–52. doi: 10.3390/antiox9080681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demiral T., Türkan I. Comparative lipid peroxidation, antioxidant defense systems and proline content in roots of two rice cultivars differing in salt tolerance. Environ Exp Bot. 2005;53:247–257. doi: 10.1016/j.envexpbot.2004.03.017. [DOI] [Google Scholar]
- 8.Tanou G., Filippou P., Belghazi M., Job D., Diamantidis G., Fotopoulos V. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 2012;72:585–599. doi: 10.1111/j.1365-313X.2012.05100.x. [DOI] [PubMed] [Google Scholar]
- 9.Filippou P., Bouchagier P., Skotti E., Fotopoulos V. Proline and reactive oxygen/nitrogen species metabolism is involved in the tolerant response of the invasive plant species Ailanthus altissima to drought and salinity. Environ Exp Bot. 2014;97:1–10. doi: 10.1016/j.envexpbot.2013.09.010. [DOI] [Google Scholar]
- 10.Wilson I.D., Neill S.J., Hancock J.T. Nitric oxide synthesis and signalling in plants. Plant, Cell Environ. 2008;31:622–631. doi: 10.1111/j.1365-3040.2007.01761.x. [DOI] [PubMed] [Google Scholar]
- 11.Gupta K.J., Fernie A.R., Kaiser W.M., van Dongen J.T. On the origins of nitric oxide. Trends Plant Sci. 2011;16:160–168. doi: 10.1016/j.tplants.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Kolbert Z., Barroso J.B., Brouquisse R., Corpas F.J., Gupta K.J., Lindermayr C. A forty year journey: the generation and roles of NO in plants. Nitric Oxide – Biol Chem. 2019;93:53–70. doi: 10.1016/j.niox.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Rockel P., Strube F., Rockel A., Wildt J., Kaiser W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot. 2002;53:103–110. doi: 10.1093/jxb/53.366.103. [DOI] [PubMed] [Google Scholar]
- 14.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 15.Arif Y., Singh P., Siddiqui H., Bajguz A., Hayat S. Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol Biochem. 2020;156:64–77. doi: 10.1016/j.plaphy.2020.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Türkan I., Demiral T. Recent developments in understanding salinity tolerance. Environ Exp Bot. 2009;67:2–9. doi: 10.1016/j.envexpbot.2009.05.008. [DOI] [Google Scholar]
- 17.Sreenivasulu N., Grimm B., Wobus U., Weschke W. Differential response of antioxidant compounds to salinity stress in salt-tolerant and salt-sensitive seedlings of foxtail millet (Setaria italica) Physiol Plant. 2000;109:435–442. doi: 10.1034/j.1399-3054.2000.100410.x. [DOI] [Google Scholar]
- 18.Parida A.K., Das A.B. Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf. 2005;60:324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Sharma A., Shahzad B., Kumar V., Kohli S.K., Sidhu G.P.S., Bali A.S. Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomolecules. 2019;9:285. doi: 10.3390/biom9070285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashraf M., Harris P.J.C. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004;166:3–16. doi: 10.1016/j.plantsci.2003.10.024. [DOI] [Google Scholar]
- 21.Kim J.K., Bamba T., Harada K., Fukusaki E., Kobayashi A. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J Exp Bot. 2007;58:415–424. doi: 10.1093/jxb/erl216. [DOI] [PubMed] [Google Scholar]
- 22.Widodo P.JH., Newbigin E., Tester M., Bacic A., Roessner U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J Exp Bot. 2009;60:4089–4103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez D.H., Szymanski J., Erban A., Udvardi M.K., Kopka J. Mining for robust transcriptional and metabolic responses to long-term salt stress: a case study on the model legume Lotus japonicus. Plant, Cell Environ. 2010;33:468–480. doi: 10.1111/j.1365-3040.2009.02047.x. [DOI] [PubMed] [Google Scholar]
- 24.Obata T., Fernie A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci. 2012;69:3225–3243. doi: 10.1007/s00018-012-1091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez D.H., Siahpoosh M.R., Roessner U., Udvardi M., Kopka J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant. 2008;132:209–219. doi: 10.1111/j.1399-3054.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 26.Tester M., Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki S., Borchert C., Deyholos M., Wang H., Brazille S., Kawai K. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Miyazaki S., Kawai K., Deyholos M., Galbraith D.W., Bohnert H.J. Temporal progression of gene expression responses to salt shock in maize roots. Plant Mol Biol. 2003;52:873–891. doi: 10.1023/A:1025029026375. [DOI] [PubMed] [Google Scholar]
- 30.Albaladejo I., Egea I., Morales B., Flores F.B., Capel C., Lozano R. Identification of key genes involved in the phenotypic alterations of res (restored cell structure by salinity) tomato mutant and its recovery induced by salt stress through transcriptomic analysis. BMC Plant Biol. 2018;18 doi: 10.1186/s12870-018-1436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai G., Xie H., Yao H., Li F., Chen X., Zhang Y. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-5839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Çakır Aydemir B., Yüksel Özmen C., Kibar U., Mutaf F., Büyük P.B., Bakır M. Salt stress induces endoplasmic reticulum stress-responsive genes in a grapevine rootstock. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0236424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oldroyd GED, Geurts R. Medicago truncatula, going where no plant has gone before. Trends Plant Sci. 2001;6:552–554. doi: 10.1016/S1360-1385(01)02153-7. [DOI] [PubMed] [Google Scholar]
- 34.Young N.D., Udvardi M. Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol. 2009;12:193–201. doi: 10.1016/j.pbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Stougaard J. Genetics and genomics of root symbiosis. Curr Opin Plant Biol. 2001;4:328–335. doi: 10.1016/S1369-5266(00)00181-3. [DOI] [PubMed] [Google Scholar]
- 36.Elmaghrabi A.M., Ochatt S., Rogers H.J., Francis D. Enhanced tolerance to salinity following cellular acclimation to increasing NaCl levels in Medicago truncatula. Plant Cell Tissue Organ Cult. 2013;114:61–70. doi: 10.1007/s11240-013-0306-2. [DOI] [Google Scholar]
- 37.Aydi S., Sassi S., Debouba M., Hessini K., Larrainzar E., Gouia H. Resistance of Medicago truncatula to salt stress is related to glutamine synthetase activity and sodium sequestration. J Plant Nutr Soil Sci. 2010;173:892–899. doi: 10.1002/jpln.200900235. [DOI] [Google Scholar]
- 38.Mhadhbi H., Fotopoulos V., Mylona P.V., Jebara M., Elarbi Aouani M., Polidoros A.N. Antioxidant gene-enzyme responses in Medicago truncatula genotypes with different degree of sensitivity to salinity. Physiol Plant. 2011;141:201–214. doi: 10.1111/j.1399-3054.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- 39.Del-Saz N.F., Florez-Sarasa I., Clemente-Moreno M.J., Mhadhbi H., Flexas J., Fernie A.R. Salinity tolerance is related to cyanide-resistant alternative respiration in Medicago truncatula under sudden severe stress. Plant Cell Environ. 2016;39:2361–2369. doi: 10.1111/pce.12776. [DOI] [PubMed] [Google Scholar]
- 40.Minotti G., Aust S.D. The requirement for iron (III) in the initiation of lipid peroxidation by iron (II) and hydrogen peroxide. J Biol Chem. 1987;262:1098–1104. doi: 10.1016/s0021-9258(19)75755-x. [DOI] [PubMed] [Google Scholar]
- 41.Velikova V., Yordanov I., Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants protective role of exogenous polyamines. Plant Sci. 2000;151:59–66. doi: 10.1016/S0168-9452(99)00197-1. [DOI] [Google Scholar]
- 42.Zhou B., Guo Z., Xing J., Huang B. Nitric oxide is involved in abscisic acid-induced antioxidant activities in Stylosanthes guianensis. J Exp Bot. 2005;56:3223–3228. doi: 10.1093/jxb/eri319. [DOI] [PubMed] [Google Scholar]
- 43.Bates L.S., Waldren R.P., Teare I.D. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 44.Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., He J., Jiang L., Wu H., Xiao Y., Liu Y. Nitric oxide production is associated with response to brown planthopper infestation in rice. J Plant Physiol. 2011;168:739–745. doi: 10.1016/j.jplph.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Wray J.L., Filner P. Structural and functional relationships of enzyme activities induced by nitrate in barley. Biochem J. 1970;119:715–725. doi: 10.1042/bj1190715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradford M. A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 48.Filippou P., Antoniou C., Fotopoulos V. The nitric oxide donor sodium nitroprusside regulates polyamine and proline metabolism in leaves of Medicago truncatula plants. Free Radic Biol Med. 2013;56:172–183. doi: 10.1016/j.freeradbiomed.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 49.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He M., Cui S., Yang X., Mu G., Chen H., Liu L. Selection of suitable reference genes for abiotic stress-responsive gene expression studies in peanut by real-time quantitative PCR. Electron J Biotechnol. 2017;28:76–86. doi: 10.1016/j.ejbt.2017.05.004. [DOI] [Google Scholar]
- 51.Gu C.S., Liu L.Q., Xu C., Zhao Y.H., Zhu X.D., Huang S.Z. Reference gene selection for quantitative real-time RT-PCR normalization in Iris lactea var. chinensis roots under cadmium, lead, and salt stress conditions. Sci World J. 2014;2014 doi: 10.1155/2014/532713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., Lu H., He Z., Wang C., Wang Y., Ji X. Selection of appropriate reference genes for quantitative real-time reverse transcription PCR in Betula platyphylla under salt and osmotic stress conditions. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0225926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 54.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 57.Alexa A, Rahnenfuhrer J. topGO: Enrichment analysis for gene ontology. 2020.
- 58.Luedemann A., Von Malotky L., Erban A., Kopka J. TagFinder: Preprocessing software for the fingerprinting and the profiling of gas chromatography-mass spectrometry based metabolome analyses. Methods Mol Biol. 2012;860:255–286. doi: 10.1007/978-1-61779-594-7_16. [DOI] [PubMed] [Google Scholar]
- 59.Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmüller E. GMD@CSB.DB: The Golm metabolome database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 60.Fernie A.R., Aharoni A., Willmitzer L., Stitt M., Tohge T., Kopka J. Recommendations for reporting metabolite data. Plant Cell. 2011;23:2477–2482. doi: 10.1105/tpc.111.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metsalu T., Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucl Acids Res. 2015;43:566–570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mhadhbi H., Fotopoulos V., Mylona P.V., Jebara M., Aouani M.E., Polidoros A.N. Alternative oxidase 1 (Aox1) gene expression in roots of Medicago truncatula is a genotype-specific component of salt stress tolerance. J Plant Physiol. 2013;170:111–114. doi: 10.1016/j.jplph.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 63.Szabados L., Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 64.Liang X., Zhang L., Natarajan S.K., Becker D.F. Proline mechanisms of stress survival. Antioxidants Redox Signal. 2013;19:998–1011. doi: 10.1089/ars.2012.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elsayed A.I., Rafudeen M.S., Golldack D. Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol. 2014;16:1–8. doi: 10.1111/plb.12053. [DOI] [PubMed] [Google Scholar]
- 66.Sengupta S., Mukherjee S., Basak P., Majumder A.L. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayat S., Hayat Q., Alyemeni M.N., Wani A.S., Pichtel J., Ahmad A. Role of proline under changing environments: a review. Plant Signal Behav. 2012;7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee H.A., Yeom S.I. Plant NB-LRR proteins: Tightly regulated sensors in a complex manner. Brief Funct Genomics. 2015;14:233–242. doi: 10.1093/bfgp/elv012. [DOI] [PubMed] [Google Scholar]
- 69.Maggio A., Raimondi G., Martino A., De Pascale S. Salt stress response in tomato beyond the salinity tolerance threshold. Environ Exp Bot. 2007;59:276–282. doi: 10.1016/j.envexpbot.2006.02.002. [DOI] [Google Scholar]
- 70.Munns R., Gilliham M. Salinity tolerance of crops - what is the cost? New Phytol. 2015;208:668–673. doi: 10.1111/nph.13519. [DOI] [PubMed] [Google Scholar]
- 71.Christou A., Manganaris G.A., Fotopoulos V. Systemic mitigation of salt stress by hydrogen peroxide and sodium nitroprusside in strawberry plants via transcriptional regulation of enzymatic and non-enzymatic antioxidants. Environ Exp Bot. 2014;107:46–54. doi: 10.1016/j.envexpbot.2014.05.009. [DOI] [Google Scholar]
- 72.Zhang F., Wang Y., Wang D. Role of nitric oxide and hydrogen peroxide during the salt resistance response. Plant Signal Behav. 2007;2:473–474. doi: 10.4161/psb.2.6.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antoniou C., Zarza X., Gohari G., Panahirad S., Filippou P., Tiburcio A.F. Involvement of polyamine metabolism in the response of Medicago truncatula genotypes to salt stress. Plants. 2021;10 doi: 10.3390/plants10020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu D., Cai S., Chen M., Ye L., Chen Z., Zhang H. Tissue metabolic responses to salt stress in wild and cultivated Barley. PLoS ONE. 2013:8. doi: 10.1371/journal.pone.0055431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helferich B, Schäfer W. Org. Synth. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2003. α-Methyl d-Glucoside. [DOI] [Google Scholar]
- 76.Ishitani M., Majumder A.L., Bornhouser A., Michalowski C.B., Jensen R.G., Bohnert H.J. Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J. 1996;9:537–548. doi: 10.1046/j.1365-313X.1996.09040537.x. [DOI] [PubMed] [Google Scholar]
- 77.Valluru R., Van den Ende W. Myo-inositol and beyond - Emerging networks under stress. Plant Sci. 2011;181:387–400. doi: 10.1016/j.plantsci.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 78.Patra B., Ray S., Richter A., Majumder A.L. Enhanced salt tolerance of transgenic tobacco plants by co-expression of PcINO1 and McIMT1 is accompanied by increased level of myo-inositol and methylated inositol. Protoplasma. 2010;245:143–152. doi: 10.1007/s00709-010-0163-3. [DOI] [PubMed] [Google Scholar]
- 79.Hu L., Zhou K., Liu Y., Yang S., Zhang J., Gong X. Overexpression of MdMIPS1 enhances salt tolerance by improving osmosis, ion balance, and antioxidant activity in transgenic apple. Plant Sci. 2020;301 doi: 10.1016/j.plantsci.2020.110654. [DOI] [PubMed] [Google Scholar]
- 80.Lorence A., Chevone B.I., Mendes P., Nessler C.L. myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004;134:1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loewus F.A., Murthy P.P.N. myo-Inositol metabolism in plants. Plant Sci. 2000;150:1–19. doi: 10.1016/S0168-9452(99)00150-8. [DOI] [Google Scholar]
- 82.Nishizawa A., Yabuta Y., Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paul M.J., Primavesi L.F., Jhurreea D., Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol. 2008;59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- 84.Figueroa C.M., Lunn J.E. A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 2016;172:7–27. doi: 10.1104/pp.16.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X., Zhang Y., Yin L., Lu J. Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma. 2017;254:957–969. doi: 10.1007/s00709-016-1005-8. [DOI] [PubMed] [Google Scholar]
- 86.Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S.P. Sensing the environment: Key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot. 2013;64:445–458. doi: 10.1093/jxb/ers354. [DOI] [PubMed] [Google Scholar]
- 87.Vaid N., Macovei A., Tuteja N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol Plant. 2013;6:1405–1418. doi: 10.1093/mp/sst033. [DOI] [PubMed] [Google Scholar]
- 88.Zhang B., Liu K., Zheng Y., Wang Y., Wang J., Liao H. Disruption of AtWNK8 enhances tolerance of Arabidopsis to salt and osmotic stresses via modulating proline content and activities of catalase and peroxidase. Int J Mol Sci. 2013;14:7032–7047. doi: 10.3390/ijms14047032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chini A., Grant J.J., Seki M., Shinozaki K., Loake G.J. Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 2004;38:810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- 90.Gholami A., De Geyter N., Pollier J., Goormachtig S., Goossens A. Natural product biosynthesis in Medicago species. Nat Prod Rep. 2014;31:356–380. doi: 10.1039/c3np70104b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in RT-qPCR analysis.
Parameters for metabolite peak annotation.
Loading values of the different metabolites for PC1 and PC2.
Expression data (fold change), statistical tests results (P-value), and annotation of genes occurring in each of the eight main clusters obtained with hierarchical clustering.