Abstract
Crithidia mellificae, a monoxenous trypanosomatid considered restricted to insects, was recently reported to infect a bat. Herein, C. mellificae has been demonstrated to have a wider range of vertebrate hosts and distribution in Brazilian biomes than once thought. Parasites isolated from haemocultures were characterized using V7V8 SSU rDNA and glyceraldehyde 3-phosphate dehydrogenase genes. Coatis (Nasua nasua) in the Cerrado; marmosets (Callithrix sp.) and bats (Carollia perspicillata, Myotis lavali, M. izecksohni, Artibeus lituratus) in the Atlantic Forest; crab-eating foxes (Cerdocyon thous) and ocelot (Leopardus pardalis) in the Pantanal biomes were infected by trypanosomatids that displayed choanomastigote forms in haemoculture in Giemsa-stained slide smears. Molecular characterization and phylogenetic inference confirmed the infection of C. mellificae in these animals. Moreover, slight differences in C. mellificae sequences were observed. Crithidia mellificae growth curves were counted at 27°C, 36°C and 37°C, and the morphotypes were able to grow and survive for up to 16 days. Serological titers for C. mellificae were observed in nonhuman primates, demonstrating that this parasite is able to induce a humoral immune response in an infected mammal. These results showed that host specificity in trypanosomatids is complex and far from understood.
Keywords: Crithidia mellificae, Monoxenous, Infection, Mammalian host, Brazilian biomes
Graphical abstract
Highlights
-
•
Crithidia mellificae infection was observed in different mammal species.
-
•
C. mellificae is a thermotolerant parasite.
-
•
C. mellificae was capable to induce humoral immune response.
-
•
Trypanosomatids host specificity is far from been known.
1. Introduction
The Kinetoplastea class (Excavata: Euglenozoa) is composed of free-living and parasitic protozoan species (Adl et al., 2019). This class includes the Trypanosomatidae family, consisting of 25 genera (d’Avila-Levy et al., 2015; Maslov et al., 2019; Kostygov et al., 2020; Lukeš et al., 2021) that include obligatory parasites of invertebrates, vertebrates, and plant hosts (Hoare, 1966; Vickerman, 1976; Lukeš et al., 2014). These species are traditionally classified according to the number of hosts that they infect during their life cycle: monoxenous species—those that have only one host, usually an invertebrate host; and dixenous species—those that require an invertebrate and a vertebrate or plant host (Hoare, 1966).
With the improvement of molecular biology tools and an increasing number of phylogenetic studies, seven subfamilies have been recognized for the Trypanosomatidae family: Leishmaniinae (Jirků et al., 2012, Kostygov and Yurchenko, 2017); Blechomonadinae (Votýpka et al., 2013); Paratrypanosomatinae (Flegontov et al., 2013); Strigomonadinae (Votýpka et al., 2014); Phytomonadinae (Yurchenko et al., 2016); Trypanosomatinae (Maslov et al., 2019) and Blastocrithidiinae (Lukeš et al., 2021). Nineteen genera are classified as monoxenous trypanosomatids (d’Avila-Levy et al., 2015; Kaufer et al., 2017; Kostygov et al., 2020; Lukeš et al., 2021): Angomononas, Blastocrithidia, Blechomonas, Crithidia, Herpetomonas, Kentomonas, Jaenimonas, Lafontella, Leptomonas, Lotmaria, Novymonas, Obuscuromonas, Paratrypanosoma, Rhynchoidomonas, Sergeia, Strigonomonas, Vickermania, Wallacemonas and Zelonia. Usually, their hosts are invertebrates from the Diptera, Hemiptera, Hymenoptera and Siphonaptera orders (Kozminsky et al., 2015). Dixenous trypanosomatids include six genera: Endotrypanum, Leishmania, Paraleishmania, Porcisia, and Trypanosoma, which infect vertebrate hosts, and Phytomonas, which infects plants (d’Avila-Levy et al., 2015; Kaufer et al., 2017). Some of the dixenous trypanosomatids are of medical or economic importance, as they cause human Chagas disease (Trypanosoma cruzi), sleeping sickness (Trypanosoma brucei) and leishmaniases (Leishmania spp.). In domestic animals, dixenous trypanosomes may cause Nagana (mainly T. brucei, T. congolensis and T. vivax), surra disease (Trypanosoma evansi), or leishmaniases (Leishmania spp.). In plants, they may cause diseases of crops (Lumsden and Evans, 1976; Podlipaev, 2001; Simpson et al., 2006; Schwelm et al., 2018).
The Crithidia genus was first described infecting Anopheles maculipennis mosquitoes, with Crithidia fasciculata as the type species (Léger, 1902). The genus includes parasites of the insect alimentary canal and is represented by choanomastigote morphology that is characterized by cell bodies of different sizes with truncated anterior and broad posterior ends, an anterior kinetoplast and a “collar” surrounding the anterior end (Olsen, 1986; Ishemgulova et al., 2017; Kaufer et al., 2017). Phylogenetic reconstructions have revealed a polyphyletic origin of the genus (Teixeira et al., 2011; Yurchenko et al., 2014), resulting in a reclassification of several trypanosomatids that were under the name Crithidia (Merzlyak et al., 2001; Yurchenko et al., 2008; Teixeira et al., 2011; Kostygov et al., 2014). Beyond Culex mosquitoes, Crithidia spp. have been described in different insect groups with worldwide distribution, including bees (Hemiptera) from the Bombus and Apis genera (Langride and McGhee, 1967; Lipa and Triggiani, 1988); reduviids (Hemiptera) from Zelus leucogrammus (Ishemgulova et al., 2017); bugs (Heteroptera) from the Leptopetalops and Largus genera (Yurchenko et al., 2009); and hoverflies (Diptera) from the Eristalis genus (Yurchenko et al., 2014). Crithidia mellificae and Crithidia bombi species are the most studied species under natural conditions since they are pathogenic to honeybees and bumblebees, causing economic losses in agriculture (Schmid-Hempel and Tognazzo, 2010; Strobl et al., 2019).
Host specificity is defined as the capability of a parasite to infect a single or a few closely related host species (narrow specificity) or a variety of host species (broad specificity) (Maslov et al., 2013). This condition is still open to debate, since new evidence has revealed that trypanosomatids are much less restricted than traditionally affirmed (Dario et al., 2017; Rangel et al., 2019, Rodrigues et al., 2019). From this perspective, some monoxenous trypanosomatids are considered nonspecific to their invertebrate hosts (Merzlyak et al., 2001; Votýpka et al., 2010; Týč et al., 2013; Kozminsky et al., 2015). Crithidia mellificae is a monoxenous trypanosomatid classically associated with honeybees (Langride and McGhee, 1967). However, recently, this parasite species was reported to infect a nectar-feeding bat (Rangel et al., 2019). In this study, we show that C. mellificae is able to parasitize other mammalian orders and demonstrate its high dispersion in nature.
2. Materials and methods
2.1. Mammal sampling
Fieldwork was conducted in two Brazilian biomes: Atlantic Forest from Rio de Janeiro (in the “Estação Biológica Fiocruz Mata Atlântica” – EFMA) and Cerrado from Campo Grande municipality (in the “Vila da Base Aérea”), and we also captured wild carnivores from the Pantanal biome (Braga, 2019). Mammal captures were performed in different periods (Supplementary Table S1). In the Atlantic Forest, bats were captured using mist nets (Zootech 9 × 3 m, 20 mm mesh). Callithrix sp. were captured in Tomahawk live traps (50 × 20 × 23 cm; EquipoFauna®). Coatis (Cerrado biome) were captured using box traps (90 × 45 × 50 cm; EquiposFauna®) baited with bacon and tinned sardines (Santos et al., 2018). Animals were anaesthetized via intramuscular injection (ketamine chloridrathe 10% and acepromazine 1% for bats and primates; tiletamine hydrochloride and zolazepan hydrochloride for carnivores). Before the blood puncture, all animals were cleared of fur in the blood collection area by a scalpel, and the area was sterilized with antiseptic soap and iodinated ethanol (70%) for blood withdrawal by cardiac puncture (bats and primates) and cephalic puncture (coati).
Appropriate biosecurity techniques and personal protective equipment were used during all procedures of collection and handling of the biological samples according to the licences of the ethics committees of the collaborating institutions: Dom Bosco Catholic University (protocol 001/2017) and Oswaldo Cruz Foundation (Licences LW-63/14, LM-6/18 and L50/16). The capture and sample collection of wild species was permitted by the Chico Mendes Institute of Biodiversity Conservation, Brazilian Environmental Agency (ICMBio - SISBIO licence numbers 5612-2, 49662-8, 19037-1 and 40968-1 for Campo Grande and Rio de Janeiro study sites, respectively), in accordance with Brazilian regulations.
2.2. Parasitological diagnosis
For parasitological diagnosis, fresh blood examination was performed by optical microscopy exam of a blood drop on a slide. In a flame safety area, blood samples (600 μl) of each mammal were inoculated in two tubes (300 μl each) containing Novy-MacNeal-Nicolle (NNN) medium with a Liver Infusion Triptose (LIT) overlay containing 10% foetal calf serum and 140 mg/ml gentamycin sulfate. Haemocultures were incubated at 27 °C in a BOD incubator and examined twice a month for up to five months. Parasite cells that eventually grew were immediately amplified in LIT medium, cryopreserved, and deposited in the Coleção de Trypanosoma de Mamíferos Silvestres, Domésticos e Vetores (COLTRYP/Fiocruz). Positive haemocultures were stained with Giemsa and observed by optical microscopy (Zeiss Axioplan microscope, Oberkochen, Germany). The amplified haemocultures were washed with phosphate-saline buffer (PBS) and centrifuged at 4000×g, and the pellets were stored at −20 °C for molecular characterization.
2.3. Molecular characterization and phylogenetic analysis
Total genomic DNA from mammalian haemocultures was extracted using a DNAse Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. To identify Trypanosomatidae species infections, DNA samples were submitted to conventional PCR for analysis of V7V8 SSU rDNA (~800 bp) and glycosomal glyceraldehyde 3-phosphate dehydrogenase (~900 bp) genes (Borghesan et al., 2013). Amplified products were visualized in a 2% agarose gel stained with SYBR® Safe DNA gel stain (Thermo Fisher, Waltham, MA, EUA). The amplified products were purified (Illustra GFX PCR DNA and gel band purification kit - GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK) and sequenced for both strands of DNA (BigDye Terminator v3.1 Cycle Sequencing Kit - Applied Biosystems, Foster City, CA, USA) on an ABI 3730 DNA sequencer available at the PDTIS/FIOCRUZ sequencing platform.
To obtain V7V8 SSU rDNA and gGAPDH consensus sequences, the sequences were assembled and edited using the SeqMan program (DNASTAR Lasergene). The consensus sequences were aligned and corrected using MegaX software (Kumar et al., 2018). For the first screening, the sequences were compared with nucleotide sequences deposited in GenBank using the BLAST (Basic Local Alignment Search Tool - https://blast.ncbi.nlm.nih.gov/Blast.cgi) algorithm.
Maximum likelihood (ML) and Bayesian inference (BI) analyses were performed to confirm C. mellificae species. For these analyses, we aligned the sequences using ClustalW available in MegaX software (Kumar et al., 2018). The best substitution models were the transition with invariant sites (TIM3+I) for V7V8 SSU rDNA and the General Time Reversible with invariant sites plus gamma distributed sites (GTR + I + G) for gGAPDH, as indicated by the corrected Akaike information criterion (AICc) score in jModelTest v.2 (Darriba et al., 2012). The ML tree reconstruction was performed in the IQ-Tree program (Nguyen et al., 2015; Chernomor et al., 2016) available on PhyloSuite v.1.2.2. For branch support, ultrafast bootstrapping (Hoang et al., 2018) was performed with 5000 replicates with 1000 maximum interactions and 0.99 minimum correlation coefficients. To validate the ultrafast bootstrap results, the SH-aLRT branch test was also applied with 5000 replicates.
Bayesian tree reconstruction was performed in Beast v2.6.2 (Bouckaert et al., 2019). The Bayesian Markov chain Monte Carlo (MCMC) method was used to assign prior C. mellificae information. The Yule model was used for tree reconstruction. Three independent runs were performed for 20 M with 5 M pre-burning and sampling every 2 M generations. Runs were converged, and the effective sample size (ESS) was calculated in TRACER v.1.6 (Rambaut et al., 2018). The parameters analysed presented an ESS higher than 500. The three runs for V7V8 SSU rDNA and gGAPDH were combined in LogCombiner after 25% exclusion (burn-in). The final tree was generated with maximum clade credibility (MCC) based on 16.878 trees (burn-in = 5625) and a 0.6 posterior probability limit in TreeAnnotator. The clade statistical support was visualized by the bootstrap values and posterior probability method in Figtree v.1.4.3. The V7V8 SSU rDNA and gGAPDH sequence data generated and analysed in this study can be found in the GenBank repository database [https://www.ncbi.nlm.nih.gov/genbank/] under the accession numbers MN879775 to MN879795 and MN913351 to MN913371, respectively (Table 1).
Table 1.
Blood sample collection, parasite isolation dates and Crithidia mellificae identification by V7V8 SSU rDNA and gGAPDH in mammalian species from the Atlantic Forest, Cerrado and Pantanal biomes.
| Coltryp ID | Host | Biome | Blood sample collection date | Parasite isolation date | GenBank accession number |
|
|---|---|---|---|---|---|---|
| V7V8 SSU rDNA | gGAPDH | |||||
| c818 | Carollia perspicillata | Atlantic Forest | 2018-09-20 | 2018-09-28 | MN879775 | MN913351 |
| c819 | Myotis lavali | Atlantic Forest | 2018-09-20 | 2018-09-28 | MN879776 | MN913352 |
| c820 | Artibeus lituratus | Atlantic Forest | 2018-09-20 | 2018-09-28 | MN879777 | MN913353 |
| c821 | Artibeus lituratus | Atlantic Forest | 2018-09-20 | 2018-09-28 | MN879778 | MN913354 |
| c822 | Myotis izecksohni | Atlantic Forest | 2018-09-21 | 2018-09-28 | MN879779 | MN913355 |
| c825 | Callithrix sp. | Atlantic Forest | 2018-10-16 | 2018-10-22 | MN879780 | MN913356 |
| c826 | Callithrix sp. | Atlantic Forest | 2018-10-16 | 2018-10-22 | MN879781 | MN913357 |
| c827 | Callithrix sp. | Atlantic Forest | 2018-10-16 | 2018-10-22 | MN879782 | MN913358 |
| c828 | Callithrix sp. | Atlantic Forest | 2018-10-16 | 2018-10-22 | MN879783 | MN913359 |
| c829 | Callithrix sp. | Atlantic Forest | 2018-10-16 | 2018-10-22 | MN879784 | MN913360 |
| c830 | Callithrix sp. | Atlantic Forest | 2018-10-16 | 2018-10-22 | MN879785 | MN913361 |
| c833 | Cerdocyon thous | Pantanal | 2018-10-18 | 2018-12-11 | MN879786 | MN913362 |
| c834 | Cerdocyon thous | Pantanal | 2018-10-20 | 2018-12-11 | MN879787 | MN913363 |
| c835 | Leopardus pardalis | Pantanal | 2018-10-24 | 2018-12-11 | MN879788 | MN913364 |
| c836 | Cerdocyon thous | Pantanal | 2018-10-31 | 2018-12-11 | MN879789 | MN913365 |
| c844 | Nasua nasua | Cerrado | 2019-01-29 | 2019-02-18 | MN879790 | MN913366 |
| c845 | Nasua nasua | Cerrado | 2019-01-29 | 2019-02-18 | MN879791 | MN913367 |
| c846 | Nasua nasua | Cerrado | 2019-01-29 | 2019-02-18 | MN879792 | MN913368 |
| c847 | Nasua nasua | Cerrado | 2019-01-23 | 2019-02-18 | MN879793 | MN913369 |
| c848 | Nasua nasua | Cerrado | 2019-01-23 | 2019-02-18 | MN879794 | MN913370 |
| c849 | Nasua nasua | Cerrado | 2019-01-23 | 2019-02-18 | MN879795 | MN913371 |
A pairwise distance matrix (PDM) for V7V8 SSU rDNA in the Crithidia genus was constructed to evaluate the distance between each species using MegaX software (Kumar et al., 2018). The Tamura-Nei parameter model plus gamma distribution among sites (TrN + G) was used for PDM analysis, as suggested by the AICc score in jModelTest v.2 (Darriba et al., 2012). The sequences used in the phylogenetic tree and PDM are listed in supplementary Table S2.
2.4. Crithidia mellificae growth curves at 27°C, 36°C and 37°C
The cryopreserved sample c828, deposited in the COLTRYP collection, was removed from nitrogen liquid, and grown in NNN medium overlayed with LIT at 27°C for three days and transferred to LIT medium for exponential phase growth. For curve growth analysis, the parasites were counted in a Neubauer chamber (BLAUBRAND, Sigma-Aldrich, Saint Louis, MO, USA) for an initial inoculum in LIT (day zero) of 3 × 104 parasites/mL. This initial inoculum value was the same as that used for Crithidia luciliae thermophila (Ishemgulova et al., 2017). The initial inoculum was seeded in three tubes (triplicate) and incubated at 27°C and 36°C. The parasites were diluted in formaldehyde when all parasites were alive or trypan blue solutions when the haemoculture presented nonviable parasite forms and counted in a Neubauer chamber (BLAUBRAND, Sigma-Aldrich, Saint Louis, MO, USA) on days 1, 2, 3, 4, 8 and 16. To observe whether growth was maintained, on day 8, a new inoculum of 3 × 104 parasites/ml was established in LIT medium in triplicate, the tubes were incubated at 27°C and 36°C, and the parasites were counted on the same days as for the first growth assay. We also determined parasite growth at 37°C from the haemoculture on the 16th day of growth at 27°C, as described above.
For each day and temperature, culture medium smear slides were set up and stained with Giemsa for morphological examination. The morphological forms were observed at 1000× magnification under an optical microscope (Zeiss Axioplan microscope, Oberkochen, Germany).
2.5. Serological tests in primates
Serological tests by indirect immunofluorescence assay (IFAT) were performed according to Camargo (1966) to evaluate whether the parasites can induce a humoral immune response and the production of antibodies against infection in primates. The antigens (T. cruzi: F90 (DTU TcI) and Y88 (DTU TcII) strains (Leishmania: L579 – L. infantum and L566 - L. braziliensis strains and the C. mellificae: LBT11071 strain) were prepared as follows: epimastigotes (T. cruzi), promastigotes (Leishmania) and choanomastigotes (C. mellificae) from eight-day cultures in LIT medium were washed three times and suspended in PBS. This final suspension, adjusted so that approximately 40 flagellates could be counted per dry high-power field, was distributed on the appropriate slides, and air-dried at room temperature, according to Lisboa and colleagues (2006). Primate sera were tested on antigen-coated slides with an IgG anti-rhesus conjugate with fluorescein isothiocyanate (Sigma-Aldrich, St Louis, MO, USA) antibody. The cut-off value for IFAT was 1:10 for all trypanosomatids tested. IFATs for T. cruzi, Leishmania and C. mellificae were not performed in carnivores and bats due to the lack of conjugated fluorescein antibodies.
3. Results
3.1. Trypanosomatid diagnosis in mammals
In this study, 72 sylvatic mammals were captured in the three biomes, including 40 bats (Artibeus [n = 14], Carollia [n = 3], Chiroderma [n = 1], Glossophaga [n = 3], Lasiurus [n = 2], Micronycteris [n = 2], Molossus [n = 1], Myotis [n = 7], Phyllostomus [n = 1], Sturnira [n = 4] and Tonatia [n = 2]), and six marmoset primates (Callithrix sp.) from the Atlantic Forest; five crab-eating foxes (Cerdocyon thous) and four ocelots (Leopardus pardalis) in the Pantanal (Braga, 2019); and 17 coatis (Nasua nasua) in the Cerrado biomes (Supplementary Table S1).
During the parasitological survey, choanomastigote forms were observed in the fresh haemoculture preparations of 21 haemocultures from the following mammalian hosts (Table 1): five [5/40] bats (Carollia perspicillata [1/3], Myotis lavali and Myotis izecksohni [2/7], Artibeus lituratus [2/14]); six marmosets [6/6]; six coatis [6/17]; three crab-eating foxes [3/5] and one ocelot [1/4] (Fig. 1). The choanomastigote forms started to be observed on haemocultures on the seventh [n = 1] and eighth [n = 4] days for bat blood samples and on the sixth day for marmoset blood samples [n = 6]. In the case of the carnivore haemoculture samples, the coati haemocultures presented choanomastigote forms on the 20th [n = 3] and 26th [n = 3] days of examination; for the crab-eating foxes and the feline from the Pantanal biome, the choanomastigote forms were observed on the 41st, 52nd, 54th and 48th days of examination, respectively.
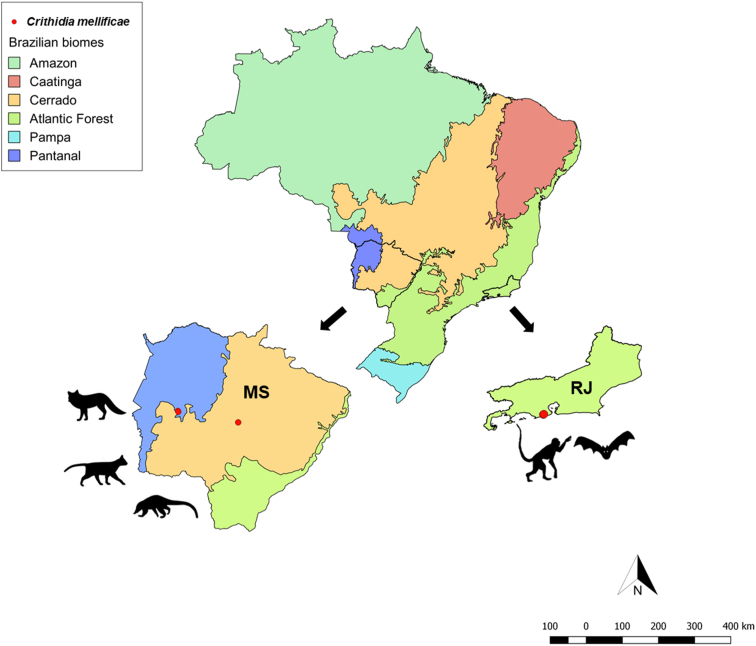
Fig. 1.
Crithidia mellificae distribution map in bats, primates, and carnivores from the Atlantic Forest (Rio de Janeiro state), Pantanal and Cerrado (Mato Grosso do Sul State) biomes. The red spots indicate mammalian C. mellificae infection in the three studied biomes: coati (Nasua nasua) in the Cerrado biome; crab-eating fox (Cerdocyon thous) and ocelot (Leopardus pardalis) in the Pantanal biome; short-tailed bat (Carollia perspicillata), insectivorous bats (Myotis lavali and Myotis izecksohni), frugivorous bat (Artibeus lituratus) and marmosets (Callithrix sp.) in the Atlantic Forest biome. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
None of these samples were positive in the fresh blood exam. The typical Crithidia choanomastigote forms observed in fresh haemoculture exams were later confirmed on Giemsa-stained slides (Fig. 2). In view of the obtained results, a suspicion was raised that the growing flagellates could be monoxenous trypanosomatids from the Crithidia genus.
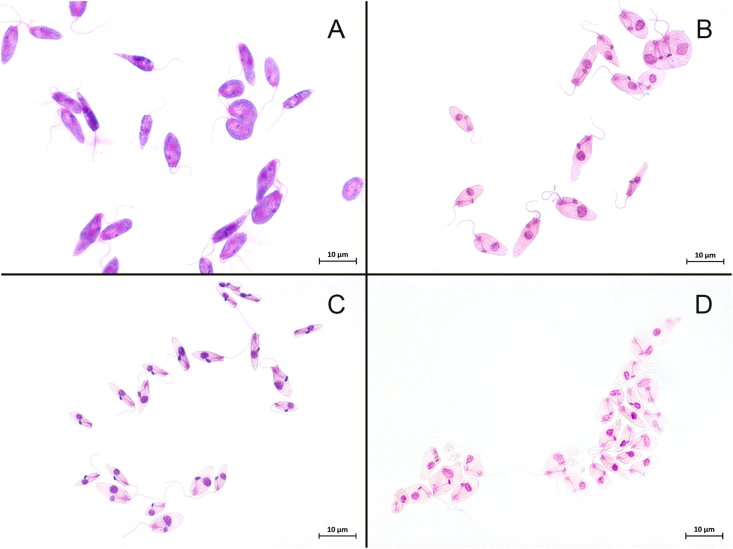
Fig. 2.
Choanomastigote forms in haemocultures from mammals stained with Giemsa. (A) and (B) choanomastigote dividing forms; (B), (C) and (D) cells in a typical collar-like extension. The kinetoplast is anterior to the nucleus and adjacent to the flagellar pocket where a single flagellum emerged.
3.2. Trypanosomatid species identification
Parasites isolated from the 21 positive haemoculture samples were subjected to DNA extraction, amplification, and sequencing of the V7V8 SSU rDNA and gGAPDH genes. In an initial screening, according to the BLAST algorithm, the 21 sequences obtained for V7V8 and gGAPDH presented similarity above 98.65% to various C. mellificae sequences deposited in GenBank. After this, phylogenetic analysis was performed, and all haemoculture isolates clustered together with parasite strains for both gene and phylogenetic inferences, in which we observed a high bootstrap value with ML analysis and a high posterior probability with BI inference (Fig. 3).
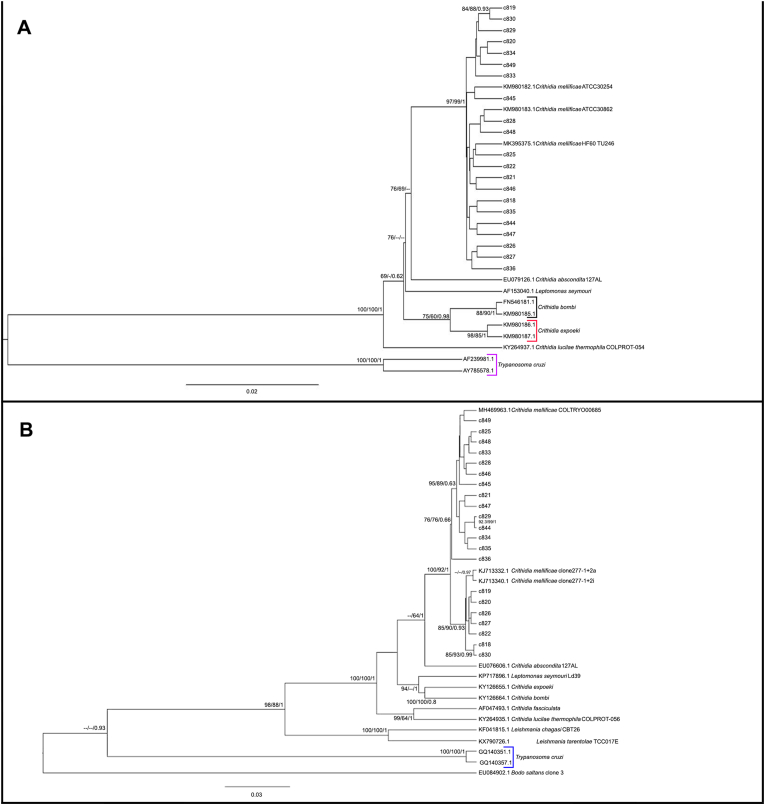
Fig. 3.
Phylogenetic analysis of V7V8 SSU rDNA and gGAPDH sequences from bat, primate, and carnivore haemocultures by maximum likelihood (ML) and Bayesian (BI) inference analyses. (A) For V7V8 SSU rDNA, the analysis was inferred using the transition-parameter model with invariant sites (TIM3 + I). (B) For gGAPDH, the analysis was inferred using the General Time Reversible with invariant plus gamma distributed sites (GTR + I + G). Maximum likelihood ultrafast and SH-aLRT bootstrap values and Bayesian posterior probabilities are shown near the nodes. The numbers at the nodes indicate support by 5000 bootstrap pseudoreplicates in the ML analysis. The scale bar shows the number of nucleotide substitutions per site. The dashes at the nodes represent bootstrap or posterior probability lower than 60 or 0.6.
By analysing in detail the phylogenetic inference, it was clear that the sequences presented genetic diversity with both genes. In the V7V8 SSU rDNA inference (Fig. 3A), samples c818 (bat) and c830 (marmoset) clustered together but separately from the other samples with significant supporting values for both reconstructions (Fig. 3A). In the gGAPDH phylogenetic inference (Fig. 3B), this genetic diversity was better observed, since two different clusters were observed between C. mellificae sequences with significant branch support: one cluster formed only with sequences originating from small mammals (reference sequence COLTRYP00685), and the other cluster formed with C. mellificae from mammals and from Apis mellifera sequences (reference sequence clones 277-1+2a and 2i).
In PDM for the V7V8 SSU rDNA gene, 11 sequences analysed in this study did not diverge from the other two C. mellificae strains (HF60 from a tabanid and ATCC30862 from Vespula squamosa), demonstrating that there was no genetic difference between these isolates according to the analysed gene, except for these ten sequences: c819, c821, c822, c825, c828, c829, c830, c833, c834, and c849 (Table 2). As demonstrated in Table 2, these sequences showed 0.001 to 0.007 divergence from honeybee, tabanid and other mammalian sequence samples. In detail, the c819, c822 and c828 sequences did not present genetic divergence between them (0.000), and the sequences with the highest genetic divergence were c829, c833 and c834 (Table 2).
Table 2.
Crithidia mellificae pairwise distance matrix for V7V8 SSU rDNA sequences from bats, carnivores, coati, and insects. The pairwise distance matrix was inferred using the Tamura-Nei-parameter model plus gamma distribution among sites (TrN + G). Other Crithidia species were also included in the analysis.
| HF60 TU246 | ATCC30862 | c825 | ATCC30254 | c849 | c819 | c822 | c828 | c821 | c830 | c829 | c833 | c834 | C. bombi | C. expoeki | C. abscondita | C. l. thermophila | C. otongatchiensis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HF60 TU246 | 0.000 | |||||||||||||||||
| ATCC30862 | 0.000 | 0.000 | ||||||||||||||||
| c825 | 0.001 | 0.001 | 0.000 | |||||||||||||||
| ATCC30254 | 0.003 | 0.003 | 0.004 | 0.000 | ||||||||||||||
| c849 | 0.003 | 0.003 | 0.004 | 0.006 | 0.000 | |||||||||||||
| c819 | 0.004 | 0.004 | 0.006 | 0.007 | 0.004 | 0.000 | ||||||||||||
| c822 | 0.004 | 0.004 | 0.006 | 0.007 | 0.004 | 0.000 | 0.000 | |||||||||||
| c828 | 0.004 | 0.004 | 0.006 | 0.007 | 0.004 | 0.000 | 0.000 | 0.000 | ||||||||||
| c821 | 0.006 | 0.006 | 0.007 | 0.008 | 0.006 | 0.001 | 0.001 | 0.001 | 0.000 | |||||||||
| c830 | 0.006 | 0.006 | 0.007 | 0.008 | 0.008 | 0.010 | 0.010 | 0.010 | 0.011 | 0.000 | ||||||||
| c829 | 0.007 | 0.007 | 0.008 | 0.010 | 0.007 | 0.003 | 0.003 | 0.003 | 0.004 | 0.013 | 0.000 | |||||||
| c833 | 0.007 | 0.007 | 0.008 | 0.010 | 0.007 | 0.003 | 0.003 | 0.003 | 0.004 | 0.013 | 0.006 | 0.000 | ||||||
| c834 | 0.007 | 0.007 | 0.008 | 0.010 | 0.007 | 0.003 | 0.003 | 0.003 | 0.004 | 0.013 | 0.003 | 0.006 | 0.000 | |||||
| C. bombi | 0.014 | 0.014 | 0.016 | 0.017 | 0.017 | 0.019 | 0.019 | 0.019 | 0.020 | 0.020 | 0.021 | 0.021 | 0.021 | 0.000 | ||||
| C. expoeki | 0.020 | 0.020 | 0.021 | 0.023 | 0.023 | 0.024 | 0.024 | 0.024 | 0.026 | 0.026 | 0.027 | 0.027 | 0.027 | 0.011–0.013 | 0.000–0.004 | |||
| C. abscondita | 0.028 | 0.028 | 0.029 | 0.031 | 0.030 | 0.032 | 0.032 | 0.032 | 0.033 | 0.033 | 0.035 | 0.035 | 0.035 | 0.028 | 0.034–0.036 | 0.000 | ||
| C. l. thermophila | 0.030 | 0.030 | 0.032 | 0.033 | 0.033 | 0.035 | 0.035 | 0.035 | 0.036 | 0.036 | 0.038 | 0.038 | 0.038 | 0.026 | 0.030–0.032 | 0.041 | 0.000 | |
| C. otongatchiensis | 0.033 | 0.033 | 0.035 | 0.036 | 0.036 | 0.038 | 0.038 | 0.038 | 0.039 | 0.039 | 0.041 | 0.041 | 0.041 | 0.028 | 0.034–0.035 | 0.035 | 0.039 | 0.000 |
3.3. Crithidia mellificae growth curves, thermal resistance, and morphological forms at different temperatures
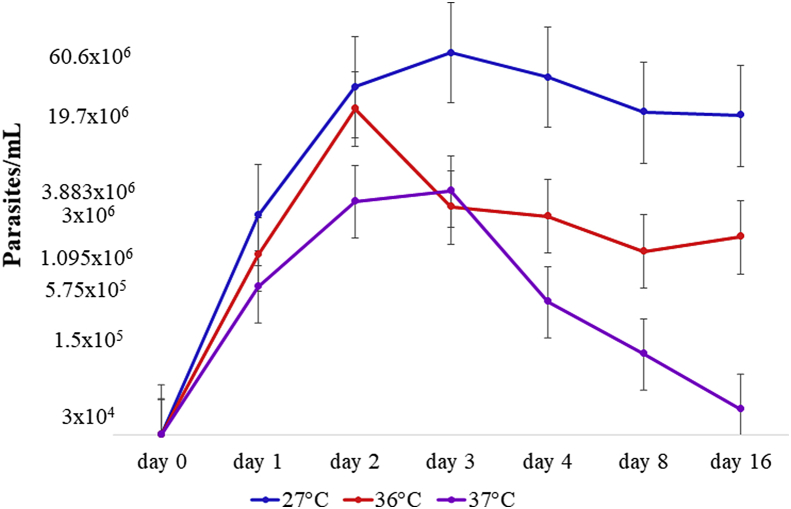
In this assay, we observed and confirmed that C. mellificae sample c828 was capable of resisting three different temperatures. Growth curves at 27°C and 36°C on day 1 (Table 3) revealed 78 and 45 times parasite growth, respectively, showing rapid parasite multiplication at both temperatures. At 36°C, the parasite presented a short log phase, reaching a maximum growth rate at day 2, and the population decreased at day 3 and remained stable until day 16 (Fig. 4). In relation to the growth at 27°C, the log phase took one day longer, reaching the maximum rate on day 3 (Fig. 4). Parasite growth started to decrease on day 4 and was stable at day 8 (Fig. 4). In contrast to the growth curve at 37°C, parasite growth was slower on day 1 than at temperatures of 27°C and 36°C (19 times – Table 3), reaching its maximum rate on day 3 (Fig. 4). The parasite growth curve rapidly declined, and parasite growth did not remain stable, with the total parasite cell/ml number decreasing in the days examined (Table 3; Fig. 4).
Table 3.
Average parasite cell count per mL (triplicate) for each temperature growth assay.
| Temperature growth | Day parasite/mL counting |
||||||
|---|---|---|---|---|---|---|---|
| day 0 | day 1 | day 2 | day 3 | day 4 | day 8 | day 16 | |
| 27°C | 3 × 104 | 2.39 × 106 | 30.5 × 106 | 60.6 × 106 | 37.3 × 106 | 18.3 × 106 | 17.2 × 106 |
| 36°C | 3 × 104 | 1.095 × 106 | 19.7 × 106 | 2.79 × 106 | 2.34 × 106 | 1.16 × 106 | 1.54 × 106 |
| 37°C | 3 × 104 | 5.75 × 105 | 3.125 × 106 | 3.833 × 106 | 4.25 × 105 | 1.5 × 105 | 5 × 104 |
Fig. 4.
Crithidia mellificae parasite growth curves at 27°C (blue line), 36°C (red line) and 37°C (purple line) from sample c828. The parasite cell number was transformed to the base 10 logarithm value (Y axis) for better curve visualization. The log phases were observed between day 1 and day 3 at 27°C and 37°C and between day 1 and day 2 at 36°C. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
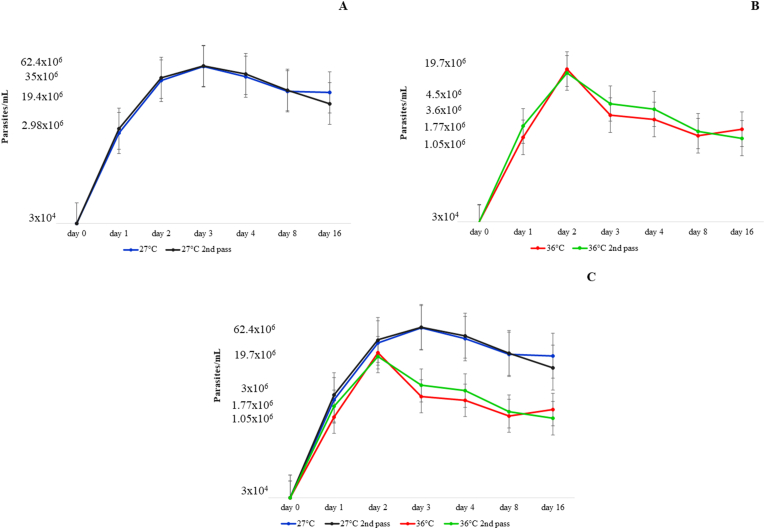
When we compared the two inoculums at 27°C and 36°C, we observed that the parasite growth curves presented the same behaviour pattern (Fig. 5): the log phase at day 2 for 36°C (Fig. 5B and C) and at day 3 at 27°C (Fig. 5A and C), and the parasite growth remained stable after day 4 (Fig. 5).
Fig. 5.
Crithidia mellificae parasite growth curves at 27°C (blue and black lines) and 36°C (red and green lines) from sample c828. The parasite cell number was transformed to the base 10 logarithm value (Y axis) for better curve visualization. The log phases were observed between day 1 and day 3 for growth at 27°C (A) and on day 1 and day 2 for growth at 36°C degrees (B). All curves were visualized together, confirming repetition in parasite growth (C). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
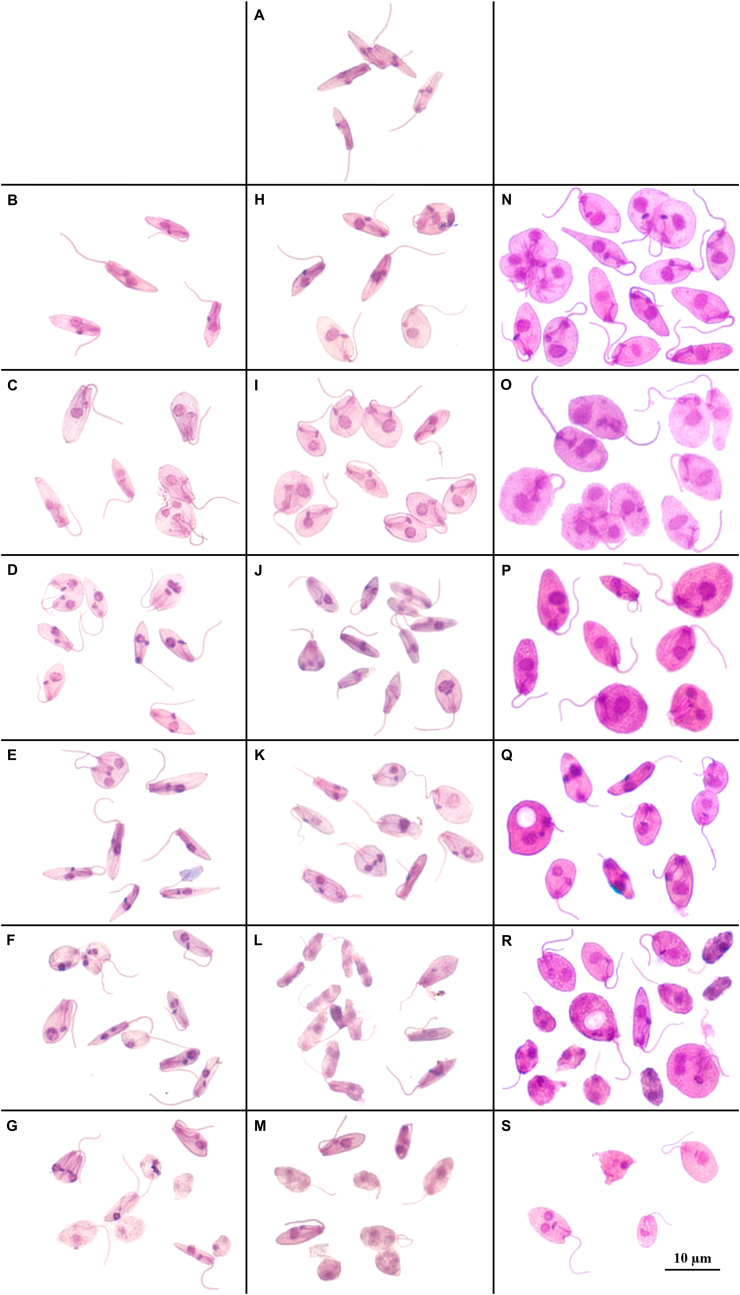
We observed at 27°C (Fig. 6B to Fig. 6G) the presence of characteristic Crithidia forms, which were elongated and thin. It was also possible to observe the presence of multiplication forms on almost every day of growth, even on day 16, when the viable parasites observed were smaller and there was a predominance of degenerate forms (Fig. 6G). For parasite growth at 36°C (Fig. 6H to M), we demonstrated the presence of rounded and large Crithidia forms, especially on day 2 (Fig. 6I), when the parasite growth peak was observed at this temperature. The morphological forms were observed after the growth peak, and a greater presence of thin than large forms was observed (Fig. 6E and J, Fig. 6K). After that, we observed both rounded larger and thinner morphologies, and on day 8 (Fig. 6L), the presence of degenerated forms was more evident. At 37°C (Fig. 6N–S), we observed a greater presence of rounded large forms since day 1 and the presence of degenerated forms on day 4. On day 16 (Fig. 6S), the predominance of the degenerated form was observed, but we observed integrity morphological forms.
Fig. 6.
Crithidia mellificae forms at different temperatures and days of growth from the c828 sample stained with Giemsa. A: C. mellificae inoculum on day zero; B to G: C. mellificae morphological forms at 27°C on day 1 (B), day 2 (C), day 3 (D), day 4 (E), day 8 (F) and day 16 (G) of cell growth; H to M: C. mellificae morphological forms at 36°C on day 1 (H), day 2 (I), day 3 (J), day 4 (K), day 8 (L) and day 16 (M) of cell growth; N to S: C. mellificae morphological forms at 37°C on day 1 (N), day 2 (O), day 3 (P), day 4 (Q), day 8 (R) and day 16 (S) of cell growth.
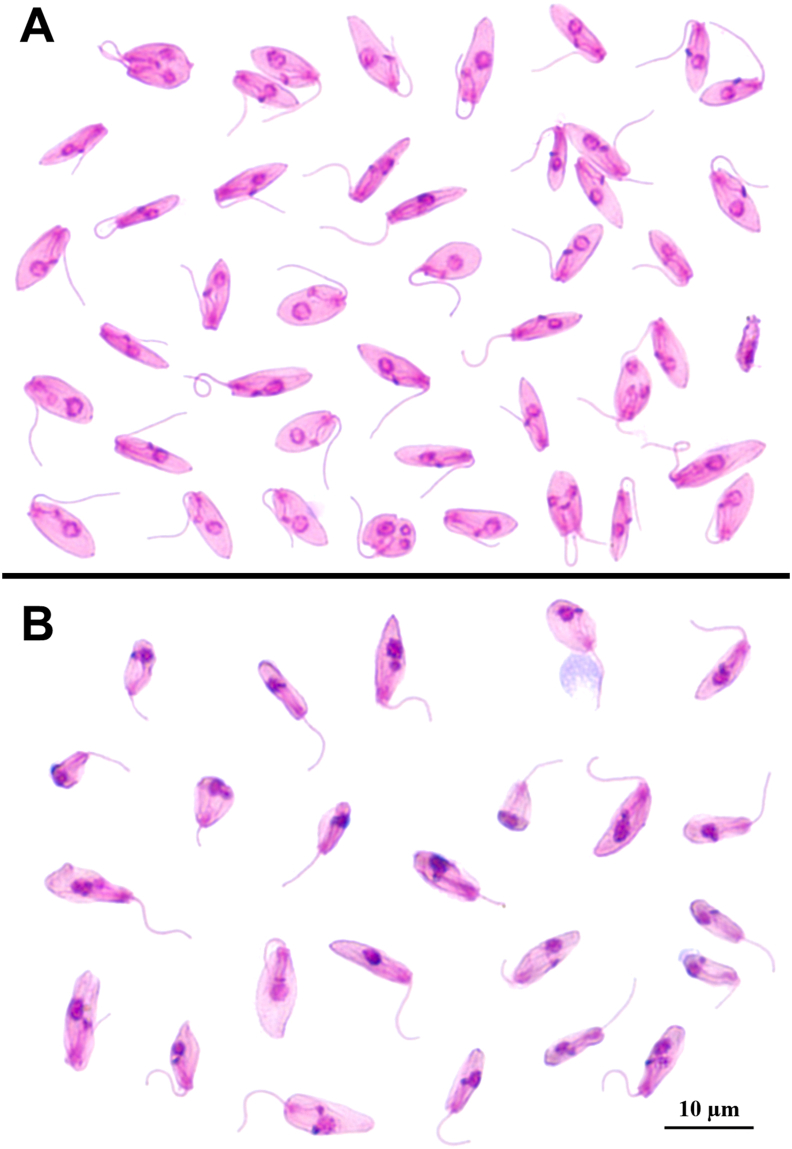
With the second inoculums at 27°C and 36°C, we observed that the growth curve peak (day 3 for 27°C and day 2 for 36°C) morphological forms were similar at 27°C (Figs. 6D and 7A), with round and thin shapes. For the 36°C second inoculum (Fig. 7B), we observed a different morphological pattern when compared to the first inoculum (Fig. 6I), in which the abundance of rounded large forms was not observed; instead, we observed thinner forms.
Fig. 7.
Crithidia mellificae forms from the second inoculum of the c828 sample at different temperatures on the peak day of growth stained with Giemsa. A: C. mellificae peak growth morphological forms on day three at 27°C; B: C. mellificae peak growth morphological forms on day two at 36°C.
3.4. Serological test for Crithidia mellificae infection in primates
In the serological analysis (IFAT) performed only for primates, the six serum samples tested presented the following serological titers for C. mellificae (Table 4): 1:10 (n = 2); 1:20 (n = 1); 1:40 (n = 2) and 1:80 (n = 1). This shows that C. mellificae was capable of inducing a humoral immune response in primates. In relation to T. cruzi antibodies, three samples presented serological titers for T. cruzi (1:10 [n = 2] and 1:20 [n = 1]), indicative of cross-reaction; however, these T. cruzi serological titers were borderline, and these samples presented higher serological titers for C. mellificae (c826 [1:40], c829 [1:40] and c830 [1:80]) (Table 4). The three other samples presented negative serological titers for T. cruzi infection. For Leishmania spp., none of the six mammalian serum samples presented serological titers showing, in this case, the absence of cross-reaction.
Table 4.
Indirect immunofluorescence assay serological titers of Trypanosoma cruzi, Leishmania sp. and Crithidia mellificae in primates from the Brazilian Atlantic Forest.
| Sample | IFAT TITLES |
||
|---|---|---|---|
| Trypanosoma cruzi | Leishmania sp. | Crithidia mellificae | |
| c825 | Negative | Negative | 1:10 |
| c826 | 1:10 | Negative | 1:40 |
| c827 | Negative | Negative | 1:20 |
| c828 | Negative | Negative | 1:10 |
| c829 | 1:10 | Negative | 1:40 |
| c830 | 1:20 | Negative | 1:80 |
4. Discussion
Classically, monoxenous trypanosomatid species have been described as having only a single host, usually invertebrates from the Diptera, Hemiptera and Siphonaptera orders (Kozminsky et al., 2015). In this study, we described the occurrence of C. mellificae in different mammalian orders from three distinct Brazilian biomes. This trypanosomatid species was first described in Apis mellifera honeybees in Australia (Langride and McGhee, 1967), but it was also observed infecting other invertebrate hosts, such as Vespula squamosa (wasp; Vespidae), Osmia cornuta (orchard bee; Megachilidae) and O. bicornis (mason bee; Megachilidae) in the United States (Schwarz et al., 2015), and haematophagous horse fly insects (Haematopota griseicoxa) in Western Africa (Votýpka et al., 2019). In addition to being a generalist insect parasite, C. mellificae has a broad geographical distribution since its occurrence was observed in invertebrate insects from Japan (Morimoto et al., 2013), Belgium (Ravoet et al., 2013), Algeria (Menail et al., 2016) and the United States (Cornman et al., 2012). This species was recently reported for the first time infecting a mammal species, a nectarivore bat species Anoura caudifer from the Atlantic Forest of Southeastern Brazil (Rangel et al., 2019).
This amazing finding represents a paradigm break. Here, we are going a step further, as we are demonstrating that Crithidia not only is able to infect other bat species (C. perspicillata, M. izecksohni, M. lavali and A. lituratus) that display different feeding habits and ecology but also infects other and unrelated wild mammal taxa. In addition to this wide diversity of mammalian hosts, the parasite is widely distributed in distinct habitats and Brazilian biomes. We obviously do not know the fate of these infections, whether they will be self-limiting and how long they will last. We know even less about the role that these mammals may eventually play in the biology of C. mellificae. However, one thing we can affirm is that the species presents a capacity to infect highly ecologically diverse mammalian species, which points to an immense ecological resilience of this parasite species.
Crithidia mellificae was demonstrated to be capable of overcoming important defence barriers, such as skin, phagocytic cells, and the complement system action of mammalian hosts, since we were able to isolate the parasite by haemoculture. None of the 21 blood samples examined in this study were positive in fresh blood smears, and we have never seen choanomastigote forms in any fresh blood test of the wild mammals we have examined in the Trypanosomatid Biology Laboratory (IOC/Fiocruz) over the years. It is rare to find parasites in fresh blood exams; even T. cruzi, a common mammalian trypanosomatid, is rarely seen in fresh blood preparations (Jansen et al., 2020). Therefore, we performed parasitological and serological diagnoses to determine infection. Additionally, parasitemia of C. mellificae-infected mammals was high enough to be detected by not sensitive but specific parasitological techniques, in this case, haemocultures. Many questions emerge from these findings, including the transmission strategies. What is certain is that the encounter frequency of insect monoxenous trypanosomatids parasitizing mammals is increasing, as has been described (Podlipaev et al., 2004; Votýpka et al., 2010; Votýpka et al., 2012a, Votýpka et al., 2012b).
Monoxenous trypanosomatids are transmitted via contamination, predation, coprophagy and necrophagy routes (Wallace, 1966; Frolov et al., 2021). Even though the infected bat species present different feeding habits, it is suggested that they may be infected by licking the bite site after a bee sting that has choanomastigote-infected faeces (Rangel et al., 2019). Primates in the genus Callithrix sp. are gummivore-insectivore animals, but they can also feed on fruits (Sussman and Kinzey, 1984; Rylands and Farias, 1993). We can speculate that possible C. mellificae transmission between this primate species could be via the ingestion of fruits contaminated by insect faeces with choanomastigote forms. Another possible route of C. mellificae infection in primates could be by the ingestion of bees trapped in gum. Concerning carnivore species, canids have an eclectic feeding habit that includes fruits and insects (Brady, 1979; Gatti et al., 2006; Ferreira et al., 2013). It can be hypothesized that these mammals could be infected by preying on insects or other animals infected by C. mellificae. On the other hand, felines are strict carnivores (Bisbal, 1986; Meza et al., 2002; Martins et al., 2008), and a plausible explanation, in this case, is that infection was due to mammalian predation. In all these situations, we hypothesized that oral transmission was the main route of C. mellificae infection.
Recently, monoxenous trypanosomatids have been reported infecting sylvatic mammals and humans, including not only immunosuppressed individuals but also immunocompetent individuals. In mammals, Crithidia sp. was reported in an Australian marsupial Bettongia penicillata (Cooper et al., 2018); Blastocrithidia sp. was reported in a bat from the USA (Hodo et al., 2016), and Angomonas deanei, Herpetomonas samuelpessoai and Leptomonas spp. were demonstrated in scent glands of Didelphis marsupialis under experimental conditions (Jansen et al., 1988). Human infection cases by monoxenous trypanosomatids Leptomonas seymouri, Crithidia sp. and Herpetomonas samuelpessoai were observed in Africa, Asia, and the American continents (Dedet et al., 1995; Pacheco et al., 1998; Boisseau-Garsaud et al., 2000; Morio et al., 2008; Ghosh et al., 2012; Kraeva et al., 2015; Ghobakhloo et al., 2018). Regarding the finding of Crithidia in immunocompetent patients in Iran (Ghobakhloo et al., 2018), Kostygov et al. (2019) did not agree with a monoxenous trypanosomatid infection of an immunocompetent host. These authors report that the infection was only possible because of the impairment of the host immune system due to coinfection with other parasites. In Brazil, a fatal human case of a Crithidia fasciculata-like infection was reported in Sergipe state (Maruyama et al., 2019). The recent encounters of monoxenous trypanosomatid infections in mammals can be justified by more discriminative diagnostic methodologies, especially molecular biology. This phenomenon is probably much more common than reported and must be taken into consideration when studying trypanosomatid infections.
According to Votýpka et al. (2012b), host switches of monoxenous trypanosomatids from their original insect hosts to new insects are likely to occur because insects present simpler defence mechanisms than mammals. This can be one of the explanations for the broad distribution of C. mellificae. Moreover, its capacity to infect mammals, as we observed, still represents a challenging question. It is important to note that the finding of C. mellificae in Haematopota insects (horse fly insects) (Votýpka et al., 2019) also demonstrates that the occurrence of Crithidia in mammals might not be occasional or accidental, since these female insects present haematophagous feeding behaviour, which reinforces the possibility of vector transmission by haematophagous insects. Previous studies reported that some monoxenous trypanosomatid species have lower host specificity and can therefore be found in different invertebrate hosts (Maslov et al., 2007; Votýpka et al., 2010, 2012a). This situation can be observed not only in monoxenous environments but also in all members from the Kinetoplastea class, even in species considered to be free-living protozoa, in which there is an increasing number of unusual host occurrences (Auty et al., 2012; Dario et al., 2017; Szőke et al., 2017).
Comparing C. mellificae SSU rDNA sequences from bats, primates and carnivores with sequences identified from Tabanidae and honeybee insects, 11 samples presented no genetic divergence. This shows that C. mellificae possibly presents a unique genetic profile and circulates between different hosts in different geographical environments. In this case, genetic diversity was not necessary to increase the parasite's host diversity. This genetic profile was competent to maintain itself in several mammal species and orders with distinct ecological profiles and is just a few steps away from suggesting that this parasite is a dixenous trypanosomatid if it cannot already be considered as such. Ten sequence samples, four from marmosets, three from bats, two from crab-eating foxes and one from coati presented genetic diversity from the other sequences (0.001 to 0.007 divergence). Although we are reporting this genetic diversity, the difference between the sequences was minimal but enough to rule out accidental contamination.
We demonstrated that C. mellificae was capable of growing and maintaining itself at different temperatures for up to 16 days. Crithidia mellificae is not the only species from the Crithidia genus capable of surviving at temperatures higher than 27°C, C. l. thermophila is capable of surviving at 34 °C (Ishemgulova et al., 2017). Therefore, we can say that C. mellificae is thermotolerant to high temperatures, which explains the occurrence of this trypanosomatid infecting different species of mammals. In the morphological forms observed at different temperatures, rounded large forms predominated at 36°C and 37°C. Most likely, the parasite passes through an adaptative shock due to this high temperature, resulting in these most observed forms at peak growth. After the peak, the thinner forms were more commonly observed, possibly because they were able to survive this event.
It was observed that C. mellificae was able to cross all innate immune barriers to establish infection and was capable of stimulating the humoral immune response, at least in primates. Specific antibodies against monoxenous trypanosomatids have already been detected in D. marsupialis serum after experimental infection by A. deanei, H. samuelpessoai, L. samueli and Leptomonas sp. by inoculation via scent gland (Jansen et al., 1988). We observed a lower/absent serological titer for T. cruzi and absence of Leishmania sp. in the same serum tested for C. mellificae antigen. Cross-reaction in serological tests involving trypanosomatids is common, mainly among T. cruzi and T. rangeli and Leishmania sp. (da Silveira et al., 2001; Caballero et al., 2007; Nieto et al., 2009). Even other taxonomically distant parasites, such as Ehrlichia canis, Toxoplasma gondii, Neospora caninum and Babesia canis, may cross-react in leishmaniasis serological tests (Zanette et al., 2014). Although we did not detect cross-reaction between C. mellificae and Leishmania sp., Garin and coworkers (2001) observed cross-reaction in serum samples from animals infected with a monoxenous trypanosomatid with enzyme-linked immunosorbent assay (ELISA). Unlike Leishmania sp., C. mellificae antibodies were capable of reacting with T. cruzi antigens, although serological titers were very low. False-positive serological results may occur since monoxenous trypanosomatids are poorly studied, and their presence in nature and other hosts may be more frequent. In addition, the use of multiple methodologies is essential to confirm trypanosomatid infections.
We can exclude laboratory contamination because the mammal capture and haemoculture exams were performed in different time periods and we do not maintain Crithidia cultures in our facilities. Therefore, these results represent important findings, breaking the concept of insect trypanosomatids being monoxenous and insect restricted. These findings also show how much is unknown of parasitic specificity, a key topic in these times of so many emerging parasitic diseases. Monoxenous trypanosomatids might be the key to understanding the origin of the parasitism phenomenon within the Trypanosomatidae family (Flegontov et al., 2013; Lukeš et al., 2014; Maslov et al., 2019). Host specificity has no sharp contour, with constant spillover and changes in hosts in nature. We can affirm that parasitic specificity within the Trypanosomatidae family is complex and far from being solved. It is important to question why monoxenous trypanosomatids have been found in mammals only over the last few years. One hypothetical explanation for this question is that concerning fundamentally digenetic trypanosomatids, many of which were recognized as pathogenic, a careful and unbiased examination has not been performed, therefore unusual forms that would eventually appear in culture media have been overlooked. Given the set of results presented herein, C. mellificae can be a generalist species of invertebrate and mammalian host species. Therefore, we can hypothesize that C. mellificae species is one step closer to becoming dixenous.
Acknowledgements
We would like to express our gratitude to Bruno Alves Silva, Pedro Cordeiro-Estrela, Andressa Fraga, and Henrique Concone, Laíza de Queiroz Viana Braga for their help in fieldwork, to Marcos Antônio dos Santos Lima e Carlos Ardé for technical support in haemoculture and Dra Samanta Cristina das Chagas Xavier for technical support in the serological methodology. We would also like to thank the Technological Development and Inputs for Health Program of the Fundação Oswaldo Cruz (PDTIS/FIOCRUZ) sequencing platform for sequencing our isolates. Special thanks to Dr Vera Bongertz for helpful comments on the English review. This study was funding by Fundação Oswaldo Cruz (Fiocruz), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) and Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul (FUNDECT). CNPq provided a PDJ fellowship (2018-2019) and FAPERJ provides a post-doctoral #10 fellowship (2019-nowadays) to MAD. CAPES provided a PNPD fellowship to FLR. RM has received financial support from FAPERJ and CNPq. HMH receives a “Bolsa de produtividade (Pq 308768/2017-5)” fellowship from CNPq and financial support from FUNDECT (03/2016 PPSUS-MS process#: 59/300.069/2017). ALRR receives a “Joven Cientista do Nosso Estado” fellowship from FAPERJ and “Bolsa de produtividade” fellowship from CNPq. AMJ receives a “Cientista do Nosso Estado” fellowship from FAPERJ and “Bolsa de produtividade” fellowship from CNPq.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2021.04.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adl S.M., Bass D., Lane C.E., Lukeš J., Schoch C.L., Smirnov A., Agatha S., Berney C., Brown M.W., Burki F., Cárdenas P., Čepička I., Chistyakova L., Campo J.d., Dunthorn M., Edvardsen B., Eglit Y., Guillou L., Hampl V., Heiss A.A., Hoppenrath M., James T.Y., Karnkowska A., Karpov S., Kim E., Kolisko M., Kudryavtsey A., Lahr D.J.G., Lara E., Gall L.L., Lynn D.H., Mann D.G., Massana R., Mitchell E.A.D., Morrow C., Park J.S., Pawlowski J.W., Powell M.J., Richter D.J., Rueckert S., Shadwick L., Shimano S., Spiegel F.W., Torruella G., Youssef N., Zlatogursky V., Zhang Q. Revision to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019;66:4–119. doi: 10.1111/jeu.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auty H., Anderson N.E., Picozzi K., Lembo T., Mubanga J., Hoare R., Fyumagwa R.D., Mable B., Hamill L., Cleaveland S., Welburn S.C. Trypanosome diversity in wildlife species from the Serengeti and Luangwa Valley ecosystems. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal F.J. Food habitats of some neotropical carnivores in Venezuela (Mammalia, Carnivora) Mammalia. 1986;50(3):329–339. [Google Scholar]
- Boisseau-Garsaud A.M., Cales-Quist D., Desbois N., Jouannelle J., Jouannelle A., Pratlong F., Dedet J.P. A new case of cutaneous infection by a presumed monoxenous trypanosomatid in the island of Martinique (French West Indies) Trans. R. Soc. Trop. Med. Hyg. 2000;94(1):51–52. doi: 10.1016/s0035-9203(00)90435-8. [DOI] [PubMed] [Google Scholar]
- Borghesan T.C., Ferreira R.C., Takata C.S., Campaner M., Borda C.C., Paiva F., Milder R.V., Teixeira M.M.G., Camargo E.P. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164:129–152. doi: 10.1016/j.protis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., Matschiner M., Mendes F.K., Müller N.F., Ogilvie H.A., du Plessis L., Popinga A., Rambaut A., Rasmussen D., Siveroni I., Suchard M.A., Wu C.H., Xie D., Zhang C., Stadler T., Drummond A.J. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15(4) doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C.A. Smithsonian Institution Press; 1979. Observations on the Behavior and Ecology of the Crab-Eating Fox (Cerdocyon Thous) pp. 161–167. [Google Scholar]
- Braga L.Q.V. Universidade Federal da Paraíba; 2019. Infecções múltiplas e a saúde de carnívoros silvestres em um agroecossistema brasileiro. [Google Scholar]
- Caballero Z.C., Sousa O.E., Marques W.P., Saez-Alquezar A., Umezawa E.S. Evaluation of serological tests to identify Trypanosoma cruzi infection in humans and determine cross-reactivity with Trypanosoma rangeli and Leishmania spp. Clin. Vaccine Immunol. 2007;14:1045–1049. doi: 10.1128/CVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo M.E. Fluorescent antibody test for the serodiagnosis of American trypanosomiasis. Technical modification employing preserved culture forms of Trypanosoma cruzi in a slide test. Rev. Soc. Bras. Med. Trop. 1966;8:227–235. [PubMed] [Google Scholar]
- Chernomor O., von Haeseler A., Minh B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C., Keatley S., Northover A., Gofton A.W., Brigg F., Lymbery A.J., Pallant L., Clode P.L., Andrew Thompson R.C. Next generation sequencing reveals widespread trypanosome diversity and polyparasitism in marsupials from Western Australia. Int. J. Parasitol. Parasites Wildl. 2018;7(1):58–67. doi: 10.1016/j.ijppaw.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman R.S., Tarpy D.R., Chen Y., Jeffreys L., Lopez D., Pettis J.S., vanEngelsdorp D., Evans J.D. Pathogen webs in collapsing honeybee colonies. PloS One. 2012;7(8) doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dario M.A., Moratelli R., Schwabl P., Jansen A.M., Llewellyn M.S. Small subunit ribosomal metabarcoding reveals extraordinary trypanosomatid diversity in Brazilian bats. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Avila-Levy C.M., Boucinha C., Kostygov A., Santos H.L., Morelli K.A., Grybchuk-Ieremenko A., Duval L., Votýpka J., Yurchenko V., Grellier P., Lukeš J. Exploring the environmental diversity of kinetoplastid flagellates in the high-throughput DNA sequencing era. Mem. Inst. Oswaldo Cruz. 2015;110(8):956–965. doi: 10.1590/0074-02760150253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira J.F., Umezawa E.S., Luquetti A.O. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 2001;17:286–291. doi: 10.1016/s1471-4922(01)01897-9. [DOI] [PubMed] [Google Scholar]
- Dedet J.P., Roche B., Pratlong F., Cales-Quist D., Jouannelle J., Benichou J.C., Huerre M. Diffuse cutaneous infection by a presumed monoxenous trypanosomatid in a patient infected with HIV. Trans. R. Soc. Trop. Med. Hyg. 1995;89:644–646. doi: 10.1016/0035-9203(95)90427-1. [DOI] [PubMed] [Google Scholar]
- Ferreira G.A., Nakano-Oliveira E., Genaro G., Lacerda-Chaves A.K. Diet of the coati Nasua nasua (Carnivora: procyonidae) in an area of woodland inserted in an urban environment in Brazil. Rev. Chil. Hist. Nat. 2013;86:95–102. [Google Scholar]
- Flegontov P., Votýpka J., Skalický T., Logacheva M.D., Penin A.A., Tanifuji G., Onodera N.T., Kondrashov A.S., Volf P., Archibald J.M., Lukeš J. Paratrypanosoma is a novel earlybranching trypanosomatid. Curr. Biol. 2013;23:1787–1793. doi: 10.1016/j.cub.2013.07.045. [DOI] [PubMed] [Google Scholar]
- Frolov A.O., Kostygov A.Y., Yurchenko V. Development of monoxenous trypanosomatids and phytomonads in insects. Trends Parasitol. 2021 doi: 10.1016/j.pt.2021.02.004. [DOI] [PubMed] [Google Scholar]
- Gatti A., Bianchi R., Rosa C.R.X., Mendes S.L. Diet of two sympatric carnivores, Cerdocyon thous and Procyon cancrivorus, in a restinga area of Espírito Santo State, Brazil. J. Trop. Ecol. 2006;22(1):227–230. [Google Scholar]
- Garin Y.J., Sulahian A., Méneceur P., Pratlong F., Prina E., Gangneux J., Dedet J.P., Derouin F. Experimental pathogenicity of a presumed monoxenous trypanosomatid isolated from humans in a murine model. J. Eukaryot. Microbiol. 2001;48(2):170–176. doi: 10.1111/j.1550-7408.2001.tb00299.x. [DOI] [PubMed] [Google Scholar]
- Ghobakhloo N., Motazedian M.H., Naderi S., Ebrahimi S. Isolation of Crithidia spp. from lesions of immunocompetent patients with suspected cutaneous leishmaniasis in Iran. Trop. Med. Int. Health. 2018;24(1):116–126. doi: 10.1111/tmi.13042. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Banerjee P., Sarkar A., Datta S., Chatterjee M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J. Clin. Microbiol. 2012;50:2774–2778. doi: 10.1128/JCM.00966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare C.A. The classification of mammalian trypanosomes. Ergeb. Mikrobiol. Immunitatsforsch. Exp. Ther. 1966;39:43–57. doi: 10.1007/978-3-662-38353-7_3. [DOI] [PubMed] [Google Scholar]
- Hodo C.L., Goodwin C.C., Mayes B.C., Mariscal J.A., Waldrup K.A., Hamer S.A. Trypanosome species, including Trypanosoma cruzi, in sylvatic and peridomestic bats of Texas, USA. Acta Trop. 2016;164:259–266. doi: 10.1016/j.actatropica.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishemgulova A., Butenko A., Kortišová L., Boucinha C., Grybchuk-Ieremenko A., Morelli K.A., Tesařová M., Kraeva N., Grybchuk D., Pánek T., Flegontov P., Lukeš J., Votýpka J., Pavan M.G., Opperdoes F.R., Spodareva V., d'Avila-Levy C.M., Kostygov A.Y., Yurchenko V. Molecular mechanisms of thermal resistance of the insect trypanosomatid Crithidia thermophila. PloS One. 2017;12(3) doi: 10.1371/journal.pone.0174165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen A.M., Carreira J.C., Deane M.P. Infection of a mammal by monogenetic insect trypanosomatids (Kinetoplastida, Trypanosomatidae) Mem. Inst. Oswaldo Cruz. 1988;83(3):271–272. doi: 10.1590/s0074-02761988000300001. [DOI] [PubMed] [Google Scholar]
- Jansen A.M., Xavier S.C.D.C., Roque A.L.R. Landmarks of the knowledge and Trypanosoma cruzi biology in the wild environment. Front. Cell. Infect. Microbiol. 2020;10:10. doi: 10.3389/fcimb.2020.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirků M., Yurchenko V.Y., Lukeš J., Maslov D.A. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J. Eukaryot. Microbiol. 2012;59:537–547. doi: 10.1111/j.1550-7408.2012.00636.x. [DOI] [PubMed] [Google Scholar]
- Kaufer A., Ellis J., Stark D., Barratt J. The evolution of trypanosomatid taxonomy. Parasites Vectors. 2017;10:287. doi: 10.1186/s13071-017-2204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostygov A.Y., Grybchuk-Ieremenko A., Malysheva M.N., Frolov A.O., Yurchenko V. Molecular revision of the genus Wallaceina. Protist. 2014;165:594–604. doi: 10.1016/j.protis.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Kostygov A.Y., Yurchenko V. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae) Folia Parasitol. 2017;64 doi: 10.14411/fp.2017.020. [DOI] [PubMed] [Google Scholar]
- Kostygov A.Y., Butenko A., Yurchenko V. On monoxenous trypanosomatids from lesions of immunocompetent patients with suspected cutaneous leishmaniasis in Iran. Trop. Med. Int. Health. 2019;24(1):127–128. doi: 10.1111/tmi.13168. [DOI] [PubMed] [Google Scholar]
- Kostygov A.Y., Frolov A.O., Malysheva M.N., Ganyukova A.I., Chistyakova L.V., Tashyreva D., Tesařová M., Spodareva V.V., Režnarová J., Macedo D.H., Butenko A., d'Avila-Levy C.M., Lukeš J., Yurchenko V. Vickermania gen. nov., trypanosomatids that use two joined flagella to resist midgut peristaltic flow within the fly host. BCM Biology. 2020;18:187. doi: 10.1186/s12915-020-00916-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminsky E., Kraeva N., Ishemgulova A., Dobáková E., Lukeš J., Kment P., Yurchenko V., Votýpka J., Maslov D.A. Host-specificity of monoxenous trypanosomatids: statistical analysis of the distribution and transmission patterns of the parasites from neotropical Heteroptera. Protist. 2015;166(5):551–568. doi: 10.1016/j.protis.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Kraeva N., Butenko A., Hlaváčová J., Kostygov A., Myškova J., Grybchuk D., Leštinová T., Votýpka J., Volf P., Opperdoes F., Flegontov P., Lukeš J., Yurchenko V. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langride D.F., McGhee R.B. Crithidia mellificae n. sp. an acidophilic trypanosomatid of the honeybee Apis mellifera. J. Protozool. 1967;14:485–487. doi: 10.1111/j.1550-7408.1967.tb02033.x. [DOI] [PubMed] [Google Scholar]
- Léger L. Sur un flagelle parasite de l'Anopheles maculipennis. Compt. Rend. Soc. Biol. 1902;54:354–356. [Google Scholar]
- Lipa J.J., Triggiani O. Crithidia bombi sp.n. a flagellated parasite of a bumblebee Bombus terrestris L. (Hymenoptera, Apidae) Acta Protozool. 1988;27:287–290. [Google Scholar]
- Lisboa C.V., Mangia R.H., Luz S.L., Kluczkovski A., Jr., Ferreira L.F., Ribeiro C.T., Fernandes O., Jansen A.M. Stable infection of primates with Trypanosoma cruzi I and II. Parasitology. 2006;133(5):603. doi: 10.1017/S0031182006000722. [DOI] [PubMed] [Google Scholar]
- Lukeš J., Skalický T., Týč J., Votýpka J., Yurchenko V. Evolution of parasitism in kinetoplastid flagellates. Mol. Biochem. Parasitol. 2014;195:115–122. doi: 10.1016/j.molbiopara.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Lukeš J., Tesařová M., Yurchenko V., Votýpka J. Characterization of a new cosmopolitan genus of trypanosomatid parasites, Obscuromonas gen. nov. (Blastocrithidiinae subfam. nov.) Eur. J. Protistol. 2021;79:125778. doi: 10.1016/j.ejop.2021.125778. [DOI] [PubMed] [Google Scholar]
- Lumsden W.H.R., Evans D.A. Academic Press; 1976. Biology of Kinetoplastida. [Google Scholar]
- Martins R., Quadros J., Mazzolli M. Hábito alimentar e interferência antrópica na atividade de marcação territorial do Puma concolor e Leopardus pardalis (Carnivora: felidae) e outros carnívoros na Estação Ecológica de Juréia-Itatins, São Paulo, Brasil. Rev. Bras. Zool. 2008;25(3):427–435. [Google Scholar]
- Maruyama S.R., de Santana A.K.M., Takamiya N.T., Takahashi T.Y., Rogerio L.A., Oliveira C.A.B., Milanezi C.M., Trombela V.A., Cruz A.K., Jesus A.R., Barreto A.S., da Silva A.M., Almeida R.P., Ribeiro J.M., Silva J.S. Non-Leishmania parasite in fatal visceral Leishmaniasis–like disease. Brazil. Emerg. Infect. Dis. 2019;25(11):2088–2092. doi: 10.3201/eid2511.181548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov D.A., Westenberger S.J., Xu X., Campbell D.A., Sturm N.R. Discovery and barcoding by analysis of Spliced Leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J. Eukaryot. Microbiol. 2007;54:57–65. doi: 10.1111/j.1550-7408.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- Maslov D.A., Votýpka J., Yurchenko V., Lukeš J. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol. 2013;29:43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Maslov D.A., Opperdoes F.R., Kostygov A.Y., Hashimi H., Lukeš J., Yurchenko V. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology. 2019;146:1–27. doi: 10.1017/S0031182018000951. [DOI] [PubMed] [Google Scholar]
- Menail A.H., Piot N., Meeus I., Smagghe G., Loucif-Ayad W. Large pathogen screening reveals first report of Megaselia scalaris (Diptera: phoridae) parasitizing Apis mellifera intermissa (Hymenoptera: apidae) J. Invertebr. Pathol. 2016;137:33–37. doi: 10.1016/j.jip.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Merzlyak E., Yurchenko V., Kolesnikov A.A., Alexandrov K., Podlipaev S.A., Maslov D.A. Diversity and phylogeny of insect trypanosomatids based on small subunit rRNA genes: polyphyly of Leptomonas and Blastocrithidia. J. Eukaryot. Microbiol. 2001;48:161–169. doi: 10.1111/j.1550-7408.2001.tb00298.x. [DOI] [PubMed] [Google Scholar]
- Meza A.D., Meyer M.E., Gonzalez C.A.L. Ocelot (Leopardus pardalis) food habits in a tropical deciduous forest of Jalisco, México. Am. Midl. Nat. 2002;148:146–154. [Google Scholar]
- Morimoto T., Kojima Y., Yoshiyama M., Kimura K., Yang B., Peng G., Kadowaki T. Molecular detection of protozoan parasites infecting Apis mellifera colonies in Japan. Environ. Microbiol. Rep. 2013;5(1):74–77. doi: 10.1111/j.1758-2229.2012.00385.x. [DOI] [PubMed] [Google Scholar]
- Morio F., Reynes J., Dollet M., Pratlong F., Dedet J.P., Ravel C. Isolation of a protozoan parasite genetically related to the insect trypanosomatid Herpetomonas samuelpessoai from a human immunodeficiency virus-positive patient. J. Clin. Microbiol. 2008;46:3845–3847. doi: 10.1128/JCM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto P.D., Boughton R., Dorn P.L., Steurer F., Raychaudhuri S., Esfandiari J., Gonçalves E., Diaz J., Malone J.B. Comparison of two immunochromatographic assays and the indirect immunofluorescence antibody test for diagnosis of Trypanosoma cruzi infection in dogs in south central Louisiana. Vet. Parasitol. 2009;165:241–247. doi: 10.1016/j.vetpar.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Olsen O.W. New York Courier Corporation; New York: 1986. Animal Parasites: Their Life Cycles and Ecology. [Google Scholar]
- Pacheco R.S., Marzochi M.C., Pires M.Q., Brito C.M., Madeira M.d.F., Barbosa-Santos E.G. Parasite genotypically related to a monoxenous trypanosomatid of dog's flea causing opportunistic infection in a HIV positive patient. Mem. Inst. Oswaldo Cruz. 1998;93(4):531–537. doi: 10.1590/s0074-02761998000400021. [DOI] [PubMed] [Google Scholar]
- Podlipaev S. The more insect trypanosomatids under study-the more diverse Trypanosomatidae appears. Int. J. Parasitol. 2001;31:648–652. doi: 10.1016/s0020-7519(01)00139-4. [DOI] [PubMed] [Google Scholar]
- Podlipaev S., Sturm N.R., Fiala I., Fernandes O., Westenberger S.J., Dollet M., Campbell D.A., Lukeš J. Diversity of insect trypanosomatids assessed from the spliced leader RNA and 5s rRNA genes and intergenic regions. J. Eukaryot. Microbiol. 2004;51(3):283–290. doi: 10.1111/j.1550-7408.2004.tb00568.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018 doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel D.A., Lisboa C.V., Novaes R.L.M., Silva B.A., Souza R.F., Jansen A.M., Moratelli R., Roque A.L.R. Isolation and characterization of trypanosomatids, including Crithidia mellificae, in bats from the Atlantic Forest of Rio de Janeiro, Brazil. PLoS Neglected Trop. Dis. 2019;13(7) doi: 10.1371/journal.pntd.0007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravoet J., Maharramov J., Meeus I., De Smet L., Wenseleers T., Smagghe G., de Graaf D.C. Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PloS One. 2013;8 doi: 10.1371/journal.pone.0072443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues Marina Silva, Lima Luciana, Xavier Samanta Cristina das Chagas, Herreira Heitor Miraglia, Rocha Fabiana Lopes, Roque André Luiz Rodrigues, Teixeira Marta M.G., Jansen Ana Maria. Uncovering Trypanosoma spp. diversity of wild mammals by the use of DNA from blood clots. Int. J. Parasitol. Parasites Wildl. 2019;14:171–181. doi: 10.1016/j.ijppaw.2019.02.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylands A.B., Farias D.S. Oxford University Press; Oxford: 1993. Marmosets and Tamarins (Systematics, Behavior, and Ecology) pp. 262–272. [Google Scholar]
- Santos F.M., de Macedo G.C., Barreto W.T.G., Oliveira-Santos L.G.R., Garcia C.M., Mourão G.M., Edith de Oliveira Porfírio G., Domenis Marino E., Rogério André M., Perles L., Elisei de Oliveira C., Braziliano de Andrade G., Jansen A.M., Miraglia Herrera H. Outcomes of Trypanosoma cruzi and Trypanosoma evansi infections on health of Southern coati (Nasua nasua), crab-eating fox (Cerdocyon thous), and ocelot (Leopardus pardalis) in the Brazilian Pantanal. PloS One. 2018;13(8) doi: 10.1371/journal.pone.0201357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel R., Tognazzo M. Molecular divergence defines two distinct lineages of Crithidia bombi (Trypanosomatidae), parasites of bumblebees. J. Eukaryot. Microbiol. 2010;57(4):337–345. doi: 10.1111/j.1550-7408.2010.00480.x. [DOI] [PubMed] [Google Scholar]
- Schwarz R.S., Bauchan G.R., Murphy C.A., Ravoet J., de Graaf D.C., Evans J.D. Characterization of two species of Trypanosomatidae from the honeybee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J. Eukaryot. Microbiol. 2015;62:567–583. doi: 10.1111/jeu.12209. [DOI] [PubMed] [Google Scholar]
- Schwelm A., Badstöber J., Bulman S., Desoignies N., Etemadi M., Falloon R.E., Gachon C.M.M.G., Legreve A., Lukeš J., Merz U., Nenarokova A., Strittmatter M., Sullivan B.K., Neuhauser S. Not in your usual Top 10: protists that infect plants and algae. Mol. Plant Pathol. 2018;19(4):1029–1044. doi: 10.1111/mpp.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson A.G., Stevens J.R., Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Strobl V., Yañez O., Straub L., Albrecht M., Neumann P. Trypanosomatid parasites infecting managed honeybees and wild solitary bees. Int. J. Parasitol. 2019;49(8):605–613. doi: 10.1016/j.ijpara.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Sussman R.W., Kinzey W.G. The ecological role of the Callitrichidae: a review. Am. J. Phys. Anthropol. 1984;64:419–449. doi: 10.1002/ajpa.1330640407. [DOI] [PubMed] [Google Scholar]
- Szőke K., Sándor A.D., Boldogh S.A., Görföl T., Votýpka J., Takács N., Estók P., Kováts D., Corduneanu A., Molnár V., Kontschán J., Hornok S. DNA of free-living bodonids (Euglenozoa: Kinetoplastea) in bat ectoparasites: potential relevance to the evolution of parasitic trypanosomatids. Acta Vet. Hung. 2017;65:531–540. doi: 10.1556/004.2017.051. [DOI] [PubMed] [Google Scholar]
- Teixeira M.M.G., Borghesan T.C., Ferreira R.C., Santos M.A., Takata C.S., Campaner M., Nunes V.L.B., Milder R.V., de Souza W., Camargo E.P. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011;162:503–524. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Týč J., Votýpka J., Klepetková H., Suláková H., Jirků M., Lukeš J. Growing diversity of trypanosomatid parasites of flies (Diptera: brachycera): frequent cosmopolitism and moderate host specificity. Mol. Phylogenet. Evol. 2013;69(1):255–264. doi: 10.1016/j.ympev.2013.05.024. [DOI] [PubMed] [Google Scholar]
- Vickerman K. Academic Press; 1976. Comparative Cell Biology of the Kinetoplastid Flagellates; pp. 35–130. [Google Scholar]
- Votýpka J., Maslov D.A., Yurchenko V., Jirků M., Kment P., Lun Z.R., Lukeš J. Probing into the diversity of trypanosomatid flagellates parasitizing insect hosts in South-West China reveals both endemism and global dispersal. Mol. Phylogenet. Evol. 2010;54:243–253. doi: 10.1016/j.ympev.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Votýpka J., Klepetková H., Yurchenko V.Y., Horák A., Lukeš J., Maslov D.A. Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist. 2012;163(4):616–631. doi: 10.1016/j.protis.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Votýpka J., Klepetková H., Jirků M., Kment P., Lukeš J. Phylogenetic relationships of trypanosomatids parasitising true bugs (Insecta: heteroptera) in sub-Saharan Africa. Int. J. Parasitol. 2012;42:489–500. doi: 10.1016/j.ijpara.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Votýpka J., Suková E., Kraeva N., Ishemgulova A., Duží I., Lukeš J. Diversity of trypanosomatids (Kinetoplastea: Trypanosomatidae) parasitizing fleas (Insecta: Siphonaptera) and description of a new genus Blechomonas gen. n. Protist. 2013;164:763–781. doi: 10.1016/j.protis.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Votýpka J., Kostygov A.Y., Kraeva N., Grybchuk-Ieremenko A., Tesařová M., Grybchuk D., Lukeš J., Yurchenko V. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist. 2014;165:825–838. doi: 10.1016/j.protis.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Votýpka J., Brzoňová J., Ježek J., Modrý D. Horse flies (Diptera: Tabanidae) of three West African countries: a faunistic update, barcoding analysis and trypanosome occurrence. Acta Trop. 2019;197:105069. doi: 10.1016/j.actatropica.2019.105069. [DOI] [PubMed] [Google Scholar]
- Wallace F.G. The trypanosomatid parasites of insects and arachnids. Exp. Parasitol. 1966;18:124–193. doi: 10.1016/0014-4894(66)90015-4. [DOI] [PubMed] [Google Scholar]
- Yurchenko V., Lukeš J., Tesarová M., Jirků M., Maslov D.A. Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus Crithidia. Protist. 2008;159:99–114. doi: 10.1016/j.protis.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Yurchenko V., Lukeš J., Jirků M., Maslov D.A. Selective recovery of the cultivation-prone components from mixed trypanosomatid infections: a case of several novel species isolated from Neotropical Heteroptera. Int. J. Syst. Evol. Microbiol. 2009;59(4):893–909. doi: 10.1099/ijs.0.001149-0. [DOI] [PubMed] [Google Scholar]
- Yurchenko V., Votýpka J., Tesarová M., Klepetková H., Kraeva N., Jirků M., Lukeš J. Ultrastructure and molecular phylogeny of four new species of monoxenous trypanosomatids from flies (Diptera: brachycera) with redefinition of the genus Wallaceina. Folia Parasitol. 2014;61:97–112. [PubMed] [Google Scholar]
- Yurchenko V., Kostygov A., Havlová J., Grybchuk-Ieremenko A., Ševčíková T., Lukeš J., Ševčík J., Votýpka J. Diversity of trypanosomatids in cockroaches and the description of Herpetomonas tarakana sp. n. J. Eukaryot. Microbiol. 2016;63:198–209. doi: 10.1111/jeu.12268. [DOI] [PubMed] [Google Scholar]
- Zanette M.F., Lima V.M., Laurenti M.D., Rossi C.N., Vides J.P., Vieira R.F.d.C., Biondo A.W., Marcondes M. Serological cross-reactivity of Trypanosoma cruzi, Ehrlichia canis, Toxoplasma gondii, Neospora caninum and Babesia canis to Leishmania infantum chagasi tests in dogs. Rev. Soc. Bras. Med. Trop. 2014;47(1):105–107. doi: 10.1590/0037-8682-1723-2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.