Highlights
-
•
Robust preparation of liposomal formulation by DELOS-susp method.
-
•
Implementation of Quality by Design methodology to liposomes preparation.
-
•
Influence of critical parameters on quality was studied through DoE analysis.
-
•
Design Space was obtained for GLA-loaded liposomes formulation.
Keywords: Quality by Design, DELOS, Scale-up, Protein-loaded liposomes, Fabry disease, α-galactosidase
Abbreviations: BCA, Bicinchoninic acid assay; Chol, Cholesterol; Chol-PEG400-RGD, Cholesterol pegylated with arginyl–glycyl–aspartic (RGD) acid peptide; CMA, Critical Material Attributes; CO2, Carbon dioxide; CoA, Certificate of Analysis; CPP, Critical Process Parameters; CQA, Critical Quality Attribute; Cryo-TEM, Cryogenic Transmission Electron Microscopy; DELOS-susp, Depressurization of an Expanded Liquid Organic Solution into aqueous solution; DLS, Dynamic Light Scattering; DMSO, Dimethyl sulfoxide; DoE, Design of Experiments; DPPC, 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine; EA, Enzymatic Activity; EE, Entrapment Efficiency; EHS, Environment, Health and Safety; EMA, European Medicines Agency; ERT, Enzyme Replacement Therapy; EtOH, Ethanol; FDA, Food and Drug Administration; fsingle, Ratio of monolayered liposomes; GLA, α-galactosidase A enzyme; H2O, Water; HPLC, High Performance Liquid Chromatography; ICH, Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use; LSD, Lysosomal storage disorders; MKC, Myristalkonium chloride; N2, Nitrogen; nanoGLA, GLA-loaded nanoliposomes; NTA, Nanoparticle Tracking Analysis; PEG, Polyethylene Glycol; PLS, Partial Least Squares; PdI, Polydispersity Index; PIC, Pressure Indicator Controller; Pw, Working pressure; QbD, Quality by Design; RGD, Arginine-Glycine-Aspartic acid; SAXS, Small-Angle X-ray Scattering; SbD, Safe by Design; SDS-PAGE, Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis; S-MLS, Static Multiple Light Scattering; TFF, Tangential Flow Filtration; TGX, Trys-Glycine eXtended; TIC, Temperature Indicator Controller; TSI, Turbiscan Stability Index; Tw, Working temperature; USP, United States Pharmacopeia; XCO2, Carbon dioxide molar fraction
Graphical Abstract
Abstract
Fabry disease is a lysosomal storage disease arising from a deficiency of the enzyme α-galactosidase A (GLA). The enzyme deficiency results in an accumulation of glycolipids, which over time, leads to cardiovascular, cerebrovascular, and renal disease, ultimately leading to death in the fourth or fifth decade of life. Currently, lysosomal storage disorders are treated by enzyme replacement therapy (ERT) through the direct administration of the missing enzyme to the patients.
In view of their advantages as drug delivery systems, liposomes are increasingly being researched and utilized in the pharmaceutical, food and cosmetic industries, but one of the main barriers to market is their scalability.
Depressurization of an Expanded Liquid Organic Solution into aqueous solution (DELOS-susp) is a compressed fluid-based method that allows the reproducible and scalable production of nanovesicular systems with remarkable physicochemical characteristics, in terms of homogeneity, morphology, and particle size. The objective of this work was to optimize and reach a suitable formulation for in vivo preclinical studies by implementing a Quality by Design (QbD) approach, a methodology recommended by the FDA and the EMA to develop robust drug manufacturing and control methods, to the preparation of α-galactosidase-loaded nanoliposomes (nanoGLA) for the treatment of Fabry disease.
Through a risk analysis and a Design of Experiments (DoE), we obtained the Design Space in which GLA concentration and lipid concentration were found as critical parameters for achieving a stable nanoformulation. This Design Space allowed the optimization of the process to produce a nanoformulation suitable for in vivo preclinical testing.
1. Introduction
One of the main challenges to produce advanced materials such as enzyme-loaded liposomes is the successful implementation of new manufacturing nanotechnologies at industrial scale. The DELOS-susp method is a compressed fluid-based technology that allows the reproducible preparation of different nanovesicular systems with remarkable physicochemical characteristics, in terms of homogeneity, morphology and particle size, as well as a high versatility to integrate different active compounds, that can lead to innovative nanomedicines with enhanced efficiency [1], [2], [3].
Lysosomal storage disorders (LSD) are diseases caused by lysosomal dysfunction usually as a consequence of deficiency of a single metabolic enzyme. In the case of Fabry disease, the lack of α-galactosidase A (GLA) enzyme causes the accumulation of glycosphingolipids leading to multiple organ pathology [4]. Enzyme replacement therapy (ERT), which is the main current treatment, exhibits several drawbacks mainly related to protein instability and low efficacy [5].
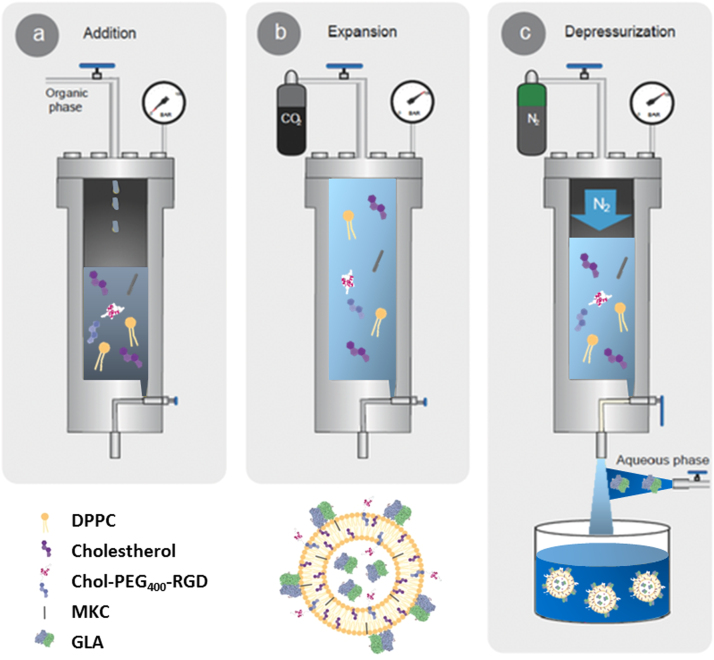
To overcome the main limitations of the current ERTs (e.g. rapid enzyme degradation, high immunogenicity, short circulation half-life, poor biodistribution, and limited efficacy among others [6]) a strategy under development consists in the encapsulation of the GLA enzyme into nanoliposomes. Such encapsulation aims to protect the enzyme, decrease the degradation and immunogenicity of GLA, and increase its plasma half-life. Moreover, the addition of a targeting peptide on the liposomal membrane can facilitate cellular up-take via peptide receptors recognition and could help to improve GLA biodistribution. Thus, GLA-loaded nanoliposomes (nanoGLA) functionalized with Arginine-Glycine-Aspartic acid (RGD) peptide and prepared by DELOS-susp, have recently been reported to increase enzymatic activity and intracellular penetration, showing the great potential of this CO2-based methodology for the simple production of protein-nanoliposome therapeutic conjugates [4], [7]. Fig. 1 shows the DELOS-susp process and the starting materials used to obtain the nanoGLA liposomal formulation.
Fig. 1.
Schematic representation of the DELOS-susp method for the preparation of GLA loaded nanoliposomes (nanoGLA). The procedure includes: a) Loading of an organic solution containing the liposome membrane components (Cholesterol, DPPC, Cholesterol-PEG400-RGD, MKC) into the high pressure vessel; b) Addition of compressed CO2 until a certain pressure to produce a CO2-expanded solution, where all membrane components remain dissolved in a single liquid phase; and c) Depressurization of the expanded solution over an aqueous solution containing the GLA enzyme at atmospheric pressure, obtaining the protein-liposomes nanoconjugates.
Figure adapted from [4].
The results obtained in earlier stages of nanoGLA development, although being biocompatible and showing interesting in vitro activity, presented some limitations regarding formulation characteristics for continuing with their translation towards the clinics. In first place, the concentration of GLA enzyme we could load into the nanoliposomes, with robust particle size and PdI, was around 20 µg/mL, far below the minimum required for reaching those doses that will allow to perform efficacy studies in mice (200 µg/mL). Besides, the enzyme entrapment efficiency in the liposomes was below 40% [4]. The addition of cationic surfactant myristalkonium chloride (MKC) into the formulation in low concentration allowed to increase the drug entrapment efficiency as well as the colloidal stability of the system [7]. However, further work was required in order to obtain a stable formulation with the proper protein concentration.
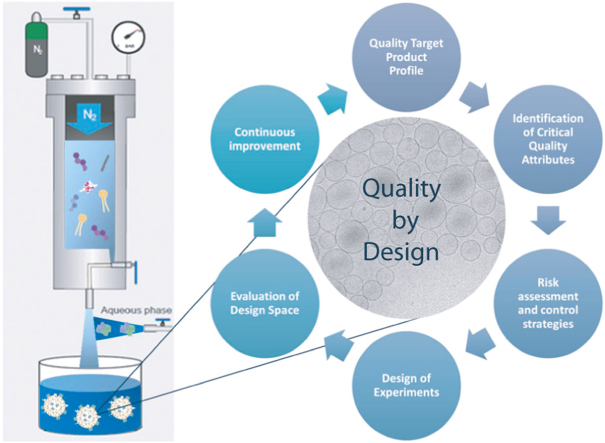
In this work we applied the Quality by Design (QbD) approach to determine formulation and process parameters that could have an important impact on nanoGLA attributes when prepared by DELOS-susp. The QbD methodology is described in Guidelines of the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) [8], [9], [10] and it is strongly encouraged by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) to develop robust drug manufacturing and control methods. Pharmaceutical QbD is a systematic approach to development that aims to ensure the quality of medicines, identifying characteristics that are critical to quality from the patient’s perspective, translating them into the product Critical Quality Attributes (CQAs), and establishing a relationship between manufacturing variables and CQAs through a Design of Experiments (DoE). The goals of the pharmaceutical QbD may include the achievement of meaningful product quality specifications as well as the reduction of product variability and defects, to increase efficiencies and to enhance root cause analysis [11].
Another systematic methodology used in pharmaceutical design is the Safe by Design (SbD) approach, which aims to eliminate possible risks to the human health and the environment at early innovation stages, i.e., during the design or planning stage [12]. SbD requires a full life cycle perspective combined with nanomaterial sustainability (environmental, social, and economic) [13] improvements, to avoid the shifting of potential risks across development stages.
In this framework, we report here the implementation of the QbD approach to develop a robust process for the preparation of nanoGLA. In the present work, the process was scaled-up from previous work [4] by a 7-fold factor, and the liposomal prototype was optimized increasing 10-times (i.e. from around 20 to 200 µg/mL) the final concentration of GLA enzyme in the nanoformulation. Its physicochemical stability was improved while meeting the defined quality specifications in order to fulfill the required therapeutic doses for advanced preclinical testing. Besides, a SbD evaluation of the nanoGLA manufacturing process by DELOS-susp is also presented.
2. Materials and methods
2.1. Materials
GLAcmycHis protein (GLA) was synthetized as explained in Appendix A. 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) was purchased from Corden Pharma (Liestal, Switzerland), cholesterol was purchased from Panreac Química (Castellar del Vallès, Spain), and myristalkonium chloride (MKC) from United States Biological (Salem, US). Cholesterol pegylated with arginyl–glycyl–aspartic acid peptide (Chol-PEG400-RGD) was synthetized as described by E. Cristóbal-Lecina et al. [14]. Ethanol (EtOH) was purchased from Scharlab (Sentmenat, Spain), dimethyl sulfoxide (DMSO) from Sigma-Aldrich (St. Louis, US), carbon dioxide (CO2) and nitrogen (N2) from Carburos Metálicos (Cornellà de Llobregat, Spain). Diafiltration column modules of 300 kDa cut-off were purchased from Repligen (Waltham, US). Ultrapure Type I water (H2O), purified using Milli-Q Advantage A10 equipment from Millipore Corporation (Burlington, US), was used for all studies.
2.2. Methods
2.2.1. NanoGLA preparation
DELOS-susp methodology was used for the nanoGLA preparation (Fig. 1). An EtOH:DMSO solution (volume ratio EtOH:DMSO 4:1) containing DPPC (from 0.86 to 3.46 mg/mL), cholesterol (from 0.30 to 1.24 mg/mL), Chol-PEG400-RGD (from 0.03 to 0.31 mg/mL), and MKC (from 0.02 to 0.07 mg/mL) at a working temperature (Tw) of 308 K was loaded into a 50 mL high pressure vessel at atmospheric pressure. The solution was, then, volumetrically expanded with compressed CO2 until a molar fraction (XCO2) of 0.55 was achieved, reaching a working pressure (Pw) of 9 MPa. The system was kept at 308 K and 9 MPa under stirring for approximately 15 min to achieve a complete homogenization and to attain thermal equilibration. A given volume of GLA stock solution at 0.28 mg/mL, measured by SDS-PAGE, was dissolved in ultrapure water to reach the desired enzyme concentration (7, 17 or 27 µg/mL). To form the nanoconjugates, the volumetrically expanded organic phase was depressurized over the aqueous solution containing the GLA. In this way, nanoGLA DELOS-susp batches of 290–500 mL were obtained. In this step, a flow of compressed N2 was used as a plunger to push down the CO2‐expanded solution from the vessel and to maintain a constant pressure of 10 MPa inside the vessel during depressurization. Average time per experiment was 60 min and the resulting dispersions of nanoconjugates were stored at 4 °C until their characterization. The process parameters that were kept constant for all experiments are listed in Appendix B.
2.2.2. Separation of the free GLA from the nanoGLA liposomal dispersion by diafiltration and concentration process
The non-conjugated GLA enzyme (with a molecular weight between 90 and 110 kDa in its dimer form) was separated from the GLA-loaded liposomes obtained by DELOS-susp using a KrossFlo Research IIi Tangential Flow Filtration (TFF) System with mPES hollow fiber modules of 300 kDa cut-off, in order to determine the GLA entrapment efficiency (EE) [4]. A volume of 10 mL of liposomal sample was added to the sample container, which was connected to the column and the buffer reservoir. Six diafiltration cycles were performed with ultrapure Type I water to eliminate the free molecules and the organic solvent present in the liposomal samples.
Concentration of the intermediate nanoGLA during the optimization step was performed using the same equipment, materials, and conditions. A volume of 50 mL of diafiltrated sample was added to the sample container and submitted to a 7.5 concentration factor, evacuating ca. 87% of water.
2.2.3. Characterization of liposomes
2.2.3.1. Mean particle size, PdI and ζ-potential
Liposomal mean size, polydispersity index (PdI) and ζ-potential were determined by Dynamic Light Scattering (DLS), using a Zetasizer Nano ZS analyzer (Malvern Instruments, Malvern, UK). The measurements were performed at 20 °C with a scattering angle of 173°, immediately after liposomes’ preparation, and 1 and 2 weeks after the production.
2.2.3.2. Turbiscan Stability Index
Turbiscan Stability Index (TSI) was determined by Static Multiple Light Scattering (S-MLS), using a Turbiscan Lab Expert (Formulaction, Toulouse, France), at 20 °C and 2 weeks after liposomes production.
2.2.3.3. Morphology
The morphology of the systems was studied using Cryogenic Transmission Electron Microscopy (Cryo-TEM). Images were obtained with a FEI Tecnai G2 12 microscope (FEI, The Netherlands) operating at 120 kV, as described in [15]. The analysis was performed 1 week after samples production.
2.2.3.4. Uni-lamellarity
Quantitative information on the liposomes degree of lamellar structure and bilayer thickness was investigated with small-angle X-ray scattering (SAXS) on an optimized NanoSTAR SAXS instrument (Bruker AXS, Karlsruhe, Germany) equipped with a liquid Ga metal jet X-ray source (Excillum AB, Sweden) and scatterless slits. The wavelength was λ = 1.34 Å and the measurements were performed 1 week after production. The data were analyzed by a model similar to the one described in [16], [17]. The model was expressed so that the fraction of bilayers presented as single-layered vesicles, fsingle, was a fit parameter.
2.2.3.5. Entrapment efficiency
The entrapment efficiency (EE) was determined by comparing the amount of enzyme encapsulated in the nanovesicles after removal of free enzyme by diafiltration (Loaded prototype) with the amount of initial GLA present in the DELOS-susp batch just after its production (Total prototype), see Eq. (1) (Entrapment efficiency calculation):
| (1) |
To detect and quantify the enzyme in each of these samples, SDS-PAGE was performed by using gels TGX Stain-Free™ FastCast™ acrylamide 12% (Bio-Rad, ref. 161-0185). To visualize the fluorescent bands, a ChemiDoc™ Touch Imaging System (Bio-Rad) was used. Samples and standards to be quantitatively compared were run in the same gel and processed as a set. Densitometric analysis of the bands were performed with the Image Lab™ software (version 5.2.1., Bio-Rad).
2.2.3.6. Enzymatic activity
The α-galactosidase specific enzymatic activity (EA) of liposomal formulations was assayed using 4-Methylumbelliferyl α-D-galactopyranoside (4-MUG, Sigma Chemical) as substrate, at a concentration of 2.46 mM in assay buffer (0.01 M acetic acid, pH 4.5) as previously described [18]. The released product (4-methylumbelliferone) was determined by fluorescence measurement (λexc = 365, λem = 450 nm) and final specific enzymatic activities were shown as 4-methylumbelliferone μmol per time and protein quantity. Measurements were performed 1 week after production of the samples.
2.2.4. Experimental design and data processing
An experimental design with 4 factors at 2 levels with 2 central points (resulting in a total of 10 experimental runs) was constructed in order to study the influence of formulation parameters (independent variables or factors) on the properties (dependent variables or responses) of nanoGLA dispersion. The selected experimental design was a Fractional Factorial design, and it was developed using Modde 12 Pro software (Umetrics, Sweden). Data fitting and calculation of statistical parameters were performed by partial least squares (PLS) method. The experimental design used in this study allowed fitting the data with a linear regression interaction model. The acceptance of the responses obtained was evaluated by means of the statistical parameters predicted by the software R2, Q2 model validity, and model reproducibility. The design space for nanoGLA was determined using Design Space Explorer option from the Optimizer module of Modde 12 Pro software.
3. Results and discussion
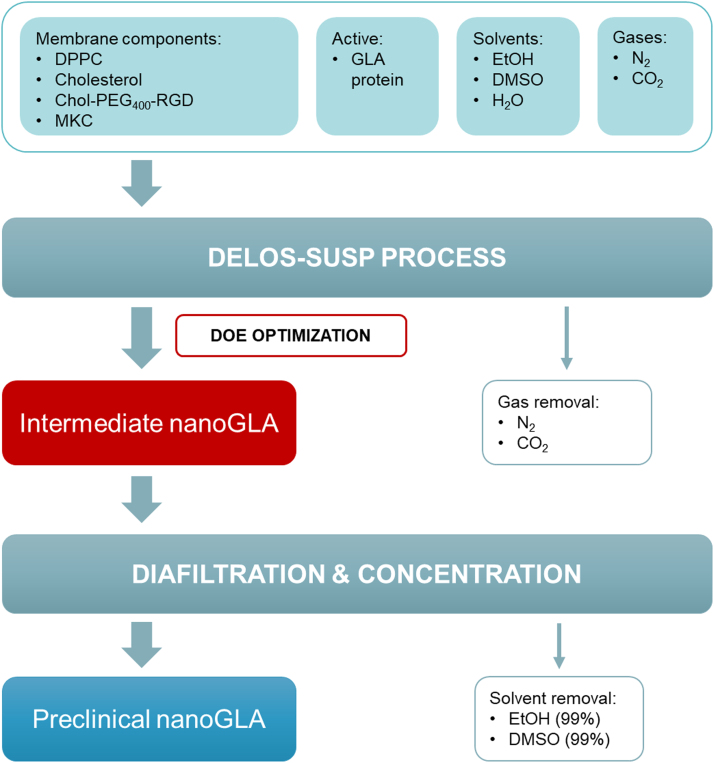
The process flow diagram of the whole nanoGLA optimized manufacturing process is presented in Fig. 2. Membrane components, solvents, and gases, as well as the GLA protein, were processed using DELOS-susp (see Section 2) obtaining an intermediate nanoGLA dispersion, whose quality was studied by QbD. Then, the optimized intermediate nanoGLA was diafiltrated and concentrated by TFF in order to remove the organic solvents and to reach the target enzyme concentration, which cannot be directly achieved in the DELOS-susp step because of the limited solubility of the liposome membrane components in the organic phase, while keeping the nanoformulation quality.
Fig. 2.
Process Flow Diagram to produce the nanoGLA. Raw materials and the materials leaving the process are also represented. In red, the intermediate nanoGLA formulation obtained by DELOS-susp that has been studied in the QbD implementation. Diafiltration and concentration processes are necessary to remove the remaining free GLA and organic solvents from the nanoformulation and to maximize the enzyme concentration in the final nanoformulation for preclinical studies, respectively.
3.1. Identification of the Critical Quality Attributes (CQAs)
During the development of liposomal drug products, the identification and pertinent characterization of CQAs of liposomal drug products is one of the main challenges from the quality point of view, together with the definition of proper control strategies. Liposomal products are complex formulations and small changes in their physicochemical attributes can have notable effects in their in vivo performance. Thus, a suitable definition of CQAs and control strategies may allow a faster and more efficient drug product development [19]. Table 1 shows the CQAs selected for QbD implementation related to the intermediate nanoGLA liposomal dispersion. The definition of the quality attributes has considered the ICH recommendations, as well as FDA and EMA guidelines [10].
Table 1.
CQAs defined for the intermediate nanoGLA liposomal dispersion obtained by DELOS-susp and their justification.
| Quality Attribute | Justification [19], [20], [21] |
|---|---|
| Macroscopic Appearance | It must be a homogeneous opalescent dispersion without sedimentation. Sedimentation could indicate poor colloidal stability. |
| Mean Particle Size | Particle size and particle size distribution are major CQAs for nanoparticle-based systems, playing an important role in determining their in vivo absorption and distribution, drug loading, drug release, and targeting ability. Therefore, robust control of particle size is one of the crucial parameters for further in vivo application of liposomal drug products. Values below 300 nm are considered acceptable for nanoGLA. |
| Polydispersity Index (PdI) | PdI reflects the heterogeneity of the particle size, indicating how wide is the particle size distribution. The lower the PdI, the higher the homogeneity of the dispersion. To meet specification, PdI should be below 0.45. |
| ζ-potential | Important parameter in the evaluation of colloidal system’s stability. Particles with a high negative or positive ζ-potential value repel each other, indicating that the colloidal system is stable. On the contrary, decreasing the ζ-potential value to nearly neutral could lead to liposomal aggregation. The liposome surface charge can also influence drug loading, cellular uptake, tissue distribution, and clearance. ζ-potential values higher than + 30 mV are considered inside the specification range for nanoGLA. |
| Particle morphology and lamellarity | Vesicles must be spheroidal and, mostly, uni-lamellar. Lamellarity can affect drug loading and release, thus, impacting the enzyme delivery. |
| GLA Entrapment Efficiency (EE) /Free drug substance | Free drug substance may have side effects and impact into pharmacokinetic profile. Besides, a high and reproducible percentage of drug entrapment could reduce manufacturing costs and increase drug concentration in the final formulation allowing greater flexibility in dosing. Depending on the pharmacokinetics, higher drug concentration can result in increased dosing intervals and hence improved patient compliance. |
| Specific Enzymatic Activity (EA) | The bioactivity of the integrated enzyme must be preserved in the nanoformulation. EA ratio to control should be higher than 0.8, referred to the EA of a commercially available GLA (agalsidase alfa, Replagal®). |
| Integration efficiency of Chol-PEG400-RGD in the vesicular membrane | The amount of targeting peptide moiety integrated in the nanoliposomal membrane must be in the proper ranges to allow the nanoGLA to interact with cells and facilitate intracellular penetration. |
| pH | pH can affect dispersion stability, drug loading and release, and cell uptake among others. A suitable pH range is from 6.0 to 7.0. |
| Dispersion stability | The intermediate formulation must be stable at least until the diafiltration and concentration step. In terms of Turbiscan stability, the TSI should be less than 10.0 at 24 h. |
| Lipid and GLA degradation products | Chemical stability of the lipid components in the liposome as well as the chemical stability of the contained drug substance is important. These will be considered at further stages of development. |
As a parenteral dosage form, liposomal products must be sterile and pyrogen-free. These parameters are not considered as CQAs at this stage of formulation development since it is an intermediate product, but they must be considered as CQAs in the final formulation.
3.2. Risk assessment and control strategies
Risk assessment is a valuable science-based process used in quality risk management that can aid in identifying which material attributes and process parameters potentially influence product CQAs [11], [22]. The potential risk factors which might influence the quality of the product were identified through risk analysis, creating the Ishikawa diagram for each CQA defined for nanoGLA (see Appendix C, where the detailed risk analysis assessment is presented). As a result, 4 formulation factors were considered relevant to be included in the subsequent DoE analysis: GLA concentration, lipid concentration, peptide content in the liposomal membrane, and ethanol concentration. The justification regarding the selection of these factors and their range of investigation is detailed in Table 2.
Table 2.
Factors included in the DoE to optimize the intermediate nanoGLA formulation, their potential impact into CQAs and the ranges of investigation.
| Factor | Potential impact into CQAs | Factor Range studied in DoE |
|---|---|---|
| GLA concentration | Protein concentration plays a key role on dispersion stability. It has been reported that increasing the loading and charge of protein will impact on size distribution and aggregation rate of the liposomal system [23]. | From 7 to 27 µg/mL of enzyme. Defined according to the viability of performing the subsequent diafiltration and concentration step up to the 200 µg/mL required for in vivo doses. |
| Lipid concentration | Besides the molar ratio between membrane components (cholesterol and phospholipid), the total lipid concentration in liposomal systems will impact on its mean size, particle distribution and stability, as well as on its loading capacity [24], [25], [26], [27]. | From 1.2 to 5.0 mg/mL, moving almost 2 mg/mL up and down of previously tested concentrations in similar nanoformulations (3.0 mg/mL). |
| Chol-PEG400-RGD molar ratio | PEGylation of liposomes improves not only the stability and circulation time, but also the passive targeting ability on tumoral tissues, through a process known as the enhanced permeation retention effect, able to improve the therapeutic effects and reduce the toxicity of encapsulated drug [28]. | From 1% to 3% mol of molar ratio in relation to the total amount of lipid components, according to previous studies. |
| EtOH concentration | Solvent concentration could have a direct impact on shape, solubility and electrostatic interactions between the liposomal bilayer and the loaded protein [29]. | From 5% to 10% v/v, folding the standard 5% v/v since higher quantities are not recommended for intravenous administration [4]. |
Although the depressurization flow rate during the DELOS-susp could be considered as a factor [30], it was not included in the present work since an accurate flow rate range variation could not be carried out in the used set-up. Due to limitations of the available in-house synthesized starting materials (GLA and Chol-PEG400-RGD), an intermediate laboratory plant was selected for performing the DoE. Thus, for this first QbD study the depressurization flow rate was kept constant at 6.3 ± 0.7 g/min because it can be controlled within narrow values where the formulation requirements are ensured. So, despite not being able to introduce the flow rate as a factor in the present study, this parameter was defined as a controlled variable that was kept constant for all experiments. QbD is based on the principle of continuous improvement, thus, by gathering knowledge during early development, it is possible to avoid potential manufacturing problems as the drug product evolves through the development pipeline. Thus, the knowledge obtained in the present study will allow to concisely define further scale-up studies. The influence of DELOS-susp depressurization flow rate variation on the nanoGLA quality will be more accurately evaluated in future studies at pilot plant scale, where it is possible to control and register this parameter. Taking the present results as the basis for refining the Design Space of the nanoGLA will ease this transition from the lab to the pilot plant.
3.3. Experimental design and development
An experimental design was constructed to study the influence of the formulation (independent variables or factors) on the physicochemical properties (dependent variables or responses) of intermediate nanoGLA obtained by DELOS-susp, as shown in Table 3. Low and high levels for each factor (Table 2) were selected based on previous experiments and studies [4], [7].
Table 3.
Matrix of experimental design. The sample codes consider the randomized order of experiments.
| Experimental run | GLA concentration (µg/mL) | Lipid concentration (mg/mL) | Chol-PEG400-RGD molar ratio to lipids (% mol) | EtOH concentration (% v/v) |
|---|---|---|---|---|
| DOE-001 | 27 | 1.2 | 1 | 10.0 |
| DOE-002 | 7 | 1.2 | 3 | 10.0 |
| DOE-003 | 17 | 3.1 | 2 | 7.5 |
| DOE-004 | 27 | 5.0 | 1 | 5.0 |
| DOE-005 | 7 | 5.0 | 3 | 5.0 |
| DOE-006 | 27 | 1.2 | 3 | 5.0 |
| DOE-007 | 17 | 3.1 | 2 | 7.5 |
| DOE-008 | 7 | 1.2 | 1 | 5.0 |
| DOE-009 | 27 | 5.0 | 3 | 10.0 |
| DOE-010 | 7 | 5.0 | 1 | 10.0 |
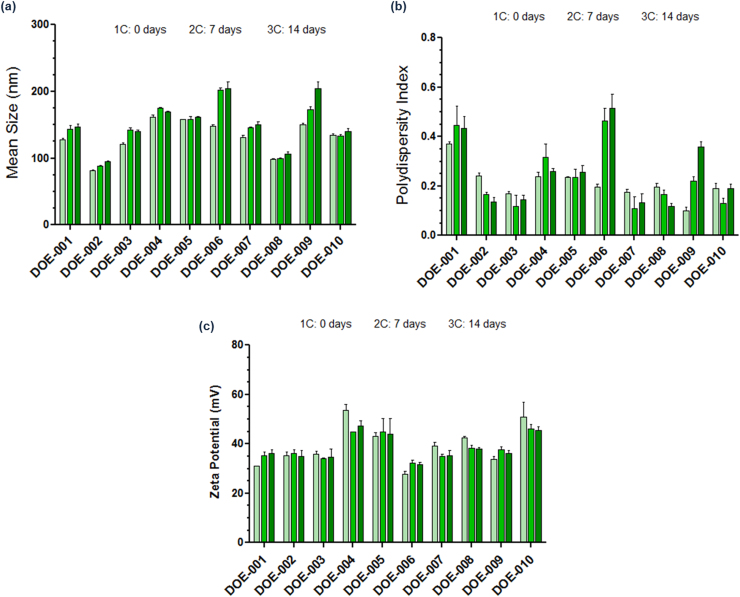
All process parameters, including depressurization flowrate at 10 g/min, were kept constant for all experiments. The responses of the experimental design were the CQAs of the intermediate nanoGLA dispersion: particle mean size, polydispersity index, ζ-potential, Turbiscan stability index, ratio of monolayered liposomes, entrapment efficiency, enzymatic activity, and pH. Furthermore, macro and microscopic appearance (i.e. particle morphology) were analyzed, but these two CQAs were not included in the design because of their qualitative nature. The matrix of the experimental design is presented in Table 3.
All experiments were produced following DELOS-susp method at the laboratory scale, as explained in Section 2. All 10 experiments were produced in 2 consecutive days.
3.4. Characterization of the intermediate nanoGLA obtained
A high variability on the stability of the samples was observed among the different experimental conditions tested. Thus, some characterizations were performed at the production day (t = 0) and followed until 14 days.
Regarding the macroscopic appearance, all DELOS-susp batches presented a homogenous, whitish, opalescent appearance without presence of sediment, except for DOE-001, DOE-006, and DOE-009.
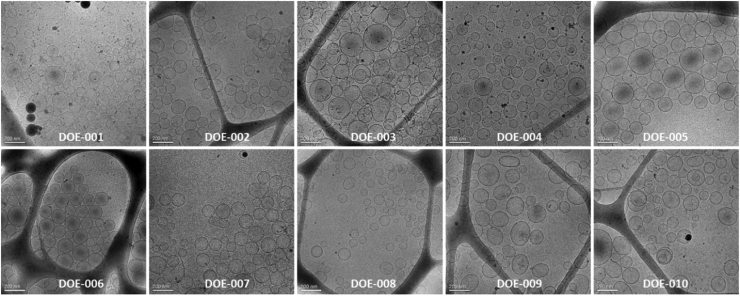
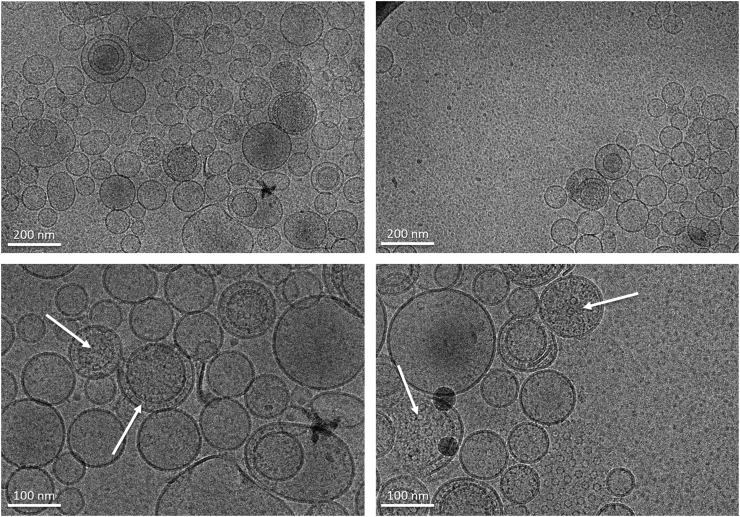
A summary of the microscopy analysis of the nanoGLA samples by cryo-TEM is presented in Fig. 3, indicating that for all experiments nanovesicles were obtained, being mostly uni-lamellar and spherical, and ranging between 100 and 200 nm, with the presence of some larger vesicles around 500 nm.
Fig. 3.
Representative cryo-TEM images of nanoGLA DoE samples at 1 week after production. Scale bar 200 nm.
Table 4 shows a summary of the characterization results of the DoE samples, which were considered for the statistical analysis. The three out of ten batches (DOE-001, DOE-006, and DOE-009) that presented small sedimentation also showed a higher TSI indicating the dispersion instability.
Table 4.
Characterization of intermediate nanoGLA CQAs after 1 week of production. Uncertainties are calculated from the standard deviation of measurement for Size, PdI, ζ-potential, and EE; and from standard error of the mean for uni-lamellarity and EA.
| Sample ID | Size (nm) | PdI | ζ-potential (mV) | TSI | Uni-lamellarity, fsingle | EE (%) | EA (ratio to control) |
|---|---|---|---|---|---|---|---|
| DOE-001a | 143 ± 6 | 0.45 ± 0.08 | 35 ± 2 | 11.5 | 0.88 ± 0.01 | 60 ± 1 | 1.7 ± 0.1 |
| DOE-002 | 88 ± 2 | 0.17 ± 0.01 | 36 ± 2 | 0.6 | 0.99 ± 0.01 | 53 ± 1 | 1.2 ± 0.0 |
| DOE-003 | 142 ± 3 | 0.12 ± 0.05 | 34 ± 0 | 1.2 | 0.95 ± 0.00 | 59 ± 1 | 1.4 ± 0.0 |
| DOE-004 | 175 ± 1 | 0.32 ± 0.05 | 45 ± 0 | 8.2 | 0.93 ± 0.00 | 45 ± 1 | 1.6 ± 0.1 |
| DOE-005 | 158 ± 4 | 0.24 ± 0.03 | 45 ± 6 | 1.9 | 0.93 ± 0.00 | 42 ± 1 | 0.8 ± 0.1 |
| DOE-006a | 202 ± 4 | 0.46 ± 0.05 | 32 ± 1 | 6.6 | 0.85 ± 0.02 | 68 ± 1 | 1.5 ± 0.0 |
| DOE-007 | 146 ± 2 | 0.11 ± 0.05 | 35 ± 1 | 1.2 | 0.99 ± 0.01 | 53 ± 1 | 1.2 ± 0.2 |
| DOE-008 | 100 ± 1 | 0.17 ± 0.02 | 38 ± 1 | 1.8 | 1.00 ± 0.01 | 84 ± 1 | 0.8 ± 0.0 |
| DOE-009a | 172 ± 5 | 0.22 ± 0.02 | 38 ± 1 | 12.0 | 0.94 ± 0.01 | 53 ± 1 | 1.1 ± 0.1 |
| DOE-010 | 134 ± 2 | 0.13 ± 0.02 | 46 ± 2 | 0.6 | 0.93 ± 0.00 | 84 ± 1 | 1.4 ± 0.1 |
DOE-001, DOE-006, and DoE-009 presented sedimentation.
Besides, samples in which aggregation and sedimentation were observed presented also higher size and PdI, and lower uni-lamellarity. Furthermore, the mean particle size and PdI of DOE-001, DOE-006, and DOE-009 significantly increased with time after production (Appendix D). On the other side, ζ-potential, EE, and EA values did not correlate with macro and microscopical characteristics of the samples.
All the formulations showed enzymatic activity, varying from 0.8 to 1.7 ratio to control referred to the enzymatic activity of a commercially available free GLA (Agalsidase alfa, Replagal® from Shire-Takeda) included in the analysis, as also shown in Table 4. These results indicate an excellent enzymatic activity in all cases, compliant with preliminary specifications.
3.5. Analysis of the influence of the factors on CQAs
The influence of the factors was studied for the rest of dependent variables or CQAs. An analysis and discussion for each quantifiable CQA is described below. From the characterized CQAs, mean particle size, ζ-potential, and uni-lamellarity could be statistically fitted and were included in the prediction model (see Appendix E for modeling and data fitting) while for the rest of attributes a bad correlation was obtained and thus a poor predictive ability of the model.
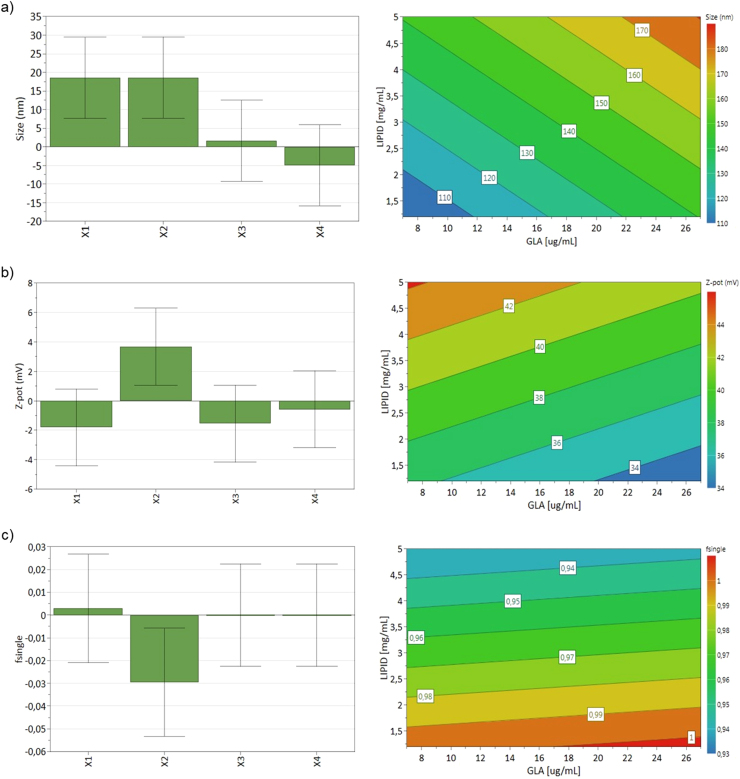
According to the data presented in Table 4, the mean particle size of the nanoGLA dispersions varied from 90 to 200 nm. These differences could be explained because of the aggregation and fusion of liposomal vesicles that could occur 1 week after preparation. The coefficients of the equation describing the influence of the factors on nanoGLA particle size are shown in Fig. 4a (left). According to this figure, the size of nanoGLA liposomes is significantly influenced by GLA concentration (X1) and the lipid concentration (X2). Thus, a significant increase in size is obtained when the concentration of GLA and lipid increase. The increase in size with the increase of GLA and lipid concentrations could be explained by the occurrence of physical aggregation of the vesicles. The influence of GLA and lipid concentration is also illustrated by the contour plot for size in Fig. 4a (right).
Fig. 4.
Left side: scaled and centered coefficients of the regression equations describing the influence of formulation parameters X1-GLA concentration, X2-lipid concentration, X3-Chol-PEG400-RGD molar ratio, X4-EtOH concentration, on the liposomes size, ζ-potential, and uni-lamellarity of nanoGLA intermediate dispersion. Right side: contour plots for the same CQAs of the nanoGLA intermediate dispersion. Molar ratio of chol-PEG400-RGD to lipid and EtOH concentration were kept constant at 2% mol and 7.5% v/v, respectively.
As shown in Table 4, ζ-potential ranged from 32 to 46 mV for non-diafiltrated liposomes. The influence of studied factors on ζ-potential is shown as coefficients of the regression equation plot in Fig. 4b (left). According to this plot, lipid concentration (X2) was the only factor that had a significant positive effect on the ζ-potential of the dispersions obtained. This could be explained because when lipid concentration increases, MKC surfactant (used as excipient in the nanoformulation) concentration also increases, giving more positive charges to the liposomal system. The influence of lipid concentration is also illustrated by the contour plot for size in Fig. 4b (right).
Monolayered liposomes fractions varied from 0.88 to 1.00, as shown in Table 4 by fsingle values. According to Fig. 4c (left), lipid concentration was the only factor affecting this CQA significantly, showing a larger difference in comparison to the rest of factors. As can be seen in Fig. 4c (right), GLA concentration had an insignificant influence on the monolayer ratio of liposomes, but it was strongly affected by the lipid concentration. The explanation can be the same as the effect of lipid concentration on vesicle size: aggregation and fusion of the vesicles when lipid concentration increases could be responsible of this behavior.
3.6. Evaluation of the design space
Among all the factors studied, GLA concentration and lipid concentration were found to have a significant influence on some of the responses. Thus, the Design Space for intermediate nanoGLA was constructed using these factors which significantly influenced the quality of the nanoformulation. On the one hand, increase of GLA and lipid concentration had a negative influence on liposomal mean particle size because of the size increase promoted by aggregation. On the other hand, as the lipid concentration increased, and, consequently, the amount of MKC surfactant was higher, ζ-potential (which is related to the electrical charge at the liposomes surface) also increased due to the formal positive charge provided by this cationic surfactant. Nevertheless, this ζ-potential increase was not correlated with a higher colloidal stability of the nanoformulation as also confirmed by an uni-lamellarity decrease. It seems that this MKC amount is not enough to stabilize such highly concentrated samples. However, higher MKC concentrations, although could improve the stability of these concentrated samples, it also could entail a toxicological concern as well as the abolishment of the enzymatic activity of the GLA, as reported by Tomsen-Melero et al. [7].
The other CQAs such as polydispersity index, TSI, GLA entrapment efficiency, and enzymatic activity could not be fitted in any reliable model because of the sedimentation of some batches and the high characterization variability, due to the use of SDS-PAGE plus TGX as a reference method for GLA quantification. Results could be improved in the future using a HPLC method for the quantification of GLA. Hence, the Design Space was determined using particle size, ζ-potential, and uni-lamellarity.
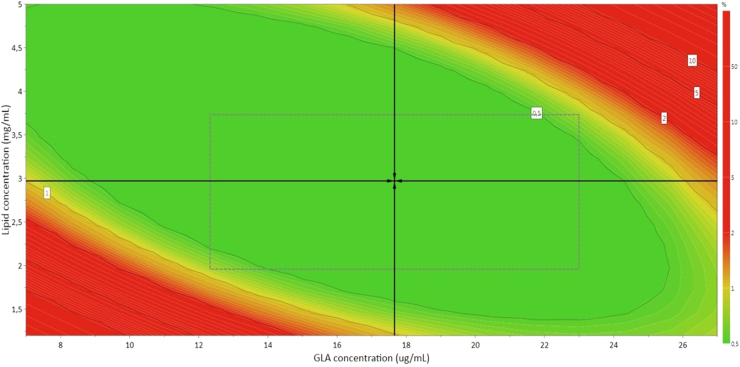
The Design Space obtained is shown as the green region in Fig. 5, and shows the combination of factors for which the nanoGLA intermediate obtained by DELOS-susp will meet the specifications in terms of CQAs, specified in Table 5, with a probability of failure less than 1%. The combination inside the design space which is pointed out by the black arrows indicates the robust setpoint (17.7 µg/mL GLA concentration, 2.97 mg/mL lipid concentration, 1.3% mol Chol-PEG400-RGD, and 6.2% v/v EtOH) corresponding to the formulation for which the prediction errors are the lowest.
Fig. 5.
The Design Space for intermediate nanoGLA liposomal dispersion that meets the specifications in terms of CQAs, expressed as the probability of failure (%). Molar ratio of chol-PEG400-RGD to lipid and EtOH concentration have been optimized in the run at 1.16% mol and 6.2% v/v, respectively.
Table 5.
The desired CQAs of nanoGLA introduced in the Design Space explorer and their values.
| Response | Criterion | Min. | Target | Max. |
|---|---|---|---|---|
| Size (nm) | Target | 50 | 150 | 250 |
| ζ-potential | Excluded | – | – | – |
| Uni-lamellarity (fsingle) | Maximize | 0.95 | 1.00 | – |
To our knowledge, there are no similar results reporting the relationship between lipid and protein concentration affecting size, ζ-potential, and uni-lamellarity in this way. We could expect that at low concentration of lipid and GLA, we would also obtain a nanoGLA within the established acceptance limits, so it could be possible that the red zone showed in Fig. 5 around the lowest concentrations belongs to a range that remained unexplored. In addition, the finding of a model which indicates that formulation will escape from the acceptance limits if both factor concentrations are high is due to aggregation effect of liposomes caused by a change of interaction between them, provoked by the high load of components in the membrane [24], [31].
3.7. Optimization of the nanoGLA formulation
The challenge was to obtain a stable nanoGLA concentrated formulation containing 200 μg/mL of GLA, suitable doses for in vivo testing. To do so, a batch of nanoGLA was prepared by DELOS-susp and diafiltrated and concentrated following the trend of the preliminary Design Space: the lipid concentration was reduced to 1.2 mg/mL so that the GLA concentration could be increased up to 35 µg/mL, obtaining an intermediate nanoGLA by DELOS-susp meeting all the CQAs.
Physicochemical properties are shown in Table 6 and Fig. 6. Particle mean size, polydispersity index, ζ-potential, and GLA concentration – all meet the requirements to go to future in vivo preclinical trials (<300 nm, ≤0.45,>30 mV, and>200 μg GLA/mL, respectively). A good colloidal stability was observed for the concentrated final nanoGLA, composed mainly by single lamellar liposomes (Fig. 6). Some structural effects were noticeable by cryo-TEM after over-concentrating the GLA-loaded liposomes. Images revealed the formation of some multilamellar complexes, where the GLA can be identified between liposomal layers as well as inside the vesicles (Fig. 6, arrows), forming structures resembling lipoplexes [32]. Interestingly, the high concentration of GLA (330 µg/mL) after the concentration process where these structures were identified was quite above the minimum required value for in vivo trials (200 µg/mL).
Table 6.
Critical Quality Attributes for optimized intermediate nanoGLA, diafiltrated and concentrated around 10-fold.
| Attributes | After DELOS-susp (Intermediate nanoGLA) | After concentration (Preclinical nanoGLA) |
|---|---|---|
| Stability | NA | >2 weeks |
| Particle mean size (nm) | 138 ± 7 | 165 ± 3 |
| Polydispersity Index | 0.38 ± 0.03 | 0.41 ± 0.02 |
| ζ-potential (mV) | 40 ± 1 | 35 ± 1 |
| Enzymatic activity (ratio to control) | 1.11 ± 0.08 | 0.93 ± 0.04 |
| GLA (µg/mL) | 34 ± 2 | 330 ± 20 |
Fig. 6.
Cryo-TEM images of optimized nanoGLA prototype after concentration. The images were acquired 2 weeks after production.
In addition, the final prototype dispersion was stable for more than 2 weeks.
3.8. Safe-by-Design implementation
The implementation of the SbD approach followed the strategy reported by [33] and was specifically tailored to complement the QbD work, to prevent hazards from the developed components and processes, and to support the redesign of technical workflows where the need is identified. Each individual process step was evaluated to identify possible exposure scenarios and safety issues.
For the used DELOS-susp methodology (see Fig. 2), four specific issues that pose a potential risk were identified:
-
1.
DMSO/EtOH in the nanoformulation
-
2.
Aerosol exposition due to organic compounds, vapors, streams from the organic phase
-
3.
Carbon dioxide poisoning
-
4.
Cold burn during lyophilization
All the identified possible risks during the DELOS-susp methodology are rated on low respectively medium risk level and are controlled (see Appendix F), thus the DELOS-susp process can be considered as safe. To fully cover the entire innovation process towards its final product application, immunogenicity, endotoxicity, and impurities of the nano-formulation will be evaluated at later stage.
4. Conclusions
The current work brings a new contribution in the preparation of GLA loaded liposomes through the successful application of the QbD approach to DELOS-susp production method. Thanks to this methodology a prototype of nanoGLA suitable for in vivo testing could be optimized. The influence of some formulation parameters was determined by using a DoE. Thus, among the formulation factors, GLA concentration and lipid concentration were the most important parameters for the quality of nanoGLA obtained by compressed fluid processing.
This study also enabled the obtaining of a preliminary design space to produce nanoGLA intermediate dispersion, in which the established quality requirements of the product are met.
Through the design space analysis, it could be predicted that for achieving a higher (10-fold), concentration of enzyme in the final formulation the concentration of lipid in the DELOS-susp step could be reduced in order to obtain a good quality product in terms of particle size and uni-lamellarity, and to avoid aggregation and sedimentation.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: J.L-C., D.P., S.Sch., M.R., I.A., J.V., S.Sa. and N.V. are inventors of patent WO/2014/001509 licensed to Biopraxis Resarch AIE. J.V., S.Sa. and N.V. are inventors of patent WO/2006/079889 owned by Nanomol Technologies SL, and stock-owners in Nanomol Technologies SL. J.M-M., J.T-M., A.F., E.G-M., J-L.C, E.C-L., D.P., M.R., S.Sch., I.A., A.S., S.Sa., J.V., N.V. and A.C. are inventors of patent application EP21382062.4.
Acknowledgments
The authors acknowledge the financial support from European Commission through H2020 program of the Smart-4-Fabry project (ID 720942).
We acknowledge financial support to our research from Instituto de Salud Carlos III, through “Acciones CIBER”. The Networking Research Center on Bioengineering, Biomaterials, and Nanomedicine (CIBER-BBN) is 2008–2011, financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. Authors acknowledge financial support from the Agencia Estatal de Investigación-Ministerio de Ciencia e Innovación through the “Severo Ochoa” Programme for Centres of Excellence in R & D (CEX2019-000917-S). This work was also financed by 1084 the Ministerio de Ciencia e Innovación (PID2019-105622RB-1085 I00). We also thank the denomination of the 1101 consolidated group from Generalitat de Catalunya: 2017-1102 SGR-1439 (M.R.) and 2017-SGR-918 (J.V.).
We also acknowledge the ICTS “NANBIOSIS”, more specifically the support from the Protein Production Platform of CIBER-BBN/IBB, at the UAB SepBioES scientific-technical service (www.nanbiosis.es/unit/u1-proteinproduction-platform-ppp/), the Soft Materials Service linked to Biomaterial Processing and Nanostructuring Unit at ICMAB-CSIC (www.nanbiosis.es/portfolio/u6-biomaterial-processing-and-nanostructuring-unit/) and the Peptide Synthesis unit at the IQAC-CSIC (www.nanbiosis.es/portfolio/u3-synthesis-of-peptides-unit/).
N. A-G. is supported by a PERIS grant from the Catalan Government (SLT006/17/270).
J. T-M. is supported by a FI-AGAUR grant from the Catalan Government and the European Social Fund (ESF-Investing in your future) of the European Union. This work has been done in the framework of the JT-M. doctorate in Materials Science of the Universitat Autònoma de Barcelona.
The technical assistance of Inbal Ionita, Ella Kesselman and Mingming Zhang from Technion - Israel Institute of Technology; and Ramon González from ICMAB-CSIC is acknowledged.
Contributor Information
Nora Ventosa, Email: ventosa@icmab.es.
Alba Córdoba, Email: acordoba@nanomol-tech.com.
Appendix A. Synthesis of GLAcmycHis
Plasmids and E. coli strains
The GLA gene was obtained from the commercial vector pReceiver-M10 (OmicsLink ORF Expression Clone, ref. EX-Q0172-M10), that encodes a cDNA version of the GLA gene (primary gene accession number: NM_000169), in which both c-myc and 6xHis tags are fused (for further detection and purification purposes) to the C-terminus.
For further steps, the pOptiVEC™-TOPO® TA Cloning Kit (Catalog Number 12744-017, Invitrogen, by Life Technologies) was used following the vendor protocol. The PCR product was cloned into the pOptiVEC™-TOPO® plasmid, and the resulting plasmid (pOptiVec-GLA) was transformed into TOP10 E. coli cells. Positive clones were selected by ampicillin and confirmed by restriction analysis. The pOptiVec-GLA plasmid was purified by “gigaprep” method, and finally linearized before transfection, as recommended by the vendor, with restriction enzyme PvuI. All plasmids were purified from their corresponding overnight bacterial cultures, using the EndoFree Plasmid Giga Kit (Qiagen, ref. 12391). For their quantification, absorbances at 260 nm (A260) and 280 nm (A280) were measured.
Mammalian cell line, transfection and selection of positive clones
For the transfection and further selection of CHO cells overexpressing the human GLA enzyme, the OptiCHO™ Express Kit (Catalog number 12745-014, Invitrogen, by Life Technologies) was used. This kit provides CHO DG44 cells, media and transfection reagent needed. DG44 dhfr- CHO cells were grown in CD DG44 medium with 8 mM L-glutamine, at 37 °C in 8% CO2 using standard techniques.
Transfection of CHO DG44 cells and selection of positive GLA-DHFR cells were performed following the vendor protocol. At 28 days post-transfection, stepwise selection for dhfr amplification was started, using two-fold increments of methotrexate hydrate (MTX, Sigma, catalog number A6770) starting at 50 nM up to 4 μM. Finally, and in order to obtain a single clone, the pool of stably transfected and 4 μM MTX-amplified cells were serially diluted and seeded at 1–2 cells per well in a 96-well plate. As suggested by vendor, to grow CHO DG44 cells under adherent conditions, the specific medium Gibco® CD-CHO-A was used. After this process, a single clone (namely CHO-DG44-GLA clone #3) was isolated and cryopreserved.
Cryopreservation of CHO-DG44-GLA clone #3
CHO-DG44-GLA clone #3 was cultured in complete CD OptiCHO medium (supplemented with L-Glutamine 8 mM) to 2 × 106 cells per milliliter and then centrifuged at 100 g for 5 min. Cell pellets were then resuspended in fresh medium containing 10% DMSO to a final cell concentration of 10 × 106 cells/mL. Cells were aliquoted into 1 mL cryovials (Nalgene) and stored in the vapor phase of liquid nitrogen.
Production and purification of extracellular GLAcmycHis protein
Cryovials of CHO-DG44-GLA clone #3 were thawed under standard procedures. Briefly, cryovial was rapidly thawed at 37 °C and resuspended in 15 mL of prewarmed complete CD OptiCHO medium. Cells were expanded by subculturing them up to the desired volume. In each passage, cells were diluted into pre-warmed complete CD DG44 Medium to give a final cell density of 2 × 105 –3 × 105 viable cells/mL. Finally, supernatant containing extracellular GLAcmycHis was harvested by centrifuging the cell culture at 14,000 rpm for 15 min. This supernatant was purified in an ÄKTA Pure system (GE Healthcare) by using an affinity chromatography column (HisTrap Excel 5 mL, Ref 17-3712-06, GE Healthcare) following the vendor protocol. Finally, the eluted protein was dialyzed in acetic buffer 0.01 M pH 5.5 and stored at −20 °C until used.
Appendix B. DELOS-susp methodology for the preparation of nanoGLA
Table B.1 summarizes the process parameters that were kept constant for all DELOS-susp experiments; and also includes the control strategies since some of them could have an impact on some CQAs. The 10 experiments carried out in this study were produced in 2 consecutive days.
Table B.1.
DELOS process parameters kept constant in all DoE experimental runs.
| Process Parameter | Value | Control |
|---|---|---|
| Working temperature, Tw | 35 °C | Heating jacket |
| High pressure stirring | 500 rpm | Magnetic stirrer controller |
| Working pressure, Pw | 9 MPa | Syringe pump for CO2 |
| Depressurization pressure in the autoclave | 10 MPa | Manometer and N2 pressure regulator |
| Collector Stirring | 300 rpm | Plate with magnetic stirrer |
| CO2 molar fraction, XCO2 | 0.55 | Solvent volume in the reactor |
| Depressurization flowrate | 10 g/min | Manually controlled due to scale, averaged |
| Collector temperature | 23 °C | Monitored, room temperature |
Appendix C. Risk analysis assessment
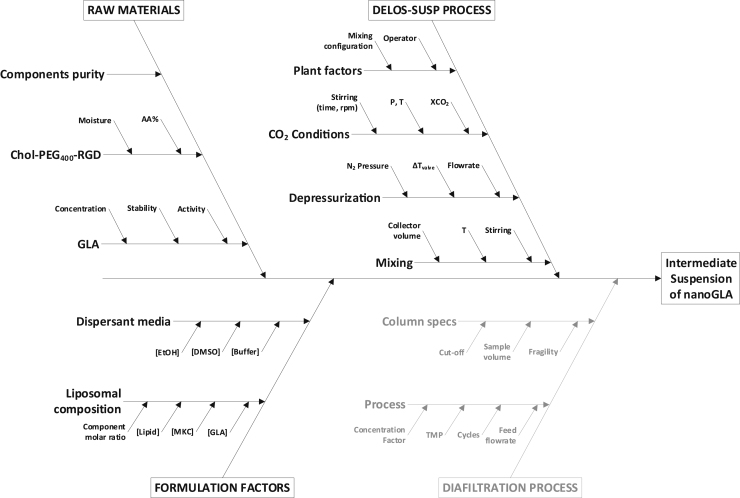
Fig C.1 represents a summary of Critical Material Attributes (CMAs) and Critical Process Parameters (CPPs) that may impact the CQAs of the intermediate nanoGLA using Ishikawa methodology.
Fig. C.1.
Ishikawa diagram illustrating the summary of CPPs and CMAs that may impact on each selected CQA of intermediate nanoGLA dispersion.
A control strategy is designed to ensure that a product of required quality will be produced consistently. These controls are based on product, formulation and process understanding and should include, at a minimum, control of the critical process parameters and material attributes. The risk assessment and the definition of control strategies serve to evaluate which CMAs and CPPs must be studied in a DoE.
In Table C.1 the risk analysis assessment of the potential CMAs and CPPs on the DELOS-susp process is presented.
Table C.1.
Risk analysis assessment and control strategies of CMAs and CPPs for intermediate nanoGLA production by DELOS-susp.
| CMA or CPP | Impact on CQA | Risk | Control strategy |
|---|---|---|---|
| Components purity | Low | Change in composition | CoA from supplier |
| Raw GLA | High | Change in morphology, nanoGLA stability an activity | CoA from supplier and check analysis by TGX, BCA or HPLC. |
| Chol-PEG400-RGD AA% | Low | Affect biological activity | CoA from supplier and check analysis |
| Chol-PEG400-RGD moisture | Low | Uni-lamellarity and morphology could be affected. | CoA from supplier and check analysis |
| EtOH concentration | High | To be evaluated in DoE. | To be included in DoE. |
| DMSO concentration | Medium | Stability, enzymatic activity | Previously defined. |
| Buffer concentration | Medium | To be studied in further steps. | To be defined in further steps of the project. |
| Membrane components molar ratio | High | Morphology, stability | Previously defined. |
| Lipid concentration | High | Entrapment efficiency, stability of the solution. | To be included in DoE. |
| Chol-PEG400-RGD concentration | High | Morphology, stability, intracellular penetration | To be included in DoE. |
| MKC concentration | High | Higher concentration, higher stability and Entrapment. | Previously defined. |
| GLA concentration | High | Stability, enzymatic activity | To be included in DoE. |
| Mixing configuration | High | Morphology, stability | Previously defined. |
| Operator | Low | Low control of the flowrate | Lab scale: Follow the standard protocol and practice on depressurization. Pilot scale: automatic control of depressurization valve. |
| Stirring of vessel | Low | Enough to obtain a single phase inside the high pressure vessel. | Previously defined, servocontrolled by high pressure stirrer. |
| Pressure of vessel | Low | Enough to obtain a liquid phase inside the high pressure vessel. | Previously defined, servocontrolled by pump and PIC. |
| Temperature of vessel | Low | Enough to obtain a liquid phase inside the high pressure vessel. | Previously defined, controlled by heating jacket and TIC. |
| CO2 molar fraction | Low | Inside the range of cosolvency. | Previously Defined and controlled by volume, P and T of the pump. |
| N2 pressure | Low | 10 bar higher than pressure of the vessel, could affect the flowrate. | Defined and controlled by a pressure regulator and PIC |
| Flowrate | High | Morphology and uni-lamellarity. To be evaluated in DoE. | To be included in DoE. |
| Collector volume | Medium | Change in flow and mixing between phases. | Previously defined. |
| Collector temperature | High | GLA degradation depends on temperature and exposition time | Previously defined. Controlled by TIC. |
| Stirring of the collector | Medium | Change in flow and mixing between phases. | Previously defined. |
Appendix D. Samples time evolution of mean particle size, PdI, and ζ-potential
Fig D.1 shows the analysis in time by DLS of the samples. As the Fig D.1. Evolution shows, the mean size of some samples increased during time, mainly explained by the aggregation of the nanovesicles, showing clearly that DOE-006 and DOE-009 were going to sediment. PdI values, remained constant except for the two samples previously mentioned. ζ-potential maintained mostly constant with time, although the decrease observed in some cases could be explained because of the change in pH during time caused by the release of dissolved CO2.
Fig. D.1.
Evolution of a) mean particle size, b) PdI, and c) ζ-potential over time during 14 days after the production of the samples.
Appendix E. Modeling and data fitting
The acceptance of the responses obtained was evaluated by means of the statistical parameters predicted by the software R2, Q2 model validity, and model reproducibility: (i) R2: it represents the percent of the variation of the response explained by the model, that is, how well the model fits the data; (ii) Q2: it is the percent of the variation of the response predicted by the model according to cross validation. It gives information about how well the model predicts new data. In Table E.1, the values of these statistical parameters are summarized, indicating if the data collected from the experimental results is fitting into a model. Only those CQAs that could be fitted (mean particle size, ζ -potential and uni-lamellarity) were included in the model.
Table E.1.
Statistical parameters and fitting for the investigated intermediate nanoGLA CQAs.
| CQA | R2 | Q2 | Fitting and model (if applies) |
|---|---|---|---|
| Mean particle size | 0.937 | 0.661 | Yes, Size = 2.0 X1 + 10.6 X2 + 11.7 X3 - 2.2 X4 + 86 |
| Polydispersity | 0.437 | 0.200 | No, a bad correlation and a poor predictive ability of the model were obtained. |
| ζ-potential | 0.791 | 0.550 | Yes, ζ-pot = −19 X1 + 2.1 X2 - 10.9 X3 - 24.5 X4 + 41 |
| TSI | 0.636 | 0.200 | No, a bad correlation and a poor predictive ability of the model were obtained. |
| Uni-lamellarity | 0.858 | 0.498 | Yes, fsingle = 3.3·10−4 X1 - 1.8·10−2 X2 - 1.1·10−7 X3 - 5.7·10−9 X4 + 1.0 |
| Entrapment efficiency | 0.420 | 0.200 | No, a bad correlation and a poor predictive ability of the model were obtained. |
| Enzymatic activity | 0.625 | 0.040 | No, it could be explained by the high measurement related variability |
Appendix F. Identified safety issues
Occupational exposure scenarios as well as environmental exposure scenarios were registered, considering different operational conditions. Table F.1 shows the obtained risk rating key for the specific activity and/or process in the DELOS-susp methodology for the preparation of the nanoGLA intermediate, and lists relevant methods to manage the possible risk and decrease its possibility to take place.
Table F.1.
Identified safety issues in relation to their severity and likelihood leads to risk rating key.
| Activity/Process | Associated Risk(s): | Severity: | Likelihood: | Method(s) to Manage the Risk: |
|---|---|---|---|---|
| The chances of that risk happening | ||||
| A list of methods you will use to minimize the chances of the risk happening | ||||
| Level of impact | ||||
| Safety issue(s) associated with the activity | ||||
| Preparation of the organic solution | EtOH/DMSO in the nanoformulation | Acceptable | Low | Related to the environment: Solvents to the non-chlorinated solvents waste container. Cholesterol, Chol-PEGn-RGD, MKC, and DPPC to the solid chemical waste container. |
| Low | ||||
| Acceptable | ||||
| Weighing solids and preparation of a solution | Dermal/occu pational safety issues during cleaning | |||
| Cleaning the laboratory material | ||||
| Related to workers: Fume hood. Mask, gloves, glasses and lab coat. | ||||
| Loading of the vessel, closing and pressurization by adding CO2 | Dermal/occupational safety issues during loading the vessel | Acceptable | Low | Related to workers: Fume hood. Mask, gloves, glasses, and lab coat. Vent and safety valves present in the equipment in case of overpressure |
| Preparation of aqueous solution | Dermal safety issues during cleaning of the material | Acceptable | Low | Related to workers: Fume hood. Mask, gloves, glasses, and lab coat. |
| Depressurization and mixing of the organic and aqueous phases | Carbon dioxide poisoning during valve opening | Tolerable | Low | Related to workers: Fume hood. Mask, gloves, glasses and lab coat. Vent and safety valves. |
| Filters for carbon dioxide exhaust expelled to the environment have to be considered in larger scales. | ||||
| Collecting the final product and storage | Cold Burn during lyophilization | Acceptable | Low | Fume hood. Mask, gloves, glasses and lab coat. |
References
- 1.N. Ventosa, J. Veciana, S. Sala, M. Cano, Patent Application EP 1843836, 2012.
- 2.Cabrera I., Elizondo E., Esteban O., Corchero J.L., Melgarejo M., Pulido D., Córdoba A., Moreno E., Unzueta U., Vazquez E., Abasolo I., Schwartz S., Villaverde A., Albericio F., Royo M., García-Parajo M.F., Ventosa N., Veciana J. Multifunctional nanovesicle-bioactive conjugates prepared by a one-step scalable method using CO2-expanded solvents. Nano Lett. 2013;13:3766–3774. doi: 10.1021/nl4017072. [DOI] [PubMed] [Google Scholar]
- 3.Grimaldi N., Andrade F., Segovia N., Ferrer-Tasies L., Sala S., Veciana J., Ventosa N. Lipid-based nanovesicles for nanomedicine. Chem. Soc. Rev. 2016;45:6520–6545. doi: 10.1039/c6cs00409a. [DOI] [PubMed] [Google Scholar]
- 4.Cabrera I., Abasolo I., Corchero J.L., Elizondo E., Gil P.R., Moreno E., Faraudo J., Sala S., Bueno D., González-Mira E., Rivas M., Melgarejo M., Pulido D., Albericio F., Royo M., Villaverde A., García-Parajo M.F., Schwartz S., Ventosa N., Veciana J. α-Galactosidase-A loaded-nanoliposomes with enhanced enzymatic activity and intracellular penetration. Adv. Healthc. Mater. 2016;5:829–840. doi: 10.1002/adhm.201500746. [DOI] [PubMed] [Google Scholar]
- 5.Alipourfetrati S., Saeed A., Norris J.M. A review of current and future treatment strategies for fabry disease: a model for treating lysosomal storage diseases. J. Pharmacol. Clin. Toxicol. 2015;3 [Google Scholar]
- 6.Solomon M., Muro S. Lysosomal enzyme replacement therapies: historical development, clinical outcomes, and future perspectives. Adv. Drug Deliv. Rev. 2017;118:109–134. doi: 10.1016/j.addr.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomsen-Melero J., Passemard S., García-Aranda N., Díaz-Riascos Z.V., González-Rioja R., Pedersen J.N., Lyngsø J., Merlo-Mas J., Cristóbal-Lecina E., Corchero J.L., Pulido D., Portnaya I., Crist E., Corchero L., Pulido D., Cámara-Sánchez P., Portnaya I., Ionita I., Schwartz S., Veciana J., Sala S., Royo M., Córdoba A., Danino D., Pedersen J.S., González-Mira E., Abasolo I., Ventosa N. Impact of chemical composition on the nanostructure and biological activity of α-galactosidase-loaded nanovesicles for fabry disease treatment!impact of chemical composition on the nanostructure and biological activity of galactosidaseloaded nanovesicles for fabry disease treatment. ACS Appl. Mater. Interfaces. 2021;13(7):7825–7838. doi: 10.1021/acsami.0c16871. [DOI] [PubMed] [Google Scholar]
- 8.International Council of Harmonisation, Q8 - Pharmaceutical Development, 2009.
- 9.International Council of Harmonisation, Q9 - Quality Risk Management, 2005.
- 10.International Council of Harmonisation, Q10 - Pharmaceutical Quality System, 2008.
- 11.Porfire A., Muntean D.M., Rus L., Sylvester B., Tomuţă I. A quality by design approach for the development of lyophilized liposomes with simvastatin. Saudi Pharm. J. 2017;25:981–992. doi: 10.1016/j.jsps.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Poel I., Robaey Z. Safe-by-design: from safety to responsibility. Nanoethics. 2017;11:297–306. doi: 10.1007/s11569-017-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purohit R., Mittal A., Dalela S., Warudkar V., Purohit K., Purohit S. Social, environmental and ethical impacts of nanotechnology. Mater. Today Proc. 2017;4:5461–5467. doi: 10.1016/j.matpr.2017.05.058. [DOI] [Google Scholar]
- 14.Cristóbal-Lecina E., Pulido D., Martin-Malpartida P., Macias M.J., Albericio F., Royo M. Synthesis of stable cholesteryl-polyethylene glycol-peptide conjugates with non-disperse polyethylene glycol lengths. ACS Omega. 2020;5:5508–5519. doi: 10.1021/acsomega.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danino D. Cryo-TEM of soft molecular assemblies. Curr. Opin. Colloid Interface Sci. 2012;17:316–329. doi: 10.1016/j.cocis.2012.10.003. [DOI] [Google Scholar]
- 16.Pabst G., Rappolt M., Amenitsch H., Laggner P. Structural information from multilamellar liposomes at full hydration: full q-range fitting with high quality X-ray data. Phys. Rev. E - Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 2000;62:4000–4009. doi: 10.1103/PhysRevE.62.4000. [DOI] [PubMed] [Google Scholar]
- 17.Hosemann R., Bagchi S.N. The interference theory of ideal paracrystals. Acta Cryst. 1952;5:612–614. [Google Scholar]
- 18.Giannotti M.I., Abasolo I., Oliva M., Andrade F., García-Aranda N., Melgarejo M., Pulido D., Corchero J.L., Fernández Y., Villaverde A., Royo M., García-Parajo M.F., Sanz F., Schwartz S. Highly versatile polyelectrolyte complexes for improving the enzyme replacement therapy of lysosomal storage disorders. ACS Appl. Mater. Interfaces. 2016;8:25741–25752. doi: 10.1021/acsami.6b08356. [DOI] [PubMed] [Google Scholar]
- 19.Kapoor M., Lee S.L., Tyner K.M. Liposomal drug product development and quality: current US experience and perspective. AAPS J. 2017;19:632–641. doi: 10.1208/s12248-017-0049-9. [DOI] [PubMed] [Google Scholar]
- 20.FDA, Liposome Drug Products Chemistry, Manufacturing, and Controls; Human Pharmacokinetics and Bioavailability; and Labeling Documentation, 2018.
- 21.European Medicine Agency . EMA/Committee Hum. Med. Prod.; 2013. Reflection Paper on the Data Requirements for Intravenous Liposomal Products Developed with Reference to an Innovator Liposomal Product; pp. 1–13. 806058/2009/Rev. 02. 44. [Google Scholar]
- 22.Xu X., Khan M.A., Burgess D.J. A quality by design (QbD) case study on liposomes containing hydrophilic API: II. Screening of critical variables, and establishment of design space at laboratory scale. Int. J. Pharm. 2012;423:543–553. doi: 10.1016/j.ijpharm.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 23.Pintre I.C., Webb S.J. Binding and Reactivity at Bilayer Membranes. 1st ed. Elsevier Ltd; 2013. [DOI] [Google Scholar]
- 24.Shaker S., Gardouh A., Ghorab M. Factors affecting liposomes particle size prepared by ethanol injection method. Res. Pharm. Sci. 2017;12:346–352. doi: 10.4103/1735-5362.213979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pamunuwa G., Karunaratne V., Karunaratne D.N. Effect of lipid composition on in vitro release and skin deposition of curcumin encapsulated liposomes. J. Nanomater. 2016;2016:1–9. doi: 10.1155/2016/4535790. [DOI] [Google Scholar]
- 26.Awad N.S., Paul V., Mahmoud M.S., Al Sawaftah N.M., Kawak P.S., Al Sayah M.H., Husseini G.A. Effect of pegylation and targeting moieties on the ultrasound-mediated drug release from liposomes. ACS Biomater. Sci. Eng. 2019;6:48–57. doi: 10.1021/acsbiomaterials.8b01301. [DOI] [PubMed] [Google Scholar]
- 27.Briuglia M.L., Rotella C., McFarlane A., Lamprou D.A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv. Transl. Res. 2015;5:231–242. doi: 10.1007/s13346-015-0220-8. [DOI] [PubMed] [Google Scholar]
- 28.Milla P., Dosio F., Cattel L. PEGylation of proteins and liposomes: a powerful and flexible strategy to improve the drug delivery. Curr. Drug Metab. 2011;13:105–119. doi: 10.2174/138920012798356934. [DOI] [PubMed] [Google Scholar]
- 29.Guner S., Oztop M.H. Food grade liposome systems: effect of solvent, homogenization types and storage conditions on oxidative and physical stability. Colloids Surf. A Physicochem. Eng. Asp. 2017;513:468–478. doi: 10.1016/j.colsurfa.2016.11.022. [DOI] [Google Scholar]
- 30.Carugo D., Bottaro E., Owen J., Stride E., Nastruzzi C. Liposome production by microfluidics: potential and limiting factors. Sci. Rep. 2016;6:1–15. doi: 10.1038/srep25876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X., Costa A., Burgess D.J. Protein encapsulation in unilamellar liposomes: High encapsulation efficiency and a novel technique to assess lipid-protein interaction. Pharm. Res. 2012;29:1919–1931. doi: 10.1007/s11095-012-0720-x. [DOI] [PubMed] [Google Scholar]
- 32.Dan N., Danino D. Structure and kinetics of lipid-nucleic acid complexes. Adv. Colloid Interface Sci. 2014;205:230–239. doi: 10.1016/j.cis.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Schimpel C., Resch S., Flament G., Carlander D., Vaquero C., Bustero I., Falk A. A methodology on how to create a real-life relevant risk profile for a given nanomaterial. J. Chem. Heal. Saf. 2018;25:12–23. doi: 10.1016/j.jchas.2017.06.002. [DOI] [Google Scholar]