Abstract
Objective: To analyze differentially expressed genes (DEGs) related to liver fibrosis, and clarify the key genes and the possible targets in the progression of liver fibrosis. Methods: Using microarray datasets, GSE38199 was extracted from Gene Expression Omnibus (GEO), and a bioinformatics method was performed to find DEGs and transcription factors related to liver fibrosis. Results: A total of 58 DEGs were screened out according to GEO2R online analysis tool, which included 49 up-regulated and 9 down-regulated genes. These DEGs were mainly involved in formation with the extracellular region and extracellular exosome through gene ontology (GO) enrichment analysis. Kyoto Encyclopedia of Gene and Genome (KEGG) pathway enrichment analysis showed that DEGs mainly participated in the PI3K-Akt signaling pathway, focal adhesion, ECM-receptor interaction, and metabolic pathways. Based on the results of the Protein-Protein Interaction (PPI) network and Molecular Complex Detection (MCODE) analysis, 9 key genes (COL1A1, FBN1, BGN, COL6A3, MMP2, FBLN5, LUM, PDGFRB, LOXL1) were screened out. A total of 30 transcription factors were found according to these 9 key genes, of which 4 transcription factors (Stat3, Trp53, NF-κB1, Sp1) were enriched. Conclusion: Stat3, Trp53, NF-κB1, and Sp1 were all related to the development of liver fibrosis, and FBLN5 might be a target for liver fibrosis.
Keywords: Liver fibrosis, differentially expressed genes, bioinformatics
Introduction
Liver fibrosis is an intermediate process of liver cell damage and inflammatory response with various causes (such as hepatitis virus infection, alcohol abuse, immune response, drug and chemical poison damage), which may lead to chronic progressive liver disease. It has been found that during the liver injury-inflammation-repair progress, quiescent hepatic satellite cells (HSCs), normally located at the space of Disse, are activated and then trans-differentiated into myofibroblasts (MFCs). These MFCs can produce a large amount of extracellular matrix (ECM) dominated by collagen and thus lead to liver fibrosis [1-3]. The imbalance of ECM deposition and degradation in the liver is a necessary stage for the development of chronic liver injury to cirrhosis, and the main reason for progression to liver cancer [4]. It has been confirmed that the activation and proliferation of HSCs is a key step in the progression of liver fibrosis, so taking effective measures to interfere with the activation process of HSCs is of great importance for treatment of fibrosis. There are several classic animal models of liver fibrosis, such as bile duct ligation and repeated injections of carbon tetrachloride (CCL4) [5]. But these models only lead to hepatocellular carcinoma when giving additional processing factors (such as a lack of choline or carcinogens). Therefore, the author established platelet-derived growth factor C (PDGF-C) transgenic mice [6].
PDGF-C is a recently discovered member of platelet derived growth factor (PDGF) ligand family. The difference between PDGF-C and PDGF-A or PDGF-B is that PDGF-A and PDGF-B are secreted by cells as bioactive dimers, while PDGF-C is a precursor polypeptide secreted in cells. By extracellular protein cleavage, PDGF-C produces active growth factor PDGF-CC [7-9]. Overexpression of PDGF-C in hepatocytes will induce liver fibrosis, hepatocyte steatosis, hepatoma, and even hepatocellular carcinoma. Therefore, PDGE-C transgenic mice could be used as a unique model to study progression from liver fibrosis to tumor.
At present, research on liver fibrosis mainly focuses on inhibition of the activation of HSCs and trans-differentiation of HSCs into MFCs and fibroblasts. TGF-β and PDGF are two major cytokines that promote activation of HSCs and ECM proliferation. Besides these, there are many other cytokines and intracellular signal transduction pathways involved in this process [10,11]. Therefore, the mechanism of occurrence and development of liver fibrosis is very complicated. If we could find more specific biomarkers concerned with liver fibrosis or related mechanisms at the molecular level, it would benefit research. The purpose of this study is to use gene chip and bioinformatic technology to find molecular markers related to liver fibrosis.
Gene chips, an important technology platform in the field of life sciences, is an effective method for screening differentially expressed genes (DEGs). However, bioinformatic technology is also required to analyze and process thousands of genes on the gene chip to obtain useful information.
Materials and methods
Microarray data information
Gene Expression Omnibus (GEO), the largest public database of gene expression profiles at present, is managed and maintained by the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). The chip data GSE38199, downloaded from GEO, contains 7 cases of PDGF-C transgenic and 9 cases of wild-type mice.
Processing and analyzing of DEG data
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), an online tool, was used to compare different data sets and analyze the GSE38199 microarray data set. DEGs between PDGF-C transgenic and wild-type mice were screened out by P < 0.05 and |logFC| > 2, then heat map analysis was performed with these DEGs on Heml (http://hemi.biocuckoo.org/down.php).
Analyzing DEGs by GO function and KEGG pathway
GO and KEGG pathway analysis were used to describe and classify a class of genes. GO refers to the basic ontology database, which can classify, define and annotate genes from three aspects: cell composition, bioinformatics process, and molecular function [12]. Moreover, KEGG can also classify genes with the same biologic function, which helps researchers to study gene expression and explore information expressed as a whole [13]. Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) database was used to identify DEGs by analyzing GO function and KEGG pathway, in which P < 0.05 was considered significant.
PPI network construction and module selection
PPI network could effectively screen out the key proteins correlated with liver fibrosis, and analyze their relationship. The internet online tool-STRING (https://string-db.org/) database is commonly used to exploring gene correlation, which contains known and predicted protein interactions [14]. In this study, a PPI network was constructed on STRING between these DEGs. The interactions between proteins with a score ≥ 0.4 were visualized using Cytoscape. The top 10 DEGs with a high degree of connectivity in the PPI network were selected as key genes of liver fibrosis.
MCODE is a plug-in of Cytoscape, which is used to find highly connected regions in PPI networks. According to the modules selected from the PPI network, GO function and KEGG pathway enrichment analysis of the most meaningful modules were carried out using DAVID, with P < 0.05 considered significant.
Searching for transcription factors
Transcriptional Regulatory Relationships Unraveled by Sentence based Text mining (TRRUST, https://www.grnpedia.org/trrust/) database records the regulatory relationships of transcription factors. It contains not only the target genes corresponding to transcription factors, but also the regulatory relationships between transcription factors [15]. In this study, transcription factors related to liver fibrosis were found by key genes in the TRRUST database.
Result
Identification of DEGs
In the study, 7 cases of PDGF-C transgenic mice and 9 cases of wild-type mice were used in this study. A total of 35557 genes were obtained from the GSE38199 data set, and 58 DEGs, including 49 up-regulated and 9 down-regulated genes, were screened out. Analysis of DEGs was carried out by heat map (Figure 1).
Figure 1.
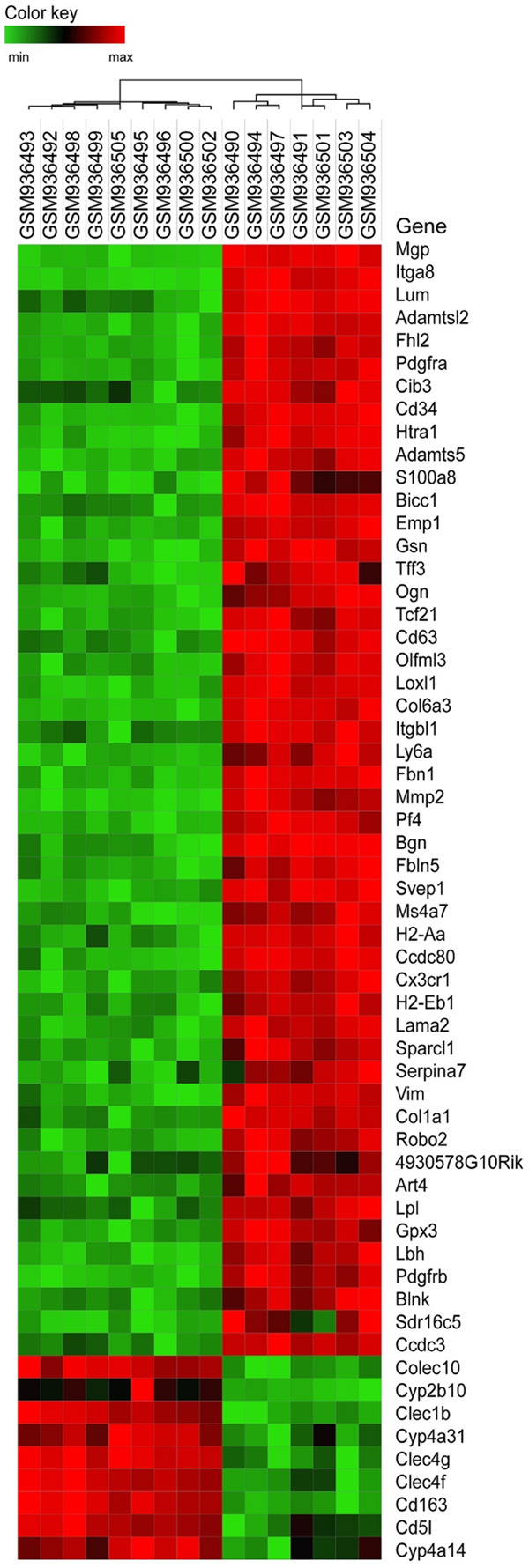
Heat map of DEGs. The heat map consists of 16×58 squares, each representing the level of gene expression in a sample, with high expression indicated by red and low expression indicated byf green.
Results of GO functional enrichment analysis
Based on DAVID analyzing GO function of DEGs, 8 GO function enrichment results were screened out (Table 1). The cytological composition analysis of these DEGs showed that most of them were involved in the composition of extracellular region, extracellular exosome, and extracellular space. Biologic processes were mainly concentrated in extracellular matrix organization, Chemotaxis, and metanephric glomerular capillary formation.
Table 1.
GO function analysis of 8 differentially expressed genes (DEG)
| Category | Term | Description | Count | P value |
|---|---|---|---|---|
| CC | GO:0005576 | extracellular region | 30 | 9.45E-17 |
| CC | GO:0031012 | extracellular matrix | 15 | 5.15E-14 |
| CC | GO:0005578 | proteinaceous extracellular matrix | 15 | 1.39E-13 |
| CC | GO:0005615 | extracellular space | 19 | 6.79E-08 |
| CC | GO:0070062 | extracellular exosome | 23 | 1.11E-06 |
| BP | GO:0030198 | extracellular matrix organization | 6 | 1.54E-05 |
| BP | GO:0006935 | Chemotaxis | 6 | 1.82E-05 |
| BP | GO:0072277 | metanephric glomerular capillary formation | 3 | 2.24E-05 |
Results of KEGG pathway enrichment analysis
KOBAS 3.0 software was used to analyze the KEGG pathway of DEGs. Significantly enriched pathways were screened out (Tables 2, 3 and Figure 2). KEGG pathway enrichment analysis showed that the up-regulated DEGs were mainly involved in PI3K-Akt signaling pathway, Focal adhesion, Human papillomavirus infection, ECM-receptor interaction, and Cell adhesion molecules. Down-regulated DEGs were mainly related to Metabolic pathways, Arachidonic acid metabolism, and Retinol metabolism.
Table 2.
KEGG pathway analysis of the top 10 up-regulated genes
| Category | Term | Description | Input number | Corrected P-Value | P value |
|---|---|---|---|---|---|
| KEGG | mmu04510 | Focal adhesion | 6 | 2.78E-05 | 3.31E-07 |
| KEGG | mmu04512 | ECM-receptor interaction | 4 | 0.000255332 | 6.69E-06 |
| KEGG | mmu04151 | PI3K-Akt signaling pathway | 6 | 0.000255332 | 9.12E-06 |
| KEGG | mmu04514 | Cell adhesion molecules (CAMs) | 4 | 0.001908865 | 9.09E-05 |
| KEGG | mmu05165 | Human papillomavirus infection | 5 | 0.002184925 | 0.000130055 |
| KEGG | mmu05205 | Proteoglycans in cancer | 4 | 0.002564466 | 0.00018317 |
| KEGG | mmu04810 | Regulation of actin cytoskeleton | 4 | 0.002581966 | 0.000222585 |
| KEGG | mmu05416 | Viral myocarditis | 3 | 0.002581966 | 0.000261389 |
| KEGG | mmu05169 | Epstein-Barr virus infection | 4 | 0.002581966 | 0.000276639 |
| KEGG | mmu04640 | Hematopoietic cell lineage | 3 | 0.002644313 | 0.000314799 |
Table 3.
KEGG pathway analysis of the top 10 down-regulated genes
| Category | Term | Description | Count | P value |
|---|---|---|---|---|
| CC | GO:0005578 | proteinaceous extracellular matrix | 5 | 3.78E-07 |
| CC | GO:0005615 | extracellular space | 4 | 0.00393979261 |
| CC | GO:0070062 | extracellular exosome | 5 | 0.01271927179 |
| CC | GO:0031012 | extracellular matrix | 2 | 0.04169655023 |
Figure 2.
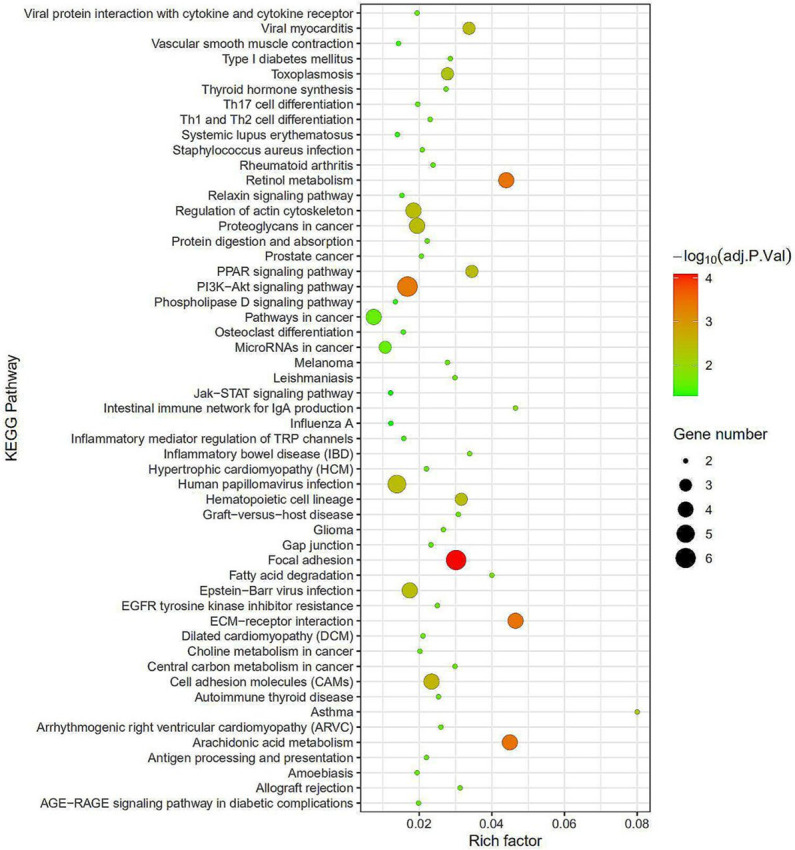
KEGG pathways enrichment analysis. The redder the bubble color indicates a smaller the P value, the larger the bubble indicates a greater the number of genes. It can be seen from the figure that the bubble on Focal adhesion is the largest, and the color is the reddest, which implies the best enrichment effect.
Analysis of DEGs by PPI network
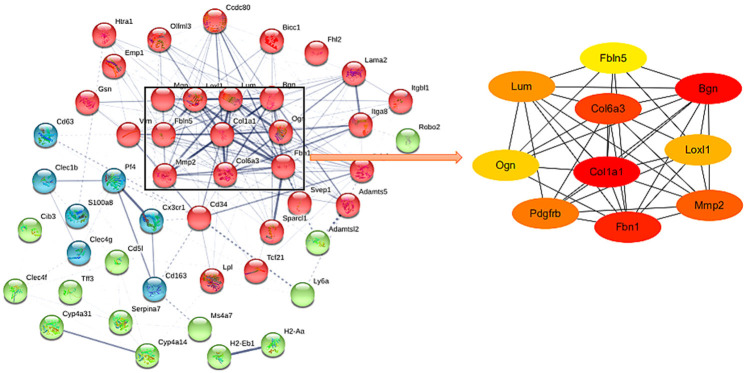
58 DEGs were inputted into STRING database to analyze molecular interaction. Then the data were imported into Cytoscape, and the top 10 Hub genes were found by plug-in cytoHubba (Figure 3).
Figure 3.
PPI network. Each node represents a relevant gene, and the right side is the top 10 Hub genes of the PPI network.
Analysis of PPI function modules
Module analysis was conducted using MCODE, and the most significant modules of DEGs were extracted from PPI network with MCODE score = 8 (Figure 4). The results of the module analysis were almost the same as the 10 Hub genes obtained by PPI network analysis. Then, KEGG pathway and GO function analysis of module genes were conducted by DAVID online analysis tool, and the results are shown in Tables 4 and 5.
Figure 4.
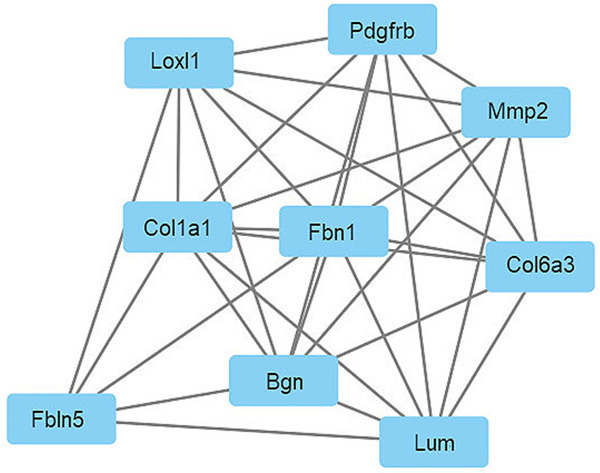
Significant modules in the PPI network with MCODE score = 8.
Table 4.
Submodule GO analysis
| Category | Term | Description | Input number | Corrected P-Value | P value |
|---|---|---|---|---|---|
| KEGG | mmu00590 | Arachidonic acid metabolism | 3 | 6.86E-06 | 1.29E-06 |
| KEGG | mmu00830 | Retinol metabolism | 3 | 6.86E-06 | 1.37E-06 |
| KEGG | mmu00071 | Fatty acid degradation | 2 | 0.000237189 | 7.12E-05 |
| KEGG | mmu03320 | PPAR signaling pathway | 2 | 0.000522879 | 0.000209151 |
| KEGG | mmu04750 | Inflammatory mediator regulation of TRP channels | 2 | 0.000877398 | 0.000438699 |
| KEGG | mmu04270 | Vascular smooth muscle contraction | 2 | 0.000885118 | 0.000531070 |
| KEGG | mmu01100 | Metabolic pathways | 3 | 0.006852979 | 0.004797085 |
| KEGG | mmu00140 | Steroid hormone biosynthesis | 1 | 0.025751782 | 0.021968794 |
| KEGG | mmu05204 | Chemical carcinogenesis | 1 | 0.025751782 | 0.023176604 |
| KEGG | mmu04625 | C-type lectin receptor signaling pathway | 1 | 0.027513751 | 0.027513751 |
Table 5.
Submodule KEGG analysis
| Category | Term | Description | Input number | Corrected P-Value | P value |
|---|---|---|---|---|---|
| KEGG | mmu04510 | Focal adhesion | 3 | 0.00035532121 | 1.95E-05 |
| KEGG | mmu05205 | Proteoglycans in cancer | 3 | 0.00035532121 | 2.15E-05 |
| KEGG | mmu04151 | PI3K-Akt signaling pathway | 3 | 0.00092345123 | 0.00010922 |
| KEGG | mmu05165 | Human papillomavirus infection | 3 | 0.00092345123 | 0.00011193 |
| KEGG | mmu04512 | ECM-receptor interaction | 2 | 0.00153386492 | 0.00025518 |
| KEGG | mmu04974 | Protein digestion and absorption | 2 | 0.00153386492 | 0.00027888 |
| KEGG | mmu04933 | AGE-RAGE signaling pathway in diabetic complications | 2 | 0.00164721576 | 0.00034940 |
| KEGG | mmu04926 | Relaxin signaling pathway | 2 | 0.00239796570 | 0.00058132 |
| KEGG | mmu05200 | Pathways in cancer | 2 | 0.03294246829 | 0.00898430 |
| KEGG | mmu05219 | Bladder cancer | 1 | 0.03776680097 | 0.01144448 |
Finding related transcription factors
In the study, 9 key genes were used for searching related transcription factors in TRRUST database. TRRUST database finally identified 4 out of 9 genes, namely COL1A1, MMP2, PDGFRB, FBLN5, and the transcription factors contained in these 4 genes are shown in Table 6. Finally, 4 transcription factors were enriched, and their enrichment is shown in Table 7.
Table 6.
Distribution of transcription factors in 9 key genes
| TF | Target | TF | Target | TF | Target |
|---|---|---|---|---|---|
| Nfic | Col1a1 | Ep300 | Mmp2 | Trp53 | Mmp2 |
| Nfkb1 | Col1a1 | Fos | Mmp2 | Smad2 | Fbln5 |
| Prdm5 | Col1a1 | Fosb | Mmp2 | Smad3 | Fbln5 |
| Rela | Col1a1 | Fosl1 | Mmp2 | Myc | Pdgfrb |
| Runx2 | Col1a1 | Jun | Mmp2 | Trp53 | Pdgfrb |
| Smad1 | Col1a1 | Junb | Mmp2 | Trp73 | Pdgfrb |
| Sox8 | Col1a1 | Jund | Mmp2 | ||
| Sp1 | Col1a1 | Nfkb1 | Mmp2 | ||
| Stat3 | Col1a1 | Snai1 | Mmp2 | ||
| Usf1 | Col1a1 | Sp1 | Mmp2 | ||
| Usf2 | Col1a1 | Srf | Mmp2 | ||
| Yy1 | Col1a1 | Stat3 | Mmp2 |
Table 7.
Distribution of 4 enriched transcription factors
| Key TF | Description | overlapped genes | P value | Q value | List of overlapped genes |
|---|---|---|---|---|---|
| Stat3 | signal transducer and activator of transcription 3 | 2 | 0.000657 | 0.00263 | Col1a1, Mmp2 |
| Trp53 | transformation related protein 53 | 2 | 0.00289 | 0.00548 | Pdgfrb, Mmp2 |
| Nfkb1 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 | 2 | 0.00411 | 0.00548 | Mmp2, Col1a1 |
| Sp1 | trans-acting transcription factor 1 | 2 | 0.00736 | 0.00736 | Mmp2, Col1a1 |
Discussion
In this study, bioinformatics was used to analyze the chip data GSE38199 in the GEO database. A total of 58 DEGs were screened out. In addition, PPI network and module analysis revealed 9 DEGs related to liver fibrosis: COL1A1, FBN1, BGN, COL6A3, MMP2, FBLN5, LUM, PDGFRB and LOXL1. 30 total transcription factors were obtained from these 9 DEGs, of which 4 transcription factors were enriched: Stat3, Trp53, NF-κB1, and Sp1.
Transcription factor specific protein 1 (Specificity protein 1, Sp1) is a nuclear transcription factor that interacts with the GC/GT box of the gene regulatory region and other proteins. It is related to initiation of transcription of many target genes. It plays a very important role in cell proliferation, apoptosis, differentiation and tumor formation [16]. Among transcription factors, Sp1 is involved in the expression of ECM genes, and regulates the expression of several genes associated with downstream targets of TGF-β. Thus it plays an important role in the progression of liver fibrosis [17]. One study showed that Sp1 Decoy ODN inhibited the activation of HSCs by down-regulating the expression of liver fibrosis-related genes, such as PDGF-BB, α-SMA, and COL1α1 [18]. It reported that COL1A1 promoter contains two Sp1 binding sites, and COL1A1 expression induced by malondialdehyde in HSCs could be blocked by the mutation of the Sp1 site and the application of Sp1 inhibitors [19]. Another experiment found that there were two Sp1 binding sites on the MMP2 promoter, and the deletion of this region would reduce 76% of the activity of MMP2 promoter. However, extracellular signals may phosphorylate Sp1 through the ERK signaling pathway to promote MMP2 expression. These results indicate that Sp1 is closely related to liver fibrosis [20].
Transcription activating factor-3 (Stat3), an important member of Stat family, participates in cellular signal transduction and transcriptional activation, and gets continuously activated in colon cancer and gastric cancer [21]. Lee has reported that Stat3 plays a key role in the occurrence and development of fibrosis in different organs, particularly in promoting fibrosis by inducing the production of ECM. While as an inhibitor of Stat3, HJC0123 can inhibit fibrosis markers in HSCs, such as type I collagen and ECM protein fibrinogen [22]. Another Stat3 inhibitor, S3I-201, improved fibrosis in preclinical models through inhibiting the activity of Stat3 by blocking Stat3 dimerization, Stat3-DNA binding, and transcription activity [23]. Stat3 is mainly involved in the biologic processes of COL1A1, such as negative regulation of cell-substrate adhesion, collagen fiber organization, and collagen biosynthetic processes. Moreover, studies have proven that the expression of activated Stat3 is positively correlated with MMP1, MMP2, and MMP10. In addition, Stat3 had a high affinity binding site in the MMP2 promoter region, which directly regulates the transcription of MMP2 [24]. The Stat3 knockout mice had a higher degree of liver fibrosis than CCl4 and 3,5-diethoxycarbon-yl-1,4-dihydrocollidine (DDC)-induced wild-type liver fibrosis models [25,26]. Moreover, activated Stat3 could stimulate liver cells to produce some unknown soluble factors, thus inhibiting the activation of HSCs and liver fibrosis. Stat3 might thus be a key signal transduction and transcription factor for regulating liver fibrosis.
NF-κB family of transcription factors, including rela (p65), NF-κB1 (p50 and p 105), NF-κB2 (p52 and p100), c-rel, and RelB, is present in almost all cell types. They can be activated by various intracellular and extracellular stimuli, such as cytokines, oxidized free radicals, and viral products [27,28]. Activated NF-κB1 enters cells and participates in various biologic processes, such as collagen fiber organization, collagen biosynthetic process, collagen catabolic process, and positive regulation of innate immune response. Rippe and Schrum have shown that NF-κB could inhibit α1(I) collagen gene expression and localized this inhibitory activity within the promoter region (within 220 bp of the transcription start site) of the α1(I) collagen gene [29].
Trp53 (Tp53 or p53), is a transcription factor and tumor suppressor, whose protein level and post-translational modification state can be changed due to cellular stress (such as hypoxia, or DNA, and spindle damage). Activation of Trp53 causes cell apoptosis while its inactivation may lead to tumors, and mutation or inactivation of Trp53 occur in more than 50% of tumors. Trp53 not only plays an important role in tumor diseases, but also participates in the development of chronic liver disease. Weng and Yang have detected mild fibrosis in the liver of adult Alb-Mcl-1-/- mice, and the loss of Trp53 exacerbated the damage of liver in Alb-Mcl-1-/- mice, along with enhanced occurrence and severity of liver fibrosis [30]. In a CCL4-induced mouse liver fibrosis model, deletion of Trp53 in HSCs increased fibroblast proliferation and ECM deposition, which finally enhanced liver fibrosis [31]. Trp53 also participates in the positive regulation of collagen biosynthetic process, platelet-derived growth factor receptor-beta signaling pathway, and platelet-derived growth factor receptor signaling pathway. Through its target gene PDGFRB, Trp53 participates in the positive regulation of HSCs activation. All these findings indicate that Trp53 plays an important role in the development of liver fibrosis.
Among these target genes, the COL1A1 encodes the largest protein connectivity, which suggests its core position in the network. Overexpression of collagen is the basic feature of liver fibrosis, and an important cause of organic pathologic change of organs. Besides, liver fibrosis is mainly characterized by abnormal accumulation of extracellular matrix components which mainly composed of type I collagen [32]. Thus, inhibiting the expression of type I collagen is one of the current strategies to treat liver fibrosis. When HSCs are activated, the expression of MMPs increases, along with the increased expression of TIMPs, the inhibitor of MMPs. If TIMPs increase too fast, the ratio of MMPs/TIMPs will change, which makes an imbalance of synthesis and degradation of ECM, and promotes the development of liver fibrosis [33,34]. During this process, the expression of MMP2 also increased significantly, which closely related to proliferation of activated HSCs. So inhibition of MMP2 activity is a choice to treat liver fibrosis. In general, PDGFRB in liver is minimally expressed, but significantly expressed in injured liver cells, thus it might have an important role in promoting fibrosis [35]. Studies have shown that one of the targets of RTK inhibitor imatinib mesylate (Gleevec) is PDGFRB, which can inhibit the activation of HSCs and reduce fibrosis [36]. At the same time, PDGFRB transcription factor Trp53 is also involved in the positive regulation of activated HSCs, which indicates that its antagonistic role against PDGFRB in anti-fibrosis has become an attractive research direction.
Liver damage and an inflammatory reaction result in excessive ECM deposition in the liver, which leads to liver fibrosis. The main components of ECM include collagen, elastase, and fibronectin [37]. However, much attention has been given to the function of collagen in liver fibrosis, and less attention to the role of elastin. As an ECM protein, FBLN5 plays an important role in composition of elastic fibers, and it has been reported that the loss of FBLN5 not only decreases tissue stiffness, but also diminishes the inflammatory response and abrogates the fibrotic phenotype in a mouse cutaneous fibrosis model [38]. ECM remodeling is related to liver fibrosis. Bracht has found that the expression of FBLN5 at the transcription and protein level increased significantly with the progression of the fibrosis stage in patients [39]. It was also noted that the expression of FBLN5 in a NAFLD group is significantly higher than in other groups. According to bioinformatic analysis, Yuan found that FBLN5 was the most up-regulated gene in NAFLD. GO analysis and KEGG pathway analysis also showed that FBLN5 was involved in ECM tissue [40]. Normally, Smad2 protein exists in the cytoplasm. When phosphorylated by the activation of TGF-β1, it will move to the nucleus, and initiate the transcription of ECM and other related genes in nucleus. After that the phosphorylated Smad2 would dephosphorylate, and return to the cytoplasm. Smad2 shuttling between the cytoplasm and nucleus would transmit a fibrogenic signal, which ultimately promotes the production of ECM proteins that accelerate liver fibrosis [41,42]. Smad3 can inhibit the activation of HSCs through the TGF-β signaling pathway and further inhibit the occurrence and development of liver fibrosis [43]. As the target gene of Smad2 and Smad3, FBLN5 plays an important role in the occurrence and development of liver fibrosis. Thus FBLN5 may be considered as a possible target gene for liver fibrosis treatment. However, how to regulate and control the relationship between FBLN5 and Smad2 and Smad3 in liver fibrosis requires further research. The process of liver fibrosis is very complicated, and usually involves changes at the gene transcription level. So research regarding transcription factors and signaling pathways related to liver fibrosis is needed in gene therapy for liver fibrosis.
Conclusions
Based on the results of PPI network analysis and module analysis related to liver fibrosis, 9 key genes were obtained, of which COL1A1, MMP2, PDGFRB and their transcription factors Stat3, Trp53, NF-κB1, and Sp1 were associated with the occurrence and development of liver fibrosis. Related studies have confirmed that COL1A1, MMP2, and PDGFRB were commonly used as targets for the treatment of liver fibrosis. Smad2 and Smad3 whose target gene is FBLN5, are closely related to the development of liver fibrosis. Thus FBLN5 appears to be a target for the treatment of liver fibrosis.
Acknowledgements
This work was partly supported by the Special Fund for the Talent of China Three Gorges University (8000303).
Disclosure of conflict of interest
None.
References
- 1.Leeming DJ, Byrjalsen I, Jiménez W, Christiansen C, Karsdal MA. Protein fingerprinting of the extracellular matrix remodelling in a rat model of liver fibrosis-a serological evaluation. Liver Int. 2013;33:439–447. doi: 10.1111/liv.12044. [DOI] [PubMed] [Google Scholar]
- 2.Ding N, Yu Ruth T, Subramaniam N, Sherman Mara H, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, Lau Sue L, Atkins Annette R, Barish Grant D, Gunton Jenny E, Liddle C, Downes M, Evans Ronald M. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang J, Mielenz D, Tomcik M, Fürnrohr BG, Scholtysek C, Dees C, Beyer C, Krönke G, Metzger D, Distler O, Schett G, Distler JH. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med. 2015;21:150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 4.Koyama Y, Brenner DA. Liver inflammation and fibrosis. J Clin Invest. 2017;127:55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiler-Normann C, Herkel J, Lohse AW. Mouse models of liver fibrosis. Z Gastroenterol. 2007;45:43–50. doi: 10.1055/s-2006-927387. [DOI] [PubMed] [Google Scholar]
- 6.Wright JH, Johnson MM, Shimizu-Albergine M, Bauer RL, Hayes BJ, Surapisitchat J, Hudkins KL, Riehle KJ, Johnson SC, Yeh MM, Bammler TK, Beyer RP, Gilbertson DG, Alpers CE, Fausto N, Campbell JS. Paracrine activation of hepatic stellate cells in platelet-derived growth factor C transgenic mice: evidence for stromal induction of hepatocellular carcinoma. Int J Cancer. 2014;134:778–788. doi: 10.1002/ijc.28421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folestad E, Kunath A, Wågsäter D. PDGF-C and PDGF-D signaling in vascular diseases and animal models. Mol Aspects Med. 2018;62:1–11. doi: 10.1016/j.mam.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Martin IV, Borkham-Kamphorst E, Zok S, van Roeyen CR, Eriksson U, Boor P, Hittatiya K, Fischer HP, Wasmuth HE, Weiskirchen R, Eitner F, Floege J, Ostendorf T. Platelet-derived growth factor (PDGF)-C neutralization reveals differential roles of PDGF receptors in liver and kidney fibrosis. Am J Pathol. 2013;182:107–117. doi: 10.1016/j.ajpath.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Campbell JS, Johnson MM, Bauer RL, Hudkins KL, Gilbertson DG, Riehle KJ, Yeh MM, Alpers CE, Fausto N. Targeting stromal cells for the treatment of platelet-derived growth factor C-induced hepatocellular carcinogenesis. Differentiation. 2007;75:843–852. doi: 10.1111/j.1432-0436.2007.00235.x. [DOI] [PubMed] [Google Scholar]
- 10.Tu X, Zhang H, Zhang J, Zhao S, Zheng X, Zhang Z, Zhu J, Chen J, Dong L, Zang Y, Zhang J. MicroRNA-101 suppresses liver fibrosis by targeting the TGFβ signalling pathway. J Pathol. 2014;234:46–59. doi: 10.1002/path.4373. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Kong LN, Huang C, Ma TT, Meng XM, He Y, Wang QQ, Li J. Hesperetin derivative-7 inhibits PDGF-BB-induced hepatic stellate cell activation and proliferation by targeting Wnt/β-catenin pathway. Int Immunopharmacol. 2015;25:311–320. doi: 10.1016/j.intimp.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2009;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2017;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JA, Suh DC, Kang JE, Kim MH, Park H, Lee MN, Kim JM, Jeon BN, Roh HE, Yu MY, Choi KY, Kim KY, Hur MW. Transcriptional activity of Sp1 is regulated by molecular interactions between the zinc finger DNA binding domain and the inhibitory domain with corepressors, and this interaction is modulated by MEK. J Biol Chem. 2005;280:28061–28071. doi: 10.1074/jbc.M414134200. [DOI] [PubMed] [Google Scholar]
- 17.Jia D, Ni YR, Zhang YQ, Rao C, Hou J, Tang HQ, Liu CB, Wu JF. SP1 and UTE1 Decoy ODNs inhibit activation and proliferation of hepatic stellate cells by targeting tissue inhibitors of metalloproteinase 1. Cell Biosci. 2016;6:31. doi: 10.1186/s13578-016-0094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Wang L, Song G, Han B. The mechanism through which octreotide inhibits hepatic stellate cell activity. Mol Med Rep. 2013;7:1559–1564. doi: 10.3892/mmr.2013.1385. [DOI] [PubMed] [Google Scholar]
- 19.García-Ruiz I, de la Torre P, Díaz T, Esteban E, Fernández I, Muñoz-Yagüe T, Solís-Herruzo JA. Sp1 and Sp3 transcription factors mediate malondialdehyde-induced collagen α1(I) gene expression in cultured hepatic stellate cells. J Biol Chem. 2002;277:30551–30558. doi: 10.1074/jbc.M203368200. [DOI] [PubMed] [Google Scholar]
- 20.Lee M, Song SU, Ryu JK, Suh JK. Sp1-dependent regulation of the tissue inhibitor of metalloproteinases-1 promoter. J Cell Biochem. 2004;91:1260–1268. doi: 10.1002/jcb.20021. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Yue P, Fletcher S, Zhao W, Gunning PT, Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem Pharmacol. 2010;79:1398–1409. doi: 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee DY, Yun SM, Song MY, Ji SD, Son JG, Kim EH. Administration of steamed and freeze-dried mature silkworm larval powder prevents hepatic fibrosis and hepatocellular carcinogenesis by blocking TGF-beta/STAT3 signaling cascades in rats. Cells. 2020;9:568. doi: 10.3390/cells9030568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Li J, Xiao W, Long J, Zhang H. The STAT3 inhibitor S3I-201 suppresses fibrogenesis and angiogenesis in liver fibrosis. Lab Invest. 2018;98:1600–1613. doi: 10.1038/s41374-018-0127-3. [DOI] [PubMed] [Google Scholar]
- 24.Tekle C, Nygren MK, Chen YW, Dybsjord I, Nesland JM, Mælandsmo GM, Fodstad Ø. B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int J Cancer. 2012;130:2282–2290. doi: 10.1002/ijc.26238. [DOI] [PubMed] [Google Scholar]
- 25.Abiko Y, Kojima T, Murata M, Tsujiwaki M, Takeuchi M, Sawada N, Mori M. Changes of tight junction protein claudins in small intestine and kidney tissues of mice fed a DDC diet. J Toxicol Pathol. 2013;26:433–438. doi: 10.1293/tox.2013-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louka ML, Ramzy MM. Involvement of fibroblast-specific protein 1 (S100A4) and matrix metalloproteinase-13 (MMP-13) in CCl4-induced reversible liver fibrosis. Gene. 2016;579:29–33. doi: 10.1016/j.gene.2015.12.042. [DOI] [PubMed] [Google Scholar]
- 27.Tergaonkar V, Correa RG, Ikawa M, Verma IM. Distinct roles of IκB proteins in regulating constitutive NF-κB activity. Nat Cell Biol. 2005;7:921–923. doi: 10.1038/ncb1296. [DOI] [PubMed] [Google Scholar]
- 28.Moynagh PN. The NF-κB pathway. J Cell Sci. 2005;118:4589. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 29.Rippe RA, Schrum LW, Stefanovic B, Solis-Herruzo JA, Brenner DA. NF-kappaB inhIbits expression of the alpha1(I) collagen gene. DNA Cell Biol. 1999;18:751–761. doi: 10.1089/104454999314890. [DOI] [PubMed] [Google Scholar]
- 30.Weng SY, Yang CY, Li CC, Sun TP, Tung SY, Yen JJ, Tsai TF, Chen CM, Chen SH, Hsiao M, Huang PH, Yang-Yen HF. Synergism between p53 and Mcl-1 in protecting from hepatic injury, fibrosis and cancer. J Hepatol. 2011;54:685–694. doi: 10.1016/j.jhep.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 31.Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh Darjus F, Bolden Jessica E, Zhao Z, Thapar V, Joyce Johanna A, Krizhanovsky V, Lowe Scott W. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kisseleva T, Brenner DA. Role of hepatic stellate cells in fibrogenesis and the reversal of fibrosis. J Gastroenterol Hepatol. 2007;22:S73–S78. doi: 10.1111/j.1440-1746.2006.04658.x. [DOI] [PubMed] [Google Scholar]
- 33.Tsukamoto H, Zhu NL, Asahina K, Mann DA, Mann J. Epigenetic cell fate regulation of hepatic stellate cells. Hepatol Res. 2011;41:675–682. doi: 10.1111/j.1872-034X.2011.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okazaki I, Noro T, Tsutsui N, Yamanouchi E, Kuroda H, Nakano M, Yokomori H, Inagaki Y. Fibrogenesis and carcinogenesis in nonalcoholic steatohepatitis (NASH): involvement of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinase (TIMPs) Cancers (Basel) 2014;6:1220–1255. doi: 10.3390/cancers6031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambrecht J, Verhulst S, Mannaerts I, Sowa JP, Best J, Canbay A, Reynaert H, van Grunsven LA. A PDGFRβ-based score predicts significant liver fibrosis in patients with chronic alcohol abuse, NAFLD and viral liver disease. EBioMedicine. 2019;43:501–512. doi: 10.1016/j.ebiom.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kocabayoglu P, Lade A, Lee YA, Dragomir AC, Sun X, Fiel MI, Thung S, Aloman C, Soriano P, Hoshida Y, Friedman SL. β-PDGF receptor expressed by hepatic stellate cells regulates fibrosis in murine liver injury, but not carcinogenesis. J Hepatol. 2015;63:141–147. doi: 10.1016/j.jhep.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du JJ, Sun JC, Li N, Li XQ, Sun WY, Wei W. β-Arrestin2 deficiency attenuates oxidative stress in mouse hepatic fibrosis through modulation of NOX4. Acta Pharmacol Sin. 2020 doi: 10.1038/s41401-020-00545-9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakasaki M, Hwang Y, Xie Y, Kataria S, Gund R, Hajam EY, Samuel R, George R, Danda D, M J P, Nakamura T, Shen Z, Briggs S, Varghese S, Jamora C. The matrix protein Fibulin-5 is at the interface of tissue stiffness and inflammation in fibrosis. Nat Commun. 2015;6:8574. doi: 10.1038/ncomms9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bracht T, Schweinsberg V, Trippler M, Kohl M, Ahrens M, Padden J, Naboulsi W, Barkovits K, Megger DA, Eisenacher M, Borchers CH, Schlaak JF, Hoffmann AC, Weber F, Baba HA, Meyer HE, Sitek B. Analysis of disease-associated protein expression using quantitative proteomics-Fibulin-5 is expressed in association with hepatic fibrosis. J Proteome Res. 2015;14:2278–2286. doi: 10.1021/acs.jproteome.5b00053. [DOI] [PubMed] [Google Scholar]
- 40.Yuan X, Sun Y, Cheng Q, Hu K, Ye J, Zhao Y, Wu J, Shao X, Fang L, Ding Y, Sun X, Shi X, Xue B. Proteomic analysis to identify differentially expressed proteins between subjects with metabolic healthy obesity and non-alcoholic fatty liver disease. J Proteomics. 2020;221:103683. doi: 10.1016/j.jprot.2020.103683. [DOI] [PubMed] [Google Scholar]
- 41.Gaarenstroom T, Hill CS. TGF-β signaling to chromatin: how Smads regulate transcription during self-renewal and differentiation. Semin Cell Dev Biol. 2014;32:107–118. doi: 10.1016/j.semcdb.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Lee H, Joung YK, Jung KH, Choi JH, Lee DH, Park KD, Hong SS. The use of low molecular weight heparin-pluronic nanogels to impede liver fibrosis by inhibition the TGF-β/Smad signaling pathway. Biomaterials. 2011;32:1438–1445. doi: 10.1016/j.biomaterials.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 43.Latella G, Vetuschi A, Sferra R, Catitti V, D’Angelo A, Zanninelli G, Flanders KC, Gaudio E. Targeted disruption of Smad3 confers resistance to the development of dimethylnitrosamine-induced hepatic fibrosis in mice. Liver Int. 2009;29:997–1009. doi: 10.1111/j.1478-3231.2009.02011.x. [DOI] [PubMed] [Google Scholar]