Abstract
Cancer stem cells (CSCs) are a cellular subpopulation accelerating cancer cell growth, invasion and metastasis and survival. After chemoradiotherapy, CSCs are enriched because of their survival advantages and lead to tumor relapse and metastasis. Elimination of CSCs is critically important for the radical treatment of human cancers. Long non-coding RNAs (lncRNAs) are a group of RNAs longer than 200 nucleotides and have no protein-coding potential. Aberrant expressions of lncRNAs are associated with human diseases including cancer. LncRNAs function as cancer biomarkers, prognostic factors and therapeutic targets. They induce cancer stemness by chromatin modification, transcriptional regulation or post-transcriptional regulation of target genes as a sponge or through assembling a scaffold complex. Several factors caused aberrant expressions of lncRNAs in CSCs such as genes mutations, epigenetic alteration and environmental stimuli. Targeting of lncRNAs has been demonstrated to significantly reverse the chemoradioresistance of CSCs. In this review, we have summarized the progress of studies regarding lncRNAs-mediated therapy resistance of CSCs and clarified the molecular mechanisms. Furthermore, we have for the first time analyzed the influences of lncRNAs on cell metabolism and emphasized the effect of tumor microenvironment on lncRNAs functions in CSCs. Overall, the thorough understanding of the association of lncRNAs and CSCs would contribute to the reversal of therapy resistance.
Keywords: Cancer stem cells, long non-coding RNAs, therapy resistance, anti-cancer targets, molecular mechanisms
Introduction
Cancer stem cells (CSCs), also known as tumor initiating cells (TICs) are a cellular subpopulation with the characteristics of self-renewal, refractory resistance to conventional therapies and potent differentiation potential [1]. CSCs often show high expressions of stemness markers, epithelial to mesenchymal transition (EMT) and survival advantages over non-CSCs. It is widely accepted that CSCs are the major cause of tumor relapse and metastasis since they survive chemoradiotherapy [2]. Furthermore, CSCs are enriched after chemoradiotherapy and render cancers cells to be more resistant [3]. Therefore, eradication of CSCs may be essential for the radical treatment of human cancers. The acquisition of cancer stemness is a very complex and heterogeneous process in which many genetic and epigenetic factors are involved.
During the past decades, the researchers have revealed that less than 2% of the human genome is transcribed into protein-coding RNAs while the rest dose not code proteins [4,5]. Long non-coding RNAs (lncRNAs) are characterized as a class of non-coding RNAs with more than 200 nucleotides and expressed mainly in the cytoplasm. Similar to mRNAs, lncRNAs are equipped with a poly-A tail and 5’ cap and Pol II transcripts. Although lncRNAs are expressed at relatively low level and show low evolutionary conservation, they play significant roles in human diseases including cancer. LncRNAs exert their physiological and pathological roles by transcriptional or post-transcriptional regulation of target genes. They are involved in cancer initiation and progression and serve as cancer biomarkers, prognostic factors and therapeutic targets [6,7]. They regulate a variety of cellular processes including cancer cell growth, metabolism, metastasis and invasion, survival and stemness [8-12]. The genes mutations, epigenetic alteration and environmental stimuli all contribute to aberrant expressions of lncRNAs in cancer cells.
In this review, we have focused on the progress of studies regarding lncRNAs-mediated therapy resistance of CSCs and analyzed their expressions, functions and action mechanisms. The signaling pathways and transcription factors regulated by lncRNAs in CSCs have been summarized. Furthermore, we have for the first time dissected the association of cell metabolism with lncRNAs-mediated therapy resistance in CSCs. We have also emphasized the effect of tumor microenvironment on lncRNAs functions in CSCs. Overall, the thorough understanding of the molecular mechanisms of lncRNAs would contribute to overcoming the chemoradioresistance of CSCs.
The aberrant expressions of lncRNAs in CSCs
Many factors lead to aberrant expressions of lncRNAs in CSCs such as genes mutations, epigenetic alteration and environmental stimuli. P53, a known tumor suppressor, was frequently mutated in human cancers. Yuechao Zhao et al reported that p53-R273H, one mutated form of p53, up-regulated the expressions of lnc273-31 and lnc273-34, which endowed colorectal cancer cells with stemness-like phenotype [13]. Some stemness markers activated the transcription of lncRNAs as transcription factors to maintain the stemness of cancer cells. SOX9, a member of SOX family, which are effective inducers for the formation of stem-like phenotypes, induced colorectal cancer (CRC) stemness by activating the transcription of lncRNA phenylalanyl-tRNA synthetase subunit alpha antisense RNA 1 (FARSA-AS1) through binding to FARSA-AS1 promoter [14]. In turn, FARSA-AS1 elevated SOX9 expression by absorbing miR-18b-5p and augmented FARSA via sequestering miR-28-5p, creating a positive feedback of SOX9 expression to accelerate tumor progression. SOX2 was demonstrated to enhance the expression of lncRNA H19, which was responsible for the progenitor property of tumor-initiating hepatocytes (TICs) in vitro and the tumorigenic potential in vivo in hepatocellular carcinoma [15]. Oct4, another stemness marker, initiated the transcriptions of lncRNAs NEAT1 and MALAT1 in lung cancer as transcription factors [16]. Inhibition of NEAT1 and MALAT1 rescued the tumor-promotion activity of Oct-4. MiRNAs, which are another common class of non-coding RNAs (ncRNAs), play their roles also through transcriptional or translational regulation of target genes. It has been demonstrated that miRNAs regulated lncRNAs expression by their direct interaction, accelerating EMT and tumor progression [17]. The death stimuli such as anti-cancer drugs or ionizing radiation also caused aberrant expressions of lncRNAs, inducing the acquisition of cancer stemness and therapy resistance. Furthermore, tumor microenvironment has important effect on the expressions and functions of lncRNAs in CSCs, which was specially reviewed in the latter part.
The functions of lncRNAs in therapy resistance of CSCs
CSCs are intrinsically chemoradioresistant due to several mechanisms such as high expressions of drug-efflux pumps, enhanced DNA repair capability and strong defense against ROS [18-22]. Surviving CSCs lead to tumour recurrence and metastasis after chemoradiotherapy. Elimination of CSCs is critically important for the complete tumor killing. The roles of CSCs in therapy resistance of human cancers were shown in Figure 1. LncRNAs were involved in therapy resistance by regulating the behaviors of CSCs, and thus become attractive anti-cancer targets in several human cancers. Junlong Zhuang et al reported that lncRNA-LET was down-regulated due to the activation of TGFβ/SMAD signaling after a long-time treatment with gemcitabine, leading to the enrichment of CSCs in bladder cancer. Their study demonstrated that down-regulation of lncRNA-LET increased the protein expression of NF90, which in turn inhibited miRNA-145 expression and promoted cancer stemness. Therefore, the enrichment of CSCs in bladder cancer induced gemcitabine resistance through the lncRNA-LET/NF90/miR-145 signaling axis [23]. In triple-negative breast cancer, lncRNA NEAT1 played an oncogenic role through increasing CD44+/CD24-, ALDH+, and SOX2+ stem cell populations and inducing chemoresistance [24]. LncRNA MALAT1 induced the resistance of CSCs to temozolomide through promoting the expressions of anti-apoptotic Bcl-2, HSP70, and inhibitors of apoptosis proteins (IAPs) family (cIAP-1, cIAP-2, XIAP and survivin) and multidrug resistance protein 1 (MRP1) in glioblastoma [25]. LncRNA AFAP1-AS1 was demonstrated to be highly expressed in human laryngeal specimens, promote cancer stemness and induce cisplatin resistance. AFAP1-AS1 negatively regulated the expression of miR-320a, resulting in overexpression of miR-320a target gene RBPJ, which activated Notch signaling pathway as a transcriptional effector [26]. LncRNA brain cytoplasmic 200 (BC200) RNA was aberrantly expressed in several human cancers. Recently, BC200 was demonstrated to be highly expressed in blood and in tumour tissues of glioblastoma patients. High expression of BC200 enhanced cancer stemness and temozolomide resistance by up-regulation of stemness related markers and multidrug resistance proteins through interfering with miR-218-5P expression. Targeting the BC200/miR218-5p signaling axis significantly improved the sensitivity to temozolomide and attenuated stemness features of glioblastoma cells [27]. The multidrug resistance (MDR) proteins, which are drug flux transporters and promote drug resistance, were often overexpressed in CSCs, resulting in the intrinsic resistance of CSCs. By up-regulating ABCB5, a known drug efflux transporter, lnc00963 conferred oral cancer cells stemness-like traits including drug resistance, increased self-renewal, invasion and colony formation ability [28].
Figure 1.
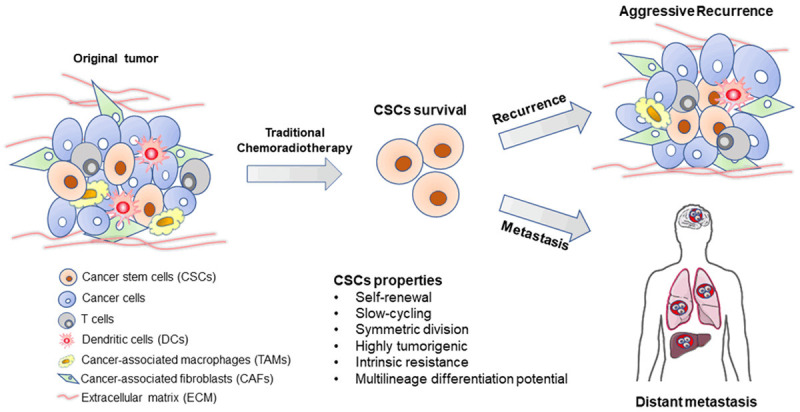
The roles of CSCs in therapy resistance of human cancers. CSCs are intrinsically resistant to chemoradiotherapy. Surviving CSCs induce tumor recurrence and metastasis after chemoradiotherapy.
Radiotherapy is used as one curative treatment modality for more than 50% cancer patients. Accumulating evidences have suggested that lncRNAs were also involved in radiotherapy resistance by regulating cancer stemness [29]. Shlomit Brodie et al reported that lncRNA TALNEC2 was overexpressed in glioblastoma (GBM) in vitro and in vivo due to E2F1-mediated transcription activation. Inhibition of TALNEC2 suppressed GBM cell proliferation and arrested cell cycle in the G1/S phase. Moreover, inhibition of TALNEC2 attenuated the self-renewal and mesenchymal transformation of GBM stem cells, enhanced radiation sensitivity and prolonged the survival of tumour-bearing mice partly through the recovery of miR-21 and miR-11 expressions [30]. Another study by Wei Yang et al also demonstrated that radiation sensitivity was improved by interfering with the expressions of lncRNAs in GBM CSCs (GSCs). They found that lincRNAp21, which is a tumour suppressor and regulates cell cycle and apoptosis in several human cancers, was down-regulated due to up-regulation of Hu antigen R (HuR) caused by miR-146b-5p down-regulation in GSCs. LincRNAp21 negatively regulated β-catenin, which is a critical activator of wnt/β-catenin signaling pathway. Decreased lincRNAp21 induced the activation of wnt/β-catenin signaling pathway, inducing the acquisition of stemness and radioresistance of GSCs. Overexpression of lincRNAp21 inhibited self-renewal of GSCs and enhanced the radiosensitivity [31]. Together, lncRNAs controlled CSCs response to chemoradiotherapy and thus the targeting of lncRNAs may be new avenues to overcoming the chemoradioresistance of CSCs.
The action mechanisms of lncRNAs in regulating therapy resistance in CSCs
Although lncRNAs have no protein-coding capability, they play their pathological roles by controlling the expressions of target genes at transcriptional or post-transcriptional levels. They activate or inhibit the transcription of target genes and regulate the activity, stability and translocation of functional proteins. They competitively bind to special RNAs or proteins as a sponge or through assembling a scaffold complex with other regulatory components to regulate target genes expressions. In CSCs, lncRNAs regulated therapy resistance mainly through controlling the expressions of genes involved in cancer stemness.
Acted as a sponge
Multidrug resistance gene 1 (MDR1) was associated with cancer cell broad-spectrum resistance to chemotherapeutic agents. LncRNA FENDRR was demonstrated to bind to the 3’ untranslated region (3’UTR) of MDR1, prevent the binding of RNA binding protein HuR to MDR1 3’UTR and thus inhibit MDR1 expression. The recovery of FENDRR expression attenuated the stemness of non-small cell lung cancer cells through down-regulating MDR1 expression [32]. In CD133+/CD166+/CD44+ colon cancer stem cells, lncRNA 1567 (LINC01567) acted as a sponge to competitively bind to miRNA-93, eliminating the inhibitory effect of miRNA-93 on target genes expressions including HDAC8, TLE4, stratifin and MSI1. Up-regulation of miRNA-93 or down-regulation of MSI1 reduced the tumorigenic role of LINC01567 [33]. Overexpression of lncRNA DLX6-AS1 was associated with cancer stemness in osteosarcoma. DLX6-AS1 competitively bound to miR-129-5p, by which the inhibitory effect of miR-129-5p on DLK1 was reversed, resulting in wnt signaling activation and wnt target genes expressions such as c-Myc, SOX2, Oct4 and Nanog [34]. In pancreatic cancer, linc-DYNC2H1-4 was overexpressed and resulted in acquired gemcitabine resistance by increasing cancer stem subpopulations. Linc-DYNC2H1-4 acted as a sponge to competitively bind to miRNA-145, which specially inhibited epithelial to mesenchymal transition and the expressions of cancer stemness markers including Lin28, Nanog, Sox2 and Oct4 [35]. Therefore, linc-DYNC2H1-4 increased pancreatic cancer stemness and gemcitabine resistance by negatively regulating miRNA-145, one tumor suppressor. Long non-coding RNA FOXD2 Adjacent Opposite Strand RNA 1 (FOXD2-AS1) was associated with cancer progression and recurrence. Xiaoli Liu et al demonstrated that high expression of FOXD2-AS1 conferred thyroid cancer cells stemness-like features and anoikis resistance in vitro. They found that FOXD2-AS1 interacted with miR-7-5p as a sponge and induced overexpression of miR-7-5p target gene telomerase reverse transcriptase (TERT). Inhibition of FOXD2-AS1 repressed thyroid cancer cells stemness-like features and reversed anoikis resistance in vitro [36]. LncRNA HOTAIR has been investigated intensively due to its close association with tumour initiation and progression. Ning Wang et al demonstrated that HOTAIR promoted the expansion of prostatic cancer stem-like cells and induced docetaxel resistance by sequestering miR-590-5p as a sponge and inducing overexpression of IL-10, which initiated the activation of STAT3 signaling [37]. LncRNA ASB16-AS1 was revealed as an inducer of gastric cancer stemness and cisplatin resistance. By acting as a sponge, ASB16-AS1 interacted with miR-3918 and miR-4676-3 and increased the expression of TRIM37, resulting in the activation of NF-κB pathway. In the meanwhile, ASB16-AS1 cooperated with ATM kinase, facilitated TRIM37 phosphorylation and further promoted the activation of NF-κB pathway. Therefore, ASB16-AS1 increased the expression of TRIM37 and improved its activity, resulting in constitutive activation of NF-κB pathway and cancer stemness [38]. In NSCLC, lncRNA DGCR5 was enriched in CSCs and responsible for the action of CSCs. DGCR5 inhibited the expression of miR-330-5p, which negatively regulated cancer stemness marker CD44 expression, by interaction with it as a sponge. Thus, overexpression of DGCR5 contributes to CSC-like traits via modulating miR-330-5p/CD44 axis in NSCLC [39].
Acted through assembling a scaffold complex
SOX2 is essential for self-renewal and conferred bladder cancer stem cells (BCSCs) chemoresistance. Low expressed in Bladder Cancer Stem cells (lncRNA-LBCS) suppressed self-renewal and chemoresistance of BCSCs through assembling a scaffold complex with heterogeneous nuclear ribonucleoprotein K (hnRNPK) and enhancer of zeste homolog 2 (EZH2), which suppressed the expression of SOX2 by inducing H3K27me3 of SOX2 promoter [40]. LncRNA Zinc2 (lncZic2) was highly expressed in liver cancer and liver CSCs and responsible for the maintainance of stemness characteristics. LncZic2 interacted with BRM/SWI2-related gene 1 (BRG1), a component of chromatin remodelling complex, and directed it to MARCKS and MARCKSL1 promoter, promoting the expressions of MARCKS/MARCKSL1. Inhibition of BRG1 or MARCKS/MARCKSL1 attenuated the self-renewal capability of liver CSCs [41]. G protein coupled receptors (GPCR) are discovered as important anti-cancer targets due to their critical regulatory roles in signal transduction [42]. Through an unbiased screening for GPCR expression, GPR107 was found to be the top GPCR expressed in liver cancer and liver CSCs. Moreover, GPR107 was responsible for CSCs self-renewal. LncGPR107, which was located neighbouring to GPR107 on the genome, recruited SRCAP complex to GPR107 promoter and initiated its transcription activation. Targeting lncGPR107-SRCAP-GPR107 axis significantly inhibited CSCs activity in liver cancer [43].
The signaling pathways and transcription factors involved in lncRNAs-mediated cancer stemness and therapy resistance
The signaling pathways
Wnt/β-catenin signaling pathway was closely associated with cancer stemness in several human cancers [44,45]. Wnt ligands bind to their receptors on cell surface such as frizzled-related family members and LRP5/6, forming an active complex to initiate the downstream signaling cascade. β-catenin is defined as a central modulator of wnt/β-catenin signaling pathway. When wnt ligands bind to their receptors, β-catenin is phosphorylated, dissociated from the APC/Axin/GSK-3β complex and translocated into the nucleus. β-catenin then interacts with TCF/LEF transcription factors to activate the transcription of target genes including those cancer stemness inducers. LncRNAs were demonstrated to induce cancer stemness through regulating wnt/β-catenin signaling pathway in several human cancers. They competitively bound to the inhibitors of wnt/β-catenin signaling pathway as a sponge or regulate the transcription activity of β-catenin through assembling a scaffold complex. In cisplatin resistant NSCLC A549 cells, lncRNA NEAT1 was overexpressed and responsible for CSCs enrichment by activation of wnt signaling pathway. Inhibition of NEAT1 restrained the stemness traits and induced apoptosis of cisplatin resistant A549 cells [46]. Another study also highlighted the involvement of wnt signaling pathway in NSCLC stemness. The authors demonstrated that lncRNA CCAT1 inhibited let-7c, one suppressor of wnt signaling pathway, resulting in the activation of wnt signaling pathway and cancer stemness in NSCLC. Down-regulation of CCAT1 or overexpression of let-7c promoted asymmetric division of NSCLC stem cells and therefore restrained cancer stemness [47]. In breast cancer, lncRNA H19 stimulated symmetric division of CSCs, resulting in their expansion by specially inhibiting let-7c, by which oestrogen receptor activated Wnt signaling was released. In turn, increased wnt signaling stimulated high expression of H19, thereby creating a positive H19/Wnt regulatory loop [48]. In colon cancer, lncRNA RBM5-AS1/LUST was enriched in CSCs. As a nuclear retained RNA, LUST directly interacts with β-catenin, stabilizes the β-catenin/TCF-4 complex and potentiates the transcription of β-catenin target genes including CMYC, CCND1, YAP1 and SGK1 [49]. LncRNA BCAR4 was highly expressed in gastric cancer tissue compared with in adjacent tissue and significantly associated with tumor size, stage and patients survival. Down-regulation of BCAR4 expression increased the sensitivity of gastric cancer cells to cisplatin by inhibition of stem cells markers such as β-catenin, Nanog, Oct3/4, Sox2, c-Myc, and Klf4 due to wnt signaling inactivation [50]. LncRNA THOR (testis-associated highly conserved oncogenic long non-coding RNA), promoted liver cancer stem cells expansion and resistance to sorafenib through activating β-catenin signaling pathway [51]. Recently, Hang-Lung Chang et al demonstrated that lncRNA MALAT1, which was overexpressed in hepatocellular carcinoma (HCC), bound to β-catenin directly to faciliate the activation of wnt/β-catenin pathway [52]. Inhibition of MALAT1 significantly repressed the nuclear translocation of β-catenin and quelled the aberrant activation of the Wnt/β-catenin. Inhibition of Wnt/β-catenin pathway attenuated caner stemness, abrogated cancerous liver cell metastasis and clonogenicity and suppressed in vivo tumor initiation and growth. Therefore, MALAT1 was an attractive molecular candidate and the therapeutic targeting of MALAT1 may constitute a novel promising anticancer strategy for HCC treatment.
In addition to wnt/β-catenin pathway, Hedgehog (Hh) signaling pathway and Notch signaling pathway are recognized as another two major stemness-related pathways. In colorectal cancer (CRC), lncRNA-cCSC1 induced cancer stemness by activation of Hh pathway. Inhibition of lncRNA-cCSC1 attenuated the self-renewal capability of colon cancer stem cells and enhanced the sensitivity to 5-fluorouracil [53]. In laryngeal carcinoma, LINC-PINT was down-regulated and associated with cancer stemness and cisplatin resistance. Down-regulation of LINC-PINT induced an increase of miR-425-5p expression while decreased the expression of PTCH1, which is a tumor suppressor protein of the Hh pathway [54]. Therefore, the authors speculated that LINC-PINT inhibited cancer stemness of laryngeal carcinoma possibly through inactivating the Hh pathway. Symmetric division is an important mechanism for CSCs expansion. Guanglin Huang et al demonstrated that down-regulation of lncRNA TUSC-7 promoted the renewal ability of lung adenocarcinoma stem cells, yielding to their symmetric division. Low expression of TUSC-7 recovered the degradation of NUMB by miR-146 and led to Notch signaling activation and the acquisition of cancer stemness [55]. Furthermore, MAPK pathway was also closely associated with cancer stemness. LncRNA H19 conferred CD133+ liver CSCs chemoresistance through activation of MAPK/Erk pathway and reduction of oxidative stress [56]. Guanqun Huang et al reported that activation of MAPK signaling pathway was involved in live CSCs. They found that lncRNA MAPK6 interacted with and recruited RNA polymerase II to MAPK6 promoter, resulting in the activation of MAPK6 transcription, which contributed to the self-renewal of live CSCs. Targeting lncRNA MAPK6 or MAPK6 could effectively eliminate live CSCs [57].
The transcription factors
Signal Transducer and Activator of Transcription (STAT) proteins are a group of transcription factors which control tumor cells growth, metastasis and survival through initiating the transcription of target genes [58]. STAT3 is phosphorylated and activated by the Janus Kinase (JAK) family, dimerized and translocated to the nucleus to initiate transcription. STAT3 activation was reported to be associated with poor prognosis of patients with lung cancer, liver cancer and renal cell carcinoma (RCC) [59-61]. STAT3 activation was also responsible for the formation of tumor spheres and the viability of CSCs in several human cancers. LncARSR promoted liver CSCs expansion and resistance to cisplatin and sorafenib by activating STAT3 signaling [62]. LncRNA PTCSC3 was defined as a tumor suppressor of thyroid cancer. Xiao-ming Wang et al reported that overexpression of PTCSC3 suppressed stem cells properties and increased the sensitivity of anaplastic thyroid cancer to doxorubicin by inhibiting STAT3 signaling and the expression of INO80, which is a positive regulator of thyroid cancer stemness [63]. SOX9, as a transcription factor, has been identified to induce cancer stemness in glioma and osteosarcoma. Furthermore, SOX9 was demonstrated to be involved in lncRNAs THOR-mediated gastric cancer stemness. THOR directly bound to SOX9 untranslated region (3’UTR), promoting SOX9 expression. Inhibition of THOR or SOX9 reduced gastric cancer stemness and enhanced cell sensitivity to cisplatin [64]. YAP is a transcription co-activator in Hippo signaling and participates in the expansion of CSCs in several human cancers. LncARSR contributed to self-renewal, tumorigenicity and metastasis of renal CSCs by physically interacting with YAP and prompting YAP nuclear translocation while LATS1-mediated YAP phosphorylation was repressed. In turn, the YAP/TEAD complex bound to lncARSR promoter and accelerated lncARSR expression, forming a positive feedback loop in renal CSCs [65]. Furthermore, YAP was demonstrated to be involved in lncRNA MALAT1 induced stemness of ovarian cancer cells. By interaction with YAP, MALAT1 inhibited its translocation from nucleus to cytoplasm, stabilized it and increased its activity, leading to the enhancement of cancer stemness and cisplatin resistance [66]. In liver cancer, MALAT1 increased the expression of YAP1 by sponging miRNA-375, which inhibited YAP1 expression through binding to its 3’-UTR. Inhibition of MALAT1 attenuated liver CSCs features including decreased sphere formation capacity and stemness genes expressions [67]. HOXA10, as a member of HOX transcription factor family, participates in several physiological and pathological processes such as tumorigenesis. Ming Shao et al demonstrated that HOXA10 was highly expressed in liver CSCs and contributed to liver CSCs self-renewal and liver tumorigenesis. LncRNA HOXA10 (lnc HOXA10), which locates closely with HOXA10 on the chromosome, was defined as a modulator of HOXA10 expression. Lnc HOXA10 recruited SNF2L, a component of the epigenetic complex NURF, which regulates chromatin remodelling and transcriptional initiation, to the promoter of HOXA10, resulting in HOXA10 overexpression. Therefore, the lncHOXA10/SNF2L/HOXA10 axis was highlighted as an attractive target to eradicate liver CSCs [68]. The signaling pathways and transcriptional factors involved in lncRNAs-mediated cancer stemness and therapy resistance were summarized in Figure 2.
Figure 2.
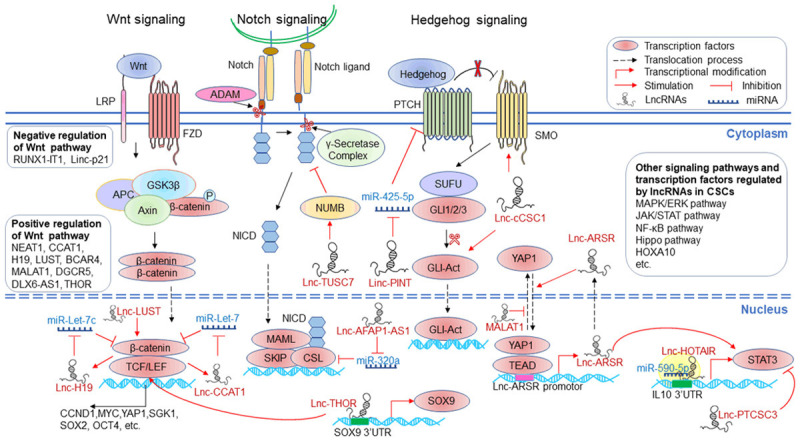
The signaling pathways and transcriptional factors involved in lncRNAs-mediated cancer stemness and therapy resistance.
The involvement of cell metabolism in lncRNAs-mediated cancer stemness and therapy resistance
Abnormal cell metabolism is an important hallmark of cancer [69]. The cell metabolism of glucose, amino acids and lipids supply energy and mass for cancer cell growth, metastasis and survival. Furthermore, abnormal cell metabolism contributes to the acquisition of cancer stemness [70,71]. Accumulating evidences have suggested that lncRNAs conferred therapy resistance by regulating CSCs metabolism. Glycolysis is a central pathway of glucose metabolism and has been demonstrated to maintain cancer stemness and induce chemoresistance [72,73]. Glycolysis is more active in CSCs compared with in non-CSCs because of increased glucose uptake and glycolytic enzyme expressions in CSCs [74-76]. Inhibition of glycolysis attenuated cancer stemness. In breast cancer, F Peng et al reported that lncRNA H19 interacted with miRNA let-7 as a competitive endogenous RNA to release hypoxia-inducible factor 1α, resulting in up-regulation of pyruvate dehydrogenase kinase 1 (PDK1), a critical glycolytic enzyme. Increased expression of PDK1 promoted glycolysis to enhance stemness in hypoxia. Depletion of H19 or PDK1 suppressed the maintainance of breast cancer stemness [77]. Another study by Fei Ma et al also highlighted the association of lncRNAs regulated cell metabolism and breast cancer stemness [78]. They found that lncRNA FGF13-AS1 was down-regulated in breast tumor tissues and inhibited glycolysis and stemness properties of breast cancer cells. By competitively interacting with IGF2BP proteins, FGF13-AS1 reduced the mRNA stability of c-Myc, which controls glycolysis by regulating glucose transporters and glycolytic enzymes. In turn, c-Myc, as a transcription factor, inhibited the transcription of FGF13-AS1, forming a negative feedback loop. Therefore, FGF13-AS1 inhibited glycolysis and breast cancer stemness by negatively regulating c-Myc through binding to IGF2BP proteins, which are RNA binding proteins and prevents c-Myc mRNA degradation. Glioblastoma multiform (GBM) is the most common brain tumor with a dismal 5-year overall survival rate [79]. Temozolomide (TMZ), an alkylating agent, is prescribed as a first-line chemotherapeutic drug for the treatment of GBM [80]. Gal Mazor et al reported that lncRNA TP73-AS1 was responsible for the induction of TMZ resistance by glioblastoma multiform cancer stem cells (gCSCs) through regulating the expressions of genes involved in metabolism, mitochondria, and nucleotide metabolism and curbing ROS levels. LncRNA TP73-AS1 also promoted the expression of ALDH1A1, a known marker of CSCs and inducer of tumor chemoresistance in GBM. Their study recovered TP73-AS1 as a prognostic factor for GBM patients receiving TMZ treatment [81]. Thus, the regulation of cell metabolism by lncRNAs played important roles in cancer stemness and therapy resistance.
The effect of tumor microenvironment on lncRNAs-mediated cancer stemness and therapy resistance
Tumor cells have close contact with their host tumor microenvironment (TME), which promotes tumor initiation and progression [82]. TME supports tumor growth, migration and invasion and cell resistance to chemoradiotherapy by inducing cancer stemness, epithelial to mesenchymal transition, regulating DNA damage and inhibiting apoptosis [83]. In this review, we have for the first time focused on the effect of TME on lncRNA regulated therapy resistance in CSCs. TME regulates the expressions of lncRNAs in tumor cells. Hypoxic is an important characteristic of TME and promotes the malignant progression of tumors such as increased cells proliferation, metastasis, therapy resistance and cancer stemness. Hypoxia/HIF-1α signaling has been shown to modulate the non-coding transcriptome including lncRNAs and miRNAs. In hepatocellular carcinoma (HCC), hypoxia-driven histone deacetylase 3 (HDAC3) promoted cancer stemness by inhibiting the expression of lncRNA RUNX1-IT1. Mechanical studies revealed that lncRNA RUNX1-IT1 directly bound to miR-632, acting as a competing endogenous RNA to facilitate the expression of the miR-632 target gene GSK-3β and suppress WNT/β-catenin pathway activation [84]. In multicellular tumor spheroids culture (MCTS) of breast cancer cell MCF-7 which mimics the traits of TME including hypoxia and acidosis, lncRNA HAL (an uncharacterized lncRNA) was detected in the quiescent stem cell population. Silencing of lncRNA HAL inhibited the proportion and function of CSCs, confirming that TME was involved in cancer stemness by regulating lncRNAs expressions [85]. Furthermore, stromal cells endowed cancer cells with CSCs features by regulating lncRNAs expressions. Our previous study demonstrated that cancer-associated fibroblasts (CAFs), one major component of TME, increased the expression of lncRNA DNM3OS, which was a critical mediator of radioresistance by inhibiting irradiation induced DNA damage while enhancing DNA damage repair in esophageal cancer cells [86]. Stromal cells secreted lncRNAs in exosomes to cancer cells [87]. In colorectal cancer (CRC), CAFs promoted the stemness and conferred resistance to oxaliplatin by secretion of lncRNA H19-loaded exosomes. Upon access to colorectal cancer cells, H19 acted as a sponge to activate the β-catenin pathway through competitively binding to miR-141, an inhibitor of CRC stemness [87]. Similar to CAFs, mesenchymal stem cells (MSCs), as another component of TME, regulate proliferation, metastasis and angiogenesis of cancer cells [88,89]. Moreover, MSCs are able to endow cancer cells with CSCs feathers. The co-culture of gastric cancer cells and MSCs induced overexpression of lncRNA HCP5, which interacted with miR-3619-5P as a sponge and up-regulated PPARG coactivator 1 alpha (PPARGC1A). Then, carnitine palmitoyltransferase 1 (CPT1) was transcriptionally activated, enhancing fatty acid oxidation (FAO), which maintains gastric cancer cells stemness [90]. Furthermore, MSCs induced stemness by reprogramming FAO in gastric cancer cells [91]. The transforming growth factor β1 (TGF-β1), which was secreted by MSCs, induced an activation of SMAD2/3 signaling upon binding to its receptors and increased lncRNA MACC1-AS1 expression in gastric cancer cells. MACC1-AS1 enhanced FAO and induced gastric cancer cells stemness and resistance to 5-FU and oxaliplatin by sequestering miR-145-5p, which is correlated with lipid metabolism and inhibits drug resistance. Tumor-associated macrophages (TAMs), as the major immunosuppressive cells in TME, were demonstrated to mediate stemness and drug resistance through the crosstalk with tumor cells. Yingnan Ye et al revealed that TAMs induced overexpression of lncRNA H19, which sequestered the tumor suppressor miR-193b, increasing MAPK1 expression to activate MAPK signaling pathway in hepatocellular carcinoma cells. Thus, the H19/miR-193b/MAPK1 axis induced stemness and epithelial to mesenchymal transition of hepatocellular carcinoma cells [92]. The effect of tumor microenvironment on lncRNAs-mediated therapy resistance in CSCs was shown in Figure 3.
Figure 3.
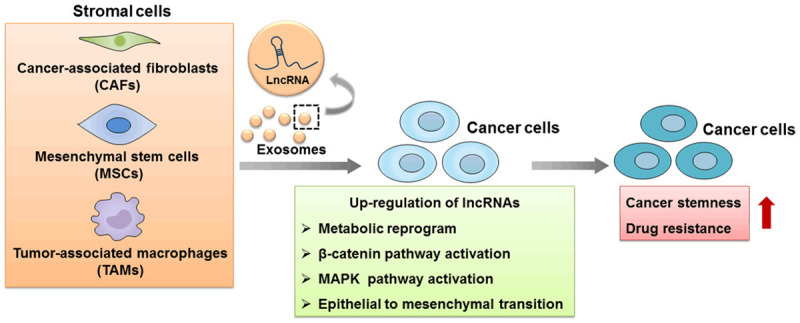
The roles of tumor microenvironment in lncRNAs-mediated cancer stemness and therapy resistance. The components of tumor microenvironment such as CAFs, MSCs and TAMs can secret lncRNAs in exosomes or up-regulate lncRNAs expressions in cancer cells. This crosstalk of tumor microenvironment and cancer cells induced metabolic reprogramming, β-catenin pathway activation, MAPK pathway activation or epithelial to mesenchymal transition, which confers cancer cells stemness and increased drug resistance.
Conclusions and perspectives
CSCs are the major obstacle for the complete tumor killing and thus lead to tumor recurrence and metastasis due to their intrinsic therapy resistance. However, there have not been feasible approaches to directly target CSCs to eliminate their influences until now [2]. LncRNAs exert their pathological roles by regulating the expressions of target genes at transcriptional or post-transcriptional levels. They served as the inducers or suppressors of cancer stemness and controlled tumor response to chemoradiotherapy as shown in Table 1. They were discovered as prognostic factors of cancer patients due to their critical roles in CSCs. Since lncRNAs are secreted and circulated in the body, they can be detected in tumor tissues as well as in body fluids such as serum, plasma, urine and saliva [7,93]. Furthermore, lncRNAs are highly sensitive and stable and therefore are suitable for the non-invasive detection to diagnose, monitor and manage cancer [7]. Ling-Yun Lin et al demonstrated that lncUEGC1 encapsulated within exosomes in the plasma was highly sensitive, stable, and may be used as a promising biomarker for early-stage gastric cancer [94]. In the clinic, lncRNA PCA3 has been routinely used as a biomarker for prostate cancer [95]. Furthermore, several clinical trials are being undergoing to evaluate the effectiveness of circulating lncRNAs as biomarkers or prognostic factors in human cancers. Therefore, the lncRNAs involved in cancer stemness may be dynamically detected by liquid biopsy to predict tumor response to chemoradiotherapy and prognosis of the patients.
Table 1.
The lncRNAs involved in cancer stemness
| Cancer type | Function | Mechanisms | Ref. | |
|---|---|---|---|---|
| LncRNA-LET | Bladder cancer | Inhibition of cancer stemness and gemcitabine resistance | Down-regulation of lncRNA-LET increased the protein expression of NF90 which in turn inhibited miRNA-145 expression and promoted cancer stemness. | [23] |
| NEAT1 | Triple-negative breast cancer | Induction of cancer stemness | NEAT1 increased CD44+/CD24-, ALDH+, and SOX2+ stem cell populations. | [24] |
| NSCLC | Induction of cancer stemness and cisplatin resistance | NEAT1 was overexpressed and activated wnt signaling pathway. | [46] | |
| AFAP1-AS1 | Laryngeal cancer | Induction of cancer stemness | AFAP1-AS1 negatively regulated the expression of miR-320a, resulting in overexpression of RBPJ and Notch signaling activation. | [26] |
| BC200 | Glioblastoma | Induction of cancer stemness and temozolomide resistance | BC200 up-regulated stemness related markers and multidrug resistance proteins through interfering with miR-218-5P expression. | [27] |
| Lnc00963 | Oral cancer | Induction of cancer stemness | By up-regulating ABCB5, a known drug efflux transporter, lnc00963 conferred oral cancer cells stemness-like traits. | [28] |
| TALNEC2 | Glioblastoma | Induction of cancer stemness and radioresistance | Inhibition of TALNEC2 inhibited cancer stemness partly through the recovery of miR-21 and miR-11 expression. | [30] |
| LincRNAp21 | Glioblastoma | Inhibition of cancer stemness and radioresistance | LincRNAp21 negatively regulates β-catenin, which is a critical activator of wnt/β-catenin signaling pathway. | [31] |
| FENDRR | Lung cancer | Inhibition of cancer stemness | FENDRR bound to 3’UTR of MDR1, prevent the binding of RNA binding protein HuR to MDR1 3’UTR and thus inhibit MDR1 expression. | [32] |
| LINC01567 | Lung cancer | Inhibition of cancer stemness | LINC01567 acted as a sponge to competitively bind to miRNA-93, increasing target genes expressions including HDAC8, TLE4, stratifin and MSI1. | [33] |
| DLX6-AS1 | Osteosarcoma | Induction of cancer stemness | DLX6-AS1 competitively bind to miR-129-5p and increased DLK1 expression, resulting in wnt signaling activation. | [34] |
| Linc-DYNC2H1-4 | Pancreatic cancer | Induction of cancer stemness and gemcitabine resistance | Linc-DYNC2H1-4 competitively bound to miRNA-145 and inhibited EMT and the expressions of cancer stemness markers. | [35] |
| FOXD2-AS1 | Thyroid cancer | Induction of cancer stemness | FOXD2-AS1 interacted with miR-7-5p as a sponge and induced overexpression of miR-7-5p target gene telomerase reverse transcriptase (TERT). | [36] |
| HOTAIR | Prostatic cancer | Induction of cancer stemness and docetaxel resistance | HOTAIR sequestered miR-590-5p as a sponge and induced overexpression of IL-10, which initiated the activation of STAT3 signaling. | [37] |
| ASB16-AS1 | Gastric cancer | Induction of cancer stemness and cisplatin resistance | ASB16-AS1 interacted with miR-3918 and miR-4676-3 and increased the expression of TRIM37. Furthermore, ASB16-AS1 cooperated with ATM kinase, facilitated TRIM37 phosphorylation and promoted the activation of NF-κB pathway. | [38] |
| DGCR5 | NSCLC | Induction of cancer stemness | DGCR5 inhibited the expression of miR-330-5p, which negatively regulated cancer stemness marker CD44 expression, by interaction with it as a sponge. | [39] |
| LBCS | Bladder cancer | Inhibition of cancer stemness | LBCS formed a scaffold complex with hnRNPK and EZH2, which suppressed SOX2 expression by mediating H3K27me3 of SOX2 promoter. | [40] |
| LncZic2 | Liver cancer | Induction of cancer stemness | LncZic2 interacted with BRG1 and directed it to MARCKS and MARCKSL1 promoter, promoting the expressions of MARCKS/MARCKSL1. | [41] |
| LncGPR107 | Liver cancer | Induction of cancer stemness | LncGPR107 recruited SRCAP complex to GPR107 promoter and initiated its transcription activation. | [43] |
| CCAT1 | NSCLC | Induction of cancer stemness | CCAT1 inhibited let-7c, one suppressor of wnt signaling pathway, resulting in the activation of wnt signaling pathway. | [47] |
| H19 | Breast cancer | Induction of cancer stemness | H19 inhibited let-7c by which oestrogen receptor activated Wnt signaling was released. | [48] |
| Liver cancer | Induction of cancer stemness and drug resistance | H19 conferred activated MAPK/Erk pathway and reduction of oxidative stress. | [56] | |
| Breast cancer | Up-regulation of cancer stemness | H19 interacted with miRNA let-7 to release HIFr1α, resulting in upregulation of pyruvate dehydrogenase kinase 1 (PDK1) and increase of glycolysis. | [77] | |
| Colorectal cancer | Up-regulation of cancer stemness | H19 in exosomes secreted by CAFs activated the β-catenin pathway via acting as a competing endogenous RNA sponge for miR-141. | [87] | |
| Hepatocellular carcinoma | Induction of cancer stemness | TAMs induced overexpression of H19 which sequestered miR-193b, increasing MAPK1 expression to activate MAPK signaling pathway. | [92] | |
| RBM5-AS1/LUST | Colon cancer | Induction of cancer stemness | LUST directly interacts with β-catenin, stabilizes the β-catenin/TCF-4 complex and potentiates the transcription of β-catenin target genes. | [49] |
| BCAR4 | Gastric cancer | Induction of cancer stemness and cisplatin resistance | BCAR4 increased the expressions of stem cells markers such as β-catenin, Nanog, Oct3/4, Sox2, c-Myc, and Klf4 due to wnt signaling activation. | [50] |
| THOR | Liver cancer | Induction of cancer stemness and sorafenib resistance | THOR promoted liver cancer stem cells expansion and resistance to sorafenib treatment through activating β-catenin signaling pathway. | [51] |
| Gastric cancer | Induction of cancer stemness and cisplatin resistance | THOR directly bound to SOX9 untranslated region (3’UTR), promoting SOX9 expression. | [64] | |
| LncRNA-cCSC1 | Colon cancer | Induction of cancer stemness and 5-fluorouracil resistance | LncRNA-cCSC1 induced cancer stemness by activation of Hh pathway. | [53] |
| LINC-PINT | Laryngeal carcinoma | Inhibition of cancer stemness | LINC-PINT inhibited cancer stemness of laryngeal carcinoma possibly through inactivating the Hedgehog pathway. | [54] |
| TUSC-7 | Lung adenocarcinoma | Induction of cancer stemness | TUSC-7 inhibited the degradation of NUMB by miR-146 and lead to Notch signaling activation. | [55] |
| LncRNA MAPK6 | Esophageal cancer | Induction of cancer stemness | LncRNA MAPK6 interacted with and recruited RNA polymerase II to MAPK6 promoter, resulting in the activation of MAPK6 transcription. | [57] |
| LncARSR | Liver cancer | Induction of cancer stemness and resistance to cisplatin and sorafenib | lncARSR promoted liver CSCs expansion and resistance to cisplatin and sorafenib by activating STAT3 signaling. | [62] |
| Renal cancer | Induction of cancer stemness | LncARSR interacted with YAP and prompted YAP nuclear translocation while repressed YAP phosphorylation. | [65] | |
| PTCSC3 | Thyroid cancer | Inhibition of cancer stemness and doxorubicin resistance | Overexpression of PTCSC3 inhibited STAT3 signaling and the expression of INO80, which is a positive regulator of thyroid cancer stemness. | [63] |
| MALAT1 | Ovarian cancer | Induction of cancer stemness and cisplatin resistance | By interaction with YAP, MALAT1 inhibited it translocation from nucleus to cytoplasm, stabilized it and increased its activity in ovarian cancer. | [66] |
| Liver cancer | Induction of cancer stemness | MALAT1 increased the expression of YAP1 by sponging miRNA-375, which inhibited YAP1 expression through binding to its 3’-UTR. | [67] | |
| Lnc HOXA10 | Liver cancer | Induction of cancer stemness | Lnc HOXA10 recruited SNF2L to the promoter of HOXA10, resulting in HOXA10 overexpression. | [68] |
| FGF13-AS1 | Breast cancer | Inhibition of cancer stemness | By competitively interacting with IGF2BP proteins, FGF13-AS1 reduced the mRNA stability of c-Myc to suppress glycolysis. | [79] |
| TP73-AS1 | Glioblastoma | Induction of cancer stemness and temozolomide resistance | TP73-AS1 regulated, mitochondria, and nucleotide metabolism, and curbed ROS levels. | [81] |
| HAL | Breast cancer | Induction of cancer stemness | HAL was induced by tumor microenvironment such as hypoxia and acidosis. | [85] |
| HCP5 | Gastric cancer | Up-regulation of cancer stemness | MSCs induced overexpression of HCP5, which interacted with miR-3619-5P and initiated the transcription of CPT1, prompting FAO. | [90] |
| MACC1-AS1 | Gastric cancer | Induction of cancer stemness and resistance to 5-FU and oxaliplatin | TGF-β1 secreted by MSCs induced MACC1-AS1 overexpression which promoted FAO by sequestering miR-145-5p. | [91] |
LncRNAs are the bridge of cancer stem cells and therapy resistance in many human cancers. Due to highly tissue-specific expression, lncRNAs are being becoming novel anti-cancer targets in cancers [96]. Their expressions and functions can be altered with RNAi technology, antisense oligonucleotides (ASOs), or small molecule inhibitors. Furthermore, gene therapy approaches may be applied for lncRNAs-targeted anti-cancer strategies. Companies such as the Curna Inc., MiNA Therapeutics Ltd. and RaNA Therapeutics Inc. are taking steps for the development of lncRNA based strategies. Therefore, it holds promise that lncRNAs based anti-cancer drugs would be developed to eliminate CSCs in the future. Although the mechanisms by which lncRNAs regulated cancer stemness are diverse in different cancers, more attention should be paid on those stemness-related signaling pathways and transcription factors due to their significant roles. Our review highlighted the important roles of wnt/β-catenin pathway, Hedgehog (Hh) signaling pathway and Notch signaling pathway in lncRNAs-mediated cancer stemness and therapy resistance. Several inhibitors of these pathways have been developed and some of them have entered clinical phases for human cancers [97-99]. Targeting these stemness-related signaling pathways may be another effective approach to eliminate cancer stemness mediated by lncRNAs. Furthermore, tumor microenvironment played essential roles in the development of cancer stemness by regulating lncRNAs expressions. The role of tumor microenvironment should not be ignored in the battle against cancer stem cells. Although many lncRNAs have been demonstrated to be attractive targets of reversing therapy resistance of CSCs, more studies are required before they are targeted in the clinical treatment of human cancers. Their structure, expression pattern and molecular mechanisms all need to be clarified. Together, we believe that lncRNAs are promising targets to overcome therapy resistance of CSCs and deserve to be further studied for their clinical significance.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 81872477), Basic and Public Welfare Research Foundation of Zhejiang Province, China (No. LGF18H160087), Medical Scientific Research Foundation of Zhejiang Province, China (No. 2018KY595) and Scientific Technology Research Foundation of Hangzhou City, Zhejiang Province, China (No. 20150733Q64, No. 20170533B93 and No. 20163501).
Disclosure of conflict of interest
None.
References
- 1.Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 2.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 3.Kuşoğlu A, Biray Avcı Ç. Cancer stem cells: a brief review of the current status. Gene. 2019;681:80–85. doi: 10.1016/j.gene.2018.09.052. [DOI] [PubMed] [Google Scholar]
- 4.ENCODE Project Consortium; Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermüller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaöz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Löytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute. Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrímsdóttir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou N, Liu G, Pan Y. Long noncoding RNA ANRIL as a novel biomarker in human cancer. Future Oncol. 2020;16:2981–2995. doi: 10.2217/fon-2020-0470. [DOI] [PubMed] [Google Scholar]
- 7.Gupta SC, Tripathi YN. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Wu Z, Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int J Immunopathol Pharmacol. 2019;33:2058738418820743. doi: 10.1177/2058738418820743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malakar P, Stein I, Saragovi A, Winkler R, Stern-Ginossar N, Berger M, Pikarsky E, Karni R. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Res. 2019;79:2480–2493. doi: 10.1158/0008-5472.CAN-18-1432. [DOI] [PubMed] [Google Scholar]
- 10.Lan T, Yuan K, Yan X, Xu L, Liao H, Hao X, Wang J, Liu H, Chen X, Xie K, Li J, Liao M, Huang J, Zeng Y, Wu H. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res. 2019;79:3220–3234. doi: 10.1158/0008-5472.CAN-18-4044. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Yu Y, Li H, Hu Q, Chen X, He Y, Xue C, Ren F, Ren Z, Li J, Liu L, Duan Z, Cui G, Sun R. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18:33. doi: 10.1186/s12943-019-0947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang X, Cai J. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8:110685–110692. doi: 10.18632/oncotarget.22161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Li Y, Sheng J, Wu F, Li K, Huang R, Wang X, Jiao T, Guan X, Lu Y, Chen X, Luo Z, Zhou Y, Hu H, Liu W, Du B, Miao S, Cai J, Wang L, Zhao H, Ying J, Bi X, Song W. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J Exp Clin Canc Res. 2019;38:379. doi: 10.1186/s13046-019-1375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T, Wu L, Ma N, Tang F, Yu Z, Jiang Z, Li Y, Zong Z, Hu K. SOX9-activated FARSA-AS1 predetermines cell growth, stemness, and metastasis in colorectal cancer through upregulating FARSA and SOX9. Cell Death Dis. 2020;11:1–15. doi: 10.1038/s41419-020-03273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Han C, Ungerleider N, Chen W, Song K, Wang Y, Kwon H, Ma W, Wu T. A transforming growth factor-beta and H19 signaling axis in tumor-initiating hepatocytes that regulates hepatic carcinogenesis. Hepatology. 2019;69:1549–1563. doi: 10.1002/hep.30153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16:104. doi: 10.1186/s12943-017-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta P, Kulkarni P, Majid S, Shahryari V, Hashimoto Y, Bhat NS, Shiina M, Deng G, Saini S, Tabatabai ZL, Yamamura S, Tanaka Y, Dahiya R. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018;17:1061–1069. doi: 10.1158/1535-7163.MCT-17-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borst P. Cancer drug pan-resistance: pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2012;2:120066. doi: 10.1098/rsob.120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 20.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–3. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 22.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y, Zheng X, Zhao X, Zhang D, Huang R, Guo H, Yan J. TGF beta 1 promotes gemcitabine resistance through regulating the LncRNA-LET/NF90/miR-145 signaling axis in bladder cancer. Theranostics. 2017;7:3053–3067. doi: 10.7150/thno.19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin VY, Chen J, Cheuk IWY, Siu MT, Ho CW, Wang X, Jin H, Kwong A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10:1–10. doi: 10.1038/s41419-019-1513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SS, Harford JB, Moghe M, Rait A, Pirollo KF, Chang EH. Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res. 2018;46:1424–1440. doi: 10.1093/nar/gkx1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Z, Xiu C, Song K, Pei R, Miao S, Mao X, Sun J, Jia S. Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. J Cell Mol Med. 2018;22:4253–4262. doi: 10.1111/jcmm.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su YK, Lin JW, Shih JW, Chuang HY, Fong IH, Yeh CT, Lin CM. Targeting BC200/miR218-5p signaling axis for overcoming temozolomide resistance and suppressing glioma stemness. Cells Basel. 2020;9:1859. doi: 10.3390/cells9081859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SP, Hsieh PL, Fang CY, Chu PM, Liao YW, Yu CH, Yu CC, Tsai LL. LINC00963 promotes cancer stemness, metastasis, and drug resistance in head and neck carcinomas via ABCB5 regulation. Cancers. 2020;12:1073. doi: 10.3390/cancers12051073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang Z, Jifu E, Guo K, Ma X, Zhang Y, Yu E. Knockdown of long non-coding RNA TINCR decreases radioresistance in colorectal cancer cells. Pathol Res Pract. 2019;215:152622. doi: 10.1016/j.prp.2019.152622. [DOI] [PubMed] [Google Scholar]
- 30.Brodie S, Lee HK, Jiang W, Cazacu S, Xiang C, Poisson LM, Datta I, Kalkanis S, Ginsberg D, Brodie C. The novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells. Oncotarget. 2017;8:31798–31814. doi: 10.18632/oncotarget.15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Yu H, Shen Y, Liu Y, Yang Z, Sun T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNAp21/beta-catenin pathway. Oncotarget. 2016;7:41505–41526. doi: 10.18632/oncotarget.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong F, Dong D, Zhang T, Xu W. Long non-coding RNA FENDRR attenuates the sternness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR. Eur J Pharmacol. 2019;853:345–352. doi: 10.1016/j.ejphar.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Mi L, Dong J, Zou J. Long intergenic non-protein-coding RNA 1567 (LINC01567) acts as a “sponge” against microRNA-93 in regulating the proliferation and tumorigenesis of human colon cancer stem cells. BMC Cancer. 2017;17:1–15. doi: 10.1186/s12885-017-3731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang RM, Tang T, Yu HM, Yao XD. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates sternness of osteosarcoma through Wnt signaling. Biochem Bioph Res Co. 2018;507:260–266. doi: 10.1016/j.bbrc.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Gao Y, Zhang Z, Li K, Gong L, Yang Q, Huang X, Hong C, Ding M, Yang H. Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis. 2017;8:e2924. doi: 10.1038/cddis.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Fu Q, Li S, Liang N, Li F, Li C, Sui C, Dionigi G, Sun H. LncRNA FOXD2-AS1 functions as a competing endogenous RNA to regulate TERT expression by sponging mir-7-5p in thyroid cancer. Front Endocrinol. 2019;10:207. doi: 10.3389/fendo.2019.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang N, Jiang Y, Lv S, Wen H, Wu D, Wei Q, Dang Q. HOTAIR expands the population of prostatic cancer stem-like cells and causes docetaxel resistance via activating STAT3 signaling. Aging (Albany NY) 2020;12:12771–12782. doi: 10.18632/aging.103188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu T, Ji K, Jin L, Zhang J, Wu X, Ji X, Fan B, Jia Z, Wang A, Liu J, Bu Z, Ji J. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-kappa B pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer. 2020;24:45–49. doi: 10.1007/s10120-020-01096-y. [DOI] [PubMed] [Google Scholar]
- 39.Wang R, Dong HX, Zeng J, Pan J, Jin XY. LncRNA DGCR5 contributes to CSC-like properties via modulating miR-330-5p/CD44 in NSCLC. J Cell Physiol. 2018;233:7447–7456. doi: 10.1002/jcp.26590. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Xie R, Gu P, Huang M, Han J, Dong W, Xie W, Wang B, He W, Zhong G, Chen Z, Huang J, Lin T. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25:1389–1403. doi: 10.1158/1078-0432.CCR-18-1656. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Liu Y, Yao L, Guo S, Gao Y, Zhu P. The long noncoding RNA lncZic2 drives the self-renewal of liver tumor-initiating cells via the protein kinase C substrates MARCKS and MARCKSL1. J Biol Chem. 2018;293:7982–7992. doi: 10.1074/jbc.RA117.001321. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Soond SM, Zamyatnin AA Jr. Targeting G protein-coupled receptors in cancer therapy. Adv Cancer Res. 2020;145:49–97. doi: 10.1016/bs.acr.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Huang G, Jiang H, Lin Y, Xia W, Luo Y, Wu Y, Cai W, Zhou X, Jiang X. LncGPR107 drives the self-renewal of liver tumor initiating cells and liver tumorigenesis through GPR107-dependent manner. J Exp Clin Canc Res. 2018;37:121. doi: 10.1186/s13046-018-0794-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang P, Xu H, Xu C, Chen A, Chen L, Zhou M, ul Haq I, Zahula X, Mariyam Z, Feng Q. NEAT1 contributes to the CSC-like traits of A549/CDDP cells via activating Wnt signaling pathway. Chem Biol Interact. 2018;296:154–161. doi: 10.1016/j.cbi.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Xu C, Xiao G, Zhang B, Wang M, Wang J, Liu D, Zhang J, Ren H, Sun X. CCAT1 stimulation of the symmetric division of NSCLC stem cells through activation of the Wnt signalling cascade. Gene Ther. 2018;25:4–12. doi: 10.1038/gt.2017.98. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Li Y, Xiao GD, Zheng XQ, Wang JC, Xu CW, Qin S, Ren H, Tang SC, Sun X. H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif. 2019;52:e12534. doi: 10.1111/cpr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Cecilia S, Zhang F, Sancho A, Li S, Aguilo F, Sun Y, Rengasamy M, Zhang W, Del Vecchio L, Salvatore F, Walsh MJ. RBM5-AS1 is critical for self-renewal of colon cancer stem-like cells. Cancer Res. 2016;76:5615–5627. doi: 10.1158/0008-5472.CAN-15-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Chunyan Q, Zhou Y, He O, Ma Y, Ga Y, Wang X. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmaco. 2017;21:4064–4070. [PubMed] [Google Scholar]
- 51.Cheng Z, Lei Z, Yang P, Si A, Xiang D, Zhou J, Hueser N. Long non-coding RNA THOR promotes liver cancer stem cells expansion via beta-catenin pathway. Gene. 2019;684:95–103. doi: 10.1016/j.gene.2018.10.051. [DOI] [PubMed] [Google Scholar]
- 52.Chang HL, Bamodu OA, Ong JR, Lee WH, Yeh CT, Tsai JT. Targeting the epigenetic non-coding RNA MALAT1/wnt signaling axis as a therapeutic approach to suppress stemness and metastasis in hepatocellular carcinoma. Cells. 2020;9:1020. doi: 10.3390/cells9041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H, Xiong Y, Peng L, Wang R, Zhang H, Fu Z. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the hedgehog signaling pathway. J Cell Biochem. 2020;121:2510–2524. doi: 10.1002/jcb.29473. [DOI] [PubMed] [Google Scholar]
- 54.Yuan Z, Xiu C, Liu D, Zhou G, Yang H, Pei R, Ding C, Cui X, Sun J, Song K. Long noncoding RNA LINC-PINT regulates laryngeal carcinoma cell stemness and chemoresistance through miR-425-5p/PTCH1/SHH axis. J Cell Physiol. 2019;234:23111–23122. doi: 10.1002/jcp.28874. [DOI] [PubMed] [Google Scholar]
- 55.Huang G, Wang M, Li X, Wu J, Chen S, Du N, Li K, Wang J, Xu C, Ren H, Tang SC, Sun X. TUSC7 suppression of notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cell. Life Sci. 2019;232:116630. doi: 10.1016/j.lfs.2019.116630. [DOI] [PubMed] [Google Scholar]
- 56.Ding K, Liao Y, Gong D, Zhao X, Ji W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;502:194–201. doi: 10.1016/j.bbrc.2018.05.143. [DOI] [PubMed] [Google Scholar]
- 57.Huang G, Jiang H, He Y, Lin Y, Xia W, Luo Y, Liang M, Shi B, Zhou X, Jian Z. LncMAPK6 drives MAPK6 expression and liver TIC self-renewal. J Exp Clin Cancer Res. 2018;37:1–11. doi: 10.1186/s13046-018-0770-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Igelmann S, Neubauer HA, Ferbeyre G. STAT3 and STAT5 activation in solid cancers. Cancers (Basel) 2019;11:1428. doi: 10.3390/cancers11101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong M, Wang J, Jiang N, Pan H, Li D. Correlation between p-STAT3 overexpression and prognosis in lung cancer: a systematic review and meta-analysis. PLoS One. 2017;12:e0182282. doi: 10.1371/journal.pone.0182282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang L, Liu JX, Zhang ZJ, Xu CZ, Zhang XN, Huang WR, Zhou DH, Wang RR, Chen XD, Xiao MB, Qu LS, Lu CH. High expression of Anxa2 and Stat3 promote progression of hepatocellular carcinoma and predict poor prognosis. Pathol Res Pract. 2019;215:152386. doi: 10.1016/j.prp.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto K, Hara T, Nakagawa T, Hirai M, Miyake H, Fujisawa M, Yano I. Association of expression levels or activation status of STAT3 with treatment outcomes of sunitinib in patients with renal cell carcinoma. Target Oncol. 2018;13:371–378. doi: 10.1007/s11523-018-0563-4. [DOI] [PubMed] [Google Scholar]
- 62.Yang C, Cai WC, Dong ZT, Guo JW, Zhao YJ, Sui CJ, Yang JM. lncARSR promotes liver cancer stem cells expansion via STAT3 pathway. Gene. 2019;687:73–81. doi: 10.1016/j.gene.2018.10.087. [DOI] [PubMed] [Google Scholar]
- 63.Wang XM, Liu Y, Fan YX, Liu Z, Yuan QL, Jia M, Geng ZS, Gu L, Lu XB. LncRNA PTCSC3 affects drug resistance of anaplastic thyroid cancer through STAT3/INO80 pathway. Cancer Biol Ther. 2018;19:590–597. doi: 10.1080/15384047.2018.1449610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song H, Xu Y, Shi L, Xu T, Fan R, Cao M, Xu W, Song J. LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed Pharmacother. 2018;108:338–346. doi: 10.1016/j.biopha.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 65.Qu L, Wu Z, Li Y, Xu Z, Liu B, Liu F, Bao Y, Wu D, Liu J, Wang A, Chu X, Sun Y, Chen C, Zhang Z, Wang L. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu X, Wang Y, Zhong W, Cheng H, Tian Z. The long non-coding RNA MALAT1 enhances ovarian cancer cell sternness by inhibiting YAP translocation from nucleus to cytoplasm. Med Sci Monitor. 2020;26:e922012. doi: 10.12659/MSM.922012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao L, Lou G, Li A, Liu Y. lncRNA MALAT1 modulates cancer stem cell properties of liver cancer cells by regulating YAP1 expression via miR-375 sponging. Mol Med Rep. 2020;22:1449–1457. doi: 10.3892/mmr.2020.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao M, Yang Q, Zhu W, Jin H, Wang J, Song J, Kong Y, Lv X. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol Cancer. 2018;17:1–12. doi: 10.1186/s12943-018-0921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 70.Sharif T, Dai C, Martell E, Ghassemi-Rad MS, Hanes MR, Murphy PJ, Kennedy BE, Venugopal C, Subapanditha M, Giacomantonio CA, Marcato P, Singh SK, Gujar S. TAp73 modifies metabolism and positively regulates growth of cancer stem-like cells in a redox-sensitive manner. Clin Cancer Res. 2019;25:2001–2017. doi: 10.1158/1078-0432.CCR-17-3177. [DOI] [PubMed] [Google Scholar]
- 71.Kamarajan P, Rajendiran TM, Kinchen J, Bermudez M, Danciu T, Kapila YL. Head and neck squamous cell carcinoma metabolism draws on glutaminolysis, and sternness is specifically regulated by glutaminolysis via aldehyde dehydrogenase. J Proteome Res. 2017;16:1315–1326. doi: 10.1021/acs.jproteome.6b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao H, Duan Q, Zhang Z, Li H, Wu H, Shen Q, Wang C, Yin T. Up-regulation of glycolysis promotes the stemness and EMT phenotypes in gemcitabine-resistant pancreatic cancer cells. J Cell Mol Med. 2017;21:2055–2067. doi: 10.1111/jcmm.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Brit J Cancer. 2016;114:1305–1312. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng W, Gentles A, Nair RV, Huang M, Lin Y, Lee CY, Cai S, Scheeren FA, Kuo AH, Diehn M. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells. 2014;32:1734–1745. doi: 10.1002/stem.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang ZF, Wang M, Xu JL, Ning YJ. Hypoxia promotes mitochondrial glutamine metabolism through HIF1 alpha-GDH pathway in human lung cancer cells. Biochem Biophys Res Commun. 2017;483:32–38. doi: 10.1016/j.bbrc.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 76.Palorini R, Votta G, Balestrieri C, Monestiroli A, Olivieri S, Vento R, Chiaradonna F. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J Cell Biochem. 2014;115:368–379. doi: 10.1002/jcb.24671. [DOI] [PubMed] [Google Scholar]
- 77.Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, Cui B, Wang HF, Zhao Y, An F, Guo T, Liu XF, Zhang L, Lv L, Lv DK, Xu LZ, Xie JJ, Lin WX, Lam EW, Xu J, Liu Q. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–1074. doi: 10.1038/onc.2017.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma F, Liu X, Zhou S, Li W, Liu C, Chadwick M, Qian C. Long non-coding RNA FGF13-AS1 inhibits glycolysis and sternness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019;450:63–75. doi: 10.1016/j.canlet.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 79.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14:434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 80.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 81.Mazor G, Levin L, Picard D, Ahmadov U, Caren H, Borkhardt A, Reifenberger G, Leprivier G, Remke M, Rotblat B. The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 2019;10:1–14. doi: 10.1038/s41419-019-1477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, Bilsland AE, Boosani CS, Chen S, Ciriolo MR, Crawford S, Fujii H, Georgakilas AG, Guha G, Halicka D, Helferich WG, Heneberg P, Honoki K, Keith WN, Kerkar SP, Mohammed SI, Niccolai E, Nowsheen S, Vasantha Rupasinghe HP, Samadi A, Singh N, Talib WH, Venkateswaran V, Whelan RL, Yang X, Felsher DW. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin Cancer Biol. 2015;35(Suppl):S199–S223. doi: 10.1016/j.semcancer.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun L, Wang L, Chen T, Shi Y, Yao B, Liu Z, Wang Y, Li Q, Liu R, Niu Y, Tu K, Liu Q. LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. Cell Death Dis. 2020;11:1–15. doi: 10.1038/s41419-020-2274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.García-Venzor A, Mandujano-Tinoco EA, Lizarraga F, Zampedri C, Krötzsch E, Salgado RM, Dávila-Borja VM, Encarnación-Guevara S, Melendez-Zajgla J, Maldonado V. Microenvironment-regulated lncRNA-HAL is able to promote sternness in breast cancer cells. Biochim Biophys Acta Mol Cell Res. 2019;1866:118523. doi: 10.1016/j.bbamcr.2019.118523. [DOI] [PubMed] [Google Scholar]
- 86.Zhang H, Hua Y, Jiang Z, Yue J, Shi M, Zhen X, Zhang X, Yang L, Zhou R, Wu S. Cancer-associated fibroblast-promoted LncRNA DNM3OS confers radioresistance by regulating DNA damage response in esophageal squamous cell carcinoma. Clin Cancer Res. 2019;25:1989–2000. doi: 10.1158/1078-0432.CCR-18-0773. [DOI] [PubMed] [Google Scholar]
- 87.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rhodes LV, Muir SE, Elliott S, Guillot LM, Antoon JW, Penfornis P, Tilghman SL, Salvo VA, Fonseca JP, Lacey MR, Beckman BS, McLachlan JA, Rowan BG, Pochampally R, Burow ME. Adult human mesenchymal stem cells enhance breast tumorigenesis and promote hormone independence. Breast Cancer Res Treat. 2010;121:293–300. doi: 10.1007/s10549-009-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weaver VM, Lelièvre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ. beta 4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1 alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11:1–17. doi: 10.1038/s41419-020-2426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, Liu Z, Yao Z, Wu Q, Liao W, Zhang S, Liu Y, Xiang Y, Liu J, Shi M. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637–4654. doi: 10.1038/s41388-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, Yu W, Xu L, Zhao Y, Yu J. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310–322. doi: 10.1016/j.canlet.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 93.Rebolho Batista Arantes LM, De Carvalho AC, Melendez ME, Carvalho AL. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev Mol Diagn. 2018;18:85–112. doi: 10.1080/14737159.2017.1404906. [DOI] [PubMed] [Google Scholar]
- 94.Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, Li BA, Cai JC, Cai WY. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soares JC, Soares AC, Rodrigues VC, Melendez ME, Santos AC, Faria EF, Reis RM, Carvalho AL, Oliveira ON Jr. Detection of the prostate cancer biomarker PCA3 with electrochemical and impedance-based biosensors. ACS Appl Mater Interfaces. 2019;11:46645–46650. doi: 10.1021/acsami.9b19180. [DOI] [PubMed] [Google Scholar]
- 96.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang S, Ivy SP. Targeting notch, hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling - are we there yet? Nat Rev Drug Discov. 2014;13:359–380. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]


