Abstract
Former clinical trials and experimental research have indicated that Interferon-gamma therapy does not achieve an ideal effect in solid tumors. Autophagy has been associated with tumor chemoresistance. The aim of this study was to explore the efficacy of Interferon-gamma and autophagy inhibitor in the combination treatment of oral squamous cell carcinoma. Interferon-gamma-induced apoptosis was evaluated by the expression of relative proteins (cleaved-PARP and caspase-3) and flow cytometry. Interferon-gamma induced autophagy was assessed by the expression of Beclin1, LC3B, and P62. The synergistic effect of interferon-gamma and autophagy inhibitor (chloroquine) was evaluated in vitro and in vivo. Interferon-gamma induced anti-proliferation, apoptosis, and autophagy in oral squamous cell carcinoma cells. Autophagy-related protein 5 was a key feature in Interferon-gamma-induced autophagy flux. Interferon-gamma and chloroquine had obvious synergistic effects on cellular growth inhibition and apoptosis promotion in oral squamous cell carcinoma cells and xenograft models. Our findings suggest that Interferon-gamma-induced autophagy plays a cellular protective role, and blocking autophagy flux can promote Interferon-gamma mediated oral squamous cell carcinoma cell apoptosis. The combination of Interferon-gamma and autophagy inhibitors represents a novel strategy for oral squamous cell carcinoma therapy.
Keywords: Oral squamous cell carcinoma, interferon-gamma, chloroquine, autophagy, apoptosis
Introduction
Oral squamous cell carcinoma (OSCC) is the most common malignant tumor in the oral and maxillofacial region, and there are approximately 300,000 new cases worldwide each year [1,2]. Despite the progress achieved in radical surgical resection with proper reconstruction and postoperative radiotherapy/chemoradiotherapy [3], the 5-year survival rate has remained at 50% to 60% [4,5], and even lower in the patients with locally advanced lesions [6]. Consequently, there is an urgent need to explore effective OSCC therapeutic strategies.
Macroautophagy (autophagy or ‘self-eating’) is a lysosome-mediated process whereby cells degrade organelles and macromolecules and recycle cellular waste [7]. Accumulating evidence suggests that autophagy can both suppress and promote tumor functions in cancer progression [8,9]. On the one hand, the initial stages of tumorigenesis can be inhibited by autophagy [10] and on the other hand, autophagy could nurture established cancers [11-13]. Yang et al. [14] revealed the mechanism of how autophagy promotes pancreatic tumor growth via p53 alternation. In research relating to OSCC, autophagy was considered as crucial for either nurturing cancer cells [15,16] or initiating programmed cell death [17,18]. Accordingly, the inhibition of autophagy may be a potential strategy for OSCC treatment.
Studies in interferon-gamma (IFNγ) have demonstrated an anti-cancer effect in several tumors, including those in colorectal, gastric, and cervical cancers [19]. Recent findings suggest that members of the IFN family synergize with traditional anti-cancer treatment for OSCC through several mechanisms, including augmenting various immune functions [20]. However, IFNγ therapy alone does not achieve an ideal effect on established cancers, which may reflect an adaptive resistance [19,21,22]. Studies of chemoresistance in brain, gastric, and ovarian cancers also suggest that autophagy plays a key role [23-25].
Herein, autophagy co-activated by IFNγ is considered to be a cancer-promoting factor, which provides a potential strategy for combined inhibition therapy. In our present study, we first demonstrate autophagy and apoptosis induced by IFNγ in OSCC cells. The ATG5 molecule was found to be necessary for IFNγ-induced autophagy. Furthermore, IFNγ and chloroquine (CQ) exhibited clear synergistic anti-cancer effects in vitro and in vivo. This study supports the combined application of autophagy inhibitors and IFNγ in the clinical therapy of OSCC.
Materials and methods
Culture system
HN4, HN6, HB96, and CAL27 cell lines were used in our study. The HB96 cell line was established from our in vitro cellular carcinogenesis model of OSCC. HN4 and HN6 cell lines were provided as a gift from the National Institutes of Health (USA). The HN4 cell line was derived from tongue squamous cell carcinoma. Cell line HN6 was established from pharyngeal squamous cell carcinoma. The tongue squamous cell carcinoma cell line CAL27 was purchased from ATCC (Manassas, VA, USA). These cell lines were cultured in complete DMEM (Gibco, Carlsbad, CA, USA), supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% glutamine. Cells were maintained in a humidified 5% CO2 atmosphere at 37°C.
Cell proliferation assay
To assess the cytotoxicity of IFNγ, OSCC cells were seeded in 96-well flat-bottom plates in triplicate at a density of 1×104 cells/ml, supplied with different concentrations IFNγ. A cell proliferation assay was performed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg/ml). The plates were incubated in a humidified incubator at 37°C for 2 h. The absorbance was measured at 450 nm.
Colony formation assay
To evaluate the influence of IFNγ on colony formation, HN4 and CAL27 cells were seeded in 24-well plates at approximately 1×103 cells/well and cultured in complete medium with 200 ng/ml IFNγ for 48 h. Then the medium was replaced by complete DMEM for 6 d. Plates were finally washed with PBS twice, fixed with 4% paraformaldehyde for 30 min, and stained with 0.5% crystal violet for 5 min. The counting of colonies was performed using Image J software.
Real-time PCR assay
The purity and concentration of the RNA were assessed by using a NanoDrop 2000/2000C spectrophotometer at wavelengths of 260/280 nm. A PrimeScript™ RT Reagent Kit (TaKaRa Biotechnology) was used to reverse transcribe RNA into cDNA. The resultant cDNA was used as a template in a TB Green® Premix Ex TaqTM Kit (TaKaRa Biotechnology) master mix and qPCR reactions were performed on a StepOnePlusTM Real-Time PCR System. The human primer sequences were as follows: GAPDH forward: 5’-CCTCTGACTTCAACAGCGAC-3’ and reverse: 5’-TCCTCTTGTGCTCTTGCTGGC-3’; ATG7 forward: 5’-AGAACATGGTGCTGGTTTCC-3’ and reverse: 5’-CATCCAGGGTACTGGGCTAA-3’; P62 forward: 5’-CCGTGAAGGCCTACCTTCTG-3’ and reverse: 5’-TCCTCGTCACTGGAAAAGGC-3’; BECN1 forward: 5’-GTGGCTTTCCTGGACTGTGT-3’ and reverse: 5’-CACTGCCTCCTGTGTCTTCA-3’; ATG5 forward: 5’-TGCAGATGGACAGTTGCACA-3’ and reverse: 5’-CCACTGCAGAGGTGTTTCCA-3’.
Western blot analysis
Proteins were extracted from OSCC cells treated with 0, 2, 20, or 200 ng/ml IFNγ for 0, 12, 24, and 48 h, or from OSCC cells stimulated with 200 nM IFNγ combined with or without CQ for 48 h. The membranes were blocked in 5% skim milk in 1× TBST (Tris-buffered saline with Tween 20) at room temperature for 1 h and then incubated with primary antibodies (β-actin, 1:1000; cleaved-PARP, 1:1000; caspase-3, 1:1000; ATG5, 1:1000; P62, 1:1000; LC3B II, 1:1000; STAT1, 1:1000) overnight at 4°C. The membranes were then incubated with fluorescent-based anti-mouse or anti-rabbit IgG secondary antibodies (7076 and 7704, Cell Signaling Technology, USA) at a 1:10000 dilution for 1 h at room temperature. Immunoreactive bands were detected using enhanced chemiluminescence. The observation and analysis of immunoreactive bands were performed using the Odyssey Infrared Imaging System (LI-COR Biosciences, USA).
Immunofluorescence analysis
OSCC cells were fixed and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for 5 min. After blocking with 1% BSA-PBS for 1 h, 0.5 mg/ml of 4’,6-diamidino-2-phenylindole was used to stain the nuclei of cells. The BioTek Cytation 3 Cell Imaging Reader was used to visualize and acquire immunofluorescence images. The number of LC3B-I/II puncta was determined by Image J software.
Cell transfection
OSCC cells were seeded into a 24-well plate according to the manufacturer’s instructions. Small interfering RNA (siRNA, 100 nM) and overexpress-NC/Atg5 (oe-NC/Atg5) plasmid were transfected using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s instructions. Treatments were administered 24 h after transfection. The sequences of the ATG5 siRNA were #1, 5’-GGAAUAUCCUGCAGAAGAATT-3’, #2, 5’-AGAAUAUAUCAGACAACGATT-3’ and #3, 5’-AUCGGAAACUCAUGGAAUATT-3’.
Immunohistochemistry
To retrieve antigen, tumor slices were heated in a water bath at 100°C with EDTA buffer (pH=10.0) for 20 min. The primary antibodies were as shown below: LC3B II (CST, Danvers, MA, USA), P62, and Ki67 (Abcam, Cambridge, MA, UK). Immunohistochemistry and image analysis were performed to assess the mean optical density for Ki67, LC3B II, and P62 in vivo.
Flow cytometry analysis
HB96, HN4, and CAL27 cells were treated with IFNγ for different durations (0, 24, and 48 h) and with different concentrations (0, 2, 20, and 200 ng/ml). OSCC cells (adherent and floating) were collected and analyzed with flow cytometry analysis using an Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA). The analysis was performed using a BD FORTASA flow cytometer (BD Biosciences, San Diego, CA, USA) and Flow Jo software.
Autophagic flux assay
HB96, HN4, and CAL27 cells were seeded into glass-bottom dishes. After 12 h, OSCC cells were transfected with the GFP-mRFP-LC3 construct. Then IFNγ (200 ng/ml) and CQ (Sigma-Aldrich) were added. The cells were washed twice in ice-cold PBS, fixed, mounted with histological mounting medium (Histomount, USA), and observed using an LSM510 confocal laser microscope (Carl Zeiss, Dresden, Germany).
Transmission electron microscopy
HB96, HN4, and Cal27 cells were fixed in 4% paraformaldehyde in 0.1 M PBS (pH=7.4) and washed in 0.1 M PBS. The cells were then fixed for 2 h with 1% OsO4 dissolved in 0.1 M PBS and dehydrated with a progressive series of ethanol (50-100%) and permeated with epoxypropane. A Poly/Bed 812 kit (Polysciences, Warrington, PA, USA) was used to embed the sample. The samples were then embedded and polymerized in an electron microscope oven (TD-700, Dosaka, Japan) at 65°C for 24 h in pure fresh resin. Sections approximately 200-250 nm thick were stained with toluidine blue (Sigma-Aldrich, T3260) and then stained twice with 6% uranyl acetate (EMS, 20 min) and lead citrate (Fisher, 10 min) for comparison. The samples were prepared and analyzed by JEM 1230 transmission electron microscopy (JEOL, Akishima, Tokyo) at 60 kV, and micrographs were obtained at ×5000 and ×20,000 magnification.
Animal study
SPF BALB/c nude mice (nu/nu, n=24, 4 weeks old, 18.81 ± 0.8 g) were purchased from Shanghai experimental animal center (Shanghai, China) and placed in the SPF facility of the Ninth People’s Hospital, Shanghai Jiao Tong University, China. All laboratory procedures were approved by the laboratory animal care and use committee of the hospital. The xenograft model of the tumor in nude mice was established with CAL27 cells. In brief, 1×106 cells were subcutaneously injected with a diameter of 5 mm, the mice were divided into four different groups (6 mice in each group) and received various treatment regimens: (a) control (0.9% saline, i.p); (b) IFNγ (10,000 IU per day, i.p); (c) CQ (30 mg/kg per day, i.p); (d) IFNγ and CQ. Tumor size was monitored twice a week. Tumor volume was calculated using the formula length × width2/2. After three weeks, the mice were sacrificed by means of overdose phenobarbital sodium and the tumor tissue was resected. Tumor tissue and organ parts were immobilized and embedded in paraffin. Tissue sections (4 mm) were stained with hematoxylin and eosin. Terminal deoxynucleotide transferase dUTP notch end marker (TUNEL) was used to detect apoptotic cells.
Statistical analysis
All data were presented as the means ± standard deviation (SD). GraphPad Prism version 7 (GraphPad Software, San Diego, CA, USA) was used to process the initial data and plot the results. Statistical analyses were performed with SPSS 13.0 software for Windows (SPSS Inc., Chicago, IL, USA). Significance between two groups was performed using Student’s t test. Significance between more than two groups was evaluated by one way analysis of variance (ANOVA). The difference was considered significant at P<0.05.
Results
IFNγ exhibited anti-proliferation activity and induces apoptosis in OSCC cells
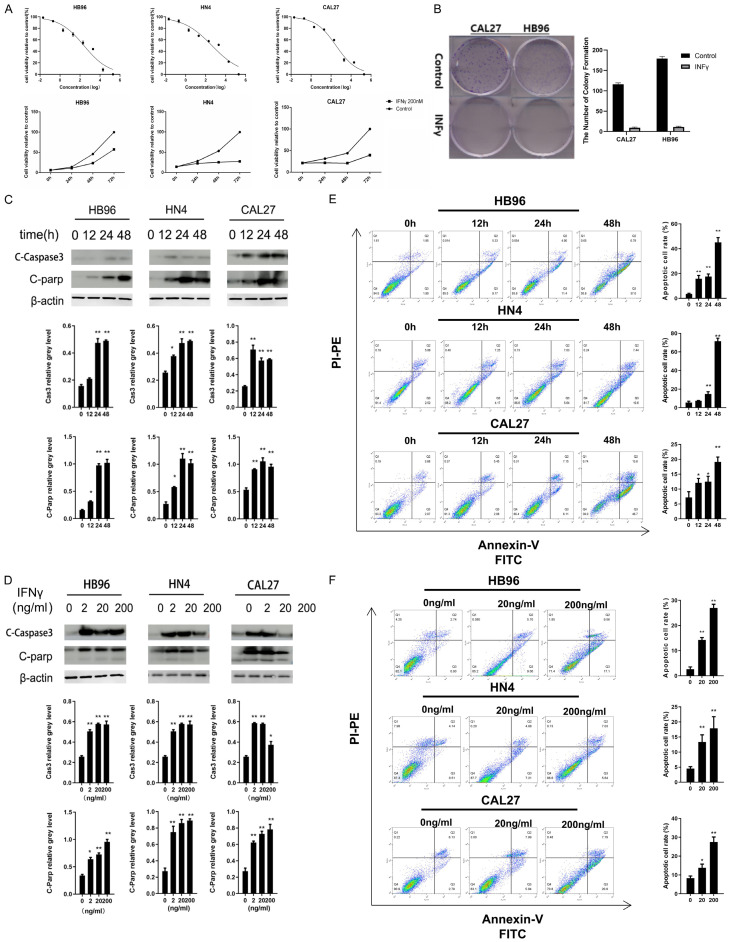
To evaluate the function of IFNγ on OSCC cell proliferation and viability, MTT and colony formation assays were performed. IFNγ exerted cytotoxicity in a time- and dose-dependent manner in HB96, HN4n, and CAL27 cells (Figure 1A). In addition, the treatment of IFNγ (200 ng/ml) for 8 days effectively attenuated the colony-forming capacity of HB96 and CAL27 cells (Figure 1B). As shown in Figure 1C and 1D, IFNγ up-regulated the cleaved-PARP and caspase-3 expression in a time-dependent manner in the HB96, HN4, and CAL27 cells. Flow cytometry assays support the above results, given that the proportion of Annexin V-positive cells increased in a time- and dose-dependent manner after IFNγ treatment (Figure 1E and 1F).
Figure 1.
IFNγ exerts cytotoxicity and induces apoptosis in HB96, HN4, and CAL27 cells. A. After the treatment of different concentrations of IFNγ within 72 h, cell viability was assessed by using an MTT assay. IC50 of IFNγ was measured in HB96, HN4, and CAL27 cells. The cells were incubated with 200 ng/ml of IFNγ or PBS for multiple time points, and then cell viability was measured using the MTT assay. B. IFNγ inhibited colony formation in CAL27 and HB96 cells. Cells were incubated in complete medium with or without IFNγ (200 ng/ml) for 7 d and stained by 0.5% crystal violet. Relative colony formation is represented in the bar graph. C. Western blot analysis showed that PARP and caspase-3 were up-regulated after 200 ng/ml IFNγ treatment for the indicated time points in the HB96, HN4, and CAL27 cells. Membranes were probed with a beta-actin antibody as a loading controL. D. Western blotting showed increased cleaved-PARP and activation of caspase-3 after treatment with indicated concentrations of IFNγ for 48 h in the HB96, HN4, and CAL27 cells. E. After IFNγ treatment (200 ng/ml) for indicated time points, apoptotic cell rates were analyzed by flow cytometry. F. HN4 and HN30 cells were treated with indicated concentrations of IFNγ for 48 h before staining with Annexin V and propidium iodide (PI), and the apoptotic rates were determined by flow cytometry. All the P values were compared with the control. *: P<0.05, **: P<0.01.
IFNγ-induced autophagy in OSCC cells
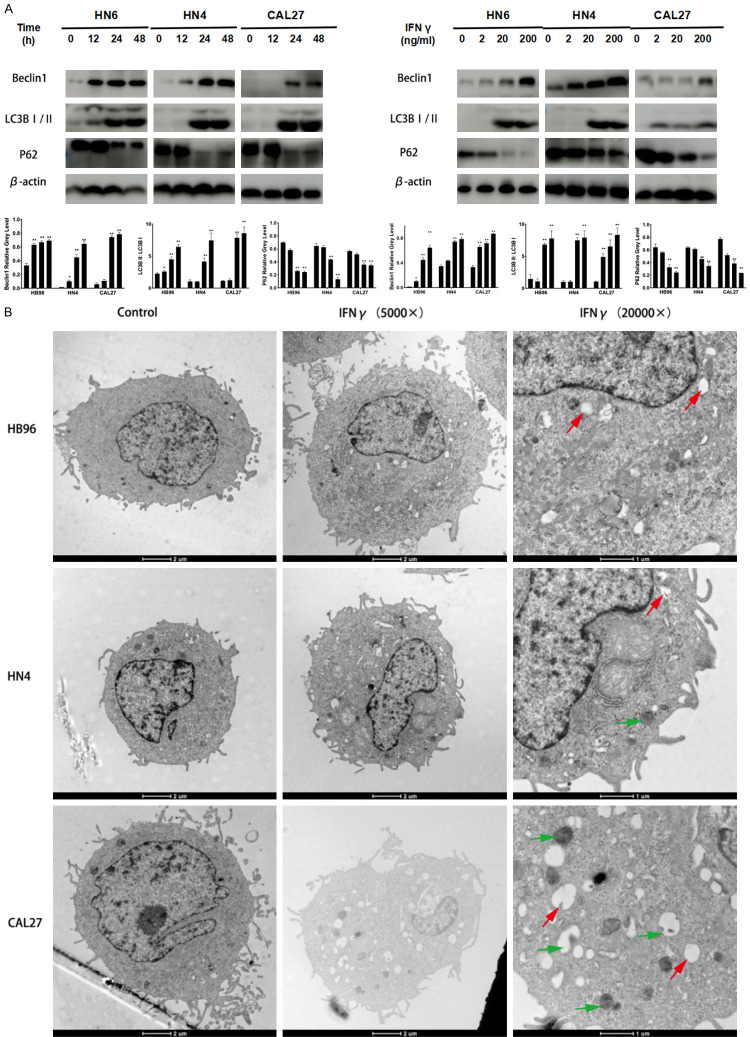
As shown in Figure 2A and 2B, Beclin1 and LC3B II expression increased in the IFNγ-treated HB96, HN4, and CAL27 cells in a time- and dose-dependent tendency. P62 level was analyzed to determine whether the autophagosome up-regulation observed after IFNγ treatment was caused by an increase of autophagic activity or a reduced turnover of autophagosomes. P62 was hydrolyzed when OSCC cells were treated with IFNγ, demonstrating IFNγ-induced autophagy. In accordance with western blot results (Figure 2A), the distribution of LC3B II puncta also increased in the IFNγ-treated OSCC cells compared with untreated control. Furthermore, we examined the morphology of the HB96, HN4, and CAL27 cells after IFNγ induction by transmission electron microscopy (TEM). As shown in Figure 2B, IFNγ activated autophagy flux by increasing the formation of the initial sequestering compartment (the phagophore), the number of autophagosomes often containing multivesicular and multilamellar structures, and autolysosomes. Herein, our data indicated that IFNγ induced autophagy in OSCC cells.
Figure 2.
IFNγ induces autophagy in OSCC cells. A. IFNγ promoted Beclin1 and LC3B II expression in a concentration- and time-dependent manner in HB96, HN4, and CAL27 cells. B. Representative transmission electron microscopy (TEM) images revealing the formation of phagophores (red arrow), autophagosomes, and autolysosomes (green arrow) after IFNγ (200 ng/ml) treatment for 24 h. All the P values were compared with the control. *: P<0.05, **: P<0.01.
IFNγ-induced autophagy via ATG5 signaling in OSCC cells
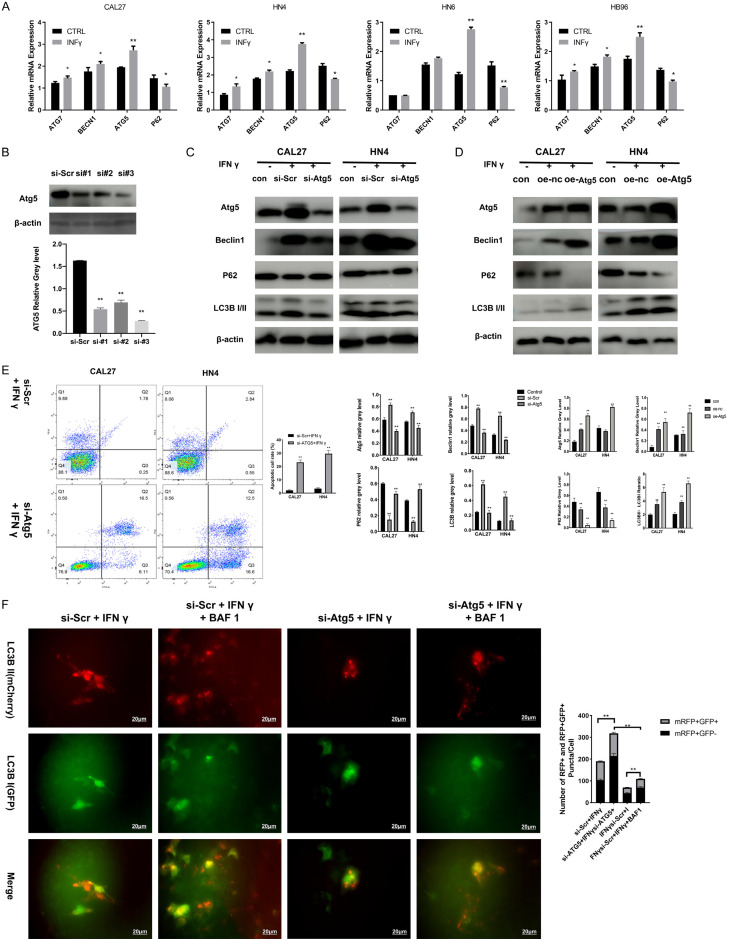
A set of autophagy-related genes (ATGs) was involved in the dynamic membrane-rearrangement reactions of autophagy. Among these genes, Beclin1, ATG5, and ATG7 represent the major regulators of the classical autophagy pathway in mammalian cells [26]. Using real-time PCR assays (Figure 3A), we found that ATG5 mRNA expression increased significantly after IFNγ treatment in HB96, HN4, and CAL27 cells. Moreover, the suppression of ATG5 expression using siRNAs (Figure 3B) in the context of IFNγ treatment (Figure 3C) decreased Beclin1 and LC3B II expression and increased P62 protein levels. The opposite result were observed when ATG5 was over expressed (Figure 3D). These results suggested that ATG5 was required for IFNγ-induced autophagy in OSCC cells. Moreover, silencing ATG5, the key regulator of autophagy, significantly enhanced the anti-cancer effects of IFNγ using FACS (Figure 3E), indicating that autophagy inhibitors might synergize with IFNγ in the treatment of OSCC. In accordance with western blot result, data of tandem fluorescent-tagged LC3B reporter plasmid (GFP-mRFP-LC3B) [25] shown a decrease autophagy flux induced by IFNγ in ATG5-downregulated HNSCC cells. However, we observed that IFNγ-induced autophagy flux was maintained when autophagosome degradation was inhibited with bafilomycin A1 (BAF1) (Figure 3F).
Figure 3.
ATG5 is necessary for IFNγ-induced autophagy in OSCC cells. A. ATGs and P62 mRNA expression level in HB96, CAL27, HN4, and HN6 cells treated with or without 200 ng/ml IFNγ was analyzed by real-time PCR assay. B. Efficiency for ATG5 gene silencing was confirmed by western blot. C. CAL27 and HN4 cells were transfected with siRNAs targeting ATG5 for 48 h and then treated with IFNγ (200 ng/ml) for 48 h. Expression of ATG5, P62, Beclin1, and LC3B II was detected using a western blot assay. D. CAL27 and HN4 cells were transfected with overexpress plasmid targeting ATG5 for 48 h and then treated with IFNγ (200 ng/ml) for 48 h. Expression of ATG5, P62, Beclin1, and LC3B II was detected using a western blot assay. Efficiency for ATG5 gene silencing was confirmed by western blot assay. E. After incubation in IFNγ (200 ng/ml) for 48 h, the apoptotic cell rate was compared between the OSCC cells transfected with ATG5 siRNA and control siRNA, which was measured by flow cytometry analysis with Annexin V and PI staining. F. ATG5-/- CAL27 and ATG5-/- HN4 cells were transfected with GFP-mRFP-LC3 construct for 24 h and then incubated in IFNγ (200 ng/ml) with or without bafilomycin A1 (200 nM) for 48 h. The merged color was observed. Red or yellow puncta of different stage LC3B were calculated by quantitative analysis and reported as the mean ± SD. All the P values were compared with the control. *: P<0.05, **: P<0.01.
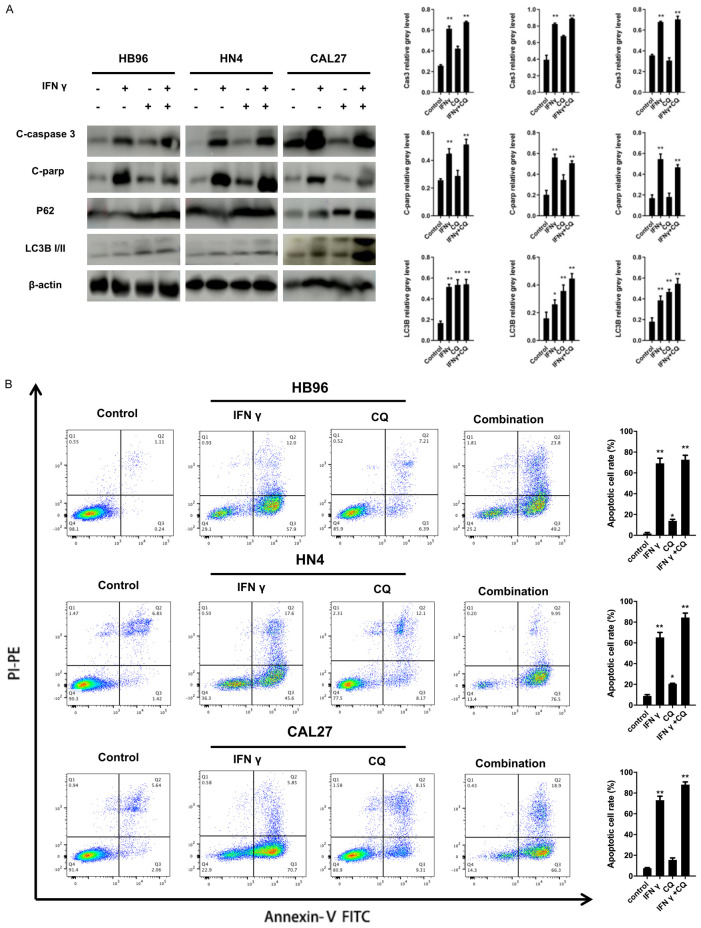
CQ inhibits IFNγ-induced autophagy and synergizes with IFNγ on anti-cancer effect in OSCC in vitro
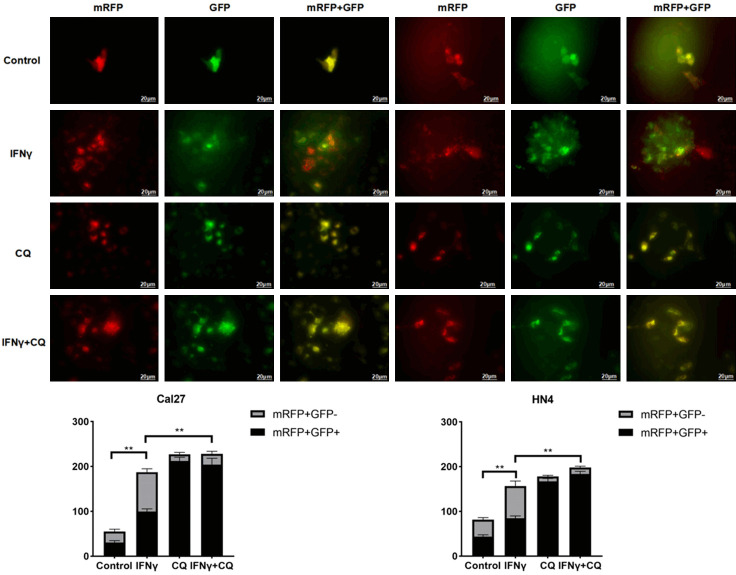
Based on previous results, CQ treatment caused the accumulation of both P62 and LC3B II. Consistent results are shown in Figure 4A. Furthermore, an autophagy flux assay (Figure 5) was performed in the HN4 and CAL27 cells treated with IFNγ using a tandem fluorescent-tagged LC3B reporter plasmid (GFP-mRFP-LC3B) [27]. The yellow fluorescence puncta represented the merging image of green and red fluorescence in autophagosomes, which indicated impaired autophagy. The red fluorescence puncta alone after fusion represented complete autophagic flux. Quantification of red (mRFP+ GFP-) and yellow (mRFP+ GFP+) puncta per cell indicates that IFNγ increased autophagy flux (red and yellow puncta). CQ resulted in the accumulation of yellow puncta (hence autophagosomes) induced by IFNγ.
Figure 4.
Synergistic effect of autophagy inhibitor (CQ) in IFNγ-induced apoptosis in OSCC cells. A. HB96, HN4, and CAL27 cells were stimulated by IFNγ (200 ng/ml) in the presence or absence of CQ (10 uM) for 48 h. Expression of apoptosis and autophagy relative protein, including caspase-3, cleaved-PARP, P62, and LC3B II was assessed by a western blot assay. Quantification of the proteins relative to beta-actin OD values is presented. B. IFNγ-induced apoptosis was synergized by CQ treatment in the HB96, HN4, and CAL27 cells, which was measured by flow cytometry analysis of Annexin V and PI staining. All the P values were compared with the control. *: P<0.05, **: P<0.01.
Figure 5.
Autophagy flux assay was performed in the HN4 and CAL27 cells treated with IFNγ (200 ng/ml for 48 h) using a tandem fluorescent-tagged LC3B reporter plasmid (GFP-mRFP-LC3B). The yellow fluorescence puncta represented the merging image of green and red fluorescence in autophagosomes, which indicated impaired autophagy. The red fluorescence puncta alone after fusion represented complete autophagic flux. Quantitative analysis of red and yellow LC3 puncta was reported as mean ± SD. All the P values were compared with the control. *: P<0.05, **: P<0.01.
CQ could also synergize IFNγ-mediated anti-cancer effects in OSCC cells. As shown in Figure 4A, compared with the IFNγ-treated group, the combined application of IFNγ and CQ significantly up-regulated the expression of caspase-3 and cleaved-PARP proteins. Flow cytometry assays support the above results (Figure 4B). The proportion of Annexin V-positive cells increased after the combination treatment. Thus, the combined application of IFNγ and CQ had a synergistic effect in OSCC cells.
Enhanced anti-cancer effect of IFNγ treatment when combined with CQ in vivo
OSCC xenografts in nude mice were used to confirm the in vitro results. As shown in Figure 6A-C, compared with the group treated with a single agent, the tumor volume in the combination treatment group was significantly smaller than in control group (P=0.0002), IFNγ (P=0.0039) and CQ group (P=0.0041), the difference of body weight of the mice was not significant among the groups. According to the TUNEL assay (Figure 6D), IFNγ up-regulated apoptosis in vivo (P=0.0071), especially when combined with CQ (P=0.0018). Furthermore, immunohistochemistry (Figure 6E) showed a reduction of Ki67, a protein relative to proliferation. In accordance with the in vitro results, CQ resulted in the accumulation of both LC3B and P62 proteins. In summary, IFNγ can induce autophagy and exhibit a synergistic effect with CQ on suppressing tumor growth in vivo.
Figure 6.
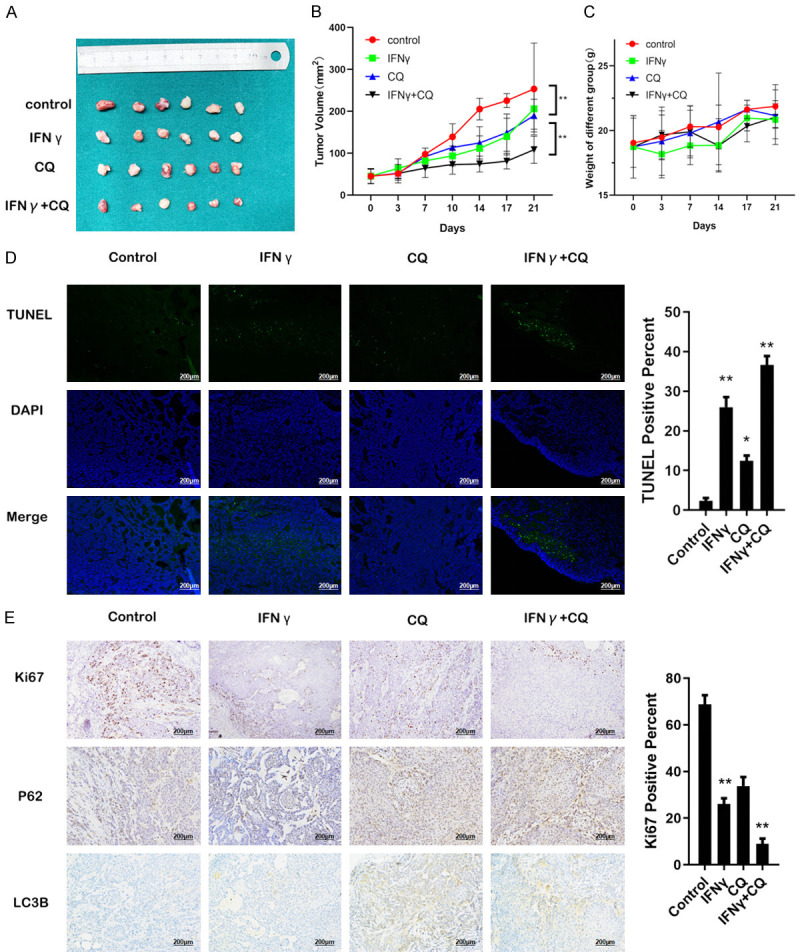
Synergistic effect of CQ and IFNγ in an OSCC tumor xenograft model. After establishing the OSCC tumor xenograft model, 24 nude mice were randomly divided into control, IFNγ, CQ, and IFNγ plus CQ groups (six mice per group). Prescription: IFNγ (10,000 IU per mouse per day, i.p), CQ (30 mg/kg per day, i.p), IFNγ plus CQ, and control (0.9% saline, i.p). All mice were sacrificed 21 days after therapy. A. Representative images of subcutaneous tumors after treatment. B. The tumor volumes were evaluated among the four groups. C. The body weight of mice was evaluated among the four groups, without significant difference. D. The percentage of TUNEL-positive cells was assessed in formalin-fixed paraffin embedding sections of tumors in each group. Magnification: ×200. E. Representative images of tumors from xenografts using HE staining and immunohistochemical staining against Ki67, P62, and LC3B. Magnification: ×200. All the P values were compared with the control. *: P<0.05, **: P<0.01.
Discussion
In this study, we demonstrated that IFNγ simultaneously induced apoptosis and autophagy, and the attenuation of autophagy also synergized the OSCC cell apoptosis mediated by IFNγ. IFNγ-induced autophagy might partly explain the limited effect of IFNγ in solid cancers. Moreover, we demonstrated a synergistic anti-cancer effect in the combination of IFNγ and CQ. This finding might provide the experimental evidence to support the clinical use of combination treatment with IFNγ and autophagy inhibitors in OSCC.
IFNγ is a crucial cytokine in anti-cancer immunity, although a pro-tumorigenic function is also found under certain circumstances [28,29], our in vitro experiment also confirmed the cytotoxicity of IFNγ and the effective suppression of colony formation in OSCC. IFNγ may up-regulate the function of immune cells (increased MHC class I expression, antigen responsive genes, and costimulatory molecules). IFNγ is also capable of inducing the apoptosis of cancer cells via different mechanisms. Notably, IFNγ, either alone or in combination with other cytokines, can induce cellular stress in selected tumor cells, leading to cell death or senescence [30-32]. Accordingly, IFNγ has been taken into clinical trials for cancer treatment [33-35]. In our study, results of western blot analysis reveal IFNγ-induced up-regulation of caspase-3 and cleaved-PARP protein expression, in a dose- and time-dependent manner in vitro. FACS also indicates increasing apoptosis in the OSCC cells, especially with Annexin V. This evidence suggests that IFNγ might be a potential agent in OSCC treatment.
However, former clinical trials of IFNγ fail to support the effectiveness of IFNγ in malignant tumors. Schiller et al. have reported a good prognosis in patients with melanoma in phase II/III clinical trials of IFNγ (Schiller et al., 1996). Unfortunately, they fail to detect the efficacious effects of IFNγ, as the response rate was only 5%, with significant side-effects. This evidence suggests that only patients with early-stage or disseminated cancer could benefit from IFNγ treatment. It seems that this phenomenon shares something in common with autophagy, which is important in tumor progression. In accordance with our hypothesis, a former study demonstrates that IFNγ induces both apoptosis and autophagy in Atf6-/- mice [36]. However, there is no evidence testifying this phenomenon in human cancer lines in vitro or in vivo. In our present study, in vitro and in vivo experimental processes were performed to verify IFNγ-induced autophagy in OSCC. Since IFNγ has not yet been approved in the therapy of most solid tumors, the autophagy activation in OSCC tissues has not been improved. Therefore, our study is the first to demonstrate that IFNγ induces autophagy in OSCC, providing a possible mechanism to explain the limited effect of IFNγ therapy in solid tumors.
Autophagy can both promote and inhibit tumor growth and the roles of autophagy vary in different contexts [37]. A cluster of ATGs is involved in the dynamic membrane-rearrangement reactions of autophagy (Bento et al., 2016). Former researches have reported that the knockdown of ATGs, such as ATG5, synergized with chemotherapy in the efficient elimination of cancer cells [38,39]. Herein, autophagy inhibition via the genetic silencing of ATG5 may contribute to the therapy of advanced cancer. According to our results, genetic silencing of ATG5 could inhibit IFNγ-mediated autophagy via the LC3B pathway. Moreover, FACS also reveals that the silencing of ATG5 synergizes IFNγ-induced apoptosis in vitro, demonstrating the cytoprotective role of autophagy during IFNγ treatment and potential strategy of OSCC treatment by IFNγ combined with an autophagy inhibitor.
Despite the complex process of autophagy and the challenges of the treatment strategy, some pharmacologic autophagy inhibitions have been used in clinical trial [37]. Bryant et al. [37] have confirmed that CQ, as an autophagy inhibitor, could promote the effect of ERK inhibitor in pancreatic cancer treatment by attenuating the resulting autophagy of ERK-inhibition. Accordingly, CQ was chosen in our study, and the results demonstrated its synergistic effect with IFNγ in anti-cancer activity. The strong evidence provided by the CQ results supports the launch of a clinical trial to assess the efficiency of IFNγ in combination with an autophagy inhibitor in OSCC. Furthermore, our present results demonstrate that CQ sensitizes the OSCC cells to enhance IFNγ induced apoptosis both in vitro and in vivo.
It is worth noting that increasing evidence has shown a positive relationship between IFNγ and PD-L1 expression. Kim et al. [41] have demonstrated that IFNγ produced by T-cells could up-regulate exosomal PD-L1 expression, which promotes tumor growth through immune escape in non-small cell lung cancer. Unfortunately, the side-effect of IFNγ in immune escaping has not been considered in this research. We are looking forward to exploring the relative results in future investigations.
In summary, we have demonstrated that IFNγ significantly induces autophagy in OSCC. The inhibition of autophagy by the autophagy inhibitor can enhance the anti-cancer effects of IFNγ in OSCC. This study provides a potential therapeutic approach to treat OSCC with combination of IFNγ and autophagy inhibitor in future.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (81972525, 81602370 and 81672660), Shanghai Municipal Education Commission (17SG18), Shanghai Municipal Commission of Health and Family Planning (2018BR41), Program of Shanghai Academic/Technology Research Leader (19XD1422300). We thank ATCC and the NIH for cell lines.
Disclosure of conflict of interest
None.
References
- 1.Kademani D. Oral cancer. Mayo Clin Proc. 2007;82:878–887. doi: 10.4065/82.7.878. [DOI] [PubMed] [Google Scholar]
- 2.Petersen PE. The World Oral Health Report 2003: continuous improvement of oral health in the 21st century - the approach of the WHO Global Oral Health Programme. Community Dent Oral Epidemiol. 2003;31:3–24. doi: 10.1046/j..2003.com122.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhao TC, Liang SY, Ju WT, Liu Y, Tan YR, Zhu DW, Zhang CP, Zhang ZY, Zhong LP. Normal BMI predicts the survival benefits of inductive docetaxel, cisplatin, and 5-fluorouracil in patients with locally advanced oral squamous cell carcinoma. Clin Nutr. 2020;39:2751–2758. doi: 10.1016/j.clnu.2019.11.037. [DOI] [PubMed] [Google Scholar]
- 4.Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52:195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 5.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 6.Zhao TC, Liang SY, Ju WT, Fu Y, Zhou ZH, Wang LZ, Li J, Zhang CP, Zhang ZY, Zhong LP. High-risk lymph node ratio predicts worse prognosis in patients with locally advanced oral cancer. J Oral Pathol Med. 2020;49:787–795. doi: 10.1111/jop.13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo JY, White E. Autophagy, metabolism, and cancer. Cold Spring Harb Symp Quant Biol. 2016;81:73–78. doi: 10.1101/sqb.2016.81.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Gene Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhammad JS, Nanjo S, Ando T, Yamashita S, Maekita T, Ushijima T, Tabuchi Y, Sugiyama T. Autophagy impairment by Helicobacter pylori-induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer. 2017;140:2272–2283. doi: 10.1002/ijc.30657. [DOI] [PubMed] [Google Scholar]
- 11.Shen S, Zhou M, Huang K, Wu Y, Ma Y, Wang J, Ma J, Fan S. Blocking autophagy enhances the apoptotic effect of 18β-glycyrrhetinic acid on human sarcoma cells via endoplasmic reticulum stress and JNK activation. Cell Death Dis. 2017;8:e3055. doi: 10.1038/cddis.2017.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong P, Zhu X, Geng Q, Xia L, Sun X, Chen Y, Li W, Zhou Z, Zhan Y, Xu D. The microRNA-423-3p-Bim axis promotes cancer progression and activates oncogenic autophagy in gastric cancer. Mol Ther. 2017;25:1027–1037. doi: 10.1016/j.ymthe.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Liu Z, Hu T, Han L, Yu S, Yao Y, Ruan Z, Tian T, Huang T, Wang M, Jing L, Nan K, Liang X. Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle. 2017;16:1053–1062. doi: 10.1080/15384101.2017.1312224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, VonHoff DD, Maitra A, Kimmelman AC. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei Y, Kansy BA, Li J, Cong L, Liu Y, Trivedi S, Wen H, Ting JP, Ouyang H, Ferris RL. EGFR-targeted mAb therapy modulates autophagy in head and neck squamous cell carcinoma through NLRX1-TUFM protein complex. Oncogene. 2016;35:4698–4707. doi: 10.1038/onc.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang I, Wang C. Inhibition of HDAC6 protein enhances bortezomib-induced apoptosis in head and neck squamous cell carcinoma (HNSCC) by reducing autophagy. J Biol Chem. 2016;291:18199–18209. doi: 10.1074/jbc.M116.717793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Zhang W, Wang Y, Liu B, Zhang W, Zhao Y, Kulkarni AB, Sun Z. Dual induction of apoptotic and autophagic cell death by targeting survivin in head neck squamous cell carcinoma. Cell Death Dis. 2015;6:e1771. doi: 10.1038/cddis.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li K, Hua K, Lin Y, Su C, Ko J, Hsiao M, Kuo M, Tan C. Inhibition of G9a induces DUSP4-dependent autophagic cell death in head and neck squamous cell carcinoma. Mol Cancer. 2014;13:172. doi: 10.1186/1476-4598-13-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Zhou C, Lu Y, Hong M, Zhang Z, Zhang Z, Chang Y, Zhang C, Li X. IFN-gamma-mediated IRF1/miR-29b feedback loop suppresses colorectal cancer cell growth and metastasis by repressing IGF1. Cancer Lett. 2015;359:136–147. doi: 10.1016/j.canlet.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee KK, Bose A, Ghosh D, Sarkar K, Goswami S, Pal S, Biswas J, Baral R. IFNalpha2b augments immune responses of cisplatin+5-fluorouracil treated tongue squamous cell carcinoma patients--a preliminary study. Indian J Med Res. 2012;136:54–59. [PMC free article] [PubMed] [Google Scholar]
- 21.Xu YH, Li ZL, Qiu SF. IFN-gamma induces gastric cancer cell proliferation and metastasis through upregulation of integrin beta3-mediated NF-kappaB signaling. Transl Oncol. 2018;11:182–192. doi: 10.1016/j.tranon.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Zhang Y, Chen L, Lu X, Li Z, Xue Y, Guan YQ. Cervical cancer HeLa cell autocrine apoptosis induced by coimmobilized IFN-gamma plus TNF-alpha biomaterials. ACS Appl Mater Interfaces. 2018;10:8451–8464. doi: 10.1021/acsami.7b18277. [DOI] [PubMed] [Google Scholar]
- 23.Pagotto A, Pilotto G, Mazzoldi EL, Nicoletto MO, Frezzini S, Pastò A, Amadori A. Autophagy inhibition reduces chemoresistance and tumorigenic potential of human ovarian cancer stem cells. Cell Death Dis. 2017;8:e2943. doi: 10.1038/cddis.2017.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei G, Luo M, Ni X, Wu J, Wang S, Ma Y, Yu J. Autophagy facilitates metadherin-induced chemotherapy resistance through the AMPK/ATG5 pathway in gastric cancer. Cell Physiol Biochem. 2018;46:847–859. doi: 10.1159/000488742. [DOI] [PubMed] [Google Scholar]
- 25.Mulcahy Levy JM, Zahedi S, Griesinger AM, Morin A, Davies KD, Aisner DL, Kleinschmidt-DeMasters BK, Fitzwalter BE, Goodall ML, Thorburn J, Amani V, Donson AM, Birks DK, Mirsky DM, Hankinson TC, Handler MH, Green AL, Vibhakar R, Foreman NK, Thorburn A. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife. 2017;6:e19671. doi: 10.7554/eLife.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burman C, Ktistakis NT. Autophagosome formation in mammalian cells. Semin Immunopathol. 2010;32:397–413. doi: 10.1007/s00281-010-0222-z. [DOI] [PubMed] [Google Scholar]
- 27.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 28.Zaidi MR. The interferon-gamma paradox in cancer. J Interferon Cytokine Res. 2019;39:30–38. doi: 10.1089/jir.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin Cancer Res. 2011;17:6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakshit S, Chandrasekar BS, Saha B, Victor ES, Majumdar S, Nandi D. Interferon-gamma induced cell death: regulation and contributions of nitric oxide, cJun N-terminal kinase, reactive oxygen species and peroxynitrite. Biochim Biophys Acta. 2014;1843:2645–2661. doi: 10.1016/j.bbamcr.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Hubackova S, Kucerova A, Michlits G, Kyjacova L, Reinis M, Korolov O, Bartek J, Hodny Z. IFNγ induces oxidative stress, DNA damage and tumor cell senescence via TGFβ/SMAD signaling-dependent induction of Nox4 and suppression of ANT2. Oncogene. 2016;35:1236–1249. doi: 10.1038/onc.2015.162. [DOI] [PubMed] [Google Scholar]
- 32.Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, Schilbach K, Essmann F, Kneilling M, Griessinger C, Ranta F, Ullrich S, Mocikat R, Braungart K, Mehra T, Fehrenbacher B, Berdel J, Niessner H, Meier F, van den Broek M, Haring HU, Handgretinger R, Quintanilla-Martinez L, Fend F, Pesic M, Bauer J, Zender L, Schaller M, Schulze-Osthoff K, Rocken M. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- 33.Creagan ET, Ahmann DL, Long HJ, Frytak S, Sherwin SA, Chang MN. Phase II study of recombinant interferon-gamma in patients with disseminated malignant melanoma. Cancer Treat Rep. 1987;71:843–844. [PubMed] [Google Scholar]
- 34.Ernstoff MS, Trautman T, Davis CA, Reich SD, Witman P, Balser J, Rudnick S, Kirkwood JM. A randomized phase I/II study of continuous versus intermittent intravenous interferon gamma in patients with metastatic melanoma. J. Clin. Oncol. 1987;5:1804–1810. doi: 10.1200/JCO.1987.5.11.1804. [DOI] [PubMed] [Google Scholar]
- 35.Kopp WC, Smith JN, Ewel CH, Alvord WG, Main C, Guyre PM, Steis RG, Longo DL, Urba WJ. Immunomodulatory effects of interferon-gamma in patients with metastatic malignant melanoma. J Immunother Emphasis Tumor Immunol. 1993;13:181–190. doi: 10.1097/00002371-199304000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Gade P, Ramachandran G, Maachani UB, Rizzo MA, Okada T, Prywes R, Cross AS, Mori K, Kalvakolanu DV. An IFN-gamma-stimulated ATF6-C/EBP-beta-signaling pathway critical for the expression of death associated protein kinase 1 and induction of autophagy. Proc Natl Acad Sci U S A. 2012;109:10316–10321. doi: 10.1073/pnas.1119273109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. 2019;33:610–619. doi: 10.1101/gad.325514.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Y, Su C, Zhao L, Shi Y. Chitosan nanoparticle-mediated co-delivery of shAtg-5 and gefitinib synergistically promoted the efficacy of chemotherapeutics through the modulation of autophagy. J Nanobiotechnology. 2017;15:28. doi: 10.1186/s12951-017-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong J, Dong X, Xiu P, Wang F, Liu J, Wei H, Xu Z, Liu F, Li T, Li J. Blocking autophagy enhances meloxicam lethality to hepatocellular carcinoma by promotion of endoplasmic reticulum stress. Cell Prolif. 2015;48:691–704. doi: 10.1111/cpr.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang G, Witkiewicz AK, Knudsen ES, Petricoin EF, Singh PK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, Der CJ. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25:628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DH, Kim H, Choi YJ, Kim SY, Lee JE, Sung KJ, Sung YH, Pack CG, Jung MK, Han B, Kim K, Kim WS, Nam SJ, Choi CM, Yun M, Lee JC, Rho JK. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp Mol Med. 2019;51:1–13. doi: 10.1038/s12276-019-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]