Abstract
Prostate cancer (PCa) is one of the major causes of cancer death among males worldwide. Our previous studies indicated that the proliferation of prostate cancer cells was reduced after BLM knockdown, however, the mechanism is still not clear. In this study, we identified a direct interaction between BLM and EZH2, which had extremely significantly positive correlations (P<0.001). In vitro, our research revealed that tumor growth was inhibited after EZH2 knockdown and that inhibition could be reversed by BLM overexpression; conversely, tumor growth was promoted after EZH2 overexpression, and promotion could be reversed by BLM knockdown. This suggests that BLM and EZH2 play important roles in the progression of prostate cancer cells. In vivo, the impact of BLM and EZH2 was investigated in mouse xenograft models, and the results showed that EZH2 could be regulated by BLM, which was consistent with our in vitro observations. Our results demonstrated that the expression of P53 is affected by the binding of BLM and EZH2 to the MDM2 promoter region. This finding indicated that EZH2 regulates the expression of MDM2 at the transcriptional level by interacting with BLM.
Keywords: BLM, EZH2, prostate cancer, prognostic, biomarker
Introduction
In 2020, the American Cancer Society (ACS) estimates the numbers of new PCa cases and deaths in the USA will reach 191,930 and 33,330, ranking 1st and 2nd out of all cancers, respectively [1]. PCa development is the result of multiple factors, including age [2], diet [3,4], and genetics [5-7]. Currently, one of the methods for the clinical diagnosis of PCa is the measurement of PSA levels in serum, but PSA is not organ-specific; rather than acting as a biomarker for PCa, the PSA level can be elevated when BPH or prostatitis occurs [8], leading to overdiagnosis [9]. Because the early clinical stage of prostate cancer is not obvious, a considerable number of patients have developed locally advanced or distant metastasis at the time of diagnosis [10,11]. Even if some patients can be relieved by castration treatment, many patients still progress to castration-resistant prostate cancer (CRPC), resulting in difficult treatment and poor effects [12,13]. Recently, many potential prognostic biomarkers for PCa have been identified, including uPA [14,15], PSCA [16,17], and ANXA3 [18], however, the prognosis of advanced PCa remains poor. Therefore, looking for PCa biomarkers and studying the underlying mechanisms for therapeutic purposes in PCa is imperative.
BLM is critical for DNA replication, repair, recombination, transcription, telomere maintenance and other cellular metabolic processes [19]. Recent studies have shown that the BLM gene is ranked first among seven high-risk PCa genes [20], and the high expression of BLM in PCa tissues is closely related to the malignant degree of the tumor [21]. Our previous studies indicated that PC3 cell proliferation was promoted after BLM overexpression, and the promotion effect was rescued by BLM knockdown [22]. We also determined that BLM enters the nucleus, requiring importin β1, RanGDP and NTF2 [23], and Larissa T et al. reported that BLM was present during nucleolar localization [24]. Protein nucleolar localization plays an important role in maintaining genomic stability and repair via the EGR1, P53, ARF, DMP1 and PTEN signaling pathways [25], which are involved in the process of tumorigenesis or apoptosis [26,27]. Therefore, BLM may play key roles in PCa development. However, the molecular mechanisms by which BLM is regulated in PCa are not entirely clear.
Our present research indicated that BLM overexpression was a poor prognostic marker for PCa patients and that overexpression of BLM can promote the proliferation and migration of prostate cancer cells. Moreover, EZH2, a member of the Polycomb-group (PcG) family [28], is involved in maintaining the transcriptional repressive state of genes over successive cell generations [29]. As a key mediator of transcriptional repression [30], EZH2 exhibits high expression in a variety of tumors [31-33], and we have identified a direct interaction between EZH2 and BLM. However, the role of the BLM and EZH2 interaction in the process of prostate cancer has not been reported. In our study, we found that both BLM and EZH2 were highly expressed and that BLM and EZH2 had extremely significantly positive correlations (P<0.001). The function of BLM is to regulate the expression of MDM2 protein through EZH2-mediated regulation to target the P53 protein.
Materials and methods
Cell culture
Human PCa cell lines (PC3, LNCap, and 22RV1) and human normal prostate cells (RWPE-1) were purchased from Zhong Qiao Xin Zhou Biotechnology Co., Ltd. (Shanghai, China). All cell lines were authenticated by Zhong Qiao Xin Zhou Biotechnology Co., Ltd. (Shanghai, China) or Viva Cell Biosciences Ltd. (Shanghai, China) via STR profiling and tested free of mycoplasma as described [34]. HEK293T cells were purchased directly from Kunming Cell Bank (Kunming, China). PC3 cells were cultured in DMEM/F12 medium (Gibco BRL, Grand Island, NY, USA), LNCap cells were cultured in RPMI 1640 (Gibco BRL, Grand Island, NY, USA) with 1% Glutamax (Life Technologies, Grand Island, NY, USA) and 1% sodium pyruvate (Life Technologies, Grand Island, NY, USA), 22RV1 cells were cultured in RPMI 1640, RWPE-1 cells were cultured in special medium (ZQ-1303, ZQXZ Bio, Shanghai, China), and HEK293T cells were cultured in DMEM/HIGH GLUCOSE (HyClone, South Logan, Utah, USA). All media were supplemented with 10% FBS (04-001-1ACS; BI, Beit HaEmek, Israel) and 1% penicillin (100 U/mL)-streptomycin (100 μg/mL) solution (SV30010; HyClone, Logan, UT, USA).
Cell viability experiments and flow cytometric analysis
Cell viability experiments were performed as previously described [35]; 2.5 × 104 PC3 cells were treated with the BLM inhibitor ML216 (Chemietek, Shanghai, MCE) at 6.25, 12.5, 25.0, 50.0, and 100.0 μM for 5 days, and trypan blue was used for counting live cells. For cell cycle experiments, cells were fixed in PBS/70% ethanol at 4°C overnight. Propidium staining (PI) was used to stain DNA. Cell cycle profile data were obtained with a Gallios flow cytometer (Beckman Coulter) and QC functionality with Levey-Jennings reports with Gallios CXP software.
For knockdown and overexpression experiments
For knockdown experiments, the siRNA pools were transiently transfected into PC3 cells with LipofectamineTM 3000 Transfection Reagent (Invitrogen, Carlsbad, CA). BLM-siRNA, two different EZH2-siRNAs, and negative control-siRNA were obtained from GenePharma Technology (Shanghai, China). The sequence of BLM-siRNA was 5’-GCUAGGAGUCUGCGUGCGAdTdT-3’ [36], the sequence of two different EZH2-siRNAs was 5’-GAGGGAAAGUGUAUGAUAAdTdT-3’ and 5’-GCUGGAAUCAAAGGAUACAdTdT-3’ [37], and the negative control-siRNA was 5’-UUCUCCGAACGUGUCACGUdTdT-3’. For stable knockdown or overexpression cell lines, PC3 cells were infected with BLM, EZH2-specific shRNA or BLM, and EZH2 overexpression lentiviral particles (5 µg/ml) (GenePharma, Shanghai). Infected cells were selected with 1 µg/ml puromycin for 4 weeks. The nucleic acid sequences of shRNA were the same as those of siRNA.
GST-pull down assay
For the GST pulldown assay, the GST and GST fusion protein pGEX-6p-1-EZH21-370 were constructed and transformed into E. coli BL21, and both cell lines were induced with 0.2 mM IPTG at 37°C for 8 h. The His-BLM fusion protein pET-32a-BLM642-1417 was induced with 0.1 mM IPTG at 17°C for 12 h. All bacterial pellets were extensively washed with 1 × PBS and high pressure, then centrifuged at 15,000 g for 10 min, and the supernatants were collected and mixed with glutathione-sepharose (Thermo Scientific) and Ni-NTA agarose (Thermo Scientific) for purification of GST, GST-EZH21-370 and His-BLM642-1417. His-BLM642-1417 protein was rotated with GST and GST-EZH21-370 at 4°C for 6 h and then added to glutathione-sepharose beads for overnight at 4°C. After centrifugation and three washes, bound proteins were eluted with 20 μl of 1 × SDS-PAGE loading buffer and processed for Western blotting.
Coimmunoprecipitation (Co-IP) assay and western blot analysis
PC3 cells were treated with lysis buffer containing 0.5% Triton X-100, 20 mM Tris-HCl (pH 8), 2 mM EDTA, 137 mM NaCl and 1 mM PMSF. The lysates were centrifuged at 15,000 × g for 10 min, and then the supernatant was collected and incubated with antibodies against endogenous proteins and purified with protein A/G beads for 24 h. The beads were washed with lysis buffer, centrifuged, and collected at 400 × g for 5 min. After the beads were boiled for 10 min, the precipitated proteins were separated and eluted in SDS-PAGE loading buffer. Western blotting was used to analyze the cell lysates and precipitated proteins. For Western blot (WB) analysis, equal amounts of cell proteins from the cells were separated and transferred. The PVDF membranes (Millipore, MMAS, USA) were blocked with 5% nonfat milk for 2 h at 37°C and then incubated with specific antibodies at 4°C overnight. Next, horseradish peroxidase (HRP)-labeled secondary antibodies were incubated for 2 h at 37°C, and the proteins were detected with a BeyoECL Plus kit (Beyotime, Shanghai). The relative amount of protein was normalized to GAPDH levels. The density of proteins was measured by ImageJ 1.8.0 software (National Institutes of Health, Bethesda, MD). The antibodies used in this experiment were as follows: BLM, EZH2, P53, MDM2, c-Caspase3, Caspase9, Bcl-2 (Abcam, Cambridge, USA), His and GST (Cwbio, Beijing, China), GAPDH (Santa Cruz, Dallas, TX, USA), Bax and Cyto.C (Proteintech, Wuhan, China).
Sliver staining and mass spectrometry
After the co-IP experiment and electrophoresis of the samples containing precipitated proteins, the protein gel was placed in stationary liquid with 50% ethanol, 40% water and 10% acetic acid on a shaker overnight at room temperature. The protein bands were observed with a silver stain kit and analyzed using MS by Genecreat (Wuhan, China).
Immunohistochemistry and hematoxylin-eosin staining
The tissue chip was purchased from Biomax. Inc. (Lot. PR807c) and contained 10 normal prostate tissues, 50 prostate adenocarcinomas, and 20 benign prostate hyperplasias. The IHC staining procedure was performed as previously described [38]. The tissue chips were incubated with anti-BLM (1:200, Abcam, Cambridge, USA) and anti-EZH2 primary antibodies (1:100; Abcam, Cambridge, USA). After washing three times, the tissue chips were incubated with HRP-conjugated anti-goat antibodies and then stained with 3,5-diaminobenzidine (DAB). The score of each tissue was calculated by multiplying the percentage of stained cells (1, weak; 2, moderate; 3, intense) by the staining value (0-5%, 0; 6-25%, 1; 26-50%, 2; 51-75%, 3; 76-100%, 4). The scores were independently generated by two pathologists (Drs. Zhu-Xue Zhang and Nan-Zhi Wen). The median IHC score was selected as the cutoff value to define high and low expression. For hematoxylin-eosin staining (HE), mouse tumor-forming subcutaneous metastatic nodules were acquired and immersed in PBS (4% paraformaldehyde), followed by routine processing [39].
Immunofluorescence
In a physiological state, PC3, LNCap and 22RV1 cells were seeded onto laser confocal microscope dishes. When cell confluence reached 70%, the cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.3% Triton X-100 for 20 min and then incubated in 5% goat serum albumin for 20 min to block nonspecific protein-protein interactions. After washing, the confocal dishes were then incubated with rabbit anti-BLM (1:200, Abcam, Cambridge, USA) and mouse anti-EZH2 antibodies (1:100, Abcam, Cambridge, USA) for 1.5 h at 37°C. After washing again, the dish was incubated with goat anti-rabbit IgG (Alexa Fluor® 594) (1:200, Abcam, Cambridge, USA) and goat anti-mouse IgG (Alexa Fluor® 488) antibodies (1:200, Abcam, Cambridge, USA) for 1 h at 37°C and stained with DAPI for 5 min at RT. Images were acquired by an Olympus IX71 Nikon imaging system (× 200).
Chromatin immunoprecipitation and deep SEQuencing (ChIP-seq)
Chromatin immunoprecipitation (ChIP) was carried out as described [40], and the PC3 cells were fixed with 1% formaldehyde and sonicated in lysis buffer (1% Triton X-100, 140 mM NaCl and 50 mM HEPES-KOH, pH 8.0). Immunoprecipitation of chromatin was performed with normal rabbit IgG in 1 × dilution buffer or rabbit anti-EZH2 antibody (Abcam, Cambridge, USA) (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1.0% Triton-X-100, and 150 mM NaCl). Immunocomplexes were incubated with A/G agarose beads at 4°C overnight, and RNA was removed by RNase A at 65°C overnight. Next, the DNA-protein mixtures were eluted, and cross-linking was reversed by adding glycine. DNA fragments were purified and used to construct a DNA library, and the concentration and purity of the DNA library were detected by a Qubit 1 × dsDNA Assay Kit (Thermo Fisher). Finally, the DNA library was sent to a company for deep sequencing when the concentration and purity reached high-throughput sequencing standards.
Dual-luciferase reporter assay
A dual-luciferase reporter assay was performed as described previously [41]. For this experiment, the primer pairs of the MDM2 promoter region were designed for a series of truncations based on the ChIP-seq results. The DNA fragments were inserted into the pGL3-basic luciferase vector. 293T cells were plated in 96-well plates, and when cell confluence reached 90%, the plasmids were transfected into 293T cells with Lipofectamine™ 3000 Transfection Reagent. The Renilla luciferase reporter pRL-TK vectors were also transiently co-transfected into 293T cells as a positive control. After 48 h, the luciferase activity was detected in a Multiskan Spectrum instrument (Synergy H4, BioTek, USA).
MTS assay and clone formation assay
Cell proliferation was detected by MTS assays (Saint-Bio, Shanghai, China). PC3 cells transfected with siRNAs or shRNAs (GenePharma, Shanghai, China) were seeded in 96-well plates at a density of 5 × 103/well. At 24 h, 48 h and 72 h, the optical density (OD) was detected using a Multiskan Spectrum instrument (Synergy H4, BioTek, USA) at 490 nm. For the clone formation assay, after PC3 cells were transfected, 500 cells per well were seeded in 6-well plates for 14 days. Next, the formative colonies were fixed with 4% formalin for 20 min and stained with crystal violet. The numbers of colonies were counted by ImageJ 1.8.0 software (National Institutes of Health, Bethesda, MD, USA).
Wound scratch assay and transwell assays
For the wound-healing assay, PC3 cells were seeded in 6-well plates until reaching confluence. Next, the PC3 cells were transfected with siRNAs or shRNAs (GenePharma, Shanghai, China), and wounds were created by using a 10 μl pipette tip. Gap images were measured at 0 h (w1) and at 24 h or 48 h (w2), and the percentage of wound closure was calculated as (w1-w2)/w1 × 100%. The Transwell migration assay was performed with BD BioCoat 9 Matrigel Invasion Chambers (BD Biosciences, Franklin Lakes, NJ, USA). A total of 3 × 104 homogeneous single-cell suspensions in 200 μl of serum-free MDED/F12 medium were added to the upper compartment of a chamber, and the lower compartment was filled with 500 μl of DMED/F12 medium containing 10% FBS. After 10 hours of incubation at 37°C, cells on the lower surface were fixed with 4% formalin for 20 min and stained with crystal violet. Cell counts were calculated by ImageJ 1.8.0 software. For the Transwell invasion assay, BD BioCoat 9 Matrigel Invasion Chambers were also used, and Matrigel was added to the upper chambers and incubated for 4 h at 37°C before filling with DMED/F12 medium. After this, other processes were the same as those in the Transwell migration assay. Each experiment was performed three times independently.
Animal model
The animal study procedures were approved by the Laboratory Animal Ethics Committee of Guizhou University. Six-week-old SPF male immune-deficient BALB/c mice from Tianqin Biotechnology Co., Ltd. (Changsha, China) were subcutaneously injected with approximately 3 × 106 cells. The tumor volumes were measured every 4 days and calculated using the equation volume = (W2 × L)/2 [42], where W and L represent width and length, respectively. After 4 weeks, the mice were sacrificed under anesthesia, and the tumor xenografts were removed and weighed and then immersed in 4% formalin for immunohistochemistry (IHC) and hematoxylin-eosin (HE) staining.
Statistical analysis
Data are presented as the mean ± SD from 3 independent experiments. Gray value of protein expression, MTS assay, clone formation assay, wound scratch assay, transwell assays and tumor weight were calculated by one-way ANOVA, tumor volume was calculated by two-way ANOVA. Correlations between BLM and EZH2 expression were analyzed by Spearman rank correlation analysis. All analysis was performed by GraphPad Prism 5.0. P values less than 0.05 were considered statistically significant.
Results
BLM was more highly expressed in PRAD than in normal cells. Co-IP and GST pulldown assays were carried out, and IF results showed that the BLM and EZH2 proteins were colocalized in cells
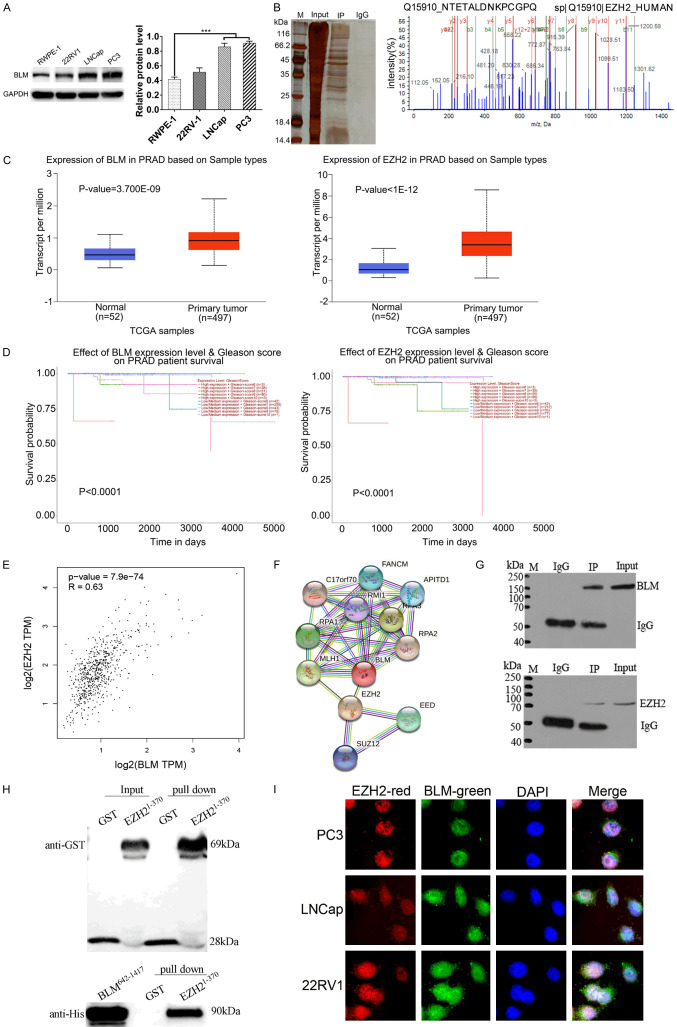
Human PCa cell lines (PC3, LNCap, and 22RV1) and human normal prostate epithelial (RWPE-1) cells were cultured, and BLM protein levels were detected by WB under physiological conditions. The results showed that PC3 cells had the highest level of BLM (Figure 1A). Therefore, PC3 cells were chosen to identify new proteins interacting with BLM in coimmunoprecipitation assays. SDS-PAGE was visualized with colloidal sliver staining (Figure 1B). Protein bands that were different from the IgG control group were sent for MS/MS analysis using MALDI-TOF/TOF, and MS/MS data were analyzed with ProteinPilotTM software using Mascot search. To select putative interacting proteins and to further explore their biological roles, we performed a literature search and selected EZH2 as a potential candidate because BLM and EZH2 are more highly expressed in cancer tissues than in normal tissues (Figure 1C). In the survival analysis of PRAD patients, the expression levels of BLM and EZH2 were significantly positively correlated with the Gleason score (Figure 1D), and EZH2 was highly correlated with the BLM protein according to the Pearson correlation statistics (Figure 1E). In addition, there may be a direct interaction of EZH2 and BLM in the STRING database (Figure 1F).
Figure 1.
BLM was highly expression in PRAD than normal cells, Co-IP and GST pull down were carried out, IF results shows that the BLM and EZH2 proteins are co-located in PC3. A. Western blot analysis of BLM expression in human normal prostate (RWPE-1) and PRAD cell lines (PC3, LNCap, 22RV1). B. BLM interaction proteins were obtained by Co-IP test under physiological conditions, IP and IgG control group were send company for MS identification. C. BLM and EZH2 mRNA levels in the PRAD TCGA dataset (normal vs. primary tumor). D. The relationship between Gleason score and mRNA expression level of BLM and EZH2 in the PRAD TCGA dataset. E. EZH2 is significantly positively correlated with BLM mRNA level in the PRAD TCGA dataset. F. Protein-protein interaction network of BLM and EZH2, the protein network was predicted using STRING database. G, H. The interaction between BLM and EZH2 was confirmed by Co-IP and GST-pull down. I. The localization of BLM and EZH2 proteins in PC3, LNCap and 22RV1 cells were detected by indirect immunofluorescence assay.
Next, to further validate the interaction between BLM and EZH2, co-IP and GST pull-down assays were performed. Co-IP assay results showed that there was a mutual connection between BLM and EZH2 (Figure 1G). Then, the recombinant plasmids pGEX-6p-1-EZH21-370, pGEX-6p-1-EZH2371-746 and pET-32a-BLM642-1417 were constructed, but only pGEX-6p-1-EZH21-370 and pET-32a-BLM642-1417 were successfully expressed in vitro for the GST pulldown assay. The results showed that there was a direct interaction between EZH21-370 and BLM642-1417 (Figure 1H). In addition, the subcellular localization of BLM and EZH2 was examined by confocal microscopy analysis. PC3, LNCap and 22RV1 cells were fixed and probed with rabbit anti-BLM and mouse anti-EZH2 antibodies followed by incubation with goat anti-rabbit IgG H&L (Alexa Fluor 488) and goat anti-mouse IgG H&L (Alexa Fluor 594)-tagged secondary antibodies. The cells were incubated with DAPI to stain the nuclei, and the obtained images indicated that BLM (green) and EZH2 (red) were colocalized in the nucleus (Figure 1I). These results together endorse the physical interaction of BLM and EZH2 in PCa cells.
The expression levels of BLM and EZH2 were investigated by IHC, and BLM is associated with EZH2 as a potential tumor marker for prostate cancer
We studied the clinicopathologic significance of BLM and EZH2 in prostate tissue chips (80 cases), which included 50 prostate adenocarcinomas, 20 benign prostate hyperplasias and 9 normal prostate tissues (1 normal tissue lost during IHC experiment). IHC analysis showed that BLM and EZH2 were both localized in the nucleus, and we discovered that patients with strong BLM staining tended to have strong EZH2 staining (Figure 2A). In addition, the expression of both BLM and EZH2 was higher in PCa tissues than in normal tissues (Figure 2B; Tables 1 and 2). The expression of BLM and EZH2 was positively correlated (r=0.291, P<0.001) by Spearman rank correlation analysis (Figure 2C). Importantly, higher expression of BLM and EZH2 was correlated with T status (P=0.030, P=0.012), clinical stage (P=0.030, P=0.012) and Gleason score (P=0.006, P=0.029) in tumor tissues but not with age, Gilson grade or NM status (P>0.05) (Table 3). The results showed that BLM was associated with EZH2 as a potential tumor marker for prostate cancer, and both were significantly different from normal tissues.
Figure 2.
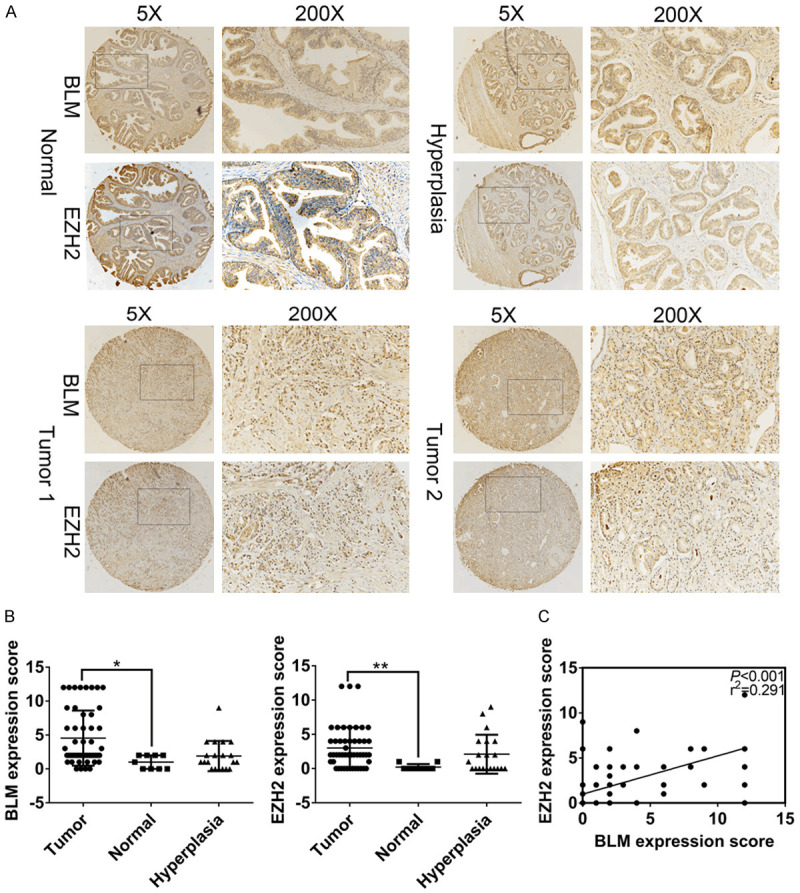
The expression levels of BLM and EZH2 were researched by IHC, BLM is associated with EZH2 as a potential tumor marker for prostate cancer. A. Representative images of BLM and EZH2 expression in Prostate adenocarcinoma, hyperplasia and ANT. B. The expression of BLM and EZH2 in 50 PCa tissues was higher than ANT, *P<0.05. C. The expression of BLM and EZH2 was positively correlated in PCa tissues.
Table 1.
The expression of BLM protein in different types of prostate tissue
| Histopathological type | N | BLM | P value (Compare with cancer tissue) | ||
|---|---|---|---|---|---|
|
| |||||
| Positive | Negative | Positive expression rate (%) | |||
| Normal tissue | 9 | 4 | 5 | 44.4 | 0.038 |
| Hyperplasia tissue | 20 | 9 | 11 | 45.0 | 0.008 |
| Cancer tissue | 50 | 40 | 10 | 80.0 | / |
Note: P value is from χ2-test.
Table 2.
The expression of EZH2 protein in different types of prostate tissue
| Histopathological type | N | EZH2 | P value (Compare with cancer tissue) | ||
|---|---|---|---|---|---|
|
| |||||
| Positive | Negative | Positive expression rate (%) | |||
| Normal tissue | 9 | 0 | 9 | 0.0 | 0.000 |
| Hyperplasia tissue | 20 | 9 | 11 | 45.0 | 0.061 |
| Cancer tissue | 50 | 35 | 15 | 70.0 | / |
Note: P value is from χ2-test.
Table 3.
Correlations between BLM, EZH2 expression and clinicopathological characteristics of PCa patients
| Clinicopathological characteristics | N | BLM | X2 | P value | KMT6 | X2 | P value | ||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Positive (%) | Negative (%) | Positive (%) | Negative (%) | ||||||
| Age (years) | 0.183 | 0.669 | 2.226 | 0.136 | |||||
| ≤69 | 22 | 17 (77.3) | 5 (22.7) | 13 (59.1) | 9 (40.9) | ||||
| >69 | 28 | 23 (82.1) | 5 (17.9) | 22 (78.6) | 6 (21.4) | ||||
| T status | 4.688 | 0.030 | 6.222 | 0.012 | |||||
| T1-T2 | 30 | 21 (70.0) | 9 (30.0) | 17 (56.7) | 13 (43.3) | ||||
| T3-T4 | 20 | 19 (95.0) | 1 (5.0) | 18 (90.0) | 2 (10.0) | ||||
| Clinical stage | 4.688 | 0.030 | 6.222 | 0.012 | |||||
| I-II | 30 | 21 (70.0) | 9 (30.0) | 17 (56.7) | 13 (43.3) | ||||
| III-IV | 20 | 19 (95.0) | 1 (5.0) | 18 (90.0) | 2 (10.0) | ||||
| Gleason Score | 7.510 | 0.006 | 4.757 | 0.029 | |||||
| <7 | 13 | 7 (53.8) | 6 (46.2) | 6 (46.2) | 7 (53.8) | ||||
| ≥7 | 37 | 33 (89.2) | 4 (10.8) | 29 (78.4) | 8 (21.6) | ||||
| Gleason Grade | 1.663 | 0.197 | 2.850 | 0.091 | |||||
| 1-3 | 21 | 15 (71.4) | 6 (28.6) | 12 (57.1) | 9 (42.9) | ||||
| 4-5 | 29 | 25 (86.2) | 4 (13.8) | 23 (79.3) | 6 (20.7) | ||||
| N-regional lymph nodes | 1.670 | 0.327 | 0.577 | 0.447 | |||||
| N0 | 44 | 34 (77.3) | 10 (22.7) | 30 (68.2) | 14 (31.8) | ||||
| N1 | 6 | 6 (100.0) | 0 (0.0) | 5 (83.3) | 1 (16.7) | ||||
| M-Distant metastasis | 0.250 | 1.000 | 0.429 | 1.000 | |||||
| M0 | 49 | 39 (79.6) | 10 (20.4) | 34 (69.4) | 15 (30.6) | ||||
| M1 | 1 | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | ||||
Note: P value is from χ2-test.
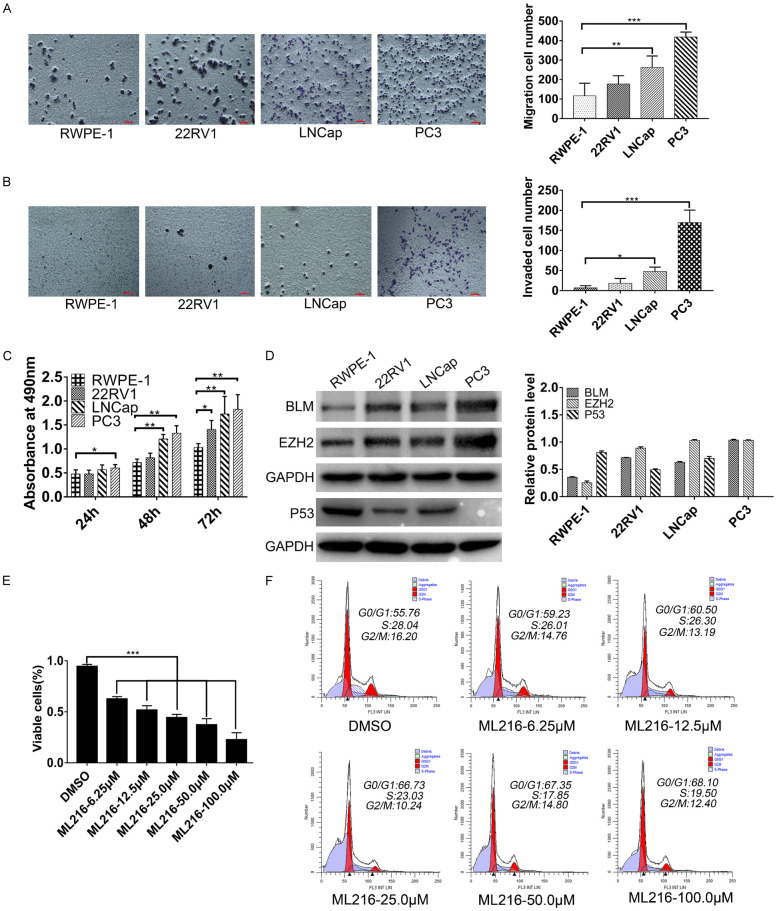
The migration, invasion and proliferation of the PRAD cell line were investigated, and BLM inhibitors reduced PC3 cell growth. We found that treatment with ML216 affected the PC3 cell cycle
To explore the malignancy of PRAD cell lines (RWPE-1, 22RV1, LNCap, PC3), we investigated their proliferation, migration, and invasion abilities. The results showed that PC3 cells had the highest degree of malignancy (Figure 3A-C), so PC3 cells were chosen for the following experiments. We further examined whether pharmacological inhibition of BLM had antitumor effects on PC3 cells in culture, and the results showed that treatment with ML216 greatly inhibited PRAD cell growth in vitro (Figure 3E). To further research whether reduced cell growth was associated with cell proliferation, cell cycle analysis was performed, and a G0/G1 block was observed (Figure 3F). As P53 is a major regulator of cell cycle progression, we detected the expression of P53 in the following experiments and found that the expression of P53 in the PC3 cell line was lower than that in normal prostate cells (RWPE-1) and other prostate cancer cell lines (22RV1 and LNCap) (Figure 3D).
Figure 3.
The migration, invasion and proliferation of PRAD cells line were investigated, BLM inhibitors reduce PC3 cells growth and found that treatment with ML216 had affected on PC3 cell cycle. A-C. The migration, invasion and proliferation ability of RWPE-1, PC3, 22RV-1 and LNcap cells was detected. Columns, mean of three independent experiments; bars, SD (n=3). *, P<0.05. D. The expressions of BLM, EZH2 and P53 in PRAD cells line were detected by Western blot, GAPDH is used as a loading control. E. ML216 treatment (5 days) inhibits PC3 cell proliferation, ***P<0.001, one-way ANOVA, n=3. F. ML216 treatment (5 days) induced PC3 cells have G0/G1 phase arrest. Cells were stained with PI and analyzed by flow cytometry. Date represented one of two independent experiments.
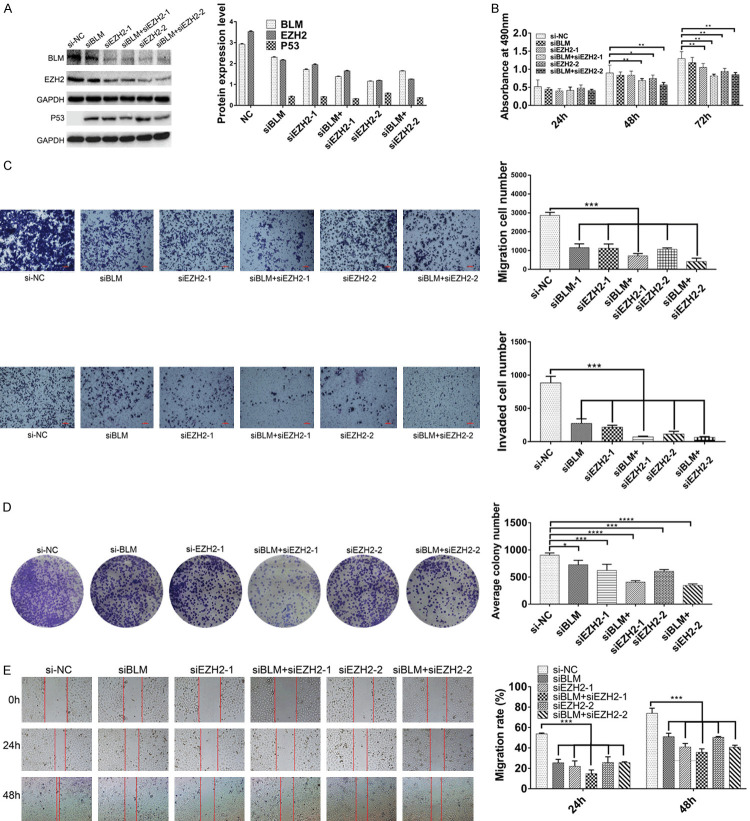
The expression of P53 was upregulated after BLM and EZH2 knockdown, and BLM may affect the P53 signaling pathway by interacting with the EZH2 protein
To further confirm whether the expression of P53 was upregulated after BLM and EZH2 knockdown in PC3 cell lines, a series of siRNAs was synthesized, and the results showed that P53 was upregulated (Figure 4A). Moreover, we investigated the effects of BLM and EZH2 knockdown on cell proliferation, migration, invasion, colony formation and wound healing (Figure 4B-E). The results showed that the degree of malignancy of PC3 cells was reduced. With both BLM and EZH2 knockdown, the degree of malignancy of PC3 cells was more obviously reduced than that of cells with BLM or EZH2 knockdown alone. Therefore, double targeting BLM and EZH2 may provide therapeutic potential for PCa treatment. Because EZH2 is a vital transcription factor, we hypothesized that BLM may affect the P53 signaling pathway by interacting with the EZH2 protein.
Figure 4.
The expression of P53 was upregulated after BLM and EZH2 knockdown, and BLM may affect the P53 signaling pathway by interacting with EZH2 protein. (A) PC3 cells were transfected by siNC, siBLM, siEZH2-1 and siEZH2-2 for 48 h, and then the expressions of BLM, EZH2 and P53 in PC3 cells were detected by Western blot, GAPDH is used as a loading control. (B) PC3 cells were treated as described in (A), and then the proliferation ability was detected after 24 h, 48 h and 72 h. Data represented one of six independent experiments. (C-E) PC3 cells were treated as described in (A), and the migration, invasion, clone formation and scratch-wound were identified. Columns, means of three independent experiments; bars, SD (n=3). *, P<0.05.
EZH2 was identified as an MDM2 promoter-binding protein, and the results were validated by dual-luciferase reporter assay
To explore the underlying molecular mechanism by which BLM and EZH2 affect P53 in PRAD cells, chromatin immunoprecipitation (ChIP) experiments were conducted on the EZH2 protein. First, PC3 cells were lysed, and ChIP grade EZH2 antibody was used for WB to verify the availability of the antibody (Figure 5A). Then, PC3 cells were crosslinked and lysed, chromatin was sheared, antibodies against EZH2 and IgG were used for immunoprecipitation, and DNA fragments of immunocomplexes were detected by DNA gel electrophoresis. After purification, a DNA library was constructed (Figure 5B), and products were sent to a sequencing company for high-throughput sequencing. Using the data produced by ChIP-seq, KEGG pathway analysis predicted several pathway networks. The top 19 scored pathway networks are presented, and the prostate cancer pathway is ranked sixth (Figure 5C). This pathway is related to cell apoptosis, proliferation, genomic stability, and survival. Interestingly, the results showed that MDM2 was included in this pathway, and EZH2 was identified as an MDM2 promoter-binding protein (Figure 5D); the binding site was found at 68,807,941 to 68,808,656 in chromosome 12. Meanwhile, many studies have shown that MDM2 can directly bind to P53 to form the MDM2-P53 complex [43-45]; therefore, the BLM/EZH2/MDM2/P53 axis may exist in PCa cells.
Figure 5.
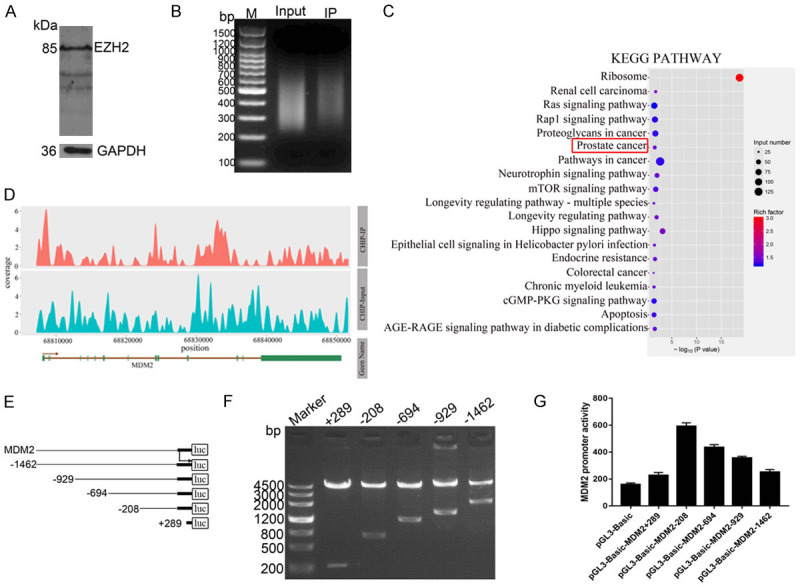
EZH2 was identified as a MDM2 promoter-binding protein, and the results was validated by luciferase reporter assay. A. PC3 cells had been lysed, ChIP grade EZH2 antibody was used for WB to verify the availability of the antibody, GAPDH is used as a positive control. B. DNA library had been constructed after immunoprecipitation. C. KEGG pathway predicted the top 19 scored pathway networks, and prostate cancer pathway is ranked the sixth. D. The map of READS on Peak associated genes, horizontal axis shows the gene location, and the vertical axis shows the depth of reads coverage. E, F. Truncated fragments of MDM2 gene promoter were designed to construct dual-fluorescence reporter plasmids, the reporter plasmids were identified by double enzyme digestion. G. Dual-luciferase reporter assay was performed to measure promoter activity of various reporter plasmids (n=3).
To further validate the transcriptional regulation mechanism between EZH2 and MDM2, a series of truncated promoter fragments of the MDM2 gene were amplified by PCR and cloned into the pGL3-basic vector to construct dual-fluorescence reporter plasmids (Figure 5E). The plasmid double enzyme digestion results of agarose gel electrophoresis showed that each truncated promoter fragment had the correct size in accordance with our designed sequence (Figure 5F). Dual-luciferase reporter assays showed that all reporter plasmids exhibited promoter activity in PC3 cells, while the pGL3-basic-MDM2-208 plasmid showed higher promoter activity (Figure 5G), indicating that the EZH2 protein could bind to the promoter region of the MDM2 gene, and the BLM/EZH2/MDM2/P53 axis may exist in PC3 cells.
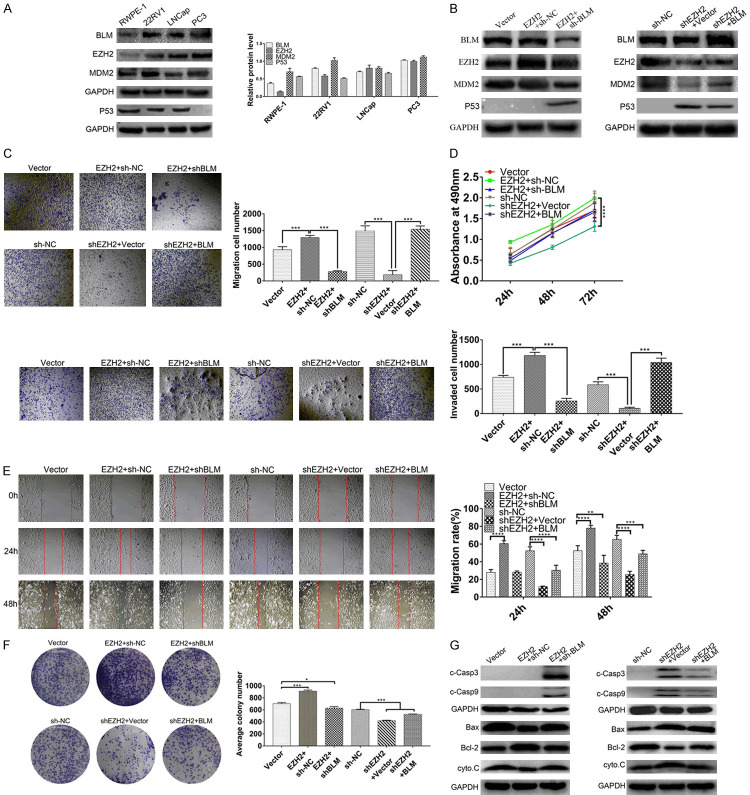
The expression of MDM2 was regulated at the transcription level via BLM interaction with EZH2 in vitro, and the BLM/EZH2/MDM2/P53 axis was identified in PC3 cells
We further evaluated whether the expression of MDM2 was regulated via BLM interaction with EZH2 and the BLM/EZH2/MDM2/P53 axis in PC3 cells. We detected the expression of BLM, EZH2, MDM2 and P53 in prostate cell lines, and we found that PC3 cells had the highest expression of BLM, EZH2 and MDM2 (Figure 6A). PC3 cells were chosen for overexpression and knockdown experiments. The first group was divided into EZH2 overexpression and EZH2 overexpression with BLM knockout. The second group was divided into EZH2 knockout and EZH2 knockout with overexpression of BLM.
Figure 6.
The expression of MDM2 was regulated at the transcription level by BLM interaction with EZH2 in vitro, and the BLM/EZH2/MDM2/P53 axis was identified in PC3 cells. (A) PRAD cell lines were analyzed by Western blot and visualized by anti-BLM, anti-EZH2, anti-MDM2 and anti-P53 antibody. GAPDH is used as a loading control. (B) PC3 cells were knockdown or overexpressed EZH2 and BLM for 48 h, and then the expressions of BLM, EZH2, MDM2 and P53 in PC3 cells were detected by Western blot, GAPDH is used as a loading control. (C-F) PC3 cells were treated as described in (B), and the migration, invasion, proliferation, scratch-wound and clone formation were identified. Columns means of three independent experiments: bars, SD (n=3). *, P<0.05. (G) PC3 cells were treated as described in (B), and the P53 signaling pathway related proteins were detected, GAPDH is used as a loading control.
In the BLM and EZH2 overexpression or knockdown experiments, the results revealed that the expression of MDM2 was increased after EZH2 overexpression, and the increased effects could be reversed by BLM knockdown (Figure 6B, Left). Then, we found that EZH2 overexpression promoted cell migration, invasion, proliferation, wound healing ability and clonogenicity, and these enhanced effects were reversed by BLM knockdown (Figure 6C-F). In the second group, WB revealed that the expression of MDM2 was inhibited after EZH2 knockdown, and the inhibitory effect was rescued by BLM overexpression (Figure 6B, Right). Similarly, we investigated the effects of EZH2 knockdown on cell proliferation, migration, invasion, clone formation and wound healing ability. The results showed that the degree of malignancy of PC3 cells was attenuated after EZH2 knockdown, and the attenuation effect was rescued by BLM overexpression (Figure 6C-F). The findings indicated that the interaction of BLM and EZH2 through MDM2 targeting of the P53 protein affects the molecular mechanism of proliferation and apoptosis of prostate cancer cells in vitro.
The interaction of BLM and EZH2 via MDM2 targeting P53 activates the TP53 signaling pathway
The BLM/EZH2/MDM2/P53 axis, which is associated with cell proliferation and metastasis, was found to be associated with the P53 signaling pathway. Western blotting was carried out, and the results showed that Bax and cyto.C levels were downregulated and Bcl-2 was increased after EZH2 overexpression, whereas EZH2 overexpression with BLM knockdown restored the protein levels of Bax and Bcl-2 (Figure 6G, Left). The levels of c-caspase3, c-caspase9, Bax and cyto.C were upregulated and Bcl-2 was decreased after EZH2 knockdown, but levels were partly restored by BLM overexpression (Figure 6G, Right), suggesting that BLM and EZH2, through MDM2 targeting of P53, activate the TP53 signaling pathway.
The expression and function of P53 were regulated by BLM and EZH2 in vivo
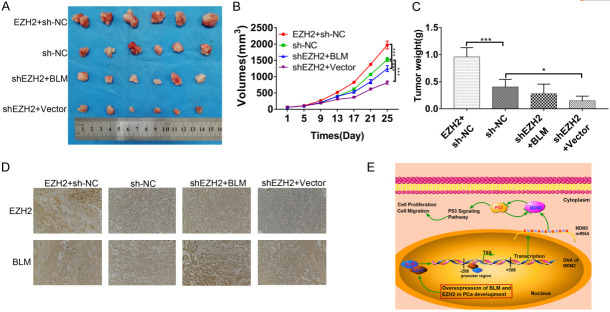
The impact of BLM and EZH2 was investigated in mouse xenograft models. PC3 cells were subcutaneously injected into the left flank of nude mice. Tumor volume was measured and recorded every 4 days. After approximately 4 weeks, the tumor xenografts were harvested, weighed, and processed for IHC staining. Our observations revealed that tumor growth was inhibited after EZH2 knockdown and that inhibition could be reversed by BLM overexpression (Figure 7A-C). In addition, the expression of BLM and EZH2 in tumor tissues presented the same variation trend as tumor growth (Figure 7D). These in vivo results were consistent with our in vitro observations and confirmed that EZH2 could be regulated by BLM in vivo. The results also indicate that BLM and EZH2 play significant roles in suppressing PC3 tumorigenicity in vivo.
Figure 7.
The expression and function of P53 were regulated by BLM and EZH2 in vivo. A. Images of PRAD tumor xenografts from each mouse (n=6 mice/group). ***, P<0.001. B. Tumor volumes, mean ± SD (n=6). ***, P<0.001. C. Tumor weights, mean ± SD (n=6). D. IHC assay was carried out for analysis the expression of BLM and EZH2 in tumor tissues. E. Proposed model for the relationship between EZH2 and BLM in PRAD development.
Discussion
PCa is a common malignant tumor and one of the greatest causes of cancer-related death in men worldwide [1]. To date, the prognosis of advanced PCa patients remains poor. Consistently, the available targeted therapies for PCa patients mainly include antiandrogen and castration [46]. At present, the main clinical indicator for the diagnosis of prostate cancer is the level of SPA in serum, but SPA is only tissue-specific, not a biomarker of prostate cancer [47,48]. Therefore, it is necessary to seek potential biomarkers for PCa that can be applied in the clinic and then research the mechanism between PCa and biomarkers.
BLM is mainly involved in physiological functions such as DNA unwinding, repair and maintenance of telomerase. In addition to the above functions, BLM may also be a potential biomarker of prostate cancer according to the literature reported recently [7,20,21]. However, the molecular mechanisms of BLM as a PCa biomarker are still not entirely clear. In this study, our results showed that BLM and EZH2 exhibited nuclear colocalization, indicating that BLM and EZH2 formed a regulatory complex in the cell nucleus and that BLM and EZH2 directly interacted. Moreover, to further demonstrate whether the expression of BLM and EZH2 in prostate cancer tissues was correlated with the malignancy of cancer cells, we examined the expression of BLM and EZH2 in PCa patients by IHC assay, Similar to most other types of malignant tumors, the expression of BLM and EZH2 in PCa was significantly increased relative to normal tissue; both BLM and EZH2 presented the same variation trends, including T status, clinical stage and Gleason score; importantly, BLM and EZH2 were significantly positively correlated in cancer tissues (P<0.001), indicating that BLM and EZH2 may have synergistic interactions in cancer tissues; and double detection of BLM and EZH2 could provide precise data for predicting the prognosis of PCa patients. ML216 is a specific inhibitor of BLM and is used on PCa cells in culture. We found that the treatment inhibited PCa cell growth in vitro. To further determine whether reduced PCa cell growth was related to cell proliferation, cell cycle analysis was performed. We observed a G0/G1 block, and then we applied DZNep, which is a specific inhibitor of EZH2 that has been reported to cause cell cycle arrest [35]. The results indicated that BLM- and EZH2-specific inhibitors could affect the cell cycle. As p53 is a major protein regulator of cell cycle progression [49], we hypothesized that BLM and EZH2, mainly via the P53 signaling pathway, affected the proliferation and migration of PCa cells.
To validate our hypothesis, siBLM or siEZH2 was transfected into PC3 cells, and we found that the expression of P53 was upregulated and that PC3 cell proliferation, clonogenicity, migration, invasion and wound healing ability were significantly attenuated. However, when BLM and EZH2 were knocked down simultaneously, the degree of cell malignancy was attenuated more obviously, so double targeting of BLM and EZH2 may provide more powerful therapeutic potential for PCa treatment. However, the above results only indicate that interference with BLM and EZH2 can affect the expression of the P53 protein. The precise molecular mechanism of the interaction between BLM and EZH2 affecting P53 expression is still unclear. Since EZH2 is an important transcription factor that can directly regulate a variety of cancer-related genes [50], we speculated that BLM may act on downstream genes through the EZH2 protein.
To further explore the downstream gene of the EZH2 target, ChIP grade EZH2 antibody was used for the ChIP assay, and the IP product was sent to the sequencing company for deep sequencing. Interestingly, according to the sequencing results, P53 was not the molecular target of EZH2 but bound to the MDM2 promoter. Meanwhile, many studies have shown that MDM2 is an oncogene, and its initiation of the synthesis of the MDM2 protein allows it to directly bind to the P53 acidic activation domain and partial transcriptional activation domain to form the P53-MDM2 complex, which can inhibit the transcriptional activation function mediated by P53 [51-54]; therefore, by regulating the expression of the MDM2 protein, EZH2 indirectly controls the expression of the P53 protein, which may regulate the proliferation of PC3 cells through the P53 signaling pathway. To further verify the mechanisms between MDM2 and EZH2, a luciferase assay was performed in PC3 cells. This suggests that EZH2 can bind to the promoter region of the MDM2 gene and then regulate the downstream P53 protein.
Then, we explored the interaction effect of BLM and EZH2 on MDM2 and P53 expression and cell malignancy degree, and we found that the promotion effect of EZH2 overexpression on MDM2 expression and cell malignancy degree was partly reversed by BLM knockdown. Conversely, the suppressive effect of EZH2 knockdown on MDM2 expression and the degree of cell malignancy could be reversed by BLM overexpression, which suggested that BLM might play a positive regulatory role in MDM2 in PCa cells. These findings illustrate that knockdown of EZH2 suppresses the binding of EZH2 to the MDM2 promoter and subsequent promoter activity; therefore, it regulates MDM2 expression at the transcriptional level by interacting with BLM. Currently, many EZH2 inhibitors are ongoing clinical trials aimed at chromosomal transcriptional regulation [55,56], but resistance to EZH2 inhibitors in solid tumors has also emerged [57]. Recently, to identify the mechanism of EZH2 drug resistance, the mechanism of action of the combined use of EZH2 and BRD4 in tumor cells has been reported [58], which provides new prospects for the application of EZH2 and BLM inhibitors in future studies. We also examined the effect of knockdown or overexpression of BLM and EZH2 on the P53 signaling pathway. We found that when EZH2 was overexpressed, proapoptotic-related proteins were downregulated, while this phenomenon was partly reversed by BLM knockdown. Conversely, when EZH2 was knocked down, proapoptotic-related proteins were upregulated, and this phenomenon was partly reversed by BLM overexpression indicating that the P53 signaling pathway is an important pathway by which BLM regulates prostate cancer. The results of the mouse xenograft models were consistent with the in vitro data.
In summary, our research demonstrated that BLM interacting with EZH2 binds the MDM2 promoter region and regulates MDM2 and P53 expression at the transcriptional level. Double targeting of BLM and EZH2 may provide promising prospects for the treatment of prostate cancer.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31860242), Scientific Research Cooperation with America and Oceania Region, Ministry of Education (LJM-2019-877) and the Young Scientific Talents Development Project of Education Department of Guizhou Province, China (QKH-KY-2018-095).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Brandt A, Bermejo JL, Sundquist J, Hemminki K. Age-specific risk of incident prostate cancer and risk of death from prostate cancer defined by the number of affected family members. Eur Urol. 2010;58:275–280. doi: 10.1016/j.eururo.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Labbe DP, Zadra G, Yang M, Reyes JM, Lin CY, Cacciatore S, Ebot EM, Creech AL, Giunchi F, Fiorentino M, Elfandy H, Syamala S, Karoly ED, Alshalalfa M, Erho N, Ross A, Schaeffer EM, Gibb EA, Takhar M, Den RB, Lehrer J, Karnes RJ, Freedland SJ, Davicioni E, Spratt DE, Ellis L, Jaffe JD, D’Amico AV, Kantoff PW, Bradner JE, Mucci LA, Chavarro JE, Loda M, Brown M. High-fat diet fuels prostate cancer progression by rewiring the metabolome and amplifying the MYC program. Nat Commun. 2019;10:4358. doi: 10.1038/s41467-019-12298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genkinger JM, Wu K, Wang M, Albanes D, Black A, van den Brandt PA, Burke KA, Cook MB, Gapstur SM, Giles GG, Giovannucci E, Goodman GG, Goodman PJ, Hakansson N, Key TJ, Mannisto S, Le Marchand L, Liao LM, MacInnis RJ, Neuhouser ML, Platz EA, Sawada N, Schenk JM, Stevens VL, Travis RC, Tsugane S, Visvanathan K, Wilkens LR, Wolk A, Smith-Warner SA. Measures of body fatness and height in early and mid-to-late adulthood and prostate cancer: risk and mortality in the pooling project of prospective studies of diet and cancer. Ann Oncol. 2020;31:103–114. doi: 10.1016/j.annonc.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgart SJ, Nevedomskaya E, Haendler B. Dysregulated transcriptional control in prostate cancer. Int J Mol Sci. 2019;20:2883. doi: 10.3390/ijms20122883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angeles AK, Bauer S, Ratz L, Klauck SM, Sultmann H. Genome-based classification and therapy of prostate cancer. Diagnostics (Basel) 2018;8:62. doi: 10.3390/diagnostics8030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledet EM, Antonarakis ES, Isaacs WB, Lotan TL, Pritchard C, Sartor AO. Germline BLM mutations and metastatic prostate cancer. Prostate. 2020;80:235–237. doi: 10.1002/pros.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gretzer MB, Partin AW. PSA markers in prostate cancer detection. Urol Clin North Am. 2003;30:677–686. doi: 10.1016/s0094-0143(03)00057-0. [DOI] [PubMed] [Google Scholar]
- 9.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 10.McLeod DG, Crawford ED, Blumenstein BA, Eisenberger MA, Dorr FA. Controversies in the treatment of metastatic prostate cancer. Cance. 1992;70:324–328. doi: 10.1002/1097-0142(19920701)70:1+<324::aid-cncr2820701321>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 11.Spiess PE, Pettaway CA, Vakar-Lopez F, Kassouf W, Wang X, Busby JE, Do KA, Davuluri R, Tannir NM. Treatment outcomes of small cell carcinoma of the prostate: a single-center study. Cancer. 2007;110:1729–1737. doi: 10.1002/cncr.22971. [DOI] [PubMed] [Google Scholar]
- 12.Mettlin C. Recent developments in the epidemiology of prostate cancer. Eur J Cancer. 1997;33:340–347. doi: 10.1016/s0959-8049(97)89003-x. [DOI] [PubMed] [Google Scholar]
- 13.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helenius MA, Savinainen KJ, Bova GS, Visakorpi T. Amplification of the urokinase gene and the sensitivity of prostate cancer cells to urokinase inhibitors. BJU Int. 2006;97:404–409. doi: 10.1111/j.1464-410X.2005.05912.x. [DOI] [PubMed] [Google Scholar]
- 15.Shariat SF, Karam JA, Walz J, Roehrborn CG, Montorsi F, Margulis V, Saad F, Slawin KM, Karakiewicz PI. Improved prediction of disease relapse after radical prostatectomy through a panel of preoperative blood-based biomarkers. Clin Cancer Res. 2008;14:3785–3791. doi: 10.1158/1078-0432.CCR-07-4969. [DOI] [PubMed] [Google Scholar]
- 16.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19:1288–1296. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Z, Liu J, Li S, Shen W. Prostate stem cell antigen mRNA expression in preoperatively negative biopsy specimens predicts subsequent cancer after transurethral resection of the prostate for benign prostatic hyperplasia. Prostate. 2009;69:1292–1302. doi: 10.1002/pros.20973. [DOI] [PubMed] [Google Scholar]
- 18.Jedinak A, Loughlin KR, Moses MA. Approaches to the discovery of non-invasive urinary biomarkers of prostate cancer. Oncotarget. 2018;9:32534–32550. doi: 10.18632/oncotarget.25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Lv H, Lv W, Shi M, Zhang M, Luan M, Zhu H, Zhang R, Jiang Y. Genome-wide haplotype association study identifies BLM as a risk gene for prostate cancer in Chinese population. Tumour Biol. 2015;36:2703–2707. doi: 10.1007/s13277-014-2893-x. [DOI] [PubMed] [Google Scholar]
- 21.Qian X, Feng S, Xie D, Feng D, Jiang Y, Zhang X. RecQ helicase BLM regulates prostate cancer cell proliferation and apoptosis. Oncol Lett. 2017;14:4206–4212. doi: 10.3892/ol.2017.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K, Xu H, Zhao J. Bloom syndrome protein activates AKT and PRAS40 in prostate cancer cells. Oxid Med Cell Longev. 2019;2019:3685817. doi: 10.1155/2019/3685817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan Z, Zhao J, Xu H, Xu H, Ji X, Chen X, Xiong J. Characterization of the nuclear import pathway for BLM protein. Arch Biochem Biophys. 2017;634:57–68. doi: 10.1016/j.abb.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Tangeman L, McIlhatton MA, Grierson P, Groden J, Acharya S. Regulation of BLM nucleolar localization. Genes (Basel) 2016;7:69. doi: 10.3390/genes7090069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsekrekou M, Stratigi K, Chatzinikolaou G. The nucleolus: in genome maintenance and repair. Int J Mol Sci. 2017;18:1411. doi: 10.3390/ijms18071411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan Y, Huang D, Chen F, Gao C, Tao T, Shi H, Zhao S, Liao Z, Lo LJ, Wang Y, Chen J, Peng J. Phosphorylation of def regulates nucleolar p53 turnover and cell cycle progression through def recruitment of calpain3. PLoS Biol. 2016;14:e1002555. doi: 10.1371/journal.pbio.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue K, Fry EA. Tumor suppression by the EGR1, DMP1, ARF, p53, and PTEN Network. Cancer Invest. 2018;36:520–536. doi: 10.1080/07357907.2018.1533965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a drosophila polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 30.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 31.Schult D, Holsken A, Siegel S, Buchfelder M, Fahlbusch R, Kreitschmann-Andermahr I, Buslei R. EZH2 is highly expressed in pituitary adenomas and associated with proliferation. Sci Rep. 2015;5:16965. doi: 10.1038/srep16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian R, Ma H, Wu Z, Zhang G, Jiao L, Miao W, Jin Q, Li R, Chen P, Shi H, Yu W. EZH2 promotes cell proliferation by regulating the expression of RUNX3 in laryngeal carcinoma. Mol Cell Biochem. 2018;439:35–43. doi: 10.1007/s11010-017-3133-7. [DOI] [PubMed] [Google Scholar]
- 33.Liu B, Pang B, Wang Q, Yang S, Gao T, Ding Q, Liu H, Yang Y, Fan H, Zhang R, Xin T, Xu G, Pang Q. EZH2 upregulation correlates with tumor invasiveness, proliferation, and angiogenesis in human pituitary adenomas. Hum Pathol. 2017;66:101–107. doi: 10.1016/j.humpath.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 34.Zhu D, Osuka S, Zhang Z, Reichert ZR, Yang L, Kanemura Y, Jiang Y, You S, Zhang H, Devi NS, Bhattacharya D, Takano S, Gillespie GY, Macdonald T, Tan C, Nishikawa R, Nelson WG, Olson JJ, Van Meir EG. BAI1 suppresses medulloblastoma formation by protecting p53 from Mdm2-mediated degradation. Cancer Cell. 2018;33:1004–1016. doi: 10.1016/j.ccell.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Zhu D, Zhang Z, Kaluz S, Yu B, Devi NS, Olson JJ, Van Meir EG. Correction: EZH2 targeting reduces medulloblastoma growth through epigenetic reactivation of the BAI1/p53 tumor suppressor pathway. Oncogene. 2020;39:1165. doi: 10.1038/s41388-019-1067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So S, Nomura Y, Adachi N, Kobayashi Y, Hori T, Kurihara Y, Koyama H. Enhanced gene targeting efficiency by siRNA that silences the expression of the Bloom syndrome gene in human cells. Genes Cells. 2006;11:363–371. doi: 10.1111/j.1365-2443.2006.00944.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Chen L, Han L, Shi Z, Zhang J, Pu P, Kang C. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer Lett. 2015;356:929–936. doi: 10.1016/j.canlet.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azevedo TT, de Faria PR, Neves LA, Do NM. Computational normalization of H&E-stained histological images: progress, challenges and future potential. Artif Intell Med. 2019;95:118–132. doi: 10.1016/j.artmed.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM, Richon VM, Kung AL, Armstrong SA. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong L, Song J, Lin X, Wei F, Zhang C, Wang Z, Zhu J, Wu S, Chen Y, Liang J, Fu X, Lu J, Zhou C, Song L. Serine-arginine protein kinase 1 promotes a cancer stem cell-like phenotype through activation of Wnt/beta-catenin signalling in NSCLC. J Pathol. 2016;240:184–196. doi: 10.1002/path.4767. [DOI] [PubMed] [Google Scholar]
- 42.Faustino-Rocha A, Oliveira PA, Pinho-Oliveira J, Teixeira-Guedes C, Soares-Maia R, Da CR, Colaco B, Pires MJ, Colaco J, Ferreira R, Ginja M. Estimation of rat mammary tumor volume using caliper and ultrasonography measurements. Lab Anim (NY) 2013;42:217–224. doi: 10.1038/laban.254. [DOI] [PubMed] [Google Scholar]
- 43.Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 44.Chen HF, Luo R. Binding induced folding in p53-MDM2 complex. J Am Chem Soc. 2007;129:2930–2937. doi: 10.1021/ja0678774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
- 46.Adamo V, Noto L, Franchina T, Chiofalo G, Picciotto M, Toscano G, Caristi N. Emerging targeted therapies for castration-resistant prostate cancer. Front Endocrinol (Lausanne) 2012;3:73. doi: 10.3389/fendo.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson IM, Ankerst DP. Prostate-specific antigen in the early detection of prostate cancer. CMAJ. 2007;176:1853–1858. doi: 10.1503/cmaj.060955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerstenbluth RE, Seftel AD, Hampel N, Oefelein MG, Resnick MI. The accuracy of the increased prostate specific antigen level (greater than or equal to 20 ng./ml.) in predicting prostate cancer: is biopsy always required? J Urol. 2002;168:1990–1993. doi: 10.1016/S0022-5347(05)64279-6. [DOI] [PubMed] [Google Scholar]
- 49.Meikrantz W, Schlegel R. Apoptosis and the cell cycle. J Cell Biochem. 1995;58:160–174. doi: 10.1002/jcb.240580205. [DOI] [PubMed] [Google Scholar]
- 50.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 52.Juven-Gershon T, Oren M. Mdm2: the ups and downs. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- 53.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 54.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremblay-LeMay R, Rastgoo N, Pourabdollah M, Chang H. EZH2 as a therapeutic target for multiple myeloma and other haematological malignancies. Biomark Res. 2018;6:34. doi: 10.1186/s40364-018-0148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morera L, Lubbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics. 2016;8:57. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibaja V, Shen F, Harari J, Korn J, Ruddy D, Saenz-Vash V, Zhai H, Rejtar T, Paris CG, Yu Z, Lira M, King D, Qi W, Keen N, Hassan AQ, Chan HM. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene. 2016;35:558–566. doi: 10.1038/onc.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang X, Yan J, Zhang M, Wang Y, Chen Y, Fu X, Wei R, Zheng XL, Liu Z, Zhang X, Yang H, Hao B, Shen YY, Su Y, Cong X, Huang M, Tan M, Ding J, Geng M. Targeting epigenetic crosstalk as a therapeutic strategy for EZH2-aberrant solid tumors. Cell. 2018;175:186–199. doi: 10.1016/j.cell.2018.08.058. [DOI] [PubMed] [Google Scholar]