Abstract
Although the classic molecular subtype of breast cancer (BRCA) has been widely used in clinical diagnosis, as a highly heterogeneous malignant tumor, the classic scheme is not enough to accurately predict the prognosis of breast cancer patients. Immune cells in the tumor microenvironment (TME) are thought to play a paramount role in tumor development and driving poor prognosis. In this study, we aimed to develop a TME-associated, immune-related signature to improve prognosis prediction of BRCA. BRCA_OURS enriched transcriptomic RNA sequencing (RNA-seq) of tumor tissue was acquired from 43 breast cancer patients before any treatment. On the immune gene profiles of 43 patients from BRCA_OURS and 932 BRCA patients from The Cancer Genome Atlas (TCGA), we identified a robust immune-related signature including one positive coefficients gene (IL-10) and other 9 genes (C14orf79, C1orf168, C1orf226, CELSR2, FABP7, FGFBP1, KLRB1, PLEKHO1, and RAC2), of which the negative coefficients suggesting higher expression were correlated with better prognosis. Based on the expression of these genes, patients were grouped into the high- and low-risk group with significant overall survival (OS) (P<0.0001). The high-risk group was likely to have inferior outcomes related to several important cancer-associated pathways, including mobilizing more Golgi vesicle-mediated transport and intensive DNA double-strand breaking, which are closely related to the infiltration of immune cells and holds the key for further growing and metastasizing. Collectively, our results highlight that the immunological value within BRCA is an essential determinant of prognostic factor. Our signature may provide an effective risk stratification tool for clinical prognosis assessment of patients with BRCA.
Keywords: Breast cancer, RNA-seq, immune-related signature, tumor microenvironment
Introduction
The incidence of BRCA has been ranked the first female malignant tumors. In China, BRCA is estimated to occupy 15% of all new cancer cases in women, and the principal consideration of cancer death in women under the age of 45 [1]. Likewise, as far as January 1, 2019, the number of new cases of invasive BRCA diagnosed reached about 3.8 million in the United States, with 150,000 women following metastasizing [2]. BRCA is a kind of heterogenous disease with various biological phenotypes, divergent treatment regimens, and prognosis. The clinicopathological characteristics such as age, molecular subtype, and AJCC stage are associated with prognosis and subsequent treatment regimen [3,4]. Although classic molecular subtypes based on the status of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) or other supplementary subtypes, such as MammaPrint [5], Oncotype DX [6], Endopredict [7] and PAM50 [8], have been widely used in clinical diagnosis and become important indicators to guide the choice of treatment options, those studies mostly always tend to focus on the tumor characteristics without considering the impact of immune cells in TME. For individualized and precise treatment, it requires a better understanding of the BRCA-specific immune microenvironment.
Recent studies on the molecular biology of cancer have put forward that beyond cancer cells, the immune cell within TME plays a pivotal role in tumorigenesis and progression biology, meanwhile it also significantly influences therapeutic response and clinical outcome [9-12]. Therefore, it is remarkably crucial to quantify the characteristics of the immune response at the tumor site and improve the understanding of the biological interaction between tumor-host immune microenvironment.
In this study, we aim to investigate the transcriptome of tumor tissue to evaluate the immune status of BRCA patients. According to the supervised clustering of specific immune cells by marker gene expression profiles, samples were divided into two distinct immune infiltration groups. Using the Lasso-Cox algorithm, we identified a robust 10-gene immune-related signature and made further replication of this signature in external cohorts. Our results showed immune-related signature was related to clinical outcomes and provided a valuable stratification tool for clinical prognosis assessment of BRCA patients.
Material and methods
Patients and sample collection of BRCA_OURS
Sample collection relied on the Cancer Hospital of the Chinese Academy of Medical Sciences (Beijing, China). Breast tissue samples (n=43) were collected from patients who had not received any treatment before and underwent surgical resection, of the molecular subtypes across Luminal A, Luminal B, Her2 enrichment, and basal-like cancers. As well, the use of patient specimens in the present study was approved by the Ethics Committee of the Cancer Hospital, Chinese Academy of Medical Sciences. Patients had signed informed consents after fully aware of the research purpose. The raw data of BRCA_OURS has been lodged in the Genome Sequence Archive (GSA) for human (accession no. HRA000272). The relevant patient characteristics were listed in Table 1.
Table 1.
Characteristics of patients in BRCA_OURS
| Characteristics | MM |
|---|---|
| Sex (male/female) | (0/43) |
| Age | 47 (29-67) |
| Molecular Subtype | |
| Luminal A | 14 |
| Luminal B | 18 |
| Her2 enrichment | 4 |
| Basal Like | 7 |
| ER status | |
| ER+ | 32 |
| ER- | 11 |
| Stage | |
| 0 | 1 |
| I | 9 |
| II | 21 |
| III | 11 |
| NA | 1 |
Characteristics of patients in BRCA_OURS (n=43).
Fresh surgical specimens were frozen at -80°C before RNA preliminaries. Tumor tissue was ground into pieces, digested in TRIzol, and total RNA was further extracted. The integrity and concentration of RNA were detected by Agilent 2100 Bioanalyzer and ND-1000 (NanoDrop Technologies). Samples of RNA integrity number (RIN) greater than 7.0 were used for subsequent sequencing.
Transcriptome sequencing and data analysis
Illumina NovaSeq6000 platform was used for transcriptome sequencing, and 150 bp paired-end readings were generated. The quality control of data was achieved by removing low-quality reads, PCR primers, adaptors, duplicates, and other contaminants. HISAT2 v2.0.5 was used to construct the index of the reference genome by using human genome build 19 (hg19) as the reference genome, as well as compare clean reads to it. Feature Counts were used to calculating the readings mapped of each gene. Transcripts per million reads (TPM) were calculated for the estimation of transcript abundance.
BRCA public datasets
The cbioportal for Cancer Genomics (https://www.cbioportal.org/datasets) was where we downloaded RNA-sequencing data directly, including TCGA and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets. The BRCA samples in TCGA_BRCA were used as a training series, while the METABRIC samples were analyzed as an external validation series. Clinical information of corresponding patients was also downloaded availably from the public cbioportal database. The samples without survival information were eliminated and a total of 2715 BRCA patients were used to analyze in the present study, with 932 patients from TCGA as training series, 1559 from METABRIC, and 224 patients from GEO, including 107 patients from GSE58812 and 117 patients from GSE88770, as the validation series. The gene expression data of the two GEO datasets were generated by the Affymetrix Human Genome U133 Plus 2.0 Array platform.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed to find the specific pathways associated with the relative risk group based on the immune-related signature on the JAVA version (http://software.broadinstitute.org/gsea/downloads.jsp).
Pathway enrichment was detected using GSEA with default parameters of 1000 random sample permutations. Those pathways with FDR<0.25 & p-value <0.01 were considered to be significantly associated [13].
Single sample gene set enrichment analysis
Single sample gene set enrichment analysis (ssGSEA) was an extension of GSEA and carried out using an R package named ‘GSVA’ [14]. Utilizing ssGSEA, we got the gene set enrichment score and evaluated immune cell infiltration of each sample according to the immune cell marker signatures [15] of 24 diverse resident immune cells. This approach quantifies the status of immune infiltration by the relative expression of specific marker genes, without isolation, preservation, and specific reagent of labeling immune cells. In this way, we obtained the immune infiltration of each sample based on the immune-specific gene expression profiling. All samples were divided into two groups according to the level of immune infiltration by the agglomeration dependent on Euclidean distance and method “ward.D2”.
Statistical analysis
All statistical analyses were implemented in the R version 3.6.2. Two groups of variables were performed by non-parameter test Wilcoxon test to assess statistical significance, as Kruskal-Wallis test was used in three groups and above. Generally, the results were considered statistically significant if p-value <0.05. The R package named ‘limma’ was used to find differential expression genes (DEGs) between two immune groups and significant DEGs were identified by the t-student test with P<0.05 and log2[FC]>1 as the cut-off. Lasso penalized Cox regression analysis was implemented by the R package of ‘glmnet’, combined least absolute shrinkage and selection operator (LASSO) and cox proportion hazard regression [16,17]. In the process of establishing the λ value of the Lasso-Cox model, lambda.1se is adopted, which contributed to the heaviest punishment and the most concise model. That exploited to reduce the dimensionality and select the most significant overall survival (OS)-associated immune-related genes to construct a prognostic signature. According to the cut-off value of the median 10-gene signature score, patients were divided into low- and high-risk groups. we performed Kaplan-Meier estimator, log-rank tests, and multivariable Cox regression using R packages of ‘survival’ and ‘survminer’. Kaplan-Meier (KM) analysis was used to assess survival outcomes. The median was used as the cut-off value to visualize the KM curves, and statistical significance was evaluated by the log-rank test. Univariate and multivariate analyses were performed by the Cox proportional hazard regression model. The Concordance index (C-index) is widely used in evaluating the prediction ability of the Cox regression model, especially the tumor prognosis model. A C-index lower than 0.70 was considered to be low accuracy, between 0.70 and 0.90 to be superior, and higher than 0.90 to be idealized accuracy. Nomogram construction and validation transformed the complex regression into a readable visual, which made the prediction model more readable and convenient for the evaluation of patients. And then bootstrapped calibration curves were plotted to the nomogram-based prediction model. We performed receiver operating characteristic (ROC) curve and time-dependent ROC analysis to visualize the survival prediction availability based on the immune-related signature risk score, AJCC stage, and subtype, while the area under the ROC curve (AUC) was used as the evaluation standard.
Results
Immune groups reflect global immune infiltration of breast cancer
To systematically assess the composition of the tumor immune microenvironment, the marker gene sets of 24 distinct leukocyte subsets were used as input, and the immune infiltration within the tumor was quantified by ssGSEA. In this way, we divided all samples of BRCA_OURS into two groups according to the immune infiltration of TME. The division represented global immune infiltration within tumor tissue (Figure 1A), including the high-infiltration and low-infiltration groups with significant differences in immune cells. The proportion of various immune cells in each sample based on immune infiltration score was visualized in Figure S1A, with T cells and related cells accounted heavily. Interestingly, we found patients in the high-immune infiltration group were with a significantly higher level of most lymphocytes and myeloid cells compared to the low-immune group (Figure 1B), especially in relative T cells, such as CD8 T cells, cytotoxic cells, and Treg (Figure S2). However, there was no obvious law of consistency between the immune infiltration and pathological stage (Figure S1B). Moreover, we observed a certain correlation between classic molecular subtypes of breast cancer with immune infiltration through an R package named ’estimate’, and the immune score of basal-like was higher than other subtypes but not statistically significant (Figure 1C). Immune signature gene sets from MSigDB were used to figure out the key immunologic signature. As Figure 1D showed, the high-immune infiltration group was endowed with a higher ration in naïve B cells/plasma cells, resting/activated CD4 T cells, and immature/mature neuron cell lines, suggesting the immune system tended to be immature and its’ anti-tumor ability decreased. Similarly, immune grouping was also performed in the BRCA_TCGA cohort (Figure S3A) and the immune cell proportion was acquired by immune cells profile in GSVA (Figure S3B). Consistent with BRCA_OURS cohort, almost all the immune cells in the high-immune group were significantly higher than the low (Figure S4). Most immune cells in the high-infiltration group of BRCA_TCGA were activated either immune promotive or immunosuppressive. These results suggested that there seemly was immune-hot but highly immunosuppressive infiltration of the high-immune group, and yet the low-immune group showed a relatively modest immune microenvironment.
Figure 1.
Immune infiltration in BRCA_OURS cohort. The Gene expression was profiled in 43 patients of BRCA_OURS (A). Boxplots of the cell compositions within TME in different immune infiltration group (B). Statistical analysis of Immune score with breast cancer molecular subtypes (C). Twenty enrichments of immunologic signature and their hazard ratios in terms of activation and inhibition of the immune function (D).
Identification of prognostic immune-related gene from the training series
DEGs between the high- and low-immune infiltration groups were identified. 3444 genes in BRCA_OURS and 3414 genes TCGA_BRCA were identified as DEGs at optimum cutoff values of P<0.05 (Figure 2A, 2B). Conducting the intersection of two datasets of DEGs, there were 932 common immune-related genes after overlapping the identified candidate genes (Figure 2C). After subjecting the common candidates of the TCGA dataset to Lasso-Cox regression analysis, a set of 10 genes was identified (Figure 3A, 3B). This prognostic immune-related signature was composed of Interleukin-10 (IL-10), one positive coefficient gene, and other 9 genes (C14orf79, C1orf168, C1orf226, CELSR2, FABP7, FGFBP1, KLRB1, PLEKHO1, and RAC2), of which the negative coefficients suggesting that their higher levels were correlated with longer survival. The specific risk-score formula was adopted based on the corresponding parameters of these 10 genes for OS prediction, as below: Risk score = (-0.114731735 * Expression of C14orf79) + (-0.019429183 * Expression of C1orf168) + (-0.049258060 * Expression of C1orf226) + (-0.055863001 * Expression of CELSR2) + (-0.028295228 * Expression of FABP7) + (-0.008174118 * Expression of FGFBP1) + (0.020753075 * Expression of IL-10) + (-0.121245004 * Expression of KLRB1) + (-0.049187024 * Expression of PLEKHO1) + (-0.003657534 * Expression of RAC2). Afterward, we calculated, ranked, and visualized the risk score of our immune-related signature for individual samples in TCGA_BRCA cohorts (Figure 3C-E).
Figure 2.
Differential expression gene high- and low-immune group. Volcano plot of BRCA_OURS (A) and BRCA_TCGA (B) test series. The black dots delimit up- and down. Red plots represent significant up genes using fold-change values and corrected p-values and blue dots represent significant down genes. A venn diagram was used to intersect the common genes between the two test series (C).
Figure 3.
The identity of 10 immune genes and risk score analysis of TCGA_BRCA test series. The tuning parameter (λ) selection in the LASSO model used 10-fold cross-validation via minimum criteria. The black solid vertical line represents the partial likelihood deviance± standard error (SE), The two vertical dotted lines are drawn at the minimum criteria and 1-SE criteria (A). LASSO coefficient profiles of the 932common immune-related genes (B). (C-E) Risk factor correlation diagram. Signature risk score distribution (C). Patients’ overall survival status and time (D). Heatmap of the immune signature expression (E). Rows represent genes, and columns represent patients. The black vertical dotted lines represent the median risk score cutoff dividing patients into low-risk and high-risk groups.
The patients’ survival of the immune-related signature in the training series
According to the risk score of 10-gene immune-related signature, samples with higher risk scores than the median were assigned to the high-risk group (n=466), as less than the median considered to be the low-risk group (n=466). Compared to the low-risk group, patients in the high-risk group had significantly shorter median OS, disease free survival (DFS), and progression free survival (PFS), respectively (log-rank test P<0.001, P=0.013, P=0.00016, respectively) (Figure 4A-C). We applied the univariable and multivariable analyses to determine whether the risk score derived from the immune-related signature was an independent prognostic factor in BRCA. Our result showed that the association of the risk score of the 10-gene immune-related signature was significant with OS and was an independent prognostic factor for BRCA patients (Figure 5A).
Figure 4.
Kaplan-Meier estimates of the overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS) of TCGA_BRCA patients using the immune signature. The Kaplan-Meier plots were used to visualize the survival probabilities for the low-risk versus a high-risk group of patients based on the cut-off of the median risk score. Kaplan-Meier curves of OS for TCGA_BRCA test series patients (A). Kaplan-Meier curves of DFS for TCGA_BRCA test series patients (B). Kaplan-Meier curves of PFS for TCGA_BRCA test series patients (C). Kaplan Meier curves of OS for METABRIC_BRCA validation series patients (D). The trick marks on the Kaplan-Meier curves represent the censored subjects. The differences between the two curves were determined by the log-rank test.
Figure 5.
Comparison of the score with prognostic clinical covariates. Multivariable Cox proportional hazards regression, analyses incorporating the risk score and known clinical characteristics, including age at diagnosis, AJCC stage, and intrinsic subtype. Multivariable analysis was performed using Cox proportional hazards regression analysis in patients of TCGA_BRCA training series (A). Multivariable analysis was performed using Cox proportional hazards regression analysis in patients of METABRIC_BRCA validation series (B).
Validation of the immune-related signature in other independent datasets
We further validated our 10-gene immune-related signature in another independent BRCA dataset from METABRIC. Similar to the training cohort, we classified patients into high-risk (n=793) and low-risk (n=766) using the median score serving as the cutoff point. Consistent with the training cohorts, patients in the two groups predicted different prognosis (log-rank test P<0.0001) (Figure 4D), with high-risk group accompanied shorter overall survival. Besides, in the multivariable Cox regression model, as a continuous variable, the risk score from the immune-related signature was significantly correlated with OS as well (Figure 5B). C-index was used to evaluate the prediction ability of the Cox regression model in GEO datasets. The C-index of GSE58812 and GSE88770 were 0.74 and 0.70 respectively, indicating a preferable accuracy in the Cox model.
Construction of nomogram based on 10-gene immune-related signature
In compliance with clinical practice, we constructed a nomogram of 3- and 5-year survival probability in TCGA_BRCA that integrated different clinical characteristics, such as age, tumor AJCC stage, and risk score from immune-related signature (Figure 6A). Meanwhile, the corresponding line-segment of the calibration plot was much closer to 45° line, suggesting better prediction and credibility of this nomogram (Figure 6B). Additionally, we compared the sensitivity and specificity of the immune signature risk score, AJCC stage, and subtype in predicting prognosis by ROC analysis and used AUC as an indicator to assess the predictive reliability among the three prognostic factors. The AUCs of time-dependent ROC curves for 1-, 3- and 5-year overall survival predictions for the immune signature were 0.757, 0.803, and 0.751, respectively (Figure 6C), indicating that the risk score had a strong predictive ability. As shown in Figure 6D, 6E, the combination of signature risk score with the AJCC stage has better efficiency than the risk score or AJCC stage alone in 3 years (0.826 versus 0.803, 0.713) or 5 years (0.784 versus 0.751, 0.686). Furthermore, the 10-gene immune-related signature risk score was obviously better than that of subtype (0.803 versus 0.544 and 0.751 versus 0.554 respectively in 3- and 5-years). In short, it was the risk score that has a strong predictive ability than the AJCC stage or subtype alone. However, the risk score combined with the AJCC stage might be the more powerful predictor in the ROC analysis.
Figure 6.
The immune signature is an independent prognostic factor in BRCA. (A, B) Nomogram construction, and validation of TCGA_BRCA series. The nomogram for predicting the probability of breast cancer patients with 3- and 5-year OS (A). The calibration curve analysis of nomogram predicting survival at 3-years (B). Nomogram-predicted OS is distributed on the x-axis; observed survival probability is distributed on the y-axis. (C-E) Receiver operating characteristic (ROC) analysis was used to analyze the sensitivity and specificity of predicting overall survival. P-values were from the comparison of the area under the ROC (AUROC) of the immune signature. ROC curve of TCGA_BRCA cohort among 1-, 3- and 5-year (C). 3-year correlation ROC curve in the TCGA_BRCA cohort for comparing the 10-gene immune-related signature risk score and clinical characteristics (D). 5-year correlation ROC curve in the TCGA_BRCA cohort for the comparison of the classification of the 10-gene immune-related signature risk score and clinical characteristics (E).
Identification of the related biological process and signaling pathways
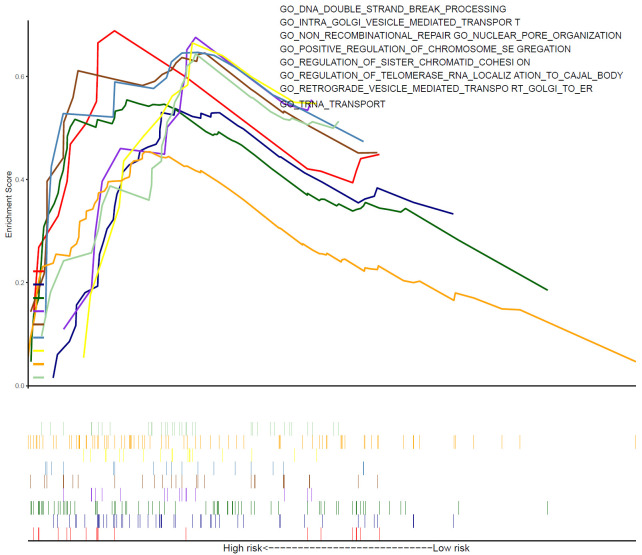
Based on the risk score of 10-gene immune-related signature, we conducted the GSEA analysis to identify the relevant biological process and signaling pathways and found a series of specific functions in the high-risk group (Figure 7). Gene ontology enrichment suggesting, the Golgi apparatus was specifically activated in the high-risk group, involving the transport and retrograde of Golgi to the endoplasmic reticulum. Perhaps, abundant Golgi apparatus activations meant providing more energy for tumor cell proliferation and metastasis. Meanwhile, in hepatocellular carcinoma (HCC), it was reported that Golgi-related protein regulates various signal transduction of EGFR/RTK to promote tumor cell growth and metastasis [18], suggesting that the activity of Golgi-related protein could promote tumor metastasis through receptor-ligand binding. In addition, there were more DNA double-strand breaking (DSB) and chromosome segregation in the high-risk group. Mis-repair of DSB, a critical DNA lesion, could lead to severe mutation, such as deletion or translocation of the chromosome [19]. As one of ten hallmarks of cancer, chromosomal instability is a genetic confusion caused by persistent errors in chromosome segregation during mitosis. An interesting study pointed out that chromosome instability can lead to DNA leakage from the nucleus of cancer cells, accompanied by a chronic inflammatory response, and eventually tumor cells spread to distant organs [20]. In addition, according to the biological processes annotated by hallmark gene sets annotation, the related pathways of the high-risk group were also enriched (Figure S5). Kinds of complements were activated, interferon-mediated inflammatory response was enhanced, and STAT-related signal pathways were activated in the high-risk group.
Figure 7.
Gene set enrichment analysis depicted biological process correlated with the high-risk group.
In order to further explore the relationship between these possible molecular mechanisms with immune infiltration in BRCA, we estimated the correlation between risk score and immune cell infiltration. The top-3 infiltrating components, respectively, with signatures for CD8 T cells, T cells, and cytotoxic cells (Figure 8). The most relevant component of immune cells, CD8 T cells, the amount of that was considered as a powerful determinant of clinical outcome and treatment response in breast cancer patients [21,22], was highly correlated with immune-related signature (r=-0.57, P<0.01), suggesting high-risk group with less CD8 T cells infiltrating and worse outcomes. The densities of cytotoxic cells in situ were significantly related to 5-year DFS and OS in patients with colorectal cancer, lower densities accompanied with worse prognosis [23], which coincided with our results that cytotoxic cells negatively correlated with risk score (r=-0.50, P<0.01). In the high-risk group, unique immune infiltration might be associated with poor prognosis of BRCA. These data suggest that the increased risk of a poor prognosis of patients in the high-risk group were likely due to their unique immune infiltration and several important cancer-associated pathways, which jointly drove tumor towards more malignant characteristics.
Figure 8.
Correlation between risk score and immune cell infiltration in TME. P-value greater than 0.05 is marked with an ‘X’.
Discussion
Current progress in RNA sequencing has expanded the understanding of the molecular pathogenesis of various tumors [24,25]. Transcriptome analysis patterns reveal the functional state of the cell and cellular behavior associated with genomic and environmental changes [26,27]. Increasing evidence shows that TME acts a critical role in breast cancer pathogenesis [28]. Given that, our study focused on exploring TME using transcriptome, and further identified an immune-related signature for predicting prognosis. The signature assessment would contribute to individualized treatment for BRCA patients.
First, taking advantage of ssGSEA, we inferred the expression and composition of leukocytes in the tumor from the expression profiles of 24 kinds of immune cells and classified them into high- and low-infiltration group. There was prominent significance in lymphocytes and myelocytes infiltration between the high- and low-infiltration group, especially in T cells. Secondly, we identified the common DEGs between BRCA_OURs and TCGA_BRCA and modeled an immune-related signature, including 10 prognostic genes by the Lasso-Cox algorithm. Additionally, we further validated our 10-gene immune-related signature in another independent BRCA dataset from METABRIC. Finally, we constructed a nomogram for clinical practice and ROC analysis was used to explore the sensitivity and specificity of our signature for survival prediction. Indeed, our study proclaimed several favorable prognostic markers. For example, as a surface marker, KLRB1 (CD161) specifically expressed in some T cell subsets, including CD8+, CD4+, and TCR γδ Τ cells, reflecting the innate immunity of organisms [29]. Future studies will help to define novel immune-associated potential implications for these signature genes.
Among the 10-gene immune-related signature, to some extent, IL-10/KLRBL/RAC2/PLEKHO1 were directly associated with tumor immunity, reflecting the characteristics of various immune cells, such as Dendritic cells (DCs) and Treg cells. For example, as an anti-inflammatory cytokine, IL-10 was broadly expressed in many immune cells and plays a vital role in the prevention and autoimmune disease [30]. On one hand, IL-10 exerted an anti-inflammatory effect by inhibiting different types of specialized antigen-presenting cells (APCs). On the other hand, IL-10 stimulation enhanced the proliferation and toxicity of CD8+ T cells [31]. The expression of IL-10 was related to several cancers, such as glioma [32], gastric carcinoma [33], and non-small cell lung cancer [34]. A related study revealed that IL-10 was expressed only in breast tumor tissue, not adjacent normal tissue, and had a significant correlation with locally advanced disease [35]. In our study, IL-10 was highly expressed in BRCA high-risk group and was found to be associated with shorter survival, which is agreed with the previous study [36]. Thus, we assumed that IL-10 might act positively in breast cancer progression and negatively regulate tumor immune microenvironment by various immune cells, such as DC and Treg. The expression of Killer Cell Lectin Like Receptor B1 (KLRB1) always is associated with superior outcome and indicates the state of some subsets of T cells and NK cells to some extent [37]. For example, KLRB was expressed in a variety of circulating lymphocytes various circulating lymphocytes, indicating its ability to produce IL-17 [38]. Recently, a study revealed the pro-inflammatory functions of KLRB+ NK cells [39]. KLRB1 was a prognostic indicator of human Esophageal Squamous Carcinoma (ESCC), with the growth of KLRB1-knockdown human ESCC cells inhibited [40]. In our study, KLRB1 was highly expressed in the low-risk group and a significant predictor of favorable survival in breast cancer. Another candidate, Rac2 Family Small GTPase (RAC2) is one of the RAC family members, restricting to be expressed in the hematopoietic system and its mutation affects the lymphocytes in different ways, including activating mutations to result in a combined immunodeficiency (CID) like phonotype or negative mutations limited to the granulocyte subsets [41]. RAC2 was also required for macrophage activation [42,43] and CCl4 treated Rac2 knockout mice confirmed that Rac2 deficiency could reduce pro-inflammatory cytokine and chemokines and inhibit fibrosis-related signals [44]. Except for immunological function, RAC2 was reported to be upregulated in NSCLC and promote quiescent cell to re-entry cell cycle [45]. In breast cancer (BC), the expression of Rac2 was significantly correlated with the survival, referring to the lower Rac2, the worse survival the patients would be [46], which was consent with our study that showed RAC2 was highly expressed in the low-risk group and regarded as a favorable survival prognostic genes. Thus, we suggested that RAC2 may play a vital role in immune cells within microenvironment communication. Pleckstrin Homology Domain Containing Family O membership 1 (PLEKHO1), also called CKIP-1, encodes a protein that specifically interacts with the CK2α subunit. A study revealed that CKIP-1 in regulating Mesenchymal stem cells (MSCs)-mediated immunomodulation, showing enhanced immunosuppressive capacity with CKIP-1 knockdown [47]. A previous study revealed that overexpression of PLEKHO1 in osteoblasts accelerated the development of inflammation in inflammatory arthritis mice [48]. PLEKHO1 promoted the growth of RCC cells maybe through Hippo and MAPK/JNK pathways [49]. In array CGH analysis, deletion of PLEKHO may be associated with the worse conditions of the BRCA1 patients [50]. Except for the aforementioned genes, others may reflect characteristics of tumor in the different microenvironment, involving proliferation and metabolism of tumor. Cordero A et al. suggested Fatty Acid Binding Proteins (FABP7) was essential for the growth of HER2+ breast cells and a potential target for the treatment of HER2+ breast cancer brain metastasis [51] and another study found that FABP7 played the oncogene role in colon cancer via MEK/ERK transduction pathway [52]. Further investigations demonstrated that FABP7 had a significantly longer breast cancer-specific survival in comparison with FABP7- basal-like breast cancer [53]. In our study, the low expression of FABP7 in the high-risk group indicates a poor prognosis of BRCA. Therefore, we agreed that FABP7 might be a critical factor that the immune microenvironment indirectly regulating tumor growth and maybe a latent target for the treatment of BRCA. The expression of Cadherin EGF LAG Seven-Pass G-Type Receptor 2 (CELSR2) protein in HCC tissue was associated with poor prognosis [54]. However, another study found that the expression of CELSR2 was not consistent in breast cancer, that is, there were strong membrane staining in benign breast epithelial cells and most lines, while a few showed a significant decrease or deletion of CELSR2 reactivity [55]. This encourages us to study the specific function of CELSR2 in BRCA in the future.
Among the 10-gene immune-related signature, some were widely studied and yet the following genes were either rarely reported or never have been investigated. For instance, C14orf79 was identified as a novel susceptibility locus for hippocampal volume near MTSU1 [56]. FYN Binding Protein2 (FYB2/C1orf79) was only found over-represented in cattle breeds [57]. C1orf226 has never been reported in any study. The functions of these genes in BRCA and other diseases are still unknown, and our findings lay a foundation for further evaluation of these genes in breast cancer and beyond.
The occurrence and development of solid tumors depend not only on the tumor cell but also closely related to the tumor microenvironment. In recent years, tumor immunity has been of great interest to researchers, especially the tumor immune microenvironment, which is mainly composed of various infiltrating immune cells and other stromal cells and cytokines secreted from them. The cells within TME are characterized by a high plasticity, rapid differentiation, and constantly changing. The tumor-infiltrating lymphocytes (TILs) are the main immune cells in the center of the tumor and is related to the adaptive immune response across cancer progression. Among these, CD8+ TILs can induce tumor cell death while Treg cells tend to inactivate CD8+ TILs. However, there was no significant correlation with prognosis both in CD8+ and Foxp3+ TILs, which maybe keep related to the subtype of breast cancer [58]. The accumulation of CD49a+ NK cells was related to poor prognosis as thought to be an inhibitory receptor [59]. The immune microenvironment is a key factor that contributes to improve or reduce survival. Surprisingly, several studies suggested that high densities of CD8+ T cells are associated with short PFS and OS [60-62]. Likewise, macrophages are the main component of immune cells within TME, and M2 phenotype is the majority, contributing to the growth and development of the invasive tumor. The density of macrophages was closely related to the prognosis of patients in many tumors, such as breast cancer, ovarian and gastric cancer [63]. Taken together, the characteristics of various immune cells within the immune microenvironment can predict the clinical outcome to different degrees. Thus, by revealing the TME, screening key immune signature gene is conducive to obtaining potential prognostic related genes.
Our study presented an immune-related signature reveals through our experimental data and currently available common tumor database across the transcriptome. This novel immune-related signature may be conducive to a more personalized prognosis prediction of BRCA patients and could be used as potential biomarkers and therapeutic targets. Our analysis suggested that this signature may involve tumor energy metabolism and chromosome replication to support the metastasis of the tumor. Further validation will be required on functional experiments and clinical trials. We believe that these genes will contribute to the clinical prediction of BRCA.
Inevitably, our study had several limitations worth noting. First, our data BRCA_OURS involved in this study were rich in sequencing data but without the prognosis information of patients. Despite this weakness, the significant and consistent correlation of the immune-related signature with survival in TCGA and METABRIC had proved the power of our prognostic signature for BRCA. Second, specific to each gene, we do not have any experimental data, nor do we study the underlying mechanism. That entails functional experiments on these valuable genes to provide further information in BRCA.
Data availability
The BRCA_OURS raw data for RNAseq has been lodged in the Genome Sequence Archive (GSA) for human, as accession number with HRA000272.
Acknowledgements
This study was supported by the PUMC Fund of the Funds for the Central Universities (NO. 3332018072), the CAMS Innovation Fund for Medical Sciences (NO. 2016-I2M-3-005), and the National Key R&D Program of China, (NO. 2018YFC1705104).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 3.Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, Díez M, Viladot M, Arance A, Muñoz M. Clinical implications of the intrinsic molecular subtypes of breast cancer. The Breast. 2015;24:S26–S35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 6.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 7.Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, Dietze O, Greil R, Jelen A, Sevelda P, Freibauer C, Müller V, Jänicke F, Schmidt M, Kölbl H, Rody A, Kaufmann M, Schroth W, Brauch H, Schwab M, Fritz P, Weber KE, Feder IS, Hennig G, Kronenwett R, Gehrmann M, Gnant M. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 8.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang R, Mao M, Lu Y, Yu Q, Liao L. A novel immune-related genes prognosis biomarker for melanoma: associated with tumor microenvironment. Aging. 2020;12:6966–6980. doi: 10.18632/aging.103054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Nirmal AJ, Regan T, Shih BB, Hume DA, Sims AH, Freeman TC. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res. 2018;6:1388. doi: 10.1158/2326-6066.CIR-18-0342. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf Anna C, Angell H, Fredriksen T, Lafontaine L, Berger A, Bruneval P, Fridman Wolf H, Becker C, Pagès F, Speicher Michael R, Trajanoski Z, Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Tibshirani RJ. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Stat Methodol. 1996;58:267–288. [Google Scholar]
- 17.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Ye QH, Zhu WW, Zhang JB, Qin Y, Lu M, Lin GL, Guo L, Zhang B, Lin ZH, Roessler S, Forgues M, Jia HL, Lu L, Zhang XF, Lian BF, Xie L, Dong QZ, Tang ZY, Wang XW, Qin LX. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30:444–458. doi: 10.1016/j.ccell.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat Res. 2017;803-805:51–55. doi: 10.1016/j.mrfmmm.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, Duran M, Pauli C, Shaw C, Chadalavada K, Rajasekhar VK, Genovese G, Venkatesan S, Birkbak NJ, McGranahan N, Lundquist M, LaPlant Q, Healey JH, Elemento O, Chung CH, Lee NY, Imielenski M, Nanjangud G, Pe’er D, Cleveland DW, Powell SN, Lammerding J, Swanton C, Cantley LC. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, Loi S. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 22.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, Earl HM, Poole CJ, Hiller L, Dunn JA, Bowden SJ, Twelves C, Bartlett JM, Mahmoud SM, Rakha E, Ellis IO, Liu S, Gao D, Nielsen TO, Pharoah PD, Caldas C. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25:1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 23.Pagès F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P, Zatloukal K, Trajanoski Z, Berger A, Fridman WH, Galon J. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 2009;27:5944–5951. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Dean DC, Hornicek FJ, Shi H, Duan Z. RNA sequencing (RNA-Seq) and its application in ovarian cancer. Gynecol Oncol. 2019;152:194–201. doi: 10.1016/j.ygyno.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Shukla S, Evans JR, Malik R, Feng FY, Dhanasekaran SM, Cao X, Chen G, Beer DG, Jiang H, Chinnaiyan AM. Development of a RNA-seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst. 2016;109:djw200. doi: 10.1093/jnci/djw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panichnantakul P, Bourgey M, Montpetit A, Bourque G, Riazalhosseini Y. RNA-Seq as a tool to study the tumor microenvironment. Methods Mol Biol. 2016;1458:311–37. doi: 10.1007/978-1-4939-3801-8_22. [DOI] [PubMed] [Google Scholar]
- 27.Henrik Heiland D, Ravi VM, Behringer SP, Frenking JH, Wurm J, Joseph K, Garrelfs NWC, Strähle J, Heynckes S, Grauvogel J, Franco P, Mader I, Schneider M, Potthoff AL, Delev D, Hofmann UG, Fung C, Beck J, Sankowski R, Prinz M, Schnell O. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun. 2019;10:2541. doi: 10.1038/s41467-019-10493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tekpli X, Lien T, Røssevold AH, Nebdal D, Borgen E, Ohnstad HO, Kyte JA, Vallon-Christersson J, Fongaard M, Due EU, Svartdal LG, Sveli MAT, Garred Ø OSBREAC. Frigessi A, Sahlberg KK, Sørlie T, Russnes HG, Naume B, Kristensen VN. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat Commun. 2019;10:5499. doi: 10.1038/s41467-019-13329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, Marchi E, Björkander S, Kang YH, Swadling L, Kurioka A, Sahgal N, Lockstone H, Baban D, Freeman GJ, Sverremark-Ekström E, Davis MM, Davenport MP, Venturi V, Ussher JE, Willberg CB, Klenerman P. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 2014;9:1075–1088. doi: 10.1016/j.celrep.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 31.Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev. 2006;212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Chen Q, Daniel V, Maher DW, Hersey P. Production of IL-10 by melanoma cells: examination of its role in immunosuppression mediated by melanoma. Int J Cancer. 1994;56:755–760. doi: 10.1002/ijc.2910560524. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Liu JW, Sun LP, Gong YH, Xu Q, Jing JJ, Yuan Y. Association of IL10 gene promoter polymorphisms with risks of gastric cancer and atrophic gastritis. J Int Med Res. 2018;46:5155–5166. doi: 10.1177/0300060518792785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vahl JM, Friedrich J, Mittler S, Trump S, Heim L, Kachler K, Balabko L, Fuhrich N, Geppert CI, Trufa DI, Sopel N, Rieker R, Sirbu H, Finotto S. Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. Br J Cancer. 2017;117:1644–1655. doi: 10.1038/bjc.2017.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee HK, Bansal VK, Nepal B, Srivastava S, Dinda AK, Misra MC. Is interleukin 10 (IL10) expression in breast cancer a marker of poor prognosis? Indian J Surg Oncol. 2016;7:320–325. doi: 10.1007/s13193-016-0512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerger A, Renner W, Langsenlehner T, Hofmann G, Knechtel G, Szkandera J, Samonigg H, Krippl P, Langsenlehner U. Association of interleukin-10 gene variation with breast cancer prognosis. Breast Cancer Res Treat. 2010;119:701–705. doi: 10.1007/s10549-009-0417-y. [DOI] [PubMed] [Google Scholar]
- 37.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, Querci V, Fambrini M, Liotta F, Levings MK, Maggi E, Cosmi L, Romagnani S, Annunziato F. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur J Immunol. 2010;40:2174–2181. doi: 10.1002/eji.200940257. [DOI] [PubMed] [Google Scholar]
- 39.Kurioka A, Cosgrove C, Simoni Y, van Wilgenburg B, Geremia A, Björkander S, Sverremark-Ekström E, Thurnheer C, Günthard HF, Khanna N, Walker LJ, Arancibia-Cárcamo CV, Newell EW, Willberg CB, Klenerman P. CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. Front Immunol. 2018;9:486. doi: 10.3389/fimmu.2018.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, Liu Y, Dong F, Liu X. Transcription/expression of KLRB1 gene as a prognostic indicator in human esophageal squamous cell carcinoma. Comb Chem High Throughput Screen. 2020;23:667–674. doi: 10.2174/1386207323666200517114154. [DOI] [PubMed] [Google Scholar]
- 41.Lougaris V, Baronio M, Gazzurelli L, Benvenuto A, Plebani A. RAC2 and primary human immune deficiencies. J Leukoc Biol. 2020;108:687–696. doi: 10.1002/JLB.5MR0520-194RR. [DOI] [PubMed] [Google Scholar]
- 42.Ceneri N, Zhao L, Young BD, Healy A, Coskun S, Vasavada H, Yarovinsky TO, Ike K, Pardi R, Qin L, Qin L, Tellides G, Hirschi K, Meadows J, Soufer R, Chun HJ, Sadeghi MM, Bender JR, Morrison AR. Rac2 modulates atherosclerotic calcification by regulating macrophage interleukin-1β production. Arterioscler Thromb Vasc Biol. 2017;37:328–340. doi: 10.1161/ATVBAHA.116.308507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi S, Singh AR, Wong SS, Zulcic M, Jiang M, Pardo A, Selman M, Hagood JS, Durden DL. Rac2 is required for alternative macrophage activation and bleomycin induced pulmonary fibrosis; a macrophage autonomous phenotype. PLoS One. 2017;12:e0182851. doi: 10.1371/journal.pone.0182851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou Y, Xiong JB, Ma K, Wang AZ, Qian KJ. Rac2 deficiency attenuates CCl(4)-induced liver injury through suppressing inflammation and oxidative stress. Biomed Pharmacother. 2017;94:140–149. doi: 10.1016/j.biopha.2017.07.074. [DOI] [PubMed] [Google Scholar]
- 45.Pei H, Guo Z, Wang Z, Dai Y, Zheng L, Zhu L, Zhang J, Hu W, Nie J, Mao W, Jia X, Li B, Hei TK, Zhou G. RAC2 promotes abnormal proliferation of quiescent cells by enhanced JUNB expression via the MAL-SRF pathway. Cell Cycle. 2018;17:1115–1123. doi: 10.1080/15384101.2018.1480217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim GE, Kim NI, Lee JS, Park MH, Kang K. Differentially expressed genes in matched normal, cancer, and lymph node metastases predict clinical outcomes in patients with breast cancer. Appl Immunohistochem Mol Morphol. 2020;28:111–122. doi: 10.1097/PAI.0000000000000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Chen JF, Yang YM, Huang XH, Dong XH, Yang HX, Cao JK, Jiang XX. CKIP-1 regulates the immunomodulatory function of mesenchymal stem cells. Mol Biol Rep. 2019;46:3991–3999. doi: 10.1007/s11033-019-04844-1. [DOI] [PubMed] [Google Scholar]
- 48.He X, Liu J, Liang C, Badshah SA, Zheng K, Dang L, Guo B, Li D, Lu C, Guo Q, Fan D, Bian Y, Feng H, Xiao L, Pan X, Xiao C, Zhang B, Zhang G, Lu A. Osteoblastic PLEKHO1 contributes to joint inflammation in rheumatoid arthritis. EBioMedicine. 2019;41:538–555. doi: 10.1016/j.ebiom.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Z, Li Q, Zhang G, Lv C, Dong Q, Fu C, Kong C, Zeng Y. PLEKHO1 knockdown inhibits RCC cell viability in vitro and in vivo, potentially by the Hippo and MAPK/JNK pathways. Int J Oncol. 2019;55:81–92. doi: 10.3892/ijo.2019.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez C, Aravena A, Tapia T, Rozenblum E, Solís L, Corvalán A, Camus M, Alvarez M, Munroe D, Maass A, Carvallo P. Different array CGH profiles within hereditary breast cancer tumors associated to BRCA1 expression and overall survival. BMC Cancer. 2016;16:219. doi: 10.1186/s12885-016-2261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordero A, Kanojia D, Miska J, Panek WK, Xiao A, Han Y, Bonamici N, Zhou W, Xiao T, Wu M, Ahmed AU, Lesniak MS. FABP7 is a key metabolic regulator in HER2+ breast cancer brain metastasis. Oncogene. 2019;38:6445–6460. doi: 10.1038/s41388-019-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma R, Wang L, Yuan F, Wang S, Liu Y, Fan T, Wang F. FABP7 promotes cell proliferation and survival in colon cancer through MEK/ERK signaling pathway. Biomed Pharmacother. 2018;108:119–129. doi: 10.1016/j.biopha.2018.08.038. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Rakha EA, Ball GR, Spiteri I, Aleskandarany M, Paish EC, Powe DG, Macmillan RD, Caldas C, Ellis IO, Green AR. The proteins FABP7 and OATP2 are associated with the basal phenotype and patient outcome in human breast cancer. Breast Cancer Res Treat. 2010;121:41–51. doi: 10.1007/s10549-009-0450-x. [DOI] [PubMed] [Google Scholar]
- 54.Xu M, Zhu S, Xu R, Lin N. Identification of CELSR2 as a novel prognostic biomarker for hepatocellular carcinoma. BMC Cancer. 2020;20:313. doi: 10.1186/s12885-020-06813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H, Groth J, Sossey-Alaoui K, Hawthorn L, Beall S, Geradts J. Aberrant expression of novel and previously described cell membrane markers in human breast cancer cell lines and tumors. Clin Cancer Res. 2005;11:4357–4364. doi: 10.1158/1078-0432.CCR-04-2107. [DOI] [PubMed] [Google Scholar]
- 56.Chung J, Wang X, Maruyama T, Ma Y, Zhang X, Mez J, Sherva R, Takeyama H Alzheimer’s Disease Neuroimaging Initiative. Lunetta KL, Farrer LA, Jun GR. Genome-wide association study of Alzheimer’s disease endophenotypes at prediagnosis stages. Alzheimers Dement. 2018;14:623–633. doi: 10.1016/j.jalz.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang F, Qu K, Chen N, Hanif Q, Jia Y, Huang Y, Dang R, Zhang J, Lan X, Chen H, Huang B, Lei C. Genome-wide SNPs and InDels characteristics of three Chinese cattle breeds. Animals (Basel) 2019;9:596. doi: 10.3390/ani9090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee KH, Kim EY, Yun JS, Park YL, Do SI, Chae SW, Park CH. The prognostic and predictive value of tumor-infiltrating lymphocytes and hematologic parameters in patients with breast cancer. BMC Cancer. 2018;18:938. doi: 10.1186/s12885-018-4832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun H, Liu L, Huang Q, Liu H, Huang M, Wang J, Wen H, Lin R, Qu K, Li K, Wei H, Xiao W, Sun R, Tian Z, Sun C. Accumulation of tumor-infiltrating CD49a(+) NK cells correlates with poor prognosis for human hepatocellular carcinoma. Cancer Immunol Res. 2019;7:1535–1546. doi: 10.1158/2326-6066.CIR-18-0757. [DOI] [PubMed] [Google Scholar]
- 60.Giraldo NA, Becht E, Vano Y, Petitprez F, Lacroix L, Validire P, Sanchez-Salas R, Ingels A, Oudard S, Moatti A, Buttard B, Bourass S, Germain C, Cathelineau X, Fridman WH, Sautès-Fridman C. Tumor-infiltrating and peripheral blood T-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin Cancer Res. 2017;23:4416–4428. doi: 10.1158/1078-0432.CCR-16-2848. [DOI] [PubMed] [Google Scholar]
- 61.Petitprez F, Fossati N, Vano Y, Freschi M, Becht E, Lucianò R, Calderaro J, Guédet T, Lacroix L, Rancoita PMV, Montorsi F, Fridman WH, Sautès-Fridman C, Briganti A, Doglioni C, Bellone M. PD-L1 expression and CD8(+) T-cell infiltrate are associated with clinical progression in patients with node-positive prostate cancer. Eur Urol Focus. 2019;5:192–196. doi: 10.1016/j.euf.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Scott DW, Chan FC, Hong F, Rogic S, Tan KL, Meissner B, Ben-Neriah S, Boyle M, Kridel R, Telenius A, Woolcock BW, Farinha P, Fisher RI, Rimsza LM, Bartlett NL, Cheson BD, Shepherd LE, Advani RH, Connors JM, Kahl BS, Gordon LI, Horning SJ, Steidl C, Gascoyne RD. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical hodgkin lymphoma. J. Clin. Oncol. 2013;31:692–700. doi: 10.1200/JCO.2012.43.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The BRCA_OURS raw data for RNAseq has been lodged in the Genome Sequence Archive (GSA) for human, as accession number with HRA000272.