Abstract
B-cell acute lymphoblastic leukemia (B-ALL) is a common type of hematologic malignancy characterized by the uncontrolled growth of immature B lymphocytes. Genomics, transcriptomics, and proteomics at different levels contribute to early diagnosis and can thereby provide better treatment for cancer. MicroRNAs (miRNAs) are conducive to the diagnosis and treatment of patients with B-ALL. Moreover, evidence suggests that runaway miRNAs and exosomes containing miRNA may be involved in the occurrence of B-ALL, which can then be used as potential biomarkers. This review summarizes the role of miRNAs in the pathogenesis, diagnosis, prognosis, and treatment of B-ALL.
Keywords: MicroRNA, exosome, biomarker, diagnosis, treatment, B-cell acute lymphoblastic leukemia
Introduction
B-ALL is an aggressive malignant tumor of the blood system, with B-lymphocyte precursor cells reproducing uncontrollably in the bone marrow. Although advances in chemotherapy have significantly improved disease outcomes in children, chemotherapy has a poor outcome in adults [1]. The recurrence of B-ALL is the leading cause of death, and the disease-related mortality rate is approximately 60%. B-ALL is a common subtype of ALL, and identification of new biomarkers for the diagnosis and classification of B-ALL may lead to improved patient prognosis and treatment [2].
miRNAs can attach to the 3’-untranslated region (UTR) of mRNA targets and inhibit their translation, thus, functioning in proliferation, differentiation, and apoptosis [3]. miRNA has been used as tumor suppressor or oncogene in leukemia [4]. Nanocarriers, a type of exosome, can carry proteins, miRNAs, and mRNAs, which can then be released to multiple cells in the body, such as immune cells, nerve cells, and tumor cells, thus participating in RNA transport and tumor metastasis [5]. Exosomes containing miRNAs (exo-miRNAs) result in the survival and progression of B-ALL cells because of their abundance and convenient access of these molecules in the body. exo-miRNAs can be used for both the diagnosis and treatment of B-ALL patients.
In this review, we describe the comprehensive relationship between miRNA and B-ALL: explaining the function of miRNAs in B-ALL as well as the role of exosomes in delivery.
miRNA and the pathogenesis of B-ALL
miRNA regulates gene expression
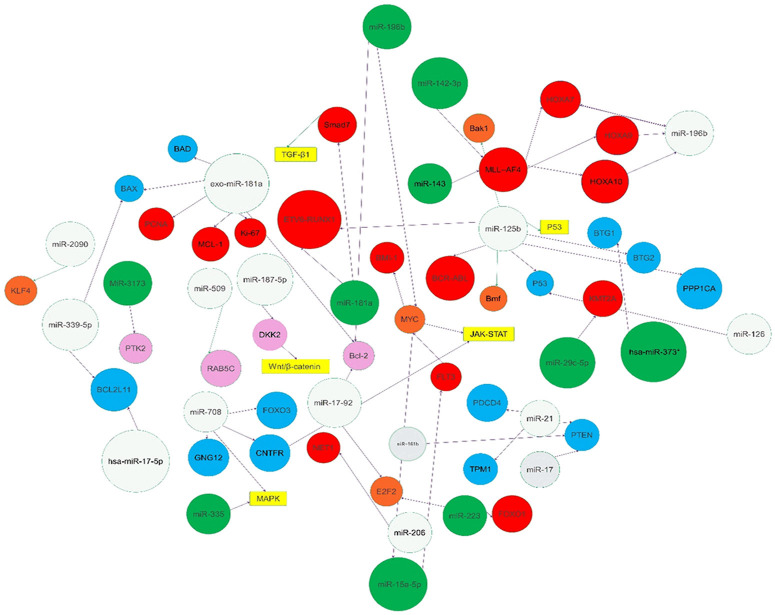
The treatment for B-ALL has improved over the past 50 years, especially in children. However, recurrence remains a threat to these patients. miRNAs act as tumor suppressors and carcinogen. Here, we summarize the latest studies that attempt to link aberrantly expressed miRNAs with their targets and describe the complex regulatory networks in B-ALL (Table 1; Figure 1).
Table 1.
Selected miRNAs involved in B-ALL pathogenesis
| MicroRNA | Expression in B-ALL | Target | Mechanism of Dysregulation | Function | Material | Sample | Ref |
|---|---|---|---|---|---|---|---|
| miR-21 | Upregulation | PDCD4, PTEN and TPM1 | Promoted cell growth, invasion, angiogenesis, and metastasis | Poorer DFS and OS. | PB or BM | 75 pediatric B-ALL and 50 controls | [6] |
| miR-339-5p | Upregulation | BCL2L11↓ | Avoided FGFR1 inactivation | Promoted cell cycle progression and an inhibition of apoptosis. | Cell line | - | [9] |
| miR-509 | Upregulation | RAB5C↓ | - | Promoted cell growth | Nalm-6 cells | - | [11] |
| miR-3173 | Downregulation | PTK2↑ | - | Promoted proliferation, migration and invasion. | PB | 135 pediatric B-ALL and 97 controls | [12] |
| miR-206 | - | NET1↑ | - | Enhanced proliferation and chemoresistance. | PB and BM | 20 pediatric B-ALL and 20 controls | [13] |
| miR-181a | Upregulation | TLR4, TLR8, IRF8 and IL6R | - | An inhibition of innate immunity and inflammation. | PB and BM smears | 9 T-ALL patients | [15] |
| miR-181a | Downregulation | Smad7↑ | TGF-β1↓ | Increase in proliferation and decrease in apoptosis; Increased diagnostic sensitivity of ALL to 100%. | PB | 23 pre-B-ALL, 5 B-ALL, 2 T-ALL | [18] |
| miR-181a | Downregulation | ETV6/RUNX1↑ | - | - | PB | PALL carrying t(12;21) translocation | [19] |
| exo-miR-181a | Upregulation | PCNA, Ki-67, MCL-1 and bcl2↑, BAD, BAX↓ | - | Upregulated proliferation, cell survival and an inhibition of apoptosis. | Cell line and serum | - | [56] |
| miR-125b, miR-17 and miR-181b | Upregulation | PPP1CA, BTG2, and PTEN↓ | Methylation of the gene promoter region | Increase in proliferation and decrease in apoptosis; Decitabine downregulates these miRNAs. | PB | 5 PALL and 7 healthy controls | [20] |
| miR-125b | Upregulation | P53, Bak1 and Bmf transcript↓ | Enhanced carcinogenicity of BCR-ABL fusion protein | Increase in proliferation and an inhibition of apoptosis. | Cell line | B-ALL with t(11;14)(q24;q32) translocation | [23] |
| miR-17~92 | Upregulation | BCL2↑ | - | An inhibition of apoptosis; An inhibition of BCL2 (e.g., ABT-737) in BCR-ABL-positive ALL cells promoted apoptosis. | Blasts and CD34 cells | 13 BCR-ABL-positive B-ALL and controls | [25] |
| sol-miR-23 | Downregulation | BCL2↑ | May promote BCR-ABL1-dependent leukemogenesis | May affect differentiation and resist to cytotoxic drugs. | BM and PB | Pre-B-ALL patients | [26] |
| miR-142-3p | Downregulation | MLL-AF4↑ | Down-regulated HOXA7, HOXA9 and HOXA10 | Promoting cell proliferation. | Cell line | B-ALL with MLL-AF4 fusion | [27] |
| miR-222, miR-339, miR-142-3p | Upregulation | miR-142-3p targets BCLAF1, LIFR, BCL2L1↓; miR-222 targets c-kit↑; miR-339 targets BCL-6 | Affected hematopoietic development | Activated cell differentiation and growth; MiR-142 was low in T-ALL and the erythroid lineages. | BM | 40 children with pre-B-ALL | [28] |
| hsa-miR-451 and hsa-miR-373* | Downregulation | AKTIP and calcium binding protein; BCL11A↓; BTG1↓ | - | Affected cell differentiation; Promoted proliferation. | BM | 40 children with pre-B-ALL | [28] |
| miR-143 | Downregulation | ERK5 and MLL-AF4↑ | Formed a methylation/miR-143/MLL-AF4 network | Promoted cell proliferation and induced apoptosis; 5-azacytidine can be used in B-ALL. | Cell line | B-ALL with MLL-AF4 fusion | [29] |
| sol-miR-6, let-7 miRNAs, miR-125b, miR-126* and miR-383 | Upregulation | - | - | - | - | TEL-AML1-positive ALL cases | [26] |
| miRNA-100/99a | Downregulation | FKBP51↑; the anti-apoptotic gene MCL1↑ | Activated IFG1R/mTOR signaling pathways; May downregulate the GR signaling pathway | Increase in proliferation and decrease in apoptosis; Mediated response to Dex in ALL cells. | BM | ALL patients with MLL-rearrangement and BCR-ABL fusion genes | [51] |
| miR-126/126*, miR-151 and miR-545 | Upregulation | Different miRNA expression patterns | - | TEL-AML1 and hyperdiploid ALL | [31] | ||
| miR-383, miR-125b, miR-100, miR-99a, let-7c | Upregulation | Higher levels of miR-125b causing vincristine-resistant express | ALL patients with TEL-AML1 fusion genes | [31] | |||
| miR-222/222*, miR-223, miR-511, miR-660 | Upregulation | Hyperdiploid ALL | [31] | ||||
| miR-10a, miR-134 and miR-214 | Downregulation | SOX2↑ | DNA hypermethylation of miR-10a | Decreased apoptosis, increased proliferation; the level of miR-10a can be reversed by DNA hypermethylation in ALL patients with MLL-rearrangement. | - | P-ALL | [31] |
| miR-196b, miR-151, miR-148a | Downregulation | c-Myc↑; Bcl-2, hTERT and AATF↑ | - | Increased proliferation and decreased apoptosis. | Cell line | - | [37] |
| miR-196b | Downregulation | - | - | - | PB or BM | Non-MLL pre-B-ALL | [32] |
| miR-196b | Upregulation | HOXA↑ | DNA hypomethylation of HOXA | Increase in proliferation. | Cells from BM or PB | MLL-rearranged pre-B ALL | [33] |
| miR-708 | Upregulation | - | - | Expressed highly in TEL-AML1, E2A-PBX1, BCR-ABL, hyperdiploid and B-other cases. | PB or BM | ALL | [32] |
| 291 differentially expressed miRNAs | miR-1246 (the most upregulated); miR-106b-5p (the most downregulated) | 19 hub TFs such as MYC, STAT5A, STAT5B, FOXO1 and HLF | Formation of feedback loops or feed-forward loops regulates genes | Regulation of gene expression and hematopoiesis; Association with 25 significantly enriched pathways. | BM | 15 patients and 10 controls | [34] |
| miR-15a-5p | Downregulation | FLT3 | Formed MYC/miR-15a-5p/FLT3 feed-forward loop | Increased proliferation. | BM | 15 patients and 10 controls | [34] |
| miR-187-5p | Upregulation | DKK2 | Activated Wnt/β-catenin signaling | Promoted cellular proliferation and inhibited apoptosis. | BM | 20 children with B-ALL and Nalm-6 cells | [38] |
| hsa-miR-124a | Downregulation | CDK6↑; phosphorylated retinoblastoma protein↑ | Cooperation with cyclinD; Promoter methylation | Increased proliferation; Predicted DFS and OS. | Cell lines | Cell lines and 353 patients | [39] |
| miR-128b | Upregulation | - | Methylation of CpG islands in miR-128b | Accurate diagnosis of ALL and AML. | BM | 18 ALL cases | [40] |
| miR-126 | Upregulation | Cdkn2aip↓ | Reduced P53 activity in HSPC; Prevented pre-BCR signaling | Evasion of senescence, cell-cycle arrest and apoptosis, and maintaining blasts. | Cell line | 17 adult patients and an experimental murine model | [44] |
| miR-148a-3p, miR-27a-3p, miR-375 | Upregulation | - | Regulating the crosstalk between transcription factors and histones | - | BM | B-ALL in CD19-CAR-T therapy | [47] |
| miR-124-3p | Upregulation | IFI44L↓ | - | Affecting inflammatory response. | Analysis of database | PALL patients | [52] |
| hsa-miR-103a-3p, hsa-miR486-3p | Downregulation | HOXA7↑, S100A10↑ | Regulating cell growth, motility, cell cycle progression and differentiation | Poor outcomes and chemoresistance. | Analysis of database | PALL patients | [52] |
miRNA, microRNA; B-ALL, B-cell acute lymphoblastic leukemia; TF, Transcription factors; HSPC, Hematopoietic stem and progenitor cells; PB, peripheral blood; BM, bone marrow; PALL, Pediatric ALL. T-ALL, T-cell acute lymphoblastic leukemia; AML, acute myeloid leukemia; DFS, disease-free survival; OS, overall survival;
refers to the star strand of the miRNA duplex that is (partly) complementary to the mature miRNA.
Figure 1.
Description of microRNAs implicated in the pathogenesis of B-cell acute lymphoblastic leukemia (B-ALL). The miRNAs studied in B-ALL can act as carcinogens or tumor suppressors. The targets, including genes, transcription factors, proteins, and signaling pathways, linked to the miRNAs are noted in the geometric figures. Increased and decreased expression of microRNAs are noted with a gray and green background, respectively. Increased and decreased expression of genes are noted with red and blue backgrounds, respectively. Transcription factors and proteins are annotated with orangey-red and pink background, respectively. Purple arrows and green arrows indicate promotion and inhibition roles, respectively.
miRNA as a carcinogen in B-ALL
As carcinogens, miRNAs can inhibit tumor suppressor genes. Many hematological tumors generally overexpress the same miRNAs, such as miR-21 and miR-155. Overexpression of miR-21 results in the acceleration of precursor B (pre-B) cells developing into malignant lymphoid phenotypes. The study indicates that miR-21 is upregulated in children with B-ALL, targeting tumor suppressor genes (PDCD4, PTEN, and TPM1) and promoting invasion, angiogenesis, and metastasis [6,7]. Medina et al. found that the overexpression of miR-155 promotes B cell proliferation, and de Yébenes et al. showed that miR-155 can serve as a key miRNA for therapeutic targets in children with hepatitis virus leukemia [7,8]. miR-339-5p is also important for B-cell precursor acute lymphoblastic leukemia (BCP-ALL) where it promotes cell cycle progression and reduces apoptosis [9]. Additionally, the upregulation of miR-708 can suppress CNTFR, NNAT, and GNG12, causing loss of control of hematopoietic differentiation in BCP-ALL [10].
miRNA as a tumor suppressor in B-ALL
miRNAs can act as tumor suppressors and negatively regulate proto-oncogenes. Overexpression of miR-509 in Nalm-6 cells increased the number of apoptotic cells. They did this by targeting RAB5C, which is important for cell growth [11]. miR-3173 inhibits PTK2 expression at the protein level and is downregulated in B-ALL. miR-3173 mimics inhibit the proliferation, migration, and invasion of B-ALL cells, while miR-3173 inhibitors do the opposite [12]. miR-206 can inhibit these effects by targeting neuroepithelial cell-transformation 1 (NET1), thus reducing the proliferation of B-ALL cells [13].
Special miRNAs in B-ALL
The same type of miRNA can have different roles in various parts of the body. For example, miR-181 regulates the differentiation of B cells, T cells, and NK cells during normal hematopoiesis [14]. However, miR-181a is one of the most controversial miRNAs in ALL with some researchers regarding miR-181a as an oncogenic factor [15], while other studies have shown miR-181a as a tumor suppressing factor. However, these may depend on the different cell backgrounds and post-target expressions [16,17]. miRNA-181a expression is significantly increased in patients with acute myeloid leukemia and T-cell acute lymphoblastic leukemia (T-ALL) through the downregulation of EGR, which produces carcinogenic effects [16]. By targeting SMAD7 and regulating TGF-β1 signaling pathways, miR-181a can suppress ALL in children. In BCP-ALL, miR-181a can effectively target the fusion protein ETV6/RUNX1, reducing its protein level, and inducing significant anti-leukemia effects [18,19]. Hence, the role of miR-181a has not been fully determined. Moreover, miR-181b tends to be a carcinogenic factor in B-ALL, which can be downregulated by decitabine through the reduced expression levels of miR-125b and miR-17 [10,20]. miR-125b can determine hematopoietic differentiation and selectively induce myeloid or lymphocytic leukemia according to expression levels and duration. Furthermore, it may regulate certain factors, such as interferon regulatory factor 4 (IRF4). In IRF4 homozygous mutant mice, miR-125b-induced B cell leukemia is significantly accelerated [21,22]. Another study showed that high levels of miR-125b shortened the incubation period of BCR-ABL-induced leukemia. Ectopic expression of miR-125b in hematopoietic embryonic hepatocytes could also induce the development of a B-ALL, a myeloproliferative neoplasm, or T-ALL mouse model [23].
miRNAs and different subtypes of B-ALL
B-ALL can be subdivided into haploid fusions, including ETV6-RUNX1 fusion, BCR-ABL1 fusion, TCF-PBX1 fusion, and MLL fusion. The first two subtypes have a better disease prognosis. The appearance of BCR-ABL fusion proteins in 3% of children with B-ALL indicates a possible poor prognosis [24]. Some miRNAs in B-ALL are dependent on BCR-ABL expression. After deleting miR-17~92 in normal lymphocyte production, accompanied by higher levels of Bim (BCL2L11), apoptosis is induced, thereby preventing the conversion of pro-B cells. miR-17~92 targeted the overexpression of BCL2 to induce apoptosis, which provides application potential for using ABT-737, a BCL2 inhibitor to cure B-ALL [25]. The decrease in sol-miR-23 levels is related to the increase in BCL2 [26]. The study found that miR-142-3p was downregulated and hence promoted B-ALL cell proliferation with MLL-AF4 fusion by targeting the HOXA7, HOXA9, and HOXA10 genes [27,28]. Various other miRNAs (miR-143, miR-223, miR-222/222*, miR-98, hsa-miR-101, miR-511, Let-7 miRNA, miR-100, miR-99a, miR-196b, and miR-708) were differentially expressed in the different types of ALL [26,29-33]. These results indicate that miRNA expression is specific to different ALL subtypes.
miRNA and the network of B-ALL gene expression
At present, the regulatory network of B-ALL gene expression is unclear. miRNAs play an important role in various physiological and pathological processes by targeting and regulating gene expression and regulating signaling pathway processes. The JAK-STAT signaling pathway, PI3K-AKT signaling pathway, cell cycle pathway, and RAS signaling pathway are associated with B-ALL pathogenesis [34]. Transcription factors (TFs) are important in biological processes. As one of the most common TFs. MYC is important in cell differentiation, proliferation, survival, and metabolism [35], and it can activate miR-17~92 clusters, thereby controlling BIM, PTEN, and p21 (CDKN1A). The rearrangement of MYC or ABL proto-oncogenes relieves the expression restriction of these key regulators, and members of the BCL-2 family transfer downstream oncogenes. Double transgenes of MYC and BCL-2 lead to excessive proliferation of immature cells and thereby promote B-ALL [36]. miR-196b functions by targeting the transcription factor MYC, thus inducing apoptosis in B-ALL [37], however, this is different for T-ALL expression. Using TF array technology and Illumina deep-sequencing, a study of microRNAs and TFs has shown that there is a MYC/miR-15a-5p/FLT3 feed-forward loop in B-ALL, which may regulate the JAK-STAT signaling pathway. This has been regarded as a vital motif in the miRNA-TF network of B-ALL. Other hub miRNAs and hub TFs were also predicted to be related to B-ALL. Among them, STAT5A, STAT5B, and MYC were associated with the JAK-STAT signaling pathway [34]. Moreover, Lou et al. first proved that uncontrolled WNT signaling can lead to B-ALL, which is regulated by the miR-187-5p-DKK2 pathway [38].
miRNAs and epigenetic regulation
The formation of acute leukemia is closely related to epigenetic regulation. Promoter methylation regulates hsa-miR-124a and can cooperate with cyclinD by targeting CDK6, thus, participating in the regulation of the cell cycle and helping to develop glucocorticoid resistance in ALL [39]. Methylation of CpG islands in miR-128b promoters leads to upregulation of miR-128, thereby resulting in the pathogenesis of high-risk pre-B-ALL [40]. In addition, miRNAs can regulate epigenetic mechanisms by affecting DNA methylation and histone changes. Researchers have found that BTG2 and PTEN are targets of miR-181b and miR-125b, this was confirmed when their expression decreased after demethylation therapy. These results suggest that demethylation drugs can be used as a first-line therapy for patients with B-ALL [20].
Effects of miRNA on hematopoietic function
Different tissues express different miRNA patterns at different stages. Various miRNAs, including miR-15a, miR-146, miR-150, miR-10a, miR-221, and miR-222 are important for normal hematopoiesis [41]. miR-155 and miR-150 affect B and T lymphocyte functions [42]. As mentioned before, miR-125b, miR-17~92, and miR-181 are important in the regulation of hematopoietic differentiation, however, their abnormal expression can lead to hematological tumors. Moreover, premature expression of miR-150 prevented the migration of pro-B cells to pre-B cells [43]. miRNA-126 regulates hematopoietic stem cell differentiation. Nucera et al. designed a mouse model with hematopoietic engineering to express miRNA-126 at all stages of differentiation. miRNA-126 expression in human progenitors and mice reduces p53 transcriptional activity by regulating p53-related targets, thus, 30% of mice developed monoclonal B-cell leukemia. The development of this type of leukemia is controlled by inhibiting miRNA-126 expression [44]. Leukemia cells can interact with the bone marrow microenvironment (BMM). BMM can reduce miR-221 expression in leukemia cells, leading to an increase in CDKN1B protein and thereby promoting cell cycle progression, increasing ALL survival, and inducing chemoresistance. By targeting the miR-221/222-p27 pathway, the residue of quiescent leukemia cells is reduced [45].
Immunomodulatory effects of miRNA
miRNAs can promote the release of mediators that activate cancer and anticancer immune activities. miRNA analysis was conducted in 19 adult and 79 pediatric patients with ALL. Immune-associated miRNAs in children and adults were significantly similar. However, researchers have observed that the level of miR-18a is low in children with ALL This is appropriate for future study because miR-18a may play a role in the better medical response in pediatric patients with ALL [46]. Chimeric antigen receptor (CAR) T-cell therapy is an important immunotherapy [47]. Zhang et al. revealed that many miRNAs, such as members of the let-7 family, are involved in the immune process during CD19-specific CAR-T therapy. miRNAs including miR-148a-3p, miR-375, and miR-27a-3p regulate the interaction between TFs and histones. The combination of histone genes with lncRNA and miRNA-TF-gene regulatory networks is necessary for these patients [48]. These findings help us to understand the mechanisms of CAR-T immunotherapy, and the effects of miRNAs on B-ALL, in cancer. Additionally, genes targeting PD-1 can also activate anticancer immunity and initiate miRNAs (including miR-424 and miR-34a) that regulate apoptosis and block the expression of PD-1 [49]. Moreover, miR-142-5p was found to suppress PD-L1 by directly binding to PD-L1 mRNA [50].
Application of comprehensive bioinformatics analysis
The identification of miRNA targets is difficult because it is challenging to locate complementary miRNA sites. Recently, many studies have used interdisciplinary methods, such as comprehensive bioinformatics analysis and open databases, to identify the correlation between potential miRNAs and disease. These studies are beneficial because they do not have the high costs and long cell experiments, thus they guide the future research direction and subsequent wet-lab experiments. One study linked miRNAs to mRNA target genes; relying on the use of publicly available data, Ramani et al. found that miRNA-100/99a inhibits B-ALL cell proliferation by targeting FKBP51 and regulating the IFG1R/mTOR signaling pathway. Furthermore, the P53 pathway is targeted by miRNAs (miR-19, miR-221, miR-515, and miR-103), suggesting that these miRNAs may have carcinogenic effects in B-ALL [51]. During early relapse of B-ALL, upregulated miR-124-3p was predicted to target the IFI44L gene. Using two predictive websites and Cytoscape data to build an mRNA-miRNA network, Huang et al. found that hsa-miR-103a-3p and hsa-miR-486-3p and their target genes change during relapse [52]. Developments in the field of bioinformatics has helped us to understand the relationship between miRNAs and their targets. Although public data and bioinformatics are useful, we are more aware of its limitations, including faults in experiments, processing data, and clinical information. Therefore, further wet-lab experiments and clinical studies involving larger number of samples are necessary. Most computer methods focus on whether the association between miRNAs and disease exists. However, new computing methods are used to predict the association between various miRNAs and diseases, and these are particularly useful for clinicians.
Role of exo-miRNA in B-ALL pathogenesis
Extracellular vesicles (EVs), including microvesicles and exosomes, are released by cells. Patel et al. confirmed that exosomes secreted from pre-B ALL cells in Ph+ ALL patients can stimulate non-growing pre-B ALL cell growth without the dependence on direct intercellular contact [53]. Exosomes containing miRNAs (exo-miRNAs) interact with the surrounding microenvironment; assisting in the development of B-ALL cells by stimulating cell proliferation and migration and altering the microenvironment, thus, inhibiting tumor immune monitoring and anti-tumor responses (Figure 2) [54]. ALL central recurrence is an important reason for the high mortality rate of patients with ALL. Only a limited number of studies have explored the role of exosomes or miRNAs in the metastasis to the central nervous system (CNS) of patients with ALL (Figure 2). Egyed et al. found that miR-181a and its specific EV subtypes in cerebrospinal fluid can predict ALL central nervous system infiltration, moreover, the sensitivity of cerebrospinal fluid miR-181a detection in the early diagnosis of CNS leukemia (90%) was significantly higher than that of conventional cytology (54.5%) [55]. Exo-miR-181a was first reported to specifically induce B lymphocyte proliferation in pediatric ALL (PALL). Moreover, miR-181a has unique and significant exosome amplification in PALL sera and leukemia cell lines. miR-181a inhibitors were successfully transfected into exosomes. Inhibiting exo-miR-181a, with the downregulation of pro-survival genes (MCL-1 and BCL2) and proliferative genes (PCNA and KI-67) as well as the inhibition of pro-apoptotic genes (BAD, BAX), can inhibit exosome-induced cell proliferation. However, inhibitors based on exosome transport-targeting miRNAs have higher safety [56].
Figure 2.
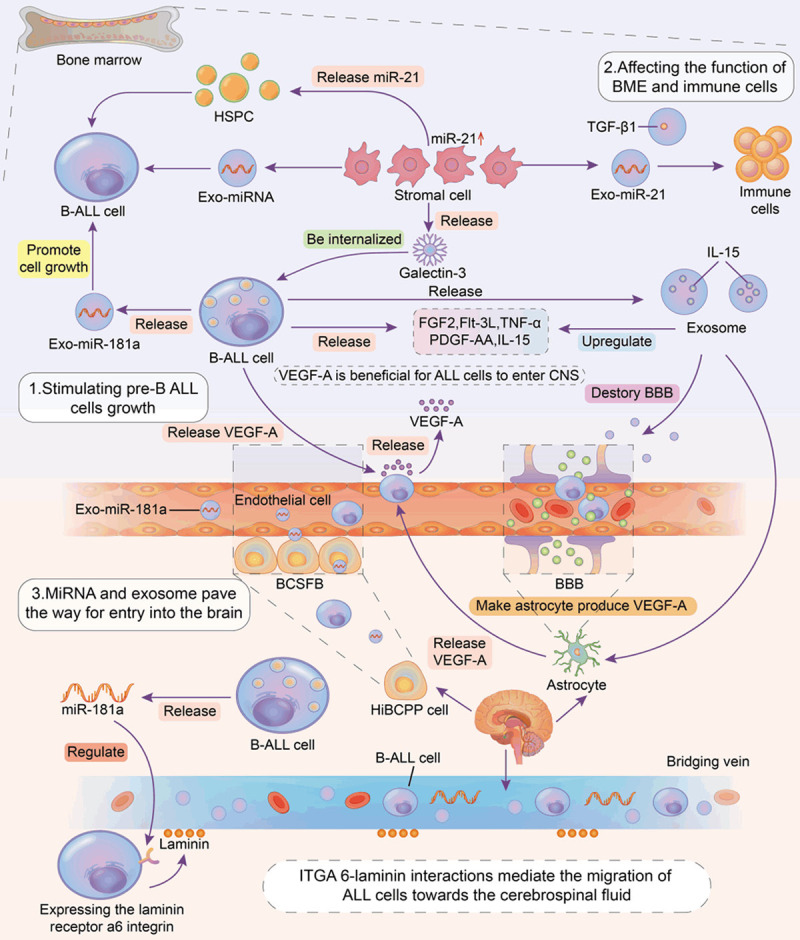
In patients with B-cell acute lymphoblastic leukemia (B-ALL), miRNAs and exosomes containing miRNAs play a key role in promoting the occurrence, progression and the central nervous system (CNS) metastasis of B-ALL. Pre-B lymphocytes, bone marrow microenvironment cells, and nervous system astrocytes can produce miRNAs and exosomes, which can mediate the bidirectional action between cells. Besides this, important cytokines and special humoral pathways also mediate the development of B-ALL cells. B-ALL cells enter the CNS through three main routes including the blood-cerebrospinal fluid barrier, blood brain barrier and bridging vein. miRNA, exo-miRNA and VEGF-A regulate these routes, thus they affect the metastasis of B-ALL cells. miRNA, exo-miRNA and VEGF-A regulate these ways, affecting the metastasis of B-ALL cells. miRNA, microRNA; Exo-miRNA, exosome containing miRNA; B-ALL, B-cell acute B-lymphoblastic leukemia; HSPC, hematopoietic stem progenitor cells; BME, bone marrow microenvironment; VEGF-A, vascular endothelial growth factor A; CNS, central nervous system; BBB, blood-brain barrier; BCSFB, Blood-cerebrospinal Fluid Barrier; HiBCPP cells, the human choroid plexus papilloma epithelial cells; ITGA 6-laminin, integrin alpha 6-laminin; EVs, extracellular vesicles.
miRNA as a biomarker in B-ALL
Mortality in ALL is increasing, this is due to a large proportion of patients dying because of a lack of biomarkers to guide treatment, assess drug resistance, and tack prognosis. Table 2 shows that miRNAs have great potential in the early diagnosis of malignancy, as well as being useful biomarkers in prognosis and drug resistance. Exosomes, a membrane-bound vesicle approximately 30-100 nm in diameter, are widely distributed in various biological body fluids [57]. Circulating miRNAs are packaged in microbubbles (e.g., exosomes), and their distribution in patients typically reflects patterns observed in tumor tissues. Their properties support their use as minimally invasive and robust biomarkers [2].
Table 2.
Application of MiRNA in Diagnosis and Treatment of B-ALL
| MicroRNA | Expression in B-ALL | Target | Mechanism of Dysregulation | Function | Material | Sample | Ref |
|---|---|---|---|---|---|---|---|
| miR-29c-5p | Downregulation | AFF1; KMT2A | Transcriptional dysregulation in cancer; Calcium signaling may be upregulated | Distinguish childhood T-ALL from B-ALL | BM | 8 B-ALL and with 8 T-ALL patients (Brazilian children) | [58] |
| miR-2909 | Upregulation | KLF4↓ | Decrease in tumor suppressor | Inhibition of apoptosis; Distinguish T-ALL and B-ALL | - | 30 pediatric B-ALL and 20 T-ALL cases and 50 controls | [59] |
| miR-21 | Upregulation | PDCD4, PTEN and TPM1 | - | Poorer DFS and OS | PB or BM | 75 pediatric B-ALL and 50 healthy controls | [6] |
| miR-151-5p, miR-451 | Downregulation | - | - | Independent prognostic markers for relapse; MiR-151 could distinguish T-ALL and B-ALL | BM | 189 pediatric ALL patients | [63] |
| miR-1290 | Upregulation | - | - | Independent prognostic markers for relapse | BM | 189 pediatric ALL patients | [63] |
| miR-101-3p, miR-1324, miR-4774-5p, miR-631, miR-922 and miR-4699 | Upregulation | - | - | - | BM | 6 early relapse and 6 prolonged remission patients and an independent set of 14 early relapse and 14 prolonged remission specimens | [64] |
| miR-708 | Upregulation | CNTFR, NNAT and GNG12↓ | Affecting hematopoietic differentiation; Activating JAK/STAT signal transduction | Increase in cell proliferation | BM | 34 pre-B-ALL cell patients and cell line | [10] |
| miR-708, miR-223 and miR-27a | miR-708 was upregulated; miR-223 and miR-27a were downregulated | E2F1, MDR1, FOXO3 | Activating cell proliferation and leukemia remission to relapse | Biomarkers for Glucocorticoid therapy response and predicting RFS | BM | B-ALL patients | [67] |
| miR-335 | Downregulation | Activating MAPK1-mediated survival of cells | DNA methylation; PRED resistance | Poorer 5-year EFS and GC resistance | BM | 56 B-ALL and 7 T-ALL patients | [68] |
| hsa-miR-17-5p | Upregulation | Bim protein levels↓ | - | GC resistance | A computational approach | PALL | [69] |
| miR-99a, miR-100, miR-125b-5p | - | - | - | Resistance to daunorubicin and vincristine | - | Pediatric B-ALL patients | [70] |
| hsa-miR-3143, hsa-miR-744-3p, hsa-miR-6503-3p, hsa-miR-1226-3p, hsa-miR-4658, hsa-miR-493-3p, hsa-miR-10-5p | Differentially expressed between LPs and LSCs | - | Affecting leukemogenesis, clone and stemness capacities | Prediction for the presence of resistant LSCs | BM | Tree adult Ph+ B-ALL patients | [71] |
| miR-3173 | Downregulation | PTK2↑ | Activated proliferation, migration and invasion | Having therapeutic potential | PB | 135 pediatric B-ALL and controls | [12] |
| miR-19b | Upregulation | - | Increase in apoptosis | miR-19b inhibitors could inhibit the proliferation | - | - | [75] |
miRNA, microRNA; B-ALL, B-cell acute B-lymphoblastic leukemia; T-ALL, T-cell acute lymphoblastic leukemia; Ph+ B-ALL, B-cell Acute lymphoblastic leukemia with chromosome positive in Philadelphia; DFS, disease-free survival; OS, overall survival; LSC, stationary leukemia stem cells; LPs, leukemia progenitor cells; GC, Glucocorticoid; EFS, event-free survival.
miRNAs function as diagnostic biomarkers
Some patients with ALL cannot be diagnosed using traditional approaches. Expression levels of certain miRNAs and overall miRNA expression profiles have shown that certain cancer types can be distinguished from each other. Mi et al. showed that ALL could be distinguished from AML based on four miRNAs (miR-128a, miR-128b, let-7b, and miR-223), with an accuracy of more than 95% [40]. The sensitivity of ALL diagnosis can be improved by 100% through miR-181a and TGF-β1 [18,31]. Other miRNAs, including miR-127 and miR-143, also have applied value in this setting [31]. Almeida et al. have used the HiSeq 2500 platform and found that miR-29c-5p is the best differentiator for children with either B-ALL or T-ALL, because its participation in calcium signaling is crucial for the fate of B cells [58]. Malik et al. found a new diagnostic/prognostic marker that can distinguish between pediatric B cells and T cells: a miR-2909-KLF4 molecular axis [59].
miRNAs function as prognostic biomarkers
Expression of miRNAs and their association with prognosis can be used as biomarkers for screening leukemia progression. Nemes et al. suggested that miR-223 and miR-128b levels could predict ALL recurrence. At diagnosis, high levels of miR-128b expression predict a good prognosis [60]. El-Khazragy et al. showed that miRNA-155a and miRNA-181a are associated with poor prognosis, with the critical value of miRNA-155a being significantly different between patients with different risk stratification, however this was not the case for miRNA-181a (P > 0.05). Low expression of miR-335 predicts poor five-year patient survival, moreover, the level of miR-335 at diagnosis was an independent prognostic indicator for treatment outcomes of PALL [61]. Labib et al. reported that upregulated miR-21 expression was associated with reduced disease-free survival (DFS) and overall survival (OS), as well as being correlated with a poor response to induction therapy in children with B-ALL [6]. Avigad et al. found that miR-451, miR-151-5p, and miR-1290 were novel biomarkers and prognostic indicators in children with BCP-ALL regardless of the treatment options. Furthermore, they detected a significant correlation between at least one of the three indicators of abnormal expression and poor prognosis (P < 0.0001). Their use may improve risk stratification and allow early treatment interventions to improve survival in at-risk patients [62]. In addition, Amankwah et al. found six miRNAs related to early relapse in children with B-ALL [63].
As briefly mentioned earlier, miR-181a and its specific subtype of EV in the serial cerebrospinal fluid could also be a predictor of central nervous system involvement in ALL [55]. Human hematological malignancy-derived exosomes have special molecular membrane markers, including TGF-β1, MHC class I polypeptide-related sequences A/B, and medulloblastoma cell markers, such as CD34, CD33, and CD117 [64]. The miRNAs contained in exosomes have a higher clinical research potential. Through monitoring B-ALL exosome contents, the efficacy of treatments can be evaluated providing more accurate means of diagnosis and evaluation of the curative effects of B-ALL.
MiRNA and treatment of B-ALL
miRNA and chemotherapy resistance and adverse reactions
Currently, there is plenty of evidence that miRNAs tend to influence treatment responses by modulating a variety of transport proteins in patients with ALL, including ATP-binding cassette (ABC) transporters [65]. Ghodousi and Rahgozar found miRNA-326 and miRNA-200c target the transporter genes ABCA3 and ABCA2, respectively, and all patients showed that a degression in miR-326 and miR-200 had new diagnostic functions [66].
Han et al. showed that miR-708 results in a poor reaction to glucocorticoids in pediatric B-ALL [67]. Ectopic expression of miR-335 increases sensitivity to prednisone [68]. Chen et al. cloned two microRNAs (hsa-miR-142-3p and hsa-miR-17-5p) and bioinformatically predicted their function in glucocorticoid resistance in PALL [69]. Some miRNAs (including miR-125b-5p, miR-99a, and miR-100) have been shown to result in daunorubicin and vincristine (VCR) resistance in pediatric B-ALL [70]. Thirty-nine percent of patients with VCR resistance had the ETV6/RUNX1 subtype. These patients showed high levels of miR-125b expression; affecting miR-125b function may enhance the chemotherapeutic response in patients [31]. The survival of stationary leukemia stem cells (LSCs) after therapy is one of the causes of relapse in patients with Ph+ ALL. A recent study identified that hsa-miR-6503-3p and hsa-miR-3143 may affect the production, cloning, and cell capacity of these subpopulations of Ph+ B-ALL and thus help develop drug-resistant LSCs [71].
Single nucleotide polymorphisms (SNPs) in miRNAs can alter the levels or functions of miRNAs. Iparraguirre et al. detected SNPs in miR-5189, miR-595, and miR-6083, which may affect the regulation of the MTX transporter genes SLC46A1, SLC19A1, and SLCO1A2, thus influencing the effect of MTX in children with B-ALL [72]. In terms of drug side-effect-related miRNA expression, this study identified two SNPs that might be involved in VCR-and neurotoxicity-related miR-3117-3p [73]. Gutierrez-Camino et al. found an association between the rs2114358 variant of miR-1206 in ALL patients with MTX-induced oral mucositis [74].
miRNA and prognosis of B-ALL patients
The role of miRNAs in tumor pathogenesis provides a new research perspective on therapeutic targets (Table 2). miRNAs regulate protein expression by altering the mRNA transition to a lower state. Changing protein levels by manipulating miRNAs is possible; however, these techniques are still in their infancy. The transfection of miRNA mimics or miRNA inhibitors into cells is the usual way to regulate miRNA expression. miR-3173 inhibits PTK2 expression at the protein level. In vitro experiments demonstrated that miR-3173 mimics inhibited proliferation, migration, and invasion in B-ALL cells, while miR-3173 inhibitors had the opposite effect. Therefore, miR-3173 shows therapeutic potential [12]. miR-206 inhibits cell proliferation and chemical resistance by targeting neuroepithelial cell-transformation 1 (NET1); this suggests the possibility of developing therapeutic interventions with this miRNA [13]. In addition, in vitro studies showed that miR-19b inhibitors could inhibit the proliferation of B cells better than T cells [75].
However, safety and degradation limit the application of miRNA mimics or miRNA inhibitors in vivo. It is important to maintain the stability of miRNA mimics or miRNA inhibitors in vivo; therefore, delivery methods need to be adequately studied [76]. miRNAs in exosomes have non-degradable stability, therefore, the application prospects of exosomes in transporting miRNAs are extensive [77]. A previous study successfully transfected miR-181a inhibitors into exosomes, which could then inhibit exo-miR-181a, thereby inhibiting the proliferation of B-ALL cells. This provides the possibility for accurate and effective treatment of B-ALL [56]. The technology and miRNA utilization of edited miRNAs need to be improved, and the possible risks of miRNA non-target effects need to be further studied. Moreover, clinical studies based on transport miRNA therapy are underway and should elucidate more data [78].
Conclusion and future perspectives
Our understanding of the molecular mechanisms underlying B-ALL remains limited. miRNAs can act as carcinogens or proto-oncogenes in different cell types. The specific mechanisms of miRNA in B-ALL disease progression and therapeutic drug resistance is not fully understood. The establishment of miRNA-mRNA interactions and signaling pathways in B-ALL helps to understand the role of miRNAs in disease occurrence, progression, epigenetic regulation, immunosuppression, and drug resistance. Since an miRNA can target many gene transcripts, it usually has related functions. Moreover, several individual transcripts can be targeted by many miRNAs that are often regulated together. Combining miRNAome data with both transcriptome and proteome integration will be the basis for a more comprehensive analysis of the consequences of disrupting the cellular structure maintained by the microRNA. However, this requires a large number of patients and appropriate healthy tissues. Therefore, interdisciplinary methods are required. Although studies on exo-miRNAs are limited, exo-miRNAs specifically regulate the pathogenesis of B-ALL, especially during the invasion of BCP-ALL cells into the CNS.
There is good research potential for disease progression biomarkers and therapeutic targets for miRNAs. Specific miRNAs help diagnose B-ALL and determine the prognosis of patients. However, their roles also require large-scale clinical trials to confirm their clinical potential. So far, most studies on miRNAs in B-ALL have focused on children. However, there are differences in the expression patterns of miRNAs between children and adults with ALL. Therefore, it is important to analyze the differences, which may help explain the poor effect of chemotherapy in adults and provide a new perspective for the choice of treatment options available for adult patients with B-ALL. Understanding the interaction between miRNA-mediated immune cells and leukemia cells is essential for the successful development of immunotherapy.
At present, exosomes can replace liposomes in transporting chemical drugs, miRNAs, and many other components, moreover, they are highly specific to target cells. Because exosomes are small, the internalization mechanism of exosomes is not clear. In the future it is necessary to determine how the exosomes are transferred in vivo and how long they maintain stability in systemic circulation, furthermore, new methods to isolate and identify exosomes must be established. EV-based clinical trials are underway, but most of them focus on solid cancer. Moreover, the Exo ReBly project (NCT03985696) is a clinical trial on Diffuse Large B-cell Lymphoma (DLBCL) aiming to explore whether there is a large number of CD20 and PD-L1 in DLBCL derivative EVs. This study will determine whether they are bait targets and whether they have strong immunosuppressive signals for rituximab antibodies, which may then lead to therapeutic resistance. As mentioned before, the killing activity of CAR-T cells in transfusion patients will still be weakened by the PD-1/PD-L1 immunosuppressive pathway. We must discover whether exosomes secreted by B-ALL leukemic cells carry PD-L1, thus decreasing CAR-T immunotherapy. This question requires further exploration, which may help to improve the effectiveness of CAR-T immunotherapy in patients with B-ALL.
In summary, studies of both miRNAs and exo-miRNAs have revealed the pathogenesis of B-ALL. Specific miRNAs and exo-miRNAs are promising biomarkers for the diagnosis and prognosis of patients with B-ALL. However, in the future it is necessary to determine how exosomes are transferred in vivo and how long they are maintained. Furthermore, disease progression biomarkers and therapeutic targets for miRNAs have good research potential.
Acknowledgements
This work was supported by the National Natural Science Foundation, P. R. China (No: 81600183).
Disclosure of conflict of interest
None.
References
- 1.Woo J, Alberti M, Tirado C. Childhood B-acute lymphoblastic leukemia: a genetic update. Exp Hematol Oncol. 2014;3:16. doi: 10.1186/2162-3619-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L, Guo Y, Zhang X, Chen J, Hu S. Blood-based circulating microRNAs are potential diagnostic biomarkers for leukemia: result from a meta-analysis. Cell Physiol Biochem. 2016;38:939–49. doi: 10.1159/000443046. [DOI] [PubMed] [Google Scholar]
- 3.Fathullahzadeh S, Mirzaei H, Honardoost M, Sahebkar A, Salehi M. Circulating microRNA-192 as a diagnostic biomarker in human chronic lymphocytic leukemia. Cancer Gene Ther. 2016;23:327–332. doi: 10.1038/cgt.2016.34. [DOI] [PubMed] [Google Scholar]
- 4.Mardani R, Jafari Najaf Abadi MH, Motieian M, Taghizadeh-Boroujeni S, Bayat A, Farsinezhad A, Gheibi Hayat SM, Motieian M, Pourghadamyari H. MicroRNA in leukemia: tumor suppressors and oncogenes with prognostic potential. J Cell Physiol. 2019;234:8465–8486. doi: 10.1002/jcp.27776. [DOI] [PubMed] [Google Scholar]
- 5.Saadatpour L, Fadaee E, Fadaei S, Nassiri Mansour R, Mohammadi M, Mousavi S, Goodarzi M, Verdi J, Mirzaei H. Glioblastoma: exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 2016;23:415–418. doi: 10.1038/cgt.2016.48. [DOI] [PubMed] [Google Scholar]
- 6.Labib HA, Elantouny NG, Ibrahim NF, Alnagar AA. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology. 2017;22:392–397. doi: 10.1080/10245332.2017.1292204. [DOI] [PubMed] [Google Scholar]
- 7.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 8.de Yébenes VG, Bartolomé-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol Rev. 2013;253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu T, Chong Y, Lu S, Wang R, Qin H, Silva J, Kitamura E, Chang CS, Hawthorn L, Cowell JK. miR-339 promotes development of stem cell leukemia/lymphoma syndrome via downregulation of the BCL2L11 and BAX proapoptotic genes. Cancer Res. 2018;78:3522–3531. doi: 10.1158/0008-5472.CAN-17-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Li D, Zhuang Y, Shi Q, Wei W, Ju X. Overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr Blood Cancer. 2013;60:2060–7. doi: 10.1002/pbc.24583. [DOI] [PubMed] [Google Scholar]
- 11.Tan YS, Kim M, Kingsbury TJ, Civin CI, Cheng WC. Regulation of RAB5C is important for the growth inhibitory effects of MiR-509 in human precursor-B acute lymphoblastic leukemia. PLoS One. 2014;9:e111777. doi: 10.1371/journal.pone.0111777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian L, Cao J, Ji Q, Zhang C, Qian T, Song X, Huang B, Tian X. The downregulation of miR-3173 in B-cell acute lymphoblastic leukaemia promotes cell invasion via PTK2. Biochem Biophys Res Commun. 2017;494:569–574. doi: 10.1016/j.bbrc.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Sun H, Zhang Z, Luo W, Liu J, Lou Y, Xia S. NET1 enhances proliferation and chemoresistance in acute lymphoblastic leukemia cells. Oncol Res. 2019;27:935–944. doi: 10.3727/096504019X15555388198071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duyu M, Durmaz B, Gunduz C, Vergin C, Yilmaz Karapinar D, Aksoylar S, Kavakli K, Cetingul N, Irken G, Yaman Y, Ozkinay F, Cogulu O. Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. Biomed Res Int. 2014;2014:967585. doi: 10.1155/2014/967585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verduci L, Azzalin G, Gioiosa S, Carissimi C, Laudadio I, Fulci V, Macino G. microRNA-181a enhances cell proliferation in acute lymphoblastic leukemia by targeting EGR1. Leuk Res. 2015;39:479–485. doi: 10.1016/j.leukres.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Fragoso R, Mao T, Wang S, Schaffert S, Gong X, Yue S, Luong R, Min H, Yashiro-Ohtani Y, Davis M, Pear W, Chen CZ. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012;8:e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nabhan M, Louka ML, Khairy E, Tash F, Ali-Labib R, El-Habashy S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene. 2017;628:253–258. doi: 10.1016/j.gene.2017.07.052. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Yen C, Pai C, Chen H, Yu S, Lin C, Hu C, Jou S, Lin D, Lin S, Lin S. A double negative loop comprising ETV6/RUNX1 and MIR181A1 contributes to differentiation block in t(12;21)-positive acute lymphoblastic leukemia. PLoS One. 2015;10:e0142863. doi: 10.1371/journal.pone.0142863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vafadar A, Mokaram P, Erfani M, Yousefi Z, Farhadi A, Elham Shirazi T, Tamaddon G. The effect of decitabine on the expression and methylation of the PPP1CA, BTG2, and PTEN in association with changes in miR-125b, miR-17, and miR-181b in NALM6 cell line. J Cell Biochem. 2019;120:13156–13167. doi: 10.1002/jcb.28590. [DOI] [PubMed] [Google Scholar]
- 21.So AY, Sookram R, Chaudhuri AA, Minisandram A, Cheng D, Xie C, Lim EL, Flores YG, Jiang S, Kim JT, Keown C, Ramakrishnan P, Baltimore D. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 2014;124:1502–1512. doi: 10.1182/blood-2014-02-553842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Khazragy N, Elshimy AA, Hassan SS, Matbouly S, Safwat G, Zannoun M, Riad RA. Dysregulation of miR-125b predicts poor response to therapy in pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018 doi: 10.1002/jcb.28017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Bousquet M, Harris M, Zhou B, Lodish H. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J. Clin. Oncol. 2017;35:975–983. doi: 10.1200/JCO.2016.70.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherr M, Elder A, Battmer K, Barzan D, Bomken S, Ricke-Hoch M, Schröder A, Venturini L, Blair HJ, Vormoor J, Ottmann O, Ganser A, Pich A, Hilfiker-Kleiner D, Heidenreich O, Eder M. Differential expression of miR-17~92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia. Leukemia. 2014;28:554–565. doi: 10.1038/leu.2013.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schotte D, Akbari Moqadam F, Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R, den Boer ML. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- 27.Dou L, Li J, Zheng D, Li Y, Gao X, Xu C, Gao L, Wang L, Yu L. MicroRNA-142-3p inhibits cell proliferation in human acute lymphoblastic leukemia by targeting the MLL-AF4 oncogene. Mol Biol Rep. 2013;40:6811–6819. doi: 10.1007/s11033-013-2798-6. [DOI] [PubMed] [Google Scholar]
- 28.Ju X, Li D, Shi Q, Hou H, Sun N, Shen B. Differential microRNA expression in childhood B-cell precursor acute lymphoblastic leukemia. Pediatr Hematol Oncol. 2009;26:1–10. doi: 10.1080/08880010802378338. [DOI] [PubMed] [Google Scholar]
- 29.Dou L, Zheng D, Li J, Li Y, Gao L, Wang L, Yu L. Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene. 2012;31:507–517. doi: 10.1038/onc.2011.248. [DOI] [PubMed] [Google Scholar]
- 30.Li XJ, Luo XQ, Han BW, Duan FT, Wei PP, Chen YQ. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J Cancer. 2013;109:2189–2198. doi: 10.1038/bjc.2013.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R, Den Boer ML. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96:703–711. doi: 10.3324/haematol.2010.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R, den Boer ML. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 33.Schotte D, Lange-Turenhout EA, Stumpel DJ, Stam RW, Buijs-Gladdines JG, Meijerink JP, Pieters R, Den Boer ML. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1675–1682. doi: 10.3324/haematol.2010.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin XC, Liu XG, Zhang YM, Li N, Yang ZG, Fu WY, Lan LB, Zhang HT, Dai Y. Integrated analysis of microRNA and transcription factor reveals important regulators and regulatory motifs in adult B-cell acute lymphoblastic leukemia. Int J Oncol. 2017;50:671–683. doi: 10.3892/ijo.2016.3832. [DOI] [PubMed] [Google Scholar]
- 35.Pinz S, Unser S, Rascle A. Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer. BMC Mol Biol. 2016;17:10. doi: 10.1186/s12867-016-0063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sochalska M, Schuler F, Weiss JG, Prchal-Murphy M, Sexl V, Villunger A. MYC selects against reduced BCL2A1/A1 protein expression during B cell lymphomagenesis. Oncogene. 2017;36:2066–2073. doi: 10.1038/onc.2016.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010;340:97–106. doi: 10.1007/s11010-010-0406-9. [DOI] [PubMed] [Google Scholar]
- 38.Lou Y, Liu L, Zhan L, Wang X, Fan H. miR-187-5p regulates cell growth and apoptosis in acute lymphoblastic leukemia via DKK2. Oncol Res. 2016;24:89–97. doi: 10.3727/096504016X14597766487753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agirre X, Vilas-Zornoza A, Jiménez-Velasco A, Martin-Subero JI, Cordeu L, Gárate L, San José-Eneriz E, Abizanda G, Rodríguez-Otero P, Fortes P, Rifón J, Bandrés E, Calasanz MJ, Martín V, Heiniger A, Torres A, Siebert R, Román-Gomez J, Prósper F. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 40.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly M, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander S, Le Beau M, Larson R, Golub T, Rowley J, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Undi R, Kandi R, Gutti R. MicroRNAs as haematopoiesis regulators. Adv Hematol. 2013;2013:695754. doi: 10.1155/2013/695754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nucera S, Giustacchini A, Boccalatte F, Calabria A, Fanciullo C, Plati T, Ranghetti A, Garcia-Manteiga J, Cittaro D, Benedicenti F, Lechman ER, Dick JE, Ponzoni M, Ciceri F, Montini E, Gentner B, Naldini L. miRNA-126 orchestrates an oncogenic program in B cell precursor acute lymphoblastic leukemia. Cancer Cell. 2016;29:905–921. doi: 10.1016/j.ccell.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Moses BS, Evans R, Slone WL, Piktel D, Martinez I, Craig MD, Gibson LF. Bone marrow microenvironment niche regulates miR-221/222 in acute lymphoblastic leukemia. Mol Cancer Res. 2016;14:909–919. doi: 10.1158/1541-7786.MCR-15-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosakhani N, Missiry ME, Vakkila E, Knuutila S, Vakkila J. Low expression of miR-18a as a characteristic of pediatric acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2017;39:585–588. doi: 10.1097/MPH.0000000000000921. [DOI] [PubMed] [Google Scholar]
- 47.Freyer CW. Tisagenlecleucel: the first CAR on the highway to remission for acute lymphoblastic leukemia. J Adv Pract Oncol. 2018;9:537–544. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q, Hu H, Chen SY, Liu CJ, Hu FF, Yu J, Wu Y, Guo AY. Transcriptome and regulatory network analyses of CD19-CAR-T immunotherapy for B-ALL. Genomics Proteomics Bioinformatics. 2019;17:190–200. doi: 10.1016/j.gpb.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, Guo X, Zhang J, Zhang Q, Zhang L, Xue Z, Li Y, Da Y, Zhao P, Zhang R. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun. 2017;488:425–431. doi: 10.1016/j.bbrc.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 51.Ramani R, Megason G, Schallheim J, Karlson C, Vijayakumar V, Vijayakumar S, Hicks C. Integrative analysis of MicroRNA-mediated gene signatures and pathways modulating white blood cell count in childhood acute lymphoblastic leukemia. Biomark Insights. 2017;12:1177271917702895. doi: 10.1177/1177271917702895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y, Li J, Chen Y, Jiang P, Wang L, Hu J. Identification of early recurrence factors in childhood and adolescent B-cell acute lymphoblastic leukemia based on integrated bioinformatics analysis. Front Oncol. 2020;10:565455. doi: 10.3389/fonc.2020.565455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel SJ, Darie CC, Clarkson BD. Exosome mediated growth effect on the non-growing pre-B acute lymphoblastic leukemia cells at low starting cell density. Am J Transl Res. 2016;8:3614–3629. [PMC free article] [PubMed] [Google Scholar]
- 54.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 55.Egyed B, Kutszegi N, Sági JC, Gézsi A, Rzepiel A, Visnovitz T, Lőrincz P, Müller J, Zombori M, Szalai C, Erdélyi DJ, Kovács GT, Semsei ÁF. MicroRNA-181a as novel liquid biopsy marker of central nervous system involvement in pediatric acute lymphoblastic leukemia. J Transl Med. 2020;18:250. doi: 10.1186/s12967-020-02415-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haque S, Vaiselbuh SR. Silencing of exosomal miR-181a reverses pediatric acute lymphocytic leukemia cell proliferation. Pharmaceuticals (Basel) 2020;13:241. doi: 10.3390/ph13090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J Cell Biol. 2013;200:367–371. doi: 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almeida RS, Costa E Silva M, Coutinho LL, Garcia Gomes R, Pedrosa F, Massaro JD, Donadi EA, Lucena-Silva N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol Oncol. 2019;37:103–112. doi: 10.1002/hon.2567. [DOI] [PubMed] [Google Scholar]
- 59.Malik D, Kaul D, Chauhan N, Marwaha RK. miR-2909-mediated regulation of KLF4: a novel molecular mechanism for differentiating between B-cell and T-cell pediatric acute lymphoblastic leukemias. Mol Cancer. 2014;13:175. doi: 10.1186/1476-4598-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nemes K, Csóka M, Nagy N, Márk Á, Váradi Z, Dankó T, Kovács G, Kopper L, Sebestyén A. Expression of certain leukemia/lymphoma related microRNAs and its correlation with prognosis in childhood acute lymphoblastic leukemia. Pathol Oncol Res. 2015;21:597–604. doi: 10.1007/s12253-014-9861-z. [DOI] [PubMed] [Google Scholar]
- 61.El-Khazragy N, Noshi MA, Abdel-Malak C, Zahran RF, Swellam M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J Cell Biochem. 2019;120:6315–6321. doi: 10.1002/jcb.27918. [DOI] [PubMed] [Google Scholar]
- 62.Avigad S, Verly IR, Lebel A, Kordi O, Shichrur K, Ohali A, Hameiri-Grossman M, Kaspers GJ, Cloos J, Fronkova E, Trka J, Luria D, Kodman Y, Mirsky H, Gaash D, Jeison M, Avrahami G, Elitzur S, Gilad G, Stark B, Yaniv I. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2016;55:328–339. doi: 10.1002/gcc.22334. [DOI] [PubMed] [Google Scholar]
- 63.Amankwah EK, Devidas M, Teachey DT, Rabin KR, Brown PA. Six candidate miRNAs associated with early relapse in pediatric B-cell acute lymphoblastic leukemia. Anticancer Res. 2020;40:3147–3153. doi: 10.21873/anticanres.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Miguel D, Basáñez G, Sánchez D, Malo PG, Marzo I, Larrad L, Naval J, Pardo J, Anel A, Martinez-Lostao L. Liposomes decorated with Apo2L/TRAIL overcome chemoresistance of human hematologic tumor cells. Mol Pharm. 2013;10:893–904. doi: 10.1021/mp300258c. [DOI] [PubMed] [Google Scholar]
- 65.Kibria G, Hatakeyama H, Harashima H. Cancer multidrug resistance: mechanisms involved and strategies for circumvention using a drug delivery system. Arch Pharm Res. 2014;37:4–15. doi: 10.1007/s12272-013-0276-2. [DOI] [PubMed] [Google Scholar]
- 66.Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;119:6024–6032. doi: 10.1002/jcb.26800. [DOI] [PubMed] [Google Scholar]
- 67.Han B, Feng D, Li Z, Luo X, Zhang H, Li X, Zhang X, Zheng L, Zeng C, Lin K, Zhang P, Xu L, Chen Y. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Genet. 2011;20:4903–4915. doi: 10.1093/hmg/ddr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan J, Jiang N, Huang G, Tay JL, Lin B, Bi C, Koh GS, Li Z, Tan J, Chung TH, Lu Y, Ariffin H, Kham SK, Yeoh AE, Chng WJ. Deregulated MIR335 that targets MAPK1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia. Br J Haematol. 2013;163:93–103. doi: 10.1111/bjh.12489. [DOI] [PubMed] [Google Scholar]
- 69.Chen H, Zhang D, Zhang G, Li X, Liang Y, Kasukurthi MV, Li S, Borchert GM, Huang J. A semantics-oriented computational approach to investigate microRNA regulation on glucocorticoid resistance in pediatric acute lymphoblastic leukemia. BMC Med Inform Decis Mak. 2018;18:57. doi: 10.1186/s12911-018-0637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luan C, Yang Z, Chen B. The functional role of microRNA in acute lymphoblastic leukemia: relevance for diagnosis, differential diagnosis, prognosis, and therapy. Onco Targets Ther. 2015;8:2903–2914. doi: 10.2147/OTT.S92470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valiollahi E, Ribera JM, Genescà E, Behravan J. Genome-wide identification of microRNA signatures associated with stem/progenitor cells in philadelphia chromosome-positive acute lymphoblastic leukemia. Mol Biol Rep. 2019;46:1295–1306. doi: 10.1007/s11033-019-04600-5. [DOI] [PubMed] [Google Scholar]
- 72.Iparraguirre L, Gutierrez-Camino A, Umerez M, Martin-Guerrero I, Astigarraga I, Navajas A, Sastre A, Garcia de Andoin N, Garcia-Orad A. MiR-pharmacogenetics of methotrexate in childhood B-cell acute lymphoblastic leukemia. Pharmacogenet Genomics. 2016;26:517–525. doi: 10.1097/FPC.0000000000000245. [DOI] [PubMed] [Google Scholar]
- 73.Gutierrez-Camino Á, Umerez M, Martin-Guerrero I, García de Andoin N, Santos B, Sastre A, Echebarria-Barona A, Astigarraga I, Navajas A, Garcia-Orad A. Mir-pharmacogenetics of vincristine and peripheral neurotoxicity in childhood B-cell acute lymphoblastic leukemia. Pharmacogenomics J. 2018;18:704–712. doi: 10.1038/s41397-017-0003-3. [DOI] [PubMed] [Google Scholar]
- 74.Gutierrez-Camino A, Oosterom N, den Hoed MAH, Lopez-Lopez E, Martin-Guerrero I, Pluijm SMF, Pieters R, de Jonge R, Tissing WJE, Heil SG, García-Orad A, van den Heuvel-Eibrink MM. The miR-1206 microRNA variant is associated with methotrexate-induced oral mucositis in pediatric acute lymphoblastic leukemia. Pharmacogenet Genomics. 2017;27:303–306. doi: 10.1097/FPC.0000000000000291. [DOI] [PubMed] [Google Scholar]
- 75.Budde H, Rau AL, Riggert J, Legler TJ. Apoptosis induction by miR-19b inhibition: does it show therapeutic potential for leukemia? Nucleosides Nucleotides Nucleic Acids. 2020;39:1073–1081. doi: 10.1080/15257770.2020.1755042. [DOI] [PubMed] [Google Scholar]
- 76.Mencía Castaño I, Curtin CM, Shaw G, Murphy JM, Duffy GP, O’Brien FJ. A novel collagen-nanohydroxyapatite microRNA-activated scaffold for tissue engineering applications capable of efficient delivery of both miR-mimics and antagomiRs to human mesenchymal stem cells. J Control Release. 2015;200:42–51. doi: 10.1016/j.jconrel.2014.12.034. [DOI] [PubMed] [Google Scholar]
- 77.Le MT, Hamar P, Guo C, Basar E, Perdigão-Henriques R, Balaj L, Lieberman J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin JA, O’Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the united kingdom MRC AML-15 trial. Blood. 2012;120:2826–2835. doi: 10.1182/blood-2012-06-435669. [DOI] [PubMed] [Google Scholar]