Abstract
Angiogenesis is a cornerstone of cancer as it allows tumors to receive oxygen and nutrients. A high level of angiogenesis within a tumor may therefore be indicative of its aggressiveness. In this study, we examined this hypothesis in gastric cancer. Gene set variation analysis was used to measure the level of angiogenesis in tumors in 1,348 gastric cancer patients using the Hallmark_angiogenesis gene set to score tumor transcriptomes. As we predicted, there was a significant correlation between angiogenesis score and expression of angiogenesis-related genes. The score moderately correlated with abundance of vessel-related stromal cells, fibroblasts and chondrocytes in the tumor microenvironment (TME). Tumors with high score had low infiltration of T helper type 1 and 2 cells but a greater infiltration of M1 macrophages and dendritic cells. They also had enriched expression of gene sets for coagulation, hypoxia, epithelial mesenchymal transition (EMT), and TGF-β signaling. High angiogenesis score was significantly associated with advanced AJCC stage and higher T- but not N-parameters in the TNM staging system. Patients with a high score also had shorter survival. In conclusion, bulk tumor transcriptome-based quantification of tumor angiogenesis using a computational algorithm may serve to identify patients with worse survival in gastric cancer.
Keywords: Angiogenesis, EMT, gastric cancer, gene set, survival, GSVA, prognostic biomarker
Introduction
Across the world, gastric cancer ranks third in cancer-related deaths [1]. At diagnosis, eighty percent of patients have unresectable or metastatic disease [2]. Despite improvements in perioperative treatment, approximately half of the patients who undergo curative intent surgery eventually relapse. Though efforts have focused on identifying prognostic biomarkers that can be applied in clinical practice to improve the poor outcome [3,4], we urgently need a predictive biomarker to help guide treatment decision-making to precisely address gastric cancer progression and metastasis.
Angiogenesis is the creation of de novo blood vessels from existing ones. Angiogenesis is a cornerstone of cancer as it paves the way for cancer progression and metastasis [5,6]. Blood vessels allow oxygen and nutrients to be delivered to tumors and also carry cells and factors that promote immune resistance. Additionally, the new blood vessels generated can act as a gateway for cancer cells to enter the blood stream and metastasize distantly [7]. Cancer and stromal cells produce angiogenic factor such as vascular endothelial growth factor and endothelial cell growth factor in several types of cancers including gastric cancer [8,9]. We have previously reported that there is an association between abundant blood vessels and both infiltration of anti-tumor immune cells and increased survival in pancreatic adenocarcinoma [10]. Therefore, it is critical to study angiogenesis not only in cell culture and in vivo animal models, but also in large human cohorts to assess its clinical relevance.
Utilizing the Gene Set Variation Analysis (GSVA) method to computationally analyze tumor transcriptomes to score them for activity of various biological processes, we have demonstrated the clinical relevance of multiple gene pathways [11-13]. We previously reported that in breast cancer patients, angiogenesis score was significantly associated with metastasis and inflammation [14]. The main advantage of scoring is that it is quantifiable using the gene expression data of the existing bulk tumors in large patient cohorts. In this study, we hypothesized that intra-tumoral angiogenesis promotes gastric cancer aggressiveness.
Materials and methods
Collection of transcriptomic and clinical data in gastric cancer patients
Transcriptome data of The Cancer Genome Atlas Stomach Adenocarcinoma project (TCGA-STAD, n = 375) was obtained through the Genomic Data Commons data portal (GDC). Clinical outcomes of the project’s patients were from the curated and filtered Pan-Cancer Clinical Data Resource of survival endpoints [15]. Normalized genomic and clinical data published by Paik et al. (GSE26253, n = 432) [16], Lee et al. (GSE26901, n = 432) [17], and Yoon et al. (GSE84437, n = 432) [18] were procured by the Gene Expression Omnibus (GEO) repository. Gene expression was log2-transformed for all analyses.
Hallmark pathway enrichment analysis
Using our previously published methodology [14], Gene Set Variation Analysis (GSVA) Bioconductor package [19] was used to measure the “HALLMARK_ANGIOGENESIS” in the MSigDB Hallmark gene sets collection [20].
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was performed using the GSEA Java software (version 4.0) with MSigDB Hallmark gene sets [21] to study the enrichment of signaling pathways between low- and high-angiogenesis score groups [22-27]. For statistical significance in GSEA analysis, we used the recommended false discovery rate (FDR) of less than 0.25.
Tumor immune microenvironment analysis
As we previously reported [29], we estimated the relative abundance of 64 types of immune and stromal cells in tumors using the xCell algorithm from their transcriptome profile [28].
Other statistical analyses
R software (version 4.0.2) was utilized for all analyses. Boxplots that are used in the figures depict medians and interquartile range (IQR). P values for group comparisons were calculated by one-way ANOVA or Fisher’s exact tests. The within-cohort median value was used to categorize the patients into low or high angiogenesis score groups. Survival curves were plotted using the Kaplan-Meier curve with log-rank test. Statistical significance was reached with P-values of less than 0.05.
Results
Angiogenesis-related genes are highly expressed in gastric cancers with high angiogenesis score
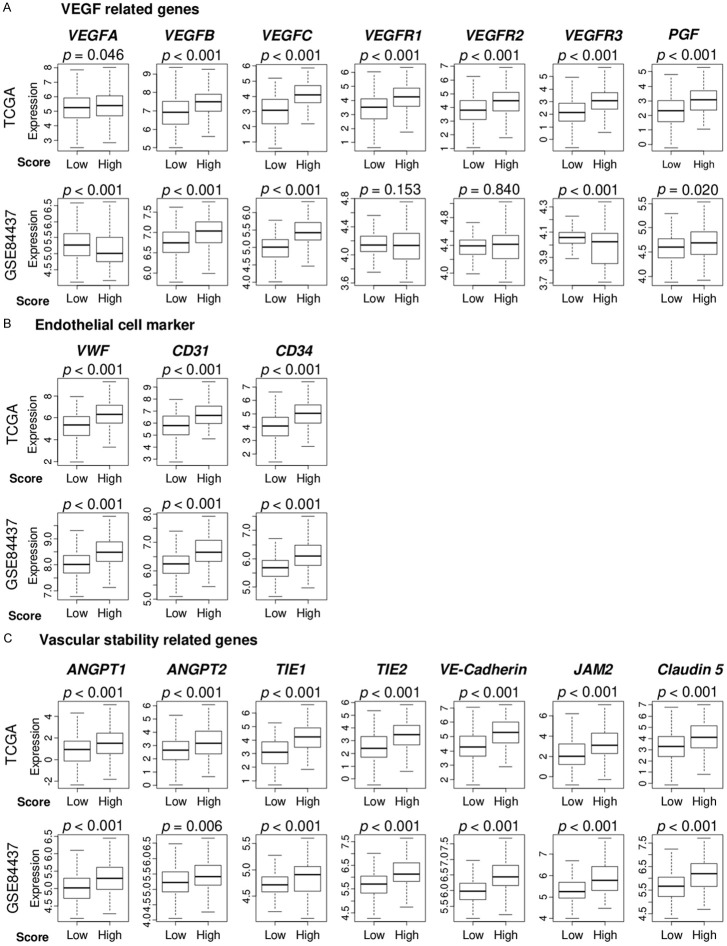
We calculated the angiogenesis score of tumors from 1,348 gastric cancer patients in multiple cohorts using the GSVA method we previously reported [14] to score tumor transcriptomes in the Hallmark angiogenesis gene set. First, to confirm that the angiogenesis score adequately captured intratumoral angiogenesis in gastric cancer, we examined the association between the score and multiple angiogenesis-related gene expression using the TCGA (n = 375) and GSE84437 (n = 432) gastric cancer cohorts. We used the median as a cutoff between the high vs low scores within cohorts because the angiogenesis score was distributed either normally or bimodally in the TCGA and GSE84437 cohorts (Figure S1). We chose VEGF-related (VEGFA, VEGFB, VEGFR1, VEGFR2, and VEGFR3), endothelial cell marker (VWF and CD31), and vascular stability-related (ANGPT1, ANGPT2, TIE1, TIE2, VE-Cadherin, JAM2, and Claudin5) genes as angiogenesis-related genes. All angiogenesis-related genes had increased expression in the high angiogenesis score group in the TCGA gastric cancer cohort (Figure 1A-C). These results were validated in the GSE84437 cohort where most genes were highly expressed in the high angiogenesis score group except for VEGFA, VEGFR1, VEGFR2 and VEGFR3 genes. There were no significant differences in the expression of VEGFR1 and VEGFR2 genes, and there was a decreased expression of VEGFA and VEGFR3 genes in the high angiogenesis score group.
Figure 1.
The angiogenesis score was associated with expressions of angiogenesis-related genes. Boxplots showing comparisons of the expression levels of (A) vascular endothelial growth factor (VEGF)-related genes, (B) endothelial cell marker-related genes; and (C) vascular stability-related genes, between low and high angiogenesis score groups in the TCGA and GSE84437 cohorts. We defined the VEGF-related genes as VEGFA, VEGFB, VEGFC, VEGFR1 (FLT1), VEGFR2 (KDR), VEGFR3 (FLT4), and PGF, and the endothelial cell marker-related genes as VWF, CD31 (PECAM1), and CD34, and vascular stability-related genes as ANGPT1, ANGPT2, TIE1, TIE2 (TEK), VE-cadherin (CDH5), JAM2 and Claudin5 (CLDN5).
Angiogenesis score correlated with fibroblasts and chondrocytes in addition to epithelial cells and pericytes in gastric cancer
Both cancer and stromal cells in gastric cancer produce several angiogenic factors [30]. Thus, we examined the association of intratumoral angiogenesis with stromal cells. To determine the fraction of stromal cells in tumors, we used the xCell algorithm [28], which estimates the abundance of 64 cell subtypes in the tumor microenvironment (TME) from bulk tumor transcriptomic data [31-35]. The angiogenesis score significantly correlated with cells that constitute vessels, namely endothelial cells, lymphatic- and microvascular-endothelial cells, and pericytes as noted in both the TCGA and GSE84437 cohorts (Figure 2). Infiltration of other stromal cells, including fibroblasts and chondrocytes, correlated with angiogenesis in gastric cancer. Furthermore, angiogenesis score correlated with fibroblast-related genes, αSMA (ACTA2) and FAPα (FAP) genes (Figure S2). These findings suggest tumors with a high angiogenesis score have a high fraction of not only vascular-related cells, but also fibroblasts and chondrocytes.
Figure 2.
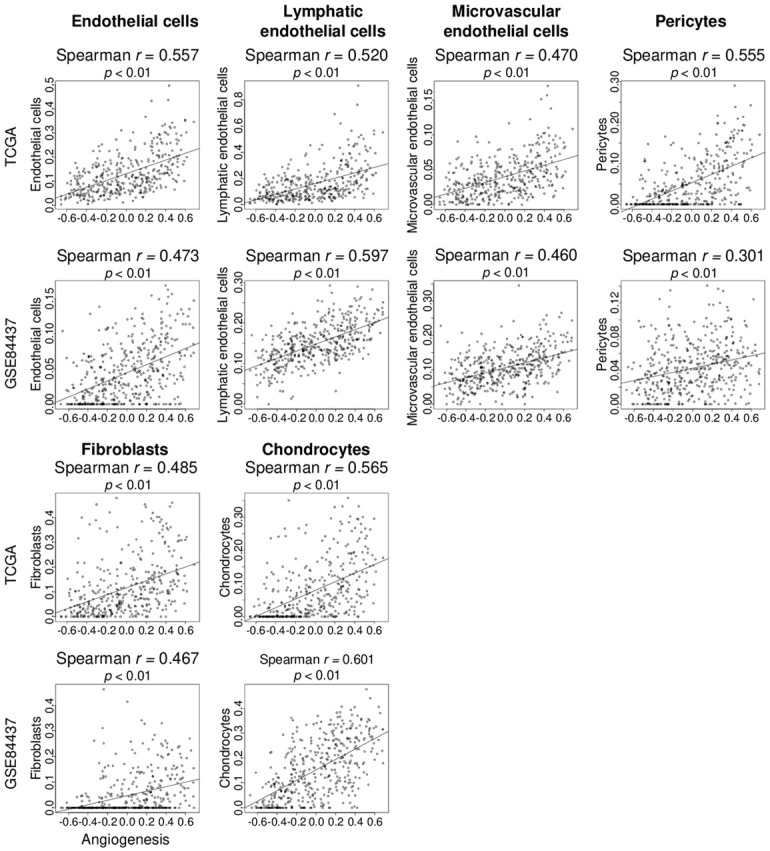
The angiogenesis score was associated with fraction of stromal cells in gastric cancer. Correlation curve of the angiogenesis score with stromal cells, including endothelial cells, lymphatic- and microvascular-endothelial cells, pericytes, fibroblasts, and chondrocytes, were estimated using the xCell algorithm in the TCGA and GSE84437 cohorts. Spearman rank correlation was used in the analysis.
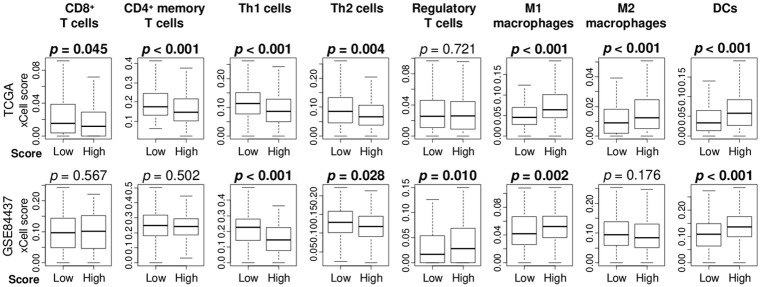
High angiogenesis score patients had low infiltration of T helper type 1 and 2 cells and high infiltration of dendritic cells and M1 macrophages
Based on the notion that the tumor immune microenvironment (TIME) is influenced by tumor angiogenesis, we studied the association between the angiogenesis score and TIME using the xCell algorithm in the TCGA and GSE84437 gastric cancer cohorts. High angiogenesis score tumors were infiltrated with a low fractions of type 1 T helper (Th1) and type 2 T helper (Th2) cells, but high fractions of dendritic cells (DC) and M1 macrophages consistently in both cohorts (Figure 3). High angiogenesis score tumors had infiltration with a low fractions of CD8+ T cells, CD4+ memory T cells, and a high fraction of M2 macrophages in the TCGA cohort, but this result was not validated in the GSE84437 cohort. Furthermore, we investigated the correlation of the angiogenesis score with inflammation-related scores (inflammatory response, TNF-α signaling, IL6/JAK/STAT3 signaling) and with immune response score (interferon (IFN)-γ response), which were calculated as the GSVA score (similarly to how the angiogenesis score was calculated). We found that angiogenesis was significantly and strongly correlated with inflammation-related pathways especially inflammatory response and IL6/STAT3 signaling, but not with immune response pathway (IFN-γ response) in both cohorts (Figure S3). This is consistent with the notion that inflammation promotes and immune response suppresses cancer.
Figure 3.
The angiogenesis score was associated with the fraction of immune cells in gastric cancer. Boxplots comparing immune cells including CD8+ T cells, CD4+ memory T cells, regulatory T cells, T helper cell type 1 (Th1), T helper cell type 2 (Th2) cells, M1 macrophages, M2 macrophages, and dendritic cells (DCs) by high and low angiogenesis score in TCGA and GSE84437 cohorts.
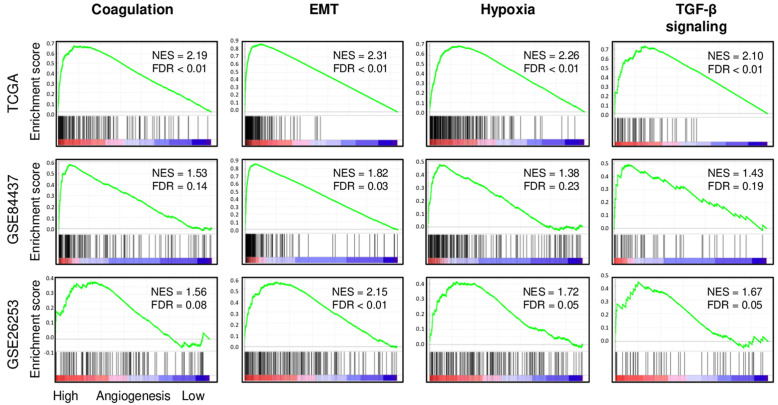
High angiogenesis score tumors significantly enriched coagulation, epithelial mesenchymal transition (EMT), hypoxia, and transforming growth factor (TGF)-β signaling gene sets
To further understand the association between abundance of angiogenesis and gastric cancer aggressiveness, Gene Set Enrichment Analysis (GSEA) was used to perform pathway analysis with Hallmark gene sets in three large cohorts (TCGA, GSE84437, and GSE26253). In all three cohorts, high angiogenesis score tumors consistently and significantly enriched coagulation, epithelial mesenchymal transition (EMT), hypoxia, and transforming growth factor (TGF)-β signaling gene sets (Figure 4). High angiogenesis score tumors also significantly enriched KEGG extracellular matrix (ECM) receptor interaction gene set consistently in all cohorts, whereas none of the Hallmark defined metabolism-related gene sets (cholesterol homeostasis, fatty acid metabolism, glycolysis, oxidative phosphorylation, and xenobiotic metabolism) were enriched to angiogenesis (Figure S4).
Figure 4.
High angiogenesis score tumor groups enriched coagulation, epithelial mesenchymal transition (EMT), hypoxia, and transforming growth factor (TGF)-β signaling gene sets in the TCGA, GSE84437, and GSE26253 cohorts. Correlation curve of coagulation, epithelial mesenchymal transition (EMT), hypoxia, and TGF-β signaling gene sets with false discovery rate (FDR) and normalized enrichment score (NES) by gene set enrichment analysis.
Aggressive American Joint committee on cancer (AJCC) stage and worse survival in gastric cancer with high angiogenesis score
As we found that angiogenesis score was related to tumor aggressiveness, we wanted to see if the abundance of intratumoral angiogenesis was also associated with more aggressive disease in gastric cancer patients. We examined the relationship between the angiogenesis score and AJCC pathological stage and T- and N-categories using gastric cancer cohorts in the TCGA (n = 375), GSE26901 (n = 109), and GSE84437 (n = 432) cohorts. As we expected, abundance of angiogenesis was associated with higher AJCC stage in both TCGA and GSE26901 cohorts (Figure 5A; P = 0.003 and P < 0.001, respectively). The angiogenesis score also correlated with the T parameter in both TCGA and GSE84437 cohorts (both P < 0.001), but not with N parameter in the TCGA cohort.
Figure 5.
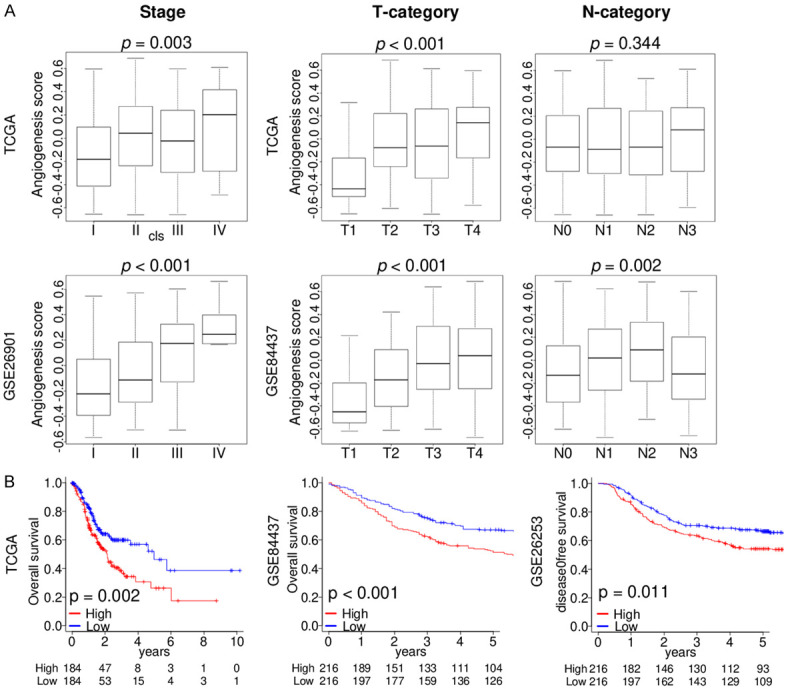
The angiogenesis score was associated with clinical cancer aggressiveness. Boxplots of the angiogenesis score by (A) AJCC stage, T- and N-category in the TCGA, GSE26901, and GSE84437 cohorts. (B) Kaplan-Meier curves of OS in the TCGA (n = 368) and GSE84437 (n = 432) cohorts, and DFS in the GSE26253 (n = 432) cohort of angiogenesis score high (red) and low (blue) in gastric cancer. AJCC, American Joint Committee on Cancer; disease-free survival, DFS; overall survival, OS.
Next, we studied how the angiogenesis score affects clinical outcome. Patients with high angiogenesis score gastric cancer had reduced overall survival in the TCGA and GSE84437 cohorts as well as worse disease-free survival in the GSE26253 (n = 432) cohort (Figure 5B; P = 0.002, P < 0.001, and P = 0.011, respectively). Our findings suggest that high compared to low angiogenesis score gastric cancer is clinically more aggressive and has worse survival.
Discussion
In this study, we used the angiogenesis score to study its association with clinical aggressiveness and as a potential biomarker in gastric cancer. Gastric cancer with high score highly expressed of multiple angiogenesis-related genes including VEGF-related, endothelial cell marker-related, and vascular stability-related genes. Furthermore, the angiogenesis score was moderately correlated with not only vessel-related stromal cells, but also fibroblasts and chondrocytes in the TME. Gastric cancer with high angiogenesis score was significantly associated with a lower infiltration of Th1, Th2 cells, and DCs, and a higher infiltration of M1 macrophages compared with low angiogenesis score tumors. Gastric cancer with a high angiogenesis score was enriched with coagulation, epithelial mesenchymal transition (EMT), hypoxia, and TGF-β signaling gene sets. Interestingly, a high angiogenesis score was significantly associated with a higher AJCC stage and T-category, but not N-category, and also associated with shorter survival.
To quantify the activity of angiogenesis pathway, we defined the angiogenesis pathway score based on the gene set variation analysis (GSVA) [19] score of the Molecular Signatures Database (MSigDB) collection [20], similar to how we previously did in breast cancer [12,36,37]. The hallmark gene sets were determined by Liberzon et al. in Cell systems [20] using computational biological methods that identified overlaps between the other gene sets in the MSigDB and retained genes that display coordinate expression. The hallmark gene sets are widely accepted and recognized as representing specific and representing specific and distinct biological functions and exhibiting consistent expression including angiogenesis. The Hallmark angiogenesis gene set is constructed of 36 genes. Biological processes such as angiogenesis is regulated by multiple genes acting in concert. Hence, using multiple genes in a shared pathway to score gene expression is a more comprehensive approach than using individual genes. Employing gene sets allows consideration of gene regulation, decreased model complexity, and greater explanatory power of prediction models. For these reasons, results were obtained for cancer biology such as survival and gene enrichment analysis in independently different cohorts, although the relationship between expressions of certain single genes, such as VEGFA and VGFR3, and angiogenesis score was not always consistent between cohorts.
Increased deposition and remodeling of the extracellular matrix with collagen crosslinking drive fibrosis in gastric cancer. Fibrosis leads to stromal stiffening which in turn promotes tumor growth, migration, and mesenchymal transition [38,39]. A dense extracellular matrix leads to poor vascularization and induces hypoxia which hinders drug delivery. In addition, a stiffer stroma and abundance of fibrosis is associated with more aggressive tumor and worse prognosis. In agreement, we found strong correlation between angiogenesis and fibroblast-related genes (αSMA (ACTA2) and FAPα (FAP)). Angiogenesis occurs when tumor and normal cells produce an imbalance of positive and negative angiogenic factors into the TME [30]. Various angiogenic factors, including VEGF and EGF, are released by cancer and stromal cells in gastric cancers [8,9]. Hence tumors with increased tissue fibrosis and stromal stiffness are more aggressive and linked with worse prognosis [40].
We demonstrated that the intratumoral angiogenesis score is correlated with fraction of fibroblasts and chondrocytes using the gene expression profile of patient cohorts. Therefore, their mechanistic roles in gastric cancer need to be elucidated by in vitro or in vivo studies.
In this study, we demonstrated that the high angiogenesis score tumor was enriched with coagulation, hypoxia, EMT, and TGF-β signaling. Our analyses did not provide information on causality; however, we speculate that elevated coagulation levels may worsen hypoxia in TME and subsequently promote angiogenesis. On the other hand, our results may implicate that high angiogenesis gastric cancer has enhanced EMT and TGF-β signaling, which are both known to worsen cancer progression and metastasis [41]. Weidner et al. [42] reported that the number and density of blood vessels correlated with metastatic frequency in invasive breast cancer. Similar observations have been obtained for gastrointestinal tumors [43,44]. We previously reported that in breast cancer, the angiogenesis score was not associated with survival [14]; however, in this study, we found that gastric cancer patients with a high angiogenesis score had worse OS and DFS. This may demonstrate the differences in clinical relevance of angiogenesis across cancer types.
Both TIME and tumor-associated macrophages (TAMs) play important roles in gastric cancer progression [45]. TAMs arise from monocytes circulating in the TME in response to chemoattractants. TAMs interact with cancer cells and play a major role in angiogenesis and tumor vascularity [46] and is thus associated with gastric cancer progression as related to depth of tumor invasion, metastasis to lymph nodes and clinical stage [47]. In this study, the fraction of infiltrated cells in the TME was quantified using the xCell algorithm. xCell algorithm allows the estimation of 64 types of immune and stromal cells in the TME from global gene expression data and is widely used throughout the world. We used the algorithm to demonstrate the clinical relevance of CD8+ T cells [35], regulatory T cells [29], and dendritic cells [31] in breast cancer. In this study, M1 macrophages were highly infiltrated in tumors with high angiogenesis score, whereas M2 macrophages were not. M1 macrophages are usually associated with anti-cancer activities, which is somewhat puzzling given our finding indicating worse survival in high angiogenesis gastric cancer. We believe this seemingly contradictory result is due to how M1 macrophage has been defined transcriptomically. In our current study, we used xCell algorithm to estimate the fraction of immune cells including M1 macrophages. In our previous study in breast cancer patients, we reported that high infiltration of M1 macrophages defined by xCell was associated with increased cancer aggressiveness and worse survival [48]. In other words, M1 macrophages defined by xCell are not associated with better survival. Although xCell algorithm is a useful tool, its limitation is that it can only compare scores between samples within the same cell type, but not between cell types due to the way it is standardized. To this end, the number of different types of cells including macrophages cannot be compared using xCell.
Furthermore, we found that the angiogenesis score was highly correlated with inflammation-related scores, including inflammatory response, TNF-α signaling, and IL6/JAK/STAT3 signaling score, but not with IFN-γ response score that reflects immune response pathway. This is consistent with the notion that angiogenesis is associated with inflammation that promotes cancer progression and not with immune response that suppresses it; thus angiogenesis is associated with poor prognosis.
Robust use of sequencing technology and mandated storage of genomic information in the public domain allow for a massive number of translational studies by many investigators including our group. Indeed, research on cancer-associated stromal cells as well as immune cells using transcriptome of a bulky tumor has become common [28,49,50]. In this current study, we demonstrated several associations between angiogenesis and clinical relevance as well as biological features. We consistently found that the angiogenesis score was a negative prognostic biomarker for gastric cancer survival in multiple large patient cohorts.
There are several limitations to this study. First, this is a retrospective study using multiple large cohorts, and the angiogenesis pathway score cannot be rigorously established as a prognostic biomarker without a prospective study. Second, detailed information on systemic treatment, which would have affected survival outcomes, was unavailable for all analyzed cohorts. Third, our study lacked experimental data demonstrating the mechanisms in which tumor angiogenesis contributes to tumor features and outcomes in gastric cancer.
In conclusion, the angiogenesis score can be a useful tool to assess intratumoral angiogenesis, which has clinical consequences in gastric cancer patients.
Acknowledgements
This work was supported by US National Institutes of Health/National Cancer Institute grant R01CA160688, R01CA250412, R37CA248018, US Department of Defense BCRP grant W81XWH-19-1-0674, as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to K.T., Grant-in-Aid for Scientific Research grants 19H03714 and 18K19576 to M.N., and US National Cancer Institute cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 2.Songun I, Putter H, Kranenbarg EM, Sasako M, van de Velde CJ. Surgical treatment of gastric cancer: 15-year follow-up results of the randomised nationwide Dutch D1D2 trial. Lancet Oncol. 2010;11:439–449. doi: 10.1016/S1470-2045(10)70070-X. [DOI] [PubMed] [Google Scholar]
- 3.Yao F, Wang Q, Wu Q. The prognostic value and mechanisms of lncRNA UCA1 in human cancer. Cancer Manag Res. 2019;11:7685–7696. doi: 10.2147/CMAR.S200436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Wang JW, Ren JY, Guo M, Guo CW, Ning SW, Yu S. Long noncoding RNAs in gastric cancer: from molecular dissection to clinical application. World J Gastroenterol. 2020;26:3401–3412. doi: 10.3748/wjg.v26.i24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Takabe K, Spiegel S. Export of sphingosine-1-phosphate and cancer progression. J Lipid Res. 2014;55:1839–1846. doi: 10.1194/jlr.R046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Y, Fu Y, Xie Q, Zhu B, Wang J, Zhang B. Anti-angiogenic agents in combination with immune checkpoint inhibitors: a promising strategy for cancer treatment. Front Immunol. 2020;11:1956. doi: 10.3389/fimmu.2020.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitadai Y, Haruma K, Sumii K, Yamamoto S, Ue T, Yokozaki H, Yasui W, Ohmoto Y, Kajiyama G, Fidler IJ, Tahara E. Expression of interleukin-8 correlates with vascularity in human gastric carcinomas. Am J Pathol. 1998;152:93–100. [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi Y, Cleary KR, Mai M, Kitadai Y, Bucana CD, Ellis LM. Significance of vessel count and vascular endothelial growth factor and its receptor (KDR) in intestinal-type gastric cancer. Clin Cancer Res. 1996;2:1679–1684. [PubMed] [Google Scholar]
- 10.Katsuta E, Qi Q, Peng X, Hochwald SN, Yan L, Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci Rep. 2019;9:1310. doi: 10.1038/s41598-018-37909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10:897–907. [PMC free article] [PubMed] [Google Scholar]
- 12.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. [Google Scholar]
- 13.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21:6708. doi: 10.3390/ijms21186708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, Park YS, Lim HY, Sohn TS, Bae JM, Choi MG, Lim DH, Min BH, Lee JH, Rhee PL, Kim JJ, Choi DI, Tan IB, Das K, Tan P, Jung SH, Kang WK, Kim S. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One. 2014;9:e90133. doi: 10.1371/journal.pone.0090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE, Park KC, Lee SH, Park JL, Park YY, Lee HS, Jang HJ, Park ES, Kim SC, Heo J, Chu IS, Jang YJ, Mok YJ, Jung W, Kim BH, Kim A, Cho JY, Lim JY, Hayashi Y, Song S, Elimova E, Estralla JS, Lee JH, Bhutani MS, Lu Y, Liu W, Lee J, Kang WK, Kim S, Noh SH, Mills GB, Kim SY, Ajani JA, Lee JS. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun. 2018;9:1777. doi: 10.1038/s41467-018-04179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon SJ, Park J, Shin Y, Choi Y, Park SW, Kang SG, Son HY, Huh YM. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC Cancer. 2020;20:314. doi: 10.1186/s12885-020-06814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, Yan L, Takabe K. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20:2655. doi: 10.3390/ijms20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20:4197. doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of microRNA-143 is associated with favorable tumor immune microenvironment and better survival in estrogen receptor positive breast cancer. Int J Mol Sci. 2020;21:3213. doi: 10.3390/ijms21093213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi H, Oshi M, Asaoka M, Yan L, Endo I, Takabe K. Molecular biological features of nottingham histological grade 3 breast cancers. Ann Surg Oncol. 2020;27:4475–4485. doi: 10.1245/s10434-020-08608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chouliaras K, Tokumaru Y, Asaoka M, Oshi M, Attwood KM, Yoshida K, Ishikawa T, Takabe K. Prevalence and clinical relevance of tumor-associated tissue eosinophilia (TATE) in breast cancer. Surgery. 2020:S0039-6060(20)30525-0. doi: 10.1016/j.surg.2020.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitadai Y. Cancer-stromal cell interaction and tumor angiogenesis in gastric cancer. Cancer Microenviron. 2010;3:109–116. doi: 10.1007/s12307-009-0032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21:5744. doi: 10.3390/ijms21165744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokumaru Y, Katsuta E, Oshi M, Sporn JC, Yan L, Le L, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of miR-34a associated with less aggressive cancer biology but not with survival in breast cancer. Int J Mol Sci. 2020;21:3045. doi: 10.3390/ijms21093045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandhi S, Elkhanany A, Oshi M, Dai T, Opyrchal M, Mohammadpour H, Repasky EA, Takabe K. Contribution of immune cells to glucocorticoid receptor expression in breast cancer. Int J Mol Sci. 2020;21:4635. doi: 10.3390/ijms21134635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 39.De Wever O, Mareel M. Role of tissue stroma in cancer cell invasion. J Pathol. 2003;200:429–447. doi: 10.1002/path.1398. [DOI] [PubMed] [Google Scholar]
- 40.Piersma B, Hayward MK, Weaver VM. Fibrosis and cancer: a strained relationship. Biochim Biophys Acta Rev Cancer. 2020;1873:188356. doi: 10.1016/j.bbcan.2020.188356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hapke RY, Haake SM. Hypoxia-induced epithelial to mesenchymal transition in cancer. Cancer Lett. 2020;487:10–20. doi: 10.1016/j.canlet.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis--correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 43.Maeda K, Chung YS, Ogawa Y, Takatsuka S, Kang SM, Ogawa M, Sawada T, Sowa M. Prognostic value of vascular endothelial growth factor expression in gastric carcinoma. Cancer. 1996;77:858–863. doi: 10.1002/(sici)1097-0142(19960301)77:5<858::aid-cncr8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi Y, Kitadai Y, Bucana CD, Cleary KR, Ellis LM. Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res. 1995;55:3964–3968. [PubMed] [Google Scholar]
- 45.Fan X, Jin J, Yan L, Liu L, Li Q, Xu Y. The impaired anti-tumoral effect of immune surveillance cells in the immune microenvironment of gastric cancer. Clin Immunol. 2020;219:108551. doi: 10.1016/j.clim.2020.108551. [DOI] [PubMed] [Google Scholar]
- 46.Ohta M, Kitadai Y, Tanaka S, Yoshihara M, Yasui W, Mukaida N, Haruma K, Chayama K. Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human gastric carcinomas. Int J Oncol. 2003;22:773–778. [PubMed] [Google Scholar]
- 47.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Okumura H, Matsumoto M, Miyazono F, Hokita S, Aikou T. Tumor-associated macrophage (TAM) infiltration in gastric cancer. Anticancer Res. 2003;23:4079–4083. [PubMed] [Google Scholar]
- 48.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, Guo AY. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) 2020;7:1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.