Radionuclide polluted environments harbor microbial species highly tolerant to these elements through mechanisms like biosorption, biotransformation, biomineralization and intracellular accumulation. The microbial‐ radionuclide interaction processes have a great potential for biotechnological applications. This review provides the state‐ of‐ the‐ art of all aspects of these investigations.

Summary
Radionuclides (RNs) generated by nuclear and civil industries are released in natural ecosystems and may have a hazardous impact on human health and the environment. RN‐polluted environments harbour different microbial species that become highly tolerant of these elements through mechanisms including biosorption, biotransformation, biomineralization and intracellular accumulation. Such microbial–RN interaction processes hold biotechnological potential for the design of bioremediation strategies to deal with several contamination problems. This paper, with its multidisciplinary approach, provides a state‐of‐the‐art review of most research endeavours aimed to elucidate how microbes deal with radionuclides and how they tolerate ionizing radiations. In addition, the most recent findings related to new biotechnological applications of microbes in the bioremediation of radionuclides and in the long‐term disposal of nuclear wastes are described and discussed.
Introduction: Radionuclides: natural/anthropogenic sources and environmental impact
Anthropogenic activities related to mining of uranium (U), atmospheric nuclear weapon tests and nuclear industry are the main sources of radionuclides in the environment (Hain et al., 2020). Radionuclides such as uranium (U), plutonium (Pu) or curium (Cm) have an adverse impact on human health and the environment. These effects are closely tied to their mobility and bioavailability, which in turn strongly depend on their speciation and physicochemical form.
Uranium is a naturally occurring radionuclide whose levels of contamination are associated with mining activities, the weathering of uranium‐containing minerals or accidental local release (Lloyd and Macaskie, 2000; Meinrath et al., 2003). The environmental impact of U is related to its chemistry (Burns 1999). Uranium occurs in four oxidation states: U(III), U(IV), U(V) and U(VI). While U(VI), the soluble, mobile and toxic U species, occurs mainly under oxic conditions, U(IV), which is insoluble, immobile and a less toxic U species, is distributed mainly under reducing conditions (Burns 1999). Plutonium is the naturally occurring chemical element with the highest atomic number (Burns 1999). Trace amounts of natural Pu are present in natural 238U deposits. Yet, the main anthropogenic sources of Pu include nuclear weapon testing (Sholkovitz 1983), accidental release (Zheng et al., 2012) and discharges from nuclear fuel reprocessing sites or nuclear power plants (Dai et al., 2005). Anthropogenic radionuclides 239Pu and 240Pu have high radiological toxicity and can persist for a long time in the environment. Curium and americium are trivalent actinides, considered as minor actinides due to their low environmental concentration, and are produced by bombarding 239Pu with neutrons and alpha particles in nuclear plants. Cm(III) can be used as a molecular probe to mimic the chemical speciation and behaviour of trivalent actinides such as 241Am due to its excellent fluorescence properties (Edelstein et al., 2006). 241Am concentrations in contaminated areas range from 4 to 10.004 Bq/kg soil (Thakur and Ward, 2019). These concentrations correspond to 1.3 x 10‐7 to 3.3 x 10‐4 µM AmIII.
The Chernobyl nuclear power plant accident in 1986 was responsible for an environmental accumulation of about 1.5 x 1014 Bq of 241Am through the decay of approximately 6x1015 Bq of 241Pu released from this plant (Thakur and Ward, 2019). This corresponds to 1181 g of 241Am and illustrates the dire need for knowledge of the behaviour of trivalent actinides in biological systems.
In the last decade, different studies were focused in analysing the structure and composition of microbial populations in radionuclide‐containing environments and to elucidate the mechanisms by which these microbes interact with these elements. In addition, the application of these studies in the field of bioremediation was largely investigated. However, few works addressed the new applications of these mechanisms in the field of nuclear industry (e.g. safety of future deep geological disposal of radioactive wastes) and in bioelectrochemical device‐based bioremediation. The present review provides insights on microbial diversity of radionuclide‐polluted environments, the mechanisms involved in microbial tolerance to radiation and interaction with radionuclides. In addition, new application of microbe–radionuclide interactions is highlighted.
Microbial diversity and activity in radionuclide‐contaminated sites
The microbial community is a sensitive indicator of environmental stress, reflecting even small changes in the geochemical composition of their microhabitat due to anthropogenic activities (Li et al., 2017; Guillot et al., 2019; Hallsworth 2019; Xiao et al., 2019). Microorganisms play a major role in element cycling as well as the weathering of rocks and sediments (Bennett et al., 2001; Jaswal et al., 2019a). They may also affect the geochemical properties of groundwater by modifying the transport of organic and inorganic contaminants (Stegen et al., 2016; Jaswal et al., 2019b).
Interest in the biodiversity and activity of microorganisms inhabiting radionuclide‐contaminated sites has increased significantly over the past 25 years. Such settings are dominated by diverse groups of microorganisms whose structure and function determine their dynamics and interactions with biotic and abiotic components (Narendrula‐Kotha and Nkongolo, 2017; Xiao et al., 2019). Culture‐independent studies provide new insights into the microbiology of radionuclides (Carlson et al., 2019; Xiao et al., 2019). To date, numerous metagenomic studies have demonstrated that contamination – including that by radionuclides – heavily impacts the structure and functions of bacterial communities (Fan et al., 2018; Hemme et al., 2010, 2015; Narendrula‐Kotha and Nkongolo, 2017; Xiao et al., 2019, Yan et al., 2016). Because radionuclides can have complex impacts on the microbial community structure, understanding the mechanisms underlying their impact will lead to a better management of microorganisms for bioremediation.
In the past few decades, the microbial diversity and activity in radionuclide‐contaminated sites have been studied exploring mine tailings, mine wastes, mining‐impacted sites and other contaminated environments containing radionuclides (Islam and Sar, 2011; Kumar et al., 2013; Radeva et al., 2013; Yan et al., 2016; Narendrula‐Kotha and Nkongolo, 2017); the composition of the bacterial communities is now known to be site‐specific and connected to concentration levels and/or different geological and physiological environmental conditions. For instance, Acidithiobacillus, Pseudomonas, Acinetobacter and Nitrosomonas were found to be abundant in various U mine waste sites in Germany (Radeva and Selenska‐Pobell, 2005; Yi and Yao, 2012), while highly abundant Sphingomonas, Acidovorax, Acinetobacter and Ralstonia may be linked to U‐contaminated radioactive waste (Akob et al., 2007; Yan et al., 2016, Jaswal et al., 2019a). Abundant and active Proteobacteria (Alpha‐, Beta‐, Delta‐ and Gammaproteobacteria), Acidobacteria, Actinobacteria, Bacteroidetes and Firmicutes have been frequently identified in radionuclide‐contaminated environments (Hemme et al., 2010; Kumar et al., 2013; Suriya et al., 2017; Jaswal et al., 2019a,2019b).
Fungi are also known to possess the metabolic capacity to overcome high environmental stresses (Rummel et al., 2014; Stevenson et al., 2017), including high uranium concentrations. However, little is known to date about fungal diversity in radionuclide‐contaminated environments and how they may tolerate such toxicity (Mumtaz et al., 2013, Narendrula‐Kotha and Nkongolo, 2017).
As an example, Carlson et al. (2019) focused on microbial communities in a contaminated aquifer in Oak Ridge, Tennessee (USA). The conditions at this site involve large gradients of pH in addition to widely varying concentrations of uranium, nitrate and many other inorganic ions including Mn2+, Al3+, Cd2+, Zn2+, Co2+ and Ni2+ (Brooks 2001; Moon et al., 2006; Carlson et al., 2019). In general, taxa frequently associated with contaminated environments can (i) reduce nitrate and heavy metals, (ii) be resistant to heavy metal toxicity and (iii) degrade polyaromatic compounds and polychlorinated biphenyls (Akob et al., 2007). Phylotypes such as Rhodanobacter, Methylobacterium, Caulobacter, Sphingomonas, Acidovorax, Acinetobacter and Ralstonia are nitrate‐reducing bacteria that tolerate acidic and low‐nutrient environments, and are highly resistant to metals. They tend to be plentiful in the microbial communities of such contaminated sites (Spain and Krumholz, 2011; Thorgersen et al., 2019), and it may be that toxic compounds select taxa typically resistant to the high concentrations of radionuclides in contaminated environments such as those reported in the RN‐contaminated aquifer in the Oak Ridge (Carlson et al., 2019).
Likewise, Hoyos‐Hernandez et al (2019) studied the bacterial community structure and functional genes in RN‐contaminated soils at Chernobyl and Fukushima. They explored the ability of microorganisms to survive in these contaminated environments and identified Proteobacteria, Acidobacteria and Actinobacteria as the most common phyla in all studied soils. In addition, the main detected taxa were those tolerant to extreme environmental conditions such as Rhodospirillales, Acetobacteraceae and Candidatus Solibacter, as well as to metals such as Acidimicrobiales (Gołębiewski et al., 2014). Verrumicrobia were also detected and previously isolated from uranium deposits (Mondani et al., 2011; Theodorakopoulos et al., 2017). Again, these authors concluded that the presence of these microorganisms would suggest some form of selection for tolerant microorganisms to uranium and heavy metal contamination.
Similarly, the highly contaminated (with uranium and other metals) sediments of the Cauvery river (India) were found to contain abundant Proteobacteria (Gamma, Alpha, Delta, Epsilon and Beta) followed by Bacteroidetes and Firmicutes (Suriya et al., 2017). In addition to confirming the impact of uranium and metal accumulation on the bacterial communities in RN‐contaminated sites, these authors highlighted specific species as bioindicators of contamination and as candidates for bioremediation strategies.
In general, bacteria are sensitive to environmental contamination, and their response to heavy metals varies. Proteobacteria, the predominant phylum common to all RN‐contaminated sites, harbour many U(VI)‐reducing genera and play a crucial role in decreasing U toxicity and bioavailability (Barns et al., 2007; Rastogi et al., 2010; Merroun et al., 2011; Kolhe et al., 2018). They can survive in oligotrophic environments, function at low water activity, adapt to metal reduction and metal resistance, and play a very important role in U immobilization in biostimulated U‐contaminated sediments or groundwater (Suzuki et al., 2003; North et al., 2004; Li et al., 2017). Members of the genera Thiobacillus (beta), Ferribacterium (beta), Pantoea (gamma), Geobacter (delta) and Pseudomonas (gamma), to name a few, are frequently detected. Others, such as Desulfomicrobium, Desulfotomaculum, Desulfovibrio, Acidovorax and Shewanella, known to reduce or immobilize U, have been detected in many RN‐contaminated sediments/soils (Rastogi et al., 2010; Spain and Krumholz, 2011). Belonging to Proteobacteria, members of the families Geobacteraceae and Shewanellaceae are able to reduce U(VI), iron(III), nitrate and elemental sulfur with electron donors such as alcohol, acetate and aromatic compounds (Richter et al., 2012; Lovley 2013). Geobacteraceae became numerically prevalent over other groups as U reduction proceeded in uranium‐contaminated sites (Lovley et al., 2011). Sulfate‐reducing bacteria such as Desulfomicrobium, Desulfotomaculum and Desulfovibrio are known to reduce U to a less soluble uraninite. The nearly ubiquitous Desulfovibrio involves the reduction in many electron acceptors (such as sulfate, nitrate and nitrite) for growth and respiration with the enzymatic reduction in chromium(VI), manganese(IV), iron(III), U(VI) and technetium(VII) (Wall and Krumholz, 2006; Zeng et al., 2019).
Acidobacteria are also frequently retrieved from radionuclide‐contaminated soils (Barns et al., 2007; Mumtaz et al., 2018). They are known to tolerate extreme conditions, including metal contamination, acidic pH and limited nutrients (Kielak et al., 2016). Among the known genera of this phylum, Geothix, capable of nitrate reduction, were found to be largely associated with Fe(III) and indirectly to U(VI) reduction under field conditions (Cardenas et al., 2008; Li et al., 2018). Bacteroidetes are known for their ecological importance as primary metabolizers of dead organic matter (Thomas et al., 2011). Their resistance to high levels of radionuclide contamination may be due to the involvement of classes such as Sphingobacteria that produce sphingolipids, signalling molecules that protect cell surfaces and functions in the face of a variety of environmental stresses (Hannun and Obeid, 2008; Mumtaz et al., 2018).
Actinobacteria also exist in uranium‐contaminated sediments and soils (Akob et al., 2007; Akob et al., 2011, Jaswal et al., 2019a); among them, the family Cellulomonadaceae (Cellulomonas) is known to reduce/immobilize U(VI) by means of phosphate precipitation and reduction based on environmental conditions (Sivaswamy et al., 2011). Arthrobacter and Microbacterium have likewise been reported in many radionuclide‐contaminated areas and are known to remove high amounts of U in heavy metal‐enriched environments (Islam and Sar, 2016 ). Moreover, Arthrobacter is well adapted to nutrient‐deprived (oligotrophic) conditions (Osman et al., 2008 ). Besides being one of the candidates used to resolve the molecular basis for resistance against uranium (Chauhan et al., 2018), Arthrobacter can account for about a third of the total soil cultivable bacterial community in highly radioactive sediments (Fredrickson et al., 2004 ).
Tolerance of microbes to radiation
In addition to their chemical toxicity, active species of radionuclides such as Pu, Np and Cm exhibit also radiotoxicity, which should be considered as one of the main criteria for screening of microbes with bioremediation potential of aqueous radioactive wastes generated by nuclear fuel reprocessing.
It is well documented that ionizing radiations lead to degradation of DNA through radiolysis or loss of electron from its structure (Ravanat and Douki, 2016) with consequent inhibition of microbe reproduction (IAEA 2017). Some microbes inhabiting radionuclides impacted environments developed different mechanisms for their survival and tolerance to ionizing radiation. For instance, Deinococcus radiodurans, the most radiation‐resistant microbes presently known, tolerate up to 15 ,000 Gy of acute ionizing radiation and 60 Gy h‐1 of chronic radiation (Daly 2006). The highly efficient radiation–protection mechanisms of Deinococcus are mediated by a combination of passive and active defence processes, (i) efficient repair of disintegrated DNA including self‐repair of DNA damage (Krisko and Radman, 2013), (ii) efficient cellular damage clearance mechanisms through hydrolysis of damaged proteins, overexpression of repair proteins, etc., and (iii) effective elimination of reactive oxygen species (ROS) (Jin et al., 2019).
Paterson‐Beedle et al. (2006) reported that members of genus Serratia tolerate high levels of Cs137, Sr90 and Co60. In addition, cells of Serratia sp. were described for their ability to precipitate high levels of hydrogen uranyl phosphate (HUP) via the activity of a highly radiostable phosphatase enzyme. The radiostability of this enzyme was increased to 100% by the incorporation of radioprotectant such as mercaptoethanol (Paterson‐Beedle et al., 2012). Malo and Ddachova (2019) reported that some fungi used melanin as radioprotector for their survival in extreme environments with high levels of ionizing radiation such as the damaged nuclear reactor at Chernobyl. In addition, these Chernobyl‐associated fungi are able to harvest usable energy from forms of ionizing radiation, phenomenon termed as radiotrophism (Dadachova and Casadevall, 2008).
Microbial interactions with radionuclides
Microorganisms are able to carry out several metabolic processes influencing the cycling of organic and inorganic species (Falkowski et al., 2008). Under certain conditions, microbially driven processes are used for biotechnological applications (Lloyd 2003). The different microbial–radionuclide interaction mechanisms include biomineralization, biosorption, biotransformation and intracellular accumulation. However, it is necessary to point out that radionuclides exhibit highly microbial species‐specific behaviour in their interactions with microorganisms (Brookshaw et al., 2012).
Biomineralization is the process by which microorganisms transform ions in solution into solid minerals as the result of cellular activities (Simkiss and Wilbur, 2012). This process is an efficient way to sequester radionuclides within relatively stable solid phases. Three different types of biomineralization processes have been described: biologically induced biomineralization, which results from indirect modification of chemical conditions such as a pH shift or redox transformations in the environment; biologically influenced biomineralization, defined as passive mineral precipitation in the presence of organic matter, such as cell surfaces or extracellular polymeric substances, whose properties influence crystal morphology and composition; and biologically controlled biomineralization, specific cellular activity that directs the nucleation, growth, morphology and final location of a mineral (Benzerara et al., 2011). Oxides, phosphates, sulfides and oxalates are the most commonly precipitated biominerals, having high metal sorption capacities and redox catalysis due to their chemical properties (Gadd and Pan, 2016), while radionuclide carbonates and hydroxides are precipitated by localized alkalinization at the cell surface (Newsome et al., 2014). U biomineralization processes have been extensively studied. They are generally based on passive U sorption by the negatively charged cell wall extracellular polymers and active secretion of phosphate groups owing to phosphatase activity, under acidic and neutral conditions (Martinez et al., 2007; Merroun and Selenska‐Pobell, 2008). Some examples of bacterial U biomineralization include Pseudomonas aeruginosa (Choudhary and Sar, 2011), Bacillus and Sphingomonas (Merroun et al., 2011; Lopez‐Fernandez et al., 2014), Gallionella (Krawczyk‐Bärsch et al., 2020) or Staphylococcus aureus (Shukla et al., 2020). Moreover, the bacterial strain Stenotrophomonas sp. Br8 mediates U(VI) immobilization under changing environmental conditions due to phosphatase enzymes (Sánchez‐Castro et al., 2020; Fig. 1). In the case of Serratia sp., biomineralization entails the activity of both alkaline and acidic phosphatases (Newsome et al., 2015; Chandwadkar et al., 2018), as well as an indigenous bentonite microbial population (Povedano‐Priego et al., 2019). Moreover, fungal U biomineralization has also been described by Aspergillus niger and Paecilomyces javanicus (Liang et al., 2015) or Saccharomyces cerevisiae (Zheng et al., 2018). In addition, phosphate mineral formation by several yeast strains may involve an organic source of phosphorus such as glycerol 2‐phosphate or phytic acid, in the presence of soluble U (Liang et al., 2016). Biomineralization processes with other radionuclides, such as caesium, strontium, technetium, plutonium or neptunium, have also been described (Macaskie et al., 1994; Macaskie and Basnakova, 1998; Hallberg and Ferris, 2004; Mitchell and Ferris, 2005; Thorpe et al., 2016, Cleary et al., 2019).
Fig. 1.
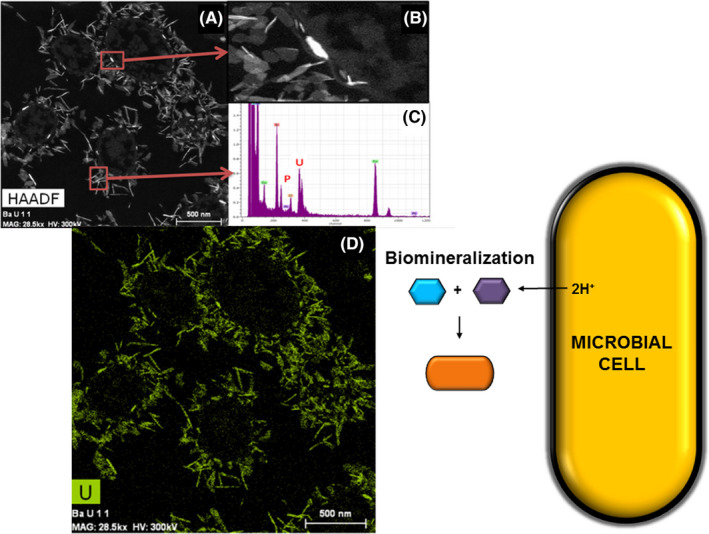
Example of uranium precipitates (1 mM) around Stenotrophomonas sp. Br8 cells from Sánchez‐Castro et al. (2020).
A. HAADF‐STEM micrograph, B. zoomed‐in view and C. EDXS spectrum of the points marked with a red square in A, and D. EDX element distribution map for U, plus schematic representation of the bioprecipitation mechanism.
Biosorption describes the metabolism‐independent sorption of radionuclides to biomass (Newsome et al., 2014). It is a complicated process influenced by parameters such as the status of the biomass (living or not living), properties of radionuclide solution chemistry or pH (Das 2012). Concretely, the biosorption of metal ions by living cells is a two‐step process. First, radionuclides accumulate at the cell surface through physical adsorption or ion exchange, and then penetrate the cell membrane of atoms or molecules of one phase, forming a solution with the second phase, mainly through chemical functional groups such as carboxyl, amine, hydroxyl, phosphate and sulfhydryl groups (Lloyd and Macaskie, 2000; Goyal et al., 2003; Newsome et al., 2014). Differences in cell wall composition among microorganisms lead to significant differences in the type and amount of metal ion binding to them (Goyal et al., 2003). U biosorption has been extensively studied in bacteria (Hu et al., 1996; Nakajima and Tsuruta, 2004; Merroun et al., 2005), fungi (Khani et al. 2005; Lopez‐Fernandez et al., 2018; Chen et al., 2020) and even in lichens (Purvis et al., 2004). An early study reported U uptake from aqueous solutions by Rhizopus arrhizus (Tsezos and Volesky, 1982). Further findings entail plutonium, americium and cerium sorption from aqueous solutions at pH 1‐2 by immobilized Saccharomyces cerevisiae (Kedari et al., 2001); the reversible and pH‐dependent biosorption of curium by yeast Rhodotorula mucilaginosa (Lopez‐Fernandez et al., 2019, Fig. 2); or strontium biosorption from aqueous solutions by several microorganisms (Ilyas et al., 2020). Moreover, caesium sorption has been described in conjunction with various microorganisms. The native and chemically modified biomass of marine algae Sargassum glaucescens and Cystoseira indica might be used as caesium biosorbents (Jalali‐Rad et al., 2004; Dabbagh et al., 2008), while Pseudomonas fluorescens and Bacillus cereus have caesium biosorption capacity (Mao et al., 2011; Kim et al., 2016). However, a recent review addressing the radiocaesium soil‐to‐plant transfer indicates that although mycorrhizal fungi can play a role in the biosorption of radiocaesium, they may not significantly contribute to its uptake by plants (Almahayni et al., 2019). The marine actinobacterial Nocardiopsis sp. and immobilized Aspergillus niger have been described as strontium biosorbent (Pan et al., 2009; Sivaperumal et al., 2018), and strontium can be biosorbed from aqueous solution by Bacillus cereus (Long et al., 2017). Thorium biosorption in living and dead cells of Streptomyces sporoverrucosus is apparently dependent on pH and ionic strength (Ding et al., 2014), while neptunium can be biosorbed by whole cells, the cell wall and extracellular polymeric substances of Shewanella algae (Deo et al., 2010).
Fig. 2.
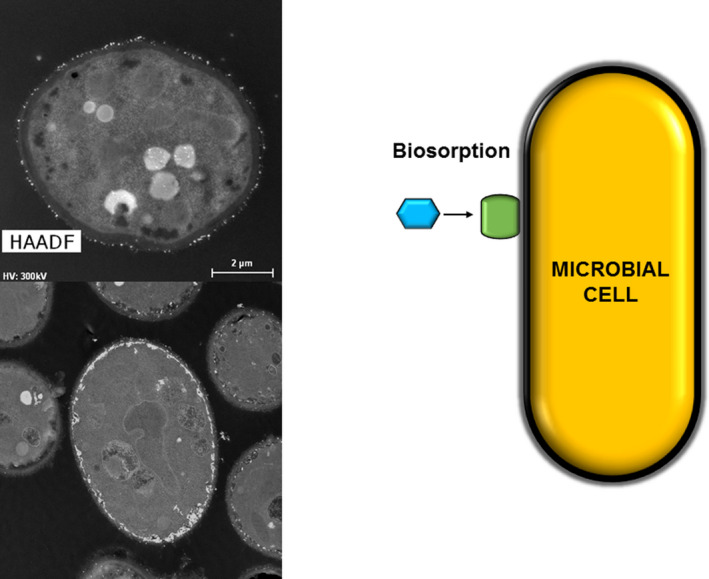
Example of curium biosorption by Rhodotorula mucilaginosa, using europium (1 mM Eu(III)) as inactive analogue of curium. High‐angle annular dark‐field images (scale bars: 2 μm) and schematic representation of the biosorption mechanism.
Biotransformations are microbial structural modifications in a chemical compound by organisms/enzyme systems that lead to the formation of molecules with relatively greater polarity (Smitha et al., 2017). These transformations, including reduction and oxidation, are useful in a wide range of biotechnological processes (Cresnar et al., 2011). A wide diversity of prokaryotes can enzymatically reduce U (Williams et al., 2013). The first case of bioreduction was described for iron reducers Geobacter metallireducens and Shewanella oneidensis, coupling their growth to the dissimilatory reduction in U (Lovley et al., 1991). Microbial cells therefore use different mechanisms to transfer electrons from the electron donor to an electron acceptor, such as c‐type cytochrome, pili, flagella or extracellular carriers to bioreduce U (Newsome et al., 2014). But not only bioreduction in this radionuclide has been described. Lloyd (2003) reviewed the microbial reduction in selenium, neptunium, plutonium and technetium. A mix of technetium and uranium was reduced by the indigenous microbial community of a nitrate‐contaminated aquifer (Istok et al., 2004). Different bacterial strains have also shown their plutonium biosorption capability, for example Sporomusa sp. cells enhanced the removal of plutonium from solution by biosorption and bioreduction (Moll et al., 2017), and the extracellular polymeric substances of Pseudomonas sp. are an effective reductant and sorbent for plutonium, influencing its redox cycling and mobility in the environment (Boggs et al., 2016). Plutonium polymers were found to bind to Desulfovibrio äspöensis biomass during the first 24 h of contact time; yet, most of the reduced plutonium dissolved from the cell envelope went back to the aqueous solution due to the weak complexing properties of this plutonium oxidation state (Moll et al., 2006). Finally, the mobility of selenium within the deep geological radioactive waste repositories was also studied, relating the bioreduction in this element in anaerobic bentonite samples using acetate as electron donor (Ruiz‐Fresneda et al., 2019, Fig. 3) and the biotransformation of amorphous selenium nanospheres to trigonal 1D nanostructures (Ruiz Fresneda et al., 2018).
Fig. 3.
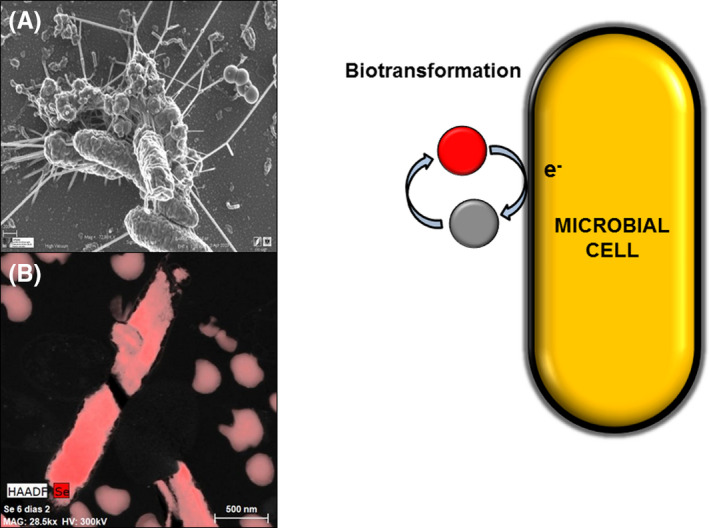
Example of selenium biotransformation by Stenotrophomonas bentonitica (2 mM Se(IV)) under anaerobic conditions.
A. VP‐SEM image showing the Se nanoparticles (scale bar: 200 nm), B. HAADF‐STEM micrograph and element distribution map of Se nanoparticles (scale bar: 500 nm) and schematic representation of the biotransformation mechanism.
Intracellular accumulation is considered to be a metabolism‐independent process. Microbial cells are able to immobilize radionuclides via different bioaccumulation mechanisms (Merroun and Selenska‐Pobell, 2008). This uptake may occur because the transported metals are similar to essential elements needed for cell functioning, so are actively taken up into the cell (Newsome et al., 2014); or due to increased membrane permeability caused for example by U toxicity (Suzuki and Banfield, 1999). Several studies describe the intracellular accumulation of U by bacteria and fungi (Fomina et al., 2007; Choudhary and Sar, 2011; Lee and Hur, 2014; Zhang et al., 2014; Gerber et al., 2018, Fig. 4). One in‐depth proteogenomic study revealed the complex cellular response to uranium in Microbacterium oleivorans, perturbing the phosphate and iron metabolic pathways by means of several transporters specifically associated with uranyl stress (Gallois et al., 2018). In addition, caesium accumulation has been demonstrated at the cell surface, and in cytoplasm and mycelia (Kato et al., 2000; Kuwahara et al., 2011; Dräxl et al., 2013; Sivaperumal et al., 2018). Further examples are the accumulation of strontium into biogenic carbonate minerals by Bacillus sp. (Horiike et al., 2017) or into calcium carbonates by cyanobacteria (Mehta et al., 2019). The bacterial accumulation of technetium (Sierra et al., 2008) or plutonium by the iron reducers Geobacter sulfurreducens and Shewanella oneidensis (Renshaw et al., 2009) has been documented, as well as the sorption of plutonium by bacteria and fungi, accumulating this radionuclide intracellularly (Lujanienė et al., 2017).
Fig. 4.
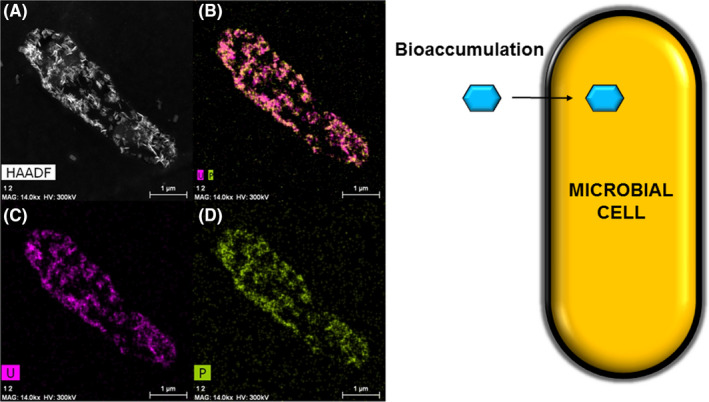
Example of intracellular accumulation of uranium (0.1 mM U(VI)) by Rhodosporidium toruloides as needle‐like structure localized at the inner cytoplasm membrane.
A–D. Micrograph and distribution analysis of uranium (purple) and phosphorus (green) from Gerber et al. (2018) and schematic representation of the bioaccumulation mechanism
Microscopic/spectroscopic/omics‐based methodologies used to study the interaction of radionuclides with microbes
A wide variety of methodologies are usually employed to characterize the physicochemical properties – morphology, size, structure, local coordination, elemental composition, chemical speciation, oxidation state, etc. – of microbial interaction products with radionuclides. The application of complementary techniques including microscopy, spectroscopy and synchrotron‐based techniques may successfully determine the microbial processes involved in these interactions. In recent years, powerful new omic‐based methodologies are revolutionizing the nature of biomolecular studies in the field of microbial–radionuclide interactions.
Fourier transform infrared (FTIR) spectroscopy
Infrared (IR) spectroscopy uses the absorption of IR radiation by the molecular bonds to identify the bond types that can absorb energy by oscillating, vibrating and rotating (Mattox 2010). An IR spectrum of a sample is generated by scanning the intensity of IR radiation before and after passage of the IR beam through the sample. The IR spectra of most materials comprise a large number of absorption bands, which originate from the interaction between discrete light quanta and mechanical motions (vibrational and rotational modes) of the molecules that are excited by the absorption of IR radiation (Naumann 2006). In FTIR spectrometers, the interference patterns of the modulated signals from interferograms are amplified, digitized, computer‐stored and transformed into a spectrum by the fast Fourier transform (FFT) algorithm (Naumann 2006; Beekes et al., 2007).
This technique is usually employed in microbiology to study the nature of functional groups from bacterial cells involved in the interaction with radionuclides and other toxic elements. IR absorption bands observed in the region between approximately 800 and 4000 cm‐1 can often be assigned to particular functional groups (Naumann 2006). These bands are sensitive to molecular‐level and structural changes (molecular interactions, membrane constitution, lipid–protein interaction, etc.) in the composition of the cellular macromolecules involved. Carboxyl, amide, hydroxyl and phosphoryl groups from Pantoea sp. TW18 and Nocardiopsis sp. 13H reportedly play an important role in the accumulation and adsorption of U(VI) and strontium (Sr+) respectively (Sivaperumal et al., 2018; Zhang et al., 2018a). In contrast, yet diffuse reflectance FTIR spectroscopy did not detect structural changes in the cellular macrocomponents of the interaction between 241Am and Photobacterium phosphoreum (Kamnev et al., 2013). This technique may also be used to investigate the nature of organic matter (proteins, lipids, polysaccharides, etc.) attached to biogenic nanoparticles of interest (Eswayah et al., 2019; Fischer et al., 2020; Ruiz‐Fresneda et al., 2020), which could affect their interactions and hence their environmental fate. For instance, the presence of absorption bands at 1659 and 1538 cm‐1 corresponding to C‐O, N‐H and C‐N vibrations attributed to amide I and amide II groups in purified selenium nanoparticles from Stenotrophomonas bentonitica suggested the presence of proteins surrounding them (Ruiz‐Fresneda et al., 2020).
X‐ray absorption spectroscopy (XAS)
XAS is a well‐established synchrotron‐based analytical technique used extensively to characterize the local chemical structure of diverse materials, including semiconductors in solid or liquid, crystalline or amorphous, bulk or nanoscale form (Schnohr and Ridgway, 2015). Under this technique, a sample of interest is bombarded with X‐rays of a given energy. Some of the X‐rays are absorbed by the atoms in the sample, inducing the excitation of a core electron (Calvin 2013). By stepping through a range of energies in this way, a spectrum is created. The sharp rise typical of XAS spectrum is generally called the edge. At energies below the edge, incident X‐rays do not have energy enough to excite electrons, while above the edge they do (Calvin 2013). The XAS spectrum is commonly divided into the extended X‐ray absorption fine structure (EXAFS) and the X‐ray absorption near‐edge structure (XANES) region. The EXAFS region comprises the oscillations above the edge and provides reliable structural information concerning the local, short‐range coordination environment and the chemical and electronic structure of specific sites within materials, including the number, chemical nature and distance of neighbouring atoms from the atomic site of interest (Ormerod, 2001). The XANES region includes peaks and other features near or on the edge, and basically provides information about the oxidation state of the atoms.
XAS revealed the impact of the bacterial strain Sporomusa sp. MT‐2.99, isolated from Mont Terri Opalinus Clay (selected as a reference material for engineered barriers in the deep geological repository system of radioactive waste), on Pu speciation through a reduction pathway (Moll et al., 2017). Since Pu can be released from nuclear waste disposal sites, the characterization of complex Pu–bacteria interactions is key for a better understanding of the fate of such toxic elements. The structural parameters of plutonium (number and chemical identities of near neighbour atoms and the average interatomic distances) helped to determine the crystalline and non‐crystalline nature of selenium nanoparticles produced by Methylococcus capsulatus and Methylosinus trichosporium (Eswayah et al., 2017). The first Se‐Se coordination shell at a bond distance of 2.35 Å reflected an amorphous nature of the nanoparticles, while the second Se‐Se shell at 3.69 Å suggested their crystalline structure. Similar results were obtained for selenium nanoparticles produced by S. bentonitica (Ruiz‐Fresneda et al., 2020). They found two Se‐Se coordination shell that would correspond to amorphous and trigonal Se, and suggested, with the help of other techniques, a transformation process from amorphous to trigonal selenium. In addition, the XANES region indicated the zero‐valent oxidation state of the nanoparticles and confirmed they are produced by a reduction process from Se (+IV). Meanwhile, EXAFS analysis of U‐bacteria interactions showed that Desulfovibrio vulgaris is able to reduce U(VI) to non‐crystalline U(IV) associated with cell‐bound phosphate groups, most probably from bacterial extracellular polymeric substances (EPS) (Stylo et al., 2015).
Other synchrotron‐based techniques such as X‐ray fluorescence (XRF) imaging have been employed to study the accumulation of radionuclides by plants through arbuscular mycorrhizal (AM) fungi. Several studies indicated the role of AM in the translocation of 238U, Cd and Zn into root tissues and in their limitation into other tissues of the plant as a defensive mechanism (Rufyikiri et al., 2002, 2003; Chen et al., 2008; Nayuki et al., 2014; Davies et al., 2015). Phytoaccumulation of the toxic metals Cd and Zn in plant and fungal structures has been demonstrated by XRF imaging analysis of mycorrhizal root sections of Lotus japonicum and Rhizophagus irregularis (Nayuki et al., 2014). The former work indicates the potential of synchrotron‐based imaging methodologies, in combination with XAS spectroscopy, for the analysis of other elements such as U in AM fungal–plant tissues.
Time‐resolved laser‐induced fluorescence spectroscopy (TRLFS) analysis
TRLFS is a useful technique for the identification of certain actinide (An) or lanthanide (Ln) species resulting from various biogeochemical processes (Collins et al., 2011). Fluorescent metal ions emit fluorescence emission spectra and decay lifetimes when irradiated with laser pulses. This phenomenon occurs as a consequence of their transition from electronically excited states to the ground state. Analysis of the emission spectra and lifetimes allows different chemical species produced by a fluorescent metal ion to be identified (Collins et al., 2011; Saito et al., 2015). TRLFS can be applied to studies of actinide speciation in aqueous and solid phases, or the coordination of metal ions adsorbed to mineral and bacterial surfaces (Collins et al., 2011).
TRLFS measurements can help determine the microbial functional groups involved in An/Ln–microorganism interaction. The complexation of Cm(III) with carboxylic functional groups from the archaeon Halobacterium noricense DSM15987T was observed using TRLFS (Bader et al., 2019). This technique also allowed identifying the association of phosphate species from this halophilic archaeon with Eu(III), the inactive analogue of Cm(III). Moll et al. (2020) demonstrated a stable interaction of Cm(III) with the protein S‐layer of Lysinibacillus sphaericus JG‐A12 by means of TRLFS. They indicated that S‐layer protein carboxyl sites could be involved in Cm(III) binding. The importance of investigating not only the interaction between cellular surfaces and actinides, but also cellular protein layers with these elements relies on the fact that proteins can highly influence their migration behaviour in the environment through the formation of colloids (Moll et al., 2020).
Microscopic techniques
The morphology and cellular location of interaction products can be analysed by means of transmission or scanning electron microscopy (TEM/SEM), equipped with energy dispersive X‐ray (EDX) for elemental composition analysis. Several high‐resolution microscopes – e.g. the high‐angle annular dark‐field scanning transmission electron microscope (HAADF‐STEM) – allow for a structural characterization of biologically and chemically produced materials by using selected‐area electron diffraction (SAED) and high‐resolution TEM combined with fast Fourier transform (FFT). Another useful system is variable pressure field emission scanning electron microscopy (VP‐FESEM), equipped with a Raman X‐ray detector, which made possible an in situ 3D structural and elemental characterization. Field emission gun environmental scanning electron microscopy (FEG‐ESEM), equipped with secondary and circular backscatter electron detectors such as ETD (Everhart‐Thornley detector) and CBS (concentric backscatter detector), provides high‐resolution imaging of surface morphology and topography, and elemental composition as a function of image contrast (Vernon‐Parry 2000; Zhang et al., 2016).
In the study of Boggs et al. (2016), Pu(V) and Pu(IV) were shown by TEM analysis to be associated with the cell walls of Pseudomonas sp. strain EPS‐1W. Wild‐type cells with bound extracellular polymeric substances (EPS) were found to sorb more efficiently than cells without EPS in the case of Pu(V). Another example is the sorption of U(VI) by microorganisms, reported widely owing to electron microscopy (Bader et al., 2018; Povedano‐Priego et al., 2019; Kolhe et al., 2020). SEM images of the yeast Yarrowia lipolytica exposed to uranyl carbonate clearly showed the apparition of uranyl deposits attached to the cell surfaces, due to biosorption and biomineralization processes (Kolhe et al., 2020). In summary, microscopic techniques provide visual evidences of the specific mechanisms involved in the microbe–radionuclide interaction, which may be very useful and improve the quality of the works in the field.
Multi‐omics
The development of multiple omic techniques – including genomics, proteomics, metabolomics and transcriptomics – is becoming a very important tool for biomolecular studies in many different fields. These high‐throughput techniques can provide a comprehensive analysis of the total pool of molecules of the same type (DNA, proteins and other metabolites) from a biological sample. Specifically, multi‐omics is leading the way towards more proficient studies and approaches in the field of bioremediation.
Genomic approaches including genome sequencing, and comparative genomics have proven useful for the identification of essential genes (key players) encoding biomolecules, mainly proteins, involved in radionuclide and heavy metal detoxification (Yan et al., 2016; Hemmat‐Jou et al., 2018, Jaswal et al., 2019a). For instance, metagenomic‐based taxonomic studies performed by Jaswal et al. (2019a) from contaminated ecosystems led to the identification and characterization of U‐resistant microorganisms and fungi belonging to Burkholderia and Penicillium species and having environmentally relevant functions. The genome is usually stable, whereas the protein expression level by the genome depends on the environmental conditions. Qualitative proteomics can systematically identify the types of proteins from a biological sample, while quantitative proteomics can determine the amount of a specific protein expressed under different environmental conditions. Proteomics therefore serves to determine the type and abundance of proteins and enzymes synthesized as a consequence of radionuclide stress response, which contributes to our understanding of the specific peptides involved in the interaction process. It is well known that c‐type cytochromes in Geobacter species promote the reduction of U(VI) to poorly soluble U(IV) as a strategy to prevent its migration through contaminated environments (Lovley et al., 2011). Recently, a c‐type cytochrome (GscA) from Geobacter sp. M18 was determined by proteomic studies to play an important role during uranium bioremediation (Yun et al., 2016).
Transcriptomics is acquiring a crucial role in bioremediation studies through the analysis of gene expression and regulation. For instance, a targeted gene expression analysis indicated the transcriptional induction of arsenate reductase genes, among others related to the ars gene cluster (arsR, arsA, acr3 and arsS) by the cells of Rhodococcus aetherivorans BCP1 when grown in the presence of As(V) at sublethal concentrations (Firrincieli et al., 2019). The latter work revealed that unique metabolic mechanisms of a Rhodococcus strain could tolerate this toxic metal. Transcriptional studies of the response of the strain Cupriavidus metallidurans CH34 to heavy metals were performed as well (Monsieurs et al., 2011). Similar expression profiles were found when the cells were exposed to different heavy metals, suggesting a complex transcriptional‐level interaction between the different metal responses.
Biotechnological applications of radionuclide microbe interactions: Bioremediation and deep geological repository
The different processes by which microbes interact with radionuclides, as described in this review, give rise to diverse biotechnological applications in the field of bioremediating radionuclide‐contaminated sites and in the implementation of deep geological disposal of radioactive wastes.
Bioremediation of radionuclide‐polluted environments
Most works conducted on bioremediation of radionuclide‐polluted environments have been focused on uranium, attempting to overcome the legacies of past U mining around the world. Western countries including France and German, having ceased their U mining activities decades ago, nowadays focus on developing effective technology for the remediation of U pollution. Conventional remediation techniques are based on the use of chemical and physical agents, entailing high costs and low contaminant specificity (Gavrilescu et al., 2009; Selvakumar et al., 2018; Zhang et al., 2018). In contrast, remediation using microorganisms, known as bioremediation, is gaining importance as a feasible and ecofriendly in situ technology for the clean‐up of environmental pollutants (Pollmann et al., 2006). Bioremediation is a process based on reducing the solubility, mobility, bioavailability and toxicity of contaminants. In the case of U, the toxicity is intimately related to its chemical speciation, which in turn depends on its oxidation state. U(IV), a highly insoluble and less toxic form of U, is present in the precipitated form as mineral uraninite (UO2) under anaerobic conditions (Nymans et al. 2006). In contrast, U(VI) species are much more soluble and toxic, and occur predominantly as mobile aqueous species under oxic conditions (Burns 1999). Under aerobic conditions, biomineralization of U(VI) phosphates is upheld as an efficient long‐term strategy to remediate U. These processes are less prone to U re‐oxidation in contrast to the product of U bioreduction, uraninite, which tends to re‐oxidize back to more soluble U (Kulkarni et al., 2013). The biomineralization of U has proven successful when using bacterial strains able to produce phosphatase (acid, neutral and alkaline phosphatase) (Hu et al., 2005; Beazley et al., 2009; Sánchez‐Castro et al., 2020; Shukla et al., 2020). Phosphatases release orthophosphates from organic phosphate substrates, leading to the formation of different U(VI)‐phosphate minerals, mainly autunite [Ca(UO2)2(PO4)2·10‐12H2O], although chernikovite [(H3O)2(UO2)2(PO4)2·6H2O] or ankoleite [K2(UO2)2(PO4)2.6(H2O)] has been also observed (Merroun and Selenska‐Pobell, 2008; Beazley et al., 2009; Salome et al., 2017). Bacteria and fungi were documented for their ability to express phosphatase activity in the presence of U to biomineralize radionuclide. The process was first reported for Citrobacter sp. by Macaskie et al. (2000), and thereafter described in several bacterial strains, including Pseudomonas, Caulobacter crescentus, Deinococcus radiodurans, Serratia, Sphingomonas and Bacillus, as well as in fungi including Aspergillus, Paecilomyces species or archaea Halobacteriium noricense DSM 15987 (Hu et al., 2005; Merroun et al., 2011; Liang et al., 2016; Tu et al., 2019). In addition, the potential utility of genetically engineered phosphatase for the bioprecipitation of uranium from alkaline solutions has been demonstrated by Nilgiriwala et al. (2008). This work reports on the cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR‐1 in E. coli strain EK4 for bioprecipitation of uranium from alkaline solutions.
In addition to the great impact of microbial phosphatase in the biomineralization of U described above, the economic costs of such bioremediation‐based technology should be taken into account. Certain strategies may reduce the costs of industrial upscaling U bioremediation technology: i) use of cheap organic phosphate substrates (e.g. organic phosphate‐containing wastes such as manure, crop residues and sludge from different treatments in sustainable waste handling systems) and ii) immobilization of phosphatase‐producing microbes on inorganic substrates (e.g. alginates).
Several international experiments were undertaken to remove U from U mining wastes in Germany, France and the USA. Recent studies have aimed to design U bioremediation technologies based on U phosphate biomineralization of contaminated former U mining sites in France (Sánchez‐Castro et al., 2017, 2020). Over 100 bacterial strains were isolated from U mill tailings pore waters in France and screened for U bioremediation potential (Sánchez‐Castro et al., 2017). The strains had to fulfil the following properties to be further investigated and applied at large scale: (i) high tolerance to U and other heavy metals (e.g. Cd, Se, Pb), (ii) high phosphatase activity and (iii) U immobilization potential through U(VI) phosphate biomineralization. A highly U‐tolerant Stenotrophomonas sp. Br8 strain – isolated from U mill tailing pore waters with high phosphatase activity – was described for its U phosphate biomineralization (Sánchez‐Castro et al., 2020).
In Germany, ongoing investigations are dedicated to remediating U‐contaminated sites in Saxony and Thuringia after U mining activities ceased. Recently, Krawczyk‐Bärsch et al. (2018) explored the U(VI) phosphate biomineralization potential of Acidovorax facilis. Species of the genus Acidovorax were widely found in U‐contaminated sites (e.g. U mine tailings) (Bondici et al., 2013).
Recently, new bioremediation technologies based on the use of redox behaviour of some microbes through exchange of electrons form cell compartments such as proteins and pili to metals and electrodes have been developed. This bioelectrochemical device‐based bioremediation technology referred to microbial fuel cell (MFC) exhibits excellent abilities to remove pollutants along with electrical power generation within the concept of circular economy (Fang and Achal, 2019). MFCs consist of a two‐chamber system where the anode chamber is occupied by bacterial cells separated from the cathode chamber by a polymeric proton exchange membrane.
Most MFCs use aqueous cathodes where water is bubbled with air to provide dissolved oxygen to electrode (Das 2020). MFCs are constructed to remove radionuclides such as uranium through the cathodic reduction reaction, while organic substrates are oxidized and serve as the carbon and electron donor at the anode (Das 2020). Electrochemically active bacteria used in the bioremediation of U through this innovative technique include Shewanella putrefaciens and Geobacter sp., directly transfer electrons from the microbe to the electrode through reduction of U(VI) to U(IV) (Kato 2015).
Impact of microbial processes in the deep geological disposal of radioactive wastes
In the past decade, a number of studies have aimed to study the impact of microbial processes on the safety of future deep geological repositories (DGR) to dispose radioactive wastes generated by the nuclear industry, U mining activity, etc. DGR is the safest option for the disposal of radioactive waste when encapsulated in metal containers surrounded by artificial (e.g. bentonite) and natural barriers (clays, granite and salt formations) (Alexander and McKinley, 2011). The microbial populations of these barriers, described as abundant, can affect the safety of the disposal system through different microbial processes: (i) transformation of clay mineralogy; (ii) corrosion of metal canisters; and (iii) mobility of radionuclides (e.g. U, Se, Cm, Pu, Am, Np) (Anderson et al., 2011).
Several studies report on the impact of microbial processes in the mobility of radionuclides in terms of DGR safety (Povedano‐Priego et al., 2019). In Spain, research has looked into the microbial processes occurring at the bentonite/radionuclide interface that may be involved in the mobility of U, Cm and Se. In the case of uranium, Lopez‐Fernandez et al. (2018) put forth a multidisciplinary approach combining X‐ray absorption and IR spectroscopy; the cells of the yeast Rhodotorula mucilaginosa BII‐R8, isolated from Spanish bentonites, sorbed mobile uranium species (U‐hydroxides and U‐hydroxo‐carbonates) from solution via a time‐dependent process initiated by the adsorption of uranium species to carboxyl groups, and then by organic phosphate groups, forming uranium complexes with a structure similar to that of meta‐autunite. This yeast strain was reported to bind Cm(III), another critical radionuclide, through phosphoryl and carboxyl groups present in bacterial cell envelopes (Lopez‐Fernandez et al., 2019). Povedano‐Priego et al. (2019) documented the immobilization of U through natural U phosphate biomineralization by bentonite bacteria in microcosms amended with glycerol‐2‐phosphate as an electron donor and organic phosphate source. In the case of selenium, a bentonite isolate from the genus Stenotrophomonas was studied to determine its impact on the mobility of this element under DGR‐relevant conditions (Ruiz Fresneda et al. (2018; Ruiz‐Fresneda 2019, 2020). Reduction of Se(IV) to Se(0) nanoparticles under aerobic and anaerobic conditions was reported. HRTEM and electron diffraction studies revealed that the structure of the Se nanoparticles depends upon their shape, while Se nanospheres are amorphous, and (a‐Se) nanowires and hexagons exhibited trigonal structures (t‐Se) due to the presence of two distinct lattice spacings of 0.37 and 0.29 nm, respectively, corresponding to the (100) and (101) planes of t‐Se. Given the low solubility of t‐Se nanostructures compared with that of a‐Se nanospheres and Se(IV), the mobility of selenium in the environment may be significantly reduced. All these studies are needed to enhance understanding of how microbial processes can influence the safety and performance of future repositories in Europe and around the world.
The above summarized laboratory‐scale radionuclide microbe interaction is the basis for future large‐scale application. However, these field bioremediation applications are still in their infancy and several challenges related to the biotic and abiotic complexity of contaminated environments (pH, presence of cations, anions, etc.) should be broadly investigated. In addition, other parameters should be taken into account in assessing field bioremediation technologies including systematic life cycle analysis take into account technical, economic and sustainability issues (Gong et al. 2018).
Conclusions and future perspectives
Despite the substantial amount of basic research and remarkable advances in microbial–radionuclide interactions summarized in this review, biotechnological applications based on such knowledge are still limited. Largely to blame is the restricted access to such contaminated environments, along with their high geochemical and biological complexity. Therefore, the application of key technologies could best be ensured through collaborative work, involving interdisciplinary teams of geologists, geochemists and microbiologists, adopting multidisciplinary approaches combining microbial ecology, radiochemistry, geochemistry, mineralogy, microscopy and spectroscopy, plus the emerging techniques based on omics, for instance. New developments are required to gain a sound understanding of the impact of microbial processes on the geochemical cycle of plutonium, neptunium, curium and radon, and subsequently design bioremediation technologies for these actinides. Another emerging application of microbes in the framework of energy and chemical wastes that calls for further study is metal cycling. There is a lack of balance between the available resources of U (natural resources; U‐containing minerals) and the industrial demand for this radionuclide. The fact that U‐tolerant microbes are able to recover radionuclides (including U) from radioactive wastes means that waste can take on an active role as raw material under the concept of circular economy. In addition, from an educational point of view, microbiologists should consider radionuclide‐contaminated environments as didactic tools and materials to attract the attention of primary and secondary scholars through the organization of excursions, seminars, etc. (McGenity et al., 2020).
Conflict of interest
None declared.
Acknowledgments
This work was supported by the ERDF‐financed grant RTI2018‐101548‐B‐I00 (80% funding by FEDER) (Ministerio de Ciencia e Innovación, Spain).
Microbial Biotechnology (2021) 14(3), 810–828
Funding Information This work was supported by the ERDF‐financed grant RTI2018‐101548‐B‐I00 (80% funding by FEDER) (Ministerio de Ciencia e Innovación, Spain).
Contributor Information
Margarita Lopez‐Fernandez, Email: margaritalopez@ugr.es.
Mohamed L. Merroun, Email: merroun@ugr.es.
References
- Akob, D.M. , Kerkhof, L. , Kuesel, K. , Watson, D.B. , Palumbo, A.V. , and Kostka, J.E. (2011) Linking specific heterotrophic bacterial populations to bioreduction of uranium and nitrate in contaminated subsurface sediments by using stable isotope probing. Appl Environ Microbiol 77: 8197–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akob, D.M. , Mills, H.J. , and Kostka, J.E. (2007) Metabolically active microbial communities in uranium‐contaminated subsurface sediments. FEMS Microbiol Ecol 59: 95–107. [DOI] [PubMed] [Google Scholar]
- Alexander, W.R. , and McKinley, L. (2011) Deep geological disposal of radioactive waste. Amsterdam, Netherlands: Elsevier. eBook ISBN: 9780080468884 [Google Scholar]
- Almahayni, T. , Beresford, N.A. , Crout, N.M. , and Sweeck, L. (2019) Fit‐for‐purpose modelling of radiocaesium soil‐to‐plant transfer for nuclear emergencies: a review. J Environ Radioact 201: 58–66. 10.1016/j.jenvrad.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Anderson, C. , Johnsson, A. , Moll, H. , and Pedersen, K. (2011) Radionuclide geomicrobiology of the deep biosphere. Geomicrobiol J 28: 540–561. [Google Scholar]
- Bader, M. , Moll, H. , Steudtner, R. , Lösch, H. , Drobot, B. , Stumpf, T. , and Cherkouk, A. (2019) Association of Eu(III) and Cm(III) onto an extremely halophilic archaeon. Environ Sci Pollut Res 26: 9352–9364. [DOI] [PubMed] [Google Scholar]
- Bader, M. , Müller, K. , Foerstendorf, H. , Schmidt, M. , Simmons, K. , Swanson, J.S. , et al. (2018) Comparative analysis of uranium bioassociation with halophilic bacteria and archaea. PLoS One 13: e0190953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns, S.M. , Cain, E.C. , Sommerville, L. , and Kuske, C.R. (2007) Acidobacteria phylum sequences in uranium‐contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl Environ Microbiol 73: 3113–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beazley, M.J. , Martinez, R.J. , Sobecky, P.A. , Webb, S.M. , and Taillefert, M. (2009) Nonreductive biomineralization of uranium (VI) phosphate via microbial phosphatase activity in anaerobic conditions. Geomicrobiol J 26: 431–441. [Google Scholar]
- Beekes, M. , Lasch, P. , and Naumann, D. (2007) Analytical applications of Fourier transform‐infrared (FT‐IR) spectroscopy in microbiology and prion research. Veterinary Microbiology 123:305–319. [DOI] [PubMed] [Google Scholar]
- Bennett, P.C. , Hiebert, F.K. , Rogers, J.R. , and Choi, W.J. (2001) Silicates, silicate weathering, and microbial ecology. Geomicrobiol J 18: 3–19. [Google Scholar]
- Benzerara, K. , Miot, J. , Morin, G. , Ona‐Nguema, G. , Skouri‐Panet, F. , and Ferard, C. (2011) Significance, mechanisms and environmental implications of microbial biomineralization. CR Geosci 343: 160–167. [Google Scholar]
- Boggs, M.A. , Jiao, Y. , Dai, Z. , Zavarin, M. , and Kersting, A.B. (2016) Interactions of plutonium with Pseudomonas sp. strain EPS‐1W and Its extracellular polymeric substances. Appl Environ Microbiol 82: 7093–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondici, Vf , Lawrence, Jr , Khan, Nh , Hill, Je , Yergeau, E. , Wolfaardt, Gm , et al. (2013) Microbial communities in low permeability, high pH uranium mine tailings: characterization and potential effects. J Appl Microbiol 114: 1671–1686. [DOI] [PubMed] [Google Scholar]
- Brooks, S.C. (2001) Waste Characteristics of the Former S‐3 Ponds and Outline of Uranium Chemistry Relevant to NABIR Field Research Center Studies. Oak Ridge: Oak Ridge National Laboratory, 21 ORNL/TM‐2001/2027, 814525. [Google Scholar]
- Brookshaw, D. , Pattrick, R. , Lloyd, J. , and Vaughan, D. (2012) Microbial effects on mineral–radionuclide interactions and radionuclide solid‐phase capture processes. Mineral Mag 76: 777–806. [Google Scholar]
- Burns, P.C. (1999) The crystal chemistry of uranium. Uranium: Miner Geochem Environ 38: 23–90. [Google Scholar]
- Calvin, S. (2013) XAFS for Everyone. Boca Raton, FL: CRC Press. ISBN 9781439878637. [Google Scholar]
- Cardenas, E. , Wu, W.‐M. , Leigh, M.B. , Carley, J. , Carroll, S. , Gentry, T. , et al. (2008) Microbial communities in contaminated sediments, associated with bioremediation of uranium to submicromolar levels. Appl Environ Microbiol 74: 3718–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, H.K. , Price, M.N. , Callaghan, M. , Aaring, A. , Chakraborty, R. , Liu, H. , et al. (2019) The selective pressures on the microbial community in a metal‐contaminated aquifer. ISME J 13: 937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandwadkar, P. , Misra, H.S. , and Acharya, C. (2018) Uranium biomineralization induced by a metal tolerant Serratia strain under acid, alkaline and irradiated conditions. Metallomics 10: 1078–1088. [DOI] [PubMed] [Google Scholar]
- Chauhan, A. , Pathak, A. , Jaswal, R. , Edwards, B. , Chappell, D. , Ball, C. , et al. (2018) Physiological and comparative genomic analysis of Arthrobacter sp. SRS‐W‐1‐2016 provides insights on niche adaptation for survival in uraniferous soils. Genes 9: 93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Hu, J. , and Wang, J. (2020) Biosorption of uranium by immobilized Saccharomyces cerevisiae . J Environ Radioact 213: 106158. [DOI] [PubMed] [Google Scholar]
- Chen, B.D. , Roos, P. , Zhu, Y.G. , and Jakobsen, I. (2008) Arbuscular mycorrhizas contribute to phyto stabilization of uranium in uranium mining tailings. J Environ Radioact 99: 801–810. [DOI] [PubMed] [Google Scholar]
- Choudhary, S. , and Sar, P. (2011) Uranium biomineralization by a metal resistant Pseudomonas aeruginosa strain isolated from contaminated mine waste. J Hazard Mater 186: 336–343. [DOI] [PubMed] [Google Scholar]
- Cleary, A. , Lloyd, J. , Newsome, L. , Shaw, S. , Boothman, C. , Boshoff, G. , et al. (2019) Bioremediation of strontium and technetium contaminated groundwater using glycerol phosphate. Chem Geol 509: 213–222. [Google Scholar]
- Collins, R.N. , Saito, T. , Aoyagi, N. , Payne, T.E. , Kimura, T. , and Waite, T.D. (2011) Applications of time‐resolved laser fluorescence spectroscopy to the environmental biogeochemistry of actinides. J Environ Qual 40: 731–741. [DOI] [PubMed] [Google Scholar]
- Cresnar, B. , and Petric, S. (2011) Cytochrome P450 enzymes in the fungal kingdom. Biochem Biophys Acta 1: 29–35. [DOI] [PubMed] [Google Scholar]
- Dabbagh, R. , Ebrahimi, M. , Aflaki, F. , Ghafourian, H. , and Sahafipour, M. (2008) Biosorption of stable cesium by chemically modified biomass of Sargassum glaucescens and Cystoseira indica in a continuous flow system. J Hazard Mater 159: 354–357. [DOI] [PubMed] [Google Scholar]
- Dadachova, E. , and Casadevall, A. (2008) Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr Opin Microbiol 11: 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, M. , Buesseler, K.O. , and Pike, S.M. (2005) Plutonium in groundwater at the 100K‐Area of the US DOE Hanford Site. J Contam Hydrol 76: 167–189. [DOI] [PubMed] [Google Scholar]
- Daly, M.J. (2006) Modulating radiation resistance: insights based on defenses against reactive oxygen species in the radio‐resistant bacterium Deinococcus radiodurans . Clin Lab Med 26: 491–504. [DOI] [PubMed] [Google Scholar]
- Das, N. (2012) Remediation of radionuclide pollutants through biosorption–an overview, CLEAN–Soil . Air Water 40: 16–23. [Google Scholar]
- Das, K.S. (2020) Microbial fuel cells: a path to green, renewable energy. In: Green Energy and Technology. Berlin, Germany: Springer, pp. 195–206. [Google Scholar]
- Davies, H.S. , Cox, F. , Robinson, C.H. , and Pittman, J.K. (2015) Radioactivity and the environment: technical approaches to understand the role of arbuscular mycorrhizal plants in radionuclide bioaccumulation. Front Plant Sci 6: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo, R.P. , Songkasiri, W. , Rittmann, B.E. , and Reed, D.T. (2010) Surface complexation of neptunium (V) onto whole cells and cell components of Shewanella alga: modeling and experimental study. Environ Sci Technol 44: 4930–4935. [DOI] [PubMed] [Google Scholar]
- Ding, C. , Feng, S. , Cheng, W. , Zhang, J. , Li, X. , Liao, J. , et al. (2014) Biosorption behavior and mechanism of thorium on Streptomyces sporoverrucosus dwc‐3. J Radioanal Nucl Chem 301: 237–245. [Google Scholar]
- Dräxl, S. , Müller, J. , Li, W.B. , Michalke, B. , Scherb, H. , Hense, B.A. , et al. (2013) Caesium accumulation in yeast and plants is selectively repressed by loss of the SNARE Sec22p/SEC22. Nat Commun 4: 1–10. [DOI] [PubMed] [Google Scholar]
- Edelstein, N.M. , Klenze, R. , Fanghänel, T. , and Hubert, S. (2006) Optical properties of Cm (III) in crystals and solutions and their application to Cm (III) speciation. Coord Chem Rev 250: 948–973. [Google Scholar]
- Eswayah, A.S. , Hondow, N. , Scheinost, A.C. , Merroun, M. , Romero‐González, M. , Smith, T.J. , and Gardiner, P.H.E. (2019) Methyl selenol as a precursor in selenite reduction to Se/S Species by methane‐oxidizing bacteria. Appl Environ Microbiol 85: e01379‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswayah, A.S. , Smith, T.J. , Scheinost, A.C. , Hondow, N. , and Gardiner, P.H. . (2017) Microbial transformations of selenite by methane‐oxidizing bacteria. Appl Microbiol Biotechnol 101: 6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski, P.G. , Fenchel, T. , and Delong, E.F. (2008) The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039. [DOI] [PubMed] [Google Scholar]
- Fan, Y.‐Y. , Li, B.‐B. , Yang, Z.‐C. , Cheng, Y.‐Y. , Liu, D.‐F. , and Yu, H.‐Q. (2018) Abundance and diversity of iron reducing bacteria communities in the sediments of a heavily polluted freshwater lake. Appl Microbiol Biotechnol 102: 10791–10801. [DOI] [PubMed] [Google Scholar]
- Fang, C. , and Achal, V. (2019) The potential of microbial fuel cells for remediation of heavy metals from soil and water‐review of application. Microorganisms 7: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firrincieli, A. , Presentato, A. , Favoino, G. , Marabottini, R. , Allevato, E. , Stazi, S.R. , et al. (2019) Identification of resistance genes and response to arsenic in rhodococcus aetherivorans BCP1. Front Microbiol 10: 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, S. , Krause, T. , Lederer, F. , Merroun, M.L. , Shevchenko, A. , Hübner, R. , et al. (2020) Bacillus safensis JG‐B5T affects the fate of selenium by extracellular production of colloidally less stable selenium nanoparticles. J Hazard Mater 384: 121146. [DOI] [PubMed] [Google Scholar]
- Fomina, M. , Charnock, J. , Hillier, S. , Alvarez, R. , and Gadd, G. (2007) Fungal transformations of uranium oxides. Environ Microbiol 9: 1696–1710. [DOI] [PubMed] [Google Scholar]
- Fredrickson, J.K. , Zachara, J.M. , Balkwill, D.L. , Kennedy, D. , Li, S.‐M.W. , and Kostandarithes, H. M. et al. (2004) Geomicrobiology of high‐level nuclear waste‐contaminated vadose sediments at the hanford site, Washington State. Appl Environ Microbiol 70: 4230–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd, G.M. , and Pan, X. (2016) Biomineralization, bioremediation and biorecovery of toxic metals and radionuclides. Milton Park, UK: Taylor & Francis.175–178. 10.1080/01490451.2015.1087603. [DOI] [Google Scholar]
- Gallois, N. , Alpha‐Bazin, B. , Ortet, P. , Barakat, M. , Piette, L. , Long, J. , et al. (2018) Proteogenomic insights into uranium tolerance of a Chernobyl's Microbacterium bacterial isolate. J Proteomics 177: 148–157. [DOI] [PubMed] [Google Scholar]
- Gavrilescu, M. , Pavel, L.V. , and Cretescu, I. (2009) Characterization and remediation of soils contaminated with uranium. J Hazard Mater 163: 475–510. [DOI] [PubMed] [Google Scholar]
- Gerber, U. , Hübner, R. , Rossberg, A. , Krawczyk‐Bärsch, E. , and Merroun, M.L. (2018) Metabolism‐dependent bioaccumulation of uranium by Rhodosporidium toruloides isolated from the flooding water of a former uranium mine. PLoS One 13: e0201903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gołębiewski, M. , Deja‐Sikora, E. , Cichosz, M. , Tretyn, A. , and Wróbel, B. (2014) 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb Ecol 67: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Y. , Zhao, D. , and Wang, D. (2018) An overview of field‐scale studies on remediation of soil contaminated with heavy metals and metalloids: technical progress over the last decade. Water Res 147: 440–460. [DOI] [PubMed] [Google Scholar]
- Goyal, N. , Jain, S. , and Banerjee, U. (2003) Comparative studies on the microbial adsorption of heavy metals. Adv Environ Res 7: 311–319. [Google Scholar]
- Guillot, E. , Hinsinger, P. , Dufour, L. , Roy, J. , and Bertrand, I. (2019) With or without trees: resistance and resilience of soil microbial communities to drought and heat stress in a Mediterranean agroforestry system. Soil Biol Biochem 129: 122–135. [Google Scholar]
- Hain, K. , Steier, P. , Froehlich, M. b , Golser, R. , Hou, X. , Lachner, J. , et al. (2020) 233 U/236 U signature allows to distinguish environmental emissions of civil nuclear industry from weapons fallout. Nat Commun 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg, R. , and Ferris, F.G. (2004) Biomineralization by gallionella. Geomicrobiol J 21: 325–330. [Google Scholar]
- Hallsworth, J.E. (2019) Microbial unknowns at the saline limits for life. Nat Ecol Evol 3: 1503–1504. [DOI] [PubMed] [Google Scholar]
- Hannun, Y.A. , and Obeid, L.M. (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9: 139–150. [DOI] [PubMed] [Google Scholar]
- Hemmat‐Jou, M.H. , Safari‐Sinegani, A.A. , Mirzaie‐Asl, A. , and Tahmourespour, A. (2018) Analysis of microbial communities in heavy metals‐contaminated soils using the metagenomic approach. Ecotoxicology 27: 1281–1291. [DOI] [PubMed] [Google Scholar]
- Hemme, C.L. , Deng, Y. , Gentry, T.J. , Fields, M.W. , Wu, L. , Barua, S. , et al. (2010) Metagenomic insights into evolution of a heavy metal‐contaminated groundwater microbial community. ISME J 4: 660–672. [DOI] [PubMed] [Google Scholar]
- Hemme, C.L. , Tu, Q. , Shi, Z. , Qin, Y. , Gao, W. , Deng, Y. , et al. (2015) Comparative metagenomics reveals impact of contaminants on groundwater microbiomes. Front Microbiol 6: 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiike, T. , Dotsuta, Y. , Nakano, Y. , Ochiai, A. , Utsunomiya, S. , Ohnuki, T. , and Yamashita, M. (2017) Removal of soluble strontium via incorporation into biogenic carbonate minerals by halophilic bacterium Bacillus sp. strain TK2d in a highly saline solution. Appl Environ Microbiol 83: e00855–00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos‐Hernandez, C. , Courbert, C. , Simonucci, C. , David, S. , Vogel, T.M. , and Larose, C. (2019) Community structure and functional genes in radionuclide contaminated soils in Chernobyl and Fukushima. FEMS Microbiol Lett 366: fnz180. [DOI] [PubMed] [Google Scholar]
- Hu, P. , Brodie, E.L. , Suzuki, Y. , McAdams, H.H. , and Andersen, G.L. (2005) Whole‐genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus . J Bacteriol 187: 8437–8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M.Z.C. , Norman, J.M. , Faison, B.D. , and Reeves, M.E. (1996) Biosorption of uranium by Pseudomonas aeruginosa strain CSU: characterization and comparison studies. Biotechnol Bioeng 51: 237–247. [DOI] [PubMed] [Google Scholar]
- IAEA (2017) IAEA Radiation Technology the Industrial Revolution Behind the Scenes. Vienna: IAEA Bulletin (March) 58–1 International Atomic Energy Agency. [Google Scholar]
- Ilyas, S. , Srivastava, R.R. , and Ilyas, N. (2020) In Strontium Contamination in the Environment, Pathak, P. , & Gupta, D. K. (eds). Berlin, Germany: Springer, pp. 65–83. ISBN 978‐3‐030‐15314‐4 [Google Scholar]
- Islam, E. , and Sar, P. (2011) Culture‐dependent and ‐independent molecular analysis of the bacterial community within uranium ore. J Basic Microbiol 51: 372–384. [DOI] [PubMed] [Google Scholar]
- Islam, E. , and Sar, P. (2016) Diversity, metal resistance and uranium sequestration abilities of bacteria from uranium ore deposit in deep earth stratum. Ecotoxicol Environ Saf 127: 12–21. [DOI] [PubMed] [Google Scholar]
- Istok, J. d , Senko, J. m , Krumholz, L. r , Watson, D. , Bogle, M. a , Peacock, A. , et al. (2004) In situ bioreduction of technetium and uranium in a nitrate‐contaminated aquifer. Environ Sci Technol 38: 468–475. [DOI] [PubMed] [Google Scholar]
- Jalali‐Rad, R. , Ghafourian, H. , Asef, Y. , Dalir, S. , Sahafipour, M. , and Gharanjik, B. (2004) Biosorption of cesium by native and chemically modified biomass of marine algae: introduce the new biosorbents for biotechnology applications. J Hazard Mater 116: 125–134. [DOI] [PubMed] [Google Scholar]
- Jaswal, R. , Pathak, A. , and Chauhan, A. (2019) Metagenomic evaluation of bacterial and fungal assemblages enriched within diffusion chambers and microbial traps containing uraniferous soils. Microorganisms 7(9): 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaswal, R. , Pathak, A. , Edwards, B. III , Lewis, R. III , Seaman, J.C. , and Stothard, P. et al. (2019b) Metagenomics‐guided survey, isolation, and characterization of uranium resistant microbiota from the savannah river Site, USA. Genes 10(5): 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M. , Xiao, A. , Zhu, L. , Zhang, Z. , Huang, H. , and Jiang, L. (2019) The diversity and commonalities of the radiation‐resistance mechanisms of Deinococcus and its up‐to‐date applications. AMB Express 9: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnev, A.A. , Tugarova, A.V. , Selivanova, M.A. , Tarantilis, P.A. , Polissiou, M.G. , and Kudryasheva, N.S. (2013) Effects of americium‐241 and humic substances on Photobacterium phosphoreum: bioluminescence and diffuse reflectance FTIR spectroscopic studies. Spectrochimica Acta ‐ Part A: Mol Biomolecul Spectrosc 100: 171–175. [DOI] [PubMed] [Google Scholar]
- Kato, S. (2015) Biotechnological aspects of microbial extracellular electron transfer. Microb Environ 30: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, F. , Kuwahara, C. , Oosone, A. , Ichikawa, T. , Terada, H. , Morita, Y. , and Sugiyama, H. (2000) Accumulation and subcellular localization of cesium in mycelia of Streptomyces lividans and a Cs tolerant strain, Streptomyces sp. TOHO‐2. J Health Sci 46: 259–262. [Google Scholar]
- Kedari, C. , Das, S. , and Ghosh, S. (2001) Biosorption of long lived radionuclides using immobilized cells of Saccharomyces cerevisiae . World J Microbiol Biotechnol 17: 789–793. [Google Scholar]
- Khani, M. , Keshtkar, A. , Meysami, B. , Zarea, M. , and Jalali, R. (2005) Biosorption of uranium from aqueous solutions by nonliving biomass of marine algae Cystoseiraical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol 71: 6308–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielak, A.M. , Barreto, C.C. , Kowalchuk, G.A. , van Veen, J.A. , and Kuramae, E.E. (2016) The ecology of acidobacteria: moving beyond genes and genomes. Front Microbiol 7: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.Y. , Jang, S.‐C. , Song, Y.H. , Lee, C.‐S. , Huh, Y.S. , and Roh, C. (2016) Screening and identification of a cesium‐tolerant strain of bacteria for cesium biosorption. Korean J Environ Biol 34: 304–313. [Google Scholar]
- Kolhe, N. , Zinjarde, S. , and Acharya, C. (2018) Responses exhibited by various microbial groups relevant to uranium exposure. Biotechnol Adv 36: 1828–1846. [DOI] [PubMed] [Google Scholar]
- Kolhe, N. , Zinjarde, S. , and Acharya, C. (2020) Impact of uranium exposure on marine yeast, Yarrowia lipolytica: insights into the yeast strategies to withstand uranium stress. J Hazard Mater 381: 121226. [DOI] [PubMed] [Google Scholar]
- Krawczyk‐Bärsch, E. , Gerber, U. , Müller, K. , Moll, H. , Rossberg, A. , Steudtner, R. , and Merroun, M. (2018) Multidisciplinary characterization of U (VI) sequestration by Acidovorax facilis for bioremediation purposes. J Hazard Mater 347: 233–241. [DOI] [PubMed] [Google Scholar]
- Krawczyk‐Bärsch, E. , Scheinost, A.C. , Rossberg, A. , Müller, K. , Bok, F. , Hallbeck, L. , et al. (2020) Uranium and neptunium retention mechanisms in Gallionella ferruginea/ ferrihydrite systems for remediation purposes. Environ Sci Pollut Res 26:23850–23860. [Google Scholar]
- Krisko, A. , and Radman, M. (2013) Biology of extreme radiation resistance: the way of Deinococcus radiodurans . Cold Spring Harbor, Perspect Biol 5(7): a012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, S. , Ballal, A. , and Apte, S.K. (2013) Bioprecipitation of uranium from alkaline waste solutions using recombinant Deinococcus radiodurans . J Hazard Mater 262: 853–861. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Nongkhlaw, M. , Acharya, C. , and Joshi, S.R. (2013) Uranium (U)‐tolerant bacterial diversity from U ore deposit of Domiasiat in North‐East India and its prospective utilisation in bioremediation. Microb Environ 28: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara, C. , Fukumoto, A. , Nishina, M. , Sugiyama, H. , Anzai, Y. , and Kato, F. (2011) Characteristics of cesium accumulation in the filamentous soil bacterium Streptomyces sp. K202. J Environ Radioact 102: 138–144. [DOI] [PubMed] [Google Scholar]
- Lee, J.‐H. , and Hur, H.‐G. (2014) Intracellular uranium accumulation by Shewanella sp. HN‐41 under the thiosulfate‐reducing condition. J Korean Soc Appl Biol Chem 57: 117–121. [Google Scholar]
- Li, X. , Meng, D. , Li, J. , Yin, H. , Liu, H. , Liu, X. , et al. (2017) Response of soil microbial communities and microbial interactions to long‐term heavy metal contamination. Environ Pollut 231: 908–917. [DOI] [PubMed] [Google Scholar]
- Li, B. , Wu, W.‐M. , Watson, D.B. , Cardenas, E. , Chao, Y. , Phillips, D.H. , et al. (2018) Bacterial community shift and coexisting/coexcluding patterns revealed by network analysis in a uranium‐contaminated site after bioreduction followed by reoxidation. Appl Environ Microbiol 84: e02885‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Csetenyi, L. , and Gadd, G.M. (2016) Uranium bioprecipitation mediated by yeasts utilizing organic phosphorus substrates. Appl Microbiol Biotechnol 100: 5141–5151. [DOI] [PubMed] [Google Scholar]
- Liang, X. , Hillier, S. , Pendlowski, H. , Gray, N. , Ceci, A. , and Gadd, G.M. (2015) Uranium phosphate biomineralization by fungi. Environ Microbiol 17: 2064–2075. [DOI] [PubMed] [Google Scholar]
- Lloyd, J.R. (2003) Microbial reduction of metals and radionuclides. FEMS Microbiol Rev 27: 411–425. [DOI] [PubMed] [Google Scholar]
- Lloyd, J.R. , and Macaskie, L.E. (2000) Bioremediation of radionuclide‐containing wastewaters. Environ Microbe‐metal Interact: Am Soc Microbiol 77: 277–327. [Google Scholar]
- Long, J. , Li, H. , Jiang, D. , Luo, D. , Chen, Y. , Xia, J. , and Chen, D. (2017) Biosorption of strontium (II) from aqueous solutions by Bacillus cereus isolated from strontium hyperaccumulator Andropogon gayanus . Process Saf Environ Prot 111: 23–30. [Google Scholar]
- Lopez‐Fernandez, M. , Fernández‐Sanfrancisco, O. , Moreno‐García, A. , Martín‐Sánchez, I. , Sánchez‐Castro, I. , and Merroun, M.L. (2014) Microbial communities in bentonite formations and their interactions with uranium. Appl Geochem 49: 77–86. [Google Scholar]
- Lopez‐Fernandez, M. , Moll, H. , and Merroun, M.L. (2019) Reversible pH‐dependent curium (III) biosorption by the bentonite yeast isolate Rhodotorula mucilaginosa BII‐R8. J Hazard Mater 370: 156–163. [DOI] [PubMed] [Google Scholar]
- Lopez‐Fernandez, M. , Romero‐González, M. , Günther, A. , Solari, P.L. , and Merroun, M.L. (2018) Effect of U (VI) aqueous speciation on the binding of uranium by the cell surface of Rhodotorula mucilaginosa, a natural yeast isolate from bentonites. Chemosphere 199: 351–360. [DOI] [PubMed] [Google Scholar]
- Lovley, D. (2013) Dissimilatory Fe(III)‐ and Mn(IV)‐Reducing Prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry. Rosenberg, E. , DeLong, E.F. , Lory, S. , Stackebrandt, E. , and Thompson, F. (eds). Berlin, Heidelberg: Springer, pp. 287–308. [Google Scholar]
- Lovley, D. , Phillips, E. , Gorby, Y. , and Landa, E. (1991) Microbial reduction of uranium. Nature 350: 413–416. [Google Scholar]
- Lovley, D.R. , Ueki, T. , Zhang, T. , Malvankar, N.S. , Shrestha, P.M. , Flanagan, K.A. , et al. (2011) Geobacter. The microbe electric's physiology, ecology, and practical applications. In: Advances in Microbial Physiology. Berlin, Germany: Springer‐Verlag, pp. 1‐100. [DOI] [PubMed] [Google Scholar]
- Lujanienė, G. , Levinskaitė, L. , Kačergius, A. , and Gavutis, M. (2017) Sorption of plutonium to bacteria and fungi isolated from groundwater and clay samples. J Radioanal Nucl Chem 311: 1393–1399. [Google Scholar]
- Macaskie, L.E. , and Basnakova, G. (1998) Microbially‐enhanced chemisorption of heavy metals: a method for the bioremediation of solutions containing long‐lived isotopes of neptunium and plutonium. Environ Sci Technol 32: 184–187. [Google Scholar]
- Macaskie, L.E. , Bonthrone, K.M. , Yong, P. , and Goddard, D.T. (2000) Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 146: 1855–1867. [DOI] [PubMed] [Google Scholar]
- Macaskie, L. , Jeong, B. , and Tolley, M. (1994) Enzymically accelerated biomineralization of heavy metals: application to the removal of americium and plutonium from aqueous flows. FEMS Microbiol Rev 14: 351–367. [DOI] [PubMed] [Google Scholar]
- Malo, M.E. , and Dadachova, E. (2019) Melanin as an energy transducer and a radioprotector in Black Fungi. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance. Tiquia‐Arashiro, S. , and Grube, M. (eds). Cham, Switzerland: Springer. [Google Scholar]
- Mao, Y. , Hu, H. , and Yan, Y. (2011) Biosorption of cesium (I) from aqueous solution by a novel exopolymers secreted from Pseudomonas fluorescens C‐2: equilibrium and kinetic studies. J Environ Sci 23: 1104–1112. [DOI] [PubMed] [Google Scholar]
- Martinez, R.J. , Beazley, M.J. , Taillefert, M. , Arakaki, A.K. , Skolnick, J. , and Sobecky, P.A. (2007) Aerobic uranium (VI) bioprecipitation by metal‐resistant bacteria isolated from radionuclide‐and metal‐contaminated subsurface soils. Environ Microbiol 9: 3122–3133. [DOI] [PubMed] [Google Scholar]
- Mattox, D.M. (2010) Substrate (“Real”) surfaces and surface modification. In: Handbook of Physical Vapor Deposition (PVD) Processing. Oxford: Elsevier, 25–72. 10.1016/B978-0-8155-2037-5.00002-2. [DOI] [Google Scholar]
- McGenity, T.J. , Gessesse, A. , Hallsworth, J.E. , Garcia Cela, E. , Verheecke‐Vaessen, C. , Wang, F. , et al. (2020) Visualizing the invisible: class excursions to ignite children’s enthusiasm for microbes. Microb Biotechnol 3:844–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, N. , Benzerara, K. , Kocar, B.D. , and Chapon, V. (2019) Sequestration of radionuclides radium‐226 and strontium‐90 by cyanobacteria forming intracellular calcium carbonates. Environ Sci Technol 53: 12639–12647. [DOI] [PubMed] [Google Scholar]
- Meinrath, A. , Schneider, P. , and Meinrath, G. (2003) Uranium ores and depleted uranium in the environment, with a reference to uranium in the biosphere from the Erzgebirge/Sachsen, Germany. J Environ Radioactivity 64: 175–193. [DOI] [PubMed] [Google Scholar]
- Merroun, M.L. , Nedelkova, M. , Ojeda, J.J. , Reitz, T. , Fernández, M.L. , Arias, J.M. , et al. (2011) Bio‐precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X‐ray absorption spectroscopic analyses. J Hazardous Materials 197: 1–10. [DOI] [PubMed] [Google Scholar]
- Merroun, M.L. , Raff, J. , Rossberg, A. , Hennig, C. , Reich, T. , and Selenska‐Pobell, S. (2005) Complexation of uranium by cells and S‐layer sheets of Bacillus sphaericus JG‐A12. Appl Environ Microbiol 71: 5532–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merroun, M.L. , and Selenska‐Pobell, S. (2008) Bacterial interactions with uranium: an environmental perspective. J Contam Hydrol 102: 285–295. [DOI] [PubMed] [Google Scholar]
- Mitchell, A.C. , and Ferris, F.G. (2005) The coprecipitation of Sr into calcite precipitates induced by bacterial ureolysis in artificial groundwater: temperature and kinetic dependence. Geochim Cosmochim Acta 69: 4199–4210. [Google Scholar]
- Moll, H. , Cherkouk, A. , Bok, F. , and Bernhard, G. (2017) Plutonium interaction studies with the Mont Terri Opalinus Clay isolate Sporomusa sp. MT‐2.99: changes in the plutonium speciation by solvent extractions. Environ Sci Pollut Res 24: 13497–13508. [DOI] [PubMed] [Google Scholar]
- Moll, H. , Lehmann, F. , and Raff, J. (2020) Interaction of curium(III) with surface‐layer proteins from Lysinibacillus sphaericus JG‐A12, Colloids and Surfaces B: Biointerfaces.110950 10.1016/j.colsurfb.2020.110950. [DOI] [PubMed] [Google Scholar]
- Moll, H. , Merroun, M. , Hennig, C. , Rossberg, A. , Selenska‐Pobell, S. , and Bernhard, G. (2006) The interaction of Desulfovibrio äspöensis DSM 10631T with plutonium. Radiochim Acta 94: 815–824. [Google Scholar]
- Mondani, L. , Benzerara, K. , Carrière, M. , Christen, R. , Mamindy‐Pajany, Y. , Février, L. , et al. (2011) Influence of uranium on bacterial communities: a comparison of natural uranium‐rich soils with controls. PLoS One 6: e25771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs, P. , Moors, H. , Van Houdt, R. , Janssen, P.J. , Janssen, A. , Coninx, I. , et al. (2011) Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals 24: 1133–1151. [DOI] [PubMed] [Google Scholar]
- Moon, J.‐W. , Roh, Y. , Phelps, T.J. , Phillips, D.H. , Watson, D.B. , Kim, Y.‐J. , and Brooks, S.C. (2006) Physicochemical and mineralogical characterization of soil‐saprolite cores from a field research site, Tennessee. J Environ Qual 35: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Mumtaz, S. , Streten, C. , Parry, D.L. , McGuinness, K.A. , Lu, P. , and Gibb, K.S. (2018) Soil uranium concentration at ranger uranium mine land application areas drives changes in the bacterial community. J Environ Radioact 189: 14–23. [DOI] [PubMed] [Google Scholar]
- Mumtaz, S. , Streten‐Joyce, C. , Parry, D.L. , McGuinness, K.A. , Lu, P. , and Gibb, K.S. (2013) Fungi outcompete bacteria under increased uranium concentration in culture media. J Environ Radioact 120: 39–44. [DOI] [PubMed] [Google Scholar]
- Nakajima, A. , and Tsuruta, T. (2004) Competitive biosorption of thorium and uranium by Micrococcus luteus . J Radioanal Nucl Chem 260: 13–18. [Google Scholar]
- Narendrula‐Kotha, R. , and Nkongolo, K.K. (2017) Bacterial and fungal community structure and diversity in a mining region under long‐term metal exposure revealed by metagenomics sequencing. Ecol Genet Genom 2: 13–24. [Google Scholar]
- Naumann, D. (2006) Infrared Spectroscopy in microbiology. In: Encyclopedia of Analytical Chemistry. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Nayuki, K. , Chen, B.D. , Ohtomo, R. , and Kuga, Y. (2014) Cellular imaging of cadmium in resin sections of arbuscular mycorrhizas using synchrotron micro X‐ray fluorescence. Microb Environ 29: 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome, L. , Morris, K. , and Lloyd, J.R. (2014) The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem Geol 363: 164–184. [Google Scholar]
- Newsome, L. , Morris, K. , and Lloyd, J.R. (2015) Uranium biominerals precipitated by an environmental isolate of Serratia under anaerobic conditions. PLoS One 10: e0132392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilgiriwala, K.S. , Alahari, A. , Rao, A.S. , and Apte, S.K. (2008) Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR‐1 for bioprecipitation of uranium from alkaline solutions. Appl Environ Microbiol 74: 5516–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North, N.N. , Dollhopf, S.L. , Petrie, L. , Istok, J.D. , Balkwill, D.L. , and Kostka, J.E. (2004) Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl Environ Microbiol 70: 4911–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman, J.L. , Marsh, T.L. , Ginder‐Vogel, M.A. , Gentile, M. , Fendorf, S. & Criddle, C. (2006) Heterogeneous response to biostimulation for U(VI) reduction in replicated sediment microcosms. Biodegradation, 17: 303–316. [DOI] [PubMed] [Google Scholar]
- Ormerod, R.M. (2001) Surface Chemistry: Electron Yield Spectroscopy. In Encyclopedia of Materials: Science and Technology. Jürgen, Buschow et al. (eds). Oxford: 9006‐9008. 10.1016/B0-08-043152-6/01623-5 [DOI] [Google Scholar]
- Osman, S. , Peeters, Z. , Duc, M.T.L. , Mancinelli, R. , Ehrenfreund, P. , and Venkateswaran, K. (2008) Effect of shadowing on survival of bacteria under conditions simulating the martian atmosphere and UV radiation. Appl Environ Microbiol 74: 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Meng, X. , Zhang, D. , and Wang, J. (2009) Biosorption of strontium ion by immobilised Aspergillus niger . Int J Environ Pollut 37: 276–288. [Google Scholar]
- Paterson‐Beedle, M. , Jeong, B.C. , Lee, C.H. , Jee, K.Y. , Kim, W.H. , Renshaw, J.C. , and Macaskie, L.E. (2012) Radiotolerance of phosphatases of a Serratia sp.: potential for the use of this organism in the biomineralization of wastes containing radionuclides. Biotechnol Bioeng 109: 1937–1946. [DOI] [PubMed] [Google Scholar]
- Paterson‐Beedle, M. , Macaskie, L.E. , Lee, C.H. , Hriljac, J.A. , Jee, K.Y. , and Kim, W.H. (2006) Utilisation of a hydrogen uranyl phosphate‐based ion exchanger supported on a biofilm for the removal of cobalt, strontium and caesium from aqueous solutions. Hydrometallurgy 83: 141–145. [Google Scholar]
- Pollmann, K. , Raff, J. , Merroun, M. , Fahmy, K. , and Selenska‐Pobell, S. (2006) Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol Adv 24: 58–68. [DOI] [PubMed] [Google Scholar]
- Povedano‐Priego, C. , Jroundi, F. , Lopez‐Fernandez, M. , Sánchez‐Castro, I. , Martin‐Sánchez, I. , Huertas, F.J. , and Merroun, M.L. (2019) Shifts in bentonite bacterial community and mineralogy in response to uranium and glycerol‐2‐phosphate exposure. Sci Total Environ 692: 219–232. [DOI] [PubMed] [Google Scholar]
- Purvis, O.W. , Bailey, E.H. , McLean, J. , Kasama, T. , and Williamson, B.J. (2004) Uranium biosorption by the lichen Trapelia involuta at a uranium mine. Geomicrobiol J 21: 159–167. [Google Scholar]
- Radeva, G. , Kenarova, A. , Bachvarova, V. , Flemming, K. , Popov, I. , Vassilev, D. , and Selenska‐Pobell, S. (2013) Bacterial diversity at abandoned uranium mining and milling sites in bulgaria as revealed by 16S rRNA genetic diversity study. Water Air Soil Pollut 224: 1748. [Google Scholar]
- Radeva, G. , and Selenska‐Pobell, S. (2005) Bacterial diversity in water samples from uranium wastes as demonstrated by 16S rDNA and ribosomal intergenic spacer amplification retrievals. Can J Microbiol 51: 910–923. [DOI] [PubMed] [Google Scholar]
- Rastogi, G. , Osman, S. , Vaishampayan, P.A. , Andersen, G.L. , Stetler, L.D. , and Sani, R.K. (2010) Microbial diversity in uranium mining‐impacted soils as revealed by high‐density 16S microarray and clone library. Microb Ecol 59: 94–108. [DOI] [PubMed] [Google Scholar]
- Ravanat, J.L. , and Douki, T. (2016) UV and ionizing radiations induced DNA damage, differences and similarities. Radiat Phys Chem 128: 92–102. [Google Scholar]
- Renshaw, J. , Law, N. , Geissler, A. , Livens, F. , and Lloyd, J. (2009) Impact of the Fe (III)‐reducing bacteria Geobacter sulfurreducens and Shewanella oneidensis on the speciation of plutonium. Biogeochemistry 94: 191–196. [Google Scholar]
- Richter, K. , Schicklberger, M. , and Gescher, J. (2012) Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl Environ Microbiol 78: 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufyikiri, G. , Thiry, Y. , and Declerck, S. (2003) Contribution of hyphae and roots to uranium uptake and translocation by arbuscular mycorrhizal carrot roots under root‐organ culture conditions. New Phytology 158: 391–399. [Google Scholar]
- Rufyikiri, G. , Thiry, Y. , Wang, L. , Delvaux, B. , and Declerck, S. (2002) Uranium uptake and translocation by the arbuscular mycorrhizal fungus, Glomus intraradices, under root‐organ culture conditions. New Phytology 156: 275–281. [DOI] [PubMed] [Google Scholar]
- Ruiz Fresneda, M.A. , Martín, J.D. , Bolívar, J.G. , Cantos, M.V.F. , Bosch‐Estévez, G. , Moreno, M.F.M. , and Merroun, M.L. (2018) Green synthesis and biotransformation of amorphous Se nanospheres to trigonal 1D Se nanostructures: impact on Se mobility within the concept of radioactive waste disposal. Environmental Science: Nano 5: 2103–2116. [Google Scholar]
- Ruiz‐Fresneda, M.A. , Eswayah, A.S. , Romero‐González, M. , Gardiner, P. , Solari, P.L. , and Merroun, M.L. (2020) Chemical and structural characterization of SeIV biotransformations by Stenotrophomonas bentonitica into Se0 nanostructures and volatiles Se species. Environmental Science: Nano. 10.1039/d0en00507j [DOI] [Google Scholar]
- Ruiz‐Fresneda, M.A. , Gomez‐Bolivar, J. , Delgado‐Martin, J. , Abad‐Ortega, M.d.M. , Guerra‐Tschuschke, I. , and Merroun, M.L. (2019) The bioreduction of selenite under anaerobic and alkaline conditions analogous to those expected for a deep geological repository system. Molecules 24: 3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel, J.D. , Beaty, D.W. , Jones, M.A. , Bakermans, C. , Barlow, N.G. , Boston, P.J. , et al. (2014) A new analysis of mars “special regions”: findings of the second MEPAG Special Regions Science Analysis Group (SR‐SAG2). Astrobiology 14: 887–968. [DOI] [PubMed] [Google Scholar]
- Saito, T. , Aoyagi, N. , and Kimura, T. (2015) Time‐resolved laser‐induced fluorescence spectroscopy combined with parallel factor analysis: a robust speciation technique for UO2 2+ . J Radioanal Nucl Chem 303(2): 1129–1132. [Google Scholar]
- Salome, K.R. , Beazley, M.J. , Webb, S.M. , Sobecky, P.A. , and Taillefert, M. (2017) Biomineralization of U (VI) phosphate promoted by microbially‐mediated phytate hydrolysis in contaminated soils. Geochim Cosmochim Acta 197: 27–42. [Google Scholar]
- Sánchez‐Castro, I. , Amador‐García, A. , Moreno‐Romero, C. , López‐Fernández, M. , Phrommavanh, V. , Nos, J. , et al. (2017) Screening of bacterial strains isolated from uranium mill tailings porewaters for bioremediation purposes. J Environ Radioact 166: 130–141. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Castro, I. , Martínez‐Rodríguez, P. , Jroundi, F. , Solari, P.L. , Descostes, M. , and Merroun, M.L. (2020) High‐efficient microbial immobilization of solved U(VI) by the Stenotrophomonas strain Br 8. Water Res 183: 116110. [DOI] [PubMed] [Google Scholar]
- Schnohr, C.S. , and Ridgway, M.C. (eds). (2015) Introduction to X‐Ray absorption spectroscopy. In: X‐Ray Absorption Spectroscopy of Semiconductors. Heildelberg:Springer Series in Optical Sciences; 355p. 10.1007/978-3-662-44362-0 [DOI] [Google Scholar]
- Selvakumar, R. , Ramadoss, G. , Menon, M.P. , Rajendran, K. , Thavamani, P. , Naidu, R. , and Megharaj, M. (2018) Challenges and complexities in remediation of uranium contaminated soils: a review. J Environ Radioact 192: 592–603. [DOI] [PubMed] [Google Scholar]
- Sholkovitz, E.R. (1983) The geochemistry of plutonium in fresh and marine water environments. Earth Sci Rev 19: 95–161. [Google Scholar]
- Shukla, S.K. , Hariharan, S. , and Rao, T.S. (2020) Uranium bioremediation by acid phosphatase activity of Staphylococcus aureus biofilms: can a foe turn a friend? J Hazard Mater 384: 121316. [DOI] [PubMed] [Google Scholar]
- Sierra, J. , Rodriguez‐Puig, D. , Soriano, A. , Mensa, J. , Piera, C. , and Vila, J. (2008) Accumulation of 99mTc‐ciprofloxacin in Staphylococcus aureus and Pseudomonas aeruginosa . Antimicrob Agents Chemother 52: 2691–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkiss, K. , and Wilbur, K.M. (2012) Biomineralization. Amsterdam, Netherlands: Elsevier. [Google Scholar]
- Sivaperumal, P. , Kamala, K. , and Rajaram, R. (2018) Biosorption of long half‐life radionuclide of strontium ion (Sr+) by marine Actinobacterium nocardiopsis sp. 13H. Geomicrobiol J 35(4): 300–310. [Google Scholar]
- Sivaswamy, V. , Boyanov, M.I. , Peyton, B.M. , Viamajala, S. , Gerlach, R. , Apel, W.A. , et al. (2011) Multiple mechanisms of uranium immobilization by Cellulomonas sp. strain ES6. Biotechnol Bioeng 108: 264–276. [DOI] [PubMed] [Google Scholar]
- Smitha, M.S. , Singh, S. , and Singh, R. (2017) Microbial bio transformation: a process for chemical alterations. J Bacteriol Mycol 4: 47–51. [Google Scholar]
- Spain, A.M. , and Krumholz, L.R. (2011) Nitrate‐reducing bacteria at the nitrate and radionuclide contaminated oak ridge integrated field research challenge site: a review. Geomicrobiol J 28: 418–429. [Google Scholar]
- Stegen, J.C. , Konopka, A. , McKinley, J.P. , Murray, C. , Lin, X. , Miller, M.D. , et al. (2016) Coupling among microbial communities, biogeochemistry and mineralogy across biogeochemical facies. Sci Rep 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, A. , Hamill, P.G. , O’Kane, C.J. , Kminek, G. , Rummel, J.D. , Voytek, M.A. , et al. (2017) Aspergillus penicillioides differentiation and cell division at 0.585 water activity. Environ Microbiol 19: 687–697. [DOI] [PubMed] [Google Scholar]
- Stylo, M. , Neubert, N. , Roebbert, Y. , Weyer, S. , and Bernier‐Latmani, R. (2015) Mechanism of uranium reduction and immobilization in desulfovibrio vulgaris biofilms. Environ Sci Technol 49: 10553–10561. [DOI] [PubMed] [Google Scholar]
- Suriya, J. , Chandra Shekar, M. , Nathani, N.M. , Suganya, T. , Bharathiraja, S. , and Krishnan, M. (2017) Assessment of bacterial community composition in response to uranium levels in sediment samples of sacred Cauvery River. Appl Microbiol Biotechnol 101: 831–841. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y. , and Banfield, J.F. (1999) Geomicrobiology of uranium. Uranium: Mineral Geochem Environ 38: 393–432. [Google Scholar]
- Suzuki, Y. , Kelly, S.D. , Kemner, K.M. , and Banfield, J.F. (2003) Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl Environ Microbiol 69: 1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, P. , and Ward, A.L. (2019) Sources and distribution of 241 Am in the vicinity of a deep geologic repository. Environ Sci Pollut Res 26: 2328–2344. [DOI] [PubMed] [Google Scholar]
- Theodorakopoulos, N. , Février, L. , Barakat, M. , Ortet, P. , Christen, R. , Piette, L. , et al. (2017) Soil prokaryotic communities in Chernobyl waste disposal trench T22 are modulated by organic matter and radionuclide contamination. FEMS Microbiol Ecol 93:fix079. [DOI] [PubMed] [Google Scholar]
- Thomas, F. , Hehemann, J.‐H. , Rebuffet, E. , Czjzek, M. , and Michel, G. (2011) Environmental and gut bacteroidetes: the food connection. Front Microbiol 2: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgersen, M.P. , Ge, X. , Poole, F.L. , Price, M.N. , Arkin, A.P. , and Adams, M.W.W. (2019) Nitrate‐utilizing microorganisms resistant to multiple metals from the heavily contaminated oak ridge reservation. Appl Environ Microbiol 85: e00896–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, C.L. , Lloyd, J.R. , Law, G.T. , Williams, H.A. , Atherton, N. , Cruickshank, J.H. , and Morris, K. (2016) Retention of 99mTc at ultra‐trace levels in flowing column experiments–insights into bioreduction and biomineralization for remediation at nuclear facilities. Geomicrobiol J 33: 199–205. [Google Scholar]
- Tsezos, M. , and Volesky, B. (1982) The mechanism of uranium biosorption by Rhizopus arrhizus . Biotechnol Bioeng 24: 385–401. [DOI] [PubMed] [Google Scholar]
- Tu, H. , Lan, T. , Yuan, G. , Zhao, C. , Liu, J. , Li, F. , et al. (2019) The influence of humic substances on uranium biomineralization induced by Bacillus sp. dwc‐2. J Environ Radioact 197: 23–29. [DOI] [PubMed] [Google Scholar]
- Vernon‐Parry, K.D. (2000) Scanning Electron Microscopy : An introduction. III‐Vs Review.13 (4), 40–44. 10.1016/S0961-1290(00)80006-X. [DOI] [Google Scholar]
- Wall, J.D. , and Krumholz, L.R. (2006) Uranium reduction. Annu Rev Microbiol 60: 149–166. [DOI] [PubMed] [Google Scholar]
- Williams, K.H. , Bargar, J.R. , Lloyd, J.R. , and Lovley, D.R. (2013) Bioremediation of uranium‐contaminated groundwater: a systems approach to subsurface biogeochemistry. Curr Opin Biotechnol 24: 489–497. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Zhang, Q. , Chen, X. , Dong, F. , Chen, H. , Liu, M. , and Ali, I. (2019) Speciation distribution of heavy metals in uranium mining impacted soils and impact on bacterial community revealed by high‐throughput sequencing. Front Microbiol 10: 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X. , Luo, X. , and Zhao, M. (2016) Metagenomic analysis of microbial community in uranium‐contaminated soil. Appl Microbiol Biotechnol 100: 299–310. [DOI] [PubMed] [Google Scholar]
- Yi, Z.‐J. , and Yao, J. (2012) Kinetic and equilibrium study of uranium (VI) adsorption by Bacillus licheniformis . J Radioanal Nucl Chem 293: 907–914. [Google Scholar]
- Yun, J. , Malvankar, N.S. , Ueki, T. , and Lovley, D.R. (2016) Functional environmental proteomics: elucidating the role of a c‐type cytochrome abundant during uranium bioremediation. ISME J 10: 310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, T. , Zhang, S. , Liao, W. , Ma, H. , Lens, P.N.L. , and Xie, S. (2019) Bacterial community analysis of sulfate‐reducing granular sludge exposed to high concentrations of uranium. J Water Supply: Res Technol‐Aqua 68: 645–654. [Google Scholar]
- Zhang, X. , Cen, X. , Ravichandran, R. , Hughes, L.A. , and Van Benthem, K. (2016) Simultaneous scanning electron microscope imaging of topographical and chemical contrast using in‐lens, in‐column, and everhart‐thornley detector systems. Microscopy and Microanalysis. 22 565–575. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Liu, H. , Song, W. , Ma, W. , Hu, W. , Chen, T. , and Liu, L. (2018a) Accumulation of U(VI) on the Pantoea sp. TW18 isolated from radionuclide‐contaminated soils. J Environ Radioact 192: 219–226. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Malhotra, S. , and Francis, A. (2014) Toxicity of ionic liquids to Clostridium sp. and effects on uranium biosorption. J Hazard Mater 264: 246–253. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Song, H. , Chen, Z. , Liu, S. , Wei, Y. , Huang, J. , et al. (2018b) Biomineralization mechanism of U (VI) induced by Bacillus cereus 12–2: the role of functional groups and enzymes. Chemosphere 206: 682–692. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Shen, Y. , Wang, X. , and Wang, T. (2018) Effect of pH on uranium (VI) biosorption and biomineralization by Saccharomyces cerevisiae . Chemosphere 203: 109–116. [DOI] [PubMed] [Google Scholar]
- Zheng, J. , Tagami, K. , Watanabe, Y. , Uchida, S. , Aono, T. , Ishii, N. , et al. (2012) Isotopic evidence of plutonium release into the environment from the Fukushima DNPP accident. Sci Rep 2: 304. [DOI] [PMC free article] [PubMed] [Google Scholar]


