Summary
The surge of SARS‐CoV‐2 has challenged health systems worldwide and efficient tests to detect viral particles, as well as antibodies generated against them, are needed. Specificity, sensitivity, promptness or scalability are the main parameters to estimate the final performance, but rarely all of them match in a single test. We have developed SCOVAM, a protein microarray with several viral antigens (spike, nucleocapsid, main protease Nsp5) as capturing probes in a fluorescence immunoassay for COVID‐19 serological testing. SCOVAM depicts IgG and IgM antibody responses against each of these proteins of 22 individuals in a single microscope slide. It detects specific IgM (0.094 μg ml‐1) and IgG (~0.017 μg ml‐1) and is scalable and cost‐effective. We validated SCOVAM by comparing with a widely used chemiluminescent commercial serological test (n = 742). SCOVAM showed twice the sensitivity and allowed following seroconversion in a single assay. By analysing the prevalence 4 months later in a subset of 76 positive sera, we still detected 93.42% of positives, almost doubling the detection of the commercial assay. The higher sensitivity of SCOVAM is especially relevant to screen sera for convalescent plasma‐based treatments, high‐throughput antibody response monitoring after vaccination or evaluation of vaccine efficiency.
We have developed a test to discriminate antibodies against SARS‐CoV‐2 in the blood serum. It detects IgM and IgG antibodies against the Spike, Nucleocapsid and Main protease of the virus, simultaneously. We printed these viral proteins on slides forming microarrays and incubated them with human sera for fluorescent detection. We established the limit of detection and an algorithm for automated diagnostics. Our test resulted twice as sensitive as a commercial test, being able to better follow antibody response through time, what will be very useful to track the epidemiology of COVID‐19 and the effectivity of the developed vaccines.
Introduction
The newly arisen severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), causing COVID‐19 infections in humans, has triggered a worldwide pandemic in 2020. Most patients display mild symptoms (fever, cough, shortness of breath), typically starting around five days after the infection, but some cases show the acute respiratory distress syndrome due to the event of a so‐called cytokine storm; a dysregulated secretion of proinflammatory cytokines that leads to multi‐organ failure, septic shock and blood clots (Song et al., 2020).
The antibody response plays an important role in the neutralization of the virus (Zost et al., 2020) and the recovery from the disease (Garcia, 2020). Virus specific immunoglobulin M (IgM) antibodies can be detected around day 7 after the onset of the symptoms, while specific IgG antibodies are detected at day 10 (Zhang et al., 2020a). Though the duration of the antibody responses has not yet fully established, it has been recently demonstrated that neutralizing antibodies could be detectable 7 months after the infection (Ripperger et al., 2020).
SARS‐CoV‐2 genome encodes, like all coronaviruses (Zumla et al., 2016), structural spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. These proteins are expressed from subgenomic mRNAs. In addition, the viral RNA is translated as replicase polyproteins pp1a and pp1ab, which are autoproteolytically processed by proteases, such as the cysteine‐like main protease Nsp5 (Mpro), allowing the release of individual functional non‐structural proteins (Jin et al., 2020; Zhang et al., 2020b). While N holds the viral RNA genome, S, E and M shape the envelope of the viral particle. In addition, S interacts with host cells via its receptor, the human angiotensin converting enzyme 2 (hACE2) (Walls et al., 2020). Composed of two subunits, S1 participates in the attachment and S2 in the fusion of the viral envelope with the host cell membrane. Within S1, the receptor binding domain (S‐RBD) is the specific peptide that binds hACE2 (Hoffmann et al., 2020). As to N, it consists of three highly conserved parts: an N‐terminal (N‐NTD) RNA‐binding domain, a C‐terminal (N‐CTD) dimerization domain and a central Ser/Arg‐rich linker (Zeng et al., 2020).
The current golden standard to identify COVID‐19 resides on the detection of the virus in the respiratory tract by specific RT‐PCR amplification of its RNA genome. However, it does not inform about a possible prior infection, detecting only active cases. Instead, antibodies remain in the serum after recovery, so that they can still be detected after elimination of the virus in negative RT‐PCR tests (Vabret, et al., 2020). Current serological tests include serum chemiluminescence immunoassays (CLIA), IgG/IgM lateral immunochromatography or other S or N protein‐based ELISA tests (Bryant et al., 2020). Lateral flow‐based methods are quick and specific, but have poor sensitivity (Krammer and Simon, 2020). In the case of ELISA or CLIA assays, it is difficult to establish a robust threshold distinguishing patients that have had mild symptoms and show a poorer antibody response, and require independent assays to determine each type of Ig response against each viral protein (Kontou et al., 2020). Thus, the development of a more accurate, robust, semi‐quantitative, multiplex and multivariant methodology would better assess the protection of a population against this virus, even more with vaccination campaigns now arriving all over the world (Pandey et al., 2020).
We have developed SCOVAM, a fluorescent multiplex microarray assay that simultaneously detects and discriminates specific IgG and IgM against key SARS‐CoV‐2 proteins, efficiently identifying positives from a blind set of verified sera. SCOVAM is based on three viral proteins, what improved the sensitivity and the specificity. Thus, SCOVAM will allow a more accurate estimation of the seroprevalence and will be advantageous to monitor the serum response against specific viral proteins after vaccination.
Results
SCOVAM detects specific key anti‐SARS‐CoV‐2 antibodies in serum quantitatively
To develop a dual‐fluorescence serological assay for detecting specific IgG and IgM to SARS‐CoV‐2, we printed several viral proteins (Table 1) in duplicates at two different concentrations on a low‐density antigen microarray, called SCOVAM (SARS‐CoV‐2 Antigen Microarray). In SCOVAM, 24 identical microarrays are printed per slide to analyse 22 samples, a positive and a negative control at a time. By testing a positive serum, we can detect and discriminate IgM and IgG antibodies against SARS‐CoV‐2 (Fig. 1). We confirmed the reproducibility of the assay regardless the position on the slide (Fig. S1A), given a variation lower than 500 RFUs (relative fluorescence units) from a total range of 65 000.
Table 1.
List of SCOVAM proteins.
| CODE | Description | Expression | Buffer | Source |
|---|---|---|---|---|
| NC1 | Nucleocapsid | Insect cells | 20 mM Tris, 500 mM NaCl, pH 8, 10% glycerol | SinoBiological. Ref.: 40588‐V08B |
| NC2 | Nucleocapsid | HEK293 cells | PBS, trehalose 10% | Acro Biosystems. Ref.: NUN‐C5227 |
| NC3 | Nucleocapsid | E. coli | Phosphate pH 8, 300 mM NaCl, imidazole | CNB‐CSIC (Spain) |
| N‐NTD | Nucleocapsid, N terminal domain (43–180) | E. coli | PBS | CRG (Spain) |
| N‐CTD | Nucleocapsid, C terminal domain (250–360) | E. coli | PBS | CRG (Spain) |
| S | Spike | HEK293 cells | PBS | CRG (Spain) |
| RBD1 | Spike, RBD domain (319–541) | Insect cells | Phosphate 20 mM, 300 mM NaCl, pH 7, 10% glycerol | SinoBiological. Ref.: 40591‐V08B2 |
| RBD2 | Spike, RBD domain (319–541) | HEK293 cells | PBS | CRG (Spain) |
| Mpro | Main protease Nsp5 | E. coli | Phosphate pH 8, NaCl 300 mM, imidazole | CNB‐CSIC (Spain) |
Highlighted in grey are the selected proteins for test evaluation.
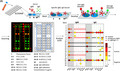
Fig. 1.
A. SCOVAM assay. Blood serum samples were incubated on 3 x 8 microarrays printed with viral proteins spike (S), nucleocapsid (N) and the main protease (Mpro). It was developed with fluorescent anti‐IgM (red) and anti‐IgG (green) antibodies. Fluorescent images were analyzed using Matlab to determine positive tests according to the relative fluorescence compared with the negative control.
B. Determination of the detection limit of the assay using known h‐IgM concentrations detected with anti‐IgM fluorescent antibodies.
C. Example of internal IgG (green) and IgM (red) calibration curves of each microarray.
D and E. A set of 89 negative (D) and 143 positive (E) sera were used to settle the threshold for positive determination (red line). Arrowheads: negative sera amongst positives, determined with SCOVAM.
To optimize the assay and avoid false positive results, we estimated the optimal serum dilution using three positive samples, previously confirmed by a CLIA assay against the N protein. We tested dilutions 1/100, 1/300, 1/900, 1/2700 and 1/8100 in three biological replicates, normalizing with a negative serum (Fig. S1B). Lower serum dilutions rendered high background signals (data not shown). We focused our analysis on those protein fragments from our panel showing a more robust response: NC2 (full length N), Mpro (main protease) and RBD1 (RBD domain of S1). The relative signal of each protein to that of the negative serum showed positive results in the three sera. In two of them, specific antibodies were detected at 1/2700 dilution, while the other serum only showed positive signals at 1/100 dilution, suggesting it as a consensus dilution for detection anti‐SARS‐CoV‐2 antibodies with SCOVAM.
SCOVAM has the advantage of being qualitative and semi‐quantitative. We estimated the concentration of IgM in serum that can be detected with SCOVAM by assaying serial dilutions of known h‐IgM concentration, as if it were serum samples, on a microarray chip printed with goat anti‐human‐IgM antibodies (Fig. 1B). From the relative fluorescence units (RFUs) on the capturing antibody spots, we calculated the limit of detection of IgM to be 4.71 ng per 50 μl of sample (94.2 ng ml‐1). Additionally, in each SCOVAM assay, we included calibration spots with known concentrations of h‐IgM and h‐IgG, to correlate the RFU of each serum sample to the number of antibody molecules (Fig. 1C).
Setting up the criteria for positive tests with SCOVAM
To establish signal thresholds to consider a serum positive for anti‐SARS‐CoV‐2 IgG antibodies with SCOVAM, we tested a set of 232 sera, 143 corresponding to patients with mild COVID‐19 symptoms (positive), and 89 to pre‐pandemic patients or blood donors (negative). The presence of anti‐SARS‐CoV‐2 antibodies had previously been verified with IgG CLIA (with protein N).
Based on the obtained signals (Fig. 1D and E, Fig. S2), we set a two‐parameter threshold: a signal to noise ratio (SNR) over three in each replicate, and a signal two times greater than the sum of the negative control and two standard deviations, for any of the selected proteins. The latter (100% threshold) was deliberately rather restrictive to avoid false positive results (Fig. 1D). Regarding the positive sera set (Fig. 1E), most positive sera presented a strong response against all proteins in SCOVAM, which coincided with the CLIA test in 141 out of 143 sera (98.6%). Considering each protein independently, protein NC2 identified 138 out of 143 positives (96.6%); protein Mpro, 121 out of 143 (84,6%); and RBD1, 127 out of 143 (88,8%). Thus, only two positive sera resulted negative for all proteins using SCOVAM and, remarkably, both showed no relevant signals for any of the proteins. Interestingly, some positive sera exhibited a strong response only against one viral protein, suggesting that some patients develop a limited but still specific response against the virus. Altogether, SCOVAM showed a specificity of 100% and a sensitivity of 98.6%, compared with the CLIA assay using the established thresholds.
To escalate, automatize the whole process, and manage the large amount of data from each 24‐sample chip, we created an algorithm (Fig. S2A) that automates sample evaluation and plots the results for an easy visualization. Considering that early screening of SARS‐CoV‐2 IgMs can improve the accuracy of epidemiological prevention and control, we also included the evaluation of the IgM response in the analysis (Fig. S2B and C). Given the unusual characteristics of IgM (Gong and Ruprecht, 2020), there is no robust test yet detecting IgM antibody with high specificity (Krammer and Simon, 2020; Wang, et al., 2020). Therefore, we established a more restrictive threshold to consider a serum to be IgM‐positive, being in this case a 300% of the sum of the negative control signal and two standard deviations. When applied to the pre‐pandemic sera, this threshold was valid for all sera for proteins Mpro and N, and 83 out of 89 sera (93.2%) when S‐RBD was considered (Fig. S2B), indicating that natural IgMs can be specific enough against SARS‐CoV‐2 in humans with no previous contact with the virus. We considered just the IgM/IgG ratio, which can hint at the stage of infection of positive patients, and established a threshold of > 1 in this ratio to consider an early‐stage infection. A particular case tested just right at the beginning of the infection (also determined by PCR) (Fig. 2A), revealed an IgM‐only response. We followed IgM and IgG dynamics through time, and 30 days after the initial extraction, IgG response clearly dominated over IgM. Interestingly, only anti‐RBD1 IgM antibodies were present in this serum, while IgGs could be detected against all proteins.
Fig. 2.
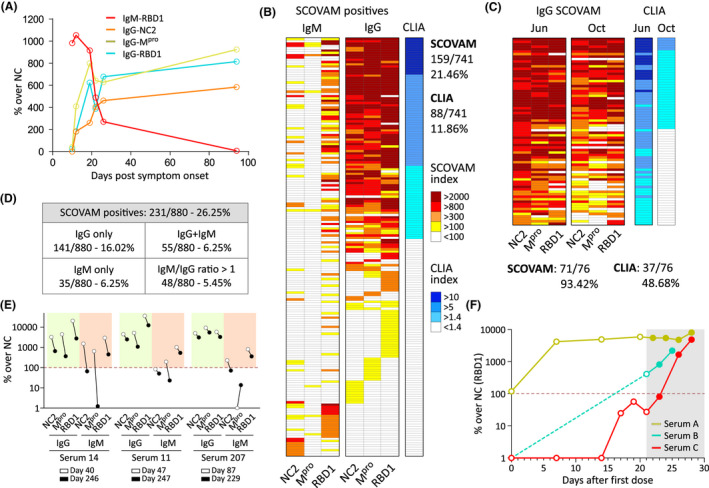
A. IgM to IgG seroconversion was followed in one individual.
B. SCOVAM validation with CLIA. The same set of sera (n = 742) were analyzed using both assays. White boxes indicate negative results.
C. A subset of sera (n = 76) from positives among the validation set (determined with the CLIA) were analyzed 4 months after the first extraction with the CLIA and SCOVAM assays.
D. Another set of sera was analysed with SCOVAM rendering similar results.
E. Serum prevalence 8 months after the infection of selected sera (n = 3) IgG and IgM signals were analyzed against the three viral proteins.
F. IgG response after vaccination with the Pfizer vaccine (RBD1 only). Serum samples were extracted at the indicated timepoints after the first inoculation. Shadowed: second dose. Dotted line: positive threshold.
Validation of SCOVAM with a random set of samples
We validated SCOVAM against the CLIA assay using 742 sera samples from voluntary donors residing in the Madrid metropolitan area (ages 25–65). Regarding SCOVAM (Fig. 2B), we detected 159 IgG‐positive sera (21.46%) and 29 with IgM/IgG > 1 (3.91%). Instead, the CLIA test identified just 88 IgG‐positive sera (11.86%). A more detailed comparison showed that some positives only detected with SCOVAM just presented antibodies against either RBD1 (2.83%), Mpro (1.35%) or NC2 (1.35%). Therefore, results suggest SCOVAM is a more sensitive and informative test compared with CLIA.
SCOVAM as a potent tool to follow SARS‐CoV‐2 seroprevalence
Being able to challenge different sera against several proteins simultaneously, SCOVAM is an excellent method to follow humoral response over time. To test the longevity of humoral immunity four months after the first extraction, we collected sera from 76 high‐titre positive donors from the validation set and tested them with SCOVAM and the commercial CLIA assay (Fig. 2C). SCOVAM identified 71 out of the 76 sera (93.42%) to be IgG‐positive against SARS‐CoV‐2, while 5 (6.58%) were under the limit of detection. Contrarily, the commercial CLIA assay detected specific antibodies in only 37 out of the 76 (48.68%).
Over the curse of the study, we have analysed additional 880 samples from a more randomized population of volunteers within central Spain (Fig. 2D). This set showed 231 (26.25%) positive sera, being 141 (16.02%) only IgG‐positive, 55 (6.25%) IgM + IgG positive, 35 (3.98%) IgM positive only, with 5.45% having IgM/IgG > 1. We also followed the seroprevalence of PCR‐positive volunteers over a longer time. These sera kept a consistent level of antibodies even over 8 months after the onset of symptoms (Fig. 2E).
Finally, in a preliminary study, we analysed sera samples (A,B,C) from three people having received the COVID‐19 vaccine (Pfizer) (Fig. 2F). While ‘B’ and ‘C’ had not been infected with SARS‐CoV‐2, ‘A’ had the infection 4 months before and was still positive for anti‐RBD1 antibodies prior to the first dose. Thus, after 7 days, serum ‘A’ already showed a specific boost of anti‐RBD1 antibodies and the second dose, given after 21 days, did not have an additional effect. Regarding serum ‘B’, it was already positive at the second dose, but this second injection boosted the response even further. That was not the case of serum ‘C’, which needed both doses to boost the signal significantly. These results point out once more to the wide variety of the humoral response against SARS‐CoV‐2 in the population.
Discussion
Most COVID‐19 serological tests focus on the specific binding of circulating antibodies to just one viral protein, which differs among those used by researchers and clinicians (Ravi et al., 2020). In this study, we developed SCOVAM, a low‐density protein microarray that allows integrated high throughput detection of IgM and IgG antibodies in human sera against representative SARS‐CoV‐2 proteins related to cell invasion (Spike, RBD1), its basic structure (Nucleocapsid, NC2) and its activity within the cells (Main protease, Mpro).
Starting from nine different purified protein fragments and production hosts, we looked for those representing better robustness, sensitivity and specificity in the detection. The full‐length nucleocapsid (N) produced in insect (NC1) or human HEK293 (NC2) cells similarly detected specific antibodies, while NC3 had a lower signal. Alternatively, we tested the N‐terminal (N‐NTD) and the C‐terminal (N‐CTD) domains separately (Peng et al., 2020). While N‐NTD robustly detected antibodies in positive sera, N‐CTD showed weaker signals in most of the sera, suggesting that N‐NTD is more immunogenic. Similarly, the main protease (Mpro), implicated in the processing of the early polypeptides of the virus (Hegyi and Ziebuhr, 2002), showed good performance, mainly in sera having high antibody titres. Full‐length S was not robustly recognized by positive sera, maybe because of an inappropriate structure when printed on the slide and/or because it is glycosylated, what may hide surface epitopes from antibody recognition (Watanabe et al., 2020; Zhao et al., 2020). Instead, RBD1 produced in insect cells robustly reacts with positive sera even at 1:8100 serum dilutions or with low antibody titres.
These results highlight the importance of using a particular viral antigen for the detection, possibly explaining the sensitivity differences between serological tests (Haselmann et al., 2020). Furthermore, they reinforce SCOVAM as a multiplex assay, which can easily be implemented with micro/nano printing technology and high‐throughput liquid handling platforms. The automatization of the assay using robotic platforms and the rapid analysis of results facilitated by the script described here, will grant a virtually automatized process, where only sampling of sera would be hand operated. The need of just micro/nano amounts of materials translates into an affordable test, estimated in below 0.5€ per assay, that could be optimized with its mass production using non‐contact nanodispensing robots, which print over 100 slides (24 assays in each) in 1 h. Alternatively, SCOVAM could be printed on the bottom of 96‐well microtiter plates, to be handled as a standard ELISA, and read with commercial two‐laser scanners.
We settled the limits that discriminate negative and positive sera for IgG, testing a set of 232 sera with SCOVAM. Though the selected threshold avoided false positive results for all proteins, the test presented two discrepancies with commercial CLIA among the positives. Those assays were repeated rendering similar results (data not shown). Given that those patients were not tested by PCR, and the CLIA has a specificity of 98%, it is feasible that these were indeed false positive results of the CLIA. Regarding IgM, its total concentration in human serum is estimated to be 500–2000 μg ml‐1. We could detect less than 0.094 μg ml‐1 with confidence, which is at least 50 times lower than the expected concentrations found in the 1/100 dilutions of the tested sera. The limit of detection of IgG cannot be tested with this assay but can be inferred to be 0.016 μg ml‐1, considering its size being 1/6 smaller than IgM. These values are consistent with the literature (Lavinder, et al., 2014), where was reported that non‐dominant blocking antibody clones are found at a concentration of 0.15–0.60 μg ml‐1.
IgM detection with SCOVAM showed variability among pre‐pandemic sera, mostly associated with protein RBD1, which showed higher sensitivity but also several false positive results, even with a stricter threshold. Conversely, variations among the positives may be related to the severity of the disease, or due to the waning observed in recent antibody surveys involving IgM (Gudbjartsson et al., 2020; Perreault et al., 2020). Thus, IgM detection should not serve as an indicator for positive determination, but rather an informative marker to identify early states of the infection in a patient.
The validation of our assay with a set of 742 sera showed that a 21.46% of them were positive with SCOVAM by June 2020. This incidence almost doubles that reported (11.50%) by a wider survey performed in the area of Madrid at that time (Pollan et al., 2020), which used two different serological tests ‐lateral flow quick test and CLIA‐, both detecting just the nucleocapsid. These discrepancies are consistent with the higher sensitivity exhibited in this work by SCOVAM compared with CLIA. Furthermore, SCOVAM detected some individuals carrying antibodies against just S‐RBD or Mpro, protein, antigens absent in most used commercial tests, which use N protein only. Again, SCOVAM exhibited nearly twice (93.42%) the sensitivity than the CLIA assay (48.68%) of a selection of positives from this set, 4 months after the first extraction. Additionally, we observed that the IgG titer was maintained for all proteins 7–8 months after the onset of symptoms, in line with recent surveys (Ripperger et al., 2020), while specific IgMs were only present against S‐RBD.
Regarding the IgM response, several volunteers from the additional set of 880 sera showing an IgM/IgG > 1 rendered a negative PCR result (not shown). This could be indicative of either the presence of natural IgMs (Gong and Ruprecht, 2020) protecting them from disease, or a delay between blood extraction and the PCR testing, during which the viral load could have been depleted, given the absence of symptoms. These results raise the question of whether natural IgM against S‐RBD could protect against the virus due to direct neutralization. Nevertheless, we detected several IgM‐only sera in PCR‐positive individuals, what allowed following the humoral response over time (Fig. 2A), indicating that changes in the IgM/IgG ratio are indeed informative of early stages of the infection.
Finally, anti‐RBD antibodies have been directly correlated with neutralizing antibodies against SARS‐CoV‐2 (Jackson et al., 2020; Zost et al., 2020). SCOVAM would be useful to screen candidate sera for treatments based on COVID‐19 convalescent plasma, tested in more than 100 clinical trials (Duan et al., 2020), and a valuable tool for the analysis and monitoring of immunized individuals after vaccination with different vaccine types. Here, we showed that individualized responses take place upon vaccination, so SCOVAM offers a unique opportunity to check the antibody pattern of vaccinated individuals. SCOVAM can also be implemented with new target antigens, variants of the virus (Weisblum et al., 2020), or even with other markers that could be relevant for the progression of the COVID illness, all‐in‐one assay.
Experimental procedures
Antigen production and purification
SCOVAM antigens are described in Table 1. Protease Nsp5 (Mpro) was produced in E. coli (patent pending). RBD1 was purchased to SinoBiological (produced in insect cells). RBD2 was produced following an established protocol (Stadlbauer et al., 2020). N‐NTD or N‐CTD sequence were inserted into pETM14, with a N‐terminal His‐tag, for expression in E. coli BL21 DE3 strain (IPTG 0.2 mM, 16 h, 18°C). Cells were resuspended in Tris 20, 250 mM NaCl, 10 mM Imidazole, 0.5% Triton‐X100 (Merck, Darmstadt, Germany) and complete mini EDTA‐free protease (Roche, Basel, Switzerland), disrupted using a French Press and centrifuged (30 min, 4°C, 30 000 g). Proteins were purified from the supernatant using Hitrap Ni‐NTA column (GE Healthcare, Boston, MA, USA) and an increasing Imidazol gradient (up to 500 mM). They were dialysed against PBS, concentrated by Vivaspin 5KD (Merck) and quantified using a NanoDrop.
Generation of SCOVAM
Antigens were diluted to 0.2, 0.1 or 0.05 mg ml‐1 in sciSPOT Protein D1 buffer (Scenion, Berlin, Germany) with 0.01% Tween20, as well as two‐fold dilutions of h‐IgM (from 40 mg ml‐1) and h‐IgG (from 20 mg ml‐1). 20 μl aliquots were distributed in a 384‐well plate as source for printing on epoxy‐activated microscope slides (Cel Associates Inc., Pearland, TX, USA) with a MicroGrid TAS II 600 (Biorobotics, Boston, MA, USA). A fluorescently labelled rabbit non‐immunized IgG fraction (10 μg ml‐1) was used as frame.
SCOVAM assay with human sera
Blood sera were obtained after written informed consent. Samples were heat‐inactivated (56ºC, 30 min) and cooled to RT. SCOVAM was blocked (Tris–HCl 0.5 M pH 8, 2% BSA, RT, 30 min), dried in a minicentrifuge and placed in an 3 × 8 microarray hybridization cassette (Arrayit, Sunnyvale, CA, USA). We incubated 50 μl of each serum (1 h, RT) on a plate shaker (600 rpm) at the indicated dilutions in 1x PBS, 0.2% Tween 20 (PBST). Wells were washed three times (PBST). For detection, wells were incubated with fluorescently labelled goat anti‐human‐IgG‐Alexa555 and goat anti‐human‐IgM‐Alexa647 (Thermo Fisher Scientific, Waltham, MA, USA) at the optimal titrated dilution (20–80 ng ml‐1) in PBST, 1% BSA. Slides were washed, dried and scanned for fluorescence in GenePix 4100A scanner at 532 and 635 nm. Fluorescence was quantified with GenePixPro (Molecular Devices, San Jose, CA, USA) and data analysed using Matlab software (Mathworks, Natick, MA, USA) (https://github.com/MolecularEcologyLab/SCOVAM). There were no missing data in the study.
To determine the detection limit of IgM, goat anti h‐IgM was printed on epoxy slides. After blocking, slides were incubated one hour with a series of five‐fold dilutions in PBST (5 μg ml‐1 to 64 ng ml‐1) of h‐IgM, and developed with goat anti‐human‐IgM‐Alexa647, as above.
Statistics
The calibration of IgM to set the limit of detection was adjusted using Graphpad Prism software (Graphad Software, San Diego, CA, USA). The curve was fitted using non‐linear hyperbolic regression and the limit of detection was selected at 150 RFUs, where the background noise is minimal. The internal calibration of the arrays was adjusted using linear regression.
Chemiluminescence immunoassays
Inactivated sera were tested using VirClia IgG monotest, Abbott SARS‐CoV‐2 IgG or Abbott SARS‐CoV‐2 IgM, detecting the nucleocapsid. VirClia IgM + IgA monotest (Vircell, Granada, Spain) detected spike and nucleocapsid. Signals were read as a relative light units (RLU) using the VirClia or the ARCHITECT system (Abbott, Abbott Park, IL, USA), with positivity cut‐off determined by the manufacturer.
Quantitative real‐time PCR assay
Clinical samples were retrieved with a nasopharyngeal swab (Deltalab, Rubí, Spain) at the Hospital Central de la Defensa ‘Gómez Ulla’ (Spain) and inactivated with 200 μl guanidine thiocyanate. From them, viral RNA was obtained using MagCore HF16 (RBC bioscience, New Taipei City, Taiwan), Nimbus Microlab Seegene (Hamilton Company, Franklin, MA, USA) or m2000 system (Abbott). Viral RNAs of E, N and the RNA‐dependent RNA‐polymerase were amplified from 200 μl of the sample using the PCR platform Allplex 2019‐nCoV (Seegene, Seoul, Republic of Korea). Thermal cycling was performed for 45 cycles in a CFX96 (Bio‐Rad Laboratories, Hercules, CA, USA).
Conflict of interests
The authors declare no competing interests.
Author contributions
DRG, MGV, MMP, MP, MSS and VP performed the experiments. DRG, JGE, MGV, MMP and VP performed the bioinformatic analysis. HR, CC, NR, YBC, PR, SZ and LE provided technical support and edited the paper. DRG and VP wrote, supervised, and edited the manuscript.
Ethical approval
The study was conducted according to the ethical requirements established by the Declaration of Helsinki. The Ethics Committee of Hospital Central de la Defensa Gómez Ulla (Madrid) approved the study.
Supporting information
Acknowledgements
We thank all voluntary blood donors, INTA’s medical service, Dr. María Mateo, chief of the Microbiology and Parasitology Service at the Hospital ‘Gómez Ulla’, and the ‘Subdirección de Coordinación y Planes’ of INTA. This work has been funded by INTA’s internal budget and the Spanish Ministry of Science and Innovation grant nos. RTI2018‐094368‐B‐I00, and MDM‐2017‐0737 (Excelencia ‘María de Maeztu’ to Centro de Astrobiología). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. DRG is funded by the ‘Programa de Atracción de Talento’ of local Government of Madrid. Centro Superior de Investigaciones Cientificas: project number 202020E079.
Microbial Biotechnology (2021) 14(3), 1228–1236
Contributor Information
David Ruano‐Gallego, Email: druano@cab.inta-csic.es.
Victor Parro, Email: parrogv@cab.inta-csic.es.
References
- Bryant, J.E. , Azman, A.S. , Ferrari, M.J. , Arnold, B.F. , Boni, M.F. , Boum, Y. , et al. (2020) Serology for SARS‐CoV‐2: Apprehensions, opportunities, and the path forward. Sci Immunol 5: eabc6347. [DOI] [PubMed] [Google Scholar]
- Duan, K. , Liu, B. , Li, C. , Zhang, H. , Yu, T. , Qu, J. , et al. (2020) Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proc Natl Acad Sci USA 117: 9490–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, L.F. (2020) Immune Response, inflammation, and the clinical spectrum of COVID‐19. Front Immunol 11: 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, S. , and Ruprecht, R.M. (2020) Immunoglobulin M: an ancient antiviral weapon ‐ rediscovered. Front Immunol 11: 1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson, D.F. , Norddahl, G.L. , Melsted, P. , Gunnarsdottir, K. , Holm, H. , Eythorsson, E. , et al. (2020) Humoral Immune response to SARS‐CoV‐2 in Iceland. N Engl J Med 383: 1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselmann, V. , Kittel, M. , Gerhards, C. , Thiaucourt, M. , Eichner, R. , Costina, V. , and Neumaier, M. (2020) Comparison of test performance of commercial anti‐SARS‐CoV‐2 immunoassays in serum and plasma samples. Clin Chim Acta 510: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi, A. , and Ziebuhr, J. (2002) Conservation of substrate specificities among coronavirus main proteases. J Gen Virol 83: 595–599. [DOI] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Kruger, N. , Herrler, T. , Erichsen, S. , et al. (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, L.A. , Anderson, E.J. , Rouphael, N.G. , Roberts, P.C. , Makhene, M. , Coler, R.N. , et al. (2020) An mRNA vaccine against SARS‐CoV‐2 ‐ preliminary report. N Engl J Med 383: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z. , Du, X. , Xu, Y. , Deng, Y. , Liu, M. , Zhao, Y. , et al. (2020) Structure of M(pro) from SARS‐CoV‐2 and discovery of its inhibitors. Nature 582: 289–293. [DOI] [PubMed] [Google Scholar]
- Kontou, P.I. , Braliou, G.G. , Dimou, N.L. , Nikolopoulos, G. , and Bagos, P.G. (2020) Antibody tests in detecting SARS‐CoV‐2 infection: a meta‐analysis. Diagnostics (Basel) 10: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer, F. , and Simon, V. (2020) Serology assays to manage COVID‐19. Science 368: 1060–1061. [DOI] [PubMed] [Google Scholar]
- Lavinder, J.J. , Wine, Y. , Giesecke, C. , Ippolito, G.C. , Horton, A.P. , Lungu, O.I. , et al. (2014) Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA 111: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S.C. , Pande, V. , Sati, D. , Upreti, S. , and Samant, M. (2020) Vaccination strategies to combat novel corona virus SARS‐CoV‐2. Life Sci 256: 117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Du, N. , Lei, Y. , Dorje, S. , Qi, J. , Luo, T. , et al. (2020) Structures of the SARS‐CoV‐2 nucleocapsid and their perspectives for drug design. EMBO J e105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault, J. , Tremblay, T. , Fournier, M.J. , Drouin, M. , Beaudoin‐Bussieres, G. , Prevost, J. , et al. (2020) Waning of SARS‐CoV‐2 RBD antibodies in longitudinal convalescent plasma samples within four months after symptom onset. Blood 136: 2588‐2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollan, M. , Perez‐Gomez, B. , Pastor‐Barriuso, R. , Oteo, J. , Hernan, M.A. , Perez‐Olmeda, M. , et al. (2020) Prevalence of SARS‐CoV‐2 in Spain (ENE‐COVID): a nationwide, population‐based seroepidemiological study. Lancet 396: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, N. , Cortade, D.L. , Ng, E. , and Wang, S.X. (2020) Diagnostics for SARS‐CoV‐2 detection: a comprehensive review of the FDA‐EUA COVID‐19 testing landscape. Biosens Bioelectron 165: 112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger, T.J. , Uhrlaub, J.L. , Watanabe, M. , Wong, R. , Castaneda, Y. , Pizzato, H.A. , et al. (2020) Orthogonal SARS‐CoV‐2 serological assays enable surveillance of low‐prevalence communities and reveal durable humoral immunity. Immunity 53: 925–933 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, P. , Li, W. , Xie, J. , Hou, Y. , and You, C. (2020) Cytokine storm induced by SARS‐CoV‐2. Clin Chim Acta 509: 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer, D. , Amanat, F. , Chromikova, V. , Jiang, K. , Strohmeier, S. , Arunkumar, G.A. , et al. (2020) SARS‐CoV‐2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 57: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret, N. , Britton, G.J. , Gruber, C. , Hegde, S. , Kim, J. , Kuksin, M. , et al. (2020) Immunology of COVID‐19: Current State of the Science. Immunity 52: 910–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A.C. , Park, Y.J. , Tortorici, M.A. , Wall, A. , McGuire, A.T. , and Veesler, D. (2020) Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 181: 281–292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Du, Q. , Guo, B. , Mu, D. , Lu, X. , Ma, Q. , et al. (2020) A method to prevent SARS‐CoV‐2 IgM false positives in gold immunochromatography and enzyme‐linked immunosorbent assays. J Clin Microbiol 58: e00375‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y. , Allen, J.D. , Wrapp, D. , McLellan, J.S. , and Crispin, M. (2020) Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science 369: 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum, Y. , Schmidt, F. , Zhang, F. , DaSilva, J. , Poston, D. , Lorenzi, J.C. , et al. (2020) Escape from neutralizing antibodies by SARS‐CoV‐2 spike protein variants. eLife 9: e61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, W. , Liu, G. , Ma, H. , Zhao, D. , Yang, Y. , Liu, M. , et al. (2020) Biochemical characterization of SARS‐CoV‐2 nucleocapsid protein. Biochem Biophys Res Commun 527: 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Nie, S. , Zhang, Z. , and Zhang, Z. (2020a) Longitudinal change of severe acute respiratory syndrome coronavirus 2 antibodies in patients with coronavirus disease 2019. J Infect Dis 222: 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Lin, D. , Sun, X. , Curth, U. , Drosten, C. , Sauerhering, L. , et al. (2020b) Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved alpha‐ketoamide inhibitors. Science 368: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, P. , Praissman, J.L. , Grant, O.C. , Cai, Y. , Xiao, T. , Rosenbalm, K.E. , et al. (2020) Virus‐receptor interactions of glycosylated SARS‐CoV‐2 spike and human ACE2 receptor. Cell Host Microbe 28: 586‐601.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost, S.J. , Gilchuk, P. , Case, J.B. , Binshtein, E. , Chen, R.E. , Nkolola, J.P. , et al. (2020) Potently neutralizing and protective human antibodies against SARS‐CoV‐2. Nature 584: 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A. , Chan, J.F. , Azhar, E.I. , Hui, D.S. , and Yuen, K.Y. (2016) Coronaviruses ‐ drug discovery and therapeutic options. Nat Rev Drug Discov 15: 327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


