Summary
Several studies have recently pointed towards an increased occurrence and prevalence of several taxa of the lactic acid bacteria (LAB) in the microbiota of the upper respiratory tract (URT) under healthy conditions versus disease. These include several species of the Lactobacillales such as Lacticaseibacillus casei, Lactococcus lactis and Dolosigranulum pigrum. In addition to physiological studies on their potential beneficial functions and their long history of safe use as probiotics in other human body sites, LAB are thus increasingly to be explored as alternative or complementary treatment for URT diseases. This review highlights the importance of lactic acid bacteria in the respiratory tract and their potential as topical probiotics for this body site. We focus on the potential probiotic properties and adaptation factors that are needed for a bacterial strain to optimally exert its beneficial activity in the respiratory tract. Furthermore, we discuss a range of in silico, in vitro and in vivo models needed to obtain better insights into the efficacy and adaptation factors specifically for URT probiotics. Such knowledge will facilitate optimal strain selection in order to conduct rigorous clinical studies with the most suitable probiotic strains. Despite convincing evidence from microbiome association and in vitro studies, the clinical evidence for oral or topical probiotics for common URT diseases such as chronic rhinosinusitis (CRS) needs further substantiation.
Several studies have recently pointed towards an increased occurrence and prevalence of several taxa of the lactic acid bacteria (LAB) in the microbiota of the upper respiratory tract (URT) under healthy conditions versus disease. This review highlights the importance of lactic acid bacteria in the respiratory tract, and their potential as topical probiotics for this body site. We focus on the potential probiotic properties and adaptation factors that are needed for a bacterial strain to optimally exert its beneficial activity in the respiratory tract.
Introduction
Inflammatory upper respiratory tract (URT) diseases such as rhinitis, and acute and chronic rhinosinusitis (CRS), impose a major burden on public health and account for significant healthcare costs (Meltzer, 2016; Hellings et al., 2017). Rhinitis is the most common URT disease defined as symptomatic inflammation of the lining of the nose that is caused by infectious agents, allergens or other factors (e.g., drugs and hormones) (Bousquet et al., 2008). Allergic rhinitis (AR) represents the most commonly encountered type of non‐infectious rhinitis, while the term rhinosinusitis is defined as inflammation of the nose and paranasal sinuses, characterized by two or more symptoms, one of which should be either nasal obstruction or nasal discharge (Fokkens et al., 2020). When the latter condition lasts for more than 12 weeks, it is defined as chronic rhinosinusitis (CRS) (Fokkens et al., 2020). These disorders have in common that inflammation, and sometimes also a disruption of the nasal epithelial barrier, underlie the pathology. In addition, a disbalance of the microbial communities or dysbiosis inhabiting the airways, the ‘airway microbiota’, has been suggested as a key factor in the pathology of these URT diseases.
Dysbiosis in URT diseases
Recent research shows that a balanced airway microbiota plays an important gatekeeper role for respiratory health, as reviewed by (Man et al., 2017), although it is yet very difficult to determine what defines a balanced microbiota. It is generally seen as the counterpart of dysbiosis. Such an imbalance in the composition and metabolic activity of our microbiota can appear in several manners, for example as a loss of beneficial microorganisms, or an excessive growth of potential pathobionts (Wilkins et al., 2019). The identification of beneficial versus potential pathobionts is not always straightforward. Bacterial genera that are commonly abundant in the URT of healthy individuals, without clear symptoms, belong to Staphylococcus, Corynebacterium, Propionibacterium, Dolosigranulum and Streptococcus species (Man et al., 2017; Kumpitsch et al., 2019). These taxa can thus be seen to contain potential beneficial strains. Pathobionts such as Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis appear to be more prevalent or abundant in URT diseases (Van Eldere et al., 2014; Duell et al., 2016; Mahdavinia et al., 2016; van den Broek et al., 2019). In addition to these classic URT pathogens, other emerging pathogens, such as Corynebacterium tuberculostearicum and Stenotrophomonas, have been suggested based on microbiome data (Abreu et al., 2012; Chalermwatanachai et al., 2018; Koeller et al., 2018). Nevertheless, it remains highly challenging to investigate whether these changes and disruptions in the microbiome are either a causality or a consequence.
Although various treatment options yet exist for CRS and AR, there is a lack of broadly applicable and effective treatment approaches (Hellings et al., 2017; Meng et al., 2019). Current options include mainly nasal irrigations with saline solutions, systemic and/or topical corticosteroid treatment, antihistamines (in AR) (Meng et al., 2019), antibiotic treatments (often macrolides because they also have anti‐inflammatory actions) and functional endoscopic sinus surgery or FESS (in CRS) (Fokkens et al., 2020). The latter surgery has the aim to clear the diseased mucosa, eliminate the infection, relieve the obstruction and restore the sinus ventilation (Piromchai et al., 2013). However, patients do not always respond to the current treatment options and patients often relapse, even after the surgery. For example, CRS patients often receive multiple antibiotic treatments, leading to several disadvantages, such as microbiota disruption and multidrug‐resistant bacteria (Szaleniec et al., 2019). Also for otitis media (OM), the most common URT infection in children, antibiotic prescription rates are high despite clinical guidelines limiting their use to strict indications (reviewed in van den Broek et al., 2019). There is thus a clear need for alternative or complementary treatment options for these highly prevalent URT diseases. In this review, we will describe the current knowledge on the potential of lactic acid bacteria as topical probiotics for the URT, with a focus on the probiotic properties and adaptation factors that are needed for a bacterial strain to exert its beneficial activity in the URT. In addition, future perspectives and challenges for URT probiotics will be addressed.
The potential of probiotics for the URT
Probiotics have been predominantly used to improve intestinal health with many promising clinical results up to date (Sanders et al., 2019). However, it is only recently that we start to explore the potential for other applications. Yet, the definition of probiotics, as formulated by an expert panel of the International Scientific Association on Probiotics and Prebiotics (ISAPP) is not limited to gut applications. Probiotics are ‘live microorganisms that, when administered in adequate amounts, confer a health benefit on the host’ (Hill et al., 2014). Most probiotics studied so far are lactic acid bacteria (LAB) (order Lactobacillales), with some of the most promising probiotic strains belonging to the Lactobacillaceae family, recently reclassified (Zheng et al., 2020). Lactobacilli are an interesting choice as probiotics, since they have a long history of safe use for over 100 years by people of all ages on a daily basis in dairy products, fermented foods and food supplements (Salvetti and O’Toole, 2017). Yet, their potential as URT probiotics is not yet widely considered.
Lactic acid bacteria have a habitat in the respiratory tract
In the recent decade, members of LAB, including Lacticaseibacillus, Dolosigranulum and Lactococcus species, have been described as part of the normal URT microbiota of healthy adults and/or children (Bogaert et al., 2011; Laufer et al., 2011; Jensen et al., 2013; Ling et al., 2013; Stearns et al., 2015). Lactobacilli are for instance detected in the nasopharynx of Chinese (Ling et al., 2013; Gan et al., 2019), Canadian (Stearns et al., 2015), and Belgian individuals (De Boeck et al., 2020) and the tonsillar crypts of children and adults (Jensen et al., 2013). The pilot study by Abreu and colleagues was one of the first that reported that certain LAB taxa, including Latilactobacillus sakei, was decreased in CRS patients, pointing towards a potential benefit for sinus health. Furthermore, they developed a murine model of sinus infection, where they demonstrated that L. sakei ATCC15521 can protect the sinus mucosa from C. tuberculostearicum‐induced pathogenesis after nasal inoculation, using goblet cell hyperplasia and mucin hypersecretion as outcome parameters (Abreu et al., 2012). These results were obtained in a not yet widely applied murine model with only small sample sizes (n = 3) and have to the best of our knowledge not yet been confirmed in larger sample sizes. In our recent microbiome comparison study between healthy controls and CRS patients, various genera of the Lactobacillaceae including Lactiplantibacillus, Latilactobacillus and Lacticaseibacillus were also found to be more prevalent and abundant in healthy controls compared with CRS patients (De Boeck et al., 2019, 2020) (Figure 1). Similarly, a lower abundance of lactobacilli was recently reported within the nasal microbiome of AR patients compared with healthy controls (Gan et al., 2020). Compared with other human body sites, such as the vagina (Petrova et al., 2015) and the gastrointestinal tract (Heeney et al., 2018), the abundances of lactobacilli in the URT (Abreu et al., 2012; Ling et al., 2013; Stearns et al., 2015; De Boeck et al., 2020), are much lower (Figure 1). Yet, this does not exclude their potentially important ecological and health‐promoting role as we will pinpoint in the next paragraphs.
Fig. 1.
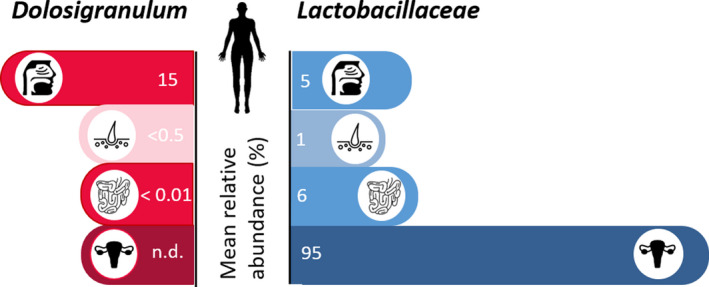
Mean relative abundances of Dolosigranulum (red bars) and Lactobacillaceae (blue bars) as part of the microbiome in different human body habitats. Based on our research (De Boeck et al., 2019,2020) and available literature (Laufer et al., 2011; Biesbroek et al., 2014a; Stearns et al., 2015; Hasegawa et al., 2017; Gan et al., 2020), Dolosigranulum and Lactobacillus are proposed as indicator taxa for health. Using the curatedMetagenomicData R‐package based on publicly available shotgun sequencing data data (Pasolli et al., 2017), their mean relative abundances were calculated in the URT, skin, gut and vagina. N.d., not detected.
In contrast to Lactobacillaceae, other LAB taxa are more prevalent in the URT. The less explored species Dolosigranulum pigrum has particularly gained interest as potential next‐generation URT probiotic, because it is found in high abundances that can range up to 50% in the healthy URT (Laufer et al., 2011; Biesbroek, Tsivtsivadze, et al., 2014; Hasegawa et al., 2017; Gan et al., 2019). It is also often more prevalent or more dominant in healthy subjects than in diseased individuals and is therefore believed to be associated with URT health (Biesbroek, Tsivtsivadze, et al., 2014; Hasegawa et al., 2016; De Boeck et al., 2019; Gan et al., 2019). Of note, based on presence and relative abundance, vaginal delivery and breastfeeding are associated with this potential health‐promoting bacterium (Biesbroek et al., 2014a; Bosch et al., 2016). In addition, a lower incidence of parental‐reported URT infections was reported in children that had higher numbers of Dolosigranulum (Biesbroek, Tsivtsivadze, et al., 2014). A lower likelihood of bronchiolitis was also observed in children with higher relative abundances of Dolosigranulum detected in their bacterial profiles (Hasegawa et al., 2017). Gan and colleagues observed a significantly higher abundance of the genus Dolosigranulum in the middle meatus of a control group (mean relative abundance of 8.52%) compared with the same sites sampled in subjects with CRS with nasal polyps (CRSwNP) (mean relative abundance < 1%) (Gan et al., 2019). Our recently published microbiome study also identified D. pigrum to be more associated with the anterior nares and nasopharynx of healthy controls compared with CRS patients (De Boeck et al., 2019). Interestingly, a higher abundance of Dolosigranulum was also observed in the nasopharyngeal microbiomes of healthy children compared to those with recurrent acute OM, suggesting that the link of Dolosigranulum with health might extend beyond the nasal cavity (Lappan et al., 2018).
Based on these microbiome studies, several taxa of the Lactobacillaceae and Dolosigranulum are thus promising URT taxa to further explore their potential as probiotics for the URT, both mechanistically and in clinical trials, which is at the moment still a rather uncharted field. However, insights from probiotic research at other human body sites such as the gut can help to define the requirements and considerations that are needed to select the most suitable strains for clinical trials.
Properties that mediate the activity of probiotics in the URT
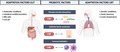
In order for a probiotic to exert beneficial health‐promoting effects in the targeted host site, it must possess an appropriate set of beneficial properties, as well as properties that help the microorganism to adapt and thrive –at least temporarily –in the target site (Lebeer et al., 2008). The first set of properties can be classified as probiotic factors because they are directly involved in the health benefits, while the second set refers more to adaptation factors. Both set of factors are important for effective probiotics. To screen for and select the most suitable probiotic candidates, it is important that the specific ecological and physiological conditions of the target site are well‐understood. Bacterial strains that colonize the URT for instance will experience different selective pressures compared with bacteria in the gastrointestinal tract. Interactions with the host cell types and mucus, specific components of the innate and adaptive respiratory immune system, exposure to medical treatments and competition with other bacterial/microbial members are different at the various body sites. Consequently, strains from the URT itself might hold specific advantages, compared with the probiotics that are already available on the market and mainly originate from the human gut or food. For example, in a study of Mårtensson et al. (2017), the effects of a LAB spray in CRS patients without nasal polyps were tested. They did not observe beneficial effects of the used probiotic spray on the disease outcomes, but could show that the spray was well‐tolerated and safe to use. It is tempting to speculate that this lack of efficacy might be linked to the fact that the LAB strains that were used in the study originate from honeybee and might be not adapted to the human URT (Mårtensson et al., 2017). While probiotic factors related to immune modulation and antimicrobial action might be more conserved across different human body habitats, adaptation factors are more determined by the specific habitat where the probiotic are applied. Figure 2 gives a schematic representation of the probiotic and adaptation factors, which we rationalized for the URT in comparison with the gastrointestinal tract based on the available evidence. In the following two paragraphs, the probiotic and adaptation factors will be discussed into more detail.
Fig. 2.
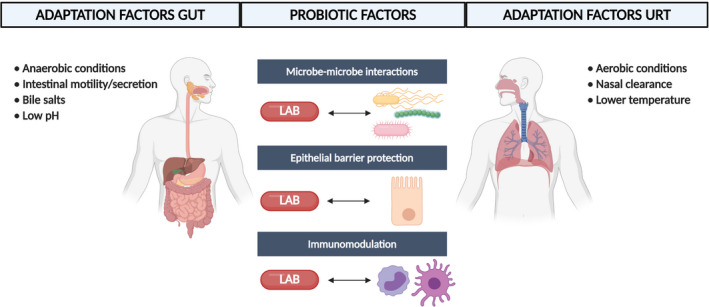
Probiotic and adaptation factors in the URT compared with the gastrointestinal tract. Probiotic factors related to antipathogenic effects, immunomodulation and barrier protection are more conserved across different human body sites, while each body site has its own unique features that determine the adaptation factors that are needed for a probiotic to be effective in that body site. Figure adapted from (Lebeer et al., 2008; Man et al., 2017) and created with BioRender.com 121 × 64 mm (300 × 300 DPI)
Adaptation factors relevant to the URT
Based on the available physiological and anatomic information on the URT (reviewed in Man et al., 2017) and how many pathogens adapt to the URT, several potential adaptation mechanisms can be envisaged for potential probiotic bacteria. First, a strong adherence capacity to the human nasal epithelium would be an important requirement for an URT probiotic, in order to be able to cope with the mechanism of nasal mucociliary clearance (Deborah and Prathibha, 2014). In addition, sufficient adherence of probiotic candidates is also of interest for competition with URT pathobionts, since adherence is often essential in their pathogenesis. S. aureus for instance produces several surface proteins, such as clumping factors B (ClfB) and fibronectin‐binding protein (FnBP), involved in adhesion to human airway epithelium (Mongodin et al., 2002; Corrigan et al., 2009). In probiotic research, knowledge on the adherence capacity of probiotic strains is mainly limited towards the gastrointestinal tract and much remains to be explored for the URT. In gastrointestinal probiotic strains, adherence was shown to be facilitated via pili or surface adhesin proteins. For instance, in the model strain Lactocaseibacillus rhamnosus GG, SpaCBA pili have been described that mediate adhesion to intestinal mucus and epithelial cells (Kankainen et al., 2009). Recently, we also demonstrated that these pili are one of the key factors promoting its adhesion to human epithelial keratinocytes and interfering with adhesion of S. aureus (Spacova et al., 2020a,2020b) showing that the SpaCBA pili are adaptation factors in other body sites than the gut. For the URT, relatively good adherence of L. rhamnosus GG to the respiratory epithelial cell line Calu‐3 and nasal epithelial cells of patients and healthy controls was observed (De Boeck et al., 2020). Yet, a particular related URT isolate, named Lacticaseibacillus casei AMBR2 was isolated that outperformed L. rhamnosus GG in adaptation and adherence to URT epithelia. This strain from a healthy nasopharynx expresses another type of fimbriae, putatively encoded by a secA2/secY2 gene cluster. The adherence capacity was also documented in vivo in healthy volunteers, as we found that L. casei AMBR2 was able to temporally colonize the URT in healthy volunteers after nasal administration (De Boeck et al., 2020).
Second, specific environmental conditions are inherent to the URT, such as higher oxygen levels, a lower temperature and higher pH compared with instance the gastrointestinal tract. The body temperature in the nasopharynx is 34°C, while this is even lower in the anterior nares (Man et al., 2017). In addition, the pH differs significantly between different human body habitats and should be taken into account. In the URT, the pH is approximately pH 6.3 in the nasal cavity and is slightly higher (pH 7) in the nasopharynx (Man et al., 2017). For comparison, in the healthy vagina, the pH is rather acidic (pH 3.8 – pH 4.3), while along the gastrointestinal tract, the pH changes drastically ranging from pH 2 in the stomach to pH 8.5 in duodenum. Finally, higher levels of oxygen (partial pressures up to 160 mmHg pO2) are encountered in the URT (Man et al., 2017), especially when compared with the anaerobic conditions of the gut. As part of the host normal cellular metabolism, the use of oxygen leads to the formation of reactive oxygen species (ROS) such as hydrogen peroxide and superoxide, that can cause oxidative stress for colonizing microbiota that are anaerobic. Many URT pathogens are known to adapt to increasing ROS associated with higher oxygen levels by different antioxidant defence mechanisms such as the resistance via catalase or superoxide dismutase enzymes (Harrison et al., 2012; Eason and Fan, 2014). For the URT strain L. casei AMBR2, a haeme‐ and manganese‐dependent catalase gene were identified and its oxygen tolerance was confirmed in vitro. Whether the presence of antioxidant defence mechanisms is common in URT probiotics is however currently underexplored. LAB are aerotolerant, so oxidative stress may not be induced in the URT. In addition, commensals like LAB may not induce the production of ROS by the host defences. Also, for D. pigrum, factors involved in its adaptation to the URT remain to be investigated.
Probiotic modes of action relevant in the URT
Probiotic modes of action are generally divided into at least three broad mechanisms of action: (i) modulating microbe‐microbe interactions, (ii) immunomodulation and (iii) epithelial barrier protection (Lebeer et al., 2008). Microbe‐microbe interactions include both inhibitory and stimulatory interactions, but for most probiotic applications, inhibition of pathogens is often the key desired property. This can be obtained in different ways, for example, via the production of antimicrobial substances or competition for limiting resources and binding sites known as competitive exclusion. Lacticaseibacillus rhamnosus GG for instance is able to interfere with the adherence of the prominent respiratory pathogen Moraxella catarrhalis to the human Calu‐3 airway epithelial cells by approximately 50% (van den Broek et al., 2018). In addition, spot antimicrobial assays showed that L. rhamnosus GG is able to inhibit the growth of M. catarrhalis (van den Broek et al., 2018). Such antimicrobial effects are also observed for URT strain L. casei AMBR2 against M. catarrhalis, H. influenzae and S. aureus (De Boeck et al., 2020). Also for D. pigrum strains, antimicrobial effects against S. aureus have been described (Brugger et al., 2020). Immunomodulatory effects can be achieved via direct interaction between probiotic microbe‐associated molecular patterns (MAMPs) and the pattern recognition receptors (PRRs) of host epithelial and immune cells on one hand (Lebeer et al., 2010, 2018), or via the release of microbial soluble factors and metabolites that trigger signaling cascades in host cells (Oelschlaeger, 2010). Finally, certain probiotic strains could enhance and/or regulate the epithelial barrier function at different body sites (Martens et al., 2018). For instance, Lactiplantibacillus plantarum MB452 has been shown to increase the expression of tight‐junction genes in vitro in intestinal epithelial cells (Anderson et al., 2010). Also in human primary keratinocytes, it has been shown that tight‐junction barrier function was enhanced by the addition of bacterial lysate from L. rhamnosus GG, Limosilactobacillus reuteri and L. plantarum (O’Neill et al., 2013). The L. casei AMBR2 isolate (De Boeck et al., 2020) was also able to restore disruption of the airway epithelial barrier in primary cells from CRSwNP patients and differentiated Calu‐3 cells (Martens et al. accepted in principle AAIR, De Rudder et al., 2020).
Future perspectives and challenges for the use of urt probiotics
Substantiating the efficacy of URT probiotics in vitro and in vivo
Despite promising evidence, based on microbiome research that lactic acid bacteria are important for URT health, confirmation of these findings in patients with URT disease are lacking and studies investigating topical application with lactobacilli are scarce. For CRS in particular, the EPOS2020 (European Position Paper on Rhinosinusitis and Nasal Polyps) steering group recently concluded, based on the small studies with limited sample sizes that are conducted up to now, that there is currently no evidence for the use of oral or topical URT probiotics as treatment option for CRS patients (Fokkens et al., 2020). It should be noted however that analysing probiotic interventions is highly challenging, and sweeping generalizations on their efficacy based on this limited amount of available studies is difficult, as also mentioned by the EPOS2020 steering group, considering the probiotic strain‐specificity and multifactorial mechanisms of action that furthermore depend on the application site (Spacova et al., 2020a,2020b). Such screenings for probiotic efficacy and safety demands a dedicated pipeline, where a combination of in silico, in vitro and in vivo models are advised. Such dedicated pipelines can help to further distinguish the role of the microbiome and potential of probiotics for CRS.
Traditional and advanced in vitro screening
Traditional in vitro screening methods are the first step to evaluate the probiotic efficacy and safety (Irina Spacova et al., 2020a,2020b). In addition, whole‐genome sequencing of probiotic candidate strains followed by detailed in silico analysis at gene level can detect potential probiotic and adaptation factors, as well as virulence and antibiotic resistance genes (Salvetti et al., 2016). State‐of‐the‐art research is increasingly focused on the development of complex human cell models (Shah et al., 2016; De Rudder et al., 2018; Xu et al., 2018) as an important alternative for simple in vitro models and animal models for transition of probiotic efficacy to the clinic. Recently, several promising polymicrobial and multicellular models for the human respiratory tract have been described (e.g., De Rudder et al., 2020). Examples of such models include organoids, which are 3D tissues in vitro that can be derived from embryonic stem cells, induced pluripotent stem cells, adult stem cells and cancer cells (Xu et al., 2018). Successful healthy organoids have for instance been developed from the human lungs (Dye et al., 2015; Chen et al., 2017; Zacharias et al., 2018) and they have been used in microbial infection studies (Quantius et al., 2016; Shen et al., 2017). However, their use to study effects of beneficial bacteria have so far mostly been limited to other body niches, such as the gut (Han et al., 2019).
Animal models
An important discussion point in probiotic and other therapeutic research remains whether or not it is necessary to perform animal studies before probiotic candidate strains can be tested in humans, especially for probiotic candidates with a well‐established safety status. The relevance and necessity of animal models can be debated, as it remains extremely challenging to translate results obtained in animal models towards humans and it requires ethical considerations for animal welfare.
Several factors should however be considered when using animal models of URT disease. In particular for CRS, the available animal models do not represent all aspects of the pathology of CRS relevant for microbiome research. Development of mouse models is especially challenging due to significant differences within their respiratory system, including sinonasal anatomy that is not representative for humans (Wenzel and Holgate, 2006; Al‐Sayed et al., 2017; Kolanjiyil et al., 2019; Lux et al., 2019). In addition, mice are obligated nose‐only breathers, whereas humans breathe via their nose and/or mouth (Kolanjiyil et al., 2019). These differences are also reflected in the microbiota that colonizes the murine and human respiratory tract, which can have a profound impact on the outcomes of probiotic interventions (Maldonado‐Gómez et al., 2016). Recently, a rabbit model for CRS was described by Cho and colleagues (Cho et al., 2017). The sinonasal anatomy of rabbits is more similar to the human anatomy compared with the anatomy of mice, and the inventors of the model also reported that some of the detected dominant bacterial taxa, for example Corynebacterium or bacteria from the order Burkholderiales and Pseudomonadales, resemble the dominant genera in humans. It should be noted that this comparison was mainly done at the phylum and order level and no other studies have investigated the similarities of the rabbit and human respiratory tract microbiota. Some higher animal models such as pigs and sheep can be considered as well, as they show more anatomical URT similarities with humans (Lux et al., 2019). The paranasal sinuses of pigs are for instance largely similar to humans, with two main complexes, that is, maxillary and frontal sinuses, and the sphenoid and lacrimal sinuses as smaller sinuses, making them a feasible model for sinonasal research (Wang et al., 2013). However, more strict ethical regulations, more expensive housing facilities and differences in microbiota composition represent the main limitations of higher animal models in URT disease research.
Overall, for next‐generation probiotics, such as the more recently suggested D. pigrum, the safety profile is less well‐known and animal studies are thus of more interest to evaluate the safety before clinical studies are performed (Cordaillat‐Simmons et al., 2020; Rouanet et al., 2020). The more traditional strains, especially strains that belong to the Lactobacillaceae, are already used on a large scale and have an established safety profile, hence animal studies might not be essential before proceeding to clinical trials in humans.
Translation to clinical studies
As described above, the transition from in vitro screening to the clinic in the probiotic field is not always straightforward. In particular for CRS and AR, clinical studies investigating the potential of topical URT probiotics are limited. The L. casei AMBR2 strain that we have tested in a nasal spray in healthy volunteers showed to be safe, and the strain was able to temporally colonize the URT, which we hypothesize can be attributed to its adaptation potential to the URT. Further application of this strain in CRS patients is thus of great interest and is currently being explored by our research group. To the best of our knowledge, only two published studies have yet investigated the effects of topical probiotic treatment in CRS patients, by using a probiotic nasal spray (Mårtensson et al., 2017) or sinus irrigations (Endam et al., 2020). The randomized, double‐blinded study by Mårtensson and colleagues investigated the previously mentioned nasal spray device with a mixture of 13 honeybee LAB (1011 CFU ml‐1) in CRSsNP patients, of which 14 out of 20 participants had previously undergone surgery, compared with placebo (Mårtensson et al., 2017). SNOT‐22 questionnaires, changes in the microbial community and inflammatory markers in nasal lavage fluids were monitored, but no differences between probiotic treatment and control groups were observed. As previously described, a possible explanation might be that the LAB strains that were used are not adapted to the URT. In the other study, CRS patients with previous undergone FESS were recruited and received intranasal irrigation with Lactococcus lactis W136 for two weeks (Endam et al., 2020). The origin of L. lactis W136 is not well‐described in the paper, but Lactococcus is often detected in URT microbiome studies in healthy subjects (Laufer et al., 2011; Pettigrew et al., 2012; De Boeck et al., 2017). Moreover, Desrosiers and colleagues postulate that the cocci form could be important, since the common nasal commensal coccus Staphylococcus epidermidis has been shown to have probiotic potential against URT pathogen S. aureus in a mouse model (Cleland et al., 2014). However, due to safety reasons, the authors mention that the use of this potential pathogenic S. epidermidis is not ideal for CRS. The nasal irrigations with L. lactis W136 were overall found to be well tolerated. During and up to two weeks after the treatment, significant improvements in symptoms and quality of life were noted based on several parameters including the sinonasal outcome test 22 (SNOT‐22). Mean SNOT‐22 scores significantly reduced from 41 at day 0 to 36 at day 28 (95% CI: 27.28–46.87). In comparison, the study of Mårtensson and colleagues also evaluated SNOT‐22 scores but did not observe significant differences between the treated groups (Mårtensson et al., 2017). Both studies thus highlight the feasibility for topical administration of LAB in the URT, but also pinpoint that proper selection of probiotic strains is essential. In addition to possible strain‐specific effects, also other considerations should be addressed, such as the dose and formulation of the probiotic strains (Broeckx et al., 2017; Kiekens et al., 2019; Jokicevic et al., 2020). According to the definition, an ‘adequate amount’ of the probiotic is needed; however, the set‐up of dose‐response curves for probiotics is difficult (Ouwehand, 2017) and clinical studies on probiotic dose‐response curves in the URT are currently lacking.
Regulatory framework
The future of probiotics in the field of human diseases is currently highly challenging due to the lack of a well‐designed regulatory framework for probiotic and microbiome‐derived products. The route of probiotic administration has a direct impact on the regulatory framework for the product. While oral probiotics with nutritional or physiological health claims are regulated by the European Food Safety Authority (EFSA), for example as functional foods, for clinical URT applications probiotics would rather be regulated as medical or pharmaceutical products in Europe by the European Medicines Agency (EMA). Such regulations typically require rigorous testing to demonstrate appropriate quality, efficacy and safety evidence of the product. Currently, the European pharmaceutical regulation is not adapted for the marketing of products containing active microorganisms. In the United States, the regulation falls under the FDA that introduced a guidance document for the use of life biotherapeutic products (LBPs) in early clinical trials (FDA, 2016). In this document, an LBP is defined as a biological product that (i) contains live organisms, such as bacteria; (ii) is applicable to the prevention, treatment or cure of a disease or condition of human beings and (iii) is not a vaccine. So the term LBPs is more used for medical claims and purposes. Changes in the current regulation are thus inevitable as the probiotic field evolves towards topical application in novel body niches, such as the URT.
Conclusion
The need for alternative therapies for CRS and other URT diseases, such as OM, is high among the general population. URT probiotics, including both the traditional Lactobacillales and next‐generation candidate probiotics (e.g., Dolosigranulum), can offer a natural solution to standard treatment options that often fail or have known side effects. Although the field is still rather uncharted compared with for instance gut probiotic research, the use of topically applied probiotics is of interest since they can broadly act on different aspects of URT diseases due to their multifactorial modes of action: microbiome restoration, antimicrobial activity, immunomodulation and barrier enhancement.
To substantiate the choice and efficacy of URT probiotics, clinical studies investigating the targeted application of probiotics in the URT are urgently needed, as well as adjusted preclinical pipelines that include complex in vitro screening models as alternative to animal models. This review aimed to emphasize that future studies should focus not only on the probiotic properties, but also on the use of probiotic isolates that are adapted to the unique environment of the URT and can thus provide temporary colonization and site‐specific beneficial effects.
Funding Information
This research was funded by a grant from the Flanders Innovation and Entrepreneurship Agency [IWT‐SBO ProCure project (IWT/50052)], by the personal grant of Ilke De Boeck 1S17916, by a grant from the University of Antwerp (IOF POC ReLACT), and by the European Research Council grant of prof. Sarah Lebeer (42/FA070500/8330).
Conflict of interest
A patent application (PCT/EP2018/057497) was filed on March 23, 2018, related to this work.
Acknowledgements
The authors want to thank the entire research group ENdEMIC of the University of Antwerp. They also want to thank the entire ENT Department of the Antwerp University Hospital and University Hospitals of Leuven. Finally, the authors want to thank all partners of the IWT‐SBO ProCure project.
Microbial Biotechnology (2021) 14(3), 859–869
References
- Abreu, N.A. , Nagalingam, N.A. , Song, Y. , Roediger, F.C. , Pletcher, S.D. , Goldberg, A.N. , and Lynch, S.V. (2012) Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med 4: 151ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Sayed, A.A. , Agu, R.U. , and Massoud, E. (2017) Models for the study of nasal and sinus physiology in health and disease: a review of the literature. Laryngoscope Investig Otolaryngol 2: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, R.C. , Cookson, A.L. , McNabb, W.C. , Park, Z. , McCann, M.J. , Kelly, W.J. , and Roy, N.C. (2010) Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol 10: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesbroek, G. , Bosch, A.A.T.M. , Wang, X. , Keijser, B.J.F. , Veenhoven, R.H. , Sanders, E.A.M. , and Bogaert, D. (2014) The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med 190: 298–308. [DOI] [PubMed] [Google Scholar]
- Biesbroek, G. , Tsivtsivadze, E. , Sanders, E. , Montijn, R. , Veenhoven, R.H. , Keijser, B.J.F. , and Bogaert, D. (2014) Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190: 1283–1292. [DOI] [PubMed] [Google Scholar]
- De Boeck, I. , van den Broek, M.F.L. , Allonsius, C.N. , Spacova, I. , Wittouck, S. , Martens, K. , et al. (2020) Lactobacilli have a niche in the human nose. Cell Rep 31: 107674. [DOI] [PubMed] [Google Scholar]
- De Boeck, I. , Wittouck, S. , Wuyts, S. , Oerlemans, E.F.M. , van den Broek, M.F.L. , Vandenheuvel, D. , et al. (2017) Comparing the healthy nose and nasopharynx microbiota reveals continuity as well as niche‐specificity. Front Microbiol 8: 2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boeck, I. , Wittouck, S. , Martens, K. , Claes, J. , Jorissen, M. , Steelant, B. , et al. (2019) Anterior nares diversity and pathobionts represent sinus microbiome in chronic rhinosinusitis. mSphere 4: e00532‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert, D. , Keijser, B. , Huse, S. , Rossen, J. , Veenhoven, R. , van Gils, E. , et al. (2011) Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6: e17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, A.A.T.M. , Levin, E. , van Houten, M.A. , Hasrat, R. , Kalkman, G. , Biesbroek, G. , et al. (2016) Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9: 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet, J. , Khaltaev, N. , Cruz, A.A. , Denburg, J. , Fokkens, W.J. , Togias, A. , et al. (2008) Allergic Rhinitis and its Impact on Asthma (ARIA) 2008*. Allergy 63: 8–160. [DOI] [PubMed] [Google Scholar]
- Broeckx, G. , Vandenheuvel, D. , Henkens, T. , Kiekens, S. , van den Broek, M.F.L. , Lebeer, S. , and Kiekens, F. (2017) Enhancing the viability of Lactobacillus rhamnosus GG after spray drying and during storage. Int J Pharm 534: 35–41. [DOI] [PubMed] [Google Scholar]
- van den Broek, M.F.L. , De Boeck, I. , Claes, I.J.J. , Nizet, V. , and Lebeer, S. (2018) Multifactorial inhibition of lactobacilli against the respiratory tract pathogen Moraxella catarrhalis . Benef Microbes 9: 429–439. [DOI] [PubMed] [Google Scholar]
- van den Broek, M.F.L. , De Boeck, I. , Kiekens, F. , Boudewijns, A. , Vanderveken, O.M. , and Lebeer, S. (2019) Translating recent microbiome insights in otitis media into probiotic strategies. Clin Microbiol Rev 15:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger, S. D. , Eslami, S. M. , Pettigrew, M. M. , Escapa, I. F. , Henke, M. M. , Kong, Y. , and Lemon, K. P. (2020) Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere 5: e00852‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalermwatanachai, T. , Vilchez‐Vargas, R. , Holtappels, G. , Lacoere, T. , Jáuregui, R. , Kerckhof, F.M. , et al. (2018) Chronic rhinosinusitis with nasal polyps is characterized by dysbacteriosis of the nasal microbiota. Sci Rep 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.‐W. , Huang, S.X. , de Carvalho, A.L.R.T. , Ho, S.‐H. , Islam, M.N. , Volpi, S. , et al. (2017) A three‐dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19: 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, D.‐Y. , Mackey, C. , Van Der Pol, W.J. , Skinner, D. , Morrow, C.D. , Schoeb, T.R. , et al. (2017) Sinus microanatomy and microbiota in a rabbit model of rhinosinusitis. Front Cell Infect Microbiol 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland, E.J. , Drilling, A. , Bassiouni, A. , James, C. , Vreugde, S. , and Wormald, P.J. (2014) Probiotic manipulation of the chronic rhinosinusitis microbiome. Int Forum Allergy Rhinol 4: 309–314. [DOI] [PubMed] [Google Scholar]
- Cordaillat‐Simmons, M. , Rouanet, A. , and Pot, B. (2020) Live biotherapeutic products: the importance of a defined regulatory framework. Exp Mol Med 52: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan, R.M. , Miajlovic, H. , and Foster, T.J. (2009) Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborah, S. , and Prathibha, K.M. (2014) Measurement of nasal mucociliary clearance. Clin Res Pulmonol 2: 1019. [Google Scholar]
- Duell, B.L. , Su, Y.C. , and Riesbeck, K. (2016) Host–pathogen interactions of nontypeable Haemophilus influenzae: from commensal to pathogen. FEBS Lett 590: 3840–3853. [DOI] [PubMed] [Google Scholar]
- Dye, B.R. , Hill, D.R. , Ferguson, M.A. , Tsai, Y.‐H. , Nagy, M.S. , Dyal, R. , et al. (2015) In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4: e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eason, M.M. , and Fan, X. (2014) The role and regulation of catalase in respiratory tract opportunistic bacterial pathogens. Microb Pathog 74: 50–58. [DOI] [PubMed] [Google Scholar]
- Van Eldere, J. , Slack, M.P.E. , Ladhani, S. , and Cripps, A.W. (2014) Non‐typeable Haemophilus influenzae, an under‐recognised pathogen. Lancet Infect Dis 14: 1281–1292. [DOI] [PubMed] [Google Scholar]
- Endam, L.M. , Alromaih, S. , Gonzalez, E. , Madrenas, J. , Cousineau, B. , Renteria, A.E. , and Desrosiers, M. (2020) Intranasal application of Lactococcus lactis W136 is safe in chronic rhinosinusitis patients with previous sinus surgery. Front Cell Infect Microbiol 10: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2016) Early Clinical Trials with Live Biotherapeutic Products: Chemistry, Manufacturing, and Control Information. Guidance for Industry. [Google Scholar]
- Fokkens, W.J. , Lund, V.J. , Hopkins, C. , Hellings, P.W. , Kern, R. , Reitsma, S. , et al. (2020) European position paper on rhinosinusitis and nasal polyps 2020. Rhinol J 1–464. [Google Scholar]
- Gan, W. , Yang, F. , Tang, Y. , Zhou, D. , Qing, D. , Hu, J. , et al. (2019) The difference in nasal bacterial microbiome diversity between chronic rhinosinusitis patients with polyps and a control population. Int Forum Allergy Rhinol 9: 582–592. [DOI] [PubMed] [Google Scholar]
- Gan, W. , Yang, F. , Meng, J. , Liu, F. , Liu, S. , and Xian, J. (2020) Comparing the nasal bacterial microbiome diversity of allergic rhinitis, chronic rhinosinusitis and control subjects. Eur Arch Oto‐Rhino‐Laryngology 1–8. [DOI] [PubMed] [Google Scholar]
- Han, X. , Lee, A. , Huang, S. , Gao, J. , Spence, J.R. , and Owyang, C. (2019) Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon‐gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microbes 10: 59–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, A. , Bakaletz, L.O. , and Munson, R.S., Jr (2012) Haemophilus influenzae and oxidative stress. Front Cell Infect Microbiol 2: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, K. , Linnemann, R.W. , Mansbach, J.M. , Ajami, N.J. , Espinola, J.A. , Petrosino, J.F. , et al. (2017) nasal airway microbiota profile and severe bronchiolitis in infants: a case‐control study. Pediatr Infect Dis J 36: 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, K. , Mansbach, J.M. , Ajami, N.J. , Espinola, J.A. , Henke, D.M. , Petrosino, J.F. , et al. (2016) Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 48: 1329‐1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney, D.D. , Gareau, G. , and Marco, M.L. (2018) Intestinal Lactobacillus in health and disease, a driver or just along for the ride ? Curr Opin Biotechnol 49: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellings, P.W. , Akdis, C.A. , Bachert, C. , Bousquet, J. , Pugin, B. , Adriaensen, G. , et al. (2017) EUFOREA Rhinology Research Forum 2016: report of the brainstorming sessions on needs and priorities in rhinitis and rhinosinusitis. Rhinology 55: 202–210. [DOI] [PubMed] [Google Scholar]
- Hill, C. , Guarner, F. , Reid, G. , Gibson, G.R. , Merenstein, D.J. , Pot, B. , et al. (2014) The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11: 506–514. [DOI] [PubMed] [Google Scholar]
- Jensen, A. , Fagö‐Olsen, H. , Sørensen, C.H. , and Kilian, M. (2013) Molecular mapping to species level of the tonsillar crypt microbiota associated with health and recurrent tonsillitis. PLoS One 8: e56418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokicevic, K. , Broeckx, G. , Vandenheuvel, D. , Boeck, I.D. , Allonsius, C.N. , Lebeer, S. , and Kiekens, F. (2020) Processing of potential upper respiratory tract probiotics by spray drying. Dry Technol. 10.1080/07373937.2020.1759083. [DOI] [Google Scholar]
- Kankainen, M. , Paulin, L. , Tynkkynen, S. , von Ossowski, I. , Reunanen, J. , Partanen, P. , et al. (2009) Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human‐ mucus binding protein. Proc Natl Acad Sci USA 106: 17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekens, S. , Vandenheuvel, D. , Broeckx, G. , Claes, I. , Allonsius, C. , De Boeck, I. , et al. (2019) Impact of spray‐drying on the pili of Lactobacillus rhamnosus GG. Microb Biotechnol 12: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeller, K. , Herlemann, D.P.R. , Schuldt, T. , Ovari, A. , Guder, E. , and Collin, M. (2018) Microbiome and culture based analysis of chronic rhinosinusitis compared to healthy sinus mucosa. Front Microbiol 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanjiyil, A.V. , Kleinstreuer, C. , Kleinstreuer, N.C. , Pham, W. , and Sadikot, R.T. (2019) Mice‐to‐men comparison of inhaled drug‐aerosol deposition and clearance. Respir Physiol Neurobiol 260: 82–94. [DOI] [PubMed] [Google Scholar]
- Kumpitsch, C. , Koskinen, K. , Schöpf, V. , and Moissl‐Eichinger, C. (2019) The microbiome of the upper respiratory tract in health and disease. BMC Biol 17: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappan, R. , Imbrogno, K. , Sikazwe, C. , Anderson, D. , Mok, D. , Coates, H. , et al. (2018) A microbiome case‐control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol 18: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer, A.S. , Metlay, J.P. , Gent, J.F. , Fennie, K.P. , Kong, Y. , and Pettigrew, M.M. (2011) Microbial communities of the upper respiratory tract and otitis media in children. MBio 2: e00245–e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer, S. , Bron, P.A. , Marco, M.L. , Van Pijkeren, J.‐P. , O’Connell Motherway, M. , Hill, C. , et al. (2018) Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol 49: 217–223. [DOI] [PubMed] [Google Scholar]
- Lebeer, S. , Vanderleyden, J. , and De Keersmaecker, S.C.J. (2008) Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 72: 728–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer, S. , Vanderleyden, J. , and De Keersmaecker, S.C.J. (2010) Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol 8: 171–184. [DOI] [PubMed] [Google Scholar]
- Ling, Z. , Liu, X. , Luo, Y. , Yuan, L. , Nelson, K.E. , Wang, Y. , et al. (2013) Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genom 14: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux, C.A. , Douglas, R.G. , Cho, D.‐Y. , Taylor, M.W. , and Biswas, K. (2019) Animal models for inflammatory mucosal disease and their potential for studying the microbiome in chronic rhinosinusitis* Animal models for studying CRS. Rhinol Online 2: 69–80. [Google Scholar]
- Mahdavinia, M. , Keshavarzian, A. , Tobin, M.C. , Landay, A.l. , and Schleimer, R.P. (2016) A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin Exp Allergy 46: 21–41. 10.1111/cea.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado‐Gómez, M.X. , Martínez, I. , Bottacini, F. , O’Callaghan, A. , Ventura, M. , van Sinderen, D. , et al. (2016) Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20: 515–526. [DOI] [PubMed] [Google Scholar]
- Man, W.H. , de Steenhuijsen Piters, W.A.A. , and Bogaert, D. (2017) The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 15: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, K. , Pugin, B. , De Boeck, I. , Spacova, I. , Steelant, B. , Seys, S.F. , et al. (2018) Probiotics for the Airways: potential to improve epithelial and immune homeostasis. Allergy 73: 1954–1963. [DOI] [PubMed] [Google Scholar]
- Mårtensson, A. , Abolhalaj, M. , Lindstedt, M. , Mårtensson, A. , Olofsson, T.C. , Vásquez, A. , et al. (2017) Clinical efficacy of a topical lactic acid bacterial microbiome in chronic rhinosinusitis: a randomized controlled trial. Laryngoscope Investig Otolaryngol 2: 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer, E.O. (2016) Allergic rhinitis. Burden of illness, quality of life, comorbidities, and control. Immunol Allergy Clin North Am 36: 235–248. [DOI] [PubMed] [Google Scholar]
- Meng, Y. , Wang, C. , and Zhang, L. (2019) Recent developments and highlights in allergic rhinitis. Allergy 74: 2320–2328. [DOI] [PubMed] [Google Scholar]
- Mongodin, E. , Bajolet, O. , Cutrona, J. , Bonnet, N. , Dupuit, F. , Puchelle, E. , and de Bentzmann, S. (2002) Fibronectin‐binding proteins of Staphylococcus aureus are involved in adherence to human airway epithelium. Infect Immun 70: 620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill, C. , Sultana, R. , McBain, A.J. , and O'Neill, C.A. (2013) Strain‐dependent augmentation of tight‐junction barrier function in human primary epidermal keratinocytes by lactobacillus and bifidobacterium lysates. Appl Environ Microbiol 79: 4887–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger, T.A. (2010) Mechanisms of probiotic actions ‐ A review. Int J Med Microbiol 300: 57–62. [DOI] [PubMed] [Google Scholar]
- Ouwehand, A.C. (2017) A review of dose‐responses of probiotics in human studies. Benef Microbes 8: 143–151. [DOI] [PubMed] [Google Scholar]
- Pasolli, E. , Schiffer, L. , Manghi, P. , Renson, A. , Obenchain, V. , and Truong, D. T. (2017) Accessible, curated metagenomic data through ExperimentHub. Nat Methods 14: 1023–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, M.I. , Lievens, E. , Malik, S. , Imholz, N. , and Lebeer, S. (2015) Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol 6: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew, M.M. , Laufer, A.S. , Gent, J.F. , Kong, Y. , Fennie, K.P. , and Metlay, J.P. (2012) Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 78: 6262–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piromchai, P. , Kasemsiri, P. , Laohasiriwong, S. , and Thanaviratananich, S. (2013) Chronic rhinosinusitis and emerging treatment options. Int J Gen Med 6: 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantius, J. , Schmoldt, C. , Vazquez‐Armendariz, A.I. , Becker, C. , El Agha, E. , Wilhelm, J. , et al. (2016) Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b‐driven epithelial repair. PLOS Pathog 12: e1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouanet, A. , Bolca, S. , Bru, A. , Claes, I. , Cvejic, H. , Girgis, H. , et al. (2020) Live biotherapeutic products, a road map for safety assessment. Front Med 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rudder, C. , Calatayud Arroyo, M. , Lebeer, S. , and Van de Wiele, T. (2018) Modelling upper respiratory tract diseases: getting grips on host‐microbe interactions in chronic rhinosinusitis using in vitro technologies. Microbiome 6: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rudder, C. , Calatayud Arroyo, M. , Lebeer, S. , and Van de Wiele, T. (2020) Dual and triple epithelial coculture model systems with donor‐derived microbiota and THP‐1 macrophages to mimic host‐microbe interactions in the human sinonasal cavities. mSphere 5: e00916‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rudder, C. , Garcia‐Tímermans, C. , De Boeck, I. , Lebeer, S. , Van de Wiele, T. , and Calatayud Arroyo, M. (2020) Lacticaseibacillus casei AMBR2 modulates the epithelial barrier function and immune response in a donor‐derived nasal microbiota manner. Sci Rep 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti, E. , and O’Toole, P.W. (2017) When regulation challenges innovation: the case of the genus Lactobacillus. Trends Food Sci Technol 66: 187–194. [Google Scholar]
- Salvetti, E. , Orrù, L. , Capozzi, V. , Martina, A. , Lamontanara, A. , Keller, D. , et al. (2016) Integrate genome‐based assessment of safety for probiotic strains: Bacillus coagulans GBI‐30, 6086 as a case study. Appl Microbiol Biotechnol 100: 4595–4605. [DOI] [PubMed] [Google Scholar]
- Sanders, M.E. , Merenstein, D.J. , Reid, G. , Gibson, G.R. , and Rastall, R.A. (2019) Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol 16: 605–616. [DOI] [PubMed] [Google Scholar]
- Shah, P. , Fritz, J.V. , Glaab, E. , Desai, M.S. , Greenhalgh, K. , Frachet, A. , et al. (2016) A microfluidics‐based in vitro model of the gastrointestinal human–microbe interface. Nat Commun 7: 11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Chen, L. , Wang, M. , Lin, D. , Liang, Z. , Song, P. , et al. (2017) Flagellar hooks and hook protein FlgE participate in host microbe interactions at immunological level. Sci Rep 7: 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spacova, I. , Dodiya, H.B. , Happel, A.‐U. , Strain, C. , Vandenheuvel, D. , Wang, X. , and Reid, G. (2020a) Future of probiotics and prebiotics and the implications for early career researchers. Front Microbiol 11: 1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spacova, I. , O’Neill, C. , and Lebeer, S. (2020b) Lacticaseibacillus rhamnosus GG inhibits infection of human keratinocytes by Staphylococcus aureus through mechanisms involving cell surface molecules and pH reduction. Benef Microbes 1: 703‐715. [DOI] [PubMed] [Google Scholar]
- Stearns, J.C. , Davidson, C.J. , McKeon, S. , Whelan, F.J. , Fontes, M.E. , Schryvers, A.B. , et al. (2015) Culture and molecular‐based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J 9: 1246–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaleniec, J. , Gibała, A. , Pobiega, M. , Parasion, S. , Składzień, J. , Stręk, P. , et al. (2019) Exacerbations of chronic rhinosinusitis— microbiology and perspectives of phage therapy. Antibiotics 8: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.C.C. , Hathorn, I. , Habib, A.‐R. , Chang, E. , and Javer, A.R. (2013) Evaluation of domestic and Yucatan swine nasal sinus anatomy as models for future sinonasal research of medications delivered by standard instruments used in functional endoscopic sinus surgery. Int Forum Allergy Rhinol 3: 150–156. [DOI] [PubMed] [Google Scholar]
- Wenzel, S. , and Holgate, S.T. (2006) The Mouse Trap. Am J Respir Crit Care Med 174: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Wilkins, L.J. , Monga, M. , and Miller, A.W. (2019) Defining dysbiosis for a cluster of chronic diseases. Sci Rep 9: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Jiao, Y. , Qin, S. , Zhao, W. , Chu, Q. , and Wu, K. (2018) Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp Hematol Oncol 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, W.J. , Frank, D.B. , Zepp, J.A. , Morley, M.P. , Alkhaleel, F.A. , Kong, J. , et al. (2018) Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Wittouck, S. , Salvetti, E. , Franz, C.M.A.P. , Harris, H.M.B. , Mattarelli, P. , et al. (2020) A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70: 2782–2858. [DOI] [PubMed] [Google Scholar]


