Summary
Bacterial colony morphology can reflect different physiological stages such as virulence or biofilm formation. In this work we used transposon mutagenesis to identify genes that alter colony morphology and cause differential Congo Red (CR) and Brilliant Blue G (BBG) binding in Shewanella algae, a marine indigenous bacterium and occasional human pathogen. Microscopic analysis of colonies formed by the wild‐type strain S. algae CECT 5071 and three transposon integration mutants representing the diversity of colony morphotypes showed production of biofilm extracellular polymeric substances (EPS) and distinctive morphological alterations. Electrophoretic and chemical analyses of extracted EPS showed differential patterns between strains, although the targets of CR and BBG binding remain to be identified. Galactose and galactosamine were the preponderant sugars in the colony biofilm EPS of S. algae. Surface‐associated biofilm formation of transposon integration mutants was not directly correlated with a distinct colony morphotype. The hybrid sensor histidine kinase BarA abrogated surface‐associated biofilm formation. Ectopic expression of the kinase and mutants in the phosphorelay cascade partially recovered biofilm formation. Altogether, this work provides the basic analysis to subsequently address the complex and intertwined networks regulating colony morphology and biofilm formation in this poorly understood species.
In this work we observed a distinct phenotype of colonies formed by the marine indigenous bacterium Shewanella algae when grown on agar plates supplemented with dyes, and we used transposon mutagenesis to identify genes contributing to colony morphotype development and biofilm formation. Microscopic inspection of the colonies revealed distinct cell morphologies, arrangements, and extracellular matrix production in the wild‐type strain S. algae CECT 5071 and key transposon integration mutants representing the diversity of morphotypes. Electrophoretic and chemical analyses of extracted biofilm extracelluar matrix components of wild‐type and mutants showed differential fingerprints between strains, with galactose and galactosamine being the preponderant monosaccharides. Our work provides a basic analysis that paves the ground towards the dissection of the complex and intertwined networks regulating colony morphology and biofilm formation in this poorly understood species.
Introduction
Microbial colony morphology and the capacity to bind dyes can serve as an indicator of a distinct state of microbial physiology and metabolism including virulence and biofilm formation. In this context, structured hydrophobic bacterial colonies of species such as Salmonella Typhimurium with a high capacity to bind the dyes Congo Red (CR) and Calcofluor White have been identified as biofilm forming and concomitantly to extensively produce amyloid fimbriae and the exopolysaccharide cellulose (Römling, 2005). Biofilms formed by Shewanella spp. have major ecological and biotechnological impact (Hau and Gralnick, 2007). Different biofilm models and an array of genetic and biochemical approaches have been used to analyze the formation, regulation and development of biofilms in some Shewanella spp. For example, batch and flow biofilm models were developed for Shewanella putrefaciens (Bagge et al., 2001), early biofilm formation of Shewanella frigidimarina was analyzed by proteomics (Linares et al., 2016), and biofilm formation under flow conditions in Shewanella oneidensis MR‐1 was assessed by transposon mutagenesis (Thormann et al., 2004). Within the genus Shewanella, S. oneidensis MR‐1 is a widely used genetic and physiological model with readily accessible gene manipulation strategies, a broad repertoire of anaerobic respiration pathways including metal oxide reduction, and diverse extracellular electron transfer capacities (Hau and Gralnick, 2007).
Species identification by the use of molecular approaches increasingly reports the marine bacterium S. algae as causing disease in humans (Vogel et al., 2000; Janda and Abbott, 2012; Martín‐Rodríguez et al., 2017, 2019). Metabolites produced by S. algae are known to participate in diverse ecological interactions (Gong et al., 2015; Singh et al., 2015; Rütschlin et al., 2017). A distinctive feature of S. algae is the production of tetrodotoxin (Nozue et al., 1992). Biofilm formation has been proposed to play a role in certain S. algae infections (Holt et al., 2005). We have shown that the morphological and nano‐mechanical properties of statically‐grown S. algae biofilms are dramatically affected by the nutrient environment (Martín‐Rodríguez et al., 2014), and we have recently demonstrated that respiration of alternative electron acceptors acts as a driver for strain‐specific induction of biofilm formation in this species (Martín‐Rodríguez et al., 2021). Besides surface‐associated biofilm formation under static or hydrodynamic conditions, the colony biofilm is another widely employed model validated for diverse bacterial species including Pseudomonas spp. (Friedman and Kolter, 2004), Bacillus spp. (Romero et al., 2010), Vibrio cholerae (Yildiz and Schoolnik, 1999) and the enterobacterial species Salmonella Typhimurium and Escherichia coli (Römling et al., 1998; Römling, 2005).
The physiology of S. algae has hardly been explored at the molecular level. In this work we initially investigated whether and to which extent a characteristic dye binding colony morphotype of S. algae on agar plates supplemented with CR and Brilliant Blue G (BBG) develops, and whether such a morphotype can be associated with a multicellular biofilm behavior with pronounced production of extracellular matrix components. To this end, a collection of over 5,000 transposon insertion mutants was screened to identify genes involved in colony morphology development in S. algae CECT 5071, and the biofilm extracellular matrix composition of the wild‐type (WT) strain and representative transposon integration mutants was analyzed using an array of techniques spanning from electrophoretic profiles to Fourier‐transform Infrared Spectroscopy (FTIR) and monosaccharide content analyses. Regulation of a different type of biofilm, namely surface‐associated biofilm formation in static broth culture, was investigated in selected transposon insertion mutants. Genes with diverse functionalities affected the colony morphotype and surface‐associated biofilm formation, including the gene coding for the sensor histidine kinase BarA which affected biofilm formation independent of catalysis.
Results
Shewanella algae shows distinct colony morphotypes on CR‐ and BBG‐supplemented agar plates
We observed a distinct colony phenotype of S. algae CECT 5071 when grown on LB agar supplemented with CR and BBG. When colonies were visualized against a black background (referred to as “top‐black view”), the colony phenotype was characterized by concentric rings of intense blue‐green coloration, a slight red outer margin, and pale rims (Fig. 1A). In contrast, a light red color of the colony was observed when visualized perpendicularly over a clear background (referred to as “top‐trans” view). Without dye supplementation to LB agar, the WT colony does not show such a pigmentation, therefore colony coloration must derive from dye binding (Fig. 1A). To confirm this observation, methanolic extracts of colonies grown on LB agar with or without CR and BBG were prepared, and the absorbance spectra were recorded and compared to those of methanolic solutions of CR and BBG (Fig. 1B). An absorbance maximum at 610 nm matching the maximum absorbance of BBG was recorded in extracts of colonies grown in the presence of the dyes, consistent with the blue‐green colony morphotype with negligible CR accumulation. Consistently, no absorbance maxima were recorded in the extracts of WT colonies grown on LB agar without dye supplementation. Next, we questioned whether this colony phenotype was exclusively observed in a single isolate or shared by a larger number of strains. To address this question, we examined the phenotypes of a collection of 23 strains of S. algae and the closely‐related species Shewanella chilikensis on CR and BBG‐supplemented LB agar. Investigation of morphotype development over 48 h showed the phenotype to be strain‐dependent, varying from blue‐green to red as a consequence of differential absorption of the dyes (Fig. 1C). This phenotype was temperature‐independent, most evident beyond 24 h of incubation at 30°C (Fig. 1C), and developed faster at a higher temperature (tested at 22, 30 and 37°C, data not shown).
Fig. 1.
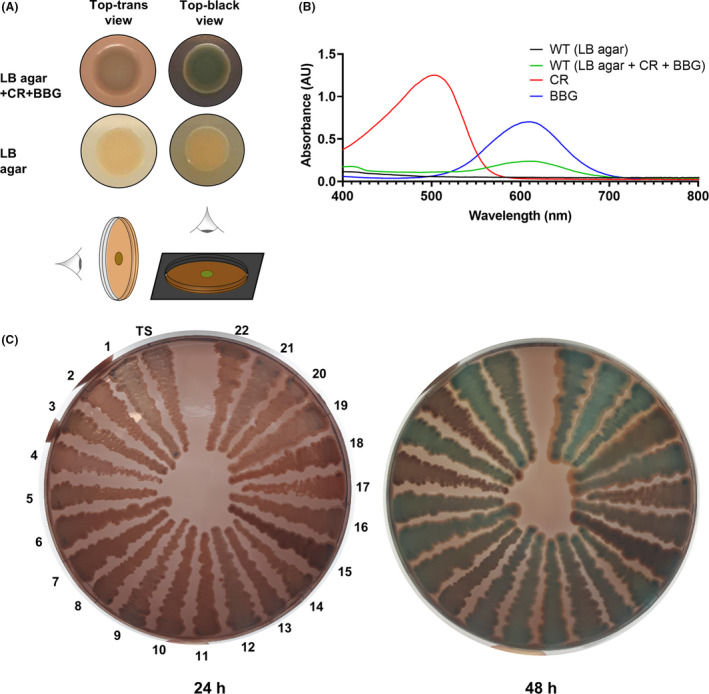
Colony biofilm phenotypes of Shewanella algae.
A. Colony morphotypes of S. algae CECT 5071 wild‐type (WT) on LB agar with or without Congo Red (CR) and Brilliant Blue G (BBG) supplementation. Top‐trans and top‐black views are shown.
B. Absorbance spectra of methanolic extracts of S. algae CECT 5071 colony biofilms grown on LB agar with or without CR and BBG supplementation. Spectra of CR and BBG solutions in methanol are also shown.
C. Phenotypes (top‐trans view) of S. algae CECT 5071 (type strain, TS) and 22 S. algae or Shewanella chilikensis strains listed in Table S1 on LB agar supplemented with CR and BBG.
Transposon mutagenesis identifies genes that alter the S. algae CECT 5071 colony morphotype and static biofilm formation
To identify genes that alter the appearance of S. algae CECT 5071 colony morphology, we performed mini‐Tn5 transposon mutagenesis. Specifically, we focused on mutants displaying either one or several of the following features: altered coloration (either more ‘white’, more ‘red’ or more ‘blue’ mutants), altered colony size, and altered colony margin. Screening of 5082 transposon insertion mutants resulted in the recovery of 43 colonies presenting an altered morphotype, for which the location of the transposon insertion site was determined (Fig. 2, Table 1). Thus, 28 disrupted genes with diverse functions as inferred by their annotations and biological pathway assignations were identified. Seven genes were found to be interrupted at distinct sites in more than one mutant. Those genes code for the type 2 secretion system (T2SS) protein GspF, a delta‐aminolevulinic acid dehydratase, the WecB/TagA/CpsF family glycosyltransferase WecG, the phosphoribosylformylglycinamidine synthase PurL, a DNA adenine methylase, and two hypothetical proteins with gene locus tags E1N14_01730 and E1N14_14870, with E1N14_01730 having 8 distinct transposon insertion sites. Of note, the colony morphotypes resulting from transposon insertion at different locations within the same gene were almost identical and therefore only one representative insertion mutant is shown in Fig. 2. Furthermore, several of the interrupted genes belonged to the same gene cluster or were located in close proximity to each other (Fig. S1).
Fig. 2.
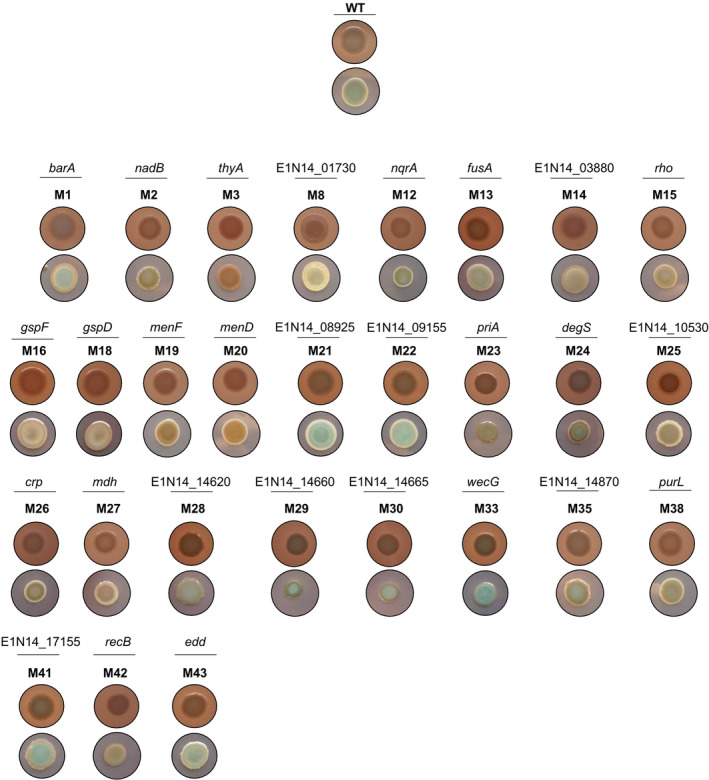
Colony morphotypes of S. algae CECT 5071 and transposon insertion mutants. Shown are the top‐trans and top‐black views of colonies of the WT strain and isogenic transposon insertion mutant derivatives. Genes without a name assigned are named after their GenBank locus tags.
Table 1.
Genes interrupted by transposon insertion in colony morphology mutants.
| Mutant(s) | Gene | Locus tag | Gene product | Biological process |
|---|---|---|---|---|
| M1 (nt. 717) | barA | E1N14_00465 | Two‐component sensor histidine kinase BarA | Transcription, Transcription regulation, Two‐component regulatory system |
| M2 (nt. 1057) | nadB | E1N14_01455 | L‐aspartate oxidase | Pyridine nucleotide biosynthesis |
| M3 (nt. 662) | thyA | E1N14_01485 | Thymidylate synthase | Nucleotide biosynthesis, Translation regulation |
|
M4 (nt. 503) M5 (nt. 513) M6 (nt. 177) M7 (nt. 77) M8 (nt. 91) M9 (nt. 119) M10 (nt. 77) M11 (nt. 544) |
E1N14_01730 | E1N14_01730 | Hypothetical protein | – |
| M12 (nt. 208) | nqrA | E1N14_01800 | Na+‐translocating NADH‐quinone reductase subunit A | Ion transport, Sodium transport, Transport |
| M13 (nt. 190) | fusA | E1N14_03520 | Elongation factor G | Protein biosynthesis |
| M14 (nt. 171) | E1N14_03880 | E1N14_03880 | Hypothetical protein | ‐ |
| M15 (nt. 499) | rho | E1N14_05040 | Transcription termination factor rho | Transcription, Transcription regulation, Transcription termination |
|
M16 (nt. 978) M17 (nt. 396) |
gspF | E1N14_05760 | T2SS protein GspF | Protein transport, Transport |
| M18 (nt. 570) | gspD | E1N14_05770 | T2SS protein GspD | Protein transport, Transport |
| M19 (nt. 217) | menF | E1N14_07510 | Isochorismate synthase | Menaquinone biosynthesis |
| M20 (nt. 1574) | menD | E1N14_08030 | 2‐Succinyl‐5‐enolpyruvyl‐6‐hydroxy‐3‐cyclohexene‐1‐carboxylase synthase | Menaquinone biosynthesis |
| M21 a | E1N14_08925 | E1N14_08925 | Type I secretion C‐terminal target domain‐containing protein | Cell communication |
| M22 (nt. 68) | E1N14_09155 | E1N14_09155 | Hypothetical protein | ‐ |
| M23 (nt. 1915) | priA | E1N14_09830 | Primosomal protein N’ | DNA replication |
| M24 (nt. 143) | degS | E1N14_10330 | Outer membrane‐stress sensor serine endopeptidase DegS | Stress response |
| M25 (nt. 61) | E1N14_10530 | E1N14_10530 | L‐sorbosone dehydrogenase | ‐ |
| M26 (nt. 79) | crp | E1N14_10795 | cAMP‐activated global transcriptional regulator CRP | Transcription, Transcription regulation |
| M27 (nt. 239) | mdh | E1N14_12485 | Malate dehydrogenase | Tricarboxylic acid cycle |
| M28 (nt. 902) | E1N14_14620 | E1N14_14620 | LPS O‐antigen length regulator | Lipopolysaccharide biosynthetic process |
| M29 (nt. 567) | E1N14_14660 | E1N14_14660 | Glycosyltransferase family 1 protein | Glycogen biosynthesis |
|
M30 (nt. 963) M31 (nt. 430) |
E1N14_14665 | E1N14_14665 | Delta‐aminolevulinic acid dehydratase | Heme biosynthesis, Porphyrin biosynthesis |
|
M32 (nt. 8) M33 (nt. 137) |
wecG | E1N14_14675 | WecB/TagA/CpsF family glycosyltransferase | Enterobacterial common antigen biosynthetic process |
|
M34 (nt. 566) M35 (nt. 297) M36 (nt. 297) M37 (nt. 90) |
E1N14_14870 | E1N14_14870 | Hypothetical protein | ‐ |
|
M38 (nt. 2499) M39 (nt. 3654) |
purL | E1N14_15175 | Phosphoribosylformylglycinamidine synthase | Purine biosynthesis |
|
M40 (nt. 19) M41 (nt. 191) |
E1N14_17155 | E1N14_17155 | DNA adenine methylase | DNA restriction‐modification system |
| M42 (nt. 618) | recB | E1N14_17705 | Exodeoxyribonuclease V subunit beta | DNA damage, DNA repair |
| M43 (nt. 1494) | edd | E1N14_19200 | Phosphogluconate dehydratase | Carbohydrate metabolism, Gluconate utilization |
Gene abbreviations, locus tag in the S. algae CECT 5071 chromosome and gene annotations are indicated. Biological processes associated with gene products are shown for annotated genes.
The open reading frame appears to be truncated (edge of a contig), therefore the transposon insertion site is ambiguous.
Expression and composition of extracellular matrix of S. algae CECT 5071 wild‐type and transposon mutants
To gain a better understanding of extracellular matrix expression and the chemical basis of the different colony biofilm phenotypes, the WT strain and mutants M1 (barA), M3 (thyA) and M8 (E1N14_01730) were selected as representatives of the phenotypes observed in our study. Thus, compared to the WT, the barA mutant M1 showed preserved BBG binding with CR binding shifted to the outmost colony rim, the thyA mutant M3 showed increased CR binding, and the E1N14_01730 mutant M8 showed negligible dye binding. Inspection of the central and peripheral areas of these colonies by Scanning Electron Microscopy (SEM) (Fig. 3) revealed cells locally producing different extracellular matrices, a hallmark of biofilm formation (Flemming and Wingender, 2010). Of note, the arrangement of cells to each other within the colony as well as their morphology differed dramatically across the selected mutants compared to the WT strain; most distinctively, the barA mutant M1 displayed extensively aligned cells and the thyA mutant M3 displayed dramatically elongated, filamented cells. Altogether, these observations show extracellular matrix production in S. algae colony communities and suggest potential differences in EPS structure and composition between WT and mutant strains as well as different levels of multicellular organization.
Fig. 3.
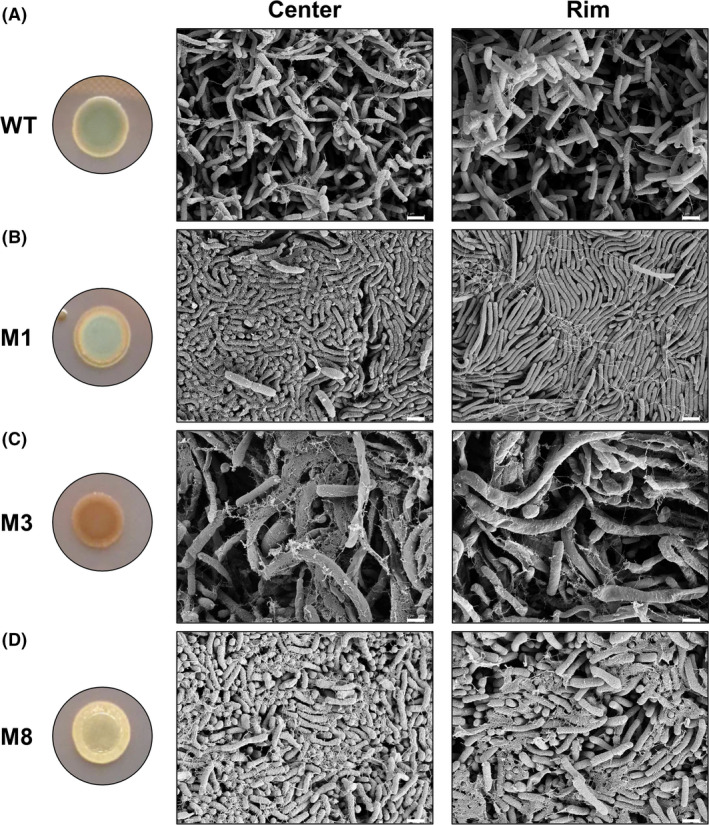
Scanning Electron Microscopy images of colony biofilms of S. algae CECT 5071 wild‐type and transposon mutant derivatives. Shown are representative images of the central and peripheral areas of colony biofilms formed by the WT strain (A) and the transposon insertion mutants M1 (barA) (B), M3 (thyA) (C), and M8 (E1N14_01730) (D) after 48 h of incubation on LB agar at 30°C. Macroscopic colony images are reproduced from Fig. 2. Scale bar = 1 µm.
As the composition of S. algae biofilm EPS is unknown, EPS was extracted from colony biofilms of the WT strain and mutants M1 (barA), M3 (thyA) and M8 (E1N14_01730) following two procedures relying on different chemistries, namely cation exchange resin (CER) extraction in PBS and formaldehyde‐NaOH extraction of PBS‐resuspended colonies, the latter reported to increase the solubility of biofilm EPS constituents (Liu and Fang, 2002). The total protein and carbohydrate content of extracted EPS was analyzed. Per unit of biomass, the EPS of M3 (thyA) contained a higher carbohydrate content than that of the WT strain, and this difference was statistically significant for the CER‐extracted EPS (Fig. 4A and C). No significant differences in total carbohydrate content were found in the EPS of mutants M1 (barA) and M8 (E1N14_01730) compared to the WT using either extraction method. The total protein content of CER‐extracted EPS of M1 (barA) and M3 (thyA) was significantly higher than that of the WT strain (Fig. 4B), whereas no significant difference with respect to the WT was recorded for the EPS of M8 (E1N14_01730). The total protein content in the formaldehyde‐NaOH EPS extract of M8 (E1N14_01730) was significantly lower than that of the WT strain, whereas no significant differences were recorded for the other two mutants with respect to the WT (Fig. 4D ). Overall, the total carbohydrate and protein content was higher in formaldehyde‐NaOH extractions than in CER extractions, consistent with previous reports (Liu and Fang, 2002).
Fig. 4.
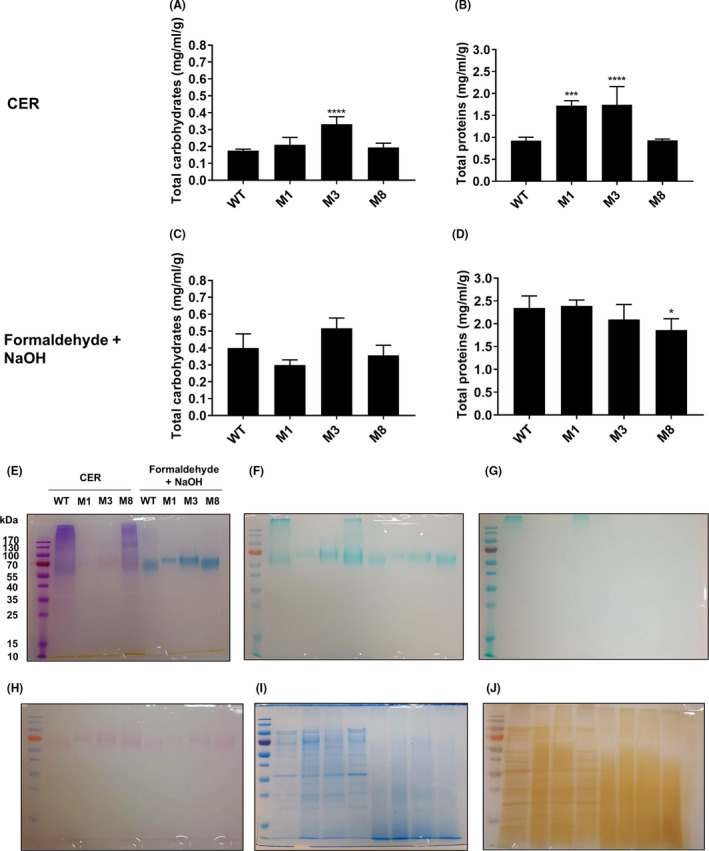
Biofilm Extracellular Polymeric Substances (EPS) analysis of colony biofilms of S. algae CECT 5071 WT and selected transposon insertion mutants. Total carbohydrate content per unit of biofilm biomass (wet weigh) (A, C) and total protein content per unit of biofilm biomass (B, D) in CER EPS extracts and formaldehyde‐NaOH EPS extracts of the WT strain and mutants M1 (barA), M3 (thyA) and M8 (E1N14_01730). The statistical significance of the differences between the WT strain and transposon insertion mutants was calculated by One‐way ANOVA followed by Dunnett’s post hoc analysis, with P = 0.05 as the threshold for significance (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). SDS‐PAGE electrophoretic profiles of extracted EPS after Stains‐All staining (E), Alcian blue staining at pH = 2.5 (F), Alcian blue staining at pH = 1.0 (G), PAS staining (H), colloidal Coomassie staining (I), and silver staining (J).
We next separated the EPS content by SDS‐PAGE followed by gel staining using an array of dyes specific to different biochemical components. All electrophoretic runs contained similar quantities of EPS, i.e. 7.5 µg of total protein per lane. Stains‐All staining (Fig. 4E) showed a smear of purple‐stained high MW components (proteoglycans) in WT and M8 (E1N14_01730) CER extracts, which gradually diffused for lower MW components under 55 kDa. Notably, discrete bands of approximately 60 kDa were observed in the EPS of the WT and mutant M8 (E1N14_01730), and of 100 kDa in the EPS of mutants M1 (barA) and M3 (thyA), irrespective of the extraction method. In CER EPS extractions, these bands stained faint pink, indicating anionic protein composition. The same bands were remarkably more prominent in formaldehyde‐NaOH extracts and stained intense blue, consistent with an identity as highly anionic proteins, with the change in global charge being presumably a consequence of the extraction method. Alcian Blue staining at pH = 2.5 (Fig. 4F) and PAS staining (Fig. 4H) revealed these components to likely be carboxylated glycoproteins. Consistently, these discrete bands were absent upon Alcian Blue staining at pH = 1.0 (Fig. 4G), which otherwise revealed sulfated components of a MW significantly higher than our detection limit, presumably of a polymeric nature, in the CER EPS extracts of the WT and M8 (E1N14_01730) strains. Staining of proteins by colloidal Coomassie (Fig. 4I) revealed a proteome in CER EPS extracts that was similar for the WT strain and the M8 (E1N14_01730) mutant, and for the M1 (barA) and M3 (thyA) mutants, respectively. Formaldehyde‐NaOH treatment, however, seemed to cause extensive protein degradation as noted by a diffuse smear in all extracts. Loading lower amounts of extracts (down to < 2.5 µg of total protein) did not contribute to the development of discrete bands (data not shown). Such profiles for each mutant and extraction method were consistent with those observed upon silver staining, which otherwise revealed additional discrete bands in CER extracts given the higher sensitivity of this method over colloidal Coomassie (Fig. 4J).
FTIR analysis of EPS and monosaccharide composition
Fingerprinting of the EPS material extracted by both the CER and formaldehyde‐NaOH treatments was performed by FTIR analysis (Fig. 5A, B, respectively). The FTIR spectra collected from two biological replicate samples were essentially identical (Fig. S2). FTIR spectra showed similar bands for the EPS material of the different strains and extraction procedures, with a broad band centered at 3250 cm‐1 corresponding to the O‐H stretching of the hydroxyl groups in polysaccharides and proteins; a triple band at 2900 cm‐1 that can be assigned to the CH2 asymmetric stretch in lipids and fatty acids; a shoulder at 1720 cm‐1 corresponding to the carbonyl (C = O) stretch of ester groups; the bands at 1650 and 1550 cm‐1 assigned to the Amide I and Amide II bands of proteins, respectively; the absorption bands at 1135 and 1050 cm−1 that usually represent the stretching vibrations of C‐O‐C and C‐O in carbohydrates and nucleic acids; and finally, a band at 850 cm‐1 that could be assigned to the C‐H bending in an aromatic moiety (Cao et al., 2011; Sardari et al., 2017; Vilaplana et al., 2015).
Fig. 5.
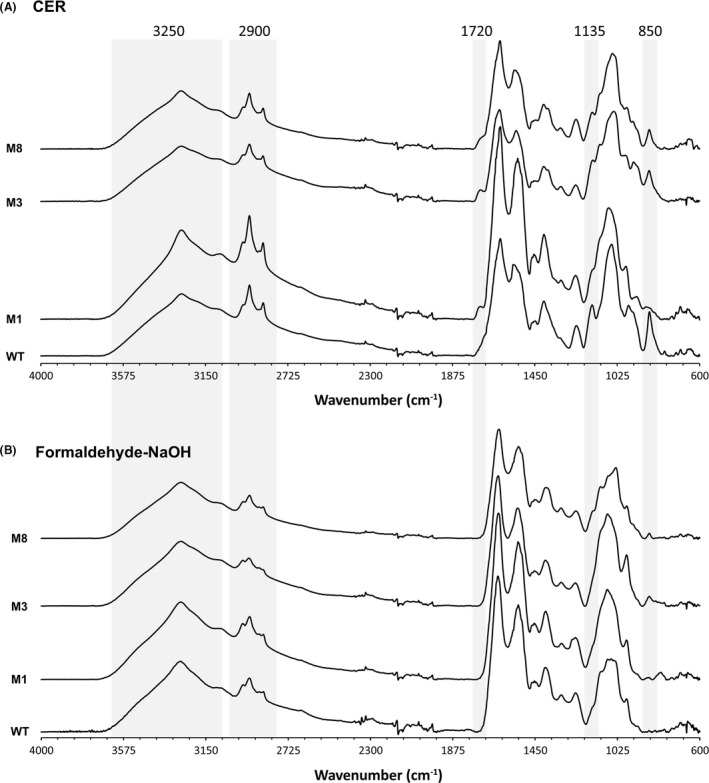
FTIR spectra of EPS extracts of S. algae WT and selected transposon insertion mutants. Spectra from two biologically replicate experiments per strain are shown for (A) CER EPS extracts (B) formaldehyde‐NaOH EPS extracts. Peaks with differences between samples are highlighted.
The CER EPS extracts showed a higher relative absorbance of the bands at 2900 and 1720 cm‐1, potentially related to the presence of lipid and fatty acid populations that might have been partially removed by the formaldehyde‐NaOH treatment. When comparing the EPS from different strains, no major differences could be observed in the FTIR spectra. The bands at 2900 cm‐1 were more prominent for the WT and M1 (barA) strains compared with M3 (thyA) and M8 (E1N14_01730) for both the CER and formaldehyde‐NaOH extracts, indicating a higher presence of lipids and fatty acids. The presence of a small shoulder at 1720 cm‐1 could be detected in the CER EPS extracts of all strains except the WT; this shoulder can be assigned to the carbonyl stretch of ester groups potentially present in acidic sugars. Differences in the area between 1300 and 900 cm‐1 could be assigned to the nucleic acid and polysaccharide content and composition in the different extraction processes and strains. Finally, the absence of the band at 850 cm‐1 in the M1 (barA) mutant CER‐extracted EPS compared with those of the WT, M3 (thyA) and M8 (E1N14_01730) strains could be attributed to the relative presence of aromatic side chains of the respective amino acids (Vilaplana et al., 2015).
The monosaccharide analysis of the EPS (Table 2) revealed the presence of galactose (Gal) and galactosamine (GalN) as the main components of the extracts, with minor amounts of glucosamine (GlcN), glucose (Glc), mannose (Man), ribose (Rib) and arabinose (Ara). The presence of other monosaccharides such as rhamnose (Rha) or fucose (Fuc) was not detected in the EPS. Interestingly, major differences were observed in the monosaccharide composition depending on the extraction procedure, with a marked abundance of Ara in the NaOH‐formaldehyde extracts compared to the CER EPS materials. In general, the WT strain showed a trend to higher GalN relative abundance compared to Gal, whereas in the M1 (barA), M3 (thyA) and M8 (E1N14_01730) mutants the Gal content was highest (Table 2).
Table 2.
Monosaccharide composition of S. algae WT and selected Tn5 integration mutants.
| Monosaccharide | CER | NaOH‐Formaldehyde | ||||||
|---|---|---|---|---|---|---|---|---|
| WT | M1 | M3 | M8 | WT | M1 | M3 | M8 | |
| Ara | 0.9 (0.3) | 0.7 (0.1) | 0.5 (0.3) | 0.2 (0.1) | 12.0 (0.2) | 16.7 (0.9) | 19.7 (0.2) | 10.9 (1.5) |
| Rib | 1.9 (1.9) | 0.2 (0.1) | 0.2 (0.0) | 0.2 (0.0) | 1.4 (0.3) | 1.4 (0.1) | 2.2 (0.4) | 1.2 (0.3) |
| Man | 2.0 (0.5) | 2.6 (0.1) | 0.8 (0.2) | 0.9 (0.2) | 0.4 (0.0) | 3.7 (2.9) | 0.4 (0.1) | 0.3 (0.0) |
| Gal | 30.1 (0.8) | 54.9 (3.2) | 37.8 (10.8) | 44.1 (8.7) | 33.4 (1.4) | 41.5 (1.3) | 37.1 (0.8) | 46.8 (1.9) |
| Glc | 8.3 (0.6) | 9.1 (0.5) | 5.1 (1.8) | 4.5 (1.5) | 2.1 (0.5) | 7.2 (1.5) | 5.6 (3.3) | 2.4 (0.5) |
| GlcN | 12.4 (0.8) | 12.4 (2.3) | 11.4 (0.3) | 11.1 (0.7) | 10.1 (0.2) | 10.6 (1.0) | 9.1 (0.5) | 8.5 (0.0) |
| GalN | 44.4 (1.9) | 20.0 (1.6) | 44.2 (9.1) | 39.1 (9.9) | 40.7 (1.2) | 18.9 (5.7) | 25.9 (2.9) | 29.9 (4.0) |
Ara, arabinose; Rib,ribose; Man, mannose; Gal, galactose; Glc, glucose; GlcN, glucosamine; GalN, galactosamine.
Values indicate %mol of neutral sugars as determined in two replicate biological extracts with three technical replicates each (standard deviation is presented in parentheses).
Static biofilm formation analyses
We next questioned whether altered binding of CR or BBG correlates with altered biofilm formation in static broth culture. To this end, the WT strain and the 43 transposon mutants were assayed for biofilm formation in MB medium. Bacterial total cell density at 600 nm, OD600, monitored during 24 h of static incubation indicated that all strains had reached stationary phase at the experimental endpoint (data not shown). Qualitative features of the growth and biofilm phenotypes of all strains are shown in Fig. S3. Thus, preferential growth at the air‐liquid interface was observed for the WT strain, with significant pellicle formation as well as biofilm formation on the walls and on the bottom of the wells. Different levels of planktonic growth and biofilm formation patterns were recorded across mutant strains and included mutants with stronger pellicle production, e.g. M42 (recB); mutants with proficient sub‐aerial biofilm formation, e.g. M32‐M33 (wecG); mutants with impaired pellicle production but preserved sub‐aerial biofilm formation capacity, e.g. M19‐M20 (menF and menD, respectively); and mutants unable to form biofilm, e.g. M15 (rho) and M21 (E1N14_08925). To account for effects derived from growth alterations in mutant strains, both biofilm formation indexes as determined by crystal violet (CV)‐stained biomass and normalized biofilm indexes as determined by the ratio between CV‐stained biomass and total cell density (OD600) are presented (Fig. 6A, B). Absolute and normalized biofilm indexes gave comparable results.
Fig. 6.
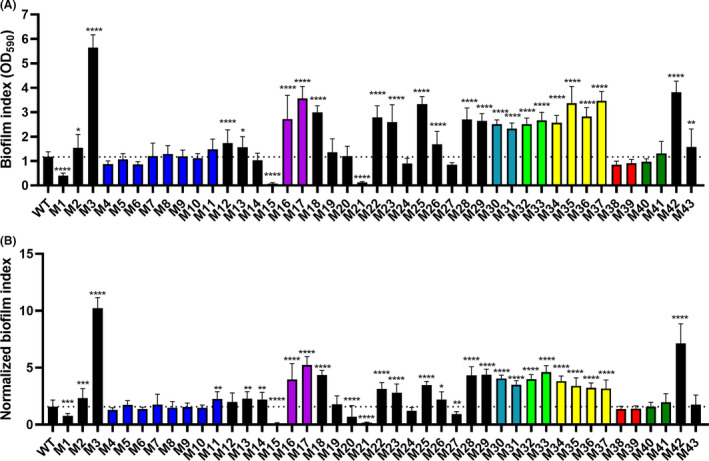
Biofilm formation in static marine broth culture of S. algae CECT 5071 WT and transposon insertion mutants.
A. Surface‐associated biofilm indexes as quantified by crystal violet staining (OD590) from at least three biological replicate cultures with seven technical replicates each. The statistical significance of the differences between the WT strain and transposon insertion mutants was calculated by One‐way ANOVA followed by Dunnett’s post hoc analysis, with P = 0.05 as the threshold for significance (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
B. Normalized biofilm indexes, as determined by the ratio between eluted CV recordings and the OD600 of the cultures before CV staining. The statistical significance of the differences between WT and mutants was calculated by One‐way ANOVA followed by Dunnett’s post hoc analysis, with P = 0.05 as the threshold for significance (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). The dotted line in panels (A) and (B) indicate WT levels of biofilm or normalized biofilm, respectively.
Altered surface‐associated biofilm formation of transposon insertion mutants with respect to the WT was frequently observed. The lack of apparent correlation between the colony and surface‐associated biofilm phenotypes suggest different components are involved in the development of either type of biofilm in different media. For example, the M3 (thyA), M19 (menF) and M20 (menD) mutants show increased CR binding, but only M3 exhibited significantly increased biofilm formation (> 5‐fold) compared to the WT strain. Likewise, transposon mutants M4‐M11 interrupting the same hypothetical gene (E1N14_01730) all displayed a colony phenotype with abrogated CR and BBG binding compared to the WT, even though their surface‐associated biofilm formation in marine broth culture as determined by CV staining was not significantly different. On the other side of the spectrum, biofilm formation was strongly impaired or almost entirely abolished in three mutants: M1 (barA), M15 (rho), and M21 (E1N14_08925). Consistently, mutants within the same gene interrupted by mini‐Tn5 insertion exhibited similar biofilm formation changes with respect to the WT strain.
Functional analysis of the S. algae CECT 5071 sensor histidine kinase BarA
Signal transduction systems integrate environmental stimuli with physiological responses, including biofilm formation. BarA is the sensor histidine kinase of the BarA‐UvrY two‐component system, catalyzing phosphorylation of the cognate response regulator UvrY via a phosphorelay cascade from the transmitter (HisKA) domain with a conserved His302 residue to a central receiver (REC) domain containing a conserved Asp718 residue, and subsequently to a C‐terminal phosphotransfer domain (Hpt) containing a conserved His861 residue (Ishige et al., 1994; Pernestig et al., 2001) (Fig. S4A). Domain structure and phosphotransfer residues are conserved in the S. algae ortholog of E. coli.
To functionally characterize the role of the BarA phosphorelay on S. algae static biofilm formation, the native barA gene or mutant derivatives harboring one or several amino acid replacements in the phosphorelay cascade were expressed from plasmid pSRK‐Gm in the barA mutant M1 showing impaired biofilm production. Of note, the empty expression vector pSRK‐Gm had a pronounced effect resulting in a higher baseline level of biofilm formation in the WT strain (Fig. S4B). Ectopic expression of barA restored biofilm formation to 40 % WT pSRK‐Gm levels.
Next, single, double, and triple amino acid replacements resulting in H306A, D729A, H860A, H306A‐D729A and H306A‐D729A‐H860A, were introduced into BarA on the complementation vector pBarA. Mutant proteins expressed in mutant M1 with the amino acid replacements D729A, H860A, H306A‐D729A and H306A‐D729A‐H860A did not significantly change biofilm formation with respect to the native protein. The only marginally statistically significant difference was recorded for cells expressing the BarA H306A mutant with respect to the native protein, yet with a presumably negligible biological significance. Of note, WT pSRK‐Gm and M1 cells expressing the native protein and mutant derivatives with single amino acid replacements exhibited cell aggregation (Fig. S4B). Cell aggregation was, though, not observed in M1 cells harboring the empty plasmid or upon expression of variants of BarA with double or triple amino acid replacements. These results evidence a substantial plasmid effect and, in addition, suggest that BarA plays a complex role in the expression of the cell aggregation and biofilm formation phenotypes.
Discussion
Regulation of colony morphology and biofilm formation are complex processes, often involving multiple, interconnected pathways. In this work, we analyzed genes contributing to the development of distinct colony morphotypes by differential CR and BBG binding in the type strain S. algae CECT 5071 as well as biofilm formation onto an abiotic surface in static broth culture. Over 5,000 transposon insertion mutants were generated, and the transposon insertion site was identified in 43 mutants showing altered colony morphology features. While our transposon screening is far from saturated, the likelihood of random integration into the S. algae CECT 5071 chromosome is supported by the retrieval of independent transposon insertions.
Targeted open reading frames included genes involved in essential processes such as nucleotide biosynthesis, DNA replication and repair, transcription, and central metabolic pathways, supporting the notion that CR and BBG colony morphology development is a multi‐factorial process deeply integrated into the physiology of S. algae. Transposon insertions in specific pathways like menaquinone biosynthesis (men mutants M19 and M20) and sodium transport (nqrA mutant M12) point to respiration as a key process regulating colony morphotypes of S. algae. Of note, many of the transposon‐interrupted genes code for hypothetical proteins of yet unknown function. For example, gene E1N14_01730 encoding a hypothetical protein that abolished BBG binding, a novel colony dye binding type, was found to be disrupted in 8 of the 43 mapped mutants and is a primary candidate for further functional studies.
Binding of specific dyes to agar‐grown colonies has been shown to be an indicator of, for example, virulence, biofilm formation and iron acquisition (Daskaleros and Payne, 1987). We do not know which physiological stage the BBG and CR binding colony morphotype reflects. To our knowledge, binding of BBG has not been associated with a certain physiological stage or surface protein expression. However, we hypothesize that an elevated ‘red’ morphotype might resemble the rdar or rugose colony biofilm morphotype of E. coli and S. Typhimurium (Römling et al., 2003; Smith et al., 2017) or the rugose morphotype of diverse Vibrio spp. including Vibrio cholerae (Yildiz and Visick, 2009) and other bacteria. Major hallmarks of a biofilm can include the expression of adhesive EPS components that induce a rough/rugose colony phenotype, an associated surface‐ and self‐aggregative phenotype, a regulation of the colony morphotype by the ubiquitous messenger cyclic di‐GMP, a substantially altered cell physiology and metabolism, and tolerance against antimicrobials and disinfectants (Römling and Balsalobre, 2012).
One of the intrinsic features of the biofilm lifestyle is the production of EPS with structural, protective and physiological functions (Flemming et al., 2007). To our knowledge, colony phenotypes have not been investigated previously in S. algae. We showed that the colony morphotype features of S. algae on CR and BBG‐supplemented agar emerge from dye binding. As such phenotypes might be associated with a different protein and carbohydrate composition of the extracellular matrix, biofilm EPS was extracted using two methods. Our biochemical analyses showed similarities between the EPS of the colony biofilms of WT and M8 (E1N14_01730) on the one hand, and of M1 (barA) and M3 (thyA) on the other hand, which were consistent in both extraction procedures. Major differences in monosaccharide content were observed between the CER and formaldehyde‐NaOH extracts, the latter showing ten‐fold higher arabinose content than the former. In both extraction procedures, galactose and galactosamine were the most abundant monosaccharides. Nevertheless, CR or BBG binding to specific, individual EPS components produced by S. algae could not be assigned. It should be noted that many biofilm EPS components such as the exopolysaccharides cellulose and curdlan, type 1 fimbriae, and amyloid fimbriae are not soluble in water and can only be depolymerized and/or extracted by specific treatment which enables their isolation and subsequent disaggregation/hydrolysis into monomeric components, such as treatment with acids or alkali (Collinson et al., 1991; Zogaj et al., 2001). It is not known whether similar insoluble polymers may be produced by S. algae. It should also be noted that CR is reported to bind as well to membrane‐integrated outer membrane proteins (Qadri et al., 1988), or soluble but firmly‐attached LPS (Budanova et al., 2018). Differential elucidation of the identity of EPS constituents of S. algae WT and mutant biofilms certainly requires additional experimentation with alternative technical approaches.
Testing selected transposon mutants in static liquid culture allowed us to identify several genes positively or negatively regulating surface‐associated biofilm formation in S. algae. Genes whose interruption increased biofilm formation included the T2SS (gsp mutants M16‐M18), which has been shown to be required for biofilm formation in enteropathogenic E. coli (Baldi et al., 2012) and Vibrio cholerae (Johnson et al., 2014). Notably, the interruption of genes involved in nucleotide biosynthesis nadB (M2) and thyA (M3) resulted in increased biofilm production, the latter causing a remarkable 5‐fold increase. The biofilm formation increase observed in the nqrA mutant M12 suggests an involvement of the electron transport chain and respiration in the regulation of this type of biofilm, a process that we have reported to be deeply involved in the regulation of biofilm formation in S. algae and E. coli (Kamal et al., 2020; Martín‐Rodríguez et al., 2020, 2021).
Among the genes whose disruption caused inhibition of surface‐associated biofilm formation, a transposon insertion in the gene with locus tag E1N14_08925 (mutant M21) practically abrogated biofilm production. Biofilm formation in the S. algae rho mutant M15 was also virtually entirely abolished, pointing to transcription termination to be involved in biofilm formation. Interruption of the hybrid sensor histidine kinase barA caused significant inhibition of biofilm formation. A study in Yersinia pseudotuberculosis showed a barA mutant to increase CR binding while surface‐associated biofilm in multiwell plates was not significantly different from that of the WT (Schachterle et al., 2018). Surprisingly, while the phosphorelay activity of BarA was not required for S. algae biofilm formation on the abiotic surface, phenotypic differences in biofilm formation patterns were recorded upon the expression of mutants in two or all three amino acids in the phosphorylation cascade. Notably, control of biofilm cohesiveness by BarA has been reported in Y. pseudotuberculosis (Schachterle et al., 2018). A limitation of our study is the strong plasmid effect, which altered dramatically the biofilm phenotypes under investigation, as well as the intrinsic difficulty of performing gene manipulation in this strain. The metabolic costs of plasmid maintenance, including significant effects on biofilm formation, are recognized (Ghigo, 2001; Ow et al., 2006; Teodósio et al., 2012) .
In summary, this work explored genes involved in the regulation of different multicellular behaviors in S. algae using a transposon mutagenesis approach. We have defined a dye binding colony model of S. algae on LB agar supplemented with CR and BBG, paving the way towards studies aiming to explore the identity of the CR and BBG binding targets and the nutrient factors affecting the development of different strain‐specific morphotypes. A first chemical characterization of the biofilm EPS of S. algae has been provided, which showed distinctive electrophoretic fingerprints of the WT strain and selected mutants. Monosaccharide analyses identified galactose and galactosamine as the main components of S. algae colony extracellular matrix. In addition, distinct cell‐cell arrangement and cell morphology was observed in mutant colonies, the biological significance still to be explored. The development of alternative genetic tools will facilitate subsequent molecular studies with S. algae.
Experimental procedures
Strains and growth conditions
The bacterial strains used in this study are listed in Table S1. Shewanella algae CECT 5071 was routinely cultured in Marine Broth (MB) or on Marine Agar at 30°C. Escherichia coli strains were routinely cultured in LB broth or on LB agar at 37°C. When required, kanamycin (50 µg ml‐1), gentamycin (50 µg ml‐1), and/or isopropyl β‐D‐1‐thiogalactopyranoside (IPTG, 500 µM) were supplemented.
Transposon mutagenesis
Plasmid pRL27 (Table S2) was mobilized from E. coli MFDpir to S. algae by bi‐parental mating (Ferrières et al., 2010). Briefly, 1‐ml aliquots of overnight cultures of both strains were washed twice with fresh LB medium containing 0.3 mM diaminopimelic acid (DAP), mixed, spotted on LB agar containing DAP, and incubated overnight at 37°C. Mating was interrupted by re‐suspending the cells in LB medium and plating on Km agar.
A total of 5082 mutants were individually picked, grown in MB, and their colony biofilm morphotype analyzed on LB agar supplemented with 40 µg ml‐1 CR and 20 µg ml‐1 BBG, on which S. algae showed the most pronounced colony morphotype. Mutants exhibiting altered colony biofilm morphotypes were preserved. Mapping of the transposon chromosomal insertion sites was conducted as previously described (Larsen et al., 2002). Briefly, genomic DNA was isolated and digested with an enzyme that does not cut within the transposon (BamHI and/or SpeI). The resulting DNA fragments containing the transposon and adjacent chromosomal DNA were self‐ligated, propagated in E. coli DH5α λpir and sequenced using primers tpnRL17–1 and tpnRL13–2 (Table S2) pointing outwards of the transposon. The genetic environment of the transposon insertion sites was reconstructed using the S. algae CECT 5071 draft genome sequence obtained as described below.
Whole‐genome sequencing
DNA was extracted from S. algae CECT 5071 using the DNeasy Blood & Tissue Kit (Qiagen). Library preparation was performed using the TruSeq Nano DNA library preparation kit (Illumina) and the libraries were subsequently sequenced on a MiSeq platform, 2 x 300 bp paired end reads. The sequencing reads were processed using the BACTpipe bacterial assembly and annotation pipeline v2.6.0 (https://github.com/ctmrbio/BACTpipe). The Whole Genome Shotgun project was deposited at the GenBank under the accession SMNR00000000 and annotation was added by the NCBI Prokaryotic Genome Annotation Pipeline
Colony biofilm morphotype analyses
The colony biofilm morphotype of S. algae WT and transposon insertion mutants was evaluated on LB agar supplemented with CR (40 µg ml‐1) and a second additive, BBG (20 µg ml‐1), to provide better contrast for the visualization of colony morphotypes (Friedman and Kolter, 2004; Cimdins and Simm, 2017). Ten µl overnight cultures were spotted on CR and BBG‐supplemented agar plates and incubated at 30°C for 48 h for phenotype recording. Extraction of colony‐bound dyes was performed by scraping and resuspending the colonies in 500 µl methanol, followed by intense vortexing and centrifugation to collect cell debris. The absorbance spectra were recorded from 200‐µl supernatant aliquots.
Assessment of biofilm formation in broth culture
Static biofilm formation assays were conducted as reported (Martín‐Rodríguez et al., 2014) with minor modifications. Briefly, fresh overnight cultures were homogenized by vigorous vortexing and diluted 1:100 in MB. Two‐hundred µl were pipetted into each well of a flat‐bottomed 96‐well plate and incubated statically at 30°C for 24 h. Total cell density was determined as OD600. The total biofilm index was determined by CV staining (0.2 %) followed by elution with 30% acetic acid and absorbance recording at 590 nm. The normalized biofilm index was calculated as the ratio between eluted CV recordings and OD600. Growth and biofilm formation patterns were documented using strip‐wells (Greiner Bio‐One, Kremsmünster, Austria, #762070). Statistical analyses were performed using GraphPad Prism software version 8.4.3 (GraphPad Software, San Diego, CA, USA).
Extraction of soluble biofilm extracellular matrix components
Extraction of soluble extracellular polymeric substances (EPS) from the colony biofilms of selected transposon mutants was performed following two previously described procedures, namely CER extraction and formaldehyde‐NaOH extraction (Liu and Fang, 2002; Jachlewski et al., 2015) with minor modifications. Thus, overnight cultures of the test strains were prepared in two biological replicates, homogenized by vigorous vortexing and used to grow colony biofilms by spotting 10 µl on LB agar plates without addition of dyes (30°C, 48 h, 50 colonies per biological replicate). Each set of 50 colonies was scraped from the agar and suspended in 10 ml PBS (pH = 7.4, 0.01 M, Medicago, Sweden) in pre‐weighed 50 ml tubes, and the biofilm biomass was recorded.
For CER extraction, 2 g of the strongly acidic resin Amberlite HPR1100 (Merck, Darmstadt, Germany) pre‐washed with 20 ml PBS (2 x 15 min) were added. The biofilm suspensions were vigorously shaken at maximal speed for 10 min using a KEBO‐Lab REAX 2000 vortex. Subsequently, the tubes were centrifuged at 4°C, 4000 rpm, for 2 h, and the supernatants were filtered (0.2 µm). The filtered supernatants were transferred to Pur‐A‐Lyzer dialysis membranes (MWCO 3.5 kDa) and dialyzed against ddH2O 2 x 1 h at room temperature and then 24 h at 4°C.
For formaldehyde‐NaOH EPS extraction, the resuspended biofilm biomass was fixed for 1 h at 4°C by addition of 60 µl formaldehyde (37 % v/v). After fixation, 800 µl 5N NaOH were added and the samples were stirred for 3 h at 4°C. Subsequently, the extracts were centrifuged for 2 h at 4°C, 4000 rpm. The supernatants were collected, filtered (0.2 µm), and dialyzed against ddH2O 2 x 1 h and then 24 h at 4°C.
Scanning electron microscopy
Prior to imaging, bacterial colonies were overlayered with 2.5% glutaraldehyde and 1% paraformaldehyde in 0.1 M phosphate buffer, pH = 7.4. After fixation, the bacterial colonies were cut out and the bulk of the supporting agar was carefully trimmed away prior to washing in ddH2O and stepwise ethanol dehydration. The colonies were finally subjected to critical‐point‐drying (Leica EM CPD 030, Wetzlar, Germany), mounted on specimen stubs using carbon adhesive tabs and sputter coated with platinum (Quorum Q150T ES). SEM images were acquired using an Ultra 55 field emission scanning electron microscope (Zeiss, Oberkochen, Germany) at 5kV and the SE2 detector.
Quantification of total carbohydrates and proteins in EPS extracts
Total carbohydrate contents per unit of biomass were determined using the phenol‐sulfuric acid method (Total Carbohydrate Assay Kit; Sigma‐Aldrich, St. Louis, MO, USA) and normalized with respect to the total biofilm biomass. Total proteins in the EMC extractions were quantified by the Lowry method after Peterson’s modification (Peterson, 1977) using a commercially‐available kit (Sigma‐Aldrich TP0300) and normalized with respect to the total colony biomass. Statistical analyses were performed using GraphPad Prism software version 8.4.3 (GraphPad Software, San Diego, CA, USA).
Electrophoretic analysis of biofilm EPS
EPS components were separated by SDS‐PAGE (12% polyacrylamide gels, 1× Tris‐Glycine‐SDS buffer, 100 V) and the gels were submitted to different staining methods to detect EPS components based on their chemical nature, as indicated below:
Stains‐all. A protocol by Lee and Cowman (Lee and Cowman, 1994) was adapted with minor modifications. Thus, after electrophoresis, gels were washed twice with ddH2O, fixed in 25% isopropanol for 3 h, washed again twice with ddH2O and stained overnight in a 0.005% (w/v) solution of Stains‐All in 50% ethanol. Gels were destained in ddH2O under light before imaging.
Alcian Blue staining. A protocol by Shori and co‐workers (Shori et al., 2001) was adapted with minor modifications. After electrophoresis, gels were washed twice with ddH2O, fixed (10% acetic acid, 25% ethanol, 4 × 15 min) (Boleij et al., 2019), washed again with ddH2O twice, and stained overnight in a Alcian Blue solution at pH = 2.5 (0.1% w/v Alcian Blue, 3% acetic acid, 0.5% v/v Tween‐20) or pH = 1.0 (0.1 % w/v Alcian Blue, 0.1 M HCl, 0.5% v/v Tween‐20). At pH = 2.5, the cationic dye Alcian Blue stains dissociated (ionic) acidic groups (Boleij et al., 2019) including carboxylated and sulfated mucins, whereas at pH = 1.0 the dye is specific to sulfated mucopolysaccharides. Gels were destained in solutions of the same composition as the respective staining solutions, except for Alcian Blue, and documented.
Periodic acid‐Schiff (PAS) staining. Glycoproteins were stained using the periodic acid‐Schiff (PAS) method with the Pierce Glycoprotein Staining Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. This method is specific for glycans bearing vicinal hydroxyl groups (Boleij et al., 2019).
Colloidal Coomassie staining. Visualization of proteins in EPS extracts was performed by colloidal Coomassie staining. Briefly, gels were washed twice with water, fixed for 1 h (40% ethanol, 10% acetic acid), washed 2x15 with water, and stained overnight with a colloidal Coomassie solution (0.1% w/v Brilliant Blue G‐250, 2% w/v 85% ortho‐phosphoric acid, 10% w/v ammonium sulfate). Excess dye was removed with 1% acetic acid before imaging.
Silver staining. EPS components in extracts were also visualized by silver staining using a commercial kit (Pierce Silver Stain Kit, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s indications. Excess stain was removed by placing the gel in a 1% hydrogen peroxide solution.
FTIR analysis
FTIR spectra were collected using a Spectrum 100 FTIR instrument (Perkin Elmer, Norwalk, CT, USA) equipped with a single reflection accessory unit (Golden Gate, Graseby Speac Ltd, Kent, UK). The data were recorded in a spectral range of 600–4000 cm‐1 at a resolution of 4.0 cm‐1 after 16 scans. The baseline correction and normalization of the recorded spectra were performed using Spectrum software (Perkin Elmer, Norwalk, CT, USA).
Monosaccharide analysis (neutral sugars and uronic acids)
Analysis of neutral sugars and uronic acids was performed by acid hydrolysis followed by chromatographic quantification of the released monosaccharides. Two biological replicates and two analytical replicates were performed per bacterial strain and extraction methodology. In brief, around 1 mg of freeze‐dried EPS material was subjected to acid hydrolysis with 2 M trifluoroacetic acid (TFA) at 121°C for 3 h (Albersheim et al., 1967). An aliquot of the acid hydrolysate was dried under N2 air, and the neutral sugars were derivatized to their alditol acetates (Blakeney et al., 1983) by reduction (using NaBH4 in 1 M NH3 at room temperature) and acetylation (with acetic anhydride in pyridine at 100°C for 60 min). The derivatized alditol acetates were purified by liquid:liquid extraction using ethyl acetate and further analyzed by gas chromatography – mass spectrometry (GC‐MS) using a HP‐6890 GC system and an HP‐5973 electron‐impact mass spectrometer (Agilent Technologies) equipped with a SP‐2380 capillary column (30 m by 0.25 mm, Supelco). Separation of the alditol acetates was performed using a temperature program from 180°C to 230°C at a rate of 1.5°C min‐1 (Mélida et al., 2013).
Site‐directed mutagenesis
Amino acid substitutions in S. algae BarA, H306A, D729A, H860A, H306A‐D729A and H306A‐D729A‐H860A, were introduced using the primers listed in Table S2 using the Q5 site‐directed mutagenesis kit (NEB) following the manufacturer’s instructions. Point mutations were confirmed by Sanger sequencing. Plasmids were propagated in Escherichia coli TOP10 and introduced in S. algae by electroporation. Electrocompetent S. algae cells were prepared by washing overnight cultures three times with 1 M sorbitol, pH = 7.2 (Corts et al., 2019).
Funding information
Stiftelsen Lars Hiertas Minne (Grant/Award Number: FO2019‐0293). Stiftelsen Längmanska Kulturfonden (Grant/Award Number: BA20‐0736). Karolinska Institutet (Grant/Award Number: 2020‐01556).
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
Study design: AJM‐R, UR. Experiments: AJM‐R, assisted by KV; FTIR and monosaccharide analyses were performed by SYT and FV. Writing – original draft: AJM‐R. Writing – review and editing: AJM‐R, FV, ÅS, UR. Funding acquisition: AJM‐R.
Supporting information
Fig. S1. Gene synteny and proximity of S. algae CECT 5071 transposon‐interrupted genes. Tn5‐disrupted genes are colored in red. Genes putatively belonging to the same operon are colored in pale yellow. Key annotations are indicated.
Fig. S2. Individual FTIR spectra of CER and formaldehyde‐NaOH EPS extracts. Data from two biological experiments are shown for CER EPS extracts, A‐D, and formaldehyde‐NaOH EPS extracts, E‐H, of the WT strain and mutants M1 (barA), M3 (thyA) and M8 (E1N14_01730).
Fig. S3. Growth and biofilm formation patterns of S. algae WT and transposon insertion mutants. Side and top views of strip‐well cultures (24 h, 30°C, MB medium) of S. algae CECT 5071 WT and isogenic transposon insertion mutant derivatives before and after crystal violet staining of biofilm biomass.
Fig. S4. Functional analysis of the hybrid sensor histidine kinase BarA of S. algae. A. Domain architecture of the BarA protein as determined by SMART (http://smart.embl‐heidelberg.de/). The location of Tn5 integration and key amino acid residues H306, D729 and H860 putatively involved in the phosphorelay cascade are indicated. B. Biofilm formation of S. algae WT and mutant M1 (barA) harboring the empty vector pSRK‐Gm plasmid, or the mutant strain complemented with the native or mutant barA derivatives cloned in pSRK‐Gm. Statistical significance of the differences are calculated from 4 biological replicates with 6 technical replicates each using One‐way ANOVA followed by Tukey’s post hoc analysis, with P = 0.05 as the threshold for significance (* P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001). Inset: side view of CV‐stained wells showing phenotypic differences upon native or mutant barA expression with respect to the WT or mutant empty vector controls.
Table S1. Bacterial strains used in this study.
Table S2. Plasmids and primers used in this study.
Acknowledgements
AJM‐R acknowledges funding from the Längmanska Kulturfonden (BA20‐0736), the Lars Hiertas Minne Foundation (FO2019‐0293) and the Karolinska Institutet Research Foundation (2020‐01556). Sequencing was performed at the Center for Translational Microbiome Research, a collaboration between SciLife Lab, Ferring Pharmaceuticals and Karolinska Institutet. The authors are grateful to Dr. K. Thorell (University of Gothenburg) for bioinformatics support and Dr. Lars Haag, Electron Microscopy Unit, Karolinska Institutet, for SEM imaging.
Microbial Biotechnology (2021) 14(3), 1183–1200
References
- Albersheim, P. , Nevins, D.J. , English, P.D. , and Karr, A. (1967) A method for the analysis of sugars in plant cell‐wall polysaccharides by gas‐liquid chromatography. Carbohydr Res 5: 340–345. [Google Scholar]
- Bagge, D. , Hjelm, M. , Johansen, C. , Huber, I. , and Gram, L. (2001) Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl Environ Microbiol 67: 2319–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi, D.L. , Higginson, E.E. , Hocking, D.M. , Praszkier, J. , Cavaliere, R. , James, C.E. , et al. (2012) The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli . Infect Immun 80: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeney, A.B. , Harris, P.J. , Henry, R.J. , and Stone, B.A. (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113: 291–299. [Google Scholar]
- Boleij, M. , Seviour, T. , Wong, L.L. , van Loosdrecht, M.C.M. , and Lin, Y. (2019) Solubilization and characterization of extracellular proteins from anammox granular sludge. Water Res 164: 114952. [DOI] [PubMed] [Google Scholar]
- Budanova, A.A. , Shirokov, A.A. , Shchyogolev, S.Y. , and Matora, L.Y. (2018) Analysis of Congo red‐induced changes in the cell surface and macrocolony structure of the bacterium Azospirillum brasilense . Microbiology (Russian Fed) 87: 60–65. [Google Scholar]
- Cao, B. , Shi, L. , Brown, R.N. , Xiong, Y. , Fredrickson, J.K. , Romine, M.F. , et al. (2011) Extracellular polymeric substances from Shewanella sp. HRCR‐1 biofilms: characterization by infrared spectroscopy and proteomics. Environ Microbiol 13: 1018–1031. [DOI] [PubMed] [Google Scholar]
- Cimdins, A. , and Simm, R. (2017. ) Semiquantitative analysis of the red, dry, and rough colony morphology of Salmonella enterica Serovar Typhimurium and Escherichia coli using congo red. Meth Mol Biol (Clifton, N.J.) 1657, 225–241. [DOI] [PubMed] [Google Scholar]
- Collinson, S.K. , Emödy, L. , Müller, K.H. , Trust, T.J. , and Kay, W.W. , et al. (1991) Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis . J Bacteriol 173: 4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corts, A.D. , Thomason, L.C. , Gill, R.T. , and Gralnick, J.A. (2019) A new recombineering system for precise genome‐editing in Shewanella oneidensis strain MR‐1 using single‐stranded oligonucleotides. Sci Rep 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskaleros, P.A. , and Payne, S.M. (1987) Congo red binding phenotype is associated with hemin binding and increased infectivity of Shigella flexneri in the HeLa cell model. Infect Immun 55: 1393–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrières, L. , Hémery, G. , Nham, T. , Guérout, A.‐M. , Mazel, D. , Beloin, C. , and Ghigo, J.‐M. (2010) Silent mischief: Bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad‐host‐range RP4 conjugative machinery. J Bacteriol 192: 6418–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming, H.‐C. , Neu, T.R. , and Wozniak, D.J. (2007) The EPS matrix: the "house of biofilm cells". J Bacteriol 189: 7945–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming, H.C. , and Wingender, J. (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Friedman, L. , and Kolter, R. (2004) Two genetic loci produce distinct carbohydrate‐rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186: 4457–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo, J.M. (2001) Natural conjugative plasmids induce bacterial biofilm development. Nature 412: 442–445. [DOI] [PubMed] [Google Scholar]
- Gong, A.‐D. , Li, H.‐P. , Shen, L. , Zhang, J.‐B. , Ai‐Bo, W. , and He, W.‐J. , et al. (2015) The Shewanella algae strain YM8 produces volatiles with strong inhibition activity against Aspergillus pathogens and aflatoxins. Front Microbiol 6: 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau, H.H. , and Gralnick, J.A. (2007) Ecology and biotechnology of the genus Shewanella . Annu Rev Microbiol 61: 237–258. [DOI] [PubMed] [Google Scholar]
- Holt, H.M. , Gahrn‐Hansen, B. , and Bruun, B. (2005) Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin Microbiol Infect 11: 347–352. [DOI] [PubMed] [Google Scholar]
- Ishige, K. , Nagasawa, S. , Tokishita, S. , and Mizuno, T. (1994) A novel device of bacterial signal transducers. EMBO J 13: 5195–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachlewski, S. , Jachlewski, W.D. , Linne, U. , Bräsen, C. , Wingender, J. , and Siebers, B. (2015) Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius . Front Bioeng Biotechnol 3: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J.M. , and Abbott, S.L. (2012) The genus Shewanella: from the briny depths below to human Pathogen. Crit Rev Microbiol 7828: 1–21. [DOI] [PubMed] [Google Scholar]
- Johnson, T.L. , Fong, J.C. , Rule, C. , Rogers, A. , Yildiz, F.H. , and Sandkvist, M. (2014) The Type II secretion system delivers matrix proteins for biofilm formation by Vibrio cholerae . J Bacteriol 196: 4245–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal, S.M. , Cimdins‐Ahne, A. , Lee, C. , Li, F. , Martín‐Rodríguez, A.J. , Seferbekova, Z. , et al. (2020) A recently isolated human commensal Escherichia coli ST10 clone member mediates enhanced thermotolerance and tetrathionate respiration on a P1 phage‐derived IncY plasmid. Mol Microbiol 115: 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, R. , Wilson, M. , Guss, A. , and Metcalf, W. (2002) Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178: 193–201. [DOI] [PubMed] [Google Scholar]
- Lee, H.G. , and Cowman, M.K. (1994) An agarose gel electrophoretic method for analysis of hyaluronan molecular weight distribution. Anal Biochem 219: 278–287. [DOI] [PubMed] [Google Scholar]
- Linares, D. , Jean, N. , Van Overtvelt, P. , Ouidir, T. , Hardouin, J. , Blache, Y. , and Molmeret, M. (2016) The marine bacteria Shewanella frigidimarina NCIMB400 upregulates the type VI secretion system during early biofilm formation. Environ Microbiol Rep 8: 110–121. [DOI] [PubMed] [Google Scholar]
- Liu, H. , and Fang, H.H.P. (2002) Extraction of extracellular polymeric substances (EPS) of sludges. J Biotechnol 95: 249–256. [DOI] [PubMed] [Google Scholar]
- Martín‐Rodríguez, A.J. , González‐Orive, A. , Hernández‐Creus, A. , Morales, A. , Dorta‐Guerra, R. , Norte, M. , et al. (2014) On the influence of the culture conditions in bacterial antifouling bioassays and biofilm properties: Shewanella algae, a case study. BMC Microbiol 14: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Rodríguez, A.J. , Martín‐Pujol, O. , Artiles‐Campelo, F. , Bolaños‐Rivero, M. , and Römling, U. (2017) Shewanella spp. infections in Gran Canaria, Spain: retrospective analysis of 31 cases and a literature review. JMM Case Reports 4: e005131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Rodríguez, A.J. , Reyes‐Darias, J.A. , Martín‐Mora, D. , González, J.M. , Krell, T. , and Römling, U. (2021) Reduction of alternative electron acceptors drives biofilm formation in Shewanella algae . npj Biofilms Microb 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Rodríguez, A.J. , Rhen, M. , Melican, K. , and Richter‐Dahlfors, A. (2020) Nitrate metabolism modulates biosynthesis of biofilm components in uropathogenic Escherichia coli and acts as a fitness factor during experimental urinary tract infection. Front Microbiol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Rodríguez, A.J. , Suárez‐Mesa, A. , Artiles‐Campelo, F. , and Römling, U. (2019) Multilocus sequence typing of Shewanella algae isolates identifies disease‐causing Shewanella chilikensis strain 6I4. FEMS Microbiol Ecol 95: fiy210. [DOI] [PubMed] [Google Scholar]
- Mélida, H. , Sandoval‐Sierra, J.V. , Diéguez‐Uribeondo, J. , and Bulone, V. (2013) Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot Cell 12: 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue, H. , Hayashi, T. , Hashimoto, Y. , Ezaki, T. , Hamasaki, K. , Ohwada, K. , and Terawaki, Y. (1992) Isolation and characterization of Shewanella alga from human clinical specimens and emendation of the description of S. alga Simidu et al.,Isolation and characterization of Shewanella alga from human clinical specimens and emendation of the description of S. alga Simidu. Int J Syst Bacteriol 42: 628–634. [DOI] [PubMed] [Google Scholar]
- Ow, D.S.W. , Nissom, P.M. , Philp, R. , Oh, S.‐K.‐W. , and Yap, M.‐G.‐S. (2006) Global transcriptional analysis of metabolic burden due to plasmid maintenance in Escherichia coli DH5α during batch fermentation. Enzyme Microb Technol 39: 391–398. [Google Scholar]
- Pernestig, A.K. , Melefors, Ö. , and Georgellis, D. (2001) Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli . J Biol Chem 276: 225–231. [DOI] [PubMed] [Google Scholar]
- Peterson, G.L. (1977) ,A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem 83: 346–356. [DOI] [PubMed] [Google Scholar]
- Qadri, F. , Abu Hossain, S. , Ciznar, I. , Haider, K. , Ljungh, A. , Wadstrom, T. , and Sack, D.A. (1988) Congo red binding and salt aggregation as indicators of virulence in Shigella species. J Clin Microbiol 26: 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, D. , Aguilar, C. , Losick, R. , and Kolter, R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA 107: 2230–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U. (2005) Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci 62: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U. , and Balsalobre, C. (2012) Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med 272: 541–561. [DOI] [PubMed] [Google Scholar]
- Römling, U. , Bokranz, W. , Rabsch, W. , Zogaj, X. , Nimtz, M. , and Tschäpe, H. (2003) Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int J Med Microbiol 293: 273–285. [DOI] [PubMed] [Google Scholar]
- Römling, U. , Sierralta, W.D. , Eriksson, K. , and Normark, S. (1998) Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28: 249–264. [DOI] [PubMed] [Google Scholar]
- Rütschlin, S. , Gunesch, S. , and Böttcher, T. (2017) One enzyme, three metabolites: Shewanella algae controls siderophore production via the cellular substrate pool. Cell Chem Biol 24: 598–604.e10. [DOI] [PubMed] [Google Scholar]
- Sardari, R.R.R. , Kulcinskaja, E. , Ron, E.Y.C. , Björnsdóttir, S. , Friðjónsson, Ó.H. , Hreggviðsson, G.Ó. , and Karlsson, E.N. (2017) Evaluation of the production of exopolysaccharides by two strains of the thermophilic bacterium Rhodothermus marinus . Carbohydr Polym 156: 1–8. [DOI] [PubMed] [Google Scholar]
- Schachterle, J.K. , Stewart, R.M. , Schachterle, M.B. , Calder, J.T. , Kang, H. , Prince, J.T. , and Erickson, D.L. (2018) Yersinia pseudotuberculosis BarA‐UvrY two‐component regulatory system represses biofilms via CsrB. Front Cell Infect Microbiol 8: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shori, D. , Kariyawasam, H. , Knight, R.A. , Hodson, M.E. , Genter, T. , Hansen, J. , et al. (2001) Sulphation of the salivary mucin MG1 (MUC‐5B) is not correlated to the degree of its sialylation and is unaffected by cystic fibrosis. Pflugers Archiv Eur J Physiol 443: S50–S54. [DOI] [PubMed] [Google Scholar]
- Singh, R.P. , Baghel, R.S. , Reddy, C.R.K. , and Jha, B. (2015) Effect of quorum sensing signals produced by seaweed‐associated bacteria on carpospore liberation from Gracilaria dura . Front Plant Sci 6: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. , Price, J. , Burby, P. , Blanco, L. , Chamberlain, J. , and Chapman, M. (2017) The production of curli amyloid fibers is deeply integrated into the biology of Escherichia coli . Biomolecules 7: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodósio, J.S. , Simões, M. , and Mergulhão, F.J. (2012) The influence of nonconjugative Escherichia coli plasmids on biofilm formation and resistance. J Appl Microbiol 113: 373–382. [DOI] [PubMed] [Google Scholar]
- Thormann, K.M. , Saville, R.M. , Shukla, S. , Pelletier, D.A. , and Spormann, A.M. (2004) Initial Phases of biofilm formation in Shewanella oneidensis MR‐1. J Bacteriol 186: 8096–8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaplana, F. , Nilsson, J. , Sommer, D.V.P. , and Karlsson, S. (2015) Analytical markers for silk degradation: comparing historic silk and silk artificially aged in different environments. Anal Bioanal Chem 407: 1433–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, B.F. , Holt, H.M. , Gerner‐Smidt, P. , Bundvad, A. , Søgaard, P. , and Gram, L. (2000) Homogeneity of Danish environmental and clinical isolates of Shewanella algae . Appl Environ Microbiol 66: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, F.H. , and Schoolnik, G.K. (1999) Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA 96: 4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, F.H. , and Visick, K.L. (2009) Vibrio biofilms: so much the same yet so different. Trends Microbiol 17: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj, X. , Nimtz, M. , Rohde, M. , Bokranz, W. , and Romling, U. (2001) The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39: 1452–1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Gene synteny and proximity of S. algae CECT 5071 transposon‐interrupted genes. Tn5‐disrupted genes are colored in red. Genes putatively belonging to the same operon are colored in pale yellow. Key annotations are indicated.
Fig. S2. Individual FTIR spectra of CER and formaldehyde‐NaOH EPS extracts. Data from two biological experiments are shown for CER EPS extracts, A‐D, and formaldehyde‐NaOH EPS extracts, E‐H, of the WT strain and mutants M1 (barA), M3 (thyA) and M8 (E1N14_01730).
Fig. S3. Growth and biofilm formation patterns of S. algae WT and transposon insertion mutants. Side and top views of strip‐well cultures (24 h, 30°C, MB medium) of S. algae CECT 5071 WT and isogenic transposon insertion mutant derivatives before and after crystal violet staining of biofilm biomass.
Fig. S4. Functional analysis of the hybrid sensor histidine kinase BarA of S. algae. A. Domain architecture of the BarA protein as determined by SMART (http://smart.embl‐heidelberg.de/). The location of Tn5 integration and key amino acid residues H306, D729 and H860 putatively involved in the phosphorelay cascade are indicated. B. Biofilm formation of S. algae WT and mutant M1 (barA) harboring the empty vector pSRK‐Gm plasmid, or the mutant strain complemented with the native or mutant barA derivatives cloned in pSRK‐Gm. Statistical significance of the differences are calculated from 4 biological replicates with 6 technical replicates each using One‐way ANOVA followed by Tukey’s post hoc analysis, with P = 0.05 as the threshold for significance (* P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001). Inset: side view of CV‐stained wells showing phenotypic differences upon native or mutant barA expression with respect to the WT or mutant empty vector controls.
Table S1. Bacterial strains used in this study.
Table S2. Plasmids and primers used in this study.


