Summary
Engineering nitrogenase in eukaryotes is hampered by its genetic complexity and by the oxygen sensitivity of its protein components. Of the three types of nitrogenases, the Fe‐only nitrogenase is considered the simplest one because its function depends on fewer gene products than the homologous and more complex Mo and V nitrogenases. Here, we show the expression of stable Fe‐only nitrogenase component proteins in the low‐oxygen mitochondria matrix of S. cerevisiae. As‐isolated Fe protein (AnfH) was active in electron donation to NifDK to reduce acetylene into ethylene. Ancillary proteins NifU, NifS and NifM were not required for Fe protein function. The FeFe protein existed as apo‐AnfDK complex with the AnfG subunit either loosely bound or completely unable to interact with it. Apo‐AnfDK could be activated for acetylene reduction by the simple addition of FeMo‐co in vitro, indicating preexistence of the P‐clusters even in the absence of coexpressed NifU and NifS. This work reinforces the use of Fe‐only nitrogenase as simple model to engineer nitrogen fixation in yeast and plant mitochondria.
Fe‐only nitrogenase components were expressed and matured in S. cerevisiae mitochondrial. The electron donor component was active as isolated and the catalytic component was ready for activation by cofactor insertion.

Introduction
Biological nitrogen fixation (BNF), the reduction of dinitrogen (N2) to ammonia (NH3), is catalysed by the nitrogenase protein, an O2‐sensitive metalloenzyme complex present in some prokaryotes called diazotrophs (Raymond et al., 2004). Three variants of nitrogenase exist in nature and are defined as the molybdenum (Mo), vanadium (V) or iron‐only (Fe‐only) nitrogenase with respect to their cofactor metal composition. All diazotrophs carry a Mo nitrogenase and some of them additionally carry alternative V or Fe‐only nitrogenase (Seefeldt et al., 2009; O’Carroll and Santos, 2011). The Mo nitrogenase is the best studied one and consists of an Fe protein (NifH) homodimer acting as electron donor and a MoFe protein (NifDK) heterotetramer that is the catalytic component. The electrons provided by NifH accumulate at the iron–molybdenum cofactor (FeMo‐co) of NifDK where N2 is reduced (Seefeldt et al., 2009). The V and Fe‐only nitrogenases also have two components, the Fe protein (AnfH) and FeFe protein (AnfDGK) for the Fe‐only nitrogenase, and the Fe protein (VnfH) and FeV protein (VnfDGK) for the V nitrogenase, but much less is known about their biosynthesis and catalytic properties. Of the three, the Fe‐only nitrogenase is considered as the simplest one because its function depends on fewer gene products than the homologous and more complex Mo and V nitrogenases (Joerger et al., 1989).
Despite BNF contributions, most reactive N in agricultural managed ecosystems is of industrial origin (Tilman et al., 2011; Foley et al., 2011). To reduce dependency of synthetic N fertilizers, mainly in cereal crops, researchers are trying to leverage biotechnology. One proposed strategy involves direct transfer of prokaryotic nitrogenase genes into cereal plants (Vicente and Dean, 2017; Burén and Rubio, 2018). In this path, Saccharomyces cerevisiae stands as unicellular, easy to culture, genetically tractable, model of eukaryotic cell. Expression of active NifH (the Fe protein component of the Mo nitrogenase) and NifB (a FeMo‐co maturating protein) in mitochondria of S. cerevisiae has proven that these extremely O2‐sensitive nitrogenase proteins can accumulate in a eukaryotic cell, representing the first steps towards engineering nitrogenase in eukaryotes (López‐Torrejón et al., 2016; Burén et al., 2017). Recent studies have also shown that both mitochondria and chloroplasts of tobacco can harbour active NifH at the end of the dark period (Eseverri et al., 2020; Jiang et al., 2021).
Engineering the Fe‐only nitrogenase is attractive because of its genetic simplicity and because availability of Mo (and V) in mitochondria and chloroplasts is not well understood. In the diazotrophic bacterium Azotobacter vinelandii, the Fe‐only nitrogenase genes are organized in a single anfHDGKOR gene cluster (anf for alternative nitrogen fixation genes) (Joerger et al., 1989). The anfH, anfD and anfK genes are equivalent to their Nif counterparts, whereas the anfG gene encodes a gamma subunit, as in contrast to the Mo nitrogenase NifDK protein the alternative nitrogenases exist as heterohexamers (VnfDGK and AnfDGK) in their mature forms. The structure of the homologous VnfDGK component of the V nitrogenase has been solved (Sippel and Einsle, 2017). While the active‐site metallocluster FeFe‐co is thought to be structurally and functionally equivalent to FeMo‐co, the N2 reduction activity of Fe‐only nitrogenase is about three times lower (Krahn et al., 2002; Harris et al., 2018). Genetic evidence indicates that the product of the NifB enzyme (a [Fe8S9C] cluster named NifB‐co) serves as biosynthetic precursor for all three nitrogenase cofactors FeMo‐co, FeV‐co and FeFe‐co (Bishop and Joerger, 1990; Curatti et al., 2007). Genetic and transcriptome analysis of A. vinelandii has suggested that the Anf system depends on VnfEN for FeFe‐co maturation (Wolfinger and Bishop, 1991; Hamilton et al., 2011). However, work by Yang and colleagues later showed that neither NifEN nor VnfEN scaffold proteins were required for the biosynthesis of FeFe‐co in Escherichia coli (Yang et al., 2014 ), although they are essential for the biosynthesis of FeMo‐co (Jacobson et al., 1989) and FeV‐co (Wolfinger and Bishop, 1991) respectively. This is consistent with studies showing that several diazotrophs lack the NifEN protein (Soboh et al., 2010) and that FeFe‐co biosynthesis can take place in its absence (Schüddekopf et al., 1993).
In addition to the structural components, nitrogenase expression and maturation in native hosts and heterologous systems require several gene products with accessory functions. In E. coli, an engineered 10‐gene cluster (anfHDGK, nifBUSV, nifF and nifJ) generated active Fe‐only nitrogenase (Yang et al., 2014 ). The products of nifJ and nifF are involved in electron donation to nitrogenase, whereas the A. vinelandii anfO and anfR genes did not seem to be strictly required. In yeast, the [Fe–S] cluster assembly proteins NifU and NifS were not necessary to produce active NifH in mitochondria (López‐Torrejón et al., 2016), suggesting that the mitochondrial [Fe–S] cluster biosynthetic machinery of yeast could support [Fe4S4] cluster insertion into NifH. On the contrary, NifU and NifS were essential to obtain NifB with a complete set of [Fe4S4] clusters in the very same system (Burén et al., 2019), emphasizing that exact genetic requirements differ among nitrogenase proteins.
In this work, the structural components of the A. vinelandii Fe‐only nitrogenase (AnfH and AnfDGK) were expressed together with the [Fe–S] cluster biosynthetic proteins NifS and NifU in S. cerevisiae and targeted to the mitochondrial matrix. Purified yeast AnfH was active as Fe protein, and enriched preparations of yeast AnfDK could be activated in vitro by the simple addition of purified FeMo‐co. Interestingly, cofactor‐deficient apo‐FeFe protein was a heterotetramer lacking the AnfG subunit. This is the first report of an engineered eukaryotic nitrogenase catalytic component ready for activation by cofactor insertion.
Results
Expression of Fe‐only nitrogenase structural components in S. cerevisiae
To synthesize Fe‐only nitrogenase components in S. cerevisiae, we generated strains expressing the A. vinelandii anfHDGK genes either alone or in combination with the [Fe–S] cluster assembly proteins NifU and NifS (Table 1 and Table S1). All genes were synthesized with codon optimization for S. cerevisiae (Fig. S1), cloned into pESC expression vectors containing the galactose‐regulated GAL1 and GAL10 promoters, and introduced into S. cerevisiae W303‐1a by transformation. Nitrogenase gene expression was induced under respiratory growth conditions with galactose as carbon source. To ensure mitochondrial protein targeting, the su9 leader sequence (Westermann and Neupert, 2000) was fused to the 5′‐end of each anf and nif gene sequence (Fig. S2). Correct translocation of each expressed component to the organelle was verified by SDS‐PAGE and immunoblot analysis of isolated mitochondria (Fig. 1). All Anf proteins were specifically targeted to the mitochondria, while a fraction of NifU and NifS proteins could also be found in the extra‐mitochondrial fraction. Fe‐only nitrogenase proteins were detected using antibodies generated to specifically recognize the AnfH, AnfD, AnfK and AnfG polypeptides. For this, AnfH, AnfD, AnfK and AnfG were individually expressed and purified from recombinant E. coli cells (Fig. S3).
Table 1.
S. cerevisiae strains expressing combinations of Anf and Nif proteins.
| Strain | Proteins expressed | Purification tags present |
|---|---|---|
| GF13 | AnfH, NifUS | His10‐AnfH |
| GF15 | AnfHDGK | His10‐AnfH, His10‐AnfD |
| GF16 | AnfHDGK, NifUS | His10‐AnfH, His10‐AnfD |
| GF17 | AnfHDGK | His10‐AnfH, TS‐AnfD |
| GF18 | AnfHDGK, NifUS | His10‐AnfH, TS‐AnfD |
| GF19 | AnfHDGK | His10‐AnfH, SS‐AnfD |
| GF20 | AnfHDGK, NifUS | His10‐AnfH, SS‐AnfD |
His10, 10 histidine tag at N‐terminal end; TS, Twin‐Strep‐tag at N‐terminal end, SS, Single Strep‐tag at the N‐terminal end.
Fig. 1.
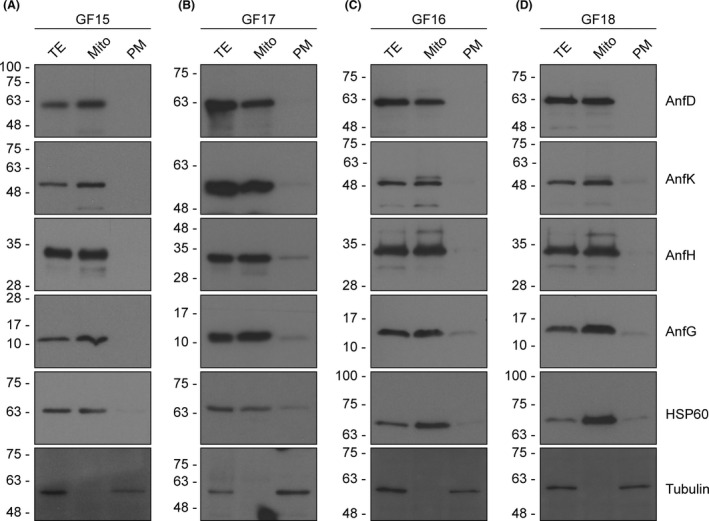
Expression and mitochondria targeting of nitrogenase proteins in S. cerevisiae. Immunoblot analysis of yAnfD, yAnfK, yAnfH and yAnfG proteins in total protein extracts (TE), in isolated mitochondria (Mito) and in ‘post‐mitochondrial supernatant’ fractions (PM) from S. cerevisiae strains GF15 (A), GF17 (B), GF16 (C) and GF18 (D). Total extracts and mitochondria isolation were prepared from aerated cultures of galactose‐induced cells. Antibodies recognizing A. vinelandii AnfD, AnfK, AnfH and AnfG proteins, HSP60 (mitochondrial marker protein), or tubulin (cytosolic marker) were used for immunoblotting analysis.
Isolation of yAnfH from mitochondria of S. cerevisiae
Mitochondria‐targeted His‐tagged Fe protein (yAnfH) was purified by anaerobic Co2+ affinity chromatography from cells of GF13, GF15 and GF16 strains (see Table 1). The cultures were grown aerobically and induced as previously reported for the yNifH protein (López‐Torrejón et al., 2016). The yAnfH protein was purified to near homogeneity from cell‐free extracts of GF15 and GF16 strains, but it was poorly produced in GF13. SDS‐PAGE analysis of a typical yAnfH purification process from GF15 is shown in Fig. 2A. Most soluble proteins eluted at imidazole concentration between 100 and 130 mM (E1), while yAnfH eluted at 130 mM imidazole (E2). Immunoblot analysis developed with antibodies against the His‐tag or AnfH indicated that the majority of soluble yAnfH produced could be recovered during purification and that it migrated as expected with no proteolytic fragments present (Fig 2B). The yAnfH protein in purified preparations was quantified by BCA and by SDS‐PAGE band densitometry using ImageJ (Fig. S4A).
Fig. 2.
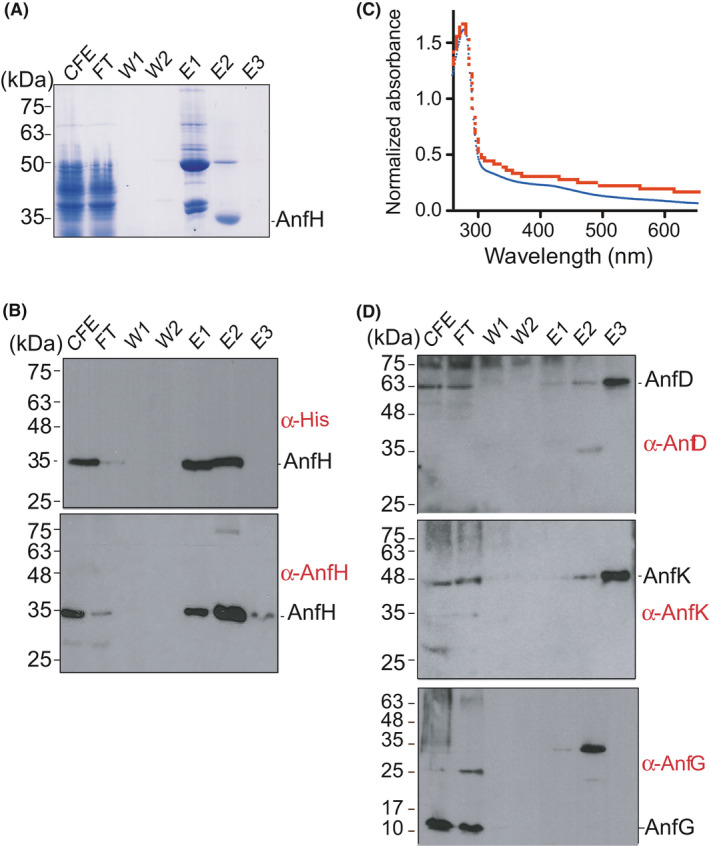
Purification of yAnfH from S. cerevisiae GF15. Coomassie‐stained SDS gel of purification fractions (A) and immunoblot analysis of the same fractions developed with antibodies against AnfH or the His‐tag (B), or with antibodies against AnfD, AnfG and AnfK (D). The 35 kDa band detected in E1 and E2 with α‐AnfG likely corresponds to unspecific recognition of an enriched polypeptide in yAnfH preparations. CFE, cell‐free extract; FT, flow‐through fraction; W1, fraction after washing with binding buffer; W2, fraction after washing with binding buffer supplemented with 80 mM imidazole; E1, E2 and E3, fractions eluted by applying a 100–175 mM imidazole gradient in binding buffer.
C, UV‐visible absorption spectra of as‐isolated yAnfH (blue) and air‐exposed yAnfH (red).
yAnfH exhibited the characteristic brown colour of nitrogenase Fe proteins. UV‐vis absorbance spectrum of as‐isolated yAnfH showed the 320 nm and 420 nm features associated with [Fe4S4] clusters (Fig. 2C) (Smith et al., 2005). Consistently, metal analysis confirmed the presence of 4 Fe atoms per yAnfH dimer. UV‐vis absorption spectral changes were observed upon exposure to air.
Because yAnfH was coexpressed with yAnfDGK in GF15 cells, the separation of both nitrogenase components during affinity chromatography was assessed by SDS‐PAGE and immunoblot. The yAnfDGK protein components were not detected by Coomassie staining (Fig. 2A). However, immunoblot analysis showed enrichment of AnfD and AnfK polypeptides in the E3 fraction containing 160 mM imidazole (Fig. 2D). The yAnfG protein was only detected in the flow‐through fraction, and therefore, it was neither stably associated with yAnfH nor with yAnfDK. The amount of yAnfDK protein in the preparations was quantified by immunoblot band densitometry using ImageJ against standards produced with E. coli purified AnfD and AnfK polypeptides (Fig. S4B).
yAnfH functions as nitrogenase Fe protein
Fe protein function of as‐isolated yAnfH protein was tested using the acetylene reduction assay after addition of pure A. vinelandii MoFe protein (AvNifDK). The unavailability of A. vinelandii FeFe protein and the functional similarity between both nitrogenases (Schneider et al., 1997; O’Carroll and Santos, 2011; Harris et al., 2018) prompted us to use AvNifDK as complementary component. Electron donation to AvNifDK was observed, where the yAnfH protein isolated from the GF15 strain yielded NifDK specific activity of 950 units (nmol of ethylene formed·min‐1·mg of NifDK‐1) when yAnfH molar concentration was 100‐fold higher than AvNifDK (Fig. 3A). Surprisingly, activity was reduced by half when yAnfH was purified from GF16 cells coexpressing NifU and NifS. Furthermore, when yAnfH was coexpressed with NifU and NifS in the absence of yAnfDGK (strain GF13), activity was 10‐fold lower compared with GF16. One explanation for this observation could be that the presence of the yAnfDGK components could stabilize yAnfH, thereby improving its solubility or functionality.
Fig. 3.
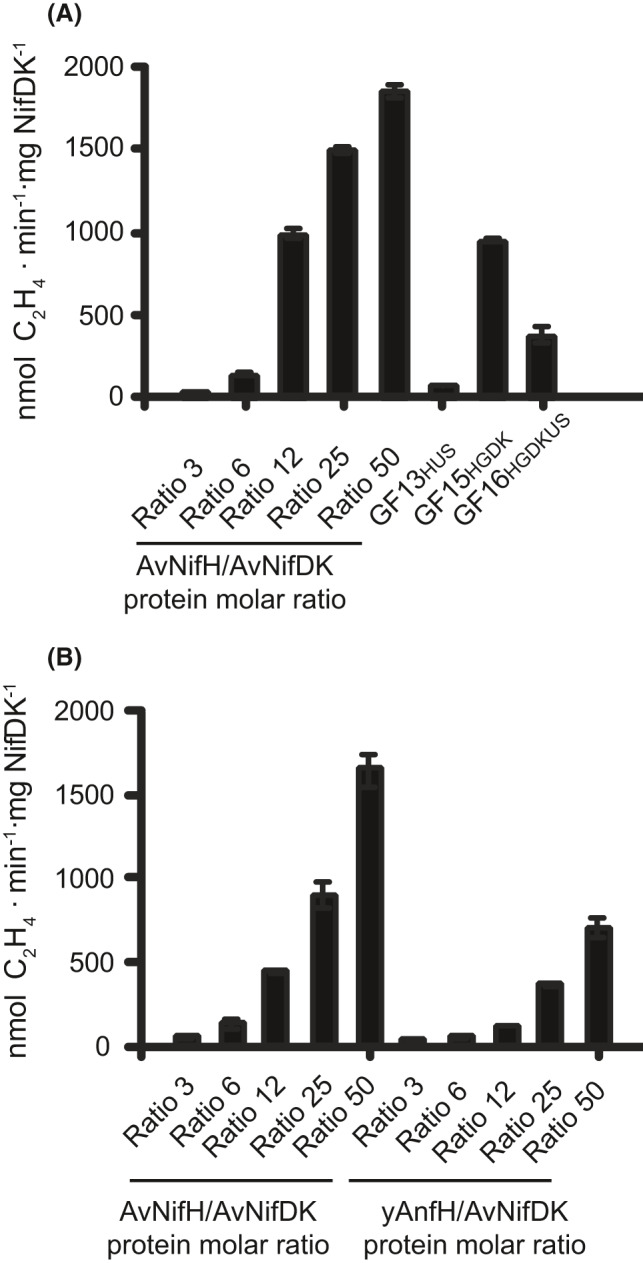
Determination of yAnfH activity.
A. Determination of Fe protein activity in yAnfH purified preparations from strains GF13HUS (coexpressing yAnfH with yNifU and yNifS), GF15HGDK (coexpressing yAnfH with yAnfDGK) and GF16 HGDKUS (coexpressing yAnfH with yAnfDGK, yNifU and yNiAS).
B. AvNifDK activity titration with yAnfH purified from GF15. AvNifDK activity titrations with AvNifH were carried out as control reactions in (A) and (B). Data represent mean ± SD (n = 6) for each yAnfH containing reaction.
Titration of AvNifDK activity with increasing amounts of yAnfH or AvNifH is shown in Fig. 3B. The specific activity of yAnfH was half compared to AvNifH along the titration consistent with a poorer interaction of AvNifDK with the non‐physiological complementary component. It is important to note that the assays in Fig. 3 compare yAnfH activity to that of AvNifH and not to the AvAnfH counterpart.
Enriched preparations of yAnfDK from mitochondria of S. cerevisiae
To improve yAnfDK protein isolation, the His‐tag at the N‐terminus of AnfD was replaced by either a Single‐ or Twin‐Strep‐tag. This strategy had previously been successful to purify NifB from yeast mitochondria (Burén et al., 2017b; Burén et al., 2019). Cells from aerated cultures of GF17 or GF19 strains coexpressing His‐tagged yAnfH were used as source of yAnfDK (Table 1). Protein preparations enriched in yAnfDK were obtained by anaerobic Strep‐Tactin affinity chromatography of soluble protein extracts, while yAnfH was simultaneously purified using anaerobic Co2+ affinity chromatography. A typical purification result from GF17 cells shown in Fig. 4. The enriched yAnfDK fraction showed light brown colour but the yAnfD and yAnfK polypeptides were not easily observed by Coomassie staining of SDS gels, although they were detected by immunoblot analysis using anti‐AnfD and anti‐AnfK antibodies (Fig. 4A). Again, yAnfDK co‐eluted as a complex devoid of yAnfG suggesting that yAnfG was either released during purification or that heterohexamer stability was low. The yAnfDK protein concentration in enriched fractions was estimated using ImageJ (Fig. S4B). Preparations of yAnfH protein purified by Co2+ affinity chromatography from GF17 soluble extracts contained more contaminants than those from GF15 cells (Fig. 4B).
Fig. 4.
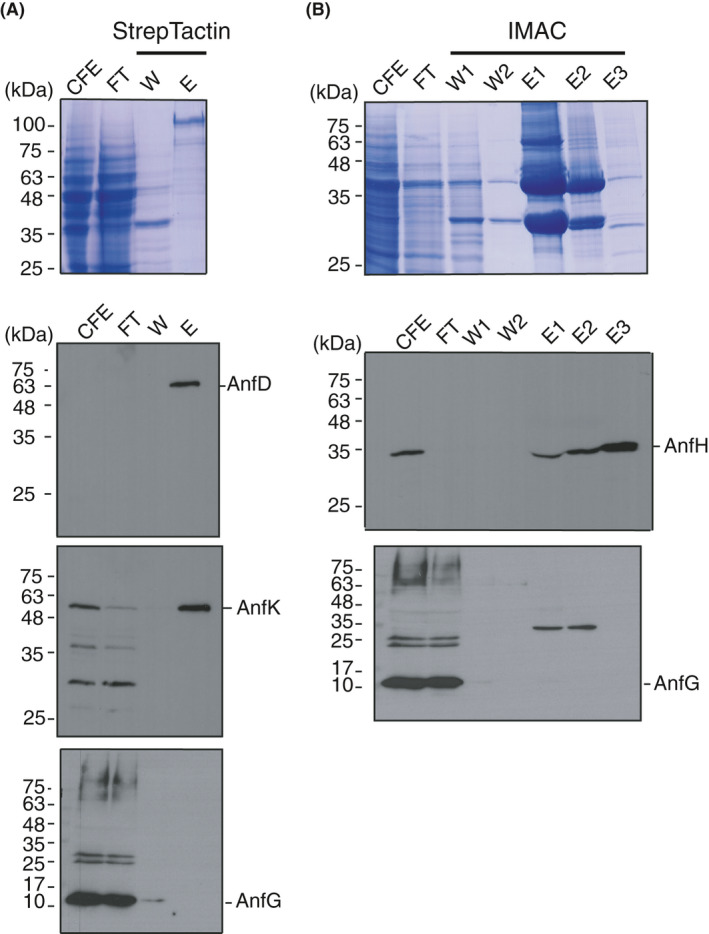
Enriched yAnfDK preparations from GF17 cells.
A. Strep‐Tactin affinity chromatography fraction analysis by SDS‐PAGE (stained with Coomassie) and by immunoblots developed with antibodies against the AnfD, AnfK and AnfG polypeptides. CFE, cell‐free extract; FT, flow‐through fraction; W, fraction after washing with binding buffer; E, fraction after elution with 50 mM biotin in binding buffer.
B. Analysis of purification fractions from Co2+ affinity chromatography (IMAC) by SDS‐PAGE (stained with Coomassie) and by immunoblots developed with antibodies against AnfH and AnfG. CFE and FT fractions as in panel (A); W1, fraction after washing with binding buffer; W2, fraction after washing with binding buffer supplemented with 80 mM imidazole; E1, E2 and E3, fractions after eluting with a 100‐175 mM imidazole gradient in binding buffer. The 35 kDa band detected in E1 and E2 with α‐AnfG likely corresponds to unspecific recognition of an enriched polypeptide in those fractions.
Activation of apo‐yAnfDK by isolated FeMo‐co
The yAnfDK protein isolated from GF15, GF17 and GF19 cells was incubated with FeMo‐co isolated from A. vinelandii. Apo‐NifDK purified from A. vinelandii was used as control of FeMo‐co reconstitution. The unavailability of FeFe‐co in our laboratory and the functional equivalence between FeFe‐co and FeMo‐co (Harris et al., 2018) were the reasons to use FeMo‐co in this assay. Previous results have shown the incorporation of FeMo‐co into apo‐AnfDGK to form a hybrid nitrogenase able to reduce acetylene to ethylene and ethane (Gollan et al., 1993). Similarly, apo‐NifDK reconstitution with FeV‐co forms a hybrid nitrogenase that can reduce acetylene to both ethylene and ethane (Smith et al., 1988). Activation of yAnfDK by FeMo‐co was then determined by the acetylene reduction assay following addition of excess AvNifH and ATP regeneration mixture (Shah and Brill, 1977; Allen et al., 1993). FeMo‐co insertion into 6 µM of yAnfDK isolated from GF17 strain resulted in reconstituted FeFe protein with specific activity of 300 nmol of ethylene formed·min‐1·mg of yAnfDK‐1 (Fig. 5). Moreover, 12 µM of yAnfDK protein isolated from GF15 and GF19 rendered 60 and 140 nmol of ethylene formed·min‐1·mg of yAnfDK‐1 respectively. The variability of specific activities shown in Fig. 5 was probably due to the different affinity tags used. These results demonstrate that the yAnfDK present in mitochondria is in fact an apo‐protein readily activated by the simple addition of active‐site cofactor.
Fig. 5.
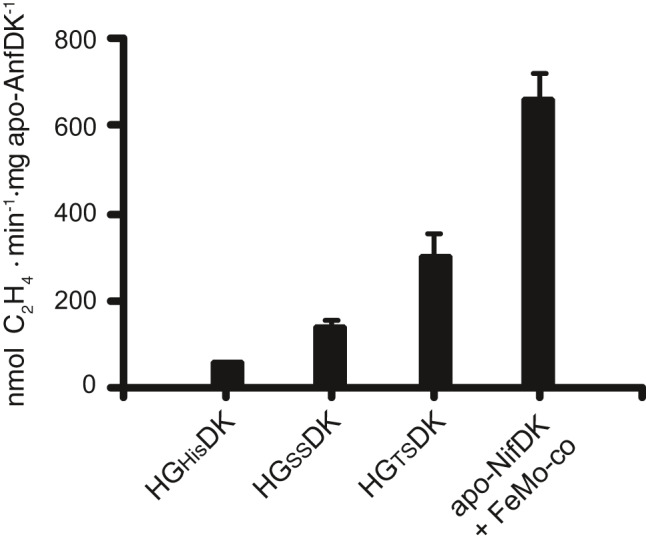
In vitro activation of apo‐yAnfDK using purified FeMo‐co. Preparations of apo‐yAnfDK with His10, Twin‐Strep (TS) or Single‐Strep (SS) purification tags at the N‐terminal end of AnfD were obtained from GF15 (expressing AnfHGHisDK), GF17 (expressing AnfHGTSDK) or GF19 (expressing AnfHGSSDK) respectively. Five µl FeMo‐co isolated from A. vinelandii was added to 6 µM of GF17 apo‐yAnfDK or 12 µM of GF15 or GF19 apo‐yAnfDK. Positive control reactions were carried with 11 µM of A. vinelandii apo‐NifDK. The activity of reconstituted yAnfDK was determined by the acetylene reduction assay after addition of AvNifH and ATP‐regenerating mixture. Data represent mean ± SD (n = 6) for each yAnfDK.
Discussion
Given the genetic simplicity of the Fe‐only nitrogenase, expression of its four structural genes anfHDGK could be sufficient to render active Fe protein and cofactor‐activatable FeFe protein. Therefore, we targeted the AnfH, AnfD, AnfG and AnfK polypeptides to the mitochondrial matrix of S. cerevisiae, with or without the A. vinelandii [Fe‐S] cluster assembly proteins NifU and NifS. The most salient result of this study is that yAnfDK produced in mitochondria could be activated in vitro by the simple addition of FeMo‐co. This activation was possible because FeFe‐co and FeMo‐co are structurally and functionally equivalent (Krahn et al., 2002; Harris et al., 2018). This activation implies that the yAnfDK complex carries the P‐clusters (or their equivalent in the FeFe protein) and only lacks the active‐site cofactor. Thus, NifU and NifS are not essential for yAnfDK P‐cluster formation in mitochondria.
Fully matured FeFe protein is a heterohexamer of AnfD2G2K2 subunit composition (Schneider and Müller, 2004; O’Carroll and Santos, 2011). However, the yAnfDK protein complex enriched from S. cerevisiae protein extracts lacked the yAnfG subunit suggesting that yAnfG was either lost during the purification process or that it was never part of a yAnfDGK complex. The complex might be unstable in yeast or require additional components for stability. One obvious component missing is the active‐site FeFe cofactor. The presence of AnfG is relevant because A. vinelandii strains with altered anfG gene expression were shown unable to grow diazotrophically albeit exhibiting acetylene reduction activity (Waugh et al., 1995). Similarly, removing anfG from the engineered Fe‐only nitrogenase in E. coli decreased acetylene reduction and eliminated N2 fixation activity (Yang et al., 2014 ).
The Fe protein activity of purified yAnfH preparations was similar to that of mitochondria‐targeted yNifH (López‐Torrejón et al., 2016), which is not surprising due to their high (61%) amino acid sequence conservation (Joerger et al., 1989). In contrast to yNifH, yAnfH maturation to render active Fe protein was independent of the ancillary protein NifM. Surprisingly, lower yAnfH Fe protein activity was observed when coexpressing NifU and NifS in addition to yAnfHDGK (strain GF16). This study confirms that mitochondria endogenous [Fe‐S] cluster assembly proteins, probably Isu1/Isu2 and Nfs1 (Lill, 2009), are able to produce [Fe4S4] clusters for yAnfH as they do for yNifH (López‐Torrejón et al., 2016). However, because NifU and NifS have been shown essential for yNifB activity (Burén et al., 2019), the native mitochondria [Fe‐S] cluster assembly machinery would not be able to replace the function of NifU and NifS in a completely engineered nitrogenase.
In summary, this study demonstrates that expression and targeting of yAnfHDGK proteins to the mitochondrial matrix of S. cerevisiae is possible and that all proteins appear to be stable. Both structural components can be purified by affinity chromatography. While the Fe protein (yAnfH) was active as‐isolated, the yAnfG subunit was not found as part of the FeFe protein, which existed as apo‐yAnfDK complex. Importantly, apo‐yAnfDK could be activated upon FeMo‐co insertion indicating that it contained the P‐clusters. Future efforts should focus on engineering FeFe‐co assembly in S. cerevisiae to obtain fully mature nitrogenase. A nitrogen‐fixing yeast could have industrial application as ammonium factory or, by coupling to glutamate dehydrogenase, a glutamate factory.
Experimental procedures
Strain growth and culture media
E. coli DH5α was used for storage and amplification of yeast expression vectors. E. coli strains were grown at 37ºC in Luria–Bertani medium supplemented with ampicillin when necessary.
Saccharomyces cerevisiae W303‐1a (MATa [leu2‐3,112 trp1‐1 can1‐100 ura3‐1 ade2‐1 his3‐11,15]) and derivative strains, constructed herein, were grown at 28ºC in flasks at 200 rpm in synthetic dropout (SD) medium composed of 1.9 g l‐1 yeast nitrogen base, 5 g l‐1 ammonium sulfate, 20 g l‐1 glucose and Kaiser dropout mixture (Kaiser et al., 1994) supplemented with 20 mg l‐1 adenine, 40 mg l‐1 tryptophan, 40 mg l‐1 histidine, 20 mg l‐1 uracil or 60 mg l‐1 leucine, depending on auxotrophic requirements. To express anf and nif genes, 20 g l‐1 galactose was added to the culture medium when glucose was consumed. In addition, the medium was supplemented with 5 g l‐1 yeast extract, 25 g l‐1 bactopeptone and 100 mM of ammonium iron (III) citrate.
DNA constructs and generation of E. coli and S. cerevisiae strains
A. vinelandii anfH, anfD, anfK and anfG genes were synthesized with codon optimization for S. cerevisiae (GenScript, Piscataway, NJ, USA) and cloned in‐frame downstream of DNA sequences encoding a Su9 mitochondria leader peptide together with either a His10 tag, a Strep‐tag or a Twin‐Strep‐tag (Westermann and Neupert, 2000). To generate GF13 strain, the Su9 encoding sequence 5’ of anfH was replaced by a Mam33 encoding sequence. Transformation of S. cerevisiae strains was carried out according to Chen et al. (1992). Generated strains are listed in Table S1.
To overproduce A. vinelandii AnfH, AnfD, AnfK and AnfG proteins in E. coli, the corresponding anfH, anfD, anfK and anfG genes were amplified by PCR and cloned into the pET16b expression vector. A. vinelandii genomic DNA was used as template, and the following oligonucleotides were used as primers for PCR (NdeI and BamHI sites included in the oligonucleotides are underlined): 5’‐AAGTCATATGACTCGTAAAGTAGCCATTTAC‐3’ and 5‐AACTGGATCCTCAGTCGGCAATACCGTACTTGAC‐3’ to amplify anfH, 5’‐ AAGTCATATGCCGCATCACGAGTTCGAGTGCAGCAAGGT‐3’ and 5’‐AACTGGATCCTCAGCCGACCTTGCGCTCGAA‐3’ to amplify anfD, 5’‐ AAGTCATATGACTTGCGAAGTCAAGGAAAAAGGG‐3’ and 5’‐AACTGGATCCTTACCAGACGTTGAGGACCCATTCC‐3’ to amplify anfK, 5’‐AAGTCATATGAGTACCGCTTCCGCCGCTGCTGTGG‐3’ and 5’‐AACTGGATCCTTAATAGTGTTTGTCGCTCA‐3’ to amplify anfG. The PCR fragments were isolated and cloned into the NdeI and BamHI sites of pET16b to generate plasmids pN2GLT19 (anfH), pN2GLT20 (anfD), pN2GLT21 (anfK) and pN2GLT22 (anfG) respectively.
Yeast and bacterial strains were stored in 20% glycerol at −80°C. Generated strains are listed in Table S1.
Rabbit polyclonal antibody production
Polyclonal anti‐AnfH, anti‐AnfD, anti‐AnfK and anti‐AnfG antibodies were generated by immunizing rabbits with the corresponding purified antigens. Recombinant AnfH, AnfD, AnfK and AnfG proteins were purified from isopropyl β‐D‐1‐thiogalactopyranoside (IPTG)‐induced cells of E. coli BL21 carrying plasmids pN2GLT19, pN2GLT20, pN2GLT21 or pN2GLT22 respectively. Purifications were performed by Co2+ affinity chromatography under anaerobic conditions (< 0.1 ppm O2) using an AKTA Prime FPLC system (GE Healthcare, Wauwatosa, WI, USA) inside an MBraun glovebox (Fig. S3). AnfD protein was recovered from inclusion bodies in E. coli BL21 (pN2GLT20) cell‐free extracts by solubilization in buffer containing 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 4 M guanidinium chloride and subsequent refolding by dialysis at 4°C for 4 hours. One mg of each pure protein was sent for antibody production (Centro de Investigaciones Biológicas CIB‐CSIC) following a 1‐rabbit 70‐day protocol with 3 antigen boosters. Anf antibodies were titrated by ELISA Díaz‐Perales et al. (2003).
Western blotting
Anf and Nif proteins were separated by SDS‐PAGE transferred to membranes and detected by immunoblotting using rabbit polyclonal antibodies (1:10,000 dilution for anti‐AnfH, and 1:3,000 dilutions for anti‐AnfD, anti‐AnfK and anti‐AnfG) or commercially available anti‐His‐tag (H‐3, sc‐8036; Santa Cruz Biotechnology, Dallas, TX, USA)‐ and anti‐Strep‐tag II (StrepMAB‐Classic, 2‐1507‐001; IBA Lifesciences, Goettingen, Germany)‐specific antibodies. Antigen–antibody complexes were visualized with goat anti‐Rabbit IgG secondary antibody–HRP (H + L, AS09 602; Agrisera, Vännäs, Sweden).
Expression of AnfH, AnfG, AnfD, AnfK, NifU and NifS in S. cerevisiae
Yeast strains were aerobically cultured according to López‐Torrejón et al. (2016) in synthetic dropout (SD) medium containing 1.9 g l‐1 yeast nitrogen base, 5 g l‐1 ammonium sulfate, 20 g l‐1 glucose and Kaiser dropout mixture (SC‐His‐Leu‐Trp‐Ura, ForMediumTM) supplemented with 20 mg l‐1 adenine and 40 mg l‐1 tryptophan, 40 mg l‐1 histidine, 20 mg l‐1 uracil or 60 mg l‐1 leucine, depending on auxotrophic requirements, in a 4 l fermentor, during 16 h at 28ºC. Then, the medium was supplemented with 5 g l‐1 of yeast extract, 25 g l‐1 of bactopeptone and 100 µM of ammonium iron (III) citrate, and culture continued until glucose was consumed, at which time 20 g l‐1 galactose and a trace element solution (13 g l‐1 CaCl2, 2.5 g l‐1 Cl2Mn, 0.5 g l‐1 ZnSO4, 1.4 g l‐1 Na2 MoO4, 1.85 g l‐1 FeCl3, 1 g l‐1 H3BO4, 0.7 g l‐1 IK in 2 M HCl) were added to induce anf and nif gene expression. Cells were harvested under anoxic conditions in the late exponential phase by centrifugation at 4ºC and 5000 rpm for 5 min. Recovered cell paste was frozen and stored into liquid N2 until used.
Growth of yeast strains for mitochondria isolations
Saccharomyces cerevisiae strains GF15, GF16, GF17 and GF18 were grown in flasks at 28°C and 200 rpm in synthetic dropout (SD) medium containing 1.9 g l‐1 yeast nitrogen base, 5 g l‐1 ammonium sulfate, 20 g l‐1 glucose and Kaiser dropout mixture (SC‐His‐Leu‐Trp‐Ura, ForMediumTM) supplemented with 20 mg l‐1 adenine, 40 mg l‐1 tryptophan and 20 mg l‐1 uracil, as previously described (López‐Torrejón et al., 2016). Protein expression was induced by replacing glucose with galactose in the above‐described SD medium, additionally supplemented with 0.1% yeast extract and 1% peptone, for 16 h. Mitochondria isolations were performed as previously described (Diekert et al., 2001). Organelle enrichment was verified using tubulin (cytosolic) and HSP60 (mitochondria) marker proteins.
His‐tagged yAnfH and yAnfDK protein purification
Saccharomyces cerevisiae cells (GF13, GF15, GF16, GF17 or GF19 strain) were re‐suspended in anaerobic buffer A containing 100 mM Tris–HCl pH 8.0, 250 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM leupeptin, 2 mM sodium dithionite (DTH) and 5 µg ml‐1 DNase I. Cells were disrupted in an Emulsiflex‐C5 homogenizer (Avestin Inc., Ottawa, ON, Canada) at 30 000 psi (2,070 bar). Cell‐free extracts (CFE) were filtered in 0.2 µm pore size filter (Nalgene Rapid‐Flow; Thermo Scientific, Waltham, MA, USA), after removing cell debris by centrifugation at 70 000 g for 1 h at 4ºC, under anaerobic conditions.
The His‐tagged yAnfH and yAnfDGK proteins were partially purified by Co2+ affinity chromatography under anaerobic conditions (< 0.1 ppm O2) using an AKTA Prime FPLC system (GE Healthcare) inside an MBraun glovebox. CFE from 150 g of cells was loaded at 2 ml min‐1 onto a HiTrap IMAC column loaded with Co2+ and equilibrated with anaerobic buffer A. After additional washing with 10 column volumes (CV) of buffer A, the proteins were eluted using a linear gradient of up to 70% elution buffer (280 mM imidazole) within 20 CV. Eluted fractions were concentrated using a Vivaspin 500 concentrator (Sartorius, Goettingen, Germany) with cut‐off pore size of 30 kDa and then desalted in PD10 desalting columns (GE Healthcare) equilibrated with anaerobic buffer B containing 50 mM Tris–HCl, pH 8, 300 mM NaCl, 5% v/v glycerol. Purified proteins were frozen and stored in liquid N2. Overall protein concentration in the preparations was determined by the BCA assay using BSA as a standard. The amount of partially purified yAnf components was estimated after gel or membrane scanning and comparison of relative intensities against known amounts of standard proteins (BSA for yAnfH and E. coli produced AnfD and AnfK for yAnfDK) using the ImageJ software (Taylor et al., 2013). Average relative densities are shown in Fig. S4.
Single‐ and Twin‐Strep‐tag yAnfDK protein purification
Saccharomyces cerevisiae GF17 and GF19 cells were re‐suspended in anaerobic buffer A containing 100 mM Tris–HCl pH 8.0, 250 mM NaCl, 1 mM PMSF, 1 mM leupeptin, 2 mM DTH and 5 µg ml‐1 DNase I. Cells were lysed in an Emulsiflex‐C5 homogenizer (Avestin Inc.) at 30 000 psi (2,070 bar). CFE was obtained by centrifugation at 70 000 g for 1 h at 4ºC under anaerobic conditions.
Single‐ or Twin‐Strep‐tagged yAnfDK proteins were purified by Strep‐Tactin Superflow XT affinity chromatography (IBA) under anaerobic conditions (< 0.1 ppm O2) using an AKTA Prime FPLC system (GE Healthcare) inside an MBraun glovebox. CFE from 200 g of cells was loaded at 2 ml min‐1 onto a Strep‐Tactin Superflow XT column equilibrated with anaerobic buffer A and washed with 10 CV with same buffer. Finally, bound protein was eluted with buffer A containing 50 mM biotin. Co‐eluted yAnfDK proteins were concentrated with a Vivaspin 500 concentrator (Sartorius) with cut‐off pore size of 30 kDa and then desalted in PD10 desalting columns (GE Healthcare) equilibrated with buffer B containing 50 mM Tris–HCl, pH 8, 300 mM NaCl, 5% v/v glycerol. Purified proteins were frozen and stored in liquid N2. Overall protein concentration in the preparations was determined by the BCA assay using BSA as a standard. The amount of partially purified yAnfDK was estimated after immunoblot membrane scanning and comparison of relative intensity against known amounts of pure AnfD and AnfK (produced in E. coli) using ImageJ software (Taylor et al., 2013). Average relative densities are shown in Fig. S4.
Fe protein activity determination
Fe protein activity of yAnfH preparations obtained from S. cerevisiae GF13, GF15, GF16, GF17 and GF19 strains was routinely analysed by the acetylene reduction assay after addition of excess pure A. vinelandii NifDK (AvNifDK) and ATP‐regenerating mixture (1.23 mM ATP, 18 mM phosphocreatine, 2.2 mM MgCl2, 3 mM DTH and 40 µg of creatine phosphokinase) in 100 mM MOPS pH 7.0 (Shah and Brill, 1973; Emerich and Burris, 1978). To titrate AvNifDK activity with yAnfH, increasing yAnfH to AvNifDK molar ratios were assayed. Positive control reactions were carried out with AvNifH and AvNifDK proteins purified from A. vinelandii cells under anaerobic conditions as described (Curatti et al., 2007).
In vitro apo‐yAnfDK activation by FeMo‐co isolated from A. vinelandii
Apo‐AvNifDK preparations were isolated under anaerobic conditions from A. vinelandii cells as described (Curatti et al., 2007). FeMo‐co was isolated in NMF from purified preparations of A. vinelandii NifDK as reported in Shah et al. (1977).
The apo‐yAnfDK activation assay was carried out by adding 5 µl of isolated FeMo‐co and incubating at 30°C for 10 min under anaerobic conditions in 100 mM MOPS pH 7.0 plus 1.2 µg ml‐1 BSA. Six or 12 µM apo‐yAnfDK (obtained from GF15, GF17 or GF19 cells) was used in reconstitution reactions. Reactions containing 11 µM of A. vinelandii apo‐NifDK were used as positive control. The resulting activation of apo‐NifDK or apo‐yAnfDK present in the reaction mixtures was analysed by the acetylene reduction assay after addition of AvNifH and ATP‐regenerating mixture (1.23 mM ATP, 18 mM phosphocreatine, 2.2 mM MgCl2, 3 mM DTH and 40 µg of creatine phosphokinase) in a 9 ml stoppered vials with Ar/C2H2 filling the headspace, at 30°C for 30 min, and then quenched by addition of 0.1 ml of 8 M NaOH.
Funding Information
This work was supported by the Bill and Melinda Gates Foundation Grant OPP1143172 to LMR.
Conflict of interest
None declared.
Author contributions
LMR and GLT designed the experiments. GLT and SB conducted the experiments. MV carried out yeast fermentations and some DNA constructs. GLT, SB and LMR analysed data and wrote the manuscript. The authors have no conflict of interest.
Supporting information
Fig. S1. DNA optimized sequences.
Fig. S2. Schematic representation of synthetic anf and nif genes in S. cerevisiae GF15, GF16 and GF18.
Fig. S3. Purification of AnfH, AnfK, AnfG and AnfD proteins for the generation of polyclonal antibodies.
Fig. S4. Quantification of yAnfH and yAnfDK proteins in partially purified fractions.
Table S1. List of strains used in this work.
Acknowledgements
We thank Jose María Buesa for help with yeast fermentations and Emilio Jiménez‐Vicente for helpful insights about the Azotobacter FeFe protein.
Microbial Biotechnology (2021) 14(3), 1073–1083
References
- Allen, R.M. , Homer, M.J. , Chatterjee, R. , Ludden, P.W. , Roberts, G.P. , and Shah, V.K. (1993) Dinitrogenase reductase‐ and MgATP‐dependent maturation of apodinitrogenase from Azotobacter vinelandii . J Biol Chem 268: 23670–23674. [PubMed] [Google Scholar]
- Bishop, P.E. , and Joerger, R.D. (1990) Genetics and molecular biology of alternative nitrogen fixation systems. Annu Rev Plant Physiol Plant Mol Biol 41: 109–125. [Google Scholar]
- Burén, S. , Jiang, X. , López‐Torrejón, G. , Echavarri‐Erasun, C. , and Rubio, L.M. (2017b) Purification and in vitro activity of mitochondria targeted nitrogenase cofactor maturase NifB. Front Plant Sci 8: 1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén, S. , Pratt, K. , Jiang, X. , Guo, Y. , Jimenez‐Vicente, E. , Echavarri‐Erasun, C. , et al. (2019) Biosynthesis of the nitrogenase active‐site cofactor precursor NifB‐co in Saccharomyces cerevisiae . Proc Natl Acad Sci USA 116: 25078–25086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén, S. , and Rubio, L.M. (2018) State of the art in eukaryotic nitrogenase engineering. FEMS Microbiol Lett 365: fnx274. 10.1093/femsle/fnx274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén, S. , Young, E.M. , Sweeny, E.A. , Lopez‐Torrejón, G. , Veldhuizen, M. , Voigt, C.A. , and Rubio, L.M. (2017a) Formation of nitrogenase NifDK tetramers in the mitochondria of Saccharomyces cerevisiae . ACS Synth Biol 6: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.M. , Chang, A.C. , Shie, B.J. , Chang, Y.H. , and Chang, N.C. (1992) Molecular cloning and characterization of a mouse alpha 2C2 adrenoceptor subtype gene. Biochim Biophys Acta 1171: 219–223. [DOI] [PubMed] [Google Scholar]
- Curatti, L. , Hernandez, J.A. , Igarashi, R.Y. , Soboh, B. , Zhao, D. , and Rubio, L.M. (2007) In vitro synthesis of the iron‐molybdenum cofactor of nitrogenase from iron, sulfur, molybdenum, and homocitrate using purified proteins. Proc Natl Acad Sci USA 104: 17626–17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz‐Perales, A. , Sanz, M.L. , García‐Casado, G. , Sánchez‐Monge, R. , García‐Selles, F.J. , Lombardero, M. , et al. (2003) Recombinant Pru p 3 and natural Pru p 3, a major peach allergen, show equivalent immunologic reactivity: a new tool for the diagnosis of fruit allergy. J Allergy Clin Immunol 111: 628–633. [DOI] [PubMed] [Google Scholar]
- Diekert, K. , de Kroon, A.I. , Kispal, G. , and Lill, R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae . Methods Cell Biol 65: 37–51. [DOI] [PubMed] [Google Scholar]
- Emerich, D.W. , and Burris, R.H. (1978) Complementary functioning of the component proteins of nitrogenase from several bacteria. J Bacteriol 134: 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eseverri, A. , López‐Torrejón, G. , Jiang, X. , Burén, S. , Rubio, L.M. , and Caro, E. (2020) Use of synthetic biology tools to optimize the production of active nitrogenase Fe protein in chloroplasts of tobacco leaf cells. Plant Biotechnol J 18: 1843–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley, J.A. , Ramankutty, N. , Brauman, K.A. , Cassidy, E.S. , Gerber, J.S. , Johnston, M. , et al. (2011) Solutions for a cultivated planet. Nature 478: 337–342. [DOI] [PubMed] [Google Scholar]
- Gollan, U. , Schneider, K. , Müller, A. , Schüddekopf, K. , and Klipp, W. (1993) Detection of the in vivo incorporation of a metal cluster into a protein. The FeMo cofactor is inserted into the FeFe protein of the alternative nitrogenase of Rhodobacter capsulatus . Eur J Biochem 215: 25–35. [DOI] [PubMed] [Google Scholar]
- Hamilton, T.L. , Ludwig, M. , Dixon, R. , Boyd, E.S. , Dos Santos, P.C. , Setubal, J.C. , et al. (2011) Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii . J Bacteriol 193: 4477–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, D.F. , Lukoyanov, D.A. , Shaw, S. , Compton, P. , Tokmina‐Lukaszewska, M. , Bothner, B. , et al. (2018) Mechanism of N2 reduction catalyzed by Fe‐nitrogenase involves reductive elimination of H2 . Biochemistry 57: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M.R. , Brigle, K.E. , Bennett, L.T. , Setterquist, R.A. , Wilson, M.S. , Cash, V.L. , et al. (1989) Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii . J Bacteriol 171: 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Payá‐Tormo, L. , Coroian, D. , García‐Rubio, I. , Castellanos‐Rueda, R. , Eseverri, A. , et al. (2021) Exploiting genetic diversity and gene synthesis to identify superior nitrogenase NifH protein variants to engineer N2‐fixation in plants. Commun Biol 4:4. 10.1038/s42003-020-01536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger, R.D. , Jacobson, M.R. , Premakumar, R. , Wolfinger, E.D. , and Bishop, P.E. (1989) Nucleotide sequence and mutational analysis of the structural genes (anfHDGK) for the second alternative nitrogenase from Azotobacter vinelandii . J Bacteriol 171: 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C. , Michaelis, S. , and Mitchell, A. (1994) Methods in Yeast Genetics: Cold Spring Harbor Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Krahn, E. , Weiss, B. , Kröckel, M. , Groppe, J. , Henkel, G. , Cramer, S. , et al. (2002) The Fe‐only nitrogenase from Rhodobacter capsulatus: identification of the cofactor, an unusual, high‐nuclearity iron‐sulfur cluster, by Fe K‐edge EXAFS and 57Fe Mössbauer spectroscopy. J Biol Inorg Chem 7: 37–45. [DOI] [PubMed] [Google Scholar]
- Lill, R. (2009) Function and biogenesis of iron‐sulphur proteins. Nature 460: 831–838. [DOI] [PubMed] [Google Scholar]
- López‐Torrejón, G. , Jiménez‐Vicente, E. , Buesa, J.M. , Hernandez, J.A. , Verma, H.K. , and Rubio, L.M. (2016) Expression of a functional oxygen‐labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat Commun 7: 11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll, I.P. , and Dos Santos, P.C. (2011) Genomic analysis of nitrogen fixation. Methods Mol Biol 766: 49–65. [DOI] [PubMed] [Google Scholar]
- Raymond, J. , Siefert, J.L. , Staples, C.R. , and Blankenship, R.E. (2004) The natural history of nitrogen fixation. Mol Biol Evol 21: 541–554. [DOI] [PubMed] [Google Scholar]
- Schneider, K. , Gollan, U. , Dröttboom, M. , Selsemeier‐Voigt, S. , and Müller, A. (1997) Comparative biochemical characterization of the iron‐only nitrogenase and the molybdenum nitrogenase from Rhodobacter capsulatus . Eur J Biochem 244: 789–800. [DOI] [PubMed] [Google Scholar]
- Schneider, K. , and Müller, A. (2004) Iron‐only nitrogenase: Exceptional catalytic, structural and spectroscopic features. Catalysts for Nitrogen Fixation. Dordrecht: Springer, pp. 281–307. [Google Scholar]
- Schüddekopf, K. , Hennecke, S. , Liese, U. , Kutsche, M. , and Klipp, W. (1993) Characterization of anf genes specific for the alternative nitrogenase and identification of nif genes required for both nitrogenases in Rhodobacter capsulatus . Mol Microbiol 8: 673–684. [DOI] [PubMed] [Google Scholar]
- Seefeldt, L.C. , Hoffman, B.M. , and Dean, D.R. (2009) Mechanism of Mo‐dependent nitrogenase. Annu Rev Biochem 78: 701–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, V.K. , and Brill, W.J. (1973) Nitrogenase IV. Simple method of purification to homogeneity of nitrogenase components from Azotobacter vinelandii . Biochim Biophys Acta 305: 445–454. [DOI] [PubMed] [Google Scholar]
- Shah, V.K. , and Brill, W.J. (1977) Isolation of an iron‐molybdenum cofactor from nitrogenase. Proc Natl Aca. Sci USA 74: 3249–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippel, D. , and Einsle, O. (2017) The structure of vanadium nitrogenase reveals an unusual bridging ligand. Nat Chem Biol 13: 956–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.D. , Jameson, G.N.L. , Dos Santos, P.C. , Agar, J.N. , Naik, S. , Krebs, C. , et al. (2005) NifS‐mediated assembly of [4Fe‐4S] clusters in the N‐ and C‐terminal domains of the NifU scaffold protein. Biochemistry 44: 12955–12969. [DOI] [PubMed] [Google Scholar]
- Smith, B.E. , Eady, R.R. , Lowe, D.J. , and Gormal, C. (1988) The vanadium‐iron protein of vanadium nitrogenase from Azotobacter chroococcum contains an iron‐vanadium cofactor. Biochem J 250: 299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboh, B. , Boyd, E.S. , Zhao, D. , Peters, J.W. , and Rubio, L.M. (2010) Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB‐co at its active site. FEBS Lett 584: 1487–1492. [DOI] [PubMed] [Google Scholar]
- Taylor, S.C. , Berkelman, T. , Yadav, G. , and Hammond, M. (2013) A defined methodology for reliable quantification of Western blot data. Mol Biotechnol 55: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman, D. , Balzer, C. , Hill, J. , and Befort, B.L. (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108: 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente, E.J. , and Dean, D.R. (2017) Keeping the nitrogen‐fixation dream alive. Proc Natl Acad Sci USA 114: 3009–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh, S.I. , Paulsen, D.M. , Mylona, P.V. , Maynard, R.H. , Premakumar, R. , and Bishop, P.E. (1995) The genes encoding the delta subunits of dinitrogenases 2 and 3 are required for Mo‐independent diazotrophic growth by Azotobacter vinelandii . J Bacteriol 177: 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, B. , and Neupert, W. (2000) Mitochondria‐targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae . Yeast 16: 1421–1427. [DOI] [PubMed] [Google Scholar]
- Wolfinger, E.D. , and Bishop, P.E. (1991) Nucleotide sequence and mutational analysis of the vnfENX region of Azotobacter vinelandii . J Bacteriol 173: 7565–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Xie, X. , Wang, X. , Dixon, R. , and Wang, Y.P. (2014) Reconstruction and minimal gene requirements for the alternative iron‐only nitrogenase in Escherichia coli . Proc Natl Acad Sci USA 111: 3718–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. DNA optimized sequences.
Fig. S2. Schematic representation of synthetic anf and nif genes in S. cerevisiae GF15, GF16 and GF18.
Fig. S3. Purification of AnfH, AnfK, AnfG and AnfD proteins for the generation of polyclonal antibodies.
Fig. S4. Quantification of yAnfH and yAnfDK proteins in partially purified fractions.
Table S1. List of strains used in this work.


