Abstract
New advances in sequencing technology and bioinformatics analysis tools have significantly supported the culture-independent analysis of complex microbial communities associated with environmental, plant, animal and human samples. However, previous work has shown that DNA extraction can have a major influence in the community profile. As such there is a constant need for new methods to efficiently and rapidly prepare and analyze DNA for microbiome research, especially in the case new and emerging technology like the Oxford Nanopore Technologies (ONT) MinION. A commercial standard was used, in triplicate, to evaluate three DNA extraction protocols, including two commercially available and one ”in-house” DNA extraction method. All DNA extractions were done as per manufacturer's instructions and prepared with the same commercial ONT 16S sample preparation kit, prior to being analysed using MinION sequencing. Eight MinION 16S datasets of this microbial reference community were obtained. Reads were initially base called and demultiplexed using ONT's Guppy™ sequencing software (version 3.2.4), filtered using NanoFilt and then classified using Usearch. A set of R scripts are presented to process sintax files generated from Usearch and produce an OTU table that can be used for further analyses. All datasets were deposited into the SRA (NCBI) database. These datasets will allow future extraction kit comparisons using MinION sequencing since a standardize laboratory process using commercially available components, such as the MinION 16S sample preparation kit, microbial reference community and extraction kits, were used. The current ONT 16S workflow making use of the Epi2me agent only provides QC metrics and the ID's of the main genera identified and does not provide any tools currently for further downstream community comparison. The analyses scripts provided in the supplementary material will thus further enable the testing of new datasets against these reference sets and provide users the ability to compare their workflows with ours, thus standardizing comparisons and workflows.
Keywords: Full length 16s meta-barcoding microbiome, Reference community standard, Long read sequencing
Specifications Table
| Subject | Biological sciences |
| Specific subject area | Targeted reference metagenomic comparison of DNA extractions |
| Type of data | Fast5 files Raw fastq files Filtered and merged fastq files Table Figure Diagram R scripts |
| How data were acquired | ONT MinION platform was used for sequencing of eight (9) 16S amplicon libraries |
| Data format | Raw and analysed |
| Parameters for data collection | DNA from a single ZymoBIOMICS™ Microbial Community Standard (Zymo, USA) was extracted with two commercially available kits, and one in-house developed method. All 16S amplicon libraries were prepared with the same sequencing preparation kit prior to sequencing according to the recommended ONT 16S Barcoding protocol. All reactions were performed in triplicate. |
| Description of data collection | DNA from the ZymoBIOMICS™ Microbial Community Standard was extracted using three methods. The two commercially available kits included a standard beat beating kit (GenElute™ Stool DNA Isolation Kit, Sigma, USA) and a kit with a host DNA removal step prior to DNA extraction (QIAamp DNA microbiome kit, Qiagen, Germany). The in-house kit used a “lyses micro tube” to extract DNA. 16S libraries were generated and multiplexed using same the ONT 16S Barcoding Kit (SQK-RAB204, ONT, UK). MinION sequencing was carried out with the aid of the MinKNOW software (ONT, UK), with the fast5 files obtained converted to fastq with the ONT's Guppy™ sequencing software (version 3.2.4). Resulting fastq sequences were analysed with available opensource software and a set of R scripts included as part of the dataset. |
| Data source location | Institution: North-West University City/Town/Region: North-West, Potchefstroom Country: South Africa GPS coordinates: Latitude: −26.7167 Longitude: 27.1000 |
| Data accessibility | Raw fast5, fastq and, filtered and merged MinION sequence data is available at NCBI under the BioProject No. PRJNA675451. SRA accession numbers: SRR13994872 - SRR13994884 (fast5), SRR13632459 - SRR13632466 (raw fastq) and SRR13011317 - SRR13011324 (filtered and merged fastq). SRA link: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA675451&o=acc_s%3Aa Supplementary material can be found on Mendeley data at the following link: https://data.mendeley.com/datasets/yhv4rsr426/1 |
Value of the Data
-
•
The work presented provides raw fastq files obtained following sequencing of a standard microbial community on the MinION nanopore sequencer along with the already filtered and merged fastq files. Additionally, a simplified, rapid workflow for the analyses of ONT 16S reads is provided with the MinION dataset to act as a standard workflow for comparison.
-
•
The data sets provided generates useful information for researchers involved in the application of long read 16S metagenomics especially those working with 16S data obtained by MinION sequencing.
-
•
The 16S sequencing data provided is of a commercially available microbial community standard of known composition, and as such, can be used to test newly developed laboratory workflows by comparing datasets with these already existing workflows needed to process the 16S data produced by the MinION platform. The workflow provided with the R scripts will enable the testing of new workflows and datasets against the data sets provided as a reference.
1. Data Description
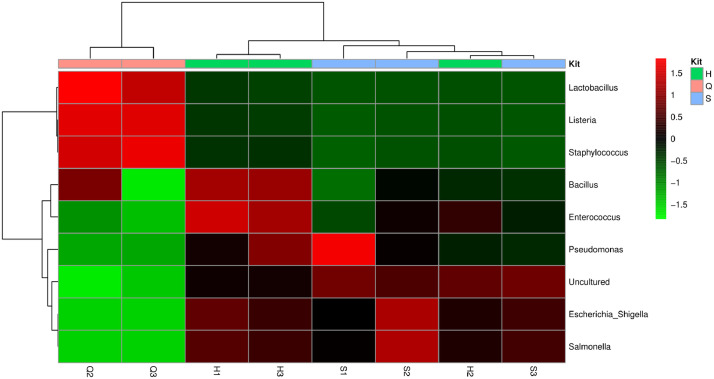
Samples for the sequencing dataset were based on a comparison study of various DNA extraction methods using a reference microbial community (ZymoBIOMICS™ Microbial Community Standard, Zymo, USA). Dataset 1 represents the obtained fast5 files and raw fastq data after sequencing by the ONT MinION 16S Barcoding Kit, as well as the filtered and merged fastq files that can be used directly with the supplied workflow of dataset 2. Dataset 2 represent a series of text files (Supplementary material) containing instructions and R script for processing of the data once quality control and demultiplexing have been done. The set of files included in the supplementary files include the instructions and R scripts (Database and sintax generation.txt, Script 1 – Multiple files.txt, Script 2-Final version.txt, Script 3-Final version.txt, Script 4-Final version.txt, Script 5-Final version.txt) applied in the workflow as depicted in Table 1. Additionally the supplementary files also contain the expected outputs (st_pre.txt, presum.txt, for_xls.txt, MA_OTU.txt, Summary_out.xlsx) when processing the supplied fastq files for each sample with the provided workflow from Table 1 as well as an example mapping file (Mappingfile_eg.txt). The MinION 16S sequencing dataset contains a total of 5, 427, 602 reads. Table 1 provides a summary of the supplied pipeline that was established for bioinformatics analysis of the long sequencing reads obtained from a single sequencing run on a MinION™. Table 2 contains a summary of the observed, and thus expected DNA quality metrics seen after extraction of the ZymoBIOMICS™ Microbial Community Standard with the 3 applied methods, while Table 3 contains a summary of the subsamples following sequencing – sample Q1 failed at PCR and thus no values were recorded. Fig. 1 represents an example of the abundance graphs that can be drawn using the excel output from step 5 the workflow described in Table 1. Figs. 2, 3 and 4 represents the expected outputs such as alpha (chao1, observed and Shannon index), beta diversity and heatmap examples generated as part of the workflow.
Table 1.
Summarised workflow steps for the simple and rapid analyses of 16S reads produced by the ONT MinION 16S barcoding.
| Step | Description | Software | Input file(s) | Output files and data |
|---|---|---|---|---|
| 1 | Install Usearch | Usearch | NA | NA |
| 2 | Database generation | Usearch | Downloaded database file (RDP 16S training set v16 (RTS)) | Database ready for classification |
| 3 | Classification | Usearch | Merged and filtered fastq files for each demultiplexed sample | .SINTAX file ready for processing using R |
| 4 | Script 1 – Pre-processing | R | .SINTAX file/files | Temporary output file for Script 2 (st_pre.txt) |
| Script 2 – Pre-processing | R | Temporary file for Script 2 (st_pre.txt) | Temporary output file for Script 3 (presum.txt) | |
| Script 3 – Pre-processing | R | Temporary file for Script 3 (presum.txt) | Temporary output file for Script 4 (for_xls.txt) | |
| 5 | Script 4 – Generate Excel summary | R | Temporary file for Script 4 (for_xls.txt) | Excel summary |
| 6 | Script 5 – Generate OUT table | R | Temporary file for Script 4 (for_xls.txt) | MA_OTU.txt |
| 7 | Import into Microbiome analyst | Web application | MA_OTU.txt Mapping file (generated by user, see example supplied in supplementary material) |
Table 2.
Summary of expected DNA quality following extraction of the ZymoBIOMICS™ Microbial Community Standard. Sample H refers to the average values obtained for the in-house kit, S refers to the GenElute™ Stool DNA Isolation Kit and Q to the QIAamp DNA microbiome kit.
| Method | Nanodrop Concentration (ng/µL) | Qubit Concentration (ng/µL) | A260/A280 | A260/A280 |
|---|---|---|---|---|
| H | 27.73 ± 0.87 c | 15.23 ± 3.72 b | 1.9 ± 0.01 a | 1.72 ± 0.08 a,b |
| S | 20.46 ± 2.02 b | 1.33 ± 0.21 a | 1.82 ± 0.03 a | 1.76 ± 0.02 b |
| Q | 12.8 ± 1.23 a | 1.97 ± 0.09 a | 1.81 ± 0.02 a | 1.4 ± 0.1 a |
a,b,c Methods sharing a common letter belong to the same group according to one way ANOVA with Tukey post-hoc test for multiple pairwise comparisons. These values do not differ significantly according to the test (p > 0.05). Values not sharing a common letter indicate a statistically significant difference in comparison to a value belonging to other groups (p < 0.05).
n = 3.
Table 3.
SRA dataset matrices, including each triplicate repeat's, raw reads, filtered reads, and percentage of surviving reads after filtering that should be expected after running the presented data analyses pipeline. Sample H refers to the in-house kit repeats, S refers to the repeats for the GenElute™ Stool DNA Isolation Kit and Q to the repeats for the QIAamp DNA microbiome kit.
| Sample | Barcode | Reads prior to processing | Reads post filtering | Percentage retained |
|---|---|---|---|---|
| H1 | 1 | 219,020 | 170,943 | 78.05% |
| H2 | 2 | 113,7251 | 759,743 | 66.81% |
| H3 | 3 | 365,529 | 279,836 | 76.56% |
| S1 | 4 | 151,1825 | 993,650 | 65.73% |
| S2 | 5 | 443,793 | 351,756 | 79.26% |
| S3 | 6 | 973,672 | 648,025 | 66.55% |
| Q1* | 7 | NA | NA | NA |
| Q2 | 8 | 523,226 | 370,027 | 70.72% |
| Q3 | 9 | 253,286 | 166,910 | 65.90% |
Sample Q1 failed at PCR.
Fig. 1.
Relative abundance chart of the bacterial genera from each sample above a 0.01% abundance cut-off. Subsamples extracted with the GenElute™ Stool DNA Isolation Kit, QIAamp DNA microbiome kit and the in-house methods have been labelled S, Q and H respectively. The expected relative abundance from the published reference community standard is labelled as Z in the figure.
Fig. 2.
An example of Alpha-diversity, measured by Observed (A), Chao1 (B) and Shannon diversity (C) indexes for the ZymoBIOMICS™ Microbial Community Standard.
Fig. 3.
An example of Principal coordinates analysis based on Bray-Curtis distances of samples extracted by various methods for the ZymoBIOMICS™ Microbial Community Standard using the presented workflow.
Fig. 4.
The Hierarchical clustering heat map based on the relative abundance of the most abundant genera classified to the genus level for ZymoBIOMICS™ Microbial Community Standard generated with the presented workflow. Individual cells in the heat map are color-coded according to the row Z-scores.
2. Experimental Design, Materials and Methods
2.1. DNA extraction methods evaluated with a commercial standard
DNA from the same ZymoBIOMICS™ Microbial Community Standard (Zymo, USA) was prepared by 3 different DNA extraction methods, with the standard being divided into 9 aliquots (75 μl at 26.7 ng/μl) prior to DNA extractions in triplicate. Commercial kits selected included both chemical lyses and mechanical shearing to ensure that DNA extraction from difficult to lyse bacteria occurred. QIAamp DNA microbiome kit (Qiagen, Germany) and GenElute™ Stool DNA Isolation Kit (Sigma, USA) were used as the commercial kits that acted as “industry controls” to which we've compared a NWU in-house cell lysis method as previously described (Mutingwende et al., 2015). Briefly 250 µl of sample was mixed with 250 µl of a proprietary lysis buffer consisting of 20 mM TCEP (Sigma-Aldrich, USA), 20 x TE (Tris–EDTA, Sigma-Aldrich, USA) and 1.5% Tween 20 (Sigma-Aldrich, USA) in a lyses micro tube (LMT). The lyses micro tube (LMT) was placed on the pre-set (95 °C and 3600 rpm) lyser device for 7 min. Bacterial cell lysis was then concurrently achieved through chemical, thermal and mechanical means (Mutingwende et al., 2015). Samples were labelled as “S” for the GenElute™ Stool DNA Isolation Kit, “Q” for the QIAamp DNA microbiome kit and “H” for the NWU in-house cell lysis method. DNA concentration was assessed using the Qubit 4 Fluorometer (Thermofisher Scientific, USA) along with the Qubit BR assay kit (Thermofisher Scientific, USA), while quality was determined by nanodrop spectrophotometry on a Nanodrop One (Thermofisher Scientific, USA), summarised in Table 2. All isolated DNA samples were stored at −20 °C until further processing.
2.2. Amplicon library and flow cell preparation
Sequencing of the ZymoBIOMICS™ Microbial Community Standard was carried out at the Agricultural Research Council's (ARC) Biotechnology Platform using the ONT 16S Barcoding Kit (SQK-RAB204) according to the ONT protocol. Polymerase chain reaction (PCR) barcoding amplification was conducted in 50 µl reactions consisting of 1 µl of 16S barcode primer, 25 µl of LongAmp Taq 2X master mix (New England Biolabs, USA), 14 µl nuclease-free water and 1 µl of template DNA (10 ng). A total of 9 samples, 3 extracted by each kit were prepared for sequencing. PCR cycling conditions were set at 95 °C for 1 min followed by 25 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 30 s, extension at 65 °C for 2 min and a final extension step of 65 °C for 5 min before holding at 4 °C. PCR products were cleaned using AMPure XP beads (Beckman Coulter, USA) and eluted in 10 µl of 10 mM Tris-HCl pH 8.0 with 50 mM NaCl. Following PCR, 1 µl of eluted sample was quantified using a Qubit fluorometer to pool the DNA barcoded libraries into an equal ratio. All barcoded libraries were pooled in the desired ratios to a total of 50–100 fmoles in 10 µl of 10 mM Tris-HCl pH 8.0 with 50 mM NaCl. Platform quality control (QC) was carried out using MinKNOW™ on a new R9.4.1 chemistry MinION™ flow cell before the flowcell was primed. In total 75 µl of sequencing mix, consisting of the DNA library, sequencing buffer and library loading beads, was prepared according to the manufacturer's instructions and added in a drop-wise fashion via the SpotOn sample port. The standard 48 h sequencing script was chosen with 1D live base calling.
2.3. Bioinformatics analysis
The raw data generated from the ZymoBIOMICS™ Microbial Community Standard was acquired as fast5 files using ONT's MinION™ sequencing software (version 19.06.8). The fast5 files were based called and de-multiplexed prior to adapter and primer sequence removal with ONT's Guppy™ sequencing software (version 3.2.4). These barcode-sorted fastq files were merged prior to further processing and renamed according to sample. Reads were filtered to remove reads with a Phred score below 7 and keep lengths between 1200 and 1500 bp using NanoFilt (https://github.com/wdecoster/nanofilt) [5]. The passing reads were processed with the bioinformatics workflow described in Table 1. The first part of the workflow (steps 1 – 3) includes the installation of Usearch, database generation and classification. Usearch takes quality controlled and demultiplexed fastq files containing ONT generated 16S reads from each sample as input and assigns taxonomy to each read using the Usearch sintax command and the selected 16S database (RDP 16S training set v16, RTS) [2]. Detailed instructions can be found in the Database and sintax generation.txt file supplied in the supplementary information file.
Following classification the generated .SINTAX files are pre-processed in step 4 with a set of R scripts. Step 4 involves pre-processing and includes the use of scripts 1 to 3. The pre-processing scripts import and merge the .SINTAX files generated for each sample during step 3, remove any unnecessary information and generate a counts table (i.e. “for_xlx.txt”, supplementary files). Following pre-processing, the script for step 5 takes the output from the final script of step 4 (Table 1) and generates an excel sheet (i.e. “Summary_out.xlsx”, supplementary file). This excel output allows easy and simple generation of relative abundance graphs at several taxonomic levels as demonstrated by Fig. 1. Finally, the scripts (step 6) take the output from step 4 (Table 1) and generate an OTU table (i.e.“MA_OTU.txt”, supplementary file). The OTU table, along with a mapping file (step 7, Table 1), can then be imported into the Microbiome analyst (https://www.microbiomeanalyst.ca/) online software suite for further evaluation which included alpha and beta diversity determination and clustering analyses [1,3]. As an example, by importing of the output file from step 6 into microbiome analyst the following standard analyses were carried out in a demonstration: Data was normalized using total sum scaling (TSS) and the alpha diversity of samples was measured by observed, chao1 and Shannon diversity indexes as demonstrated with Fig. 2. Additionally, beta-diversity was determined and visualized using principal coordinate analysis plots (PCoA) based on Bray–Curtis distances, and compared using the nonparametric analysis of similarities (ANOSIM) test (Fig. 3). Finally, a heatmap (Fig. 4) of the most abundant genera classified to the genus level was generated using complete hierarchical clustering by Euclidian distance [1,3,6]. Detailed descriptions and directions for the use of each script and links for all software used and packages required in R, are provided in the supplementary material within the R scripts.
2.4. Statistical analysis
The influences of each extraction method on the DNA quantity (yield) and quality (A260/A280 and A260/A230) was evaluated with ANOVA (one-way analysis of variance) and Tukey post hoc test for multiple pairwise comparisons [4]. This statistical testing was carried out using statistica (v13.1) (Statsoft, Inc, USA) and visualised in GraphPad Prism (v8) (GraphPad, Inc., USA).
Ethics Statement
This study was approved by the North-West University Health Research Ethics Committee (NWU-HREC) of the faculty of health sciences, Ethics number: NWU-00,127–18-A1.
CRediT Author Statement
B.C. Mann: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization; J.J. Bezuidenhout: Conceptualization, Methodology, Software, Writing - Review & Editing, Supervision; A.F. Grobler: Conceptualization, Writing - Review & Editing, Resources, Project administration, funding acquisition; Z.H. Swanevelder: Conceptualization, Writing - Review & Editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This manuscript was written with the support of funding provided by the DSI/NWU Preclinical Drug Development Platform and HANKS TB diagnostics (Pty) Ltd.
References
- 1.Dhariwal A., Chong J., Habib S., King I.L., Agellon L.B., Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic. Acids Res. 2017;45 doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgar E.C. Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ. 2018;6:e4652. doi: 10.7717/peerj.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong J., Liu P., Zhou G., Xia J. Using microbiome analyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 4.Corcol N., Österlund T., Sinclair L., Eiler A., Kristiansson E., Backhaus T., Eriksson K.M. Comparison of four DNA extraction methods for comprehensive assessment of 16S rRNA bacterial diversity in marine biofilms using high-throughput sequencing. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnx139. [DOI] [PubMed] [Google Scholar]
- 5.Kai S., Matsuo Y., Nakagawa S., Kryukov K., Matsukawa S., Tanaka H., Iwai T., Imanishi T., Hirota K. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION™ nanopore sequencer. FEBS Open Bio. 2019;9:548–557. doi: 10.1002/2211-5463.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia Y., Sun J. Hypothesis Testing and Statistical Analysis of Microbiome. Genes dis. 2017;4:138–148. doi: 10.1016/j.gendis.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]