Abstract
Background
Organoid technology has recently emerged as a powerful tool to assess drug sensitivity of individual patient tumors in vitro. Organoids may therefore represent a new avenue for precision medicine, as this circumvents many of the complexities associated with DNA- or transcriptional-profiling.
Materials and methods
The SENSOR trial was a single-arm, single-center, prospective intervention trial to evaluate the feasibility of patient-derived organoids to allocate patients for treatment with off-label or investigational agents. The primary endpoint was an objective response rate of ≥20%. Patients underwent a biopsy for culture before commencing their last round standard of care. Organoids were exposed to a panel of eight drugs and patients were treated after progression on standard-of-care treatment and when a clear signal of antitumor activity was identified in vitro.
Results
Sixty-one patients were included and we generated 31 organoids of 54 eligible patients. Twenty-five cultures were subjected to drug screening and 19 organoids exhibited substantial responses to one or more drugs. Three patients underwent treatment with vistusertib and three with capivasertib. Despite drug sensitivity of organoids, patients did not demonstrate objective clinical responses to the recommended treatment.
Conclusions
Organoid technology had limited value as a tool for precision medicine in this patient population because a large fraction of patients could not undergo treatment or because the recommended treatment did not elicit an objective response. We identified several essential parameters, such as the culture success rate, clinical deterioration of patients during standard of care, and rational design of drug panels that need to be accounted for in organoid-guided clinical studies.
Key words: colorectal cancer, precision medicine, experimental treatment, clinical trial, tumor organoids, drug screening
Highlights
-
•
The first prospective clinical trial that leverages tumor organoids to guide experimental treatment decisions.
-
•
Clinical implementation of tumor-organoid-guided treatment is challenging.
-
•
Patients that received organoid-informed treatment did not experience clinical benefit.
-
•
Organoid drug screening can distinguish differential drug responses in identical genetic genotypes.
Introduction
The paradigm of personalized medicine revolves around allocating the right treatment to the right patient. As a consequence, most newly developed anticancer drugs usually have a defined, genomics-based target. The implementation of personalized medicine in daily clinical practice has led to variable successes in the treatment of cancer patients.1, 2, 3, 4 Early experiences have identified several important challenges, including the tissue-specificity of cancer drivers, variants of unknown significance (VUS), and the context-dependency of genomic aberrations among the thousands found in cancer genomes.5 These issues currently limit efficient use of new anticancer drugs and inspired us to explore other means (beyond genomics) to improve precision medicine.3,6
Organoid technology allows culturing, expansion, and drug screening of patients' individual tumors.7 Organoids provide a morphological and genetic representation of a patient's tumor and several studies demonstrated that in vitro drug responses mirrored the clinical responses of the patient.8,9 Organoids are therefore widely recognized as a novel opportunity to test a long-standing concept in the field of precision oncology: treatment based on individualized, ex vivo drug screening of patient tumor cells.6,10, 11, 12, 13, 14, 15 As such, many groups are now pursuing organoid-based drug screening of patient tumor cells to guide clinical decision-making.11,12,16 Here, we present the first formal, prospective intervention trial, the Selecting Cancer Patients for Treatment Using Tumor Organoids (SENSOR) trial, designed to evaluate the potential and feasibility of treating patients based on their in vitro organoid drug response profile.
Materials and methods
Study design
The SENSOR study was an open-label, single-center, prospective, feasibility study (NL50400.031.14; https://www.clinicaltrialsregister.eu/ctr-search/trial/2014-003811-13/NL/) at The Netherlands Cancer Institute. The study was approved by the ethical review board of The Netherlands Cancer Institute and was designed in accordance with the Declaration of Helsinki for medical research involving human subjects, Dutch law, and good clinical practice. The study objective was to evaluate the feasibility of organoids to allocate patients for treatment with specific targeted agents and the primary endpoint of the study was an objective response rate (ORR) of ≥20% according to Response Evaluation Criteria in Solid Tumors (RECIST) 1.1.17 Secondary and exploratory endpoints were to identify potential biomarkers of response, to identify mechanisms of primary and secondary resistance, and to determine whether standard-of-care (SOC) treatment-induced changes of the drug response profile in pre- and post-treatment organoids. Metastatic colorectal cancer (CRC) patients without curative treatment options were accrued at The Netherlands Cancer Institute before start of their last SOC treatment and referred by their treating physicians. Patients underwent pre-treatment biopsies before start of their last round SOC treatment and second biopsy before start with experimental treatment to control for potential effect of intermittent SOC treatment. Treatment was provided for 5 patients per drug (vistusertib, capivasertib, selumetinib, and gefitinib) by AstraZeneca and for 10 patients per drug (palbociclib, axitinib, gedatolisib, and glasdegib) by Pfizer. Eligibility criteria were an Eastern Cooperative Oncology Group (ECOG) performance status of ≤2; measurable disease according to RECIST 1.1 criteria17; histologic tumor biopsy feasible; age >18 years. Patients with a life expectancy of <3 months due to tumor progression under SOC, with symptomatic brain or leptomeningeal metastasis, with other malignancies within the last 5 years, or with a known HIV infection, and pregnant/nursing women were excluded from study participation. Written informed consent was provided before any study-specific procedures or assessments.
Statistical analysis
RECIST response and progression-free survival will be presented for the evaluable patient population. Adverse events will be presented separately for each safety population. As this is a feasibility study, there is no formal sample size calculation. We planned preliminary analysis of a cohort when at least five patients were treated. All statistical tests were carried out as two-tailed in GraphPad Prism V7.03. P values < 0.05 were considered statistically significant.
Patient material processing and organoid culture
Patient-derived organoids (PDOs) for the decision model were derived from van de Wetering et al. 2015, our personal biobank, or our prior studies.13,18 For patients included in the trial, we used one or two 18-gauge tumor biopsies for both organoid culture and DNA sequencing. Biopsies were collected and processed as previously described in Weeber et al. 2015 and Dijkstra et al. 2018.9,18 In short, biopsies were collected by a trained radiologist and collected on phosphate-buffered salt (PBS)-wetted, gauze pads. Upon completion of biopsy procedure, tissue was immediately transferred to a sterile 15-ml tube containing Advanced Dulbecco's Modified Eagles Medium with Nutrient Mixture F-12 Hams (Ad-DF) (Invitrogen; #12634), supplemented with 1% penicillin/streptomycin (Invitrogen; #15140-122), 1% HEPES (Invitrogen; #15630-056), and 1% GlutaMAX (Invitrogen; #35050) (Ad-DF+++). Tubes containing fresh biopsies were kept at 4°C and processed within 24 h to ensure viability of the tissue. Biopsies were mechanically dissociated with needles to cell clumps, washed with Ad-DF+++, and cultured in colorectal cancer (CRC) medium as described previously.8,18 PDOs were expanded into master and working biobanks, and PDOs from working biobanks were used for drug sensitivity testing, typically passage four or lower. Cultures were checked for mycoplasma contamination every month using the MycoAlert Mycoplasma Detection Kit (Lonza). As part of quality control, PDOs were authenticated using a TaqMan-based SNP array targeting 26 single-nucleotide polymorphisms (SNPs) (Hartwig Medical Foundation). PDOs with identity scores <0.9 (tumor versus blood) were discarded.19 In case of eligibility and consent of the patient, cultures were also used in context of the tumor organoids: feasibility to predict sensitivity to treatment in cancer patients (TUMOROID) study, of which the goal was to determine whether clinical response of patients to SOC treatment correlate with in vitro drug sensitivity.13
Drug screening
All drug screens were carried out in duplicate by an independent researcher, blinded for all genetic (and clinical) data of patients. In case of discrepant results, the given drug was repeated a third time. Screens were carried out as described previously by Ooft et al. 2019.13 Organoids were mechanically and enzymatically dissociated to cell clumps in TrypLE (Gibco, #12604-013) for 5-10 min, filtered for sizes <40 μm, and re-plated to allow for formation of organoids over the course of 4 days. After 4 days, organoids were collected, incubated with 2 mg/ml dispase II (Sigma #D4693) for 15 min to remove Geltrex, and counted using a hemocytometer and trypan blue. Organoids were resuspended in 1 : 2 Ad-DF+++ : Geltrex at a concentration of 20 organoids/μl. Five μl/well of the suspension was dispensed in clear-bottomed, white-walled 96-well plates (Corning, #3707) using an automated repeat pipet and overlaid with 200 μl CRC medium. Read-outs were carried out at day 0 (‘baseline’) and at day 6 in the positive control (10 μM phenylarsine oxide), negative control, and the drug-treated wells. Quantification of cell viability was done by replacing medium with 50 μl CellTiter-Glo 3D (Promega, #G9681) mixed with 50 μl Ad-DF+++ according to manufacturer instructions on an Infinite 200 Pro plate reader (Tecan Life Sciences). Compounds were provided by AstraZeneca and Pfizer, dissolved in di-methylsulfoxide (DMSO), and plated using a Tecan D300e digital dispenser. The choice of concentrations used in drug screening was based on the maximum concentration (Cmax) found in patients and concentrations 1-2 orders of magnitude lower20, 21, 22, 23: 0.137, 0.249, and 2.49 μM vistusertib; 0.37, 0.825, and 3.7 μM capivasertib; 0.041, 0.26, and 2.6 μM selumetinib; 0.0531, 0.098, and 0.531 μM gefitinib; 0.0303, 0.147, and 0.303 μM palbociclib; 0.000815, 0.0163, and 0.163 μM axitinib; 0.0069, 0.0407, and 0.407 μM gedatolisib; 0.1, 0.8, and 8.0 μM glasdegib. Organoid drug sensitivity was calculated using growth-rate inhibition metrics (GR; scale 1 to −1). A score of 1 represents identical growth to non-treated condition, 0 represents a size identical to day 0 (no growth), and anything <0 represents a degree of cytotoxicity after 6 days of exposure to the respective drug.
Study treatment
Organoid sensitivity, as defined by GR <0.1 (described in more detail in results), to one of the experimental compounds rendered patients eligible to treatment with that compound. In case patients were subsequently treated, additional inclusion and exclusion criteria were applied and an additional written consent form was obtained. The inclusion and exclusion criteria, study-related procedures and assessments, and written consent were specific for each experimental treatment regimen. Disease assessment with computed tomography (CT) or magnetic resonance imaging (MRI) was carried out at baseline and every two treatment cycles (2 months). Tumor measurements and treatment response were assessed according to RECIST 1.1.17 Treatment was continued up until disease progression or withdrawal. Adverse events were registered and graded by the investigator according to the Common Terminology Criteria for Adverse Events (CTCAE version 4.0) until 30 days after discontinuation of study treatment. Patients were treated with vistusertib or capivasertib according to investigator brochure at the recommended phase II dose: both orally and twice daily, at a dose of 480 mg in 28-day cycles using an intermittent dosing schedule (4 days on/3 days off) for vistusertib and 125 mg in 28-day cycles using an intermittent dosing schedule (2 days on/5 days) off for capivasertib.
DNA sequencing
Part of the biopsied material for the trial was used for routine clinical or cancer panel sequencing (Illumina TruSeq; ABL1; AKT1; ALK; APC; ATM; BRAF; CDH1; CDKN2A; CSF1R; CTNNB1; EGFR; ERBB2; ERBB4; FBXW7; FGFR1; FGFR2; FGFR3; FLT3; GNA11; GNAQ; GNAS; HNF1A; HRAS; ADH1; JAK2; JAK3; KDR; KIT; KRAS; MET; MLH1; MPL; NOTCH1; NPM1; NRAS; PDGFRA; PIK3CA; PTEN; PTPN11; RB1; RET; SMAD; SMARCB1; SMO; SRC; STK11; TP53; VHL) for samples T2, T4, T7, T10-15, P2, P3, P7-10, P14-16, P18, P20, and P22-24 or whole-genome sequencing (WGS) by the Hartwig Medical Foundation (HMF) for patients T9, P1, P4-6, P11-13, P17, P19, and P25. Both libraries were prepared according to manufacturer's instructions (targeted sequencing: FC-130-1008; WGS: TruSeq Nano LT; FC-121-4001-3) and sequenced on Illumina MiSeq (panel) or HiSeqX paired-end 2 × 150 basepairs (WGS) platform. Analysis of the targeted panel was carried out with Somatic Variant Caller v1.3 (Illumina). Analysis of the WGS data by the HMF was carried out using their standard pipeline.
Results
Development of a treatment decision model
Five Food and Drug Administration (FDA)-approved compounds and three drugs in advanced phase of clinical development were included in the study. This drug panel, made available by AstraZeneca and Pfizer, consisted of vistusertib (mTOR), capivasertib (AKT), selumetinib (MEK), gefitinib (EGFR), palbociclib (CDK4/6), axitinib (VEGFR), gedatolisib (PI3K/mTOR), and glasdegib (SMO). The panel was designed to target both frequent and sporadic genetic events in metastatic CRC, such as mutations or amplifications in EGFR (gefitinib), MEK (selumetinib), AKT (capivasertib), or SMO (glasdegib).24 To determine what would qualify as ‘sensitive’ or ‘resistant’, we amended our previously developed drug screening protocol and screened a dedicated pilot cohort of organoids to construct a decision model for patient treatment in the SENSOR trial.13 The cohort consisted of 16 organoids, of which the genetic and clinical data are summarized in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100103. We exposed all 16 organoids for 6 days to three concentrations per drug. The concentrations used in drug screening were guided by pharmacokinetic data from the investigator brochures or the published phase I studies of each drug.20, 21, 22, 23 In order to use patient-relevant concentrations in the drug screens, we used the maximum drug concentration found in patients (Cmax) and concentrations 1-2 orders of magnitude lower.20, 21, 22, 23 Drug effects were quantified using GR metrics.25
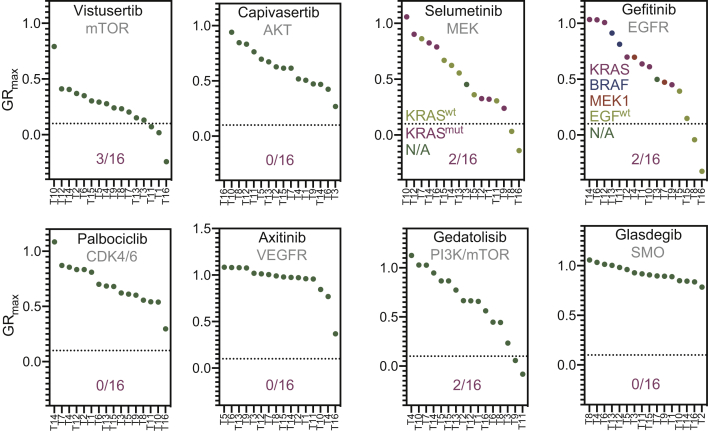
As expected, drug response data demonstrated a dose-dependent pattern, in which the highest concentration (GRmax) generally elicited a substantial effect (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100103). We decided that effects below 0.1 at these concentrations would be sufficiently stringent to qualify as in vitro hits, as this would identify cultures in which we observe near-cytostatic effects at physiologically achievable concentrations. This criterium resulted in hits on four drugs across the cohort targeting various oncogenic signaling nodes in CRC: vistusertib (mTOR), selumetinib (MEK), gefitinib (EGFR), and gedatolisib (PI3K/mTOR) (Figure 1).24
Figure 1.
Development of a decision model based on drug screening of 16 organoids.
The cohort of 16 organoids and their response to the GRmax concentration of each drug after 6 days. Effects were calculated using GR metrics. 1 = no effect, 0 = no growth, and <0 = a certain amount of cytotoxicity (up to −1). A cut-off of 0.1 was set, represented by the dotted line, discriminating between what was considered a hit versus no hit. All organoids were plotted on the x-axis and sorted from resistant to sensitive. All drug names were plotted at the top of each graph and the target(s) below. At the bottom of each graph we noted the number of virtual hits per drug. For selumetinib and gefitinib, additional, color-coded information on KRAS or the EGF-pathway status was added per organoid line.
GR, growth-rate corrected metric; N/A, not available.
To test whether our defined threshold would identify potentially eligible patients, we assessed the predictive value for two known biomarker-drug combinations. Because epidermal growth-factor receptor (EGFR) inhibition is solely efficacious in epidermal growth-factor (EGF)-pathway wild-type (EGFwt) tumors and not in EGF-pathway mutated (EGFmut) tumors, we expected that gefitinib in combination with our threshold can discriminate between these two genotypes. When we applied GR <0.1 as cut-off for potential responders, none of the EGFmut organoids were classified as a hit. Analogous to the variable response rate of EGFwt patients to the anti-EGFR monoclonal antibodies cetuximab and panitumumab, we observed a subset (2/4) of EGFwt organoids responding to gefitinib treatment. This suggests that our model could not only discriminate between EGFwt and EGFmut organoids but also identify differential responses within a genomically identical genotype (EGFwt).26 To further test the ability of our model to exclude non-responders and prevent overtreatment, we also correlated organoids with gain-of-function mutations in KRAS (KRASmut) and the effects of selumetinib. Preclinical evidence demonstrated that there is no benefit for selumetinib in KRASmut tumors, and we therefore expected this to be reflected in our organoid drug sensitivity data too.27,28 Seven organoids harbored KRAS mutations (T10, T12, T14, T16, T2, T1, and T9), which were all classified as resistant by our pipeline in line with (pre)clinical data. These data suggest that, at least for gefitinib and selumetinib, organoid drug screening excludes genotypes that are known to be resistant to treatment (EGF/KRASmut; gefitinib/selumetinib). Conversely, organoids displayed differential responses within a genetically similar genotype-drug combination (EGFwt; gefitinib), of which only a subset is known to respond well in the clinic to EGFR inhibition.26 We therefore concluded that organoids can be used as a treatment selection step in addition to purely genomics-based approaches.26
Prospective treatment of patients with metastatic colorectal cancer
Following validation of the decision model, we included patients of whom organoids were screened for responsiveness to our drug panel (Figure 2A). All patients were included before they received their last SOC treatment line (irinotecan-based therapy or anti-EGFR mABs), while all had progressed on one or two prior lines of SOC treatment (containing 5-fluoruracil, with or without oxaliplatin and/or irinotecan) according to the SOC for palliative treatment of patients with metastatic CRC (Figure 2A).
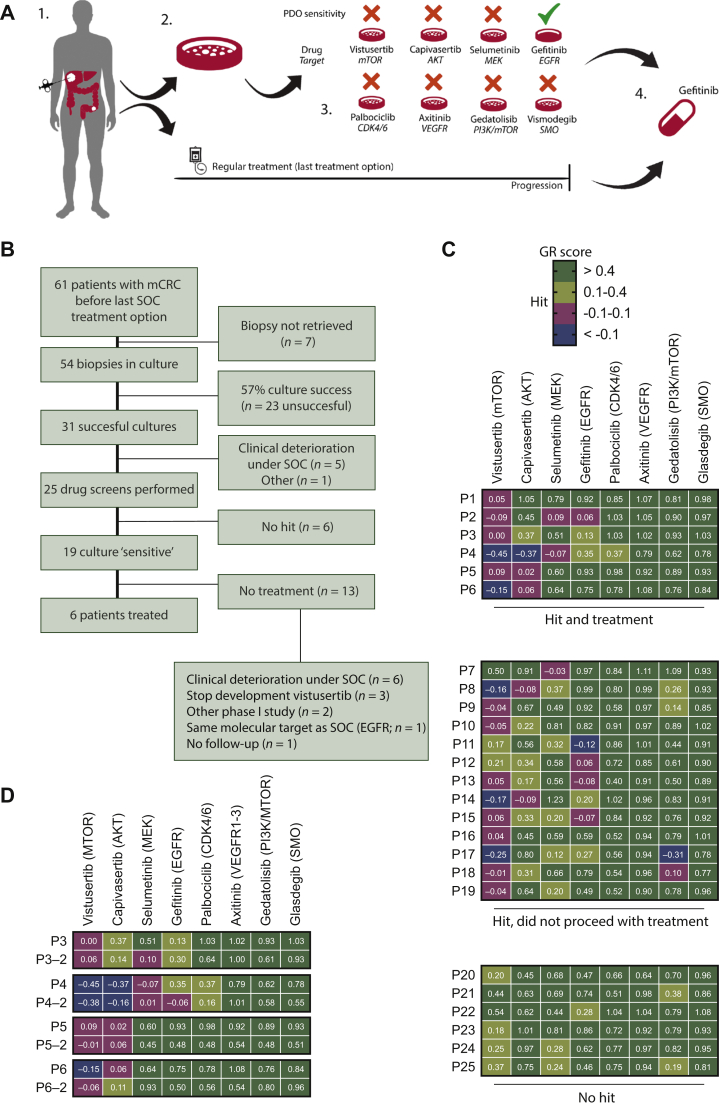
Figure 2.
Enrolment and organoid drug profiling of patients in the SENSOR trial.
(A) Outline of the SENSOR trial. Patients undergo a biopsy of a metastatic lesion for generation of organoids before start of their last line standard-of-care (SOC) treatment. 1. Patients underwent biopsy of a metastatc lesion. Part of the biopsies were also used for DNA sequencing. 2. Organoid cultures are generated from biopsies and frozen in master and working biobanks at low passage. 3. Organoids are profiled for their response to eight FDA-approved or investigational drugs. Patients received treatment when organoid drug response was qualified as a hit after two repeated experiments. In case organoids displayed sensitivity towards multiple drugs, patients received drug treatment with the strongest hit. 4. Patients received the drug identified in the drug screen after progression on SOC or went off study when no treatment option was available. Patient underwent a mandatory second biopsy before start of experimental treatment, which was used for a second confirmatory organoid culture. These were subjected to the same drug screen as identified in step 3 to control for potential change in drug sensitivity due to intermittent SOC treatment. (B) Flow chart on inclusion and dropout of patients in the SENSOR trial. (C) Heatmap with drug screening result of all organoids. Organoids were profiled for their drug response to the drug concentrations identified in Figure 1 (GRmax) and the average of two or, in the case of discrepant results, three independent replicates is given in the heatmap per drug and organoid. At the top, hits in six organoids were identified that subsequently led to treatment of the respective patients. The hits in the middle 13 organoids did not lead to treatment due to various reasons stated at the bottom of the flowchart. In the bottom six organoids, no hits were identified. (D) Patients that started treatment underwent a second biopsy after progression on SOC (represented by P#-2), and organoids were re-screened to control for potential shifts in drug sensitivity.
GR, growth-rate corrected metric; mCRC, metastatic colorectal cancer; SOC, standard of care.
Sixty-one patients were included in the trial, of which 54 underwent a successful biopsy procedure, as the biopsy failed to obtain tumor material in seven cases (Figure 2B; baseline characteristics of patients intended to treat are presented in Table 1).
Table 1.
Baseline characteristics of all included patients, patients with a successful organoid culture and subsequent drug screen, and patients that received an organoid-informed treatment, in comparison with the clinical characteristics in the pilot set. Information not available for a = 7, b = 9, c = 4, d = 6, e = 12, f = 1. For 7 patients, no tissue for organoid culture was retrieved (g)
| All patients |
Drug screen |
Treated patients |
Pilot set |
|
|---|---|---|---|---|
| n = 62 | n = 25 | n = 6 | n = 16 | |
| Median age (range), years | 59.5 (26-78) | 57 (30-73) | 57 (51-65) | N/A |
| Male : female n : n | 35 : 26 | 11 : 14 | 3 : 3 | N/A |
| WHO, n (%) | a | |||
| 0 | 34 (61%) | 14 (56%) | 3 (50%) | N/A |
| 1 | 20 (36%) | 10 (40%) | 3 (50%) | |
| 2 | 1 (2%) | 1 (4%) | 0 | |
| Localization of primary, n (%) | ||||
| Colon | 34 (55%) | 16 (64%) | 3 (50%) | 10 (63%) |
| Rectum | 14 (23%) | 5 (20%) | 3 (50%) | 2 (13%) |
| Rectosigmoid | 2 (3%) | 2 (8%) | 0 | 1 (6%) |
| Colorectal NOS | 12 (19%) | 2 (8%) | 0 | 3 (19%) |
| Differentiation, n (%) | c | d | ||
| Well/moderately | 38 (72%) | 14 (67%) | 5 (83%) | 8 (80%) |
| Poorly | 12 (23%) | 6 (29%) | 1 (17%) | 1 (10%) |
| Mucinous | 3 (6%) | 1 (5%) | 0 | 1 (10%) |
| Undifferentiated | 0 | 0 | 0 | 0 |
| Biopsied lesion, n (%) | g | |||
| Liver | 28 (51%) | 18 (72%) | 3 (50%) | 6 (38%) |
| Primary | 0 | 0 | 0 | 7 (44%) |
| Lymph node | 11 (20%) | 5 (20%) | 2 (33%) | 1 (6%) |
| Peritoneum | 5 (9%) | 1 (4%) | 0 | 0 |
| Other | 12 (22%) | 1 (4%) | 1 (17%) | 1 (6%) |
| Microsatellite status, n (%) | e | f | ||
| MSI | 1 (2%) | 1 (4%) | 0 | 2 (13%) |
| MSS | 49 (98%) | 24 (96%) | 6 (100%) | 13 (87%) |
| Prior lines of (chemo)therapy | ||||
| 0 | 20 (32%) | 2 (8%) | 0 | 12 (75%) |
| 1 | 25 (40%) | 11 (44%) | 2 (33%) | 2 (13%) |
| ≥2 | 17 (27%) | 12 (48%) | 4 (67%) | 2 (13%) |
The remaining 54 biopsies were taken into culture and organoids were generated as previously described.9,13,18 We obtained successful cultures for 31 patients, resulting in a culture success rate of 57% (31/54 biopsies). Twenty-three cultures failed due to either low epithelial/tumor cell content in the biopsy (n = 14), infection (n = 5), unknown (n = 3), or quality control (n = 1). Six patients dropped out before culture was successful, of which five clinically deteriorated during their last line of SOC treatment. For these six patients, we did not perform drug screening. In total, 25 drug screens were carried out for patients whose clinical and genetic parameters are presented in Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100103. Expansion of organoids from needle biopsies and drug screening was generally carried out within 10 weeks of the biopsy and finished before the first evaluation of treatment response of SOC. Repeats were highly reproducible with a median Pearson's r correlation between repeats of 0.937 (Supplementary Figure S1A and B, available at https://doi.org/10.1016/j.esmoop.2021.100103). In vitro drug sensitivity to at least one drug was found for 19 patients, resulting in a patient-to-drug matching rate of 31% (19/61 patients). We identified hits to vistusertib (n = 16), capivasertib (n = 5), selumetinib (n = 3), gefitinib (n = 5), and gedatolisib (n = 2) (Figure 2B). Of 19 patients eligible for study treatment (i.e. in vitro drug sensitivity), 6 patients started treatment based on the organoid assay. The other 13 patients did not start treatment mostly due to clinical deterioration under SOC (n = 6). For three patients, we identified vistusertib as a potential hit, but when the drug did not show superiority to everolimus in a randomized phase II study, AstraZeneca deprioritized further development of this drug.29 We therefore also decided against further use of this drug. Of two patients enrolled in other phase I studies (n = 2), one patient had an identical target of intermittent and experimental treatment (panitumumab/gefitinib targeting EGFR; n = 1) and another patient did not receive follow-up (Figure 2B).
Analogous to our test cohort, we also analyzed genotype drug-response relationships in the patient cohort. Drug sensitivity of P11 and P13 to gefitinib coincided with EGFwt status, as found earlier in our test cohort (Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2021.100103; Figure 1). In contrast, all other organoids harbored kinase gain-of-function mutations downstream of EGFR and did not respond to gefitinib in vitro (P12 harbored an ARAFD24N VUS located outside the kinase domain; Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2021.100103). We also assessed the effect of KRAS mutations and response to selumetinib, as in our test cohort (Figure 1). In line with (pre)clinical studies and the test cohort, all except one KRAS mutant culture in our patient cohort did not respond to selumetinib (Figure S2B).27,28 The exception, P7, harbored an oncogenic KRASG13D mutation in the context of an EGFRG724V VUS, located in the kinase domain. These complex genotype-drug relationships uncovered by organoid drug screening can therefore provide data complementary to DNA sequencing.
P1-3 started treatment with vistusertib and P4-6 with capivasertib after they underwent a second post-SOC biopsy for organoid culture. Treatment-related and non-treatment-related adverse effects are described in Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2021.100103. Post-SOC organoid cultures of four patients were successfully generated and could be re-screened to control for potential interference with drug sensitivity by intermittent SOC treatment. P3 retained sensitivity to vistusertib, and P4 and P5 remained sensitive to capivasertib (Figure 2D). P6 had a marginal decrease in sensitivity to capivasertib (GR = 0.106 post-SOC versus GR = 0.05 pre-SOC; Supplementary Figure S2C, available at https://doi.org/10.1016/j.esmoop.2021.100103).
Of the six patients that started treatment, P1 had a radiologically stable disease at the first evaluation but presented with neurological symptoms. The presence of symptomatic brain metastases was subsequently radiologically confirmed (Table 2 and Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2021.100103). Also, cancer embryonic antigen (CEA; a surrogate tumor marker) levels did increase during treatment. P2 showed disease stabilization of both tumor masses at the first response evaluation but had progressive disease 2 months later. P3 (vistusertib) and P6 (capivasertib) did not reach first evaluation due to clinical progression (symptomatic brain metastasis and intestinal obstruction with peritoneal carcinomatosis, which was unrelated to study treatment), despite a substantial decrease in CEA levels during treatment of P6. Both P4 and P5 showed disease progression at the first evaluation (Table 2). Notably, both vistusertib and capivasertib do not cross the blood-brain barrier. In conclusion, we did not observe durable clinical responses for organoid-informed treatment decisions.
Table 2.
Prospective, organoid-informed treatment of patients in the SENSOR trial
| Drug | Patient | Baseline |
First response evaluation |
End of treatment |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Sum of RECIST lesions in mm | Biopsied lesion in mm | CEA (ng/ml) | Sum of RECIST lesions in mm (% relative to baseline) | Biopsied lesion in mm (% relative to baseline) | CEA (ng/ml) | Days after start treatment | Reason for end of treatment | ||
| Vistusertib | P1 | 101 | 30 | 253 | 107 (+6%) | 29 (−3%) | 1165 | 58 | New symptomatic brain metastases |
| P2 | 110 | 21 | 648 | 123 (+12%) | 22 (+5%) | 727 | 107 | Progressive disease | |
| 132 (+20%)a | 26 (+24%)a | 1119a | |||||||
| P3 | 300 | 9 | 923 | 351 (+17%) | 11 (+22%) | 652 | 57 | New lesions at response evaluation | |
| Capivasertib | P4 | 67 | 91 (+35%) | 52 | Progressive disease | ||||
| P5 | 46 | 22 | 47 (+2%) | 25 (+14%) | 20 | Clinical progression | |||
| P6 | 234 | 81 | 359 | 245 (+5%) | 89 (+10%) | 514 | 20 | New symptomatic brain metastases | |
Response to experimental treatment was evaluated using RECIST 1.1 every 2 months. When available, the response in the biopsied lesion and levels of CEA (in ng/ml) were also recorded. Clinical disease progression was observed for patients P5 and P6 before the first response evaluation. The tumor measurements and CEA levels at first response evaluation correspond with the CT scan and blood withdrawal for clinical purposes, on day 20 after start of treatment of both patients.
CEA, cancer embryonic antigen; CT, computed tomography.
Second evaluation.
Because we observed limited clinical benefit in the six patients that underwent treatment, we decided to perform an unplanned interim analysis to evaluate the feasibility of organoid-guided precision medicine in this patient population. We concluded there was substantial dropout (55/61 patients), mostly because of unsuccessful cultures (n = 23) and disease progression (n = 11). Together with the absence of objective responses so far, we decided there was insufficient basis to continue the trial.
Discussion
In this feasibility study we did not succeed in establishing organoids as a means to improve response rates to off-label or investigational drugs. The major factor that prohibited testing of organoids as a predictive tool for every individual patient was the culture success of 57%, which was slightly lower than our and others' prior success rate of 63%-71%.9,11,13 This was also the main reason we did not pursue extension of the study to non-small-cell lung cancer (NSCLC) as stated in our initial study protocol. We found that, among other difficulties, the success rate of establishment for NSCLC is only 17%.30
In our study we attempted to establish cultures of all included patients because patients had exhausted all other SOC treatment opportunities. As a consequence, we lowered the bar for biopsies that were used to initiate an organoid culture compared with our other studies where we noted that the quantity of tumor material (number and size of the biopsies and tumor/epithelial cell content) were important indicators of culture success.11,31,32 Pre-selection of biopsies that contain sufficient tumor cells (cellularity 2+) by a pathologist, as done by us and others (Vlachogiannis et al.), is an important logistical and technical lesson learned from our prospective intervention clinical study.11 However, even when we include this additional step, the current success rates of 57%-71% for the establishment of mCRC organoids from biopsies remains a challenge.9,11,13 We therefore underpin that further optimization of culture conditions from biopsies of metastatic lesions is critical for the clinical applicability of organoids. As an alternative to biopsies with high cell content, other studies demonstrated that tumor resections can be a source of material, as this increases the success rate and shortens time to establishment.14,15,32 However, patients with metastatic cancer generally do not undergo this type of surgery.
Secondly, organoids were expanded and subjected to the drug screen parallel to the last line of SOC treatment. Unfortunately, given the advanced stages of the disease of our patients, 11 patients deteriorated during SOC treatment or were considered to have progressive disease at the first evaluation after 8-10 weeks. Stricter patient selection at time of inclusion (i.e. earlier stages of treatment, better performance status) would potentially improve the likelihood for patients to start experimental treatment. An important finding is that there were no major shifts in drug sensitivity after SOC treatment in four paired biopsies, suggesting that the intermittent (chemo)therapy in our trial design does not alter drug sensitivity and is not an underlying factor for the lack of responses. This design can provide additional testing time and confirms our initial assumption.
Ideally we would like to test a large panel of (experimental) drugs and select the ones with the strongest propensity to kill the organoids. However, the larger the number of drugs or combinations thereof, the more organoids are needed for adequate testing which can further delay treatment. Here, we have chosen to select a limited set of drugs and defined activity on the basis of single-agent activity. With improved culture conditions (leading to more organoids) it may become feasible to test a broader panel of drugs and also include combinatorial approaches to increase the chance of clinical benefit for patients.4 We foresee that additional DNA sequencing and other (pre)clinical data, such as organoids established from clinical responders and/or preliminary evidence of activity in basket trials, can significantly contribute to more streamlined design of drug (combination) panels and robust prediction models.2,11
A more streamlined design of the drug panel can also be achieved by pre-screening a pilot cohort with an expanded number of drugs or drug combinations. Our drug panel contained both drugs for which some activity was expected (vistusertib, selumetinib, gefitinib), while for others, there was limited clinical evidence available for mCRC (palbociclib, glasdegib). This set-up allowed us to test whether organoids can be leveraged to identify potential treatments beyond the expected EGF/insulin growth-factor (IGF)-targeting drugs. Unfortunately, we observed little activity of these drugs in the pilot cohort and in the patient cohort. Therefore, it may be beneficial to test an expanded number of drugs or drug combinations in the pilot cohort and continue with a set that demonstrates activity in a reasonable fraction of cultures (>10%) to increase the odds of finding active drugs for enrolled patients. It could also allow for a more stringent cut-off, as the number of options will be larger and treating physicians can propose the most promising treatment.
The employed threshold for in vitro hits (0.1) is another limitation of our study. It still allows (very) limited in vitro growth, which could explain the ongoing, slow growth observed in most patients (Table 1). Using a more stringent cut-off could potentially improve the specificity of organoid drug screening. Also, assays that measure cell death or apoptosis can be considered, as these are considered to be more conservative criteria of in vitro response. It should be noted, though, that it is not clear whether the outcomes of these assays correlate with clinical responses of patients, which we and others have demonstrated for CellTiter-Glo.10, 11, 12, 13, 14, 15
The limited number of patients does not allow firm conclusions on whether organoid in vitro sensitivity predicts clinical response in vivo. P2 showed disease stabilization at the first evaluation, but this clinical benefit was not durable; as for the stable disease of P1, no clinical responses (e.g. partial or complete responses) were observed. Surprisingly, even when organoids predicted a strong effect, for instance, P4 and the FDA-approved drug capivasertib (GR = −0.35; Supplementary Figure S2C, available at https://doi.org/10.1016/j.esmoop.2021.100103), the patient progressed rapidly. Although still anecdotal, it suggests organoids might not be universally predictive as we suggested earlier and warrants careful design of drug panels.13
In conclusion, the culture success rate, the ineligibility of patients due to clinical deterioration during last line SOC treatment and the limited clinical efficacy of the proposed treatments are challenges for the use of organoids for precision medicine. Our experiences may pave the way for future trials in this area of precision medicine.
Acknowledgements
We acknowledge all members of the Voest lab, Rene Bernards, Daniel Peeper, Piet Borst, Hans Clevers, Silvia Boj, Marc van de Wetering, and Rene Overmeer for useful discussions and advice. We acknowledge Stef van Lieshout and Ewart de Bruijn for performing WGS and profiling of organoids and Suzanne van der Kolk, Judith Westra, and Sandra Visser for help with patient inclusion. We thank all The Netherlands Cancer Institute core facilities, with special thanks to Robotics and Screening, Molecular Pathology, and Cryogenic storage for their contribution and help. We also acknowledge the NKI Department of Pharmacy and Pharmacology for the logistics and Manon Winter (Pfizer), Colin Glover and Peter Mortimer (both AstraZeneca) for their input and discussion. Furthermore, we thank all participating centers, medical oncologists, and patients for contributing to the study. R-Spondin-producing cells were a kind gift from C. Kuo (Stanford).
Funding
This work was supported by grants from the Koningin Wilhelmina Fonds [grant numbers NKI2015-7732, HUBR2014-7006] to EEV and the NWO gravitation program (2012-2022) to EEV. Vistusertib, capivasertib, selumetinib, and gefitinib for in vitro and clinical use were provided by AstraZeneca. Palbociclib, axitinib, gedatolisib, and glasdegib were provided by Pfizer.
Disclosure
EEV is the medical director of The Netherlands Cancer Institute and legally responsible for all contracts with AstraZeneca and Pfizer. AstraZeneca and Pfizer provided compounds for drug screening and patient treatment. All other authors have declared no conflicts of interest.
Data sharing
Reagents used under material transfer agreement (MTA) are Wnt-3a, Noggin (Hans Clevers, Hubrecht Institute), and R-spondin-1 producer lines (Calvin Kuo, Stanford). DNA sequencing data for T1, T5, T6, and T8 is published in the manuscript by Wetering et al. Cell, 2015.8 WGS data is published and deposited by Priestley et al. Nature, 2019.33 Deposition of targeted-sequencing data and distribution of organoids and deposition of DNA sequencing data in publicly available databases are regulated by the informed consent that participants to this study signed. All materials and data on a per-patient level can be obtained through the Institutional Review Board of The Netherlands Cancer Institute (IRB@nki.nl). All other materials used in this study are freely or commercially available.
Supplementary data
References
- 1.Le Tourneau C., Delord J.P., Goncalves A. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 2.van der Velden D.L., Hoes L.R., van der Wijngaart H. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature. 2019;574(7776):127–131. doi: 10.1038/s41586-019-1600-x. [DOI] [PubMed] [Google Scholar]
- 3.Friedman A.A., Letai A., Fisher D.E., Flaherty K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15(12):747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sicklick J.K., Kato S., Okamura R. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019;25(5):744–750. doi: 10.1038/s41591-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman D.M., Taylor B.S., Baselga J. Implementing genome-driven oncology. Cell. 2017;168(4):584–599. doi: 10.1016/j.cell.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letai A. Functional precision cancer medicine – moving beyond pure genomics. Nat Med. 2017;23(9):1028–1035. doi: 10.1038/nm.4389. [DOI] [PubMed] [Google Scholar]
- 7.Sato T., Stange D.E., Ferrante M. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 8.van de Wetering M., Francies H.E., Francis J.M. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weeber F., van de Wetering M., Hoogstraat M. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112(43):13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs N., de Ligt J., Kopper O. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1-2):373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Vlachogiannis G., Hedayat S., Vatsiou A. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (New York, NY) 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiriac H., Belleau P., Engle D.D. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8(9):1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ooft S.N., Weeber F., Dijkstra K.K. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11(513):eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y., Xu X., Yang L. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26(1):17–26.e6. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Ganesh K., Wu C., O'Rourke K.P. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019;25(10):1607–1614. doi: 10.1038/s41591-019-0584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiriac H., Plenker D., Baker L.A., Tuveson D.A. Organoid models for translational pancreatic cancer research. Curr Opin Genet Dev. 2019;54:7–11. doi: 10.1016/j.gde.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenhauer E.A., Therasse P., Bogaerts J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer (Oxford, England: 1990) 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Dijkstra K.K., Cattaneo C.M., Weeber F. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174(6):1586–1598.e12. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang-Chu M.M., Yu M., Haverty P.M. Human biosample authentication using the high-throughput, cost-effective SNPtrace(TM) system. PLoS One. 2015;10(2):e0116218. doi: 10.1371/journal.pone.0116218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu B., Dean E., Puglisi M. First-in-human pharmacokinetic and pharmacodynamic study of the dual m-TORC 1/2 inhibitor AZD2014. Clin Cancer Res. 2015;21(15):3412–3419. doi: 10.1158/1078-0432.CCR-14-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro G.I., Bell-McGuinn K.M., Molina J.R. First-in-human study of PF-05212384 (PKI-587), a small-molecule, intravenous, dual inhibitor of PI3K and mTOR in patients with advanced cancer. Clin Cancer Res. 2015;21(8):1888–1895. doi: 10.1158/1078-0432.CCR-14-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerji U., Camidge D.R., Verheul H.M. The first-in-human study of the hydrogen sulfate (Hyd-sulfate) capsule of the MEK1/2 inhibitor AZD6244 (ARRY-142886): a phase I open-label multicenter trial in patients with advanced cancer. Clin Cancer Res. 2010;16(5):1613–1623. doi: 10.1158/1078-0432.CCR-09-2483. [DOI] [PubMed] [Google Scholar]
- 23.Wagner A.J., Messersmith W.A., Shaik M.N. A phase I study of PF-04449913, an oral hedgehog inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2015;21(5):1044–1051. doi: 10.1158/1078-0432.CCR-14-1116. [DOI] [PubMed] [Google Scholar]
- 24.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hafner M., Niepel M., Chung M., Sorger P.K. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 2016;13(6):521–527. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahabreh I.J., Terasawa T., Castaldi P.J., Trikalinos T.A. Systematic review: anti-epidermal growth factor receptor treatment effect modification by KRAS mutations in advanced colorectal cancer. Ann Intern Med. 2011;154(1):37–49. doi: 10.7326/0003-4819-154-1-201101040-00006. [DOI] [PubMed] [Google Scholar]
- 27.Janne P.A., van den Heuvel M.M., Barlesi F. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA. 2017;317(18):1844–1853. doi: 10.1001/jama.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C., Hobor S., Bertotti A. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7(1):86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 29.Schmid P., Zaiss M., Harper-Wynne C. Fulvestrant plus vistusertib vs fulvestrant plus everolimus vs fulvestrant alone for women with hormone receptor-positive metastatic breast cancer: the MANTA phase 2 randomized clinical trial. JAMA Oncol. 2019;5(11):1556–1563. doi: 10.1001/jamaoncol.2019.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dijkstra K.K., Monkhorst K., Schipper L.J. Challenges in establishing pure lung cancer organoids limit their utility for personalized medicine. Cell Rep. 2020;31(5):107588. doi: 10.1016/j.celrep.2020.107588. [DOI] [PubMed] [Google Scholar]
- 31.Kodack D.P., Farago A.F., Dastur A. Primary patient-derived cancer cells and their potential for personalized cancer patient care. Cell Rep. 2017;21(11):3298–3309. doi: 10.1016/j.celrep.2017.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M., Mun H., Sung C.O. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nature Commun. 2019;10(1):3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priestley P., Baber J., Lolkema M.P. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575(7781):210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.