Summary
Tissue imaging in 3D using visible light is limited and various clearing techniques were developed to increase imaging depth, but none provides universal solution for all tissues at all developmental stages. In this review, we focus on different tissue clearing methods for 3D imaging of heart and vasculature, based on chemical composition (solvent-based, simple immersion, hyperhydration, and hydrogel embedding techniques). We discuss in detail compatibility of various tissue clearing techniques with visualization methods: fluorescence preservation, immunohistochemistry, nuclear staining, and fluorescent dyes vascular perfusion. We also discuss myocardium visualization using autofluorescence, tissue shrinking, and expansion. Then we overview imaging methods used to study cardiovascular system and live imaging. We discuss heart and vessels segmentation methods and image analysis. The review covers the whole process of cardiovascular system 3D imaging, starting from tissue clearing and its compatibility with various visualization methods to the types of imaging methods and resulting image analysis.
Subject areas: Imaging Methods in Chemistry, Optical Imaging, Biology Experimental Methods
Imaging Methods in Chemistry; Optical Imaging; Biology Experimental Methods
Introduction
Recent advances in tissue clearing technology have enabled rapid and comprehensive cellular analyses of entire organs in the whole-mount preparations by a combination of state-of-the-art technologies of optical imaging and image processing. All tissue clearing methods seek to homogenize the refractive index (RI) of a sample by removing, replacing, or modifying some of its components. Incorporating a tissue clearing step in standard immunostaining protocols could open the door to routine high-resolution imaging of a few hundred microns deeper into samples with confocal microscopes and objectives that are quite commonly available in research environment around the world.
Determining the best method for a given application is difficult owing to proliferation of new techniques with trade-offs between speed, cost, complexity, conservation of protein-based fluorescence, compatibility with immunostaining, and other criteria (Ariel, 2017). The plethora of clearing protocols suggests that it is unlikely there will be a universal one for all applications, so trying several might be necessary to achieve the best results for particular samples. Most tissue cleaning techniques are optimized for the nervous tissue, which differs significantly from the heart specimens. Brain tissue has high fat content and relative lack of autofluorescence unlike the heart tissue. Similar to the central nervous system, the cardiovascular system represents a complex three-dimensional (3D) structure; thus, availability of a non-destructive method of high-resolution 3D imaging without specimen distortion is highly desirable. The possibility to track selected structures (myocyte bundles, vessels, nerves) in 3D is beneficial for our understanding of normal and pathological anatomy.
Some selected methods can assist visualization of, for example, the vasculature, the structure of the myocardium, GFP-tagged (green fluorescent protein-tagged) ion channels, and proteins labeled by immunochemistry (Matryba et al., 2019). A variety of methods exists to clear tissue samples, including hydrophobic, hydrophilic, and hydrogel-based tissue clearing methods (Richardson and Lichtman, 2015). Tissue clearing and staining (e.g., with fluorescent dyes, fluorescently labeled antibodies, or nucleic acid probes) are usually conducted with the following steps: (1) fixation, (2) permeabilization, (3) staining (if applicable), (4) bleaching (optional), (5) adjusting of RI using tissue clearing, and (f) RI matching, and imaging.
Tissue clearing methods optimized for heart and vessel imaging
Here we summarize the tissue clearing methods, which were published for use on the developing cardiovascular system. These methods were tested and used for visualization of embryonic heart and vasculature.
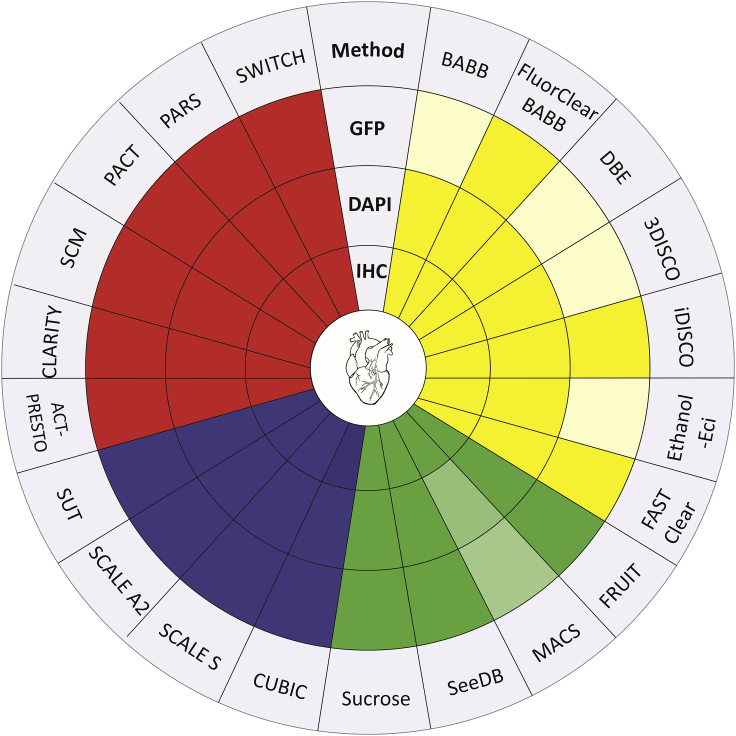
Tissue clearing techniques can be used for various purposes and, therefore, can be divided into four main categories: solvent-based, simple immersion, hyperhydration, and hydrogel embedding (Richardson and Lichtman, 2015; Silvestri et al., 2016) (Table 1). Each of these distinct methods has advantages and disadvantages based on many independent factors. When planning an experiment, it is necessary to consider limitations of each method (see below) and determine the most efficient and effective approach for the tissue of interest (Figure 1).
Table 1.
Tissue clearing methods used on heart tissue—abbreviations and names
| Method name | Full name of the method | Citation | |
|---|---|---|---|
| Solvent based | BABB | Benzyl-Alcohol:Benzyl-Benzoate | Zucker et al., (1998) |
| FluorClear BABB | Fluorophore preservation Clearing Benzyl-Alcohol:Benzyl-Benzoate | Schwarz et al. (2015); Foster et al. (2019) | |
| DBE | Phenylmethoxymethylbenzene (Dibenzyl Ether) | Becker et al., (2012) | |
| 3DISCO | Three-Dimensional Imaging of Solvent-Cleared Organs | Molbay et al., (2021) | |
| iDISCO | Immunolabeling-enabled three-Dimensional Imaging of Solvent-Cleared Organs | Renier et al., (2016) | |
| Ethanol-Eci | Ethanol-ethyl-3-phenylprop-2-enoate (Ethyl Cinnamate [ECi]) | Klingberg et al. (2017) | |
| FASTClear | Free of Acrylamide SDS-based Tissue Clearing | Liu et al., (2017) | |
| Simple immersion | FRUIT | Fructose and Urea | Hou et al., (2015) |
| MACS | MXDA-based Aqueous Clearing System | Zhu et al. (2020) | |
| SeeDB | See Deep Brain | Ke et al. (2013); Murray et al. (2015) | |
| Sucrose | Sucrose | Tsai et al., (2009) | |
| Hyperhydratation | CUBIC | Clear, Unobstructed Brain Imaging Cocktails and computational analysis | Susaki et al. (2014); Susaki et al., (2015) |
| SCALE S | SCALE—name, not abbreviation | Hama et al. (2015) | |
| SCALE A2 | SCALE—name, not abbreviation | Hama et al. (2015) | |
| SUT | Scheme Update on tissue Transparency | Wang et al. (2018); Matryba et al. (2019) | |
| Hydrogel embedding | ACT-PRESTO | Active Clarity Technique-Pressure Related Efficient and Stable Transfer of macromolecules into Organs technique | Lee et al. (2016) |
| Clarity | Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/in situ-hybridization-compatible Tissue hYdrogel | Chung et al. (2013); Tomer et al. (2014) | |
| SCM | Simplified Clarity Method | Sung et al. (2016); Matryba et al. (2019) | |
| PACT | PAssive Clarity Technique | Yang et al., (2014); Neckel et al., (2016) | |
| PARS | Perfusion-assisted Agent Release in Situ | Yang et al., (2014) | |
| SWITCH | System-Wide control of Interaction Time and kinetics of Chemicals | Murray et al. (2015) |
Figure 1.
Overview of tissue clearing methods used on heart
Tissue clearing methods are divided into four groups based on their clearing principle. Solvent-based (yellow), simple immersion (green), hyperhydration (purple), and hydrogel embedding (red). Applicability of each method is marked in a dark color: GFP, method preserves natural GFP fluorescence; IHC, method can be used with immunohistochemistry; DAPI, method is suitable for DAPI/Hoechst nuclei staining. Light color, method is not suitable for this methodology. To summarize, most of the methods used on heart clearing are applicable for many techniques.
Previous reviews addressed various aspects of tissue clearing methods. Silvestri et al. (2016) reviewed tissue clearing methods and their applications for various methods of analysis. The review by Vigouroux et al. (2017) addressed more practical aspects and hands-on methodology in tissue clearing. Gomez-Gaviro et al. (2020) reviewed in detail the application of various tissue clearing methods on various types of tissue. However, the main focus of tissue clearing methods is in neuroscience and brain development. The majority of original papers and reviews in tissue clearing as well as 3D brain CUBIC-atlas come from this field (Murakami et al., 2018; Tian et al., 2021; Ueda et al., 2020; Vigouroux et al., 2017). However, the number of publications discussing tissue clearing methods on heart is limited (Kolesova et al., 2016; Nehrhoff et al., 2016; Yokoyama et al., 2017), even though the field of heart and vasculature development has its avid audience.
Each method is known mainly by its abbreviation; therefore, an overview of the abbreviations and methods published for cardiovascular tissue is summarized in Table 1. Table 2 summarizes the methods used for cardiovascular tissue clearing, providing the key ingredients of each method; length of the clearing process, which is an important parameter to consider when choosing a particular tissue clearing method; and the main publication describing protocol modifications for heart and vascular tissues.
Table 2.
Heart and heart vasculature tissue clearing methods
| Method name | Key components | Clearing time | Protocol modification for heart or heart vasculature | |
|---|---|---|---|---|
| Solvent based | BABB | Benzoic Acid/Benzyl Benzoate | Days | Miller et al. (2005); Kolesova et al. (2016); Ivins et al., 2016 |
| FluorClear BABB | Tert-butanol/BABB | Hours | Epah et al. (2018) | |
| DBE | DiBenzylEther | Hours | Kolesova at al.,2016 | |
| 3DISCO | Dichlormethane/DBE | Hours-Days | Belle et al. (2017); Epah et al. (2018) | |
| iDISCO | Dichlormethane/DBE | Hours-Days | Belle et al. (2017); Epah et al. (2018), Goodyer et al. (2019) | |
| Ethanol-Eci | Ethyl-3-phenylprop-2-enoate | Days | Klingberg et al. (2017) | |
| FASTClear | THF/DBE | 45 min | Perbellini et al. (2017) | |
| Simple immersion | FRUIT | Fructose/Triglycerol/Urea | Days | Xu et al. (2019a), 2019b |
| MACS | MXDA/Sorbitol | Days | Xu et al. (2019a), 2019b; Zhu et al. (2020) | |
| SeeDB | Fructose/Thioglycerol | Week | Tainaka et al. (2014) | |
| Sucrose | Sucrose | 1 day | Tsai et al., (2009) | |
| Hyperhydratation | CUBIC | 4M Urea/50% Sucrose | Days | Kolesova et al. (2016); Nehrhoff et al. (2016); Tainaka et al. (2014) |
| SCALE S | 4M Urea/Sorbitol | Days-weeks | Li et al. (2016); Matryba et al. (2019) | |
| SCALE A2 | 4M Urea/10% Glycerol | Weeks | Kolesova et al. (2016) | |
| SUT | Urea/SDS | Days | Wang et al. (2018); Matryba et al. (2019) | |
| Hydrogel embedding | ACT-PRESTO | Hydrogel/SDS | Days | Kim et al. (2016) |
| Clarity | FocusClear/80% Glycerol | Days | Kolesova et al. (2016) | |
| SCM | Hydrogel | Weeks | Sung et al. (2016); Matryba et al. (2019) | |
| PACT | Hydrogel/SDS | Days-Weeks | Treweek et al. (2015), Xu et al. (2019a), 2019b | |
| PARS | Hydrogel/SDS | Days | Treweek et al. (2015) | |
| SWITCH | SDS/Thioglycerol | Weeks | Murray et al. (2015) |
Solvent-based clearing techniques such as BABB and DISCO are the most popular methods of the cardiac optical clearing. These methods are relatively inexpensive, and samples can be cleared in a matter of days. Solvent-based methods are commonly used together with antibody staining. Whole-mount immunohistochemical staining followed by clearing with BABB was used to visualize 3D myocardial or vascular architecture (Ivins et al., 2016; Miller et al., 2005). Limiting factors for the BABB method are mainly incomplete clearing (for thick/older hearts) and specimen autofluorescence. The main inconvenience is that BABB does not preserve GFP fluorescence. Therefore, FluoClearBABB clearing was developed. The main difference is that either 1-propanol or tert-butanol is used during dehydration and a basic pH is maintained throughout the procedure. This method minimizes optical distortions and preserves the majority of GFP fluorescence (Schwarz et al., 2015). It is also effective at clearing heart tissue specimens while preserving fluorescent immunolabeling (Epah et al., 2018). However, the GFP fluorescence of the specimen, when using THF-DBE/dichlormethane or FluoClearBABB lasts only a few days (Richardson and Lichtman, 2015). To summarize, BABB and THF methods are used mainly to study cardiovascular system marked with antibodies, but its use for studying GFP fluorescence is time limited.
The next commonly used method for heart tissue clearing is DISCO. DISCO can be successfully combined with immunohistochemistry and 3DISCO was used to study vasculature development of the human heart by immunostaining for smooth-muscle-specific alpha-actin (SMA) (Belle et al., 2017; Epah et al., 2018). iDISCO + tissue clearing was used for 3D reconstruction of the mouse heart conduction system (Goodyer et al., 2019). Like BABB, DISCO does not preserve GFP fluorescence or preserves the fluorescence only for a very short time. In 3DISCO, a rapid decline in endogenous fluorescence signals during the tissue clearing and storage procedure with a half-life of approximately 1–2 days has been reported (Pan et al., 2016). To address this limitation, a recently developed method, FDISCO, achieved a higher level of fluorescence preservation by temperature and pH condition adjustments (Qi et al., 2019).
The FASTClear protocol was specifically designed for cardiac tissue. FASTClear does not induce structural or microstructural distortion and can be combined with immunostaining to identify the micro- and macrovascular networks (Perbellini et al., 2017). Ethanol-Ethyl-3-phenylprop-2-enoate (Ethyl Cinnamate or Ethanol-ECi) clearing was also used on heart tissue and is suitable for immunohistochemistry (CD31 antibody) but was also reported to preserve EYFP (Enhanced Yellow Fluorescent Protein) fluorescence (Klingberg et al., 2017). For summary, see Figure 1.
Simple immersion tissue clearing
These conventional tissue clearing reagents are mostly based on sugars and have been adapted to some of the clearing protocols. The best known is the SeeDB protocol, which maintains GFP fluorescence and also fluorescence from immunohistochemical staining, as well as heart sample volume (Susaki et al., 2014). One of the SeeDB-derived clearing methods is termed FRUIT, and it utilizes a cocktail of fructose and urea. The FRUIT solution has low viscosity, and may be used for clearing via arterial perfusion, which is impossible to achieve with a saturated fructose solution. Arterial perfusion with tissue clearing solution significantly improves the tissue clearing and shortens tissue clearing time (Hou et al., 2015). However, arterial perfusion with tissue clearing reagents has not been performed yet on heart tissue, and we assume that arterial perfusion would improve tissue clearing especially in older hearts. The FRUIT method was also applied to clear intact hearts (Xu et al., 2019b), but in comparison with the other tissue clearing methods (CUBIC and PACT) it can achieve only partial tissue transparency (similar to SCALE).
Another advantage of FRUIT compared with SeeDB is its compatibility with lipophilic tracers (DiI) (Hou et al., 2015), which was demonstrated on the brain. Another method from this group that was successfully used together with lipophilic dyes is MACS, a rapid aqueous clearing method based on m-xylylenediamine (MXDA). This method achieved high transparency of intact organs with a short incubation time and showed ideal compatibility with multiple probes, especially lipophilic dyes. Besides being used for brain and spleen, MACS tissue clearing was used also to acquire heart vasculature images labeled with DiI (Zhu et al., 2020). From few studies it is confirmed that these methods are suitable for cardiovascular development studies; however, they are not commonly used.
Hyperhydratation
These common tissue clearing methods are based on hydrophilic substances, and they expand the tissue volume. They are commonly used for heart tissue clearing as they preserve GFP fluorescence and are compatible with immunostaining. The SCALE is in primarily an aqueous solution of urea, which is compatible with immunostaining (Azaripour et al., 2016) and preserves the GFP signal (Hama et al., 2015). Several improvements of the SCALE method are available: ScaleS, ScaleA2. However, only few studies have used SCALE on the heart tissue, and details about the difference among SCALE methods on heart tissue have yet to be further explored. SCALE clearing was used to clear superficial layers of the embryonic heart (Kolesova et al., 2016) and to perform single cell tracing and analysis of the whole heart during early cardiac development (Li et al., 2016). A drawback of this method is the long incubation time up to months (Keller and Dodt, 2012), or with shorter incubation, tissue is not cleared in its whole depth (Kolesova et al., 2016). CUBIC is a modification of SCALE. It contains only nontoxic water-soluble chemicals (Tainaka et al., 2014); however, sale of one of its components (Quadrol) is monitored as it is a component for explosive fabrication and therefore can be challenging to obtain (authors' experience). The method has been commonly used for heart tissue clearing (Kolesova et al., 2016; Nehrhoff et al., 2016; Yokoyama et al., 2017). The main advantage of CUBIC over the other clearing methods is that CUBIC constituents (Quadrol) effectively elute endogenous chromophores (mainly heme) and thus reduce myocardial and blood autofluorescence (Susaki et al., 2014; Tainaka et al., 2014). The CUBIC tissue clearing protocol can be used for embryonic and adult mouse hearts (Kolesova et al., 2016) and enables one to observe in detail structures such as coronary arteries, fine structure of the ventricular trabeculae, and pectinate muscles. According to the original description, CUBIC is also suitable for tissue clearing after immunolabeling and preserves the fluorescent antibody signal (Tainaka et al., 2014). The main disadvantage is still a long clearing period for larger samples. This significant limitation was recently overcome with a modification named SUT (Wang et al., 2018). SUT is an effective method for clearing and imaging of cardiac microstructures in whole hearts from different species. Wang et al. (2018) accelerated the protocol even further by using electrophoresis to shorten the time of antibody penetration.
Hydrogel embedding
These tissue clearing methods are based on preserving the tissue structure by embedding the sample in hydrogel and subsequent removal of lipids. CLARITY (Chung et al., 2013) is based on the concept that lipids have to be removed to clear tissue (using acrylamide-based hydrogels built from within, and linked to, the tissue). CLARITY is commonly used to clear many organs (Lee et al., 2014) including embryonic and adult heart tissue (Kolesova et al., 2016; Sereti et al., 2018) as it preserves GFP and other types of fluorescence. The optimized CLARITY protocol termed a Simplified Clarity Method (SCM) was applied also on heart samples and achieved full transparency of hearts within similar time as CLARITY. The speed of the clearing can be accelerated by increasing the temperature to 42°C (Sung et al., 2016). CLARITY also has been modified as PACT (passive CLARITY technique) and PARS (Perfusion-Assisted agent Release in Situ (Treweek et al., 2015) or SWITCH (System-Wide control of Interaction Time and kinetics of CHemical) (Murray et al., 2015). PACT-PARS uses tissue-hydrogel hybrids to stabilize tissue biomolecules during selective lipid extraction, resulting in enhanced clearing efficiency. The PACT tissue clearing method is suitable for heart tissue and preserves the GFP fluorescence (authors' unpublished data). SWITCH accelerates clearing using high temperatures with the addition of sodium sulfite as an antibrowning agent, and it synchronizes labeling reactions. However, these methods are more expensive and also very slow. The rapid and highly reproducible ACT-PRESTO (Active Clarity Technique-Pressure Related Efficient and Stable Transfer of macromolecules into Organs) method clears tissues or the whole body within one day while preserving tissue architecture and GFP fluorescence. Moreover, ACT-PRESTO is compatible with conventional immunolabeling methods and expedites antibody penetration into thick specimens by applying pressure (Lee et al., 2016). ACT-PRESTO was successfully applied to clearing mouse heart tissue (Kim et al., 2016). For an overview see Figure 1.
Tissue clearing and GFP preservation in the heart
The fast development and usage of tissue clearing techniques relates to the rapid development of imaging methods, such as confocal microscopy and light sheet microscopy. Together these methods can be used to reconstruct the 3D anatomy of the tissue (Costantini et al., 2019).
One of the main model organisms in biological research is the mouse model, with its wide array of possible genetical modification including insertion of fluorescently labeled proteins (e.g., GFP) as reporter genes. The first developed tissue clearing methods used hydrophobic compounds, which dehydrate the tissue and dehydration quench the introduced fluorescent proteins (i.e., GFP). To avoid this phenomenon, various hydrophilic (water-based) clearing solutions were developed. However, water-based methods have had lower tissue clearing performance (Silvestri et al., 2016), while preserving the fluorescence. Figures 2C and 2D illustrate preserving GFP fluorescence in embryonic heart of different stages (ED10.5 and ED14.5) by the CUBIC tissue clearing method. Besides using a hydrophilic clearing solution, another approach to maintain fluorescence is a tissue transformation method involving embedding the tissue in acrylamide gel—CLARITY (Chung et al., 2013).
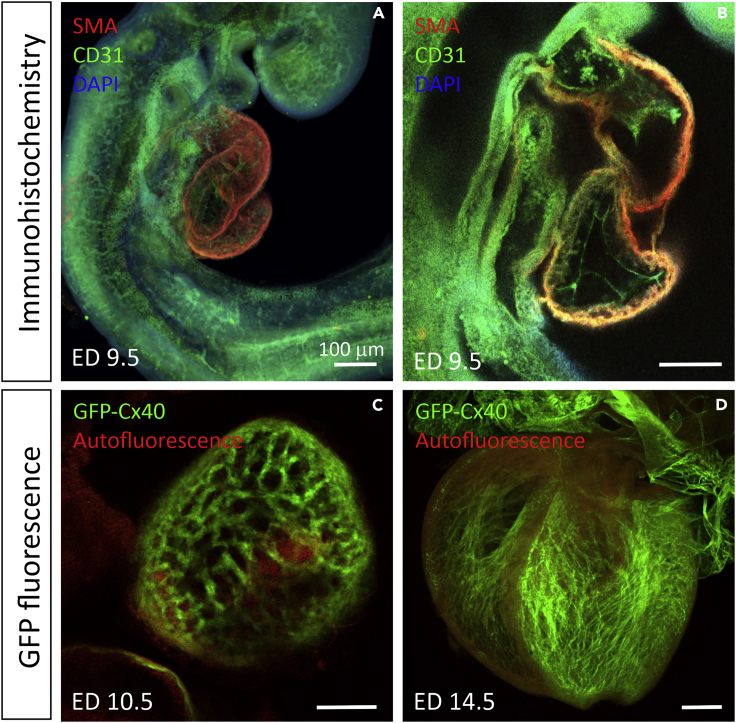
Figure 2.
Examples of immunostaining and preserving GFP fluorescence with tissue clearing on the developing mouse heart
(A and B) Immunohistochemistry combined with BABB on ED 9.5 mouse embryo, smooth muscle actin antibody (SMA) in red labeling the myocardium, CD31 (PECAM-1) in green staining the endocardium (B), and DAPI nuclear staining (not very distinct at this low magnification) in blue.
(C and D) Preservation of natural GFP fluorescence with CUBIC tissue clearing on ED 10.5 (C) and ED 14.5 (D) in mouse Connexin 40 - GFP (Cx40-GFP) embryo hearts with superimposed autofluorescence in red. Ventricular trabeculae and atria are positive for Cx40 at these stages. Autofluorescent blood is present in the ventricles (C). All images were captured with confocal microscopy. Scale bar represents 100 μm in all figures.
Detailed analysis of the fluorescence preservation by various clearing methods was performed by Xu and colleagues (Xu et al., 2019b ) on intestinal tissues. They confirmed that most aqueous clearing methods performed better in fluorescence preservation than organic solvent-based ones. On measuring signal to background ratio after tissue clearing, they found that the best method to preserve fluorescence was FRUIT, followed by ScaleS and SeeDB. However, CUBIC and PACT preserved similar levels of fluorescence as 3DISCO and uDISCO, which are the organic hydrophobic clearing agents. They did not include CLARITY and its variations in their testing. Even though their study included heart tissue, the part about fluorescence preservation was performed on intestine, which may react differently from the dense, highly vascularized heart tissue. Therefore, further analysis of 3DISCO, uDISCO, and FRUIT on cardiac tissue is needed. An overview is summarized in Figure 1.
Only few studies have directly analyzed compatibility of tissue clearing and fluorescence preservation in the adult or embryonic heart tissue. In our previous study (Kolesova et al., 2016) we used CLARITY, SCALE, CUBIC, and DBE clearing methods and compared their GFP fluorescence preservation ability in embryonic hearts (illustrated in Figures 2C, 2D and 3B with CUBIC tissue clearing on Cx40:GFP trabeculae and coronary vasculature in embryonic hearts). We found that CUBIC cleared the tissue to a deeper level compared with SCALE and therefore was more suitable for analysis of intact hearts. However, although SCALE also preserved the GFP signal, it only cleared the superficial layers of the heart, which may be sufficient for studies of the great vessels of the coronary vasculature (Kolesova et al., 2016). The difference between SCALE and CUBIC was more obvious in postnatal hearts, where CUBIC effectively cleared all the way through the hearts, whereas SCALE did not (Shaikh Qureshi et al., 2016). Another study tested the ability of CUBIC to preserve various fluorescent signals in the heart tissue (Nehrhoff et al., 2016). They found that CUBIC clearing can also preserve other fluorescent proteins such as TdTomato and GFP in 750-μm-thick heart sections.
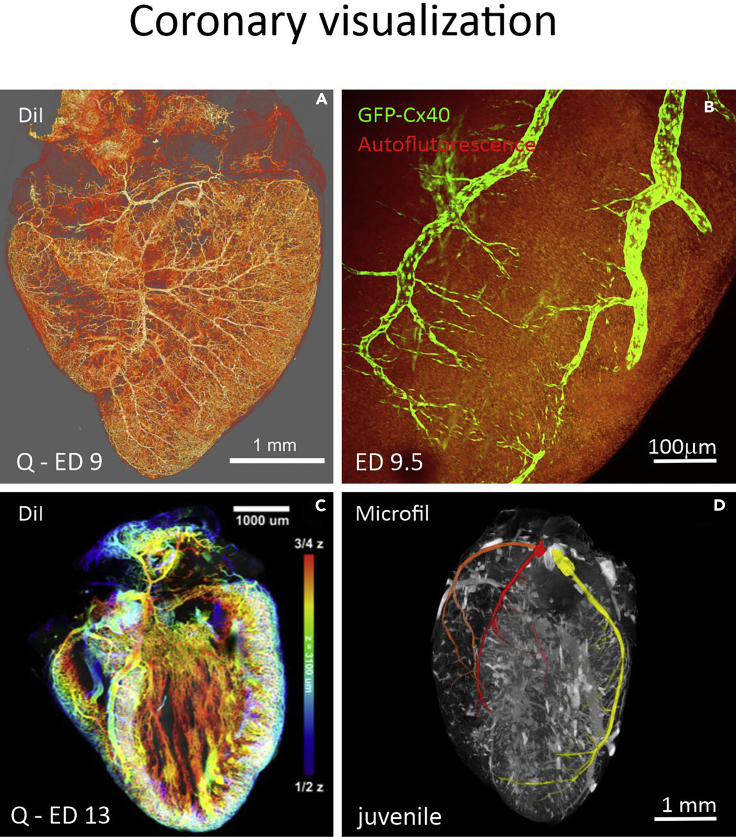
Figure 3.
Coronary vasculature visualization in the developing hearts
(A) Vasculature was injected with DiI in a quail ED9 embryo.
(B) Coronary arteries were visualized on heart surface of an ED18.5 Cx40-GFP mouse embryo cleared with CUBIC. Imaged on a confocal microscope.
(C) Analysis of the DiI injected heart with coronary vessels pseudocolored to indicate depth within the ventricular wall, ED13 quail embryo.
(D) Juvenile mouse heart with coronary vasculature injected with Microfil and main coronary arteries indicated with color: Right coronary artery (orange) and its branch, septal artery (red), and left coronary artery (yellow). Imaged on a micro-CT scanner.
CLARITY also has been shown to preserve many fluorescent proteins such as GFP, mCherry, mOrange, and Cerulean in heart tissue (Sereti et al., 2018). Also, modified CLARITY protocols have been used to analyze cardiac tissue. SUT (Scheme Update on tissue Transparency, combination of CUBIC and CLARITY) has been used to analyze fibrotic healing in myocardial infarction (Wang et al., 2018). SCM (Simplified CLARITY Method) has been used to analyze heart tissue (Sung et al., 2016); however, better results were obtained when the blood was washed from the heart prior to fixation as decolorizing of heme using aminoalcohol treatment reduced YFP fluorescence in heart samples (Sung et al., 2016).
Tissue clearing and preserving the nuclei staining (DAPI/Hoechst) fluorescence
Nuclei staining and visualization are important parts of tissue imaging and heart imaging. Like with other visualization methods only some tissue clearing techniques are compatible with nuclear staining. However, little is published on this topic and most of the published studies worked with DAPI or Hoechst nuclear staining, while a study focusing on compatibility of the fluorescence from far red staining TOPRO and DRAQ5, or any other nuclear staining, with optical clearing is missing.
The combination of DAPI (4′,6-diamidino-2-phenylindole) and a fluorescently labeled antibody is a powerful cytochemical strategy for concurrent visualization of DNA and individual proteins in the cell (Kapuscinski, 1995). DAPI could be used in quantification of nuclei in the heart tissue culture, in dissociated cardiomyocytes, and other heart cells (Tarnowski et al., 1991). DAPI staining was successfully applied in whole-mount heart (Sandell et al., 2018; Xu et al., 2019b), for cardiomyocytes orientation (Lapierre-Landry et al., 2020) or DNA content or ploidy. However, fluorescence in DAPI is very easy to damage by breaking its binding bonds and thus decreasing the signal-to-background ratio (SBR). Therefore, not every clearing method is compatible with DAPI.
Generally, the fluorescence preservation of solvent-based methods is limited owing to harsh dehydration or delipidation, while the aqueous clearing methods have been developed for the purpose of better fluorescence preservation. By comparing several clearing methods, such as CUBIC or SCALE (Xu et al., 2019b), the most suitable methods for clearing DAPI-labeled samples while still maintaining a strong SBR appear to be SeeDB (Ke et al., 2013) and SCALE on brain tissue. SeeDB and SCALE maintain an SBR between 68.3% and 68.7%, whereas other methods decrease the SBR to less than 50.0% or even quench fluorescence more substantially, such as CUBIC or PACT (Xu et al., 2019b). However, the SeeDB has not yet been tested in other organs, such as the heart, and we think that future testing would be beneficial.
The presence of red blood cells, especially in non-mammalian model organism, where even mature red blood cells remain nucleated, could make tissue clearing while preserving nuclei staining more difficult in the heart. It is crucial to ensure complete washing off of the blood cells in the sample prior to cross-linking with paraformaldehyde. This critical step must be done very carefully, because red blood cells lyse and their accumulation and lysis may lead to hemoglobin polymerization and therefore to increased autofluorescence (Sung et al., 2016).
In addition to the mentioned methods, a recent protocol, SUT-CLARITY, has been designed specifically for the adult mouse heart, and therefore, it also preserves the tissue for DAPI staining and immunohistochemistry (Wang et al., 2018). The SUT-CLARITY could also be used for coronary vascularization imaging in different kind of mammalian hearts, such as in mice or rats (Wang et al., 2018). Overall, the SUT-CLARITY is a very promising method, and it should be tested not only in adult vertebrate hearts but also in embryonic hearts during development (summarized in Figure 1).
Tissue clearing and heart and vessel immunolabeling
Visualization of various proteins within whole-mount immunohistochemistry is another well-known and well-established method used in biology. Tissue clearing can be combined with various labeling techniques such as fluorescent immunohistochemistry and in situ hybridization (Tainaka et al., 2016). One of the disadvantages of these techniques might be the insufficient antibody (or probe) tissue penetration during the whole mount staining protocol and therefore limited imaging in depths.
Compared with preserving natural fluorescence (GFP), immunohistochemistry on heart tissue is compatible with a broader array of tissue clearing techniques and especially with many hydrophobic tissue clearing methods. From the hydrophobic tissue clearing methods, clearing with BABB (benzyl-alcohol:benzyl-benzoate) was successfully used in tissue clearing of endocardial painting with fluorescently labeled poly-L-lysine for the purpose of screening for congenital heart defects (Miller et al., 2005). Similarly, Ivins et al., 2016 used BABB for clearing whole-mount heart samples of coronary vasculature labeled with endothelial marker CD31 in mouse. BABB was found to preserve the fluorescent signal in hearts for several weeks without loss of the signal (Ivins et al., 2016). Figures 2A and 2B illustrate the compatibility of BABB tissue clearing with immunohistochemistry for CD31 vascular and endocardial marker and SMA (smooth muscle actin) in early embryonic hearts (Vrbacky et al., 2016)
Organic solvent (hydrophobic) tissue clearing methods were adapted to fit the needs of immunohistochemical staining. The 3DISCO was used to image the whole mount of early human embryo, to build a 3D atlas of human development. Embryos were stained with various antibodies to visualize the coronary vasculature, cardiac nerves, and other heart structures (Belle et al., 2017). The iDISCO+ was successfully used not only for whole-mount immunohistochemistry but also for assessment of the transcriptional profiles from the mouse cardiac conduction system using fluorescence RNA in situ hybridization (Goodyer et al., 2019).
Another family of tissue clearing methods that enables immunohistochemical staining and in situ hybridization includes CLARITY and its modifications (Chung et al., 2013; Tomer et al., 2014). An updated method, SUT, was developed and successfully used for clearing adult mouse cardiac tissue stained immunohistochemically with several cardiac markers (Cardiac Troponin I, Alpha Tubulin, Collagen I, Vimentin, and Actin) to produce a 3D reconstruction model of their expression (Wang et al., 2018). Many authors working on cardiac tissues encountered the problem of slow or insufficient antibody diffusion into the dense cardiac tissue during immunohistochemical staining. Therefore, Wang et al. (2018) developed EAL (electrophoretic antibody labeling) that can be used to achieve faster antibody labeling by electric force, which significantly reduces antibody incubation time from days to hours and can be combined with SUT clearing. Compared with the PACT reagent (passive CLARITY), SUT can achieve higher transparency and less protein loss. The electrophoretic field was also used on other tissues, resulting in faster clearing and enhanced antibody staining (Kim et al., 2015). Besides using electrophoretic fields the antibody penetration can be enhanced by applying pressure as in ACT-PRESTO (Active Clarity Technique Pressure Related Efficient and Stable Transfer of macromolecules into Organs technique; Lee et al., 2016). This method with increased pressure shortens the tissue clearing time and improves immunolabeling and preserves at the same time endogenous fluorescent proteins. These methods apply an electrophoretic field or increased pressure, which also improves the antibody penetration and staining, so important in whole-mount heart labeling (summarized in Figure 1).
Myocardium visualization with autofluorescence and tissue clearing
Unlike other organs, heart tissue contains highly autofluorescent myocardium. Myocardial autofluorescence is often used to visualize the whole heart anatomy, shape, and anomalies (Ding et al., 2018). The myocardium is highly autofluorescent owing to sarcomeric proteins and myoglobin, which have a tetrapyrol in the heme. Myocardial autofluorescence can be useful to visualize cardiomyocyte orientation at various depths of the heart. Combined with tissue clearing method it enables 3D reconstruction and orientation analysis of myocardial fibers orientation in the whole heart. This makes tissue autofluorescence an accessible and noninvasive tool for characterization of cardiomyocytes architecture (Lee et al., 2018).
The naturally high level of autofluorescence in the myocardium is usually considered a disadvantage of cardiac tissue as it may compete with fluorescence from immunohistochemistry in whole-mount specimens. To prevent obscuring of signal with tissue autofluorescence, spectral shift can be applied. Fluorescent signal from the blue/green channel can be moved to the red/far-red region of the spectrum, where myocardium autofluorescence is lower (e.g., GFP can be visualized with antibody staining detected with Cy5 fluorescence) and therefore the signal becomes more prominent. The blue/green region of the spectrum may then be used for anatomical annotation using autofluorescence (Miller et al., 2005). For many applications that are based on expressing fluorescent proteins (particularly GFP), antibody amplification and spectral shifting of the signal may be critical for obtaining good results (Buffinton et al., 2013). However, Figure 4 illustrates that visualization of superficial structures, such as the heart conduction system on the inner surface of ventricle, is not obscured by high myocardial autofluorescence and they can be visualized without spectral shift. GFP is visualized in green channel and autofluorescence in red channel to illustrate the anatomical context.
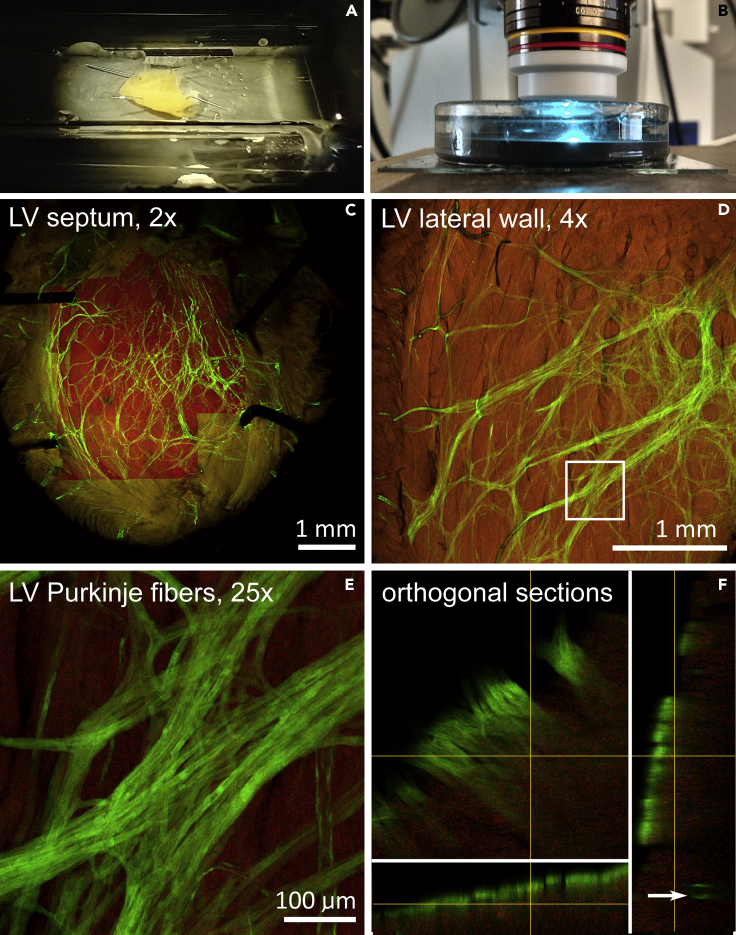
Figure 4.
Workflow for 3D imaging in adult mouse heart
(A) Custom made imaging chamber with pinned embryonic heart for 2x lens imaging. Glass tubes border the chamber and support the glass coverslip.
(B) Imaging of the heart on an upright confocal microscope with immersion objective lens 25×; the lens is immersed in a chamber filled up with SCALE with the heart specimen pinned at the bottom.
(C) Field of view using 2x lens, allowing visualization of the entire mouse heart (postnatal day 30) without image stitching. The highlighted region was imaged also by the 10× objective to obtain a high-resolution 3D dataset of the left bundle branch. Cx40-GFP (green), tissue autofluorescence (red).
(D and E) Left ventricular lateral wall imaged with 4x dry lens for overview of the Purkinje network on Cx40-GFP mouse (D) with one region (boxed area) imaged with 25x ScaleView lens (E).
Depth of imaging could be appreciated on orthogonal views below, allowing clear distinction between subendocardial Purkinje fibers (solid bands) and more deeply located coronary artery branch (arrow) (F).
Other highly fluorescent structures are red blood cells. Red blood cells autofluorescence can be reduced by using tissue clearing protocols that clear hemoglobin, such as CUBIC (Kolesova et al., 2016). CUBIC was also reported to clear the hemoglobin from cardiac tissue; however, it is recommended to wash blood from the heart prior to fixation as decolorization of heme using aminoalcohol treatment reduced YFP fluorescence in heart samples (Sung et al., 2016). Therefore, hemoglobin tissue clearing methods not only improve transparency of the tissue but also limit blood autofluorescence.
Red blood cells autofluorescence can help to generally locate the main coronary vessels (especially veins) as the blood cells usually remain in the venous system. This can be used for the purpose of imaging museum samples, for example (Sedmera et al., 2003), where immunohistochemistry staining is not possible. Similarly, in medium-sized vessels autofluorescence permits distinguishing veins from arteries as blood remains predominantly in the venous system (authors’ observations).
Perfusion methods for heart vessel imaging and tissue clearing
The heart is a complex organ, and therefore it is supplied by a rich vascular network as illustrated in Figures 3A and 3C, where embryonic coronary vasculature was injected with contrast fluorescent dye. The vasculature of the heart is commonly imaged with tools like those used to visualize vasculature in other organs. Coronary vasculature can be visualized with immunohistochemistry using various endothelial (CD31 in mouse [Cavallero et al., 2015]; QH1 in quail [Kattan et al., 2004]) smooth muscle markers (SMA, smooth muscle actin), as reviewed in Sharma et al. (2017). Alternatively, one can use various mouse strains expressing fluorescent protein with these markers, reviewed in Red-Horse et al. (2010). Figure 2B illustrates the expression of CD31 in heart endocardium. An interesting method of vasculature visualization, which was successfully used within cardiac muscle WGA (wheat germ agglutinin) lectin staining. WGA binds to all plasma membranes, but as endothelial cells are much thinner and smaller than cardiomyocytes, membranous staining is more intense in coronary vasculature. In addition, it binds strongly to the extracellular matrix, highlighting the arterial adventitia and regions of perivascular fibrosis in the heart. This method was successfully used for cardiac vessels quantification and analysis (Bensley et al., 2016).
Besides these common methods, another advantage of the vasculature is that it can be injected with various substances to facilitate viewing. First, vasculature was imaged based on vessel perfusion by a contrast agent observable in visible spectrum. For example, valve morphogenesis as well as inadequate coronary tree development under hypoxic condition was studied using india ink (Hu et al., 2009; Nanka et al., 2008; Wang et al., 2009). Even though tissue clearing was not applied to these samples, we showed that clearing of india ink-injected hearts should improve image quality in a bright field microscopy (Kolesová et al., 2018). Another possibility is using latex/carbon black or perfusion gelatin mixed with various dyes to identify the vessels (Hasan et al., 2012; Maeda et al., 1998).
Vasculature also can be visualized by injection of various lectins (specific carbohydrate-binding proteins) that are fluorescently labeled. Lectins are species specific and are used mainly to study avian vascular development (as the chicken embryos are lacking the endothelial marker such as PECAM-1 found in the mouse or QH1 marker in quail). These have been applied in numerous studies of the vasculature (increased permeability, vascular subtype identification, etc.) and were summarized by Jilani and colleagues (Jilani et al., 2003). Visualization and imaging of the lectins is more vessels specific, if lectins are injected into the vasculature. Intravascular lectin injection make lectins binding more vessel specific compared to their application on histological sections (Jilani et al., 2003).
When combined with the clearing methods, the 3D vascular network can be imaged in detail. After 3DISCO clearing single endothelial cell integration into capillaries in the spheroid-based matrigel plug assay was measured using ultramicroscopy (Epah et al., 2018). Tissue clearing also enables vessels imaging deeper into organs. Study of brain microvasculature showed that after intravenous lectin tomato-FITC injection microvessels down to a depth of 2 mm after CLARITY optical clearing (Lagerweij et al., 2017) could be imaged. Using CLARITY in this study lead to a tissue expansion of approximately 1.59x of the original tissue diameters, whereas the iDISCO procedure resulted in shrinkage of approximately 0.7x. In addition, authors stated that, in iDISCO processed samples, the CD31 staining was hampered by a low signal-to-noise ratio (Lugo-Hernandez et al., 2017; Qi et al., 2019).
Other methods that can be applied to various species heart studies is the injection of fluorescently labeled dextran. This approach was used for imaging heart and vessel development in various species. To image mouse vessel fluorescein isothiocyanate (FITC)-conjugated dextran was successfully combined with BABB (Bryson et al., 2011). Using tetramethylrhodamine-dextran injection, the mechanical stability of the developing perlecan-deficient mouse hearts was analyzed (Sasse et al., 2008). Furthermore, fluorescently labeled dextran proved its compatibility with intravital cardiovascular imaging in zebrafish (Sarkar and Schmued, 2012; Zhang et al., 2002). These studies showed that, solvent-based methods such as BABB and DISCO, and CLARITY clearing are suitable for heart vasculature studies and compatible with antibody labeling, lectins or dextran labeling.
Other methods of vasculature visualization use well-known fluorescent dye such as long-chain carbocyanine dyes (DiI, DiO, DiD, and DiD) commonly used for membrane staining. These lipophilic substances show intense fluorescence when incorporated into cell membranes. Their intense fluorescence with little bleaching when exposed to the excitation light make carbocyanines a powerful tool for vessel visualization (Honig and Hume 1989). Carbocyanines can be successfully combined with various tissue clearing methods (Carrillo et al., 2018; Hou et al., 2015; Jensen and Berg, 2016; Lapierre-Landry et al., 2020) (Figures 3A and 3C).
A common use of carbocyanine dyes is for in vivo cell tracing by staining the cell membrane, as carbocyanines have low level of toxicity (Nagyova et al., 2014; Lankford et al., 2018). By labeling cell clusters with DiI and DiO, the tissue dynamic during early heart tube formation was visualized, showing convergent extension as a key mechanism in the heart tube elongation (Kidokoro et al., 2018).
DiI labeling also found its application in studies of vasculature development. Direct Dil perfusion has been described by Li et al. (2008) to incorporate the dye to the endothelial cell membranes and thus rapidly and reliably visualize the inner surface of the vasculature of a hollow organ such as the eye. However, the majority of publications using DiI to visualize vessels did not use any tissue clearing method (Nagyova et al., 2014; Lankford et al., 2018). Recently, DiI was used to visualize coronary microvasculature and to quantify vessel density and orientation even in whole ventricles of middle gestational age quail hearts. Moreover, in combination with nuclear staining of cardiomyocytes (with DAPI), the mutual cardiomyocyte and coronary microvasculature orientation and alignment was assessed and quantified (Lapierre-Landry et al., 2020). Moreover, not only the vessels could be determined by Dil perfusion, but also lateral diffusion helps to stain membrane structures that are not in direct contact with the DiI solution and with vessel structures with minimal or no lumen. Thus the angiogenic sprouts and pseudopodial processes of angiogenic endothelial cells could be imaged as well (Li et al., 2008).
The advantage of Dil labeling lies in its compatibility with other labeling methods. DiI vessels visualization can be combined with DAPI nuclear staining (Lapierre-Landry et al., 2020) as well as with subsequent immunohistochemical analysis such as fluorescence in situ hybridization or electron microscopy (Li et al., 2008). One drawback we found is that DiI fluorescence is so intense that it may be excited by other wavelengths and bleed into many channels, which limits observations of the other fluorophore staining. In addition to other carbocyanines, DiR might be useful for in vivo imaging and tracing owing to the effective transmission of infrared light through cells and tissues and low level of autofluorescence in the infrared range (Lankford et al., 2018).
In addition to the above-mentioned approaches based on vessel endothelial cells labeling, the gelatin hydrogel containing FITC-conjugated albumin method stains the entire blood vessel (Di Giovanna et al., 2018; Lugo-Hernandez et al., 2017). The high molecular weight of albumin prevents the marker from crossing blood vessel walls, which ensures the confinement of the fluorescent signal within the blood vessels. Since the fluorescent marker and the gel comprise proteins, they are both retained in the hydrogel matrix during the procedure. In comparison with lectin-FITC endothelial labeling, staining of the blood vessel lumen improved the signal-to-noise ratio and provided clear demarcation of blood vessels imaged by light-sheet fluorescence microscopy (LSFM). This approach in combination with LSFM and CLARITY clearing allowed whole mouse brain vasculature reconstruction down to the capillary level (Di Giovanna et al., 2018). In combination with another clearing procedure, 3DISCO, it enabled detailed analysis of microvessels in the whole ischemic mouse brain (Lugo-Hernandez et al., 2017).
Other perfusion methods for heart vessel imaging with no need of tissue clearing
Tissue clearing is the method necessary for imaging in the visible spectrum, as the tissue has to be transparent for visible light. However, biological tissue including the heart can be visualized in other wavelengths. A commonly used method in heart visualization is whole-mount imaging with X-rays using a micro-CT scanner. Recent advantages in imaging and enormous improvement in resolution have made micro-CT suitable to visualize in great details even the tiny embryonic hearts. As micro-CT can image whole-organ morphology, it is commonly used to study pathological phenotypes and gene mutations involved in the cardiovascular development, like persistent truncus arteriosus and aberrant origins of the coronary arteries (Degenhardt Karl et al., 2010; Ermakova et al., 2018).
Micro-CT is of limited value in imaging of the cardiac vasculature as there is an insufficient contrast in tissue saturation between vessels and the surrounding tissue. However, these limits can be overcome by injecting contrast agents into the coronary vasculature. The most commonly used injecting media, which are visible in micro-CT imaging, are hydrophobic plastic resins such as Microfil (Microfil, Flow Tech, Carver, MA), PU4ii (vasQtec, Zurich, Switzerland), mAngiofil (Fumedica AG, Muri, Switzerland), and XlinCA (University of Zurich, Switzerland) (Bohuslavova et al., 2019; Ghanavati et al., 2014; Le et al., 2020; Schaad et al., 2017). Figure 3D shows a juvenile heart injected with Microfil and imaged on micro-CT. A novel method was recently developed with cross-linkable polymeric contrast agent called XlinCA, which combines the reliable perfusion of the hydrophilic angiography contrast agents with the permanent retention and contrast of the hydrophobic vascular casting resins. Good water solubility and sufficient viscosity ensure homogeneous filling without the contrast agent escaping out of the vasculature (Le et al., 2020).
With transcardially injected contrast agent (in vivo), the vasculature of the entire animal can be visualized and the vasculature the various organs (heart, brain, pancreas, even placenta) can be analyzed. However, the organ is typically excised and ex vivo perfused (Bohuslavova et al., 2019; Junaid et al., 2017; Schambach et al., 2010; Zagorchev et al., 2010). Micro-CT and vascular casting were successfully used to measure volumetric parameters of cardiac development, aortic arch morphogenesis, and outflow tract as well as atrioventricular valve formation (Bharadwaj et al., 2012; Butcher et al., 2007; Hu et al., 2009; Wang et al., 2009; Yalcin et al., 2011). Detailed analysis (down to 4 μm) of the development of the aberrant vascular system can be performed (Bohuslavova et al., 2019; Le et al., 2020). Pathologies found using this approach included abnormal coronary artery origin and branching caused by impaired Hif-1a signaling, coronary vasculature defects in the time-restricted Fog2 knockout, or defect vascular patterning due to affected Notch signaling (Proweller et al., 2007; Bohuslavova et al., 2019; Zhou at al. 2009).
Injected resins do not contain larger particles, and together with the high-resolution modality of micro-CT, the vasculature could be tracked down to the level of capillaries (Schürmann et al., 2015). Moreover, it was applied to study mechanisms of collateral vessel growth (He et al., 2016; Zhang et al., 2019). Moreover, the technical challenges such as timely restricted polymerization and the necessity for adequate perfusion pressure make study at the level of capillaries using micro-CT quite challenging (Le et al., 2020). Considering developmental studies, imaging of neonatal and embryonic vascular network could be limited owing to the fragility of the immature vessel wall and its intolerance of high perfusion pressure.
It should be noted that a limitation of all the above perfusion techniques is that they cannot differentiate between arterial and venous parts of the vascular system. Differentiation is possible only if the vessels segmentation is done manually based on the course of the vessel. For better differentiation of arteries and veins, the techniques need to be followed up with detecting specific markers. Even without artery/vein distinction, 3D high resolution reconstruction of all mentioned perfusion techniques provides important spatial visualization and allows precise analysis of vessel density and arrangement.
Tissue clearing, tissue size changes, and expansion microscopy
Various tissue clearing techniques either expand or shrink the heart tissue (Kolesova et al., 2016) depending on the reagent compositions and clearing mechanism as well as on the characteristics of the tissue or organ. The changes in sample size can sometimes be advantageous; for example, shrinking associated with most dehydration-based protocols allows use of higher-power objectives with correspondingly higher numerical aperture, improving the Z-resolution. Most tissue clearing methods claim isotropic shrinkage or expansion; hence, linear size deformation is used to calibrate samples. However, further detailed work to determine if the expansion/shrinkage is indeed isotropic is required (Xu et al., 2019b). On the other hand, more elaborate tissue expansion techniques were developed, based on tissue expansion during tissue clearing, and enabled study of tissues in greater detail using regular imaging methods.
Expansion microscopy (ExM) (Chen et al., 2015) is a novel, fluorescence imaging technique, which allows 3D nanoscale imaging of specimens on a conventional fluorescence microscope (Faulkner et al., 2020). This unique technique physically expands preserved cells and tissues, mostly brain, isotropically via a chemical process, so that 3D nanoscale resolution imaging of specimens becomes possible on common, fast, diffraction-limited microscopes (Alon et al., 2019; Chen et al., 2015; Gao et al., 2017; Murakami et al., 2018). In expansion microscopy, hydrogel embedding is performed with a polyelectrolyte gel of cross-linked sodium polyacrylate, which swells substantially when exposed to water (Alon et al., 2019) to physically magnify the specimen ≥100-fold in volume (Gao et al., 2017). The ExM techniques are conducted mainly with slices approximately 50–200 μm thick (Tillberg et al., 2016; Wassie et al., 2019) as well as with tissue culture (Chen et al., 2015) or single cell observation (Alon et al., 2019; Chen et al., 2015; Gao et al., 2017; Murakami et al., 2018). Yet, as a result, ExM is suited for rapid super-resolution imaging in cells, and also it has now been validated across a range of cell and tissue types and even whole organs, such as the brain (Migliori et al., 2018; Tillberg et al., 2016), pancreas, spleen, and lung (Wassie et al., 2019), and larval zebrafish (Freifeld et al., 2017). Clearing methods, which go hand in hand with ExM, are CUBIC and CLARITY (Freifeld et al., 2017; Wassie et al., 2019).
In the heart, just a few pilot studies exist on expansion microscopy in comparison with other organs such as the brain. However, there are several experiments with heart tissue slices for studying the vasculature and coronary vessels (Yokoyama et al., 2017) and also for observing whole-mount heart vasculature (Nehrhoff et al., 2016), which broaden the knowledge of the heart itself even at the nanoscale level.
Moreover, ExM could be combined with other super-resolution techniques including structured illumination microscopy (SIM) (Halpern et al., 2017), stimulated emission depletion (STED) microscopy (Gao et al., 2018), and stochastic optical reconstruction microscopy (STORM) (Xu et al., 2019a). This method could allow more detailed observations of the different kinds of nanostructures, e.g., kinetochore structures in cells undergoing mitosis (Chozinski et al., 2016) or transverse filaments spanning sister chromatids (Cahoon et al., 2017). Cahoon et al. (2017) were even able to map how different proteins dimerize and interact with one another, by labeling different proteins in different locations and then performing expansion microscopy. Moreover, DNA and RNA could be observed and visualized via ExM as well (Chen et al., 2016). These studies show expansion microscopy as a powerful tool for higher-resolution microscopy and therefore gaining new information. Its impact in the field of heart and vasculature studies is promising.
Although ExM provides us more resolution, the tissue shrinkage (Pan et al., 2016) should decrease sample size to enable fitting the sample into small imaging chambers of the microscopes and to reduce the disadvantage of short working distances of the high numerical aperture objective lenses. Hence sample shrinkage is beneficial for imaging large-volume samples in the whole-mount setting such as with light sheet microscopy (Xu et al., 2019b).
Intentional tissue shrinkage has not yet been performed on heart and vasculature studies, even though the heart is a good candidate organ for tissue shrinkage. uDISCO tissue clearing (Pan et al., 2016) resulted in high-quality images; however, shrinkage allows imaging of 2× to 3× larger volumes in the same setup without apparent loss of resolution. These new methods developed from tissue clearing will enable the collection of new information in the field of normal and abnormal heart and vasculature development as well as in studying pathologies that are applicable to human cardiovascular medicine.
Live imaging of transparent tissues in cardiovascular development
Some of the developmental biology model species and methods are already transparent enough to enable direct observation without tissue clearing. Here, we would like to briefly summarize these models and methods and their impact on the study of heart and vasculature development. The great advantage of these vertebrate models is that direct in vivo observations of developmental processes can be made and captured.
Live imaging is the study of living cells using time-lapse microscopy. It is used for obtaining a better understanding of biological function through the study of cellular dynamics (Cooper and Bakal, 2017). It requires simultaneous incubation of cells in stress-free conditions while imaging is being performed. There are several factors that must be considered when choosing imaging conditions such as phototoxicity, photobleaching, and tracking ease. These are all related to imaging frequency and illumination intensity (Stephens and Allan, 2003). However, live imaging is a promising method in heart morphogenesis studies.
The ideal model for using live imaging protocols is zebrafish, commonly used for studying cardiac morphogenesis as well as hemodynamics (Boselli and Vermot, 2016; Forouhar et al., 2006; Mellman et al., 2012; Taylor et al., 2019; Truong et al., 2011). The zebrafish eggs are externally fertilized; embryos are nearly transparent, providing optical access to the earliest stages of cardiogenesis (Stainier et al., 1996); and many GFP-labeled transgenic strains have been derived (D'Amico et al., 2007; Lawson and Weinstein, 2002; Nguyen et al., 2008). In using high-speed microscopy, optical sections through 26-h postfertilization Tg(gata1:GFP) zebrafish hearts expressing GFP in blood cells, endocardium, and myocardium could be reconstructed into four-dimensional datasets (Liebling et al., 2005). There are many useful zebrafish mutations, which could be used as models for studying heart morphogenesis, e.g., Tg(flk1:EGFP) (Beis et al., 2005), Tg(gata1:dsRed) (Traver et al., 2003), Tg(myl7:HRASEGFP) (D'Amico et al., 2007), Tg(gata1:GFP), or Tg(cmlc2:GFP). Life imaging of heart tube formation and contribution of primary and secondary heart field was published by Ivanovitch et al. (2017).
Other vertebrate models that are transparent and can be used for in vivo analysis are adult Medaka fish. Also, early avian embryos are transparent and can be used for in ovo imaging of early heart and vasculature development (Lansford and Rugonyi, 2020). However, the number of genetically modified (GFP) lines is limited in avians (Chapman et al., 2005). Also tissue cultures are transparent enough to permit in vivo imaging, i.e., mouse tissue cultures (Yue et al., 2020).
In vivo imaging is a powerful tool to study hemodynamics of the embryonic heart (Forouhar et al., 2006), and improvements of microscopical techniques are bringing higher resolution and imaging depth to live imaging of transparent model organisms or tissue culture.
Recently, developing mouse heart was successfully live imaged with vertical LSFM while growing in culture. Cardiomyocyte nuclei were imaged and then traced during their further development. This demanding method enabled the reconstruction of migration and orientation of cell division in various cell populations of the developing heart (Yue et al., 2020).
Tissue clearing and microscopic methods for embryonic heart and vessel imaging
Optical microscopy is a widely used tool for non-destructive and minimally invasive observations of experimental structures. Fluorescence microscopy allows the detection of structures based on immunolabeling or genetic targeting by fluorophores. Although various optical microscopic methods enable 3D reconstruction, the requirement for high degree of transparency is usually provided by tissue clearing.
Single-photon confocal laser scanning laser microscopy (CLSM) is a well-established method in embryology allowing detailed observation of surface as well as deep structures with the possibility of subsequent 3D reconstruction. Using CLSM, the role of genetic mutations, epigenetic factors, as well as cell-cell interactions in cardiovascular development could be studied. To improve the depth of imaging, various clearing protocols, such as BABB, CUBIC, CLARITY, SCALE, have been used (Bryson et al., 2011; Kolesova et al., 2016; Zhao et al., 2015). However, owing to limited field of view, it is not suitable for detailed imaging of large heart structures. Imaging depth and clarity can be further improved by two-photon excitation (Miller et al., 2005). Unlike one-photon microscopy, in two-photon laser scanning microscopy two photons are absorbed by the label at virtually the same instant. It also uses longer-wavelength photons, which are lower energy and penetrate more deeply, creating less tissue damage while imaging farther into the sample (Wu et al., 2017). These properties overcome other limitations of conventional single-photon imaging modalities, such as image blurriness due to out-of-focus fluorescence, relatively limited penetration depth, phototoxicity, photoactivation, and photobleaching (Cahalan et al., 2002; van Zandvoort et al., 2004). Moreover, TPLSM provides a combination of high resolution and high penetration depth modality with the possibility of intravital imaging (Wu et al., 2017).
Optical projection tomography (OPT) is an optical imaging technique that measures the 3D distribution of the absorption of the fluorochromes (Sharpe et al., 2002). In cardiovascular studies, OPT was successfully used in combination with BABB clearing to quantify infarction size in adult mouse hearts (Zhao et al., 2015). It was also used for 3D imaging of embryonic working as well as conduction myocardium development (Kolesova et al., 2016). Using a mouse model expressing Cx40:GFP and CUBIC the embryonic coronary tree was successfully reconstructed (Kolesova et al., 2016). Compared with single-photon confocal microscopy OPT allows acquisition of data with proper morphological and spatial information of whole specimens, up to several millimeters in diameter and thus making it possible to resolve macroscopic anatomical structures (Kolesova et al., 2016). In contrast, the resolution of confocal microscope is much higher (up to 200 nm/pixel), which provides high-resolution insight into subtle structures of specimens, albeit in a more restricted field of view (Kolesova et al., 2016).
3D light sheet fluorescence microscopy (LSFM) of labeled cells uses plane illumination of the sample and detects the fluorescence perpendicular to the illumination axis (Epah et al., 2018). Various clearing procedures such as 3DISCO (3D imaging of solvent-cleared organs), iDISCO, uDISCO (ultimate 3DISCO), BABB, Ethanol-ECi (ethyl cinnamate), or CUBIC (Ertürk et al., 2012; Renier et al., 2016; Pan et al., 2016; Schwarz et al., 2015; Klingberg et al., 2017; Nojima et al., 2017) are compatible with LSFM. A recently published review by Ueda et al. (2020) discussed the importance of LSFM imaging for brain and large samples imaging, which enables acquisition of large volumes in real time. LSFM has not been used for imaging of developing heart as intensively as for brain, but clearly imaging heart by LSFM will be beneficial as is the case for brain tissue. After tissue clearing LSFM was successfully used for ex vivo vasculature imaging in embryos (Lowe et al., 2015) with limited application in the adult tissue (Epah et al., 2018; Ertürk et al., 2012; Lowe et al., 2015). Moreover, LSFM has also been described to image intravital angiogenesis in translucent animals like zebrafish larvae expressing protein-based fluorescent molecules (Fei et al., 2016; Kugler et al., 2019).
Epah at al. (2018) tested ultramicroscopy and micro-CT modality for vascular imaging. Ultramicroscopy is the method of virtual sectioning of the sample on the light-sheet microscope, used to achieve high-resolution images. They showed that, although ultramicroscopy is ideal for imaging and especially quantifying capillary networks and arterioles, larger vascular structures are easier and faster to quantify and visualize using micro-CT (Epah et al., 2018). Moreover, ultramicroscopy has been reported to achieve resolution sufficient for visualization of single endothelial cell integration into capillaries in the spheroid-based Matrigel plug assay (Epah et al., 2018).
Another cardiovascular imaging method is optical coherence tomography (OCT). Although OCT could be used for vascular imaging of embryonic tissue, the major disadvantage represents the limited depth of penetration, which allows analysis only of a relatively small part of the tissue. This importantly limits 3D vascular imaging. This obstacle could be overcome by improving tissue transparency through optical clearing methods bringing the advantages of a larger visualized area and microvascular mapping in large tissue volumes. Two recent studies described such protocols. The first one combined a novel OCT contrast agent with clearing method using a modified Scale/CUBIC-1 solution (Liu et al., 2019). The strong OCT signal was ensured by filling the vasculature with a specialized OCT contrast agent, while its leakage was prevented by rapid cross-linking (Liu et al., 2019). Using this method, microvascular abnormalities (chaotic alignment, reduced vessel density) have been demonstrated in quail embryo heart model of fetal alcohol syndrome (Liu et al., 2019). Furthermore, not only heart vasculature density but also differences in local tortuosity in the quail heart after ethanol exposure have been demonstrated (Liu et al., 2019). The second protocol used OCT and OCT angiography in optically cleared (BABB) murine embryos (Choi et al., 2020). This study confirmed that OCT is a feasible, rapid, and high-quality tool for phenotyping transgenic mouse embryos.
Besides the above-mentioned methods, further intravital methods, but used without tissue clearing, include micro magnetic resonance angiography or recently developed photoacoustic tomography (PAT). PAT works on laser pulse-induced ultrasound signals through the photoacoustic effect. As hemoglobin provides the strongest photoacoustic signal for imaging in the visible and near-infrared regions, PAT should be ideal for high-resolution in vivo imaging of vasculature (Shan et al., 2020). Since the reported depth of imaging is up to 4 cm, even deeper vessels in the embryo could be easily visualized (Wang et al., 2016). Thus, PAT allowed measurements of a rapid decrease in vessel diameter and density in the murine fetal brain within minutes after maternal ethanol consumption (Shan et al., 2020).
Another challenge in imaging of cardiovascular system is visualization of its development in 3D. This is particularly true for the delicate structures that are difficult to distinguish based on simple observation, such as cardiac conduction system (CCS). The facts that the CCS myocardium shares similar morphology as the working ones and represents a rapidly developing structure make its differentiation particularly challenging. Various markers were established to better understand its growth (reviewed by van Eif et al., 2018). The first attempts of 3D reconstruction of the developing CCS were based on in situ hybridizations or immunohistochemistry on serial sections (Aanhaanen et al., 2010; Jensen et al., 2012; Sizarov et al., 2011). However, the high number of images was required for the 3D reconstruction and lower level of spatial resolution was achieved. To image the CCS subpopulation within the heart with improved spatiotemporal resolution, advanced projection modalities in combination with genetically targeted models and optical clearing techniques provided a new possibility. The contribution of various cell lineages to the final CCS morphology was studied using genetic fate mapping (Choquet et al., 2020; Mohan et al., 2018). Employing the model expressing Cx40:GFP, whole ventricular conduction system up to Purkinje fibers was reconstructed in BABB cleared heart (Miquerol et al., 2010). To visualize the CCS myocardium in specific developmental periods we compared various imaging modalities in the Cx40:GFP model. We showed that OPT and SPIM enabled complete 3D images of the whole CUBIC cleared embryonic hearts up to late stages and allowed a general overview of the developed CCS at the macroscopic level. CLSM combined with clearing procedure preserving GFP (CUBIC, SCALE) enabled detailed analysis of the CCS at the cellular level.
Notes on specimen chambers for the heart imaging
Unlike the standardized specimen slides, imaging of larger 3D objects usually requires construction of a specific chamber meeting the following criteria: (1) it must be large enough for mounting the specimen and yet fit into the imaging instrument (typically a microscope stage). This is not trivial, as the objectives with high numerical aperture required to obtain good z-resolution are typically of large diameter and with short working distance (Figure 4A; (2) it must be resistant to the media used (e.g., BABB dissolves some plastic and glues); (3) it must provide a clear illumination/imaging path, especially challenging for techniques using different excitation and emission pathways such as SPIM or OPT; specialized optical grade cuvettes and thin, transparent specimen tubes allowing for sample rotation in the media are provided by the manufactures (Walter et al., 2010), but the mounting and movements of the specimen in the imaging media are not simple. The chamber design for confocal imaging including multiphoton is dictated mainly by configuration of particular microscope (upright or inverted). Although dry or immersion objectives can be successfully used on both (for the inverted microscope, glass bottom Petri dishes are recommended), the use of dipping lenses (for Scale or similar solutions) with high numerical aperture and working distance is restricted to the upright systems (Figure 4B).
Our design for upright confocal microscope is based on a deep glass Petri dish filled with black silicone. For dry lenses, we embed two pieces of glass tubing to provide a surface for mounting of coverslip; a schematic design of this chamber was published previously (Miller et al., 2005). For dipping lenses, a dish with a large diameter and depth is critical. For detailed imaging of adult and embryonic mouse and avian hearts we used the 25×, immersion, 1.0 NA ScaleView lens. In combination with standard 4× and 10× objectives these lenses enable overview as well as detailed imaging of the heart (as illustrated in Figure 4, with an overview of the whole interventricular septum in Figure 4C and in a detailed view of Purkinje fibers in Figures 4D, E, F).
Tissue clearing and heart and vessels segmentation and image analysis
Image analysis is the subsequent step after the tissue clearing and imaging. Tissue clearing significantly simplifies image segmentation of the heart or the vessels in the process of the image analysis. In cleared tissue locating and identifying (segmentation) of the heart parts and vessels can be done automatically by thresholding, especially when heart or vasculature are visualized with a specific marker (e.g., Khoradmehr et al., 2019; Lapierre-Landry et al., 2020). Image segmentation is used for 3D visualization and 3D animations of heart. Visualization of the brain is on the top, with 3D brain atlas, rendered at a single-cell resolution (Murakami et al., 2018). Developing human heart innervation was visualized in 3D by (Belle et al., 2017).
However, when the tissue clearing is not possible, the image segmentation techniques help to obtain high-resolution images. In live heart imaging, especially using LSFM, segmentation methods are rapidly advancing. In zebrafish embryos they were used to study volumetric changes and cardiac cycle and its changes in pathological situation in vivo (Fei et al., 2016; Packard et al., 2017) and in cardiac zebrafish regeneration (Baek et al., 2018). A special field of image segmentation is vasculature segmentation, where vessels can be segmented and skeletonized and branching points established, and 3D vasculature network can then be reconstructed (Park et al., 2015; Corliss et al., 2019; Kennel et al., 2018). As a next step, the blood flow can be computed and modeled (Kennel et al., 2020).
Advances in light sheet imaging of the cardiac system and its analysis are summarized in the review by Ding et al. (2018). In the last 2 years segmentation analysis made progress in applying Deep learning image analysis on LSCM-generated images (Ding et al., 2021). This technique was developed and applied to obtain information about volumetric changes and cardiac function of zebrafish (Akerberg et al., 2019) and cardiac architecture and mechanics (Ding et al., 2021). Deep learning was also successfully used on segmentation of brain microvasculature and analysis of vessels density in various brain areas (Todorov et al., 2020) (Veith and B Baker, 2020).
Rapid improvement is also seen in the field of cardiac surgery image segmentation, where Deep learning is used for segmenting-identifying coronary architecture in CT coronary angiography (Xian et al., 2020; Yang et al., 2019). Even before applying Deep learning methods, segmentation methods were used to identify the main vessels and heart chambers when performing an open heart surgery (Al-Surmi et al., 2014; Ecabert et al., 2011). Segmentation methods applied onto heart and coronary vasculature and its imaging methods in patients are summarized in Chen et al. (2020).
On a smaller scale, the segmentation can be performed in 3D to recognize cells, or filamentous structures as small vessels. On confocal microscope, the main limiting factor in image segmentation is the loss of the signal with increasing depth in the sample, which can be improved by tissue clearing or by microscope acquisition settings. Also, further image processing such as deconvolution can be applied (Li et al., 2007). Vessels can be segmented (Cebasek et al., 2010) also on the whole embryonic heart and reconstructed in 3D model of coronary vasculature (Lapierre-Landry et al., 2020). This reconstructed 3D model of the vasculature can be further analyzed. Whole-embryo vascular network was recently segmented and analyzed (Hahn et al., 2020). Embryonic trabecular network can be also segmented and reconstructed (Olejnickova et al., 2018) and used for modeling of electrical impulse propagation.
Nuclei of the heart can be also segmented and analyzed. The overall orientation of the nuclei can be assessed by Fourier transform to assess orientation of the myocardium in various depths and regions of the ventricular wall (Lapierre-Landry et al., 2020). Alternatively, individual nuclei can be segmented (gradual flow tracking method) for further analysis such as cell counting or measurements of percentages of cells with certain specific markers (e.g., proliferation or apoptosis) (Li et al., 2007). Nuclei segmentation was performed on quail hearts (Lapierre-Landry et al., 2020) as well as on zebrafish and Caenorhabditis elegans (Li et al., 2007) with high accuracy.
From acquired 3D models, besides the various characteristics of myocardium or microvasculature orientation, advanced analysis can be performed. Computational modeling has been used to compute the blood flow within the vasculature, and data have been already published for brain (Blinder et al., 2013; Kirst et al., 2020) and placenta (Plitman Mayo, 2018). In the brain, even the oxygen transport was modeled (Gagnon et al., 2016). However, computing of the blood flow in developing coronary microvasculature has not yet been performed. In computation of coronary microvasculature, the pattern of vessels was analyzed post myocardial infarction and compared with normal microvasculature (Gkontra et al., 2018). Recently the computational model for heart perfusion areas was published (Di Gregorio et al., 2021).
However, modeling in the heart focuses predominantly on modeling the ventricular contraction and its motion analysis, usually in adult patients using MRI (Creswell et al., 1992), CT, or ultrasonography and their variations (Frangi et al., 2001; Park et al., 2015). Project of Cardiac atlas analyzed data from normal and pathological human hearts and modeled several heart parameters such as ejection fraction, diastolic volume, sphericity, conicity, longitudinal shortening, and wall thickness (Fonseca et al., 2011). Image analysis and computation are bringing the acquired data to a new level and enable to gain more and detailed information about the heart and coronary vasculature.
Conclusion and future directions and perspectives in embryonic heart and vasculature tissue clearing and imaging
Tissue clearing protocols have provided us with a plethora of new information, which would be hard to obtain from serial sectioning approaches even with the use of the episcopic imaging (Weninger et al., 2006). First, with proper execution of the clearing and imaging protocol, it results in faster data collection without production of artifacts inherent to sectioning (compression) and their alignment and registrations. This makes tracking of fine structures (e.g., blood vessels) much easier in 3D. A variety of developed approaches attests to the fact that there is no “gold standard” that would outperform all the alternatives. Some techniques are more suitable for prenatal specimens, distinct by their higher water content, less pronounced tissue autofluorescence and lower lipid concentration. Others (intravascular contrast injection) are suitable for whole animal or whole organ imaging of the vasculature using both visible light and X-rays. Again, the ability to obtain distortion-free images of the entire vascular bed presents a major advance in comparison with the tedious and time-consuming 3D reconstruction of the vascular bed from serial sections.
The necessity of future development of these strategies is obvious, as their advantages are clear and with the biggest limitations being the expense of the instrument or limited availability. This is the case of multiphoton imaging, which improves depth of imaging in all the confocal applications and significantly limits photobleaching, making the working distance of the objective lens the major limitation. For clearing techniques, increased popularity is evident in protocols that are faster, water based, using only non-toxic substances and resulting in minimal tissue volume changes. In developmental studies, emphasis is being put on live imaging approaches that allow repeated imaging of the same specimen over time; for these purposes, using very young stages or those with sufficient transparency (including specific mutations) is the way forward. Translational potential is possible through the standardization of 3D clearing protocols and adoption for human pathology samples, which can result in faster and more accurate diagnoses (e.g., whole-mount imaging of lymph nodes, tumor vascularization, or definition of tumor border zone).
Future perspectives for tissue clearing of heart tissue are in steadily improving imaging, collection of large datasets, and detailed data analysis. The heart would deserve similarly detailed imaging and analysis, which is currently done for the brain. Future will be also in combination with other visualizing and analyzing methods used on developing hearts, as physiological analysis of heart activation and conduction system analyzed by optical mapping. Image analysis also could be combined with advanced mathematical modeling, as in brain samples illustrated with blood flow modeling in brain capillaries (Kennel et al., 2020; Perdikaris et al., 2016). For heart research the blood flow modeling in coronary vasculature would move the field significantly.
Tissue clearing is the method that is opening the broad field of whole heart imaging and 3D analysis and moving biological imaging to a new level.
Acknowledgments
We would like to thank Dr. Yehe Liu for providing the images of coronary vasculature injection, pseudocolored for depth (Figure 3C); Dr. Martin Bartoš for imaging micro-CT specimens (Figure 3D); Tom and Lihi Gidor for acquiring images (Figures 4D–4F) and Matěj Kočka for collecting Figure 4C; and Prof. Michiko Watanabe and Dr. Maryse Lapierre-Landry for providing the comments to the text and proofreading.
We would also like to thank Ms. Blanka Topinková, Jarmila Svatůňková, Eva Zábrodská, and Mr. Patrik Štych for their excellent technical assistance.
Funding: This study was supported by grants from the Ministry of Education, Youth and Sports of the Czech Republic (PROGRES-Q38/LF1, INTER-COST LTC17023, and LM2015062 Czech-BioImaging) CZ.02.1.01/0.0/0.0/16_013/0001775 Modernization and support of research activities of the national infrastructure for biological and medical imaging Czech-BioImaging funded by OP RDE; Grant Agency of the Czech Republic, Czechia 18-03207S, 18-03461S, and 21-03847S; Grant Agency of Charles University, Czechia, 1456217. Institutional funding was provided through the Czech Academy of Sciences, Czechia, RVO: 67985823.
Author contributions
H.K., V.O., A.K., M.G., D.S. prepared the manuscript. H.K., V.O., D.S. prepared the figures.
Declaration of interests
The authors declare no competing or financial interests.
References
- Aanhaanen W.T.J., Mommersteeg M.T.M., Norden J., Wakker V., Vries C.de G., Anderson R.H., Kispert A., Moorman A.F.M., Christoffels V.M. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction Axis of the mouse heart. Circ. Res. 2010;107:728–736. doi: 10.1161/CIRCRESAHA.110.222992. [DOI] [PubMed] [Google Scholar]
- Akerberg A.A., Burns C.E., Burns C.G., Nguyen C. Deep learning enables automated volumetric assessments of cardiac function in zebrafish. Dis. Model. Mech. 2019;12:dmm040188. doi: 10.1242/dmm.040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S., Huynh G.H., Boyden E.S. Expansion microscopy: enabling single cell analysis in intact biological systems. FEBS J. 2019;286:1482–1494. doi: 10.1111/febs.14597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Surmi A., Wirza R., Mahmod R., Khalid F., Dimon M.Z. A new human heart vessel identification, segmentation and 3D reconstruction mechanism. J. Cardiothorac. Surg. 2014;9:161. doi: 10.1186/s13019-014-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel P. A beginner’s guide to tissue clearing. Int. J. Biochem. Cell Biol. 2017;84:35–39. doi: 10.1016/j.biocel.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaripour A., Lagerweij T., Scharfbillig C., Jadczak A.E., Willershausen B., Van Noorden C.J.F. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog. Histochem. Cytochem. 2016;51:9–23. doi: 10.1016/j.proghi.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Baek K.I., Ding Y., Chang C.-C., Chang M., Sevag Packard R.R., Hsu J.J., Fei P., Hsiai T.K. Advanced microscopy to elucidate cardiovascular injury and regeneration: 4D light-sheet imaging. Prog. Biophys. Mol. Biol. 2018;138:105–115. doi: 10.1016/j.pbiomolbio.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Jährling N., Saghafi S., Weiler R., Dodt H.-U. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beis D., Bartman T., Jin S.-W., Scott I.C., D’Amico L.A., Ober E.A., Verkade H., Frantsve J., Field H.A., Wehman A.J.D. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Belle M., Godefroy D., Couly G., Malone S.A., Collier F., Giacobini P., Chédotal A. Tridimensional visualization and analysis of early human development. Cell. 2017;169:161–173.e12. doi: 10.1016/j.cell.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Bensley J.G., De Matteo R., Harding R., Black M.J. Three-dimensional direct measurement of cardiomyocyte volume, nuclearity, and ploidy in thick histological sections. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep23756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj K.N., Spitz C., Shekhar A., Yalcin H.C., Butcher J.T. Computational fluid dynamics of developing avian outflow tract heart valves. Ann. Biomed. Eng. 2012;40:2212–2227. doi: 10.1007/s10439-012-0574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinder P., Tsai P.S., Kaufhold J.P., Knutsen P.M., Suhl H., Kleinfeld D. The cortical angiome: an interconnected vascular network with noncolumnar patterns of blood flow. Nat. Neurosci. 2013;16:889–897. doi: 10.1038/nn.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohuslavova R., Cerychova R., Papousek F., Olejnickova V., Bartos M., Görlach A., Kolar F., Sedmera D., Semenza G.L., Pavlinkova G. HIF-1α is required for development of the sympathetic nervous system. Proc. Natl. Acad. Sci. U S A. 2019;116:13414–13423. doi: 10.1073/pnas.1903510116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boselli F., Vermot J.J.M. Live imaging and modeling for shear stress quantification in the embryonic zebrafish heart. Methods. 2016;94:129–134. doi: 10.1016/j.ymeth.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Bryson J.L., Coles M.C., Manley N.R. A method for labeling vasculature in embryonic mice. J. Vis. Exp. 2011:e3267. doi: 10.3791/3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffinton C.M., Faas D., Sedmera D. Stress and strain adaptation in load-dependent remodeling of the embryonic left ventricle. Biomech. Model. Mechanobiol. 2013;12:1037–1051. doi: 10.1007/s10237-012-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher J.T., Sedmera D., Guldberg R.E., Markwald R.R. Quantitative volumetric analysis of cardiac morphogenesis assessed through micro-computed tomography. Dev. Dyn. 2007;236:802–809. doi: 10.1002/dvdy.20962. [DOI] [PubMed] [Google Scholar]
- Cahalan M.D., Parker I., Wei S.H., Miller M.J. Two-photon tissue imaging: seeing the immune system in a fresh light. Nat. Rev. Immunol. 2002;2:872–880. doi: 10.1038/nri935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon C.K., Yu Z., Wang Y., Guo F., Unruh J.R., Slaughter B.D., Hawley R.S. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U S A. 2017;114:E6857–E6866. doi: 10.1073/pnas.1705623114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo M., Chuecos M., Gandhi K., Bednov A., Moore D.L., Maher J., Ventolini G., Ji G., Schlabritz-Loutsevitch N. Optical tissue clearing in combination with perfusion and immunofluorescence for placental vascular imaging. Medicine (Baltimore) 2018;97:e12392. doi: 10.1097/MD.0000000000012392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallero S., Shen H., Yi C., Lien C.-L., Kumar S.R., Sucov H.M. CXCL12 signaling is essential for maturation of the ventricular coronary endothelial plexus and establishment of functional coronary circulation. Dev. Cell. 2015;33:469–477. doi: 10.1016/j.devcel.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebasek V., Erzen I., Vyhnal A., Janacek J., Ribaric S., Kubinova L. The estimation error of skeletal muscle capillary supply is significantly reduced by 3D method. Microvasc. Res. 2010;79:40–46. doi: 10.1016/j.mvr.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Chapman S.C., Lawson A., MacArthur W.C., Wiese R.J., Loechel R.H., Burgos-Trinidad M., Wakefield J.K., Ramabhadran R., Mauch T.J., Schoenwolf G.C. Ubiquitous GFP expression in transgenic chickens using a lentiviral vector. Development. 2005;132:935–940. doi: 10.1242/dev.01652. [DOI] [PubMed] [Google Scholar]
- Chen F., Tillberg P.W., Boyden E.S. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wassie A.T., Cote A.J., Sinha A., Alon S., Asano S., Daugharthy E.R., Chang J.-B., Marblestone A., Church G.M. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Qin C., Qiu H., Tarroni G., Duan J., Bai W., Rueckert D. Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med. 2020;7:25. doi: 10.3389/fcvm.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.J., Maga A.M., Kim E.S., Wang R.K. A feasibility study of OCT for anatomical and vascular phenotyping of mouse embryo. J. Biophotonics. 2020;13:e201960225. doi: 10.1002/jbio.201960225. [DOI] [PubMed] [Google Scholar]
- Choquet C., Kelly R.G., Miquerol L. Nkx2-5 defines distinct scaffold and recruitment phases during formation of the murine cardiac Purkinje fiber network. Nat. Commun. 2020;11:5300. doi: 10.1038/s41467-020-19150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chozinski T.J., Halpern A.R., Okawa H., Kim H.-J., Tremel G.J., Wong R.O., Vaughan J.C. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Bakal C. Accelerating live single-cell signalling studies. Trends Biotechnol. 2017;35:422–433. doi: 10.1016/j.tibtech.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Corliss B.A., Mathews C., Doty R., Rohde G., Peirce S.M. Methods to label, image, and analyze the complex structural architectures of microvascular networks. Microcirculation. 2019;26:e12520. doi: 10.1111/micc.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini I., Cicchi R., Cicchi R., Silvestri L., Vanzi F., Vanzi F., Pavone F.S., Pavone F.S. In-vivo and ex-vivo optical clearing methods for biological tissues: review. Biomed. Opt. Express. 2019;10:5251–5267. doi: 10.1364/BOE.10.005251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creswell L.L., Wyers S.G., Pirolo J.S., Perman W.H., Vannier M.W., Pasque M.K. Mathematical modeling of the heart using magnetic resonance imaging. IEEE Trans. Med. Imaging. 1992;11:581–589. doi: 10.1109/42.192695. [DOI] [PubMed] [Google Scholar]
- D’Amico L., Scott I.C., Jungblut B., Stainier D.Y. A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr. Biol. 2007;17:252–259. doi: 10.1016/j.cub.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Degenhardt Karl, Wright Alexander C., Debra H., Arun P., Epstein Jonathan A. Rapid 3D phenotyping of cardiovascular development in mouse embryos by micro-CT with iodine staining. Circ. Cardiovasc. Imaging. 2010;3:314–322. doi: 10.1161/CIRCIMAGING.109.918482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanna A.P., Tibo A., Silvestri L., Müllenbroich M.C., Costantini I., Allegra Mascaro A.L., Sacconi L., Frasconi P., Pavone F.S. Whole-brain vasculature reconstruction at the single capillary level. Sci. Rep. 2018;8:12573. doi: 10.1038/s41598-018-30533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Ma J., Langenbacher A.D., Baek K.I., Lee J., Chang C.-C., Hsu J.J., Kulkarni R.P., Belperio J., Shi W. Multiscale light-sheet for rapid imaging of cardiopulmonary system. JCI Insight. 2018;3:e121396. doi: 10.1172/jci.insight.121396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio S., Fedele M., Pontone G., Corno A.F., Zunino P., Vergara C., Quarteroni A. A computational model applied to myocardial perfusion in the human heart: From large coronaries to microvasculature. Journal of Computational Physics. 2021;424:109836. doi: 10.1016/j.jcp.2020.109836. [DOI] [Google Scholar]
- Ding Y., Gudapati V., Lin R., Fei Y., Song S., Chang C.-C., In K., Wang Z., Roustaei M., Kuang D. Saak transform-based machine learning for light-sheet imaging of cardiac trabeculation. IEEE Trans. Biomed. Eng. 2021;68:225–235. doi: 10.1109/TBME.2020.2991754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecabert O., Peters J., Walker M.J., Ivanc T., Lorenz C., von Berg J., Lessick J., Vembar M., Weese J. Segmentation of the heart and great vessels in CT images using a model-based adaptation framework. Med. Image Anal. 2011;15:863–876. doi: 10.1016/j.media.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Epah J., Pálfi K., Dienst F.L., Malacarne P.F., Bremer R., Salamon M., Kumar S., Jo H., Schürmann C., Brandes R.P. 3D imaging and quantitative analysis of vascular networks: a comparison of ultramicroscopy and micro-computed tomography. Theranostics. 2018;8:2117–2133. doi: 10.7150/thno.22610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova O., Orsini T., Gambadoro A., Chiani F., Tocchini-Valentini G.P. Three-dimensional microCT imaging of murine embryonic development from immediate post-implantation to organogenesis: application for phenotyping analysis of early embryonic lethality in mutant animals. Mamm. Genome. 2018;29:245–259. doi: 10.1007/s00335-017-9723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A., Becker K., Jährling N., Mauch C.P., Hojer C.D., Egen J.G., Hellal F., Bradke F., Sheng M., Dodt H.-U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Faulkner E.L., Thomas S.G., Neely R.K. An introduction to the methodology of expansion microscopy. Int. J. Biochem. Cell Biol. 2020;124:105764. doi: 10.1016/j.biocel.2020.105764. [DOI] [PubMed] [Google Scholar]
- Fei P., Lee J., Packard R.R.S., Sereti K.-I., Xu H., Ma J., Ding Y., Kang H., Chen H., Sung K. Cardiac light-sheet fluorescent microscopy for multi-scale and rapid imaging of architecture and function. Sci. Rep. 2016;6:22489. doi: 10.1038/srep22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca C.G., Backhaus M., Bluemke D.A., Britten R.D., Chung J.D., Cowan B.R., Dinov I.D., Finn J.P., Hunter P.J., Kadish A.H. The Cardiac Atlas Project—an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. 2011;27:2288–2295. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar A.S., Liebling M., Hickerson A., Nasiraei-Moghaddam A., Tsai H.-J., Hove J.R., Fraser S.E., Dickinson M.E., Gharib M. The embryonic vertebrate heart tube is a dynamic suction pump. Science. 2006;312:751–753. doi: 10.1126/science.1123775. [DOI] [PubMed] [Google Scholar]
- Foster D.S., Nguyen A.T., Chinta M., Salhotra A., Jones R.E., Mascharak S., Titan A.L., Ransom R.C., da Silva O.L., Foley E., Briger E., Longaker M.T. A Clearing Technique to Enhance Endogenous Fluorophores in Skin and. Soft Tissue. Sci Rep. 2019;9:15791. doi: 10.1038/s41598-019-50359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangi A.F., Niessen W.J., Viergever M.A. Three-dimensional modeling for functional analysis of cardiac images: a review. IEEE Trans. Med. Imaging. 2001;20:2–25. doi: 10.1109/42.906421. [DOI] [PubMed] [Google Scholar]
- Freifeld L., Odstrcil I., Förster D., Ramirez A., Gagnon J.A., Randlett O., Costa E.K., Asano S., Celiker O.T., Gao R. Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proc. Natl. Acad. Sci. U S A. 2017;114:E10799–E10808. doi: 10.1073/pnas.1706281114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon L., Smith A.F., Boas D.A., Devor A., Secomb T.W., Sakadžić S. Modeling of cerebral oxygen transport based on in vivo microscopic imaging of microvascular network structure, blood flow, and oxygenation. Front. Comput. Neurosci. 2016;10:82. doi: 10.3389/fncom.2016.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Maraspini R., Beutel O., Zehtabian A., Eickholt B., Honigmann A., Ewers H. Expansion stimulated emission depletion microscopy (ExSTED) ACS Nano. 2018;12:4178–4185. doi: 10.1021/acsnano.8b00776. [DOI] [PubMed] [Google Scholar]
- Gao R., Asano S.M., Boyden E.S. Q&A: expansion microscopy. BMC Biol. 2017;15:50. doi: 10.1186/s12915-017-0393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanavati S., Lerch J.P., Sled J.G. Automatic anatomical labeling of the complete cerebral vasculature in mouse models. Neuroimage. 2014;95:117–128. doi: 10.1016/j.neuroimage.2014.03.044. [DOI] [PubMed] [Google Scholar]
- Gkontra P., Norton K.-A., Żak M.M., Clemente C., Agüero J., Ibáñez B., Santos A., Popel A.S., Arroyo A.G. Deciphering microvascular changes after myocardial infarction through 3D fully automated image analysis. Sci. Rep. 2018;8:1854. doi: 10.1038/s41598-018-19758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gaviro M.V., Sanderson D., Ripoll J., Desco M. Biomedical applications of tissue clearing and three-dimensional imaging in health and disease. iScience. 2020;23:101432. doi: 10.1016/j.isci.2020.101432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W.R., Beyersdorf B.M., Paik D.T., Tian L., Li G., Buikema J.W., Chirikian O., Choi S., Venkatraman S., Adams E.L. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 2019;125:379–397. doi: 10.1161/CIRCRESAHA.118.314578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A., Bode J., Alexander A., Karimian-Jazi K., Schregel K., Schwarz D., Sommerkamp A.C., Krüwel T., Abdollahi A., Wick W. Large-scale characterization of the microvascular geometry in development and disease by tissue clearing and quantitative ultramicroscopy. J. Cereb. Blood Flow Metab. 2020 doi: 10.1177/0271678X20961854. 271678X20961854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern A.R., Alas G.C., Chozinski T.J., Paredez A.R., Vaughan J.C. Hybrid structured illumination expansion microscopy reveals microbial cytoskeleton organization. ACS Nano. 2017;11:12677–12686. doi: 10.1021/acsnano.7b07200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Hioki H., Namiki K., Hoshida T., Kurokawa H., Ishidate F., Kaneko T., Akagi T., Saito T., Saido T., Miyawaki A. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- Hasan M.R., Herz J., Hermann D.M., Doeppner T.R. Visualization of macroscopic cerebral vessel anatomy--a new and reliable technique in mice. J. Neurosci. Methods. 2012;204:249–253. doi: 10.1016/j.jneumeth.2011.11.024. [DOI] [PubMed] [Google Scholar]
- He L., Liu Q., Hu T., Huang X., Zhang H., Tian X., Yan Y., Wang L., Huang Y., Miquerol L. Genetic lineage tracing discloses arteriogenesis as the main mechanism for collateral growth in the mouse heart. Cardiovasc. Res. 2016;109:419–430. doi: 10.1093/cvr/cvw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig M.G., Hume R.I. Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing. Trends Neurosci. 1989;12:340–341. [PubMed] [Google Scholar]
- Hou B., Zhang D., Zhao S., Wei M., Yang Z., Wang S., Wang J., Zhang X., Liu B., Fan L. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Christensen D.A., Agrawal A.K., Beaumont C., Clark E.B., Hawkins J.A. Dependence of aortic arch morphogenesis on intracardiac blood flow in the left atrial ligated chick embryo. Anat. Rec. 2009;292:652–660. doi: 10.1002/ar.20885. [DOI] [PubMed] [Google Scholar]
- Ivanovitch K., Temiño S., Torres M. Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis. Elife. 2017;6:e30668. doi: 10.7554/eLife.30668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B., Boukens B.J.D., Postma A.V., Gunst Q.D., van den Hoff M.J.B., Moorman A.F.M., Wang T., Christoffels V.M. Identifying the evolutionary building blocks of the cardiac conduction system. PLoS One. 2012;7:e44231. doi: 10.1371/journal.pone.0044231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivins S., Roberts C., Vernay B., Scambler P.J. Analysis of coronary vessels in cleared embryonic hearts. J. Vis. Exp. 2016:54800. doi: 10.3791/54800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.H.R., Berg R.W. CLARITY-compatible lipophilic dyes for electrode marking and neuronal tracing. Sci. Rep. 2016;6:32674. doi: 10.1038/srep32674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilani S.M., Murphy T.J., Thai S.N.M., Eichmann A., Alva J.A., Iruela-Arispe M.L. Selective binding of lectins to embryonic chicken vasculature. J. Histochem. Cytochem. 2003;51:597–604. doi: 10.1177/002215540305100505. [DOI] [PubMed] [Google Scholar]
- Junaid T.O., Bradley R.S., Lewis R.M., Aplin J.D., Johnstone E.D. Whole organ vascular casting and microCT examination of the human placental vascular tree reveals novel alterations associated with pregnancy disease. Sci. Rep. 2017;7:4144. doi: 10.1038/s41598-017-04379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuscinski J. DAPI: a DNA-specific fluorescent probe. Biotech. Histochem. 1995;70:220–233. doi: 10.3109/10520299509108199. [DOI] [PubMed] [Google Scholar]
- Kattan J., Dettman R.W., Bristow J. Formation and remodeling of the coronary vascular bed in the embryonic avian heart. Dev. Dyn. 2004;230:34–43. doi: 10.1002/dvdy.20022. [DOI] [PubMed] [Google Scholar]
- Ke M.-T., Fujimoto S., Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Keller P.J., Dodt H.-U. Light sheet microscopy of living or cleared specimens. Curr. Opin. Neurobiol. 2012;22:138–143. doi: 10.1016/j.conb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Kennel P., Dichamp J., Barreau C., Guissard C., Teyssedre L., Rouquette J., Colombelli J., Lorsignol A., Casteilla L., Plouraboué F. From whole-organ imaging to in-silico blood flow modeling: a new multi-scale network analysis for revisiting tissue functional anatomy. PLoS Comput. Biol. 2020;16:e1007322. doi: 10.1371/journal.pcbi.1007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennel P., Teyssedre L., Colombelli J., Plouraboué F. Toward quantitative three-dimensional microvascular networks segmentation with multiview light-sheet fluorescence microscopy. JBO. 2018;23:086002. doi: 10.1117/1.JBO.23.8.086002. [DOI] [PubMed] [Google Scholar]
- Khoradmehr A., Mazaheri F., Anvari M., Tamadon A. A Simple Technique for Three-Dimensional Imaging and Segmentation of Brain Vasculature U sing Fast Free-of-Acrylamide Clearing Tissue in Murine. Cell J. 2019;21:49–56. doi: 10.22074/cellj.2019.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro H., Yonei-Tamura S., Tamura K., Schoenwolf G.C., Saijoh Y. The heart tube forms and elongates through dynamic cell rearrangement coordinated with foregut extension. Development. 2018;145:dev152488. doi: 10.1242/dev.152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Ahn H.H., Sun W., Rhyu I.J. Electrophoretic tissue clearing and labeling methods for volume imaging of whole organs. AM. 2016;46:134–139. [Google Scholar]
- Kim S.-Y., Cho J.H., Murray E., Bakh N., Choi H., Ohn K., Ruelas L., Hubbert A., McCue M., Vassallo S.L. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. U SA. 2015;112:E6274–E6283. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst C., Skriabine S., Vieites-Prado A., Topilko T., Bertin P., Gerschenfeld G., Verny F., Topilko P., Michalski N., Tessier-Lavigne M., Renier N. Mapping the fine-scale organization and plasticity of the brain vasculature. Cell. 2020;180:780–795.e25. doi: 10.1016/j.cell.2020.01.028. [DOI] [PubMed] [Google Scholar]
- Klingberg A., Hasenberg A., Ludwig-Portugall I., Medyukhina A., Männ L., Brenzel A., Engel D.R., Figge M.T., Kurts C., Gunzer M. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J. Am. Soc. Nephrol. 2017;28:452–459. doi: 10.1681/ASN.2016020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesová H., Bartoš M., Hsieh W.C., Olejníčková V., Sedmera D. Novel approaches to study coronary vasculature development in mice. Dev Dyn. 2018;247:1018–1027. doi: 10.1002/dvdy.24637. [DOI] [PubMed] [Google Scholar]
- Kolesova H., Capek M., Radochova B., Janacek J., Sedmera D. Comparison of different tissue clearing methods and 3D imaging techniques for visualization of GFP-expressing mouse embryos and embryonic hearts. Histochem. Cell. Biol. 2016;146:141–152. doi: 10.1007/s00418-016-1441-8. [DOI] [PubMed] [Google Scholar]
- Kugler E., Plant K., Chico T., Armitage P. Enhancement and segmentation Workflow for the developing zebrafish vasculature. J. Imaging. 2019;5:14. doi: 10.3390/jimaging5010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerweij T., Dusoswa S.A., Negrean A., Hendrikx E.M.L., de Vries H.E., Kole J., Garcia-Vallejo J.J., Mansvelder H.D., Vandertop W.P., Noske D.P. Optical clearing and fluorescence deep-tissue imaging for 3D quantitative analysis of the brain tumor microenvironment. Angiogenesis. 2017;20:533–546. doi: 10.1007/s10456-017-9565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankford K.L., Arroyo E.J., Nazimek K., Bryniarski K., Askenase P.W., Kocsis J.D. Intravenously delivered mesenchymal stem cell-derived exosomes target M2-type macrophages in the injured spinal cord. PLoS One. 2018;13:e0190358. doi: 10.1371/journal.pone.0190358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansford R., Rugonyi S. Follow me! A tale of avian heart development with comparisons to mammal heart development. J. Cardiovasc. Dev. Dis. 2020;7:8. doi: 10.3390/jcdd7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre-Landry M., Kolesova H., Liu Y., Watanabe M., Jenkins M.W. Three-dimensional alignment of microvasculature and cardiomyocytes in the developing ventricle. Sci. Rep. 2020;10:14955. doi: 10.1038/s41598-020-71816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N.D., Weinstein B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Le N.A., Kuo W., Müller B., Kurtcuoglu V., Spingler B. Crosslinkable polymeric contrast agent for high-resolution X-ray imaging of the vascular system. Chem. Commun. (Camb.) 2020;56:5885–5888. doi: 10.1039/c9cc09883f. [DOI] [PubMed] [Google Scholar]
- Lee E., Choi J., Jo Y., Kim J.Y., Jang Y.J., Lee H.M., Kim S.Y., Lee H.-J., Cho K., Jung N. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Park J.-H., Seo I., Park S.-H., Kim S. Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung, and intestine. BMC Dev. Biol. 2014;14:48. doi: 10.1186/s12861-014-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-E., Nguyen C., Yoon J., Chang H.-J., Kim S., Kim C.H., Li D. Three-dimensional cardiomyocytes structure revealed by diffusion tensor imaging and its validation using a tissue-clearing technique. Sci. Rep. 2018;8:6640. doi: 10.1038/s41598-018-24622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu T., Tarokh A., Nie J., Guo L., Mara A., Holley S., Wong S.T. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 2007;8:40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Song Y., Zhao L., Gaidosh G., Laties A.M., Wen R. Direct labeling and visualization of blood vessels with lipophilic carbocyanine dye DiI. Nat. Protoc. 2008;3:1703–1708. doi: 10.1038/nprot.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Jingjing, Miao L., Shieh D., Spiotto E., Li J., Zhou B., Paul A., Schwartz R.J., Firulli A.B., Singer H.A. Single-cell lineage tracing reveals that oriented cell division contributes to trabecular morphogenesis and regional specification. Cell Rep. 2016;15:158–170. doi: 10.1016/j.celrep.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebling M., Forouhar A.S., Gharib M., Fraser S.E., Dickinson M.E. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J. Biomed. Opt. 2005;10:054001. doi: 10.1117/1.2061567. [DOI] [PubMed] [Google Scholar]
- Liu Y., Broberg M.C.G., Watanabe M., Rollins A.M., Jenkins M.W. SLIME: robust, high-speed 3D microvascular mapping. Sci. Rep. 2019;9:893. doi: 10.1038/s41598-018-37313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.K.L., Lai H.M., Chang R.C.-C., Gentleman S.M. Free of acrylamide sodium dodecyl sulphate (SDS)-based tissue clearing (FASTClear): a novel protocol of tissue clearing for three-dimensional visualization of human brain tissues. Neuropathol Appl Neurobiol. 2017;43:346–351. doi: 10.1111/nan.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K.L., Finney B.A., Deppermann C., Hägerling R., Gazit S.L., Frampton J., Buckley C., Camerer E., Nieswandt B., Kiefer F., Watson S.P. Podoplanin and CLEC-2 drive cerebrovascular patterning and integrity during development. Blood. 2015;125:3769–3777. doi: 10.1182/blood-2014-09-603803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Hernandez E., Squire A., Hagemann N., Brenzel A., Sardari M., Schlechter J., Sanchez-Mendoza E.H., Gunzer M., Faissner A., Hermann D.M. 3D visualization and quantification of microvessels in the whole ischemic mouse brain using solvent-based clearing and light sheet microscopy. J. Cereb. Blood Flow Metab. 2017;37:3355–3367. doi: 10.1177/0271678X17698970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Hata R., Hossmann K.A. Differences in the cerebrovascular anatomy of C57black/6 and SV129 mice. Neuroreport. 1998;9:1317–1319. doi: 10.1097/00001756-199805110-00012. [DOI] [PubMed] [Google Scholar]
- Matryba P., Kaczmarek L., Gołąb J. Advances in ex situ tissue optical clearing. Laser Photon. Rev. 2019;13:1800292. [Google Scholar]
- Mellman K., Huisken J., Dinsmore C., Hoppe C., Stainier D.Y. Fibrillin-2b regulates endocardial morphogenesis in zebrafish. Dev. Biol. 2012;372:111–119. doi: 10.1016/j.ydbio.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Migliori B., Datta M.S., Dupre C., Apak M.C., Asano S., Gao R., Boyden E.S., Hermanson O., Yuste R., Tomer R. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. 2018;16:57. doi: 10.1186/s12915-018-0521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.E., Thompson R.P., Bigelow M.R., Gittinger G., Trusk T.C., Sedmera D. Confocal imaging of the embryonic heart: how deep? Microscop. Microanal. 2005;11:216–223. doi: 10.1017/S1431927605050464. [DOI] [PubMed] [Google Scholar]
- Miquerol L., Moreno-Rascon N., Beyer S., Dupays L., Meilhac S.M., Buckingham M.E., Franco D., Kelly R.G. Biphasic development of the mammalian ventricular conduction system. Circ. Res. 2010;107:153–161. doi: 10.1161/CIRCRESAHA.110.218156. [DOI] [PubMed] [Google Scholar]
- Mohan R.A., Mommersteeg M.T.M., Domínguez J.N., Choquet C., Wakker V., Vries C.de G., Boink G.J.J., Boukens B.J., Miquerol L., Verkerk A.O., Christoffels V.M. Embryonic Tbx3+ cardiomyocytes form the mature cardiac conduction system by progressive fate restriction. Development. 2018;145:dev167361. doi: 10.1242/dev.167361. [DOI] [PubMed] [Google Scholar]
- Molbay M., Kolabas Z.I., Todorov M.I., Ohn T.-L., Ertürk A. A guidebook for DISCO tissue clearing. Mol Syst Biol. 2021;17:e9807. doi: 10.15252/msb.20209807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T.C., Mano T., Saikawa S., Horiguchi S.A., Shigeta D., Baba K., Sekiya H., Shimizu Y., Tanaka K.F., Kiyonari H. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 2018;21:625–637. doi: 10.1038/s41593-018-0109-1. [DOI] [PubMed] [Google Scholar]
- Murray E., Cho J.H., Goodwin D., Ku T., Swaney J., Kim S.-Y., Choi H., Park Y.-G., Park J.-Y., Hubbert A. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagyova M., Slovinska L., Blasko J., Grulova I., Kuricova M., Cigankova V., Harvanova D., Cizkova D. A comparative study of PKH67, DiI, and BrdU labeling techniques for tracing rat mesenchymal stem cells. In Vitro Cell. Dev. Biol. Anim. 2014;50:656–663. doi: 10.1007/s11626-014-9750-5. [DOI] [PubMed] [Google Scholar]
- Nanka O., Krizova P., Fikrle M., Tuma M., Blaha M., Grim M., Sedmera D. Abnormal myocardial and coronary vasculature development in experimental hypoxia. Anat. Rec. (Hoboken) 2008;291:1187–1199. doi: 10.1002/ar.20738. [DOI] [PubMed] [Google Scholar]
- Neckel P.H., Mattheus U., Hirt B., Just L., Mack A.F. Large-scale tissue clearing (PACT): Technical evaluation and new perspectives in immunofluorescence, histology, and ultrastructure. Sci Rep. 2016;6:34331. doi: 10.1038/srep34331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrhoff I., Bocancea D., Vaquero J., Vaquero J.J., Ripoll J., Desco M., Gómez-Gaviro M.V. 3D imaging in CUBIC-cleared mouse heart tissue: going deeper. Biomed. Opt. Express. 2016;7:3716–3720. doi: 10.1364/BOE.7.003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C.T., Lu Q., Wang Y., Chen J.-N. Zebrafish as a model for cardiovascular development and disease. Drug Discov. Today Dis. Models. 2008;5:135–140. doi: 10.1016/j.ddmod.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima S., Susaki E.A., Yoshida K., Takemoto H., Tsujimura N., Iijima S., Takachi K., Nakahara Y., Tahara S., Ohshima K. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci. Rep. 2017;7:9269. doi: 10.1038/s41598-017-09117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejnickova V., Sankova B., Sedmera D., Janacek J. Trabecular architecture determines impulse propagation through the early embryonic mouse heart. Front. Physiol. 2018;9:1876. doi: 10.3389/fphys.2018.01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard R.R.S., Baek K.I., Beebe T., Jen N., Ding Yichen, Shi F., Fei P., Kang B.J., Chen P.-H., Gau J. Automated segmentation of light-sheet fluorescent imaging to characterize experimental doxorubicin-induced cardiac injury and repair. Sci. Rep. 2017;7:8603. doi: 10.1038/s41598-017-09152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Cai R., Quacquarelli F.P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., Plesnila N., Dichgans M., Hellal F., Ertürk A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- Park O.K., Kwak J., Jung Y.J., Kim Y.H., Hong H.-S., Hwang B.J., Kwon S.-H., Kee Y. 3D light-sheet fluorescence microscopy of cranial neurons and vasculature during zebrafish embryogenesis. Mol. Cells. 2015;38:975–981. doi: 10.14348/molcells.2015.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbellini F., Liu A.K.L., Watson S.A., Bardi I., Rothery S.M., Terracciano C.M. Free-of-Acrylamide SDS-based Tissue Clearing (FASTClear) for three dimensional visualization of myocardial tissue. Sci. Rep. 2017;7:5188. doi: 10.1038/s41598-017-05406-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdikaris P., Grinberg L., Karniadakis G.E. Multiscale modeling and simulation of brain blood flow. Phys. Fluids. 2016;28:021304. doi: 10.1063/1.4941315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitman Mayo R. Advances in human placental biomechanics. Comput. Struct. Biotechnol. J. 2018;16:298–306. doi: 10.1016/j.csbj.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proweller A., Wright A.C., Horng D., Cheng L., Lu M.M., Lepore J.J., Pear W.S., Parmacek M.S. Notch signaling in vascular smooth muscle cells is required to pattern the cerebral vasculature. Proc. Natl. Acad. Sci. U S A. 2007;104:16275–16280. doi: 10.1073/pnas.0707950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Yu T., Xu J., Wan P., Ma Y., Zhu J., Li Y., Gong H., Luo Q., Zhu D. FDISCO: advanced solvent-based clearing method for imaging whole organs. Sci. Adv. 2019;5:eaau8355. doi: 10.1126/sciadv.aau8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K., Ueno H., Weissman I.L., Krasnow M.A. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N., Adams E.L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A.E., Kadiri L., Umadevi Venkataraju K., Zhou Y. Mapping of brain activity by automated volume Analysis of immediate early genes. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D.S., Lichtman J.W. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L., Inman K., Trainor P. DAPI staining of whole-mount mouse embryos or fetal organs. Cold Spring Harb. Protoc. 2018;2018:prot094029. doi: 10.1101/pdb.prot094029. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Schmued L. In vivo administration of fluorescent dextrans for the specific and sensitive localization of brain vascular pericytes and their characterization in normal and neurotoxin exposed brains. Neurotoxicology. 2012;33:436–443. doi: 10.1016/j.neuro.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Sasse P., Malan D., Fleischmann M., Roell W., Gustafsson E., Bostani T., Fan Y., Kolbe T., Breitbach M., Addicks K. Perlecan is critical for heart stability. Cardiovasc. Res. 2008;80:435–444. doi: 10.1093/cvr/cvn225. [DOI] [PubMed] [Google Scholar]
- Schaad L., Hlushchuk R., Barré S., Gianni-Barrera R., Haberthür D., Banfi A., Djonov V. Correlative imaging of the murine hind limb vasculature and muscle tissue by MicroCT and light microscopy. Sci. Rep. 2017;7:41842. doi: 10.1038/srep41842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach S.J., zag Bag S., Groden C., Schilling L., Brockmann M.A. Vascular imaging in small rodents using micro-CT. Methods. 2010;50:26–35. doi: 10.1016/j.ymeth.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Schürmann C., Gremse F., Jo H., Kiessling F., Brandes R.P. Micro-CT technique is well suited for documentation of remodeling processes in murine carotid arteries. PLoS One. 2015;10:e0130374. doi: 10.1371/journal.pone.0130374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M.K., Scherbarth A., Sprengel R., Engelhardt J., Theer P., Giese G. Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS One. 2015;10:e0124650. doi: 10.1371/journal.pone.0124650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmera D., Misek I., Klima M., Thompson R.P. Heart development in the spotted dolphin (Stenella attenuata) Anat. Rec. A. Discov. Mol. Cell. Evol. Biol. 2003;273:687–699. doi: 10.1002/ar.a.10086. [DOI] [PubMed] [Google Scholar]
- Sereti K.-I., Nguyen N.B., Kamran P., Zhao P., Ranjbarvaziri S., Park S., Sabri S., Engel J.L., Sung K., Kulkarni R.P. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat. Commun. 2018;9:754. doi: 10.1038/s41467-018-02891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh Qureshi W.M., Miao L., Shieh D., Li J., Lu Y., Hu S., Barroso M., Mazurkiewicz J., Wu M. Imaging cleared embryonic and postnatal hearts at single-cell resolution. J. Vis. Exp. 2016;7:54303. doi: 10.3791/54303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Zhao Y., Jiang S., Jiang H. In-vivo hemodynamic imaging of acute prenatal ethanol exposure in fetal brain by photoacoustic tomography. J. Biophotonics. 2020;13:e201960161. doi: 10.1002/jbio.201960161. [DOI] [PubMed] [Google Scholar]
- Sharma B., Chang A., Red-Horse K. Coronary artery development: progenitor cells and differentiation pathways. Annu. Rev. Physiol. 2017;79:1–19. doi: 10.1146/annurev-physiol-022516-033953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J., Ahlgren U., Perry P., Hill B., Ross A., Hecksher-Sørensen J., Baldock R., Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Silvestri L., Costantini I., Sacconi L., Pavone F.S. Clearing of fixed tissue: a review from a microscopist’s perspective. JBO. 2016;21:081205. doi: 10.1117/1.JBO.21.8.081205. [DOI] [PubMed] [Google Scholar]
- Sizarov A., Ya J., de Boer B.A., Lamers W.H., Christoffels V.M., Moorman A.F.M. formation of the building plan of the human heart: morphogenesis, growth, and differentiation. Circulation. 2011;123:1125–1135. doi: 10.1161/CIRCULATIONAHA.110.980607. [DOI] [PubMed] [Google Scholar]
- Stainier D., Fouquet B., Chen J.-N., Warren K.S., Weinstein B.M., Meiler S.E., Mohideen M., Neuhauss S., Solnica-Krezel L., Schier A.F. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Stephens D.J., Allan V.J. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- Sung K., Ding Y., Ma J., Chen H., Huang V., Cheng M., Yang C.F., Kim J.T., Eguchi D., Di Carlo D. Simplified three-dimensional tissue clearing and incorporation of colorimetric phenotyping. Sci. Rep. 2016;6:30736. doi: 10.1038/srep30736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T.M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Kubota S.I., Suyama T.Q., Susaki E.A., Perrin D., Ukai-Tadenuma M., Ukai H., Ueda H.R. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Tainaka K., Kuno A., Kubota S.I., Murakami T., Ueda H.R. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev. Biol. 2016;32:713–741. doi: 10.1146/annurev-cellbio-111315-125001. [DOI] [PubMed] [Google Scholar]
- Tarnowski B.I., Spinale F.G., Nicholson J.H. DAPI as a useful stain for nuclear quantitation. Biotech. Histochem. 1991;66:297–302. [PubMed] [Google Scholar]
- Taylor J.M., Nelson C.J., Bruton F.A., Baghbadrani A.K., Buckley C., Tucker C.S., Rossi A.G., Mullins J.J., Denvir M.A. Adaptive prospective optical gating enables day-long 3D time-lapse imaging of the beating embryonic zebrafish heart. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-019-13112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Yang Z., Li X. Tissue clearing technique: recent progress and biomedical applications. J. Anat. 2021;238:489–507. doi: 10.1111/joa.13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg P.W., Chen F., Piatkevich K.D., Zhao Y., Yu C.-C., Jay), English B.P., Gao L., Martorell A., Suk H.-J. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov M.I., Paetzold J.C., Schoppe O., Tetteh G., Shit S., Efremov V., Todorov-Völgyi K., Düring M., Dichgans M., Piraud M. Machine learning analysis of whole mouse brain vasculature. Nat. Methods. 2020;17:442–449. doi: 10.1038/s41592-020-0792-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R., Ye L., Hsueh B., Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D., Paw B.H., Poss K.D., Penberthy W.T., Lin S., Zon L.I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Treweek J.B., Chan K.Y., Flytzanis N.C., Yang B., Deverman B.E., Greenbaum A., Lignell A., Xiao C., Cai L., Ladinsky M.S. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 2015;10:1860–1896. doi: 10.1038/nprot.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong T.V., Supatto W., Koos D.S., Choi J.M., Fraser S.E. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- Tsai P.S., Kaufhold J.P., Blinder P., Friedman B., Drew P.J., Karten H.J., Lyden P.D., Kleinfeld D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J Neurosci. 2009;29:14553–14570. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H.R., Ertürk A., Chung K., Gradinaru V., Chédotal A., Tomancak P., Keller P.J. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 2020;21:61–79. doi: 10.1038/s41583-019-0250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eif V.W.W., Devalla H.D., Boink G.J.J., Christoffels V.M. Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol. 2018;15:617–630. doi: 10.1038/s41569-018-0031-y. [DOI] [PubMed] [Google Scholar]
- van Zandvoort M., Engels W., Douma K., Beckers L., Oude Egbrink M., Daemen M., Slaaf D.W. Two-photon microscopy for imaging of the (atherosclerotic) vascular wall: a proof of concept study. J. Vasc. Res. 2004;41:54–63. doi: 10.1159/000076246. [DOI] [PubMed] [Google Scholar]
- Veith A., B Baker A. A rapid, nondestructive method for vascular network visualization. Biotechniques. 2020;69:443–449. doi: 10.2144/btn-2020-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux R.J., Belle M., Chédotal A. Neuroscience in the third dimension: shedding new light on the brain with tissue clearing. Mol. Brain. 2017;10:33. doi: 10.1186/s13041-017-0314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbacky M., Kovalcikova J., Chawengsaksophak K., Beck I.M., Mracek T., Nuskova H., Sedmera D., Papousek F., Kolar F., Sobol M. Knockout of Tmem70 alters biogenesis of ATP synthase and leads to embryonal lethality in mice. Hum. Mol. Genet. 2016;25:4674–4685. doi: 10.1093/hmg/ddw295. [DOI] [PubMed] [Google Scholar]
- Walter T., Shattuck D.W., Baldock R., Bastin M.E., Carpenter A.E., Duce S., Ellenberg J., Fraser A., Hamilton N., Pieper S. Visualization of image data from cells to organisms. Nat. Methods. 2010;7:S26–S41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Wang Y., Wang W., Luo D., Chitgupi U., Geng J., Zhou Y., Wang L., Lovell J.F., Xia J. Deep tissue photoacoustic computed tomography with a fast and compact laser system. Biomed. Opt. Express. 2016;8:112–123. doi: 10.1364/BOE.8.000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dur O., Patrick M.J., Tinney J.P., Tobita K., Keller B.B., Pekkan K. Aortic arch morphogenesis and flow modeling in the chick embryo. Ann. Biomed. Eng. 2009;37:1069–1081. doi: 10.1007/s10439-009-9682-5. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang Jie, Fan G., Zhao H., Wang X., Zhang Jing, Zhang P., Wang W. Imaging transparent intact cardiac tissue with single-cell resolution. Biomed. Opt. Express. 2018;9:423. doi: 10.1364/BOE.9.000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassie A.T., Zhao Y., Boyden E.S. Expansion microscopy: principles and uses in biological research. Nat. Methods. 2019;16:33–41. doi: 10.1038/s41592-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger W.J., Geyer S.H., Mohun T.J., Rasskin-Gutman D., Matsui T., Ribeiro I., Costa L.da F., Izpisúa-Belmonte J.C., Müller G.B. High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat. Embryol. 2006;211:213–221. doi: 10.1007/s00429-005-0073-x. [DOI] [PubMed] [Google Scholar]
- Wu Z., Rademakers T., Kiessling F., Vogt M., Westein E., Weber C., Megens R.T.A., van Zandvoort M. Multi-photon microscopy in cardiovascular research. Methods. 2017;130:79–89. doi: 10.1016/j.ymeth.2017.04.013. [DOI] [PubMed] [Google Scholar]
- Xian Z., Wang X., Yan S., Yang D., Chen J., Peng C. Main coronary vessel segmentation using deep learning in smart medical. Math. Probl. Eng. 2020:8858344. doi: 10.1155/2020/8858344. [DOI] [Google Scholar]
- Xu H., Tong Z., Ye Q., Sun T., Hong Z., Zhang L., Bortnick A., Cho S., Beuzer P., Axelrod J. Molecular organization of mammalian meiotic chromosome axis revealed by expansion STORM microscopy. Proc. Natl. Acad. Sci. U S A. 2019;116:18423–18428. doi: 10.1073/pnas.1902440116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Ma Y., Yu T., Zhu D. Quantitative assessment of optical clearing methods in various intact mouse organs. J. Biophotonics. 2019;12:e201800134. doi: 10.1002/jbio.201800134. [DOI] [PubMed] [Google Scholar]
- Yalcin H.C., Shekhar A., McQuinn T.C., Butcher J.T. Hemodynamic patterning of the avian atrioventricular valve. Dev. Dyn. 2011;240:23–35. doi: 10.1002/dvdy.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Kweon J., Roh J.-H., Lee J.-H., Kang H., Park L.-J., Kim D.J., Yang H., Hur J., Kang D.-Y. Deep learning segmentation of major vessels in X-ray coronary angiography. Sci. Rep. 2019;9:16897. doi: 10.1038/s41598-019-53254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Treweek J.B., Kulkarni R.P., Deverman B.E., Chen C.-K., Lubeck E., Shah S., Cai L., Gradinaru V. Single-Cell Phenotyping within Transparent Intact Tissue Through Whole-Body Clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T., Lee J.-K., Miwa K., Opthof T., Tomoyama S., Nakanishi H., Yoshida A., Yasui H., Iida T., Miyagawa S. Quantification of sympathetic hyperinnervation and denervation after myocardial infarction by three-dimensional assessment of the cardiac sympathetic network in cleared transparent murine hearts. PLoS One. 2017;12:e0182072. doi: 10.1371/journal.pone.0182072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Zong W., Li X., Li J., Zhang Y., Wu R., Liu Y., Cui J., Wang Q., Bian Y. Long-term, in toto live imaging of cardiomyocyte behaviour during mouse ventricle chamber formation at single-cell resolution. PLoS One. 2020;15:e0226791. doi: 10.1038/s41556-020-0475-2. [DOI] [PubMed] [Google Scholar]
- Zagorchev L., Oses P., Zhuang Z.W., Moodie K., Mulligan-Kehoe M.J., Simons M., Couffinhal T. Micro computed tomography for vascular exploration. J. Angiogenes. Res. 2010;2:7. doi: 10.1186/2040-2384-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chalothorn D., Faber J.E. Collateral vessels have unique endothelial and smooth muscle cell phenotypes. Int. J. Mol. Sci. 2019;20:3608. doi: 10.3390/ijms20153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.G., Zhang L., Tsang W., Soltanian-Zadeh H., Morris D., Zhang R., Goussev A., Powers C., Yeich T., Chopp M. Correlation of VEGF and angiopoietin expression with disruption of blood-brain barrier and angiogenesis after focal cerebral ischemia. J. Cereb. Blood Flow Metab. 2002;22:379–392. doi: 10.1097/00004647-200204000-00002. [DOI] [PubMed] [Google Scholar]
- Zhao X., Wu J., Gray C.D., McGregor K., Rossi A.G., Morrison H., Jansen M.A., Gray G.A. Optical projection tomography permits efficient assessment of infarct volume in the murine heart postmyocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 2015;309:H702–H710. doi: 10.1152/ajpheart.00233.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Kong S.W., Hu Y., Campbell P.H., McGowan F.X., Ackerman K.G., Wu B., Zhou B., Tevosian S.G., Pu W.T. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J. Clin. Invest. 2009;119:1462–1476. doi: 10.1172/JCI38723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yu T., Li Y., Xu J., Qi Y., Yao Y., Ma Y., Wan P., Chen Z., Li X. MACS: rapid aqueous clearing system for 3D mapping of intact organs. Adv. Sci. (Weinh) 2020;7:1903185. doi: 10.1002/advs.201903185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker, R.M., Hunter, S., Rogers, J.M., 1998. Confocal laser scanning microscopy of apoptosis in organogenesis-stage mouse embryos. Cytometry 33, 348–354. 10.1002/(sici)1097-0320(19981101)33:3%3C348::aid-cyto9%3E3.0.co;2-c. [DOI] [PubMed]