Graphical abstract
Keywords: Metabolomics, Mass spectrometry, Multiblock PCA, Type 2 Diabetes, Biomarkers
Abbreviations: LC/MS, liquid chromatography mass spectrometry; GC/MS, gas chromatography mass spectrometry; CE/MS, capillary electrophoresis mass spectrometry; PCA, principal component analysis; SD, Sprague Dawley; ZDF, Zucker diabetic fatty; QC, quality control; CV, coefficient of variation; UPLC, ultra-performance liquid chromatography; ESI, electrospray ionization; TG, triacylglycerol; TCA, tricarboxylic acid. CoA, coenzyme A; AMP, adenosine monophosphate; FABP, fatty acid-binding protein; PPAR, peroxisome proliferator-activated receptor
Highlights
-
•
“Multiblock metabolomics” elucidates the global metabolic network in a whole body.
-
•
“Multiblock metabolomics” combines LC/MS-based metabolomics with multiblock PCA.
-
•
“Multiblock metabolomics” highlights and elicits organ-specific metabolism.
-
•
TGs with less unsaturated fatty acids were highly accumulated in the diabetic liver.
Abstract
Principal component analysis (PCA) is a useful tool for omics analysis to identify underlying factors and visualize relationships between biomarkers. However, this approach is limited in addressing life complexity and further improvement is required. This study aimed to develop a new approach that combines mass spectrometry-based metabolomics with multiblock PCA to elucidate the whole-body global metabolic network, thereby generating comparable metabolite maps to clarify the metabolic relationships among several organs. To evaluate the newly developed method, Zucker diabetic fatty (ZDF) rats (n = 6) were used as type 2 diabetic models and Sprague Dawley (SD) rats (n = 6) as controls. Metabolites in the heart, kidney, and liver were analyzed by capillary electrophoresis and liquid chromatography mass spectrometry, respectively, and the detected metabolites were analyzed by multiblock PCA. More than 300 metabolites were detected in the heart, kidney, and liver. When the metabolites obtained from the three organs were analyzed with multiblock PCA, the score and loading maps obtained were highly synchronized and their metabolism patterns were visually comparable. A significant finding in this study was the different expression patterns in lipid metabolism among the three organs; notably triacylglycerols with polyunsaturated fatty acids or less unsaturated fatty acids showed specific accumulation patterns depending on the organs.
1. Introduction
Comprehensive analysis “omics” is a new approach to address life complexity [1]. Metabolomics clarifies disease-associated biochemical activities using noninvasive samples such as blood and urine [2], [3]. The most significant issue for metabolomics research is handling enormous amount of data to uncover the underlying cellular messages. However, since the data obtained from a single organ is large, the data collected from multiple organs (or multiple regions) becomes considerable to handle effectively [4], [5]. Classical pathway analysis is limited in eliciting latent biological functions as detected metabolites are usually assigned to already known metabolic pathways; consequently, unknown functions or relations are rarely discovered [6].
PCA has been widely used in metabolomic analysis to identify underlying factors or to visualize the relationships between biomarkers [7]. However, its ability is limited in addressing complex and considerable metabolomics data. PLS-DA and OPLS-DA, the extension models of PCA, are powerful tools to discover significant biomarkers distinguishing two or more groups [8], [9]; however, these tools are disadvantageous in overviewing metabolic expression patterns or elucidating the relationship between biomarkers. Several metabolomic studies with PCA have been conducted to uncover domain-specific metabolism using multiple tissues [10]; however, the small metabolic changes in some domains are often obscured by the large metabolic changes in the other domains. Since the fundamental function of PCA is to simplify data while retaining data variation [7], conventional PCA is limited in highlighting minor changes when handling several block data. Recently, several PCA methods dealing with multiple blocks have been developed to improve the interpretation of complicated events [11], [12], [13], [14], [15], [16], [17], [18], of which multiblock PCA has been used for metabolomics [19], [20]. However, these have been only to reduce the complexity of multiple data; therefore, multiblock PCA has not yet addressed the issue of domain-specific metabolism.
Here, we propose “multiblock metabolomics” to elucidate whole-body metabolism by combining capillary electrophoresis mass spectrometry (CE/MS) [21] and LC/MS metabolomics with multiblock PCA. This approach was evaluated by analyzing the heart, kidney, and liver of a type 2 diabetes rat model, Zucker diabetic fatty (ZDF) model [22]. This murine model has a mutation in the leptin receptor, accompanied by enhanced β-cell destruction and impaired glucose homeostasis. Since the metabolism of this model has been analyzed using urine, plasma, or a single organ [23], multiple organ analysis will provide new insights regarding the entire metabolism in diabetic rats. SD rats were used as a control; these are widely used as a control for diabetic studies using the ZDF model [24], [25], [26], [27].
2. Materials and methods
2.1. Algorithm for multiblock PCA strategies
The multiblock PCA program was generated following Hassani’s method [16]. The procedure is briefly summarized below.
2.1.1. Preparation
Matrix Xb, composed of N (samples) × Kb (metabolites), was individually prepared for the heart (b = 1), kidney (b = 2), and liver (b = 3). The sample number N was 12, including SD rats (n = 6) and ZDF rats (n = 6), which were common in the heart, kidney, and liver; however, the number of detected metabolites (Kb) changed depending on the organ. Unit-variance scaling and zero-average centering of the matrix were conducted for all Xb values.
2.1.2. Initialization
An arbitrary starting global score vector t (N × 1) was chosen.
2.1.3. Computation of block loadings (for b = 1, 2, 3)
All blocks of Xb were regressed using the t global score to obtain the block loading vector pb (Kb × 1).
2.1.4. Computation of block scores for b = 1, 2, 3
2.1.5. Computation of global score and block weights
All block scores were combined to the T global score matrix (N × b).
The T global score matrix was regressed by the global score vector t, resulting in the global weight vector w (b × 1) as the regression coefficient.
2.1.6. Normalization
The global weights were normalized to length one, and a new global score vector, t, was calculated.
This algorithm iterates until convergence was achieved. Deflation was performed with the block loading pab, and the residual matrix Xa+1b was calculated as follows: The second component was calculated using Xa+1b (where a is the number of components).
As with conventional PCA, the block score vector t of multiblock PCA represents the similarity of samples (SD or ZDF individuals), and the block loading vector p of multiblock PCA represents the similarity of metabolites.
2.2. Materials
Ammonium acetate was obtained from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan), and methanol and acetonitrile were purchased from Kanto Chemical Corporation (Tokyo, Japan). We purchased 1379 authentic standards listed in Chemicals.xlsx in the Supplementary Data to identify metabolites. Standards were dissolved in Milli-Q water (Millipore, Bedford, MA, USA), 0.1 mol/L acetic acid (Kanto Chemical Corporation), 0.1 mol/L ammonium hydroxide (FUJIFILM Wako Pure Chemical Corporation), or 50% methanol aqueous solution to obtain stock solutions.
2.3. Samples
The heart, liver, kidney, and plasma of eight-week-old male ZDF (n = 6) and SD rats (n = 6) were obtained from KAC (Kyoto, Japan). All breeding and experimental procedures were performed according to the following Japanese animal welfare laws: “Act on Welfare and Management of Animals” (1973); “Standards relating to the Care and Keeping and Reducing Pain of Laboratory Animals” (Ministry of the Environment, Japan, 2006); and “Basic guidance about animal experiment in the Health, Labour and Welfare Ministry competent organization” (enacted by the Ministry of Health, Labour and Welfare, Japan, 2006).
2.4. Preparation of rat tissues
Approximately 100 mg of tissue was transferred to tissue disruptor tubes supplied by Yasui Kikai (Osaka, Japan) and shaken with iron cones cooled in liquid nitrogen. Tissue powders were suspended with 1 mL of water and then mixed with 2 mL of methanol and chloroform, respectively. After mixing using a shaker for 15 min, samples were centrifuged at 1000 × g for 10 min. Supernatants were transferred to 15-mL Falcon tubes and dried under a nitrogen stream at 40 °C [28]. The lower layers were transferred to LC vials and analyzed via LC/MS (lipid). Dried residues of the supernatants were dissolved with a 200-µL of 10% acetonitrile and 90% water solution, and then analyzed using CE/MS and LC/MS (hilic). Twelve samples were analyzed twice for each of the platforms: CE/MS, LC/MS (hilic), and LC/MS (liquid) analyzed by both positive and negative polarities. The averages of the two datasets were obtained as expression values. Quality control (QC) was prepared by pooling 12 samples and was analyzed for every six measurements. Five QCs were analyzed in each run. The coefficient of variance (CV%) of every metabolite was calculated using five QCs, and metabolites with more than 50% CV were omitted from the data table.
2.5. CE/MS-metabolomics
CE/MS experiments were performed using an Agilent CE capillary electrophoresis system (Agilent Technologies, Waldbronn, Germany) and an Agilent 6520 QTOF system (Agilent Technologies, Palo Alto, CA, USA). Cationic separation was performed in fused-silica capillaries (50 µm i.d. 100 cm total length) filled with 1 mol/L formic acid [28], and anionic separation was conducted with 20 mmol/L ammonium acetate and 20 mmol/L ammonium formate (pH 10) [29] as the electrolyte. Sample solutions were injected at 50 mbar for 10 s, and a voltage of 30 kV was applied. The capillary temperature was not controlled (at room temperature), and the temperature of the sample tray was maintained at 4 °C using an external thermostatic cooler. The sheath liquid (methanol/water, 50% v/v) was delivered at 8 µL/min. ESI-TOF-MS was conducted in both positive (cationic) and negative (anionic) ion modes. The capillary voltage was set to 3 kV, and the flow rate of nitrogen gas (heater temperature 300 °C) was set to 10 psi. Exact mass data were acquired over a 60–1200 m/z range in the full scan mode.
2.6. LC/MS- hilic mode
LC/MS (hilic) analyses were performed using an Agilent 1290 series UPLC system equipped with a 6520 quadrupole TOF system (Agilent Technologies, Palo Alto, CA, USA) operated by the Masshunter Workstation B.04.01 software. The analytical column was a CAPCELL PAK C18 IF 2.0 mm I.D. × 50 mm, 2 µm (Osaka Soda, Osaka, Japan). The mobile phase consisted of two solvents: eluent A was water/ammonium acetate (5 mmol/L), and eluent B was acetonitrile. Metabolites were eluted at a flow rate of 0.2 mL/min at 40 °C with a linear gradient of 10–100% of eluent B over 10 min with a further 5 min hold at 100% of solvent B. The injection volume was 10 μL. Mass spectrometric analysis was performed in both positive and negative ionization modes with a scan rate of 2 spectra/s, mass range of 60–1200 (m/z), capillary voltage of 3500 V, and fragmentor setting of 120 V. The pressure of the nebulizer was 40 psi, the gas temperature was 350 °C, and the continuous gas flow was 8 L/min [30], [31], [32].
2.7. LC/MS-lipid mode
LC/MS (lipid) experiments were performed using an Agilent 1290 series UPLC system equipped with a 6220 TOF system (Agilent Technologies, Palo Alto, CA, USA) operated by the Masshunter Workstation B.04.01 software. Lipids were separated using Agilent EC-C18 Poroshell columns (2.1 × 5 mm and 2.1 × 150 mm) with a particle size of 2.7 μm and pore size of 120 Å. The mobile phase comprised solvent A (5% water, 47.5% acetonitrile, and 47.5% methanol) and solvent B (10 mmol/L ammonium acetate in 1% water and 99% isopropyl alcohol) [33]. The gradient cycle began with a solvent composition of 90% solvent A and 10% solvent B and reached a solvent composition of 55% solvent B within 10 min. The injection volume was 1 μL. The mass spectrometer was operated in both positive and negative modes with a mass range of 50–2000 m/z, and the capillary voltage was set to 4000 V. The nebulizing gas pressure was 40 psi, and the dry gas flow rate was 8 L/min at 350 °C.
2.8. Data analysis
Raw data obtained from the mass spectrometer were converted to CSV-type using a mass Hunter Export (Agilent Technologies). The converted CSV data comprised m/z, retention time, and intensity. Data processing was performed in the following sequence: 1) peak picking (identifying peak positions and areas), 2) bias correction of retention time and m/z, 3) peak alignment, 4) noise reduction, 5) bias correction of peak intensity, and 6) peak identification. All processes were performed using the Marker analysis [34]. All peak areas were divided by the area of the internal standard to avoid injection-volume and mass-spectrometry detector sensitivity bias among multiple measurements. Furthermore, peaks were normalized by wet tissue weights. Peak areas with a signal-to-noise ratio of less than three were converted to zero. Then, isotopic, fragment, and adduct ions were eliminated, and the peak datasets were compared across the samples and aligned according to both m/z and retention time. Noise peaks in the samples were recognized by comparison with blank preparation samples. Peaks detected in samples were identified based on the matched m/z values and the normalized retention times of the 1379 standards purchased from reagent manufacturers (“Chemicals.xlsx in Supplementary Data); all standards were previously analyzed using the same platforms. When several metabolite candidates were simultaneously identified, MS/MS analysis was carried out to confirm the structure by matching the fragmentation patterns with those of the candidates. Although all metabolites were quantified separately, the sum of the quantified values of several isomers, such as citrate and isocitrate, were counted as a single marker due to low peak separation. Recovery and stability of each metabolite was not assessed. The multiblock PCA program was developed with reference to Hassani’s architecture [16] using Excel VBA.
3. Results
3.1. Overview of the metabolome analysis of the heart, kidney, and liver of ZDF rats
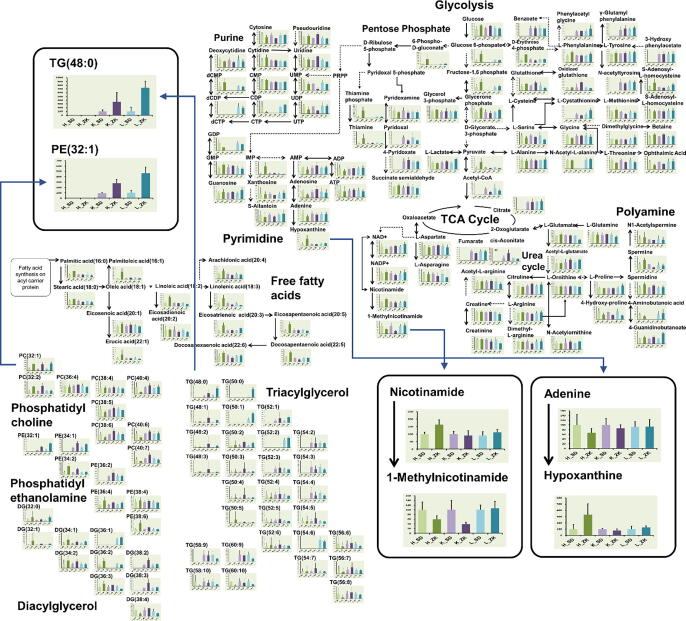
Metabolomics data acquired from CE/MS and LC/MS (hilic and lipid modes) were integrated into a data table for each organ. In total, 334, 442, and 363 metabolites were detected in the heart, kidney, and liver, respectively, of which 203 were common amongst the three organs (Figure S1A). To overview the ZDF rat metabolism, 203 metabolites were analyzed using classical metabolomic approaches, that is, correlation, volcano plot, metabolic pathway, and heat map analyses. The correlation analysis was to understand the similarity of metabolic expression patterns among the three organs. Positive correlations represent similar metabolic patterns between the two organs, whereas negative correlations indicate dissimilar metabolism. Correlation coefficients calculated from 12 samples (six SD rats and six ZDF rats) for the 203 common metabolites between the two organs (heart vs. kidney, kidney vs. liver, and liver vs. heart) are shown in Figure S1B. These results showed that the metabolism patterns of the liver and kidney were relatively similar (70% of the correlation coefficients were positive), whereas the positive correlation ratios for heart vs. liver and heart vs. kidney were 52% and 43%, respectively, suggesting that metabolism in heart was different from the others. Several methylated compounds, including methylimidazoleacetic acid and 1-methyladenosine, were highly correlated among the three organs (Figure S1B). Both these metabolites are mostly detected in urine as end products of metabolism, suggesting that end products will exhibit a high correlation between multiple organs. Volcano plots highlight metabolites that significantly increase or decrease in concentration in each organ [35]. This approach revealed a significant increase in linoleate levels (18:2n6) and a decrease in UDP-galactose levels in ZDF rat hearts (Figure S1C). These metabolite behaviors are related to the inhibition of glycolysis and enhanced lipid metabolism in rat hearts during diabetes development [36]. Volcano plot analysis facilitates the discovery of new biomarkers; however, this method is limited in revealing biological mechanisms as it does not show the relationships among these metabolites. Metabolic pathway analysis provides an overview of metabolic dynamism between SD and ZDF rats, and between their organs (Fig. 1). The most significant advantage of this approach is the pursuit of critical points that affect metabolism. For example, the concentration of 1-methylnicotinamide in ZDF hearts decreased remarkably even though levels of the precursor (nicotinamide) increased compared with those in control SD rats, implying that the reaction from nicotinamide to 1-methylnicotinamide was inhibited by some diabetic factors in ZDF rats. Similarly, the hypoxanthine concentration in ZDF hearts increased significantly, whereas that of the precursor adenine decreased suggesting that this metabolism was promoted in ZDF rat hearts. Levels of triacylglycerols (TGs) containing short fatty acid esters were increased in ZDF rats, whereas those of long fatty acid esters decreased. This meant that fatty acid beta oxidation was promoted in ZDF rats resulting in increased TG levels with short fatty acids. Metabolite pathway analysis is effective when these opposite changes are observed before and after a reaction, indicating that enzyme activities change at these points. However, as shown in Fig. 1, the levels of many metabolites randomly and independently changed regardless of their precursor or post-product behaviors, which implied that unknown feedback mechanisms existed in the metabolism. Heatmap analysis can reveal which metabolic fields are most influenced by diabetes [37]. Here, there were significant increases in free fatty acid and phosphatidylcholine content in the heart and kidney of ZDF rats (Figure S2). Although heatmap analysis was useful for overviewing metabolism, it was ill-suited to elucidate minor biological changes.
Fig. 1.
Metabolite pathway analysis. Pathways of glycolysis, pentose phosphate, tricarboxylic acid (TCA), purine, and lipid metabolism are shown as bar graphs: light green [H_SD], SD heart; green [H_ZK], ZDF heart; light purple [K_SD], SD kidney; purple [K_ZK], ZDF kidney; light blue [L_SD], SD liver; and dark blue [L_ZK], ZDF liver. Opposite behaviors between the precursor and product (e.g., nicotinamide to 1-methylnicotinamide or adenine to hypoxanthine in the heart) are highlighted with magnified bar graphs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Multiblock metabolomics
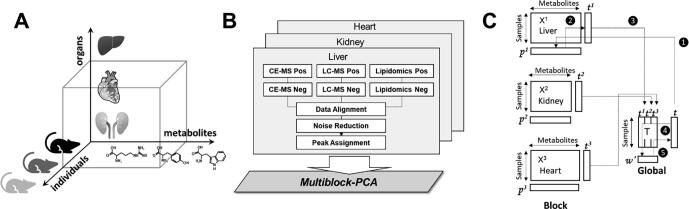
Traditional approaches provide a rough understanding of metabolic changes in each organ caused by diabetes; however, they are limited in their ability to clarify complex metabolic networks between organs. Multiblock metabolomics aims to elucidate the underlying metabolic relationships among several organs that these traditional approaches cannot achieve. The data structure of the multiblock metabolomics is shown in Fig. 2A. In contrast to conventional metabolomics, data structures become three-dimensional (individuals × metabolites × organs). The multiblock metabolomic scheme is shown in Fig. 2B. After combining CE/MS, LC/MS (hilic mode), and LC/MS (lipid mode) into one sheet, metabolites were identified based on elution times and m/z of standard metabolites. The data tables obtained for the heart, kidney, and liver were analyzed using the multiblock PCA algorithm (Fig. 2C). As with conventional PCA, block score vector t of multiblock PCA represented the similarity of samples (SD or ZDF individuals), and block loading vector p of multiblock PCA represented the similarity of metabolites.
Fig. 2.
Multiblock metabolomics scheme. (A) Multiblock metabolomics requires a three-dimensional data structure, metabolites, individual samples, and organs. (B) Metabolomics data obtained from CE/MS and LC/MS (hilic and lipid modes) were merged into one data table. After noise reduction, peaks were identified based on the matched m/z values and normalized retention times of the corresponding standard compounds. This process was repeated for the heart, kidney, and liver, and three data matrices were integrated using multiblock PCA. (C) Multiblock PCA architecture: ❶All blocks of X1,2,3 were regressed by an arbitrary global score t to obtain the block loadings p1,2,3. ❷The block scores t1,2,3 were calculated with the normalized block loadings p1,2,3 using the following equation: tb = Xbpb where b = 1, 2, 3. ❸All block scores were combined to a global score matrix T. ❹The global score matrix T was regressed by the global score vector t, resulting in the global weights. ❺ Global weights were normalized to length one and a new global score vector t was then calculated.
3.3. Comparison of multiblock-PCA and solo-PCA
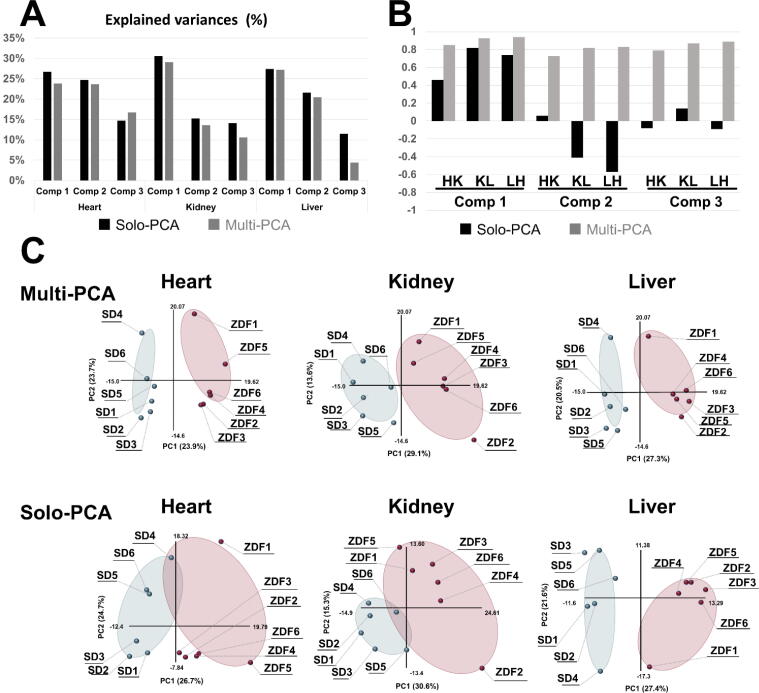
We analyzed the metabolomic data obtained from the heart, kidney, and liver using multiblock and solo-PCA to compare their performances. Solo-PCA was performed individually for each organ using conventional PCA architecture. Explained variance of the model indicates the degree of extracted information where values close to 100% indicate high reliability. The explained variances of the first component in multiblock PCA were 2.8%, 1.5%, and 0.1% less than those of solo-PCA for the heart, kidney, and liver, respectively (Fig. 3A). The explained variances of solo-PCA are theoretically larger than those of multiblock PCA; however, the degree of losses in multiblock PCA was limited. The block scores (t values) of multiblock PCA obtained from the heart, kidney, and liver were nearly equal, whereas those of solo-PCA were unequal, especially in the second and third components (Figure S3). Symmetries of block scores between two organs were evaluated using inner products for t vectors with the following equation: cos θ p,q = tpt·tq / (||tp||·||tq||), where p or q = heart, kidney, or liver. These values range between − 1 and 1, where values closer to 1 indicate high symmetry. The symmetries of t scores in multiblock PCA were evidently much higher than those of solo-PCA, indicating that the block p loadings of each organ calculated by multiblock PCA were also comparable (Fig. 3B). Solo-PCA maximizes the variance of each dataset regardless of the other dataset; thus, axis orientations were set independently from the dataset of other organs. However, the orientations of multiblock PCA depended on the other organs, and the axis orientations showed nearly identical directions. The block score plots with the first and second components generated by multiblock PCA were highly synchronized among the three organs, whereas those of solo-PCA did not (Fig. 3C).
Fig. 3.
Comparison of multiblock PCA and solo PCA. Multiblock and solo-PCA were performed with the metabolomic data of the heart, kidney, and liver. (A) The explained variances (%) are indicated by black (solo) and gray (multiblock) bars for the first three components. (B) The cos θ values of the t block scores in the solo and multiblock PCAs are indicated by black and gray bars, respectively. HK: cos θ between heart and kidney, KL: cos θ between kidney and liver, LH: cos θ between liver and heart. (C) The tb block scores of the first and second components are plotted for the multiblock and solo PCAs for the three organs. Six SD rats and six ZDF rats are plotted as light blue and red solid circles in the scatter plots. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Elucidation of metabolism with multiblock PCA loadings
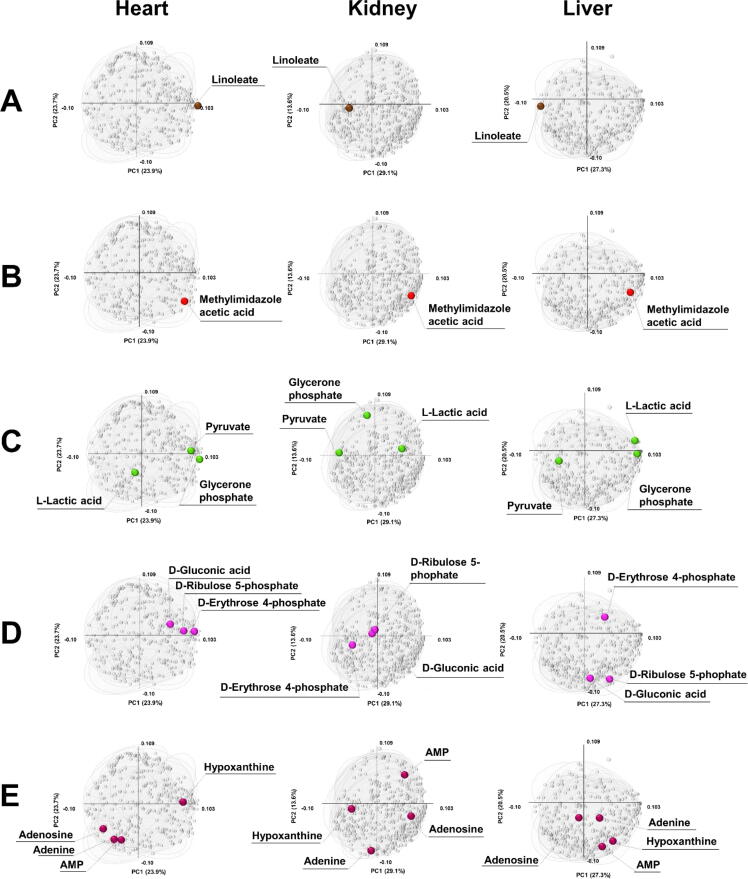
The multiblock PCA generates block scores and loading values among the target organs. Clear separations of ZDF and SD groups were observed in the score maps given by multiblock PCA (Fig. 3C), with the ZDF group located on the right side (positive side of the first component) for all organs. These observations mean that the metabolites plotted on the right side of the loading maps increase in the ZDF group as the block score and loading maps are synchronized. The axes of each component in the three organs showed the same directions; therefore, the similarity or dissimilarity of metabolite expression patterns in the three organs were comparable.
Linoleate content (18:2n6), which significantly increased in the ZDF rat heart but decreased in the ZDF rat liver in volcano plot analysis (Figure S1C), was located on the right side of the heart and the left side of the liver in the block loading plots (Fig. 4A). This indicated that the linoleate (18:2n6) expression pattern was completely opposite in the heart and liver, and multiblock PCA clearly identified these metabolite patterns. Methylimidazoleacetic acid, which showed high correlations between the three organs (Figure S1B), was located in the same direction (lower right side) in the three block loading maps (Fig. 4B), indicating that metabolites with the same expression pattern were in the same direction of the block-loading maps. Glycerone phosphate is a major metabolite of glycolysis and is generated from D-fructose 1,6-bisphosphate and then transformed to pyruvate via glyceraldehyde 3-phosphate. Pyruvate further reacts with CoA to generate acetyl-CoA, or is transformed to L-lactate under anaerobic conditions. Fig. 4C illustrates the expression patterns of glycerone phosphate, pyruvate, and L-lactate in the three organs. Glycerone phosphate was located on the right side of both the heart and liver maps, suggesting that glycolysis in ZDF rats was disrupted by diabetic factors and that glycerone phosphate accumulated in the ZDF rat heart and liver. Pyruvate and L-lactate were also located on the right side of the heart and liver maps, respectively, a result of sluggish tricarboxylic acid metabolism; low-level oxygen supply increases L-lactate content in the liver more than that in the heart. D-erythrose 4-phosphate, D-ribulose 5-phosphate, and D-gluconic acid, which are metabolites of the pentose phosphate pathway, are also important for glucose metabolism. These were located on the right side of the heart map, implying that the pentose phosphate pathway metabolism was stagnant in the heart, whereas these phenomena were not observed in the kidney and liver (Fig. 4D). In metabolic pathway analysis (Fig. 1), there was a significant increase in hypoxanthine levels and a decrease in adenine levels in the heart, which implied that adenine aminohydrolase activity in the ZDF rat heart was promoted. When adenine and hypoxanthine were plotted on the loading maps, hypoxanthine was located at a symmetric position to adenine with its precursor AMP and adenosine, which indicated that the reaction from adenine to hypoxanthine was promoted (Fig. 4E).
Fig. 4.
Simultaneous multiorgan analysis with multiblock PCA. Scattered block loading plots generated by the multiblock PCA for the heart, kidney, and liver were aligned horizontally: linoleate (A), methylimidazoleacetic acid (B), glycolysis (C), pentose phosphate pathway (D), and AMP-related metabolites (E). All metabolites are plotted as light gray circles to clarify the relative locations.
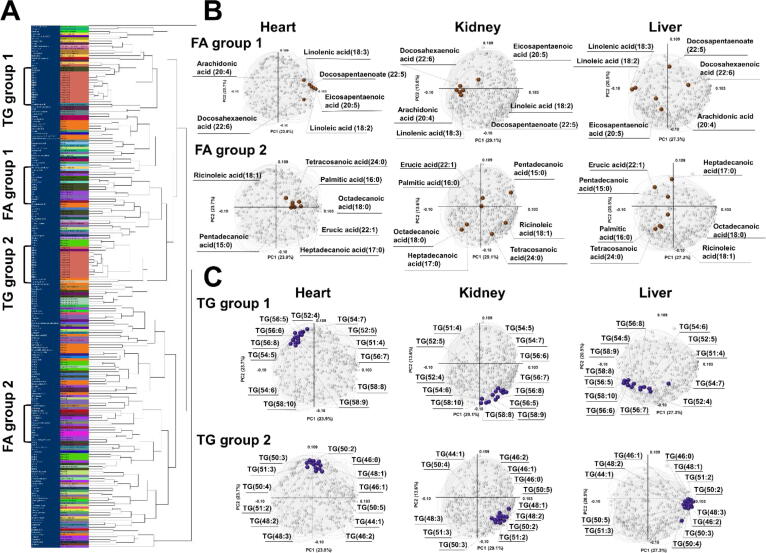
3.5. Clustering multiblock PCA loadings
Clustering multiblock loading p values help with the understanding of small differences in metabolite expression patterns among organs. Clustering was performed for multiblock loading p values with metabolite categories, such as amino acids, bile acids, fatty acids, and triacylglycerol (Fig. 5A). The results revealed that metabolites in the same categories were not always classified into the same clusters. The fatty acid group was divided into two major groups: highly unsaturated fatty acids (FA group 1) and less unsaturated fatty acids (FA group 2). FA group 1 included arachidonic acid (20:4), docosapentaenoic acid (22:5), and docosahexaenoic acid (22:6), and FA group 2 included palmitic acid (16:0) and stearic acid (octadecanoic acid) (18:0). FA group 1 was located on the right side of the heart block loading map (Fig. 5B), which meant that the unsaturated fatty acid levels increased significantly in the ZDF rat hearts compared with those in the SD rat hearts. The FA group 2 was also located on the right side of the heart map; however, this was closer to the origin, which indicated that the increases of fatty acids belonging to FA group 2 was less striking compared with those in the FA group 1. Contrastingly, FA group 1 in the kidney was located on the left side of the map, indicating that unsaturated fatty acid expression patterns in the kidney were completely different from those in the heart. Triacylglycerols were also classified into two groups: those with long and highly unsaturated fatty acids (TG group 1), and those with short and less unsaturated fatty acids (TG group 2). TG group 1 included TG (54:5), TG (56:6), and TG (58:9), and TG group 2 comprised TG (44:1), TG (46:1), TG (48:1), etc. Both TG groups 1 and 2 were located at the top of the heart map, whereas both groups were located at the bottom of the kidney map, indicating that TG expression patterns in the heart and kidney were opposite (Fig. 5C). TG group 1 in the heart was shifted slightly to the left compared to TG group 2, indicating that the levels of TGs with long and highly unsaturated fatty acids in the heart decreased slightly in ZDF rats compared with SD rats. TG group 2 was concentrated on the right side of the liver map, whereas TG group 1 was on the left side, indicating that TG group 2 was highly accumulated in the ZDF liver, even though TG group 1 was not.
Fig. 5.
Combination of cluster analysis and multiblock PCA. (A) The multiblock PCA block loading p values of 203 commonly detected metabolites in the three organs were classified by cluster analysis based on their metabolite categories. (B) Free fatty acids belonging to FA groups 1 and 2 are plotted as brown solid circles in the heart, kidney, and liver block loading maps. (C)Triacylglycerols belonging to TG groups 1 and 2 are plotted with violet solid circles for the first and second component loading maps in the heart, kidney, and liver. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Here, we propose multiblock metabolomics that combines mass spectrometry-based metabolomics with multiblock PCA to elucidate metabolic networks among multiple organs. Since organs have intrinsic roles and mutually interact in an individual organism, single-organ analysis is limited in understanding an entire body mechanism. A notable discovery of this study was to reveal organ-specific lipid metabolism, levels of TGs with long and polyunsaturated fatty acids decreased significantly, whereas those of TGs containing saturated or less unsaturated fatty acids increased in the ZDF rat liver. Although previous studies have shown TG levels in the liver of diabetic models to increase compared to non-diabetic models [39], our study revealed that TG accumulation patterns changed depending on the TG class and organs. These findings suggest that TG accumulation is related to fatty acid-binding proteins (FABPs) that specifically localize in each organ. FABPs are categorized into several families, including liver-FABP (FABP1), heart-FABP (FABP3), and brain-FABP, and they are involved in the binding, transport, and metabolism of long-chain fatty acids [38]. FABPs are controlled by peroxisome proliferator-activated receptors (PPARs), which bind to fatty acids or hydrophobic agonists. Wang et al. demonstrated the relationship between PPAR-γ activity and FABP expression using a c-γ agonist, and revealed that the expression of FABP1 and FABP3, controlled by PPAR-γ, was completely independent [40]. This finding supports our results on the different TG accumulation patterns among the heart, kidney, and liver (Fig. 5C). Jönsson et al. observed an increase of saturated and less unsaturated fatty acids, and a decrease of polyunsaturated fatty acids in the serum of ZDF rats [41]. This phenomenon was also observed in the TGs of ZDF rat liver in our study; however, it was limited to the liver and was not observed in the other organs. Kubota et al. investigated rice bran to improve diabetic conditions using ZDF rats and revealed that some proteins in rice bran induced β-oxidation and alleviated the diabetic condition [42]. They also revealed a significant increase of pyruvate in the liver during the improvement; this phenomenon was also observed in our study (if SD is considered an improved state, Fig. 4C). However, this phenomenon was only observed in the liver, and the expression pattern in the heart was opposite.
5. Conclusion
In this study, we developed multiblock metabolomics that combined mass spectrometry-based metabolomics and multiblock PCA. The biggest advantage of multiblock metabolomics is the generation of highly synchronized loading maps that can visually compare metabolic expression patterns among multiple organs. We revealed organ-specific TG accumulation in a ZDF rat model using this platform. We conclude that metabolism patterns differed depending on the organs, and investigating multiple organs is necessary to understand entire body metabolism.
Author contributions
K. T. organized the study and developed multiblock metabolomics. C. H. and T. K. analyzed the metabolites and improved the system. K. S. and K. I. interpreted the biological phenomena.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to thank Yuko Miyazaki for preparing metabolite standards. We would also like to thank Sadayo Iijima for supporting this project. This work was supported by LSI Medience Corporation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.015.
Contributor Information
Kazuhiro Tanabe, Email: tanabe.kazuhiro@mp.medience.co.jp.
Chihiro Hayashi, Email: hayashi.chihiro@ma.medience.co.jp.
Tomoko Katahira, Email: katahira.tomoko@ma.medience.co.jp.
Katsuhiko Sasaki, Email: sasaki.katsuhiko@mh.medience.co.jp.
Ko Igami, Email: igami.ko@mu.medience.co.jp.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou W., Sailani M.R., Contrepois K., Zhou Y., Ahadi S. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature. 2019;569:663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burrage L.C., Thistlethwaite L., Stroup B.M., Sun Q., Miller M.J. Untargeted metabolomic profiling reveals multiple pathway perturbations and new clinical biomarkers in urea cycle disorders. Genet Med. 2019;21:1977–1986. doi: 10.1038/s41436-019-0442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hocher B. Adamski J Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol. 2017;13:269–284. doi: 10.1038/nrneph.2017.30. [DOI] [PubMed] [Google Scholar]
- 4.Carneiro T.J., Araújo R., Vojtek M., Gonçalves-Monteiro S., Diniz C. Multi-organ NMR metabolomics to assess in vivo overall metabolic impact of cisplatin in mice. Metabolites. 2019;9(11):279. doi: 10.3390/metabo9110279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyagi R., Rana P., Gupta M., Khan A.R., Bhatnagar D. Differential biochemical response of rat kidney towards low and high doses of NiCl2 as revealed by NMR spectroscopy. J Appl Toxicol. 2013;33:134–141. doi: 10.1002/jat.1730. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino D, Kawata K, Kunida K, Hatano A, Yugi K, et al. Trans-omic analysis reveals ROS-dependent pentose phosphate pathway activation after high-frequency electrical stimulation in C2C12 myotubes. iScience 2020;23(10):8. [DOI] [PMC free article] [PubMed]
- 7.Jolliffe I.T. Principal Component Analysis. Springer Series. Statistics. 2002 [Google Scholar]
- 8.Hayashi M., Matsuo K., Tanabe K., Ikeda M., Miyazawa M. Comprehensive Serum Glycopeptide Spectra Analysis (CSGSA): A Potential New Tool for Early Detection of Ovarian Cancer. Cancers. 2019;11(5):591. doi: 10.3390/cancers11050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuo K., Tanabe K., Hayashi M., Ikeda M., Yasaka M. Utility of Comprehensive Serum Glycopeptide Spectra Analysis (CSGSA) for the Detection of Early Stage Epithelial Ovarian Cancer. Cancers. 2020;12(9):2374. doi: 10.3390/cancers12092374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi W.T., Tosun M., Jeong H.H., Karakas C., Semerci F. Metabolomics of mammalian brain reveals regional differences. BMC Syst Biol. 2018;12:127. doi: 10.1186/s12918-018-0644-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westherhuis J.A., Kourti T., Macgregor J.F. Analysis of multiblock and hierarchical PCA and PLS models. J Chemom. 1998;12:301–321. [Google Scholar]
- 12.Worley B., Powers R. A sequential algorithm for multiblock orthogonal projections to latent structures. Chemom Intell Lab Syst. 2015;149:3339. doi: 10.1016/j.chemolab.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenenhaus M., Tenenhaus A., Groenen P.J.F. Regularized generalized canonical correlation analysis: A framework for sequential multiblock component methods. Psychometrika. 2017;82:737–777. doi: 10.1007/s11336-017-9573-x. [DOI] [PubMed] [Google Scholar]
- 14.Scotti L., Ferreira E.I., Silva M.S., Scotti M.T. Chemometric studies on natural products as potential inhibitors of the NADH oxidase from Trypanosoma cruzi using the VolSurf approach. Molecules. 2010;15:7363–7377. doi: 10.3390/molecules15107363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall D.D., Lei S., Worley B., Huang Y., Garcia-Garci A. Combining DI-ESI-MS and NMR datasets for metabolic profiling. Metabolomics. 2015;11:391–402. doi: 10.1007/s11306-014-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassani S., Hanafi M., Qannari E.M., Kohler A. Deflation strategies for multi-block principal component analysis revisited. Chemom Intell Lab Syst. 2013;120:154–168. [Google Scholar]
- 17.Lee D.S., Vanrolleghem P.A. Adaptive consensus principal component analysis for on-line batch process monitoring. Environ Monit Assess. 2004;92:119–135. doi: 10.1023/b:emas.0000014498.72455.18. [DOI] [PubMed] [Google Scholar]
- 18.Eide I., Westad F. Automated multivariate analysis of multi-sensor data submitted online: Real-time environmental monitoring. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0189443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y., Goodacre R. Multiblock principal component analysis: an efficient tool for analyzing metabolomics data which contain two influential factors. Metabolomics. 2012;8:S37–S51. [Google Scholar]
- 20.Surowiec I., Skotare T., Sjögren R., Gouveia-Figueira S., Orikiiriza J. Joint and unique multiblock analysis of biological data - multiomics malaria study. Faraday Discuss. 2019;218:268–283. doi: 10.1039/c8fd00243f. [DOI] [PubMed] [Google Scholar]
- 21.Sasak K., Sagawa H., Suzuki M., Yamamoto H., Tomita M. Metabolomics platform with capillary electrophoresis coupled with high-resolution mass spectrometry for plasma analysis. Anal Chem. 2019;91:1295–1301. doi: 10.1021/acs.analchem.8b02994. [DOI] [PubMed] [Google Scholar]
- 22.Cabello-Olmo M., Oneca M., Torre P., Sainz N., Moreno-Aliaga M.J. A fermented food product containing lactic acid bacteria protects ZDF rats from the development of type 2 diabetes. Nutrients. 2019;11:2530. doi: 10.3390/nu11102530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedemann M.S., Hermansen K., Pedersen S., Bach Knudsen K.E. Resistant starch but not enzymatically modified waxy maize delays development of diabetes in Zucker diabetic fatty rats. J Nutr. 2017;147:825–834. doi: 10.3945/jn.116.243899. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X., Rougée L.R.A., Bedwell D.W., Cramer J.W., Mohutsky M.A. Difference in the Pharmacokinetics and Hepatic Metabolism of Antidiabetic Drugs in Zucker Diabetic Fatty and Sprague-Dawley Rats. Drug Metab Dispos. 2016;44(8):1184–1192. doi: 10.1124/dmd.116.070623. [DOI] [PubMed] [Google Scholar]
- 25.Pal A., Rhoads D.B., Tavakkoli A. Portal milieu and the interplay of multiple antidiabetic effects after gastric bypass surgery. Am J Physiol Gastrointest Liver Physiol. 2019;316(5):G668–G678. doi: 10.1152/ajpgi.00389.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dantas-Ferreira R.F., Raingard H., Dumont S., Schuster-Klein C., Guardiola-Lemaitre B. Melatonin potentiates the effects of metformin on glucose metabolism and food intake in high-fat-fed rats. Endocrinol Diabetes Metab. 2018;1(4) doi: 10.1002/edm2.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vammen L., Rahbek S., Secher N., Povlsen J.A., Jessen N. Type 2 diabetes mellitus worsens neurological injury following cardiac arrest: an animal experimental study. Intensive Care Med Exp. 2018;6(1):23. doi: 10.1186/s40635-018-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soga T., Baran R., Suematsu M., Ueno Y., Ikeda S. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 29.Nogami C., Sawada H. Positional and geometrical anionic isomer separations by capillary electrophoresis-electrospray ionization-mass spectrometry. Electrophoresis. 2005;26:1406–1411. doi: 10.1002/elps.200410122. [DOI] [PubMed] [Google Scholar]
- 30.Ambati C.R., Vantaku V., Donepudi S.R., Amara C.S., Ravi S.S. Measurement of methylated metabolites using liquid chromatography-mass spectrometry and its biological application. Anal Methods. 2019;11(1):49–57. doi: 10.1039/C8AY02168F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan B., Zhao D., Du R., Kshatriya D., Bello N.T. A highly sensitive ultra-high performance liquid chromatography/tandem mass spectrometry method with in-source fragmentation for rapid quantification of raspberry ketone. J Food Drug Anal. 2019;27:778–785. doi: 10.1016/j.jfda.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murugesu S., Ibrahim Z., Ahmed Q.U., Uzir B.F., Yusoff N.I.N. Identification of α-glucosidase inhibitors from Clinacanthus nutans leaf extract using liquid chromatography-mass spectrometry-based metabolomics and protein-ligand interaction with molecular docking. J Pharm Anal. 2019;9(2):91–99. doi: 10.1016/j.jpha.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi H., Ogiso H., Taguchi R. Qualitative and quantitative analyses of phospholipids by LC-MS for lipidomics. Methods Mol Biol. 2009;579:287–313. doi: 10.1007/978-1-60761-322-0_15. [DOI] [PubMed] [Google Scholar]
- 34.Tanabe K., Kitagawa K., Kojima N., Iijima S. Multifucosylated alpha-1-acid glycoprotein as a novel marker for hepatocellular carcinoma. J Proteome Res. 2016;15:2935–2944. doi: 10.1021/acs.jproteome.5b01145. [DOI] [PubMed] [Google Scholar]
- 35.Chen J.J., Wang S.J., Tsai C.A., Lin C.J. Selection of differentially expressed genes in microarray data analysis. Pharmacogenomics J. 2007;7:212–220. doi: 10.1038/sj.tpj.6500412. [DOI] [PubMed] [Google Scholar]
- 36.Zhao L., Dong M., Xu C., Zheng H., Wei T. Identification of energy metabolism changes in diabetic cardiomyopathy rats using a metabonomic approach. Cell Physiol Biochem. 2018;48:934–946. doi: 10.1159/000491960. [DOI] [PubMed] [Google Scholar]
- 37.Jones D.R., Wu Z., Chauhan D., Anderson K.C., Peng J. A nano ultra-performance liquid chromatography-high resolution mass spectrometry approach for global metabolomic profiling and case study on drug-resistant multiple myeloma. Anal Chem. 2014;86:3667–3675. doi: 10.1021/ac500476a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuhashi M., Hotamisligil G.S. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fishman S., Muzumdar R.H., Atzmon G., Ma X., Yang X. Resistance to leptin action is the major determinant of hepatic triglyceride accumulation in vivo. FASEB J. 2007;21(1):53–60. doi: 10.1096/fj.06-6557com. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z., Yue Y.X., Liu Z.M., Yang L.Y., Li H. Genome-wide analysis of the FABP gene family in liver of chicken (Gallus gallus): Identification, dynamic expression profile, and regulatory mechanism. Int J Mol Sci. 2019;20(23):5948. doi: 10.3390/ijms20235948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jönsson T.J., Schäfer H.L., Herling A.W., Brönstrup M. A metabolome-wide characterization of the diabetic phenotype in ZDF rats and its reversal by pioglitazone. PLoS ONE. 2018;13(11) doi: 10.1371/journal.pone.0207210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubota M., Watanabe R., Hosojima M., Saito A., Sasou A. Rice bran protein ameliorates diabetes, reduces fatty liver, and has renoprotective effects in Zucker diabetic fatty rats. J Funct Foods. 2020;70 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.