Graphical abstract
In this paper, we mainly concluded the Hi-C data analysis methods from genome 3D structure, A/B compartment, TADs, and loops detection, how to apply the visualization technology into the identification of chromosome feature structure, and correlation analysis among multi-omics data about human cancer and cell differentiation.
Keywords: Visualization, Hi-C, Omics data, Chromosome structure, A/B compartment, TADs, Loop
Abstract
With the development of 3C (chromosome conformation capture) and its derivative technology Hi-C (High-throughput chromosome conformation capture) research, the study of the spatial structure of the genomic sequence in the nucleus helps researchers understand the functions of biological processes such as gene transcription, replication, repair, and regulation. In this paper, we first introduce the research background and purpose of Hi-C data visualization analysis. After that, we discuss the Hi-C data analysis methods from genome 3D structure, A/B compartment, TADs (topologically associated domain), and loop detection. We also discuss how to apply genome visualization technologies to the identification of chromosome feature structures. We continue with a review of correlation analysis differences among multi-omics data, and how to apply Hi-C and other omics data analysis into cancer and cell differentiation research. Finally, we summarize the various problems in joint analyses based on Hi-C and other multi-omics data. We believe this review can help researchers better understand the progress and applications of 3D genome technology.
1. Introduction
With the completion of the Human Genome Project and the progress of other model organisms' genome projects, how to deal with the massive amount of molecular biology information is a considerable challenge. The existing multi-omics research is divided into genomics, transcriptomics, proteomics, epigenomics, and other omics research.
Transcriptomics studies how the same genome can result in different cell types and how gene expression is regulated. Among the genome analysis “omics” technologies, RNA-Seq [1] can be used to identify genes in the genome or to identify which genes are active at a specific time point, and read counts can be used to simulate relative gene expression levels accurately.
Epigenes are the genome's supporting structure, including protein and RNA binders, alternative DNA structures, and chemical modifications of DNA. Among technologies used to study epigenes, MNase-seq [2], DNase-Seq [3], ATAC-Seq [4], FAIRE-Seq [5], [6] are all used to study open chromatin regions and determine chromatin accessibility by detecting transcription factor (TF) footprints. ChIP-seq [7] to study the interaction between proteins and DNA in nuclei. Hi-C [8], [9] is a technology to analyze the spatial structure of chromatin in cells, quantifying the number of interactions between genomic loci that are adjacent in 3D space, but may be separated by many nucleotides in the linear genome (in this paper, chromatin interactions may be just the product of the random ligation of two DNA fragments detected by the Hi-C experiment, may be an interaction of chromatin segments mediated by proteins, etc.).
In recent years, multi-omics data analyses about diseases and cell differentiation appeared as shown in Table 1 and Table 2. Gene structure changes can lead to different diseases, for example, Holoprosencephaly (a forebrain disease caused by mutations in the SBE2 enhancer element [10]); PPD2 (polydactyly of a triphalangeal thumb caused by mutations in the ZRS enhancer [11]) and adenocarcinoma of the lung (caused by duplication of MYC gene enhancer [12]). In addition to multi-omics data analysis, in 2015, Ya Guo et al., [13] used CRISPR technology to invert the CTCF site, which changed the genome topology and enhancer/promoter functions. In each of these diseases, the underlying genetic defect could not have been identified without the use of multi-omics technologies and analyses, so understanding the relationship between gene structure changes and gene expression, combined with gene-editing technology, is expected to treat various genetic diseases. Hi-C technology, as the basis for studying genome structure, is fundamental among these technologies. So, this paper will concentrate on the analysis of Hi-C with other omics data.
Table 1.
Multi-omics datasets for human cancer and disease.
| Cancer/disease | Cell line | Sequencing method | Data ID/data link | Reference |
|---|---|---|---|---|
| liver, lung tumours, breast pancreas and lymphoma samples | GM12878 | WGS, Hi-C | GSE63525 | [35] |
| breast cancer, colorectal cancer, lung cancer | GM06990, K562 | (SNP)-arrays, Hi-C | GSE19399,GSE18199, GSE18350 | [36] |
| melanoma, prostate cancer, lung cancer, leukaemia | GM06990 | SNVs, SNPs, Hi-C | GSE18199 | [37] |
| cardiovascular disease | HCASMCs | ATAC-seq, RNA-Seq, Hi-C | GSE101498 | [38] |
| the Crohn’s disease | T cells | ATAC-seq, RNA-Seq, Hi-C | GSE101498 | [38] |
| the celiac disease | intestinal T cell | ATAC-seq, RNA-Seq, Hi-C | GSE101498 | [38] |
| IDH mutant gliomas | IMR90, NHEK, KBM7, K562, HUVEC, HMEC, GM12878 | ChIP-seq, RNA-seq, Hi-C, DNA methylation quantification |
GSE70991 | [39] |
| Pan-cancer analysis | 3T3, HCC-15, CR, HCT116, IMR90 cell line | Hi-C | http://cancergenome.nih.gov/ | [40] |
aEach column denotes the key properties of multi-omics data analysis with Hi-C technology for cancer and other diseases. ‘Cancer/disease column’ denotes the cancer or disease’s name, ‘cell line’ column denotes the cell line that analyzed, ‘sequencing method’ column denotes the sequencing methods that were used, ‘data ID/data link’ column denotes the availability of sequencing data, among them, data ID GSEXXX can be searched in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), ‘reference’ column denotes the references where the data acquisition and analysis were described. Readers are referred to these papers for further information.
Table 2.
Multi-omics datasets for cell differentiation.
| Species | Cell line | Sequencing method | Data ID | Reference |
|---|---|---|---|---|
| Homo sapiens | Embryonic stem cell | Hi-C, RNA-seq, Chip-seq | GSE116862 | [41] |
| Hi-C, RNA-seq | GSE105028 | [42] | ||
| Hi-C, RNA-seq | GSE86821 | [43] | ||
| ChIP-seq, Dnase-HiC | GSE90680 | [44] | ||
| Hi-C, HiChIP, ChIP-seq, ATAC-seq, eU-Seq | GSE105028 | [42] | ||
| Hi-C,RNA-seq and ATAC-seq | GSE106687 | [45] | ||
| Hi-C | GSE107148 | |||
| Hi-C | GSE86821 | [43] | ||
| Hi-C | GSE52457 | |||
| Mus musculus | mESC | Capture Hi-C | GSE124698 | [46] |
| ATAC-seq, Hi-C,RNA-seq, ChIP-seq | GSE115933 | [47] | ||
| RNA-seq, ChIP-seq, Hi-C and Promoter Capture Hi-C | GSE100835 | [48] | ||
| Hi-C, RNA-seq | GSE89520 | |||
| ChIP-seq,Hi-C | GSE95533 | [49] | ||
| ChIP-seq,RNA-seq, ATAC-seq, WGBS and Hi-C | GSE138102 | [50] | ||
| Hi-C | GSE153884 | |||
| ChIP-seq,Hi-C and 5C | GSE156868 | [51] | ||
| In situ Hi-C | GSE118911 | [52] | ||
| ChIP-seq,ATAC-seq,Hi-ChIP,Hi-C | GSE113339 | |||
| RNA-seq,ChIP-seq, Xist CHART-seq, and in situ Hi-C | GSE116413 | [53] | ||
| Hi-C | GSE119805 | [54] | ||
| HiC,STEM-seq and RNA-seq | GSE109344 | [55] | ||
| ChIP-seq,RNA-seq, Hi-C | GSE119697 | [56] | ||
| Capture Hi-C | GSE114619 | [46] | ||
| Chip-Seq,Gro-Seq,Mnase-Seq, ATAC-seq and Hi-C | GSE82144 | [57], [58] | ||
| Hi-C | GSE110061 | |||
| Hi-C | GSE125656 | [59] | ||
| Hi-C | GSE146001 | [60] | ||
| Hi-C, RNA-seq | GSE118263 | [61] | ||
| Hi-C | GSE133246 | [62] | ||
| Hi-C | GSM4386021 | |||
| PLAC-seq. Hi-C, mRNA-seq and ChIP-seq | GSE146449 | [63] | ||
| Hi-C | GSE119347 | |||
| Hi-C | GSE124342 | [64] | ||
| Hi-C | GSE59027 | [65] | ||
| Hi-C, ChIP-Seq, RNA-Seq, DNase-Hypersensitivity | GSE72164 | [66] | ||
| DNA SPRITE, RNA-DNA SPRITE | GSE114242 | [67] | ||
| Hi-C | GSE130723 | |||
| Hi-C | GSE130725 | [68] | ||
| Hi-C, RNA-seq | GSE136307 | |||
| Hi-C | GSE152918 | [69] |
aEach column denotes the key properties of multi-omics data analysis of cell differentiation using Hi-C technology. ‘Species’ denotes the species’ name, ‘cell line’ column denotes the cell line that was analyzed, ‘sequencing method’ column denotes the sequencing methods that were used, ‘data ID’ column denotes the availability of the sequencing data, data ID GSEXXX can be searched in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), ‘reference’ column denotes the references where the data were published. Readers are encouraged to seek out these papers for further information.
Many authors have written relevant reviews on Hi-C data in the past ten years, mainly divided into three types: Hi-C data fundamental technical analysis, Hi-C structure analysis method, and explanations of applications of Hi-C data. The first case mainly focused on developing 3C technology [8], [14], [15], [16], [17], [18], [19], [20], [21], [22] and fundamental analysis methods [23], [24], [25]. Some reviews summarize the various hierarchical analysis methods based on Hi-C [26], [27], [28], [29], [30], [31], while the others summarize the different hierarchical structures of the 3D genome applications to human disease development [31], [32], [33], [34]. This review will focus on Hi-C technology's basic principles and the multi-level chromosome structures that can be identified based on Hi-C technology: overall structure, A/B compartment, TAD, and loop. In addition, we will briefly introduce how to combine Hi-C data with other epigenomic data and transcriptome data to study the relationship with an understanding of human diseases. Finally, we will give examples to illustrate the application of multi-omics joint analyses to provide ideas for researchers who have just started 3D genome research.
2. Demand for Hi-C data visualization analysis
2.1. Principle theory of Hi-C
The most commonly used Hi-C experiment was proposed by Erez Lieberman Aiden as follows [9]: (1) formaldehyde cross-linking so that the spatially adjacent chromatin fragments are covalently connected; (2) restriction enzymes digestion to cut the genome and the use of biotin to label the cut ends; (3) use of DNA ligase to ligate the cut ends and create chimeric molecules; (4) purify and break DNA chimeric molecules, and isolate DNA fragments with the biotin tag; (5) sequence both ends of the fragments of the DNA library; (6) construct the chromatin interaction matrix by counting the number of chimeric molecules between any two regions of the genome.
Using Hi-C technology, we obtain raw sequency data, but if other visualization methods are needed, the following steps are required: 1) Perform linker trimming processing on raw data to obtain valid sequencing data; 2) Obtain comparison file (.sam format) by double-end sequence alignment to the reference genome; 3) Read alignment file and process it into a matrix, tuple or .hic format; 4) use normalization algorithms like KR [70], ICE [71] or others [72], [73], [74], [75], [76], [77], [78], [79] to normalize the data. After the above experiment and data processing steps, we obtain the Hi-C contact matrix. With the Hi-C data and other omics data, we can explore the chromosomes' architecture and study the relationship between chromosome structure and transcriptional expression.
2.2. Hi-C enhancement
In the past few years, the 3D genome analysis methods have rapidly improved, and a large amount of data appeared, but the current resolution of most Hi-C data ranges from 25 kb to 1 Mb. Some high-resolution Hi-C data (range from 1 kb to 10 kb) are only available in a few tissues or cell lines, which affects our analysis of structures at kilobase pair (kb) resolution. But the higher the data resolution is, the deeper the sequencing depth required and the greater the expense is. So, how to map existing low-resolution Hi-C data to high-resolution Hi-C data has become a hot spot in the past five years.
Many authors used the deep learning framework to enhance the resolution of Hi-C data in recent years. In 2018, Yan Z et al. [80] developed HiCPlus, a method based on a super-resolution convolutional neural network (SRCNN). This algorithm, which can infer from low-resolution Hi-C data, is highly similar to the original matrix, a high-resolution Hi-C matrix, using only 1/16 of the original sequence reads. In 2019, Tong L et al. [81], [82] developed two new calculation methods to enhance the resolution of Hi-C data: HiCNN, based on a 54-layer convolutional neural network, and HiCNN2, inclusive of three different deep convolutional neural network architectures. Liu Q et al. [83] proposed hicGAN to enhance low-resolution Hi-C data through Generative Adversarial Networks (GAN). Same as hicGAN [83], in 2020, Hong, Hao et al. [84] developed the DeepHiC method, which can reproduce high-resolution Hi-C data from down-sampled reads as low as 1%. Zhilan L et al. [85] developed SRHiC based on the ResNet and WDSR model. They improved the Res-block in ResNet to increase the network's nonlinearity and learning ability. Simultaneously, a small convolution kernel is used multiple times to reduce the contact matrix’s size instead of using a large convolution kernel at once. This method has a strong generalization ability.
2.3. Hi-C data analysis
Many methods can be used to analyze Hi-C data, such as principal component analysis, interaction network analysis, heat maps, etc., to analyze Hi-C data:
1). cis–trans analysis to determine the quality of the Hi-C library:
Generally, the cis/trans ratio in high-quality Hi-C experiments is between 40 and 60 [86].
2). chromatin interactions visualization using heat maps: a) Whole-genome interactive heatmap; b) Interaction analysis between chromosomes; c) Interaction analysis within chromosomes as shown in Fig. 1.
Fig. 1.
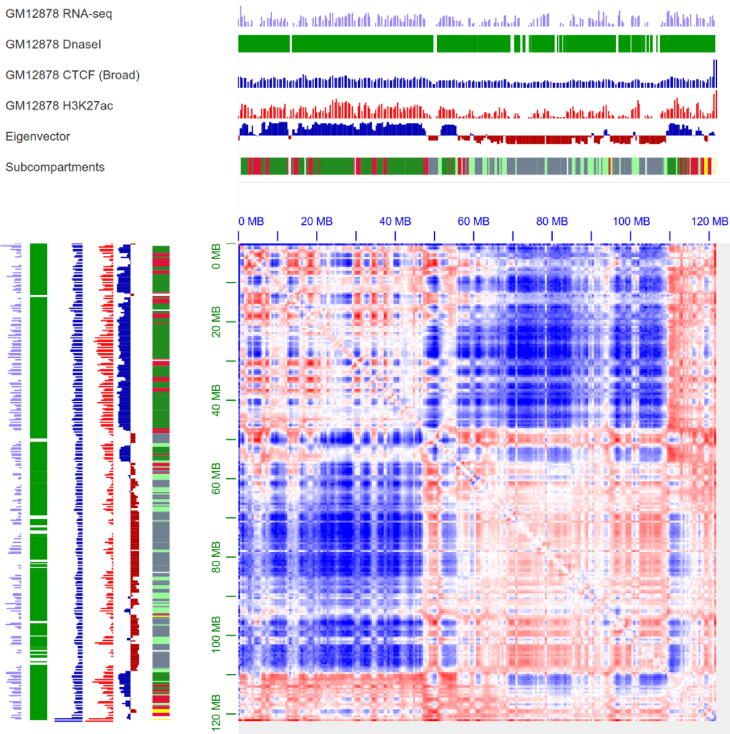
A/B compartment visualization with annotation. From top to bottom, the following visualization is shown: RNA-seq, Dnasel, CTCF (Broad), H3K27ac annotation, eigenvector, subcompartments and Hi-C contact heatmap. For Hi-C contact heatmap, we choose to show observed/expected balanced Hi-C data to visualize A/B compartment with 500 kb resolution, blue means A compartment, red means B compartment, combined with RNA-seq, Dnasel and H3K27ac, we can see, the blue region had higher gene expression and higher signals of h3k27ac, dnasel and CTCF. (Note: this Figure is drawn using the juicer box tool [87], Data Source: Rao and Huntley et al. [18] Cell GM12878 Hi-C in situ chr1: 0 MB-120 MB). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3). Structural analysis: a) 3D structure visualization for the whole/local chromatin, available tools to reconstruct structure can be seen in Table 3; b) compartments analysis to find the open/closed regions as Fig. 1 shows, available tools to find A/B compartments can be seen in Table 3; c) TADs detection to find CTCF histone as shown in Fig. 2, available tools to detect TAD boundaries can be seen in Table 3; d) loop calling mediated by CTCF and other proteins, available tools to find loops can be seen in Table 3.
Table 3.
Tools to analyze Hi-C data at different structural levels.
| Tool | Function/algorithm/download link | Resolution | Reference |
|---|---|---|---|
| loop Detection | loop detection | 1 ~ 10 kb | |
| HiCCUPS | [18] | ||
| HOMER | [115] | ||
| GOTHiC | [116] | ||
| Fit-Hi-C | [117] | ||
| HiC-DC | [118] | ||
| SIP | [119] | ||
| cLoops/cDBSCAN | [120] | ||
| Mustache | [121] | ||
| Chicago | [122] | ||
| PSYCHIC | [123] | ||
| diffHiC | differential analysis | [124] | |
| FIND | [125] | ||
| HICcompare | [126] | ||
| TADs Detection | ~40 kb | ||
| HMM | Directionality Index | [127] | |
| DP | Dynamic programming | [128] | |
| HicSeg | Two-dimensional segmentation | [129] | |
| Arrowhead | Arrow matrix | [18] | |
| insulation score | Insulation Square Analysis | [130] | |
| DHDF | Cluster-based | [131] | |
| TopDom | IdentifyTD, evaluate quality | [132] | |
| TADtree | hierarchical TADs | [133] | |
| TADs_Identification | Spectral identification | [134] | |
| IC-Finder | Hierarchical clustering | [135] | |
| MrTADFinder | network modularity based | [136] | |
| 3DNetMod | network modularity based | [137] | |
| HiTAD | domain-based alignment | [138] | |
| rGAMP | Gaussian Mixture model and Proportion test | [139] | |
| HiCDB | local relative, insulation metric | [140] | |
| deDoc | graph structural entropy | [141] | |
| tadbit | breakpoint detection algorithm | ||
| TADBoundaryDectector | deepLearning-based | [142] | |
| EAST | Haar-based algorithm | [143] | |
| TADBD | Haar-based algorithm | [144] | |
| TADCompare | Differential TADs | [145] | |
| TADpole | hierarchy of TADs in intra-chromosomal interaction matrices | [146] | |
| SpectralTAD | Spectral cluster | [147] | |
| ClusterTAD | an unsupervised machine learning approach | [148] | |
| Matryoshka | cluster | [149] | |
| A/B compartment | |||
| PCA | A/B compartment | 100 kb | |
| HOMER | [115] | ||
| juicebox | [87] | ||
| CscoreTools | https://github.com/scoutzxb/CscoreTool | ||
| HiCPro | http://github.com/nservant/HiC-Pro | [151] | |
| 3D structure | |||
| contact-based | |||
| Gen3D | adaptation, simulated annealing, and genetic algorithm | 200 kb | [152] |
| MOGEN | Gradient ascent | 200 kb-1 Mb | [153] |
| GEM | manifold learning | 1 Mb | [114] |
| GEM-FISH | polymer model | 5 kb | [154] |
| SuperRec | multidimensional scaling | 100 kb | [155] |
| distance-based | |||
| AutoChrom3D | considering the sequencing depth | 8 kb | [73] |
| ChromSDE | semi-definite embedding approach | 500 kb-1 Mb | [103] |
| ShRec3D | Short-path algorithm | 3–150 kb | [156] |
| FisHiCal | SMACOF algorithm | 1 Mb | [100] |
| MBO | manifold optimization | unknow | [107] |
| InfMod3DGen | Gradient ascent | unknow | [104] |
| 3D-GNOME | Markov chain, Simulated annealing | 1–2 Mb | [93] |
| Chromosome3D | Simulated annealing | 500 kb-1 Mb | [97] |
| LorDG | lorentzian objective function | 500 kb-1 Mb | [111] |
| HSA | Multi-track modeling, Markov chain, Simulated annealing | 25 kb-1 Mb | [157] |
| miniMDS | Hierarchical modeling | 10–100 kb | [108] |
| TADbit | https://github.com/3DGenomes/tadbit | unknow | [94] |
| mdsga | genetic algorithm | unknow | [95] |
| ShRec3+ | two-step algorithm | 1 Mb | [112] |
| 3DMax | maximum likelihood algorithm | 1 Mb | [88] |
| Hierarchical3DGenome | Hierarchical modeling | 1–5 kb | [101] |
| EVR | Error-Vector Resultant | unknow | [99] |
| ShNeigh | Gaussian formula | unknow | [158] |
| Probability-Based | |||
| BAC, BACH-MIX | Bayesian Inference | 40 kb | [159] |
| pastis | multidimensional scaling | 100 kb-1 Mb | [90] |
| tRex | Monte Carlo sampling etc. | 1 Mb | [92] |
| PGS | simulated annealing | 50 kb-1 Mb | [159] |
| SIMBA3D | Bayesian Estimation | [160] | |
| CHROMSTRUCT 4 | Monte Carlo sampling | [161] | |
| online tools | |||
| NDB | https://ndb.rice.edu/ | [162] | |
| Csynth | https://csynth.org | [163] | |
| GSDB | sysbio.rnet.missouri.edu/3dgenome/GSDB | [164] | |
| 3D-GNOME 2.0 | 3dgnome.cent.uw.edu.pl/ | [165] | |
| 3DGD | http://3dgd. biosino.org/ | [166] | |
| 3DIV | http://3div.kr/ | [167] | |
| 3DGB | http://3dgb.cs.mcgill.ca/ | [168] |
aEach column denotes the key properties of available tools to analyze Hi-C data at different structural levels. ‘Tools’ denotes availability of open-source software for a method. ‘Function/algorithm/download link’ column denotes Function, algorithms used by a method or download link for access,’ Resolution’ column denotes the resolution of Hi-C data described in the published method’s, ‘Reference’ column denotes the references where the methods were published.
Fig. 2.
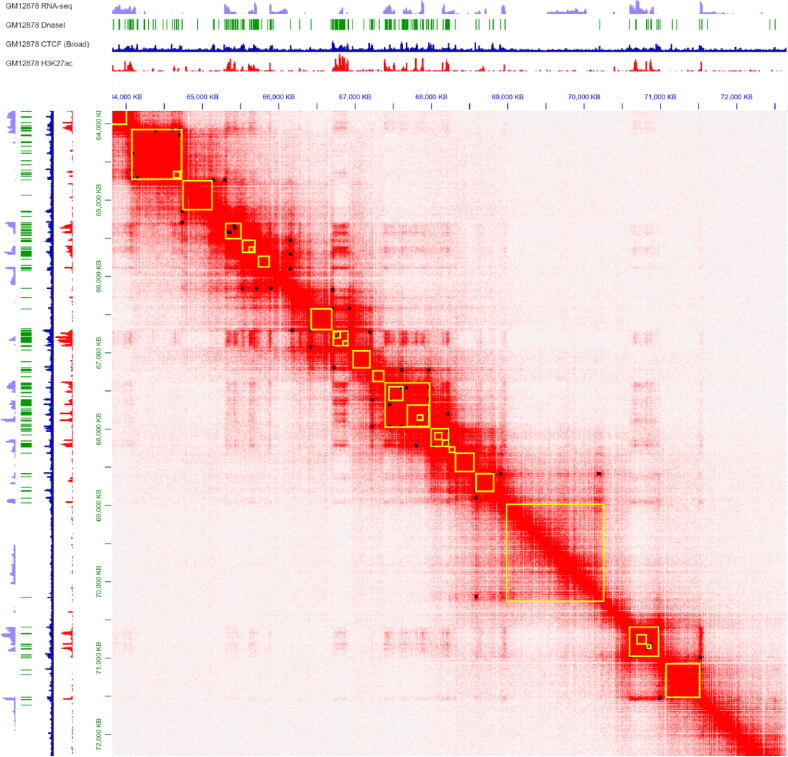
TADs and loops visualization with annotation. From top to bottom, the following visualization is shown: RNA-seq, Dnasel, CTCF (Broad), H3K27ac annotation and Hi-C contact heatmap. For the Hi-C contact heatmap, Squares of contact frequency along with the diagonal (yellow squares) indicate the TADs, peaks (black points) in the contact heatmap indicate the chromatin loops. (Note: this Figure is drawn using the juicer box tool [87], Data Source: Rao and Huntley et al. [18] Cell GM12878 Hi-C in situ chr1: 63,820,000–72,460,000). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Analysis methods for multi-omics data
As shown in Table 4, researchers can perform the following analyses according to their actual research goals: chromatin feature structure identification, correlation analysis among different samples, and Hi-C multi-omics joint analysis. We can analyze RNA-seq, ChIP-Seq, and ATAC-seq data using visualization methods such as box plots, scatter plots, heat maps, and volcano plots to do the following studies: 1) distribution of sequencing reads on the whole genome; 2) statistical information on the enrichment area of sequencing data (Peak); 3) difference analysis of multiple samples; and 4) motif identification.
Table 4.
Common tools for the joint analysis of omics datasets.
| Tools | Function/algorithm | Omics data | References |
|---|---|---|---|
| MACS | peak calling | ChIP-seq/RNA-seq/ATAC-seq | [169] |
| ChIPseeker | peak annotation | ChIP-seq | [170] |
| HOMER | peak calling and Motif analysis | ChIP-seq/ATAC-seq | [115] |
| Calculate ChIP-Seq expression near TSS | ChIP-seq | ||
| BEDtools | Extracting promoter sequences | ChIP-seq | [171] |
| RNA-seq coverage analysis | RNA-seq | ||
| SeqSite | detect transcription factor (TF) | ChIP-seq | [172] |
| EdgeR | Peak comparisons | ChIP-seq/ATAC-seq | [173] |
| DESeq2 | Peak comparisons | ChIP-seq/RNA-seq/ATAC-seq | [174] |
| DiffBind | Peak comparisons | ChIP-seq/ATAC-seq | [175] |
| methyKit | DNA methylation analysis | WGBS | [176] |
| MethGo | genomic and epigenomic analyses | WGBS/RRBS | [177] |
| GATK | variant analysis | WGS | [178] |
| BGI Online | variant analysis | WGS/RNA-seq | |
| SAMtools | variant analysis | WGS/RNA-seq | [179] |
| limma | differential expression | RNA-seq | [180] |
| Cufflinks | RNA-Seq analysis workflow | RNA-seq | [181] |
| RNA-Cocktail | RNA-Seq analysis workflow | RNA-seq | [182] |
| topGO | GO/KEGG enrichment | RNA-seq | |
| DAVID | GO/KEGG enrichment | RNA-seq | [183], [184] |
| KOBAS | GO/KEGG enrichment | RNA-seq | [185] |
aEach column denotes the key properties of available tools to analyze other omics data. ‘Tools’ denotes availability of open-source software for a method. ‘Function/algorithm/’ column denotes the primary function, algorithms used by a method. ’Omics data’ column denotes the omics data that serves as the input for the tool. ‘Reference’ column denotes the references where the methods were published.
3. Identification of chromatin structure
3.1. 3D visualization of chromosomes
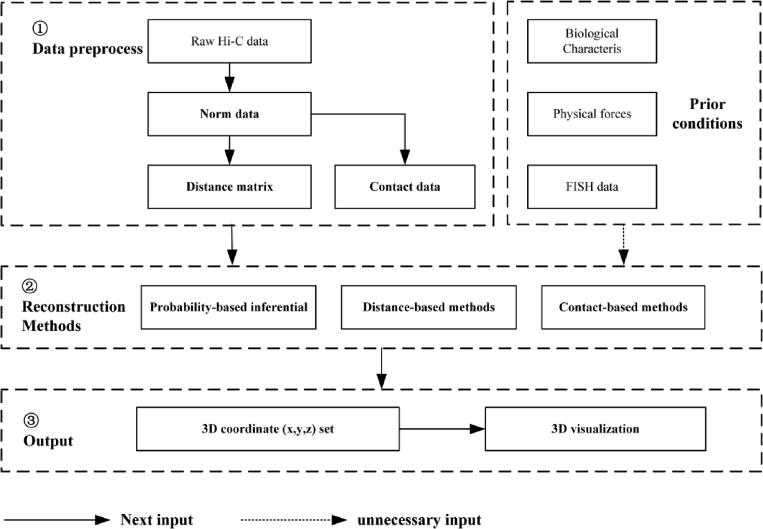
To better understand the relationship between chromosome structure and function, many researchers have reconstructed the 3D structures of chromosomes based on population Hi-C data at different resolutions, or on single-cell Hi-C data. Using the process shown in Fig. 3, before modeling, we need to obtain clean data by normalizing the raw Hi-C data [70], [71], [72], [73], [74], [75], [76], [77], [78], [79]. These methods can be classified as probability-based, distance-based or contact-based.
Fig. 3.
Process of simulating chromosomes’ 3D structure. First, data is preprocessed to get distance matrix or contact data, which is used as the second step’s input. Second, we could choose or not choose some prior conditions (such as biological characteristics, physical forces, and FISH data) to help us build chromosome reconstruction models. In the past ten years, most researchers chose to reconstruct chromosomes by probability-based inferential, distance-based or contact-based methods. At the end of the analysis, we will have a 3D coordinate set for visualization.
1) Probability-based: some researchers assumed the contact counts of Hi-C data follow a normal distribution [88], [89] or Poisson distribution [90], [91], [92] and designed a transfer function between the distribution intensity and spatial distance to infer a genome structure.
2) Distance-based: most reconstruction methods are based on the principle that the frequency of chromatin interaction is inversely proportional to the bin distance. Researchers [73], [88], [89], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109] determined the chromosomes structures based on optimization functions, such as semi-definite embedding [103], [113], manifold-based [107], [114], simulated annealing [97], Lorentzian objective function [111], maximum likelihood function [88], and shortest path [112].
3) Contact-based: some researchers set the biological characteristics and physical forces of chromosomes as a priori conditions [89], [94], [106], [109], [114], [152], [153], [154], [159], [186], [187], [188], [189], and directly use contact data to optimize the chromosome structure of contacts. Gen3d [152] reconstructs chromosomes with an adaptive, simulated annealing and genetic algorithm. MOGEN [153] uses the optimized scoring function to convert the Hi-C intrachromosomal and interchromosomal contact data into a 3D conformation set by satisfying as many contacts with high probability as possible. TADbit [94], GEM [114], and GEM-FISH [153] reconstruct spatial organization of chromosomes by combining their conformational energy.
The above methods can be used at different resolutions. At Mb-500 kb resolution, we can choose these methods [73], [89], [90], [91], [93], [94], [96], [98], [102], [103], [104], [105], [107], [109], [152], [153], [156], [187], [190], [191], [192], [193], [194] to observe the overall chromosome; at 5 kb resolution, we can choose the following methods [88], [95], [101], [108], [110], [112], [114], [154], [195] to observe the spatial structure of the whole chromosome or the local three-dimensional structure.
3.2. A/B compartment
In 2009, Erez Lieberman-Aiden, et al. [8] found the A/B compartments using principal component analysis. As shown in Fig. 1, the Hi-C heatmap region can be divided into A and B compartments, corresponding to the positive (the blue region) and negative (the read region) parts of the principal eigenvector. Through the study of gene expression levels, histone modifications, and DNase enzyme hypersensitivity sites corresponding to positive and negative regions, they found that in the regions with positive eigenvalue, there are more genes, and the corresponding gene expression levels are relatively high. The signal of the Dnase-sensitive DNA site is also relatively high. These characteristics indicate that these regions are more open and accessible, and the region of transcriptional activation is defined as the A compartment, which corresponds to the open chromatin region; on the contrary, the B compartment corresponds to the closed chromatin region.
In the next few years, most researchers verified the relationships between the structures of the A/B compartments and their functional characteristics by predicting chromosome A/B compartments [196], [197], [198], other researchers developed tools to analyze the A/B compartments, such as HiC-Pro [151] and CscoreTool[150].
3.3. TADs detection
In 2012, the concept of TADs was first proposed by E. P. Nora et al [200] and Dixon et al. [127], to explain the squares in the Hi-C matrix diagonal. They thought the interaction frequency within the TADs was significantly higher than the interaction frequency between two adjacent regions. In 2019, E. D. Wit [201] gave a definition of TADs that considers the mechanisms that shape them: TADs are an emergent property of an underlying biological mechanism, i.e. loop extrusion or compartmentalization and are dynamic genomic regions rather than a static structural feature of the genome. As shown in Fig. 2, a heat map is used to perform TADs boundaries identification analysis. The heat map indicates the chromatin interaction at 10 kb resolution. The genome interaction map is a symmetric matrix, so the information on both sides of the diagonal is equal in Fig. 2. As shown in this figure, let’s just see the upper right corner of heat map, the interaction intensity changes from weak to strong, which is indicated by the color of the cell changing from white to red. We can see some small triangular regions appear on the bottom edge repeatedly, which are depicted in red, indicating that the interaction frequency between chromatin fragments within these regions is high, and the frequency of interaction between adjacent triangular regions is lower. In this heatmap, these regions (yellow boxes in Fig. 2) are called TADs.
In the past ten years, most researchers identified the TADs by extracting one-dimensional features from the two-dimensional interaction matrix for segmentation, or by using the clustering algorithm. Regarding the first method: in 2012, to identify TADs in chromatin, DI (directionality index proposed by J. R. Dixon et al. [127]) was used to quantify the degree of bias in upstream or downstream interactions of genomic regions. By determining DI in the genome, we can determine the location of TAD boundaries in the genome. In the next ten years, many authors continued to improve the TADs recognition algorithm. Dynamic programming [128] has been used to reveal the TADs hierarchical structure. In addition two-dimensional segmentation [129], insulation score [130], laplacian graph clustering method [134], hierarchical clustering [137], unsupervised machine learning methods [148], the modular concept of network science [136], Gaussian Mixture model [139], local relative insulation metrics and multi-scale aggregation methods [140], have all been developed to detect contact domain boundaries. Based on structural information theory, a method called deDoc [141] proposed a solution to predict the structure of high-resolution TADs from low-resolution data.
3.4. Loop calling
People often refer to chromatin interactions as chromatin loops. But there are subtle differences between the two concepts. The chromatin loop is a circular structure formed by folding and wrapping chromatin due to protein and other mediation, the chromatin interaction may only be the product of the random connection of two DNA fragments detected by 3C-based experiments. Loops bring promoters and enhancers to closely together in space to regulate gene expression. With dynamic changes of the loop structures, such as new formation or disappearance, genes’ regulation will be affected to a certain extent [18] [202].
The chromatin loop can be identified by constructing Hi-C maps with a resolution of less than 5 kb. In 2014, S. S. Rao et al. [18] identified the positions of chromatin loops by using HiCCUPS (integrated into the developed juicer [87]) to search for pairs of loci, whose pixels with higher contact frequency than typical pixels in their neighborhood. (These pixels are defined as “peaks” in the Hi-C contact matrix and the corresponding pair of loci are called “peak loci”). As shown by the black mark in Fig. 2, we used the juicerbox software to mark the loops. Many other tools to find loops, such as HOMER [203] and GOTHiC [116], are available and are listed in Table 1.
4. Applications of integrated omics data analysis
As described in section 3 and shown in Fig. 4, we can combine Hi-C data of different resolutions with other omics data regarding epigenetic regulation at different structural levels and gene expression analysis.
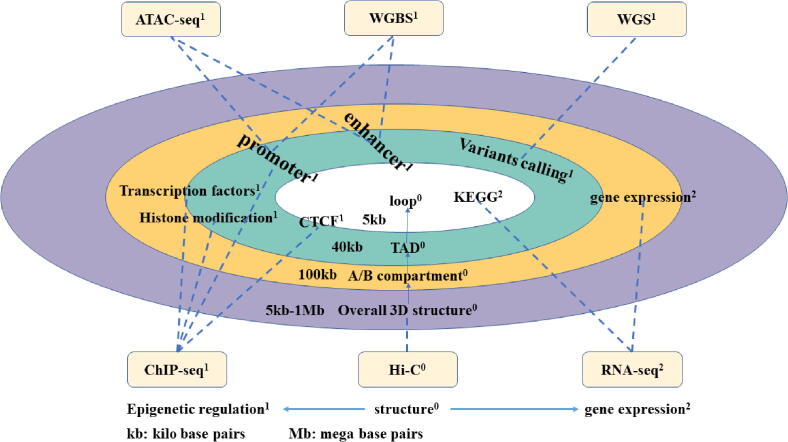
Fig. 4.
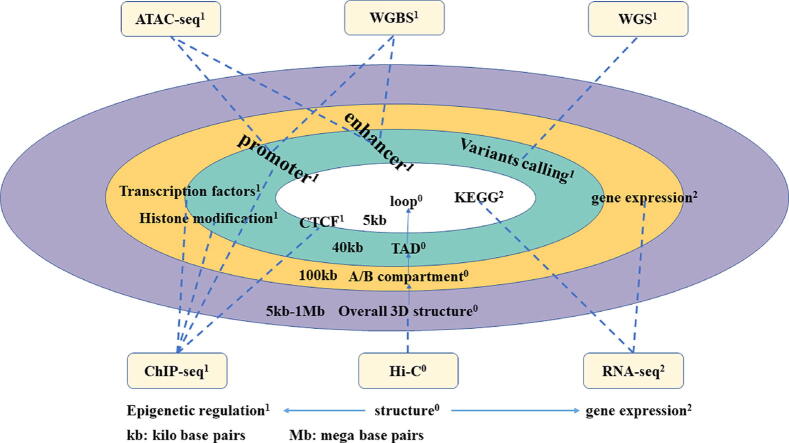
Analysis of multi-omics data. The superscripts 0, 1, and 2 represent the structure, epigenetic regulation, and expression related sequencing technology and corresponding analysis methods. From the loop level, we can do KEGG pathway and CTCF analysis; from the TAD level, we can use ChIP-seq, ATAC-seq, and WGBS to detect transcription factor sites or do variant calling and histone modification analysis; from the A/B compartment level, we can use ChIP-seq, ATAC-seq, WGBS, and RNA-seq to detect promoter, enhancer, transcription factor sites or conduct gene expression analysis.
4.1. Multi-omics data analysis at the loop level
As discussed in subsection 3.4, we can identify whether the two ends of chromatin loops are regulatory elements and gene loci by determining the locations of loops, so as to obtain a list of genes specifically regulated by different loops [18],[202]. As can be seen in Fig. 4, analyses from a variety of data sets such as DNase-seq, ChIP-seq, RNAseq or ATAC-seq may be combined to draw a whole-genome loop model. Comparing loops of multiple samples, finding loops that have changed at the genome-wide level, and using RNA-Seq to count the expression of related genes, can helps explain the relationship between loops and the potential differences in transcription regulation among different samples.
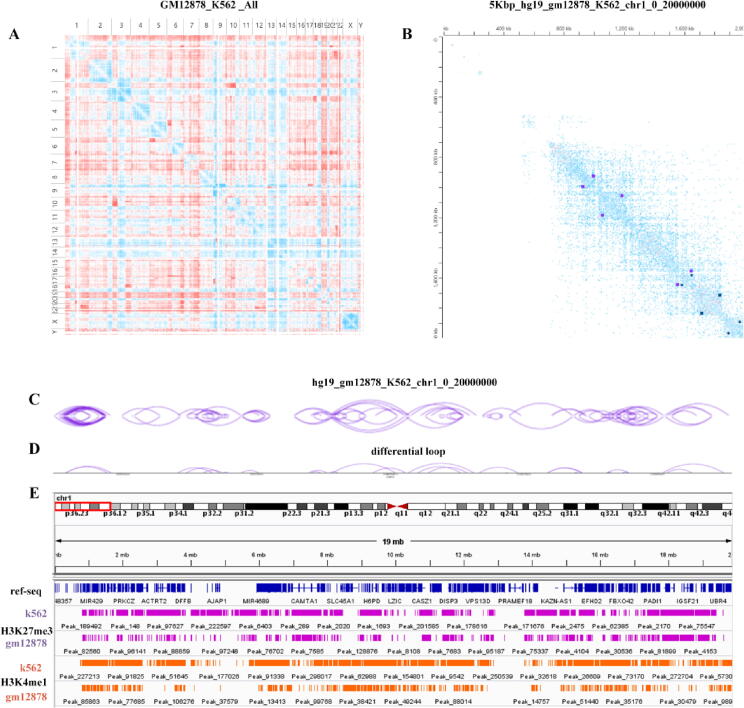
As shown in Fig. 5, Fig. 5(A) represents the differential interaction map of the whole genome map of GM12878 and K562 cells, Fig. 5(B) represents the differential interaction map of the two cell lines GM12878 and K562 in the region of chromosome 1: 0:2,000,000 bp and the corresponding identification of differential loops, Fig. 5(C, D) uses arc diagrams to visualize the interaction loop’s location and corresponding differential loops of the GM12878 and K562 cell lines in the 0:2,000,000 bp of chromosome 1. After statistically analyzing the differential loops between samples, we find loops that have changed at the genome-wide level, and use RNA-Seq to do differential expression analysis, which helps explain the relationship between loops and gene transcription regulation among different samples.
Fig. 5.
Differential analysis of data from the GM12878 and K562 cell lines. A. Differential heatmap of all chromosomes between the GM12878 lymphocyte line and the K562 cell line. The red color in the figure indicates sites with stronger interactions in the GM12878 cell line than the K562 cell line, and the blued ones indicate weaker interactions in the GM12878 cell line than in the K562 cell line. B. Differential heatmap of chromosome 1 between the GM12878 lymphocyte line and the K562 cell line. The points on the diagonal lines are the identified loops. The dark purple points represent the loops of GM12878, and the dark blue points represent the loops of K562. C-D. The loops and differential loops’ location of the GM12878 lymphocyte line and the K562 cell line chromosome 1 from 0 to 20,000,000 bp. Each arc indicates chromatin interaction from the start site to the end site. E. ChIP-Seq data is used to visualize the H3K27me3 and H3K4me1 histone modification peaks in the GM12878 and K562 cell lines, chromosome 1 range of 0–20,000,000 bp. (note: GM12878 Hi-C in situ and K562 Hi-C in situ data is from reference [18], A-D are drawn with the juicer box tool [87], and the G is drawn with the IGV tool [204]). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Let’s give an example: Rao et al. [18] found 9448 loops in the GM12878 cell line, of which 2854 loops are related to known promoter-enhancer functions. The expression of the gene's promoter with a loop was significantly higher than without a loop. For example, there is a loop in the GM12878 cell line, which is connected to the SELL promoter and a distal enhancer SELP, where the gene transcription is turned on, and the expression is increased. However, there is no loop in the same location in the IMR90 cell line, and the gene is not expressed. Greenwald, et al. [202] used a Hi-C experiment to generate high-resolution chromatin loops of pancreatic islets in three samples, as well as ATAC-seq, and ChIP-seq data to identify the target genes of pancreatic islet enhancers. Finally, these loops were annotated with target genes of islet enhancer, which shows that enhancer looping is correlated with islet-specific gene expression.
4.2. Multi-omics data analysis on TADs level
As section 3.3 has described, although TADs are statistical constructs rather than structural components of the 3D genome, TADs are an emergent property of an underlying biological mechanism, i.e. Loop extrusion or compartmentalization [201], if TADs boundaries are destroyed, the loops structure may change. so, analyzing differential TADs boundaries along with many other omics data as described below can help to understand the relationship between changes in loops/compartments and their functions.
1) Differential TADs boundaries region gene expression can be analyzed by using RNA-seq and Hi-C data, as described in reference [205].
2) Differential TADs boundaies and CNV identification and analysis of gene expression in related regions by using RNA-seq, WGS and Hi-C data can be done as in references [206], [207].
3) Analysis of the distribution of transcription factors and binding sites, histone modifications in differential TADs’ boundaries region by using ChIP-seq and Hi-C data, can be performed as in references [208], [209].
For example, in 2017, Rubin et al. used Hi-C and ChIP-seq data [209] to jointly analyze the interaction patterns of enhancers and promoters throughout the genome during the differentiation of isolated and cultured human primary keratinocytes. They confirmed two types of enhancers-promoter interactions: one is a ‘gaining’ interaction, which is enhanced during differentiation and is consistent with the enhancer obtaining H3K27ac activation marker; the other is a ‘stable’ interaction with enhancers constitutively marked by H3K27ac. Furthermore, these two interactions were not detected in pluripotent cells, suggesting that this lineage-specific chromatin structure was established in precursor cells and remodeled during terminal differentiation.
In reference [207], the authors performed Hi-C, whole-genome sequencing (WGS), ChIP-seq, and RNA-seq on two multiple aneuploidy myeloma (MM) cell lines to study the 3D genome structure of multiple myeloma (MM) and its relationship with genomic variation and gene expression. The authors found that the average interaction count inside each CNV block was positively correlated with its copy number, which indicates that raw interaction counts in cancer Hi-C data are biased by CNVs. This suggests that we can detect CNV by inferring the interaction counts of Hi-C data. Similarly, combining Hi-C and WGS data can improve the detection of translocations. The CNV breakpoints and TAD boundaries significantly overlapped. Compared with normal B cells, the number of TADs in MM increased by 25%, the average size of TADs was smaller, and about 20% of the genomic regions switched their chromatin A/B compartments type.
4.3. Multi-omics data analysis on the A/B compartment level
On the A/B compartment level, RNA-seq, ChIP-seq, ATAC-seq, WGS, or WGBS data is analyzed with Hi-C data as follows. For example, biological changes in myocardial cells are a mainly cause of heart failure. M. Rosagarrido et al. [210] found that this problematic cell function failure results from gene expression changes and is affected by transcription factors and chromatin remodeling enzymes. In reference [210], a chromatin conformation study of myocardial cell lines induced by load stimulation and mouse myocardial cell lines lacking CTCF function was conducted with Hi-C and RNA-seq. The analysis explores the effect of the entire genome structure on heart failure—generally, changes in A/B compartments correlated with gene expression. A change from the A compartment to the B compartment correlated with the down-regulation of gene expression, while a change from the B compartment to the A compartment correlated with the up-regulation of gene expression. This study's transcriptome data showed that most genes in regions with changed A/B compartments occurred in diseased cell lines, and the regulation of the expression of the sera changed, including the activation of some pathogenic marker genes.
Reference [211] used mouse embryonic stem cells to comprehensively study the effects on the three-dimensional structure (Hi-C) and on the chromatin accessibility (ATAC-seq), caused by the knockout of the methyltransferase complex subunit MLL2. Authors also studied alterations in protein modification (ChIP-seq), and gene expression levels (RNA-seq) casued by the MLL2 knockout. They found that the deletion of MLL2 increased the Polycomb complex's occupation, reshaped long-distance gene interaction and histone modification, reduced the transcription levels of critical genes, and ultimately caused abnormal embryonic development.
5. Conclusion
Overall, the Hi-C and other omics data analysis methods have appeared in different ways to help researchers understand the relationship between function and genome structure. However, there are still many improvements that could be made to the Hi-C analysis methods.
For the 3D structure analysis, although we have many methods to simulate the 3D structure of the genome, we still face many obstacles: (1) How can we simulate the genome structure of at a resolution even higher than 1 kb? Maybe we can apply deep learning methods to 3D structure simulation; (2) How can we improve microscope resolution to see the genome structure at kb resolution, to verify the accuracy of the three-dimensional simulated structure? Maybe we can use image super resolution technology to enhance the images from microscope; (3) How can we verify the accuracy of the 3D reconstructed model, and not just compare with FISH (Fluorescence in situ hybridization) data? (4) How can we detect TADs or loops when obtaining 3D structures in order to understand the relationship between structure and function?
Many methods were proposed to detect TADs and loops, but how can we detect TADs or loops from low-resolution Hi-C data? One way is by detecting them from enhanced Hi-C data. How can we detect them more accurately, faster and with fewer parameters? These problems still need to be solved.
There are many problems to deal with in order to observe an accurate genome structure, we still have additional methods to help us understand the relationship between function and genome structure using multi-omics analysis, for example, CRISPR/CAS9 genome editing technology. Ultimately these approaches will help us to develop cancer treatments and accelerate drug development.
Author contributions
All authors contributed to the conception and writing of this manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work has been supported by the National Key R&D Program of China [2018YFB0704301] and Scientific and Technological Innovation Foundation of Shunde Graduate School, USTB [BK20BF009]. Funding for open access charge: Department of Computer Science and Technology, Advanced Innovation Center for Materials Genome Engineering, University of Science and Technology Beijing. We thank all members of Dr. Zhang’s laboratories, the Department of Computer Science and Technology at the University of Science and Technology Beijing, to help collect all data related to visualization. The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and review services.
References
- 1.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui K, Zhao K. Genome-Wide Approaches to Determining Nucleosome Occupancy in Metazoans Using MNase-Seq. Methods in molecular biology (Clifton, N.J.), 2012, 833: 413-419. [DOI] [PMC free article] [PubMed]
- 3.Song L, Crawford G E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor Protocols, 2010, 2010(2): pdb. prot5384. [DOI] [PMC free article] [PubMed]
- 4.Buenrostro J.D., Wu B., Chang H.Y. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protocols Mol Biol. 2015;109(1) doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giresi P.G., Kim J., Mcdaniell R.M. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giresi P.G., Lieb J.D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48(3):233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt D., Wilson M.D., Spyrou C. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48(3):240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman-Aiden E., Van Berkum N.L., Williams L. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Berkum N.L., Lieberman-Aiden E., Williams L. Hi-C: a method to study the three-dimensional architecture of genomes. J Visualized Experiments: JoVE. 2010;39 doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong Y., El-Jaick K., Roessler E. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133(4):761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 11.Lettice L.A., Heaney S.J., Purdie L.A. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X., Choi P.S., Francis J.M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016 doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Xu Q., Canzio D. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162(4):900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker J., Rippe K., Dekker M. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 15.Simonis M., Klous P., Splinter E. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on-chip (4C) Nat Genet. 2006;38(11):1348. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 16.Nagano T., Lubling Y., Stevens T.J. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma W., Ay F., Lee C. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2014;12(1):71. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao S.S., Huntley M.H., Durand N.C. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beagrie R.A., Scialdone A., Schueler M. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543(7646):519. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Z., Li G., Wang Z. BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat Commun. 2017;8(1):1622. doi: 10.1038/s41467-017-01754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramani V., Deng X., Qiu R. Massively multiplex single-cell Hi-C. Nat Methods. 2017;14(3):263. doi: 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens T.J., Lando D., Basu S. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544(7648):59. doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Wit E., De Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekker J., Marti-Renom M.A., Mirny L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14(6):390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackay K., Kusalik A. Computational methods for predicting 3D genomic organization from high-resolution chromosome conformation capture data. Brief Funct Genomics. 2020;19(4):292–308. doi: 10.1093/bfgp/elaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Cheng L.G., Hongyu Zhang, Yijun Ruan. Reconstruction of three-dimensional structures of chromatin and its biological implications. ResearchGate. 2014;44(8):794–802. [Google Scholar]
- 27.Chang P., Gohain M., Yen M.R. Computational methods for assessing chromatin hierarchy. Comput Struct Biotechnol J. 2018;16:43–53. doi: 10.1016/j.csbj.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diament A., Tuller T. Modeling three-dimensional genomic organization in evolution and pathogenesis. Semin Cell Dev Biol. 2019;90:78–93. doi: 10.1016/j.semcdb.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Kong S., Zhang Y. Deciphering Hi-C: from 3D genome to function. Cell Biol Toxicol. 2019;35(1):15–32. doi: 10.1007/s10565-018-09456-2. [DOI] [PubMed] [Google Scholar]
- 30.Oluwadare O., Highsmith M., Cheng J. An overview of methods for reconstructing 3-D chromosome and genome structures from Hi-C data. Biol Proced Online. 2019;21:7. doi: 10.1186/s12575-019-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Li G. Advances in technologies for 3D genomics research. Sci China Life Sci. 2020;63(6):811–824. doi: 10.1007/s11427-019-1704-2. [DOI] [PubMed] [Google Scholar]
- 32.Babu D., Fullwood M.J. 3D genome organization in health and disease: emerging opportunities in cancer translational medicine. Nucleus. 2015;6(5):382–393. doi: 10.1080/19491034.2015.1106676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans S.A., Horrell J., Neretti N. The three-dimensional organization of the genome in cellular senescence and age-associated diseases. Semin Cell Dev Biol. 2019;90:154–160. doi: 10.1016/j.semcdb.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumm A., Duan Z. Understanding the 3D genome: Emerging impacts on human disease. Semin Cell Dev Biol. 2019;90:62–77. doi: 10.1016/j.semcdb.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vera B., Kaise Mutational biases drive elevated rates of substitution at regulatory sites across cancer types. PLoS Genet. 2016 doi: 10.1371/journal.pgen.1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.] De, Subhajyoti, Michor, et al. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nature Biotechnology, 2011. [DOI] [PMC free article] [PubMed]
- 37.Schuster-Bckler B., Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488(7412):504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 38.Mumbach MR, Satpathy AT, Boyle EA, et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements, 2017. [DOI] [PMC free article] [PubMed]
- 39.Flavahan W.A., Drier Y., Liau B.B. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weischenfeldt J., Dubash T., Drainas A.P. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet. 2016 doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Li T., Preissl S. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet. 2019;51(9) doi: 10.1038/s41588-019-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyu X., Rowley M.J., Corces V.G. Architectural proteins and pluripotency factors cooperate to orchestrate the transcriptional response of hESCs to temperature stress. Other. 2018;71(6) doi: 10.1016/j.molcel.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freire-Pritchett P., Schoenfelder S., Várnai C. Global reorganisation of $extit{cis}$-regulatory units upon lineage commitment of human embryonic stem cells. eLife Sciences. 2017;6 doi: 10.7554/eLife.21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battle S.L., Jayavelu N.D., Azad R.N. Enhancer chromatin and 3D genome architecture changes from naive to primed human embryonic stem cell states. Stem Cell Rep. 2019;12(5) doi: 10.1016/j.stemcr.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertero A., Fields P.A., Ramani V. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nature Commun. 2019;10(1) doi: 10.1038/s41467-019-09483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sima J., Chakraborty A., Dileep V. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell. 2018 doi: 10.1016/j.cell.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nature Structural & Molecular Biology, 2018. [DOI] [PubMed]
- 48.Comoglio, Federico, Park, et al. Thrombopoietin signaling to chromatin elicits rapid and pervasive epigenome remodeling within poised chromatin architectures, 2018. [DOI] [PMC free article] [PubMed]
- 49.Siersbk R., Madsen J.G.S., Javierre B.M. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte. Differentiation. 2017 doi: 10.1016/j.molcel.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Q., Ang J.Y.J., Lee A.Y. G9a plays distinct roles in maintaining DNA methylation, retrotransposon silencing, and chromatin looping. Cell Reports. 2020;33(4) doi: 10.1016/j.celrep.2020.108315. [DOI] [PubMed] [Google Scholar]
- 51.Nora E.P., Caccianini L., Fudenberg G. Molecular basis of CTCF binding polarity in genome folding. Nature Commun. 2020;11(1) doi: 10.1038/s41467-020-19283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fulco C.P., Nasser J., Jones T.R. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet. 2019;51(12):1664–1669. doi: 10.1038/s41588-019-0538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C.Y., Colognori D., Sunwoo H. PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nature Commun. 2019;10(1) doi: 10.1038/s41467-019-10755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alavattam KG, Maezawa S, Sakashita A, et al. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nature Structural & Molecular Biology. [DOI] [PMC free article] [PubMed]
- 55.Wang Y., Wang H., Zhang Y. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol Cell. 2019;73(3):547. doi: 10.1016/j.molcel.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 56.Rosario B.C.D., Kriz A.J., Rosario A.M.D. Exploration of CTCF post-translation modifications uncovers Serine-224 phosphorylation by PLK1 at pericentric regions during the G2/M transition. Elife Sci. 2019;8 doi: 10.7554/eLife.42341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The Energetics and Physiological Impact of Cohesin Extrusion. Cell, 2018.
- 58.Guideng Alex, Yick-Lun Epigenetic silencing of miR-125b is required for normal B-cell development. Blood J Am Soc Hematol. 2018;131(17):1920–1930. doi: 10.1182/blood-2018-01-824540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong S., Li Q., Zhang G. Exonuclease combinations reduce noises in 3D genomics technologies. Nucl Acids Res. 2020;8:8. doi: 10.1093/nar/gkaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M., Zhu Q., Li C. Chromatin architecture reorganization in murine somatic cell nuclear transfer embryos. Nature Commun. 2020;11(1) doi: 10.1038/s41467-020-15607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du Z., Zheng H., Kawamura Y.K. Polycomb group proteins regulate chromatin architecture in mouse oocytes and early embryos. Mol Cell. 2019;77(4) doi: 10.1016/j.molcel.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Ng A.P., Coughlan H.D., Hediyeh-Zadeh S. An Erg-driven transcriptional program controls B cell lymphopoiesis. Nature Commun. 2020;11(1) doi: 10.1038/s41467-020-16828-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kubo N., Ishii H., Xiong X. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat Struct Mol Biol. 2021:1–10. doi: 10.1038/s41594-020-00539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mclaughlin K., Flyamer I.M., Thomson J.P. DNA methylation directs polycomb-dependent 3D genome re-organization in naive pluripotency. Cell Reports. 2019;29(7):1974–1985.e6. doi: 10.1016/j.celrep.2019.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraser J., Ferrai C., Chiariello A.M. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol. 2015;11(12):852. doi: 10.15252/msb.20156492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joshi O., Wang S.Y., Kuznetsova T. Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell. 2015;17(6):748–757. doi: 10.1016/j.stem.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 67.Trinh S Q N O B T a P J S E D M L a S P B V. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. cell, 2017, 174(3): 744-757. [DOI] [PMC free article] [PubMed]
- 68.Kurup Jt H Z, Jin W, Kidder Bl. H4K20me3 methyltransferase SUV420H2 shapes the chromatin landscape of pluripotent embryonic stem cells. Development, 2020, 147. [DOI] [PMC free article] [PubMed]
- 69.Zhang C, Xu Z, Yang S, et al. tagHi-C Reveals 3D Chromatin Architecture Dynamics during Mouse Hematopoiesis - ScienceDirect. [DOI] [PubMed]
- 70.Knight P.A., Ruiz D. A fast algorithm for matrix balancing. IMA J Numerical Anal. 2013;33(3):1029–1047. [Google Scholar]
- 71.Imakaev M., Fudenberg G., Mccord R.P. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9(10):999. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung M.-S., Down T.A., Latorre I. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucl Acids Res. 2011;39(15):e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng C., Fu L.-Y., Dong P.-F. The sequencing bias relaxed characteristics of Hi-C derived data and implications for chromatin 3D modeling. Nucl Acids Res. 2013;41(19):e183. doi: 10.1093/nar/gkt745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu T., Wang Z. scHiCNorm: a software package to eliminate systematic biases in single-cell Hi-C data. Bioinformatics. 2018;34(6):1046–1047. doi: 10.1093/bioinformatics/btx747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yaffe E., Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 2011;43(11):1059. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 76.Cournac A., Marie-Nelly H., Marbouty M. Normalization of a chromosomal contact map. BMC Genomics. 2012;13(1):436. doi: 10.1186/1471-2164-13-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teytelman L., Özaydın B., Zill O. Impact of chromatin structures on DNA processing for genomic analyses. PLoS ONE. 2009;4(8) doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu M., Deng K., Selvaraj S. HiCNorm: removing biases in Hi-C data via Poisson regression. Bioinformatics. 2012;28(23):3131–3133. doi: 10.1093/bioinformatics/bts570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Servant N., Varoquaux N., Heard E. Effective normalization for copy number variation in Hi-C data. BMC Bioinf. 2018;19(1):313. doi: 10.1186/s12859-018-2256-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y., An L., Xu J. Enhancing Hi-C data resolution with deep convolutional neural network HiCPlus. Nat Commun. 2018;9(1):750. doi: 10.1038/s41467-018-03113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tong L, Zheng W. HiCNN: a very deep convolutional neural network to better enhance the resolution of Hi-C data[J]. Bioinformatics (Oxford, England), 2019, 35(21). [DOI] [PMC free article] [PubMed]
- 82.Liu T., Wang Z. HiCNN2: enhancing the resolution of Hi-C data using an ensemble of convolutional neural networks. Genes. 2019;10(11):862. doi: 10.3390/genes10110862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Q., Lv H., Jiang R. hicGAN infers super resolution Hi-C data with generative adversarial networks. Bioinformatics (Oxford, England) 2019;35(14):i99–i107. doi: 10.1093/bioinformatics/btz317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hong H., Jiang S., Li H. DeepHiC: a generative adversarial network for enhancing Hi-C data resolution. PLoS Comput Biol. 2020;16(2) doi: 10.1371/journal.pcbi.1007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhilan L, Zhiming D. SRHiC: A Deep Learning Model to Enhance the Resolution of Hi-C Data. Front Genetics, 2020, 11. [DOI] [PMC free article] [PubMed]
- 86.Lajoie B.R., Dekker J., Kaplan N. The Hitchhiker's guide to Hi-C analysis: practical guidelines. Methods. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durand N.C., Shamim M.S., Machol I. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Systems. 2016;3(1):95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oluwadare O., Zhang Y., Cheng J. A maximum likelihood algorithm for reconstructing 3D structures of human chromosomes from chromosomal contact data. BMC Genomics. 2018;19(1):161. doi: 10.1186/s12864-018-4546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanizawa H., Iwasaki O., Tanaka A. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucl Acids Res. 2010;38(22):8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Varoquaux N., Ay F., Noble W.S. A statistical approach for inferring the 3D structure of the genome. Bioinformatics. 2014;30(12):i26–i33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu M., Deng K., Qin Z. Bayesian inference of spatial organizations of chromosomes. PLoS Comput Biol. 2013;9(1) doi: 10.1371/journal.pcbi.1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park J., Lin S. Impact of data resolution on three-dimensional structure inference methods. BMC Bioinf. 2016;17(1):70. doi: 10.1186/s12859-016-0894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Szalaj P., Michalski P.J., Wróblewski P. 3D-GNOME: an integrated web service for structural modeling of the 3D genome. Nucl Acids Res. 2016;44(W1):W288–W293. doi: 10.1093/nar/gkw437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Serra F., Baù D., Goodstadt M. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kapilevich V., Seno S., Matsuda H. Chromatin 3D reconstruction from chromosomal contacts using a genetic algorithm. IEEE/ACM Trans Comput Biol Bioinf. 2018;16(5):1620–1626. doi: 10.1109/TCBB.2018.2814995. [DOI] [PubMed] [Google Scholar]
- 96.Mei H., Ma Y., Wei Y. The design space of construction tools for information visualization: a survey. J Visual Languages Comput. 2017;44:120–132. [Google Scholar]
- 97.Adhikari B., Trieu T., Cheng J. Chromosome3D: reconstructing three-dimensional chromosomal structures from Hi-C interaction frequency data using distance geometry simulated annealing. BMC Genomics. 2016;17(1):886. doi: 10.1186/s12864-016-3210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fraser J., Rousseau M., Shenker S. Chromatin conformation signatures of cellular differentiation. Genome Biol. 2009;10(4):R37. doi: 10.1186/gb-2009-10-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hua K-J, Ma B-G. EVR: Reconstruction of Bacterial Chromosome 3D Structure Using Error-Vector Resultant Algorithm. bioRxiv, 2018: 401513. [DOI] [PMC free article] [PubMed]
- 100.Shavit Y., Hamey F.K., Lio P. FisHiCal: an R package for iterative FISH-based calibration of Hi-C data. Bioinformatics. 2014;30(21):3120–3122. doi: 10.1093/bioinformatics/btu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trieu T, Oluwadare O, Cheng J. Hierarchical Reconstruction of High-Resolution 3D Models of Human Chromosomes. bioRxiv, 2018: 415810 [DOI] [PMC free article] [PubMed]
- 102.Zou C., Zhang Y., Ouyang Z. HSA: integrating multi-track Hi-C data for genome-scale reconstruction of 3D chromatin structure. Genome Biol. 2016;17:40. doi: 10.1186/s13059-016-0896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Z, Li G, Toh K-C, et al. Inference of spatial organizations of chromosomes using semi-definite embedding approach and Hi-C data. Annual international conference on research in computational molecular biology, 2013: 317-332.
- 104.Wang S., Xu J., Zeng J. Inferential modeling of 3D chromatin structure. Nucl Acids Res. 2015;43(8) doi: 10.1093/nar/gkv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Szałaj P., Tang Z., Michalski P. An integrated 3-dimensional genome modeling engine for data-driven simulation of spatial genome organization. Genome Res. 2016 doi: 10.1101/gr.205062.116. gr. 205062.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carstens S., Nilges M., Habeck M. Inferential structure determination of chromosomes from single-cell Hi-C data. PLoS Comput Biol. 2016;12(12) doi: 10.1371/journal.pcbi.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paulsen J., Gramstad O., Collas P. Manifold Based Optimization for Single-Cell 3D Genome Reconstruction. PLoS Comput Biol. 2015;11(8) doi: 10.1371/journal.pcbi.1004396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rieber L., Mahony S. miniMDS: 3D structural inference from high-resolution Hi-C data. Bioinformatics. 2017;33(14):i261–i266. doi: 10.1093/bioinformatics/btx271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Duan Z., Andronescu M., Schutz K. A three-dimensional model of the yeast genome. Nature. 2010;465(7296):363. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hua K.-J., Ma B.-G. EVR: reconstruction of bacterial chromosome 3D structure models using error-vector resultant algorithm. BMC Genomics. 2019;20(1):1–10. doi: 10.1186/s12864-019-6096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trieu T., Cheng J. 3D genome structure modeling by Lorentzian objective function. Nucl Acids Res. 2016;45(3):1049–1058. doi: 10.1093/nar/gkw1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li J., Zhang W., Li X. 3D Genome Reconstruction with ShRec3D+ and Hi-C Data. IEEE/ACM Trans Comput Biol Bioinform. 2018;15(2):460–468. doi: 10.1109/TCBB.2016.2535372. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Z., Li G., Toh K.-C. 3D chromosome modeling with semi-definite programming and Hi-C data. J Comput Biol. 2013;20(11):831–846. doi: 10.1089/cmb.2013.0076. [DOI] [PubMed] [Google Scholar]
- 114.Zhu G., Deng W., Hu H. Reconstructing spatial organizations of chromosomes through manifold learning. Nucl Acids Res. 2018;46(8):e50. doi: 10.1093/nar/gky065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sven Heinz. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mifsud B., Tavares-Cadete F., Young A.N. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47(6):598. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 117.Ay F., Bailey T.L., Noble W.S. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014 doi: 10.1101/gr.160374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carty M., Zamparo L., Sahin M. An integrated model for detecting significant chromatin interactions from high-resolution Hi-C data. Nat Commun. 2017;8:15454. doi: 10.1038/ncomms15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rowley M J, Poulet A, Nichols M H, et al. Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. Elegans to mammals. Genome Research, 2020, 30(3): gr.257832.119. [DOI] [PMC free article] [PubMed]
- 120.Cao Y., Chen Z., Chen X. Accurate loop calling for 3D genomic data with cLoops. Bioinformatics. 2019;36(3) doi: 10.1093/bioinformatics/btz651. [DOI] [PubMed] [Google Scholar]
- 121.Ardakany A.R., Gezer H.T., Lonardi S. Mustache: multi-scale detection of chromatin loops from Hi-C and micro-C maps using scale-space representation. Genome Biol. 2020;21(1):256. doi: 10.1186/s13059-020-02167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Putna NH, O'connell BL, Stites JC, et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Research, 2016. [DOI] [PMC free article] [PubMed]
- 123.Ron G., Globerson Y., Moran D. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun. 2017;8(1):2237. doi: 10.1038/s41467-017-02386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aaron T.L., Lun diffHic: a Bioconductor package to detect differential genomic interactions in Hi-C data. BMC Bioinf. 2015 doi: 10.1186/s12859-015-0683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Djekidel M.N., Chen Y., Zhang M.Q. FIND: difFerential chromatin INteractions Detection using a spatial Poisson process. Genome Res. 2018 doi: 10.1101/gr.212241.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stansfield J.C., Cresswell K.G., Vladimirov V.I. HiCcompare: An R-package for joint normalization and comparison of HI-C datasets. BMC Bioinf. 2018;19(1):279. doi: 10.1186/s12859-018-2288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dixon J.R., Selvaraj S., Yue F. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Filippova D., Patro R., Duggal G. Identification of alternative topological domains in chromatin. Algorithms Mol Biol. 2014;9(1):14. doi: 10.1186/1748-7188-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lévy-Leduc C., Delattre M., Mary-Huard T. Two-dimensional segmentation for analyzing Hi-C data. Bioinformatics. 2014;30(17):i386–i392. doi: 10.1093/bioinformatics/btu443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Crane E., Bian Q., Mccord R.P. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523(7559):240. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Y., Li Y., Gao J. A novel method to identify topological domains using Hi-C data. Quantitative Biol. 2015;3(2):81–89. [Google Scholar]
- 132.Shin H., Shi Y., Dai C. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucl Acids Res. 2015;44(7):e70. doi: 10.1093/nar/gkv1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weinreb C., Raphael B.J. Identification of hierarchical chromatin domains. Bioinformatics. 2015;32(11):1601–1609. doi: 10.1093/bioinformatics/btv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen J., Hero Iii A.O., Rajapakse I. Spectral identification of topological domains. Bioinformatics. 2016;32(14):2151–2158. doi: 10.1093/bioinformatics/btw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Haddad N., Vaillant C., Jost D. IC-Finder: inferring robustly the hierarchical organization of chromatin folding. Nucl Acids Res. 2017;45(10):e81. doi: 10.1093/nar/gkx036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yan K.-K., Lou S., Gerstein M. MrTADFinder: a network modularity based approach to identify topologically associating domains in multiple resolutions. PLoS Comput Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Norton H.K., Emerson D.J., Huang H. Detecting hierarchical genome folding with network modularity. Nat Methods. 2018;15(2) doi: 10.1038/nmeth.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X.-T., Cui W., Peng C. HiTAD: detecting the structural and functional hierarchies of topologically associating domains from chromatin interactions. Nucl Acids Res. 2017;45(19):e163. doi: 10.1093/nar/gkx735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu W., He B., Tan K. Identifying topologically associating domains and subdomains by Gaussian Mixture model And Proportion test. Nat Commun. 2017;8(1):535. doi: 10.1038/s41467-017-00478-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen F., Li G., Zhang M.Q. HiCDB: a sensitive and robust method for detecting contact domain boundaries. Nucl Acids Res. 2018;46(21):11239–11250. doi: 10.1093/nar/gky789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li A., Yin X., Xu B. Decoding topologically associating domains with ultra-low resolution Hi-C data by graph structural entropy. Nat Commun. 2018;9(1):3265. doi: 10.1038/s41467-018-05691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Henderson J., Ly V., Olichwier S. Accurate prediction of boundaries of high resolution topologically associated domains (TADs) in fruit flies using deep learning. Nucl Acids Res. 2019;47(13):e78. doi: 10.1093/nar/gkz315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Abbas Roayaei Ardakany S L. Efficient and Accurate Detection of Topologically Associating Domains from Contact Maps. 17th International Workshop on Algorithms in Bioinformatics (WABI 2017), 2017.
- 144.Lyu H., Li L., Wu Z. TADBD: a sensitive and fast method for detection of typologically associated domain boundaries. Biotechniques. 2020;69(1) doi: 10.2144/btn-2019-0165. [DOI] [PubMed] [Google Scholar]
- 145.Cresswell K.G., Dozmorov M.G. TADCompare: an R package for differential and temporal analysis of topologically associated domains. Front Genet. 2020;11(158) doi: 10.3389/fgene.2020.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Soler-Vila P., Cuscó P., Farabella I. Hierarchical chromatin organization detected by TADpole. Nucl Acids Res. 2020;48(7):e39. doi: 10.1093/nar/gkaa087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cresswell K.G., Stansfield J.C., Dozmorov M.G. SpectralTAD: an R package for defining a hierarchy of topologically associated domains using spectral clustering. BMC Bioinf. 2020;21(1) doi: 10.1186/s12859-020-03652-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oluwadare O., Cheng J. ClusterTAD: an unsupervised machine learning approach to detecting topologically associated domains of chromosomes from Hi-C data. BMC Bioinf. 2017;18(1):480. doi: 10.1186/s12859-017-1931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Malik L, Patro R. Rich Chromatin Structure Prediction from Hi-C Data, 2017: 184-193. [DOI] [PubMed]
- 150.Xiaobin Z, Yixian Z. CscoreTool: Fast Hi-C Compartment Analysis at High Resolution. Bioinformatics(9): 9. [DOI] [PMC free article] [PubMed]
- 151.Servant N., Varoquaux N., Lajoie B.R. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16(1):259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nowotny J., Ahmed S., Xu L. Iterative reconstruction of three-dimensional models of human chromosomes from chromosomal contact data. BMC Bioinf. 2015;16(1):338. doi: 10.1186/s12859-015-0772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Trieu T., Cheng J. MOGEN: a tool for reconstructing 3D models of genomes from chromosomal conformation capturing data. Bioinformatics. 2015;32(9):1286–1292. doi: 10.1093/bioinformatics/btv754. [DOI] [PubMed] [Google Scholar]
- 154.Abbas A., He X., Niu J. Integrating Hi-C and FISH data for modeling of the 3D organization of chromosomes. Nat Commun. 2019;10(1):2049. doi: 10.1038/s41467-019-10005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang Y., Liu W., Lin Y. Large-scale 3D chromatin reconstruction from chromosomal contacts. BMC Genomics. 2019;20(Suppl 2):186. doi: 10.1186/s12864-019-5470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lesne A., Riposo J., Roger P. 3D genome reconstruction from chromosomal contacts. Nat Methods. 2014;11(11):1141. doi: 10.1038/nmeth.3104. [DOI] [PubMed] [Google Scholar]
- 157.Zou C., Zhang Y., Ouyang Z. HSA: integrating multi-track Hi-C data for genome-scale reconstruction of 3D chromatin structure. Genome Biol. 2016;17(1):40. doi: 10.1186/s13059-016-0896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li F.Z., Liu Z.E., Li X.Y. Chromatin 3D structure reconstruction with consideration of adjacency relationship among genomic loci. BMC Bioinf. 2020;21(1):17. doi: 10.1186/s12859-020-03612-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hua N., Tjong H., Shin H. Producing genome structure populations with the dynamic and automated PGS software. Nat Protoc. 2018;13(5):915. doi: 10.1038/nprot.2018.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rosenthal M, Bryner D, Huffer F, et al. Bayesian Estimation of 3D Chromosomal Structure from Single Cell Hi-C Data. bioRxiv, 2018: 316265. [DOI] [PMC free article] [PubMed]
- 161.Caudai C., Salerno E., Zoppe M. CHROMSTRUCT 4: A Python Code to Estimate the Chromatin Structure from Hi-C Data. Ieee-Acm Transactions on Computational Biology and Bioinformatics. 2019;16(6):1867–1878. doi: 10.1109/TCBB.2018.2838669. [DOI] [PubMed] [Google Scholar]
- 162.Contessoto V.G., Cheng R.R., Hajitaheri A. The Nucleome Data Bank: web-based resources to simulate and analyze the three-dimensional genome. Nucl Acids Res. 2020 doi: 10.1093/nar/gkaa818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Todd S, Todd P, Mcgowan S J, et al. CSynth: An Interactive Modelling and Visualisation Tool for 3D Chromatin Structure. Bioinformatics (Oxford, England), 2020. [DOI] [PMC free article] [PubMed]
- 164.Oluwadare O, Highsmith M, Turner D, et al. GSDB: a database of 3D chromosome and genome structures reconstructed from Hi-C data (vol 21, 60, 2020). BMC Mol Cell Biol, 2020, 21(1). [DOI] [PMC free article] [PubMed]
- 165.Wlasnowolski M., Sadowski M., Czarnota T. 3D-GNOME 2.0: a three-dimensional genome modeling engine for predicting structural variation-driven alterations of chromatin spatial structure in the human genome. Nucl Acids Res. 2020;48(W1):W170–W176. doi: 10.1093/nar/gkaa388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li C., Dong X., Fan H. The 3DGD: a database of genome 3D structure. Bioinformatics. 2014;30(11):1640–1642. doi: 10.1093/bioinformatics/btu081. [DOI] [PubMed] [Google Scholar]
- 167.Kim K., Jang I., Kim M. 3DIV update for 2021: a comprehensive resource of 3D genome and 3D cancer genome. Nucl Acids Res. 2020 doi: 10.1093/nar/gkaa1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Butyaev A, Mavlyutov R, Blanchette M, et al. 3DGB: A Low-Latency, Big Database System and Browser for Storage, Querying and Visualization of 3D Genomic Data. [DOI] [PMC free article] [PubMed]
- 169.Yong, Zhang, Tao, et al. Model-based Analysis of ChIP-Seq (MACS)[J]. Genome Biology, 2008. [DOI] [PMC free article] [PubMed]
- 170.Guangchuang Y, Li-Gen W, Qing-Yu H. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics(14): 2382-2383. [DOI] [PubMed]
- 171.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang X, Zhang X. Pinpointing transcription factor binding sites from ChIP-seq data with SeqSite. Bmc Systems Biology, 2011, 5(2): S3. [DOI] [PMC free article] [PubMed]
- 173.Robinson M.D., Mccarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Anders S. Differential gene expression analysis based on the negative binomial distribution. J Marine Technol Environ. 2009;2(2) [Google Scholar]
- 175.R. Stark G B. Diffbind differential binding analysis of chip-seq peak data, 2014.
- 176.Mason A a M K L S F G M F a M C. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol, 2015. [DOI] [PMC free article] [PubMed]
- 177.Liao W.W., Yen M.R., Ju E. MethGo: a comprehensive tool for analyzing whole-genome bisulfite sequencing data. BMC Genomics. 2015;16(Suppl 12):S11. doi: 10.1186/1471-2164-16-S12-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mckenna A.H.M., Banks E. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li H., Handsaker B., Wysoker A. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kerr Kathleen M. Linear models for microarray data analysis: hidden similarities and differences. J Comput Biol J Comput Mol Cell Biol. 2003;10(6):891–901. doi: 10.1089/106652703322756131. [DOI] [PubMed] [Google Scholar]
- 181.Trapnell C., Roberts A., Goff L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sahraeian S.M.E., Mohiyuddin M., Sebra R. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8(1):59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 185.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 2011, 39(Web Server issue): 316-22. [DOI] [PMC free article] [PubMed]
- 186.Mateos-Langerak J, Bohn M, De Leeuw W, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci, 2009, 106(10): 3812-3817. [DOI] [PMC free article] [PubMed]
- 187.Kalhor R., Tjong H., Jayathilaka N. Solid-phase chromosome conformation capture for structural characterization of genome architectures. Nat Biotechnol. 2012;30(1):90. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Paulsen J., Sekelja M., Oldenburg A.R. Chrom3D: three-dimensional genome modeling from Hi-C and nuclear lamin-genome contacts. Genome Biol. 2017;18(1):21. doi: 10.1186/s13059-016-1146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Paulsen J., Ali T.M.L., Collas P. Computational 3D genome modeling using Chrom3D. Nat Protoc. 2018;13(5):1137. doi: 10.1038/nprot.2018.009. [DOI] [PubMed] [Google Scholar]
- 190.Rousseau M., Fraser J., Ferraiuolo M.A. Three-dimensional modeling of chromatin structure from interaction frequency data using Markov chain Monte Carlo sampling. BMC Bioinf. 2011;12(1):414. doi: 10.1186/1471-2105-12-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Baù D., Sanyal A., Lajoie B.R. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 2011;18(1):107. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Meluzzi D., Arya G. Recovering ensembles of chromatin conformations from contact probabilities. Nucl Acids Res. 2013;41(1):63–75. doi: 10.1093/nar/gks1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tjong H, Li W, Kalhor R, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proceedings of the National Academy of Sciences, 2016, 113(12): E1663-E1672. [DOI] [PMC free article] [PubMed]
- 194.Trieu T., Cheng J. Large-scale reconstruction of 3D structures of human chromosomes from chromosomal contact data. Nucl Acids Res. 2014;42(7):e52. doi: 10.1093/nar/gkt1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang Y., Liu W., Lin Y. Large-scale 3D chromatin reconstruction from chromosomal contacts. BMC Genomics. 2019;20(2):186. doi: 10.1186/s12864-019-5470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fortin J.-P., Hansen K.D. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 2015;16(1):180. doi: 10.1186/s13059-015-0741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dong P., Tu X., Chu P.Y. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol Plant. 2017;10(12):1497. doi: 10.1016/j.molp.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 198.Miura H, Poonperm R, Takahashi S, et al.: Practical Analysis of Hi-C Data: Generating A/B Compartment Profiles: Methods and Protocols, 2018: 221-245. [DOI] [PubMed]
- 200.Nora E.P., Lajoie B.R., Schulz E.G. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wit E.D. TADs as the caller calls them. J Mol Biol. 2019;432(3) doi: 10.1016/j.jmb.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 202.Greenwald W.W., Chiou J., Yan J. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat Commun. 2019;10(1):2078. doi: 10.1038/s41467-019-09975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Heinz S., Benner C., Spann N. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Robinson J.T., Thorvaldsdottir H., Winckler W. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Barutcu A.R., Lajoie B.R., Mccord R.P. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol. 2015;16(1):214. doi: 10.1186/s13059-015-0768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Taberlay P.C., Achinger-Kawecka J., Lun A.T. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. 2016;719 doi: 10.1101/gr.201517.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu P., Li T., Li R. 3D genome of multiple myeloma reveals spatial genome disorganization associated with copy number variations. Nat Commun. 2017;8(1):1937. doi: 10.1038/s41467-017-01793-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hug C.B., Grimaldi A.G., Kruse K. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169(2):216. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 209.Rubin A.J., Barajas B.C., Furlan-Magaril M. Lineage-specific dynamic and pre-established enhancer–promoter contacts cooperate in terminal differentiation. Nat Genet. 2017 doi: 10.1038/ng.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rosagarrido M., Chapski D.J., Schmitt A.D. High resolution mapping of chromatin conformation in cardiac myocytes reveals structural remodeling of the epigenome in heart failure. Circulation. 2017;136(17):1613–1625. doi: 10.1161/CIRCULATIONAHA.117.029430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mas G., Blanco E., Ballare C. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat Genet. 2018;50(10):1452–1462. doi: 10.1038/s41588-018-0218-5. [DOI] [PubMed] [Google Scholar]