Abstract
This paper reviews the technologies that have been invented in the last few years on high-throughput phenotyping, imaging, screens, and related techniques using microfluidics. The review focuses on the technical challenges and how microfluidics can help to solve these existing problems, specifically discussing the applications of microfluidics to multicellular model organisms. Some of the challenges include handling multicellular organisms in an efficient manner, and controlling the microenvironment and precise manipulation of the local conditions to allow the phenotyping, screening, and imaging of the small animals. Not only does microfluidics have the proper length scale for manipulating these biological entities, but automation has been demonstrated with these systems, and more importantly the ability to deliver stimuli or alter biophysical/biochemical conditions to the biological entities with good spatial and temporal controls. In addition, integration with and interfacing to other hardware/software allows quantitative approaches. We will include several successful examples of microfluidics solving these high-throughput problems. The paper will also highlight other applications that can be developed in the future.
Introduction
Microfluidics has been a field that develops tools for biology and medicine for the last two decades or so [1–5]. The success stories so far include DNA analysis, sensors, and micro total analysis. More complex maneuvers have also been demonstrated with cells or populations of cells in controlled microenvironments for a variety of potential applications. Recently, multicellular organisms such as worms (C. elegans), flies (Drosophila melanogaster), and zebrafish (Danio rerio) have emerged as additional biological systems that can benefit from the technological development of microfluidics and related technologies.
The fields of genetics, cell biology and drug screen have long been using small multicellular organisms such as C. elegans, Drosophila melanogaster, and Danio rerio. This is because of the genetic tractability, ease of genetic or molecular manipulation, and the cost-effectiveness as compared to larger/higher organisms. In addition, most of these organisms are transparent, at least in a large part of the developmental stages, and numerous imaging, optical, or pharmacological tools have been developed. There are also many human disease models available in these model organisms, making them ideal for studying fundamental disease mechanisms, disease progression, and performing drug screening. Compared to cell culture, these small organisms provide an in vivo environment for the particular problems of interest. Although experimentally extremely useful, manipulating these organisms in the laboratory has not been subjected to many engineering innovations, much less being used in conjunction with microfluidics. Part of the reason is in the complexity of handling them as well as the complexity of the data generated from the associated experiments.
The benefits of microfluidics in manipulating these organisms, however, are numerous, and they mirror the benefits for microfluidics in other biological fields. First, the size scales of the microsystems are of the same orders as the organisms of interest, in the range of tens to hundreds of microns. This makes the parts of the microsystem more compatible than their macro counterparts, which are spatulas, hair, pipettes, and tubes, sometimes much bigger than the biological entities they are handling. For example, innovative MEMS devices have been used advantageously for force sensing and manipulation of model organsisms [12–14]. Further, the maneuvers are also easier in microfluidics. With moving parts, flowing fluids, or other passive mechanisms, microsystems can be used to align samples with a particular orientation as compared to hand-manipulations with the macroscopic objects such as a spatula. Second, at the micro scale it is much easier to control the sample environment, which includes temperature, dissolved gas, nutrients, and stimuli, as compared to at the macro scale. This is in part due to the laminar nature of the flow at micro scale, and the efficient mass and energy transfer (often in the form of heat transfer). The micro length scale gives rise to the small transport resistance. For instance, dissolved gases can diffuse across tens of microns (of fluids or polymer membrane materials) in a matter of a few seconds in a completely predictable manner. Additionally, small conduits and small devices generally correspond to small thermal mass, which give rise to quick temperature rise and fall time. Laminar flow, as in the case of cellular or molecular analysis, can be exploited for delivering reagents that are spatially segregated without having a physical membrane between streams. Other benefits of micro systems include the ability to automate: recent efforts have been successful in automation and streamlining analysis including image-based analysis in some instances. Because genetic systems often require large number of (different) samples, it is often advantageous to have automated analysis that can speed up the analysis. Standardized and computerized analysis also reduces human bias and improved the quantitation in the analysis. An automated commercial system, COPAS (Union Biometrica), is capable of performing high speed imaging along the length of the C. elegans, and Drosophila and zebrafish embryos, and has had a large impact on the model organism community [15]. It is anticipated that microfluidics as a complementary technology will eventually allow routine high-throughput multi-dimensional and time-series imaging of organisms in addition to complex manipulations such as laser ablation.
To push the frontier of using microsystems for multicellular organisms to truly benefit genetics and therapeutic screens, a few challenges need to be overcome. First, because of the large number requirement for these types of studies, the micro systems have to be robust over long periods of time; the practicality of running a system thousands to millions of times is very different from running a one-time assay. Second, because some of these systems need to recover live samples (such as in a genetic screen), not just the information gathered from assaying a biological system, the chips need to be gentle and be able to sustain the organisms’ physiology (and growth in some cases). Third, many genetic, cell biology studies, and drug screens are morphometric studies, largely using fluorescent markers or reporters. For reasons stated earlier, phenotypical analyses also benefit from quantitation and automation. Therefore the integrated system, the control scheme, the analyses, and the hardware system all have to be compatible and work well with the existing optical microscopy setups.
This review will focus on demonstrated systems for handling multicellular organisms for complex biological analyses, and point towards areas of future research.
Microfluidics Enabling Controlled Experimental Conditions and Environment for Multicelluar Organisms
Controlling Geometry and Mechanical Environment
One benefit of microfluidics is the ability to control precisely the environment surrounding an organism in multiple ways in order to design more accurate experiments to elucidate biological mechanisms. This is one of the central components in most successful microfluidic designs for cell biology and analytical sciences [2, 4, 5]. Two of the primary ways this can be done is by modifying the geometry of the device so that how the organism interacts with it affects its behavior, and by directly controlling the fluid flow in a spatial and temporal manner. Using the well-established soft lithographic fabrication techniques, principles of laminar flow and knowledge of heat and mass transfer, researchers can precisely regulate the microenvironment surrounding the micro-organisms (Drosophila, zebrafish and C. elegans). In contrast to conventional bench-top experiments where the microenvironment surrounding an organism can vary dramatically between experiments, on-chip experiments allow researchers far more precise control. One example is controlling the physical properties surrounding the organism, through the modification of the geometry and material stiffness for studying sensory behavior and locomotion. For example, C. elegans moves by crawling in a sinusoidal manner and generating thrust for forward or backwards motion. This process relies on both the material and geometric properties surrounding the organism. Pillar arrays were used to control, modulate, and observe the locomotion patterns of C. elegans [16, 17]. By altering the pillar size and pillar-to-pillar distance, crawling velocities can be modulated. These microstructured devices are easy to make using PDMS or agar replica-molding from a master, and offer a simple and inexpensive method to investigate how geometry and material properties of an organism’s microenvironment affect its locomotion. These devices are now in the position to allow for complex analyses of behavior genetics and mechanosensory or chemosensory biology.
Controlling Chemical Environment and Mass Transport
Besides mechanical cues, multicellular organisms sense complex chemical cues in order to survive and reproduce, and thus the ability to directly control and selectively expose organisms to chemical cues is of great importance for sensory biology. Chemotaxis assays are commonly performed in macro-scale environments (e.g. using an agar plate with odors or tastants spotted in one part of the plate), but the standard techniques offer limited control of concentration and gradient quality, and can result in a large degree of uncertainty when behavior is of interest. By capitalizing on laminar flow, small diffusion time scales, and spatial confinement, it is possible to improve standard assays using microfluidics to make more controlled gradients with faster temporal control. In an early work, Gray et al. probed the ability of C. elegans to sense molecular oxygen [7]. A stable and repeatable gradient of dissolved oxygen was created by flowing nitrogen and air (21% oxygen) through opposite ends of a PDMS device as “sink” and “source” for the gradient (Fig. 1).
Figure 1.
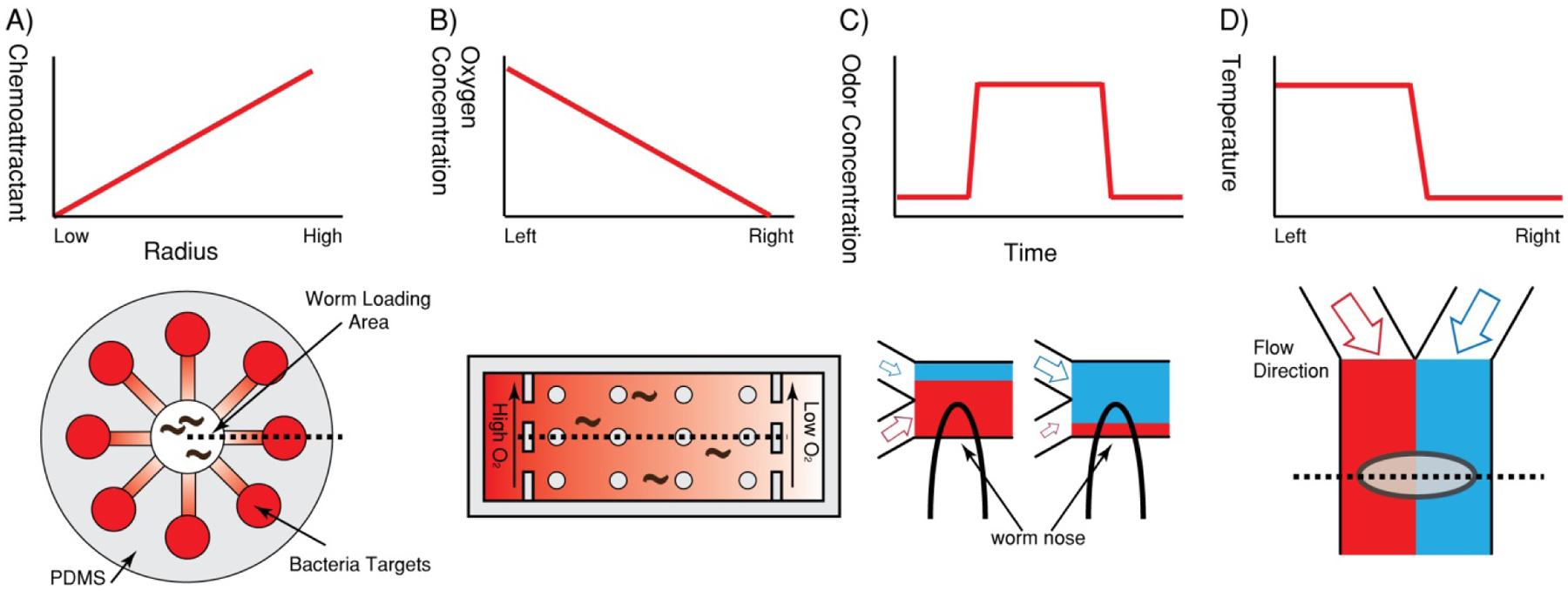
Well-defined and well-controlled mass and thermal diffusion to manipulate the microenvironment for multicellular organisms. (A) Creating an (attractive or repellent) odor gradients of different types of bacteria. Worms placed at the center of the device experience mixed odors and chemotax towards attractive odour sources [6]. (B) Creating gas-phase oxygen gradients with gases of two different concentrations of oxygen as “source” and “sink”. Aerotaxis behavior can be observed in such a device [7]. (C) Creating temporal chemical gradient using laminar flow. This is used to study chemosensory neurons in C. elegans [8]. (D) Creating temperature gradients using hot and cold flow streams in laminar flow. This is used to study the effect of temperature on development in fruit fly embryos [9–11].
Another example of handling gasses and establishing odor gradients for longer-term experiments is the maze olfactory learning assay [6]. By combining spatial or geometric restrictions with the constraints these designs placed on the diffusion of chemical signals has facilitated studies to probe C. elegans olfactory chemotactic response to pathogenic bacteria. This device consisted of a central chamber connected to eight channels leading to open chambers where different strains of pathogenic bacteria were spotted (Fig. 1). The worms were then placed in the central chamber where they experienced multiple chemical cues from the various bacteria used. By observing the choices of the worms, olfactory learning of trained and naive, wild-type or mutant C. elegans can be measured; this further allows the dissection of the genetic pathways and neural circuitry of olfactory learning. Compared to an open-plate conventional chemotactic assay, this olfactory maze allows for directional cues (i.e. not complete mixing of all the odors), which is critical in allowing for the “multiple-choices” instead of the simple traditional two-choice assays.
In addition to creating gas gradients, microfluidic technologies also lend themselves to fast and repeatable switching between dissolved gasses of constant concentrations. PDMS has a relatively high permeability to oxygen as well as other gasses. Using a simple two-layer PDMS device Zimmer et al. [18] imaged neuron activities (calcium transients using calcium-sensitive fluorescent proteins) in response to temporal step gradients of oxygen [18]. In switching of oxygen concentrations, it was found that the dissolved oxygen equilibrated in 5–10 sec after switching and this allowed for both steps-up and -down in concentrations and the recording of the neuronal response in a highly repeatable manner. PDMS gaseous permeability has also been utilized by other researchers for immobilization of C. elegans (which will be discussed later in this review).
The exploitation of microfluidic laminar flow as applied to multicellular model organisms can be best seen in the work by Chronis et al. [8] (Fig. 1). In this work the researchers wished to deliver a chemical stimulus across the tip of the nose of C. elegans (where many sensory neurons have exposed ciliated processes) and record neuronal activities measured by calcium transients. The stimuli needed to be delivered with both precise spatial and temporal control. In a microfluidic device, the worm was loaded into a channel with a slightly smaller cross-section than that of the animal to restrict movement; the animal was held in place by positive pressure and then stimulated by the odors of interest. The temporal control of the simulation was achieved by opening or closing the flow to the control side channels, which also maintains the overall volumetric flow rate and thus stabilizing the pressure. To ensure that no mixing occurred between the buffer and stimulus flow streams, a relatively high flow rate of buffer and stimulus was used (i.e. high Peclet number for mass diffusion).
Fakhoury et al. was able to use a simple microfluidic device for the analysis of drug affects on Drosophila embryo development [19]. A Y-channel design was used to introduce one or two drugs into the main channel. In this main channel, a number of embryos were immobilized using interfacial tension created by oil, water/alcohol and SAM-modified surfaces, and a single or combination of drugs was flowed over the embryos. The constant flow of drug insured a consistent concentration, and the small channel dimensions allowed for minimal reagent consumption. Again, a set of relatively simple to fabricate and use devices has allowed researchers to probe the response of an organism on time and spatial scales not possible using macro methods while using smaller amount of reagents.
Controlling Temperature and Heat Transfer
Besides controlling mass transport, laminar flow can also be used for controlling heat transfer and thus temperature distributions in microfluidic devices. Luchetta et al. and others used a simple Y-shaped PDMS microfluidic device in which one inlet contained a warm buffer solution and the other a cold solution to control the developmental rate in different parts of live Drosophila embryos [9, 10, 19] (Fig. 1). Because of the laminar flow and the relatively large flow rates (resulting in high Peclet numbers for heat transfer, analogous the mass diffusion scenario discussed earlier), little thermal diffusion occurred across the two streams and a sharp temperature step was created across the Drosophila embryo. By visualizing the number of nuclei on the two halves of the embryo that were exposed to the two different temperatures, the researchers were able to discern developmental differences in the same embryo due to temperature effects and begin to understand how embryos control such important processes. This work was later expanded with the parallelization of many fly embryos in a temperature step gradient to further increase the throughput of such assays [11].
Thermal control in microfluidics has also been used to immobilize the nematode C. elegans [20, 21]. C. elegans has very small thermal mass: an adult animal of ~10−5 J/K and L1 larva of ~10−7 J/K; in other words, to raise the temperature by 10K, 0.1 mJ of heat is needed for an adult animal and 1 μJ for an L1 animal. Due to this extremely small thermal mass, C. elegans can be cooled or warmed very rapidly (practically instantaneously) by controlling the surrounding temperature. Chung et al. incorporated a cooling channel into the devices, relying on thermal diffusion through a thin PDMS membrane (also of small thermal mass) between the temperature controlling channel and the worm channel to achieve rapid immobilization. The animals were instantaneously immobilized as soon as they entered the cooling region at ~4 °C, and instantaneously mobilized as soon as they left the region to warm back to room temperature.
These examples demonstrate that through microfluidic technology, it is possible to precisely and repeatedly control the environment for a variety of behavioral, neural, and developmental investigations in multicellular organisms. The level of spatial and temporal control these groups have achieved would have been significantly harder (if not impossible) using macroscopic methods and the field of microfluidics have opened the door to many previously unattainable studies.
Unique Challenges in Manipulating Multicellular Organisms in Microfluidics
Although microscopic in length scale, multicellular organisms pose additional changes when manipulating them in microfluidics for the following reason: (1) multicellular organisms are capable of rapid and sometimes unpredictable locomotion, (2) their size variation as a function of age, development, or genetic make-up can be significant, in contrast to the relative uniformity of sizes of cell lines, and (3) the thickness of the specimen and the presence of multiple tissue types and organs can present significant optical challenges. Recent development of new devices and strategies has begun to address these issues.
When working with the embryos of multicellular organisms, one of the pressing concerns is the ability to precisely position and orient them. For example, positioning a large number of embryos in well-ordered arrays and orient them in a predetermine manner is a prerequisite for automating high-throughput microinjection or image processing [19, 22–26]. Due to the size of the embryos, there is considerable diffraction and optical distortion when acquiring images with particular orientations. To obtain high quality data, the embryos are required to be precisely aligned. However, this manipulation (of the small embryos of Drosophila or zebrafish for example) done manually is very difficult due to the delicate structures and the fact that they can be easily damaged during the process. To address this issue, Cornell et al. developed a microstructure that contained a dense array of precisely machined U-shaped grooves on a block of stainless steel [26]. The Drosophila embryos are brushed into the grooves with a paintbrush and aligned in the U-shaped groove that is slightly wider than the embryo diameter. Although this method requires manual adjustments to improve alignment, this simple micro-groove structure enables high throughput as many rows can be aligned at once.
Another group (Bernstein et al.) reported a simple and passive way of positioning Drosophila embryos using array of microfabricated gold pads on an oxidized silicon substrate [22] (Fig. 2). When a thin layer of oil is deposited on the array, only hydrophobic gold pad is covered with oil. Because the oil pad acts as an adhesive, the embryos are self-assembled on the ordered array of the gold pads and properly oriented as well. With this fluidic self-assembly technique, the researchers achieved a high success rate, with only < 5 % of embryos misplaced.
Figure 2.
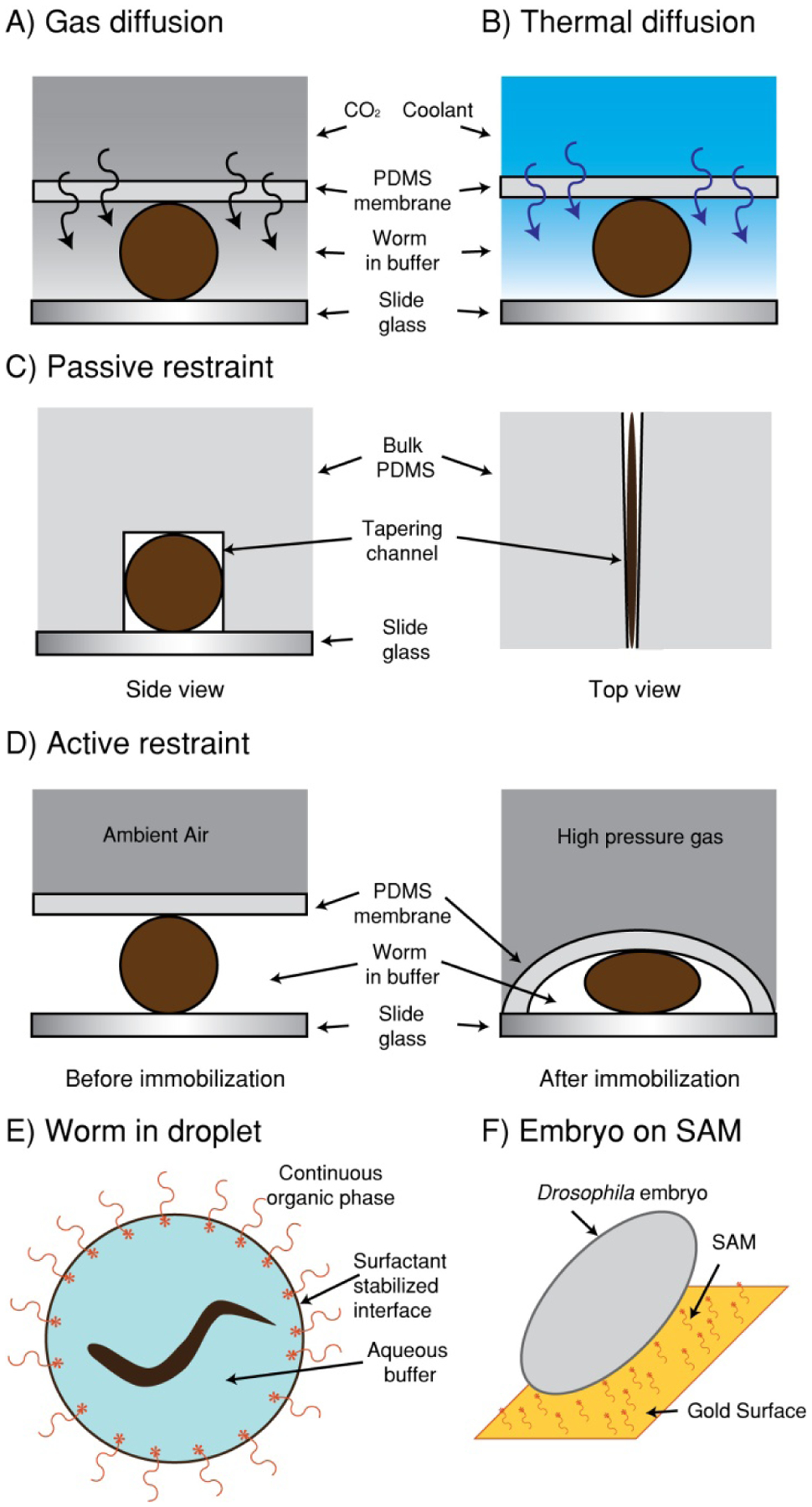
Microfluidic methods used to directly manipulate multicellular organisms. (A-B) Methods of immobilizing C. elegans using the principles of mass and thermal diffusion. (A) The creation of a gradient across a thin membrane of PDMS allow CO2 to diffuse and immobilize the animal[37]. (B) Using the same principle with thermal diffusion to cool the animals to ~4 °C[21]. (C-D) A common method of immobilizing C. elegans is to restrict the range of motion either passively or actively. (C) Passive restriction involves flowing the animals into a channel slightly smaller than the animal cross section to prevent movement[27]. (D) Active restriction uses a two-layer device, wherein a pressurized membrane deforms around the worm to prevent movement[34, 37]. (E-F) Lipids have been used to manipulate animals. (E) Single animals can be isolated by surrounding a small amount of an aqueous buffer containing an animal or embryo with an organic continuous phase [30, 31, 53]. (F) Using SAM-on–gold surface modification, it is possible to precisely align embryos for microinjections [23].
Researchers using zebrafish commonly perform microinjection to insert transgenic plasmids, or RNAi, which poses many of the same problems with handling Drosophila embryos. For zebrafish embryo microinjection, Wang et al. developed an embryo positioning device that consists of evenly-spaced through holes connected to a vacuum source via a backside channel [25]. The negative pressure through a well-defined microstructure successfully positioned the embryos within a few seconds without damaging them. Embryos positioned outside of the through-holes can be easily flushed away from the device since negative holding pressure is applied only to embryos located directly on the through holes. Using the device with full automation, the researchers demonstrated microinjection with a high throughput and a high success rate.
Microstructure design is also utilized for positioning live C. elegans for imaging. Hulme et al. developed a device that consists of an array of tapering channels [27, 28] (Fig. 2). Once a worm enters a channel with a cross-section similar to that of the worm, the hydrodynamic resistance of the channel increases dramatically, simultaneously reducing the flow rate through that channel and increasing it through the vacant channels. This process automatically distributes the individual worms into each channel by the passive pressure changes. Using this passive mechanism the researchers demonstrated rapid positioning of a large number of worms (over 100 worms in less than 15 min).
Another microfluidic technology that has allowed precise manipulation of model organisms is the use droplets for encapsulating the organisms [29–31] (Fig. 2). Microfluidic devices, when its micro structure, surface properties, and the flow rates are properly controlled, enable formation of monodispersed droplets at high speeds [32, 33]. By distributing the multicellular organisms in the aqueous phase, individual droplets can be used as containers for encapsulating the individual organisms. For instance, Clausell et al. developed a droplet-based microfluidic platform that generates droplets encapsulating C. elegans [30]. In this study, biocompatible surfactants used in droplet formation and the gas-permeable PDMS device allow long-term culture of C. elegans in droplets. They demonstrated that the worms remain fully viable for several days and proliferate in droplets containing E. coli OP 50 as a food source. This system could be used in high-throughput biochemical screens. Funfak et al. adopted the droplet technology to study development of the zebrafish embryos [29]. They generated aqueous plugs containing zebrafish embryo and observed the development of the embryos until hatching time. This method could be a promising technique to study the effects of drugs or toxic substances on single individuals. Microfluidic technology not only enables generating droplets containing the organisms, it also allows precise manipulation of the droplets. Shi et al. developed a system that allows droplet generation, transportation, and immobilization in a single device [31]. The researchers demonstrated the encapsulation of C. elegans into a parallel series of nanolitre-volume droplets and immobilization of them in a droplet trap array. Using the microdevice they investigated the behavioral response of C. elegans to a neurotoxin, 1-methyl-4-phenylpyridinum. This microdevice allows easy handling of individual worms without mechanical injury.
When it comes to handling small model organisms, the most unique advantage conferred by microfluidic technology is integration of functional components. On-chip functional components could enable sophisticated level of control that is otherwise impossible to achieve using macro-scale methods. For example, in order to image C. elegans, one needs to immobilize samples with anesthetics, manually mount the samples on a slide glass, locate each individual, and then image them. For phenotypical screening and laser ablation purposes, additional processes are required, and these include locating target neurons, laser firing, and rescuing the worms by sliding the coverslip off and picking the worms using a “worm pick”, which is often a small platinum wire. This painstaking manual handling not only significantly limits the experimental throughput, but also increases noise due to variation in sample-to-sample handling.
To address this limitation, three groups independently developed multilayer PDMS devices that can rapidly route, load, and immobilize live C. elegans for high-resolution imaging and laser ablation [20, 21, 34–39] (Fig. 2). These microdevices are similar in utilizing intricate sequences of on-chip PDMS valves [40, 41] to control a buffer suspension of worms, while differing in key mechanisms. Each group explored a unique approach for single worm loading. Zeng and Rohde et al. developed two-step suction mechanism whereby a single worm is first captured by a single suction channel while the remaining un-trapped worms are flushed away [34, 36]. The captured worm is then transferred from the single suction channel to an array of suction channels on the opposing wall. This is done by manually actuating on-chip valves. Guo et al. utilized a set of side manipulation channels located at each end of the imaging/surgery region to manually position a worm in the region [35]. Both of these multi-step processes require additional image acquisition, analysis, and valve actuation to coordinate the activities on-chip. An alternative to these designs is by Chung et al., which uses self-regulated loading mechanism and passive positioning mechanism using partially closable on-chip valves [20, 21]. This particular design completely eliminates the need to have (manual or image-based) feedback control of the sample loading to ensure one and only one worm is loaded at a time, and thus ensures robust and consistent operation of the chip.
Once a worm is loaded, it needs to be completely immobilized because, unlike embryos, C. elegans has high motility that can cause blurring of image or incorrect laser ablation. Various groups have developed different strategies to achieve immobilization. While Hulme et al. [27] and Allen et al. [39] uses simple geometric constraint, Guo et al. [35] and Zeng et al. [36] used the elastomeric properties of PDMS and positive pressure to physically restrain the animal. The latter groups integrated a thin PDMS membrane, similar to an on-chip valve [41], over the channel where the worm is loaded. By pressurizing the membrane, and deforming it around the animal they were able to mechanically restrict the worm’s movement reversibly. Both groups demonstrated immobilization of worm’s body movement and successfully performed femtosecond-laser microsurgery.
Two other approaches for immobilization are explored by Chung et al. and Chokshi et al. [20, 21, 37] (Fig. 2). The two technologies take advantage of the rapid heat and mass transfer in microsystems. As the size of microstructure is reduced, the surface area-to-volume ratio increases, which allows the microsystem to reach steady state rapidly. As reviewed earlier, Chung et al. used an integrated temperature control channel to locally cool the loaded worm for immobilization. The small thermal mass of C. elegans and large surface area-to-volume ratio of the microchannel results in nearly instantaneous immobilization of the body as well as stopping the pharyngeal pumping, which is critical in cell laser ablation. Analogously, Chokshi et al. use rapid mass diffusion of gas through PDMS membranes to create a high CO2 micro-environment [37]. Although more time for immobilization than the other methods is required (within 1~2 min), this technique proved sufficient and is an alternative for long-term immobilization (1–2 hours).
To further widen the possibilities of many biological studies, researchers can precisely manipulate the organisms in high-throughput manner by orchestrating the functional components. Several of the most interesting categories of biological studies include the ability to image and sort multicellular organisms, and the ability to conduct genetic (or pharmacological) screens. Although sorting samples from a collection (or sometimes referred to as a “library”) of small entities such as bacteria, organelles, or mammalian cells has been well demonstrated [30, 42–48], sorting multicellular organisms is nontrivial due to the size, and the active motion of multicellular organisms. On the other hand, sorting is of great interest to biologists, because the precise handling and control of organisms has significant labor-saving potential, and could reduce the time spent performing genetic screens from months to days. To sort for mutants of interest (or animals treated with different drugs for instance), they must be detected, imaged (or otherwise evaluated), and then sorted by opening specific valves. The specific sequence and fluid handling can vary significantly depending on the device design and manipulation [21, 34, 38].
Intrinsic to the promise of the microfluidic field is the potential to optimize designs to achieve a significantly higher throughput. In dealing with multicellular organisms, this has been done by creating arrays, or parallelization, and by optimizing the control process so as to minimize the number of steps and variation involved in a serial process. The creation of arrays, as in dealing with single-cell systems, offers the ability to inspect large numbers of different culture conditions or a single-animal cultures. Two such approaches mentioned earlier achieve the parallelization: single animals were either trapped in aqueous droplets surrounded by an oil medium with these droplets introduced to an array device where flow forced them to occupy single chambers [31], or were cultured and trapped in a narrowing microfluidic channel in parallel [27, 28]. Analogously optimized processes are also critical to increasing throughput in serial operations. Ans example of this is Chung et al. [21] where the authors used the shape of the animal and hydrodynamic forces to reliably position the animal in the channel. This reduces the number of active steps, and allows reliable imaging without moving the stage. All of these approaches take advantages of the different hydrodynamic resistances of an open chamber/channel versus a filled one. This is advantageous as compared to when active components are employed because of its simplicity and the consequent robustness.
Towards Robust and Automated Microfluidic Systems for Multicellular Organism Research: Off-chip and Systemic Considerations
Systemic, or off-chip, components are central to the robust operations of microfluidic chips, although this is less emphasized in microfluidic literature. Well engineered off-chip components and software are critical to creating an ease of operation sufficient to allow microfluidic devices to tackle practical biological problems of interest. The following are important areas to consider when designing a microfluidic system: image analysis and signal processing, automation (through error handling and valve control), incorporation of appropriate microsurgery laser tools, and lab-to-chip interface methods. There are currently an abundance of biological problems that require significant advances in modern technology before being solved, but without the creation of a comprehensive system solution, microfluidic devices would fail to have the significant impact on the biological community that could otherwise be achieved.
Autonomous, or semi-autonomous operation requires devices to be integrated with computer controls [21, 28, 38]. Typically this entails off-chip solenoid valves actuated by a digital control board, electronically controlled pressure regulators, a computer controlled image acquisition system, and potentially an x-y-z stage. Automation, however, requires significantly more than computer control. Extensive error-handling is required since the size of a multicellular organism can vary dramatically, and when coupled with motility and the inevitable problem of debris, can result in device failure. Creating a fully automated system requires a collaborative effort with computer science, electronics, and robotics. Although the requirements for automated microfluidic systems used with multicellular organisms differs little from those used with particles or single-cells, commercially available systems that integrate with microfluidics are not available at present, and custom-made systems are typically used.
Along with automated routines, it is important to perform image analysis in a robust manner and potentially automated decision making for screens and to perform other operations based on the images, such as laser ablations. The primary promises of engineering to biology are the ability for increased throughput and control and reduce bias. Increased control in the realm of microfluidics comes not just from the ability to precisely manipulate the environment on a microscale but from the ability to position samples and acquire images in a rapid, highly repeatable manner. The images acquired during an experiment can be stored for manual analysis later on; however, many high-impact applications such as microsurgery and genetic screening benefit greatly from the real-time decision-making to achieve their full potentials. In these scenarios images must be processed in order to identify specific features of interest such as cells, axons/dendrites, or even synapses. When using sufficiently bright fluorescent reporters this can be achieved by a series of intensity thresholding and simple morphological operations [21, 28, 34]. In the case of screening, the processed images can be classified using statistical learning methods to separate different classes of animals [21]. In the case of monitoring long-term behavior of samples, e.g. motility and life-span [28], image processing routines facilitate the experiments and standardize data analysis. The use of alternative imaging modalities, such as brightfield, Ca2+ or DIC, can potentially make the processing more difficult, but can still be tackled using computer science tools.
Microfluidic systems that meet some of the highest impact biological needs, such as laser microsurgery or microinjection, also require additional specialized hardware. Laser microsurgery on model organisms has been extremely useful since it was first popularized for killing specific cells [49] and studying development. Using a low-powered, nano-second laser allows researchers to selectively ablate cells and study the behavioral response when a lesion is created in the network. Taking advantage of the precise control and higher throughput, this has recently been demonstrated using an automated microfluidic system [20]. Additionally, the utility of femtosecond lasers (although considerably more expensive) has allowed researchers to dramatically limit the amount of energy absorbed by surrounding tissues and to ablate axons or dendrites to study axonal regeneration. Two groups have demonstrated the ability to immobilize C. elegans in a microfluidic device and cut axons [35, 36, 50]. One group reported that the use of microfluidic immobilization resulted in significantly more rapid axonal regeneration when compared to conventional methods using anesthetics [35]. Another area of interest is the injection of Drosophila and zebrafish embryos with RNAi for large-scale genetic screens [24–26, 51, 52]. Using microneedles developed by conventional silicon MEMS fabrication coupled with a microfluidic device, the ability to inject large numbers of embryos with RNAi has been demonstrated.
Most microfluidic devices are currently created and operated in engineering labs where the difficulty of setting up and replacing devices is secondary to creating novel designs. As such, most microfluidic devices are painstakingly set up for each experiment by connecting small pieces of tubing to holes cored into the PDMS device. Not only is this a time consuming bottleneck, but it creates a failure point and limits the ability to use standard lab equipment such as micro-pipettes or liquid-handlers in conjunction with the microfluidic systems created. In the near future, one could expect that interfaces capable of meeting the needs of many labs and applications can be developed and commercially marketed. This would not only speed up the development time for new microfluidic systems, but reduce the early adopter cost to biology labs interested in utilizing systems already published.
Summary and Future Outlook
Although genetic screens of multicellular model organisms are an important part of modern biology, the every-day common methods with which scientists manipulate these organisms are still labor intensive and time-consuming. Microfluidic engineering presents an excellent opportunity in making an impact in these fields for high-throughput, high-content screens. To best accomplish this, understanding the needs of the biology communities is crucial, as is the ability to harness the unique advantages conferred by microfluidics in terms of manipulating flow and the transport of mass, energy, and momentum. One must also consider the unique challenges in handling multicellular organisms and the integration of off-chip components.
In particular we believe that the future of microfluidics for multicellular organisms will focus on standardization, inreased scale of experimentation, and automation. For instance, many large-scale experiments such as genetic screen (in developmental biology) or drug screens for particular disease models (e.g. Alzheimer’s disease) will benefit from automation and high-throughput. The many laboratories performing related experiments and laboratories mining the large-scale data sets for genetic or genomic studies will also want to be able to compare data gathered from different experiments and from different laboratories, making standardization an absolute necessity. As microfluidics becomes more common in biological laboratories, we expect the end-users in the future to be able to purchase a single system containing valves, controllers, and software that can handle many different devices. We also expect that many relevant biological experiments will need to be performed in longer time scales; therefore the ability to culture organisms longer term would be of tremendous interest. In addition, many questions in immunology and ecology are best addressed by co-culturing mutliple species; there will be needs for engineering solutions for materials, fabrication, design, protocol optimization, and system integration. We look forward to many exciting works yet to come, and the impacts these engineered microsystems will have in fundamental genetics and disease studies, systems biology, and pharmaceutical developments.
Notes and references
- 1.deMello AJ, Control and detection of chemical reactions in microfluidic systems. Nature, 2006. 442(7101): p. 394–402. [DOI] [PubMed] [Google Scholar]
- 2.El-Ali J, Sorger PK, and Jensen KF, Cells on chips. Nature, 2006. 442(7101): p. 403–411. [DOI] [PubMed] [Google Scholar]
- 3.Psaltis D, Quake SR, and Yang CH, Developing optofluidic technology through the fusion of microfluidics and optics. Nature, 2006. 442(7101): p. 381–386. [DOI] [PubMed] [Google Scholar]
- 4.Whitesides GM, The origins and the future of microfluidics. Nature, 2006. 442(7101): p. 368–373. [DOI] [PubMed] [Google Scholar]
- 5.Yager P, et al. , Microfluidic diagnostic technologies for global public health. Nature, 2006. 442(7101): p. 412–418. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Lu H, and Bargmann CI, Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature, 2005. 438(7065): p. 179–184. [DOI] [PubMed] [Google Scholar]
- 7.Gray JM, et al. , Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature, 2004. 430(6997): p. 317–322. [DOI] [PubMed] [Google Scholar]
- 8.Chronis N, Zimmer M, and Bargmann CI, Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nature Methods, 2007. 4: p. 727–731. [DOI] [PubMed] [Google Scholar]
- 9.Lucchetta EM, et al. , Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature, 2005. 434(7037): p. 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchetta EM, Munson MS, and Ismagilov RF, Characterization of the local temperature in space and time around a developing Drosophila embryo in a microfluidic device. Lab on a Chip, 2006. 6(2): p. 185–190. [DOI] [PubMed] [Google Scholar]
- 11.Dagani GT, et al. , Microfluidic self-assembly of live Drosophila embryos for versatile high-throughput analysis of embryonic morphogenesis. Biomedical Microdevices, 2007. 9(5): p. 681–694. [DOI] [PubMed] [Google Scholar]
- 12.Doll JC, et al. , SU-8 force sensing pillar arrays for biological measurements. Lab on a Chip, 2009. 9(10): p. 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SJ, Goodman MB, and Pruitt BL, Analysis of nematode mechanics by piezoresistive displacement clamp. Proceedings of the National Academy of Sciences of the United States of America, 2007. 104: p. 17376–17381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin JH and Wheeler AR, Maze exploration and learning in C-elegans. Lab on a Chip, 2007. 7(2): p. 186–192. [DOI] [PubMed] [Google Scholar]
- 15.Dupuy D, et al. , Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nature Biotechnology, 2007. 25: p. 663–668. [DOI] [PubMed] [Google Scholar]
- 16.Lockery SR, et al. , Artificial dirt: Microfluidic substrates for nematode neurobiology and behavior. Journal of Neurophysiology, 2008. 99(6): p. 3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S, et al. , Enhanced Caenorhabditis elegans locomotion in a structured microfluidic environment. PLoS One, 2008. 3(6): p. e2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmer M, et al. , Neurons Detect Increases and Decreases in Oxygen Levels Using Distinct Guanylate Cyclases. Neuron, 2009. 61(6): p. 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fakhoury JR, Sisson JC, and Zhang XJ, Microsystems for controlled genetic perturbation of live Drosophila embryos: RNA interference, development robustness and drug screening. Microfluidics and Nanofluidics, 2009. 6(3): p. 299–313. [Google Scholar]
- 20.Chung K and Lu H, Automated high-throughput cell microsurgery on-chip. Lab on a Chip, 2009. 9(19): p. 2764–2766. [DOI] [PubMed] [Google Scholar]
- 21.Chung KH, Crane MM, and Lu H, Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nature Methods, 2008. 5(7): p. 637–643. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein RW, et al. Characterization of fluidic microassembly for immobilization and positioning of Drosophila embryos in 2-D arrays. 2004: Elsevier Science Sa. [Google Scholar]
- 23.Zhang XJ, et al. , Microoptical characterization and modeling of positioning forces on drosophila embryos self-assembled in two-dimensional arrays. Journal of Microelectromechanical Systems, 2005. 14(5): p. 1187–1197. [Google Scholar]
- 24.Zappe S, et al. , Automated MEMS-based Drosophila embryo injection system for high-throughput RNAi screens. Lab on a Chip, 2006. 6(8): p. 1012–1019. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, et al. , A Fully Automated Robotic System for Microinjection of Zebrafish Embryos. PLoS ONE, 2007. 2(9): p. e862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornell E, et al. , Automating fruit fly Drosophila embryo injection for high throughput transgenic studies. Review of Scientific Instruments, 2008. 79(1): p. 013705. [DOI] [PubMed] [Google Scholar]
- 27.Hulme SE, et al. , A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab on a Chip, 2007. 7(11): p. 1515–1523. [DOI] [PubMed] [Google Scholar]
- 28.Hulme SE, et al. , Lifespan-on-a-chip: microfluidic chambers for performing lifelong observation of C. elegans. Lab on a Chip, 2010. 10(5): p. 589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funfak A, et al. , Micro fluid segment technique for screening and development studies on Danio rerio embryos. Lab on a Chip, 2007. 7(9): p. 1132–1138. [DOI] [PubMed] [Google Scholar]
- 30.Clausell-Tormos J, et al. , Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chemistry & Biology, 2008. 15(5): p. 427–437. [DOI] [PubMed] [Google Scholar]
- 31.Shi WW, et al. , Droplet-based microfluidic system for individual Caenorhabditis elegans assay. Lab on a Chip, 2008. 8(9): p. 1432–1435. [DOI] [PubMed] [Google Scholar]
- 32.Link DR, et al. , Geometrically Mediated Breakup of Drops in Microfluidic Devices. Physical Review Letters, 2004. 92(5): p. 54503. [DOI] [PubMed] [Google Scholar]
- 33.Utada AS, et al. , Monodisperse double emulsions generated from a microcapillary device. Science, 2005. 308(5721): p. 537–541. [DOI] [PubMed] [Google Scholar]
- 34.Rohde CB, et al. , Microfluidic system for on-chip high-throughput whole-animal sorting and screening at subcellular resolution. PNAS, 2007. 104(35): p. 13891–13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo SX, et al. , Femtosecond laser nanoaxotomy lab-on-achip for in vivo nerve regeneration studies. Nature Methods, 2008. 5(6): p. 531–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng F, Rohde CB, and Yanik MF, Sub-cellular precision on-chip small-animal immobilization, multi-photon imaging and femtosecond-laser manipulation. Lab on a Chip, 2008. 8(5): p. 653–656. [DOI] [PubMed] [Google Scholar]
- 37.Chokshi TV, Ben-Yakar A, and Chronis N, CO2 and compressive immobilization of C. elegans on-chip. Lab on a Chip, 2009. 9(1): p. 151–157. [DOI] [PubMed] [Google Scholar]
- 38.Crane MM, Chung K, and Lu H, Computer-enhanced high-throughput genetic screens of C. elegans in a microfluidic system. Lab on a Chip, 2009. 9(1): p. 38–40. [DOI] [PubMed] [Google Scholar]
- 39.Allen PB, et al. , Single-synapse ablation and long-term imaging in live C. elegans. Journal of Neuroscience Methods, 2008. 173(1): p. 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quake SR and Scherer A, From micro- to nanofabrication with soft materials. Science, 2000. 290(5496): p. 1536–1540. [DOI] [PubMed] [Google Scholar]
- 41.Unger MA, et al. , Monolithic microfabricated valves and pumps by multilayer soft lithography. Science, 2000. 288(5463): p. 113–116. [DOI] [PubMed] [Google Scholar]
- 42.Fu AY, et al. , A microfabricated fluorescence-activated cell sorter. Nature Biotechnology, 1999. 17(11): p. 1109–1111. [DOI] [PubMed] [Google Scholar]
- 43.Lu H, et al. , A microfabricated device for subcellular organelle sorting. Analytical Chemistry, 2004. 76(19): p. 5705–5712. [DOI] [PubMed] [Google Scholar]
- 44.Vahey MD and Voldman J, An Equilibrium Method for Continuous-Flow Cell Sorting Using Dielectrophoresis. Anal. Chem, 2008. 80(9): p. 3135–3143. [DOI] [PubMed] [Google Scholar]
- 45.Chabert M and Viovy JL, Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells. Proceedings of the National Academy of Sciences, 2008. 105(9): p. 3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu X, et al. , Marker-specific sorting of rare cells using dielectrophoresis. Proceedings of the National Academy of Sciences of the United States of America, 2005. 102(44): p. 15757–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lou X, et al. , Micromagnetic selection of aptamers in microfluidic channels. Proceedings of the National Academy of Sciences, 2009. 106(9): p. 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toner M and Irimia D, Blood-on-a-chip. Annu. Rev. Biomed. Eng, 2005. 7: p. 77–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bargmann CI and Avery L, Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol, 1995. 48: p. 225–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohde C, et al. , High-throughput in vivo genetic and drug screening using femtosecond laser nano-surgery, and microfluidics. Conf Proc IEEE Eng Med Biol Soc, 2008. 2008: p. 2642. [DOI] [PubMed] [Google Scholar]
- 51.Lu Z, et al. , A micromanipulation system with dynamic force-feedback for automatic batch microinjection. Journal of Micromechanics and Microengineering, 2007. 17(2): p. 314–321. [Google Scholar]
- 52.Zhang XJ, et al. , Microoptical characterization of piezoelectric vibratory microinjections in Drosophila embryos for genome-wide RNAi screen. Journal of Microelectromechanical Systems, 2006. 15(2): p. 277–286. [Google Scholar]
- 53.Son SU and Garrell RL, Transport of live yeast and zebrafish embryo on a droplet (“digital”) microfluidic platform. Lab on a Chip, 2009. 9(16): p. 2398–2401. [DOI] [PubMed] [Google Scholar]


