Summary
This protocol describes the detailed procedures for utilizing human pluripotent stem cells (hPSCs) for pancreatic progenitor and hepatic differentiation, followed by the application of hPSC-derived cells in a luciferase reporter-based assay to study gene regulation. The generated hPSC-derived cells have been shown to achieve morphologies and gene expression profiles specific to their differentiated cell types, and subsequent luciferase assay has been shown to effectively elucidate the role of disease-relevant gene variants. Therefore, this protocol provides a valuable approach for pancreatic and liver disease modeling.
For complete details on the use and execution of this protocol, please refer to Ng et al. (2019).
Subject areas: Cell culture, Cell-based Assays, Molecular Biology, Gene Expression, Stem Cells, Cell Differentiation
Graphical Abstract

Highlights
-
•
Protocol generates pancreatic and hepatic progenitors from human pluripotent stem cells
-
•
Describes use of luciferase reporter assay to study gene regulation in hPSC-derived cells
-
•
Enables the study of gene regulation during pancreatic and liver development
-
•
Provides a valuable approach for modeling pancreatic and liver diseases
This protocol describes the detailed procedures for utilizing human pluripotent stem cells (hPSCs) for pancreatic progenitor and hepatic differentiation, followed by the application of hPSC-derived cells in a luciferase reporter-based assay to study gene regulation. The generated hPSC-derived cells have been shown to achieve morphologies and gene expression profiles specific to their differentiated cell types, and subsequent luciferase assay has been shown to effectively elucidate the role of disease-relevant gene variants. Therefore, this protocol provides a valuable approach for pancreatic and liver disease modeling.
Before you begin
Preparation of coated plates, media solutions, growth factors, and small molecules
Prepare all coated plates, media, and stock solutions before use. This includes coated culture plates, cell culture media, growth factors and small molecules used for differentiation, and DNA plasmids used for transfections. Refer to the Materials and Equipment section on guidelines and storage of reagents and materials. A complete list of reagents and materials can be found in the key resources table.
Culture of human pluripotent stem cells in preparation for differentiation
Timing: up to 2 weeks
Before setting up human pluripotent stem cells (hPSCs) for differentiation, culture the hPSCs under feeder-free conditions, or on feeder cells (such as CF-1 Mouse Embryonic Fibroblast [MEF] feeder cells). Note that there will be variations in this protocol for hPSCs grown in feeder-free media versus feeder-based culture.
-
1.For feeder-free culture:
-
a.Prior to seeding of cells, pre-coat surfaces of culture plates with 0.1% gelatin and FBS with reference to instructions stated in the “Preparation of Gelatin/FBS-containing Medium-Coated Plates for Feeder-free hPSC Culture” subsection; Materials and Equipment section in this paper.
-
b.Passage hPSCs using ReLeSR™, once a week when confluent according to the manufacturer’s instructions. Generally, a confluent well of a 6-well plate of hPSCs can be seeded into a 100 mm pre-coated culture plate.
-
c.Maintain hPSCs by changing fresh TeSR™-E8™ or mTeSR™1 medium daily.
-
d.Culture hPSCs in a 37°C, 5% CO2 humidified incubator.
-
a.
-
2.Alternatively, for feeder-based culture:
-
a.Prior to seeding of cells, pre-coat surfaces of culture plates with 0.1% gelatin and seed CF-1 MEF feeder cells at a density of 1–2 million cells per 6-well plate or 100 mm plate in FBS-containing medium. The feeder cells will be ready for use on the next day.
-
b.Passage hPSCs once a week when confluent by manual picking of individual colonies. Generally, a confluent well of a 6-well plate of hPSCs can be passaged into a 100 mm pre-coated culture plate.
-
c.Maintain hPSCs by changing fresh hPSC culture medium daily. The composition of hPSC culture medium used for feeder-based hPSC culture can be found in the “hPSC Culture Medium” subsection; Materials and Equipment section of this paper.
-
d.Culture hPSCs in a 37°C, 5% CO2 humidified incubator.
-
a.
Note: The scale of the differentiation experiment determines the number of confluent 100 mm hPSC culture plates required on the day of setup.
CRITICAL: The homogeneity of undifferentiated hPSCs during setup affects the efficiency of the subsequent differentiation. A plate of hPSC culture observed visually to contain more than 10% differentiated cells should not be used as starting material for differentiation.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Activin A | STEMCELL Technologies | Cat # 78001.2 |
| Advanced RPMI 1640 | Thermo Fisher Scientific (Gibco) | Cat # 12633020 |
| All-trans retinoic acid (RA) | Wako | Cat #186-01114 |
| B-27 Supplement (50×), Minus Vitamin A | Thermo Fisher Scientific (Gibco) | Cat # 12587010 |
| Bone morphogenetic factor 4 (BMP4) | Miltenyi Biotec | Cat # 130-111-168 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat # A9418 |
| CHIR99021 | Singlab Technologies | Cat # 4423 |
| Collagenase type IV | Thermo Fisher Scientific (Invitrogen) | Cat # 17104019 |
| DAPT | Abcam | Cat # AB120633 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat # D2650 |
| Dispase in DMEM/F12 | STEMCELL Technologies | Cat # 7923 |
| DMEM/F-12, GlutaMAX™ Supplement | Thermo Fisher Scientific (Gibco) | Cat # 10565018 |
| DMEM/high glucose with L-glutamine, sodium pyruvate | Hyclone Laboratories Inc. | Cat # SH30243.01 |
| Dulbecco’s phosphate buffered saline (DPBS), 1×, without calcium, magnesium, cell culture grade | Hyclone Laboratories Inc. | Cat # SH30028.02 |
| Fibroblast growth factor 2 (FGF2) | Miltenyi Biotec | Cat # 130-093-840 |
| Fibroblast growth factor 10 (FGF10) | Miltenyi Biotec | Cat # 130-093-850 |
| Gelatin from porcine skin | Sigma-Aldrich | Cat # G1890 |
| HCMTM Hepatocyte Culture Medium BulletKitTM | Lonza | Cat # CC-3198 |
| Hepatocyte growth factor (HGF) | Sigma-Aldrich | Cat # H9661 |
| Knockout serum replacement (KOSR) | Thermo Fisher Scientific (Gibco) | Cat # 10828028 |
| L-Glutamine | Thermo Fisher Scientific (Invitrogen) | Cat # 25030149 |
| LY294002 | LC Labs | Cat # L-7962 |
| MEM-Non-Essential Amino Acids (MEM-NEAA) Solution (100×) | Thermo Fisher Scientific (Gibco) | Cat # 11140050 |
| mTeSR™1 | STEMCELL Technologies | Cat # 85850 |
| Na2HPO4 | Tocris Bioscience | Cat # 3153 |
| NaH2PO4 | Sigma-Aldrich | Cat # S3139 |
| Nicotinamide | Sigma-Aldrich | Cat # N0636 |
| Oncostatin M (OSM) | Mitenyi Biotec | Cat # 130-093-976 |
| Opti-MEM™ Reduced Serum Medium | Thermo Fisher Scientific (Gibco) | Cat # 31985062 |
| Penicillin/streptomycin | Thermo Fisher Scientific (Gibco) | Cat # 15140122 |
| Phosphoric acid, ACS reagent, ≥85 wt. % in H2O | Sigma-Aldrich | Cat # 695017 |
| ReLeSR™ Human Pluripotent Stem Cell Passaging Reagent | STEMCELL Technologies | Cat # 05872 |
| Research Grade Fetal Bovine Serum South American Origin (FBS) | Hyclone Laboratories Inc. | Cat # SV30160.03 |
| Rho kinase inhibitor Y-27632 | STEMCELL Technologies | Cat # 72308 |
| Sodium hydroxide ACS reagent, ≥97.0%, pellets | Sigma-Aldrich | Cat # 221465 |
| TeSR™-E8™ | STEMCELL Technologies | Cat # 05990 |
| TrypLE™ Express Enzyme (1×), Phenol Red | Thermo Fisher Scientific (Gibco) | Cat # 12605010 |
| Water, Cell Culture Grade | Hyclone Laboratories Inc. | Cat # SH30529.02 |
| Critical commercial assays | ||
| Dual-Luciferase® Reporter 1000 Assay System | Promega | Cat # E1980 |
| FuGENE® 6 Transfection Reagent | Promega | Cat # E2691, E2692 and E2693 |
| Experimental models: Cell lines | ||
| CF-1 MEF Feeder Cells | Lonza | Cat #GSC-6201G |
| H9 (WA09) Human Embryonic Stem Cells (hESCs) | WiCell | N/A |
| hiPSC Lines (iN904-1A/B/C, iN904-2, iN904-7A/B/C, iN904-13A/B) | Adrian Teo Laboratory | N/A |
| Recombinant DNA | ||
| pCDH-HNF4A | Adrian Teo Laboratory | N/A |
| pGL4.10-APOBp | Adrian Teo Laboratory | N/A |
| pRL-TK Renilla | Promega | Cat # E2241 |
| Other | ||
| Axygen® 1.7 mL MaxyClear Snaplock Microcentrifuge Tube | Axygen, Inc. | Cat # MCT-175-C |
| Corning® 100 mm TC-treated Culture Dish | Corning Inc. | Cat # 430167 |
| Costar® 6-well Clear TC-treated Multiple Well Plates, Individually Wrapped, Sterile | Corning Inc. | Cat # 3516 |
| Costar® 24-well Clear TC-treated Multiple Well Plates, Individually Wrapped, Sterile | Corning Inc. | Cat # 3524 |
| Duran® Laboratory Bottles, with Caps, Capacity 500 mL, Blue PP Screw Cap and Pouring Ring | Duran | Cat # Z305197 |
| Falcon® 15 mL High Clarity PP Centrifuge Tube, Conical Bottom, with Dome Seal Screw Cap, Sterile | Corning Inc. | Cat # 352096 |
| Falcon® 50 mL High Clarity PP Centrifuge Tube, Conical Bottom, Sterile | Corning Inc. | Cat # 352070 |
| Nalgene™ Rapid-Flow™ Sterile Single Use Vacuum Filter Units | Thermo Fisher Scientific (Nalgene) | Cat # 566-0020 |
| SPL 5 mL Serological Pipette | SPL Life Sciences | Cat # 91005 |
| SPL 10 mL Serological Pipette | SPL Life Sciences | Cat # 91010 |
| SPL 25 mL Serological Pipette | SPL Life Sciences | Cat # 91025 |
| SPL Cell Strainer, PP, 70 μm, Sterile, Individually Wrapped | SPL Life Sciences | Cat # 93070 |
| Syringe Filter 0.20 μm Blue Rim (50) | Sigma-Aldrich | Cat # 16534-K |
Materials and equipment
Coating of tissue culture plates
Making of 0.1% Gelatin Solution
-
•
Measure 5 g of gelatin powder from porcine skin.
-
•
Pour the gelatin powder into a bottle of 500 mL cell culture grade water.
-
•
Warm the mixture in a 37°C water bath until all the gelatin dissolves to obtain a pre-dissolved 1% gelatin solution.
-
•
Filter sterilize using a vacuum filtration system through a 0.2 μm filter unit.
Note: It takes 1–3 h on average for the gelatin to be completely dissolved.
-
•
Add 50 mL of sterile pre-dissolved 1% gelatin solution to 450 mL of cell culture grade water to obtain a 0.1% gelatin solution.
Note: 0.1% gelatin solution can be stored at 15°C–25°C for 6 months up to a year.
FBS-containing medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM High Glucose with GlutaMAX Supplement, Pyruvate | N/A | 445 mL |
| Heat-inactivated fetal bovine serum (FBS) | 10% | 50 mL |
| MEM-NEAA | 1% | 5 mL |
| Total | N/A | 500 mL |
Note: FBS-containing medium can be stored at 4°C for up to a month.
Preparation of Gelatin/FBS-containing Medium-Coated Plates for Feeder-free hPSC Culture
-
•
Add 2 mL of 0.1% gelatin per well of a 6-well plate or 7 mL per 100 mm tissue culture dish. Ensure that the gelatin solution covers the entire surface of the well/plate.
-
•
Leave the coated plates in a 37°C, 5% CO2 humidified incubator for 24 h.
Note: Coating with gelatin is recommended for a minimum of 15 min. In general, coating with gelatin is carried out for 24 h.
-
•
The next day, remove the gelatin solution from the coated wells/plates.
-
•
Add 2 mL of FBS-containing medium per well of a 6-well plate or 7 mL per 100 mm tissue culture plate.
-
•
Leave the coated plates in a 37°C, 5% CO2 humidified incubator for 48 h.
Note: Coat plates with FBS-containing medium for a minimum of 48 h up to a maximum of 2 weeks.
hPSC culture medium
For a feeder-free culture of hPSCs, use mTeSR™1 or TeSR™-E8™ medium from STEMCELL Technologies.
Alternatively, for feeder-based hPSC culture, follow the medium composition stated below:
hPSC culture medium for feeder-based culture
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F12 with GlutaMAX supplement | N/A | 395 mL |
| KOSR | 20% | 100 mL |
| MEM-NEAA | 1% | 5 mL of stock |
| FGF2 100 ng/μL | 10 ng/mL | 50 μL |
| Total | N/A | 500 mL |
Note: hPSC culture medium can be stored at 4°C for up to a month.
Dispase and collagenase solution
Dispase solution
Note: Dispase is available at a concentration of 1 U/mL in DMEM/F-12 from the manufacturer. It is recommended to store dispase at −20°C for short-term storage or −80°C for long-term storage.
Collagenase solution
-
•
Dissolve 5 g of Collagenase IV in 500 mL of DPBS to make a stock solution of 10 mg/mL.
-
•
Filter-sterilize the solution through a 0.2 μm filter unit.
-
•
Prepare aliquots of 5 mL in 50 mL cell culture tubes.
-
•
Store at −20°C for short-term storage or −80°C for long-term storage.
-
•
Before use, add 45 mL of DPBS to the 5 mL of Collagenase IV solution in the 50 mL tube to obtain a final volume of 50 mL at a concentration of 1 mg/mL.
Dispase/collagenase (1:1) solution
To make 50 mL of dispase/collagenase (1:1) solution, mix 25 mL of 1 U/mL dispase and 25 mL of 1 mg/mL collagenase solution together in a 50 mL tube.
Note: Dispase/collagenase solution can be stored at 4°C for up to a month.
Growth factors and small molecules
Here, the preparation of stock solutions of various growth factors and small molecules used during differentiation is described. For all growth factors and small molecules, centrifuge vials before opening and avoid vortexing of the contents. To avoid repeated freeze-thawing, it is recommended to aliquot all reconstituted growth factors and small molecules into multiple 1.7 mL micro-centrifuge tubes and to store the aliquots at −80°C for 6 months up to a year unless otherwise stated. Note that all growth factors and small molecules are to be prepared under sterile conditions.
Preparation of 7.5% bovine serum albumin (BSA) used for reconstituting growth factors
-
•
Dissolve 3.75 g of BSA in 50 mL of 1× cell culture grade DPBS in a 37°C water bath until it is fully dissolved.
-
•
Filter-sterilize the 7.5% BSA through a 0.2 μm syringe filter.
Preparation of Activin A at a stock concentration of 50 ng/μL
-
•
Mix 1000 μg of Activin A with 18 mL of 1× cell culture grade DPBS and 2 mL of 7.5% BSA to obtain 20 mL of Activin A at 50 ng/μL stock concentration.
Preparation of CHIR99021 at a stock concentration of 3 mM
-
•
Mix 10 mg of CHIR99021 with 7.163 mL of DMSO to obtain 7.163 mL of CHIR99021 at 3 mM stock concentration.
Preparation of LY294002 at a stock concentration of 10 mM
-
•
Mix 50 mg of LY294002 with 16.268 mL of DMSO to obtain 16.268 mL of LY294002 at 10 mM stock concentration.
Preparation of FGF2 at a stock concentration of 100 ng/μL
-
•
Mix 10 μg of FGF2 with 100 μL of cell culture grade water to obtain 100 μL of FGF2 at 100 ng/μL stock concentration.
Preparation of all-trans retinoic acid at a stock concentration of 100 mM
-
•
Mix 50 mg of all-trans retinoic acid with 1.66 mL of DMSO to obtain 1.66 mL of all-trans retinoic acid at 100 mM stock concentration.
Preparation of nicotinamide at a stock concentration of 1 M
-
•
Mix 12.21 g of nicotinamide with 100 mL of cell culture grade water to obtain 100 mL of nicotinamide at 1 M stock concentration.
-
•
Filter-sterilize 1 M Nicotinamide through a 0.2 μm syringe filter.
Preparation of DAPT at a stock concentration of 20 mM
-
•
Mix 50 mg of DAPT with 5.78 mL of DMSO to obtain 5.78 mL of DAPT at 20 mM stock concentration.
Preparation of BMP4 at a stock concentration of 50 ng/μL
-
•
Mix 25 μg of BMP4 with 450 μL of 1× cell culture grade DPBS and 50 μL of 7.5% BSA to obtain 500 μL of BMP4 at 50 ng/μL stock concentration.
Preparation of FGF10 at a stock concentration of 100 ng/μL
Note: FGF10 is soluble in 5 mM sodium phosphate. Before preparing 5 mM sodium phosphate, prepare 1 M Na2HPO4 and 1 M NaH2PO4 first.
-
•
To make 1 M Na2HPO4, dissolve 14.2 g of Na2HPO4 in 100 mL of cell culture grade water.
-
•
Next, to make 1 M NaH2PO4, dissolve 12 g of NaH2PO4 in 100 mL of cell culture grade water.
-
•
Then, mix 387 μL of 1 M Na2HPO4 and 113 μL of 1 M NaH2PO4 with 99.5 mL of cell culture grade water to obtain 100 mL of 5 mM sodium phosphate buffer.
-
•
Adjust the pH of the buffer to 7.4 by adding either phosphoric acid to decrease pH or sodium hydroxide to increase pH.
-
•
Filter sterilize using a vacuum filtration system through a 0.2 μm filter unit.
-
•
Finally, to make 100 ng/μL FGF10, dissolve 25 μg of FGF10 in 250 μL of filter-sterilized 5 mM sodium phosphate buffer.
Preparation of OSM at a stock concentration of 30 ng/μL
-
•
Mix 30 μg of OSM with 900 μL of 1× cell culture grade DPBS and 100 μL of 7.5% BSA to obtain 1 mL of OSM at 30 ng/μL stock concentration.
Preparation of HGF at a stock concentration of 50 ng/μL
-
•
Mix 25 μg of HGF with 450 μL of 1× cell culture grade DPBS and 50 μL of 7.5% BSA to obtain 500 μL of HGF at 50 ng/μL stock concentration.
Differentiation media used for differentiation
Basal differentiation media
Note: All basal differentiation media are to be prepared under sterile conditions.
RPMI/2% B27 differentiation medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | N/A | 485 mL |
| B-27 Supplement (50×), Minus Vitamin A | 2% | 10 mL |
| MEM-NEAA | 1% | 5 mL of stock |
| Total | N/A | 500 mL |
Note: RPMI/2% B27 differentiation medium can be stored at 4°C for up to a month.
Hepatocyte basal medium
Prepare the medium according to manufacturer’s specifications stated in the HCMTM Hepatocyte Culture Medium BulletKitTM from Lonza.
Note: Hepatocyte basal medium can be stored at 4°C for a duration as specified by the manufacturer.
Step-by-step method details
Pancreatic progenitor and hepatic differentiation
Methods to differentiate hPSCs to pancreatic progenitor and/or hepatic cells have been reported extensively in the past few years (Kroon, et al., 2008; Rashid, et al., 2010; Hannan, et al., 2013; Pagliuca, et al., 2014; Russ, et al., 2015; Teo, et al., 2015; Teo, et al., 2016). This protocol describes the detailed procedures for in vitro pancreatic progenitor and hepatic differentiation from hPSCs (Ng, et al., 2019). Note that the first 6 days (Days -2 to 4) of both differentiation protocols are identical, and the protocols deviate after the induction of the definitive endoderm (DE) (Tables 1 and 2). The specification of cells to the DE stage is crucial as these cells are responsible for the formation of organs such as the pancreas, liver, and intestines in vivo (Zorn & Wells, 2007). Once DE cells are generated, they are then directed towards either the pancreatic or hepatic lineage through the introduction of various differentiation media, growth factors, and small molecules at different time points. An overview of the specific growth factors and small molecules added at specific time points in this protocol is demonstrated in Tables 1 and 2.
Table 1.
Overview of the pancreatic progenitor (PP) differentiation timeline, the final concentrations of growth factors and small molecules used
| Day | Cell type | Final concentration of growth factors and small molecules |
|---|---|---|
| D-2 | hPSCs | N/A |
| D-1 | N/A | |
| D0 | hPSCs | Activin A 100 ng/mL + CHIR99021 3 μM + LY294002 10 μM |
| D1 | N/A | |
| D2 | N/A | |
| D3 | Definitive Endoderm | Activin A 50 ng/mL |
| D4 | N/A | |
| D5 | Anterior Definitive Endoderm | FGF2 50 ng/mL + RA 3 μM + Nicotinamide 10 mM |
| D6 | Primitive Gut Tube | N/A |
| D7 | FGF2 50 ng/mL + RA 3 μM + Nicotinamide 10 mM | |
| D8 | N/A | |
| D9 | N/A | |
| D10 | Posterior Foregut | FGF2 50 ng/mL + RA 3 μM + Nicotinamide 10 mM + DAPT 20 μM |
| D11 | N/A | |
| D12 | N/A | |
| D13 | FGF2 50 ng/mL + RA 3 μM + Nicotinamide 10 mM + DAPT 20 μM | |
| D14 | Pancreatic Progenitors | N/A |
Table 2.
Overview of the hepatic differentiation timeline, the final concentration of growth factors and small molecules used
| Day | Cell type | Final concentration of growth factors and small molecules |
|---|---|---|
| D-2 | hPSCs | N/A |
| D-1 | N/A | |
| D0 | hPSCs | Activin A 100 ng/mL + CHIR99021 3 μM + LY294002 10 μM |
| D1 | N/A | |
| D2 | N/A | |
| D3 | Definitive Endoderm |
Activin A 50 ng/mL |
| D4 | N/A | |
| D5 | Anterior Definitive Endoderm |
Activin A 50 ng/mL |
| D6 | BMP4 20 ng/mL + FGF10 10 ng/mL | |
| D7 | BMP4 20 ng/mL + FGF10 10 ng/mL | |
| D8 | N/A | |
| D9 | Hepatoblasts | BMP4 20 ng/mL + FGF10 10 ng/mL |
| D10 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D11 | N/A | |
| D12 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D13 | N/A | |
| D14 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D15 | N/A | |
| D16 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D17 | N/A | |
| D18 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D19 | N/A | |
| D20 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D21 | N/A | |
| D22 | OSM 30 ng/mL + HGF 50 ng/mL | |
| D23 | N/A | |
| D24 | Hepatocyte-like cells | OSM 30 ng/mL + HGF 50 ng/mL |
Setup on day 2
Harvesting hPSCs grown in feeder-free culture
Timing: 30 min
-
1.This step describes the setting up of hPSCs grown in feeder-free culture on a 100 mm culture plate two days prior to the start of differentiation. Here, ReLeSR™ is used to dissociate hPSCs into evenly-sized cell aggregates. It is recommended to split one confluent 100 mm plate of hPSCs into a full 6-well plate for differentiation.
-
a.Retrieve hPSCs cultured in a 100 mm tissue culture dish and observe hPSCs under the microscope.
 CRITICAL: It is recommended to harvest hPSCs when they are grown to 80%–90% confluency. It is also important to check that there are less than 10% differentiated cells on the plate before proceeding.
CRITICAL: It is recommended to harvest hPSCs when they are grown to 80%–90% confluency. It is also important to check that there are less than 10% differentiated cells on the plate before proceeding. -
b.Aspirate the hPSC culture medium and wash cells once with 3 mL of DPBS. Aspirate the DPBS.
-
c.Add 1 mL of ReLeSR™ to the hPSCs, ensure that the ReLeSR™ solution covers the surface of the entire plate.
-
d.After 45 s of incubation at 15°C–25°C, aspirate the ReLeSR™.
-
e.Immediately place the cell culture dish into a 37°C humidified incubator for an additional 2.5 min.
-
f.After 2.5 min, gently wash the plate once with 1 mL of DPBS by dispensing the DPBS slowly at the side of the plate without dislodging the cells. Aspirate the DPBS.
-
g.Add 3 mL of TeSR™-E8™ or mTeSR™1 medium to the plate, and firmly tap the side of the plate to dislodge the cells. Transfer detached cell aggregates to a 15 ml tube.
-
h.Rinse the remaining cells on the plate with another 3 mL of TeSR™-E8™ or mTeSR™1 medium and transfer the cells to the same 15 mL tube.
-
i.Top up the cell mixture with 3 mL of TeSR™-E8™ or mTeSR™1 medium and, pipette up and down once using a 5 mL serological pipette.
-
j.Seed 1.5 mL of cells in each well of a coated 6-well plate.Note: 6-well plates are coated as per instructions stated in the “Preparation of Gelatin/FBS-containing Medium-Coated Plates for Feeder-free hPSC Culture” subsection; Materials and Equipment section in this paper.
 CRITICAL: Ensure that the cells are evenly distributed in the wells by rocking the plate back and forth.
CRITICAL: Ensure that the cells are evenly distributed in the wells by rocking the plate back and forth. -
k.Incubate cells in a 37°C, 5% CO2 humidified incubator for two days to allow cells to attach and recover.
 CRITICAL: During the two days of recovery, to minimize disturbance to the cells, it is not recommended to change the mTeSR™1 medium.
CRITICAL: During the two days of recovery, to minimize disturbance to the cells, it is not recommended to change the mTeSR™1 medium.
-
a.
Harvesting hPSCs grown in feeder-based culture
Timing: 30 min
-
2.This step describes the setting up of hPSCs grown in feeder-based culture two days prior to the start of differentiation. Here, hPSCs are harvested by enzymatic means using dispase/collagenase mixed in a 1:1 ratio, followed by gentle mechanical scraping to generate dissociated cell aggregates. It is recommended to split confluent hPSCs grown on a 100 mm plate into a full 6-well plate for differentiation.
-
a.Warm dispase/collagenase (1:1) solution in a 37°C water bath.
-
b.Retrieve hPSCs cultured in a 100 mm tissue culture dish and observe hPSCs under the microscope.
 CRITICAL: It is recommended to harvest hPSCs when they are grown to 80%–90% confluency. It is also important to check that there are less than 10% differentiated cells on the plate before proceeding.
CRITICAL: It is recommended to harvest hPSCs when they are grown to 80%–90% confluency. It is also important to check that there are less than 10% differentiated cells on the plate before proceeding. -
c.Aspirate the hPSC culture medium and wash cells once with 3 mL of DPBS. Aspirate the DPBS.
-
d.Add 3 mL of warmed dispase/collagenase (1:1) solution to the hPSCs, ensuring that the solution covers the surface of the entire plate.
-
e.Incubate the cells for 3 min at 37°C. This step facilitates the subsequent dislodging of hPSCs but not the feeder cells.
-
f.Remove the dispase/collagenase solution and wash the cells gently with 3 mL DPBS. Aspirate the DPBS.
-
g.Add 3 mL of hPSC culture medium to the cells.
-
h.Use a 5 mL serological pipette to scrape cells off the plate mechanically, from left to right, up to down, in a horizontal and vertical straight-line fashion, until the entire area of the plate has been scraped mechanically. Collect dissociated cell clusters into a 15 mL tube. The action of cell-scraping in two perpendicular orientations allows the hPSCs to be cut into small evenly-sized pieces.Note: The aim of this step is to cut hPSC colonies into small evenly-sized clusters while minimizing damage to the cells.
-
i.Rinse the remaining cells on the plate with another 3 mL of hPSC medium and transfer the cells to the same 15 mL tube.
-
j.Pass the harvested cell clusters through an inverted 70 μm cell strainer to deplete dislodged feeder cells.
-
k.Insert the cell strainer into a 50 mL tube and rinse the strainer with 9 mL of fresh mTeSR™1 medium to collect the trapped hPSC clusters.
-
l.Gently resuspend the cell mixture using a serological pipette and seed 1.5 mL of cells into each well of a coated 6-well plate.Note: 6-well plates are coated as per instructions stated in the “Preparation of Gelatin/FBS-containing Medium-Coated Plates for Feeder-free hPSC Culture” subsection; Materials and Equipment section in this paper.
 CRITICAL: Ensure that the cells are evenly distributed in the wells by rocking the plate back and forth.
CRITICAL: Ensure that the cells are evenly distributed in the wells by rocking the plate back and forth. -
m.Incubate cells in a 37°C, 5% CO2 humidified incubator for two days to allow cells to attach and recover.
 CRITICAL: During the two days of recovery, to minimize disturbance to the cells, it is not recommended to change the culture medium.
CRITICAL: During the two days of recovery, to minimize disturbance to the cells, it is not recommended to change the culture medium.
-
a.
Pancreatic progenitor (PP) differentiation after day 2 setup
Timing: 10 min per day
-
3.This step describes the pancreatic progenitor differentiation process after day -2 setup.Note: The cell types generated from day -2 to day 14, the specific growth factors and small molecules to be added on the stipulated days are summarized in Table 1.Days 0 to 2 of the PP differentiation process aim to differentiate hPSCs into DE cells.
-
a.On day 0, aspirate the hPSC culture medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 2 μL of 50 ng/μL Activin A, 1 μL of 3 mM CHIR99021 and 1 μL of 10 mM LY294002 per well of a 6-well plate.Days 3 to 5 of the PP differentiation process aim to differentiate DE cells into anterior definitive endoderm (ADE) cells.
-
b.On day 3, aspirate old differentiation medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 1 μL of 50 ng/μL Activin A per well of a 6-well plate.Days 5 to 9 of the PP differentiation process aim to differentiate ADE cells into primitive gut tube cells within one day, and then to posterior foregut within the next four days.
-
c.On day 5, aspirate old differentiation medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 0.5 μL of 100 ng/μL FGF2, 0.3 μL of 10 mM RA and 10 μL of 1 M Nicotinamide per well of a 6-well plate.
-
d.Repeat the previous step on day 7.Days 10 to 13 of the PP differentiation process aim to differentiate posterior foregut cells into PP cells (Figure 1).
-
e.On day 10, aspirate old differentiation medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 0.5 μL of 100 ng/μL FGF2, 0.3 μL of 10 mM RA, 10 μL of 1 M Nicotinamide and 1 μL of 20 mM DAPT per well of a 6-well plate.
-
f.Repeat the previous step on day 13.
-
g.On day 14, PP cells are ready for harvesting.
-
a.
Figure 1.
Representative images from the PP differentiation time course
Scale bar, 200 μm.
Hepatic differentiation after day 2 setup
Timing: 10 min per day
-
4.This step describes the hepatic differentiation process after day -2 setup.Note: The cell types generated from day -2 to day 25, the specific growth factors and small molecules to be added on stipulated days are summarized in Table 2.Days 0 to 2 of the hepatic differentiation process aim to differentiate the hPSCs into DE cells.
-
a.On day 0, aspirate the hPSC culture medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 2 μL of 50 ng/μL Activin A, 1 μL of 3 mM CHIR99021 and 1 μL of 10 mM LY294002 per well of a 6-well plate.Days 3 to 5 of the hepatic differentiation process aim to differentiate DE cells into ADE cells.
-
b.On day 3, aspirate old differentiation medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 1 μL of 50 ng/μL Activin A per well of a 6-well plate.
-
c.Repeat the previous step on day 5.Days 6 to 9 of the hepatic differentiation process aim to differentiate ADE cells into hepatoblasts.
-
d.On day 6, aspirate old differentiation medium from the wells and replace with 1 mL of RPMI/2% B27 differentiation medium with 0.4 μL of 50 ng/μL BMP4 and 0.1 μL of 100 ng/μL FGF10 per well of a 6-well plate.
-
e.Repeat previous step on days 7 and 9.Lastly, days 10 to 24 of the hepatic differentiation process aim to differentiate hepatoblasts into hepatocyte-like cells (Figure 2).
-
f.On day 10, aspirate old differentiation medium from the wells and replace with 1 mL of hepatocyte basal medium with 1 μL of 30 ng/μL OSM and 1 μL of 50 ng/μL HGF per well of a 6-well plate.
-
g.Repeat the previous step every alternate day from day 12 onwards.
-
h.On day 25, hepatocyte-like cells are ready for harvesting.
-
a.
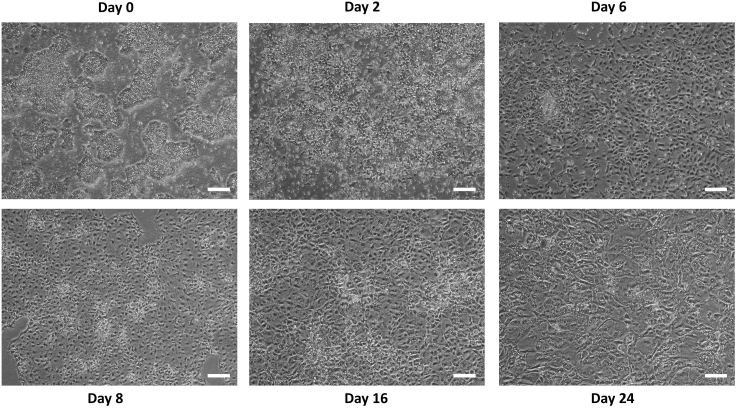
Figure 2.
Representative images from the hepatic differentiation time course
Scale bar, 200 μm.
Gene regulatory assay
The pancreatic and hepatic cells generated from the directed differentiation of hPSCs are useful as cell models for the study of gene regulation during development. This protocol describes the use of these hPSC-derived cells in a gene regulation assay, specifically a luciferase reporter-based transcriptional assay. The cells at any stage of differentiation can be harvested for the subsequent assays, depending on the developmental time point of interest.
Cell harvesting and FuGENE® 6 transfection
Timing: 3 days
-
5.
Collect hPSC-derived cells at an appropriate time point. For example, if gene regulation at the PP stage (Day 14) is of interest, cells should be harvested on Day 11.
-
6.
Aspirate differentiation medium from the wells and wash cells once with DPBS. Aspirate the DPBS.
-
7.
Add 0.5 mL of TrypLE to each well of a 6-well plate and incubate at 37°C for 3–4 min.
Note: The length of incubation time with TrypLE should be optimized for the specific cell type to ensure that cells are not subjected to over dissociation.
-
8.
Add 1.5 mL of basal differentiation medium (without any growth factors) to each well and collect all the cells into a 50 mL tube.
-
9.
Rinse each well with additional 1 mL of basal differentiation medium and transfer the remaining cells to the same 50 mL tube.
-
10.
Centrifuge cells at 280 g for 3 min.
-
11.
Remove the supernatant and resuspend cells in an appropriate volume of freshly-prepared differentiation medium with growth factors and small molecules that are required for the relevant time point.
-
12.
Count the cells and seed 300,000 cells in each well of a 24-well plate (250–500 μL volume).
Note: Seed the cells at an appropriate density to ensure that cells are of 50%–60% confluency at the point of transfection with FuGENE® 6.
-
13.
Culture cells in the incubator for 24 h to allow for cell attachment and recovery.
-
14.
The next day, before transfection of cells, replace the media with 250 μL per well of freshly-prepared differentiation medium with growth factors and small molecules that are required for the relevant time points.
-
15.
Prepare DNA and transfection mix as shown in Table 3 below:
-
16.
For each well of a 24-well plate, make up the transfection mix by adding 4 μl of FuGENE® 6 Transfection reagent to 46 μl of Opti-MEM Reduced Serum Medium, according to the manufacturer’s instructions (based on a DNA: FuGENE® 6 ratio of 1:4).
-
17.
Prepare sufficient volume in a master mix for the transfection of triplicate wells per condition as shown in Table 3.
-
18.
Add the DNA mix to the FuGENE® 6 transfection mix, flick to mix and let the reaction stand for 15 min at 15°C–25°C.
-
19.
Add the DNA-transfection mix dropwise to each well of cells.
-
20.
Harvest the cells for the luciferase assay 48 h (or 72 h) after transfection.
Note: It is not necessary to change media after transfection, except for the replacement of differentiation medium during the relevant time points.
Table 3.
Components and quantity of DNA mix
| Component | Quantity per well | Quantity per triplicate wells (3×) |
|---|---|---|
| Luciferase reporter plasmid (such as pGL4.10 vector containing the promoter of interest) | 490 ng | 1.47 μg |
| Overexpression plasmid (such as pCDH vector containing the protein-coding sequence of interest) | 500 ng | 1.5 μg |
| pRL-TK Renilla Vector (internal control reporter vector) | 10 ng | 30 ng |
| Total | 1 μg | 3 μg |
Luciferase assay
Timing: 1 h
-
21.
Dilute an appropriate amount of 5× Passive Lysis Buffer (PLB) with four volumes of distilled water, depending on the number of samples to be collected for the luciferase assay. For each well of a 24-well plate, prepare 100 μl of 1× PLB. Therefore, for a full 24-well plate of cells, 2.4 mL of 1× PLB is required.
-
22.
Reconstitute LAR II and the Stop & Glo reagents according to the manufacturer’s instructions. For example, to prepare 10 mL of fresh 1× Stop and Glo solution, add 200 μl of 50× Stop & Glo substrate to 9.8 mL of Luciferase Assay Buffer before the assay.
-
23.
Aspirate differentiation medium from wells.
-
24.
Wash cells once in 250 μL of DPBS per well. Aspirate the DPBS.
-
25.
Add 100 μl of 1× PLB per well of a 24-well plate.
-
26.
Gently rock the plate for 15–20 min at 15°C–25°C to facilitate passive cell lysis.
Note: The plate of samples may be frozen and stored at −20°C for a few days until the luciferase assay is to be performed. For longer-term storage, store plate at −80°C.
-
27.
Centrifuge plate at 195 g for 3 min at 4°C to collect the cell debris at the bottom of the plate.
-
28.
To perform the luciferase assay, transfer 20 μl of the clear supernatant (do not collect any visible cell debris) into each well of a flat-bottomed, white 96-well assay plate. For technical duplicates, transfer 20 μl of the clear supernatant twice into two separate wells.
-
29.
Dispense 100 μl of LAR II to each well using a multi-channel pipette or an appropriate automated injector system.
-
30.
Measure firefly luciferase activity immediately using a suitable luminescence microplate reader.
-
31.
Dispense 100 μl of 1× Stop & Glo solution to each well using a multi-channel pipette or an appropriate automated injector system.
-
32.
Measure Renilla luciferase activity immediately using a suitable luminescence microplate reader.
-
33.
To analyze data, subtract background luminescence detected in a non-transfected control sample from all firefly or Renilla luciferase activity results. Next, divide luciferase reporter activity by the respective Renilla luciferase activity for each sample to obtain normalized luciferase activity data.
CRITICAL: Both LAR II and Stop & Glo reagents are light-sensitive. Therefore, ensure that the reagents and plates containing samples are protected from light.
Expected outcomes
Pancreatic progenitor differentiation
Figure 1 shows the expected differentiation of hPSCs into PP cells over 14 days utilizing a previously-established protocol (Teo, et al., 2015; Teo, et al., 2016). hPSCs that were dissociated into evenly-sized clumps on day -2 were used as starting cell colonies for pancreatic differentiation on day 0. At day 5, cells have migrated outwards and formed cobblestone-shaped cells that are characteristic of DE cells. By day 10, pancreatic cell fate specification resulted in uniform, distinct cell clusters that continue to proliferate and give rise to PP cells at day 14.
Hepatic differentiation
Figure 2 shows the expected differentiation of hPSCs into hepatic cells over 24 days utilizing a previously-established protocol with slight modifications (Hannan, et al., 2013). hPSCs that were dissociated into evenly-sized clumps on day -2 were used as starting cell colonies for hepatic differentiation on day 0. At day 2, hPSCs are migrating outwards and are starting to display a morphology resembling DE cells. It is typical to observe high amounts of cell death at this point. By day 6, uniform, cobblestone-shaped cells are observed upon successful DE formation. At day 8, DE cells are undergoing hepatic cell fate commitment under the influence of BMP4 and FGF10, and are differentiated into hepatoblasts. At day 16, hepatic progenitor cells are undergoing hepatocyte maturation under the influence of OSM and HGF. On day 24, hepatocyte-like cells with multi-nuclei and distinct canaliculated borders are formed.
Molecular analyses such as RT-qPCR for target gene expression, western blot or flow cytometry for protein expression, and gene regulatory assays such as those described in our protocol may be performed to study specific stages of both pancreatic and hepatic differentiation processes.
Luciferase assay
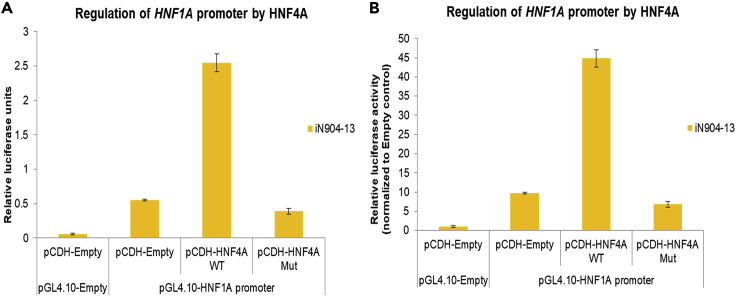
Figure 3 illustrates the use of luciferase assay to study the regulation of the HNF1A promoter by transcription factor HNF4A in day 8 hepatoblasts differentiated from a healthy donor iPSC line iN904-13 (Ng, et al., 2019). In the presence of only the HNF1A promoter-containing luciferase reporter plasmid, a 10-fold upregulation of luciferase activity is seen due to endogenous activation of the HNF1A promoter. In the presence of overexpressed HNF4A wild type (WT), an approximate 45-fold upregulation of luciferase activity is seen due to strong activation of the HNF1A promoter by HNF4A protein. On the other hand, the loss-of-function HNF4A mutant was unable to upregulate luciferase activity. These results highlight the potential application of this protocol to investigate disease-relevant genetic variants and/or disease-relevant hPSC-derived cells.
Figure 3.
Luciferase assay showing HNF1A promoter regulation by transcription factor HNF4A
Luciferase assay results showing the regulation of the HNF1A promoter by HNF4A wild type (WT) and a loss-of-function mutant (Mut) in hiPSC-derived hepatoblasts, plotted based on (A) relative luciferase units or (B) relative luciferase units normalized to the Empty vector control. Data shown is obtained from one experiment with each condition in triplicate samples.
Limitations
This protocol describes the generation of hPSC-derived cells for downstream gene regulatory assays. As hPSC differentiation is inherently a heterogeneous process, the use of hPSC-derived cells for molecular experiments may result in high variability especially across different cell lines and across multiple independent differentiation experiments. The materials and methods described in Part I of this protocol are limited to the generation of hPSC-derived PPs and hepatic cells in a monolayer culture. To determine the quality of cells obtained from each round of differentiation, additional cell characterization will need to be performed, such as the evaluation of cell stage-specific markers through gene expression or flow cytometry analysis. Furthermore, if functionally mature cells, suspension cells or other cell types are required, the use of alternative protocols will be needed, together with subsequent optimization of cell dissociation and seeding for the molecular assay.
Another limitation relates to the potentially low transfection efficiency of certain cell types of interest, that may result in luciferase activity readings that are too low to be meaningfully interpreted.
Troubleshooting
Problem 1
At step 1, the quality of cells after the setup (hPSCs from feeder-free or feeder-based cultures) plays a critical role in influencing the outcome of differentiation. For feeder-free cultures, inappropriate use of ReLeSR™ or other dissociation reagents that have not been optimized for the particular cell line may result in poor cell viability or a high level of differentiation. For feeder-based cultures, ensure that the feeders are depleted and are not carried over to the setup. Mechanical scraping may result in poor cell viability, a high level of differentiation, or high variability in the size of the seeded cell clusters. Poor quality in the starting hPSC material will compromise the success of the differentiation.
Potential solution
Dissociation based on ReLeSR™ or reagents need to be optimized for the particular cell line. When performing mechanical scraping, scrape in parallel lines and in a clean swift motion to minimize physical cell damage. Minimize the duration of cell processing during setup to maintain the quality of undifferentiated hPSCs. If hPSCs are not confluent enough or display more than 10% differentiated areas, discard the cells and restart the experiment.
Problem 2
At step 1, poor attachment of hPSCs and hPSC-derived cells or even cell death may occur after harvesting and seeding them in coated plates prior to differentiation and transfection.
Potential solution
Optimize the type, concentration and duration of coating media that is most suited for the cell type of interest. Add 10 μM of Rho-kinase inhibitor Y-27632 to the hPSC culture medium after harvesting and seeding the cells to increase cell survival rate and adhesion. After incubation of cells in 37°C, 5% CO2 humidified incubator for 24 h, aspirate hPSC medium with Y-27632 and replace with fresh hPSC culture medium without Y-27632 to minimize unintended effects of Y-27632 on differentiation.
Problem 3
At step 2, high cell death may be observed on day 3 after DE generation in both PP and hepatic differentiation, resulting in significant loss of cells.
Potential solution
One potential cause for high cell death is the presence of the PI3K inhibitor LY294002. Decrease and re-optimize the concentration of LY294002 added at the start of differentiation to reduce cell death. Provide fresh change of differentiation medium midway to promptly remove dead cells. Cells at higher confluency or more wells of cells during the start of differentiation may also be prepared, to make up for the projected cell loss.
Problem 4
At step 13 during the preparation of the DNA mix for transfection, the volume of DNA added to each well may vary due to differences in concentrations of plasmids or manual pipetting inconsistencies. This may give rise to variability in results for both the firefly luciferase reporter plasmid and the Renilla luciferase plasmid control.
Potential solution
Plasmids in use can be made up to identical concentrations to ensure the same volume of DNA is used across all conditions. Plasmids should also be stored in small aliquots to minimize freeze-thawing. As far as possible, master mixes for the DNA and transfection reagents should be made up for all conditions requiring the same components.
Problem 5
At steps 23 and 24, cells might not be completely or efficiently lysed by 1× PLB during the stipulated 15–20 min of incubation.
Potential solution
After step 24, scrape the cells from the 24-well plate using a 200 μl pipette tip. Freeze the plate of samples at −20°C or −80°C for at least one hour before performing the luciferase assay.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Adrian Kee Keong Teo (ateo@imcb.a-star.edu.sg; drainteo@gmail.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate and/or analyze any new datasets.
Acknowledgments
A.K.K.T. is supported by the Institute of Molecular and Cell Biology (IMCB), A∗STAR, NMRC OFYIRG16may014, A∗STAR ETPL Gap Funding ETPL/18-GAP005-R20H, NHG-KTPH SIG/14033, Lee Foundation Grant SHTX/LFG/002/2018, NMRC OF-LCG/DYNAMO, Skin Innovation Grant SIG18011, FY2019 SingHealth Duke-NUS Surgery Academic Clinical Programme Research Support Programme Grant, Precision Medicine and Personalised Therapeutics Joint Research Grant 2019, Industry Alignment Fund – Industry Collaboration Project (IAF-ICP) I1901E0049, and the 2nd A∗STAR-AMED Joint Grant Call 192B9002. N.H.J.N. is supported by NMRC OFYIRG18may040 and A∗STAR 1st CDA.
Author contributions
Conceptualization, N.H.J.N. and A.K.K.T; investigation, L.S.T., R.S.E.C., and N.H.J.N.; writing – original draft, L.S.T., R.S.E.C., and N.H.J.N.; writing – review & editing, L.S.T., R.S.E.C., N.H.J.N., and A.K.K.T.; funding acquisition, A.K.K.T.
Declaration of interests
N.H.J.N. and A.K.K.T. are co-founders of BetaLife Pte Ltd.
References
- Hannan N.R.F., Segeritz C.-P., Touboul T., Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Ng N.H.J., Jasmen J.B., Lim C.S., Lau H.H., Krishnan V.G., Kadiwala J., Kulkarni R.N., Ræder H., Vallier L., Hoon S., Teo A.K.K. HNF4A haploinsufficiency in MODY1 abrogates liver and pancreas differentiation from patient-derived induced pluripotent stem cells. iScience. 2019;16:192–205. doi: 10.1016/j.isci.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid S.T., Corbineau S., Hannan N., Marciniak S.J., Miranda E., Alexander G., Huang-Doran I., Griffin J., Ahrlund-Richter L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 2010;120(9):3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ H.A., Parent A.V., Ringler J.J., Hennings T.G., Nair G.G., Shveygert M., Guo T., Puri S., Haataja L., Cirulli V. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K.K., Lau H.H., Valdez I.A., Dirice E., Tjora E., Raeder H., Kulkarni R.N. Early developmental perturbations in a human stem cell model of MODY5/HNF1B pancreatic hypoplasia. Stem Cell Rep. 2016;6:357–367. doi: 10.1016/j.stemcr.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K.K., Tsuneyoshi N., Hoon S., Tan E.K., Stanton L.W., Wright C.V., Dunn N.R. PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Rep. 2015;4:578–590. doi: 10.1016/j.stemcr.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn A.M., Wells J.M. Molecular basis of vertebrate endoderm development. Int. Rev. Cytol. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate and/or analyze any new datasets.