Summary
Hypoxia is known to stimulate mitochondrial reactive oxygen species (mROS) in cells. Here, we present a detailed protocol to detect mROS using MitoSOX staining in live cells under normoxia and hypoxia. Flow cytometry allows sensitive and reliable quantification of mROS by FlowJo software. We optimized several aspects of the procedure including hypoxic treatment, working concentrations of the staining buffer, and quantitative analyses. Here, we use HepG2 cells, but the protocol can be applied to other cell lines.
For complete details on the use and execution of this protocol, please refer to Yang et al. (2020).
Subject areas: Cell Biology, Flow Cytometry/Mass Cytometry, Cell-based Assays, Molecular/Chemical Probes
Graphical abstract
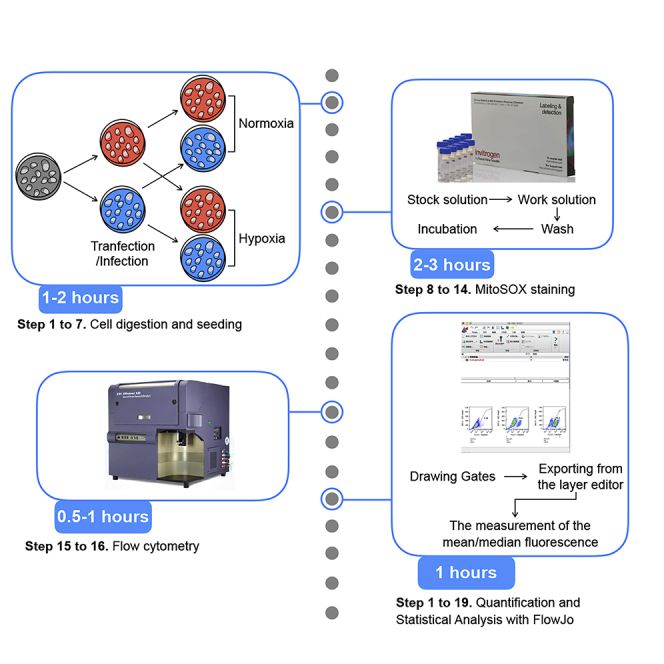
Highlights
-
•
Detailed protocol to detect mitochondrial ROS in cell lines
-
•
MitoSOX staining in normoxic and hypoxic live cells
-
•
Flow cytometry allows quantification and statistical analysis
Hypoxia is known to stimulate mitochondrial reactive oxygen species (mROS) in cells. Here, we present a detailed protocol to detect mROS using MitoSOX staining in live cells under normoxia and hypoxia. Flow cytometry allows sensitive and reliable quantification of mROS by FlowJo software. We optimized several aspects of the procedure including hypoxic treatment, working concentrations of the staining buffer, and quantitative analyses. Here, we use HepG2 cells, but the protocol can be applied to other cell lines.
Before you begin
Timing: 30 min/step
-
1.
Provide a sufficient supply of carbon dioxide to maintain concentrations inside the incubator at 5%. Prepare sufficient nitrogen gas to achieve hypoxic condition (1% O2, 5% CO2, 94% N2).
-
2.
Make sure that there is sufficient DMEM medium and PBS buffer for cell culture of HepG2 and dissolving reagents.
-
3.
All medium or buffers should be put into 37°C incubator for preheating at least half an hour in advance. Temperature change may stimulate cells, leading to incorrect results of mROS quantification.
-
4.
At least 24 h in advance, the DMEM medium and PBS buffer should be placed in the hypoxic incubator, so that the oxygen concentration in the medium and buffers reaches hypoxic levels (hypoxia-pretreated medium).
-
5.
Calculate the amount of MitoSOX required for the experiment to make sure that there is sufficient MitoSOX to quantify mROS production. For example, for each 6 well plate prepared for testing, 30 nmol MitoSOX (6 mL 5 μM) is needed.
CRITICAL: All MitoSOX reagent should be stored at −20°C and be protected from light. The working solution should be stored in 4°C.
CRITICAL: MitoSOX is easily oxidized, so avoid contact with air.
CRITICAL: MitoSOX is a derivative of ethidium bromide and has toxicity. Operate with care and wear lab clothes and disposable gloves.
-
6.
HepG2 cells should be passaged at log phase, during which the cells are actively dividing, healthy and ≥ 90% viable. As passaging cells too late can lead to overcrowding, apoptosis and senescence, influencing the result of quantification of mROS.
-
7.
HepG2 cells will show different cell proliferation kinetics during the log phase and it is therefore the doubling time of the cells should be determined before the experiment.
CRITICAL: Ensure that all flow cytometry equipment is set up in advance, since MitoSOX is a live-cell dye and mROS is a stress response mediator, the stained cells should immediately be examined by flow cytometry.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| MitoSOX | Thermo Fisher | Cat #M36008 |
| Trypsin-EDTA (0.25%) | Thermo Fisher | Cat #25200056 |
| Opti-MEMTM | Thermo Fisher | Cat #31985070 |
| LipofectamineTM 3000 | Thermo Fisher | Cat #L3000015 |
| Hanks’s balanced salt solution (with Ca2+ & Mg2+) | Beyotime | Cat #C0219 |
| 1 × PBS buffer | Sangon Biotech | Cat #B540626 |
| HepG2 | ATCC cell lines | Cat #HB-8065 |
| DMEM basic (1×) | Gibco | Cat #C11995500BT |
| DMSO | Sigma | Cat #41639-100ML |
| Software and algorithms | ||
| FlowJoTM 10 | BD Biosciences | https://www.flowjo.com |
| GraphPad Prism | GraphPad | https://www.graphpad.com |
| Other | ||
| Countstar® Bio Tech system | Countstar | https://aberinstruments.com/ |
| BD LSRFortessa™ Flow Cytometer | BD Biosciences | https://www.bdbiosciences.com/ |
| IV5100FL inverted fluorescence microscope | Olympus Imaging | https://olympus-imaging.cn |
| Tri-gas CO2 incubator (150i) | Thermo Fisher | https://www.thermofisher.com/ |
Note: Hypoxic incubator is absolutely required. MitoSOX is a critical reagent.
Step-by-step method details
Cell digestion and seeding—day 1
Timing: 1–2 h
Timing: 10 min for steps 1 and 2
Timing: 10 min for step 3
Timing: 10 min for step 4
Timing: 20 min for step 5
Timing: 30 min for step 6
-
1.
Passage cells at log phase during which the cells are actively dividing, healthy and ≥ 90% viable. If the cells are adherent cells, culture the cells to achieve a dense monolayer.
-
2.
Discard the culture medium, gently shake the cell bottle, and discard the dead cells or aging cells with poor adherence.
-
3.
Add preheated trypsin-EDTA (0.25%) to the cells and hold at almost 18°C–25°C until cells become shrunk and round (after almost 3 to 5 min).
-
4.
Neutralize trypsin-EDTA by adding the preheated complete medium until almost half of the cells become suspended when tapping the edge of the culture bottle with your hand. The volume of preheated complete medium was twice that of trypsin-EDTA.
-
5.
Count cells using hemocytometer or other automated cell counters and cell analyzers such as the CountStar system.
-
6.Seed the cells into 6-well plate at a density of 3 to 7 × 105 cells/well, depending on the doubling time. Add medium to a final volume of 2 mL/well. Incubate the cells in a normoxic CO2 incubator for 12 h.
 CRITICAL: This protocol certainly does not only apply to 6-well plate. If the cells are relatively slow to grow, or if the users could not afford 6-well plate cell counts, it is suggested to choose other plates, but these plates should be used in proportion to the number of cells. To our knowledge, the number of cells required for each sample point to achieve meaningful measurement should be no less than 7 × 104 in 24-well plate.Alternatives: After the cells attached, plasmid or siRNA transfection may be performed if the expression of a gene needs to be modulatedNote: Lipo3000 is not the only transfection agents but our preferred agents, other transfection methods can also be used.
CRITICAL: This protocol certainly does not only apply to 6-well plate. If the cells are relatively slow to grow, or if the users could not afford 6-well plate cell counts, it is suggested to choose other plates, but these plates should be used in proportion to the number of cells. To our knowledge, the number of cells required for each sample point to achieve meaningful measurement should be no less than 7 × 104 in 24-well plate.Alternatives: After the cells attached, plasmid or siRNA transfection may be performed if the expression of a gene needs to be modulatedNote: Lipo3000 is not the only transfection agents but our preferred agents, other transfection methods can also be used.-
a.Change the cell culture media to 1750 μL fresh media with FBS per well.
-
b.Dilute 2.5 μg plasmid in 125 μL Opti-MEMTM media within a 1.5 mL tube (Tube 1).
-
c.Add 5 μL P3000TM from LipofectamineTM 3000 Kit into Tube 1.
-
d.Dilute 3.75 μL LipofectamineTM 3000 in 125 μL Opti-MEMTM media in another 1.5 mL tube (Tube 2).
-
e.Incubate at almost 18°C–25°C for 10 min.
-
f.Add the plasmid-p3000TM mixture in Tube 1 to Tube 2.
-
g.Add plasmid-Lipofectamine 3000-p3000TM mixture into the well.
 CRITICAL: Viral infections including human immunodeficiency virus result in increased ROS production (Schwarz, 1996), which is essential to viral replication (Cheng et al., 2014). We observed this phenomenon in cells infected with control lentivirus that is still unstable to quantify mROS. Under these conditions, cells under normoxic conditions exhibit an abnormally high level of mROS, just like hypoxic cells. We therefore recommend using cells that have been stably passaged for at least three generations.
CRITICAL: Viral infections including human immunodeficiency virus result in increased ROS production (Schwarz, 1996), which is essential to viral replication (Cheng et al., 2014). We observed this phenomenon in cells infected with control lentivirus that is still unstable to quantify mROS. Under these conditions, cells under normoxic conditions exhibit an abnormally high level of mROS, just like hypoxic cells. We therefore recommend using cells that have been stably passaged for at least three generations.
-
a.
-
7.
Divide the cells into the hypoxic group (1% O2, 5% CO2, 94% N2) and the normoxic group (21% O2, 5% CO2, 74% N2) and place them in corresponding incubator for the time as required by the experiment. The photos of incubator settings can be seen as Figure 1. The approximate incubation time to induce mROS under the hypoxic condition could be detected between 10 min to 72 h.
CRITICAL: During hypoxic treatment, the frequency and duration of door openings should be limited as much as possible. Otherwise, intermittent hypoxia may cause the level of mROS to be unstable and uneven, thus leading to general underestimation of mROS. If multiple door openings are necessary, consider using a hypoxia incubator chamber that is self-contained and sealed inside a normal CO2 incubator. This equipment has advantages for allowing the cells to return to a stable hypoxic environment as soon as possible if the door is opened.
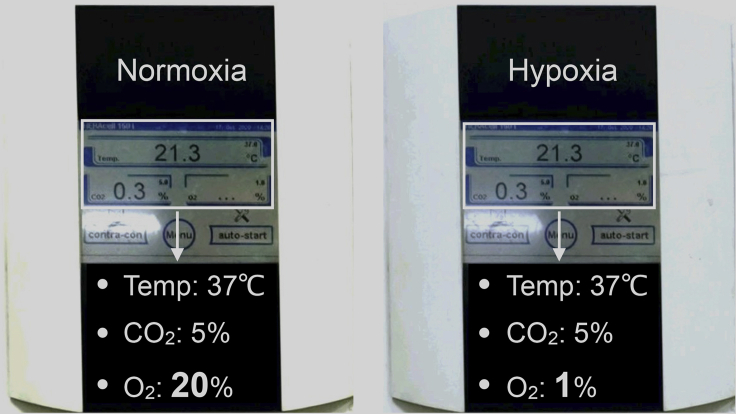
Figure 1.
Photos of normoxic settings and hypoxic settings
Normoxic settings in incubator are as follows: Temp: 37°C, CO2: 5%, and O2: 21% (Left); Hypoxic settings in incubator are as follows: Temp: 37°C, CO2: 5%, and O2: 1% (Right).
MitoSOX staining and flow cytometry—day 4
Timing: 2–3 h
Timing: 10 min for step 8
Timing: 10 min for step 9
Timing: 20 min for step 10
Timing: 20 min for step 11
Timing: 10 min for step 12
Timing: 20 min for step 13
Timing: 20 min for step 14
Timing: 30 min for steps 15 and 16
-
8.
Observe the (transfected) cells under a (fluorescence) microscope to estimate confluence (and the transfected efficiency).
-
9.
Prepare 5 mM MitoSOX reagent stock solution. Each MitoSOX vial should be placed at almost 18°C–25°C before opening. After centrifugation at 1000 g of the vial for 1 min, dissolve the contents (50 μg) of one vial of MitoSOX in 13 μL of dimethylsulfoxide (DMSO) to make a 5 mM MitoSOX reagent stock solution.
Note: We recommend that DMSO should use molecular biology grade.
-
10.
Collect normoxic cells with 1 mL preheated medium without serum or preheated Hanks’ balanced salt solution (HBSS) into flow cytometry tubes. Collect hypoxic cells with 1 mL hypoxia-pretreated medium (See the 4th step in before you begin) into flow cytometry tubes. If the cells are adherent cells, digest cells with preheated trypsin-EDTA for 3–5 min and washed cells 3 times with preheated medium/HBSS or hypoxia-pretreated medium during which cells should centrifugated each time at 400 g for 3 min at almost 18°C–25°C .
-
11.
Count cells using hemocytometer or other automated cell counters and cell analyzers, and adjust cells to a concentration of 1–5 × 106 cells per 1 mL in the corresponding medium.
CRITICAL: The cell concentration should be in this range, and specific value of concentration can be determined according to the recommendation from the users’ flow cytometry.
-
12.
Add the 1 μL of MitoSOX stock solution to each cell suspension to make a 5 μM MitoSOX reagent working solution and shake gently.
CRITICAL: The concentration of the MitoSOX reagent working solution should not exceed 5 μM, as concentrations exceeding 5 μM can produce cytotoxic effects, including altered mitochondrial morphology and redistribution of fluorescence to nuclei and the cytosol. It is important to note that too many cells lead to a deficiency of the dye relative to each cell and thus low fluorescence. We recommend that cells should be diluted at a density of 5 × 106 cells/mL before staining.
CRITICAL: The remaining working solution can be stored at 4 °C in a refrigerator, protected from light for almost one week.
CRITICAL: Prepare one tube of cells as a negative control by adding the same amount of DMSO.
-
13.
Incubate the cells for 20 min in a normoxic incubator or hypoxic incubator, protected from light.
CRITICAL: The incubation of cells with MitoSOX should last 15 min to 30 min at 37°C. The methods also required that the buffer is preheated to 37°C, otherwise the staining will not be sufficient.
-
14.
Wash cells gently three times with preheated medium/HBSS or hypoxia-pretreated medium as soon as possible. During washing, cells should centrifugated each time at 400 g for 3 min at almost 18°C –25°C . Prepare cells for flow cytometry.
-
15.
The expression of MitoSOX is quantified in the PE channel by flow cytometry.
CRITICAL: Compensation can be calculated using single-stained cells if the cells are labeled with FITC or other fluorescence excited at 510 nm and detected at 580 nm.
CRITICAL: If the cells are detected under sterile conditions, cells can also be sorted for cell culture.
CRITICAL: An optimal sequence for sample measurement would be from low ROS to high, such as from normoxic samples to hypoxic samples. The recommended positive control samples can be the cells that are treated with lasting hypoxia, virus like lentivirus, or ROS-activating inhibitors like retenone. The recommended negative control samples can be the cells under normal culturing (21% O2, 5% CO2).
-
16.
Perform data analysis using FlowJo software.
Expected outcomes
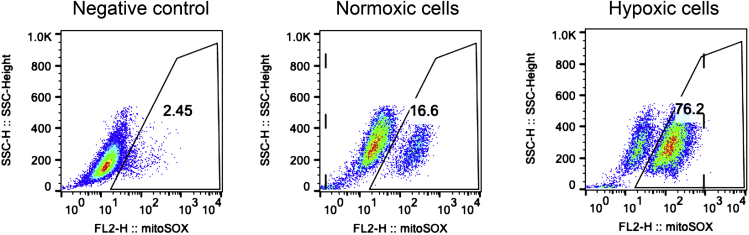
Successful MitoSOX staining not only shows the cells that is stained with DMSO (without MitoSOX) have the least population (<3%) as the negative control, but also demonstrates a clear difference between normoxic and hypoxic cells (Figure 2).
Note: We found that the range of the percentage of mitoSOX-positive cells under normoxia is approximately 10% to 30% (not including activated T cells), and under hypoxia the range is approximately 70% to 100%.
Figure 2.
Flow cytometry analysis of the negative control, normoxic, and hypoxic cells
HepG2 cells were cultured in normoxic (Middle) and hypoxic (right) incubator for 3 days. And then the normoxic and hypoxic cells were digested, washed with pre-heated buffer and hypoxia-pretreated medium, respectively. They were suspended by each medium at a concentration of 5 μM MitoSOX and incubated for 20 min, while part of normoxic HepG2 cells was separated to be treated with DMSO for 20 min (left, Negative control). Post washing, all the cells were performed with flow cytometry analysis immediately.
Quantification and statistical analysis
The mean/median fluorescence can be analyzed to quantify MitoSOX (mROS) using FlowJo analysis software .
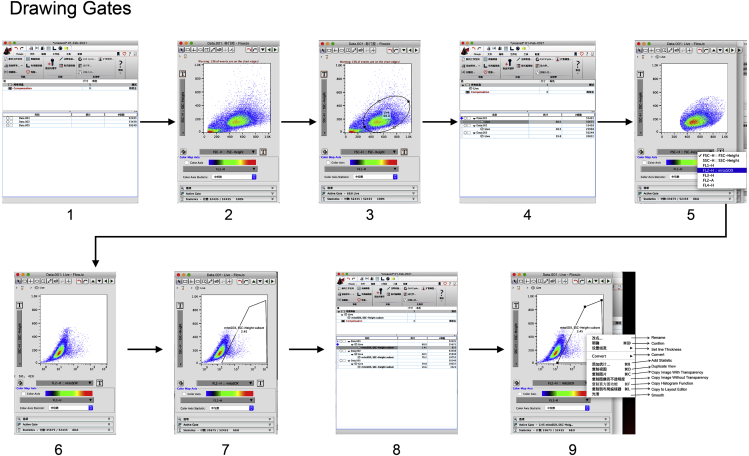
Drawing gates (Figure 3)
-
1.
Start Flowjo software and drag the data such as fcs format to the analysis column.
-
2.
Double click icon of any data in the data list, the pop-up window showing FSC as horizontal axis and SSC as vertical axis will appear.
-
3.
Draw a loose gate that excludes debris found at the very bottom left corner of the FSC/SSC plots.
-
4.
Apply this gating to any sample by drawing icon of the “live” in the data list to icon of “all samples”.
-
5.
Double click icon of the “live” of the sample that was not stained with mitoSOX as negative control in the data list, and change the horizontal axis into “FL2-H:: mitoSOX”.
-
6.
The pop-up window showing “FL2-H:: mitoSOX” as horizontal axis will appear.
-
7.
Draw a gate that excludes mitoSOX-negative cells and includes mitoSOX-positive cells.
-
8.
Apply this gating to any sample by drawing icon of the “mitoSOX” in the data list to icon of “live” under “all samples”.
-
9.
Right click the pop-up window and select “Copy to layout editor”.
Figure 3.
Draw gates by using a negative control
Drag all data to FlowJo (step 1), open the negative control in sample list (step 2), draw a loose gate that excludes debris (step 3), draw a gate that excludes mitoSOX-negative cells and includes mitoSOX-positive cells (step 7), and apply the gating to all samples (step 8).
Exporting from the layout editor (Figure 4)
-
10.
The auto pop-up window will show the Layout Editor to create graphical reports.
-
11.
Double click the graph with the Layout Editor, customize the look of the overlay graph that best highlights your data.
-
12.
The interface of the overlay graph will display the image post customization. Click “Batching” button to create a table report.
-
13.
The in layout batch will display the same graph for sets of samples.
-
14.
Change the order or delete the overlay graph as you wish.
-
15.
Click “Export Image” to save the image.
Figure 4.
Export graphics from the layout editor
Customize the look of any overlay graph (step 11), create a table report (step 12), change the order or delete the overlay graph (step 13 or 14), and save the image by clicking “Export Image” (step 15).
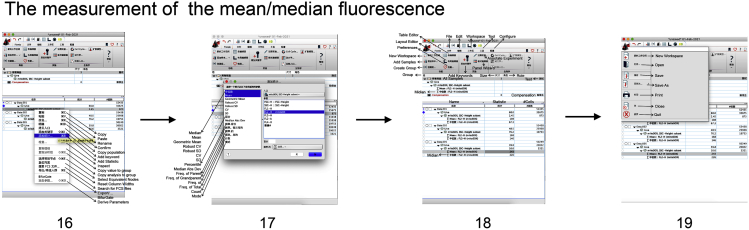
The measurement of the mean/median fluorescence (Figure 5)
-
16.
Back to the pop-up window in the step 9, right click the icon of “mitoSOX” of any sample in the data list.
-
17.
In the new pop-up window showing “Add statistics”, click successively “mean” and “median”, “FL2-H:: mitoSOX”, and “Add”.
-
18.
Apply the mean/median statistics to all sample by drawing icon of the “∑ Mean: FL2-H (mitoSOX)” and “∑ Median: FL2-H (mitoSOX)”in the data list to icon of “mitoSOX, SSC-Height subset” under “ all samples”.
-
19.
Save all procedure throughout mitoSOX analysis as .wsp format.
Figure 5.
Measure the mean/median fluorescence
Select the icon of “mitoSOX” of any sample (step 16), click successively “mean” and “median”, “FL2-H:: mitoSOX”, and “Add” (step 17), apply the mean/median statistics to all sample, record all the mean/median values, and save all procedure (steps 18 and 19).
Limitations
Reactive oxygen species (ROS) are mainly produced by mitochondria, including free radicals such as superoxide anion (O2⋅−), hydroxyl radical (OH⋅−), and hydrogen peroxide (H2O2) (De Biasi et al., 2016). Among ROS, the predominant mROS is O2⋅−, which causes a cascade of other ROS by superoxide dismutase (Idelchik et al., 2017). Our measurement relies on the concept that the MitoSOX, which is oxidized by O2⋅− generated in the mitochondria of live cells, elicits a fluorescence response proportional to the superoxide concentration (Kauffman et al., 2016; Robinson et al., 2008). Therefore, the total mROS are not precisely measured; the detected value can reflect mROS generation but will be lower than the actual value.
As a cationic molecule, MitoSOX uptake into mitochondria may contribute to direct mROS generation by depolarizing the mitochondrial membrane potential (ΔΨm), so that the quantified values post-analysis are higher than the real values (Smith et al., 2011). However, a study showed that mROS generated in this case was miniscule compared with the mROS produced by hypoxic stress (Ghanian et al., 2018). MitoSOX uptake into mitochondria may contribute to direct mROS generation by depolarizing the mitochondrial membrane potential (ΔΨm), so that the quantified values post-analysis are higher than the real values (Smith et al., 2011). However, a study showed that mROS generated in this case was miniscule compared with the mROS produced by hypoxic stress (Ghanian et al., 2018). It is worth noting that some probes may reflect extracellular signals that mask mitochondria signals, since they can be readily oxidized by multiple extracellular and intracellular oxidation processes (Kalyanaraman, 2020). The detection of mitochondrial superoxide in cells remains experimentally challenging and the interpretation contentious.
MitoSOX uptake increases 10-fold for every 60 mV increase in ΔΨm (Kalbacova et al., 2003; Porteous et al., 2010). A decrease in ΔΨm upon using uncoupler and ETC inhibitors could impede the uptake of MitoSOX (Kalbacova et al., 2003). Therefore, the fluorescence intensities presented could be an underestimate for the levels of the ROS in the inhibited and uncoupled ETC. If the mROS levels are too high and therefore decrease mitochondrial membrane potential, mitoSOX may underestimate ROS levels. It is worth noting that cell digestion might affect mitoSOX uptake through ΔΨm for two reasons. First, digestion is to break down the adhesive proteins on cell surface into simpler forms, so it is an exothermic reaction and power is generated in the digestion phase. This thermal energy might act as a stress perceived by cell membrane and subsequently as a signal transduce into mitochondria altering ΔΨm, leading to changes of probe uptake. Second, digestion is to make cells from cell connection and cell adhesion to single cell suspensions, so the digested cells lost their connection between them and the connection between cell and cell matrix. The missing connections might be sensed by mitochondria that always are sensitive to external stimuli, further leading to alteration of probe uptake by the change of ΔΨm . Therefore, digestion may contribute to ΔΨm, leading to alteration of mitoSOX uptake. In the process of digestion, 3–5 min is appropriate in order to avoid over digestion.
There are some alternative methods to detect mitochondrial redox state. For example, ratiometric biosensor mito-roGFP can be used(Vevea et al., 2013) . Sara De Biasi et al has described a new protocol that can take advantage of multi-laser (De Biasi et al., 2016), polychromatic flow cytometry to analyze simultaneously hydrogen peroxide and mitochondrial superoxide in PBMC cells, which displayed a higher loss of mitochondrial membrane potential and derived from patients with Down Syndrome. Kristine M. Robinson also has reported that exact quantitation of mROS is fraught with difficulties (Robinson et al., 2006), and therefore HPLC methods quantifying the relative amount of hydroxylated and nonhydroxylated products should be used in conjunction with fluorescence experiments when possible.
Troubleshooting
Problem 1
No difference of mROS levels between normoxic and hypoxic groups.
Potential solution
Mitochondrial ROS increased rapidly under hypoxia, and the difference could be detected within 10 min (Yang et al., 2020). However, if there is no difference between the two groups, the two groups should be compared with the negative control group that was not subjected with mitoSOX staining. Specifically, in the first case, the level of mROS in the two groups was similar to the negative control group, which suggests that the two groups of cells did not complete staining, and the solution should refer to the problem 2. In the second case, the level of mROS in the two groups was between the negative control and the positive control, but there was no difference. It is possible that the problem that no detectable difference in mROS between normoxic and hypoxic groups is due to the time of hypoxia treatment or the conditions of mitoSOX staining. For example, it should be considered whether the hypoxic incubator need to be repaired, such as oxygen sensor, or whether different concentrations of mitoSOX were used when the normoxic and hypoxic cells were staining. In the third case, the mROS of the two groups was similar to that of the positive control group, refer to problem 3.
Problem 2
NO mitoSOX-positive cells in both normoxic and hypoxic groups.
The possible reasons could be incorrect usage of mitoSOX. For example, the concentration of mitoSOX is too low to stain most cells due to the wrong configuration of working solution; the incubation conditions are not consistent; during incubation the working medium is cold, almost 4°C; or the normoxic group have hereditary mitochondrial damage, such as Rho0 cells (Chandel and Schumacker, 1999), which lack critical respiratory chain catalytic subunits.
Problem 3
Both normoxic and hypoxic groups showing high level of mROS.
This indicates that mROS in normoxic group increased abnormally, and there is a false positive signal caused by non-hypoxic factors. For example, when normoxic cells are infected with lentivirus, they will exhibit an abnormally high level of mROS, just like hypoxic cells; or some small molecule inhibitors, such as rotenone (Won et al., 2015), antimycin A (Choi and Lee, 2011), can also induce rapid elevation of mROS.
Problem 4
Intro-Normoxia groups show large variability.
Potential solution
Mitochondrial ROS varies depending on cell types. We once detected mROS under normoxia in five different cell types (data not shown), the results showed that the order of mROS from high to low is as follows: activated mouse T cells, Hep-3B, HepG2, Huh7, and 293T. As far as one cell type is concerned, if there is large variability in intra-group, it should be considered that the process during cell culture appears contamination, because bacterial contamination in an individual sample is likely to affect the level of mROS production, contributing to large variation within intra-group. Further, once a part of cells in normoxia has just been transfected with plasmids or infected with virus like lentivirus, which led to elevation of mROS. We recommend detecting mROS when the cells have been stably passaged for at least three generations.
Problem 5
Intra-hypoxia group show large variability.
Potential solution
Hypoxia is known to stimulate elevated mROS, but the potential to elevate varies depending on cell types. That is to say, it may be normal for different cell types to show large variability in intra-hypoxia group post treatment with hypoxia. In terms of single cell type, large variability might be caused by the failure of hypoxic treatment of some cells or the failure of mROS elevation in intra-hypoxia group. First, it might be caused by the different treatment conditions of some cells in the group, for example, part of intra-hypoxia cells is treated in a damaged hypoxic incubator, inner which the oxygen sensitive electrode needs to be maintained or replaced. We have had this situation before. Second, some of cells in the intra-hypoxia group are treated with different small molecule inhibitors, which will be able to inhibit mROS generation, for example, some of NADPH oxidase inhibitors (Tang et al., 2018) and NecroX-5 (Kim et al., 2014). Third, some of cells in the intra-hypoxia group are treated with ethidium bromide to induce loss of mitochondrial DNA, which will lead to non-responsiveness to hypoxia of mROS. In general, large variability in intra-hypoxia group should be considered from the three aspects above.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hanshuo Yang (yhansh@scu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any unique datasets or code.
Acknowledgments
This project was supported by grants from the Natural Science Funding of China (81171956, 81772605, and 81572402) and the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC18008). We thank Gabrielle White Wolf, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author contributions
Conceptualization, H.Y. and Y.Y.; investigation, Y.Y., G.Z., T.Y., J.G., and L.X.; writing – original draft, Y.Y., G.Z., and H.Y.; writing – review & editing, H.Y. and G.Z.; funding acquisition, H.Y.; supervision, H.Y.
Declaration of interests
The authors declare no competing interests.
References
- Chandel N.S., Schumacker P.T. Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Lett. 1999;454:173–176. doi: 10.1016/s0014-5793(99)00783-8. [DOI] [PubMed] [Google Scholar]
- Cheng M.L., Weng S.F., Kuo C.H., Ho H.Y. Enterovirus 71 induces mitochondrial reactive oxygen species generation that is required for efficient replication. PLoS One. 2014;9:e113234. doi: 10.1371/journal.pone.0113234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.M., Lee Y.S. Mitochondrial defects and cytotoxicity by antimycin A on cultured osteoblastic MC3T3-E1 cells. Food Chem. Toxicol. 2011;49:2459–2463. doi: 10.1016/j.fct.2011.04.037. [DOI] [PubMed] [Google Scholar]
- De Biasi S., Gibellini L., Bianchini E., Nasi M., Pinti M., Salvioli S., Cossarizza A. Quantification of mitochondrial reactive oxygen species in living cells by using multi-laser polychromatic flow cytometry. Cytometry A. 2016;89:1106–1110. doi: 10.1002/cyto.a.22936. [DOI] [PubMed] [Google Scholar]
- Ghanian Z., Konduri G.G., Audi S.H., Camara A.K.S., Ranji M. Quantitative optical measurement of mitochondrial superoxide dynamics in pulmonary artery endothelial cells. J. Innov. Opt. Health Sci. 2018;11:1750018. doi: 10.1142/S1793545817500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idelchik M., Begley U., Begley T.J., Melendez J.A. Mitochondrial ROS control of cancer. Semin. Cancer Biol. 2017;47:57–66. doi: 10.1016/j.semcancer.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbacova M., Vrbacky M., Drahota Z., Melkova Z. Comparison of the effect of mitochondrial inhibitors on mitochondrial membrane potential in two different cell lines using flow cytometry and spectrofluorometry. Cytometry A. 2003;52:110–116. doi: 10.1002/cyto.a.10031. [DOI] [PubMed] [Google Scholar]
- Kalyanaraman B. Pitfalls of reactive oxygen species (ROS) measurements by fluorescent probes and mitochondrial superoxide determination using mitoSOX. In: Berliner L.J., Parinandi N.L., editors. Measuring Oxidants and Oxidative Stress in Biological Systems. Springer; 2020. pp. 7–9. [PubMed] [Google Scholar]
- Kauffman M.E., Kauffman M.K., Traore K., Zhu H., Trush M.A., Jia Z., Li Y.R. MitoSOX-based flow cytometry for detecting mitochondrial ROS. React. Oxyg. Species (Apex) 2016;2:361–370. doi: 10.20455/ros.2016.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.R., Kim D.I., Kim S.H., Lee H., Lee K.S., Cho S.H., Lee Y.C. NLRP3 inflammasome activation by mitochondrial ROS in bronchial epithelial cells is required for allergic inflammation. Cell Death Dis. 2014;5:e1498. doi: 10.1038/cddis.2014.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous C.M., Logan A., Evans C., Ledgerwood E.C., Menon D.K., Aigbirhio F., Smith R.A., Murphy M.P. Rapid uptake of lipophilic triphenylphosphonium cations by mitochondria in vivo following intravenous injection: implications for mitochondria-specific therapies and probes. Biochim. Biophys. Acta. 2010;1800:1009–1017. doi: 10.1016/j.bbagen.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Robinson K.M., Janes M.S., Beckman J.S. The selective detection of mitochondrial superoxide by live cell imaging. Nat. Protoc. 2008;3:941–947. doi: 10.1038/nprot.2008.56. [DOI] [PubMed] [Google Scholar]
- Robinson K.M., Janes M.S., Pehar M., Monette J.S., Ross M.F., Hagen T.M., Murphy M.P., Beckman J.S. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl. Acad. Sci. U S A. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K.B. Oxidative stress during viral infection: a review. Free Radic. Biol. Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- Smith R.A., Hartley R.C., Murphy M.P. Mitochondria-targeted small molecule therapeutics and probes. Antioxid. Redox Signal. 2011;15:3021–3038. doi: 10.1089/ars.2011.3969. [DOI] [PubMed] [Google Scholar]
- Tang P., Dang H., Huang J., Xu T., Yuan P., Hu J., Sheng J.F. NADPH oxidase NOX4 is a glycolytic regulator through mROS-HIF1alpha axis in thyroid carcinomas. Sci. Rep. 2018;8:15897. doi: 10.1038/s41598-018-34154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea J.D., Alessi Wolken D.M., Swayne T.C., White A.B., Pon L.A. Ratiometric biosensors that measure mitochondrial redox state and ATP in living yeast cells. J. Vis. Exp. 2013;77:50633. doi: 10.3791/50633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J.H., Park S., Hong S., Son S., Yu J.W. Rotenone-induced impairment of mitochondrial electron transport chain confers a selective priming signal for NLRP3 inflammasome activation. J. Biol. Chem. 2015;290:27425–27437. doi: 10.1074/jbc.M115.667063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang G., Guo F., Li Q., Luo H., Shu Y., Shen Y., Gan J., Xu L., Yang H. Mitochondrial UQCC3 modulates hypoxia adaptation by orchestrating OXPHOS and glycolysis in hepatocellular carcinoma. Cell Rep. 2020;33:108340. doi: 10.1016/j.celrep.2020.108340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.