Abstract
Notch receptors maintain skeletal homeostasis. NOTCH1 and 2 have been studied for their effects on bone remodeling. Although NOTCH3 plays a significant role in vascular physiology, knowledge about its function in other cellular environments, including bone, is limited. The present study was conducted to establish the function of NOTCH3 in skeletal cells using models of Notch3 misexpression. Microcomputed tomography demonstrated that Notch3 null mice did not have appreciable bone phenotypes. To study the effects of the NOTCH3 activation in the osteoblast lineage, BGLAP-Cre or Dmp1-Cre transgenics were crossed with RosaNotch3 mice, where the NOTCH3 intracellular domain is expressed following the removal of a loxP-flanked STOP cassette. Microcomputed tomography demonstrated that BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice of both sexes exhibited an increase in trabecular bone and in connectivity, with a decrease in cortical bone and increased cortical porosity. Histological analysis revealed a decrease in osteoclast number and bone resorption in trabecular bone and an increase in osteoclast number and void or pore area in cortical bone of RosaNotch3 mice. Bone formation was either decreased or could not be determined in Cre;RosaNotch3 mice. NOTCH3 activation in osteoblasts inhibited Alpl (alkaline phosphatase) and Bglap (osteocalcin) and induced Tnfsf11 (RANKL) and Tnfrsf11b (osteoprotegerin) mRNA, possibly explaining the trabecular bone phenotype. However, NOTCH3 induced Tnfsf11 and suppressed Tnfrsf11b in osteocytes, possibly explaining the cortical porosity. In conclusion, basal NOTCH3 is dispensable for skeletal homeostasis, whereas activation of NOTCH3 in osteoblasts/osteocytes inhibits osteoclastogenesis and bone resorption in cancellous bone but increases intracortical remodeling and causes cortical porosity.
Keywords: osteocyte, osteoblast, Notch receptor, bone, osteoclast, cortical porosity
Abbreviations: α-MEM, α-minimum essential medium; BGLAP, bone gamma-carboxyglutamate protein; BMM, bone marrow–derived macrophage; Dmp1, dentin matrix protein 1; FBS, fetal bovine serum; HA, hydroxyapatite; Hes, Hairy Enhancer of Split; Hey, Hes-related with YRPW motif; μCT, microcomputed tomography; MMRC, Mutant Mouse Resource and Research Center; NRR, negative regulatory region; NICD, Notch intracellular domain; qRT-PCR, quantitative RT-PCR; RANKL, receptor activator of nuclear factor-κB; TRAP, tartrate-resistant acid phosphatase
Notch receptors (NOTCH1 to 4) determine cell fate and function in a variety of cell lineages including those present in bone tissue (1, 2, 3, 4). Consequently, Notch receptors have emerged as major contributors to the regulation of skeletal development and homeostasis. Interactions of specific regions of the extracellular domain of Notch with ligands of the Jagged and Delta-like families lead to the exposure of the negative regulatory region (NRR) and its cleavage by ADAM metalloproteases and the γ-secretase complex (5, 6). The proteolytic cleavage results in the release of the Notch intracellular domain (NICD) and Notch activation (7). The NICD translocates to the nucleus where it forms a complex with recombination signal-binding protein for Ig of κ (RBPJκ) and mastermind-like (MAML) (8, 9, 10). Canonical Notch activation results in the transcription of members of the Hairy Enhancer of Split (Hes) and Hes-related with YRPW motif (Hey) family of transcription factors (11, 12). Although Notch receptors are activated following interactions with their ligands, a degree of basal activation is possible, particularly in the case of NOTCH3 (13).
Although the four Notch receptors share structural and functional properties, each receptor retains its own identity. This is in part because of their preferential expression in certain cellular environments, interactions with specific Notch ligands, and various degrees of basal activation (3). Notch1, Notch2, and Notch3 and low levels of Notch4 mRNA are expressed by skeletal cells (3, 14, 15). Notch1, Notch2, and Notch3 are expressed in cells of the osteoblast lineage, but only Notch1 and Notch2 are present in cells of the myeloid/osteoclast lineage, where they have direct effects on osteoclast differentiation (3, 16, 17). NOTCH1 inhibits osteoclast differentiation, whereas NOTCH2 induces osteoclastogenesis by direct actions on the osteoclast lineage and by inducing receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL) in the osteoblast lineage (14, 16, 17, 18). Notch3 is not expressed in the myeloid lineage, but it has the potential to induce osteoclastogenesis by enhancing the production of RANKL in osteoblasts/osteocytes (16). Notch receptors inhibit osteoblast differentiation (3). Although NOTCH1 and NOTCH2 have been studied extensively for their effects on bone remodeling, less is known regarding the role of NOTCH3 in skeletal physiology.
Recently, we created a mouse model harboring a Notch3 mutation designed to reproduce the functional outcome of lateral meningocele syndrome and termed Notch3em1Ecan (synonym Notch3tm1.1Ecan) (16, 19). Notch3em1Ecan mice exhibit a modest NOTCH3 gain of function, osteopenia, and high bone remodeling indicating that NOTCH3 plays a role in skeletal metabolism. However, to establish the function of NOTCH3 in bone, it is important to define the consequences of wildtype NOTCH3 activation on the skeleton and to determine the skeletal phenotype of mice harboring a Notch3 inactivation.
In the present work, we attempted to define the role of NOTCH3 by establishing the consequences of its misexpression on bone remodeling. For this purpose, we obtained Notch3 null and RosaNotch3 mice, where a loxP-flanked STOP cassette was cloned into the Rosa26 locus upstream of sequences coding for the Notch3 NICD (20). Because Notch3 is expressed by osteoblasts/osteocytes, RosaNotch3 mice were crossed with transgenics expressing Cre under the control of the bone gamma-carboxyglutamate protein (BGLAP, osteocalcin) or the dentin matrix protein 1 (Dmp1) promoter (21, 22). The phenotype of mice misexpressing NOTCH3 was established by microcomputed tomography (μCT) and histomorphometry and possible mechanisms involved were explored.
Results
General characteristics of Notch3 null mice
To study the consequences of the inactivation of Notch3 in the skeleton, we obtained global Notch3 null mice from the Mutant Mouse Resource and Research Centers (MMRC) and examined their phenotype following intercrosses of heterozygous Notch3−/+ mice. Although the intent was to compare Notch3−/− with littermate sex-matched wildtype controls, the yield of the expected genotype (25%) and sex-matching made littermate comparisons not always possible, so that it was necessary to compare mice from different but related litters. Notch3−/− mice had a healthy appearance, and their weight and femoral length were comparable with those of control mice (Fig. 1). Notch3 mRNA levels in tibiae from Notch3−/− mice were undetectable.
Figure 1.
Weight, femoral length, and identification of Notch3 null alleles by genotyping of tail DNA and mRNA levels in bone extracts.A, body weight and (B) femoral length of 1- and 4-month-old Notch3−/− (closed circles) and sex-matched controls (open circles). Bars represent means and ranges SD; n = 7–9 control and n = 5–10 Notch3−/− at 1 month of age, and n = 6–10 control and n = 6 Notch3−/− at 4 months of age. C, tail DNA was obtained from Notch3−/− and control mice for genotyping, and (D) total RNA was obtained from tibiae from Notch3−/− (closed circles) and sex- and age-matched control (open circles) mice. ∗Significantly different between Notch3−/− and controls by unpaired t test.
Skeletal microarchitecture of Notch3 null mice
Femoral microarchitecture, determined by μCT, of 1- and 4-month-old male Notch3−/− mice revealed no differences in trabecular or cortical bone when compared with control sex-matched mice of the same age (Table 1, Fig. 2). μCT of 1-month-old female mice revealed no alterations in either cancellous or cortical bone when compared with control mice (Table 1). Femoral bone microarchitecture of 4-month-old female Notch3−/− mice demonstrated a modest decrease in trabecular thickness and density of material in cancellous bone and a mild decrease in cortical bone thickness and area.
Table 1.
Femoral microarchitecture assessed by microcomputed tomography of 1- and 4-month-old Notch3 null male and female mice and sex- and age-matched controls
| Males | 1 Month |
4 Months |
||
|---|---|---|---|---|
| Control | Notch3−/− | Control | Notch3−/− | |
| Distal femur trabecular bone | n = 6 | n = 5 | n = 9 | n = 5 |
| Bone volume/total volume (%) | 7.1 ± 1.5 | 6.9 ± 0.7 | 9.1 ± 2.8 | 8.9 ± 0.8 |
| Trabecular separation (μm) | 250 ± 21 | 258 ± 28 | 222 ± 11 | 216 ± 16 |
| Trabecular number (1/mm) | 4.1 ± 0.3 | 3.9 ± 0.3 | 4.5 ± 0.2 | 4.6 ± 0.3 |
| Trabecular thickness (μm) | 31 ± 2 | 31 ± 1 | 39 ± 6 | 37 ± 2 |
| Connectivity density (1/mm3) | 208 ± 27 | 209 ± 17 | 159 ± 20 | 184 ± 33 |
| Structure model index | 2.4 ± 0.1 | 2.4 ± 0.1 | 2.2 ± 0.4 | 2.2 ± 0.1 |
| Density of material (mg HA/cm3) | 759 ± 26 | 747 ± 32 | 871 ± 13 | 856 ± 22 |
| Femoral midshaft cortical bone | n = 7 | n = 5 | n = 10 | n = 6 |
| Bone volume/total volume (%) | 97.0 ± 1.7 | 95.8 ± 4.6 | 99.9 ± 0.1 | 99.9 ± 0.0 |
| Porosity (%) | 3.0 ± 4.7 | 4.2 ± 4.6 | 0.1 ± 0.16 | 0.1 ± 0.0 |
| Cortical thickness (μm) | 77 ± 3 | 71 ± 5 | 171 ± 10 | 167 ± 7 |
| Total area (mm2) | 1.4 ± 0.1 | 1.3 ± 0.2 | 1.7 ± 0.2 | 1.6 ± 0.1 |
| Bone area (mm2) | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.7 ± 0.1 | 0.7 ± 0.1 |
| Periosteal perimeter (mm) | 4.1 ± 0.1 | 4.0 ± 0.3 | 4.6 ± 0.2 | 4.5 ± 0.1 |
| Endocortical perimeter (mm) | 3.6 ± 0.1 | 3.5 ± 0.2 | 3.5 ± 0.2 | 3.4 ± 0.1 |
| Density of material (mg HA/cm3) | 927 ± 32 | 905 ± 58 | 1188 ± 20 | 1179 ± 40 |
| p Moment of inertia (mm4) | 0.126 ± 0.02 | 0.103 ± 0.02 | 0.321 ± 0.07 | 0.288 ± 0.04 |
| Females | 1 Month |
4 Months |
||
|---|---|---|---|---|
| Control | Notch3−/− | Control | Notch3−/− | |
| Distal femur trabecular bone | n = 9 | n = 9 | n = 7 | n = 5 |
| Bone volume/total volume (%) | 6.4 ± 0.9 | 6.8 ± 1.2 | 4.1 ± 0.6 | 3.3 ± 1.0 |
| Trabecular separation (μm) | 282 ± 26 | 268 ± 27 | 311 ± 34 | 332 ± 40 |
| Trabecular number (1/mm) | 3.6 ± 0.3 | 3.8 ± 0.4 | 3.3 ± 0.4 | 3.0 ± 0.4 |
| Trabecular thickness (μm) | 32 ± 1 | 32 ± 2 | 35 ± 3 | 31 ± 1a |
| Connectivity density (1/mm3) | 179 ± 24 | 193 ± 44 | 81 ± 19 | 76 ± 22 |
| Structure model index | 2.3 ± 0.1 | 2.3 ± 0.2 | 2.8 ± 0.1 | 2.8 ± 0.2 |
| Density of material (mg HA/cm3) | 767 ± 14 | 767 ± 22 | 877 ± 14 | 850 ± 26a |
| Femoral midshaft cortical bone | n = 9 | n = 10 | n = 7 | n = 5 |
| Bone volume/total volume (%) | 98.2 ± 0.8 | 97.9 ± 1.6 | 99.9 ± 0 | 99.8 ± 0.1 |
| Porosity (%) | 1.8 ± 0.8 | 2.1 ± 1.6 | 0.1 ± 0 | 0.2 ± 0.1 |
| Cortical thickness (μm) | 80 ± 6 | 77 ± 6 | 180 ± 6 | 170 ± 5a |
| Total area (mm2) | 1.4 ± 0.2 | 1.3 ± 0.1 | 1.6 ± 0.2 | 1.6 ± 0.1 |
| Bone area (mm2) | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.74 ± 0.05 | 0.69 ± 0.02a |
| Periosteal perimeter (mm) | 4.2 ± 0.3 | 4.0 ± 0.1 | 4.5 ± 0.2 | 4.4 ± 0.1 |
| Endocortical perimeter (mm) | 3.6 ± 0.4 | 3.5 ± 0.1 | 3.4 ± 0.3 | 3.3 ± 0.2 |
| Density of material (mg HA/cm3) | 932 ± 23 | 930 ± 38 | 1206 ± 25 | 1171 ± 30 |
| p Moment of inertia (mm4) | 0.119 ± 0.02 | 0.111 ± 0.02 | 0.303 ± 0.06 | 0.270 ± 0.03 |
Microcomputed tomography was performed in distal femurs for trabecular bone and midshaft for cortical bone from 1- and 4-month-old Notch3 null mice and age- and sex-matched controls. Values means ± SD.
Significantly different between control and Notch3−/−, p < 0.05 by unpaired t test.
Figure 2.
Representative microcomputed tomography of femurs from (A) 1- and (B) 4-month-old male and female control and Notch3−/−mice. A sagittal cut is shown for cancellous bone, and a cross-sectional cut at mid-diaphysis is shown for cortical bone. Bar in left lower corner scale 1 mm.
Bone marrow stromal cell cultures from Notch3 null mice
To determine whether NOTCH3 had a direct effect in cells of the osteoblast lineage, bone marrow stromal cells from Notch3 null and control mice were isolated and cultured to confluence (5–7 days) prior to study (day 0). The inactivation of Notch3 did not result in appreciable changes in the Notch target genes Hey1, Hey2, and HeyL or in the expression of Notch1, Notch2, and Notch4 when compared with control cultures (Fig. 3). mRNA levels of Alpl and Bglap (osteocalcin) were stable throughout the culture period, and Alpl, Bglap, Tnfsf11 (RANKL), and Tnfrsf11b (osteoprotegerin) were not significantly different between Notch3−/− mice and controls. In addition, there was a modest increase in mineralized nodules as control cultures progressed but no difference in the formation of mineralized nodules in cultures from Notch3 null mice when compared with controls (Fig. 3).
Figure 3.
Bone marrow stromal cells harvested from tibiae of 5-week-old Notch3−/−(closed circles) and control wildtype mice (open circles) were cultured for 21 days following confluence (Day 0).A, total RNA was extracted and gene expression determined by qRT-PCR. Data are expressed as Notch3, Notch1, Notch2, Notch4, Hey1, Hey2, HeyL, Alpl, Bglap, Tnfsf11 (RANKL), and Tnfrsf11b (osteoprotegerin) copy number corrected for Rpl38 expression. Bars represent means and ranges SD; n = 4 biological replicates. Alpl and Bglap in control and experimental cultures were not statistically different between 7, 14, and 21 days and day 0 by two-way ANOVA. ∗Significantly different between Notch3−/− and controls, p < 0.05. B, representative alizarin red staining of mineralized nodules in control (top) or Notch3−/− (bottom) cultures.
Osteocyte-enriched cells from Notch3 null mice
Because Notch3 is prominently expressed by osteocytes and because these cells play a major role in the regulation of bone remodeling through the RANKL/osteoprotegerin axis, we examined for the expression of Notch target genes and Tnfsf11 and Tnfrsf11b in osteocyte-rich preparations from Notch3−/− and control mice (15, 23, 24). The inactivation of Notch3 did not alter the expression of the Notch target genes Hey1, Hey2, HeyL, or Hes1 and did not have an effect on Tnfsf11 (RANKL) or Tnfrsf11b (osteoprotegerin) mRNA levels compared with control wildtype cells (not shown).
General characteristics of RosaNotch3 mice
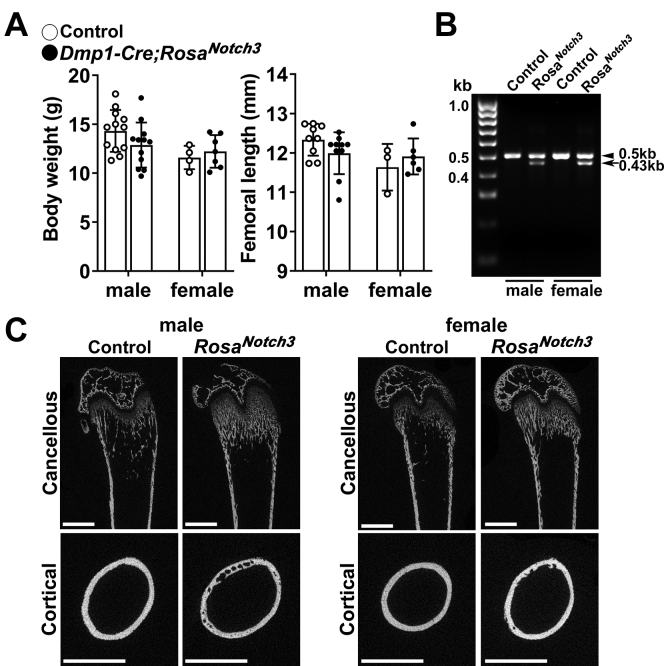
To induce the conditional activation of NOTCH3 in cells of the osteoblast lineage, homozygous RosaNotch3 mice were mated with heterozygous BGLAP-Cre or Dmp1-Cre mice so that approximately half of the pups would express NOTCH3 NICD (Cre+;RosaNotch3) and approximately half would serve as controls (RosaNotch3). Cre-mediated recombination of loxP sites flanking the STOP cassette was documented in extracts from tibiae of 1-month-old mice. The general appearance of BGLAP-Cre;RosaNotch3 or Dmp1-Cre;RosaNotch3 was normal, and their weight and femoral length were not significantly different from those of control littermates (Figs. 4 and 5).
Figure 4.
Weight, femoral length, validation of loxP recombination, and representative microcomputed tomography images of 1-month-old male and female BGLAP-Cre;RosaNotch3mice.A, body weight and femoral length of 1-month-old BGLAP-Cre;RosaNotch3 (closed circles) and sex-matched littermate controls (open circles). Bars represent means and ranges SD; n = 9 control males and n = 12 control females and n = 6 (males) and 7 (females) BGLAP-Cre;RosaNotch3. B, DNA extracted from tibiae of BGLAP-Cre;RosaNotch3 and control mice before and following loxP recombination by Cre under the control of the BGLAP promoter is shown. A 504-bp band is detected for the allele not recombined, and a 432-bp band is detected for the recombined allele. C, representative microcomputed tomography of femurs from 1-month-old male and female control and BGLAP-Cre;RosaNotch3 mice. A sagittal cut is shown for cancellous bone, and a cross-sectional cut at mid-diaphysis shown for cortical bone. Bar in left lower corner scale 1 mm.
Figure 5.
Weight, femoral length, validation of loxP recombination, and representative microcomputed tomography images of 1-month-old male and female Dmp1-Cre;RosaNotch3mice.A, body weight and femoral length of 1-month-old Dmp1-Cre;RosaNotch3 (closed circles) and sex-matched littermate controls (open circles). Bars represent means and ranges SD; n = 3 or 4 (females) and 9 or 12 (males) control and n = 6 or 7 (females) and 10 or 12 (males) Dmp1-Cre;RosaNotch3. B, DNA extracted from tibiae of Dmp1-Cre;RosaNotch3 and control mice before and following loxP recombination by Cre under the control of the Dmp1 promoter is shown. A 504-bp band is detected for the allele not recombined, and a 432-bp band is detected for the recombined allele. C, representative microcomputed tomography of femurs from 1-month-old male and female control and Dmp1-Cre;RosaNotch3 mice. A sagittal cut is shown for cancellous bone and a cross-sectional cut at mid-diaphysis is shown for cortical bone. Bar in left lower corner scale 1 mm.
Skeletal microarchitecture and histomorphometry of RosaNotch3 mice
In contrast to the absence of a phenotype in Notch3−/− mice, the expression of the NOTCH3 NICD in mature osteoblasts or in osteocytes caused a significant 1.7- to 2.4-fold increase in cancellous bone volume/tissue volume in BGLAP-Cre;RosaNotch3 or a 1.5- to 1.8-fold increase in Dmp1-Cre;RosaNotch3 mice (Tables 2 and 3, Figs. 4 and 5) The higher trabecular bone volume in BGLAP-Cre;RosaCre and Dmp1-Cre;RosaNotch3 was mostly observed in the distal metaphysis and was associated with a pronounced increase in connectivity density and a lower structure model idex indicating a tendency toward plate-like trabeculae. The trabecular and cortical bone of the distal metaphyses of BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice were not clearly distinguishable, suggesting defective or delayed bone corticalization (25). At mid-diaphysis, cortical bone volume and cortical thickness were decreased and the cortex appeared porous in BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice of both sexes (Tables 2 and 3, Figs. 4 and 5). Polar moment of inertia was decreased in Dmp1-Cre;RosaNotch3 mice, suggesting decreased bone strength, although the effect was significant only in male mice, and it was not observed in BGLAP-Cre;RosaNotch3 mice (26). There were no major sex differences in the phenotype observed, and skeletal microarchitecture was affected similarly in male and female BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice. There was a distortion of the normal skeletal microarchitecture in both cortical and cancellous compartments of experimental mice. However, periosteal perimeter, width, and anteroposterior length of the metaphysis of BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice were not different from controls (Tables 2 and 3).
Table 2.
Femoral microarchitecture and dimensions assessed by microcomputed tomography of 1-month-old BGLAP-Cre;RosaNotch3 male and female mice and sex-matched littermate controls
| Trabecular and cortical bone | Males |
Females |
||
|---|---|---|---|---|
| Control n = 9 | Cre+/−;RosaNotch3 n = 6 | Control n = 12 | Cre+/−;RosaNotch3 n = 7 | |
| Distal femur trabecular bone | ||||
| Bone volume/total volume (%) | 13.0 ± 2.6 | 22.4 ± 3.9b | 9.7 ± 1.6 | 23.5 ± 7.2b |
| Trabecular separation (μm) | 167 ± 15 | 146 ± 17b | 186 ± 18 | 170 ± 61 |
| Trabecular number (1/mm) | 6.0 ± 0.5 | 7.5 ± 1.0b | 5.4 ± 0.5 | 7.3 ± 2.3b |
| Trabecular thickness (μm) | 34 ± 3 | 35 ± 1 | 31 ± 2 | 37 ± 2b |
| Connectivity density (1/mm3) | 369 ± 83 | 852 ± 180b | 297 ± 71 | 942 ± 429b |
| Structure model index | 1.9 ± 0.3 | 0.7 ± 0.3b | 2.2 ± 0.2 | 0.4 ± 0.2b |
| Density of material (mg HA/cm3) | 825 ± 22 | 862 ± 18b | 808 ± 28 | 840 ± 27b |
| Distal femur dimensions | ||||
| Periosteal perimeter (mm) | 6.3 ± 0.2 | 6.4 ± 0.3 | 6.0 ± 0.7 | 6.3 ± 0.2 |
| Anteroposterior length (mm) | 1.8 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.1 |
| Mediolateral width (mm) | 2.7 ± 0.1 | 2.6 ± 0.1 | 2.7 ± 0.1 | 2.5 ± 0.1 |
| Femoral midshaft cortical bone | ||||
| Bone volume/total volume (%) | 98.0 ± 1.3 | 47.5 ± 4.5b | 98.8 ± 0.6 | 53.1 ± 8.2b |
| Porosity (%) | 2.0 ± 1.3 | 52.5 ± 4.5b | 1.2 ± 0.6 | 46.9 ± 8.2b |
| Cortical thickness (μm) | 95 ± 6 | 36 ± 3b | 95 ± 5 | 36 ± 5b |
| Total area (mm2) | 1.6 ± 0.1 | 2.0 ± 0.2b | 1.5 ± 0.1 | 1.8 ± 0.2b |
| Bone area (mm2)a | 0.43 ± 0.03 | 0.37 ± 0.09 | 0.40 ± 0.03 | 0.30 ± 0.06b |
| Periosteal perimeter (mm) | 4.4 ± 0.1 | 5.0 ± 0.3b | 4.3 ± 0.2 | 4.8 ± 0.2b |
| Endocortical perimeter (mm) | 3.7 ± 0.1 | 4.0 ± 0.2b | 3.7 ± 0.1 | 4.0 ± 0.1b |
| Density of material (mg HA/cm3) | 1018 ± 38 | 934 ± 11b | 1011 ± 39 | 916 ± 43b |
| p Moment of inertia (mm4) | 0.183 ± 0.02 | 0.182 ± 0.04 | 0.166 ± 0.025 | 0.157 ± 0.045 |
Microcomputed tomography was performed in distal femurs for trabecular bone and midshaft for cortical bone from 1-month-old BGLAP-Cre;RosaNotch3 mice and sex-matched littermate controls.
Segmented bone. Values means ± SD.
Significantly different between control and Cre+/−;RosaNotch3, p < 0.05 by unpaired t test.
Table 3.
Femoral microarchitecture and dimensions assessed by microcomputed tomography of 1-month-old Dmp1-Cre;RosaNotch3 male and female mice and sex-matched littermate controls
| Trabecular and cortical bone | Males |
Females |
||
|---|---|---|---|---|
| Control n = 12 | Cre+/−;RosaNotch3 n = 11 | Control n = 4 | Cre+/−;RosaNotch3 n = 7 | |
| Distal femur trabecular bone | ||||
| Bone volume/total volume (%) | 11.3 ± 1.8 | 17.1 ± 3.4b | 9.6 ± 0.9 | 14.4 ± 3.3b |
| Trabecular separation (μm) | 170 ± 5 | 152 ± 13b | 182 ± 13 | 162 ± 33 |
| Trabecular number (1/mm) | 5.9 ± 0.2 | 7.0 ± 0.5b | 5.6 ± 0.3 | 6.6 ± 1.3 |
| Trabecular thickness (μm) | 31 ± 3 | 31 ± 3 | 29 ± 1 | 29 ± 2 |
| Connectivity density (1/mm3) | 384 ± 52 | 962 ± 217b | 395 ± 52 | 908 ± 341b |
| Structure model index | 2.1 ± 0.2 | 1.4 ± 0.5b | 2.1 ± 0.1 | 1.6 ± 0.3b |
| Density of material (mg HA/cm3) | 785 ± 22 | 799 ± 22 | 776 ± 7 | 768 ± 20 |
| Distal femur dimensions | ||||
| Periosteal perimeter (mm) | 6.1 ± 0.2 | 6.1 ± 0.2 | 5.7 ± 0.4 | 6.2 ± 0.1 |
| Anteroposterior length (mm) | 1.8 ± 0.2 | 1.7 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.1 |
| Mediolateral width (mm) | 2.5 ± 0.1 | 2.5 ± 0.1 | 2.4 ± 0.1 | 2.4 ± 0.1 |
| Femoral midshaft cortical bone | ||||
| Bone volume/total volume (%) | 96.8 ± 2.4 | 85.3 ± 6.5b | 97.2 ± 1.7 | 81.3 ± 9.0b |
| Porosity (%) | 3.2 ± 2.4 | 14.7 ± 6.5b | 2.8 ± 1.7 | 18.7 ± 9.0b |
| Cortical thickness (μm) | 88 ± 8 | 55 ± 11b | 80 ± 7 | 48 ± 8b |
| Total area (mm2) | 1.4 ± 0.1 | 1.4 ± 0.1 | 1.3 ± 0.1 | 1.4 ± 0.1 |
| Bone area (mm2)+ | 0.38 ± 0.03 | 0.292 ± 0.06b | 0.32 ± 0.02 | 0.26 ± 0.01b |
| Periosteal perimeter (mm) | 4.2 ± 0.1 | 4.2 ± 0.2 | 4.0 ± 0.1 | 4.2 ± 0.1 |
| Endocortical perimeter (mm) | 3.6 ± 0.1 | 3.7 ± 0.1 | 3.4 ± 0.2 | 3.7 ± 0.1b |
| Density of material (mg HA/cm3) | 955 ± 32 | 917 ± 17b | 927 ± 31 | 895 ± 19 |
| p Moment of inertia (mm4) | 0.151 ± 0.02 | 0.120 ± 0.03b | 0.111 ± 0.01 | 0.103 ± 0.01 |
Microcomputed tomography was performed in distal femurs for trabecular bone and midshaft for cortical bone from 1-month-old Dmp1-Cre;RosaNotch3 mice and sex-matched littermate controls.
Segmented bone. Values means ± SD.
Significantly different between control and Cre+/−;RosaNotch3, p < 0.05 by unpaired t test.
Bone histomorphometry was conducted in male and female mice and sex-matched controls. In accordance with the more pronounced microarchitectural phenotype in BGLAP-Cre;RosaNotch3 mice, changes in histomorphometric parameters were more evident in this cohort than in Dmp1-Cre;RosaNotch3 mice. Histomorphometric analysis of cancellous femoral bone (in methyl methacrylate embedded sections) confirmed the increase in bone volume and trabecular number in BGLAP-Cre;RosaNotch3 mice (Table 4). This was secondary to a decrease in bone resorption since osteoclast number was significantly reduced in BGLAP-Cre;RosaNotch3 mice and eroded surface was significantly decreased in BGLAP-Cre;RosaNotch3 male mice (Table 4). Safranin O and fast green staining revealed cartilage remnants in cancellous bone (Fig. 6). Analysis of decalcified bone samples (embedded in paraffin) confirmed a decrease in tartrate-resistant acid phosphatase (TRAP)-positive multinucleated osteoclasts in cancellous bone from male and female BGLAP-Cre;RoseNotch3 mice. The number of osteoclasts/bone perimeter (1/mm) decreased from (means ± SD; n = 4) 5.1 ± 1.2 in control to 2.5 ± 1.6 (p < 0.05) in BGLAP-Cre;RosaNotch3 male mice and from 7.8 ± 2.1 in control to 2.3 ± 1.2 (p < 0.05) in BGLAP-Cre;RosaNotch3 female mice. A decrease in osteoblast number was noted in BGLAP-Cre;RosaNotch3 mice that reached statistical significance in female mice, and osteoblasts had a fibroblast-like appearance. Calcein/demeclocycline labels did not condense or were missing, not allowing the measurement of mineral apposition rate or mineralizing surface in BGLAP-Cre;RosaNotch3 male mice (Fig. 6). Bone formation rate was reduced in female mice albeit not significantly. In contrast to the results observed in cancellous bone, cross-sectional analysis of cortical bone revealed an increase in void or pore area and TRAP-positive osteoclast number in male and female BGLAP-Cre;RosaNotch3 mice demonstrating increased intracortical remodeling. An increase in TRAP-positive osteoclast number also was noted on the endocortical surface of BGLAP-Cre;RosaNotch3 male mice indicating enhanced resorption in cortical bone contrasting the suppression of bone resorption in cancellous bone (Table 4, Fig. 7).
Table 4.
Cancellous and cortical bone histomorphometry of 1-month-old BGLAP-Cre;RosaNotch3 male and female mice and sex-matched RosaNotch3 control littermates
| Cancellous bone | Males |
Females |
||
|---|---|---|---|---|
| Control n = 6 | Cre;RosaNotch3 n = 5 | Control n = 6 | Cre;RosaNotch3 n = 4 | |
| Static histomorphometry | ||||
| Bone volume/tissue volume (%) | 17.2 ± 3.8 | 47.9 ± 11.2a | 9.5 ± 3.4 | 42.1 ± 0.9a |
| Trabecular separation (μm) | 201 ± 44 | 39 ± 13a | 373 ± 139 | 55 ± 23a |
| Trabecular number (1/mm) | 4.3 ± 0.9 | 13.9 ± 3.1a | 2.7 ± 0.9 | 11.6 ± 3.9a |
| Trabecular thickness (μm) | 41 ± 7 | 36 ± 9 | 35 ± 5 | 37 ± 4 |
| Osteoblast surface/bone surface (%) | 19.1 ± 5.2 | 14.1 ± 3.6 | 20.3 ± 3.0 | 15.9 ± 5.8 |
| Osteoblasts/bone perimeter (1/mm) | 15.8 ± 5.5 | 10.4 ± 2.0 | 17.4 ± 2.6 | 11.9 ± 4.1a |
| Osteoid surface/bone surface (%) | 0.9 ± 0.9 | 0.2 ± 0.1 | 0.9 ± 0.7 | 0.2 ± 0.2 |
| Osteocytes (mm2) | 941 ± 118 | 919 ± 107 | 1083 ± 200 | 805 ± 50a |
| Osteoclast surface/bone surface (%) | 7.1 ± 1.6 | 3.5 ± 1.0a | 9.3 ± 3.3 | 3.8 ± 1.2a |
| Osteoclasts/bone perimeter (1/mm) | 2.9 ± 0.6 | 1.5 ± 0.4a | 3.9 ± 1.2 | 1.6 ± 0.3a |
| Eroded surface/bone surface (%) | 2.9 ± 0.8 | 1.2 ± 0.3a | 2.1 ± 0.9 | 1.6 ± 0.4 |
| Dynamic histomorphometry | ||||
| Mineral apposition rate (μm/day) | 2.1 ± 0.5 | ND | 1.7 ± 0.3 | 1.5 ± 0.3 |
| Mineralizing surface/bone surface (%) | 1.7 ± 0.7 | ND | 2.4 ± 1.3 | 0.8 ± 0.6b |
| Bone formation rate (μm3/μm2/day) | 0.034 ± 0.01 | ND | 0.041 ± 0.03 | 0.012 ± 0.01 |
| Cortical bone | Males |
Females |
||
|---|---|---|---|---|
| Control | Cre;RosaNotch3 | Control | Cre;RosaNotch3 | |
| Static histomorphometry | n = 5 | n = 5 | n = 3 | n = 3 |
| Cortical thickness (μm) | 127 ± 9 | 83 ± 14a | 107 ± 10 | 84 ± 16 |
| Periosteal perimeter (mm) | 5.6 ± 0.6 | 7.4 ± 1.9 | 5.5 ± 0.06 | 6.0 ± 0.87 |
| Endocortical perimeter (mm) | 4.8 ± 0.5 | 8.0 ± 2.1a | 5.1 ± 0.2 | 5.9 ± 3.0 |
| Void/pore perimeter (mm) | 0.5 ± 0.8 | 7.3 ± 3.9a | 0.8 ± 0.7 | 4.9 ± 2.6b |
| Void/pore area (mm2) | 0.006 ± 0.009 | 0.12 ± 0.06a | 0.004 ± 0.004 | 0.07 ± 0.06 |
| Void/pore area/bone area (mm2) | 0.008 ± 0.01 | 0.22 ± 0.08a | 0.008 ± 0.007 | 0.16 ± 0.09a |
| Osteoclasts/bone area (mm2) | 7.1 ± 10.3 | 124.6 ± 41.5a | 8.6 ± 7.4 | 110 ± 41a |
| Osteocytes/bone area (mm2) | 1207 ± 59 | 1207 ± 71 | 1310 ± 38 | 1176 ± 24a |
| Endocortical surface | ||||
| Static histomorphometry | n = 4 | n = 4 | n = 3 | n = 3 |
| Osteoclast surface/bone surface (%) | 7.3 ± 1.7 | 15.4 ± 4.5a | 10.6 ± 2.4 | 15.1 ± 6.2 |
| Osteoclast number/perimeter (1/mm) | 4.2 ± 0.5 | 8.0 ± 1.6a | 5.5 ± 0.8 | 7.9 ± 2.7 |
| Eroded surface/bone surface (%) | 6.3 ± 3.1 | 8.6 ± 1.0 | 6.4 ± 3.0 | 6.0 ± 1.6 |
| Dynamic histomorphometry | n = 3 | n = 3 | n = 3 | n = 4 |
| Mineral apposition rate (μm/day) | 1.15 ± 0.33 | 0.91 ± 0.29 | 0.97 ± 0.18 | 0.87 ± 0.20 |
| Mineralizing surface/bone surface (%) | 14.9 ± 8.7 | 7.9 ± 4.2 | 9.3 ± 5.2 | 9.3 ± 5.6 |
| Bone formation rate (μm3/μm2/day) | 0.18 ± 0.14 | 0.08 ± 0.06 | 0.08 ± 0.03 | 0.09 ± 0.06 |
Histomorphometry was carried out on sagittal sections of distal femurs or TRAP-stained and unstained cross sections at the femoral mid-diaphysis from 1-month-old male and female BLGAP-Cre;RosaNotch3 mice and sex-matched control littermates. Values are means ± S.D.
Significantly different between control and Cre;RosaNotch3, p < 0.05 by unpaired t test. ND, not determined.
p < 0.055.
Figure 6.
Representative cancellous bone histomorphometry of femurs from 1-month-old male and female BGLAP-Cre;RosaNotch3mice. Sections embedded in methyl methacrylate were deplasticized and stained with toluidine blue (two upper panels; scale bars 100 and 50 μm); arrows point to fibroblast-like cells or with safranin O and fast green (middle upper panel; scale bar, 200 μm). Bones embedded in paraffin were processed and stained with TRAP (red-pink) and hematoxylin (blue-purple) (fourth panel from top: scale bar, 100 μm). Calcein (green) and demeclocycline (orange) labeling of methyl methacrylate embedded sections is shown in the lower panel (scale bar, 50 μm).
Figure 7.
Representative cortical bone histomorphometry at mid-diaphysis of femurs from 1-month-old male and female BGLAP-Cre;RosaNotch3mice. Bones embedded in paraffin were processed and stained with TRAP (red-pink) and hematoxylin (blue-purple) (upper panel; scale bar, 50 μm). Frozen bones were sectioned to demonstrate calcein (green) and demeclocycline (orange) labeling (lower panel; scale bar, 20 μm). The arrows indicate voids or pores in intracortical bone of male and female BGLAP-Cre;RosaNotch3 mice.
Bone histomorphometric analysis of cancellous bone of Dmp1-Cre;RosaNotch3 male, but not female mice, demonstrated similar, although less pronounced, changes than those observed in BGLAP-Cre;RosaNotch3 mice (Table 5). Osteoclast number and bone formation were reduced. Analysis of TRAP-positive multinucleated cells in paraffin-embedded sections revealed a nonsignificant ∼20% to 25% decrease in male and female Dmp1-Cre;RosaNotch3 mice compared with controls (not shown). Cortical histomorphometry could be performed in male Dmp1-Cre;RosaNotch3 mice only due to the limited number of female mice available for study and revealed a substantial increase in cortical TRAP-positive osteoclasts and void or pore area (over 10-fold), although it did not reach statistical significance owing to variability in the results (Table 5). An increase in osteoclast number was found in the endocortical surface of Dmp1-Cre;RosaNotch3 male mice.
Table 5.
Cancellous and cortical bone histomorphometry of 1-month-old Dmp1-Cre;RosaNotch3 male and female mice and sex-matched RosaNotch3 control littermates
| Cancellous bone | Males |
Females |
||
|---|---|---|---|---|
| Control n = 6 | Cre;RosaNotch3 n = 8 | Control n = 3 | Cre;RosaNotch3 n = 7 | |
| Static histomorphometry | ||||
| Bone volume/tissue volume (%) | 14.5 ± 3.2 | 20.3 ± 5.0a | 9.0 ± 1.8 | 9.9 ± 3.9 |
| Trabecular separation (μm) | 203 ± 40 | 122 ± 40a | 317 ± 31 | 306 ± 138 |
| Trabecular number (1/mm) | 4.3 ± 0.9 | 7.0 ± 1.8a | 2.9 ± 0.3 | 3.4 ± 1.3 |
| Trabecular thickness (μm) | 33 ± 8 | 29 ± 3a | 31 ± 8 | 29 ± 3 |
| Osteoblast surface/bone surface (%) | 18.2 ± 2.1 | 20.7 ± 4.6 | 21.6 ± 5.0 | 26.0 ± 7.1 |
| Osteoblasts/bone perimeter (1/mm) | 15.0 ± 2.0 | 16.9 ± 4.9 | 20.2 ± 7.9 | 23.1 ± 6.6 |
| Osteoid surface/bone surface (%) | 0.5 ± 0.7 | 0.2 ± 0.1a | 1.4 ± 1.2 | 0.2 ± 0.2a |
| Osteocytes/bone area (mm2) | 1042 ± 73 | 1027 ± 96 | 1179 ± 150 | 1117 ± 161 |
| Osteoclast surface/bone surface (%) | 8.6 ± 1.3 | 5.0 ± 0.9a | 12.2 ± 2.8 | 10.9 ± 3.9 |
| Osteoclasts/bone perimeter (1/mm) | 3.3 ± 0.5 | 2.1 ± 0.3a | 5.1 ± 1.3 | 4.4 ± 1.5 |
| Eroded surface/bone surface (%) | 2.9 ± 0.9 | 2.1 ± 0.5 | 4.0 ± 1.2 | 3.8 ± 1.4 |
| Dynamic histomorphometry | ||||
| Mineral apposition rate (μm/day) | 2.0 ± 0.4 | 1.7 ± 0.2 | 1.8 ± 0.4 | 2.0 ± 0.9 |
| Mineralizing surface/bone surface (%) | 1.0 ± 0.3 | 0.4 ± 0.2a | 1.9 ± 1.8 | 0.8 ± 0.6 |
| Bone formation rate (μm3/μm2/day) | 0.022 ± 0.018 | 0.007 ± 0.003a | 0.068 ± 0.041 | 0.017 ± 0.015a |
| Cortical bone | Males |
|
|---|---|---|
| Control | Cre;RosaNotch3 | |
| Static histomorphometry | n = 6 | n = 5 |
| Cortical thickness (μm) | 127 ± 11 | 103 ± 22 |
| Periosteal perimeter (mm) | 5.2 ± 0.7 | 4.6 ± 0.9 |
| Endocortical perimeter (mm) | 4.3 ± 0.7 | 4.5 ± 1.2 |
| Void/pore perimeter (mm) | 0.3 ± 0.4 | 3.1 ± 3.3 |
| Void/pore area (mm2) | 0.002 ± 0.002 | 0.029 ± 0.02 |
| Void/pore area/bone area (mm2) | 0.003 ± 0.003 | 0.05 ± 0.04b |
| Osteoclasts/bone area (mm2) | 1.4 ± 2.0 | 32.3 ± 32.2 |
| Osteocytes/bone area (mm2) | 1094 ± 74 | 1135 ± 181 |
| Endocortical surface | ||
| Static histomorphometry | n = 6 | n = 6 |
| Osteoclast surface/bone surface (%) | 7.1 ± 3.6 | 11.2 ± 4.1 |
| Osteoclast number/perimeter (1/mm) | 3.5 ± 1.7 | 6.5 ± 2.7a |
| Eroded surface/bone surface (%) | 3.9 ± 2.6 | 7.2 ± 3.1a |
| Dynamic histomorphometry | n = 5 | n = 4 |
| Mineral apposition rate (μm/day) | 0.91 ± 0.25 | 0.58 ± 0.38 |
| Mineralizing surface/bone surface (%) | 7.0 ± 4.8 | 7.6 ± 5.9 |
| Bone formation rate (μm3/μm2/day) | 0.070 ± 0.06 | 0.058 ± 0.04 |
Histomorphometry was carried out on sagittal sections of distal femurs or TRAP-stained and unstained cross sections at the femoral mid-diaphysis from 1-month-old Dmp1-Cre;RosaNotch3 mice and sex-matched control littermates. Values are means ± S.D.
Significantly different between control and Cre;RosaNotch3, p < 0.05 by unpaired t test.
p < 0.055.
Gene expression profile in bones from RosaNotch3 mice
To explore mechanisms that may explain the phenotype of BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice, tibiae were analyzed for changes in gene expression. Transcript levels for the Notch3 NICD and its target genes Hey1, Hey2, and HeyL were increased in tibiae from BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice confirming activation of Notch signaling in skeletal cells (Figs. 8 and 9). Notch activation caused an increase in Tnfrsf11b (osteoprotegerin) and Tnfsf11 (RANKL) expression. The induction of Tnfrsf11b was more pronounced than that of Tnfsf11 in tibiae from BGLAP-Cre;RosaNotch3 mice. BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice exhibited a marked suppression of the Wnt antagonists Sost (encoding sclerostin) and Dickkopf 1 (Dkk1) mRNA and a modest not statistically significant increase in the Wnt target gene Wisp1 suggesting enhanced Wnt signaling.
Figure 8.
Total RNA was extracted from tibiae from BGLAP-Cre;RosaNotch3(closed circles) and age-matched littermate control (open circles) mice, and gene expression was determined by qRT-PCR. Data are expressed as Notch3NICD, Hey1, Hey2, HeyL, Dkk1, Sost, Wisp1, Tnfsf11 (RANKL), and Tnfrsf11b (osteoprotegerin) copy number corrected for Rpl38. Bars represent means and ranges SD; n = 8. Data are derived from biological replicates. ∗Significantly different between BGLAP-Cre;RosaNotch3 and controls, p < 0.05 by unpaired t test.
Figure 9.
Total RNA was extracted from tibiae from Dmp1-Cre;RosaNotch3(closed circles) and age-matched littermate control (open circles) mice, and gene expression was determined by qRT-PCR. Data are expressed as Notch3NICD, Hey1, Hey2, HeyL, Dkk1, Sost, Wisp1, Tnfsf11 (RANKL), and Tnfrsf11b (osteoprotegerin) copy number corrected for Rpl38. Bars represent means and ranges SD; n = 7. Data are derived from biological replicates. ∗Significantly different between Dmp1-Cre;RosaNotch3 and controls, p < 0.05 by unpaired t test.
Bone marrow stromal cell cultures from RosaNotch3 mice
To determine the direct effects of NOTCH3 in cells of the osteoblast lineage, bone marrow stromal cells from BGLAP-Cre;RosaNotch3 and control mice were isolated and cultured for 21 days. BGLAP-Cre;RosaNotch3 cells displayed an expected increase in Notch3-NICD mRNA and protein by immunoblot, confirming the overexpression of the NICD (Fig. 10). Activation of NOTCH3 resulted in a sustained increase in the Notch target genes Hey1, Hey2, and HeyL as well as an induction of Tnfsf11 (RANKL) and Tnfrsf11b (osteoprotegerin). Sost mRNA was expressed at low levels in control cultures and was suppressed further in BGLAP-Cre;RosaNotch3 cells. Dkk1 expression was suppressed and the Wnt target gene Wisp1 was increased suggesting enhanced Wnt signaling. Despite this effect, osteoblastogenesis was suppressed by the activation of NOTCH3 and there was a decrease in mineralized nodule formation and osteocalcin expression in cultures from BGLAP-Cre;RosaNotch3 mice (Fig. 10). In addition, a transient decrease in Alp1 was observed.
Figure 10.
Bone marrow stromal cells harvested from tibiae of 5-week-old BGLAP-Cre;RosaNotch3(closed circles) and control littermate mice (open circles) were cultured for 21 days following confluence (Day 0).A and C, total RNA was extracted and gene expression determined by qRT-PCR. Data are expressed as Notch3-Nicd, Hey1, Hey2, HeyL, Alpl, Bglap, Dkk1, Sost, Wisp1, Tnfsf11 (RANKL), and Tnfrsf11b (osteoprotegerin), copy number corrected for Rpl38. Bars represent means and ranges SD; n = 4. Data are derived from technical replicates. ∗Significantly different between BGLAP-Cre;RosaNotch3 and controls, p < 0.05 by two-way ANOVA. #Significantly different from time 0 by two-way ANOVA. B, immunoblot demonstrating the presence of NOTCH3 intracellular domain (N3ICD). D, representative alizarin red staining of mineralized nodules in control (top) or BGLAP-Cre;RosaNotch3 cultures (bottom).
Osteocyte-enriched cells from RosaNotch3 mice
To explore the direct effect of NOTCH3 in osteocytes, osteocyte-rich preparations from BGLAP-Cre;RosaNotch3 mice were obtained and analyzed. The expression of Notch3-NICD transcripts and the expression of the Notch target genes Hey1, Hey2, and HeyL were increased, whereas mRNA levels for Sost and Dkk1 were suppressed; Wisp1 mRNA was not changed (Fig. 11). In contrast to the results obtained in intact tibiae and bone marrow stromal cells, there was an induction of Tnfsf11 (RANKL) but a marked suppression in Tnfrsf11b (osteoprotegerin) mRNA levels in osteocytes from BGLAP-Cre;RosaNotch3 mice (Fig. 11).
Figure 11.
Total RNA isolated from osteocyte-enriched cells from BGLAP-Cre;RosaNotch3(closed circles) and control (open circles) littermate mice was extracted, and gene expression was determined by qRT-PCR. Data are expressed as Notch3-Nicd, Hey1, Hey2, HeyL, Dkk1, Sost, Wisp1 Tnfsf11 (RANKL), and Tnfrsf11b (osteoprotegerin) copy number corrected for Rpl38. Bars represent means and ranges SD; n = 4. Data are derived from biological replicates. ∗Significantly different between BGLAP-Cre;RosaNotch3 and controls, p < 0.05 by unpaired t test.
Cocultures of osteoblasts from RosaNotch3 mice and bone marrow–derived macrophages
To determine the contributions of RosaNotch3 osteoblasts to osteoclastogenesis, bone marrow–derived macrophages (BMMs) from wildtype mice were cocultured with either bone marrow stromal cells from BGLAP-Cre;RosaNotch3 or Dmp1-Cre;RosaNotch3 and control mice or with osteoblasts from BGLAP-Cre;RosaNotch3 and control mice. Cocultures of bone marrow stromal cells from control or experimental mice with BMMs resulted in the formation of TRAP-positive cells but not in multinucleation, and the cells were limited in number (data not shown). In contrast, multinucleated TRAP-positive cells were induced in BMMs whether they were cultured in the presence of wildtype or BGLAP-Cre;RosaNotch3 osteoblasts. Moreover, osteoblasts from BGLAP-Cre;RosaNotch3 mice enhanced osteoclastogenesis from (means ± SD; n = 4) 45 ± 11 osteoclasts in control to 126 ± 19 (p < 0.05) osteoclasts in RosaNotch3 cocultures. The results demonstrate an osteoblast-dependent increase in osteoclast differentiation in cells overexpressing NOTCH3. The effect is congruent with the increased RANKL and decreased osteoprotegerin expression observed in osteocyte-rich cultures from BGLAP-Cre;RosaNotch3 mice.
Discussion
In the present study, we explored the consequences of the Notch3 misexpression in the skeleton. The inactivation of Notch3 did not result in significant or meaningful changes in bone microarchitecture of either male or female mice studied at a young age or following maturation. These results would indicate that, under basal conditions, NOTCH3, in contrast to NOTCH1 and NOTCH2, is not required for skeletal homeostasis (27, 28, 29). The inactivation of Notch3 did not cause a decrease in Notch target genes in cells of the osteoblast lineage explaining why neither the in vitro cellular nor the in vivo skeletal phenotype was altered by the Notch3 inactivation. Although NOTCH3 regulates RANKL expression in the osteoblast lineage, in accordance with the absence of changes in Notch signaling, the expression of Tnfsf11 was not affected in either bone marrow stromal cells or osteocytes from Notch3 null mice (16). There were no changes in gene markers of osteoblast differentiation in bone marrow stromal cells from Notch3 null mice compared with wildtype cells, although the cells were studied after they had undergone a degree of osteoblast differentiation since they formed mineralized nodules and expressed Alpl and Bglap (osteocalcin) during the initial phases of the experiment, which was started approximately a week following the isolation of cells. The lack of a skeletal phenotype in Notch3 null mice may be related to a low level of basal expression or of activation in bone cells or to a possible compensation by the actions of NOTCH1 and NOTCH2. However, this did not require their overexpression since Notch1, Notch2, and Notch4 transcript levels were not increased in cells from Notch3 null mice. The absence of a Notch3 null phenotype does not mean that NOTCH3 does not play a role in skeletal remodeling under conditions of Notch activation or in the pathogenesis of skeletal disorders associated with a gain-of-NOTCH3 function. Indeed, the expression of the active NOTCH3 NICD in mature osteoblasts and in osteocytes caused a pronounced skeletal phenotype manifested by increased cancellous bone volume and high cortical porosity.
The cancellous bone phenotype observed following the activation of NOTCH3 in osteoblasts and osteocytes can be explained by an induction of osteoprotegerin, leading to an inhibition of bone resorption and decreased remodeling of cancellous bone. The phenotype was suggestive of osteopetrosis since cartilage remnants were found in cancellous bone. An additional mechanism that might have contributed to the phenotype of mice overexpressing NOTCH3-NICD is the activation of Wnt signaling secondary to the suppression of the Wnt antagonists sclerostin and Dkk1 (30). Wnt signaling induces osteoblastogenesis and has a less recognized but important inhibitory effect on osteoclastogenesis and bone resorption secondary to an increase in osteoprotegerin expression by cells of the osteoblastic lineage and to direct effects of Wnt on osteoclast precursors (31, 32, 33, 34, 35). However, the inhibitory effects of NOTCH3 on osteoblast differentiation in vitro and bone formation in vivo prevailed over the potential stimulatory effects of Wnt. This is in accordance with previous work demonstrating that Notch inhibits osteoblastogenesis and opposes Wnt signaling and Wnt effects in cells of the osteoblast lineage (36, 37, 38).
The effect of NOTCH3-NICD was bone compartment specific, and it caused increased porosity of cortical bone associated with increased intracortical number of osteoclasts and remodeling as demonstrated by an increase in void or pore area. An increase in endocortical surface osteoclasts also was noted. There was a decrease in polar moment of inertia in Dmp1-Cre;RosaNotch3 male mice indicating impaired bone strength (26). Cortical bone is a tissue rich in osteocytes, and in contrast to the induction of osteoprotegerin in osteoblast cultures, NOTCH3-NICD inhibited osteoprotegerin in osteocyte-rich cell preparations possibly explaining the cortical porosity and enhanced intracortical remodeling. Of interest, RANKL was induced both in bone marrow stromal cells, where its actions could be counteracted by an increase in osteoprotegerin, and in osteocytes where its osteoclastogenic actions prevailed. An additional mechanism responsible for the high cortical porosity could be impaired or incomplete formation of cortical bone or corticalization since both Bglap and Dmp1 are expressed late during embryogenesis and could have induced NOTCH3 (39, 40). Impaired corticalization is suggested by the severity of the cortical bone phenotype and the fact that cortical and trabecular bone were not clearly distinguishable at the distal metaphysis (25). Corticalization is the process by which trabecular bone coalesces at the metaphyseal cortex as longitudinal growth occurs (41, 42). Several genes have been shown to be critical for corticalization, including Sp7 (Osterix), Clcn7, and Socs3 (25, 41, 43, 44). It is not known whether Notch signaling interacts with any of the genes.
The increase in cancellous bone volume and in cortical porosity following the activation of NOTCH3 are similar to those reported following the supraphysiological activation of NOTCH1 in osteoblasts and osteocytes (29, 45). However, impaired corticalization is not observed in mouse models harboring Notch2 or Notch3 mutations that result in modest increments in Notch signaling (16, 17). This would indicate that supraphysiological levels of Notch signaling are necessary to impair corticalization.
Previously, we reported a mouse model (Notch3em1Ecan or Notch3tm1.1Ecan) of lateral meningocele syndrome presenting with NOTCH3 gain of function due to stabilization of NOTCH3 and osteopenia (16, 46). The bone loss was attributed to increased bone resorption due to an induction of RANKL by cells of the osteoblast lineage. Although the cancellous bone phenotype is in contrast to the one manifested by either the BGLAP-Cre;RosaNotch3 or the Dmp1-Cre;RosaNotch3 mouse model described in the present work, the three models of NOTCH3 overexpression presented with cortical bone loss. RANKL is induced in osteoblasts and osteocytes following modest (Notch3em1Ecan) as well as pronounced (BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3) levels of NOTCH3 activation, whereas the regulation of osteoprotegerin appears to be dependent on the cell context and degree of Notch activation (45). Indeed, osteoprotegerin is not induced in Notch3em1Ecan mice explaining their RANKL-dependent resorptive phenotype, is induced in RosaNotch3 bone marrow stromal cells, possibly explaining the gain in cancellous bone, and is suppressed in RosaNotch3 osteocytes possibly explaining the cortical bone porosity and osteopenia of mice overexpressing NOTCH3-NICD. Cocultures of bone marrow stromal cells with BMMs did not yield the formation of mature osteoclasts from either control or RosaNotch3 cells. Cocultures of osteoblasts with BMMs resulted in the formation of mature osteoclasts, and osteoclastogenesis was increased by osteoblasts from BGLAP-Cre;RosaNotch3 mice. The results are congruent with the increased RANKL and suppressed osteoprotegerin expression observed in osteocytes and could explain the enhanced intracortical remodeling. This is not surprising since the cells are of the same lineage and vary in degree of cell maturation. NOTCH3 overexpression inhibited cancellous bone formation, an effect that could represent direct effects of NOTCH3 on osteoblasts or a degree of decreased bone remodeling in this bone compartment. The phenotype observed in BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 mice was secondary to the exposure of skeletal tissue to supraphysiological levels of Notch activation. This may be relevant to pathological states of Notch signal activation but probably not to a physiological state of bone remodeling.
Because cells of the myeloid/osteoclast lineage do not express Notch3 mRNA, NOTCH3 is capable of modulating osteoclastogenesis only by indirect mechanisms regulating the expression of RANKL and osteoprotegerin in the osteoblast lineage (16, 47). This is in contrast to NOTCH1 and NOTCH2, which are expressed in the myeloid lineage and have direct, as well as indirect, effects on osteoclastogenesis (14, 17, 18, 48, 49), albeit those of NOTCH1 are inhibitory, whereas those of NOTCH2 are stimulatory.
The present observations may suggest a more important function of NOTCH1 and NOTCH2 than NOTCH3 in skeletal homeostasis since mouse models of gain or loss of function of both receptors exhibit skeletal phenotypes (17, 27, 28, 45). We reported that the dual inactivation of Notch1 and Notch2 in Sp7 (Osterix)-expressing osteoblast precursors or in Dmp1-expressing osteocytes increases cancellous bone, and others reported that the inactivation of Notch2 in osteoblasts causes increased bone mass indicating a negative role of NOTCH1 and NOTCH2 in skeletal homeostasis (27, 28, 29). Similarly, the inactivation of Notch1 and Notch2 in the limb bud causes lengthening of the growth plate and an increase in cancellous bone (50).
Notch1 null mice die during development owing to widespread cellular death, and hypomorphic Notch2 alleles cause perinatal death due to vascular and renal defects, whereas Notch3 and Notch4 null mice develop normally and mutant adults are viable and fertile (51, 52, 53, 54, 55). The present findings are in line with previous work demonstrating that NOTCH3 plays an important function in vascular cells and is required for arterial identity and maturation but it is not essential for bone development (54).
The approach to downregulate Notch signaling has been diverse; however, the approaches are often not specific (56, 57, 58). To target specific Notch receptors, antibodies to the NRR of NOTCH1, NOTCH2, and NOTCH3 have been developed (59, 60). Targeting the NRR prevents cleavage and, therefore, activation of Notch receptors, making it ideal for the specific neutralization of individual Notch isoforms. Previously, we have shown that anti-NOTCH3 NRR antibodies reverse the osteopenia of Notch3em1Ecan mice. However, anti-NOTCH3 NRR antibodies did not influence the skeletal phenotype of wildtype mice or the behavior of their skeletal cells in vitro demonstrating that the prevention of NOTCH3 activation in wildtype cells is without consequences (47). These observations are congruent with the present findings indicating that the basal expression of NOTCH3 is dispensable for skeletal homeostasis.
In conclusion, NOTCH3 activation in osteoblasts/osteocytes increases cancellous bone volume by decreasing bone resorption but causes high cortical porosity by increasing intracortical remodeling. Notch3 inactivation does not alter bone architecture indicating that basal NOTCH3 activation is dispensable for skeletal homeostasis.
Experimental procedures
Notch3 null mice
Notch3tm1.1(KOMP)Vlcg mice were created in a C57BL/6N genetic background and were obtained from MMRC; project VG18699 (University of California). Notch3tm1(KOMP)Vlcg mice in a C57BL/6N genetic background that contained a reporter-tagged null allele were bred at MMRC with Gt(ROSA)26Sortm1(ACTB-cre,-EGFP)Ics/TCP in a C57BL/6N genetic composition to remove the β-actin promoter and the neomycin selection cassette to create Notch3tm1.1(KOMP)Vlcg mice (61). Frozen sperm from Notch3tm1.1(KOMP)Vlcg was obtained from MMRC and used for the fertilization of C57BL/6N females at the Center for Mouse Genome Modification at UConn Health. Mice were studied in a C57BL/6N genetic composition following heterozygous intercrosses for the generation of Notch3−/− and control wildtype sex-matched littermates. Because the yield of the various sex-matched genotypes was modest, litters from various dams had to be pooled so that the study of littermates was not always possible. Genotypes were determined by polymerase chain reaction (PCR) analysis of tail DNA using specific primers (Integrated DNA Technologies; Table 6).
Table 6.
Primers used for allele identification
| Allele | Strand | Sequence | Amplicon size (bp) |
|---|---|---|---|
| Genotyping | |||
| BGLAP transgene | Forward | 5'-CAAATAGCCCTGGCAGAT-3' | 300 |
| Reverse | 5'-TGATACAAGGGACATCTTCC-3' | ||
| Dmp1 transgene | Forward | 5'-CCCGCAGAACCTGAAGATG-3' | 534 |
| Reverse | 5'-GACCCGGCAAAACAGGTAG-3' | ||
| Notch3 | Forward | 5'-GAGGCCCAAGGAATCGAGAC-3' | 296 |
| Reverse | 5'-ATGAGACGTTTTCTCCGAGTTCAG-3' | ||
| Notch3−/− | Forward | 5'-ACTTGCTTTAAAAAACCTCCCACA-3' | 849 |
| Reverse | 5'-CTCCCAAATGTCCCCTGACC-3' | ||
| RosaNotch3 | Forward | 5'-CCTCCTGGCTTCTGAGGAC-3' | Wt (Reverse 1) = 333 Floxed (Reverse 2) = 504 |
| Reverse 1 | 5'-CTCCGAGGCGGATCACAAGC-3' | ||
| Reverse 2 | 5'-CTCGTCCTGCAGTTCATTCA-3' | ||
| LoxP Recombination | |||
| RosaNotch3 | Forward | 5'-CCTCCTGGCTTCTGAGGAC-3' | Absent LoxP recombination (Reverse 1) = 504 Present LoxP recombination (Reverse 2) = 432 |
| Reverse 1 | 5'-CTCGTCCTGCAGTTCATTCA-3' | ||
| Reverse 2 | 5'-ACCTCCCCCATCAGACTCTC-3' | ||
RosaNotch3 mice
RosaNotch3 mice (Notch3_ICD_YPFTG) were obtained from Dr Spyros Artavanis Tsakonas (Harvard University) and Dr Silvia Fre (Institut Curie) in a C57BL/6 genetic background (20). In these mice, the Rosa26 locus is targeted with a DNA construct encoding NOTCH3-NICD, preceded by a neo-STOP cassette flanked by loxP sites, cloned downstream exon 1 of the Rosa26 gene. Expression of the NICD from the targeted Rosa26 locus occurs following the excision of the STOP cassette by Cre recombination of loxP sequences. To study the activation of NOTCH3 in osteoblasts and in osteocytes, homozygous RosaNotch3 mice were mated with either BGLAP-Cre+/− (Tg(BGLAP-Cre)/Clem/J, Jackson 019509) or Dmp1-Cre+/− (Tg(Dmp1-Cre)1 Jqfe/Bwd_, Jackson 023047) transgenics to create Cre+/−;RosaNotch3 experimental and RosaNotch3 littermate controls (21, 22). Male and female experimental and control mice were compared at 1 month of age. Genotyping of BGLAP-Cre, Dmp1-Cre transgenics and RosaNotch3 mice was carried out by PCR in tail DNA extracts (Table 6). Deletion of the loxP-flanked STOP cassette by Cre recombinase was documented by PCR in DNA from tibiae using specific primers, and the induction of NOTCH3 NICD and Notch target gene expression in tibiae was documented by quantitative reverse transcription (RT)-PCR in tibiae.
Studies were approved by the Institutional Animal Care and Use Committee of UConn Health.
Microcomputed tomography
Femoral microarchitecture was determined using a μCT instrument (Scanco μCT 40, Scanco Medical AG), which was calibrated at periodic intervals with a manufacturer provided phantom (62, 63). Femurs from control and experimental mice were scanned in 70% ethanol at high resolution, energy level of 55 peak kilovoltage, intensity of 145 μA, and integration time of 200 ms as reported (16, 17). A total of 100 slices at midshaft and 80 (for BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 and controls) or 160 (for Notch3−/− and controls) slices at the distal metaphysis were acquired at an isotropic voxel size of 216 μm3 and a slice thickness of 6 μm and chosen for analysis. Cancellous bone volume fraction (bone volume/total volume) and microarchitecture were evaluated starting ∼1.0 mm proximal from the femoral condyles. Contours were manually drawn every 10 slices, a few voxels away from the endocortical boundary, to define the region of interest for analysis, whereas the remaining slice contours were iterated automatically. Total volume, bone volume, bone volume fraction, trabecular thickness, trabecular number, connectivity density, structure model index, and material density were measured in trabecular regions using a Gaussian filter (σ = 0.8) and defined thresholds. A threshold of 240 permil equivalent to 355.5 mg of hydroxyapatite (HA)/cm3 for 1-month-old and a threshold of 260 permil equivalent to 399.1 mg of HA/cm3 for 4-month-old Notch3−/− and wildtype mice and a threshold of 250 permil equivalent to 377 mg HA/cm3 for BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 and control mice were used (62, 63). For analysis of cortical bone, contours were iterated across 100 slices along the cortical shell of the femoral midshaft, excluding the marrow cavity. Analysis of bone volume/total volume, porosity, cortical thickness, total cross-sectional and cortical bone area, segmented bone area, periosteal and endosteal perimeter, and material density, exclusive of cortical pores, and mean polar moment inertia were conducted using a Gaussian filter (σ = 0.8, support = 1) with a threshold of 390 permil equivalent to 682.9 mg of HA/cm3 for 1-month-old Notch3−/−, BGLAP-Cre;RosaNotch3 and Dmp1-Cre;RosaNotch3 and control mice or a threshold of 400 permil equivalent to 704.7 mg of HA/cm3 for 4-month-old Notch3−/− and control mice.
Bone histomorphometry
Bone histomorphometry was carried out in 1-month-old mice injected with calcein 20 mg/kg and demeclocycline 50 mg/kg at a 2-day interval and sacrificed 2 days after demeclocycline administration. For cancellous bone analysis, femurs were dissected, fixed in 70% ethanol, and embedded in methyl methacrylate. Femurs were sectioned at a thickness of 5 μm along the sagittal plane on a Microm microtome (Richards-Allan Scientific) and stained with 0.1% toluidine blue. Static and dynamic parameters of bone morphometry were measured in a defined area between 0.35 and 2.16 mm from the growth plate at a magnification of 100x using an OsteoMeasure morphometry system (Osteometrics). Stained sections were used to draw bone tissue and to measure trabecular separation, number and thickness, and osteoid and eroded surface, as well as to count osteoblast and osteoclast number. To assess for the presence of cartilage remnants in cancellous bone, sections were stained with safranin O and fast green. To assess for the presence of TRAP-positive multinucleated cells, bones were decalcified in 14% ethylenediaminetetraacetic acid for 14 days and embedded in paraffin, and 7-μm sections were stained for the presence of TRAP and counterstained with hematoxylin and analyzed at a 100x magnification using OsteoMeasureXP software. Mineralizing surface per bone surface and mineral apposition rate were measured on unstained sections visualized under UV light and a triple diamidino-2-phenylindole/fluorescein/Texas red set long pass filter, and bone formation rate was calculated (64). For cortical histomorphometry, femurs were cut through the mid-diaphysis and a half was frozen and 7-μm sections were cut using a cryostat (Leica CM1950, Leica Biosystems) to assess dynamic parameters of bone histomorphometry. The other half was decalcified and stained for the presence of TRAP as described for cancellous bone, and stained sections were used to count cell numbers and estimate intracortical remodeling.
Bone marrow stromal cell cultures
Bone marrow stromal cells were obtained from tibiae dissected from 5-week-old Notch3−/− or BGLAP-Cre;RosaNotch3 and control mice. Bone marrow cells were isolated by centrifugation following the removal of the epiphyseal ends and recovered in α-minimum essential medium (α-MEM, Life Technologies). Cells were seeded at a density of 1.25 x 106 cells/cm2 in α-MEM supplemented with 15% heat-inactivated fetal bovine serum (FBS) and grown in an incubator in an atmosphere of 5% CO2 at 37 °C. Cells in suspension were removed by replacing the culture medium 48 h after seeding, and adherent cells were considered bone marrow stromal cells (65). Approximately 5 to 7 days later, adherent cells were digested with trypsin and grown to confluence in α-MEM containing 10% FBS and then switched to α-MEM supplemented with 10% FBS, 100 μg/ml ascorbic acid, and 5 mM β-glycerophosphate (all from Sigma-Aldrich) (66, 67, 68). Culture plates were stained with Alizarin red for visualization of mineralized nodules, or cell extracts were obtained for RNA determinations.
Osteocyte-enriched cell preparations
Osteocyte-enriched cells were obtained from 1-month-old Notch3−/− or BGLAP-Cre;RosaNotch3 and control mice following a modification of a previously described method (48, 69). Tibiae were removed aseptically from 1-month-old experimental and control mice. Tibiae were dissected free from surrounding tissues, the proximal epiphyseal end was excised, and the bone marrow was removed by centrifugation. The distal epiphysis was excised, and bones were digested for 20 min at 37 °C with type II bacterial collagenase pretreated with N-α-tosyl-l-lysyl-chloromethyl ketone hydrochloride and subsequently exposed to 5 mM EDTA for 20 min at 37 °C. The resulting osteocyte-enriched cortical bones were extracted for RNA determinations or cultured overnight in Dulbecco's modified Eagle's medium supplemented with 10% FBS prior to RNA extraction (48, 70).
Osteoblast-enriched cells and cocultures with bone marrow–derived macrophages
To obtain osteoblast-enriched cells, parietal bones from 3- to 5-day-old BGLAP-Cre;RosaNotch3 and littermate wildtype mice were exposed to 1.2 units/ml Liberase TL (Sigma-Aldrich) for 20 min at 37 °C, and cells were extracted in five consecutive reactions (48, 71). Cells from the last three digestions were pooled and seeded at a density of 10,000 cells/cm2 and cultured in Dulbecco’s modified Eagle’s medium supplemented with nonessential amino acids (both from Thermo Fisher Scientific), 20 mM Hepes, 100 μg/ml ascorbic acid (both from Sigma-Aldrich), and 10% heat-inactivated fetal bovine serum (Atlanta Biologicals) in a humidified 5% CO2 incubator at 37 °C. BMMs from wildtype mice were isolated by flushing of the marrow as described (48, 49). Cells were centrifuged, and the sediment was suspended in α-MEM (Thermo Fischer Scientific) in the presence of 10% FBS and recombinant human macrophage colony stimulating factor at 30 ng/ml as described (48, 49). To determine the contribution of osteoblast-derived factors to osteoclast formation, bone marrow stromal cells or calvarial osteoblast-enriched cells from BGLAP-Cre;RosaNotch3 and wildtype control littermates were seeded at a density of 15,700 cells/cm2 in α-MEM in the presence of BMMs from wildtype mice at a density of 47,000 cells/cm2 and cultured in the presence of 1,25-dihydroxyvitamin D3 at 10 nM (16). Cultures were carried out until the formation of multinucleated TRAP-positive cells was achieved. TRAP enzyme histochemistry was conducted using a commercial kit (Sigma-Aldrich), in accordance with the manufacturer’s instructions, as reported (48). TRAP-positive cells containing three or more nuclei were considered osteoclasts.
Quantitative reverse transcription–polymerase chain reaction
Total RNA was extracted from cells with the RNeasy kit (Qiagen) and from homogenized tibiae or osteocyte-enriched preparations with the micro RNeasy kit (Qiagen), in accordance with manufacturer's instructions, as reported (17, 48, 72, 73). The integrity of the RNA from tibiae and osteocyte-rich fragments was assessed by microfluidic electrophoresis on an Experion instrument (Bio-Rad), and only RNA with a quality indicator number equal to or higher than 7.0 was used for subsequent analysis. Equal amounts of RNA were reverse transcribed using the iScript RT-PCR kit (Bio-Rad) and amplified in the presence of specific primers (Integrated DNA Technologies) (Table 7) with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) at 60 °C for 35 cycles. Transcript copy number was estimated by comparison with a serial dilution of cDNA for Alpl (encoding for alkaline phosphatase from American Type Tissue Culture Collection (ATCC), Bglap (encoding for osteocalcin; from J. Lian, University of Vermont), Dkk1 (from Thermo Fisher Scientific), Hes1 (from ATCC), Hey1 and Hey2 (both from T. Iso, Gunma University), HeyL (from D. Srivastava, Gladstone Institute of Cardiovascular Disease or Dharmacon), Notch2 and Sost (from Thermo Fisher Scientific), Notch1 (from J.S. Nye), Notch4 (from Y. Shirayoshi), Tnfsf11 (encoding for RANKL from Source BioScience), Tnfrsf11b (encoding for osteoprotegerin from ATCC), or Wisp1 (from ATCC) (74, 75, 76, 77, 78). Notch3 copy number was estimated by comparison with a serial dilution of ∼100–base pair synthetic DNA template (Integrated DNA Technologies) cloned into pcDNA3 as described, and Notch3 NICD was estimated by comparison with a CMV/N31CHA plasmid (Addgene 47618) (15, 79, 80). Amplification reactions were conducted in a CFX96 qRT-PCR detection system (Bio-Rad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed as copy number corrected for Rpl38 (from ATCC) (81).
Table 7.
Primers used for qRT-PCR
| Gene | Strand | Sequence | GenBank accession number |
|---|---|---|---|
| Alpl | Forward | 5'-TGGTATGGGCGTCTCCACAGTAACC-3' | NM_007431 |
| Reverse | 5'-CTTGGAGAGGGCCACAAAGG-3' | ||
| Bglap | Forward | 5'-GACTCCGGCGCTACCTTGGGTAAG-3' | NM_001037939 |
| Reverse | 5'-CCCAGCACAACTCCTCCCTA-3' | ||
| Dkk1 | Forward | 5'-CCCTCCCTTGCGCTGAAGATGAGGAGT-3' | NM_010051 |
| Reverse | 5'-CGCTTTCGGCAAGCCAGAC-3' | ||
| Hes1 | Forward | 5′-ACCAAAGACGGCCTCTGAGCACAGAAAGT-3′ | NM_008235 |
| Reverse | 5′-ATTCTTGCCCTTCGCCTCTT-3′ | ||
| Hey1 | Forward | 5′-ATCTCAACAACTACGCATCCCAGC-3′ | NM_010423 |
| Reverse | 5′-GTGTGGGTGATGTCCGAAGG-3′ | ||
| Hey2 | Forward | 5′-AGCGAGAACAATTACCCTGGGCAC-3′ | NM_013904 |
| Reverse | 5′-GGTAGTTGTCGGTGAATTGGACCT-3′ | ||
| HeyL | Forward | 5′-CAGTAGCCTTTCTGAATTGCGAC-3′ | NM_013905 |
| Reverse | 5′-AGCTTGGAGGAGCCCTGTTTC-3′ | ||
| HeyLa | Forward | 5'-TCCTCACCCGTCAGA-3′ | NM_013905 |
| Reverse | 5'-AGGCACCATGTAACTCA-3′ | ||
| Notch1 | Forward | 5'-GTCCCACCCATGACCACTACCCAGTTC-3′ | NM_008714 |
| Reverse | 5'-GGGTGTTGTCCACAGGGGA-3′ | ||
| Notch2 | Forward | 5'-TGACGTTGATGAGTGTATCTCCAAGCC-3′ | NM_010928 |
| Reverse | 5'-GTAGCTGCCCTGAGTGTTGTGG-3′ | ||
| Notch3 | Forward | 5′-CCGATTCTCCTGTCGTTGTCTCC-3′ | NM_008716 |
| Reverse | 5′-TGAACACAGGGCCTGCTGAC-3′ | ||
| Notch3NICD | Forward | 5′-CATCCTTATTTGACCCCGTC-3′ | NM_008716 |
| Reverse | 5′-TGGCATTGGTAGCAGTTG-3′ | ||
| Notch4 | Forward | 5′-CCAGCAGACAGACTACGGTGGAC-3′ | NM_010929 |
| Reverse | 5′-GCAGCCAGCATCAAAGGTGT-3′ | ||
| Rpl38 | Forward | 5′-AGAACAAGGATAATGTGAAGTTCAAGGTTC-3′ | NM_001048057; NM_001048058; NM_023372 |
| Reverse | 5′-CTGCTTCAGCTTCTCTGCCTTT-3′ | ||
| Sost | Forward | 5′-AGGAATGATGCCACAGAGGTC-3′ | NM_024449 |
| Reverse | 5′-CTGGTTGTTCTCAGGAGGAGGCTC-3′ | ||
| Tnfrsf11b | Forward | 5′-CAGAAAGGAAATGCAACACATGACAAC-3′ | NM_008764 |
| Reverse | 5′-GCCTCTTCACACAGGGTGACATC-3′ | ||
| Tnfsf11 | Forward | 5′-TATAGAATCCTGAGACTCCATGAAAAC-3′ | NM_011613 |
| Reverse | 5′-CCCTGAAAGGCTTGTTTCATCC-3′ | ||
| Wisp1 | Forward | 5′-TCCAGGAGTTAAGTGATTTGCTCA-3′ | NM_018865 |
| Reverse | 5′-CATGTTACATGACACTGGGCTTC-3′ |
GenBank accession numbers identify transcript recognized by primer pairs.
Used for experiment shown in Figure 3.
Immunoblotting
Bone marrow stromal cells from control or BGLAP-Cre;RosaNotch3 mice were extracted in buffer containing 25 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5% glycerol, 1 mm EDTA, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture (all from Sigma-Aldrich). Quantified total cell lysates (35 μg of total protein) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis in 8% or 10% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore). The blots were probed with anti-NOTCH3 (ab23426) (Abcam) and anti-β-Actin (3700) antibodies (Cell Signaling Technology) and exposed to anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase (Sigma-Aldrich) and incubated with a chemiluminescence detection reagent (Bio-Rad). Chemiluminescence was detected on a ChemiDoc XSR+ molecular imager (Bio-Rad) with Image Lab software (version 6.0.1), and the amount of protein in individual bands was quantified (49).
Statistics
Data are expressed as individual sample values, and means ± SD. All data represent biological replicates except for stromal cell cultures shown in Figure 10 and BMM/osteoblast coculture experiments, which represent technical replicates. Quantitative RT-PCR (qRT-PCR) values were derived from two technical replicates of biological replicates as indicated in the text and legends. Statistical differences were determined by unpaired Student's t test for pairwise comparisons or two-way analysis of variance for multiple comparisons with Holm–Šidák post hoc analysis.
Data availability
Data not shown will be shared upon request to Ernesto Canalis at canalis@uchc.edu.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article. Dr Zanotti is currently employed by Dyne Therapeutics.
Acknowledgments
The authors thank S. Artavaris Tsakonas and S. Fre for Notch3_ICD_YPETG mice, J. Lian for Bglap cDNA, T. Iso for Hey1 and Hey2 cDNAs, D. Srivastava for HeyL cDNA, J. S. Nye for Notch1 cDNA, Y. Shirayoshi for Notch4 cDNA, Madga Mocarska for technical assistance, and Mary Yurczak for secretarial support.
Author contributions
E. C. conceived the work, obtained funding, designed experiments, analyzed data, wrote the manuscript; S. Z. designed and conducted experiments, edited the manuscript; L. S. conducted experiments; T. E. conducted experiments; and J. Y. designed and conducted experiments, edited the manuscript.
Funding and additional information
This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases AR072987 (E. C.) and AR076747 (E. C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Qi-Qun Tang
Footnotes
Present address for Stefano Zanotti: Dyne Therapeutics, Waltham, MA, 02451.
References
- 1.Fortini M.E. Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Siebel C., Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 3.Canalis E. Notch in skeletal physiology and disease. Osteoporos.Int. 2018;29:2611–2621. doi: 10.1007/s00198-018-4694-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanotti S., Canalis E. Notch signaling and the skeleton. Endocr. Rev. 2016;37:223–253. doi: 10.1210/er.2016-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Irizarry C., Carpenter A.C., Weng A.P., Pear W.S., Aster J.C., Blacklow S.C. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon W.R., Zimmerman B., He L., Miles L.J., Huang J., Tiyanont K., McArthur D.G., Aster J.C., Perrimon N., Loparo J.J., Blacklow S.C. Mechanical Allostery: Evidence for a Force Requirement in the proteolytic activation of Notch. Dev. Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeter E.H., Kisslinger J.A., Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 8.Kovall R.A. More complicated than it looks: Assembly of Notch pathway transcription complexes. Oncogene. 2008;27:5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- 9.Nam Y., Sliz P., Song L., Aster J.C., Blacklow S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 10.Wilson J.J., Kovall R.A. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 11.Iso T., Kedes L., Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T., Kageyama R. Expression dynamics and functions of Hes factors in development and diseases. Curr. Top Dev. Biol. 2014;110:263–283. doi: 10.1016/B978-0-12-405943-6.00007-5. [DOI] [PubMed] [Google Scholar]
- 13.Choy L., Hagenbeek T.J., Solon M., French D., Finkle D., Shelton A., Venook R., Brauer M.J., Siebel C.W. Constitutive NOTCH3 signaling Promotes the growth of basal Breast Cancers. Cancer Res. 2017;77:1439–1452. doi: 10.1158/0008-5472.CAN-16-1022. [DOI] [PubMed] [Google Scholar]
- 14.Bai S., Kopan R., Zou W., Hilton M.J., Ong C.T., Long F., Ross F.P., Teitelbaum S.L. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J. Biol. Chem. 2008;283:6509–6518. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- 15.Zanotti S., Canalis E. Parathyroid hormone inhibits Notch signaling in osteoblasts and osteocytes. Bone. 2017;103:159–167. doi: 10.1016/j.bone.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canalis E., Yu J., Schilling L., Yee S.P., Zanotti S. The lateral meningocele syndrome mutation causes marked osteopenia in mice. J. Biol. Chem. 2018;293:14165–14177. doi: 10.1074/jbc.RA118.004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canalis E., Schilling L., Yee S.P., Lee S.K., Zanotti S. Hajdu Cheney mouse mutants exhibit osteopenia, increased osteoclastogenesis and bone resorption. J. Biol. Chem. 2016;291:1538–1551. doi: 10.1074/jbc.M115.685453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukushima H., Nakao A., Okamoto F., Shin M., Kajiya H., Sakano S., Bigas A., Jimi E., Okabe K. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol. Cell Biol. 2008;28:6402–6412. doi: 10.1128/MCB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gripp K.W., Robbins K.M., Sobreira N.L., Witmer P.D., Bird L.M., Avela K., Makitie O., Alves D., Hogue J.S., Zackai E.H., Doheny K.F., Stabley D.L., Sol-Church K. Truncating mutations in the last exon of NOTCH3 cause lateral meningocele syndrome. Am. J. Med. Genet. A. 2015;167A:271–281. doi: 10.1002/ajmg.a.36863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafkas D., Rodilla V., Huyghe M., Mourao L., Kiaris H., Fre S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Xuan S., Bouxsein M.L., von Stechow D., Akeno N., Faugere M.C., Malluche H., Zhao G., Rosen C.J., Efstratiadis A., Clemens T.L. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y., Xie Y., Zhang S., Dusevich V., Bonewald L.F., Feng J.Q. DMP1-targeted Cre expression in odontoblasts and osteocytes. J. Dent. Res. 2007;86:320–325. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 23.Xiong J., O'Brien C.A. Osteocyte RANKL: New insights into the control of bone remodeling. J. Bone Miner. Res. 2012;27:499–505. doi: 10.1002/jbmr.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dallas S.L., Prideaux M., Bonewald L.F. The osteocyte: An endocrine cell and more. Endocr. Rev. 2013;34:658–690. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker E.C., Truong K., McGregor N.E., Poulton I.J., Isojima T., Gooi J.H., Martin T.J., Sims N.A. Cortical bone maturation in mice requires SOCS3 suppression of gp130/STAT3 signalling in osteocytes. eLife. 2020;9:e56666. doi: 10.7554/eLife.56666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagi C.M., Hanson N., Andresen C., Pero R., Lariviere R., Turner C.H., Laib A. The use of micro-CT to evaluate cortical bone geometry and strength in nude rats: Correlation with mechanical testing, pQCT and DXA. Bone. 2006;38:136–144. doi: 10.1016/j.bone.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 27.Zanotti S., Canalis E. Notch1 and Notch2 expression in osteoblast precursors regulates femoral microarchitecture. Bone. 2014;62:22–28. doi: 10.1016/j.bone.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yorgan T., Vollersen N., Riedel C., Jeschke A., Peters S., Busse B., Amling M., Schinke T. Osteoblast-specific Notch2 inactivation causes increased trabecular bone mass at specific sites of the appendicular skeleton. Bone. 2016;87:136–146. doi: 10.1016/j.bone.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Canalis E., Adams D.J., Boskey A., Parker K., Kranz L., Zanotti S. Notch signaling in osteocytes Differentially regulates cancellous and cortical bone remodeling. J. Biol. Chem. 2013;288:25614–25625. doi: 10.1074/jbc.M113.470492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke H.Z., Richards W.G., Li X., Ominsky M.S. Sclerostin and dickkopf-1 as therapeutic targets in bone diseases. Endocr. Rev. 2012;33:747–783. doi: 10.1210/er.2011-1060. [DOI] [PubMed] [Google Scholar]
- 31.Canalis E. Wnt signalling in osteoporosis: Mechanisms and novel therapeutic approaches. Nat. Rev. Endocrinol. 2013;9:575–583. doi: 10.1038/nrendo.2013.154. [DOI] [PubMed] [Google Scholar]
- 32.Holmen S.L., Zylstra C.R., Mukherjee A., Sigler R.E., Faugere M.C., Bouxsein M.L., Deng L., Clemens T.L., Williams B.O. Essential role of beta-catenin in postnatal bone acquisition. J. Biol. Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 33.Wei W., Zeve D., Suh J.M., Wang X., Du Y., Zerwekh J.E., Dechow P.C., Graff J.M., Wan Y. Biphasic and dosage-dependent regulation of osteoclastogenesis by beta-catenin. Mol. Cell Biol. 2011;31:4706–4719. doi: 10.1128/MCB.05980-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albers J., Keller J., Baranowsky A., Beil F.T., Catala-Lehnen P., Schulze J., Amling M., Schinke T. Canonical Wnt signaling inhibits osteoclastogenesis independent of osteoprotegerin. J. Cell Biol. 2013;200:537–549. doi: 10.1083/jcb.201207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otero K., Shinohara M., Zhao H., Cella M., Gilfillan S., Colucci A., Faccio R., Ross F.P., Teitelbaum S.L., Takayanagi H., Colonna M. TREM2 and beta-catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J. Immunol. 2012;188:2612–2621. doi: 10.4049/jimmunol.1102836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sciaudone M., Gazzerro E., Priest L., Delany A.M., Canalis E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology. 2003;144:5631–5639. doi: 10.1210/en.2003-0463. [DOI] [PubMed] [Google Scholar]
- 37.Deregowski V., Gazzerro E., Priest L., Rydziel S., Canalis E. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone Morphogenetic protein signaling. J. Biol. Chem. 2006;281:6203–6210. doi: 10.1074/jbc.M508370200. [DOI] [PubMed] [Google Scholar]
- 38.Zanotti S., Smerdel-Ramoya A., Stadmeyer L., Durant D., Radtke F., Canalis E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149:3890–3899. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de C.B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 40.Feng J.Q., Huang H., Lu Y., Ye L., Xie Y., Tsutsui T.W., Kunieda T., Castranio T., Scott G., Bonewald L.B., Mishina Y. The Dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. J. Dent. Res. 2003;82:776–780. doi: 10.1177/154405910308201003. [DOI] [PubMed] [Google Scholar]
- 41.Moon Y.J., Yun C.Y., Choi H., Kim J.R., Park B.H., Cho E.S. Osterix regulates corticalization for longitudinal bone growth via integrin beta3 expression. Exp. Mol. Med. 2018;50:1–11. doi: 10.1038/s12276-018-0119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cadet E.R., Gafni R.I., McCarthy E.F., McCray D.R., Bacher J.D., Barnes K.M., Baron J. Mechanisms responsible for longitudinal growth of the cortex: Coalescence of trabecular bone into cortical bone. J. Bone Joint Surg. Am. 2003;85:1739–1748. doi: 10.2106/00004623-200309000-00013. [DOI] [PubMed] [Google Scholar]
- 43.Neutzsky-Wulff A.V., Sims N.A., Supanchart C., Kornak U., Felsenberg D., Poulton I.J., Martin T.J., Karsdal M.A., Henriksen K. Severe developmental bone phenotype in ClC-7 deficient mice. Dev. Biol. 2010;344:1001–1010. doi: 10.1016/j.ydbio.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 44.Cho D.C., Brennan H.J., Johnson R.W., Poulton I.J., Gooi J.H., Tonkin B.A., McGregor N.E., Walker E.C., Handelsman D.J., Martin T.J., Sims N.A. Bone corticalization requires local SOCS3 activity and is promoted by androgen action via interleukin-6. Nat. Commun. 2017;8:806. doi: 10.1038/s41467-017-00920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canalis E., Parker K., Feng J.Q., Zanotti S. Osteoblast lineage-specific effects of Notch activation in the skeleton. Endocrinology. 2013;154:623–634. doi: 10.1210/en.2012-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canalis E. The skeleton of lateral meningocele syndrome. Front Genet. 2020;11:620334. doi: 10.3389/fgene.2020.620334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J., Siebel C.W., Schilling L., Canalis E. An antibody to Notch3 reverses the skeletal phenotype of lateral meningocele syndrome in male mice. J. Cell. Physiol. 2020;235:210–220. doi: 10.1002/jcp.28960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanotti S., Yu J., Sanjay A., Schilling L., Schoenherr C., Economides A.N., Canalis E. Sustained Notch2 signaling in osteoblasts, but not in osteoclasts, is linked to osteopenia in a mouse model of Hajdu-Cheney syndrome. J. Biol. Chem. 2017;292:12232–12244. doi: 10.1074/jbc.M117.786129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J., Canalis E. The Hajdu Cheney mutation sensitizes mice to the osteolytic actions of tumor necrosis factor alpha. J. Biol. Chem. 2019;294:14203–14214. doi: 10.1074/jbc.RA119.009824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H.M., Teitelbaum S.L., Ross F.P., Kopan R., Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swiatek P.J., Lindsell C.E., del Amo F.F., Weinmaster G., Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 52.McCright B., Gao X., Shen L., Lozier J., Lan Y., Maguire M., Herzlinger D., Weinmaster G., Jiang R., Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128:491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- 53.Domenga V., Fardoux P., Lacombe P., Monet M., Maciazek J., Krebs L.T., Klonjkowski B., Berrou E., Mericskay M., Li Z., Tournier-Lasserve E., Gridley T., Joutel A. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krebs L.T., Xue Y., Norton C.R., Sundberg J.P., Beatus P., Lendahl U., Joutel A., Gridley T. Characterization of Notch3-deficient mice: Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis. 2003;37:139–143. doi: 10.1002/gene.10241. [DOI] [PubMed] [Google Scholar]
- 55.Krebs L.T., Xue Y., Norton C.R., Shutter J.R., Maguire M., Sundberg J.P., Gallahan D., Closson V., Kitajewski J., Callahan R., Smith G.H., Stark K.L., Gridley T. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 56.Ryeom S.W. The cautionary tale of side effects of chronic Notch1 inhibition. J. Clin. Invest. 2011;121:508–509. doi: 10.1172/JCI45976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J.S., Schroeter E.H., Schrijvers V., Wolfe M.S., Ray W.J., Goate A., Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 58.Duggan S.P., McCarthy J.V. Beyond gamma-secretase activity: The multifunctional nature of presenilins in cell signalling pathways. Cell. Signal. 2016;28:1–11. doi: 10.1016/j.cellsig.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 59.Li K., Li Y., Wu W., Gordon W.R., Chang D.W., Lu M., Scoggin S., Fu T., Vien L., Histen G., Zheng J., Martin-Hollister R., Duensing T., Singh S., Blacklow S.C. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y., Cain-Hom C., Choy L., Hagenbeek T.J., de Leon G.P., Chen Y., Finkle D., Venook R., Wu X., Ridgway J., Schahin-Reed D., Dow G.J., Shelton A., Stawicki S., Watts R.J. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 61.Birling M.C., Dierich A., Jacquot S., Herault Y., Pavlovic G. Highly-efficient, fluorescent, locus directed cre and FlpO deleter mice on a pure C57BL/6N genetic background. Genesis. 2012;50:482–489. doi: 10.1002/dvg.20826. [DOI] [PubMed] [Google Scholar]
- 62.Bouxsein M.L., Boyd S.K., Christiansen B.A., Guldberg R.E., Jepsen K.J., Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 63.Glatt V., Canalis E., Stadmeyer L., Bouxsein M.L. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J. Bone Miner. Res. 2007;22:1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- 64.Dempster D.W., Compston J.E., Drezner M.K., Glorieux F.H., Kanis J.A., Malluche H., Meunier P.J., Ott S.M., Recker R.R., Parfitt A.M. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2013;28:2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bianco P., Gehron R.P. Marrow stromal stem cells. J. Clin. Invest. 2000;105:1663–1668. doi: 10.1172/JCI10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zanotti S., Smerdel-Ramoya A., Canalis E. Reciprocal regulation of Notch and nuclear factor of activated T-cells (NFAT) c1 transactivation in osteoblasts. J. Biol. Chem. 2011;286:4576–4588. doi: 10.1074/jbc.M110.161893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zanotti S., Smerdel-Ramoya A., Canalis E. Nuclear factor of activated T-cells (Nfat)c2 inhibits Notch signaling in osteoblasts. J. Biol. Chem. 2013;288:624–632. doi: 10.1074/jbc.M112.340455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanotti S., Kalajzic I., Aguila H.L., Canalis E. Sex and genetic factors determine osteoblastic differentiation potential of murine bone marrow stromal cells. PLoS One. 2014;9:e86757. doi: 10.1371/journal.pone.0086757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Halleux C., Kramer I., Allard C., Kneissel M. Isolation of mouse osteocytes using cell fractionation for gene expression analysis. Methods Mol. Biol. 2012;816:55–66. doi: 10.1007/978-1-61779-415-5_5. [DOI] [PubMed] [Google Scholar]
- 70.Canalis E., Schilling L., Zanotti S. Effects of sex and Notch signaling on the osteocyte cell Pool. J. Cell. Physiol. 2017;232:363–370. doi: 10.1002/jcp.25433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Canalis E., Zanotti S., Smerdel-Ramoya A. Connective tissue growth factor is a target of Notch signaling in cells of the osteoblastic lineage. Bone. 2014;64:273–280. doi: 10.1016/j.bone.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nazarenko I., Pires R., Lowe B., Obaidy M., Rashtchian A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002;30:2089–2195. doi: 10.1093/nar/30.9.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nazarenko I., Lowe B., Darfler M., Ikonomi P., Schuster D., Rashtchian A. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 2002;30:e37. doi: 10.1093/nar/30.9.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lian J., Stewart C., Puchacz E., Mackowiak S., Shalhoub V., Collart D., Zambetti G., Stein G. Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1143–1147. doi: 10.1073/pnas.86.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iso T., Sartorelli V., Chung G., Shichinohe T., Kedes L., Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol. Cell Biol. 2001;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakagawa O., Nakagawa M., Richardson J.A., Olson E.N., Srivastava D. HRT1, HRT2, and HRT3: A new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev. Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 77.Nye J.S., Kopan R., Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 78.Shirayoshi Y., Yuasa Y., Suzuki T., Sugaya K., Kawase E., Ikemura T., Nakatsuji N. Proto-oncogene of int-3, a mouse Notch homologue, is expressed in endothelial cells during early embryogenesis. Genes Cells. 1997;2:213–224. doi: 10.1046/j.1365-2443.1997.d01-310.x. [DOI] [PubMed] [Google Scholar]
- 79.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 80.Lardelli M., Williams R., Mitsiadis T., Lendahl U. Expression of the Notch 3 intracellular domain in mouse central nervous system progenitor cells is lethal and leads to disturbed neural tube development. Mech. Dev. 1996;59:177–190. doi: 10.1016/0925-4773(96)00589-8. [DOI] [PubMed] [Google Scholar]
- 81.Kouadjo K.E., Nishida Y., Cadrin-Girard J.F., Yoshioka M., St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data not shown will be shared upon request to Ernesto Canalis at canalis@uchc.edu.