Abstract
Objective
Glucose-dependent insulinotropic polypeptide (GIP) and Glucagon-like peptide-1 (GLP-1) are incretin hormones that exert overlapping yet distinct actions on islet β-cells. We recently observed that GIP, but not GLP-1, upregulated islet expression of Transcription Factor 7 (TCF7), a gene expressed in immune cells and associated with the risk of developing type 1 diabetes. TCF7 has also been associated with glucose homeostasis control in the liver. Herein we studied the relative metabolic importance of TCF7 expression in hepatocytes vs. islet β-cells in mice.
Methods
Tcf7 expression was selectively inactivated in adult mouse hepatocytes using adenoviral Cre expression and targeted in β-cells using two different lines of insulin promoter-Cre mice. Glucose homeostasis, plasma insulin and triglyceride responses, islet histology, hepatic and islet gene expression, and body weight gain were evaluated in mice fed regular chow or high fat diets. Tcf7 expression within pancreatic islets and immune cells was evaluated using published single cell RNA-seq (scRNA-seq) data, and in islet RNA from immunodeficient Rag2−/−Il2rg−/− mice.
Results
Reduction of hepatocyte Tcf7 expression did not impair glucose homeostasis, lipid tolerance or hepatic gene expression profiles linked to control of metabolic or immune pathways. Similarly, oral and intraperitoneal glucose tolerance, plasma insulin responses, islet histology, body weight gain, and insulin tolerance were not different in mice with targeted recombination of Tcf7 in insulin-positive β-cells. Surprisingly, islet Tcf7 mRNA transcripts were not reduced in total islet RNA containing endocrine and associated non-endocrine cell types from Tcf7βcell−/− mice, despite Cre-mediated recombination of islet genomic DNA. Furthermore, glucose tolerance was normal in whole body Tcf7−/− mice. Analysis of scRNA-seq datasets localized pancreatic Tcf7 expression to islet progenitors during development, and immune cells, but not within differentiated islet β-cells or endocrine lineages within mature islets. Moreover, the expression of Tcf7 was extremely low in islet RNA from Rag2−/−Il2rg−/− mice and, consistent with expression within immune cells, Tcf7 was highly correlated with levels of Cd3g mRNA transcripts in RNA from wild type mouse islets.
Conclusions
These findings demonstrate that Tcf7 expression is not a critical determinant of glucose homeostasis in mice. Moreover, the detection of Tcf7 expression within islet mRNA is attributable to the expression of Tcf7 RNA in islet-associated murine immune cells, and not in islet β-cells.
Keywords: Islets, Liver, Glucose, Immune cells, Transcription factors, Knockout mice
Graphical abstract
Highlights
•Reduction of hepatocyte Tcf7 does not impair glucose homeostasis.
•Targeting beta cell Tcf7 using insulin-promoter-Cre does not reduce islet Tcf7 expression.
•RNA-seq localizes pancreatic Tcf7 to islet progenitors and lymphocytes.
•Tcf7 expression is markedly reduced in islet RNA from Rag2−/−Il2rg−/− mice.
1. Introduction
The control of blood glucose requires extensive communication between multiple organs, including the central nervous system, liver, muscle, adipose tissue, and islets, coupling continuous glucose sensing to regulatory mechanisms that modify glucose production and disposal. Although multiple tissues and mechanisms contribute to this process, the islets of Langerhans represent an essential glucoregulatory organ, coupling small changes in glycemia in the fasted vs. fed states to coordinate regulation of insulin and glucagon secretion. The importance of islet cells for glucose control is further supported by human genetic studies that establish a dominant role for genetic variation within genes expressed in pancreatic islets as an important determinant of the susceptibility to type 2 diabetes (T2D) [1].
Although glucose is an independent and critical signal for β-cell and α-cell function, gut hormones secreted in response to food ingestion amplify glucose-dependent pathways controlling insulin and glucagon secretion. The gut hormone Glucose-dependent Insulinotropic Polypeptide (GIP) is secreted rapidly from enteroendocrine K cells in the proximal gut following meal ingestion, and potentiates glucose-dependent insulin secretion, consistent with its description as an incretin hormone. Pharmacological GIP administration also modulates α-cell function, enhancing glucagon secretion under conditions of hypoglycemia, but not hyperglycemia [2]. These islet actions of GIP are physiologically important, as whole-body inactivation [3] or selective disruption of GIP action in murine β-cells [4] impairs incretin action and β-cell function. Consistent with these findings, transient blockade of GIP activity in humans using a GIP Receptor (GIPR)-selective antagonist similarly impairs glucose homeostasis [5].
A second incretin hormone, Glucagon-Like Peptide-1 (GLP-1), also potentiates glucose-dependent insulin secretion in animals and humans [6]. GLP-1 further inhibits glucagon secretion, delays gastric emptying, and reduces food intake, actions conserved in people with T2D [7]. Collectively, these actions supported the development of multiple GLP-1 receptor agonists for the treatment of T2D. Moreover, there remains intense interest in understanding whether dual GIPR-GLP-1R co-agonism, as exemplified by the activity of tirzepatide, represents a promising new approach for the treatment of T2D, obesity, and non-alcoholic steatohepatitis [8,9].
The resurgent interest in understanding the biological activity of GIP-based co-agonists [8], has fostered studies directed at comparing how GIP vs. GLP-1 act via their structurally-related receptors to produce overlapping yet distinct complementary actions. Within the islets, analysis of incretin action has demonstrated that GLP-1 activates β-cell signal transduction via both Gq and Gs, whereas GIP only activates Gs [10]. Differential incretin control of β-cell function may also reside at the level of gene transcription. Transcription factor 7 (TCF7) is a GIP-regulated transcription factor recently postulated to have a metabolic role in liver and β-cells [4,11]. Our previous work showed that Tcf7 expression is markedly reduced in islets from diabetic db/db mice and humans with T2D [4]. Indeed, TCF7 expression has been detected by several groups, using RNA-seq, in mouse and human islet RNA, and relative islet TCF7 expression correlates with HbA1c in humans [12,13]. Moreover, we demonstrated that GIP, but not GLP-1, activates Tcf7/TCF7 expression in murine and human islets, whereas knockdown or genetic inactivation of Tcf7 expression increased the susceptibility to apoptotic β-cell injury [4]. Intriguingly, genetic variation within the TCF7 locus is also linked to the development of type 1 diabetes [14,15].
Interpreting the putative importance of TCF7 for metabolic homeostasis is complicated by findings that changes in both hepatic and islet Tcf7 expression are linked to control of glucose homeostasis. Indeed, reduction of hepatic miR-22-3p expression leads to enhanced Tcf7 expression, associated with reduced expression of genes controlling gluconeogenesis and decreased hepatic glucose output in mice [11]. To resolve the competing actions of TCF7 in liver vs. islet β-cells, we have now examined the phenotypes of mice with genetic targeting of Tcf7 in hepatocytes vs. β-cells. Surprisingly, reduction of hepatocyte Tcf7 expression had no impact on glucose homeostasis. Moreover, we were unable to detect dysregulated glucose homeostasis or meaningful reduction of islet Tcf7 expression in Tcf7fl/fl mice expressing Cre-recombinase under the control of the insulin gene promoter. Consistent with findings implying that islet cell-associated Tcf7 expression is not attributable to β-cells, islets from Rag2−/−Il2rg−/− mice, that are devoid of T-cells, B-cells and natural killer cells, have markedly reduced Tcf7 expression. Collectively, we conclude that TCF7 does not play an important role in murine glucose homeostasis, and attribute localization of Tcf7 expression in isolated islet-associated cells predominantly to non-endocrine immune cells.
2. Material and methods
2.1. Animals
Mice were housed in the Toronto Centre for Phenogenomics under a 12-h light/dark cycle and fed a regular chow diet (RCD – 18% kcal fat, 2018 Harlan Teklad, Mississauga, ON, Canada) or a high-fat diet (HFD – 45% kcal fat, 35% kcal carbohydrate, 0.05% wt/wt cholesterol, D12451i, Research Diets, New Brunswick, NJ, USA). To induce hepatosteatosis and metabolic dysfunction, mice were fed a High Fat/High Fructose/High Cholesterol (HF/HFr/HC) diet (40% kcal Fat, 20% kcal Fructose, 2% wt/wt Cholesterol, D09100301, Research Diets) [16]. All mice had ad libitum access to water and food, unless otherwise noted. Animal experiments were approved by the Animal Care and Use Subcommittee at the Toronto Centre for Phenogenomics, Mt. Sinai Hospital. Mice were fasted for 5 h and gently restrained but not anesthetized during metabolic tests. Mice were euthanized by CO2 inhalation at the end of study and blood was collected through cardiac puncture. Tissues were snap frozen in liquid nitrogen and stored at −80 °C until the time of analysis.
Floxed Tcf7 (Tcf7tm1a (EUCOMM)Wtsi) mice (Tcf7fl/fl) were obtained from Dr. Hai-Hui Xue, at the University of Iowa. The LoxP sites flank exon 4, and Cre recombinase-mediated excision of these sequences results in a nonsense frame-shift mutation [17]. To generate hepatocyte-specific Tcf7 knockout mice (Tcf7Hep−/−), Tcf7fl/fl were injected (i.v.) with 1.5 × 1011 genome copies per mouse of AAV8.TBG.PI.eGFP.WPRE.bGH (AAV-GFP; control virus) or AAV8.TBG.PI.Cre.rBG (AAV-Cre; Cre driven by Thyroxine Binding Globulin promoter resulting in specific expression in and targeting of hepatocytes, and not the non-parenchymal cells in the liver) [18]. Both AAV8 constructs were obtained from the University of Pennsylvania Vector Core Lab. Wild-type (WT) mice injected with AAV-Cre and Tcf7fl/fl mice injected with AAV-GFP viral particles were used as controls. All viral injections were administered in 8–10 week old mice. Mice were then transferred to HF/HFr/HC diet (2 weeks post-injection) for a total of 12 weeks. Metabolic phenotyping was done after 1 week on RCD, and after 4 and 12 weeks on the HF/HFr/HC diet and mice were euthanized ∼20 weeks post AAV injection (Supplementary Fig. 1A).
To generate Tcf7βcell−/−mice, Tcf7fl/fl mice were crossed with B6.Cg-Tg (Ins1-cre/ERT)1Lphi/J mice, expressing a tamoxifen-inducible Cre driven by the mouse insulin promoter (MIP-Cre; obtained from Dr. Louis Philipson's lab) [19] or C57BL/6J mice x CBA/J F2-Tg (Ins2-cre)23Herr, expressing constitutive Cre recombinase under the Rat Insulin II promoter (RIP-Cre; obtained from Dr. Pedro Luis Herrera) [20]. To induce the expression of MIP-Cre, six-week-old mice were injected intraperitoneally with tamoxifen (40 mg/kg) for five consecutive days. Whole-body Tcf7 knockout mice (Tcf7−/−) were provided by Dr. Hans Clevers [21]. Whenever possible, we performed experiments in all four genotypes (WT, Cre, Tcf7fl/fl and Tcf7βcell−/−mice). All mice were fed a RCD and metabolic analyses performed at 8, 18 and 35 weeks of age. To induce metabolic dysfunction, mice were fed a HFD starting at 8 weeks of age, for a total of 20 weeks, and metabolic phenotyping was performed at 10 and 20 weeks post HFD feeding. Mice were sacrificed at ∼30 weeks of age. Immunodeficient, C; 129S4-Rag2tm1.1Flv Il2rgtm1.1Flv/J (Rag2−/−Il2rg−/−) mice [22] were obtained from Jackson Laboratories (Stock #014593).
2.2. Glucose, insulin, and lipid tolerance tests
Mice were administered a gavage of glucose (1.5 mg/kg) orally (oral glucose tolerance test [oGTT]) or intraperitoneally (ipGTT) after a 5 h fast. For insulin tolerance tests (ITT), mice were fasted for 5 h and injected intraperitoneally with human insulin (Humalog, Lilly) at a dose of 0.5 U/kg for RCD fed mice and 0.7 U/kg for HFD fed mice. Blood glucose levels were measured in tail vein blood at the indicated time points with a handheld glucometer (Contour glucometer, Bayer Healthcare, Toronto, Canada). For oral lipid tolerance tests, mice were given an oral gavage of 200 μL of olive oil after a 5-h fast, and tail vein blood was collected at 0, 1, 2, and 3 h post gavage in heparin-coated capillary tubes to measure plasma triglyceride (TG) levels.
2.3. Body weight and body composition
Body weights were measured at regular time-points and body composition was assessed using EchoMRI quantitative nuclear magnetic resonance (Echo Medical Systems, Houston, TX).
2.4. Blood collection and metabolic assays
Tail-vein blood samples were collected in Lithium-heparin coated capillary Microvette tubes (Sarstedt, Numbrecht, Germany) and plasma isolated by centrifugation (13,000 rpm at 4 °C, 5 min). Blood samples were collected during metabolic tests at indicated time points. Plasma insulin levels were analyzed using an Ultrasensitive Insulin ELISA kit (Alpco Diagnostics) and plasma TG levels were measured using an enzymatic assay (#11877771 216, Roche Diagnostics) and the calibrator 464–01601 (Wako, Mountain View, CA).
2.5. Mouse hepatocyte isolation and non-parenchymal cell (NPC) fractions
Mouse hepatocytes were isolated as described previously [23] and non-parenchymal cell (NPC) fractions were purified as described [18,24]. Both fractions were used for RNA analysis.
2.6. Mouse pancreatic islet isolation
Mice were sacrificed by CO2 inhalation, and the pancreas was distended by injecting collagenase type V solution (0.8 mg/mL in HBSS, Sigma–Aldrich) through the pancreatic duct (after clamping the bile duct) as previously described [25]. The pancreas was then excised and digested for 10–12 min at 37 °C, followed by vigorous shaking and subsequent washes with cold RPMI (with 0.025% BSA). Islets were then separated by a Histopaque gradient, and handpicked under a microscope, for RNA isolation.
2.7. Genomic DNA isolation and PCR
For genotyping, genomic DNA was isolated by heating ear punches in 50 mM NaOH at 95 °C for 10 min and then adding 10% volume of 1 M Tris pH 8.0. Genomic DNA from other mouse tissue samples was isolated using the DNeasy Blood and Tissue kit (Qiagen), according to the manufacturer's protocol. Polymerase chain reaction (PCR) was performed using primers for the Tcf7 floxed allele (Forward primer: agctgagcccctgttgtaga, Reverse primer: caacgagctgggtagaggag) using standard PCR.
2.8. Gene expression analysis
Total RNA was isolated from tissue by homogenizing samples in Tri Reagent (Molecular Research Center, OH, USA) using a TissueLyser II system (Qiagen, Germantown, MD, USA). For pancreatic islets, RNA was isolated using an RNeasy Mini Kit (Qiagen), because of small sample size and low yield of RNA. First strand cDNA synthesis was carried out using DNase I treated total RNA (0.25–2 μg), SuperScript III and random primers (Thermo Fisher). Quantitative PCR (qPCR) for quantifying gene expression levels was carried out on a QuantStudio System, using TaqMan Gene expression Master Mix and Assays (Thermo Fisher Scientific). Gene expression was analyzed using the 2-ΔΔCt method and expression levels were normalized to Tbp, as indicated in Figure legends. qPCR primer assay ID numbers (Thermo Fisher Scientific) are provided in Supplementary Table 1.
2.9. Histological analysis
Mouse tissues were isolated and fixed in formalin for 24 h, paraffin-embedded, sectioned and slide mounted. Slides were deparaffinised through xylenes and a series of graded alcohol solutions and taken to water. Antigen retrieval was performed using HIER with Tris–EDTA buffer (pH 9) or Citrate buffer (min 6) for 7 min. Endogenous peroxidase activity was blocked in Bloxall reagent (Agilent) for 12 min. Non-specific antibody binding was blocked using Vector ImmPRESS blocking reagent (Cat # MP-7401, Vector Laboratories) for 20 min at room temperature. Sections were then incubated for 1 h at room temperature in either Rabbit anti- TCF1/TCF7 (C63D9) (Cat # 2203, Cell Signaling Technology) diluted at 1:150 in antibody diluent (Agilent) or Rabbit Anti-Insulin (Cat # ab181547, Abcam) diluted to 1:30,000. Sections were then incubated in ImmPRESS peroxidase Polymer Anti-Rabbit IgG reagent (Cat # MP-7401, Vector Laboratories) for 30 min at room temperature. Positive immunoreactive staining was visualized using Steady DAB/Plus (Cat # ab103723, Abcam). Mayer's hematoxylin was used as a counterstain prior to dehydrating and mounting coverslips. Liver sections were stained with hematoxylin and eosin using standard protocols. Immunohistochemistry, hematoxylin and eosin staining, and slide scanning were performed by the Pathology Services facility at the Lunenfeld-Tanenbaum Research Institute. β-cell area in the pancreatic sections was quantified by positive pixel count analysis using Aperio ImageScope Viewer Software (Leica Biosystems). Weak positive pixels were discounted to minimize the effects of non-specific staining.
2.10. Single-cell RNA sequencing data analysis
Published single-cell RNA sequencing (scRNA-seq) data matrices were obtained as follows. Adult mouse islet data were obtained from GSE84133 [26] and GSE109774 [27]. Embryonic mouse pancreas data spanning E12.5, E14.5, E15.5, E17.5, and E18.5 were obtained from GSE101099 [28] and GSE120522 [29]. Isolated human islet data were obtained from E-MTAB-5061 [30], GSE81547 [31] and GSE124742 [32]. Human embryonic stem cell (hESC)-derived β cell data spanning stage 0 to stage 7 of differentiation were obtained from GSE143783 [33]. All matrices were processed in Seurat 3.2.2 [34]. Low quality mouse cells and human cells (defined by mitochondria transcripts >5% and >10% of total transcripts, respectively) were filtered out, and filtered data were scaled using SCTransform and visualized by UMAP (uniform manifold approximation and projection).
2.11. Western blot analysis
Total protein lysates were prepared from frozen thymus tissue extracts from Tcf7+/+ and Tcf7−/− mice using RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS) containing protease inhibitor cocktail (Complete mini Protease inhibitor cocktail, Roche). Forty micrograms of protein per lane were resolved on a 10% SDS-PAGE gel and transferred to nitrocellulose membranes. Membranes were blocked with 5% skim milk for 1 h at room temperature and probed with TCF7 primary antibody at 1:1000 dilution (TCF1/TCF7 (C63D9) Rabbit mAb #2203, Cell Signaling Technology) overnight at 4 °C. After incubating with secondary antibody (1:2000 dilution) for 1 h at room temperature, immunoreactive bands were detected using SuperSignal West Pico Chemiluminescent assay (Thermo Fisher Scientific) in a ChemiDoc XRS + imager (Bio-Rad). GAPDH (#2118, Cell Signaling Technology) was used as a loading control.
2.12. Statistical methods
Results are expressed as mean ± standard deviation (SD), except for the area under the curve (AUC) where data is expressed as mean ± Standard Error of the Mean (SEM). Statistical analysis was performed using Graph Pad Prism 9 (San Diego, CA, USA). Comparisons were made by i) one-way ANOVA followed by Tukey post hoc, ii) two-way ANOVA followed by a Sidak post hoc, or iii) unpaired Student's t tests, where applicable. For correlation analysis, Pearson's correlation coefficients, r, and confidence intervals were calculated. P values were computed using two-tailed t-tests and goodness of fit was assessed by R2 values, calculated using linear regression. Statistically significant differences are indicated as ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001.
3. Results
3.1. Hepatocyte-specific deletion of Tcf7 in adult mice does not perturb glucose metabolism or insulin tolerance
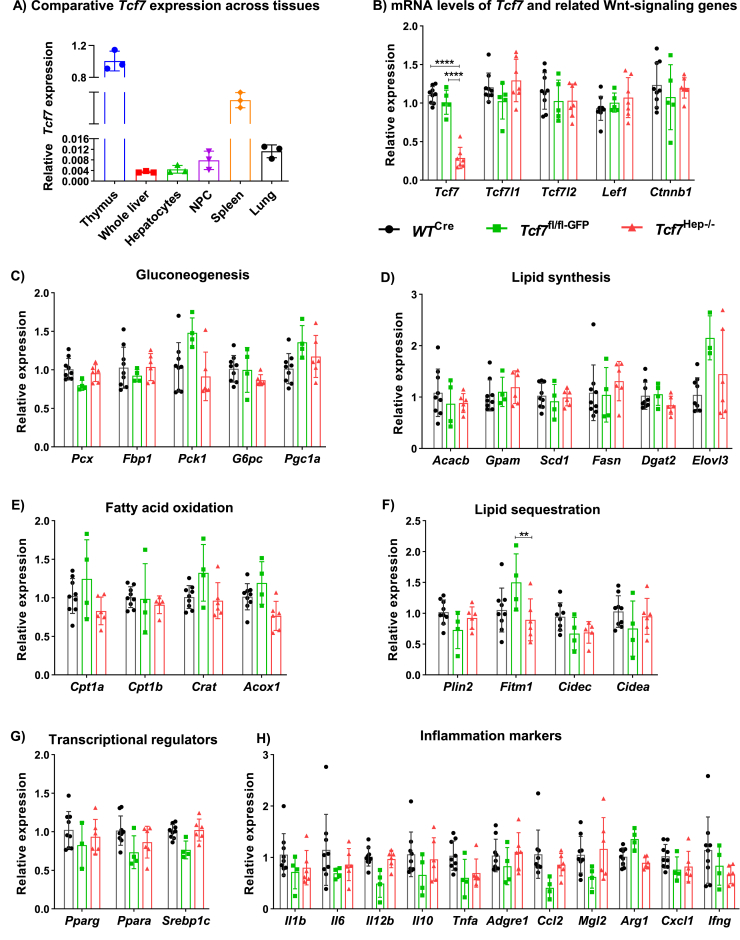
Previous studies examining Tcf7 biology within the liver correlated miR-22-3p-regulated Tcf7 expression with control of hepatic gluconeogenesis in diabetic db/db mice [11]. Given the central importance of hepatocytes for control of gluconeogenesis, we investigated if endogenous hepatocyte Tcf7 plays a direct role in regulating liver and whole-body glucose metabolism. To generate mice with selective inactivation of Tcf7 in hepatocytes (Tcf7Hep−/−), we injected adeno-associated virus (AAV) expressing Cre recombinase driven by the thyroxine-binding globulin (Tbg) promoter (AAV-Tbg-Cre) into Tcf7fl/fl mice as previously described [18,35]. For controls, we injected AAV-Tbg-Cre into WT mice (WTCre) or AAV-Tbg-GFP into Tcf7fl/flmice (Tcf7fl/fl-GFP) (Figure 1A and Fig. S1A). Levels of Tcf7 mRNA transcripts were 70% lower in RNA from whole liver and isolated hepatocytes from Tcf7Hep−/− vs. control mice, however Tcf7 expression in the NPC liver fraction, as well as in the spleen and thymus, was not reduced (Figure 1B). Oral and intraperitoneal glucose tolerance was unchanged in Tcf7Hep−/− mice on a RCD several days after AAV-induced reduction of hepatocyte Tcf7 expression (Figure 1C,D). Moreover, plasma insulin levels in response to oral glucose (Fig. S1B) and insulin tolerance after exogenous insulin administration (Figure 1E) were not different in mice with hepatocyte-selective reduction of Tcf7 expression.
Figure 1.
Hepatocyte specific deletion of Tcf7 does not perturb glucose homeostasis. A) Schematic representation of Cre-LoxP strategy to generate hepatocyte-specific Tcf7 knockout (Tcf7Hep−/−) using adeno-associated virus (AAV) expressing Cre recombinase driven by the thyroxine-binding globulin (Tbg) promoter in eight-week-old mice. B)Tcf7 mRNA expression (relative to Tbp) in whole liver, isolated hepatocytes, isolated liver non-parenchymal cell (NPC) fraction, spleen, and thymus tissue (n = 3 per group) sampled from Floxed Tcf7 mice (Tcf7fl/fl) injected with either AAV-Tbg-GFP (green, Tcf7fl/fl-GFP) or AAV-Tbg-Cre (red, Tcf7Hep−/−) and after 12 weeks of HF/HFr/HC diet. Glucose excursion in 5-h fasted mice during C) Oral glucose tolerance test (oGTT, n = 9–10 per group), D) Intraperitoneal glucose tolerance (ipGTT, n = 4 per group), and E) Insulin tolerance test (ITT, n = 4–5 per group) after 1–2 weeks of AAV injection in RCD-fed mice (black, WTCre; green, Tcf7fl/fl-GFP; red, Tcf7Hep−/−). Glucose excursion in 5-h fasted mice during F) oGTT (n = 8–9 per group), G) ipGTT (n = 7–9 per group), and H) ITT (n = 6–9 per group) after 4 weeks on HF/HFr/HC diet. Glucose excursion in 5-h fasted mice during I) oGTT (n = 7–9 per group, J) ipGTT (n = 7–9 per group), and K) ITT (n = 7–9 per group) after a total of 12 weeks on HF/HFr/HC diet (and a total of 14–16 weeks after AAV injection). Insets depict area under the curve (AUC) analysis of glucose excursions during different metabolic tests. Data are represented as mean ± standard deviation (SD) except for AUC analysis, where data are represented as mean ± standard error of the mean (SEM). Statistical significance was calculated using Student's t-test (panel B) or One-way ANOVA with Tukey's correction for multiple comparisons (panel C–K). ∗p ≤ 0.05, ∗∗∗p ≤ 0.001.
We subsequently examined the contribution of hepatocyte Tcf7 to glucose homeostasis after HF/HFr/HCD feeding for 4 and 12 weeks (Fig. S1A). Four weeks of HF/HFr/HC diet feeding did not impair glucose tolerance or the glycemic response to exogenous insulin in Tcf7Hep−/− vs. control mice (Figure 1F–H). Moreover, glucose-stimulated plasma insulin levels were not different after 4 weeks of HF/HFr/HC diet feeding (Fig. S1C). After prolonged exposure to the HF/HFr/HC diet for 12 weeks, Tcf7Hep−/− exhibited similar body weight and adiposity compared to littermate control mice (Figs. S1D and E) and the glycemic responses to oral and intraperitoneal glucose or insulin were not different in Tcf7Hep−/− vs. control mice (Figure 1I–K). Mice fed a HF/HFr/HC diet exhibited higher levels of i) fasting glucose, ii) basal and glucose-stimulated plasma insulin levels, and iii) glycemic excursions during oGTT, compared to mice fed a RCD (Figs. S1F–H). However, plasma insulin levels were comparable across all three genotypes during the oGTT (Fig. S1I). Taken together, these data show that reduced Tcf7 expression in hepatocytes does not impair glucose metabolism in RCD- or HF/HFr/HC diet-fed mice.
3.2. Hepatocyte-specific deletion of Tcf7 does not alter expression of genes regulating metabolism or inflammation
Basal levels of hepatic Tcf7 expression were lower relative to corresponding mRNA levels in the thymus, spleen, and lung tissue, organs enriched for immune cell populations (Figure 2A). Tcf7 mRNA levels were reduced in whole liver from Tcf7Hep−/− mice (Figure 2B); however, there were no changes in the mRNA levels of several related transcription factors including Tcf7l1, Tcf7l2 Lef1 or β-catenin (Ctnnb1) in the liver of Tcf7Hep−/− mice. We subsequently examined whether selective hepatocyte depletion of Tcf7 dysregulated the expression of genes important for gluconeogenesis or lipid metabolism. The relative expression of genes important for gluconeogenic or lipid metabolism pathways was not different in Tcf7Hep−/− mice after 20 weeks of HF/HFr/HC diet (Figure 2C–G). Consistent with these findings, TG excursion after oral olive oil gavage was similar in Tcf7Hep−/− mice (Fig. S1J) and plasma TG levels were similar across genotypes in fasted or re-fed animals (Fig. S1K). Organ weights (normalized to body weight) were comparable across genotypes (Fig. S1L). Although exposure to a HF/HFr/HC diet induces hepatic inflammation [16], no differences in levels of liver mRNA transcripts corresponding to genes acting within inflammatory pathways were observed in Tcf7Hep−/− mice (Figure 2H). Histological analysis of the liver revealed similar levels of hepatic fat deposition and histological appearance after HF/HFr/HC feeding in Tcf7Hep−/− vs. control mice (Fig. S1M). Moreover, we did not observe any compensatory changes in the relative expression of Tcf7, Tcf7l1, Tcf7l2, Lef1 and Ctnnb1 in adipose tissue (inguinal and epididymal fat depots, Figs. S2A and S2B) from Tcf7Hep−/− mice. Collectively, these findings show that the loss of hepatic Tcf7 does not impair transcriptional programs controlling glucose or lipid homeostasis.
Figure 2.
Gene expression analysis in tissues from Tcf7Hep−/− mice fed a HF/HFr/HC diet for 12 weeks. A) Comparative mRNA levels of Tcf7 (relative to Tbp) in different tissues from Tcf7Hep+/+ mice (n = 3). B) mRNA levels (relative to Tbp) of Tcf7, Tcf7l1, Tcf7l2, Lef1, and Ctnnb1 in whole liver tissue from WTCre (black), Tcf7fl/fl-GFP (green) or, Tcf7Hep−/− (red) mice (n = 5–9 per group). (C–H) mRNA levels (relative to Tbp) of genes related to C) Gluconeogenesis, D) Lipid synthesis, E) Fatty acid oxidation, F) Lipid sequestration, G) Transcriptional regulation and H) Inflammation in liver after AAV-Tbg-GFP or AAV-Tbg-Cre injection in mice fed a HF/HFr/HC diet for 12 weeks post AAV injection (black, WTCre, n = 9; green, Tcf7fl/fl-GFP n = 4; red, Tcf7Hep−/−, n = 6). Data are presented as mean ± SD and analyzed using One-way ANOVA with Tukey's correction for multiple comparisons for the indicated groups. ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001.
3.3. Mice with targeting of Tcf7 in adult β-cells exhibit normal glucose homeostasis
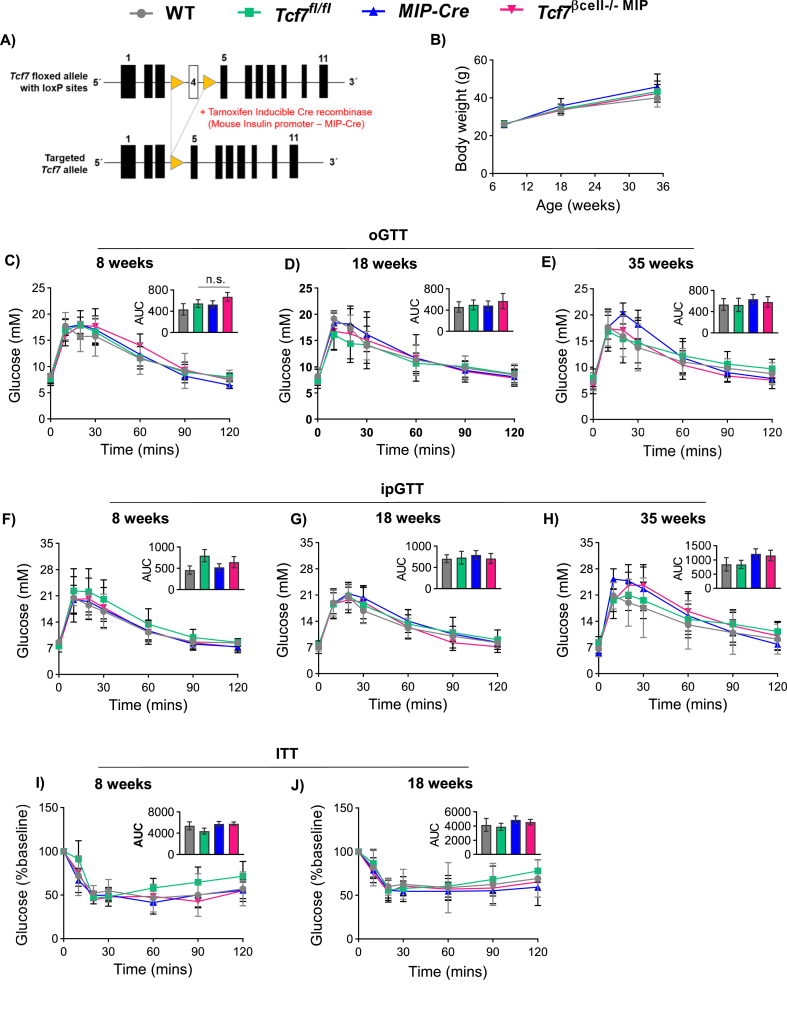
We previously observed reduced levels of Tcf7 expression in islet RNA isolated from diabetic db/db mice and from humans with T2D [4]. Moreover, Tcf7−/− mice exhibited impaired glucose tolerance and defective glucose-stimulated insulin secretion, leading us to question whether loss of Tcf7 impairs β-cell function. Accordingly, we used mice expressing tamoxifen-inducible Cre recombinase under the control of the mouse insulin promoter (MIP Cre) to target Tcf7 in adult pancreatic β-cells of Tcf7βcell−/− MIP mice analyzed from 8 to 35 weeks of age (Figure 3A and Fig. S3A). Tcf7βcell−/−MIP and control mice exhibited similar body weights, body composition (Figure 3B, Fig. S3B), and comparable responses to oral or intraperitoneal glucose at 8, 18 and 35 weeks of age on a RCD (Figure 3C–H). Similarly, the plasma insulin responses to oral glucose were not different in Tcf7βcell−/−MIP mice (Figs. S3C–E). Furthermore, the glycemic response to exogenous insulin assessed after 8 and 18 weeks of RCD feeding was not different (Figure 3I,J), consistent with similar sensitivity to insulin and comparable β-cell area across genotypes (Figs. S3F–G). Organ weights were also similar in Tcf7βcell−/−MIP vs. control mice (Fig. S3H).
Figure 3.
Targeting of β-cell Tcf7 does not impair glucose tolerance. A) Schematic representation of Cre-LoxP strategy to generate pancreatic β-cell specific Tcf7 knockout (Tcf7βcell−/−MIP). Mice expressing Tamoxifen-inducible Cre driven by the mouse insulin promoter (MIP-Cre) were mated with Floxed-Tcf7 (Tcf7fl/fl) mice, giving rise to four genotypes (WT, grey; Tcf7fl/fl, green; MIP-Cre, blue; Tcf7βcell−/−MIP, pink). B) Mouse body weights compared across genotypes at 8, 18, and 35 weeks of age on chow diet (n = 7–12 per group). (C–J) Metabolic phenotyping at 8, 18, and 35 weeks of age in mice belonging to all four genotypes. Glucose excursions in 5-h fasted mice during oGTT at C) 8 weeks (n = 7–12 per group), D) 18 weeks (n = 11–15 per group), and E) 35 weeks (n = 9–10 per group) of age. Glucose excursions in 5-h fasted mice during intraperitoneal glucose tolerance tests (ipGTT) at F) 8 weeks (n = 7–12 per group), G) 18 weeks (n = 7–9 per group), and H) 35 weeks (n = 6–9 per group) of age. Glucose excursions in 5-h fasted mice during insulin tolerance tests (ITT) at I) 8 weeks (n = 5–7 per group), and J) 18 weeks of age (n = 6–8 per group). Insets depict AUC analysis. Data are presented as mean ± SD, except AUC graphs which are presented as mean ± SEM, and analyzed using one-way ANOVA with Tukey's correction for multiple comparisons.
To independently verify the results obtained using MIP-Cre for targeting of Tcf7 in β-cells, we generated Tcf7βcell−/− RIP mice using an alternate constitutive Cre driver, RIP-Cre (Fig. S4A), expressed earlier (from E13-E15) during β-cell development [36]. Consistent with our findings using MIP-Cre (Figure 3), Tcf7βcell−/− RIP mice did not reveal any phenotypic differences vs. control animals (WT and RIP-Cre). Specifically, Tcf7βcell−/− RIP mice displayed comparable body weights, glucose tolerance (oral), and plasma insulin levels (fasting and in response to oral glucose) at 8, 18, and 35 weeks of age (Figs. S4B–H). Analysis of pancreatic histology revealed comparable β-cell area in Tcf7βcell−/−RIP vs. RIP-Cre mice (Figs. S4I–J). In conclusion, attempts to inactivate β-cell expression of Tcf7 do not impair β-cell function in RCD-fed Tcf7βcell−/− RIP mice.
3.4. High fat diet feeding does not differentially impair glucose or insulin tolerance in Tcf7βcell−/− MIP vs. control mice
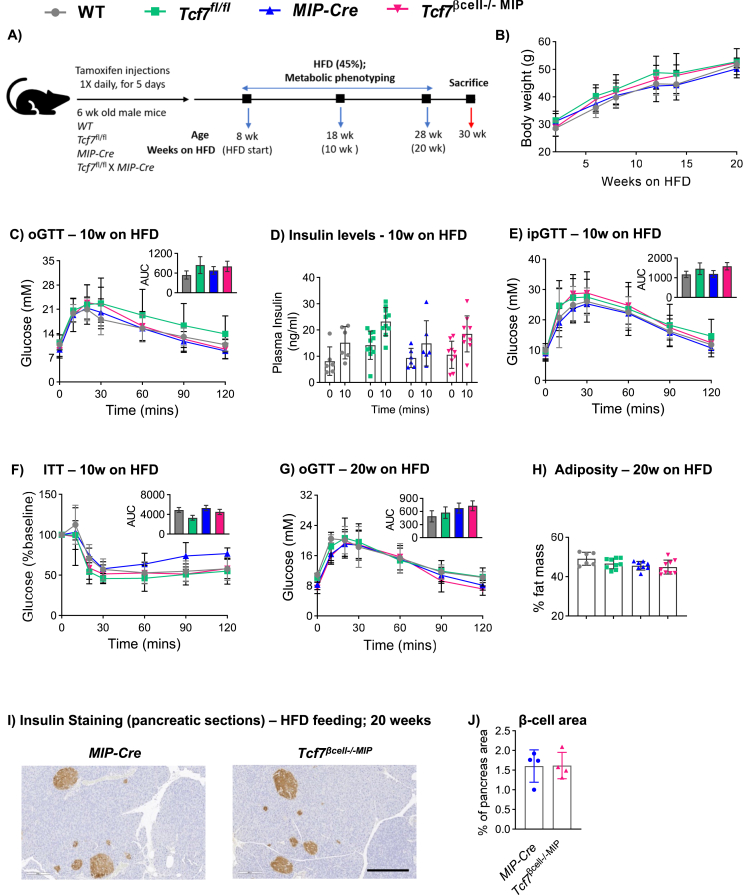
To examine the putative importance of Tcf7 for β-cell function under conditions of metabolic stress, we fed Tcf7βcell−/− MIP mice and littermate controls a HFD for 22 weeks (Figure 4A). Tcf7βcell−/− MIP mice gained weight to a similar extent as controls (Figure 4B). After 10 weeks of HFD feeding, glucose excursions during oGTT or ipGTT were similar and plasma insulin levels (fasting and in response to oral glucose) were comparable in Tcf7βcell−/− MIP vs. littermate control mice (Figure 4C–E). Consistent with exposure to a diabetogenic diet, mice on the HFD displayed elevated fasting glucose levels, increased plasma insulin levels following a glucose challenge, and elevated AUC glucose during glucose tolerance testing compared to mice fed a RCD (Figs. S5A–C). Furthermore, glucose levels were similar across genotypes during insulin tolerance testing (Figure 4F). Moreover, no glycemic differences were observed after 20 weeks of HFD feeding (Figure 4G), and body adiposity (Figure 4H), organ weights (Fig. S5D) and β-cell area (Figure 4I–J) were not different.
Figure 4.
High fat diet feeding does not impair glucose or insulin tolerance in Tcf7βcell−/−MIP mice. A) Schematic representation of the experimental design. B) Body weight during the course of high fat diet (HFD) feeding starting at eight weeks of age. C) Glucose excursion and D) Plasma insulin levels at 0 and 10 min after oral gavage with glucose during an oral glucose tolerance test (oGTT; n = 7–12 per group) after 10 weeks of HFD feeding. E) ipGTT (n = 6–12 per group) and F) ITT (5–10 per group) after 10 weeks of HFD feeding. G) oGTT (n = 6–9/group), H) Body composition (percent fat mass) measured by MRI, I) Representative photomicrographs of pancreas sections showing insulin immunostaining in islets (scale bar: 200 μm; 10× magnification), and J) β-cell area after 20 weeks of HFD feeding (n = 4 per group). Insets depict AUC analysis. Data are presented as mean ± SD, except AUC graphs that are presented as mean ± SEM. Statistical significance was determined using one-way ANOVA with Tukey's correction for multiple comparisons (panel C, E, F–H), two-way ANOVA (panel D) or Student's t-test (panel J).
3.5. Whole-body Tcf7 knockout (Tcf7−/−) mice exhibit normal glucose homeostasis
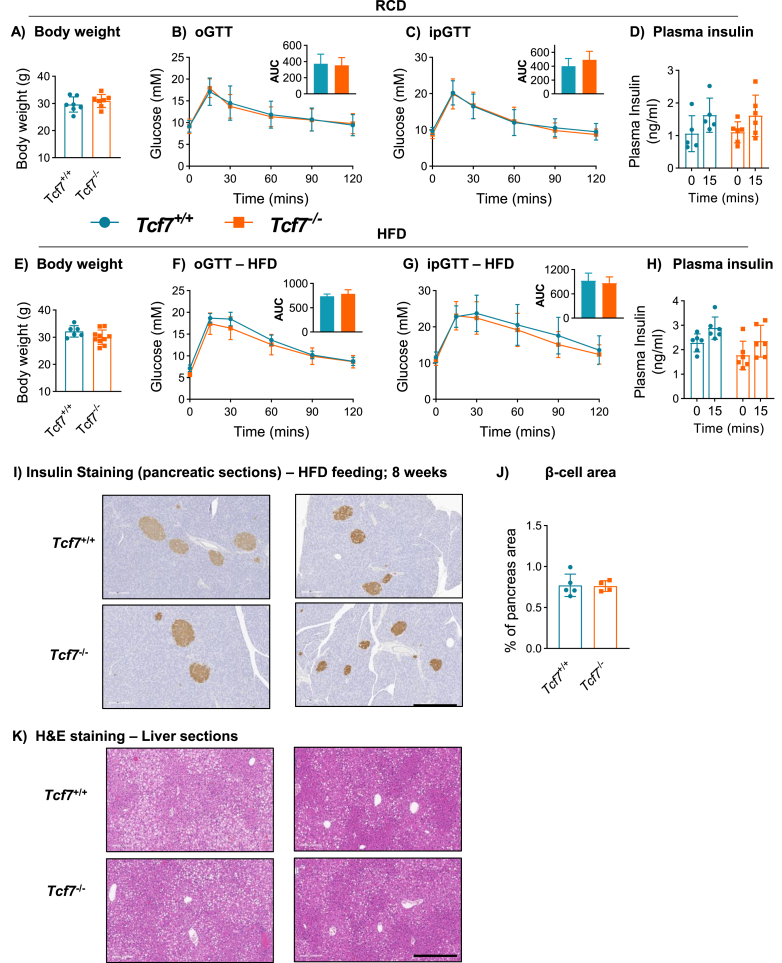
Given the surprisingly normal metabolic phenotypes of mice with targeting of Tcf7 in hepatocytes or β-cells, we re-examined the metabolic phenotype of new cohorts of whole body Tcf7−/− mice. Adult Tcf7−/− mice displayed normal body weight, glucose tolerance (either oral or intraperitoneal) and insulin levels at 18 weeks of age on a RCD (Figure 5A–D), and after 4–6 weeks on a HFD (Figure 5E–H). Furthermore, histological analysis of pancreatic sections did not reveal differences in β-cell area (Figure 5I,J) and liver histology was not grossly different in Tcf7−/− vs. Tcf7+/+ mice (Figure 5K). Taken together, the available data does not support the previous hypothesis that loss of Tcf7 dysregulates glucose homeostasis [4].
Figure 5.
Whole-body Tcf7 knockout (Tcf7−/−) mice do not exhibit dysregulated glucose tolerance. (A–D) Metabolic phenotyping of Tcf7−/− mice fed a regular chow diet (RCD). A) Body weight and glycemic excursion during B) Oral glucose tolerance (oGTT, n = 7 per group) and C) Intraperitoneal glucose tolerance test (ipGTT, n = 5–8 per group) in overnight fasted 18 week-old male Tcf7+/+ and Tcf7−/− mice. D) Plasma insulin levels at 0 and 15 min after glucose challenge during the ipGTT. (E–H) Metabolic phenotyping of Tcf7−/− mice fed a high fat diet (HFD) for 4–6 weeks. E) Body weight and glycemic excursion during F) oGTT, (n = 6–11 per group) and G) ipGTT (n = 7–11 per group) in overnight fasted 12–14 week-old male Tcf7+/+ and Tcf7−/− mice. H) Plasma insulin levels at 0 and 15 min after glucose challenge during the ipGTT. I) Representative photomicrographs of pancreas sections showing insulin immunostaining within islets and J) Quantification of β-cell area (n = 4–5 per group). K) H&E staining in liver sections (n = 4–5 per group) from Tcf7+/+ and Tcf7−/− mice fed a HFD for 8 weeks (scale bar: 500 μm; 4× magnification). Insets depict AUC analysis. Data are presented as mean ± SD except AUC graphs, which are presented as mean ± SEM, and analyzed using Student's t-test.
3.6. Gene expression analysis in pancreatic islets isolated from Tcf7βcell−/− MIP mice
To understand the failure to detect metabolic phenotypes in Tcf7βcell−/− MIP mice, we quantified Tcf7 expression in different tissues, including isolated islets. Levels of islet Tcf7 expression were much lower relative to levels of Tcf7 mRNA in the thymus, spleen, lung, and liver (Figure 6A). Moreover, levels of insulin (Ins2), glucagon (Gcg), chromogranin A (Chga), GLP-1 receptor (Glp1r) and GIP receptor (Gipr) mRNA transcripts were not different in islets from Tcf7βcell−/− MIP mice (Figure 6B). Surprisingly, levels of islet Tcf7 mRNA transcripts were not reduced in Tcf7βcell−/−MIP mice, despite evidence for Cre-mediated recombination of genomic DNA selectively in islets and not in the thymus, spleen, lung, or liver (Figure 6A,C). Consistent with these data, Tcf7 expression was also not reduced in islet RNA of Tcf7βcell−/−RIP mice (Fig. S4K).
Figure 6.
Gene expression analysis in mouse tissues and isolated pancreatic islets. A) mRNA levels of Tcf7 (relative to Tbp) in tissues and B) mRNA levels of islet-specific markers in isolated pancreatic islets from Tcf7βcell−/−MIP and control mice (n = 5–6 per group). C) Recombination of genomic DNA in pancreatic islets, but not in other Tcf7-expressing tissues (thymus, spleen, lung and liver), in Tcf7βcell−/− vs. control mice (n = 5–6 per group). D) UMAP plots for the expression of Tcf7 and various pancreas markers in 3,758 adult mouse islet cells from published single cell RNAseq data sets. Expression data are shown in a log2 scale. Data are presented as mean ± SD and analyzed using Student's t-test.
To further interrogate the cell type(s) within the pancreas that might express Tcf7, we analyzed published single-cell RNA-seq data (Figure 6D). Within the adult mouse pancreas, different pancreatic cell populations were represented in UMAP plots defined by endocrine cell (Ins1, Gcg, Sst, and Ppy), immature endocrine cell (Ghrl), acinar/exocrine cell (Cpa1), ductal cell (Krt19), and endothelial cell markers (Pecam1) (Figure 6D). UMAP plots were also generated for Tcf7 and Cd3g (a T-cell marker). Weak Tcf7 expression was detected in acinar cells, ductal cells and endothelial cells, whereas stronger Tcf7 signals overlapped primarily with Cd3g in T-cells. No Tcf7 signals were observed within endocrine cells of the adult mouse pancreas.
In scRNA-seq data of embryonic mouse pancreata (E12.5 – E18.5), weak Tcf7 signals were localized to Sox9High/Pdx1+/Ptf1aLow bipotent trunk cells (Figs. 7A and S6A) [37]. Tcf7 mRNA was also detected in pancreatic endocrine progenitors marked by Neurog3 expression [38]; however, it was not expressed in hormone-positive endocrine cell lineages. Cd3g+ T-cells expressed high levels of Tcf7 (Figure 7A) and some diffuse expression was evident in Fstl+ mesenchymal cells during pancreatic development.
Figure 7.
Tcf7 expression in mouse embryonic pancreatic cells. A) UMAP plots for the expression of Tcf7 and various embryonic pancreas markers in 60,322 embryonic mouse pancreatic cells spanning E12 to E18.5 from previously published single cell RNA-seq data sets. B) A violin plot for the expression of Tcf7 across various embryonic mouse pancreatic cell types. All expression data are shown in a log2 scale.
Violin plots (Figure 7B) show low expression of Tcf7 in at least some early pancreatic progenitor populations, but not in differentiated islet cell lineages, and the expression was predominantly localized to lymphocytes, mesenchymal cells, and other non-endocrine cell lineages (Figure 7B). Referring to the human endocrine pancreas scRNA-seq data, TCF7 was expressed at very low levels, precluding accurate assignment to a specific cell lineage (Fig. S6B). In contrast, TCF7 expression was detectable in pancreatic cells derived from human embryonic stem cells (hESC) at various stages of differentiation, including NEUROG3+ pancreatic endocrine progenitors (Figs. S6C and S6D).
Next, we analyzed TCF7 expression in pancreatic islets using a validated antibody [39]. We verified the specificity of the TCF7 antibody by Western blotting using protein lysates from thymus tissue isolated from Tcf7+/+ and Tcf7−/− mice (Fig. S7A). Consistent with RNA-seq data from mouse pancreas, we could not detect immunoreactive TCF7 cells within pancreatic islet endocrine cells from C57BL/6 WT mice (Figure 8A), whereas TCF7-immunopositive cells were detected in immune cells within the spleen and thymus tissue (Fig. S7B).
Figure 8.
TCF7 protein is not detectable in pancreatic islets of WT mice and Tcf7 mRNA levels are markedly reduced in pancreatic islets isolated from immunodeficient Rag2−/−Il2rg−/− mice. A) Representative images of immunohistochemical staining with insulin or TCF7 antibodies in pancreatic serial sections from C57BL/6 WT female mice (scale bar: 100 μm; 20 × magnification). B)Tcf7 and C) T-cell marker, Cd3g mRNA levels (relative to Tbp) in islets isolated from either Rag2+/+Il2rg+/+ or Rag2−/−Il2rg−/− mice (females; n = 7 per group). D) Correlation analysis (Pearson's Correlation coefficient, r = 0.987; r2 value = 0.975; 95% Confidence interval = 0.916–0.998) between Tcf7 and Cd3g expression levels in islets isolated from WT (Rag2+/+Il2rg+/+ females; n = 7) mice. E) mRNA levels of Islet-specific gene markers (relative to Tbp) in Rag2+/+Il2rg+/+ or Rag2−/−Il2rg−/− mice. Data are presented as mean ± SD and analyzed using Student's t-test - ∗∗p ≤ 0.01.
Because Tcf7 is highly expressed within immune cell populations including T-cells [40], we hypothesized that the Tcf7 mRNA transcripts detectable in mouse and human [4] islet preparations might arise predominantly from contaminating immune cells. Accordingly, we analyzed islet RNA from immunodeficient Rag2−/−Il2rg−/− mice, which lack T and B lymphocytes, and natural killer cells. Consistent with this hypothesis, levels of Tcf7 mRNA transcripts were markedly reduced in islet RNA isolated from Rag2−/−Il2rg−/− mice (Figure 8B). In keeping with the absence of T-cells in Rag2−/−Il2rg−/− mice, the expression of Cd3g was extremely low in islet RNA (Figure 8C). Remarkably, the levels of Tcf7 and Cd3g mRNAs were highly correlated in islets from WT mice, with a Pearson's coefficient, r equal to 0.987 (Figure 8D). No differences in levels of islet mRNA transcripts were observed in Rag2−/−Il2rg−/− mice (Figure 8E). Taken together, these findings support the hypothesis that the major source of Tcf7 in islet preparations is islet-associated immune cells, and not β-cells.
4. Discussion
Our interest in the potential role of Tcf7 in the islets stemmed from observations that Tcf7 (and Lef1) expression was reduced in islet RNA isolated from Giprβcell−/− mice [4]. Moreover, GIP directly increased Tcf7/TCF7 mRNA transcripts in the INS-1 β-cell line, as well as in primary cultures of human islets, the latter experiments carried out independently in a separate laboratory [4]. Furthermore, islet Tcf7/TCF7 mRNA transcripts were lower in db/db mice, and relatively decreased in islet RNA isolated from older human subjects as well as from islet RNA from donors with T2D [4]. Taken together with findings of glucose intolerance in Tcf7−/− mice, these findings, combined with independent reports describing islet TCF7 expression [12,13], supported a potential role for islet TCF7 in the control of glucose homeostasis.
Attribution of glucoregulatory actions of TCF7 specifically to β-cells is complicated by studies linking liver Tcf7 expression to the control of lipid and glucose homeostasis. Downregulation of TCF7 in HepG2 cells increased the expression of gluconeogenic enzymes and levels of Tcf7 RNA and protein were reduced in the liver of db/db mice, in association with increased expression of Pck1 and G6pc [11]. Nevertheless, these experiments interrogating changes in TCF7 were carried out indirectly, by manipulating expression of miR-22-3p, a microRNA that downregulates hepatic Tcf7 expression. Hence, a direct relationship between altered TCF7 expression and the control of gluconeogenesis had not been established. Herein we tested the hypothesis that selective downregulation of hepatic Tcf7 expression would be sufficient to perturb glucose homeostasis. Surprisingly, we did not detect impairment in oral or intraperitoneal glucose tolerance, plasma triglycerides, lipid tolerance, or changes in insulin sensitivity approximated by assessment of insulin tolerance in mice with ∼70% reduction of levels of hepatocyte Tcf7 mRNA transcripts. Moreover, hepatic gene expression profiles for mRNAs encoding proteins regulating glucose production, lipid metabolism, or inflammation, were not different in Tcf7Hep−/− mice. Taken together, the current evidence does not support a physiological role for endogenous hepatocyte Tcf7 expression in the control of hepatocyte glucose or lipid metabolism.
Despite previous detection of TCF7 expression in INS-1 cells, as well as in RNA isolated from mouse and human islets [4], we were unable to detect abnormal glucose homeostasis in 2 different lines of mice generated to inactivate Tcf7 expression in β-cells. In hindsight, this was not surprising, as we did not observe reduction of islet Tcf7 expression in Tcf7βcell−/− mice. This latter finding prompted us to reassess the possibility that Tcf7 was not expressed in the majority of β-cells. Interrogation of TCF7 expression using published scRNA-seq databases demonstrated that Tcf7/TCF7 could be detected within islet progenitors in the developing murine pancreas, as well as in pancreatic endocrine cell precursors arising from guided hESC differentiation. Nevertheless, the majority of differentiated mouse and human β-cells did not contain mRNA transcripts corresponding to Tcf7/TCF7. Hence, the detection of Tcf7 expression in mouse islets was likely not a reflection of Tcf7 expression within islet endocrine cells, but rather a reflection of expression in non-endocrine cells associated with islets.
Consistent with this premise, Tcf7 expression was localized to a subset of Cd3g+ cells within murine islets (Figure 6D), in agreement with identification of T lymphocytes as the cell type with the highest relative expression of Tcf7. Furthermore, Tcf7 expression was markedly reduced in islets from Rag2−/−Il2rg−/− mice, in agreement with the predominant localization of islet cell-associated Tcf7 expression to immune cells. The representation and proportions of non-endocrine immune cells within isolated islets have been described in mouse, non-human primate and human studies [26,[41], [42], [43]], with T lymphocytes being the predominant immune cell type detected within islets from donors without diabetes [44]. Indeed, Baron et al. detected 4 distinct immune cell populations, tissue-resident macrophages, mast cells, B cells, and cytotoxic T cells associated with mouse and human islets [26]. Not surprisingly, the proportion of immune cells is increased in the pancreas from islet autoantibody + donors and individuals with type 1 diabetes [43]. The majority of studies analyzing islet-associated cells in humans assessed donor islet cells from older individuals; hence, much less is known about the representation of islet-associated immune cell populations in islets from children or young adult donors.
Our previous studies employing INS-1 cells and human islets demonstrated that gain and loss of GIPR signaling regulate Tcf7/TCF7 expression [4]. It is likely that the expression of Tcf7 in an immortalized cell line reflects a more immature β-cell gene expression profile in the insulin-producing cell line, which often expresses neuropeptide Y, gastrin and related markers characteristic of fetal or immature islet development [45,46]. Indeed, scRNA-seq data demonstrate expression of Tcf7 in multiple cell lineages in the embryonic mouse pancreas, as well as in early progenitors during the course of hESC differentiation towards pancreatic cells.
Surprisingly, we were unable to replicate our previous findings [4] of glucose intolerance in whole body Tcf7−/− mice. At present, we do not have a satisfactory explanation for the failure to replicate these findings several years later. Although the reasons for different metabolic phenotypes in Tcf7−/− mice studied several years apart are unclear, this might possibly reflect changes in the bacterial colonization of our animal facility over time, perhaps impacting systemic inflammation and insulin resistance.
The GIP receptor is expressed in the bone marrow, and within cells arising from the myeloid and lymphoid lineages, including macrophages and T cells [47,48]. Moreover, gain and loss of GIPR signaling is associated with dysregulated immune responses to pro-inflammatory stimuli involving regulation of the TLR and Notch pathways in bone marrow cells [48]. Hence, it seems likely that our previous findings demonstrating GIP-dependent regulation of TCF7 expression in human islet cultures [4] might be consistent with the actions of GIP on islet-associated immune cells or a small subset of mouse β-cells that express a functional GIPR. Taken together, these findings suggest that TCF7 is not a direct regulator of β-cell, hepatocyte, or whole body glucose homeostasis in mice, implying that the previously described association of genetic variation within the TCF7 gene and type 1 diabetes [14,15] likely reflects actions of TCF7 within the immune system, and neither β-cells nor hepatocytes.
Author contributions
KK, CKW, and DJD designed the experiments, analyzed the data, and wrote and/or reviewed the manuscript. KK, CKW, LB, JB, SF, BP, XC, and DM carried out the experiments and reviewed/edited the manuscript. DJD secured funding for the studies and is the guarantor of the data.
Acknowledgments
D.J.D. is supported in part by a Novo Nordisk, Banting and Best Diabetes Center (BBDC) Chair in incretin biology, the Mt. Sinai Hospital Novo Nordisk Foundation Fund in regulatory peptides, and CIHR Foundation grant 154,321. Mt. Sinai Hospital receives support for studies of incretin biology in the Drucker lab from Novo Nordisk. CKW was supported by a fellowship from the BBDC and JLB was supported by a fellowship from Diabetes Canada. SF was supported by the Hospital for Sick Children Clinician Scientist Training Program. The Graphical Abstract was created using BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101213.
Conflict of interest
DJD receives consulting or speaking honoraria from Eli Lilly Inc., Forkhead Inc. Intarcia, Kallyope, Merck, and Novo Nordisk, within the past 12 months for advisory boards and lectures related to incretin biology. None of the other authors have competing interests. Preclinical studies of incretin biology in the Drucker lab are supported in part by grants to Mt. Sinai Hospital from Novo Nordisk and Pfizer Inc. Following completion of these studies, BP became a full-time employee of Roche Canada Inc.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Vinuela A., Varshney A., van de Bunt M., Prasad R.B., Asplund O., Bennett A. Genetic variant effects on gene expression in human pancreatic islets and their implications for T2D. Nature Communications. 2020;11(1):4912. doi: 10.1038/s41467-020-18581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen M.B., Calanna S., Holst J.J., Vilsboll T., Knop F.K. Glucose-dependent insulinotropic polypeptide: blood glucose stabilizing effects in patients with type 2 diabetes. Journal of Clinical Endocrinology Metabolism. 2014;99(3):E418–E426. doi: 10.1210/jc.2013-3644. [DOI] [PubMed] [Google Scholar]
- 3.Miyawaki K., Yamada Y., Yano H., Niwa H., Ban N., Ihara Y. Glucose intolerance caused by a defect in the entero-insular axis: a study in gastric inhibitory polypeptide receptor knockout mice. Proceedings of the National Academy of Sciences of the U S A. 1999;96(26):14843–14847. doi: 10.1073/pnas.96.26.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell J.E., Ussher J.R., Mulvihill E.E., Kolic J., Baggio L.L., Cao X. TCF1 links GIPR signaling to the control of beta cell function and survival. Nature Medicine. 2016 doi: 10.1038/nm.3997. 2284–90. [DOI] [PubMed] [Google Scholar]
- 5.Gasbjerg L.S., Helsted M.M., Hartmann B., Jensen M.H., Gabe M.B.N., Sparre-Ulrich A.H. Separate and combined glucometabolic effects of endogenous glucose-dependent insulinotropic polypeptide and glucagon-like peptide 1 in healthy individuals. Diabetes. 2019;68(5):906–917. doi: 10.2337/db18-1123. [DOI] [PubMed] [Google Scholar]
- 6.Drucker D.J., Habener J.F., Holst J.J. Discovery, characterization, and clinical development of the glucagon-like peptides. Journal of Clinical Investigation. 2017;127(12):4217–4227. doi: 10.1172/JCI97233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller T.D., Finan B., Bloom S.R., D'Alessio D., Drucker D.J., Flatt P.R. Glucagon-like peptide 1 (GLP-1) Molecular Metabolism. 2019:3072–3130. doi: 10.1016/j.molmet.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frias J.P., Nauck M.A., Van J., Kutner M.E., Cui X., Benson C. Efficacy and safety of LY3298176, a novel dual GIP and GLP-1 receptor agonist, in patients with type 2 diabetes: a randomised, placebo-controlled and active comparator-controlled phase 2 trial. Lancet. 2018;392(10160):2180–2193. doi: 10.1016/S0140-6736(18)32260-8. [DOI] [PubMed] [Google Scholar]
- 9.Baggio L.L., Drucker D.J. Glucagon-like peptide-1 receptor co-agonists for treating metabolic disease. Molecular Metabolism. 2020 doi: 10.1016/j.molmet.2020.101090. 101090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oduori O.S., Murao N., Shimomura K., Takahashi H., Zhang Q., Dou H. Gs/Gq signaling switch in beta cells defines incretin effectiveness in diabetes. Journal of Clinical Investigation. 2020;130(12):6639–6655. doi: 10.1172/JCI140046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaur K., Vig S., Srivastava R., Mishra A., Singh V.P., Srivastava A.K. Elevated hepatic miR-22-3p expression impairs gluconeogenesis by silencing the wnt-responsive transcription factor Tcf7. Diabetes. 2015;64(11):3659–3669. doi: 10.2337/db14-1924. [DOI] [PubMed] [Google Scholar]
- 12.Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L., Taneera J. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proceedings of the National Academy of Sciences of the U S A. 2014;111(38):13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benner C., van der Meulen T., Caceres E., Tigyi K., Donaldson C.J., Huising M.O. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15620 doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erlich H.A., Valdes A.M., Julier C., Mirel D., Noble J.A. Evidence for association of the TCF7 locus with type I diabetes. Genes and Immunity. 2009;10(Suppl) doi: 10.1038/gene.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble J.A., White A.M., Lazzeroni L.C., Valdes A.M., Mirel D.B., Reynolds R. A polymorphism in the TCF7 gene, C883A, is associated with type 1 diabetes. Diabetes. 2003;52(6):1579–1582. doi: 10.2337/diabetes.52.6.1579. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan S., Ke J.Y., Pellizzon M.A. Targeted nutrient modifications in purified diets differentially affect nonalcoholic fatty liver disease and metabolic disease development in rodent models. Current Developments in Nutrition. 2020;4(6):nzaa078. doi: 10.1093/cdn/nzaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinke F.C., Yu S., Zhou X., He B., Yang W., Zhou B. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nature Immunology. 2014;15(7):646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varin E.M., Mulvihill E.E., Beaudry J.L., Pujadas G., Fuchs S., Tanti J.F. Circulating levels of soluble dipeptidyl peptidase-4 are dissociated from inflammation and induced by enzymatic DPP4 inhibition. Cell Metabolism. 2019;29(2):320–334. doi: 10.1016/j.cmet.2018.10.001. e325. [DOI] [PubMed] [Google Scholar]
- 19.Tamarina N.A., Roe M.W., Philipson L. Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic beta-cells. Islets. 2014;6(1) doi: 10.4161/isl.27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera P.L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127(11):2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 21.Ioannidis V., Beermann F., Clevers H., Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nature Immunology. 2001;2(8):691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 22.Song J., Willinger T., Rongvaux A., Eynon E.E., Stevens S., Manz M.G. A mouse model for the human pathogen Salmonella typhi. Cell Host & Microbe. 2010;8(4):369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flock G., Baggio L.L., Longuet C., Drucker D.J. Incretin receptors for glucagon-like peptide 1 and glucose-dependent insulinotropic polypeptide are essential for the sustained metabolic actions of vildagliptin in mice. Diabetes. 2007;56(12):3006–3013. doi: 10.2337/db07-0697. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs S., Yusta B., Baggio L.L., Varin E.M., Matthews D., Drucker D.J. Loss of Glp2r signaling activates hepatic stellate cells and exacerbates diet-induced steatohepatitis in mice. JCI Insight. 2020;5(8) doi: 10.1172/jci.insight.136907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamont B.J., Li Y., Kwan E., Brown T.J., Gaisano H., Drucker D.J. Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. Journal of Clinical Investigation. 2012;122(1):388–402. doi: 10.1172/JCI42497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell System. 2016;3(4):346–360. doi: 10.1016/j.cels.2016.08.011. e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabula Muris C., Overall C., Logistical C., Organ C., Processing, Library P. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562(7727):367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrnes L.E., Wong D.M., Subramaniam M., Meyer N.P., Gilchrist C.L., Knox S.M. Lineage dynamics of murine pancreatic development at single-cell resolution. Nature Communications. 2018;9(1):3922. doi: 10.1038/s41467-018-06176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krentz N.A.J., Lee M.Y.Y., Xu E.E., Sproul S.L.J., Maslova A., Sasaki S. Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Reports. 2018;11(6):1551–1564. doi: 10.1016/j.stemcr.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segerstolpe A., Palasantza A., Eliasson P., Andersson E.M., Andreasson A.C., Sun X. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enge M., Arda H.E., Mignardi M., Beausang J., Bottino R., Kim S.K. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171(2):321–330. doi: 10.1016/j.cell.2017.09.004. e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camunas-Soler J., Dai X.Q., Hang Y., Bautista A., Lyon J., Suzuki K. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metabolism. 2020;31(5):1017–1031 e1014. doi: 10.1016/j.cmet.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng C., Xi J., Li H., Cui J., Gu A., Lai S. Single-cell lineage analysis reveals extensive multimodal transcriptional control during directed beta-cell differentiation. Nature Metabolism. 2020;2(12):1443–1458. doi: 10.1038/s42255-020-00314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Z., Miller R.A., Patel R.T., Chen J., Dhir R., Wang H. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nature Medicine. 2012;18(6):934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue J., Scotti E., Stoffel M. CDK8 regulates insulin secretion and mediates postnatal and stress-induced expression of neuropeptides in pancreatic beta cells. Cell Reports. 2019;28(11):2892–2904. doi: 10.1016/j.celrep.2019.08.025. e2897. [DOI] [PubMed] [Google Scholar]
- 37.Lynn F.C., Smith S.B., Wilson M.E., Yang K.Y., Nekrep N., German M.S. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proceedings of the National Academy of Sciences of the U S A. 2007;104(25):10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proceedings of the National Academy of Sciences of the U S A. 2000;97(4):1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emmanuel A.O., Arnovitz S., Haghi L., Mathur P.S., Mondal S., Quandt J. TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4(+)CD8(+) thymocytes. Nature Immunology. 2018;19(12):1366–1378. doi: 10.1038/s41590-018-0254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Wetering M., Oosterwegel M., Dooijes D., Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. The EMBO Journal. 1991;10(1):123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffey L.C., Berman D.M., Willman M.A., Kenyon N.S. Immune cell populations in nonhuman primate islets. Cell Transplantation. 2009;18(10):1213–1222. doi: 10.3727/096368909X12483162196728. [DOI] [PubMed] [Google Scholar]
- 42.Butcher M.J., Hallinger D., Garcia E., Machida Y., Chakrabarti S., Nadler J. Association of proinflammatory cytokines and islet resident leucocytes with islet dysfunction in type 2 diabetes. Diabetologia. 2014;57(3):491–501. doi: 10.1007/s00125-013-3116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddy S., Zeng N., Al-Diery H., Jung D., Yeu C., Joret M.O. Analysis of peri-islet CD45-positive leucocytic infiltrates in long-standing type 1 diabetic patients. Diabetologia. 2015;58(5):1024–1035. doi: 10.1007/s00125-015-3519-6. [DOI] [PubMed] [Google Scholar]
- 44.Radenkovic M., Uvebrant K., Skog O., Sarmiento L., Avartsson J., Storm P. Characterization of resident lymphocytes in human pancreatic islets. Clinical and Experimental Immunology. 2017;187(3):418–427. doi: 10.1111/cei.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waeber G., Thompson N., Waeber B., Brunner H.R., Nicod P., Grouzmann E. Neuropeptide Y expression and regulation in a differentiated rat insulin-secreting cell line. Endocrinology. 1993;133(3):1061–1067. doi: 10.1210/endo.133.3.8396008. [DOI] [PubMed] [Google Scholar]
- 46.Brand S.J., Wang T.C. Gastrin gene expression and regulation in rat islet cell lines. Journal of Biological Chemistry. 1988;263(32):16597–16603. [PubMed] [Google Scholar]
- 47.Mantelmacher F.D., Fishman S., Cohen K., Pasmanik Chor M., Yamada Y., Zvibel I. Glucose-dependent insulinotropic polypeptide receptor deficiency leads to impaired bone marrow hematopoiesis. The Journal of Immunology. 2017;198(8):3089–3098. doi: 10.4049/jimmunol.1601441. [DOI] [PubMed] [Google Scholar]
- 48.Pujadas G., Varin E.M., Baggio L.L., Mulvihill E.E., Bang K.W.A., Koehler J.A. The gut hormone receptor GIPR links energy availability to the control of hematopoiesis. Mol Metab. 2020 doi: 10.1016/j.molmet.2020.101008. 39101008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.