Abstract
In normal individuals, pituitary somatotrophs optimise body composition by responding to metabolic signals from leptin. To identify mechanisms behind the regulation of somatotrophs by leptin, we used Cre-LoxP technology to delete leptin receptors (LEPR) selectively in somatotrophs and developed populations purified by fluorescence-activated cell sorting (FACS) that contained 99% somatotrophs. FACS-purified, Lepr-null somatotrophs showed reduced levels of growth hormone (GH), growth hormone-releasing hormone receptor (GHRHR), and Pou1f1 proteins and Gh (females) and Ghrhr (both sexes) mRNAs. Pure somatotrophs also expressed thyroid-stimulating hormone (TSH) and prolactin (PRL), both of which were reduced in pure somatotrophs lacking LEPR. This introduced five gene products that were targets of leptin. In the present study, we tested the hypothesis that leptin is both a transcriptional and a post-transcriptional regulator of these gene products. Our tests showed that Pou1f1 and/or the Janus kinase/signal transducer and activator of transcription 3 transcriptional regulatory pathways are implicated in the leptin regulation of Gh or Ghrhr mRNAs. We then focused on potential actions by candidate microRNAs (miRNAs) with consensus binding sites on the 3’ UTR of Gh or Ghrhr mRNAs. Somatotroph Lepr-null deletion mutants expressed elevated levels of miRNAs including miR1197-3p (in females), miR103-3p and miR590-3p (both sexes), which bind Gh mRNA, or miRNA-325-3p (elevated in both sexes), which binds Ghrhr mRNA. This elevation indicates repression of translation in the absence of LEPR. In addition, after detecting binding sites for Musashi on Tshb and Prl 3’ UTR, we determined that Musashi1 repressed translation of both mRNAs in in vitro fluc assays and that Prl mRNA was enriched in Musashi immunoprecipitation assays. Finally, we tested ghrelin actions to determine whether its nitric oxide-mediated signalling pathways would restore somatotroph functions in deletion mutants. Ghrelin did not restore either GHRH binding or GH secretion in vitro. These studies show an unexpectedly broad role for leptin with respect to maintaining somatotroph functions, including the regulation of PRL and TSH in subsets of somatotrophs that may be progenitor cells.
Keywords: Cre-LoxP, ghrelin, growth hormone, growth hormone releasing hormone, leptin, leptin receptors, miRNAs, mouse, mRNAs, Musashi, post-transcriptional regulation, prolactin, somatotroph, thyroid stimulating hormone, transcriptional activators, translational regulation
1 |. INTRODUCTION
Growth hormone (GH) secretion from anterior pituitary somatotrophs stimulates skeletal growth before puberty and, under normal conditions, GH maintains an optimal body composition throughout life.1 GH synthesis and secretion are stimulated by growth hormone-releasing hormone (GHRH) and amplified by ghrelin.2–4 Somatotrophs thus determine growth and body size, lipoprotein metabolism, muscle insulin-like growth factor-1 expression, and insulin sensitivity.5 To perform these vital functions, somatotrophs receive extrinsic signals concerning the metabolic and nutritional state of the body,6 including different adipokines, especially leptin.7 Leptin deficiency causes a severe reduction in somatotroph functions.8–12 Most somatotrophs express leptin receptors13,14 and leptin treatment restores GH secretion and Ghrhr mRNA levels (but not hypothalamic GHRH) in leptin-deficient, ob/ob mice, suggesting direct actions of leptin at the pituitary level.15 Similarly, we have shown that as little as 10 pg mL−1 leptin given to cultured pituitary cells from fasted rats restored the number of somatotrophs immunolabelled for GH, which had been reduced in response to the fasting-induced low serum leptin.16
To identify the mechanism by which leptin acts upon somatotrophs, we selectively deleted leptin receptors (LEPR) in somatotrophs (Lepr-null). These mutant mice exhibited sex-specific GH deficiency17,18 and metabolic dysfunction.17,19 Serum GH was reduced along with numbers of immunolabelled GH cells (> 60% in males or > 41% in females).17 However, in situ hybridisation studies showed that male mutants had normal numbers of cells bearing Gh mRNA.20 These findings led to our early hypothesis suggesting that actions by leptin may be post-transcriptional.
To test this hypothesis and investigate potential post-transcriptional regulators unique to somatotrophs, we next used Cre-LoxP technology with mice bearing somatotroph- specific Cre-GH crossed with mice bearing a Cre-reporter transgene to develop somatotrophs purified by fluorescence-activated cell sorting (FACS). Proof of purity was based on the detection of differentiated somatotrophs expressing GH immunolabelling and GHRH receptor (GHRHR) binding in 98%−99% of the FACS fraction.21 Surprisingly, however, our assays of the somatotroph and non-somatotroph fractions detected thyroid stimulating hormone-beta (TSH) and prolactin (PRL) in the purified somatotrophs, which indicated multihormonal expression by a population that was 99% GH cells. Furthermore, we observed that levels of TSH and PRL proteins (but not Tshb and Prl mRNAs) were reduced only in pure Lepr-null somatotrophs from females.21 We therefore hypothesised that leptin may play a role in maintaining this subset of somatotrophs via post-transcriptional pathways.
This hypothesis was strengthened further by tests showing that levels of the transcription factor Pou1f1 (Pit1) proteins (but not mRNAs) were also reduced in pure somatotrophs from Lepr-null females and that leptin treatment increased Pou1f1 protein, but not mRNA levels.21 This is significant because Pou1f1 is a transcription factor required by all of the leptin targets identified in our studies (Tshb, Gh, Prl and Ghrhr). Collectively, these findings suggested a role for leptin in the regulation of multiple gene products in somatotrophs, either directly via post-transcriptional regulation of TSH or PRL, or indirectly via regulation of Pou1f1.
Because of the diversity of the gene products, we hypothesised that leptin stimulates somatotrophs via multiple signalling pathways. To test this hypothesis, we initiated additional studies aiming to identify the pathways involved in mediating the action of leptin upon somatotrophs. This report confirms our hypothesis and presents findings that show evidence for leptin regulation of the different gene products via both transcriptional and post-transcriptional signalling pathways, including novel regulators and candidate microRNAs (miRNAs).
2 |. MATERIALS AND METHODS
2.1 |. Mutant and control animal models
LEPR deletion mutant mice bearing CreGH and Lepr(fl/fl exon1) (Lepr-null) were bred to the N4/N5 generations as described previously.17,18 All animal use and care protocols were approved annually by the Institutional Animal Care and Use Committee of the University of Arkansas for Medical Sciences.
Normal, wild-type mice (FVB.P129 hybrids) were also used for all experiments involving control cultures. In addition, we bred mice bearing Cre-GH to a Cre-reporter mouse line to create a line of mice with fluorescent somatotrophs. This was a mouse line bearing floxed tdTomato-eGFP driven by a membrane promoter. In the presence of Cre-recombinase, the tdTomato is deleted, leaving enhanced green fluorescent protein (eGFP) to be driven by the membrane promoter. This model is fully described elsewhere.21
2.2 |. Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), mRNA and miRNA sample preparation and amplification
Whole pituitaries were collected from 3 month-old male and female controls and deletion mutants, total RNA was isolated and target transcripts were analysed as described previously. The assay of Prl mRNA (NM_011164.1, forward: GGCCATCTTGGAGAAGTGTG, reverse: ACAGATTGGCAGAGGCTGAA) and Ghrhr mRNA (NM_00 1003685.1, forward: ACCCGTATCCTCTGCTTGCT, reverse: AGGTGTTG TTGGTCCCCTCT) was normalised to peptidylprolyl isomerase A22 (Ppia, cyclophilin, NM_008907.1
forward: TGGTCTTTGGGAAGGTGAAAG, reverse: TGTCCAC AGTCGGAAATGGT.) Spectrophotometric analysis confirmed RNA purity (A260/A280 = 1.8–2.0 and A260/A230 > 1.7) and qPCR single product was confirmed by melting curve analysis. miRNAs (miRNAs) were analysed using the TaqMan and TaqMan Advanced MicroRNA systems (Applied Biosystems, Foster City, CA, USA) and the miR103, miR325-3p, miR590 and miR1197 Taqman primers (all normalised to U6 snRNA). qRT-PCR reactions utilised the quantstudio 12k flex system (Applied Biosystems, Life Technologies, Grand Island, NY, USA) using the recommended protocols for each system (TaqMan or Power SYBR Green) and relative expression values were determined using quantstudio 12k flex, version 1.0 and the ΔΔCT method.
2.3 |. Pituitary cell stimulation and serum analysis
Pituitary cell dispersion, culture and leptin stimulation protocols have been described previously.17,18,20 A subset of experiments used pituitary pieces stimulated with leptin (1–100 nmol L−1) for 3 hours followed by extraction of proteins or mRNA as described previously.23 For stimulation with GHRH and ghrelin, pituitaries were collected and dispersed from control and somatotroph Lepr-null mutant mice as described previously and plated 12 000 cells per well on poly-d-lysine coated glass coverslips and allowed to grow overnight in Dulbecco’s modified Eagle’s medium (DMEM) in a 5% CO2 incubator at 37°C.20 Cells were exposed to GHRH (1 or 3 nmol L−1) or a combination of GHRH (1 or 3 nmol L−1) and 10 nmol L−1 acylated ghrelin for 3 hours. GHRH and ghrelin were diluted in DMEM with 1:200 protease cocktail. Leptin was not used in these ghrelin-stimulation studies as a result of the deletion of the leptin receptor.
For tests of inhibitors, cells from control mice were treated with 3 μmol L−1 actinomycin D, 1 μmol L−1 STAT3 inhibitor peptide (STAT3i; Millipore, Burlington, MA, USA) or 1 nmol L−1 nitric oxide inhibitor, l-NG-nitro arginine methyl ester (l-NAME) for 1 hour prior to stimulation with 10 nmol L−1 leptin or 10 nmol L−1 ghrelin for 3 hours. After treatment, cells were fixed, as above, and immunolabelled for GH, as described20 previously and/or for biotinylated GHRH (Bio-GHRH) binding.24 Serum analysis of pituitary proteins utilised the Luminex LX200 (Luminex Corp, Austin, TX, USA) xPO-NENT 3.1 with the Millipore MAP Multiplex kits (Millipore), as described previously.19
2.4 |. Immunocytochemistry for GH and affinity cytochemistry for GHRHR
For the immunolabelling, the primary antibody to rat GH was supplied by the NIDDK Hormone Distribution Program (A. Parlowe, University of California, Harbor Medical Center, Los Angeles, CA, USA) and diluted 1;80 000–1:100 000 for most of the experiments. However, for immunofluorescence, the dilution was 1:30 000–1:80 000 anti-rGH. Absorption controls have been used to test specificity of this antibody, showing that the addition of 10 μg mL−1 iodination preparation rat GH neutralised the immunolabelling of plastic-embedded pituitary sections when added to 1:15 000 anti-rGH.25 Similarly, 100 ng mL−1 GH neutralised the immunolabelling of cells in culture when added to 1:80 000 anti-rGH.26 The second antibody is from Vector Laboratories (Burlingame, CA, USA), made in a goat against rabbit immunoglobulin (Ig)G. The antibody was biotinylated and thus detected by Streptavidin peroxidase as described previously.20
For immunofluorescence, we used the Dylight 488 anti-rabbit IgG (Vector Laboratories), diluted to 1:100.
We have further validated the GH antibody, showing dual labelling for Gh mRNA and GH proteins in the same cells.26 Most recently, this antibody was used along with an enzyme immunoassay (EIA) to validate the purification of somatotrophs, correlating the enrichment in GH protein and Gh mRNA levels with the counts of cells immunolabelled for GH showing 99% somatotrophs.21 We also consistently show parallel reductions in somatotroph stores and pituitary GH content in our transgenic model, Lepr-null mice, detected by both EIA and immunolabelling.18,21 This constitutes biological validation of the immunolabelling in the present study.
Finally, further validation is seen with dual labelling for GHRH receptors and GH antigens, which detect differentiated somatotrophs. This protocol, validated in 1999,24 is applied to living pituitary cells from both control and deletion mutant mice plated on coverslips. Cells were treated with 1–3 nmol L−1 Bio-GHRH for 15 minutes at 37°C and then fixed with 2% glutaraldehyde. Fixed cells were then labelled with avidin-biotin peroxidase complex (ABC kitl Vector Laboratories), and the peroxidase was detected by black diamino-benzidine substrate followed by GH immunocytochemistry as described previously.24 The specificity of the binding to Bio-GHRH has been validated by competing non-biotinylated analogues.24 In the present study, we further validate the labelling for Bio-GHRH with an EIA for GHRHR proteins. Both assays will show reductions in GHRHR in the deletion mutant model.
2.5 |. RNA immunoprecipitation of endogenous Musashi1-mRNA complex
Musashi1 was immunoprecipitated from pituitary cell lysate (Magna-RIP; MilliporeSigma, Burlington, MA, USA; 17-700) from six adult control (FVB) mixed-cycle females with Musashi1 antibody (Abcam, Cambridge, MA, USA; ab52865) or control rabbit IgG (Millipore; 12-370) as described previously.23,27 Results represent the average of two experiments (n = 4) and are expressed as fold-enrichment compared to rabbit IgG control with Ppia subtracted. Musashi1 immunoprecipitation was confirmed by western blotting as described in our previous study.23
2.6 |. Luciferase mRNA translation reporter assays
The 154-bp murine Prl mRNA 3’ UTR (NM_011164) and the terminal 300 bp of the Tshb mRNA (NM_009432) were synthesised separately as geneblock fragments (IDT). For the Prl 3’ UTR, the fragment was engineered with a 5’ SalI site and 3’ XhoI site and cloned into SalI/XhoI digested pmirGLO (Promega, Madison, WI, USA) dual Firefly Luciferase (FLuc) RNA reporter and control Renilla Luciferase plasmid, and designated pmirGLO Prl UTR. For Tshb the terminal 300 bp contained some coding sequence, the stop codon and the 72-bp 3’ UTR. The sequence was engineered to add a 5’ SacI followed by a NheI site, the stop codon was replaced with a NheI site and a XhoI site was placed at the 3’ end. The full-length fragment was cloned into SacI/XhoI digested pmirGLO and subsequently digested with NheI to delete the upstream coding region, leaving only the entire 72-bp 3’ UTR, gel-purified, religated and designated pmirGLO Tshb UTR. NIH3T3 cells (American Type Culture Collection, Manassas, VA, USA) were co-transfected with the pmirGLO Prl or Tshb UTR plasmids along with either a plasmid expressing a Musashi1-eGFP fusion protein (Msi1-AA-eGFP) or the RNA-binding mutant protein (Msi1-bm-eGFP) or control protein (the empty vector (encoding the eGFP) moiety alone, as described previously.27–29 The MSI1-AA protein is mutated in two sites of regulatory phosphorylation to ensure continued repression of target mRNAs27 and the negative control Msi1-bm mutant has attenuated RNA target binding activity. Luciferase activity was determined in quadruplicate after 24 hours, using the Dual-Luciferase Reporter Assay System (Promega) and Turner Biosystems luminometer (Promega) in accordance with the manufacturer’s instructions. Data are expressed as relative luciferase activity (Firefly Luciferase/Renilla Luciferase, FF/R) in arbitrary units. All experiments were repeated three times in separate experiments.
2.7 |. Statistical analysis
All experiments included at least three samples per group and power analyses have been described in recent studies.18,19 Statistical tests were conducted using prism (GraphPad Software Inc., San Diego, CA, USA) and one- two- or three-way ANOVA followed by Tukey’s, Bonferroni’s or Fisher’s LSD post-hoc tests or Student’s t test with Welch’s correction. P < .05 was considered statistically significant. The results of the statistical analyses (sample size, F, t, df and probability) are indicated as appropriate.
3 |. RESULTS
3.1 |. Loss of LEPR in somatotrophs reduces levels of GHRH receptors
We determined the impact of the ablation of all isoforms of LEPR in somatotrophs on GHRHR binding, detected cytochemically by Bio-GHRH binding to dispersed cultured cells from whole pituitaries.24 Figure 1A and B illustrate the dramatic reduction in numbers of cells labelled for Bio-GHRH and immunolabelled for GH in the somatotroph Lepr-null mutants (Figure 1B) compared to controls (Figure 1A). Figure 1C shows the counts of the cytochemically identified cells in Figure 1A and B and also shows the significant decrease in numbers of cells with GHRHR binding sites in whole pituitary cell cultures from mutants. In addition, Figure 1C shows the results of the ghrelin treatment, in which we treated additional coverslips bearing these cultures of dispersed pituitary cells for 3 hours with 10 nmol L−1 ghrelin. Figure 1C shows that ghrelin alone stimulated an increased number of cells with GHRHR in control cultures, although it had no restorative effect on cultures from mutant mice.
FIGURE 1.
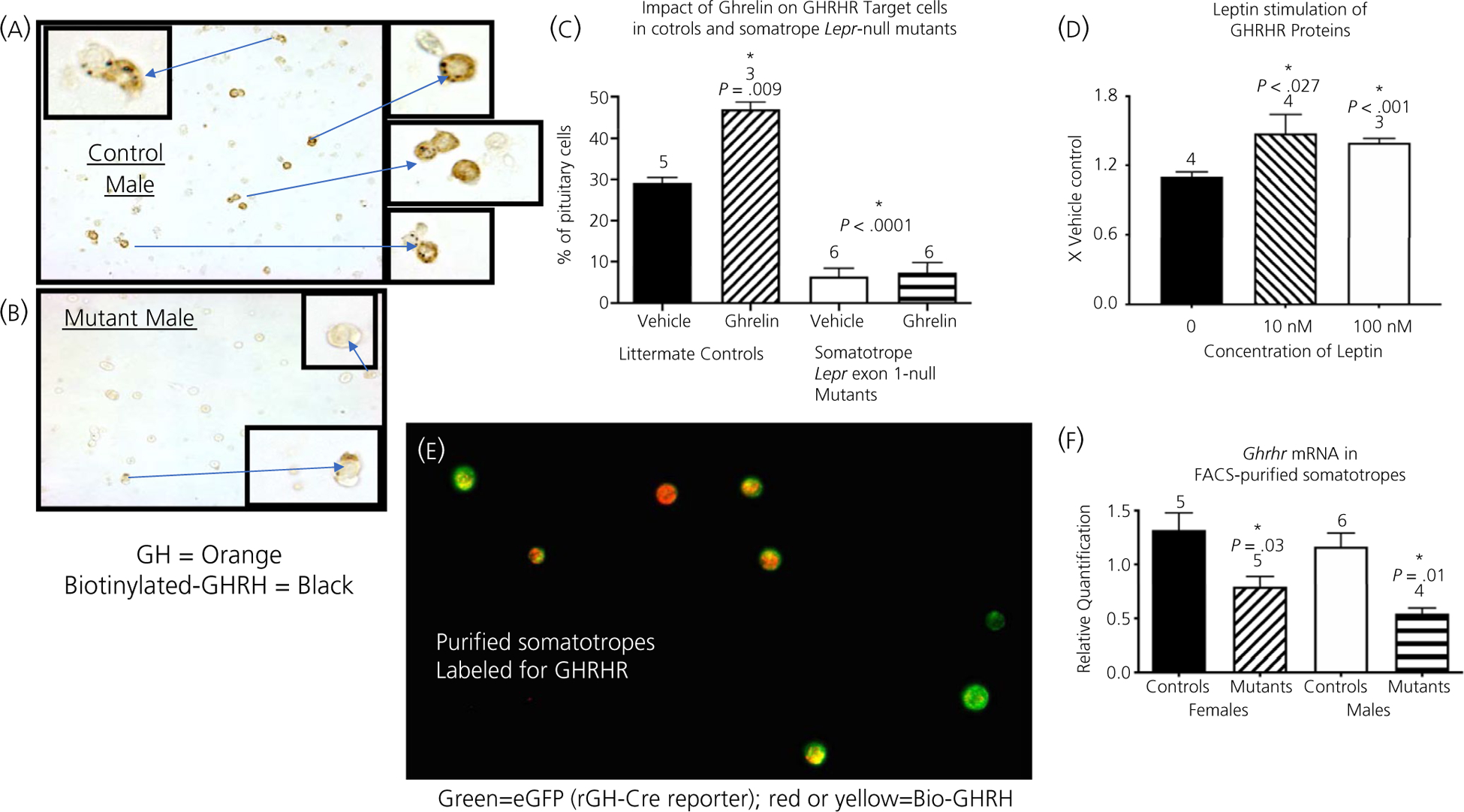
Growth hormone-releasing hormone receptor (GHRHR) reduction in the Lepr-null pituitary. A,B, Assay of one-day cultures of dispersed whole pituitary cells from adult control mice (A) or mutant mice (B); biotinylated GHRH (Bio-GHRH) (black) and GH immunolabelling (orange). C, Counts of cells labelled for Biotinylated GHRH from the whole pituitary cultures in (A,B), adding the results from the counts of cells stimulated with ghrelin. D, GHRHR enzyme immunoassay of protein extracts from pituitary pieces stimulated with leptin. E, Cytochemical detection of Bio-GHRH in purified somatotrophs from control, dioestrous females; Bio-GHRH is detected by the red fluorescence marker, Dylight 594, somatotrophs are detected by enhanced green fluorescent protein (eGFP). Cre-reporter was floxed tdTomato, eGFP. Cytoplasmic overlap in expression is detected by yellow/orange fluorescence. F, mRNA extracts from purified somatotrophs (from male and female mice) were assayed for Ghrhr mRNA in purified somatotrophs. C, Statistical analysis. *Significantly different from vehicle control as tested by two-way ANOVA followed by Tukey’s post-hoc test. In control cultures, ghrelin stimulated a significant increase in cells with GHRHR (F1,14 = 9.012, P = .009); leptin receptors (LEPR)-null mutants had a significant decrease in cells with GHRHR (F1,14 = 141.3, P < .0001). Interaction, F1,14 = 7.616, P = .015. D, One-way ANOVA followed by Bonferroni’s post-hoc test, F2,8 = 6.09, P = .025; n = 4 samples per group. F, *Significant by two-way ANOVA followed by Tukey’s post-hoc test; source of variation is controls vs mutants, F1,16 = 21.87, P = .0003; Males vs Females, F1,16 = 2.728, P = .1181. Interaction, F1,16 = 0.164, P = .691. FACS, fluorescence-activated cell sorting
Having determined that GHRHR was reduced in the somatotrophs Lepr-null mice, we next investigated whether GHRHR was a leptin target. We treated pituitary pieces from male mice for 3 hours with 10 or 100 nmol L−1 leptin and assayed protein extracts of these pieces for GHRHR. Figure 1D shows that leptin treatment at 10 or 100 nmol L−1 resulted in a significant increase in GHRHR. The results for females are similar to those obtained in males (data not shown).
Finally, as part of our analysis of purified somatotrophs, we used Bio-GHRH to identify GHRHR binding in our FACS-purified populations. A major objective was to determine whether these purified cells were differentiated somatotrophs, defined by their expression of GHRHR. We were particularly interested in the somatotroph population, which showed evidence for multihormonal expression in progenitor cells. Figure 1E shows that almost all of the somatotrophs, labelled green for eGFP by a Cre-reporter transgene, also expressed red fluorescence labelling for Bio-GHRH. Thus, almost all (99%) of the somatotrophs are differentiated in that they bind Bio-GHRH. Note that the level and distribution of bio-GHRH varied from cell-to-cell, with an overlap in fluorescence (red and green) indicated by yellow label.
We then determined whether Ghrhr mRNA levels were changed in Lepr-null somatotrophs. Pure somatotrophs from control and somatotroph LEPR null mutants were collected and mRNA extracted for qPCR analysis of Ghrhr mRNA levels. Figure 1F shows that both male and female mutants exhibit a decrease in Ghrhr mRNA in somatotrophs.
3.2 |. Leptin regulates via transcriptional pathways
This reduced Ghrhr mRNA (Figure 1F) along with the reduced Gh mRNA reported in previous studies21 pointed to leptin-dependent transcriptional activation, potentially via the Janus kinase-signal transducer and activator of transcription-signal transducer and activator of transcription (JAK-STAT) pathway and mediated via phosphorylated STAT3 (pSTAT3). We tested this hypothesis utilising two transcriptional inhibitors in our in vitro studies with dispersed pituitary cultures: (i) STAT3i, a highly selective, potent blocker of STAT3 activation and STAT3-dependent Src transformation (Millipore) and (ii) actinomycin D (ActD), which non-specifically inhibits transcription by binding DNA at the transcription initiation complex.30 We observed that STAT3i did not have an effect on the basal numbers of GH-containing somatotrophs, although it did efficiently inhibit the leptin-mediated increase in number of somatotrophs (Figure 2A and B). Similarly, ActD did not impact basal levels of somatotrophs, although it prevented the leptin stimulation of somatotrophs (Figure 2C).
FIGURE 2.
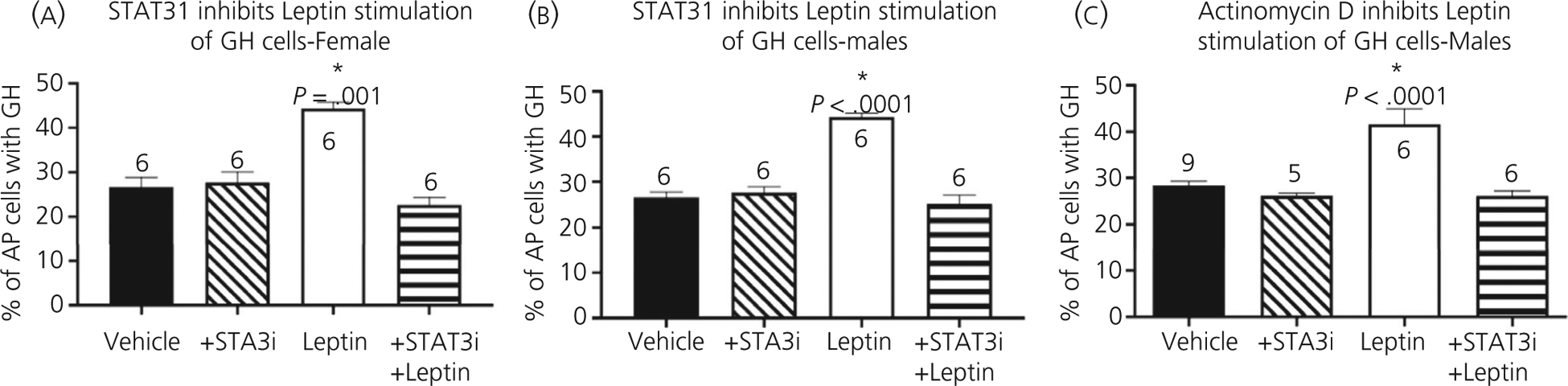
Transcriptional control of leptin-stimulated GH-containing cells. Dispersed pituitary cell cultures from adult control mice (24 hours) treated with transcriptional inhibitors (1 hour) prior to leptin stimulation (3 hour). Statistical analysis by two-way ANOVA followed by Tukey’s post-hoc test. In all cases, the source of variation came only from treatment with leptin, resulting in a higher number of growth hormone (GH) cells: (A) F1,16 = 27.49, P < .0001; (B) F1,20 = 30.16, P < .0001; (C) F1,22 = 14.34, P = .001. Interaction: (A) F1,16 = 75.41, P < .001; (B) F1,20 = 53.32, P < .001; (C) F1,22 = 14, P = .001
3.3 |. Leptin signals via post-transcriptional mechanisms
Our previous studies showed that Lepr-null mutant males have a reduction in GH protein levels despite no reduction in numbers of cells bearing Gh mRNA identified by in situ hybridisation,20 which suggested post-transcriptional repression of mRNA translation. Consistent with regulation at the level of mRNA translational control, an in silico analysis of the 105-nucleotide murine Gh mRNA 3’ UTR (NM_008117) identified predicted binding sites31 for several regulatory miRNAs. We therefore tested the expression of these candidate miRNAs by qPCR on mRNA extracts from control and mutant mice, reasoning that up-regulation would signify miRNA repression in the absence of LEPR in somatotrophs. Figure 3 shows that the certain miRNAs are increased in the somatotroph Lepr-null pituitary: miR-1197-3p (increased in female mutants only, Figure 3A) and miR103-3p and miR-590-3p (increased in both male and female mutants, Figure 3B and C). We also confirmed that miR-590-3p was specifically up-regulated in FACS-purified somatotrophs from Lepr-null females (Figure 3D). Finally, we further identified increased levels of miR325-3p, which targets the Ghrhr mRNA 3’ UTR (NM_001003685), in both male and female mutant pituitaries (Figure 3E). Collectively, these data suggest that leptin signalling may be required to de-repress miRNA-mediated control Gh or Ghrhr mRNA translation. We hypothesise that, because they are elevated, these candidate miRNAs may have exerted translational repression on the Gh and/or Ghrhr mRNAs.
FIGURE 3.
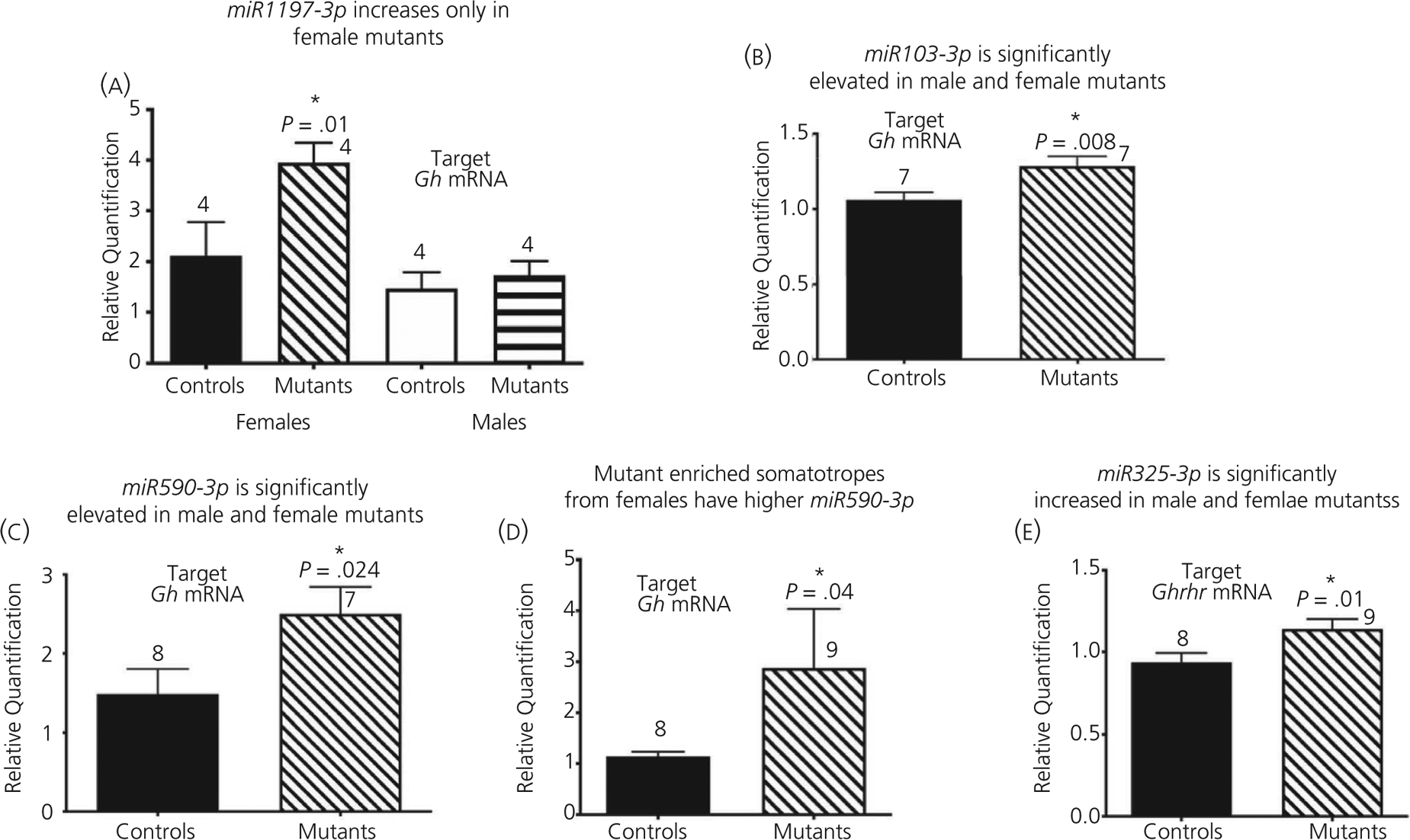
Pituitary changes in microRNA (miRNA) levels in Lepr-null pituitary. A-C,E, Pituitary miRNA from adult mice. A-C, miRNAs that potentially target Gh mRNA. D, miR590-3p in purified somatotrophs from females. E, miRNA that potentially target Ghrhr mRNA (combined males and females). *Significant. A, Two-way ANOVA followed by Fisher’s least significant difference post-hoc test showed a higher expression in mutant females than control females or males from any group. The source of variation comes from controls vs mutants, F1,10 = 6.063, P = .03; Males vs females, F1,10 = 11.36, P = .007. Interaction, F1,10 = 3.423, P = .094. B-E, Student’s t test: (B) t = 2.818 df = 11, P = .008, (C) t = 2.187 df = 12, (D) t = 2.04, df = 6, P = .04, (E) t = 2.446, df = 15, P = .01; n is indicated
In addition to the identified miRNA consensus sites in the Gh and Ghrhr mRNAs, our in silico analysis identified consensus binding sites for the mRNA translation protein Musashi (termed Musashi binding elements [MBEs]) in the regulatory 3’ UTRs of Tshb, Prl, and Pou1f1 mRNAs (the Gh and Ghrhr 3’ UTRs do not contain MBEs). The Musashi family of RNA-binding proteins has been implicated in maintenance of the stem and progenitor cells that are required for adult tissue repair and regeneration.32,33 Our ongoing studies similarly support a role for Musashi with respect to mediating adult pituitary cell function. Musashi has been shown to play a critical role in supporting stem and progenitor cell function33 by repressing the translation of target mRNAs that encode proteins required for cell maturation.34–36 To assess the ability of Musashi to exert translational repression via the Tshb and Prl 3’ UTRs, we utilised the firefly luciferase mRNA translation reporter (Fluc) assay. Figure 4A and B show that expression of Musashi (Msi1-AA) exerts translational repression of the Tshb 3’ UTR (reduced by 42%) and the Prl 3’ UTR (reduced by 20%). By contrast, a mutant form of Musashi1 with diminished RNA binding (Msi1-bm) did not exert translational repression of the Prl 3’ UTR and shows attenuated repression of the Tshb 3’ UTR reporter mRNA (Figure 4A and B). We further determined that the Prl mRNA is specifically associated with endogenous Musashi1 in immunoprecipitation assays of control pituitaries (Figure 4C).
FIGURE 4.
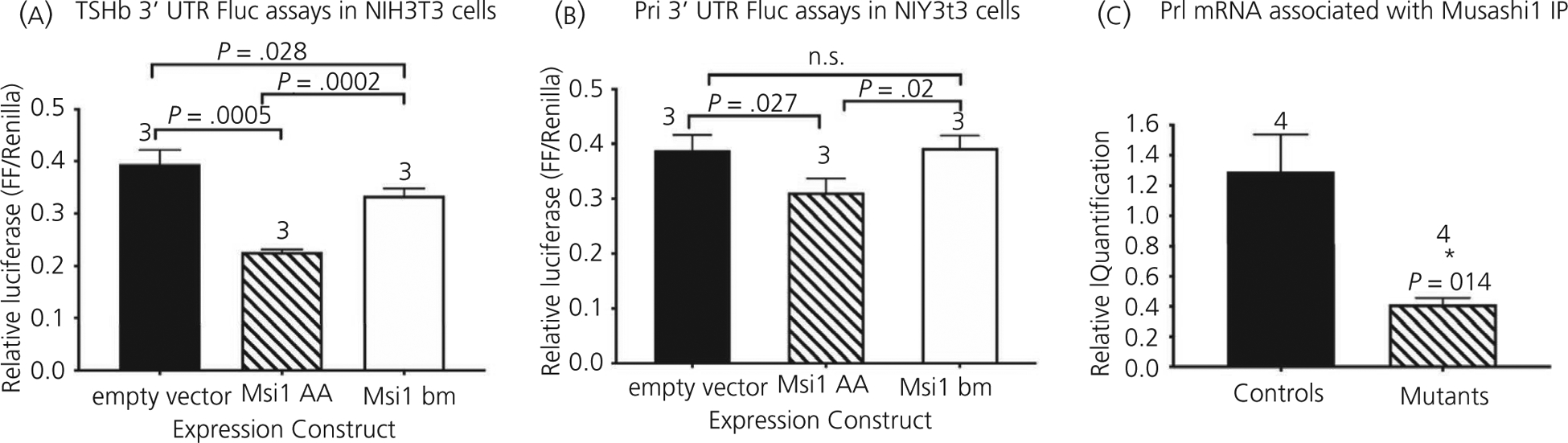
RNA luciferase reporter assay and Musashi co-association. A,B, Fluc assays for Tshb mRNA 3’ UTR (A) or Prl mRNA 3’ UTR (B). C, MSI1 immunoprecipitation assay and Prl mRNA enrichment. *Significant differences. Statistical analysis by ANOVA followed by Bonferroni’s post hoc test to detect differences between means. A, F3,6 = 65.51, P < .0001. B, F3,6 = 9.95, P = .012. C, Significance by Student’s t test with Welch’s correction. t = 2.781, df = 3.00, P = .034; n is indicated. NS, not significant
3.4 |. Ghrelin does not restore deficiencies in Lepr-null somatotrophs
Finally, after elucidating the signalling pathways used by leptin in the regulation of somatotrophs, we initiated studies of another somatotroph secretagogue, ghrelin, aiming to determine whether its unique signalling pathways would restore secretion or the number of GH-storing somatotrophs in the population. We thus performed in vitro studies on pituitary cultures from control and somatotroph Lepr exon 1-null mutant mice. The design of these studies was similar to those performed on the somatotroph Lepr-exon 17 null mutant mice.20 Figure 5A shows the GH secretory response to GHRH, in the presence or absence of 10 nmol L−1 ghrelin. As expected, ghrelin amplified secretion of GH when added with 1 or 3 nmol L−1 GHRH to control cultures, although ghrelin alone did not stimulate GH secretion. By contrast, ghrelin did not promote GH secretion in response to GHRH in the somatotroph Lepr-null pituitary cultures (Figure 5A). Notably, the GH-secretory response to GHRH by the mutant somatotrophs in the presence of ghrelin was lower than basal levels (no ghrelin stimulation) in control cultures. Figure 5B shows that the counts of somatotrophs immunolabelled for GH in control pituitaries are increased in the presence of GHRH with or without ghrelin. The mutant pituitary cell population does not exhibit increased numbers of GH cells in response to GHRH alone; however, we were able to see a modest stimulation in the presence of ghrelin (Figure 5B), although, as noted, these somatotrophs did not secrete more GH (Figure 5A).
FIGURE 5.
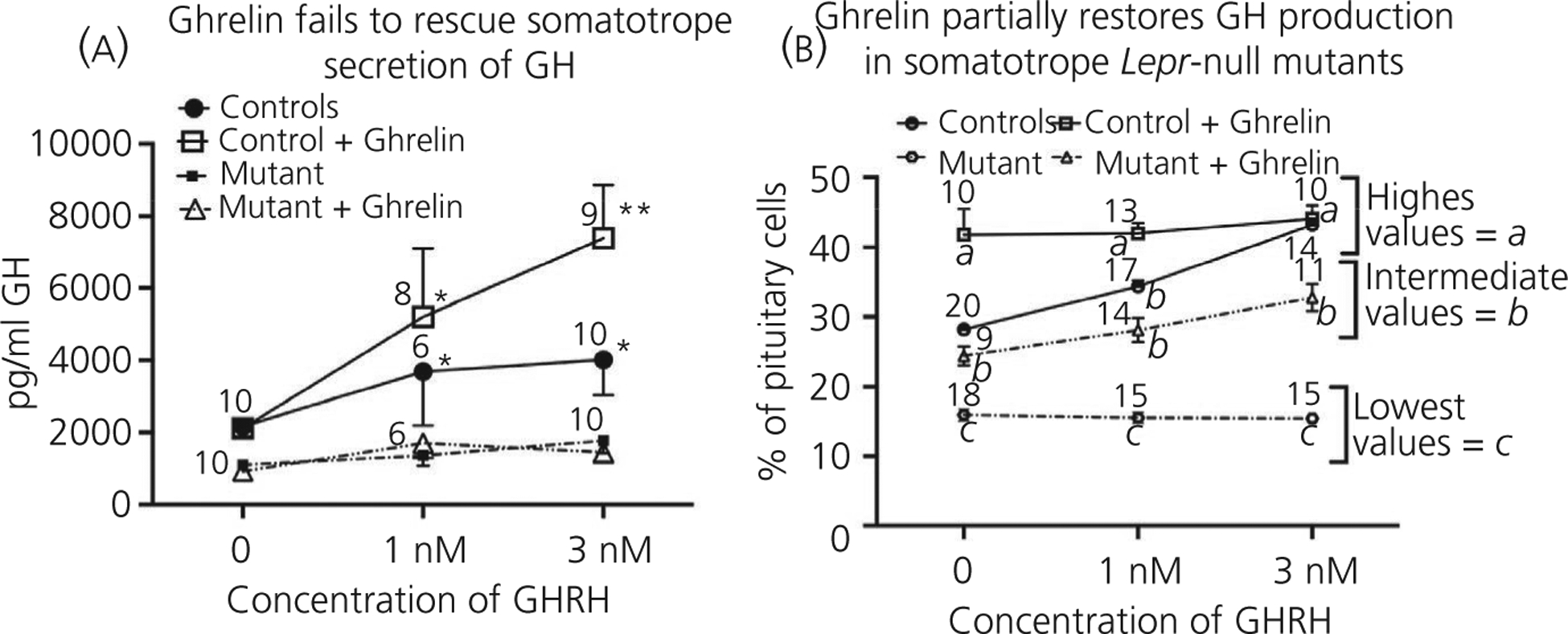
Growth hormone (GH) responses to GH-releasing hormone (GHRH) and ghrelin in Lepr-null pituitary. A, Dispersed whole pituitary cells from adult control mice and the Lepr-null model treated with 0–3 nmol L−1 GHRH (3 hours) in the absence or presence of 10 nmol L−1 ghrelin (+ ghrelin). (B) Immunolabelled GH cells were counted in fields from the experiment shown in (A). Statistical analysis: (A) analysis by three-way ANOVA followed by Tukey’s post-hoc test. The source of variation comes from comparing control vs mutant, with all mutant responses lower than control responses, F1,85 = 2.017, P < .0001 The second source of variation comes from the different doses of GHRH, F2,85 = 4.791, P = .01. Tukey’s post-hoc test identified the following different groups (designated by asterisks): **values higher than all others; *intermediate values higher than 0 nmol L−1 GHRH. (B) Analysis by three-way ANOVA followed by Tukey’s post-hoc test. The sources of variation are the dose of GHRH, F2,157 = 19.58, P < .0001; GHRH only compared to GHRH + ghrelin, F1,157 = 158.2, P < .0001; and control vs mutant, F1,157 = 443.9, P < .0001. Additional sources of variation included GHRH only vs GHRH + ghrelin × control vs mutant, F1,157 = 11.61, P = .0008 and dose of GHRH × GHRH only vs GHRH + ghrelin × control vs mutant, F2,157 = 14.10, P < .0001. Tukey’s post-hoc test identified three groups that are significantly different from one another: a, highest values; b, intermediate values; c, lowest values. These are shown on the graph; n is indicated
Finally, to identify the mechanism activated by ghrelin in control pituitaries, we tested the inhibitor of the nitric oxide pathway, l-NAME in our in vitro culture of control pituitary cells treated with ghrelin or leptin. The endpoint was a change, if any, in the number of immunolabelled GH cells, which tests the well-established stimulatory effect of leptin or ghrelin on GH stores. The percentages of GH-containing pituitary cells increased from 28 ± 1% to 47 ± 2% (P < .0001) in the presence of ghrelin and the addition of l-NAME inhibited the stimulatory effects of ghrelin (P < .0001) (Figure 6A). By contrast, Figure 6B shows that l-NAME did not inhibit leptin stimulation of GH-containing control pituitary cells, indicating that leptin acts independently of the nitric oxide pathway on somatotrophs. Similarly, ActD had no effect on ghrelin stimulation of GH cells (Figure 6C).
FIGURE 6.
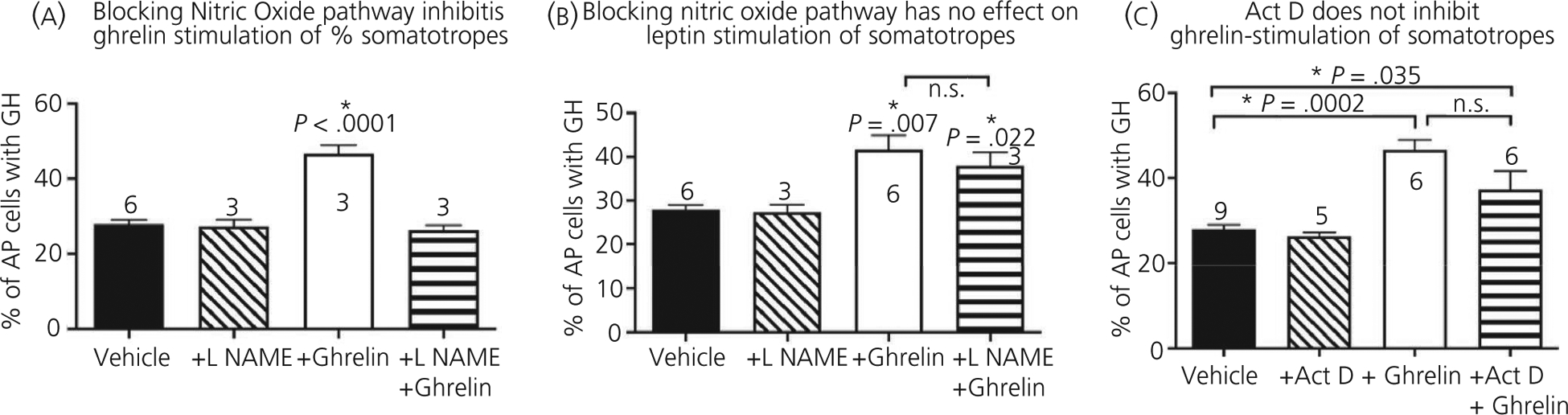
Signalling and transcription mechanisms in pituitary. Dispersed whole pituitary cells from adult control mice were stimulated with ghrelin (A) or leptin (B) for 3 hours after pretreatment with the nitric oxide signalling inhibitor, l-NG-nitro arginine methyl ester (l-NAME). (C) Actinomycin (D) pretreatment prior to ghrelin stimulation. *Significantly different from vehicle control levels as tested by two-way ANOVA followed by Tukey’s post-hoc test. A, Significance with or without l-NAME, F1,11 = 42.43, P < .0001 or vehicle vs ghrelin, F1,11 = 30.03, P = .0002. Interaction, F1,11 = 37.21, P < .0001. n is indicated. B, *Significance as tested by two-way ANOVA followed by Tukey’s post-hoc tests. B, N effect of l-NAME, F1,14 = 0.62, P = .444; and significant increases following leptin, F1,14 = 19.55, P = .0006. Interaction, F1,14 = 0.297, P = .594. C, *Significance tested by two-way ANOVA followed by Tukey’s post-hoc tests. Ghrelin stimulates more growth homrone (GH) cells, F1,11 = 44.61, P < .0001. Interaction, F1,11 = 2.980, P = .112. Actinomycin D (ActD) has no impact on control or ghrelin stimulation and high levels of GH cells are maintained following ghrelin treatment (compared to vehicle controls), F1,11 = 6.134, P = .03. AP, anterior pituitary
4 |. DISCUSSION
Our studies are directed at the identification of the cellular mechanisms behind the GH deficiency seen in Lepr-null mutants in which LEPR was functionally ablated in somatotrophs. Overall, we have observed that a loss of LEPR results in a significant reduction in GH secretion levels, GH protein stores, Gh mRNA levels (reduced in females only), GHRHR binding, Ghrhr mRNA levels and Pou1f1 protein levels.17–21 We have determined that these effects of the loss of leptin are mediated via control at both the transcriptional and post-transcriptional levels.
When addressing the role of leptin in transcriptional control, we noted that a reduction in levels of the transcription factor Pou1f1 would potentially affect transcription of both the Gh and Ghrhr genes. Furthermore, our studies utilising the STAT3 inhibitor indicate that leptin stimulation of GH stores is mediated via the transcriptional JAK/STAT pathway. This observation is consistent with findings with somatotroph tumor lines,37 showing that STAT3 specifically binds the rat Gh promoter and activates Gh transcription.
However, the JAK/STAT3 pathway is not implicated in the regulation of Ghrhr mRNA and our evidence suggests that the leptin-stimulated transcription of Ghrhr mRNA may be mediated via Pou1f1, which we find is reduced in the female deletion mutants, and up-regulated by leptin in vitro.21
When addressing the role of leptin in mRNA translation control, we noted that activation of the pSTAT3-signalling pathway has been linked to down-regulation of target miRNAs in breast cancer38 and leptin-dependent relief of miRNA-mediated translational control has been observed in adipocytes and in hepatic cells.39 We thus hypothesised that translation of the Gh and Ghrhr mRNAs may also be controlled by miRNAs in a leptin-dependent manner. Consistent with this hypothesis, our studies showed that miRNAs targeting Gh and Ghrhr mRNAs (miR1197-3p, miR103-3p, miR590-3p and miRNA-325-3p) are elevated in the absence of leptin signals. Our ongoing studies will determine the role of these identified miRNAs with respect to mediating the somatotroph response to leptin signalling, whereas our unbiased miRNA sequencing approach may identify additional leptin-sensitive miRNAs in somatotrophs.
Our studies indicate that RNA binding and the translational regulatory protein Musashi comprise another translation control mechanism that may be used by leptin. We have identified Musashi binding elements in the regulatory 3’ UTR of the Tshb, Prl and Pou1f1 mRNAs and our ongoing studies indicate that the MBEs are active and functional. Future studies will determine whether leptin impacts the repression of Tshb and Prl translation by Musashi.
Interestingly, our studies indicate that transcriptional and translational effects of leptin may also specifically target multihormonal somatotrophs. We reported that some of the TSHβ and PRL proteins are detected in a subpopulation of pure somatotrophs, which is a novel finding that may reflect the presence of a pool of progenitor cells.21 These findings correlate well with the recent analysis of single cell RNA-sequencing, which reported clusters of cells bearing Tshb mRNA overlapping clusters of cells expressing Prl and Gh mRNAs.40 The observed reduced number of somatotrophs with co-expressed GH, TSHβ and PRL in the Lepr-null mutants may indicate a role for leptin in the regulation and maintenance of this multihormonal/progenitor pool.
Our studies showing that GHRHR expression in somatotroph Lepr exon 1-null mice cannot be rescued by ghrelin is in contrast with our previous study of somatotroph Lepr-exon 17 bearing mice, which reported that ghrelin partially restored GHRH-mediated GH secretion in vitro.20 This points to the heightened severity of the deletion of all isoforms of LEPR in somatotrophs.17 Furthermore, with respect to our counts of GH-immunolabelled cells, ghrelin treatment restored GH stores to control basal levels in both types of mutants. However, the fact that GH secretion from the somatotroph Lepr exon 1 mutant somatotrophs remained significantly lower than that from control somatotrophs indicates that leptin has non-redundant roles with ghrelin with respect to mediating somatotroph secretion.
Furthermore, we have discovered that, even though ghrelin increases GHRH sensitivity normally in somatotrophs by increasing GHRHR (Figure 1), ghrelin is not able to recover the lost sensitivity to GHRH seen in the mice lacking leptin receptors (Figure 5). This may be impactful in cases of energy insufficiency following the consequential reduction in leptin signalling. Indeed, we showed previously that GHRHR binding was reduced in fasted rats, correlating with reduced serum leptin and stores of GH.16 However, our recently submitted studies of fasted male mice address this condition and show that mRNA levels of Ghrhr rise following fasting, despite the reduction in leptin. This highlights the need to discover additional regulators of Ghrhr mRNA, which may compensate for the loss in leptin signalling.
These studies demonstrate the multiple mechanisms by which leptin controls somatotroph function in response to nutritional and metabolic status. Leptin optimises somatotroph functions via both transcriptional and post-transcriptional pathways and also targets the subpopulation of multihormonal somatotrophs, suggesting a novel role for leptin in the regulation of somatotroph progenitor cells. Ongoing studies in our laboratories aim to elucidate these mechanisms further.
ACKNOWLEDGEMENTS
These studies were funded by National Institutes of Health R01HD059056 (GVC), 1R01DK113776-01 and R01HD093461 (GVC, AM, and MM) and R01HD087057 (GVC and AM); NIGMS P20 GM103425 and P30GM11070 (Dr Edgar Garcia-Rill)
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Number: R01HD059056, R01HD093461 and R01HD087057; National Institute of General Medical Sciences, Grant/Award Number: P20 GM103425 and P30GM11070; National Institute of Diabetes and Digestive and Kidney Diseases, Grant/Award Number: 1R01DK113776-01
Footnotes
The peer review history for this article is available at https://publons.com/publon/10.1111/jne.12883
CONFLICT OF INTERESTS
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Buzi F, Bontempelli AM, Alberti D, et al. Growth, insulin-like growth factor I (IGF-I), and IGF-binding proteins 1 and 3 in children with severe liver disease before and after liver transplantation: a longitudinal and cross-sectional study. Pediatr Res. 1998;43:478–483. [DOI] [PubMed] [Google Scholar]
- 2.Osterstock G, Escobar P, Mitutsova V, et al. Ghrelin stimulation of growth hormone-releasing hormone neurons is direct in the arcuate nucleus. PLoS One. 2010;5:e9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luque RM, Kineman RD, Park S, et al. Homologous and heterologous regulation of pituitary receptors for ghrelin and growth hormone-releasing hormone. Endocrinology. 2004;145:3182–3189. [DOI] [PubMed] [Google Scholar]
- 4.Kineman RD, Luque RM. Evidence that ghrelin is as potent as growth hormone (GH)-releasing hormone (GHRH) in releasing GH from primary pituitary cell cultures of a nonhuman primate (Papio anubis), acting through intracellular signaling pathways distinct from GHRH. Endocrinology. 2007;148:4440–4449. [DOI] [PubMed] [Google Scholar]
- 5.Farhy LS, Bowers CY, Veldhuis JD. Model-projected mechanistic bases for sex differences in growth hormone regulation in humans. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1577–R1593. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez-Borrego MC, Gahete MD, Martinez-Fuentes AJ, et al. Multiple signaling pathways convey central and peripheral signals to regulate pituitary function: Lessons from human and non-human primate models. Mol Cell Endocrinol. 2018;4634–4722. [DOI] [PubMed] [Google Scholar]
- 7.Sarmento-Cabral A, Peinado JR, Halliday LC, et al. Adipokines (Leptin, Adiponectin, Resistin) differentially regulate all hormonal cell types in primary anterior pituitary cell cultures from two primate species. Sci Rep. 2017;7:743537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isozaki O, Tsushima T, Miyakawa M, Demura H, Seki H. Interaction between leptin and growth hormone (GH)/IGF-I axis. Endocr J. 1999;46(Suppl):S17–S24. [DOI] [PubMed] [Google Scholar]
- 9.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim MH, Skoda RC. Defective STAT signaling by the leptin receptor in diabetic mice. Proc Natl Acad Sci U S A. 1996;93:6231–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carro E, Seoane LM, Senaris R, Casanueva FF, Dieguez C. Leptin increases in vivo GH responses to GHRH and GH-releasing peptide-6 in food-deprived rats. Eur J Endocrinol. 2000;142:66–70. [DOI] [PubMed] [Google Scholar]
- 11.Carro E, Pinilla L, Seoane LM, et al. Influence of endogenous leptin tone on the estrous cycle and luteinizing hormone pulsatility in female rats. Neuroendocrinology. 1997;66:375–377. [DOI] [PubMed] [Google Scholar]
- 12.Asada N, Takahashi Y, Honjo M. Effects of 22K or 20K human growth hormone on lipolysis, leptin production in adipocytes in the presence and absence of human growth hormone binding protein. Horm Res. 2000;54:203–207. [DOI] [PubMed] [Google Scholar]
- 13.Sone M, Osamura RY. Leptin and the pituitary. Pituitary. 2001;4:15–23. [DOI] [PubMed] [Google Scholar]
- 14.Sone M, Nagata H, Takekoshi S, Osamura RY. Expression and localization of leptin receptor in the normal rat pituitary gland. Cell Tissue Res. 2001;305:351–356. [DOI] [PubMed] [Google Scholar]
- 15.Luque RM, Huang ZH, Shah B, Mazzone T, Kineman RD. Effects of leptin replacement on hypothalamic-pituitary growth hormone axis function and circulating ghrelin levels in ob/ob mice. Am J Physiol Endocrinol Metab. 2007;292:E891–E899. [DOI] [PubMed] [Google Scholar]
- 16.Crane C, Akhter N, Johnson BW, et al. Fasting and glucose effects on pituitary leptin expression: is leptin a local signal for nutrient status? J Histochem Cytochem. 2007;55:1059–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allensworth-James M, Odle AK, Haney A, Childs GV. Sex differences in somatotrope dependency on leptin receptors in young mice: Ablation of LEPR causes severe growth hormone deficiency and abdominal obesity in males. Endocrinology. 2015;156:3253–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Childs GV, Akhter N, Haney A, et al. The somatotrope as a metabolic sensor: deletion of leptin receptors causes obesity. Endocrinology. 2011;152:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhter N, Odle AK, Allensworth-James ML, et al. Ablation of leptin signaling to somatotropes: changes in metabolic factors that cause obesity. Endocrinology. 2012;153:4705–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syed M, Cozart M, Haney AC, et al. Ghrelin restoration of function in vitro in somatotropes from male mice lacking the Janus kinase (JAK)-binding site of the leptin receptor. Endocrinology. 2013;154:1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odle A, Allensworth-James M, Akhter N, et al. A Sex-Dependent Tropic Role for Leptin In The Somatotrope As A Regulator of POU1F1 and POU1F1-dependent Hormones. Endocrinology. 2016;157:3958–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouadjo K, Nishida Y, Cadrin-Girard J, Yoshiioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genom. 2007;8:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odle AK, Benes H, Melgar Castillo A, et al. Association of Gnrhr mRNA with the stem cell determinant musashi: a mechanism for leptin-mediated modulation of GnRHR expression. Endocrinology. 2018;159:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Childs GV, Unabia G, Miller BT, Collins TJ. Differential expression of gonadotropin and prolactin antigens by GHRH target cells from male and female rats. J Endocrinol. 1999;162:177–187. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahim SN, Moussa SM, Childs GV. Morphometric studies of rat anterior pituitary cells after gonadectomy: correlation of changes in gonadotropes with the serum levels of gonadotropins. Endocrinology. 1986;119:629–637. [DOI] [PubMed] [Google Scholar]
- 26.Childs GV, Unabia G, Rougeau D. Cells that express luteinizing hormone (LH) and follicle-stimulating hormone (FSH) beta-subunit messenger ribonucleic acids during the estrous cycle: the major contributors contain LH beta, FSH beta, and/or growth hormone. Endocrinology. 1994;134:990–997. [DOI] [PubMed] [Google Scholar]
- 27.Arumugam K, MacNicol MC, Wang Y, et al. Ringo/cyclin-dependent kinase and mitogen-activated protein kinase signaling pathways regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J Biol Chem. 2012;287:10639–10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacNicol AM, Hardy LL, Spencer HJ, MacNicol MC. Neural stem and progenitor cell fate transition requires regulation of Musashi1 function. BMC Dev Biol. 2015;15:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci U S A. 1985;82:5328–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horisawa K, Imai T, Okano H, Yanagawa H. The Musashi family RNA-binding proteins in stem cells. Biomol Concepts. 2010;1:59–66. [DOI] [PubMed] [Google Scholar]
- 33.Fox RG, Park FD, Koechlein CS, Kritzik M, Reya T. Musashi signaling in stem cells and cancer. Annu Rev Cell Dev Biol. 2015;31:249–267. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai T, Tokunaga A, Yoshida T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. [DOI] [PubMed] [Google Scholar]
- 37.Zhou C, Jiao Y, Wang R, Ren S-G, Wawrosky K, Melmed S. STAT3 upregulation in pituitary somatotroph adenomas induces growth hormone hypersecretion. J Clin Invest. 2015;125:1692–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L, Chen C, Shi M, et al. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene. 2013;32:5272–5282. [DOI] [PubMed] [Google Scholar]
- 39.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. [DOI] [PubMed] [Google Scholar]
- 40.Cheung LYM, George AS, McGee SR, et al. Single-Cell RNA sequencing reveals novel markers of male pituitary stem cells and hormone-producing cell types. Endocrinology. 2018;159:3910–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]


