Abstract
Arthrinium has a widespread distribution occurring in various substrates (e.g., air, soil debris, plants, lichens, marine algae and even human tissues). It is characterized by the basauxic conidiogenesis in the asexual morph, with apiospores in the sexual morph. In this study, seventeen isolates of Arthrinium were collected in China. Based on their morphology and phylogenetic characterization, four new species (A. biseriale, A. cyclobalanopsidis, A. gelatinosum, and A. septatum) are described and seven known species (A. arundinis, A. garethjonesii, A. guizhouense, A. hydei, A. neosubglobosa, A. phyllostachium and A. psedoparenchymaticum) are identified, of which the sexual morph of three species (A. guizhouense, A. phyllostachium and A. psedoparenchymaticum) and asexual morph of A. garethjonesii are reported for the first time. The detailed descriptions, illustrations and comparisons with related taxa of these new collections are provided. Phylogenetic analyses of combined ITS, LSU, TUB2, and TEF sequence data support their placements in the genus Arthrinium and justify the new species establishments and identifications of known species.
Keywords: 4 new taxa, asexual-sexual morphs, multi-genes, phylogeny, taxonomy
Introduction
The genus Arthrinium Kunze belongs to the family Apiosporaceae, which was introduced by Hyde et al. (1998) and typified by the genus Apiospora (Ellis, 1971; Seifert et al., 2011; Hyde et al., 2020). Arthrinium is the largest genus within Apiosporaceae and it has a widespread distribution on variety of hosts, 77 species have been recorded by Species Fungrom (March, 2021). Classification of Arthrinium was primarily based on conidial shape, conidiophores, sterile cells, and the presence of setae. Some morphologically different taxa (genera) grouped with Arthrinium when use the molecular data (Crous and Groenewald, 2013). Thus, those characteristics could be not fully inferred about the phylogenetic relationships for Arthrinium (Crous and Groenewald, 2013; Wang et al., 2018; Pintos et al., 2019). Except for being reported as saprobes (Agut and Calvo, 2004; Crous and Groenewald, 2013; Dai et al., 2016, 2017; Jiang et al., 2018; Wang et al., 2018; Jiang et al., 2019, 2020; Luo et al., 2019; Pintos et al., 2019; Yan et al., 2019; Mapook et al., 2020; Senanayake et al., 2020a; Tang et al., 2020), the species of Arthrinium also includes phytopathogenic fungi, for instance, A. arundinis causing brown culm streak of Phyllostachys praecox, A. phaeospermum causing culm rot on Phyllostachys viridis and cutaneous infections of humans (Rai, 1989; Zhao et al., 1990; Martínez-Cano et al., 1992; Mavragani et al., 2007; Crous and Groenewald, 2013; Li et al., 2016; Wang et al., 2018).
we are carrying out the survey of fungal diversity in Karst formations of the Asian region, and many new taxa are described in last few years (Li et al., 2016; Chen et al., 2017, 2020; Zhang et al., 2017, 2019; Feng et al., 2019; Liu et al., 2019; Dissanayake et al., 2020b). In this study, seventeen Arthrinium-like were collected in Guizhou and Guangdong province, China and can be recognized as eleven Arthrinium species based on morphological characters and phylogeny inferred from the multi-gene sequences data (ITS, LSU, TUB2, and TEF) analyses, which four new species (A. biseriale, A. cyclobalanopsidis, A. gelatinosum, and A. septatum) and seven known species (A. arundinis, A. garethjonesii, A. guizhouense, A. hydei, A. neosubglobosa, A. phyllostachium and A. psedoparenchymaticum) are introduced and identified, respectively. The aim of this study is to describe these new taxa with detailed descriptions and illustrations, and also provide their phylogenetic relationships within Arthrinium based on multi-gene analysis.
Materials and Methods
Sample Collection, Morphological Studies and Isolation
Samples were collected from Guizhou and Guangdong Province in China. Fungal fruiting bodies were examined by using stereomicroscope (Motic SMZ 168). Free hand sections of fungal structures were mounted in water for microscopic studies and photomicrography. Images were taken by using a Nikon ECLIPSE Ni compound microscope fitted with a Canon EOS 70D digital camera. All measurements were taken by using Tarosoft Image Frame Work software (IFW) (Liu et al., 2010), and photo plates were processed with Adobe Photoshop CS6 software (Adobe Systems, United States).
The single spore isolation followed the method described in Senanayake et al. (2020b). Parts of morphological descriptions were based on sporulated cultures on WA (Water Agar) at room temperature (ca. 25°C). Type specimens were deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (HKAS), Kunming, China and Guizhou Academy of Agriculture sciences Herbarium (GZAAS). Pure cultures were deposited in China General Microbiological Culture Collection Center (CGMCC) and Guizhou Culture Collection (GZCC). Faces of Fungi1 number is obtained as described in the paper by Jayasiri et al. (2015), and the new taxa are registered in Index Fungorum (2021).
DNA Extraction, PCR Amplification and Sequencing
Fungal mycelia were scraped from the pure culture which were growing on PDA (Potato Dextrose Agar) for one week at 25°C in dark. DNA was extracted by using Ezup Column Fungi Genomic DNA Purification Kit (Sangon Biotech, China) from fresh fungal mycelia, but some were extracted directly from fruiting bodies by using Forensic DNA Kit (Omega Bio-Tek, China). Four gene regions, large subunit rDNA (LSU), internal transcribed spacer (ITS), beta-tubulin (TUB2) and the translation elongation factor 1-alpha (TEF) gene were amplified by the primer pairs LR0R and LR5 (Vilgalys and Hester, 1990), ITS5 and ITS4, T1 (O’Donnell and Cigelnik, 1997) and Bt2b (Glass and Donaldson, 1995), EF1-728F and EF-2 (O’Donnell et al., 1998; Carbone and Kohn, 1999), respectively. Polymerase chain reaction (PCR) was carried out in 25 μL reaction volume containing 12.5 μL 2 × PCR Master Mix (Sangon Biotech, China), 9.5 μL ddH2O, 1 μL of each primer and 1 μL DNA template. The annealing temperatures were adjusted to 56°C for ITS, LSU and TUB2, and 55°C for TEF. PCR products were sent to sequence at Sangon Biotech Co., Ltd., China. The PCR products were examined using 1.2% agarose electrophoresis gel stained with ethidium bromide. PCR products were purified and sequenced by Sangon Biotech (Shanghai) Co., Ltd., China. New generated nucleotide sequences were submitted in GenBank (Table 1).
TABLE 1.
GenBank accession numbers of species included in the phylogenetic study.
| Species | Strain no. | GenBank Accession Numbers |
|||
| LSU | ITS | TEF | TUB2 | ||
| Arthrinium acutiapicum | KUMCC 20–0209 | MT946338 | MT946342 | MT947359 | MT947365 |
| A. acutiapicum | KUMCC 20–0210 | MT946339 | MT946343 | MT947360 | MT947366 |
| A. aquaticum | MLFU 18–1628 | MK835806 | MK828608 | N/A | N/A |
| A. arundinis | CBS 106.12 | KF144927 | KF144883 | KF145015 | KF144973 |
| A. arundinis | CBS 114316 | KF144928 | KF144884 | KF145016 | KF144974 |
| A. arundinis | GZCC 20–0116 | MW478899 | MW481720 | MW522952 | MW522968 |
| A. aureum | CBS 244.83 | KF144935 | AB220251 | KF145023 | KF144981 |
| A. balearicum | CBS 145129 | MK014836 | MK014869 | MK017946 | MK017975 |
| A. bambusae | LC7107 | N/A | KY494719 | KY705117 | KY705187 |
| A. bambusae | LC7106 | KY494794 | KY494718 | KY806204 | KY705186 |
| A. bambusicola | MFLUCC 20–0144 | MW173087 | MW173030 | MW183262 | N/A |
| A. biseriale | CGMCC 3.20135 | MW478885 | MW481708 | MW522938 | MW522955 |
| A. biseriale | GZCC 20–0099 | MW478886 | MW481709 | MW522939 | MW522956 |
| A. biseriale | GZCC 20–0100 | MW478887 | MW481710 | MW522940 | MW522957 |
| A. camellia-sinensis | LC8181 | KY494837 | KY494761 | KY705157 | KY705229 |
| A. camellia-sinensis | LC5007 | KY494780 | KY494704 | KY705103 | KY705173 |
| A. caricicola | CBS 145127 | MK014838 | MK014871 | MK017948 | MK017977 |
| A. chinense | CFCC53036 | N/A | MK819291 | MK818545 | MK818547 |
| A. chinense | CFCC53037 | N/A | MK819292 | MK818546 | MK818548 |
| A. chromolaenae | MFLUCC 17-1505 | MT214436 | MT214342 | N/A | N/A |
| A. cyclobalanopsidis | CGMCC 3.20136 | MW478892 | MW481713 | MW522945 | MW522962 |
| A. cyclobalanopsidis | GZCC 20–0103 | MW478893 | MW481714 | MW522946 | MW522963 |
| A. descalsii | CBS 145130 | MK014837 | MK014870 | MK017947 | MK017976 |
| A. dichotomanthi | LC8175 | KY494831 | KY494755 | KY705151 | kY705223 |
| A. dichotomanthi | LC4950 | KY494773 | KY494697 | KY705096 | KY705167 |
| A. esporlense | CBS 145136 | MK014845 | MK014878 | MK017954 | MK017983 |
| A. euphorbiae | IMI 285638b | AB220335 | AB220241 | N/A | AB220288 |
| A. gaoyouense | CFCC 52301 | N/A | MH197124 | MH236793 | MH236789 |
| A. gaoyouense | CFCC 52302 | N/A | MH197125 | MH236794 | MH236790 |
| A. garethjonesii | KUMCC16–0202 | KY356091 | KY356086 | N/A | N/A |
| A. garethjonesii | GZCC 20–0115 | MW478894 | MW481715 | MW522947 | N/A |
| A. gelatinosum | KHAS 11962 | MW478888 | MW481706 | MW522941 | MW522958 |
| A. gelatinosum | GZAAS 20–0107 | MW478889 | MW481707 | MW522942 | MW522959 |
| A. guizhouense | LC5318 | KY494784 | KY494708 | KY705107 | KY705177 |
| A. guizhouense | LC5322 | KY494785 | KY494709 | KY705108 | KY705178 |
| A. guizhouense | GZCC 20–0114 | MW478895 | MW481716 | MW522948 | MW522964 |
| A. gutiae | CBS 135835 | KR149063 | KR011352 | KR011351 | KR011350 |
| A. hispanicum | IMI 326877 | AB220336 | AB220242 | N/A | AB220289 |
| A. hydei | CBS 114990 | KF144936 | KF144890 | KF145024 | KF144982 |
| A. hydei | KUMCC 16–0204 | KY356092 | KY356087 | N/A | N/A |
| A. hydei | GZCC 20–0113 | MW478900 | MW481721 | MW522953 | N/A |
| A. hyphopodii | KUMCC 16–0201 | KY356093 | KY356088 | N/A | N/A |
| A. hyphopodii | MFLUCC15–0003 | N/A | KR069110 | N/A | N/A |
| A. hysterinum | CBS 145134 | MK014843 | MK014876 | N/A | N/A |
| A. hysterinum | ICMP 6889 | MK014841 | MK014874 | MK017951 | MK017980 |
| A. ibericum | CBS 145137 | MK014846 | MK014879 | MK017955 | MK017984 |
| A. italicum | CBS 145138 | MK014847 | MK014880 | MK017956 | MK017985 |
| A. japonicum | IFO 30500 | AB220356 | AB220262 | N/A | AB220309 |
| A. japonicum | IFO 31098 | AB220358 | AB220264 | N/A | AB220311 |
| A. jatrophae | MMI 00052 | N/A | JQ246355 | N/A | N/A |
| A. jatrophae | AMH–9557 | N/A | JQ246355 | N/A | N/A |
| A. jiangxiense | LC4577 | KY494769 | KY494693 | KY705092 | KY705163 |
| A. jiangxiense | LC4578 | KY494770 | KY494694 | KY705093 | KY705164 |
| A. kogelbergense | CBS 113332 | KF144937 | KF144891 | KF145025 | KF144983 |
| A. kogelbergense | CBS 113333 | KF144938 | KF144892 | KF145026 | KF144984 |
| A. locuta-pollinis | LC11683 | N/A | MF939595 | MF939616 | MF939622 |
| A. longistromum | MFLUCC 11–0481 | KU863129 | KU940141 | N/A | N/A |
| A. longistromum | MFLUCC 11–0479 | KU863130 | KU940142 | N/A | N/A |
| A. malaysianum | CBS 102053 | KF144942 | KF144896 | KF145030 | KF144988 |
| A. malaysianum | CBS 251.29 | KF144943 | KF144897 | KF145031 | KF144989 |
| A. marii | CBS 497.90 | KF144947 | AB220252 | KF145035 | KF144993 |
| A. mediterranei | IMI 326875 | AB220337 | AB220243 | N/A | AB220290 |
| A. minus | CBS 145131 | MK014839 | MK014872 | MK017949 | MK017978 |
| A. mytilomorphum | DAOM 214595 | N/A | KY494685 | N/A | N/A |
| A. garethjonesii | KUMCC 18–0192 | MK070898 | MK070897 | N/A | N/A |
| A. neosubglobosa | KUMCC 16–0203 | KY356095 | KY356090 | N/A | N/A |
| A. neosubglobosa | JHB006 | KY356094 | KY356089 | N/A | N/A |
| A. neosubglobosa | GZAAS 20–0099 | MW478901 | MW481705 | MW522954 | MW522969 |
| A. obovatum | LC8177 | KY494833 | KY494757 | KY705153 | KY705225 |
| A. obovatum | LC4940 | KY494772 | KY494696 | KY705095 | KY705166 |
| A. ovatum | CBS 115042 | KF144950 | KF144903 | KF145037 | KF144995 |
| A. paraphaeospermum | MFLUCC 13–0644 | KX822124 | KX822128 | N/A | N/A |
| A. phaeospermum | CBS 114314 | KF144951 | KF144904 | KF145038 | KF144996 |
| A. phaeospermum | CBS 114315 | KF144952 | KF144905 | KF145039 | KF144997 |
| A. phragmites | CPC 18900 | KF144956 | KF144909 | KF145043 | KF145001 |
| A. phyllostachium | MFLUCC 18–1101 | MH368077 | MK351842 | MK340918 | MK291949 |
| A. phyllostachium | GZCC 20–0111 | MW478896 | MW481717 | MW522949 | MW522965 |
| A. phyllostachium | GZCC 20–0112 | MW478897 | MW481718 | MW522950 | MW522966 |
| A. piptatheri | CBS 145149 | MK014860 | MK014893 | MK017969 | N/A |
| A. pseudoparenchymaticum | LC7234 | KY494819 | KY494743 | KY705139 | KY705211 |
| A. pseudoparenchymaticum | LC8173 | KY494829 | KY494753 | KY705149 | KY705221 |
| A. pseudoparenchymaticum | GZCC 20–0117 | MW478898 | MW481719 | MW522951 | MW522967 |
| A. pseudorasikravindrae | KUMCC 20–0208 | N/A | MT946344 | MT947361 | MT947367 |
| A. pseudorasikravindrae | KUMCC 20–0211 | N/A | MT946345 | MT947362 | MT947368 |
| A. pseudosinense | CPC 21546 | KF144957 | KF144910 | KF145044 | N/A |
| A. pseudosinense | CBS 135459 | N/A | KF144910 | KF145044 | N/A |
| A. pseudospegazzinii | CBS 102052 | KF144958 | KF144911 | KF145045 | KF145002 |
| A. pterospermum | CBS 123185 | KF144959 | KF144912 | N/A | KF145003 |
| A. pterospermum | CPC 20193 | KF144960 | KF144913 | KF145046 | KF145004 |
| A. puccinioides | CBS 145150 | MK014861 | MK014894 | MK017970 | MK017998 |
| A. puccinioides | CBS 549.86 | AB220347 | AB220253 | N/A | AB220300 |
| A. qinlingense | CFCC 52303 | N/A | MH197120 | MH236795 | MH236791 |
| A. qinlingense | CFCC 52304 | N/A | MH197121 | MH236796 | MH236792 |
| A. rasikravindrae | CBS 145152 | MK014863 | MK014896 | MK017971 | MK017999 |
| A. rasikravindrae | LC7115 | KY494797 | KY494721 | KY705118 | KY705189 |
| A. rasikravindrae | NFCCI 2144 | N/A | JF326454 | N/A | N/A |
| A. sacchari | CBS 212.30 | KF144963 | KF144917 | KF145048 | KF145006 |
| A. sacchari | CBS 301.49 | KF144962 | KF144916 | KF145047 | KF145005 |
| A. saccharicola | CBS 191.73 | KF144966 | KF144920 | KF145051 | KF145009 |
| A. saccharicola | CBS 463.83 | KF144968 | KF144921 | KF145053 | KF145011 |
| A. septatum | CGMCC 3.20134 | MW478890 | MW481711 | MW522943 | MW522960 |
| A. septatum | GZCC 20–0109 | MW478891 | MW481712 | MW522944 | MW522961 |
| A. serenense | IMI 326869 | AB220344 | AB220250 | N/A | AB220297 |
| A. setostromum | KUMCC 19–0217 | MN528011 | MN528012 | MN527357 | N/A |
| A. sporophleum | CBS 145154 | MK014865 | MK014898 | MK017973 | MK018001 |
| A. subglobosum | MFLUCC 11–0397 | KR069113 | KR069112 | N/A | N/A |
| A. subroseum | LC7292 | KY494828 | KY494752 | KY705148 | KY705220 |
| A. thailandicum | MFLUCC 15–0199 | KX986111 | KU940146 | N/A | N/A |
| A. thailandicum | MFLUCC 15–0202 | KU863133 | KU940145 | N/A | N/A |
| A. trachycarpum | CFCC 53038 | N/A | MK301098 | MK303396 | MK303394 |
| A. trachycarpum | CFCC 53039 | N/A | MK301099 | MK303397 | MK303395 |
| A. urticae | IMI 326344 | AB220339 | AB220245 | N/A | N/A |
| A. vietnamese | IMI 99670 | KX986111 | KX986096 | N/A | KY019466 |
| A. xenocordella | CBS478.86 | KF144970 | KF144925 | KF145055 | KF145013 |
| A. xenocordella | CBS 595.66 | KF144971 | KF144926 | N/A | N/A |
| A. yunnanum | DDQ 00281 | KU863136 | KU940148 | N/A | N/A |
| A. yunnanum | MFLU 15–0002 | N/A | KU940147 | N/A | N/A |
| Seiridium phylicae | CPC 19962 | NG 042759 | LT853092 | LT853189 | LT853239 |
| Seiridium phylicae | CPC 19965 | KC005809 | LT853093 | LT853190 | LT853240 |
The newly generated sequence is shown in bold. AMH: Ajrekar Mycological herbarium, Pune, Maharashtra, India; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Microbiological Culture Collection Center, Beijing, China; CPC: Culture collection of Pedro Crous, housed at the Westerdijk Fungal Biodiversity Institute; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; DDQ: D.Q. Dai; GZAAS: Guizhou Academy of Agricultural Sciences herbarium, China; GZCC: Guizhou Culture Collection, China; HKAS: Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica; ICMP: International Collection of Microorganisms from Plants, New Zealand; IFO: Institute for Fermentation, Osaka, Japan; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; JHB: H.B. Jiang; KUMCC: Culture collection of Kunming Institute of Botany, Yunnan, China; LC: Working collection of Lei Cai, housed at CAS, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NFCCI: National Fungal Culture Collection of India.
Phylogenetic Analyses
Phylogenetic analyses were performed based on ITS, LSU, TUB2 and TEF sequence data. The related strains of Apiosporaceae (Table 1) used for analysis were referred to BLAST2 results and relevant publications (Wang et al., 2018; Jiang et al., 2019, 2020; Pintos et al., 2019; Tang et al., 2020). Sequences were obtained from GenBank and aligned using MAFFT v. 7 (Katoh and Standley, 2013). Manual adjustment was also performed when it is necessary by using BioEdit v. 7.0 (Hall, 1999). The alignment of sequences data used in analyses is deposited in TreeBASE under the accession number S27728. The analyses of maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) were carried out as detailed in Dissanayake et al. (2020a) and programs used including PAUP v.4.0b 10 (Hillis and Bull, 1993; Swofford, 2002), raxmlGUI v. 1.3 (Silvestro and Michalak, 2012), and MrBayes v3.1.2 (Rannala and Yang, 1996; Huelsenbeck and Ronquist, 2001; Zhaxybayeva and Gogarten, 2002; Nylander, 2004). Trees were visualized with FigTree v1.4.2 (Rambaut, 2012) and the layout was edited using Adobe Illustrator CS6.
Results
Phylogeny
To determine the phylogenetic placement of the new collections in this study, the combined ITS, LSU, TUB2 and TEF data set comprised 119 taxa with Seiridium phylicae (CPC 19962 and CPC 19965) as the outgroup taxa. The concatenated alignment comprises 2,770 characters (ITS: 1–635; LSU: 636–1,454; TUB2: 1,455–2,300; TEF: 2,301–2,770) including gaps, of which 1,361 characters were constant, and 1,279 characters are parsimony informative and 130 are parsimony uninformative. Maximum likelihood, maximum parsimony and Bayesian analyses were performed, respectively, and presented consistent topologies. The best scoring RAxML tree (Figure 1) is obtained with a final likelihood value of -27544.434044. Estimated base frequencies were as follows: A = 0.236829, C = 0.253015, G = 0.251523, T = 0.258634; substitution rates AC = 1.154254, AG = 2.738844, AT = 1.073582, CG = 0.896658, CT = 4.134885, GT = 1.000000; The gamma distribution shape parameter alpha is equal to 0.332641 and the Tree-Length equal to 3.795226.
FIGURE 1.
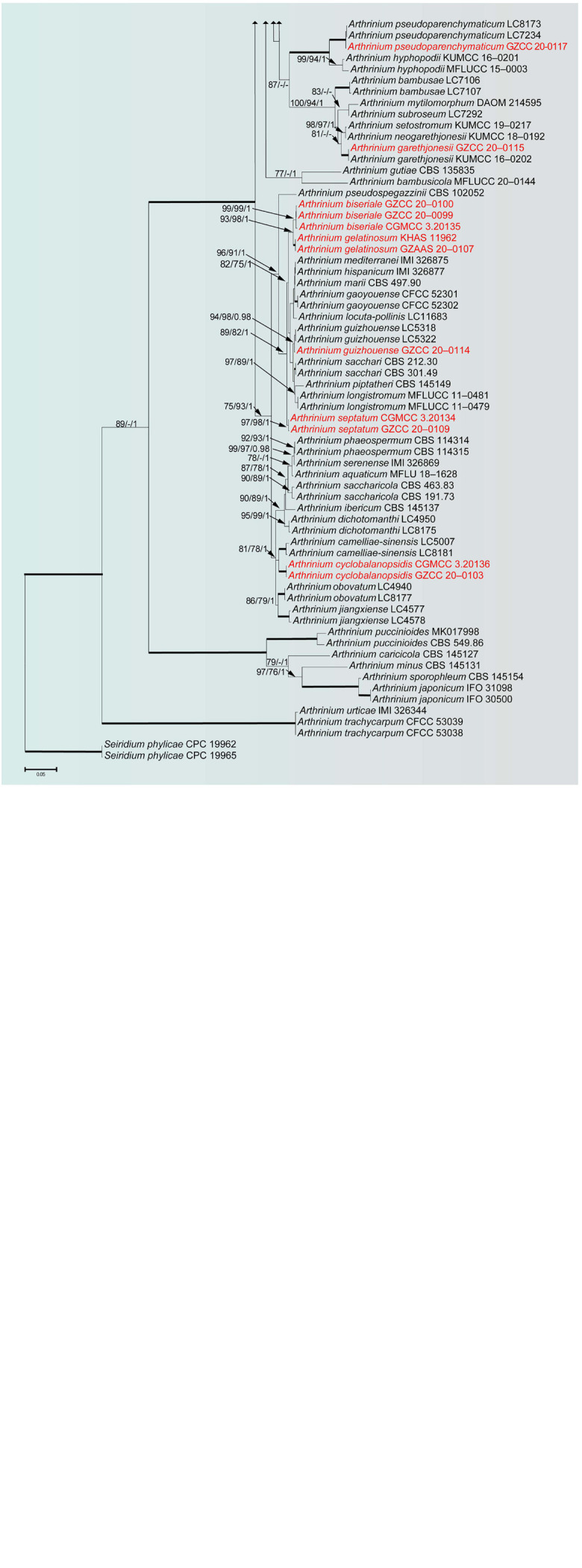
RAxML tree of representatives based on a combined dataset of ITS, LSU, TUB2 and TEF sequences. Bootstrap support values for ML, MP (≥75%) and Bayesian (≥0.95) are given at the nodes (ML/MP/BI). Branches with ML, MP and BI equal 100, 100 and 1 are in bold. The tree is rooted with Seiridium phylicae (CPC 19962 and CPC 19965). New strains are shown in red.
Phylogenetic analyses showed that our newly collected seventeen taxa clustered into eleven clades and can be recognized as seven known species (Arthrinium arundinis, A. garethjonesii, A. guizhouense, A. hydei, A. neosubglobosa, A. phyllostachium, A. psedoparenchymaticum) and four new species (A. biseriale, A. cyclobalanopsidis, A. gelatinosum, A. septatum) (Figure 1).
Taxonomy
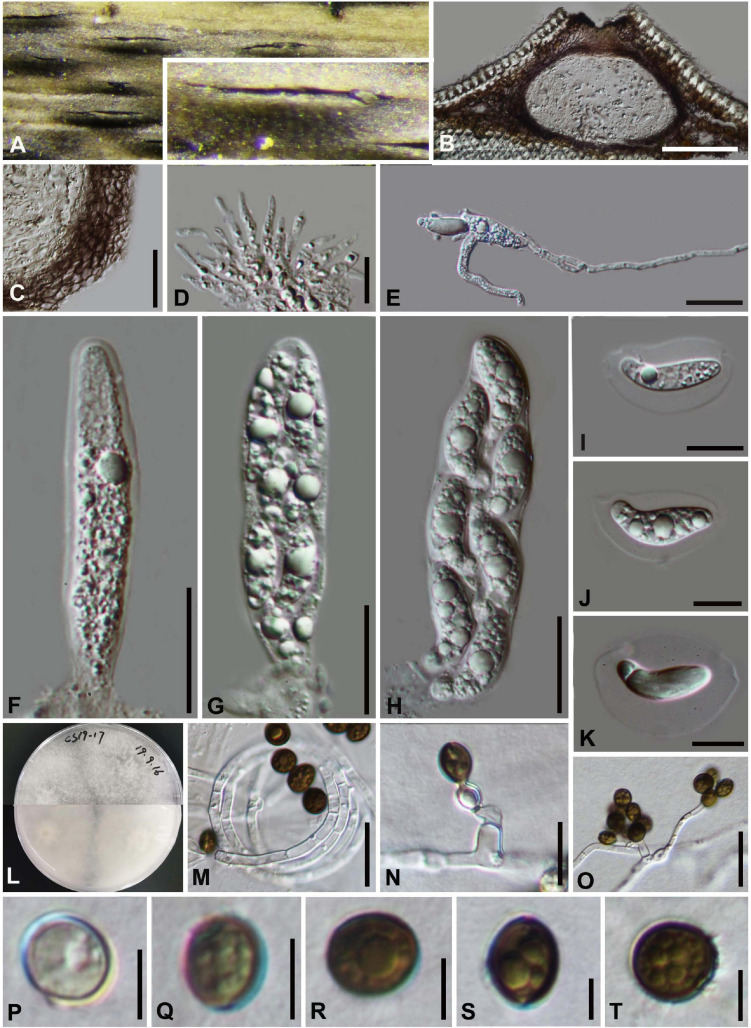
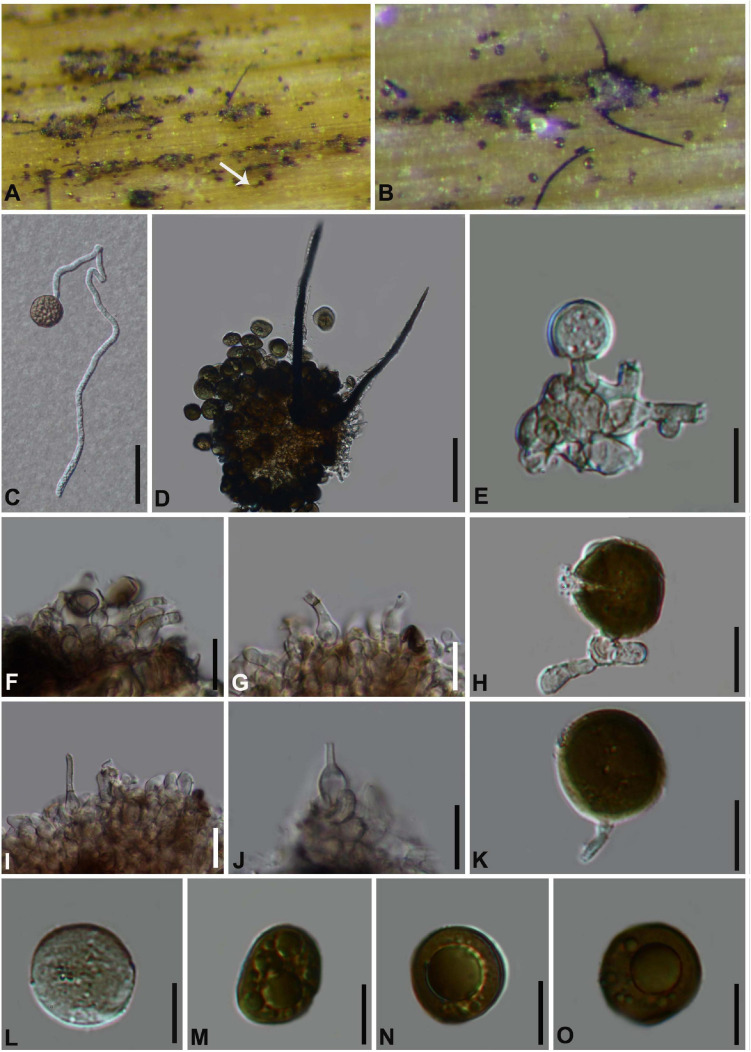
Arthrinium biseriale Y. Feng and Z.Y. Liu, sp. nov. Figure 2
FIGURE 2.
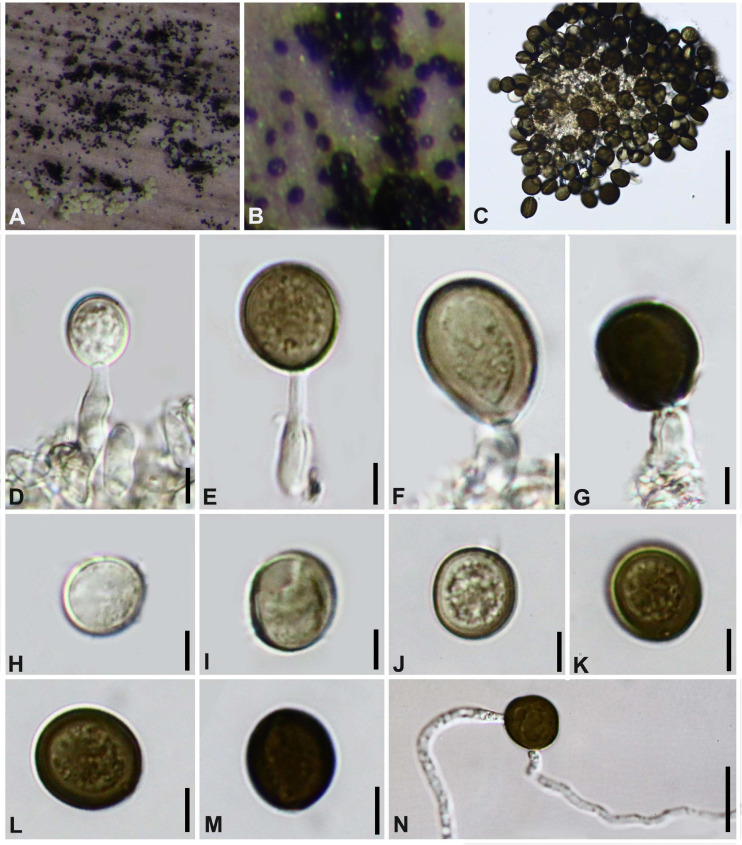
Arthrinium biseriale (HKAS 111961, holotype). (A) Appearance of stromata on bamboo host. (B) Vertical section of stroma. (C) Peridium. (D) Paraphyses. (E) Germinating ascospore. (F–H) Asci. (I–K) Ascospore. (L) Culture. (M) Hyphae. (N,O) Conidiophore and conidiogenous cells. (P–T) Conidia. Scale bars: (B) = 50 μm. (C) = 20 μm. (D) = 10μm. (E–H) = 20 μm. (I–K) = 10 μm. (M) = 20 μm. (N) = 10 μm. (O) = 20 μm. (P–T) = 5 μm.
Index Fungorum number: IF558136
Facesoffungi number: FoF 09569
Etymology: The epithet refers to the ascospores are arranged in two rows in the ascus.
Holotype: HKAS 111961
Saprobic on dead bamboo culms, forming black, lenticular spots on the host surface, with stromata breaking through raised cracks with black center, and merge with each other with age, forming an erumpent black mass visible at the naked eye. Sexual morph: Stromata scattered to gregarious, immersed to erumpent, later becoming superficial, dark brown to black, fusiform, forming a slit-like opening at the apex, multi-loculate, membranous, with a periphysate ostiole. Ascomata 122–153 μm high, 138–207 μm diam, arranged in rows, dark brown to black. Peridium 11–19 μm wide, composed of several layers of dark brown to hyaline cells of textura angularis. Hamathecium 3–4 μm wide, comprising dense, hyaline, septa paraphyses. Asci 84–116 μm × 18–25 μm ( = 97 μm × 21 μm, n = 20), 8-spored, unitunicate, clavate, apically rounded, with an indistinct pedicel. Ascospores 22–28 μm × 7–11 μm ( = 25 μm × 9 μm, n = 30), biseriate, fusiform, curved at the bottom, obtuse at both ends, slightly wider in the middle, hyaline, 1-septate, constricted at the septum, mostly curved at the lower cell, rarely straight, with a large upper cell and a small lower cell, smoothwalled. The lower cell has 1-guttulate, the upper cell has 1–3 big guttulate in the middle surrounded by multiple small guttulate with a shallow 4–7 μm thick gelatinous sheath in the early. Growing to a later stage, the guttulate filled the entire spore, and the gelatinous sheath dissolves easily. Asexual morph: On WA, Hyphae 2.5–6.0 μm diam, hyaline, branched, septate, some curled in a ring structure. Conidiophores 12.0–44.0 μm × 2.5–5.0 μm ( = 20.0 μm × 3.5 μm, n = 20), straight or flexuous, smooth, thin-walled, unbranched, hyaline to pale brown, cylindrical, cyathiform, having transverse septa, often reduced to conidiogenous cells. Conidiogenous cells 5.0–22.0 μm × 2.5–5.0 μm ( = 10.0 μm × 3.5 μm, n = 20), integrated, hyaline to pale brown, doliiform to ampulliform, or lageniform. Conidia 7–9 μm long (n = 30), brown, smooth in surface view, and 7–11 μm long (n = 30), lenticular, with a paler equatorial slit in side view, globose to ellipsoid with many guttules.
Culture characters
Ascospores germinated on WA within 24 h and germ tubes produced from middle and lower end. Colonies fast grown on PDA at 25°C, reached 7 cm in 7 days at 25°C. Colonies evenly tiled, with a large number of aerial hyphae, white, velvety, thin, gray-white on the reverse side and dirty white in the center.
Materials examined
China, Guizhou Province, Chishui City, Zhuhai National Forest Park, on dead culms of bamboo, 10 July 2019, Yao Feng, CS19-17 (HKAS 111961, holotype; GZAAS 20–0102, isotype), ex-type living cultures, CGMCC 3.20135 = GZCC 20–0101. Ibid., on dead base of the bamboo stem, 10 July 2019, Yao Feng, CS 19-25 (GZAAS 20–0101), living culture, GZCC 20–0100. Ibid., on dead branch of bamboo, 11 July 2019, Ya-Ya Chen, CS 013 (GZAAS 20–0100), living culture, GZCC 20–099.
Notes
Three strains representing Arthrinium biseriale clustered in a well-supported clade which are closely related to A. gelatinosum, but phylogenetically distinct and can be recognized as two different species (99% sequence similarity in ITS; 99% in TEF; 98% in TUB2). Morphologically, Arthrinium biseriale has smaller stromata (122–153 μm × 138–207 μm vs. 144–199 μm × 184–214 μm) and the spores of A. biseriale are more curved than those of A. gelatinosum.
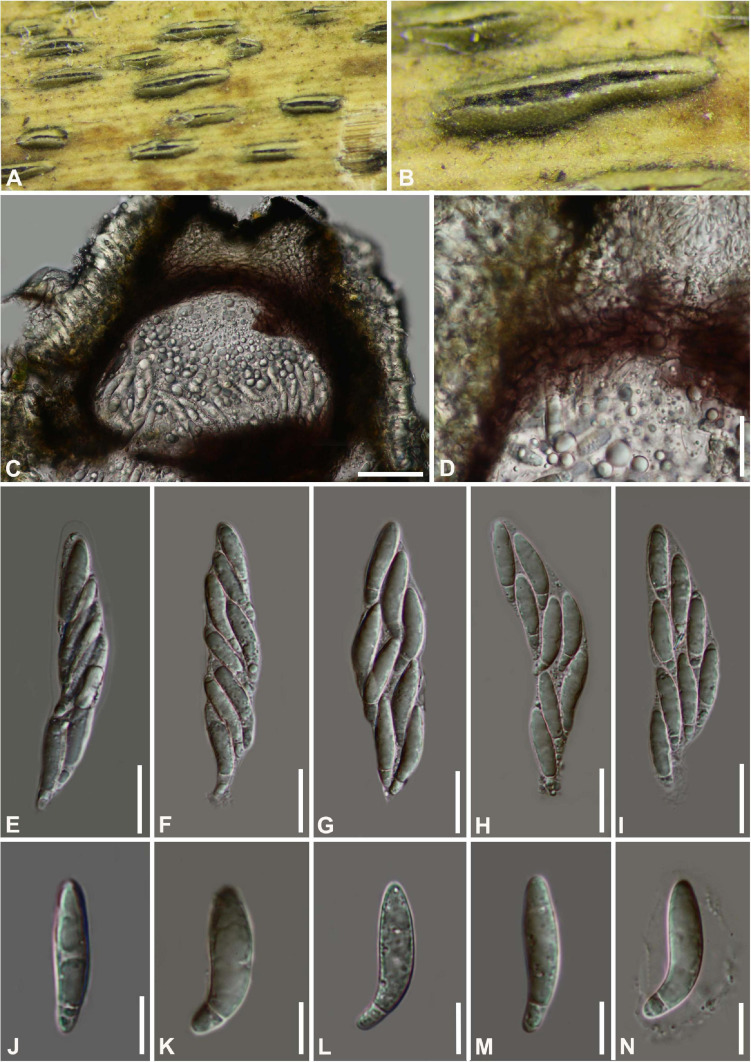
Arthrinium gelatinosum Y. Feng and Z.Y. Liu, sp. nov. Figure 3
FIGURE 3.
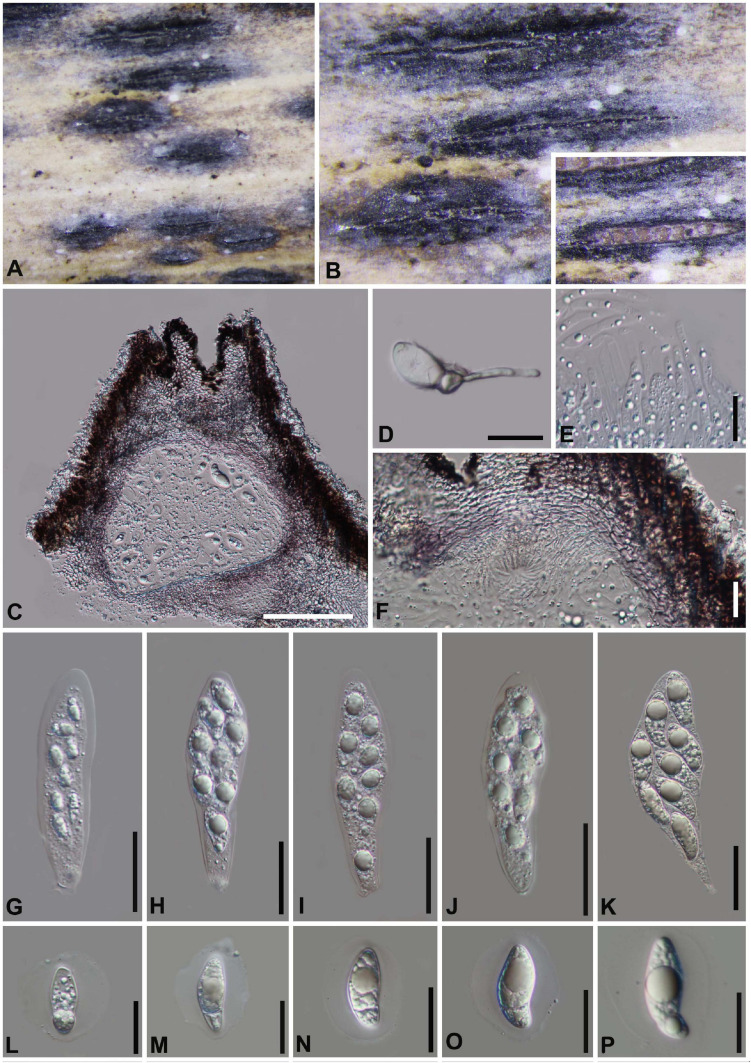
Arthrinium gelatinosum (HKAS 111962, holotype). (A,B) Appearance of stromata on bamboo host. (C) Vertical section of stroma. (D) Peridium. (E–I) Asci. (J–N) Ascospore. Scale bars: (C) = 50 μm. (D–I) = 20 μm. (J–N) = 10 μm.
Index Fungorum number: IF558137
Facesoffungi number: FoF 09570
Etymology: The epithet refers to the ascospore surrounded by gelatinous sheath.
Holotype: HKAS 111962
Saprobic on dead bamboo culms, forming black, lenticular spots on the host surface, with stromata breaking through raised cracks with black center. Sexual morph: Stromata solitary to gregarious, immersed to erumpent, fusiform, with long axis broken at the top by one cracks. Ascomata 144–199 μm high μm × 184–214 μm wide, uniseriate or irregularly arranged beneath stromata, pseudothecial, black, globose to subglobose with a flattened base. Peridium composed of 5 or 6 layers of brown cells arranged in textura angularis, with a conspicuous perfusate ostiole. Hamathecium paraphyses hyphae-like. Asci 85–121 μm × 15–24 μm ( = 100 μm × 17 μm, n = 20), 8-spored, unitunicate, clavate, apically rounded, broadly cylindrical, with an indistinct pedicel. Ascospores (27−) 28-31 (−32) μm × 6–8 μm ( = 30 μm × 7 μm, n = 30), apiosporic, clavate to fusiform with narrowly rounded ends, composed of a large guttate and small guttate, hyaline, smooth-walled, surrounded by a gelatinous sheath. Growing to a later stage, the guttate filled the entire spore, and the gelatinous sheath dissolves easily. Asexual morph: undetermined.
Materials examined
China, Guizhou Province, Chishui City, Zhuhai National Forest Park, on dead culms of bamboo, 10 July 2019, Yao Feng, CS19-32 (HKAS 111962, holotype; GZAAS20–0108, isotype). Ibid., Chishui National Scenic Area, on dead branch of bamboo, 10 July 2019, Yao Feng, CS 19–29 (GZAAS 20–0107).
Notes
Two taxa representing Arthrinium gelatinosum cluster in a well-supported lineage (ML/MP/BI = 93/98/1, Figure 1), which is a sister to A. biseriale, and they are phylogenetically distinct species.
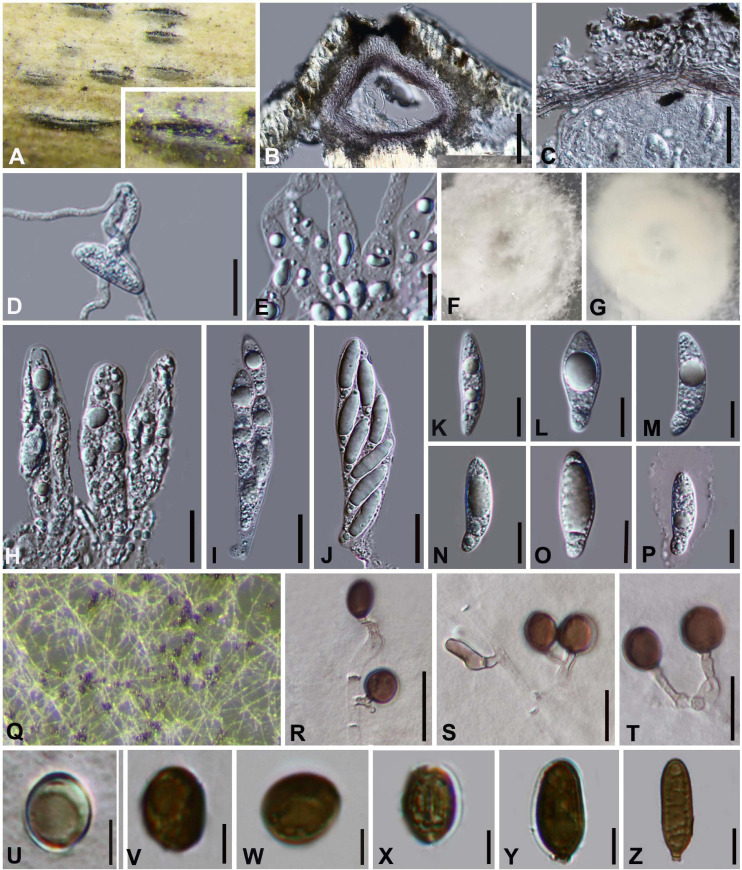
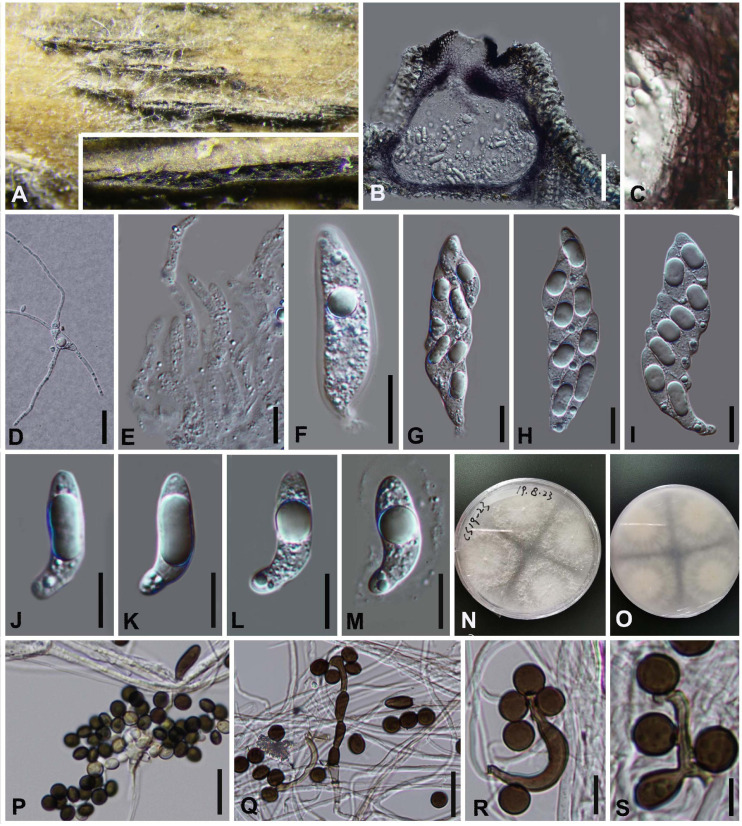
Arthrinium septatum Y. Feng and Jian K. Liu, sp. nov. Figure 4
FIGURE 4.
Arthrinium septatum (HKAS 111960, holotype). (A) Appearance of stromata on bamboo host. (B) Vertical section of stroma. (C) Peridium. (D) Germinating ascospore. (E) Paraphyses. (F,G) Culture. (H–J) Asci. (K–P) Ascospore. (Q) Colony on PDA producing conidia masses. (R–T) Conidiophore and conidiogenous cells. (U–X) Conidia. (Y,Z) Sterile cell. Scale bars: (B) = 50 μm. (C,D) = 25 μm. (E) = 10 μm. (H–J) = 20 μm. (K–P) = 10 μm. (R–T) = 10 μm. (U–Z) = 5 μm.
Index Fungorum number: IF558138
Facesoffungi number: FoF 09571
Etymology: The epithet refers to the septate conidiophore.
Holotype: HKAS 111960
Saprobic on dead bamboo culms. Sexual morph: Stromata scattered to gregarious, immersed to erumpent, initially breaks through a black spot on the host, later visible as black, raised, lenticular or dome-shaped, and will grow into a linear shape at the later stage of growth. Ostiolate, with the long axis broken at the top revealing the ostioles of pseudothecia. Ascomata 88–195 μm high × 160–185 μm wide ( = 140 μm × 173 μm, n = 10), arranged in rows, brown to dark brown, subglobose with a flattened base. Peridium with several layers of cells arranged in textura angularis, with a conspicuous ostiole 50–90 μm in diameter, periphysate. Hamathecium paraphyses hyphae-like, septate, hyaline. Asci 75–104 μm × 17–26 μm ( = 91 μm × 20 μm, n = 20), 8-spored, clavate, cylindrical, apically rounded, with distinct pedicel. Ascospores (24−) 25–30 (−32) μm × (6−) 8–10 (−11) μm ( = 29 μm × 9 μm, n = 30), biseriate, broad fusiform to cylindrical, with a large upper cell and a small lower cell, hyaline, 1-septate, constricted at septum, slightly curved, smooth-walled, with many guttules, with a large guttule at the center of large upper cell, with a distinct gelatinous sheath. Asexual morph: On PDA, Hyphae 2–4 μm in diameter, hyaline, branched, septate. Conidiophores 12.0–63.0 × 2–5 ( = 313 μm × 3 μm, n = 20), straight or flexuous, smooth, thin-walled, septate, hyaline to light brown, cylindrical, sometimes reduced to conidiogenous cells. Conidiogenous cells 4.0–18.0 × 1.5–4.0 ( = 10.0 μm × 2.5 μm, n = 20), solitary on hyphae, integrated, branched, ampuliform, cylindrical, hyaline to brown. Conidia 8–11 (−13) μm long (n = 30), brown, smooth, guttulate, globose to ellipsoid in surface view. and (8−) 9–13 (−14) μm long (n = 30), lenticular with a paler equatorial slit in side view. Sterile cells (13−) 14–21 (−27) μm × 5–7 (−9) μm elongated, mixed among conidia.
Culture characteristics
Ascospores germinated on WA within 24 h. Colonies on PDA reached 8 cm in 7 days at 25 °C, flat, aerial mycelium white. The hyphae in the center are cottony, dense, and there is a thin circle of hyphae at the edge. Reverse grayish white with a dirty white patch.
Materials examined
China, Guizhou Province, Chishui City, Zhuhai National Forest Park, on dead culms of bamboo, 10 July 2019, Yao Feng, CS19-8 (HKAS 111960, holotype; GZAAS 20–0109, isotype), ex-type living culture CGMCC 3.20134 = GZCC20–0108. Ibid., on dead branch of bamboo, 11 July 2019, Ya-Ya Chen, CS 025 (GZAAS 20–0111), living culture GZCC 20–0109.
Notes
Two isolates, representing Arthrinium septatum, grouped in a well-supported clade and appear to be distinct from other Arthrinium species phylogenetically (Figure 1). Arthrinium septatum resembles to A. biseriale in having biseriate, broad fusiform to cylindrical ascospores and cylindrical, clavate asci. However, Arthrinium septatum differs from A. biseriale by having smaller stromata (160–185 μm diam vs. 138–207 μm diam) and asci (75–104 × 17–26 μm vs. 84–116 μm × 18–25 μm).
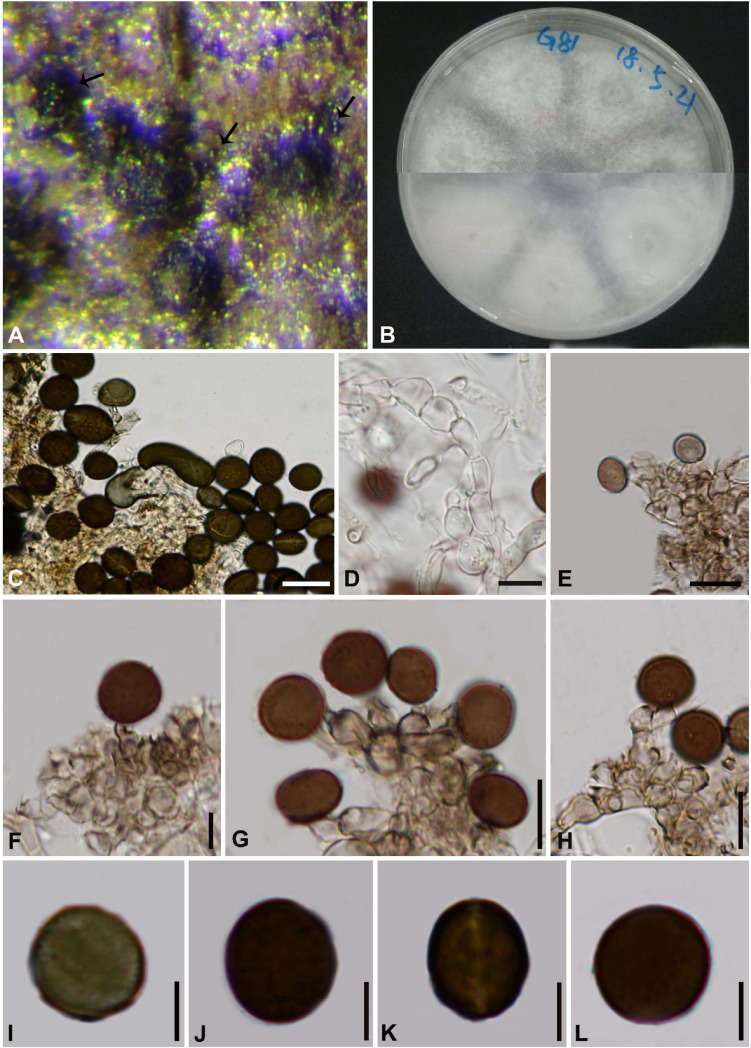
Arthrinium cyclobalanopsidis Y. Feng and Jian K. Liu, sp. nov. Figure 5
FIGURE 5.
Arthrinium cyclobalanopsidis (HKAS 111963, holotype) (A) Conidia masses on host. (B) Culture. (C) Sterile cell mixed among conidia. (D) Hyphae of chain structure. (E–H) Conidiogenous cells and conidia. (I–L) Conidia. Scale bars: (C–H) = 10 μm. (I–L) = 5 μm.
Index Fungorum number: IF558139
Facesoffungi number: FoF 09572
Etymology: The epithet “cyclobalanopsidis” refers to the host plant, Cyclobalanopsidis glauca (Thunb.) Oerst.
Holotype: HKAS 111963
Saprobic on Cyclobalanopsidis glauca (Thunb.). Sexual morph: Undetermined. Asexual morph: On PDA, Hyphae 2.5–5.5 μm in diameter, hyaline, septate, branched with chain structure. Conidiophores reduced to the conidiogenous cells. Conidiogenous cells 6.0–19.0 μm × 2.5–7.0 μm ( = 11.0 μm × 4.5 μm, n = 20), aggregated in clusters on hypha, pale brown, ampuliform or cylindrical. Conidia 8–12 μm long (n = 30), brown, smooth, globose to ellipsoid in surface view, and 10–14 μm long (n = 30), lenticular, with a paler equatorial slit in side view. Sterile cells elongated, rolled up, sometimes mixed among conidia.
Culture characteristics
Conidia germinated on WA within 12 h. Sporulated on PDA, Colonies flat, margin circular, fluffy, sparse, white, with dirty white patches in center, reverse white, with sparse aerial mycelium, reached 8 cm in 7 days at 25°C.
Material examined
China, Guizhou Province, Qianxinan Buyi and Miao Autonomous Prefecture, Ceheng County, on Leaf of cyclobalanopsidis glauca (Thunb.) Oerst., 13 May 2018, Yao Feng, G81 (HKAS 111963, holotype; GZAAS 20–0096, isotype), ex-type living cultures, CGMCC 3.20136 = GZCC 20–0102. Ibid., 20 Oct. 2019, Yao Feng, G82 (GZAAS 20–0097), living culture GZCC 20–0103.
Notes
Two isolates, representing Arthrinium cyclobalanopsidis, cluster together with A. camelliae-sinensis which was introduced by Wang et al. (2018) from Camellia sinensis (Figure 1). Arthrinium cyclobalanopsidis can be distinguished from A. camelliae-sinensis (567/572 in ITS; 390/414 in TEF; 715/748 in TUB2). Morphologically, Arthrinium cyclobalanopsidis resembles to A. camelliae-sinensis in having similar conidia (8–12 μm × 10–14 μm vs. 9.0–13.5 μm × 7.0–12.0 μm), but can be distinguished by its relatively longer conidiogenous cells (6.0–19.0 μm vs. 4.0–9.5 μm).
Arthrinium garethjonesii D.Q. Dai and H.B. Jiang, Mycosphere 7 (9): 1337 (2017). Figure 6
FIGURE 6.
Arthrinium garethjonesii (GZAAS 20–0117) (A,B) Appearance of sporodochia on bamboo host. (C) Germinating conidia. (D) Conidia with setae. (E–K) Conidiogenous cells and conidia. (L–O) Conidia. Scale bars: (C) = 30 μm. (D) = 50 μm. (E) = 20 μm. (F,G) = 10 μm. (H) = 5 μm. (I,J) = 10 μm. (K–O) = 5 μm.
Saprobic on dead bamboo branch. Sexual morph: See Dai et al. (2016). Asexual morph: Sporodochia on host with hair-like setae, also grow in the gaps of the perithecia and scatter on the surface of the perithecia, black. Conidiophores reduced to conidiogenous cells. Conidiogenous cells (5−) 6–19 (−20) μm × (2−) 3–5 (−7) μm ( = 11 μm × 4 μm, n = 20), aggregated in black sporodochia, hyaline to pale brown, smooth, ampulliform. Conidia (14–) 16–19 (–20) μm diam, brown, smooth, granular, globose to subglobose in surface view, and (16−) 17–22 (−23) μm diam, with pale equatorial slit in side view.
Culture characteristics
Conidia germinated on WA within 12 h, colonies fast growing on PDA, reached 8 cm in 7days at 25°C, fluffy, circular, dense, raised at center, white, reverse reddish. Hyphae 2–4 μm diam, hyaline to pale brown, branched, septate.
Material examined
China, Guizhou Province, Chishui City, Zhuhai National Forest Park, on dead branches of bamboo, 10 July 2019, Yao Feng, CS19-9 (GZAAS 20–0117); living culture GZCC 20–0115.
Notes
Arthrinium garethjonesii was originally described by Dai et al. (2016) based on the sexual morph from dead bamboo culms (HKAS 96289) collected from Yunnan Province, China. Our phylogenetic result (Figure 1) indicates that our collection is identical to Arthrinium garethjonesii and we report its asexual morph for the first time in this study.
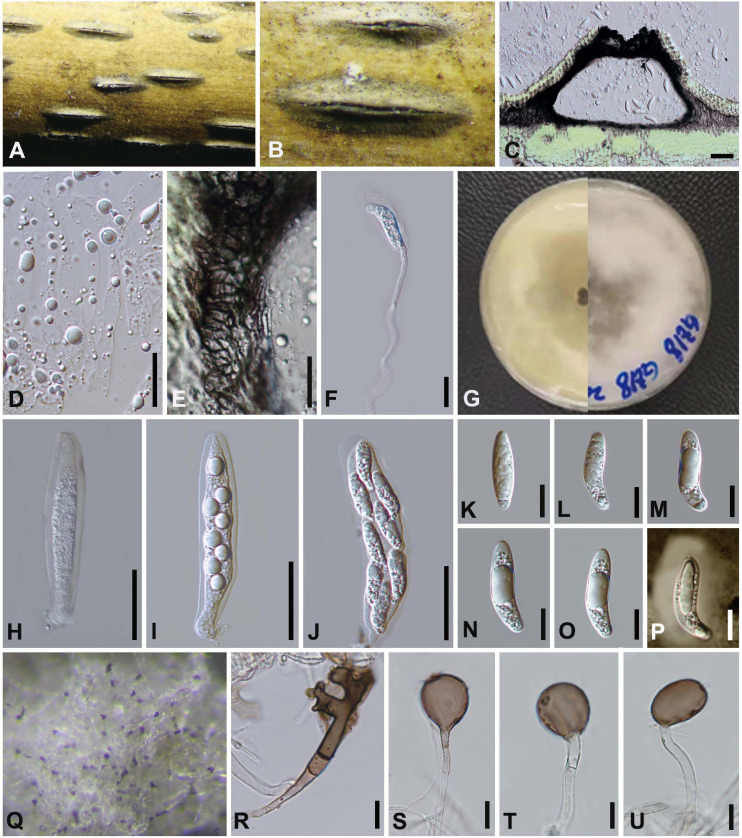
Arthrinium guizhouense M. Wang and L. Cai, Mycokeys 34: 13 (2018). Figure 7
FIGURE 7.
Arthrinium guizhouense (GZAAS 20–0114). (A) Appearance of stromata on bamboo host. (B) Vertical section of stroma. (C) Peridium. (D–F) Asci. (G) Germinating ascospore. (H,I) Culture. (J–O) Ascospore. (P) Colony on WA producing conidia masses. (Q,R) Conidiogenous cells giving rise to conidia. (S–V) Conidia. (W) Sterile cell. Scale bars: (B,C) = 50 μm. (D–F) = 30 μm. (G) = 50 μm. (J–O) = 20 μm. (Q,R) = 10 μm. (S–W) = 5 μm.
Saprobic on dead bamboo culms, forming black, lenticular spots on the host surface, stromata breaking through raised cracks with black center. Sexual morph: Stromata solitary to gregarious, immersed to erumpent, fusiform, with long axis broken at the top by one cracks. Ascomata 188–220 μm high × 170–200 μm wide, uniseriate or irregularly arranged beneath stromata, pseudothecial, black, globose to subglobose. Peridium composed of 5 or 6 layers of brown cells arranged in textura angularis, Hamathecium paraphyses 3–5 μm, hyaline, hyphae-like, septate. Asci (80−) 94–106 (−107) μm × (20−) 21–23 (−24) μm, 8-spored, clavate, apically rounded, broadly cylindrical, with an indistinct pedicel. Ascospores (24−) 25–32 (−33) μm × (6−) 7–9 (−10) μm ( = 31 μm × 8 μm, n = 30), apiosporic, clavate to fusiform with narrowly rounded ends, composed of a large upper cell and small lower cell, hyaline, smooth-walled, surrounded an inconspicuous gelatinous sheath. Asexual morph: Hyphae 2.5–7.5 μm diam, hyaline, branched, septate. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–12 μm × 2–5 μm ( = 8 μm × 3 μm, n = 20), erect, aggregated in clusters on hyphae, pale brown, smooth, subglobose, ampulliform or doliiform. Conidia 5–8 μm long (n = 30), dark brown to black, smooth, globose or subglobose, and 6–8 μm long (n = 30), lenticular, with a paler equatorial slit in side view. Sterile cells elongated, rolled up, sometimes mixed among conidia.
Culture characteristics
On PDA, colonies very fast, reached 8 cm in 8 days at 25°C, velvety, circular, with regular edge, middle densely and raised white, dense at above the from margin, aerial mycelia, surface initially white, became grayish and reverse white.
Material examined
China, Guangdong Province, Guangzhou City, on decaying bamboo culms, 03 Sep. 2019, Yao Feng, GZ 23 (GZAAS 20–0114); living culture GZCC 20–0114.
Notes
Arthrinium guizhouense was introduced from air in a karst cave (asexual morph was provided from the culture) in Guizhou province, China by Wang et al. (2018). In this study, one collection was found as saprobe on bamboo in Guangzhou, China and it is identified as A. guizhouense based on the phylogeny and morphology evidences. In addition, our new collection provides the sexual morph which only the asexual morph was illustrated by Wang et al. (2018) and Senanayake et al. (2020a).
Arthrinium phyllostachium C.L. Yang, X.L. Xu and K.D. Hyde, Phytotaxa 406 (2): 102 (2019). Figure 8
FIGURE 8.
Arthrinium phyllostachium (GZAAS 20–0112) (A) Appearance of stromata on bamboo host. (B) Vertical section of stroma. (C) Peridium. (D) Germinating ascospore. (E) Paraphyses. (F–I) Asci. (J–M) Ascospore. (N,O) Culture. (P–S) Conidiogenous cells and conidia. Scale bars: (B) = 50 μm. (C) = 10 μm. (D) = 20 μm. (E) = 20 μm. (F–I) = 30 μm. (J–M) = 10 μm. (P,Q) = 20 μm. (R–S) = 10 μm.
Saprobic on dead bamboo culms, Sexual morph: Stromata scattered to gregarious, immersed to erumpent, later becoming superficial, dark brown to black, fusiform, forming a slit-like opening at the apex, with stromata breaking through raised cracks with black center. Ascomata 135–185 μm high, 157–215 μm diam, multi-loculate, with a periphysate ostiole, arranged in rows, clustered, gregarious, ampulliform, dark brown to black. Peridium 20–25 μm wide, composed of several layers of dark brown to brown cells of textura angularis. Hamathecium 3–5 μm wide, comprising dense, hyaline, septate paraphyses, hyphae-like. Asci 66–98 × 17–27 ( = 85 μm × 21 μm, n = 20), 8-spored, unitunicate, clavate, apedicellate, apically rounded. Ascospores 29–34 μm × 7–10 μm ( = 32 μm × 9 μm, n = 30), biseriate, elliptical, hyaline, 1-septate, constricted at the septum, mostly curved at the lower cell, rarely straight, with a large upper cell and a small lower cell, smooth-walled, with a shallow gelatinous sheath. Asexual morph: On WA, Hyphae 1.5–4.0 μm in diameter, hyaline, septate, branched. Conidiophores reduced to the conidiogenous cells. Conidiogenous cells (8−) 9–28 (−31.5) μm × (1.5−) 2–4 (−6) μm ( = 20 μm × 3 μm, n = 20), aggregated in clusters on hypha or solitary, erect, ampuliform or cylindrical, arising holoblastically from vegetative hyphae, monoblastic, or polyblastic, sympodial, terminal, cylindrical to clavate, ampulliform, hyaline, smooth, thin-walled. Conidia 5–8 μm long (n = 30), brown, smooth, globose to ellipsoid in surface view, and 7–9 μm long (n = 30), lenticular, with a paler equatorial slit in side view. Sterile cells light brown, elongated, and occasionally irregularly angled.
Culture characters
Ascospores germinated on WA within 24 h and germ tubes produced from Upper, middle and lower end. Colonies fast growed on PDA at 25°C, flat, spreading, the middle is white, smooth, the edge surface tapetum, gray-white, indistinct aerial hyphae, the reverse side is yellowish in the middle, and the edge is grayish white.
Materials examined
China, Guizhou Province, Chishui City, Zhuhai National Forest Park, on dead culms of bamboo, 10 July 2019, Yao Feng, CS 19-23 (GZAAS 20–0112), living culture GZCC 20–0111; Ibid., Chishui National Scenic Area, on dead culms of bamboo, 10 July 2019, Ya-Ya Chen, CS004 (GZAAS 20–0113), living culture GZCC 20–0112.
Notes
Arthrinium phyllostachium was introduced by Yang et al. (2019) based on the asexual morph and phylogeny analyses. It was collected from culms of Phyllostachys heteroclada (Poaceae) in China (Yang et al., 2019). The phylogenetic results showed that our new collections are identical to A. phyllostachium, Yang et al. (2019) only provided the asexual morph and the sexual morph is given in this study.
Arthrinium pseudoparenchymaticum M. Wang and L. Cai, Mycokeys 34 (1): 17 (2018), Figure 9
FIGURE 9.
Arthrinium pseudoparenchymaticum (GZAAS 20–0115) (A,B) Appearance of stromata on bamboo host. (C) Vertical section of stroma. (D) Paraphyses. (E) Peridium. (F) Germinating ascospore. (G) Culture. (H–J) Asci. (K–O) Ascospore. (P) Ascospore in Indian ink and present clear gelatinous sheath. (Q) Colony on WA producing conidia masses. (R) Dentate conidia. (S–U) Conidiogenous cells giving rise to conidia. Scale bars: (C) = 50 μm. (D–F) = 20 μm. (H–J) = 30 μm. (K–P) = 15 μm. (R–U) = 15 μm.
Saprobic on dead bamboo culms, forming black, lenticular spots on the host surface, with stromata breaking through raised cracks with black center. Sexual morph: Ascomata 187–242 μm high × 242–373 μm wide, uniseriate, or irregularly arranged beneath stromata, black, globose to subglobose with a flattened base. Peridium composed of 5–7 layers of brown cells arranged in textura angularis, with a conspicuous perfusate ostiole. Hamathecium paraphyses 4–8 μm, hyphae-like, septa hyaline. Asci (95−) 107–110 (−133) μm × 23–25 (−27) μm, 8-spored, broadly cylindrical, clavate or subglobose, apically rounded, with an indistinct pedicel. Ascospores (35–) 35–43 (–44) × (10–) 11–13 μm ( = 41 × 12, n = 30), apiosporic, clavate to fusiform with narrowly rounded ends, composed of a large upper cell and small lower cell, hyaline, smooth-walled, surrounded by a gelatinous sheath. Asexual morph: On WA, Hyphae 1.5–4 μm diam, hyaline to pale brown, branched, septate. Conidiophore extends from the vegetative hypha, up to 60 μm long. Conidiogenous cells 10.0–40.0 μm × 3.0–6.0 μm, scattered in clusters on hyphae, smooth, unbranched, hyaline to pale yellow, smooth, erect, subcylindrical. Conidia 17–27 μm × 17–21 μm ( = 23 μm × 19 μm, n = 30), pale to light brown, smooth, globose to subglobose, sometimes lobed or dentate, polygonal or irregular in surface view.
Culture characteristics
Ascospores germinating on WA within 24 h and germ tubes produced from upper. Colonies fast growing on PDA at 25°C, under 12 h light/12 h dark, cottony, circular, sparse, raised, with irregular edge, white in center.
Material examined
China, Guangdong Province, Guangzhou City, on decaying bamboo culms, 3 Sep. 2019, Yao Feng, GZ18 (GZAAS 20–0115), living culture GZCC20–0117.
Notes
Arthrinium pseudoparenchymaticum was introduced by Wang et al. (2018) based on the asexual morph characters and phylogeny analyses. It was originally collected from bamboo in China (Wang et al., 2018). In this study, a fresh specimen was collected and it is identical to A. pseudoparenchymaticum (Figure 1), both sexual and asexual morphs were described and illustrated (Figure 9).
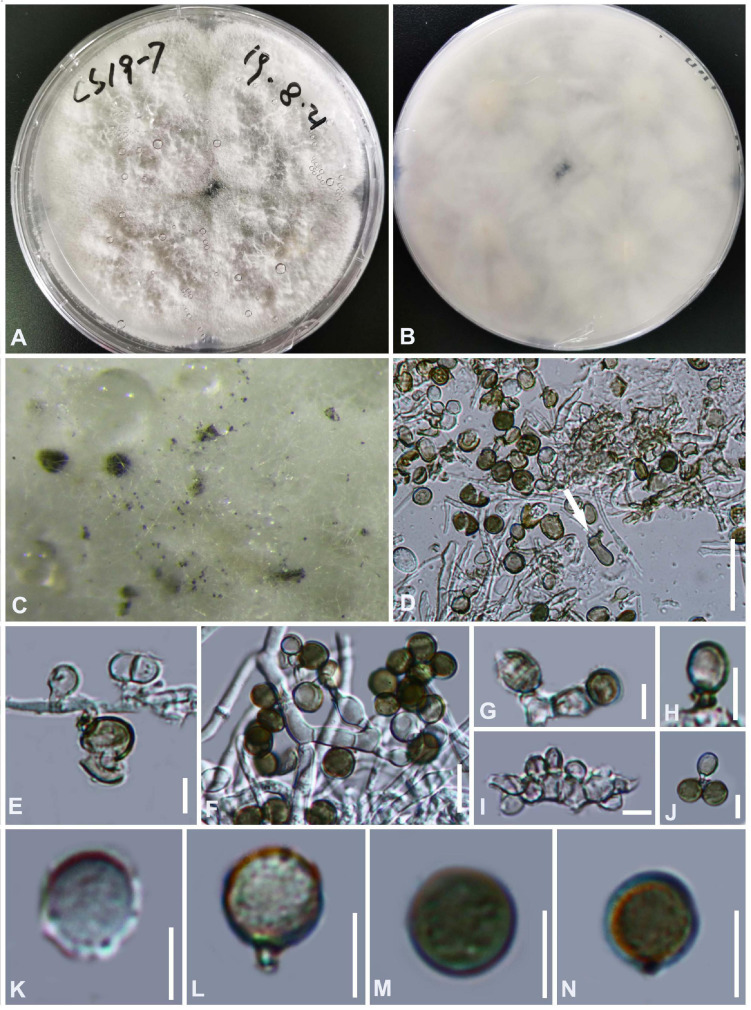
Arthrinium arundinis (Corda) Dyko and B. Sutton, Mycotaxon 8: 119 (1979). Figure 10.
FIGURE 10.
Arthrinium arundinis (GZAAS 20–0116) (A,B) culture. (C) Colonies on culture. (D) Sterile cell mixed among conidia. (E–J) Conidiogenous cells and conidia. (K–N) Conidia. Scale bars: (D) = 50 μm. (E) = 5 μm. (F) = 10 μm. (G–N) = 5 μm.
Saprobic on dead bamboo culms. Asexual morph: On PDA, Hyphae 2–3 μm diam, consisting of smooth, hyaline, branched, septate. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–8 μm × 2–5 μm, aggregated in clusters on hyphae, pale brown, smooth, ampulliform. Conidia (4–) 5–6 (–7) μm, brown, smooth, globose in surface view, and (4−) 5–7 (−8) μm diam, lenticular with pale equatorial slit in side view. Sterile cells at times intermingled among conidia. Sexual morph: Undetermined.
Culture characteristics
The colony is flat, cotton-like, thick and dense, with sparse aerial mycelia. The surface of PDA is white and the reverse side is grayish white.
Materials examined
China, Guizhou Province, Chishui City, Zhuhai National Forest Park, on dead culms of bamboo, 10 July 2019, Yao Feng, CS 19-7 (GZAAS 20–0116), living culture GZCC 20–0116.
Notes
Our collection clusters together with the isolates of Arthrinium arundinis (Figure 1) and its morphology lines up with the type species. Therefore we identify it as Arthrinium arundinis.
Arthrinium hydei Crous, IMA Fungus 4 (1): 142 (2013). Figure 11
FIGURE 11.
Arthrinium hydei (GZAAS 20–0098) (A,B) Colonies on substrate. (C–F) Conidiogenous cells and conidia. (G–M) Conidia. (N) Germinating conidia. Scale bars: (C) = 50 μm. (D–M) = 5 μm. (N) = 25 μm.
Saprobic on bamboo leaves. Asexual morph: Colonies on the host punctiform, pulvinate, blackish brown. Conidiophores pale brown, smooth, transversely septate, subcylindrical. Conidiogenous cells 5–15 × 3–6 μm, brown, smooth, subcylindrical to doliiform to lageniform. Conidia (13−) 14–19 (−20) μm diam in surface view, brown, roughened, globose, and 15–20 μm diam, lenticular with pale equatorial slit in side view. Sexual morph: Undetermined.
Culture characteristics
Colonies flat, spreading, with sparse aerial mycelium. On PDA surface and reverse pale luteous. Mycelium consisting of smooth, hyaline to pale brown, branched, septate, 2.0–4.5 μm diam hyphae.
Materials examined
China, Guizhou Province, Guiyang City, Baihua Lake, on bamboo leaves, 20 April 2018, Yao Feng, 67 (GZAAS 20–0098), living culture GZCC20–0113.
Notes
This collection is identified as Arthrinium hydei based on both morphological characters and molecular data. Crous and Groenewald (2013) originally described A. hydei based on the asexual morph from a culture (CBS 114990) which was isolated from bamboo culms in Hong Kong, China; we found this species in Guizhou from the substrate in nature with its asexual morph.
Arthrinium neosubglobosa D.Q. Dai and H.B. Jiang, Mycosphere 7 (9): 1337 (2017) Figure 12.
FIGURE 12.
Arthrinium neosubglobosa (GZAAS 20–0099). (A,B) Appearance of stromata on bamboo host. (C) Vertical section of stroma. (D) Germinating ascospore. (E) Paraphyses. (F) Peridium. (G–K) Asci. (L–P) Ascospore. Scale bars: (C) = 100 μm. (D–F) = 20 μm. (G–K) = 30 μm. (L–P) = 20 μm.
Saprobic on dead bamboo culms. Sexual morph: Stromata scattered to gregarious, superficial to raised, with a slit-like opening, dark brown to black, naviculate, with black papillate ostiole, multi-loculate. Ascomata 205–328 μm high, 168–345 μm, perithecial, arranged in a row, immersed in stromata, later becoming erumpent through host surface to superficial, obpyriform to ampulliform, dark brown, membranous. Ostiole raised from center of Ascomata, internally lined with periphyses. Peridium 4 layers, outer layer composed of dark brown, cells of textura prismatica, inner layer thin, with hyaline cells of textura angularis. Hamathecium 4.0–5.5 μm wide, comprising dense paraphyses, indistinctly aseptate, unbranched, not anastomosing, filamentous, clustered embedded in gelatinous matrix. Asci 80–119 μm × 20–37 μm ( = 97 μm × 28 μm, n = 20), 8-spored, unitunicate, clavate, with a short pedicel, apically rounded. Ascospores 25–36 μm × 11–15 μm ( = 29 μm × 13 μm, n = 30), 2-seriate, elliptical, hyaline, 1-septate, constricted at the septum, mostly curved at the lower cell, rarely straight, with a large upper cell and a small lower cell, smoothwalled, 1-guttulate, with a shallow 8–12 μm thick gelatinous sheath. Asexual morph: Undetermined.
Materials examined
China, Guizhou Province, Zunyi City, Daozhen County, on dead culms of bamboo, 15 August 2018, Yao Feng, DZ22 (GZAAS 20–0099).
Notes
Arthrinium neosubglobosa was introduced by Dai et al. (2016) based on the sexual morph characters and phylogeny analyses. The pure culture was attempted by single spore isolation and the DNA was extracted directly from the fruiting body, the new collection is identified as A. neosubglobosa based on the phylogeny (Figure 1) and morphology evidences.
Discussion
Arthrinium species have been reported from many hosts, includes hive-stored pollen lichens, marine algae, soil debris, gut of insects and nodules of human skin (Sharma et al., 2014; Crous et al., 2015, Senanayake et al., 2015; Wijayawardene et al., 2017, Zhao et al., 2018), it can be concluded that Arthrinium is ecologically diverse. Bamboo, as one of the most reported host, is a gramineous plant integrating economy and ornamental value (Gratani et al., 2008; Kelchner and Bamboo Phylogeny Group, 2013; Dai et al., 2016, 2017; Jiang et al., 2018, 2019, 2020; Wang et al., 2018; Yang et al., 2019), there is more than 115 genera with approximately 1,450 species. According to incomplete statistics, more than 1,100 species of fungi on bamboo were reported (Hyde et al., 2002a, b; Dai et al., 2016, 2017; Senanayake et al., 2020a; Tang et al., 2020; Wijesinghe et al., 2020). It is of great significance to excavate and identify the fungi on bamboo.
Several studies have shown (Crous and Groenewald, 2013; Dai et al., 2016, Dai et al., 2017; Wang et al., 2018; Yang et al., 2019) that it is difficult to identify the Arthrinium species solely rely on morphology and the multi-gene phylogenetic analyses are needed in the identification and classification of Arthrinium. The morphology of conidia is variable which can be depending on the period of incubation on different habitats, for example, A. biseriale, A. gelatinosum and A. septatum, are very similar in morphology, but molecular data distinguish them into different species; Our collection A. pseudoparenchymaticum differs from the type specimen (LC 8173) in the morphology of conidiophores, the size of conidiogenous cells is also different, while the molecular data supported them as the same species. These results are in agreement with the previous observations and publications (Crous and Groenewald, 2013; Dai et al., 2016, Dai et al., 2017; Wang et al., 2018, Yang et al., 2019). In addition, as a high diverse group, it is also difficult to distinguish species within Arthrinium by only using ITS and LSU gene regions, and the protein genes (TEF and TUB2) are not available for many species in the genus which bring potential problem once the new or existing taxa are introduced and identified, respectively. For example the absent of the protein genes (TEF and TUB2) of Arthrinium garethjonesii would bring the troubles in identification of Arthrinium setostromum as it is hard to confirm whether they are same species or not as they are identical in ITS and LSU gene regions, as well as the close phylogenetic relationship (Figure 1). It would be necessary to provide protein genes when new taxa are introduced in these well-study and diverse groups.
Data Availability Statement
The data presented in the study can be found in the Genbank. The accession numbers of the sequences deposited in GenBank are ITS: MW481705—MW481721; LSU: MW478885–MW478901; TEF: MW522938–MW522954; and TUB: MW522955–MW522969.
Author Contributions
YF and J-KL: conceptualization. YF and Y-YC: methodology. YF, C-GL, and J-KL: formal analysis. YF, Y-YC, M-MX, and J-KL: resources. YF: writing—original draft preparation. C-GL, Z-YL, and J-KL writing—review and editing. Z-YL and J-KL: supervision. All authors approved to publish the version of final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ning-Guo Liu, Sheng-Nan Zhang, Jing Yang, Qiu-Jiu Shang, and Milan C. Samarakoon for their suggestions on phylogeny in this study.
Funding. This study was supported by the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou province (Grant No. U1812401); the Research of Featured Microbial Resources and Diversity Investigation in Southwest Karst area (Project No. 2014FY120100); and the Guizhou Graduate Research Funding: Taxonomic and Phylogenetic studies of microfungi from Bamboo in Guizhou (Qian Jiao He YJSCXJH [2019] 022).
References
- Agut M., Calvo M. A. (2004). In vitro conidial germination in Arthrinium aureum and Arthrinium phaeospermum. Mycopathologia 157 363–367. 10.1023/B:MYCO.0000030432.08860.f3 [DOI] [PubMed] [Google Scholar]
- Carbone I., Kohn L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chen Y. Y., Dissanayake A., Liu Z. Y., Liu J. K. (2020). Additions to Karst Fungi 4: Botryosphaeria spp. associated with woody hosts in Guizhou province, China including B. guttulata sp. nov. Phytotaxa 454 186–202. 10.11646/phytotaxa.454.3.2 [DOI] [Google Scholar]
- Chen Y. Y., Maharachchikumbura S. S. N., Liu J. K., Hyde K. D., Nanayakkara R. R., Zhu G. S., et al. (2017). Fungi from asian karst formations I. Pestalotiopsis photinicola sp.nov., causing leaf spots of Photinia serrulata. Mycosphere 8 103–110. 10.5943/mycosphere/8/1/9 [DOI] [Google Scholar]
- Crous P. W., Groenewald J. Z. (2013). A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4 133–154. 10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P. W., Wingfield M. J., Le Roux J. J., Richardson D. M., Strasberg D., Shivas R. G., et al. (2015). Fungal planet description sheets: 371-399. Persoonia 35 264–327. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D. Q., Jiang H. B., Tang L. Z., Bhat D. J. (2016). Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7 1332–1345. 10.5943/mycosphere/7/9/7 [DOI] [Google Scholar]
- Dai D. Q., Phookamsak R., Wijayawardene N. N., Li W. J., Bhat D. J., Xu J. C., et al. (2017). Bambusicolous fungi. Fung. Divers. 82 1–105. [Google Scholar]
- Dissanayake A. J., Bhunjun C. S., Maharachchikumbura S. S. N., Liu J. K. (2020a). Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11 2652–2676. 10.5943/mycosphere/11/1/18 [DOI] [Google Scholar]
- Dissanayake A. J., Chen Y. Y., Liu J. K. (2020b). Unravelling diaporthe species associated with woody hosts from karst formations (Guizhou) in China. J. Fungi 6:251. 10.3390/jof6040251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. B. (1971). Dematiaceous Hyphomycetes. Kew: Commonwealth Mycological Institute, 608. [Google Scholar]
- Feng Y., Zhang S. N., Liu Z. Y. (2019). Tremateia murispora sp. nov. (Didymosphaeriaceae, Pleosporales) from Guizhou, China. Phytotaxa 416 79–87. 10.11646/phytotaxa.416.1.10 [DOI] [Google Scholar]
- Glass N. L., Donaldson G. C. (1995). Development ofprimer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratani L., Crescente M. F., Varone L., Fabrini G., Digiulio E. (2008). Growth pattern and photosynthetic activity of different bamboo species growing in the Botanical Garden of Rome. Flora Morphol. Distrib. Funct. Ecol. Plants 203 77–84. 10.1016/j.flora.2007.11.002 [DOI] [Google Scholar]
- Hall T. A. (1999). “BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT,” in Nucleic Acids Symposium Series. 95–98.10780396 [Google Scholar]
- Hillis D. M., Bull J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde K. D., Fröhlich J., Taylor J. E. (1998). Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50 21–80. [Google Scholar]
- Hyde K. D., Norphanphoun C., Maharachchikumbura S. S. N., Bhat D. J., Jones E. B. G., Bundhun D., et al. (2020). Refined families of Sordariomycetes. Mycosphere 11 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Hyde K. D., Zhou D., Dalisayl T. (2002a). Bambusicolous fungi: a review. Fung. Divers. 9 1–14. [Google Scholar]
- Hyde K. D., Zhou D., McKenzie E., Ho W., Dalisay T. (2002b). Vertical distribution of saprobic fungi on bamboo culms. Fungal Divers. 11 109–118. [Google Scholar]
- Jayasiri C. S., Hyde K. D., Ariyawansa H. A., Bhat J., Buyck B., Cai L., et al. (2015). The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jiang H. B., Hyde K. D., Doilom M., Karunarathna S. C., Xu J. C., Phookamsak R. (2019). Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. Asia. J. Mycol. 2 254–268. 10.5943/ajom/2/1/16 [DOI] [Google Scholar]
- Jiang N., Li J., Tian C. M. (2018). Arthrinium species associated with bamboo and reed plants in China. Fungal Syst. Evol. 2 1–9. 10.3114/fuse.2018.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Liang Y. M., Tian C. M. (2020). A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia 72 77–83. 10.12905/0380 [DOI] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelchner S. A., and Bamboo Phylogeny Group (2013). Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 67 404–413. 10.1016/j.ympev.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Li B. J., Liu P. Q., Jiang Y., Weng Q. Y., Chen Q. H. (2016). First report of culm rot caused by Arthrinium phaeospermum on Phyllostachys viridis in China. Plant Dis. 100 1013–1013. 10.1094/PDIS-08-15-0901-PDN [DOI] [Google Scholar]
- Liu J. K., Chomnunti P., Cai L., Phookamsak R., Chukeatirote E., Jones E. B. G., et al. (2010). Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia 62 261–276. [Google Scholar]
- Liu N. G., Bhat D. J., Hyde K. D., Liu J. K. (2019). Conioscypha tenebrosa sp. nov. (Conioscyphaceae) from China and notes on Conioscypha species. Phytotaxa 413 159–171. 10.11646/phytotaxa.413.2.5 [DOI] [Google Scholar]
- Luo Z., Hyde K. D., Liu J. K., Maharachchikumbura S. S. N., Jeewon R., Bao D. F., et al. (2019). Freshwater Sordariomycetes. Fungal Divers. 99 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Mapook A., Hyde K. D., McKenzie E. H. C., Gareth J. E. B., Bhat D. J., Jeewon R., et al. (2020). Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 101 1–175. 10.1007/s13225-020-00444-8 [DOI] [Google Scholar]
- Martínez-Cano C., Grey W. E., Sands D. C. (1992). First report of Arthrinium arundinis causing kernel blight on barley. Plant Dis. 76 1077. 10.1094/PD-76-1077B [DOI] [Google Scholar]
- Mavragani D. C., Abdellatif L., McConkey B., Hamel C., Vujanovic V. (2007). First report of damping-off of durum wheat caused by Arthrinium sacchari in the semi-arid Saskatchewan fields. Plant Dis. 91:469. 10.1094/pdis-91-4-0469a [DOI] [PubMed] [Google Scholar]
- Nylander J. A. A. (2004). MrModeltest v2. Program Distributed by the Author. Uppsala: Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K., Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- O’Donnell K., Kistler H. C., Cigelnik E., Ploetz R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 95 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos A., Alvarado P., Planas J., Jarling R. (2019). Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. Hosts. Mycokeys 49 15–48. 10.3897/mycokeys.49.32115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M. K. (1989). Mycosis in man due to Arthrinium phaeospermum var. indicum. First case report. Mycoses 32 472–475. 10.1111/j.1439-0507.1989.tb02285.x [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012). ). FigTree, Version 1.4. 2. Edinburgh: University of Edinburgh. [Google Scholar]
- Rannala B., Yang Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Seifert K., Morgan-Jones G., Gams W., Kendrick B. (2011). The Genera of Hyphomycetes.[CBS Biodiversity Series 9]. Utrecht: CBS-KNAW Fungal Biodiversity Centre. [Google Scholar]
- Senanayake I. C., Bhat J. D., Cheewangkoon R., Xie N. (2020a). Bambusicolous Arthrinium species in guangdong province. China. Front. Microbiol. 11:602773. 10.3389/fmicb.2020.602773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake I. C., Maharachchikumbura S. S., Hyde K. D., Bhat J. D., Jones E. G., McKenzie E. H., et al. (2015). Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 73 73–144. 10.1007/s13225-015-0340-y [DOI] [Google Scholar]
- Senanayake I. C., Rathnayake A. R., Marasinghe D. S., Calabon M. S., Gentekaki E., Lee H. B., et al. (2020b). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Sharma R., Kulkarni G., Sonawane M. S., Shouche Y. S. (2014). A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience 55 118–123. 10.1016/j.myc.2013.06.004 [DOI] [Google Scholar]
- Silvestro D., Michalak I. (2012). raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Swofford D. L. (2002). PAUP∗: Phylogenetic Analysis Using Parsimony (∗and Other Methods). Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Tang X., Goonasekara I. D., Jayawardena R. S., Jiang H. B., Li J. F., Hyde K. D., et al. (2020). Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in northern Thailand. Biodivers. Data J. 8:e58755. 10.3897/BDJ.8.e58755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tan X. M., Liu F., Cai L. (2018). Eight new Arthrinium species from China. MycoKeys 34 1–24. 10.3897/mycokeys.34.24221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene N. N., Hyde K. D., Rajeshkumar K. C., Hawksworth D. L., Madrid H., Kirk P. M., et al. (2017). Notes for genera: ascomycota. Fungal Divers. 86 1–594. 10.1007/s13225-017-0386-0 [DOI] [Google Scholar]
- Wijesinghe S. N., Wang Y., Camporesi E., Wanasinghe D. N., Boonmee S., Hyde K. D. (2020). A new genus of Bambusicolaceae (Pleosporales) on Corylus avellana (Fagales) from Italy. Biodivers. Data J. 8:e55957. 10.3897/BDJ.8.e55957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Jiang N., Liang L. Y., Yang Q., Tian C. M. (2019). Arthrinium trachycarpum sp. nov. from Trachycarpus fortunei in China. Phytotaxa 400 203–210. 10.11646/phytotaxa.400.3.7 [DOI] [Google Scholar]
- Yang C. L., Xu X. L., Dong W., Wanasinghe D. N., Liu Y. G., Hyde K. D. (2019). Introducing Arthrinium phyllostachium sp. nov. (Apiosporaceae, Xylariales) on Phyllostachys heteroclada from Sichuan Province, China. Phytotaxa 406 091–110. 10.1127/nova_hedwigia/2019/0526 [DOI] [Google Scholar]
- Zhang J. F., Liu J. K., Hyde K. D., Chen Y. Y., Liu Y. X., Liu Z. Y. (2017). Two new species of Dyfrolomyces (Dyfrolomycetaceae, Dothideomycetes) from karst landforms. Phytotaxa 313:267. 10.11646/phytotaxa.313.3.4 [DOI] [Google Scholar]
- Zhang J. F., Liu J. K., Jeewon R., Wanasinghe D. N., Liu Z. Y. (2019). Fungi from Asian Karst formations III. Molecular and morphological characterization reveal new taxa in Phaeosphaeriaceae. Mycosphere 10 202–220. 10.5943/mycosphere/10/1/3 [DOI] [Google Scholar]
- Zhao Y. M., Deng C. R., Chen X. (1990). Arthrinium phaeospermum causing dermatomycosis, a new record of China. Acta Mycol. Sin. 9 232–235. [Google Scholar]
- Zhao Y. Z., Zhang Z. F., Cai L., Peng W. J., Liu F. (2018). Four new filamentous fungal species from newly-collected and hivestored bee pollen. Mycosphere 9 1089–1116. 10.5943/mycosphere/9/6/3 33332223 [DOI] [Google Scholar]
- Zhaxybayeva O., Gogarten J. P. (2002). Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genom. 3:4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in the study can be found in the Genbank. The accession numbers of the sequences deposited in GenBank are ITS: MW481705—MW481721; LSU: MW478885–MW478901; TEF: MW522938–MW522954; and TUB: MW522955–MW522969.