Abstract
Antimicrobial resistance (AMR) is a complex global health issue and will push twenty-four million people into extreme poverty by 2030, risking the sustainable development goals (SDGs) 2, 3, 6, 9, 12, and 17 if not addressed immediately. Humans, animals, and the environment are the reservoirs that contribute and allow AMR to propagate in interconnected ecosystems. The emergence of antibiotic-resistant bacteria and antibiotic-resistant genes in the water environment has become an important environmental health issue. One of the major influencers from environment sector is the pharmaceutical industry which is growing globally to meet the ever-increasing demand of antibiotics, especially in low- and middle-income countries. The pharmaceutical effluent has a mix of large concentrations of antibiotics and antibiotic resistance genes, and these sites act as hotspots for environmental contamination and the spread of AMR. Inadequate treatment of the effluent and its irresponsible disposal leads to unprecedented antibiotic contamination in the environment and their persistent presence in the environment significantly modulates the bacterial genomes’ expression that is responsible for increase and spread of AMR. However, not much interventions are suggested in the National Action Plan developed on AMR by many countries. There are no regulations across the globe till date for the level of antibiotic residues in pharmaceutical effluent for the growing pharmaceutical industry. This review put together the work done showing several detrimental effects of the antimicrobial residues in the pharmaceutical effluent which leads to rise in development of AMR. The environment risk approach and need to have indicators to measure environment risk is a way forward for all countries engage in antibiotic manufacturing. Overall, efforts to address the problem are isolated and fragmented. Policymakers, regulators, manufacturers, researchers, civil society, and the community need to collaborate so that antibiotics are produced sustainably and continue to stay effective in treating bacterial infections.
Keywords: Antibiotics, Antimicrobial resistance, Environment, National Action Plan, Pharmaceutical effluent, Pharmaceutical industry, Effluent treatment
Introduction
Antibiotics are modern-day miracle medicines that have saved lives since the discovery of penicillin in the twentieth century. Effective antibiotics are the cornerstone of modern-day medicine-enabling organ transplants, surgical prophylaxis, protecting newborn lives from sepsis, and warding off infections during cancer chemotherapy. Unfortunately, the excessive and indiscriminate use of antibiotics in the globally interconnected ecosystems has led to enormous rise in antimicrobial resistance (AMR). The concern has been recognized among the top ten public health threats by the World Health Organization (WHO 2020). AMR is defined as the microorganisms’ ability to resist the therapeutic agents used to treat them, exposure to which transforms them into “superbugs,” and thus makes antimicrobials ineffective in the treatment of infectious diseases caused by such superbugs (Gandra and Kotwani 2019). It is a complex global health issue with an immense impact on the economy and development. It imposes a substantial burden on the global economy—by 2050, approximately four hundred forty-four million people would be affected (O'Neill 2016) due to the inability to treat the infections. A 2017 World Bank study projected that AMR would be responsible for decreasing global Gross Domestic Product (GDP) by 1.1 and 3.8 percentage points between 2017 and 2050 (Jonas Olga et al. 2017). Ninety of the 244 indicators for Sustainable Development Goals (SDGs) are relevant for AMR (IACG 2018), with 6 SDGs being of particular relevance (SDG 2, 3, 6, 9, 12, and 17).
In interconnected ecosystems, human, animal (including food safety), and environment sectors are the key to the development and transmission of AMR. Their involvement is essential to slow its spread as suggested by the U.N. Tripartite (WHO, FAO (Food and Agriculture Organization of the United Nations), and the OIE (Office International des Epizooties, World Organization for Animal Health (OIE)). The Global Action Plan (GAP) on AMR (2015) addresses AMR in the one health paradigm. Following the GAP, most countries have developed their National Action Plan (NAP) on AMR by 2017 as promised in the World Health Assembly 2015. However, a stark limitation in the GAP-AMR is the limited direction provided in addressing AMR in the environment sector. Therefore, in most of the NAP-AMR, the role of environmental sector and activities for the environment sector, especially treatment and disposing of the effluents containing antibiotic residue, is not stressed. With greater recognition of the environmental aspects of AMR, the United Nations Environment Program (UNEP) in 2018 joined this collaboration which is now called the “Tripartite Plus” (FAO, OIE, WHO, UNEP) (Tripartite Plus Recent Update and Memorandum of Understanding on Antimicrobial Resistance 2018) and aimed to address AMR holistically. Hence, the present review article was synthesized to impress upon the policymakers and other stakeholders involved in operationalizing and prioritizing the activities of NAP-AMR across the globe the importance of environment sector, especially the effluent discharge from pharmaceutical industry manufacturing antibiotics.
Inappropriate use of antibiotics in humans (in hospitals, communities) and animals (including livestock, poultry, aquaculture) and the utilization of animal waste from antibiotic fed animals, like poultry as manure or fertilizer in fields, also act as a source of introducing antibiotics and antibiotic-resistant genes (ARGs) in the soil and water bodies (Meersche et al. 2020) which are part of the environment. The evident sources contributing to AMR’s environmental role include agricultural and poultry farms, effluents from pharmaceutical industries and healthcare facilities, and expired or unused antibiotics disposed of by household and community (Khurana and Sinha 2019).There are numerous scientific studies that demonstrate the presence of antibiotic-resistant bacteria and the presence of antibiotic residues and resistance genes in the environment (Kumar and Kotwani 2017; Tamhankar and Lundborg 2019; Singer et al. 2016). However, there is a paucity of studies that address ways to tackle this problem in the environment (Franco et al. 2009; Larsson et al. 2018).
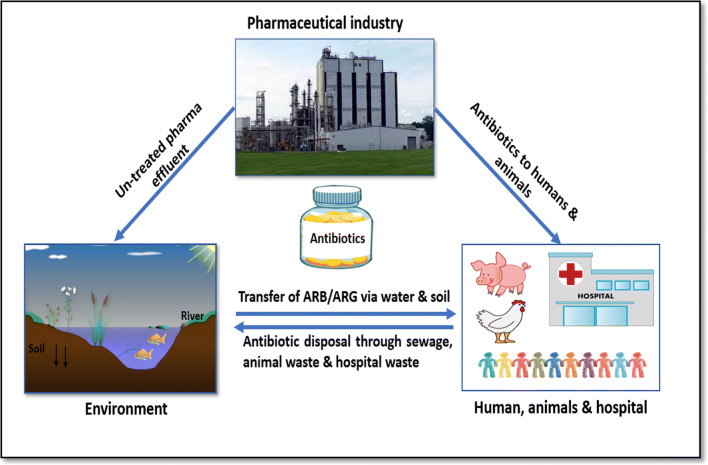
The much growing pharmaceutical industry particularly has a critical role as it acts as a reservoir for antibiotics and antibiotic resistance genes (ARGs) in the environment within the one health paradigm (Fig. 1). While it is critical to improve access to much-needed antibiotics for treating infections for humans and animals, antibiotic residue containing pharmaceutical effluent also contributes to AMR development.
Fig. 1.
The interconnection between the Pharmaceutical industry, environment, humans and animals
In this context, the current review was done with the aim to synthesize and put together the work done till recently to bring attention of policymakers and regulators to have regulations for effluent discharge from pharmaceutical industry to curb the development of AMR. The broad areas covered in this review article are as follows: (1) characterize the AMR challenge due to the pharmaceutical effluent discharge from antibiotic manufacturing industries, (2) discuss the adverse effects of antibiotic residues on the ecosystem, (3) need to have indicators to measure environment risk, and (4) the current initiatives taken by stakeholders to address the antibiotic residues from manufacturing units to curb the role of pharmaceutical effluent in decreasing AMR.
Pharmaceutical industry effluent contributions to AMR in the environment
The ever-increasing demand for life-saving antibiotics has led to large-scale manufacturing to improve access to antibiotics, especially in low- and middle-income countries (LMICs). LMICs, like Bangladesh, Brazil, India, Malaysia, Nigeria, Thailand, Philippines, and Vietnam (WHO 2011; Babatunde et al. 2014; Rader and Langer 2018), have strengthened and expanded their manufacturing capacities. The pharmaceutical industry is expected to be worth USD 1.57 trillion by 2023. The pharmaceutical market for the Asia Pacific region is expected to be the second largest (24.07%) by 2023 (Global Pharmaceuticals Industry Analysis and Trends 2019).
Broadly, the pharmaceutical industry comprises of (i) active pharmaceutical ingredient (API) manufacturing units that manufacture the raw material or an intermediate for the antibiotic and (ii) formulations or finished pharmaceutical product (FPP) manufacturing units that combine the active ingredient to produce a final pharmaceutical product (Nahar 2020). Over the past three decades, India and China have emerged as the source of APIs for the pharmaceutical sector. While China is the principal supplier of APIs, India has established itself as the hub for processing the APIs into finished pharmaceutical products or formulations (Arnum 2013; Gandra et al. 2017).
The increase in antibiotic manufacturing capacity has caused a consequent rise in the industry’s pharmaceutical effluent. The effluents generated from both types of antibiotic manufacturing units (API and FPP) contain antibiotic residues. Still, significantly higher amounts of residues are expected in the effluent from the API manufacturing units. Since the effluent from pharmaceutical manufacturing sites (especially API manufacturing plants) contain a concentrated amount of API and intermediates in the effluent, these sites act as hotspots for environmental contamination and the spread of AMR. The pharmaceutical effluent is a cocktail of the highest concentrations of antibiotics and antibiotic resistance genes (ARGs) detected in the environment (Larsson 2014; Bengtsson-Palme and Larsson 2016) when compared to discharges from agriculture sources, sewage and other human sources. Moreover, treating the effluent at source in the pharmaceutical plant itself is a feasible and economically viable solution to address AMR in the environment from pharma manufacturing.
With a half-life of a few hours to a hundred days, antibiotics persist in the environment (Ji et al. 2010), and in fact, can be described as a persistent organic contaminant due to frequent and extensive use of antibiotics and uninterrupted emissions (Hamscher et al. 2002). When discharged from the plant effluents into the environment, antimicrobial residues have several detrimental effects on the environment microbiome at different concentration levels. Even at subinhibitory concentrations, the susceptible bacterial cells continue growing at a reduced growth rate and antibiotics act as signal molecules, shifting bacterial gene expression and gene transfer, virulence, biofilm formation and quorum sensing, and the modulation of gene activity. The lowest antibiotic level that drives the selection of a resistant mutant over the wild type cells is determined as the Minimal Selective Concentration (MSC), which is usually estimated to range between 1/4 and 1/230 of the minimum inhibitory concentration (MIC) values( Bengtsson-Palme and Larsson 2016; Gullberg et al. 2011).MSC is a more sensitive metric to reflect the selective pressures at lower antibiotic concentrations than MICs (Sandegren 2014) but difficult to measure since the composition and structure of the bacterial community and the antibiotic residues differ significantly in environmental and laboratory settings.
Long-term exposure of environmental bacterial strains to low concentrations of antibiotics significantly modulates the bacterial genomes' expression. Studies have found that the sub-inhibitory concentrations of antimicrobials induce the bacterial SOS repair system, increasing the frequency of genome mutation, overexpression of certain genes, gene transfer and creation of mobile genetic elements which include those responsible for antimicrobial resistance (Couce and Blazquez 2009; Blázquez et al. 2012). Exposure to sub-inhibitory doses of erythromycin, sulfamethoxazole, or roxithromycin results in biofilm formation in P. aeruginosa strains (Bruchmann et al. 2013). A survey (1999-2018) of sub-inhibitory concentrations of antibiotics in environmental samples reported that the environmental concentrations of antibiotics in many samples fall into the MSC range and are likely to influence bacterial ecology and trigger the selection of antibiotic resistant bacterial cells (Chow et al. 2021).
Studies have confirmed antibiotic residues in effluents, municipal wastewaters, surface waters, and groundwater around the pharmaceutical manufacturing sites (Deschamps et al. 2012; Tong et al. 2020). A few studies showing presence of huge quantities of antibiotics in surface water, drinking water, pharmaceutical effluent and in the effluent of wastewater treatment plant is presented in Table 1. Several studies (Verlicchi et al. 2012; Rizzo et al. 2013; Xu et al. 2015) have indicated that wastewater treatment plants can be critical sources for releasing antibiotic-resistant bacteria (ARBs) into the environment and can be catastrophic if treated inadequately, a common occurrence in LMICs. .Recent studies have shown that the environment polluted with waste from antibiotic manufacturing units could be an important antibiotic resistance reservoir (Bengtsson-Palme et al. 2014). Studies have identified that the untreated effluent discharge from API or FPP units can lead to the emergence of hotspots unless these are adequately assessed and controlled by the industry. Studies conducted in Bangladesh, India, Indonesia, and Thailand have reported antibiotic residues in aquatic water and fish (Impens et al. 2003; Swapna et al. 2012; Bakar et al. 2013). Reports on the presence of APIs in water from pharmaceutical industries in the European Union (Cardoso et al. 2014), the USA (Phillips et al. 2010), India, and elsewhere indicate that concentrations have reached higher levels when wastewater discharges are not sufficiently treated by the industries (Larsson et al. 2007; Larsson 2014).
Table 1.
Studies on presence of antibiotics in surface water, drinking water, pharmaceutical effluent, and in the effluent of wastewater treatment plant
| Class of antibiotics | Type of antibiotic | Origin and concentration | References |
|---|---|---|---|
| β-Lactams | Lincomycin | Surface water 248.9 ng/L | (Calamari et al. 2003) |
| Amoxicillin | Raw water 1500ng/L | (Mahmood et al. 2019) | |
| Aminoglycosides | Gentamicin | Sewage plant export 1300 ng/L | (Loffler and Ternes 2003) |
| Macrolides | Erythromycin-H2O | Surface water 1700 ng/L | (Kolpin et al. 2002) |
| Azithromycin | Wastewater treatment plant effluent 1577.3 ng/L | (Rodriguez-Mozaz et al. 2020) | |
| Clarithromycin | Surface water 260 ng/L | (Hirsch et al. 1999) | |
| Wastewater treatment plant effluent 346.8 ng/L | (Rodriguez-Mozaz et al. 2020) | ||
| Roxithromycin | Surface water 560 ng/L | (Hirsch et al. 1999) | |
| Quinolones | Ciprofloxacin | Sewage plant export 260 ng/L | (Kostich et al. 2014) |
| Surface water 185 ng/L | (Bai et al. 2014) | ||
| Pharma Effluent 28 X 106–31 X 106 ng/l | (Larsson et al. 2007) | ||
| Wastewater treatment plant effluent 1435.5 ng/L | (Rodriguez-Mozaz et al. 2020) | ||
| Raw water 1270 ng/L | (Mahmood et al. 2019) | ||
| Norfloxacin | Surface water 208 ng/L | (Xu et al. 2007) | |
| Pond 152.31 ng/L | (Chen et al. 2018) | ||
| Pharma Effluent39 X 104–42 X 104 ng/L | (Larsson et al. 2007) | ||
| Ofloxacin | Surface water 89 ng/L | (Xu et al. 2007) | |
| Sewage plant export 210 ng/L | (Guerra et al. 2014) | ||
| Pond 15.98 ng/L | (Chen et al. 2018) | ||
| Pharma Effluent15 X 104–16 X 104 ng/L | (Larsson et al. 2007) | ||
| Wastewater treatment plant effluent 613 ng/L | (Rodriguez-Mozaz et al. 2020) | ||
| Enrofloxacin | Livestock farm export 680 ng/L | (Andrieu et al. 2015) | |
| Underground water 3 ng/L | (Tong et al. 2009) | ||
| Levofloxacin | Raw water 414 ng/L | (Mahmood et al. 2019) | |
| Sulfonamides | Sulfamethoxazole | Surface water 1900 ng/L | (Kolpin et al. 2002) |
| Underground water 470 ng/L | (Hirsch et al. 1999) | ||
| Wastewater treatment plant effluent 220.9 ng/L | (Rodriguez-Mozaz et al. 2020) | ||
| Sulfamethizole | Surface water 130 ng/L | (Kolpin et al. 2002) | |
| Sulfamethazine | Surface water 4660 ng/L | (Wei et al. 2011) | |
| Underground water 160 ng/L | (Hirsch et al. 1999) | ||
| Sulfadoxine | Surface water 460 ng/L | (Wei et al. 2011) | |
| Sulfamonomethoxine | Pond 21.92 ng/L | (Chen et al. 2018) | |
| Sulfaphenazole | Pond 147.95 ng/L | (Chen et al. 2018) | |
| Sulfapyridine | Wastewater treatment plant effluent 583.6 ng/L | (Rodriguez-Mozaz et al. 2020) | |
| Tetracyclines | Tetracycline | Underground water 3.8 ng/L | (Tong et al. 2009) |
| Oxytetracycline | Surface water 340 ng/L | (Kolpin et al. 2002) | |
| Chlortetracycline | Surface water 690 ng/L | (Kolpin et al. 2002) |
A study conducted in 7 European countries (Portugal, Spain, Ireland, Cyprus, Germany, Finland, and Norway) detected 17 antibiotics, i.e., ciprofloxacin, ofloxacin, enrofloxacin, orbifloxacin (Fluroquinoloes class of drugs are used to treat gram harmful abdominal infections, pyelonephritis, and as second-line treatment for typhoid fever as resistance is increasing); azithromycin, clarithromycin (Macrolides class of drugs used to treat typhoid, Rickettsial infections), sulfapyridine (used for skin infections); sulfamethoxazole, trimethoprim (used in the treatment of typhoid; pneumonia and diarrhea); nalidixic acid (used to treat urinary tract infections (UTIs)), pipemidic acid, oxolinic acid, cefalexin (antibiotic prophylaxis before surgeries, diabetic foot ), clindamycin (Clostridium difficile infections, skin and soft tissue infections, Pneumocystis carinii infections); metronidazole (diarrheas and UTIs), ampicillin (for Acinetobacter spp., susceptible typhoid strains, pre-surgery prophylaxis); and tetracycline (used to treat infections of the skin, urinary tract and sexually transmitted diseases), in the final effluent of wastewater treatment plant (Rodriguez-Mozaz et al. 2020). A similar study conducted in Vietnam detected higher concentrations of sulfonamides and quinolones class of antibiotics in the effluent of pharmaceutical plants (Thai et al. 2018). Other studies from Korea (Sim et al. 2011), Taiwan (Lin et al. 2008; Lin and Tsai 2009), and Pakistan (Khan et al. 2013) also substantiated the presence of high concentrations of APIs in the manufacturing discharges. Furthermore, some reports identified few pharmaceutical companies, including formulation units, in the USA (Phillips et al. 2010) and Europe (Norwegian Environment Agency 2005; Babic et al. 2007) as a source of antibiotic pollution, with significant concentrations of APIs in treated effluents reaching milligram per liter.
In 2008, a manufacturing unit in China was ordered to suspend activities and install an appropriate waste treatment plant after reports that the factory had furtively disposed off its waste in a 50-ha pit. Additionally, the effluent was being discharged through irrigation trenches connecting to the Yellow River (Shanghai Daily 2012). Another study in China indicated that the levels of oxytetracycline, from a manufacturing unit that apparently treated its waste, were still significant (Li et al. 2008). An analysis conducted in 2016 (Pharma Compass 2016, a trade publication) identified two critical dangers in China’s antibiotic supply chain—first, the threat of antibiotic resistance from pharma effluents and second, the lack of compliance with Good Manufacturing Practices (GMP).
Propagation of resistance through the environment
The environment plays a crucial role in the emergence and transmission of AMR determinants and pathogenic bacteria. The pharmaceutical effluent is released into the environment via soil or water (Rees 2020), further establishing soil and the aquatic environment as reservoirs for antibiotic residues and vectors for resistance genes (UNEP 2017).
Resistance to antibiotics can be disseminated through vertical gene transfer (VGT; i.e., parent to next-generation) as well as by horizontal gene transfer (HGT; i.e., the transference of genetic material, such as ARGs, from cell to cell). HGT can occur between live bacterial cells (conjugation): via a bacteriophage (transduction) or through absorption of extracellular DNA (transformation) within the cell (Vikesland et al. 2019). Both HGT and VGT are known to coexist in the natural environment (Li et al. 2019).
Selection and propagation of antibiotic-resistant genes (ARGs) in the environment are aided by the presence of not just antimicrobials (including antibiotics) but also the presence of heavy metals, biocides (disinfectants), and other natural chemicals and xenobiotics such as solvents like toluene which gives rise to antibiotic-resistant bacteria (ARB). Cross-resistance and co-resistance to each of these hazards lead to co-selection of genes (Singer et al. 2016), promote the sharing of antibiotic-resistant genes (ARGs), mobile genetic elements (MGEs), plasmids, and virulence factors mediated through horizontal transfer (Fletcher 2015) with other bacteria present in the environment (Larsson et al. 2007; Kraemer et al. 2019). The MGEs carry the genes that encode antibiotic resistance and are present in a wide variety of bacterial population. The cross phylogenetic boundaries are characteristic of HGT. The presence of diverse MGEs carrying antibiotic resistance genes makes HGT the main cause of spreading antibiotic resistance in the environment (Huijbers et al. 2015).
The selective resistance potential of bacteria varies between different species and different pharmaceutical compounds (Larsson et al. 2018). The ARBs carrying a newly evolved ARGs can subsequently spread between patients epidemically (López-Causapé et al. 2013); examples of bacteria of human health relevance include vancomycin-resistant Enterococci (VRE), methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum beta-lactamase enzymes (ESBLs) producing E. coli, Carbapenem-resistant Enterobacteriacae (CRE) (Vikesland et al. 2019), and multi-resistant Pseudomonads (Pareek et al. 2015). These resistant bacteria are the key contributors to the increasing burden of healthcare-associated infections (HAIs): infections that are acquired while receiving treatment for medical or surgical conditions in a healthcare setting. Some of the common healthcare-associated infections include catheter-related urinary tract infections, bloodstream infections related to intravenous cannulas and other devices, postoperative wound infections, and ventilator-associated pneumonia. Carbapenem-resistant Enterobacteriacae can cause resistant urinary tract infections, bloodstream infections, and ventilator-associated pneumonia and intra-abdominal abscesses which have a 50% mortality rate even in a developed country setting like the USA (CDC 2019a, b). ESBL E. coli causes urinary tract infections and severe infections in the blood stream and central nervous system. Since E. coli is a common colonizer of the normal gut microbiota, increasing prevalence of ESBL-producing strains is particularly worrisome (Brolund 2014). MRSA infections are commonly seen in the elderly, those admitted in ICUs, having extended stay in a healthcare facility, or undergoing an invasive medical procedure. MRSAs lead to infections of the skin and invasive disease in the presence of indwelling devices (intravascular lines, urinary catheter, endotracheal or tracheostomy tube, gastrostomy feeding tube) (Best Practice Guidelines Nova Scotia 2012). VRE infections are estimated to cause 54,500 infections among hospitalized patients and 5400 estimated deaths in the USA (CDC AR Threats 2019). Persons at risk include those who have been previously treated with antibiotics, including vancomycin, for long duration, those hospitalized or undergoing any surgical procedures, or having a medical device in situ, and those with weakened immune systems, such as patients in intensive care units, or in cancer or transplant wards.
Thus, with improved life expectancy and human survival, the need to control HAIs and ensure successful treatment outcomes in healthcare without any resistant microbial infections makes it imperative to consider the environment a high-interest factor to study the evolution and dissemination of antibiotic resistance.
Impacts of antibiotic residues on the ecosystem
Evolutionary effect on the microbial population
Antibiotic pollution impacts the evolutionary dynamics of the microbial population leading to a heterogeneous phenotypic response to antibiotics. The occurrence of antibiotic residues in the microbial ecosystem may lead to genetic or mutational alterations in normally sensitive bacteria. This allows the bacteria to survive and grow further as ARB that carry ARGs (Martinez 2009).Antibiotic exposure to the microbial community influences the developmental characteristics of the microbes in multiple ways such as genome reconstruction mainly in the mobile genetic elements (MGEs) of the microbes, physiology of the microbiome, and triggering expression of a shock-response system in the microbes (Martínez 2017). Diverse bacterial populations present different degrees of resistance to antibiotics due to altered gene expression or critical physiological traits under the exerted selective pressure (Sanchez-Romero and Casadesus 2013). Bacterial populations exposed to low concentrations of antibiotics can lead to bacterial lineages with increased phenotypic and genotypic diversity through the activation of an intrinsic survival response. It results in an increased mutation rate throughout the genome (Foster 2007), and via the direct mutagenic effect on the DNA (Andersson and Hughes 2014). MIC is the minimum concentration of an antibiotic that can completely inhibit the growth of a bacterial strain after an incubation period of 24 h. In many instances, environmental concentrations close to, or exceeding, the MICs of certain antibiotics have been measured in site linked to pollution from pharmaceutical production facilities (Larsson 2014) even though concentrations below the MICs can select for resistant bacteria (Andersson and Hughes 2012).
A study reported that most of the antibiotics either bind directly to the transcription factor or via other regulatory mechanisms such as riboswitches and quorum sensing altering the gene expression of various genes (Goh et al. 2002; Davies et al. 2006) and resulting in elevated phenotypic variability and in some cases increased virulence (Kraemer et al. 2019). Similarly, other studies have found structural alterations in the microbial community due to the presence of antibiotics in soil. This results in loss of biomass and a reduction in microbial activity including nitrification, denitrification, and respiration (Westergaard et al. 2001; Cycon et al. 2019). Moreover, antibiotics can also affect the enzymatic activity of the bacteria, including dehydrogenases, phosphatases, and ureases, which are considered important indicators of soil activity (Cycon et al. 2019). Thus, this consequently brings loss of key microbial taxonomy that is critical for ecological roles.
Adverse effects on the non-microbial population in the ecosystem
The implication of antibiotic pollution is not limited to the microbial population only. It is been shown that low concentrations of streptomycin and erythromycin impact the survival and behaviors of micro-invertebrates such as Daphnia magna (Flaherty and Dodson 2005) and Artemia under in vitro conditions (Migliore et al. 1997).
Strong implications can be even seen in the aquatic ecosystem where the animals are chronically exposed to antibiotic pollutants (Liu et al. 2018). Adverse effects of the presence of antibiotics in the aquatic environment, especially on fish have been reported (Corcoran et al. 2010). One of the effects seen was the suppression of the fish immune system by the presence of tetracycline (0.1–50 μg/L) in the environment (Grondel et al. 1985). A study has demonstrated that the sub-inhibitory concentrations of macrolides induces malformation of the yolk sac edema, uninflated swim bladder, and influences the movement frequency of the embryo in zebrafish (Danio rerio)(Wang et al. 2014). Similarly, norfloxacin and sulfamethoxazole at a concentration of 200μg/L have also been reported to have an adverse effect on the growth and reproduction rate of zebrafish (Danio rerio) (Yan et al. 2016). Antibiotics such as fluoroquinolones and tetracyclines can also affect the development and behavior, potentially via altering the expression of multiples genes in zebrafish (Danio rerio) (Zhang et al. 2016).
In addition to affecting the microbial population and the aquatic ecosystem, antibiotic pollution even has a negative impact on vertebrates such as amphibians, reptiles, birds, and mammals. It leads to growth inhibition, disordered locomotion behavior, cytotoxicity, and teratogenesis (Liu et al. 2018). A study conducted on Xenopus tropicalis found that the antibiotic residue in the environment is lethal to the amphibians. The study showed that exposure of tetracycline resulted in decreased body length, pericardial edema, and other malformations (Liu et al. 2018).
Adverse effects on the human health
Varying levels of antibiotics that have been found in surface and waste water (between 0.01 and 1.0 μg/L) allow for interactions between environmental bacteria and contaminating bacteria from sewage and wastewater treatment plants and are the potential medium for transfer of novel antibiotic resistance mechanisms from environmental bacteria to human and animal commensals or pathogens (or vice versa) (Aslan et al. 2018; Manaia et al. 2016). Presence of antibiotics in drinking water may persist for months and many are not completely removable by traditional disinfection technologies in drinking water treatment (Ben et al. 2019). Key classes of antibiotics that have been found in drinking water include quinolones, chloramphenicols, sulfonamides, and macrolides appeared with ciprofloxacin exhibiting the highest detected concentration up to 679.7 ng/L (Wang et al. 2010). Many studies showing presence of various classes of antibiotics present in surface water, drinking water, pharmaceutical effluent, and in the effluent of wastewater treatment plant are tabulated in Table 1.
Wastewater treatment plants (WWTPs) have a key role in the spread of antibiotic resistance (Bouki et al. 2013; Rizzo et al. 2013) by allowing bacteria from different environments and sources to interact and exchange genes and are major sources of ARB and ARGs that are released into the environment (Alexander et al. 2020; Karkman et al. 2017). The risk of transfer of ARGs to pathogens increases in environments with a high fecal load and associated fecal bacteria (Huijbers et al. 2019).
The antibiotic residues in the environment create selection pressure on the environmental microbiome, generating environmental ARB and ARGs which can be transferred to humans (Forsberg et al. 2012; Manaia 2017). Environmental ARGs can be transmitted to humans by either pathogenic ARB or human commensal ARB, both of which have the ability to colonize and proliferate in human body (Li et al. 2015; Manaia 2017; Vaz-Moreira et al. 2014). While commensal bacteria are otherwise not harmful, acquisition of ARGs from the environment enables them to transfer the virulent genes to human microbiota (Serwecinska 2020). The pathogenic ARBs could potentially penetrate into tissues and cause acute immune reaction and infectious disease (Ben et al. 2019). Among the Gram-negative bacteria, Pseudomonas, Acinetobacter, and Enterobacteriacae have been prioritized by the WHO as bacteria for which there is an urgent need to develop new effective drugs. CRE, VRE, MRSA, and ESBL E. coli as described in the earlier section (propagation of resistance) are responsible for the rise in HAIs and have been identified as the priorities for action by the WHO and the Centre for Disease Control and prevention (CDC), USA. Gaps in knowledge exist for the role of the environment in transmission of resistance in these bacteria because only a few studies have confirmed the presence of ESBL, MRSA, and VRE producers in the environment. However, even their presence in the environment is alarming as they can act as a reservoir of such resistance and such genes can be transferred into human pathogens (Czekalski et al. 2015; Chen et al. 2015, 2017; Zielinski et al. 2020).
Environment risk assessment approach: need for indicators to measure progress
Considering the negative impacts of antibiotic residues in the environment, the need to assess the environmental risk becomes necessary. This requires both science and policy inputs. At present, there is a lack of consensual scientific benchmark and surveillance indicators among all stakeholders to measure progress on the one hand and a lack of cohesive partnership at policy level to establish progress in addressing the role of pharma effluent on AMR in the environment.
Globally, neither the WHO, which lays out the standards for Good Manufacturing Practices (GMP) for pharmaceutical manufacturing, nor the national regulatory authorities have established legal regulatory requirements for antibiotic emission control at pharma manufacturing sites (though India has released draft norms). Moreover, pharmaceutical (including antibiotic) manufacturing production and supply chain (guided by intentions to decrease costs and maximize profits) is scattered across the globe for different manufacturing steps and is messy with the involvement of several agencies (Topp et al. 2018). There is lack of transparency which hinders the establishment of benchmarks for each step and fix responsibility on a particular actor in one stage.
The environmental risk assessment process for a specific pharmaceutical compound is complex due to the inadequate availability of data. It depends on overall usage, its persistence, fate, transport, and eco-toxicity of the compound in the environment. The environmental risk can be determined by the minimum inhibitory concentration (MIC), predicted environmental concentration (PEC), and predicted no-effect concentration (PNEC) for antibiotics (Peake et al. 2016).
MIC, as defined earlier, is estimated in clinically relevant bacteria, and few bacterial indicators such as Acinetobacter, Burkholderia, Stenotrophomonas, Neisseria, Enterococcus, Staphylococcus, Streptococcus, and Haemophilus have been suggested to assess MICs of antibiotics (Patel 2017). The MICs are well documented in the database of the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2013). The PEC value is the concentration of chemical compound present in the environment. It is based on modelling and data derived from known environmental factors such as the ability of the compound to be degraded (persistence), the ability of the compound to penetrate into the biological membranes and accumulate inside the flora and fauna (transport), and the ability for organisms in the environment to metabolize and detoxify the pharmaceutical, nullifying any detrimental effects arising from the bioaccumulation of the pharmaceutical (Peake et al. 2016). The PNEC value is the concentration of pharmaceuticals at which it does not exert any adverse pharmacological effect on any organism.
Bengtsson-Palme et al in 2016 suggested a methodology for deriving PNECs by extrapolating the MIC data from the EUCAST database and included an additional safety factor to derive PNECs for antimicrobial resistance (Bengtsson-Palme and Larsson 2016). These values are derived using eco-toxicity testing data. A PEC to PNEC ratio of < 1 implies no significant risk associated with the environmental presence of the pharmaceutical. A ratio ≥ 1 suggests that further characterization of environmental risk or management of the pharmaceutical is necessary (Peake et al. 2016). This data can be used as a comprehensive reference by the industry and the regulating bodies for formulating standards.
The pharma industry has come up with target (PNEC values) for more than 100 antibiotics to be used as a benchmark for self-evaluation. Efforts are needed so that pharma companies can be incentivized to establish better production systems that are sustainable, document antibiotic pollution control efforts, and provide incentives such as tax subsidies for exchangeable products tax subsidy (Bengtsson-Palme et al. 2018). Industry can also be encouraged to leverage technology of pollution control in procurement processes, and in benchmarking schemes for drug companies (Access to Medicines Foundation 2017; Bengtsson-Palme et al. 2018).
Stakeholders efforts to address antibiotic residues in pharmaceutical effluent
Addressing the contribution of the pharmaceutical industry to AMR requires key stakeholders to work together. This includes the policymakers, national regulators, pharmaceutical industry, researchers and academics, civil society, U.N. organizations, and trade organizations.
At the global level, the U.N. informal Interagency Task Team on Sustainable Procurement in the Health Sector (SPHS 2020) was established in May 2012 in Copenhagen, Denmark, for effectively engaging with suppliers and manufacturers to promote environmentally and socially responsible procurement of health commodities to save lives sustainably and protect the planet (United Nations (U.N.) Interagency Task Team on Sustainable Procurement in the Health Sector 2020).
As part of the Good Manufacturing Practices (GMPs), the WHO has drafted standards for the antibiotic manufacturing pharmaceutical industries and the inspectors from regulatory authorities for all member states (WHO 2019) to include environmental risk of AMR from the pharma effluent. Since resistant microbes do not respect borders, globally national regulatory authorities (NRA) and U.N. bodies need to ensure uniform global regulatory standards to address the role of pharmaceutical effluent in propagating AMR.
Assessing the environmental impact of a pharmaceutical product is challenging as the manufacturing supply chain is complex with various stages of production outsourced to suppliers in diversely regulated markets. Regulations in the USA and European Union (E.U.) primarily focus on drug safety through the GMP but do not currently oblige companies to put in place environmental safeguards when producing their drugs. Currently, there is no legally binding national or international environmental legislation or directives for pharmaceutical companies on how to treat and dispose of their wastes in an environmentally friendly way (Turner 2020). Moreover, there are no statutory GMP regulations for active pharmaceutical ingredients in either the E.U. or the USA. However, many countries are taking an active role and formulating guidelines for pharmaceutical effluent treatment with regard to the antibiotic residue.
Among the high-income countries (HICs), the European Union (E.U.) has recently formulated a tailored risk assessment for medicinal products including antibiotics (EMA 2018). Under this tailored risk assessment for each medicinal product, both a risk assessment and a specific hazard assessment for persistent bio-accumulative and toxic (PBT) properties are required at the time of licensure. The risk assessment can be done in phases, and the PBT assessment identifies the intrinsic properties of the active substance which have long-term risk on the environment. Particularly for antibiotics, an additional targeted effect assessment should be performed for the aquatic compartment. Further, the tailored risk assessment for antibiotics focuses on the effects of antibiotics on lower trophic levels including bacteria, algae, and aquatic invertebrates as they are sensitive for antibiotics (EMA 2018). Among LMICs, India is the first country to have prepared draft regulatory standards for the pharmaceutical industry. The Ministry of Environment Forest and Climate Change (MoEFCC) through its technical arm, the Central Pollution Control Board (CPCB), has drafted standards for PNEC values for antibiotic residue in industrial effluents (Leo 2020; Ministry of Environment, Forest, and Climate change 2020). The draft standards released in January 2020 specify limits of effluents released from pharmaceutical manufacturing plants though the recommendation still needs to be notified as a law. In China, the government encourages its pharmaceutical industry and promotes zero discharge of pharmaceuticals into the environment though there are no standards for antibiotics residue levels in the discharged effluent.
Within the pharma industry too, there is greater recognition of the need for action leading to two key initiatives—The Pharmaceutical Supply Chain Initiative (PSCI) and the Industry Declaration and Antibiotic Manufacturing Framework as part of Roadmap on AMR. In 2016, some of the leading pharmaceutical companies signed the Antibiotic Manufacturing Framework as part of the Roadmap on AMR led by the AMR Industry Alliance. The roadmap was adopted at the United Nations High-Level Meeting on AMR in 2016 at the World Economic Forum in Davos. This framework aims to ensure adequate control of effluent emissions from the manufacturing units (AMR Industry Alliance 2018). Among the measures proposed was calculating the amount of API discharge by the industries into the environment, called the predicted-no-effect concentration (PNEC) or antibiotic concentrations below which there are no adverse effects on the environment and the risk of developing resistance is minimal (Lee and Choi 2019). These values are expected to be protective of ecological species and incorporate assessment factors consistent with standard environmental risk methodologies (Brandt et al. 2015).
Besides these efforts at the global level, national governments, and industry, civil society groups have also taken a lead to address the issue. The Access to Medicine Foundation aims to improve access on the medicines and produces the AMR Benchmark report. This assesses the pharma companies engaged in Research & Development, managing the waste generated from the manufacturing process, ensuring appropriate access and stewardship where the potential and responsibility to restrict AMR is high. Benchmarking recognizes the company’s positive endeavors towards AMR which may inspire other companies to join (Antimicrobial Resistance Benchmark Report 2020).
Another initiative is the Responsible Antibiotic Manufacturer’s Platform (RAMP): a global collaborative forum bringing together responsible procurers, regulators, and companies to co-create the business-case for sustainably produced antibiotics (SIWI 2020). The initiative aims to make sustainable manufacturing the norm for the whole antibiotics industry and has initiated pilot projects.
Conclusion
The discovery of antibiotics was a revolutionary milestone in the field of medical science to treat infections and improve human life expectancy and living conditions. However, inappropriate use of antibiotics has led to sharp increase in antimicrobial resistance. AMR is a threat to global public health and the economy with significant implications on the successful treatment of infections in humans and animals. The spread of ARB and of ARGs, if left unchecked, could make antibiotics ineffective with devastating consequences on human and veterinary healthcare systems (WHO 2020). AMR is a cross-cutting public health issue encompassing the interface between humans, animals, and the environment and this is known as one-health involvement. Taking one-health approach, the WHO has prepared GAP-AMR and many countries (mainly LMICs) have developed their NAP-AMR aligning it with GAP. However, the involvement, recognition, and interventions for environmental sector are not as prominent as human or animal sector. Moreover, one of the important and growing factors is the discharge of antibiotics from pharmaceutical manufacturing plants which propagates antibiotic resistance and contaminates the ecological environment, emerging as an area of concern. Several studies highlight that bacteria in the environment acquire resistance through horizontal and vertical gene transfer, making the environment both a source and a reservoir for AMR. Tackling the discharge of pharmaceutical waste containing a concentrated amount of antibiotics at source in the pharmaceutical plants will be valuable to address the environmental burden of AMR in the ecosystem. Moreover, the lack of specific standardized methods and standards for treating the antibiotic effluent and the lack of awareness about the industrial contribution to AMR is one of the root causes of the rampant emergence of AMR in the environment. So far, there have been limited and fragmented approaches from the stakeholders to address the problem, and robust initiatives are the urgently required. Since many countries are revising their NAP-AMR or will be doing the same in the near future, it will be good if more activities and interventions targeted for environment reservoir of AMR and for effluent discharge are included. Sustainable antibiotics manufacturing can be adopted to reduce the release of antibiotics to the environment to the levels that, according to the best available knowledge, do not trigger AMR. Policymakers, regulators, manufacturers, researchers, civil society, and the community need to collaborate so that antibiotics are produced sustainably and continue to stay effective in treating bacterial infections. In true sense, a multi-sectoral and multi-dimensional “One Health” approach needs to be adopted by the leading organizations in the world and in the countries to curb AMR.
Abbreviations
- AMR
antimicrobial resistance
- API
active pharmaceutical ingredient
- ARB
antibiotic resistant bacteria
- ARG
antibiotic resistant gene
- CDC
Centers for Disease Control, USA
- CPCB
Central Pollution Control Board
- E.U.
European Union
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- FAO
Food and Agriculture Organization of the United Nations
- FPP
formulations or finished pharmaceutical product
- GAP
Global Action Plan
- GMP
good manufacturing practice
- HAI
healthcare-associated infections
- HGT
horizontal gene transfer
- LMIC
low- and middle-income countries
- MIC
minimum inhibitory concentration
- MGE
mobile genetic elements
- MoEFCC
Ministry of Environment Forest and Climate Change
- MSC
minimum selective concentration
- NRA
national regulatory authority
- OIE
Office International des Epizooties, World Organization for Animal Health
- PBT
persistent bio-accumulative and toxic
- PEC
predicted environmental concentration
- PNEC
predicted-no-effect concentration
- PSCI
pharmaceutical supply chain initiative
- RAMP
Responsible Antibiotic Manufacturer Platform
- UNEP
United Nation Environment Program
- VGT
vertical gene transfer
- WHO
World Health Organization
- WWTP
waste water treatment plants
Author contribution
AK and JJ conceived the idea of this paper and A.K. developed the format of the paper. D.K. searched the references and drafted the paper with inputs from A.K. A.K and JJ critically evaluated the manuscript. All authors have reviewed and approved the final version for submission.
Funding
DK was Junior Research Fellow on a Department of Biotechnology (DBT), Government of India (BT/IN/Indo-UK/AMR/04/AK/2018-19) funded project, and this review article is based on literature review done for the research work in the project. The funding agency had no role in the article’s design or concept, data collection, interpretation of data, and writing the manuscript. The views expressed are those of the authors alone and do not necessarily reflect those of the project funders.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Acess to medicine foundation (2017) 2017 Methodology for the 2018 Access to Medicine Index Available at: https://accesstomedicineindex.org/media/atmi/2017-Methodology-2018-Access-to-Medicine-Index.pdf. Accessed 22 Apr 2021
- Alexander J, Hembach N, Schwartz T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci Rep. 2020;10:1–9. doi: 10.1038/s41598-020-65635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMR Industry Alliance (2018) Progress Report. Geneva (C.H.): IFPMA. Available at: https://www.amrindustryalliance.org/progress-report/. Accessed Sept 26 2020
- Andersson DI, Hughes D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist Updat. 2012;15(3):162–172. doi: 10.1016/j.drup.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 2014;12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- Andrieu M, Rico A, Phu TM, Huong DTT, Phuong NT, Brink PJVD. Ecological risk assessment of the antibiotic enrofloxacin applied to Pangasius catfish farms in the Mekong Delta, Vietnam. Chemosphere. 2015;119:407–414. doi: 10.1016/j.chemosphere.2014.06.062. [DOI] [PubMed] [Google Scholar]
- Antimicrobial Resistance Benchmark (2020) Available at: https://accesstomedicinefoundation.org/media/uploads/downloads/5e270aa36821a_Antimicrobial_Resistance_Benchmark_2020.pdf. Accessed on Sept 24 2020
- Arnum PV (2013) The Weaknesses and Strengths of the Global API Market. PTSM: Pharmaceutical Technology Sourcing and Management 9. Available at: https://www.pharmtech.com/view/weaknesses-and-strengths-global-api-market. Accessed 22 Apr 2021
- Aslan A, Cole Z, Bhattacharya A, and Oyibo O (2018) Presence of antibiotic-resistant Escherichia coli in wastewater treatment plant effluents utilized as water reuse for irrigation. Water 10. 10.3390/w10060805
- Babatunde AI, Bamgbola EP, Oyelola OT (2014) The effect of pharmaceutical effluents on the quality of groundwater: a case study of Ikeja industrial area of Lagos, Nigeria. Int J Med Res Health Sci 4. http://ijsk.org/uploads/3/1/1/7/3117743/1_environmental_pollution.pdf
- Babic S, Mutavdzic D, Asperger D, Horvat AJM, Kastelan-Macan M. Determination of veterinary pharmaceuticals in production wastewater by HPTLC-videodensitometry. Chromatographia. 2007;65:105–110. doi: 10.1365/s10337-006-0109-2. [DOI] [Google Scholar]
- Bai Y, Meng W, Xu J, Zhang Y, Guo C. Occurrence, distribution and bioaccumulation of antibiotics in he river Liao River Basin in China. R Soc Chem. 2014;16:586–593. doi: 10.1039/c3em00567d. [DOI] [PubMed] [Google Scholar]
- Bakar MA, Morshed AJM, Islam F, Karim R. Screening of Chloramphenicol Residues in Chickens and Fish in Chittagong City of Bangladesh. Bangl J Vet Med. 2013;11:173–175. doi: 10.3329/bjvm.v11i2.19144. [DOI] [Google Scholar]
- Ben Y, Fu C, Hu M, Liu L, Wong MH, Zheng C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: a review. Environ Res. 2019;169:483–493. doi: 10.1016/j.envres.2018.11.040. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Larsson DGJ. Concentration of antibiotics predicted to select for resistant bacteria:Proposed limit for environmental regulation. Environ Int. 2016;86:140–149. doi: 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ. Shotgun metagenomics reveals a wide array ofantibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol. 2014;5:648. doi: 10.3389/fmicb.2014.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ. Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? J Clean Prod. 2018;171:137–146. doi: 10.1016/j.jclepro.2017.09.247. [DOI] [Google Scholar]
- Best Practice Guidelines for Reducing Transmission of Antibiotic Resistant Organisms (AROs) In Acute &Long Term Care Settings, Home Care & Prehospital Care (2012) Infection Prevention and Control Nova Scotia (IPCNS), Department of Health and Wellness. Available at: https://ipc.gov.ns.ca/sites/default/files/BestPracticesGuidelineforAROsAcuteandLTCSettings.pdf. Accessed 22 Apr 2021
- Blázquez J, Couce A, Rodríguez-Beltrán J, Rodríguez-Rojas A. Antimicrobials as promoters of genetic variation. Curr Opin Microbiol. 2012;15:561–569. doi: 10.1016/j.mib.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Bouki C, Venieri D, Diamadopoulos E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: a review. Ecotoxicol Environ Saf. 2013;91:1–9. doi: 10.1016/j.ecoenv.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Brandt KK, Amezquito A, Backhaus T, Boxall A, et al. Ecotoxicological assessment of antibiotics: a call for improved consideration of microorganisms. Environ Int. 2015;85:189–205. doi: 10.1016/j.envint.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Brolund A (2014) Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect Ecol Epidemiol 4. 10.3402/iee.v4.24555 [DOI] [PMC free article] [PubMed]
- Bruchmann J, Kirchen S, Schwartz T. Sub-inhibitory concentrations of antibiotics and wastewater influencing biofilm formation and gene expression of multi-resistant Pseudomonas aeruginosa wastewater isolates. Environ Sci Pollut Res Int. 2013;20:3539–3549. doi: 10.1007/s11356-013-1521-4. [DOI] [PubMed] [Google Scholar]
- Calamari D, Zuccato E, Castiglioni S, Bagnati R, Fanelli R. Strategic survey of therapeutic drugs in the rivers Po and Lambro in northern Italy. Environ Sci Technol. 2003;37:1241–1248. doi: 10.1021/es020158e. [DOI] [Google Scholar]
- Cardoso O, Porcher JM, Sanchez W. Factory-discharged pharmaceuticals could be a relevant source of aquatic environment contamination: review of evidence and need for knowledge. Chemosphere. 2014;115:20–30. doi: 10.1016/j.chemosphere.2014.02.004. [DOI] [PubMed] [Google Scholar]
- CDC (2019a) Information on Diseases and Organisms in Healthcare Settings, Healthcare Associated Infections ,National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Healthcare Quality Promotion (DHQP). Available at: https://www.cdc.gov/ncezid/dhqp/index.html. Accessed 22 Apr 2021
- CDC (2019b) Antibiotic Resistance Threats in the United States. U.S. Department of Health and Human Services, CDC, Atlanta Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 22 Apr 2021
- Chen B, Hao L, Guo X, Wang N, Ye B. Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ Sci Pollut Res. 2015;22:13950–13959. doi: 10.1007/s11356-015-4636-y. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yu D, He S, Ye H, Zhang L, Wen Y, Chen S. Prevalence of antibiotic-resistant Escherichia coli in drinking water sources in Hangzhou city. Front Microbiol. 2017;8:1133. doi: 10.3389/fmicb.2017.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Liu S, Zhang M, Li S, Wang J (2018) Comparison of the occurrence of antibiotic residues in two rural ponds: implication for ecopharmacovigilance. Environ Monit Assess 190. 10.1007/s10661-018-6883-0 [DOI] [PubMed]
- Chow LKM, Ghaly TM, Gillings MR. A survey of sub-inhibitory concentrations of antibiotics in the environment. J Environ Sci. 2021;99:21–27. doi: 10.1016/j.jes.2020.05.030. [DOI] [PubMed] [Google Scholar]
- Corcoran J, Winter MJ, Tyler CR. Pharmaceuticals in the aquatic environment: a critical review of the evidence for health effects in fish. Crit Rev Toxicol. 2010;40:287–304. doi: 10.3109/10408440903373590. [DOI] [PubMed] [Google Scholar]
- Couce A, Blazquez J. Side effects of antibiotics on genetic variability. FEMS Microbiol Rev. 2009;33:531–538. doi: 10.1111/j.1574-6976.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- Cycon M, Mrozik A, Piotrowska-Seget Z. Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front Microbiol. 2019;10:338. doi: 10.3389/fmicb.2019.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czekalski N, Sigdel R, Birtel J, Matthews B, Bürgmann H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ Int. 2015;81:45–55. doi: 10.1016/j.envint.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Deschamps E, Vasconcelos O, Lange L, Donnici C, Silva M, Sales J. Management of Effluents and Waste from Pharmaceutical Industry in Minas Gerais, Brazil. Braz J Pharm Sci. 2012;48:727–736. doi: 10.1590/s1984-82502012000400017. [DOI] [Google Scholar]
- EMA (2018) Guideline on the Environmental Risk Assessment of Medicinal Products for Human Use (Draft). Available at: https://www.ema.europa.eu/en/documents/scientific-guideline/draft-guideline-environmental-risk-assessment-medicinal-products-human-use-revision-1_en.pdf. Accessed 22 Apr 2021
- EUCAST (2013) [EUCAST] European Committee on Antimicrobial Susceptibility Testing. 2013. Available at: http://www.eucast.org/. Accessed on Oct 15 2020
- Flaherty CM, Dodson SI. Effects of pharmaceuticals on Daphnia survival, growth, and reproduction. Chemosphere. 2005;61:200–207. doi: 10.1016/j.chemosphere.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Fletcher S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ Health Prev Med. 2015;20:243–252. doi: 10.1007/s12199-015-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco BE, Martínez MA, Sánchez Rodríguez MA, Wertheimer AI. The determinants of the antibiotic resistance process. Infect Drug Resist. 2009;2:1–11. [PMC free article] [PubMed] [Google Scholar]
- Gandra S, Kotwani A. Need to improve availability of “access” group antibiotics and reduce the use of “watch” group antibiotics in India for optimum use of antibiotics to contain antimicrobial resistance. J Pharm Pol Pract. 2019;12:20. doi: 10.1186/s40545-019-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandra S, Joshi J, Trett A, Lamkang AS, Laxminarayan R (2017) Scoping report on antimicrobial resistance in India. Center for Disease Dynamics, Economics & Policy, Washington, DC. https://cddep.org/wp-content/uploads/2017/11/AMR-INDIA-SCOPING-REPORT.pdf. Accessed 22 Apr 2021
- Global Pharmaceuticals Industry Analysis and Trends 2023 (2019) https://www.reportlinker.com/p05750669/?utm_source=GNW. Accessed on 01 Oct 2020
- Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondel JL, Gloudemans AG, Van Muiswinkle WB. The influence of antibiotics on the immune system II. Modulation of fish leukocyte responses in culture. Vet Immunol Immunopathol. 1985;9:251–260. doi: 10.1016/0165-2427(85)90075-3. [DOI] [PubMed] [Google Scholar]
- Guerra P, Kim M, Shah A, Alaee M, Smyth SA. Occurrence and fate of antibiotic, analgesic/anti-inflammatory, and anti-fungal compounds in five wastewater treatment processes. Sci Total Environ. 2014;473:235–243. doi: 10.1016/j.scitotenv.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamscher G, Sczesny S, Höper H, Nau H. Determination of persistent tetracycline residues in soil fertilized with liquid manure by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. Anal Chem. 2002;74:1509–1518. doi: 10.1021/ac015588m. [DOI] [PubMed] [Google Scholar]
- Hirsch R, Ternes T, Haberer K, Kratz KL. Occurrence of antibiotics in the aquatic environment. Sci Total Environ. 1999;225:109–118. doi: 10.1016/S0048-9697(98)00337-4. [DOI] [PubMed] [Google Scholar]
- Huijbers PMC, Blaak H, De Jong MCM, Graat EAM, Vandenbroucke-Grauls CMJE, Husman AMR. Role of the environment in the transmission of antimicrobial resistance to humans:a review. Environ Sci Technol. 2015;49:11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- Huijbers PMC, Flach C, Larsson DGJ. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ Int. 2019;130:104880. doi: 10.1016/j.envint.2019.05.074. [DOI] [PubMed] [Google Scholar]
- IACG (2018) AMR Indicators and their relevance to the global indicator framework for SDGs and targets for the 2030 Agenda for sustainable development. Available at: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/AMR_SDG_indicators_analysis_slides.pdf. Accessed 22 Apr 2021
- Impens S, Reybroeck W, Vercammen J. Screening and confirmation of chloramphenicol in shrimp tissue using ELISA in combination with GC-MS2 and LC-MS2. Anal Chim Acta. 2003;483:153–163. doi: 10.1016/S0003-2670(02)01232-1. [DOI] [Google Scholar]
- Ji K, Kho Y, Park C, Paek D, Ryu P, Paek D, Kim M, Kim P, Choi K. Influence of water and food consumption on inadvertent antibiotics intake among general population. Environ Res. 2010;110:641–649. doi: 10.1016/j.envres.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Jonas Olga B, Irwin Alec, Berthe Franck Cesar Jean, Le Gall Francois G, Marquez Patricio V (2017)Drug-resistant infections : a threat to our economic future (Vol. 2) : final report (English). HNP/Agriculture Global Antimicrobial Resistance Initiative Washington, D.C.: World Bank Group. http://documents.worldbank.org/curated/en/323311493396993758/final-report. Accessed 22 Apr 2021
- Karkman A, Do TT, Walsh F, Virta MPJ. Antibiotic-resistance genes in wastewater. Trends Microbiol. 2017;26:220–228. doi: 10.1016/j.tim.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Khan GA, Berglund B, Khan KM, Lindgren PE, Fick J. Occurrence and abundance of antibiotics and resistance genes in rivers, canal and near drug formulation facilities: a study in Pakistan. PLoS One. 2013;8:e62712. doi: 10.1371/journal.pone.0062712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana A, Sinha R (2019) Tackling AMR Starts with the environment. Sanit Environ:78–80
- Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999−2000: a national reconnaissance. Environ Sci Technol. 2002;36:1202–1211. doi: 10.1021/es011055j. [DOI] [PubMed] [Google Scholar]
- Kostich MS, Batt AL, Lazorchak JM. Concentrations of prioritized pharmaceuticals in effluents from 50 large wastewater treatment plants in the U.S. and implications for risk estimation. Environ Pollut. 2014;184:354–359. doi: 10.1016/j.envpol.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Kraemer SA, Ramachandran A, Perron GG. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7:180. doi: 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Kotwani A. Rising threat of antimicrobial resistance: judicious use of antibiotics is the way forward. Indian J Chest Dis Allied Sci. 2017;59:65–166. [Google Scholar]
- Larsson DGJ. Pollution from drug manufacturing: review and perspectives. Philos Trans R Soc Lond Ser B Biol Sci. 2014;369:20130571. doi: 10.1098/rstb.2013.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson DGJ, Pedro CD, Paxeus N. Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J Hazard Mater. 2007;148:751–755. doi: 10.1016/j.jhazmat.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Larsson DGJ, Antoine A, Johan B, Kristian K, Maria A, Husman DR, Fagerstedt P, et al. Critical knowledge gaps and research needs related to the environmental imensions of antibiotic resistance. Environ Int. 2018;117:132–138. doi: 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- Lee D, Choi K. Comparison of regulatory frameworks of environmental risk assessments for human pharmaceuticals in E.U., USA, and Canada. Sci Total Environ. 2019;671:1026–1035. doi: 10.1016/j.scitotenv.2019.03.372. [DOI] [Google Scholar]
- Leo L (2020) Check effluents for antibiotic residue: government to pharma firms [Internet]. Livemint., 2020. Available at: https://www.livemint.com/news/india/govt-urges-pharma-cos-to-be-vigilant-about-pollution-at-manufacturing-pl. Accessed 22 Apr 2021
- Li D, Yang M, Hu J, et al. Determination and fate of oxytetracycline and related compounds in oxytetracycline production wastewater and receiving river. Environ Toxicol Chem. 2008;27:80–86. doi: 10.1897/07-080.1. [DOI] [PubMed] [Google Scholar]
- Li X, Atwill ER, Antaki E, Applegate O, Bergamaschi B, Bond RF, Chase J, Ransom KM, Samuels W, Watanabe N, Harter T. Fecal indicator and pathogenic bacteria and their antibiotic resistance in alluvial groundwater of an irrigated agricultural region with dairies. J Environ Qual. 2015;44:1435–1447. doi: 10.2134/jeq2015.03.0139. [DOI] [PubMed] [Google Scholar]
- Li B, Qiu Y, Song Y, Lin H, Yin H. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ Int. 2019;131:105007. doi: 10.1016/j.envint.2019.105007. [DOI] [PubMed] [Google Scholar]
- Lin AY, Tsai YT. Occurrence of pharmaceuticals in Taiwan’s surface waters: impact of waste streams from hospitals and pharmaceutical production facilities. Sci Total Environ. 2009;407:3793–3802. doi: 10.1016/j.scitotenv.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Lin AY, Yu TH, Lin CF. Pharmaceutical contamination in residential, industrial, and agricultural waste streams: risk to aqueous environments in Taiwan. Chemosphere. 2008;74:131–141. doi: 10.1016/j.chemosphere.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Liu L, Wu W, Zhang J, Lv P, Xu L, Yan Y. Progress of research on the toxicology of antibiotic pollution in aquatic organisms. Acta Ecol Sin. 2018;38:36–41. doi: 10.1016/j.chnaes.2018.01.006. [DOI] [Google Scholar]
- Loffler D, Ternes TA. Analytical method for the determination of the aminoglycoside gentamicin in hospital wastewater via liquid chromatography electrospraytandem mass spectrometry. J Chromatogr A. 2003;1000:583–588. doi: 10.1016/s0021-9673(03)00059-1. [DOI] [PubMed] [Google Scholar]
- López-Causapé C, Rojo-Molinero E, Mulet X, Cabot G, Moyà B, Figuerola J, Togores B, Pérez JL, Oliver A. Clonal dissemination, emergence of mutator lineages and antibiotic resistance evolution in pseudomonas aeruginosa cystic fibrosis chronic lung infection. PLoS One. 2013;8:e71001. doi: 10.1371/journal.pone.0071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood AR, Al-haideri HH, Hassan FM. Detection of antibiotics in drinking water treatment plants in Baghdad City, Iraq. Adv Public Health. 2019;2019:1–10. doi: 10.1155/2019/7851354. [DOI] [Google Scholar]
- Manaia CM. Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 2017;25(3):173–181. doi: 10.1016/j.tim.2016.11.014. [DOI] [PubMed] [Google Scholar]
- Manaia CM, Macedo G, Fatta-Kassinos D, Nunes OC. Antibiotic resistance in urban aquatic environments: can it be controlled? Appl Microbiol Biotechnol. 2016;100:1543–1557. doi: 10.1007/s00253-015-7202-0. [DOI] [PubMed] [Google Scholar]
- Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez JL (2017) Effect of antibiotics on bacterial populations: a multi-hierarchical selection process. F1000Research 6. 10.12688/f1000research.9685.1 [DOI] [PMC free article] [PubMed]
- Meersche TVD, Rasschaert G, Nest TV, Haesebrouck F, Herman L et al (2020) Longitudinal screening of antibiotic residues, antibiotic resistance genes and zoonotic bacteria in soils fertilized with pig manure. Environ Sci Pollut Res:28016–28029. 10.1007/s11356-020-09119-y [DOI] [PubMed]
- Migliore L, Civitareale C, Brambilla G, Dojmi Di Delupis G. Toxicity of several important agriculturalantibiotics to Artemia. Water Res. 1997;31:1801–1806. doi: 10.1016/S0043-1354(96)00412-5. [DOI] [Google Scholar]
- Ministry of Environment, Forest & Climate Change (2020) Available at: http://moef.gov.in/wp-content/uploads/2020/01/finalization.pdfAvailable at: http://moef.gov.in/wp-content/uploads/2020/01/finalization.pdf. Accessed on 26 Mar 2020
- Nahar S (2020) Understanding How The Indian Pharmaceutical Industry Works – Part 2. https://www.alphainvesco.com/blog/understanding-how-the-indian-pharmaceutical-industry-works-part-2/. Accessed on 05 Oct 2020
- Norwegian Environment Agency (2005) http://www.miljodirektoratet.no/no/Nyheter/Nyheter/Old-klif/2005/September/Nye_antibiotikautslipp_fra_Alpharma_i_Oslo/. Accessed on 27 Sept 2020
- O'Neill J (2016) The Review on Antimicrobial Resistance-Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available at: https://amr-review.org/sites/default/files/160518_Finalpaper_withcover.pdf. Accessed 22 Apr 2021
- Pareek S, Mathur N, Singh A, Nepalia A. Antibiotics in the environment: a review. Int J Curr Microbiol App Sci. 2015;4:278–285. [Google Scholar]
- Patel JB (2017) Performance standards for antimicrobial susceptibility testing. Clinical and laboratory standards institute
- Peake BM, Braund R, Tong AYC, Tremblay LA (2016) Impact of pharmaceuticals on the environment. In: The life-cycle of pharmaceuticals in the environment. Woodhead Publisher, pp 109–152
- Pharma Compass (2016) An Inconvenient Truth about Chinese Drug Manufacturing Available at : http://www.pharmacompass.com/radio-compass-blog/an-inconve-nient-truth-about-chinese-drug-manufacturing. Accessed on 20 Sept 2020
- Phillips PJ, Smith SG, Kolpin DW, Zaugg SD, Buxton HT, Furlong ET, Esposito K, Stinson B. Pharmaceutical formulation facilities as sources of opioids and other pharmaceuticals to wastewater treatment plant effluents. Environ Sci Technol. 2010;44:4910–4916. doi: 10.1021/es100356f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader RA, Langer ES (2018) Worldwide Biopharmaceutical Manufacturing Capacity Analysis: Growth Continues Across the Board, Bioprocess International. Available at: https://bioprocessintl.com/business/economics/worldwide-biopharmaceutical-manufacturing-capacity-analysis-growth-continues-across-the-board/. Accessed 22 Apr 2021
- Rees V (2020) Limiting antibiotic manufacturing discharge in Indian wastewater (2020) Available at: https://www.europeanpharmaceuticalreview.com/article/115074/limiting-antibiotic-manufacturing-discharge-in-indian-wastewater/. Accessed on 29 Sept 2020
- Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ. 2013;447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S, Vaz-Moreira I, Varela Della Giustina S, Llorca M, Barceló D, Schubert S, Berendonk TU, Michael-Kordatou I, Fatta-Kassinos D, Martinez JL, Elpers C, Henriques I, Jaeger T, Schwartz T, Paulshus E, O'Sullivan K, Pärnänen KMM, Virta M, Do TT, Walsh F, Manaia CM. Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ Int. 2020;140:105733. doi: 10.1016/j.envint.2020.105733. [DOI] [PubMed] [Google Scholar]
- Sanchez-Romero MA, Casadesus J. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Natl Acad Sci U S A. 2013;111:355–360. doi: 10.1073/pnas.1316084111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L. Selection of antibiotic resistance at very low antibiotic concentrations. Ups J Med Sci. 2014;119:103–107. doi: 10.3109/03009734.2014.904457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwecinska L. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health: a review. Water. 2020;12:3313. doi: 10.3390/w12123313. [DOI] [Google Scholar]
- Shanghai Daily (2012) H.K. Drug maker accused of destroying lake. Available at: https://archive.shine.cn/nation/HK-drug-maker-accused-of-destroying-lake/shdaily.shtml. Accessed on 25 Sept 2020
- Sim W-J, Lee J-W, Lee E-S, Shin S-K, Hwang S-R, Oh J-E. Occurrence and distribution of pharmaceuticals in wastewater from households, livestock farms, hospitals and pharmaceutical manufactures. Chemospere. 2011;82:179–186. doi: 10.1016/j.chemosphere.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Singer AC, Shaw H, Rhodes V, Hart A (2016) Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front Microbiol 7. 10.3389/fmicb.2016.01728 [DOI] [PMC free article] [PubMed]
- SIWI (2020) New initiative to tackle antimicrobial resistance in manufacturing. Available at: New initiative to tackle antimicrobial resistance in manufacturing - Stockholm International Water Institute (siwi.org). Accessed 22 Apr 2021
- SPHS (2020) UN informal interagency task team on sustainable procurement in the global health sector. Available at: UN informal Interagency Task Team on Sustainable Procurement in the Health Sector (savinglivesustainably.org). Accessed 22 Apr 2021
- Swapna KM, Rajesh R, Lakshmanan PT. incidence of antibiotic residues in farmed shrimps from the southern states of India. Indian J GeoMar Sci. 2012;41:344–347. [Google Scholar]
- Tamhankar AJ, Lundborg CS (2019) Antimicrobials and antimicrobial resistance in the environment and its remediation: a global one health perspective. Int J Environ Res Public Health 16. 10.3390/ijerph16234614 [DOI] [PMC free article] [PubMed]
- Thai P, Ky L, Binh V, Nhung P, Nhan P, Hieu N, et al. Occurrence of antibiotic residues and antibiotic-resistant bacteria in effluents of pharmaceutical manufacturers and other sources around Hanoi, Vietnam. Sci Total Environ. 2018;645:393–400. doi: 10.1016/j.scitotenv.2018.07.126. [DOI] [PubMed] [Google Scholar]
- Tong L, Li P, Zhu K. Analysis of veterinary antibiotic residues in swine wastewater and environmental water samples using optimized SPE-LC/MS/MS. Chemosphere. 2009;74:1090–1097. doi: 10.1016/j.chemosphere.2008.10.051. [DOI] [PubMed] [Google Scholar]
- Tong L, Qin L, Guan C, Wilson ME, Li X, Cheng D, Ma J, Liu H. Antibiotic resistance gene profiling in response to antibiotic usage and environmental factors in the surface water and groundwater of Honghu Lake, China. Environ Sci Pollut Res Int. 2020;27:31995–32005. doi: 10.1007/s11356-020-09487-5. [DOI] [PubMed] [Google Scholar]
- Topp E, Larsson J, Miller D, Van den Eede C, Virta M (2018) Antimicrobial resistance and the environment: assessment of advances, gaps and recommendations for agriculture, aquaculture and pharmaceutical manufacturing. FEMS. Microb Ecol 94. 10.1093/femsec/fix185 [DOI] [PubMed]
- Tripartite Plus Recent Update and Memorandum of Understanding on Antimicrobial Resistance (2018) https://www.who.int/antimicrobial-resistance/interagency-coordinationgroup/Tripartite_Plus_update_info_session_011018.pdf?ua=1. Accessed on 01 Oct 2020
- Turner S (2020) Antimicrobial resistance: is pharmaceutical pollution creating superbugs? Available at: https://www.pharmaceutical-technology.com/features/antimicrobial-resistance-superbugs/. Accessed on 26 Sept 2020
- UNEP (2017) Emerging Issues of Environmental Concern. United Nations Environment Programme, Nairobi. Frontiers. Available at: https://wedocs.unep.org/bitstream/%0Ahandle/20.500.11822/22255/Frontiers_2017_EN.pdf. Accessed on 01 Oct 2020
- United Nations (U.N.) Interagency Task Team on Sustainable Procurement in the Health Sector (SPHS). Available at https://savinglivesustainably.org/who-we-are. Accessed on 18 Dec 2020
- Vaz-Moreira I, Nunes OC, Manaia CM. Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome.FEMS. Microbiol Rev. 2014;38(4):761–778. doi: 10.1111/1574-6976.12062. [DOI] [PubMed] [Google Scholar]
- Verlicchi P, Al Aukidy M, Zambello E. Occurrence of pharmaceutical compounds in urban wastewater: removal, mass load and environmental risk after a secondary treatment—a review. Sci Total Environ. 2012;429:123–155. doi: 10.1016/j.scitotenv.2012.04.028. [DOI] [PubMed] [Google Scholar]
- Vikesland P, Garner E, Gupta S, Kang S, Maile-Moskowitz A, Zhu N. Differential drivers of antimicrobial resistance across the world. Acc Chem Res. 2019;52(4):916–924. doi: 10.1021/acs.accounts.8b00643. [DOI] [PubMed] [Google Scholar]
- Wang Q-JY, Mo C-H, Li Y-W, Gao P, Tai Y-P, Zhang Y, Ruan Z-L, Xu J-W. Determination of four fluoroquinolone antibiotics in tap water in Guangzhou and Macao. Environ Pollut. 2010;158:2350–2358. doi: 10.1016/j.envpol.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Wang H, Che B, Duan A, Mao J, Dahlgren RA, Zhang M, Zhang H, Zeng A, Wang X. Toxicity evaluation of β-diketone antibiotics on the development of embryo-larval zebrafish (Danio rerio) Environ Toxicol. 2014;29:1134–1146. doi: 10.1002/tox.21843. [DOI] [PubMed] [Google Scholar]
- Wei R, Ge F, Huang S, Chen M, Wang R. Ocurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere. 2011;82:1408–1414. doi: 10.1016/j.chemosphere.2010.11.067. [DOI] [PubMed] [Google Scholar]
- Westergaard K, Müller AK, Christensen S, Bloem J, Sørensen SJ. Effects of tylosin as a disturbance onthe soil microbial community. Soil Biol Biochem. 2001;33:2061–2071. doi: 10.1016/S0038-0717(01)00134-1. [DOI] [Google Scholar]
- WHO (2011) Local Production and Access to Medicines in Low- and Middle-Income Countries. Available at: https://www.who.int/phi/publications/Local_Production_Literature_Review.pdf. Accessed on 29 Sept 2020
- WHO (2019) Environmental Aspects of Good Manufacturing Practices: Points to Consider for Manufacturers and Inspectors in the Prevention of Antimicrobial Resistance. Available at: https://www.who.int/medicines/areas/quality_safety/quality_assurance/qas19_802_environmental_aspects_of_GMP.pdf?ua=1. Accessed on 01 Oct 2020
- WHO (2020) Antimicrobial Resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed on 28 Sept 2020
- Xu W, Zhang G, Li XD. Occurrence and elimination of antibiotics at four sewage treatment plants in the Pearl River Delta (PRD), South China. Water Res. 2007;41:4526–4534. doi: 10.1016/j.watres.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Xu J, Xu Y, Wang H, Guo C, Qiu H, He Y, Zhang Y, Li X, Meng W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere. 2015;119:1379–1385. doi: 10.1016/j.chemosphere.2014.02.040. [DOI] [PubMed] [Google Scholar]
- Yan Z, Lu G, Ye Q, Liu J. Long-term effects of antibiotics, norfloxacin, and sulfamethoxazole, in a partial life-cycle study with zebrafish (Danio rerio): effects on growth, development, and reproduction. Environ Sci Pollut Res. 2016;23:18222–18228. doi: 10.1007/s11356-016-7018-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Yin X, Shi M, Dahlgren RA, Wang H. Toxicity assessment of combined fluoroquinolone and tetracycline exposure in zebrafish (Danio rerio) Environ Toxicol. 2016;31:736–750. doi: 10.1002/tox.22087. [DOI] [PubMed] [Google Scholar]
- Zielinski W, Korzeniewska E, Harnisz M, Hubeny J, Buta M, Rolbiecki D. The prevalence of drug-resistant and virulent Staphylococcus spp. in a municipal wastewater treatment plant and their spread in the environment. Environ Int. 2020;143:105914. doi: 10.1016/j.envint.2020.105914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.