Abstract
In this paper, we compare and evaluate different testing protocols used for automatic COVID-19 diagnosis from X-Ray images in the recent literature. We show that similar results can be obtained using X-Ray images that do not contain most of the lungs. We are able to remove the lungs from the images by turning to black the center of the X-Ray scan and training our classifiers only on the outer part of the images. Hence, we deduce that several testing protocols for the recognition are not fair and that the neural networks are learning patterns in the dataset that are not correlated to the presence of COVID-19. Finally, we show that creating a fair testing protocol is a challenging task, and we provide a method to measure how fair a specific testing protocol is. In the future research we suggest to check the fairness of a testing protocol using our tools and we encourage researchers to look for better techniques than the ones that we propose.
Keywords: Convolutional neural networks, Covid-19, Covid-19 diagnosis, X-Ray images
1. Introduction
COVID-19 is a new coronavirus that spread in China and then in the rest of the world in 2020 and became a serious health problem worldwide [1], [2], [3]. This virus infects the lungs and causes potentially deadly respiratory syndromes [4]. The diagnosis of COVID-19 is usually performed by Real Time Polymerase Chain Reaction (RT-PCR) [5]. Recently, many researchers attempted to automatically diagnose COVID-19 using x-ray images [6]. Chest x-ray image classification is not a new problem in artificial intelligence. Convolutional neural networks have already reached very high performances in the diagnosis of lung diseases [7]. The recent publication of new small dataset of COVID-19 x-ray and CT images encouraged many researchers to apply the same techniques using these new data [8], [9], [10], [11]. Medical research already showed that pneumonia caused by COVID-19 seems to be different from a radiologist perspective [12]. Most of the papers dealing with COVID-19 classification report very high performances in this task. However, Cohen et al. [13] experimented the limits of the generalization of x-ray images classification, due to the fact that the network might learn features that are specific of the dataset more than the ones that are specific of the disease.
In this paper, we test if this is the case for most of the testing protocols used for COVID-19 classification at the moment. We downloaded four chest x-ray datasets and ran multiple tests to see whether a neural network could predict the source dataset of an image. This would be a serious problem in this case, since all COVID-19 samples come from only one dataset in most papers, hence a classifier trained to distinguish COVID-19 might actually have learnt to classify the source dataset. In order to do this, we trained AlexNet [14] to detect the source dataset of an image whose center was turned to black. In this way, we delete the lungs from the image, or at least most of the lungs, hence it is impossible for the network to learn anything on the disease detection task. We find that, if the training and the test set contain images that come from the same dataset, AlexNet can distinguish them with a confidence that is much higher than the one reported in tasks like pneumonia detection. Hence, if one does not pay enough attention to the testing protocol, the reported results might be very misleading.
To sum up, we show that every result dealing with the COVID-19 dataset in [15] should also contain a baseline model that detects the source dataset, to understand the amount of information that actually come from the lungs area. Our consideration actually do not only apply to this particular case. One must always be careful at merging more dataset and using different labels for each one of those datasets.
The structure of the paper is the following: first of all, we summarize some of the papers dealing with COVID-19 classification. After that, we describe the datasets that we use in our experiments and explain the details of our testing protocols. Finally, we run the experiments and show which testing protocols are the most suitable for COVID-19 classification.
2. Related work
Chest x-ray classification is not new in deep learning. Many datasets have been released [7,16] and neural networks trained on those dataset report high performances. As an example, Pranav et al. [7] report a 0.76 ROC-AUC for the pneumonia vs. healthy classification task on [16].
We are not the first to report potential biases in chest x-ray image classification [17,18]. Recently, Cohen et al. [13] expressed some concerns on the real world applications of the automatic classification of x-ray images. They tried to train Densenet [19] on different chest x-ray datasets and showed that the network performance dropped when it was trained on a dataset and tested on a another one.
Since the publication of the COVID-19 dataset [15] by Cohen et al., many researchers tried to classify those images and created a test by merging this dataset with other chest x-ray image datasets. We shall now describe some of them and report their testing protocols.
Narin et al. [9] created a small dataset with 50 COVID-19 cases coming from Cohen repository and 50 heathy cases coming from Kaggle (https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia). They used a 5 fold cross validation to train and test ResNet-50 [20] and obtained an accuracy of 98%. Apostolopoulos et al. [11] combined Cohen repository and many other sources to create a larger dataset containing 224 images of COVID-19, 700 of pneumonia and 504 negative. They tested VGG [21] on these data using 10 fold cross validation and obtained 93.48% accuracy, although on an unbalanced dataset. Wang et al. [22] trained Covid-Net, a new architecture introduced in their paper. They used a large dataset with 183 cases of COVID-19: 5538 of Pneumonia and 8066 are healthy subjects. They included several sources for their data. They extracted a test set with 100 images of pneumonia and of healthy lungs, and only 31 of COVID-19. In their code they explicitly mention that there is no patient overlap between the test and the training set, which is very important in problems like this. They reach a 92% accuracy. Hemdan et al. [23] trained Covidx-Net, a VGG19 [21] network on a dataset made by 50 images, half of which came from the Cohen repository. Their testing protocol was 5 fold cross validation. Pereira et al. [8] trained their model on a dataset created by merging different datasets. From every dataset, they only extracted images belonging to specific classes (one source dataset for COVID-19, one for healthy lungs, one for pneumonia,…). In their work they introduced the idea of a hierarchical classification and reached a 0.89 F1-Score.
Karim et al. [23] proposed a method to classify COVID-19 based on an explainable neural network. They used an enlarged version of the dataset used in [22], but they also add new images due to the fact that in the original dataset the healthy samples were pediatric scans. This might have led the network in [22] to learn how to classify the age of a patient more than its health status, and highlights once again the need for a fair testing protocol. An interesting protocol was tested in [24], where the authors managed to collect COVID-19 images that did not belong to the Cohen repository and used them as the test set.
Castiglioni et al. [25] proposed a completely different protocol using a dataset that they collected and that is not public. They used 250 COVID-19 and 250 healthy images for training, and used an independent test set of 74 positive and 36 negative samples. They trained an ensemble of 10 ResNets and achieved a ROC-AUC of 0.80 for the classification task. Its performance is much worse than the other ones reported in the literature, however, they used both anteroposterior and posteroanterior projections and they do not suffer of the dataset recognition problem that we highlight in this paper.
Tabik et al. [26] proposed the COVID Smart Data based Network (COVID-SDNet) methodology, which classifies COVID-19 Images according to the severity of the disease. Besides, they also show that high performances reported in the literature might be biased and propose a new and independent dataset that might help to design better experiments.
In order to understand the relevant features in COVID-19 classification, Karim et al. [27] proposed a method that highlights class-discriminating regions using gradient-guided class activation maps(Grad-CAM++) and layer-wise relevance propagation(LRP), helping to understand whether the context of a scan was useful for classification.
3. Datasets
In this paper we used four different datasets which are publicly available online. We shall now describe them. In Table 1 , we report the main traits of the various datasets.
Table 1.
Summary of the 4 Datasets.
| Dataset | COV | NIH | CHE | KAG |
|---|---|---|---|---|
| Covid Samples | 144 | 0 | 0 | 0 |
| Healthy Samples | 0 | 84,312 | 16,627 | 1583 |
| Images Resolution | Not Fixed | 1024 × 1024 | Not Fixed | Not Fixed |
3.1. NIH dataset
The Chestx-ray8 dataset [16] was released by the National Institute of Health and is one of the largest public labelled datasets in this field, which contains 108,948 images of 32,717 different patients, classified into 8 different categories, potentially overlapping. It was labelled using natural language processing techniques on the radiologists annotations. We refer to this dataset as NIH. We plot some samples in Fig 1 .
Fig. 1.
Samples from the NIH dataset.
3.2. CHE dataset
Irvin et al. collected and labelled Chexpert [28], a large dataset containing 224,316 chest radiographs of 65,240 patients divided into 14 classes. The strength of this new dataset is that the labeling tool based on natural language processing obtains higher performances than the one in Chestx-ray8. However, its test set is not publicly available, hence we use the validation set instead. For our purposes, this makes no difference, since we are not comparing our work to any previous papers. We refer to this dataset as CHE. We show some of the scans in Fig. 2 .
Fig. 2.
Samples from the CHE dataset.
3.3. KAG dataset
In 2017 Dr. Paul Mooney started a competition on Kaggle on viral and bacterial pneumonia classification (https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia/version/2).
It contained 5863 pediatric images, hence it is very different from the other datasets. We refer to this dataset as KAG. Some samples can be seen in Fig. 3 .
Fig. 3.
Samples from the KAG dataset.
3.4. COV dataset
Our source of COVID-19 images is the repository made available by Cohen et al. [15], which is the main source of most papers dealing with COVID-19. In the moment we are writing, it contains 144 images of frontal x-ray images of patients potentially positive to COVID-19. Metadata are available for every sample, containing the patient ID and, most of the times, the location and other notes that contain the reference to the doctor that uploaded the images. We refer to this dataset as COV. Some of the samples are in Fig. 4 .
Fig. 4.
Samples from the COV dataset.
4. Methods
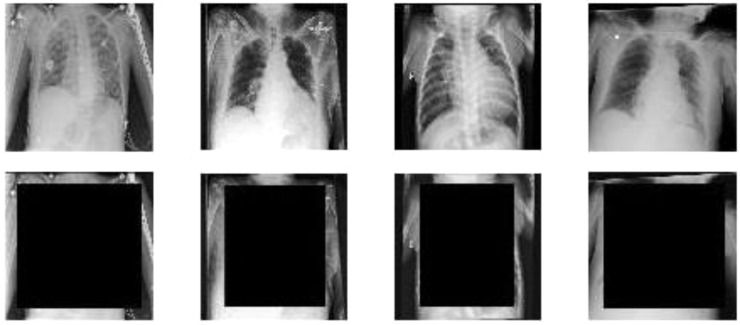
We made two different experiments. In both cases our training and test sets consist in combinations of the four datasets that we introduced in the previous section. The images were preprocessed by resizing them so that their smallest dimension was equal to 360, then a square of fixed size was turned to black in the center of the image. In our experiments we used a size of 240, 270 and 300. To give an intuition, if we the input image is a square, a 270 black square would cover the 56% of the pixels, while, if the largest dimension is the 50% larger than the shortest, which is quite an upper bound to the difference between dimensions, it would cover the 37% of the pixels. This preprocessing is made in order to create a square as small as possible, that still covers most of the lungs. The images in the dataset are more or less centered, especially in the horizontal axis. Besides, the two dimensions of the lungs are often comparable, hence they more or less fit in a square. From this observation it follows that, most of the times, the side pixels of the larger dimension do not usually contain part of the lungs, hence they can be kept in the preprocessed image. However, that is just our hypothesis, hence we shall also try a different image preprocessing later. Scans were always resized to squares of size 227 to be fed into AlexNet. The original and the transformed samples can be seen in Fig. 5 . It is clear that most of the lungs are hidden in our datasets, hence we can assume that we removed nearly all the information about the health status of the patient. We only considered the samples whose labels were Pneumonia, No Finding and COVID-19, except for the test set of Chexpert, since it was too small. We applied this preprocessing to all test and training images.
Fig. 5.
Original and transformed samples from the 4 datasets, 300 sized black square (Left to right: COV, NIH, CHE, KAG).
As the test set of Chexpert, we use all the images belonging to all the classes in the validation set. Since we wanted to detect the dataset and we removed the lungs from the images, we considered this a safe protocol. It is worth noticing that all datasets except COV have training data and test data, while COV does not have this distinction. Hence, we divided COV into 11 folds for cross validation. This number was not set in advance, but it was the result of the constraints that the folds must satisfy. We shall now describe those constraints. We randomly divided the COV dataset using two different protocols. The easiest one avoids patient overlap among folds. We refer to this protocol as PAT-OUT. The second one exploits the information in the metadata so that all the scans uploaded by the same doctor are in the same fold. We refer to this protocol as DOC—OUT. The information about the uploads is not complete and we cannot be 100% sure to do this. However, scans with no metadata about the location of the patient are in the same fold. However, most scans have metadata about the location and the doctor. We choose to do this to avoid that a network learns to recognize a specific hospital or X-Ray machine. Although this does not ensure the protocol to be unbiased, this is what this paper is about: stating that it is very hard to create a fair protocol in this field, and trying to propose the best one based on our experiments. In the DOC—OUT protocol we also required that every fold contained more than 10 samples, except for the last one. The minimum number of samples for the PAT-OUT protocol was set to 13 in order to obtain 11 folds as in the DOC—OUT protocol.
In both experiments, we fine-tuned AlexNet with a learning rate of 0.0001, except for the last fully connected layer, whose learning rate was 0.0002. We trained the network for 12 epochs with a mini-batch size of 64. We apply standard data augmentation applying random vertical flipping, random translations in [−5,5] and random rotations in [−5,5]. Data augmentation was very important because there were not many samples in the COV dataset. We always train the networks using 10 folds of COV and subsets of NIH, CHE and KAG, since they are very large. In all the experiments, for every COV sample, there are two samples of the other training datasets.
In our first experiment, we merge the training sets of NIH, CHE and KAG and 10 folds of COV and train AlexNet to recognize the source dataset of the images. Hence, we have 4 different labels. The test set is made by the test sets of NIH, CHE and KAG and the remaining fold of COV. We test this protocol 11 times, one for every fold of COV. We then merge the results obtained by every fold. This means that every sample not belonging to COV is tested 11 times, making the test set even more unbalanced than it already was. This is why the only metric we could use to test our models was class vs. class ROC-AUC over all the predictions of the 11 networks, since it is indifferent, on average, to the multiplicity of a sample in the test set. We only use the DOC—OUT protocol in this experiment. We ran this test three times. The first one with a black square of size 300. The second with a black square of size 270. In the third one we used a 240 sized square, but we also preprocess the image by applying contrast limited adaptive histogram equalization [29] and by cropping the image by cutting its upper and lower part so that the height of the new image is between the 90% and the 100% of its width. We use this preprocessing because the network could be able to deduce the original dataset by the proportion of the images. Besides, the contrast of the CHE dataset looks much larger than the contrast of the other images. In order to avoid confusion with the black square preprocessing, we called this procedure equalization. We show 4 equalized images in Fig. 6 . We refer to this experiment as dataset recognition.
Fig. 6.
Images form the four datasets (COV, NIH, CHE, KAG) after equalization.
In our second experiment, we implement a protocol which is similar to the one proposed by Cohen in [13]. We choose a dataset among NIH, CHE and KAG to be left out from the training set and to be used in the test set. Then, we train AlexNet on the training sets of two datasets among NIH, CHE and KAG and on 10 folds of COV, and we test it on the test set of the left-out dataset and on the remaining fold of COV. We repeat this for every fold of COV and using both PAT-OUT and DOC—OUT protocols. The labels of the samples are COV and non-COV. Again, we evaluated the ROC-AUC of the classification task, as we did for dataset recognition. We repeat this experiment three times, one for every choice of the left-out dataset. We refer to this experiment as COV recognition. It is worth mentioning that also in this experiment we covered the images with the black square.
5. Results
In Table 1 we report the results of the dataset recognition with a 300 square. Since the dataset is highly unbalanced, we only evaluate the ROC-AUC of the binary classifications and the confusion matrices. The best outcome would be to get a 0.5 ROC-AUC, which would mean that the two dataset cannot be distinguished by our model. However, one can see that AlexNet is very capable of recognizing the dataset without using the lungs.
The lowest ROC-AUC value is reached for the COV vs. NIH classification, but it is still 0.92. This is not surprising if one looks at the Fig. 1, Fig. 2, Fig. 3, Fig. 4. The samples in the different datasets seem to have very specific features. In particular, the images in the CHE dataset seem to have a strong contrast, which is probably recognized by the AlexNet even when the center of the images is set to black. In Table 2 we report the confusion matrix of the task. In a single row there are the samples belonging to the corresponding dataset, while on the columns there are the samples which are predicted to belong to that dataset. We can see that all datasets are accurately predicted. The only dataset that is not very well recognized is COV, and the large AUCs in the binary classifications in tasks involving COV seem to depend more on the fact that a dataset different from COV is hardly ever classified as COV.
Table 2.
Confusion Matrix of the Dataset Recognition Task – 300 – No Equalization.
| Dataset | COV | NIH | CHE | KAG |
|---|---|---|---|---|
| COV | 106,911 | 4197 | 3434 | 34 |
| NIH | 9 | 342 | 11 | 1 |
| CHE | 33 | 13 | 88 | 10 |
| KAG | 53 | 131 | 206 | 6474 |
In Table 3, we report the result of the same experiments using a square of size 270. We can see that there is not much difference with the previous experiment. As it is expected, the larger amount of information allows AlexNet to perform even better than in the 300 sized square case. This can also be seen in the confusion matrix reported in Table 4.
Table 3.
Roc-Auc Of The Dataset Recognition Task – 270 – No Equalization.
| Dataset | NIH | CHE | KAG |
|---|---|---|---|
| COV | 0.9283 | 0.9871 | 0.9937 |
| NIH | —– | 0.9995 | 0.9998 |
| CHE | —– | —– | 0.9997 |
Table 4.
Confusion Matrix of the Dataset Recognition Task – 270 – No Equalization.
| Dataset | COV | NIH | CHE | KAG |
|---|---|---|---|---|
| COV | 110,802 | 2111 | 1592 | 71 |
| NIH | 1 | 353 | 9 | 0 |
| CHE | 30 | 12 | 90 | 12 |
| KAG | 46 | 87 | 108 | 6623 |
In Table 5 we report the results of the classification using equalization and a square of size 240. Although the square is smaller than in the previous cases, recall that the upper and lower parts of image are cut, hence there are as more black pixels than the ones on the square. Some examples can be seen in Fig. 6. In Table 6 we can see that the confusion matrix is more promising than in the previous cases. In particular, the COV dataset seems to be confused with the other datasets most of the times.
Table 5.
Roc-Auc of the Dataset Recognition Task – 240 – With Equalization.
| Dataset | NIH | CHE | KAG |
|---|---|---|---|
| COV | 0.9210 | 0.9565 | 0.9741 |
| NIH | —– | 0.9899 | 0.9905 |
| CHE | —– | —– | 0.9974 |
Table 6.
Confusion Matrix of the Dataset Recognition Task – 240 – With Equalization.
| Dataset | COV | NIH | CHE | KAG |
|---|---|---|---|---|
| COV | 94,564 | 13,422 | 5195 | 1395 |
| NIH | 1 | 352 | 10 | 0 |
| CHE | 32 | 40 | 59 | 13 |
| KAG | 218 | 253 | 95 | 6298 |
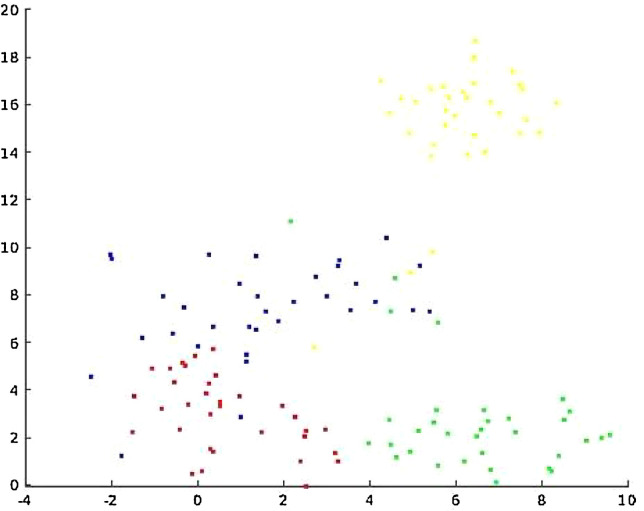
In Table 7, we report the three ROC-AUCs of the COV recognition experiment. We can see that the values are much lower than the ones of the previous experiments. Besides, there is not much evidence that the DOC—OUT protocol performs better than the PAT-OUT protocol. It might seem unexpected that KAG vs. COV performs worse than random. Probably this is due to the fact that KAG is much different from NIH and CHE that were used as non COV samples in the training set. This can be seen in Fig. 7 .
Table 7.
Roc-Auc of the Covid Dataset Recognition Task With Equalization.
| Protocol | Leave-out NIH | Leave-out CHE | Leave-out KAG |
|---|---|---|---|
| DOC—OUT | 0.68 | 0.62 | 0.36 |
| PAT-OUT | 0.68 | 0.62 | 0.42 |
Fig. 7.
t-SNE of the last hidden layer of a finetuned Alexnet. Red is NIH, green is CHE, yellow is KAG and blue is COV.
We must also report that in this protocol nearly all samples are labelled as COV.
Our hypothesis is that the COV dataset does not have particular features to be learnt, while the other datasets do. We show this in Fig. 7, where we plotted the output of the last hidden layer of AlexNet using t-distributed stochastic neighbor embedding (t-SNE) [30]. The network that we used is the one trained for dataset recognition with a 300 square. This might be due to the fact that COV is a repository of images uploaded by doctors from all over the world.
We can see that COV is more similar to the other datasets than those datasets are among themselves. This also explains why the Leave-out KAG protocol performs worse than random. If a model is trained to recognize the red and green points as non-COV and the blue points as COV, it is clear that some blue points might be confused with a red or green point, but yellow points will always be classified as blue points.
The objective of this paper is suggesting a fair testing protocol for COVID-19 classification. Our experiments show that the difference between the datasets is so large that building a fair protocol with the dataset that we considered might be very hard. One solution would be to find a dataset whose features are similar to the ones in COV. Otherwise, one can find an effective preprocessing (hopefully better than our equalization) that deletes the dataset-dependent features. We showed that it is possible to delete some of them and making the dataset classification harder, however we are far from an unbiased protocol.
Other datasets are available for chest x-ray recognition, hence one can apply our techniques to validate the use of any other repository which is available to them. We must also point out that in our experiments we removed nearly all the information about the health status of the lungs, but we also removed a large portion of the information about the dataset in general. In other words, two datasets might be distinguishable because of features that appear in the center of the images, but have nothing to do with the health status of the patient. Hence, our experiments set a minimum ability of a classifier to recognize the datasets. To the best of our knowledge, we are the first to address this problem for COVID-19 x-ray images. From the COV recognition experiment on might deduce that leaving a dataset out to be used a test set could be beneficial. However, this might be very hard in practice since the data cloud of the left-out dataset might be very far from the other non Covid datasets, as it happens in Fig. 7.
6. Conclusion
In this paper we discussed the validity of the usual testing protocols in most papers dealing with the automatic diagnosis of COVID-19. We showed that these protocols might be biased and learn to predict features that depend more on the source dataset than they do on the relevant medical information. We also suggested some solutions to find a new testing protocol and a method to evaluate its biasness. To the best of our knowledge, we are the first to provide such a metric. As future work, we plan to look for more efficient methods for dataset recognition and to create new experiments to test the biasness of testing protocols for X-Ray image classification. Besides, we plan to create new image processing techniques that might reduce the inter-dataset differences.
Table 1a.
Roc-Auc of the Dataset Recognition Task – 300 – No Equalization.
| Dataset | NIH | CHE | KAG |
|---|---|---|---|
| COV | 0.9212 | 0.9652 | 0.9898 |
| NIH | —– | 0.9956 | 0.9997 |
| CHE | —– | —– | 0.9989 |
CRediT authorship contribution statement
Gianluca Maguolo: Conceptualization, Methodology, Software, Writing - review & editing. Loris Nanni: Conceptualization, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.C.D.C. COVID, R. Team Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12–March 16, 2020. MMWR Morb. Mortal Wkly. Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020 doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.E. Mahase, Coronavirus: covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate, (2020). [DOI] [PubMed]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O. Gozes, M. Frid-Adar, H. Greenspan, P.D. Browning, H. Zhang, W. Ji, A. Bernheim, E. Siegel, Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis, ArXiv Prepr. ArXiv2003.05037. (2020).
- 7.P. Rajpurkar, J. Irvin, K. Zhu, B. Yang, H. Mehta, T. Duan, D. Ding, A. Bagul, C. Langlotz, K. Shpanskaya, others, Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning, ArXiv Prepr. ArXiv1711.05225. (2017).
- 8.R.M. Pereira, D. Bertolini, L.O. Teixeira, C.N. Silla Jr, Y.M.G. Costa, COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios, ArXiv Prepr. ArXiv2004.05835. (2020). [DOI] [PMC free article] [PubMed]
- 9.A. Narin, C. Kaya, Z. Pamuk, Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks, ArXiv Prepr. ArXiv2003.10849. (2020). [DOI] [PMC free article] [PubMed]
- 10.J. Zhang, Y. Xie, Y. Li, C. Shen, Y. Xia, COVID-19 Screening on Chest X-ray Images Using Deep Learning based Anomaly Detection, ArXiv Prepr. ArXiv2003.12338. (2020).
- 11.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J.P. Cohen, M. Hashir, R. Brooks, H. Bertrand, On the limits of cross-domain generalization in automated X-ray prediction, ArXiv Prepr. ArXiv2002.02497. (2020).
- 14.Krizhevsky A., Sutskever I., Hinton G.E. Adv. Neural Inf. Process. Syst. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 15.J.P. Cohen, P. Morrison, L. Dao, COVID-19 image data collection, ArXiv Prepr. ArXiv2003.11597. (2020).
- 16.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 17.Baltruschat I.M., Nickisch H., Grass M., Knopp T., Saalbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Yao, J. Prosky, B. Covington, K. Lyman, A strong baseline for domain adaptation and generalization in medical imaging, ArXiv Prepr. ArXiv1904.01638. (2019).
- 19.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 20.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conf. Comput. Vis. Pattern Recognit. 2016. Deep Residual Learning for Image Recognition; pp. 770–778. [Google Scholar]
- 21.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, ArXiv Prepr. ArXiv1409.1556. (2014).
- 22.L. Wang, A. Wong, Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images, ArXiv Prepr. ArXiv2003.09871. (2020). [DOI] [PMC free article] [PubMed]
- 23.E.E.-D. Hemdan, M.A. Shouman, M.E. Karar, Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images, ArXiv Prepr. ArXiv2003.11055. (2020).
- 24.L.O. Hall, R. Paul, D.B. Goldgof, G.M. Goldgof, Finding COVID-19 from Chest X-rays using Deep Learning on a Small Dataset, ArXiv Prepr. ArXiv2004.02060. (2020).
- 25.I. Castiglioni, D. Ippolito, M. Interlenghi, C.B. Monti, C. Salvatore, S. Schiaffino, A. Polidori, D. Gandola, C. Messa, F. Sardanelli, Artificial intelligence applied on chest X-ray can aid in the diagnosis of COVID-19 infection: a first experience from Lombardy, Italy, MedRxiv. (2020).
- 26.S. Tabik, A. Gómez-R\’\ios, J.L. Mart\’\in-Rodr\’\iguez, I. Sevillano-Garc\’\ia, M. Rey-Area, D. Charte, E. Guirado, J.L. Suárez, J. Luengo, M.A. Valero-González, others, COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on Chest X-Ray images, ArXiv Prepr. ArXiv2006.01409. (2020). [DOI] [PMC free article] [PubMed]
- 27.M. Karim, T. Döhmen, D. Rebholz-Schuhmann, S. Decker, M. Cochez, O. Beyan, others, DeepCOVIDExplainer: explainable COVID-19 Predictions Based on Chest X-ray Images, ArXiv Prepr. ArXiv2004.04582. (2020).
- 28.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K. Proc. AAAI Conf. Artif. Intell. 2019. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison; pp. 590–597. [Google Scholar]
- 29.Zuiderveld K. Contrast limited adaptive histogram equalization. Graph. Gems. 1994:474–485. [Google Scholar]
- 30.van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]