Abstract
Purpose
The known epithelial ovarian cancer (EOC) susceptibility genes account for less than 50% of the heritable risk of ovarian cancer suggesting that other susceptibility genes exist. The aim of this study was to evaluate the contribution to ovarian cancer susceptibility of rare deleterious germline variants in a set of candidate genes.
Methods
We sequenced the coding region of 54 candidate genes in 6385 invasive EOC cases and 6115 controls of broad European ancestry. Genes with an increased frequency of putative deleterious variants in cases versus controls were further examined in an independent set of 14 135 EOC cases and 28 655 controls from the Ovarian Cancer Association Consortium and the UK Biobank. For each gene, we estimated the EOC risks and evaluated associations between germline variant status and clinical characteristics.
Results
The ORs associated for high-grade serous ovarian cancer were 3.01 for PALB2 (95% CI 1.59 to 5.68; p=0.00068), 1.99 for POLK (95% CI 1.15 to 3.43; p=0.014) and 4.07 for SLX4 (95% CI 1.34 to 12.4; p=0.013). Deleterious mutations in FBXO10 were associated with a reduced risk of disease (OR 0.27, 95% CI 0.07 to 1.00, p=0.049). However, based on the Bayes false discovery probability, only the association for PALB2 in high-grade serous ovarian cancer is likely to represent a true positive.
Conclusions
We have found strong evidence that carriers of PALB2 deleterious mutations are at increased risk of high-grade serous ovarian cancer. Whether the magnitude of risk is sufficiently high to warrant the inclusion of PALB2 in cancer gene panels for ovarian cancer risk testing is unclear; much larger sample sizes will be needed to provide sufficiently precise estimates for clinical counselling.
Keywords: cancer: endocrine, genetic epidemiology
Introduction
Rare, predicted deleterious variants in multiple genes have been shown to be associated with a moderate to high risk of epithelial ovarian cancer (EOC). These include the DNA double stand break repair genes BRCA1,1 BRCA,2 BRIP1,3 RAD51C, and RAD51 4, and the mismatch repair genes MSH2 and MSH6.5 6 ANKRD11, FANCM, PALB2 and POLE have recently been reported as possible susceptibility genes.7–9 Multiple common variants conferring weaker risk effects have also been identified,10–17 some of which modify EOC risk in carriers of more highly penetrant gene mutations.18 19
EOC is heterogeneous with five main histotypes: high-grade serous (HGSOC), low-grade serous, endometrioid, clear cell and mucinous ovarian cancer. These have different clinical characteristics and outcomes and are characterised by different germline and somatic genetic changes that result in the perturbation of different molecular pathways. For example, germline mutations in DNA double break repair genes predispose to HGSOC while germline mutations in mismatch repair genes increase risk of the endometrioid and clear cell histotypes.6
The known susceptibility alleles account for less than 50% of the excess familial risk of ovarian cancer, suggesting that other susceptibility genes and alleles exist.15 The unexplained genetic component of risk is likely to be made up of a combination of common genetic variants conferring weak effects and uncommon alleles conferring weak to moderate relative risks (less than 10-fold).
The aim of this study was to identify additional ovarian cancer susceptibility genes using case-control sequencing of candidate genes identified through various approaches including their known function in pathways that are associated with ovarian cancer development and from whole exome sequencing studies (WES) of ovarian cancer cases that have identified putative deleterious mutations in genes not previously evaluated for EOC risk.
Material and methods
Selection of candidate genes
Genes based on known biological function
As several EOC susceptibility genes are involved in DNA double-strand break repair and Fanconi anaemia (FA),8 we selected genes involved in these pathways. FA is a rare genetic disease characterised by chromosomal instability, hypersensitivity to DNA crosslinking agents, defective DNA repair, severe bone marrow failure, cancer susceptibility and many congenital defects. To date, 22 FA genes have been identified, of which eight have previously been evaluated in ovarian cancer case-control studies:3 4 6 8 FANCD1, FNACJ (BRIP1), FANCL, FANCN (PALB2), FANCM, FANCO (RAD51C), FANCS (BRCA1) and FANCV (MAD2L2). We selected nine FA genes not previously studied in ovarian cancer: FANCA, FANCB, FANCC, FANCD2, FANCE, FANCG, FANCI, FANCP (SLX4), FANCW. We also included FANCN (PALB2), which has been studied previously in ovarian cancer3 9 20–22 but its association with EOC risk is equivocal. Eight candidate genes involved in other aspects of DNA repair were also included: ALKBH3, CHEK2, GTF2H4, POLE, POLK, RDM1 and XRCC1.
Genes from whole exome sequencing studies (WES)
Twelve genes (BUB1B, C5orf28, C6, DNAJB4, EXO1, LIG4, MKNK2, MMRN1, PARP1, RAD52, SMC1A and SNRNP200) were selected from WES analysis of EOC cases where putative deleterious (truncating) mutations were identified at a greater frequency in cases compared with publicly available WES data from controls reported by the NHLBI GO Exome Sequencing Project and The Exome Aggregation Consortium databases (http://exac.broadinstitute.org). Germline WES data for EOC cases were available for 412 HGSOC cases from the Cancer Genome Atlas ovarian cancer study; 513 ovarian cancer cases from an Australian case series 6; 97 familial non-BRCA1/BRCA2 ovarian cancer cases from Gilda Radner Familial Ovarian Cancer Registry and 54 ovarian cancer cases from the UK Familial Ovarian Cancer Registry.
Four genes from these WES studies (GANC, KNTC1, PSG6 and UPK2) were selected because more than one family member diagnosed with ovarian cancer from 10 familial cases carried the same truncating mutation in one of these genes.
Finally, 21 genes were selected from analyses of several other unpublished EOC WES studies (personal communications) where the frequency of truncating mutations was greater in cases compared with controls. These genes were ANAPC2, CNKSR1, DUOX1, FBXO10, NAT10, OSGIN1, PAK4, PHF20L1, PIK3C2G, PTGER3, PTX3, RAD54B, RECQL, RIPK3, RNASEL, SMG5, SPHK1, SULT1C2, UHRF2, WNT5A and ZFHX3.
Study subjects
We used case-control data from targeted sequencing, exome and array-based genotyping.
Targeted sequencing
We included 5914 EOC cases and 5479 controls of European ancestries from 19 studies—13 case-control studies, 1 familial ovarian cancer study from Poland, 2 clinical trials and 3 case-only studies (online supplementary table 1).14 HGSOC cases were preferentially plated out for sequencing where possible.
jmedgenet-2019-106739supp001.xlsx (424.6KB, xlsx)
Exome sequencing
We extracted data on the 54 candidate genes from 829 case and 913 controls from two ovarian cancer case-control studies (MDA23–25 and NCO14) for which whole exome sequence data were available (online supplementary table 1).
Variants from genotyping array data
For genes that reached nominal significance in the combined analysis of the targeted sequencing and exome sequencing data, we extracted genotypes of any deleterious variants included on the OncoArray and UK Biobank Axiom Array. These two arrays were used to genotype up to 18 936 controls and 13 288 cases from the Ovarian Cancer Association Consortium (OCAC),15 9725 controls and 858 cases from UK Biobank GWAS (https://www.ukbiobank.ac.uk/), respectively. Samples overlapped with the sequencing studies were excluded from the analysis.
All studies had ethics committee approval, and all participants provided informed consent.
Sequencing methods
Target sequence enrichment followed by sequencing was performed on the coding sequence and splice-sites of ALKBH3, ANAPC2, BUB1B, C5ORF28, C6, CHEK2, CNKSR1, DNAJB4, DUOX1, EXO1, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCG, FANCI, FBXO10, GANC, GTF2H4, KNTC1, LIG4, MKNK2, MMRN1, NAT10, OSGIN1, PAK4, PALB2, PARP1, PHF20L1, PIK3C2G, POLE, POLK, PSG6, PTGER3, PTX3, RAD52, RAD54B, RDM1, RECQL, REV3L, RIPK3, RNASEL, SLX4, SMC1A, SMG5, SNRNP200, SPHK1, SULT1C2, UHRF2, UPK2, WNT5A, XRCC1 and ZFHX3. The target sequence was identified from the NCBI Reference Sequence Database48.48 Fluidigm access arrays as previously described.6 A total of 1663 amplicons were designed to cover the 159 kb target region. Libraries were sequenced using 150 bp paired-end sequencing on the Illumina HiSeq4000 or HiSeq2500.
Sequencing reads were demultiplexed and then aligned against the human genome reference sequence (hg19) using the Burrows-Wheeler Aligner.26 The Genome Analysis Toolkit27 was used for base quality-score recalibration, local indel realignment and variant calling. Finally, ANNOVAR28 was used for variant annotation. Variants were called if (1) genotype information was available from a chip genotype for that sample or (2) the variants were presented in more than one amplicon or (3) read depth ≥15 and alternate allele frequency ≥40% or (4) read depth ≥100 and alternate allele frequency ≥25%. These thresholds were defined using the results from sequencing of positive controls with known variants and genotype information from chip array genotyping of overlapping samples.
We excluded 356 cases and 269 controls because <80% of the target sequence bases had a read depth of at least 15. The average percentage coverage of the genes at 15X read depth ranged from 64% to 99% (online supplementary table 2). The mean sequencing depth for these genes ranged from 130 (IQR 104–152) to 432 (IQR 364–492). Concordance for 111 duplicate pairs was 98% (7384 concordant variants out of total 7572 variants called).
For the exome sequencing, sonication fragmentation was used to fragment DNA samples. Fragments with an average size of 200 bp were selected to generate libraries for sequencing. Agilent SureSelect Clinical Research Exome (CRE) v1 was used for exome enrichment and sequencing was performed on an Illumina HiSeq 4000 using 2×150 bp paired-end reads. Cutadapt (https://doi.org/10.14806/ej.17.1.200) was used to locate and remove residual adapters in reads. FLASH (Fast Length Adjustment of SHort reads)29 was used to merge the overlapped paired-end reads into one read, using default parameters. Reference genome alignment and joint genotype calling according to a pipeline described in Yu et al.30 The coding sequences and splice sites of all 54 genes were extracted. Fifty-three genes with 100% average coverage at 10X were included in the analysis. GTF2H4 was excluded from the analysis, as the average coverage was only 43%.
Deleterious variants were defined as those predicted to result in protein truncation (frameshift indel, splice site, nonsense mutations and start loss) or predicted to be deleterious and/or likely deleterious by Clinvar.31 Any exonic single nucleotide variants within 3 bp of the exon-intron boundary and any intronic variants within 20 bp of the exon-intron boundary at the 5-prime end, and 6 bp at the 3-prime end, were evaluated using the software MaxEntScan to identify those most likely to disrupt splicing.32 Variants with a MaxEntScan score that decreased by more than 40% compared with the reference sequence and having a reference sequence score ≥3 were considered deleterious. Sequencing alignments were confirmed by visual inspection using the Integrative Genomic Viewer.33
Statistical methods
Risk estimation and genotype-phenotype analyses
We used a simple burden test for association between deleterious variants and ovarian cancer risk on a gene-by-gene basis. The burden test was based on unconditional logistic regression adjusted for country (Australia, Denmark, German, Poland, the UK and the USA) and sequencing method (targeted sequencing or exome sequencing). ORs and associated 95% CI were calculated.
Missense variant analyses
We also identified multiple rare (minor allele frequency <1%) missense variants that have an unknown functional effect on the protein. We used the rare admixture likelihood burden test34 to test these variants for association. We excluded any missense variants classified as deleterious and classified the remaining variants by whether or not they are predicted to have a damaging effect on protein function by two out of three prediction tools—SIFT (score <0.05),35 polyphen-236 (classified as probably damaging or damaging) and Provean37 (score≤−2.5). Subjects with a missense variant call rate less than 80% and variants with a call rate less than 80% or with genotype frequencies inconsistent with Hardy-Weinberg equilibrium (p<10–5) were excluded.
Results
Germline deleterious mutations in ovarian cancer cases and controls
Sequencing results were available for 6385 EOC cases and 6115 controls after quality control analysis. The characteristics of these individuals by study are summarised in online supplementary table 1. Most EOC cases were serous histotype (n=6304, 98.7%), of which 5951 were the HGSOC histotype (93.2%).
We identified 629 unique, putative-deleterious variants (online supplementary table 3) in 1051 ovarian cancer cases (967 high-grade serous histotype) and 964 controls. There was a nominally significant higher frequency of mutations in cases compared with controls for POLK, PALB2 and SLX4 and a lower frequency of mutations in cases compared with controls for FBXO10 (table 1). The associated ORs are shown in table 1—for POLK, PALB2 and SLX4 the effect size was slightly larger for HGSOC. The frequency of deleterious variants in the other genes was similar in cases compared with controls (online supplementary table 4). Given the evidence for association of multiple FA genes with EOC risk, we also carried out a burden test to compare the frequency of deleterious variants in any of the eight genes which were not significantly associated with ovarian cancer risks individually (FANCA, FANCB, FANCC, FANCD2, FANCE, FANCG, FANCI and FANCL). A combined analysis will have greater power if multiple genes were associated but the effect sizes too small to detect individually. There was no significant difference in the frequency of deleterious variants in cases (96/6184, 1.6%) and controls (85/6089, 1.4%) (p=0.50).
Table 1.
Frequency of mutations and estimated risk of EOC in candidate genes (p<0.05) from targeted sequencing and exome sequencing
| Set* | Histotype | Gene | Controls | Cases | Or (95% CI) | P value | ||
| No. | % | No. | % | |||||
| TS | Overall | POLK | 9 | 0.17 | 29 | 0.52 | 3.04 (1.43 to 6.43) | 0.0037 |
| PALB2 | 6 | 0.12 | 19 | 0.34 | 3.10 (1.23 to 7.78) | 0.016 | ||
| SLX4 | 4 | 0.08 | 13 | 0.23 | 3.08 (1.00 to 9.48) | 0.0049 | ||
| FBXO10 | 9 | 0.17 | 3 | 0.053 | 0.30 (0.08 to 1.11) | 0.071 | ||
| Non-carrier | 5174 | 99.5 | 5492 | 98.8 | ||||
| HGSOC | POLK | 9 | 0.17 | 27 | 0.53 | 3.17 (1.48 to 6.79) | 0.003 | |
| PALB2 | 6 | 0.12 | 18 | 0.35 | 3.30 (1.30 to 8.38) | 0.012 | ||
| SLX4 | 4 | 0.08 | 13 | 0.25 | 3.51 (1.13 to 10.9) | 0.029 | ||
| FBXO10 | 9 | 0.17 | 3 | 0.059 | 0.32 (0.09 to 1.18) | 0.086 | ||
| Non-carrier | 5174 | 99.5 | 5062 | 98.8 | ||||
| ES | Overall | POLK | 7 | 0.77 | 6 | 0.72 | 0.94 (0.32 to 2.82) | 0.92 |
| PALB2 | 2 | 0.22 | 3 | 0.36 | 1.65 (0.28 to 9.93) | 0.58 | ||
| SLX4 | 0 | 0 | 2 | 0.24 | NA | |||
| FBXO10 | 1 | 0.11 | 0 | 0 | ||||
| Non-carrier | 903 | 98.9 | 818 | 98.7 | ||||
| HGSOC | POLK | 7 | 0.77 | 6 | 0.72 | 0.94 (0.32 to 2.82) | 0.92 | |
| PALB2 | 2 | 0.22 | 3 | 0.36 | 1.66 (0.28 to 9.94) | 0.58 | ||
| SLX4 | 0 | 0 | 2 | 0.24 | NA | |||
| FBXO10 | 1 | 0.11 | 0 | 0 | NA | |||
| Non-carrier | 903 | 98.9 | 817 | 98.7 | ||||
*TS: targeted sequencing; ES: exome sequencing.
EOC, epithelial ovarian cancer; HGSOC, high-grade serous ovarian cancer; OCAC, Ovarian Cancer Association Consortium.
Validation analyses in ovarian cancer case-control studies
We also evaluated risk associations between deleterious variants in POLK, PALB2, and SLX4 with EOC risk based on germline genotyping data for 13 277 EOC cases and 18 930 controls from OCAC and for 858 EOC cases and 9725 controls and from UK Biobank. For OCAC samples, data were available for six deleterious non-monomorphic variants in PALB2; for UK Biobank samples, data were available for seven PALB2 and one POLK deleterious variants (table 2, list of variants in online supplementary table 5).
Table 2.
Frequency of mutations and estimated risk of EOC in candidate genes for validation chip genotyping data
| Set* | Histotype | Gene | Controls | Cases | OR (95% CI) | P value | ||
| No. | % | No. | % | |||||
| OCAC | Overall | PALB2 | 6 | 0.03 | 11 | 0.08 | 2.10 (0.74 to 5.94) | 0.16 |
| Non-carrier | 18 930 | 99.97 | 13 277 | 99.9 | ||||
| HGSOC | PALB2 | 6 | 0.03 | 6 | 0.097 | 3.48 (1.10 to 11.1) | 0.035 | |
| Non-carrier | 18 930 | 99.97 | 6168 | 99.9 | ||||
| Biobank | Overall | PALB2 | 11 | 0.11 | 3 | 0.35 | 3.12 (0.87 to 11.2) | 0.081 |
| POLK | 29 | 0.30 | 2 | 0.23 | 0.78 (0.19 to 3.29) | 0.74 | ||
| Non-carrier | 9685 | 99.6 | 853 | 99.4 | ||||
| HGSOC† | PALB2 | 11 | 0.11 | 1 | 0.28 | 2.49 (0.32 to 19.4) | 0.38 | |
| POLK | 29 | 0.30 | 1 | 0.28 | 0.92 (0.12 to 6.74) | 0.93 | ||
| Non-carrier | 9685 | 99.6 | 361 | 99.4 | ||||
*OCAC: OCAC sample genotype on the OncoArray; Biobank: genotype from UK Biobank Axiom Array.
†Information on tumour grade was not available for UK Biobank cases, all the serous cases in UK Biobank were assumed to be HGSOC.
EOC, epithelial ovarian cancer.
In OCAC case-control analyses, PALB2 variants showed a non-significant increased risk of EOC (OR 2.10, 95% CI 0.74 to 5.94, p=0.16). The strength of this association increased when the analysis was restricted to 6181 HGSOC cases (OR 3.48, 95% CI 1.10 to 11.1, p=0.035). In UK Biobank, we observed a weak association for PALB2 mutations with EOC risk (OR 3.12, 95% CI 0.87 to 11.2, p=0.081). There was no evidence of risk association for mutations in POLK (table 2).
We then performed a meta-analysis by combining the targeted sequencing, WES and chip genotyping data. Taken together, putative deleterious mutations were associated with increased risk for PALB2 (OR 2.60, 95% CI 1.45 to 4.64; p=0.0013), POLK (OR 1.77, 95% CI 1.07 to 2.93; p=0.026) and SLK4 (OR 3.37, 95% CI 1.17 to 9.70, p=0.024) and decreased risk for FBXO10 (95% CI 0.07 to 1.00; p=0.049). After stratifying cases by histological subtype, the estimated risks were higher for HGSOC for PALB2 (OR 3.01, 95% CI 1.59 to 5.68; p=0.00068), POLK (OR 1.99, 95% CI 1.15 to 3.43; p=0.014) and SLK4 (OR 3.92, 95% CI 1.33 to 11.5; p=0.013).
We used an approximate Bayes factor to calculate the Bayes false discovery probability (BFDP) described by Wakefield38 for PALB2, SLX4, POLK and FBXO10 based on several different priors and assuming that the associated risk is unlikely to be greater than an OR of 4 (table 3). The evidence for association of PALB2 was strong with a BFDP of less than 15% when the prior on the alternative hypothesis is 0.1. The nominally significant associations for the other three genes are likely to be false positives.
Table 3.
Bayes false discovery probability for the associations reported for PALB2, SLX4, POLK and FBXO10 from the meta-analysis of all the available data
| Gene | Histotype | OR (95% CI) | P value | Prior probability | ||
| 0.1 | 0.05 | 0.01 | ||||
| PALB2 | HGSC | 3.01 (1.59 to 5.68) | 0.00068 | 0.14 | 0.26 | 0.65 |
| SLX4 | HGSC | 3.92 (1.33 to 11.5) | 0.013 | 0.65 | 0.80 | 0.95 |
| POLK | HGSC | 1.99 (1.15 to 3.43) | 0.014 | 0.65 | 0.80 | 0.95 |
| FBXO10 | Overall | 0.27 (0.07 to 1.00) | 0.026 | 0.75 | 0.86 | 0.97 |
Predicting the functional impact of missense coding variants
Combining the whole exome and targeted sequencing data, we identified 5265 unique missense variants with minor allele frequency less than 1% in the 54 genes (online supplementary table 6). We used the in silico software programs SIFT, Polyphen-2 and Provean to evaluate the predicted impact of these variants on protein function for each gene. Of the 5265 variants, 2111 were classified as ‘deleterious’ based on at least 2 out of 3 of these classifiers. We found weak evidence for association with increased EOC risk for rare missense variants in DUOX1 and PAK4 using burden testing (p=0.015 and 0.025, respectively) (online supplementary table 7); for DUOX1, the strength of this association improved when the analyses were restricted to the HGSOC histotype (p=0.0061). When we performed the same analyses for 1493 very rare variants (MAF<0.001), we observed significant association for missense variants in DUOX1 and FANCE (p=0.015 and 0.034, respectively).
Discussion
We have evaluated the association between putative deleterious variants in 54 genes with the risk of HGSOC through a combination of whole exome and targeted sequencing analysis in 5951 cases and 6115 controls of broad European ancestries. We found evidence for four genes—PALB2, POLK, SLX4 and FBXO10—associated with HGSOC risk. Association analysis in an additional 14 135 ovarian cancer cases and 28 655 controls genotyped through OCAC and the UK Biobank provided further support for PALB2 as a HGSOC susceptibility gene.
The probability that a genetic association deemed statistically significant is a false positive depends on the prior of the null hypothesis and the power of the study to detect an effect size plausible under the alternative hypothesis. We calculated Wakefield’s BFDP38 based on several different priors to further evaluate the likelihood that PALB2, POLK, SLX4 and FBXO10 are EOC susceptibility genes. If we assume the prior on the alternative to be 1 in 10 or 1 in 20, the BFDPs for the association of deleterious variants in PALB2 with HGSOC are 0.14 and 0.26, respectively. These moderately strong priors are reasonable given the evidence for the association from previously published studies.20 Two studies have reported nominally significant associations for PALB2 with OR 4.4 (95% CI 2.1 to 9.1)20 and (2.87, 95% CI 1.61 to 4.74).21 Kotsopoulos and colleagues reported an increased risk that was not significant (OR, 4.55, 95% CI 0.76 to 27) and, in a subset of the samples included in this study, we also found a non-significant increase in risk (OR 3.2, 95% CI 0.86 to 12).3
It is possible that cryptic population structure could cause spurious association in these data. Principal component analysis is one approach to reducing the risk of such bias, but there are too few common variants in the regions covered by the targeted sequencing panel to do a principal component analysis and chip genotyping data that would be required for such an analysis is not available for all the samples. Adjusting for country of origin and restricting the analysis to samples from individuals of broad European ancestries should reduce any problem with population stratification.
We lacked the statistical power to identify susceptibility genes conferring relative risks of less than 2 (figure 1). Our use of targeted sequencing and a definition of deleterious variants as those that likely truncate the protein product will have probably underestimated the true prevalence of deleterious variants in these genes. Incomplete coverage of each gene will have missed some small indels and single nucleotide variants. Amplicon based sequencing will also miss large deletions and rearrangements, which are relatively common in some genes.39 40 Finally, any functional mutations in the non-coding region of these genes will have been missed.41
Figure 1.
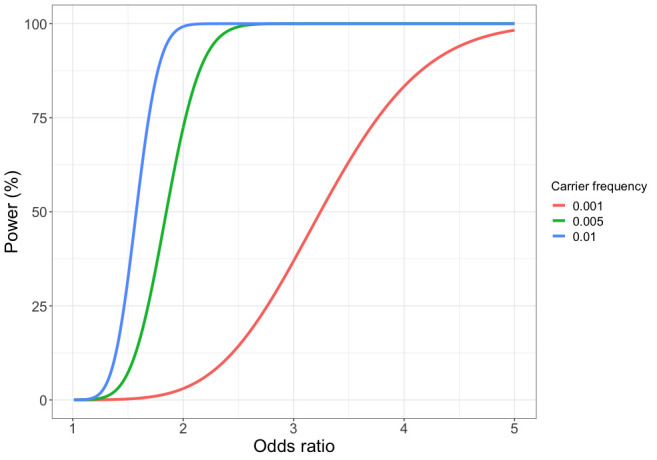
Power to detect association for 5951 cases and 6385 controls at a Type I error rate of 0.0001 by deleterious variant carrier frequency and effect size (OR).
Some commercial gene-panel tests for hereditary breast-ovarian cancer already include PALB2. However, whether there is clinical utility in testing unaffected women for deleterious mutations in PALB2 is not clear given the uncertainties in the risk estimates for this gene. There is no consensus over the risk threshold at which preventative surgery should be offered; many cancer genetics clinics in the UK will refer women if their predicted lifetime risk of EOC is greater than 10%. Others have suggested that the risk threshold should be lower given the low risk nature of the intervention; prophylactic surgery has been shown to be cost-effective for women at a lifetime risk of 5%. Recent updates to the US National Comprehensive Cancer Network Guidelines recommend considering risk reducing salpingo-oophorectomy in carriers of moderate risk genes if the lifetime risk of such mutation carriers exceeds 2.6%. Based on our data and population data for ovarian cancer incidence in England and Wales in 2016, the cumulative risk of ovarian cancer by age 80 for a carrier of a deleterious PALB2 mutation is 3.2% (figure 2). Thus, a woman carrying a PALB2 deleterious mutation would be eligible for prophylactic surgery. However, the CIs for this estimate range from 1.8% to 5.7%. Very large, well-designed case-control studies will be required to provide more precise, unbiased estimates of risk suitable for clinical counselling.
Figure 2.
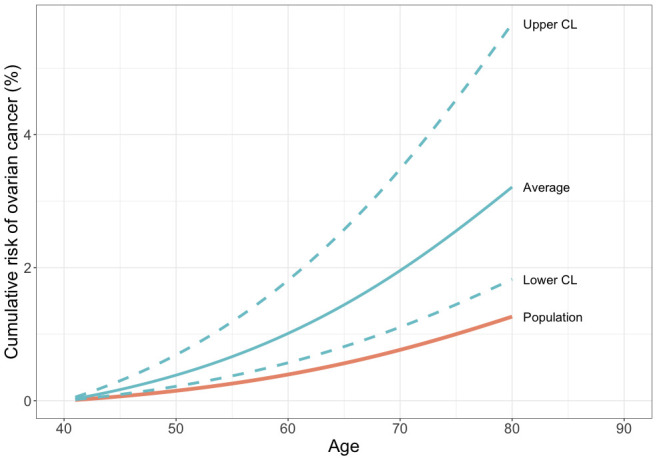
Estimated cumulative risk (%) of ovarian cancer in a PALB2 deleterious variant carrier compared with population risks for England and Wales, 2016.
In summary, we have found relatively strong evidence that deleterious germline mutations in PALB2 are associated with a moderate increase in the risk of HGSOC with weak evidence for POLK, SLX4 and FBXO10. Mutations in the other 50 genes we tested are unlikely to contribute meaningfully to genetic predisposition to HGSOC. This study highlights the importance of large sample sizes needed to obtain risk estimates with the precision necessary for clinical use.
Acknowledgments
We thank all the study participants who contributed to this study and all the researchers, clinicians and technical and administrative staff who have made possible this work. In particular, we thank: the clinical and scientific collaborators listed at www.aocstudy.org/ (AOCS) and opalstudy.qimrberghofer.edu.au (OPL).
Footnotes
HS and EMD contributed equally.
SR, SG and PP contributed equally.
Contributors: HS planned the study, co-ordinated the sequencing, collated the results and drafted the manuscript. EDM carried out the analysis of the sequencing data. JT carried out the biostatistical analyses. GC-T, DDB, NT, JB, TG, KH, AP, EVO, JD, HRH, MAR, MD, TD, NVB, FM, KM, KO, RN, BYK, JLe, AJe, SKK, EH, CL, Miquel Angel Pujana, JC, RV, SJW, MH, CH, DL, XW, YY, JBP, DAL, JMS, AB, PMW, CC, JG, AJa, JLu, JA, CJK, IC, UM, CLP, AHW and AdF were responsible for data and sample collection for the contributing studies. MJR co-ordinated the collection of the OCAC phenotype data. Maria Intermaggio and PH carried out the targeted sequencing. IGC, EG, SR, SG and PP planned the study.
Funding: American Cancer Society: SIOP-06-258-01-COUN. Cancer Councils of New South Wales, Victoria, Queensland, South Australia and Tasmania and Cancer Foundation of Western Australia: Multi-State Applications 191, 211 and 182. Cancer Institute NSW: 12/RIG/1-17, 15/RIG/1-16. Cancer Research UK: C490/A10119, C490/A10124, C490/A16561, Cambridge Cancer Centre. Kræftens Bekæmpelse: 94 222 52. Medical Research Council: MR_UU_12023. U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute: K07CA080668, K22 CA193860, MO1RR000056, P50CA159981, R01CA112523, R01CA122443, P30CA15083, P50CA136393, R01CA76016, R01CA126841, R01CA178535, R01CA188943, R01CA61107, R01CA87538, R01CA95023. U.S. Department of Health and Human Services, National Institutes of Health, National Center for Advancing Translational Sciences: UL1TR000124. National Health and Medical Research Council of Australia 199600, 310670, 400281, 400413, 628903, APP1025142. National Institutes of Health Research: Cambridge Biomedical Research Centre, University College London Hospitals Biomedical Research Centre. The Eve Appeal: UKOPS Study. U.S. Army Medical Research and Materiel Command: DAMD17-01-1-0729, DAMD17-02-1-0669, DAMD17-02-1-06. U.S Department of Defense Ovarian Cancer Research Program: W81XWH-07-0449. The University of Cambridge has received salary support in respect of PDPP from the NHS in the East of England through the Clinical Academic Reserve. SG is a recipient of the Barth Family Chair in Cancer Genetics.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available from the authors on request.
References
- 1. Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugenstrano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barrett JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994;266:66–71. 10.1126/science.7545954 [DOI] [PubMed] [Google Scholar]
- 2. Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995;378:789–92. 10.1038/378789a0 [DOI] [PubMed] [Google Scholar]
- 3. Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward J, Philpott S, Anderson C, Edlund CK, Conti D, Harrington P, Barrowdale D, Bowtell DD, Alsop K, Mitchell G, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Sieh W, McGuire V, Lester J, Bogdanova N, Dürst M, Hillemanns P, Odunsi K, Whittemore AS, Karlan BY, Dörk T, Goode EL, Menon U, Jacobs IJ, Antoniou AC, Pharoah PDP, Gayther SA, AOCS Study Group, Ovarian Cancer Association Consortium . Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst 2015;107. 10.1093/jnci/djv214. [Epub ahead of print: 27 Aug 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, Edlund CK, Conti D, Harrington P, Fraser L, Philpott S, Anderson C, Rosenthal A, Gentry-Maharaj A, Bowtell DD, Alsop K, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Høgdall E, Høgdall CK, Jensen A, Kjaer SK, Lubiński J, Huzarski T, Jakubowska A, Gronwald J, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Odunsi K, Goode EL, Menon U, Jacobs IJ, Gayther SA, Pharoah PDP. Contribution of germline mutations in the Rad51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol 2015;33:2901–7. 10.1200/JCO.2015.61.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pal T, Permuth-Wey J, Sellers TA. A review of the clinical relevance of mismatch-repair deficiency in ovarian cancer. Cancer 2008;113:733–42. 10.1002/cncr.23601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song H, Cicek MS, Dicks E, Harrington P, Ramus SJ, Cunningham JM, Fridley BL, Tyrer JP, Alsop J, Jimenez-Linan M, Gayther SA, Goode EL, Pharoah PDP. The contribution of deleterious germline mutations in BRCA1, BRCA2 and the mismatch repair genes to ovarian cancer in the population. Hum Mol Genet 2014;23:4703–9. 10.1093/hmg/ddu172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu Q, Zhang J, Chen Y, Hu Q, Shen H, Huang R-Y, Liu Q, Kaur J, Long M, Battaglia S, Eng KH, Lele SB, Zsiros E, Villella J, Lugade A, Yao S, Liu S, Moysich K, Odunsi KO. Whole-Exome sequencing of ovarian cancer families uncovers putative predisposition genes. Int J Cancer 2020;146:2147–55. 10.1002/ijc.32545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dicks E, Song H, Ramus SJ, Oudenhove EV, Tyrer JP, Intermaggio MP, Kar S, Harrington P, Bowtell DD, Group AS, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Piskorz A, Goranova T, Kent E, Siddiqui N, Paul J, Crawford R, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Sieh W, McGuire V, Lester J, Odunsi K, Whittemore AS, Bogdanova N, Dürst M, Hillemanns P, Karlan BY, Gentry-Maharaj A, Menon U, Tischkowitz M, Levine D, Brenton JD, Dörk T, Goode EL, Gayther SA, Pharoah DPP. Germline whole exome sequencing and large-scale replication identifies FANCM as a likely high grade serous ovarian cancer susceptibility gene. Oncotarget 2017;8:50930–40. 10.18632/oncotarget.15871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, Dunning AM, Redman J, Scarth J, Plaskocinska I, Luccarini C, Shah M, Pooley K, Dorling L, Lee A, Adank MA, Adlard J, Aittomaki K, Andrulis IL, Ang P, Barwell J, Bernstein JL, Bobolis K, Borg A, Blomqvist C, Claes KBM, Concannon P, Cuggia A, Culver JO, Damiola F, de Pauw A, Diez O, Dolinsky JS, Domchek SM, Engel C, Evans DG, Fostira F, Garber J, Golmard L, Goode EL, Gruber SB, Hahnen E, Hake C, Heikkinen T, Hurley JE, Janavicius R, Kleibl Z, Kleiblova P, Konstantopoulou I, Kvist A, Laduca H, ASG L, Lesueur F, Maher ER, Mannermaa A, Manoukian S, McFarland R, McKinnon W, Meindl A, Metcalfe K, Mohd Taib NA, Moilanen J, Nathanson KL, Neuhausen S, PS N, Nguyen-Dumont T, Nielsen SM, Obermair F, Offit K, Olopade OI, Ottini L, Penkert J, Pylkas K, Radice P, Ramus SJ, Rudaitis V, Side L, Silva-Smith R, Silvestri V, Skytte AB, Slavin T, Soukupova J, Tondini C, Trainer AH, Unzeitig G, Usha L, van Overeem Hansen T, Whitworth J, Wood M, Yip CH, Yoon SY, Yussuf A, Zogopoulos G, Goldgar D, Hopper JL, Chenevix-Trench G, Pharoah P, George SHL, Balmana J, Houdayer C, James P, El-Haffaf Z, Ehrencrona H, Janatova M, Peterlongo P, Nevanlinna H, Schmutzler R, Teo SH, Robson M, Pal T, Couch F, Weitzel JN, Elliott A, Southey M, Winqvist R, Easton DF, Foulkes WD, Antoniou AC, Tischkowitz M. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol 2019:JCO1901907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Vergote I, Lambrechts S, Despierre E, Risch HA, González-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guénel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, Brauch H, Garcia-Closas M, Hillemanns P, Winqvist R, Dürst M, Devilee P, Runnebaum I, Jakubowska A, Lubinski J, Mannermaa A, Butzow R, Bogdanova NV, Dörk T, Pelttari LM, Zheng W, Leminen A, Anton-Culver H, Bunker CH, Kristensen V, Ness RB, Muir K, Edwards R, Meindl A, Heitz F, Matsuo K, du Bois A, Wu AH, Harter P, Teo S-H, Schwaab I, Shu X-O, Blot W, Hosono S, Kang D, Nakanishi T, Hartman M, Yatabe Y, Hamann U, Karlan BY, Sangrajrang S, Kjaer SK, Gaborieau V, Jensen A, Eccles D, Høgdall E, Shen C-Y, Brown J, Woo YL, Shah M, Azmi MAN, Luben R, Omar SZ, Czene K, Vierkant RA, Nordestgaard BG, Flyger H, Vachon C, Olson JE, Wang X, Levine DA, Rudolph A, Weber RP, Flesch-Janys D, Iversen E, Nickels S, Schildkraut JM, Silva IDS, Cramer DW, Gibson L, Terry KL, Fletcher O, Vitonis AF, van der Schoot CE, Poole EM, Hogervorst FBL, Tworoger SS, Liu J, Bandera EV, Li J, Olson SH, Humphreys K, Orlow I, Blomqvist C, Rodriguez-Rodriguez L, Aittomäki K, Salvesen HB, Muranen TA, Wik E, Brouwers B, Krakstad C, Wauters E, Halle MK, Wildiers H, Kiemeney LA, Mulot C, Aben KK, Laurent-Puig P, Altena AM, Truong T, Massuger LFAG, Benitez J, Pejovic T, Perez JIA, Hoatlin M, Zamora MP, Cook LS, Balasubramanian SP, Kelemen LE, Schneeweiss A, Le ND, Sohn C, Brooks-Wilson A, Tomlinson I, Kerin MJ, Miller N, Cybulski C, Henderson BE, Menkiszak J, Schumacher F, Wentzensen N, Le Marchand L, Yang HP, Mulligan AM, Glendon G, Engelholm SA, Knight JA, Høgdall CK, Apicella C, Gore M, Tsimiklis H, Song H, Southey MC, Jager A, den Ouweland AMW, Brown R, Martens JWM, Flanagan JM, Kriege M, Paul J, Margolin S, Siddiqui N, Severi G, Whittemore AS, Baglietto L, McGuire V, Stegmaier C, Sieh W, Müller H, Arndt V, Labrèche F, Gao Y-T, Goldberg MS, Yang G, Dumont M, McLaughlin JR, Hartmann A, Ekici AB, Beckmann MW, Phelan CM, Lux MP, Permuth-Wey J, Peissel B, Sellers TA, Ficarazzi F, Barile M, Ziogas A, Ashworth A, Gentry-Maharaj A, Jones M, Ramus SJ, Orr N, Menon U, Pearce CL, Brüning T, Pike MC, Ko Y-D, Lissowska J, Figueroa J, Kupryjanczyk J, Chanock SJ, Dansonka-Mieszkowska A, Jukkola-Vuorinen A, Rzepecka IK, Pylkäs K, Bidzinski M, Kauppila S, Hollestelle A, Seynaeve C, Tollenaar RAEM, Durda K, Jaworska K, Hartikainen JM, Kosma V-M, Kataja V, Antonenkova NN, Long J, Shrubsole M, Deming-Halverson S, Lophatananon A, Siriwanarangsan P, Stewart-Brown S, Ditsch N, Lichtner P, Schmutzler RK, Ito H, Iwata H, Tajima K, Tseng C-C, Stram DO, van den Berg D, Yip CH, Ikram MK, Teh Y-C, Cai H, Lu W, Signorello LB, Cai Q, Noh D-Y, Yoo K-Y, Miao H, Iau PT-C, Teo YY, McKay J, Shapiro C, Ademuyiwa F, Fountzilas G, Hsiung C-N, Yu J-C, Hou M-F, Healey CS, Luccarini C, Peock S, Stoppa-Lyonnet D, Peterlongo P, Rebbeck TR, Piedmonte M, Singer CF, Friedman E, Thomassen M, Offit K, Hansen TVO, Neuhausen SL, Szabo CI, Blanco I, Garber J, Narod SA, Weitzel JN, Montagna M, Olah E, Godwin AK, Yannoukakos D, Goldgar DE, Caldes T, Imyanitov EN, Tihomirova L, Arun BK, Campbell I, Mensenkamp AR, van Asperen CJ, van Roozendaal KEP, Meijers-Heijboer H, Collée JM, Oosterwijk JC, Hooning MJ, Rookus MA, van der Luijt RB, Os TAM, Evans DG, Frost D, Fineberg E, Barwell J, Walker L, Kennedy MJ, Platte R, Davidson R, Ellis SD, Cole T, Bressac-de Paillerets B, Buecher B, Damiola F, Faivre L, Frenay M, Sinilnikova OM, Caron O, Giraud S, Mazoyer S, Bonadona V, Caux-Moncoutier V, Toloczko-Grabarek A, Gronwald J, Byrski T, Spurdle AB, Bonanni B, Zaffaroni D, Giannini G, Bernard L, Dolcetti R, Manoukian S, Arnold N, Engel C, Deissler H, Rhiem K, Niederacher D, Plendl H, Sutter C, Wappenschmidt B, Borg A, Melin B, Rantala J, Soller M, Nathanson KL, Domchek SM, Rodriguez GC, Salani R, Kaulich DG, Tea M-K, Paluch SS, Laitman Y, Skytte A-B, Kruse TA, Jensen UB, Robson M, Gerdes A-M, Ejlertsen B, Foretova L, Savage SA, Lester J, Soucy P, Kuchenbaecker KB, Olswold C, Cunningham JM, Slager S, Pankratz VS, Dicks E, Lakhani SR, Couch FJ, Hall P, Monteiro ANA, Gayther SA, Pharoah PDP, Reddel RR, Goode EL, Greene MH, Easton DF, Berchuck A, Antoniou AC, Chenevix-Trench G, Dunning AM, Australian Cancer Study, Australian Ovarian Cancer Study, Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab), Gene Environment Interaction and Breast Cancer (GENICA), Swedish Breast Cancer Study (SWE-BRCA), Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON), Epidemiological study of BRCA1 & BRCA2 Mutation Carriers (EMBRACE), Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) . Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 2013;45:371–84. 10.1038/ng.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai Y-Y, Weidhaas J, Paik D, Van Den Berg DJ, Stram DO, Pearce CL, Wu AH, Brewster W, Anton-Culver H, Ziogas A, Narod SA, Levine DA, Kaye SB, Brown R, Paul J, Flanagan J, Sieh W, McGuire V, Whittemore AS, Campbell I, Gore ME, Lissowska J, Yang HP, Medrek K, Gronwald J, Lubinski J, Jakubowska A, Le ND, Cook LS, Kelemen LE, Brooks-Wilson A, Brook-Wilson A, Massuger LFAG, Kiemeney LA, Aben KKH, van Altena AM, Houlston R, Tomlinson I, Palmieri RT, Moorman PG, Schildkraut J, Iversen ES, Phelan C, Vierkant RA, Cunningham JM, Goode EL, Fridley BL, Kruger-Kjaer S, Blaeker J, Hogdall E, Hogdall C, Gross J, Karlan BY, Ness RB, Edwards RP, Odunsi K, Moyisch KB, Baker JA, Modugno F, Heikkinenen T, Butzow R, Nevanlinna H, Leminen A, Bogdanova N, Antonenkova N, Doerk T, Hillemanns P, Dürst M, Runnebaum I, Thompson PJ, Carney ME, Goodman MT, Lurie G, Wang-Gohrke S, Hein R, Chang-Claude J, Rossing MA, Cushing-Haugen KL, Doherty J, Chen C, Rafnar T, Besenbacher S, Sulem P, Stefansson K, Birrer MJ, Terry KL, Hernandez D, Cramer DW, Vergote I, Amant F, Lambrechts D, Despierre E, Fasching PA, Beckmann MW, Thiel FC, Ekici AB, Chen X, Johnatty SE, Webb PM, Beesley J, Chanock S, Garcia-Closas M, Sellers T, Easton DF, Berchuck A, Chenevix-Trench G, Pharoah PDP, Gayther SA, Australian Ovarian Cancer Study Group, Australian Cancer Study (Ovarian Cancer), Ovarian Cancer Association Consortium . Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet 2010;42:880–4. 10.1038/ng.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, Widschwendter M, Vierkant RA, Larson MC, Kjaer SK, Birrer MJ, Berchuck A, Schildkraut J, Tomlinson I, Kiemeney LA, Cook LS, Gronwald J, Garcia-Closas M, Gore ME, Campbell I, Whittemore AS, Sutphen R, Phelan C, Anton-Culver H, Pearce CL, Lambrechts D, Rossing MA, Chang-Claude J, Moysich KB, Goodman MT, Dörk T, Nevanlinna H, Ness RB, Rafnar T, Hogdall C, Hogdall E, Fridley BL, Cunningham JM, Sieh W, McGuire V, Godwin AK, Cramer DW, Hernandez D, Levine D, Lu K, Iversen ES, Palmieri RT, Houlston R, van Altena AM, Aben KKH, Massuger LFAG, Brooks-Wilson A, Kelemen LE, Le ND, Jakubowska A, Lubinski J, Medrek K, Stafford A, Easton DF, Tyrer J, Bolton KL, Harrington P, Eccles D, Chen A, Molina AN, Davila BN, Arango H, Tsai Y-Y, Chen Z, Risch HA, McLaughlin J, Narod SA, Ziogas A, Brewster W, Gentry-Maharaj A, Menon U, Wu AH, Stram DO, Pike MC, Beesley J, Webb PM, Chen X, Ekici AB, Thiel FC, Beckmann MW, Yang H, Wentzensen N, Lissowska J, Fasching PA, Despierre E, Amant F, Vergote I, Doherty J, Hein R, Wang-Gohrke S, Lurie G, Carney ME, Thompson PJ, Runnebaum I, Hillemanns P, Dürst M, Antonenkova N, Bogdanova N, Leminen A, Butzow R, Heikkinen T, Stefansson K, Sulem P, Besenbacher S, Sellers TA, Gayther SA, Pharoah PDP, Wellcome Trust Case-Control Consortium, Australian Cancer Study (Ovarian Cancer), Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium (OCAC) . A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet 2010;42:874–9. 10.1038/ng.668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, Lin H-Y, Chen YA, Tsai Y-Y, Qu X, Ramus SJ, Karevan R, Lee J, Lee N, Larson MC, Aben KK, Anton-Culver H, Antonenkova N, Antoniou AC, Armasu SM, Bacot F, Baglietto L, Bandera EV, Barnholtz-Sloan J, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Cai Q, Campbell I, Chang-Claude J, Chanock S, Chenevix-Trench G, Cheng JQ, Cicek MS, Coetzee GA, Cook LS, Couch FJ, Cramer DW, Cunningham JM, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dörk T, du Bois A, Dürst M, Easton DF, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher DA, Flanagan JM, Garcia-Closas M, Gentry-Maharaj A, Giles GG, Glasspool RM, Gonzalez-Bosquet J, Goodman MT, Gore M, Górski B, Gronwald J, Hall P, Halle MK, Harter P, Heitz F, Hillemanns P, Hoatlin M, Høgdall CK, Høgdall E, Hosono S, Jakubowska A, Jensen A, Jim H, Kalli KR, Karlan BY, Kaye SB, Kelemen LE, Kiemeney LA, Kikkawa F, Konecny GE, Krakstad C, Kjaer SK, Kupryjanczyk J, Lambrechts D, Lambrechts S, Lancaster JM, Le ND, Leminen A, Levine DA, Liang D, Lim BK, Lin J, Lissowska J, Lu KH, Lubiński J, Lurie G, Massuger LFAG, Matsuo K, McGuire V, McLaughlin JR, Menon U, Modugno F, Moysich KB, Nakanishi T, Narod SA, Nedergaard L, Ness RB, Nevanlinna H, Nickels S, Noushmehr H, Odunsi K, Olson SH, Orlow I, Paul J, Pearce CL, Pejovic T, Pelttari LM, Pike MC, Poole EM, Raska P, Renner SP, Risch HA, Rodriguez-Rodriguez L, Rossing MA, Rudolph A, Runnebaum IB, Rzepecka IK, Salvesen HB, Schwaab I, Severi G, Shridhar V, Shu X-O, Shvetsov YB, Sieh W, Song H, Southey MC, Spiewankiewicz B, Stram D, Sutphen R, Teo S-H, Terry KL, Tessier DC, Thompson PJ, Tworoger SS, van Altena AM, Vergote I, Vierkant RA, Vincent D, Vitonis AF, Wang-Gohrke S, Palmieri Weber R, Wentzensen N, Whittemore AS, Wik E, Wilkens LR, Winterhoff B, Woo YL, Wu AH, Xiang Y-B, Yang HP, Zheng W, Ziogas A, Zulkifli F, Phelan CM, Iversen E, Schildkraut JM, Berchuck A, Fridley BL, Goode EL, Pharoah PDP, Monteiro ANA, Sellers TA, Gayther SA, Australian Cancer S, Australian Cancer Study, Australian Ovarian Cancer Study, Consortium of Investigators of Modifiers of BRCA1/2 . Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun 2013;4:1627. 10.1038/ncomms2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pharoah PDP, Tsai Y-Y, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E, Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow W-H, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dörk T, du Bois A, Dürst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao Y-T, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Høgdall E, Høgdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubiński J, Lundvall L, Lurie G, Massuger LFAG, Matsuo K, McGuire V, McLaughlin JR, Menon U, Modugno F, Moysich KB, Nakanishi T, Narod SA, Ness RB, Nevanlinna H, Nickels S, Noushmehr H, Odunsi K, Olson S, Orlow I, Paul J, Pejovic T, Pelttari LM, Permuth-Wey J, Pike MC, Poole EM, Qu X, Risch HA, Rodriguez-Rodriguez L, Rossing MA, Rudolph A, Runnebaum I, Rzepecka IK, Salvesen HB, Schwaab I, Severi G, Shen H, Shridhar V, Shu X-O, Sieh W, Southey MC, Spellman P, Tajima K, Teo S-H, Terry KL, Thompson PJ, Timorek A, Tworoger SS, van Altena AM, van den Berg D, Vergote I, Vierkant RA, Vitonis AF, Wang-Gohrke S, Wentzensen N, Whittemore AS, Wik E, Winterhoff B, Woo YL, Wu AH, Yang HP, Zheng W, Ziogas A, Zulkifli F, Goodman MT, Hall P, Easton DF, Pearce CL, Berchuck A, Chenevix-Trench G, Iversen E, Monteiro ANA, Gayther SA, Schildkraut JM, Sellers TA, Australian Cancer S, Australian Cancer Study, Australian Ovarian Cancer Study Group . Gwas meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet 2013;45:362–70. 10.1038/ng.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, Dennis J, Pirie A, Riggan MJ, Chornokur G, Earp MA, Lyra PC, Lee JM, Coetzee S, Beesley J, McGuffog L, Soucy P, Dicks E, Lee A, Barrowdale D, Lecarpentier J, Leslie G, Aalfs CM, Aben KKH, Adams M, Adlard J, Andrulis IL, Anton-Culver H, Antonenkova N, Aravantinos G, Arnold N, Arun BK, Arver B, Azzollini J, Balmaña J, Banerjee SN, Barjhoux L, Barkardottir RB, Bean Y, Beckmann MW, Beeghly-Fadiel A, Benitez J, Bermisheva M, Bernardini MQ, Birrer MJ, Bjorge L, Black A, Blankstein K, Blok MJ, Bodelon C, Bogdanova N, Bojesen A, Bonanni B, Borg Åke, Bradbury AR, Brenton JD, Brewer C, Brinton L, Broberg P, Brooks-Wilson A, Bruinsma F, Brunet J, Buecher B, Butzow R, Buys SS, Caldes T, Caligo MA, Campbell I, Cannioto R, Carney ME, Cescon T, Chan SB, Chang-Claude J, Chanock S, Chen XQ, Chiew Y-E, Chiquette J, Chung WK, Claes KBM, Conner T, Cook LS, Cook J, Cramer DW, Cunningham JM, D'Aloisio AA, Daly MB, Damiola F, Damirovna SD, Dansonka-Mieszkowska A, Dao F, Davidson R, DeFazio A, Delnatte C, Doheny KF, Diez O, Ding YC, Doherty JA, Domchek SM, Dorfling CM, Dörk T, Dossus L, Duran M, Dürst M, Dworniczak B, Eccles D, Edwards T, Eeles R, Eilber U, Ejlertsen B, Ekici AB, Ellis S, Elvira M, Eng KH, Engel C, Evans DG, Fasching PA, Ferguson S, Ferrer SF, Flanagan JM, Fogarty ZC, Fortner RT, Fostira F, Foulkes WD, Fountzilas G, Fridley BL, Friebel TM, Friedman E, Frost D, Ganz PA, Garber J, García MJ, Garcia-Barberan V, Gehrig A, Gentry-Maharaj A, Gerdes A-M, Giles GG, Glasspool R, Glendon G, Godwin AK, Goldgar DE, Goranova T, Gore M, Greene MH, Gronwald J, Gruber S, Hahnen E, Haiman CA, Håkansson N, Hamann U, Hansen TVO, Harrington PA, Harris HR, Hauke J, Hein A, Henderson A, Hildebrandt MAT, Hillemanns P, Hodgson S, Høgdall CK, Høgdall E, Hogervorst FBL, Holland H, Hooning MJ, Hosking K, Huang R-Y, Hulick PJ, Hung J, Hunter DJ, Huntsman DG, Huzarski T, Imyanitov EN, Isaacs C, Iversen ES, Izatt L, Izquierdo A, Jakubowska A, James P, Janavicius R, Jernetz M, Jensen A, Jensen UB, John EM, Johnatty S, Jones ME, Kannisto P, Karlan BY, Karnezis A, Kast K, Kennedy CJ, Khusnutdinova E, Kiemeney LA, Kiiski JI, Kim S-W, Kjaer SK, Köbel M, Kopperud RK, Kruse TA, Kupryjanczyk J, Kwong A, Laitman Y, Lambrechts D, Larrañaga N, Larson MC, Lazaro C, Le ND, Le Marchand L, Lee JW, Lele SB, Leminen A, Leroux D, Lester J, Lesueur F, Levine DA, Liang D, Liebrich C, Lilyquist J, Lipworth L, Lissowska J, Lu KH, Lubinński J, Luccarini C, Lundvall L, Mai PL, Mendoza-Fandiño G, Manoukian S, Massuger LFAG, May T, Mazoyer S, McAlpine JN, McGuire V, McLaughlin JR, McNeish I, Meijers-Heijboer H, Meindl A, Menon U, Mensenkamp AR, Merritt MA, Milne RL, Mitchell G, Modugno F, Moes-Sosnowska J, Moffitt M, Montagna M, Moysich KB, Mulligan AM, Musinsky J, Nathanson KL, Nedergaard L, Ness RB, Neuhausen SL, Nevanlinna H, Niederacher D, Nussbaum RL, Odunsi K, Olah E, Olopade OI, Olsson H, Olswold C, O'Malley DM, Ong K-R, Onland-Moret NC, Orr N, Orsulic S, Osorio A, Palli D, Papi L, Park-Simon T-W, Paul J, Pearce CL, Pedersen IS, Peeters PHM, Peissel B, Peixoto A, Pejovic T, Pelttari LM, Permuth JB, Peterlongo P, Pezzani L, Pfeiler G, Phillips K-A, Piedmonte M, Pike MC, Piskorz AM, Poblete SR, Pocza T, Poole EM, Poppe B, Porteous ME, Prieur F, Prokofyeva D, Pugh E, Pujana MA, Pujol P, Radice P, Rantala J, Rappaport-Fuerhauser C, Rennert G, Rhiem K, Rice P, Richardson A, Robson M, Rodriguez GC, Rodríguez-Antona C, Romm J, Rookus MA, Rossing MA, Rothstein JH, Rudolph A, Runnebaum IB, Salvesen HB, Sandler DP, Schoemaker MJ, Senter L, Setiawan VW, Severi G, Sharma P, Shelford T, Siddiqui N, Side LE, Sieh W, Singer CF, Sobol H, Song H, Southey MC, Spurdle AB, Stadler Z, Steinemann D, Stoppa-Lyonnet D, Sucheston-Campbell LE, Sukiennicki G, Sutphen R, Sutter C, Swerdlow AJ, Szabo CI, Szafron L, Tan YY, Taylor JA, Tea M-K, Teixeira MR, Teo S-H, Terry KL, Thompson PJ, Thomsen LCV, Thull DL, Tihomirova L, Tinker AV, Tischkowitz M, Tognazzo S, Toland AE, Tone A, Trabert B, Travis RC, Trichopoulou A, Tung N, Tworoger SS, van Altena AM, Van Den Berg D, van der Hout AH, van der Luijt RB, Van Heetvelde M, Van Nieuwenhuysen E, van Rensburg EJ, Vanderstichele A, Varon-Mateeva R, Vega A, Edwards DV, Vergote I, Vierkant RA, Vijai J, Vratimos A, Walker L, Walsh C, Wand D, Wang-Gohrke S, Wappenschmidt B, Webb PM, Weinberg CR, Weitzel JN, Wentzensen N, Whittemore AS, Wijnen JT, Wilkens LR, Wolk A, Woo M, Wu X, Wu AH, Yang H, Yannoukakos D, Ziogas A, Zorn KK, Narod SA, Easton DF, Amos CI, Schildkraut JM, Ramus SJ, Ottini L, Goodman MT, Park SK, Kelemen LE, Risch HA, Thomassen M, Offit K, Simard J, Schmutzler RK, Hazelett D, Monteiro AN, Couch FJ, Berchuck A, Chenevix-Trench G, Goode EL, Sellers TA, Gayther SA, Antoniou AC, Pharoah PDP, AOCS study group, EMBRACE Study, GEMO Study Collaborators, HEBON Study, KConFab Investigators, OPAL study group . Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet 2017;49:680–91. 10.1038/ng.3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, Kobel M, Consortium P, Ziogas A, Zheng W, Yang HP, AH W, Wozniak EL, Woo YL, Winterhoff B, Wik E, Whittemore AS, Wentzensen N, Weber RP, Vitonis AF, Vincent D, Vierkant RA, Vergote I, Van Den Berg D, Van Altena AM, Tworoger SS, Thompson PJ, Tessier DC, Terry KL, Teo SH, Templeman C, Stram DO, Southey MC, Sieh W, Siddiqui N, Shvetsov YB, Shu XO, Shridhar V, Wang-Gohrke S, Severi G, Schwaab I, Salvesen HB, Rzepecka IK, Runnebaum IB, Rossing MA, Rodriguez-Rodriguez L, Risch HA, Renner SP, Poole EM, Pike MC, Phelan CM, Pelttari LM, Pejovic T, Paul J, Orlow I, Omar SZ, Olson SH, Odunsi K, Nickels S, Nevanlinna H, Ness RB, Narod SA, Nakanishi T, Moysich KB, Monteiro AN, Moes-Sosnowska J, Modugno F, Menon U, McLaughlin JR, McGuire V, Matsuo K, Adenan NA, Massuger LF, Lurie G, Lundvall L, Lubinski J, Lissowska J, Levine DA, Leminen A, Lee AW, ND L, Lambrechts S, Lambrechts D, Kupryjanczyk J, Krakstad C, Konecny GE, Kjaer SK, Kiemeney LA, Kelemen LE, Keeney GL, Karlan BY, Karevan R, Kalli KR, Kajiyama H, BT J, Jensen A, Jakubowska A, Iversen E, Hosono S, Hogdall CK, Hogdall E, Hoatlin M, Hillemanns P, Heitz F, Hein R, Harter P, Halle MK, Hall P, Gronwald J, Gore M, Goodman MT, Giles GG, Gentry-Maharaj A, Garcia-Closas M, Flanagan JM, Fasching PA, Ekici AB, Edwards R, Eccles D, Easton DF, Durst M, du Bois A, Dork T, Doherty JA, Despierre E, Dansonka-Mieszkowska A, Cybulski C, Cramer DW, Cook LS, Chen X, Charbonneau B, Chang-Claude J, Campbell I, Butzow R, Bunker CH, Brueggmann D, Brown R, Brooks-Wilson A, Brinton LA, Bogdanova N, Block MS, Benjamin E, Beesley J, Beckmann MW, Bandera EV, Baglietto L, Bacot F, Armasu SM, Antonenkova N, Anton-Culver H, Aben KK, Liang D, Wu X, Lu K, Hildebrandt MA. Australian ovarian cancer study G, Australian cancer S, Schildkraut JM, sellers TA, Huntsman D, Berchuck a, Chenevix-Trench G, Gayther SA, Pharoah PD, Laird PW, Goode El, Pearce Cl. epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun 2013;4:1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, Anton-Culver H, Chang-Claude J, Cramer DW, DiCioccio R, Dörk T, Goode EL, Goodman MT, Schildkraut JM, Sellers T, Baglietto L, Beckmann MW, Beesley J, Blaakaer J, Carney ME, Chanock S, Chen Z, Cunningham JM, Dicks E, Doherty JA, Dürst M, Ekici AB, Fenstermacher D, Fridley BL, Giles G, Gore ME, De Vivo I, Hillemanns P, Hogdall C, Hogdall E, Iversen ES, Jacobs IJ, Jakubowska A, Li D, Lissowska J, Lubiński J, Lurie G, McGuire V, McLaughlin J, Medrek K, Moorman PG, Moysich K, Narod S, Phelan C, Pye C, Risch H, Runnebaum IB, Severi G, Southey M, Stram DO, Thiel FC, Terry KL, Tsai Y-Y, Tworoger SS, Van Den Berg DJ, Vierkant RA, Wang-Gohrke S, Webb PM, Wilkens LR, Wu AH, Yang H, Brewster W, Ziogas A, Houlston R, Tomlinson I, Whittemore AS, Rossing MA, Ponder BAJ, Pearce CL, Ness RB, Menon U, Kjaer SK, Gronwald J, Garcia-Closas M, Fasching PA, Easton DF, Chenevix-Trench G, Berchuck A, Pharoah PDP, Gayther SA, Vivo D, Hogdall E, AH W, Australian Cancer S, Australian Cancer (Ovarian) Study, Australian Ovarian Cancer Study Group, Ovarian Cancer Association Consortium . A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet 2009;41:996–1000. 10.1038/ng.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, Spindler TJ, Lin YG, Pejovic T, Bean Y, Li Q, Coetzee S, Hazelett D, Miron A, Southey M, Terry MB, Goldgar DE, Buys SS, Janavicius R, Dorfling CM, van Rensburg EJ, Neuhausen SL, Ding YC, Hansen TV, Jonson L, Gerdes AM, Ejlertsen B, Barrowdale D, Dennis J, Benitez J, Osorio A, Garcia MJ, Komenaka I, Weitzel JN, Ganschow P, Peterlongo P, Bernard L, Viel A, Bonanni B, Peissel B, Manoukian S, Radice P, Papi L, Ottini L, Fostira F, Konstantopoulou I, Garber J, Frost D, Perkins J, Platte R, Ellis S, Embrace GAK, Schmutzler RK, Meindl A, Engel C, Sutter C, Sinilnikova OM, Collaborators GS, Damiola F, Mazoyer S, Stoppa-Lyonnet D, Claes K, De Leeneer K, Kirk J, Rodriguez GC, Piedmonte M, O'Malley DM, de la Hoya M, Caldes T, Aittomaki K, Nevanlinna H, Collee JM, Rookus MA, Oosterwijk JC. Consortium of Investigators of modifiers of B, BRCA. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet 2015;47:164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramus SJ, Kartsonaki C, Gayther SA, Pharoah PD, Sinilnikova OM, Beesley J, Chen X, McGuffog L, Healey S, Couch FJ, Wang X, Fredericksen Z, Peterlongo P, Manoukian S, Peissel B, Zaffaroni D, Roversi G, Barile M, Viel A, Allavena A, Ottini L, Papi L, Gismondi V, Capra F, Radice P, Greene MH, Mai PL, Andrulis IL, Glendon G, Ozcelik H, Ocgn TM, Gerdes AM, Kruse TA, Cruger D, Jensen UB, Caligo MA, Olsson H, Kristoffersson U, Lindblom A, Arver B, Karlsson P, Stenmark Askmalm M, Borg A, Neuhausen SL, Ding YC, Nathanson KL, Domchek SM, Jakubowska A, Lubinski J, Huzarski T, Byrski T, Gronwald J, Gorski B, Cybulski C, Debniak T, Osorio A, Duran M, Tejada MI, Benitez J, Hamann U, Rookus MA, Verhoef S, Tilanus-Linthorst MA, Vreeswijk MP, Bodmer D, Ausems MG, van Os TA, Asperen CJ, Blok MJ, Meijers-Heijboer HE, Hebon E, Peock S, Cook M, Oliver C, Frost D, Dunning AM, Evans DG, Eeles R, Pichert G, Cole T, Hodgson S, Brewer C, Morrison PJ, Porteous M, Kennedy MJ, Rogers MT, Side LE, Donaldson A, Gregory H, Godwin A, Stoppa-Lyonnet D, Moncoutier V, Castera L, Mazoyer S, Barjhoux L, Bonadona V, Leroux D, Faivre L, Lidereau R, Nogues C, Bignon YJ, Prieur F, Collonge-Rame MA, Venat-Bouvet L, Fert-Ferrer S, Collaborators GS, Miron A, Buys SS, Hopper JL, Daly MB, John EM, Terry MB, Goldgar D, Bcfr HT, Jonson L, Ejlertsen B, Agnarsson BA, Offit K, Kirchhoff T, Vijai J, Dutra-Clarke AV, Przybylo JA, Montagna M, Casella C, Imyanitov EN, Janavicius R, Blanco I, Lazaro C, Moysich KB, Karlan BY, Gross J, Beattie MS, Schmutzler R, Wappenschmidt B, Meindl A, Ruehl I, Fiebig B, Sutter C, Arnold N, Deissler H, Varon-Mateeva R, Kast K, Niederacher D, Gadzicki D, Caldes T, de la Hoya M, Nevanlinna H, Aittomaki K, Simard J, Soucy P, kConFab I, Spurdle AB, Holland H, Chenevix-Trench G, Easton DF, Antoniou AC. Consortium of Investigators of modifiers of B. genetic variation at 9p22.2 and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst 2011;103:105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, Bernards SS, Casadei S, Yi Q, Burger RA, Chan JK, Davidson SA, Mannel RS, DiSilvestro PA, Lankes HA, Ramirez NC, King MC, Swisher EM, Birrer MJ. Inherited mutations in women with ovarian carcinoma. JAMA Oncol 2016;2:482–90. 10.1001/jamaoncol.2015.5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lilyquist J, LaDuca H, Polley E, Davis BT, Shimelis H, Hu C, Hart SN, Dolinsky JS, Couch FJ, Goldgar DE. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls. Gynecol Oncol 2017;147:375–80. 10.1016/j.ygyno.2017.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kotsopoulos J, Sopik V, Rosen B, Fan I, McLaughlin JR, Risch H, Sun P, Narod SA, Akbari MR. Frequency of germline PALB2 mutations among women with epithelial ovarian cancer. Fam Cancer 2017;16:29–34. 10.1007/s10689-016-9919-z [DOI] [PubMed] [Google Scholar]
- 23. Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, Matullo G, Seminara D, Yoshida T, Saeki N, Andrew AS, Dinney CP, Czerniak B, Zhang Z-feng, Kiltie AE, Bishop DT, Vineis P, Porru S, Buntinx F, Kellen E, Zeegers MP, Kumar R, Rudnai P, Gurzau E, Koppova K, Mayordomo JI, Sanchez M, Saez B, Lindblom A, de Verdier P, Steineck G, Mills GB, Schned A, Guarrera S, Polidoro S, Chang S-C, Lin J, Chang DW, Hale KS, Majewski T, Grossman HB, Thorlacius S, Thorsteinsdottir U, Aben KKH, Witjes JA, Stefansson K, Amos CI, Karagas MR, Gu J. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet 2009;41:991–5. 10.1038/ng.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu X, Hildebrandt MA, Ye Y, Chow W-H, Gu J, Cunningham S, Zhao H, Hawk ET, Wagar E, Rodriguez A, Hamilton SR. Cohort profile: the MD anderson cancer patients and survivors cohort (MDA-CPSC). Int J Epidemiol 2016;45:713–713f. 10.1093/ije/dyv317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hassan MM, Bondy ML, Wolff RA, Abbruzzese JL, Vauthey J-N, Pisters PW, Evans DB, Khan R, Chou T-H, Lenzi R, Jiao L, Li D. Risk factors for pancreatic cancer: case-control study. Am J Gastroenterol 2007;102:2696–707. 10.1111/j.1572-0241.2007.01510.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Magoč T, Salzberg SL. Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu Y, Hu H, Bohlender RJ, Hu F, Chen J-S, Holt C, Fowler J, Guthery SL, Scheet P, Hildebrandt MAT, Yandell M, Huff CD. XPAT: a toolkit to conduct cross-platform association studies with heterogeneous sequencing datasets. Nucleic Acids Res 2018;46:e32. 10.1093/nar/gkx1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44:D862–8. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 2004;11:377–94. 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
- 33. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tyrer JP, Guo Q, Easton DF, Pharoah PDP. The admixture maximum likelihood test to test for association between rare variants and disease phenotypes. BMC Bioinformatics 2013;14:177. 10.1186/1471-2105-14-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812–4. 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:7.20.1–7.20.41. 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015;31:2745–7. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 2007;81:208–27. 10.1086/519024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duraturo F, Cavallo A, Liccardo R, Cudia B, De Rosa M, Diana G, Izzo P. Contribution of large genomic rearrangements in Italian Lynch syndrome patients: characterization of a novel alu-mediated deletion. Biomed Res Int 2013;2013:1–7. 10.1155/2013/219897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramus SJ, Harrington PA, Pye C, DiCioccio RA, Cox MJ, Garlinghouse-Jones K, Oakley-Girvan I, Jacobs IJ, Hardy RM, Whittemore AS, Ponder BAJ, Piver MS, Pharoah PDP, Gayther SA. Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum Mutat 2007;28:1207–15. 10.1002/humu.20599 [DOI] [PubMed] [Google Scholar]
- 41. Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet 2017;136:1093–111. 10.1007/s00439-017-1809-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2019-106739supp001.xlsx (424.6KB, xlsx)
Data Availability Statement
Data are available from the authors on request.


