Abstract
Background
The mechanisms underlying the obesity-cancer relationship are incompletely understood. This study aimed to characterise metabolic signatures of greater body size and to investigate their association with two obesity-related malignancies, endometrial and colorectal cancers, and with weight loss within the context of an intervention study.
Methods
Targeted mass spectrometry metabolomics data from 4326 participants enrolled in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort and 17 individuals from a single-arm pilot weight loss intervention (Intercept) were used in this analysis. Metabolic signatures of body size were first determined in discovery (N = 3029) and replication (N = 1297) sets among EPIC participants by testing the associations between 129 metabolites and body mass index (BMI), waist circumference (WC), and waist-to-hip ratio (WHR) using linear regression models followed by partial least squares analyses. Conditional logistic regression models assessed the associations between the metabolic signatures with endometrial (N = 635 cases and 648 controls) and colorectal (N = 423 cases and 423 controls) cancer risk using nested case-control studies in EPIC. Pearson correlation between changes in the metabolic signatures and weight loss was tested among Intercept participants.
Results
After adjustment for multiple comparisons, greater BMI, WC, and WHR were associated with higher levels of valine, isoleucine, glutamate, PC aa C38:3, and PC aa C38:4 and with lower levels of asparagine, glutamine, glycine, serine, lysoPC C17:0, lysoPC C18:1, lysoPC C18:2, PC aa C42:0, PC ae C34:3, PC ae C40:5, and PC ae C42:5. The metabolic signature of BMI (OR1-sd 1.50, 95% CI 1.30–1.74), WC (OR1-sd 1.46, 95% CI 1.27–1.69), and WHR (OR1-sd 1.54, 95% CI 1.33–1.79) were each associated with endometrial cancer risk. Risk of colorectal cancer was positively associated with the metabolic signature of WHR (OR1-sd: 1.26, 95% CI 1.07–1.49). In the Intercept study, a positive correlation was observed between weight loss and changes in the metabolic signatures of BMI (r = 0.5, 95% CI 0.06–0.94, p = 0.03), WC (r = 0.5, 95% CI 0.05–0.94, p = 0.03), and WHR (r = 0.6, 95% CI 0.32–0.87, p = 0.01).
Conclusions
Obesity is associated with a distinct metabolic signature comprising changes in levels of specific amino acids and lipids which is positively associated with both colorectal and endometrial cancer and is potentially reversible following weight loss.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-021-01970-1.
Keywords: Metabolomics, Obesity, Weight loss, Cancer
Background
Obesity is an important risk factor for at least 13 different types of cancer [1]. It is estimated that 3.9% (~ 550,000 cases in 2012) of the total worldwide cancer burden is related to obesity, while 5.7% (~ 800,000 cases in 2012) is attributed to both obesity and type 2 diabetes [1, 2]. Experimental and molecular epidemiologic studies indicate important roles for dysregulated sex hormone metabolism, adipose tissue-derived inflammation, and alterations in insulin signalling in mediating the adiposity and cancer associations [3, 4]. However, it is likely that other, as of yet unidentified, biological pathways may also underlie these relationships. Further, it is not clear whether weight loss promotes changes in the metabolic pathways linking obesity and cancer development, and ultimately whether it lowers cancer risk.
Metabolomics is an established technology for the identification of metabolic changes and biomarkers for understanding pathophysiological processes, through simultaneous measurement of multiple metabolites in human biofluids or tissues [5, 6]. Metabolomic profiling has the potential to identify specific metabolic phenotypes that are associated with cancer and to provide insights into the mechanistic pathways involved in cancer development [7–9].
To date, a limited number of epidemiological studies have identified metabolic and biochemical pathways that are significantly altered in obesity [10–12]. In a previous analysis in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort, a metabolic signature of high body mass index (BMI) was positively associated with hepatocellular carcinoma risk (HCC) and was found to mediate much of the association between measured BMI and HCC [13]. However, to date, no study has linked metabolic signatures reflecting other anthropometric measures of obesity, such as waist circumference (WC) and waist-to-hip ratio (WHR), to obesity-related cancer development. Similarly, very few studies have investigated the biochemical pathways altered during weight loss [14, 15]. The Intercept study, a pilot intervention promoting weight loss through meal replacement diet among individuals with obesity, was one of the first studies to demonstrate potential cancer-relevant changes in colorectal tissue following substantial weight loss [16].
In this analysis, we identified metabolic signatures associated with greater body sizes as determined by BMI, WC, and WHR, and then investigated their association with risk of colorectal and endometrial cancers—two malignancies strongly linked to obesity and metabolic dysfunction [1]—in EPIC. We also explored the extent to which weight loss modified these metabolic signatures within a single-arm weight loss intervention study.
Methods
European Prospective Investigation into Cancer and Nutrition
EPIC is a multicentre cohort of 521,330 participants who were recruited between 1991 and 2000, predominantly from the general populations of 10 European countries (Denmark, France, Germany, Greece, Italy, the Netherlands, Norway, Spain, Sweden, and the UK) [17, 18]. The current study used data from all EPIC countries apart from Greece.
Anthropometric characteristics were measured by trained observers using standardised methods [18]. Body weight was measured in all centres by electronic digital scales, with participants wearing only light underwear and after voiding the bladder. Height was measured to the nearest 0.1 cm using a flexible anthropometer [19]. The exceptions were Oxford, France, and Norway where these measures were self-reported; however, they were shown to be valid for identifying associations in epidemiological studies [20, 21]. Assessed weight and height (measured and self-reported) were used to calculate body mass index (BMI) defined as weight in kilogrammes divided by height in metres squared (kg/m2). Waist circumference (WC) was measured either at the narrowest torso circumference or at the midpoint between the lower ribs and iliac crest. At baseline, questionnaires were used to collect information on demographics, behavioural factors including dietary intakes, and medical information. Physical activity levels were estimated using a questionnaire focused on past-year physical activity in occupational, leisure, and household domains and classified according to the validated Cambridge physical activity index [22]. Validated country/centre-specific dietary questionnaires were used to obtain information on dietary intake including total energy, dietary fibre, fish and shellfish, meat, and processed meat intake.
Incident cancer cases were identified using cancer registries in Norway, Sweden, UK, Spain, Italy, the Netherlands, and Denmark. For France and Germany, incident cancer cases were identified during follow-up from a combination of sources including cancer and pathology centres, health insurance records, and active follow-up of study subjects. All countries followed a detailed protocol for the collection and standardisation of clinical and pathological data on each cancer site [23–26]. Cancer cases were defined using the tenth revision of the International Classification of Diseases (ICD-10) (ICD-10) and the second revision of the International Classification of Diseases for Oncology (ICDO-2). In the current analysis, we focused on two malignancies strongly linked to obesity and metabolic dysfunction [1], colorectal (C180-209) and endometrium (C540-549), more specifically on type I (endometrioid type) endometrial tumours, and on colon cancer.
Study participants
For the derivation of the metabolic signature, we used existing data from 4326 participants who had been selected as matched control (non-cancer) participants in four separate case–control studies nested within EPIC (breast, kidney, liver and prostate cancer studies). These control participants were selected as they had both metabolomics and anthropometric data available. These data were randomly split into a discovery set (N = 3029) and a replication set (N = 1297), for the validation of the metabolic signatures. The derived metabolic signatures associated with greater body size were then investigated in relation to colorectal and endometrial cancers using data from two case-control studies nested within EPIC comprising 423 colorectal cancer cases and 423 matched controls and 635 endometrial cancer cases and 648 matched controls. For the colorectal case-control study, participants were matched on study recruitment centre, sex, age at blood collection, time of blood collection, and fasting status. Similar matching criteria were applied to the endometrial case-control study, but also included menopausal status, and for premenopausal women, phase of menstrual cycle. All participants included in the current study self-reported being free of diabetes and not using hormone replacement therapy at baseline (women only).
Intercept weight loss pilot intervention
The Intercept study was a single-arm pilot study testing the effect of weight loss among obese individuals on biomarkers of colorectal cancer risk measured in serum and colorectal tissue biopsies. The study was registered on the ISRCTN registry as ISRCTN35702367. Twenty-six participants (21–57 years old) with obesity (BMI ≥ 30 kg/m2) were recruited via advertisements placed around University College London between July 2013 and July 2014. Briefly, participants followed an 8-week liquid weight-loss diet programme (810 cal per day) based on formula diet products (Cambridge Weight Plan™, Northants., UK). A detailed description of the study and primary results has been published elsewhere [16]. The programme was carried out by trained researchers, who also offered support and advice on behaviour change techniques on a weekly basis. At the end of the 8 weeks, 4 weeks of additional support were provided to help participants with meal reintroduction and weight loss maintenance. Pre- and post-intervention measures of weight, height, and waist circumference were collected, and two 20-ml fasting blood (serum) samples were taken by a research nurse at each time point.
Study participants
Data from 17 participants from the Intercept pilot intervention with metabolomics data measured in serum samples collected pre- and post-intervention were included to assess the association between the derived body size-related metabolic signatures and weight loss.
Laboratory analysis
Plasma (EPIC) and serum (Intercept) metabolites from all study populations were assayed using the targeted AbsoluteIDQ p180 kit (BIOCRATES Life Sciences AG, Innsbruck, Austria) on the liquid chromatography mass spectrometry (LC/MS) platform at IARC, Lyon, France. The exception were the plasma metabolites from the endometrial case-control study, which were assayed at Imperial College London, UK, using the same methodology. Amino acids and biogenic amines were separated by liquid chromatography before injection into the mass spectrometer, while flow injection analysis was used for glycerophospholipids, hexoses, acylcarnitines, and sphingolipids. Metabolites with inter-batch or intra-batch coefficients of variation (CVs) larger than 20% for analytical replicates and with more than 20% missing data were excluded. For the included metabolites, measurements below the limit of detection (LOD) or quantification (LOQ) were set to half the batch-specific LOD or LOQ, respectively. When all of the samples within the same batch had measurements below the LOD and LOQ, half of the lowest measured concentration detected for that metabolite across all other batches was imputed. All metabolites above the highest calibration standard were given the highest value obtained in the sample. For the metabolites with up to 20% missing data, the median value was imputed. In the EPIC discovery and replication sets, 129 metabolites were included. For the colorectal, endometrial and Intercept data sets 129, 124, and 128 metabolites were included, respectively (Additional file 1: Table S1).
C-peptide was assayed in serum samples from a sample of EPIC participants by enzyme-linked immunosorbent assay by Mercodia (Sylveniusgatan, Sweden) as previously described [27]. The mean intra-batch and inter-batch coefficients of variation were 6.69% and 5.75%, respectively, for C-peptide concentration of 5 ng/ml [27].
Statistical analysis
Descriptive analyses were performed for sociodemographic, behavioural and blood sampling-related variables for each study population (discovery and replication sets, colorectal and endometrial nested case-control studies and Intercept pilot intervention). Within each study population, metabolite levels were log-transformed (natural logarithm) and Z-standardised.
The Principal Component Partial R-square (PC-PR2) method [28] was performed in each study population to estimate the contribution to total variability in metabolite levels attributed to anthropometric variables and other factors such as subjects’ characteristics and technical aspects of the samples. In the EPIC discovery set, lifestyle (physical activity, smoking status and dietary intake), fasting status, sex, age at blood collection, and batch/study variables all together explained 32% of the total variability in metabolite levels (Additional file 1: Figure S1, Figure S2 and Figure S3). The main contributions of variability were batch/sub-study (8%) and country (13%). Regarding the anthropometrics, BMI, WC, and WHR explained 1.34%, 1.36%, and 1.10% of the total variability, respectively. Similar variability was observed in the replication set and in the colorectal and endometrial case-control studies. In the Intercept study, the variability of the changes in metabolites over 8 weeks were mainly explained by percentage of weight loss (32%), while baseline BMI, WC, and WHR explained 15%, 17%, and 13%, respectively (Additional file 1: Figure S4).
In all study populations, the metabolite levels (Z-standardised and log-transformed) were transformed into residuals of linear models with country and sex as independent variables (when possible) and random intercepts for analytical batches (nested within studies, when relevant).
Metabolic signatures of greater body sizes
In the discovery and replication sets, the residuals were used as dependent variables in linear regression models testing confounder-adjusted associations with log-transformed BMI, WC, and WHR. Models were adjusted for a set of a priori-defined covariates that included age at blood collection (continuous), fasting status at blood collection (< 3 h/3–6 h/> 6 h/unknown), education, smoking status at recruitment (current/former/never/unknown), physical activity index (inactive/moderately inactive/moderately active/active/unknown), height, and daily intake of energy, red and processed meat products, fish and shellfish, fibre, and alcohol (all continuous). In the discovery analysis, false discovery rate (FDR) adjustment of p values was applied using the Benjamini–Hochberg method (q values < 0.05 were considered statistically significant), as it has greater power to detect real differences when compared to the Bonferroni approach and reduces the risk that potentially relevant metabolites might be missed in the discovery phase. Then, in the replication phase, in which we eliminate the non-relevant metabolites and we focus on a smaller set of metabolites (only those that were significant in the discovery analysis), we used a more conservative approach, that is, Bonferroni correction (in order to only select the metabolites that are most highly statistically significantly associated with the outcome). Separate models were run for each exposure, i.e. BMI, WC, and WHC. In the discovery set, the PC-PR2 method was performed again using residuals of each metabolite in which its association with body size was validated. Also in the discovery set, residuals of each metabolite in which its association with body size was validated were included as multivariate predictors of greater body size in the PLS regression, a multivariate method that achieves dimensionality reduction [29]. The metabolic signatures were predicted in the replication set and correlated to BMI, WC, and WHR in the entire sample as well as separately by sex (Pearson correlation was applied).
Metabolic signatures of greater body sizes and cancer risk
The metabolic signatures of greater body size were predicted in all participants from each nested case-control study using the residuals for each metabolite. Conditional logistic regression models were applied to assess the associations between the metabolic signatures of greater body size and risk for colorectal and endometrial cancers. The multivariable model included adjustment for age at blood collection, fasting status at blood collection, education, smoking status at recruitment, physical activity index, height, and daily intake of energy, red and processed meat products, fish and shellfish, fibre, and alcohol. For colorectal cancer, the models were further adjusted for fibre and calcium intake, while for endometrial cancer the models were further adjusted for menopause status (premenopausal, perimenopausal, and postmenopausal), age at first menstrual period, age at first full-term pregnancy, hormone therapy and oral contraceptive use. In an attempt to investigate whether the metabolic signatures were able to predict cancer risk beyond individuals’ body size, the multivariate models were further adjusted for BMI and residuals of the linear regression of WC on BMI and the residuals of the linear regression of WHR on BMI and WC. We have also further adjusted the models for C-peptide level, a valid marker of insulin secretion, as a possible confounder between the associations of metabolic signatures and cancer risk. Further adjustment for cancer grade and stage was also conducted. Additionally, we investigated the association between the log-transformed anthropometric variables and cancer risk. Correlations between the metabolic signatures and BMI, WC, and WHR among colorectal and endometrial controls were also tested. Finally, sensitivity analyses were performed by excluding cases of cancer diagnosed during the first two years of each participant’s follow-up to minimise/limit reverse causality bias.
Metabolic signatures of greater body size and weight loss
In the intercept pilot intervention, paired t tests were employed to determine which metabolites were significantly altered during the intervention. Changes in metabolite levels post-versus pre-intervention were calculated and were further transformed into residuals of linear models with sex as independent variable. The metabolic signatures of greater body size were predicted in all participants for the Intercept study using the residuals for metabolite changes and described by tertiles of weight loss. This approach allowed us to assess changes in the metabolic signatures as it provides results similar to calculating the difference between the metabolic signature at follow-up and at baseline. Pearson correlations between changes in metabolic signatures of body sizes and percentage of weight loss were assessed. Additionally, linear regression models tested associations between residuals for metabolite changes and weight loss adjusted for age and residuals for anthropometric variables. Statistical tests were two-sided and P values < 0.05 were considered statistically significant. All analyses were performed in Stata 15 and R 3.6.3 statistical software. A flow-diagram with the main methodological steps undertaken in the current study is presented in supplementary material (Additional file 1: Figure S5).
Results
Sociodemographic and behavioural characteristics of participants in each study population are presented in Table 1. The majority of the participants in the EPIC discovery and replication sets were men (~ 70%), while in the colorectal case-control (~ 60%), endometrial case-control (100%), and Intercept (~ 65%) studies, most participants were women. In the EPIC discovery set, around 36% of participants were normal weight and 64% were overweight or obese and similar characteristics were observed in the other EPIC sets. Participants were followed-up by a mean of 10.9 (SD 5.2) and 12.1 (SD 4.9) years in the colorectal and endometrial case-control studies, respectively. Cases and controls showed similar characteristics. In the Intercept pilot intervention, all participants were obese and lost at least 10% of their initial body weight (mean = 13.4 kg).
Table 1.
Sociodemographic, lifestyle, dietary and blood-sampling related characteristics of participants in the study populations
| European Prospective Investigation into Cancer and Nutrition | Intercept* | ||||||
|---|---|---|---|---|---|---|---|
| Discovery | Replication | Colorectal cancer | Endometrial cancer | ||||
| Cases | Controls | Cases | Control | ||||
| (n = 3029) | (n = 1297) | (n = 423) | (n = 423) | (n = 635) | (n = 648) | (n = 17) | |
| Age, mean (SD) | 55.5 (8.1) | 55.8 (8.2) | 55.7 (7.8) | 55.8 (7.9) | 53.96 (7.9) | 53.99 (7.9) | 37.5 (9.7) |
| Sex, n (%) | |||||||
| Women | 964 (31.8) | 431 (33.2) | 257 (60.8) | 253 (59.9) | 635 (100) | 648 (100) | 11 (64.7) |
| Educational level, n (%) | |||||||
| University or higher | 674 (22.2) | 296 (22.8) | 62 (14.6) | 65 (15.3) | 116 (18.2) | 92 (14.2) | 13 (76.4) |
| Body mass index (kg/m2), mean (SD) | 26.45 (3.7) | 26.47 (3.7) | 27.1 (4.5) | 26.51 (3.6) | 28 (5.4) | 25.85 (4.2) | 34.2 (3.8) |
| Categories of BMI, n (%) | |||||||
| Normal weight | 1101 (36.3) | 467 (36.0) | 142 (33.6) | 141 (33.3) | 210 (33.1) | 315 (48.6) | – |
| Overweight | 1456 (48.1) | 619 (47.7) | 182 (43.0) | 216 (51.0) | 226 (35.6) | 239 (36.8) | 1 (5.8) |
| Obese | 472 (15.6) | 211 (16.3) | 99 (23.4) | 66 (15.6) | 199 (31.3) | 94 (14.5) | 16 (94.2) |
| Waist circumference (cm), mean (SD) | 90.7 (12.0) | 90.4 (12.1) | 89.6 (13.3) | 87.8 (11.5) | 85.7 (12.1) | 81.5 (10.5) | 102.8 (8.0) |
| Waist-hip ratio (cm), mean (SD) | 0.89 (0.09) | 0.89 (0.09) | 0.87 (0.09) | 0.86 (0.1) | 0.81 (0.06) | 0.80 (0.06) | 0.86 (0.7) |
| Height (cm), mean (SD) | 169.0 (8.8) | 168.9 (8.9) | 164.3 (9.1) | 163.8 (9.4) | 160.2 (6.6) | 160.2 (6.8) | 170.6 (7.9) |
| Physical activity, n (%) | |||||||
| Inactive | 679 (22.4) | 296 (22.8) | 133 (31.4) | 122 (28.8) | 95 (14.9) | 86 (13.3) | NA |
| Moderately inactive | 1026 (33.9) | 463 (35.7) | 183 (43.2) | 175 (41.4) | 189 (29.7) | 175 (27.0) | NA |
| Moderately active | 670 (22.1) | 278 (21.4) | 63 (14.9) | 71 (16.8) | 18 (2.8) | 13 (2.0) | NA |
| Active | 597 (19.7) | 237 (18.3) | 44 (10.4) | 54 (12.7) | 115 (18.1) | 126 (19.4) | NA |
| Smoking status, n (%) | |||||||
| Never smoker | 1224 (40.4) | 518 (39.9) | 198 (46.8) | 222 (52.5) | 82 (12.9) | 111 (17.1) | NA |
| Current smoker | 703 (23.2) | 299 (23.0) | 90 (21.3) | 77 (18.2) | 417 (65.6) | 396 (61.1) | NA |
| Meat intake (g/d), mean (SD) | 86.5 (55.2) | 88.9 (59.5) | 76.7 (81.7) | 77.6 (44.4) | 634.7 (378.8) | 646.2 (376.4) | NA |
| Fish intake (g/d), mean (SD) | 37.1 (37.2) | 35.3 (33.7) | 36.3 (32.4) | 37.9 (36.5) | 523.9 (316.5) | 509.0 (312.9) | NA |
| Fibre intake (g/d), mean (SD) | 23.7 (8.1) | 23.3 (7.8) | 22.9 (7.6) | 23.3 (8.0) | NA | ||
| Alcohol intake (g/d), mean (SD) | 3.1 (1.3) | 3.1 (1.27) | 2.9 (1.4) | 2.7 (1.3) | 3.1 (1.2) | 3.1 (1.2) | NA |
| Energy intake (g/d), mean (SD) | 2253 (654.7) | 2255 (643.0) | 2221 (829.0) | 2187 (627.9) | 661 (383.9) | 667 (380.1) | NA |
| Fasting status, n (%) | |||||||
| Non-fasting | 1351 (44.6) | 601 (46.3) | 23 (5.4) | 23 (5.4) | 99 (15.6) | 105 (16.2) | 17 (100) |
| Sub-study, n (%) | |||||||
| Breast cancer controls | 752 (24.8) | 332 (25.6) | n/a | n/a | n/a | n/a | n/a |
| Kidney cancer controls | 346 (11.4) | 134 (10.3) | n/a | n/a | n/a | n/a | n/a |
| Hepatobiliary cancer controls | 151 (4.9) | 65 (5.0) | n/a | n/a | n/a | n/a | n/a |
| Prostate cancer controls | 1780 (58.7) | 766 (59.0) | n/a | n/a | n/a | n/a | n/a |
*Intercept pilot weight loss intervention. NA, not available; n/a, not applicable
Metabolic signatures of greater body sizes
In the discovery set, BMI, WC, and WHR were statistically significantly (after FDR-adjustment; degrees of freedom = 2949) associated with levels of 89, 94, and 75 metabolites, respectively (Fig. 1). In the replication phase, after Bonferroni correction, 47% (N = 42), 42% (N = 40), and 21% (N = 16) of these metabolites were associated with BMI, WC, and WHR, respectively (Additional file 1: Table S2). As shown in Table 2, a total of 16 metabolites were associated with all three measures, and 23 were associated with both BMI and WC. Considering the metabolites with the strongest association with the three anthropometric measures, higher BMI, WC, and WHR were associated with higher levels of valine, phosphatidylcholine diacyl (PC aa) C38:3, and lower levels of lysophosphatidylcholine acyl (LysoPC) C18:2. PC-PR2 was performed using residuals of each metabolite in which its association with body size was validated and showed that the anthropometric variables explained over 3% of their total variability (Additional file 1: Figure S6).
Fig. 1.
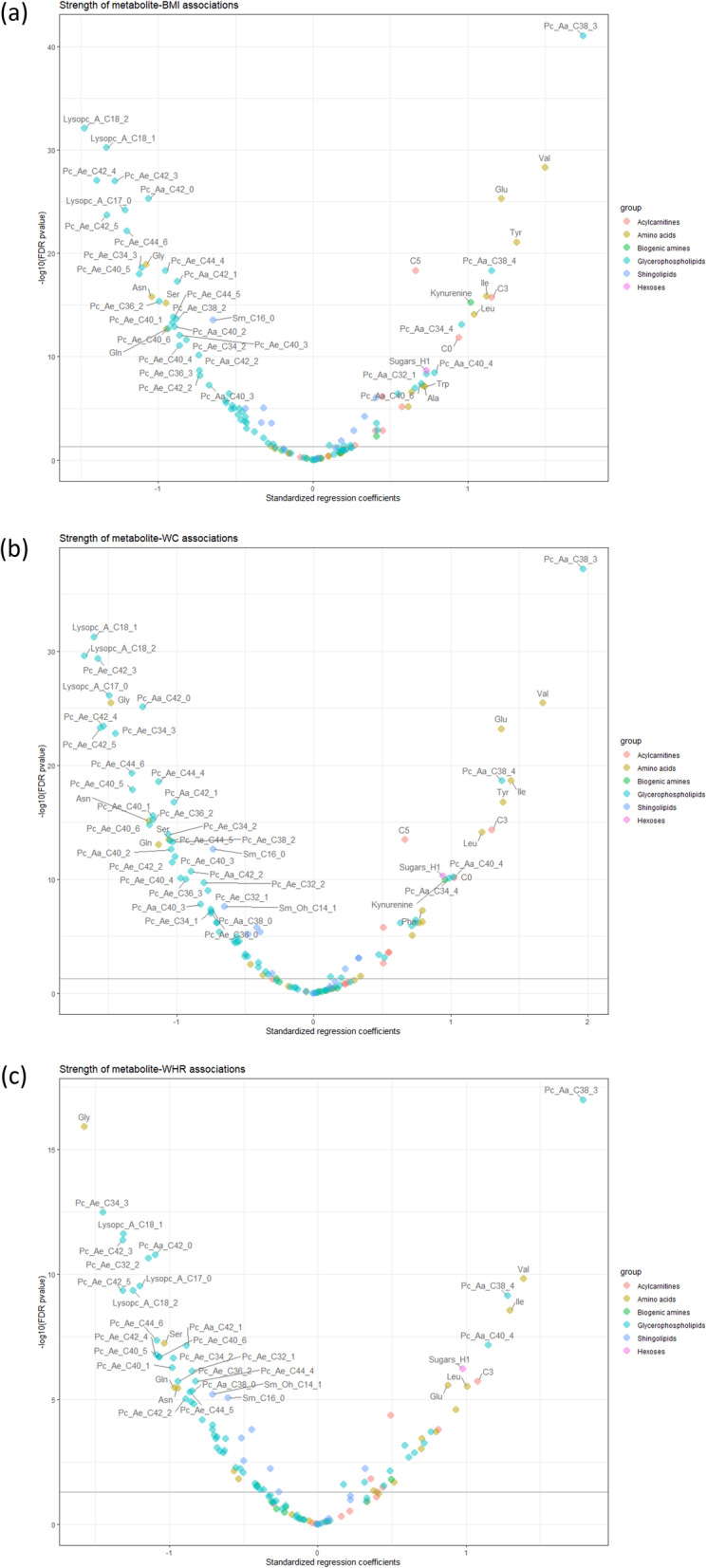
Smile plot with associations between metabolites with BMI, WC and WHR in the discovery set. a BMI, b WC, and c WHR. Smile plot with FDR (false discovery rate method) q values. Analysis using residuals from Z and Log transformed metabolites with fixed effect for country and sex and random effect for batches nested within studies. Models were adjusted for age at blood collection, fasting status at blood collection, smoking status at recruitment, Cambridge physical activity index, height, and daily intake of energy, red and processed meat, fish and shellfish, fibre, and alcohol. The metabolites above the horizontal line showed a significant association with the anthropometric measure (p < 0.05)
Table 2.
Metabolites significantly associated with each anthropometric variable in the discovery and replication sets
| N | Metabolites | Association with: | |||||
|---|---|---|---|---|---|---|---|
| BMI | WC | WHR | |||||
| β (95% CI) | p* | β (95% CI) | p* | β (95% CI) | p* | ||
| 1 | Asparagine | − 1.02 (− 1.25; − 0.79) | < .001 | − 1.11 (− 1.38; − 0.84) | < .001 | − 0.80 (− 1.15; − 0.45) | < .001 |
| 2 | Glutamine | − 0.83 (− 1.05; − 0.60) | < .001 | − 0.92 (− 1.18; − 0.65) | < .001 | − 0.69 (− 1.03; − 0.35) | < .001 |
| 3 | Glutamate | 1.03 (0.83; 1.23) | < .001 | 1.08 (0.84; 1.32) | < .001 | 0.64 (0.33; 0.95) | < .001 |
| 4 | Glycine | − 1.01 (− 1.24; − 0.79) | < .001 | − 1.31 (− 1.57; − 1.04) | < .001 | − 1.40 (− 1.74; − 1.05) | < .001 |
| 5 | Isoleucine | 1.04 (0.80; 1.27) | < .001 | 1.24 (0.95; 1.52) | < .001 | 1.03 (0.66; 1.39) | < .001 |
| 6 | Serine | − 0.93 (− 1.15; − 0.71) | < .001 | − 1.02 (− 1.29; − 0.76) | < .001 | − 1.04 (− 1.38; − 0.69) | < .001 |
| 7 | Valine | 1.35 (1.12; 1.58) | < .001 | 1.49 (1.22; 1.77) | < .001 | 1.17 (0.82; 1.53) | < .001 |
| 8 | LysoPC a C17:0 | − 1.14 (− 1.35; − 0.92) | < .001 | − 1.31 (− 1.56; − 1.05) | < .001 | − 0.82 (− 1.15; − 0.48) | < .001 |
| 9 | LysoPC a C18:1 | −1.36 (− 1.57; − 1.16) | < .001 | − 1.51 (− 1.75; − 1.27) | < .001 | − 0.97 (− 1.28; − 0.65) | < .001 |
| 10 | LysoPC a C18:2 | − 1.47 (− 1.68; − 1.25) | < .001 | − 1.59 (− 1.85; − 1.33) | < .001 | − 1.00 (− 1.34; − 0.66) | < .001 |
| 11 | PC aa C38:3 | 1.69 (1.46; 1.92) | < .001 | 1.85 (1.57; 2.12) | < .001 | 1.62 (1.27; 1.98) | < .001 |
| 12 | PC aa C38:4 | 1.06 (0.83; 1.30) | < .001 | 1.26 (0.98; 1.54) | < .001 | 1.20 (0.84; 1.56) | < .001 |
| 13 | PC aa C42:0 | − 0.22 (− 0.27; − 0.17) | < .001 | − 0.26 (− 0.32; − 0.20) | < .001 | − 0.23 (− 0.31; − 0.15) | < .001 |
| 14 | PC ae C34:3 | − 1.26 (− 1.49; − 1.03) | < .001 | − 1.7 (− 1.97; − 1.42) | < .001 | − 1.77 (− 2.12; − 1.42) | < .001 |
| 15 | PC ae C40:5 | − 1.06 (− 1.29; − 0.83) | < .001 | − 1.24 (− 1.51; − 0.97) | < .001 | − 0.99 (− 1.34; − 0.64) | < .001 |
| 16 | PC ae C42:5 | − 1.13 (− 1.35; − 0.91) | < .001 | − 1.34 (− 1.6; − 1.08) | < .001 | − 1.14 (− 1.48; − 0.8) | < .001 |
| 17 | Acylcarnitine C0 | 0.85 (0.61; 1.09) | < .001 | 0.92 (0.64; 1.21) | < .001 | – | – |
| 18 | Acylcarnitine C3 | 0.27 (0.14; 0.41) | < .001 | 0.34 (0.18; 0.49) | < .001 | – | – |
| 19 | Acylcarnitine C5 | 0.69 (0.51; 0.86) | < .001 | 0.63 (0.42; 0.83) | < .001 | – | – |
| 20 | Leucine | 1.09 (0.85; 1.33) | < .001 | 1.23 (0.95; 1.52) | < .001 | – | – |
| 21 | Phenylalanine | 0.78 (0.55; 1.01) | < .001 | 0.88 (0.61; 1.15) | < .001 | – | – |
| 22 | Tyrosine | 1.23 (0.98; 1.47) | < .001 | 1.22 (0.92; 1.51) | < .001 | – | – |
| 23 | Kynurenine | 1.10 (0.87; 1.33) | < .001 | 0.96 (0.68; 1.23) | < .001 | – | – |
| 24 | PC aa C32:1 | 0.58 (0.36; 0.81) | < .001 | 0.46 (0.20; 0.73) | 0.001 | – | – |
| 25 | PC aa C34:4 | 0.57 (0.32; 0.81) | <. 001 | 0.44 (0.15; 0.73) | 0.004 | – | – |
| 26 | PC aa C38:0 | − 0.46 (− 0.68; − 0.24) | <. 001 | − 0.62 (− 0.88; − 0.36) | < .001 | – | – |
| 27 | PC aa C40:2 | − 0.11 (− 0.17; − 0.05) | < .001 | − 0.14 (− 0.21; − 0.08) | < .001 | – | – |
| 28 | PC aa C40:4 | 0.61 (0.36; 0.85) | < .001 | 0.78 (0.50; 1.07) | < .001 | – | – |
| 29 | PC aa C40:6 | 0.73 (0.49; 0.96) | < .001 | 0.76 (0.49; 1.04) | < .001 | – | – |
| 30 | PC aa C42:1 | − 0.12 (− 0.15; − 0.09) | < .001 | − 0.15 (− 0.18; − 0.11) | < .001 | – | – |
| 31 | PC aa C42:2 | − 0.56 (− 0.72; − 0.40) | < .001 | − 0.61 (− 0.80; − 0.42) | < .001 | – | – |
| 32 | PC ae C32:1 | − 0.57 (− 0.78; − 0.37) | < .001 | − 0.9 (− 1.14; − 0.66) | < .001 | – | – |
| 33 | PC ae C34:2 | − 0.92 (− 1.14; − 0.71) | < .001 | − 1.24 (− 1.49; − 0.98) | < .001 | – | – |
| 34 | PC ae C36:2 | − 1.12 (− 1.35; − 0.90) | < .001 | − 1.32 (− 1.59; − 1.05) | < .001 | – | – |
| 35 | PC ae C36:3 | −0.87 (− 1.10; − 0.65) | < .001 | − 1.15 (− 1.42; − 0.88) | < .001 | – | – |
| 36 | PC ae C38:2 | − 1.05 (− 1.27; − 0.82) | < .001 | − 1.19 (− 1.46; − 0.92) | < .001 | – | – |
| 37 | PC ae C40:6 | − 0.89 (− 1.12; − 0.65) | < .001 | − 1.11 (− 1.38; − 0.83) | < .001 | – | – |
| 38 | PC ae C44:4 | − 1.05 (− 1.24; − 0.86) | < .001 | − 1.11 (− 1.34; − 0.89) | < .001 | – | – |
| 39 | SM C16:0 | − 0.58 (− 0.74; − 0.42) | < .001 | − 0.64 (− 0.83; − 0.45) | < .001 | – | – |
| 40 | PC ae C44:6 | − 0.64 (− 0.87; − 0.42) | < .001 | – | – | – | – |
| 41 | SM C18:1 | 0.49 (0.34; 0.64) | < .001 | – | – | – | – |
| 42 | Hexoses | 0.65 (0.44; 0.87) | < .001 | – | – | – | – |
| 43 | PC aa C32:2 | – | – | 0.45 (0.19; 0.72) | 0.002 | – | – |
Analysis using residuals from Z and Log transformed metabolites with fixed effect for country and sex and random effect for batches nested within study. The multivariable model included additional adjustment for height, physical activity, smoking status, education level, alcohol consumption, dietary intakes of total energy, red and processed meats, fish and shellfish, and fibre, age at blood collection and fasting status. *P value refers to FDR correction
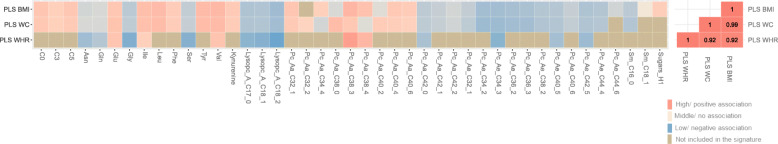
PLS regression analysis defined metabolic signatures of BMI, WC, and WHR (Fig. 2). The metabolites with the greatest contribution to the metabolic signature of BMI were PC aa C38:3 (loading = 0.19), valine (loading = 0.18), tyrosine (loading = 0.17), lysoPC C18:2 (loading = − 0.25), phosphatidylcholine acyl-alkyl (PC ae) C34:2 (loading = − 0.25), and PC ae C34:3 (loading = − 0.25). Similar results were observed for the metabolic signatures of the other anthropometric measures (Additional file 1: Table S3). In the replication set, the metabolic signatures of greater body sizes showed a moderate Pearson correlation with BMI (r = 0.48, 95% CI 0.44–0.53, p < 0.001), WC (r = 0.39, 95% CI 0.34–0.43, p < 0.001), and WHR (r = 0.28, 95% CI 0.24–0.33, p < 0.001). Similar correlations were found among CRC and endometrial cancer controls and for men and women (Additional file 1: Table S4). Additionally, the metabolic signatures of BMI, WC, and WHR explained ~ 23%, ~ 16%, and ~ 8% of the variability of their respective anthropometric variable, among CRC and endometrial cancer controls.
Fig. 2.
Pearson correlation between the PLS scores of BMI, WC, and WHR and their loadings
Metabolic signatures of greater body size and cancer risk
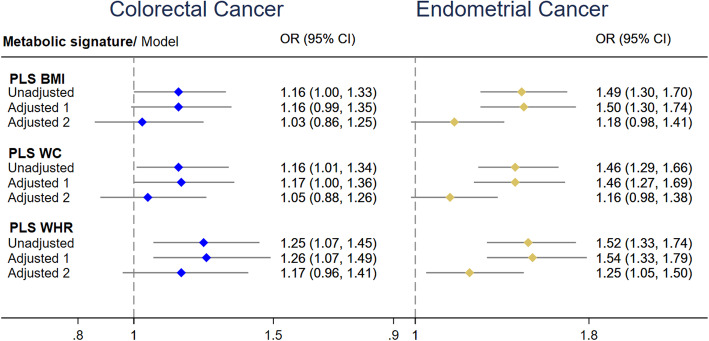
As shown in Fig. 3, endometrial cancer was positively associated with the metabolic signature of BMI (odds ratio per 1-standard deviation [OR1-sd] 1.50, 95% CI 1.30–1.74), WC (OR1-sd 1.46, 95% CI 1.27–1.69), and WHR (OR1-sd 1.54, 95% CI 1.33–1.79) after adjustment for the main confounders. Similarly, colorectal cancer was positively associated with the metabolic signature of WHR (OR1-sd: 1.26, 95% CI 1.07–1.49), but borderline statistically significantly with the metabolic signatures of BMI (OR1-sd: 1.16, 95% CI 0.99–1.35) and WC (OR1-sd: 1.17, 95% CI 1.00–1.36). The association between endometrial cancer and the metabolomic signature of WHR remained statistically significant even after adjustment for anthropometric measures of obesity. We also tested the effect of further adjusting the models for C-peptide and cancer grade and staging; however, the results remained similar to those without these adjustments (Additional file 1: Table S5). Similar results were also found when analyses were repeated removing the cancer cases reported in the first two years of follow-up (results not shown).
Fig. 3.
Association of colorectal and endometrial cancers with the metabolic signatures of obesity. ORs and 95% CIs by 1-SD change. Adjusted model 1 was adjusted for height, physical activity, smoking status, education level, consumption of alcohol, total energy, red and processed meats, fish and shellfish, age at blood collection, and fasting status. For endometrial cancer, model 1 was further adjusted for menopause status, hormonal therapy, oral contraceptive use, age at first menstrual period, and age at first full-term pregnancy, while for colorectal cancer, model 1 was further adjusted for fibre and calcium intake. Model 2 included the adjustments from model 1 plus anthropometric measures
Metabolic signatures of greater body size and weight loss
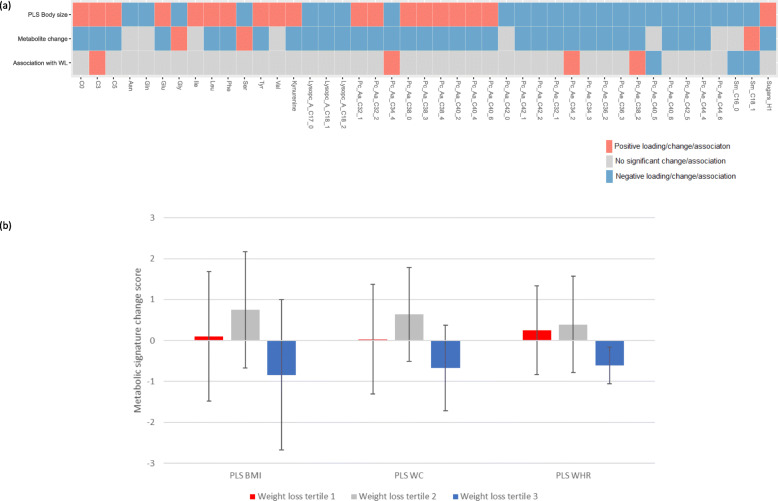
The percentage of weight loss was positively correlated with changes in the metabolic signatures of BMI (r = 0.5, 95% CI 0.06–0.94, p = 0.03), WC (r = 0.5, 95% CI 0.05–0.94, p = 0.03), and WHR (r = 0.6, 95% CI 0.32–0.87, p = 0.01). Participants in the third tertile of weight loss (mean = 16.5 kg) showed the greatest reduction of the PLS scores for BMI, WC, and WHR compared to those in the first (mean = 10.6 kg) and second (mean = 12.7 kg) tertiles (Fig. 4). There were statistically significant changes in 105 measured metabolites during the intervention (Additional file 1: Table S6). As shown in Fig. 4, there were significant decreases in the serum levels of acylcarnitines C0, C3, C5, glutamate, leucine, phenylalanine, tyrosine, kynurenine, PCs aa C32:1, C32:2, C38:0, C38:3, C38:4, C40:2, C40:4, C40:6, and hexoses and an increase in the serum levels of glycine, serine, and sphingomyelin C18:1 following the intervention. All these changes were consistent with the metabolic signatures of greater body sizes. However, the association between changes in these metabolites and weight loss were mainly not significant (Additional file 1: Table S7 and Figure S7), which may reflect the small sample size (N = 17).
Fig. 4.
Metabolomics signatures of obesity, metabolite changes, and weight loss in the Intercept pilot intervention. a Comparison of the loadings for the metabolomics signatures of greater body sizes, changes in metabolites, and association between changes in metabolites and weight loss. b Means and standard deviations for metabolomics signatures of greater body sizes by tertiles of weight loss
Measured anthropometry and cancer risk
Analysis of the association between the three measures of obesity and cancer risk showed that greater BMI and WC were significantly associated with increased risk of endometrial and colorectal cancers, while greater WHR was only associated with increased risk of colorectal cancer (Additional file 1: Figure S8). When the models were further adjusted to their respective metabolic signature, all associations were attenuated, suggesting a partial mediating role of the metabolic signatures in these relationships (Additional file 1: Figure S8).
Discussion
In this analysis which used data from a large prospective cohort, we identified metabolic signatures of BMI, WC, and WHR that were positively associated with colorectal and endometrial cancer risk. Further, the metabolomics signature of WHR predicted endometrial cancer risk beyond measured body fatness. In an exploratory analysis using data from a diet-induced weight loss intervention study, we found a positive association between weight loss and changes in the metabolic signatures of greater body sizes.
The metabolic signature of greater BMI, WC, and WHR was represented by higher levels of valine, isoleucine, glutamate, PC aa C38:3, PC aa C38:4 and lower levels of asparagine, glutamine, glycine, serine, lysoPC C17:0, lysoPC C18:1 and lysoPC C18:2, PC aa C42:0, PC ae C34:3, PC ae C40:5, and PC ae C42:5. Other metabolites were also shown to be relevant for the metabolic signatures of BMI and WC only, including leucine, phenylalanine, tyrosine, kynurenine, C0, C3, C5, SM C16:0, SM C18:1, and 17 glycerophospholipids. Additionally, many of these metabolites changed significantly after the Intercept weight loss intervention, reinforcing their association with body weight.
In line with previous studies, the current analysis showed associations between greater body size and the amino acids valine, leucine, isoleucine, tyrosine, glutamate, and the biogenic amine kynurenine [10, 11, 13]. However, this study also provided strong evidence for the associations between body weight and the amino acids phenylalanine, asparagine, glutamine, and glycine in EPIC, which has been supported by other metabolomics studies [30–32]. Systematic reviews of case-control studies examining the association of serum concentrations of metabolites and cancer diagnosis reported that tyrosine and phenylalanine are associated with both colorectal and endometrial cancers, and valine and glutamate with endometrial cancer only [33, 34]. These amino acids have been reported to be associated with insulin resistance and impaired insulin secretion [35], key factors of cancer and obesity pathogenesis [36, 37]. Valine, as well as other branched chain amino acids (BCAAs), also plays an important role in activating the mechanistic/mammalian target of rapamycin (mTOR) axis, a signalling pathway associated with cell growth, proliferation, and survival [38], important features of cancer development. Imbalances in the biogenic amine kynurenine metabolism have also been implicated in cancer development [39, 40].
Consistent with other studies, the metabolic signature of obesity reflected lipid dysregulation [10, 11, 30, 41, 42], such as higher levels of diacyl PCs and lower levels of acyl-alkyl PCs and lysoPCs. Some of these lipid alterations have also been associated with cancer risk, for example, LysoPC C18:1 and C18:2 have been reported to be downregulated in colorectal cancer patients [43]. LysoPCs are important cell-signalling molecules and their downregulation may reflect pathophysiological changes in cancer development [43]. Recent prospective studies have also reported lower levels of PC ae C34:2, C36:2, C36:3, and C38:2 to be associated with breast cancer risk [8]. The acyl-alkyl PCs seem to have antioxidant properties and when downregulated may increase reactive oxygen species (ROS) generation, promoting oxidative stress and oncogenic DNA defects [44]. Increased ROS has also been linked to many metabolic alterations, such as insulin resistance [45], decreases in adiponectin, and increased expression of pro-inflammatory cytokines including TNFα and IL-6 [46], all potential markers of obesity and cancer development. Regarding the alterations in diacyl PCs, the metabolites PC aa 32:1 and PC aa C38:3 are of particular interest as they have been associated with diabetes and cardiovascular diseases [47, 48], although not yet to cancer. The role of these lipid metabolites is still not clear and remains to be further investigated.
The metabolic signature of greater WHR included metabolites that were also significantly associated with BMI and WC and may reflect overall greater body size. Importantly, it was positively associated with endometrial cancer risk regardless of the individual’s body fatness. This suggests the metabolic signature of WHR was potentially able to differentiate individuals with similar body size but different metabolic health status. These results corroborate other research studies indicating that metabolic alterations which typically accompany obesity, such as insulin resistance and hyperinsulinemia, may be more relevant risk factors for some cancers than adiposity per se [49–53].
Exploratory analysis of data from the Intercept study showed that changes in the metabolic signatures of greater body sizes were positively correlated with the percentage of weight loss. Consistent with previous intervention studies [31, 54], the Intercept weight loss intervention promoted reductions in levels of amino acids and biogenic amines that have been consistently positively associated with obesity and cancer risk, such as tyrosine, phenylalanine, glutamate, and kynurenine, suggesting reduction in cancer risk in individuals with obesity who lose weight. The intervention also decreased levels of PC aa C38:3, PC aa C38:4, and increased levels of glycine and serine, metabolites that have been strongly associated with greater body sizes and that may also be potentially linked to cancer risk. In support of this, primary results from Intercept showed that weight loss in individuals with obesity was associated with improvement in insulin levels and reduction in Ki-67 expression in colorectal tissue, an established marker of cell proliferation [16].
To our knowledge, this is one of the largest studies examining the associations of greater body size and metabolic profiles, and the first to relate these signatures to colorectal and endometrial cancer risk. Strengths of this study include the assessment of numerous behavioural factors and anthropometric measures in EPIC, allowing us to conduct a comprehensive analysis of the associations between greater body sizes and metabolites and to control for potential confounding factors. Additionally, in the current study, we were able to validate the association of metabolites and greater body sizes in a replication set. The analyses of the association of changes in the metabolic signatures of greater body sizes and weight loss in the Intercept intervention study are also novel. However, since anthropometric variables have been previously associated to cancer risk in EPIC, an external validation of the metabolic signatures is needed. Differences in sex distribution and fasting status between the samples may have also affected the results. Another potential limitation of this study was the use of targeted metabolomics data only, in which a set of metabolites that are known a priori are measured. Untargeted metabolomics could provide a more comprehensive view of the metabolic perturbations associated with obesity and cancer; however, such data are currently not available for this cohort. Nevertheless, the findings from the current targeted analysis provide some interesting clues as to the specific metabolic perturbations that accompany obesity and that could be associated with cancer development. The small sample size and exploratory nature of the Intercept intervention study are also limitations. Additionally, the Intercept sample was composed mainly of women, younger, and relatively healthy participants with obesity, and it lacked a control group and physical activity measures. Therefore, replication of the results of this study within a large randomised controlled trial is needed. In addition, the EPIC colorectal case-control sample was relatively small, limiting the possibilities of conducting stratified analysis by sex and cancer subsite. An additional limitation could be that metabolites were assayed in plasma samples of EPIC participants and but in serum samples of Intercept participants. Furthermore, despite the prospective design of EPIC and Intercept, we cannot rule out potential reverse causation since some metabolites may have been altered by underlying subclinical carcinogenic processes. To tackle this issue, we conducted sensitivity analyses excluding cases recorded in the first 2 years of follow-up, and similar results were obtained. Finally, although we excluded participants with diagnosed diabetes, we lacked data on other comorbidities such as hypertension and cardiovascular disease which may have impacted the assayed metabolic pathways and which might represent potential confounding factors. Future studies should aim to better understand the impact of such comorbidities on the metabolic disturbances underlying the association between obesity and cancer.
Conclusions
Obesity is associated with a distinct metabolic signature comprising changes in levels of specific amino acids and lipids which is positively associated with both colorectal and endometrial cancer and is potentially reversible following weight loss. These findings may offer insights into the pathophysiological mechanisms underlying the obesity-cancer relation. Further, by measuring a specific panel of metabolites, it may be possible to identify strata of the population at higher risk for obesity-related cancers. Future studies should aim to further explore the impact of obesity on the metabolome using, for example, untargeted metabolomics which could uncover additional pathways that may be relevant for cancer development.
Supplementary Information
Additional file 1: Supplementary information. Table S1. Number of metabolites included in each study population; Table S2. Metabolites associated with each anthropometric variable in the replication set; Table S3. Metabolites with the greatest contribution for each metabolic signature; Table S4. Correlations between the metabolic signatures and their corresponding anthropometric measure; Table S5. Metabolites that changed significantly from baseline to follow-up in the Intercept; Table S6. Metabolites significantly associated with weight loss in the Intercept; Table S7. Metabolites significantly associated with weight loss in the Intercept; Figure S1. Overall Rpartial2 and weighted Rpartial2 for each covariate and BMI in the discovery set; Figure S2. Overall Rpartial2 and weighted Rpartial2 for each covariate and WC in the discovery set; Figure S3. Overall Rpartial2 and weighted Rpartial2 for each covariate and WHR in the discovery set; Figure S4. Overall Rpartial2 and weighted Rpartial2 for each covariate and weight loss in the Intercept; Figure S5. Association of colorectal and endometrial cancers with the metabolic signatures further adjusted to C-peptide; Figure S6. Association of colorectal and endometrial cancers with the anthropometric measures of obesity; Figure S7. Smile plot of the associations between metabolites with weight loss in the Intercept.
Acknowledgements
The authors would like to thank the EPIC and Intercept study participants and staff for their valuable contribution to this research. The authors would also like to thank Mr. Bertrand Hemon for his support in preparing the databases.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Abbreviations
- BMI
Body mass index
- EPIC
European Prospective Investigation into Cancer and Nutrition
- HCC
Hepatocellular carcinoma risk
- LOD
Limit of detection
- LOQ
Limit of quantification
- LysoPC
Lysophosphatidylcholine acyl
- PC aa
Phosphatidylcholine diacyl
- PC ae
Phosphatidylcholine acyl-alkyl
- WC
Waist circumference
- WHR
Waist-to-hip ratio
Authors’ contributions
MJG, AT, RK, MBS, SP, and MJ (Johansson) are principal investigators of EPIC centres and are responsible for the ongoing follow-up and management of the EPIC cohort. The Intercept pilot study was conceptualised and designed by the late Professor Jane Wardle. RJB contributed to the conceptualisation, design, and led the running of Intercept, in collaboration with MJG. The laboratory analyses were performed by AG and DA with assistance from SR and AS. The statistical analyses were performed by NK with assistance from MJG, VV, and PF. This article was written by NK, VV, NM, RJB, JAR, NA, EHVR, JAS, PF, MJG, and KKT, taking into account the comments and suggestions from SR, KBB, CA, AHR, HS, JMH, AT, JH, BBM, AG, DA, KA, MBS, AKH, GM, SP, RK, RTF, BVG, LD, AS, HCK, RCT, MJ (Jenab), and MJ (Johansson). All co-authors had the opportunity to comment on the analysis and interpretation of the findings and approved the final version for publication. All authors read and approved the final manuscript.
Funding
EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany; Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada; PI13/01162 to EPIC-Murcia), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, and the Catalan Institute of Oncology (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (UK). The Intercept study was supported by a grant from Cancer Research UK (C1418/A14133). Meal replacement products were supplied by the Cambridge Weight Plan. RJB is supported by Yorkshire Cancer Research Academic Fellowship funding. Additionally, the work reported in this paper was undertaken during the tenure of a Postdoctoral Fellowship from the International Agency for Research on Cancer to NK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
EPIC data and biospecimens are available for investigators who seek to answer important questions on health and disease in the context of research projects that are consistent with the legal and ethical standard practices of IARC/WHO and the EPIC Centres. The primary responsibility for accessing the data belongs to IARC and the EPIC centres. Access to materials from the EPIC study can be requested by contacting epic@iarc.fr.
Declarations
Ethics approval and consent to participate
All EPIC study participants provided written informed consent and the study was approved by the ethics committee at the International Agency for Research on Cancer (IARC) and the local ethics centres. The Intercept study was approved by the London–Harrow Research Ethics Committee (reference: 13/LO/0080) and all study participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K, et al. Body fatness and cancer - viewpoint of the IARC Working Group. N Engl J Med. 2016;375(8):794–798. doi: 10.1056/NEJMsr1606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson-Stuttard J, Zhou B, Kontis V, Bentham J, Gunter MJ, Ezzati M. Worldwide burden of cancer attributable to diabetes and high body-mass index: a comparative risk assessment. Lancet Diab Endocrinol. 2018;6(6):E6–E15. doi: 10.1016/S2213-8587(18)30150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Flanagan C, Bowers L, Allott EH, Hursting SD. Molecular and metabolic mechanisms underlying the obesity-cancer link. Lyon: International Agency for Research on Cancer; 2018. [PubMed] [Google Scholar]
- 4.Murphy N, Jenab M, Gunter MJ. Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat Rev Gastroenterol Hepatol. 2018;15(11):659–670. doi: 10.1038/s41575-018-0038-1. [DOI] [PubMed] [Google Scholar]
- 5.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35(Database):D521–D5D6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbons H, Brennan L. Metabolomics as a tool in the identification of dietary biomarkers. Proc Nutr Soc. 2017;76(1):42–53. doi: 10.1017/S002966511600032X. [DOI] [PubMed] [Google Scholar]
- 7.Farshidfar F, Weljie AM, Kopciuk KA, Hilsden R, McGregor SE, Buie WD, et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115(7):848–857. doi: 10.1038/bjc.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.His M, Viallon V, Dossus L, Gicquiau A, Achaintre D, Scalbert A, Ferrari P, Romieu I, Onland-Moret NC, Weiderpass E, Dahm CC, Overvad K, Olsen A, Tjønneland A, Fournier A, Rothwell JA, Severi G, Kühn T, Fortner RT, Boeing H, Trichopoulou A, Karakatsani A, Martimianaki G, Masala G, Sieri S, Tumino R, Vineis P, Panico S, van Gils CH, Nøst TH, Sandanger TM, Skeie G, Quirós JR, Agudo A, Sánchez MJ, Amiano P, Huerta JM, Ardanaz E, Schmidt JA, Travis RC, Riboli E, Tsilidis KK, Christakoudi S, Gunter MJ, Rinaldi S. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019;17(1):178. doi: 10.1186/s12916-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troisi J, Sarno L, Landolfi A, Scala G, Martinelli P, Venturella R, di Cello A, Zullo F, Guida M. Metabolomic signature of endometrial cancer. J Proteome Res. 2018;17(2):804–812. doi: 10.1021/acs.jproteome.7b00503. [DOI] [PubMed] [Google Scholar]
- 10.Bachlechner U, Floegel A, Steffen A, Prehn C, Adamski J, Pischon T, et al. Associations of anthropometric markers with serum metabolites using a targeted metabolomics approach: results of the EPIC-potsdam study. Nutr Diabetes. 2016;6(6). 10.1038/nutd.2016.23. [DOI] [PMC free article] [PubMed]
- 11.Carayol M, Leitzmann MF, Ferrari P, Zamora-Ros R, Achaintre D, Stepien M, Schmidt JA, Travis RC, Overvad K, Tjønneland A, Hansen L, Kaaks R, Kühn T, Boeing H, Bachlechner U, Trichopoulou A, Bamia C, Palli D, Agnoli C, Tumino R, Vineis P, Panico S, Quirós JR, Sánchez-Cantalejo E, Huerta JM, Ardanaz E, Arriola L, Agudo A, Nilsson J, Melander O, Bueno-de-Mesquita B, Peeters PH, Wareham N, Khaw KT, Jenab M, Key TJ, Scalbert A, Rinaldi S. Blood metabolic signatures of body mass index: a targeted metabolomics study in the EPIC cohort. J Proteome Res. 2017;16(9):3137–3146. doi: 10.1021/acs.jproteome.6b01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aleksandrova K, Egea Rodrigues C, Floegel A, Ahrens W. Omics biomarkers in obesity: novel etiological insights and targets for precision prevention. Curr Obes Rep. 2020;9(3):219–230. doi: 10.1007/s13679-020-00393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assi N, Thomas DC, Leitzmann M, Stepien M, Chajes V, Philip T, et al. Are metabolic signatures mediating the relationship between lifestyle factors and hepatocellular carcinoma risk? Results from a nested case-control study in EPIC. Cancer Epidemiol Biomarkers. 2018;27(5):531–540. doi: 10.1158/1055-9965.EPI-17-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SH, Crosslin DR, Haynes CS, Nelson S, Turer CB, Stevens RD, Muehlbauer MJ, Wenner BR, Bain JR, Laferrère B, Gorroochurn P, Teixeira J, Brantley PJ, Stevens VJ, Hollis JF, Appel LJ, Lien LF, Batch B, Newgard CB, Svetkey LP. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia. 2012;55(2):321–330. doi: 10.1007/s00125-011-2356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes TI, Geloneze B, Pareja JC, Calixto AR, Ferreira MM, Marsaioli AJ. Blood metabolome changes before and after bariatric surgery: a (1) H NMR-based clinical investigation. Omics. 2015;19(5):318–327. doi: 10.1089/omi.2015.0009. [DOI] [PubMed] [Google Scholar]
- 16.Beeken RJ, Croker H, Heinrich M, Obichere A, Finer N, Murphy N, Goldin R, Guppy NJ, Wilson R, Fisher A, Steptoe A, Gunter MJ, Wardle J. The impact of diet-induced weight loss on biomarkers for colorectal cancer: an exploratory study (INTERCEPT) Obesity. 2017;25:S95–S101. doi: 10.1002/oby.21984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slimani N, Kaaks R, Ferrari P, Casagrande C, Clavel-Chapelon F, Lotze G, Kroke A, Trichopoulos D, Trichopoulou A, Lauria C, Bellegotti M, Ocké MC, Peeters PHM, Engeset D, Lund E, Agudo A, Larrañaga N, Mattisson I, Andren C, Johansson I, Davey G, Welch AA, Overvad K, Tjønneland A, van Staveren W, Saracci R, Riboli E. European Prospective Investigation into Cancer and Nutrition (EPIC) calibration study: rationale, design and population characteristics. Public Health Nutr. 2002;5(6b):1125–1145. doi: 10.1079/PHN2002395. [DOI] [PubMed] [Google Scholar]
- 18.Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, Charrondière UR, Hémon B, Casagrande C, Vignat J, Overvad K, Tjønneland A, Clavel-Chapelon F, Thiébaut A, Wahrendorf J, Boeing H, Trichopoulos D, Trichopoulou A, Vineis P, Palli D, Bueno-de-Mesquita HB, Peeters PHM, Lund E, Engeset D, González CA, Barricarte A, Berglund G, Hallmans G, Day NE, Key TJ, Kaaks R, Saracci R. European prospective investigation into cancer and nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5(6b):1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 19.Klipstein-Grobusch K, Georg T, Boeing H. Interviewer variability in anthropometric measurements and estimates of body composition. Int J Epidemiol. 1997;26(Suppl 1):S174–S180. doi: 10.1093/ije/26.suppl_1.S174. [DOI] [PubMed] [Google Scholar]
- 20.Spencer EA, Appleby PN, Davey GK, Key TJ. Validity of self-reported height and weight in 4808 EPIC-Oxford participants. Public Health Nutr. 2002;5(4):561–565. doi: 10.1079/PHN2001322. [DOI] [PubMed] [Google Scholar]
- 21.Skeie G, Mode N, Henningsen M, Borch KB. Validity of self-reported body mass index among middle-aged participants in the Norwegian Women and Cancer study. Clin Epidemiol. 2015;7:313–323. doi: 10.2147/CLEP.S83839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wareham NJ, Jakes RW, Rennie KL, Schuit J, Mitchell J, Hennings S, Day NE. Validity and repeatability of a simple index derived from the short physical activity questionnaire used in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Public Health Nutr. 2003;6(4):407–413. doi: 10.1079/PHN2002439. [DOI] [PubMed] [Google Scholar]
- 23.Bergmann MM, Bussas U, Boeing H. Follow-up procedures in EPIC-Germany--data quality aspects. European Prospective Investigation into Cancer and Nutrition. Ann Nutr Metab. 1999;43(4):225–234. doi: 10.1159/000012789. [DOI] [PubMed] [Google Scholar]
- 24.Bergmann MM, Noethlings U, Eisinger B, Streller B, Quehl A, Walter D, et al. [The importance of the common cancer registry for the identification of cancer cases in the EPIC Potsdam-study -- results of the first record linkage]. Gesundheitswesen (Bundesverband der Arzte des Offentlichen Gesundheitsdienstes (Germany)). 2004;66(8–9):475–81. [DOI] [PubMed]
- 25.Miller AB, Altenburg HP, Bueno-de-Mesquita B, Boshuizen HC, Agudo A, Berrino F, Gram IT, Janson L, Linseisen J, Overvad K, Rasmuson T, Vineis P, Lukanova A, Allen N, Amiano P, Barricarte A, Berglund G, Boeing H, Clavel-Chapelon F, Day NE, Hallmans G, Lund E, Martinez C, Navarro C, Palli D, Panico S, Peeters PHM, Quirós JR, Tjønneland A, Tumino R, Trichopoulou A, Trichopoulos D, Slimani N, Riboli E. Fruits and vegetables and lung cancer: findings from the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2004;108(2):269–276. doi: 10.1002/ijc.11559. [DOI] [PubMed] [Google Scholar]
- 26.Pischon T, Lahmann PH, Boeing H, Friedenreich C, Norat T, Tjonneland A, et al. Body size and risk of colon and rectal cancer in the European Prospective Investigation Into Cancer and Nutrition (EPIC) J Natl Cancer Inst. 2006;98(13):920–931. doi: 10.1093/jnci/djj246. [DOI] [PubMed] [Google Scholar]
- 27.Jenab M, Riboli E, Cleveland RJ, Norat T, Rinaldi S, Nieters A, Biessy C, Tjønneland A, Olsen A, Overvad K, Grønbæk H, Clavel-Chapelon F, Boutron-Ruault MC, Linseisen J, Boeing H, Pischon T, Trichopoulos D, Oikonomou E, Trichopoulou A, Panico S, Vineis P, Berrino F, Tumino R, Masala G, Peters PH, van Gils CH, Bueno-de-Mesquita HB, Ocké MC, Lund E, Mendez MA, Tormo MJ, Barricarte A, Martínez-García C, Dorronsoro M, Quirós JR, Hallmans G, Palmqvist R, Berglund G, Manjer J, Key T, Allen NE, Bingham S, Khaw KT, Cust A, Kaaks R. Serum C-peptide, IGFBP-1 and IGFBP-2 and risk of colon and rectal cancers in the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2007;121(2):368–376. doi: 10.1002/ijc.22697. [DOI] [PubMed] [Google Scholar]
- 28.Fages A, Ferrari P, Monni S, Dossus L, Floegel A, Mode N, Johansson M, Travis RC, Bamia C, Sánchez-Pérez MJ, Chiodini P, Boshuizen HC, Chadeau-Hyam M, Riboli E, Jenab M, Elena-Herrmann B. Investigating sources of variability in metabolomic data in the EPIC study: the Principal Component Partial R-square (PC-PR2) method. Metabolomics. 2014;10(6):1074–1083. doi: 10.1007/s11306-014-0647-9. [DOI] [Google Scholar]
- 29.Assi N, Fages A, Vineis P, Chadeau-Hyam M, Stepien M, Duarte-Salles T, Byrnes G, Boumaza H, Knüppel S, Kühn T, Palli D, Bamia C, Boshuizen H, Bonet C, Overvad K, Johansson M, Travis R, Gunter MJ, Lund E, Dossus L, Elena-Herrmann B, Riboli E, Jenab M, Viallon V, Ferrari P. A statistical framework to model the meeting-in-the-middle principle using metabolomic data: application to hepatocellular carcinoma in the EPIC study. Mutagenesis. 2015;30(6):743–753. doi: 10.1093/mutage/gev045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JY, Park JY, Kim OY, Ham BM, Kim HJ, Kwon DY, Jang Y, Lee JH. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS) J Proteome Res. 2010;9(9):4368–4375. doi: 10.1021/pr100101p. [DOI] [PubMed] [Google Scholar]
- 31.Tulipani S, Griffin J, Palau-Rodriguez M, Mora-Cubillos X, Bernal-Lopez RM, Tinahones FJ, Corkey BE, Andres-Lacueva C. Metabolomics-guided insights on bariatric surgery versus behavioral interventions for weight loss. Obesity (Silver Spring) 2016;24(12):2451–2466. doi: 10.1002/oby.21686. [DOI] [PubMed] [Google Scholar]
- 32.Park S, Sadanala KC, Kim EK. A metabolomic approach to understanding the metabolic link between obesity and diabetes. Mol Cells. 2015;38(7):587–596. doi: 10.14348/molcells.2015.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffone A, Troisi J, Boccia D, Travaglino A, Capuano G, Insabato L, Mollo A, Guida M, Zullo F. Metabolomics in endometrial cancer diagnosis: a systematic review. Acta Obstet Gynecol Scand. 2020;99(9):1135–1146. doi: 10.1111/aogs.13847. [DOI] [PubMed] [Google Scholar]
- 34.Hashim NAA, Ab-Rahim S, Suddin LS, Saman MSA, Mazlan M. Global serum metabolomics profiling of colorectal cancer. Mol Clin Oncol. 2019;11(1):3–14. doi: 10.3892/mco.2019.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vangipurapu J, Stancakova A, Smith U, Kuusisto J, Laakso M. Nine amino acids are associated with decreased insulin secretion and elevated glucose levels in a 7.4-year follow-up study of 5,181 Finnish men. Diabetes. 2019;68(6):1353–1358. doi: 10.2337/db18-1076. [DOI] [PubMed] [Google Scholar]
- 36.Siddik MAB, Shin AC. Recent progress on branched-chain amino acids in obesity, diabetes, and beyond. Endocrinol Metab. 2019;34(3):234–246. doi: 10.3803/EnM.2019.34.3.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez AV, Pasupuleti V, Benites-Zapata VA, Thota P, Deshpande A, Perez-Lopez FR. Insulin resistance and endometrial cancer risk: a systematic review and meta-analysis. Eur J Cancer. 2015;51(18):2747–2758. doi: 10.1016/j.ejca.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 38.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mezrich JD, Fechner JH, Zhang XJ, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185(6):3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang SC, Fanidi A, Ueland PM, Relton C, Midttun O, Vollset SE, Gunter MJ, Seckl MJ, Travis RC, Wareham N, Trichopoulou A, Lagiou P, Trichopoulos D, Peeters PHM, Bueno-de-Mesquita HB, Boeing H, Wientzek A, Kuehn T, Kaaks R, Tumino R, Agnoli C, Palli D, Naccarati A, Aicua EA, Sánchez MJ, Quirós JR, Chirlaque MD, Agudo A, Johansson M, Grankvist K, Boutron-Ruault MC, Clavel-Chapelon F, Fagherazzi G, Weiderpass E, Riboli E, Brennan PJ, Vineis P, Johansson M. Circulating biomarkers of tryptophan and the kynurenine pathway and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2014;23(3):461–468. doi: 10.1158/1055-9965.EPI-13-0770. [DOI] [PubMed] [Google Scholar]
- 41.Tulipani S, Palau-Rodriguez M, Minarro Alonso A, Cardona F, Marco-Ramell A, Zonja B, et al. Biomarkers of morbid obesity and prediabetes by metabolomic profiling of human discordant phenotypes. Clin Chim Acta. 2016;463:53–61. doi: 10.1016/j.cca.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn T, Floegel A, Sookthai D, Johnson T, Rolle-Kampczyk U, Otto W, et al. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14(1):13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao ZW, Xiao YJ, Elson P, Tan HY, Plummer SJ, Berk M, Aung PP, Lavery IC, Achkar JP, Li L, Casey G, Xu Y. Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25(19):2696–2701. doi: 10.1200/JCO.2006.08.5571. [DOI] [PubMed] [Google Scholar]
- 44.Porporato PE, Filigheddu N, Bravo-San Pedro JM, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28(3):265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urakawa H, Katsuki A, Sumida Y, Gabazza EC, Murashima S, Morioka K, Maruyama N, Kitagawa N, Tanaka T, Hori Y, Nakatani K, Yano Y, Adachi Y. Oxidative stress is associated with adiposity and insulin resistance in men. J Clin Endocrinol Metab. 2003;88(10):4673–4676. doi: 10.1210/jc.2003-030202. [DOI] [PubMed] [Google Scholar]
- 46.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114(12):1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semba RD, Gonzalez-Freire M, Moaddel R, Sun K, Fabbri E, Zhang P, Carlson OD, Khadeer M, Chia CW, Salem N, Jr, Ferrucci L. Altered plasma amino acids and lipids associated with abnormal glucose metabolism and insulin resistance in older adults. J Clin Endocr Metab. 2018;103(9):3331–3339. doi: 10.1210/jc.2018-00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Floegel A, Stefan N, Yu ZH, Muhlenbruch K, Drogan D, Joost HG, Fritsche A, Haring HU, Hrabe de Angelis M, Peters A, Roden M, Prehn C, Wang-Sattler R, Illig T, Schulze MB, Adamski J, Boeing H, Pischon T. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunter MJ, Xie XH, Xue XN, Kabat GC, Rohan TE, Wassertheil-Smoller S, Ho GYF, Wylie-Rosett J, Greco T, Yu H, Beasley J, Strickler HD. Breast cancer risk in metabolically healthy but overweight postmenopausal women. Cancer Res. 2015;75(2):270–274. doi: 10.1158/0008-5472.CAN-14-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy N, Cross AJ, Abubakar M, Jenab M, Aleksandrova K, Boutron-Ruault MC, et al. A nested case-control study of metabolically defined body size phenotypes and risk of colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC) Plos Med. 2016;13(4):e1001988. doi: 10.1371/journal.pmed.1001988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabat GC, Kim MY, Lee JS, Ho GY, Going SB, Beebe-Dimmer J, Manson JAE, Chlebowski RT, Rohan TE. Metabolic obesity phenotypes and risk of breast cancer in postmenopausal women. Cancer Epidemiol Biomark Prev. 2017;26(12):1730–1735. doi: 10.1158/1055-9965.EPI-17-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang XY, Margolis KL, Hendryx M, Rohan TE, Groessl EJ, Thomson CA, Kroenke CH, Simon MS, Lane D, Stefanick M, Luo J. Metabolic phenotype and risk of colorectal cancer in normal-weight postmenopausal women. Cancer Epidemiol Biomark Prev. 2017;26(2):155–161. doi: 10.1158/1055-9965.EPI-16-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore LL, Chadid S, Singer MR, Kreger BE, Denis GV. Metabolic health reduces risk of obesity-related cancer in Framingham study adults. Cancer Epidemiol Biomarkers Prev. 2014;23(10):2057–2065. doi: 10.1158/1055-9965.EPI-14-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rangel-Huerta OD, Pastor-Villaescusa B, Gil A. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics. 2019;15(6):93. doi: 10.1007/s11306-019-1553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary information. Table S1. Number of metabolites included in each study population; Table S2. Metabolites associated with each anthropometric variable in the replication set; Table S3. Metabolites with the greatest contribution for each metabolic signature; Table S4. Correlations between the metabolic signatures and their corresponding anthropometric measure; Table S5. Metabolites that changed significantly from baseline to follow-up in the Intercept; Table S6. Metabolites significantly associated with weight loss in the Intercept; Table S7. Metabolites significantly associated with weight loss in the Intercept; Figure S1. Overall Rpartial2 and weighted Rpartial2 for each covariate and BMI in the discovery set; Figure S2. Overall Rpartial2 and weighted Rpartial2 for each covariate and WC in the discovery set; Figure S3. Overall Rpartial2 and weighted Rpartial2 for each covariate and WHR in the discovery set; Figure S4. Overall Rpartial2 and weighted Rpartial2 for each covariate and weight loss in the Intercept; Figure S5. Association of colorectal and endometrial cancers with the metabolic signatures further adjusted to C-peptide; Figure S6. Association of colorectal and endometrial cancers with the anthropometric measures of obesity; Figure S7. Smile plot of the associations between metabolites with weight loss in the Intercept.
Data Availability Statement
EPIC data and biospecimens are available for investigators who seek to answer important questions on health and disease in the context of research projects that are consistent with the legal and ethical standard practices of IARC/WHO and the EPIC Centres. The primary responsibility for accessing the data belongs to IARC and the EPIC centres. Access to materials from the EPIC study can be requested by contacting epic@iarc.fr.