Abstract
Wastewater-based epidemiology is currently being utilized to monitor the dissemination of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), on a population scale. The detection of SARS-CoV-2 in wastewater is highly influenced by methodologies used for its isolation, concentration and RNA extraction. Although various viral concentration methods are currently employed, including polyethylene glycol (PEG) precipitation, adsorption-extraction, ultracentrifugation and ultrafiltration, to our knowledge, none of these methods have been standardized for use with a variety of wastewater matrices and/or different kits for RNA extraction and quantification. To address this, wastewater with different physical characteristics was seeded with gamma-irradiated SARS-CoV-2 and used to test the efficiency of PEG precipitation and adsorption-extraction to concentrate the virus from three physiochemically different wastewater samples, sourced from three distinct wastewater plants. Efficiency of viral concentration and RNA extraction was assessed by reverse-transcriptase polymerase chain reaction and the recovery yields calculated. As co-purification of inhibitors can be problematic for subsequent detection, two commonly used commercial master mixes were assessed for their sensitivity and efficiency to detect two SARS-CoV-2 target nucleocapsid (N) gene sequences. Recovery rates varied greatly between wastewater matrices and concentration methods, with the highest and most reproducible recovery rates (46.6–56.7%) observed when SARS-CoV-2 was precipitated with PEG and detected by the Luna® Universal master mix. The adsorption-extraction method was less effective (0–21.7%). This study demonstrates that PEG precipitation is the more robust method, which translates well to varying wastewater matrices, producing consistent and reproducible recovery rates. Furthermore, it is compatible with different kits for RNA extraction and quantitation.
Keywords: Wastewater-based epidemiology, COVID-19, Virus recovery rates, Wastewater matrix, Viral extraction method, PCR detection
Graphical abstract
1. Introduction
Approximately a year after the first detection of the coronavirus disease 2019 (COVID-19) over 125 million people worldwide are known to have been infected and deaths due to this disease are rapidly approaching a staggering 3 million (WHO, 2020 - https://covid19.who.int/). COVID-19 is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a single stranded, enveloped RNA zoonotic coronavirus (Abdel-Moneim and Abdelwhab, 2020; Wu et al., 2020a; Zhou et al., 2020).
The ability of SARS-CoV-2 to spread rapidly and the difficulty in containing the virus are the two main factors that differentiate COVID-19 from the two previous coronavirus outbreaks, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), and also from Ebola hemorrhagic fever. With an incubation period of 2 to 14 days (Lauer et al., 2020), SARS-CoV-2 can be spread by asymptomatic or presymptomatic individuals; the only way of diagnosing an infection is by performing a test on clinical specimens. This then must be followed by quarantine and contact tracing. However, in the midst of an outbreak it is often not logistically possible to perform enough tests to sufficiently measure dissemination. Hence, in most countries, initial efforts were focused on individuals that were symptomatic, have travelled or have been in contact with a known case of COVID-19.
Since SARS-CoV-2 can be shed through faeces, wastewater-based epidemiology (WBE) (Daughton, 2020) has been utilized to assess the presence of the virus in wastewater (Aguiar-Oliveira et al., 2020; Ahmed et al., 2021; Gonzalez et al., 2020). It is now being used as a potential warning sign of infections in an area, with the information gathered on a population scale serving to lead to interventions being launched without the need to test every individual in a catchment. This information could be utilized as a predictive tool for immediate intervention, for example to focus clinical screening of a specific geographical area. However, as the virus is usually present in low numbers in wastewater, viral particles require concentration prior to nucleic acid extraction. Various viral concentration methods have been utilized throughout this pandemic to concentrate SARS-CoV-2 from wastewater including polyethylene glycol precipitation (PEG) (Kumar et al., 2020; Perez-Cataluna et al., 2021; Philo et al., 2021; Wu et al., 2020b), a two-phase (PEG-dextran) separation method (La Rosa et al., 2020b), adsorption-extraction using electronegative membranes (Ahmed et al., 2020a; Haramoto et al., 2020), ultracentrifugation (Wurtzer et al., 2020), ultrafiltration (Ahmed et al., 2020a; Medema et al., 2020; Nemudryi et al., 2020; Philo et al., 2021) and aluminum hydroxide adsorption-extraction (Randazzo et al., 2020). Although a comparison of concentration methods has been carried out (Ahmed et al., 2020c; Perez-Cataluna et al., 2021; Philo et al., 2021; Rusiñol et al., 2020), questions remain unanswered. Subtle, but important differences in the methods used, along with different RNA extraction methods has led to significant variations in the final viral yields recovered. Many of the studies early in the pandemic did not utilize actual SARS-CoV-2, instead opting for surrogates (Ahmed et al., 2020c; Randazzo et al., 2020). In addition, upon review of current WBE SARS-CoV-2 studies, it was revealed that very few of these (Kumar et al., 2020; Torii et al., 2021) have reported on the physiochemical properties of the wastewater being analyzed. The wastewater matrix itself is a key component of WBE and can differ greatly depending on the catchment. Differences in organic material, total dissolved solids and pH can influence virus survival and subsequently the detection of the SARS-CoV-2 RNA in wastewater (Geller et al., 2012; Ye et al., 2016). For example, at a pH greater than 6, RNA is more susceptible to alkaline hydrolysis (Wozniak et al., 2020), thus the stability and subsequent detection of viral RNA in highly alkaline wastewater may be compromised. As such, it is imperative that a method is found which is sufficiently robust to overcome matrix variability. In addition, the method must be feasible and non-onerous in its execution, especially during a pandemic when resources are scarce. As concentration is a pivotal step in the ongoing detection of the virus from wastewater, a consensus method is thus essential to maximize viral concentration, minimize viral loss and reduce coprecipitation of inhibitory substances present in wastewater which may interfere with downstream qPCR analysis.
Following concentration, efficient extraction of viral RNA presents the next hurdle in the successful detection of the SARS-CoV-2 virus from wastewater. Even when using commercial kits tailored to the recovery of RNA from wastewater and faecal samples, the recovery rate can vary greatly, depending on the kit. Factors such as bead and buffer composition and wash procedures may affect both the yield and purity of extracted nucleic acids (Petrich et al., 2006). Despite this, reviews of the current SARS-CoV-2 detection methods primarily focus on the viral concentration methods from wastewater but lack a comparison of genomic extraction methods (Farkas et al., 2020; La Rosa et al., 2020a).
Wastewater carries many potential environmental PCR inhibitors. Hence, concentration of viral particles may also lead to the copurification and concentration of inhibitors which can greatly reduce the sensitivity of qPCR. Presence of nearby industry and pollution levels can all impact the survival of the SARS-CoV-2, as well as the type and load of inhibitors found within the water. A large-scale study investigating qPCR inhibitors present in sewage, surface, ground, drinking water and agricultural runoff samples revealed that 34% of 3193 samples investigated would have resulted in false negative or under-represented viral concentrations (Gibson et al., 2012). Studies have also shown that inhibitors may not affect all qPCR primers and probes equivalently and can lead to the over and under representation of certain viral targets within a sample (da Silva AK et al., 2007). Recent reports highlight that not all SARS-CoV-2 genomic targets are equally detected by RT-qPCR (Medema et al., 2020; Randazzo et al., 2020). The authors of these studies suggest using a combination of primer sets against multiple genomic targets are required for valid detection.
This study aimed to establish an optimized, robust method for detecting SARS-CoV-2 in wastewater. Wastewater samples collected from three different regional South Australian treatment plants were examined to account for the physiochemical differences inherent to each sample. Two of these, plant 1 and 2, were used for method development and the third for validation of the optimized method.
A gamma-irradiated SARS-CoV-2, was used to evaluate the effectiveness of the methods trialed in this study. For comparison, we also assessed the suitability of the commercially available AccuPlex™ SARS-CoV-2 reference material. This engineered virus is a non-infectious and replication deficient alphavirus, carrying Centers for Disease Control and Prevention (CDC) and WHO targeted SARS-CoV-2 consensus sequences.
Recovery rates were determined using different combinations and permutations of two concentration/extraction methods, two RNA extraction kits and two commercial RT-qPCR master mixes. From this data, an optimized pipeline for SARS-CoV-2 isolation and detection in a variety of different wastewater matrices was constructed, that is easily adaptable for use in different laboratories and can serve as a potential early warning to supplement and guide clinical screening.
2. Materials and methods
2.1. Collection of wastewater samples
Influent wastewater samples were collected from three different South Australian regional catchments, as part of wastewater testing for the SARS-CoV-2. Site 1 and 3 are mainly commercial and residential catchments, whilst site 2 included light industry. Grab samples were collected three times during the day at plants 1 and 2. The daily samples were combined for each site and frozen before transport to the laboratory. Plant 3 (serving ~10,000 people) was equipped with an auto-sampler, which collected time proportional 24-h composite samples. Samples were frozen before transport to the laboratory.
Wastewater parameters including pH, oxidation-reduction potential (ORP), electrical conductivity (EC), total dissolved solids (TDS) and salinity were measured using a Hanna multiparameter probe (model HI19895, Hanna Instrument, USA). Total suspended solids (TSS) were determined by filtration of 100 ml of wastewater through a pre-weighed 0.2 μm pore size and 47 mm diameter membrane (Millipore, Cork, Ireland), and measured by weighing the recovered solids after being dried at 103 °C for 16 h.
2.2. Materials
A plasmid harboring a cloned SARS-CoV-2 nucleocapsid (N) gene was used as a positive control for all RT-qPCR assays. The AccuPlex™ SARS-CoV-2 reference material (SeraCare, USA), was used as a proxy for the virus. Comprising of a non-replicative recombinant alphavirus, the AccuPlex™ SARS-CoV-2 reference material carries partial SARS-CoV-2 genome sequences from 5 different regions including the ORF1a, RdRp, spike, envelope and nucleocapsid region. Finally, the gamma-irradiated SARS-CoV-2 virus was obtained from The Victorian Infectious Diseases Reference Laboratory (VIDRL) and used throughout this study to evaluate viral concentration and recovery methods.
2.3. Sample preparation
Wastewater samples, determined to be free of the SARS-CoV-2, were used for method development in this study. Verification of wastewater was carried out by viral concentration using both the PEG precipitation and the adsorption-extraction method, followed by RNA extraction and detection by RT-qPCR targeting the N1 and N2 gene segments.
Unseeded wastewater from each plant was initially tested for the presence SARS-CoV-2 using the methods described in this study. Aliquots of 100 ml of wastewater were then seeded with 5 μl (approximately 1 × 104 viral particles) of the gamma-irradiated SARS-CoV-2 virus or 150 μl (equivalent to 750 viral copies) of the AccuPlex™ SARS-CoV-2 reference material and incubated for 2 h with shaking at 4 °C, before being processed by either the PEG or adsorption-extraction method.
2.4. Virus concentration
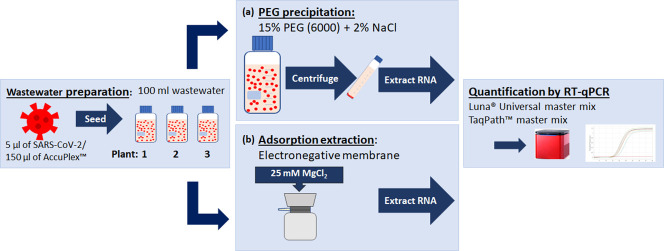
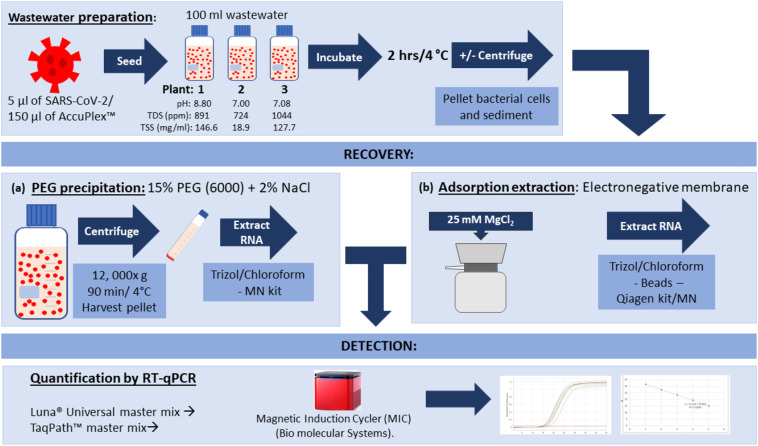
SARS-CoV-2 seeded wastewater samples were used in PEG precipitation of the virus and adsorption-extraction following the methods set out in Fig. 1 .
Fig. 1.
Wastewater (WW) preparation, virus concentration, extraction and detection methods trialed in this study. Of the methods shown, the PEG precipitation protocol was most successful in the recovery of SARS-CoV-2. (Qiagen kit; RNeasy PowerWater, MN kit; NucleoSpin™ RNA Virus).
2.4.1. PEG precipitation
PEG precipitation in combination with NaCl, which acts as a co-precipitant, have been used to purify viruses and act by altering their solubility causing precipitation (Farkas et al., 2018; Fouladvand et al., 2020; Vajda, 1978). This method was used in this study with modification. SARS-CoV-2 seeded wastewater samples of 100 ml were centrifuged at 5000×g for 30 min at 4 °C to pellet bacteria, sediment, and large particles. Virus in the clarified wastewater was precipitated overnight by gentle agitation at 4 °C with 15% of polyethylene glycol 6000 (PEG6000) and 2% NaCl. The precipitated virus was recovered in a pellet by centrifugation at 12,000×g for 90 min at 4 °C. To ensure the collection of viral particles that may have deposited on the side of the tube following the removal of the supernatant, the samples were centrifuged for an additional 5 min in a swinging-bucket rotor at 5000×g. The final pellet was then resuspended in 100 μl of phosphate buffered saline (PBS, pH 7.5).
To assess recovery efficiency of both the liquid and solid phase of the wastewater, the initial centrifugation step was omitted, and PEG precipitation carried out on the entire 100 ml of seeded wastewater. The precipitated virus was then extracted following the steps outlined above and recovery efficiencies compared.
2.4.2. Adsorption-extraction
The adsorption-extraction method, modified from previously described methods (Ahmed et al., 2020c; Ikner et al., 2012) was used to concentrate both the SARS-CoV-2 and the AccuPlex™ reference material from wastewater samples. Negatively charged MF-Millipore membrane filters (cat. no. HAWP-047-00; Millipore, Cork, Ireland), with a 0.45 μm pore size and 47 mm diameter were used with a vacuum pump. To aid adsorption to the electronegative membranes, MgCl2 was added to the wastewater to a final concentration of 25 mM before being filtered. Filters were also pre-treated with 50 ml of 25 mM MgCl2 prior to sample filtration. A volume of 100 ml of SARS-CoV-2 seeded wastewater and MgCl2 was then filtered through the membrane for viral adsorption. Membranes were placed in a 5 ml PowerWater DNA Bead Tube (Qiagen) containing garnet beads.
To evaluate viral recovery rates from the liquid fraction of wastewater, solids were removed by centrifugation at 5000×g for 30 min at 4 °C, prior to filtration. Recoveries from both the total liquid and solid phase of the wastewater and just the liquid phase were then compared.
Finally, as ultrafiltration and ultracentrifugation have also been utilized in the concentration of SARS-CoV-2 from wastewater (Ahmed et al., 2020c; Medema et al., 2020; Wurtzer et al., 2020), they were trialed in this study following the protocol outlined in Table S1.
2.5. RNA extraction
2.5.1. PEG precipitation
Viral RNA was extracted using a combination of the Trizol/phenol extraction method (Chomczynski and Sacchi, 1987) and the NucleoSpin™ RNA Virus (Macherey-Nagel™) extraction kit. Briefly, 1 ml of Trizol (Invitrogen, USA) and 200 μl of chloroform (Sigma, Australia) were added to the resuspended pellet obtained by PEG6000 precipitation. Following a 2-min incubation step, the sample was centrifuged at 12,000 ×g for 15 min at 4 °C. The aqueous phase was transferred to a new microcentrifuge tube and an equal volume of ethanol added. Total RNA binding was then carried out using the NucleoSpin™ RNA Virus column and RNA eluted with 30 μl of RNase-free water.
2.5.2. Adsorption-extraction
Viral RNA was extracted directly from the membrane using a modified Trizol/phenol method in combination with the RNeasy PowerWater (Qiagen) or the NucleoSpin™ RNA Virus (Macherey-Nagel™) kits. Changes to the standard RNeasy PowerWater Kit (Qiagen) protocol, included the addition of 1.4 ml of Trizol instead of Solution 1 and β-mercaptoethanol directly to the 5 ml PowerWater DNA Bead Tube, which was then vortexed at maximum speed for 10 min using the Vortex-Genie® 2 Vortex. Tubes were then centrifuged at 4000×g for 1 min and the viral RNA containing Trizol sample transferred to fresh microcentrifuge tube. Following the addition of chloroform, centrifugation step and addition of ethanol to the aqueous layer (as outlined above), the sample was transferred to a RNeasy PowerWater Kit (Qiagen) Spin Filter or the NucleoSpin™ RNA Virus (Macherey-Nagel™) column for RNA binding and washing. RNA was eluted in 30 μl of RNase-free water.
2.6. RT-qPCR analysis
The primer/probe sets used in this study, as listed by the CDC (USA) (2020), targeted different regions of the nucleocapsid (N) gene; the N1 forward (5’-GAC CCC AAA ATC AGC GAA AT-3′) and reverse (5’-TCT GGT TAC TGC CAG TTG AAT CTG-3′) primers and the N1 probe (6FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1); the N2 forward (5′-TTA CAA ACA TTG GCC GCA AA-3′) and reverse (5’-GCG CGA CAT TCC GAA GAA-3′) primers and the N2 probe (6FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1). Primers and probes were obtained from Sigma (NSW, Australia). RT-qPCR assays for SARS-CoV-2 were performed using the TaqPath™ 1-Step Multiplex Master Mix (No ROX) (Thermo-Fisher Scientific) and the Luna® Universal Probe One-Step RT-qPCR Kit (New England Biolabs). All RT-qPCR assays were performed using a magnetic induction rotor-based platform, the Magnetic Induction Cycler (MIC) (Bio molecular Systems).
-
(i)
TaqPath™ reactions. RT-qPCR assays were performed in a 10 μl qPCR reaction volume containing 2.5 μl of TaqPath™ master mix, 2 μl of RNA template, 400 nM of forward and reverse primer, and 400 nM of FAM-BHQ1 labelled oligonucleotide probe.
The thermal cycling conditions of the qPCR assays were as follows: Initial incubation at 25 °C for 2 min, then 53 °C for 10 min, and 95 °C for 2 min. This was followed by 40 cycles of 3 s at 95 °C, 30 s at 60 °C.
-
(ii)
Luna® Universal reactions. RT-qPCR assays were performed in a 10 μl qPCR reaction volume containing 5 μl of Luna® Universal master mix, 0.5 μl of Luna RT Enzyme Mix, 2 μl of RNA template, 400 nM of forward and reverse primer, and 400 nM of FAM-BHQ1 labelled oligonucleotide probe.
The thermal cycling conditions of the qPCR assays were as follows: Initial incubation at 55 °C for 10 min, then 95 °C for 1 min. This was followed by 40 cycles of 10 s at 95 °C,30 s at 60 °C.
2.7. RT-qPCR inhibition
Potential RT-qPCR inhibition was investigated using RNA extracts obtained from unseeded wastewater, confirmed by RT-qPCR to be free of the SARS-CoV-2, extracted by both the PEG precipitation and the adsorption-extraction method. Genomic material was extracted using a combination of the Trizol/chloroform method and the RNeasy PowerWater Kit (Qiagen) or the NucleoSpin™ RNA Virus (Macherey-Nagel™) kit. Resulting RNA extracts were assayed with and without the addition of the N plasmid control, and the cycle threshold (Ct) scores compared to a sample containing the same volume of the N plasmid control added to RNAse-free water.
2.8. Recovery rate quantification
Gamma irradiated SARS-CoV-2 used throughout this study was supplied at a concentration of Ct 19. However, as the virus was nonviable it was difficult to accurately determine copy numbers. Copy number and recovery rate estimates of the SARS-CoV-2 were thus based on the AccuPlex™ reference material, which came with a known copy number of 5000/ml. Standard curves were generated in triplicate using two-fold serial dilutions of the AccuPlex™ reference material for both the N1 and N2 primer/probe sets, using both the TaqPath™ and Luna® Universal master mixes. Similarly, a two-fold serial dilution of SARS-CoV-2 was also performed in triplicate, starting from approximately 1 × 104 virus particles. The copy number of the SARS-CoV-2 was calculated based on a comparison between the resulting SARS-CoV-2 and AccuPlex™ standard curves.
2.9. Statistical analysis
Data analysis was performed using Microsoft Office 365 ProPlus (Microsoft Corp) and the GraphPad software (Prism 8.3, La Jolla, Ca. USA). A paired t-test applied to two groups of experimental data was used to assess statistical significance for the comparison between the qPCR master mixes, the comparison between methods and kits used an unpaired t-test. Two-tailed p-values were calculated, and a p-value of <0.05 was considered statistically significant.
3. Results
3.1. Wastewater analysis
To ascertain that the PEG based detection method used in this study would be suitable for different wastewater samples without the need for extensive optimization by individual laboratories, influent wastewater samples used for method development were collected from two different regional treatment plants. These plants are operated by different authorities and are also in distinct geographical locations. The physiochemical characteristics of the wastewater samples differed substantially (Table 1 ). Wastewater from plant 1 consistently presented with a higher pH, ranging from 8.8 to 9.3, high visual turbidity and high TDS. Plant 2 wastewater, which carried a minimal load of TDS, presented with low turbidity and ranged in pH from 7 to 7.4.
Table 1.
Physiochemical characteristics of plant 1, 2 and 3 wastewater samples.
| Parameters | Plant 1 | Plant 2 | Plant 3 |
|---|---|---|---|
| pH | 8.8–9.3 | 7.0–7.4 | 7.08 |
| ORP (mV) | 100.2 | 141.9 | 133.4 |
| EC (μS/cm) | 1782 | 1449 | 2008 |
| TDS (ppm) | 891 | 724 | 1044 |
| Salinity (PSU) | 0.9 | 0.72 | 1.06 |
| TSS (mg/ml) | 146.6 | 18.9 | 127.7 |
3.2. Concentration and extraction of SARS-CoV-2 from wastewater samples
Recovery rates of deactivated SARS-CoV-2 from seeded wastewater samples were compared for two different concentration/extraction methods (a) PEG precipitation and, (b) membrane adsorption-extraction (Fig. 1).
3.2.1. PEG precipitation
Variations to the PEG precipitation approach were investigated in this study and the efficiency of this method verified after RNA extraction and detection using either Luna® Universal or TaqPath™ master mix. Significantly higher recovery rates were obtained (p < 0.05) for both plants when the Luna® Universal master mix was employed (avg. 56.7% ± 2.5 for plant 1 and 46.6% ± 7.5 for plant 2) compared to TaqPath™ (avg. 37.2% ± 6.7 for plant 1 and 27.5% ± 5.4 for plant 2) (Fig. 2 ).
Fig. 2.
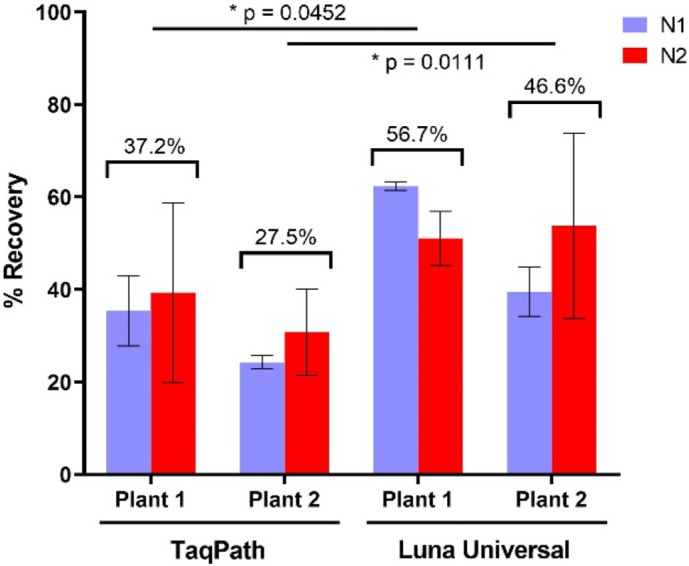
The mean recovery rates of the gamma-irradiated SARS-CoV-2 from wastewater. Virus material was concentrated from wastewater samples using PEG6000 precipitation, followed by extraction using the NucleoSpin™ RNA Virus kit and detection by either the Luna® Universal Probe One-Step RT-qPCR Kit or the TaqPath™ 1-Step Multiplex Master Mix kit as indicated. Values are means of three triplicates and error bars indicate standard deviation. P values for the paired t-test comparing the RT-qPCR master mixes are also presented.
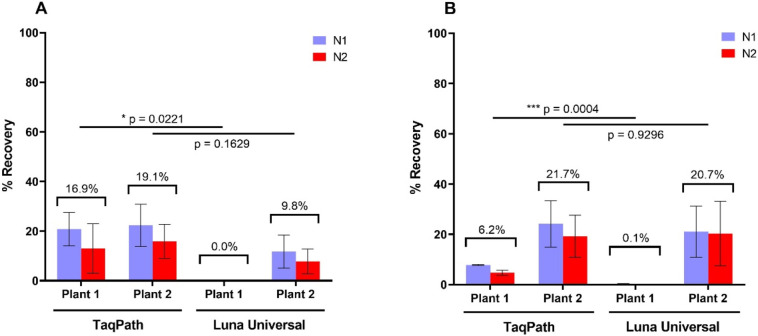
3.2.2. Adsorption-extraction
Initial attempts at recovering the seeded SARS-CoV-2 virus from wastewater in this study, using the adsorption-extraction method resulted in very low recovery rates (< 1.0%). As previously used, the effect of lowering the pH of the wastewater matrices to pH 3.5 on viral adsorption to the membrane was also initially evaluated (Ahmed et al., 2020a). However, this proved to have a negative effect on yield and so was not continued. A crucial consideration to ensure high recovery rates was the addition of Trizol and chloroform as first steps in the extraction protocol (Amsalu et al., 2020). In addition to the NucleoSpin™ RNA Virus kit the RNeasy PowerWater kit, which is designed for RNA extraction from water, was also investigated for RNA extraction efficiency. Different permutations and combinations of RNA extraction kits and PCR polymerase master mixes were investigated using wastewater sourced from both wastewater plants. Recovery rates from plant 2 wastewater showed little variability between kits when coupled with the TaqPath™ master mix (avg. of 20.4%) (Fig. 3A and B). However, a combination of the NucleoSpin™ RNA Virus kit and the Luna® Universal master mix resulted in reduced recovery rates of <10%. Greater variability was observed for plant 1 where <0.1% was detected with the Luna® Universal master mix after extraction using either kit (Fig. 3A and B). This was most likely due to the presence of PCR inhibitors which were co-purified during this extraction protocol. These results demonstrated that the TaqPath™ master mix may be less prone to inhibition as recovery rates of 16.9% for NucleoSpin™ and 6.2% for RNeasy PowerWater kit were observed.
Fig. 3.
The mean recovery rates of the gamma-irradiated SARS-CoV-2 from wastewater. Virus material was concentrated from wastewater samples using the adsorption-extraction method followed by RNA extraction using the (A) NucleoSpin™ MN RNA Virus kit and (B) RNeasy PowerWater Kit. Subsequently, detection of SARS-CoV-2 was performed using the TaqPath™ 1-Step Multiplex Master Mix or the Luna® Universal Probe One-Step RT-qPCR Kit as indicated. Values are means of three triplicates and error bars indicate standard deviation. P values for the paired t-test comparing the RT-qPCR master mixes are also presented.
3.3. Particle associated virus – liquid/solid fraction analysis
As viral association with solids in wastewater has previously been reported (Hejkal et al., 1981; Templeton et al., 2008), PEG precipitated samples are routinely pre-spun to remove solids before precipitating with PEG. Therefore, the possible loss of sample adhered to this solid fraction was investigated by performing PEG precipitation on whole wastewater without the pre-spinning step, as well as extracting RNA directly from the pre-spun pellet. However, low recovery yields (<2%) were observed in either of the solids from pre-spin pellets (Table 2 ). For plant 2 wastewater, SARS-CoV-2 recovery rates were similar when PEG precipitation was carried out on whole wastewater or with the removal of solids (avg. 41.1% whole and 46.6% with pre-spin). However, the SARS-CoV-2 could not be detected from plant 1 wastewater without the inclusion of the pre-spin step. Wastewater from this plant contained a high load of dissolved solids and was found to have a high pH ranging from 8.80 to 9.35.
Table 2.
SARS-CoV-2 recovery rates from the liquid and/or solid wastewater phase following PEG6000 precipitation, adsorption-extraction and viral RNA extraction by the NucleoSpin™ RNA Virus (Macherey-Nagel™).
| Average % (N1 & N2) |
|||
|---|---|---|---|
| TaqPath™ | Luna® Universal | ||
| PEG precipitation | Whole wastewater | ||
| Plant 1 | 8.91 | – | |
| Plant 2 | 8.69 | 41.1 | |
| Supernatant after initial pre-spin | |||
| Plant 1 | 37.65 | 56.7 | |
| Plant 2 | 27.49 | 46.62 | |
| Pellet after initial pre-spin | |||
| Plant 1 | 1.58 | 0.99 | |
| Plant 2 | 0.94 | 0.62 | |
| Adsorption-extraction | Whole wastewater | ||
| Plant 1 | 16.9 | – | |
| Plant 2 | 19.11 | 9.78 | |
| Supernatant after initial pre-spin | |||
| Plant 1 | 0.01 | – | |
| Plant 2 | 0.73 | 0.9 | |
The adsorption-extraction method is routinely performed on whole wastewater without the removal of solids (Schmitz et al., 2016; Sherchan et al., 2020; Tandukar et al., 2020). Here, a pre-spin and removal of the solid fraction led to significantly reduced recovery rates of the virus (Table 2).
3.4. Detection of the AccuPlex™ RNA in wastewater samples
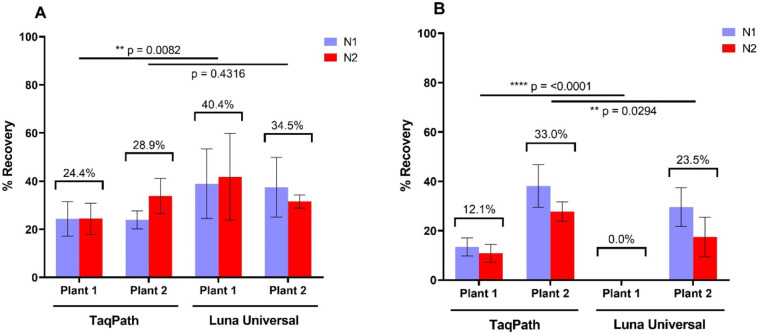
As it can be difficult to obtain deactivated SARS-CoV-2 which can be used as an internal control, confirmation of viral extraction methods using the commercially available AccuPlex™ reference material was investigated. The adsorption-extraction concentration and PEG6000 precipitation methods were thus trialed using AccuPlex™. Following extraction using the Trizol/chloroform adapted viral RNA extraction method along with the same combination of the PCR master mixes trialed for the recovery of the SARS-CoV-2, it was observed that the PEG6000 precipitation method was more efficient at concentrating the alphavirus (Fig. 4A and B). Detection for AccuPlex™ for plant 1 was on avg. 40.4% ± 2.0 and plant 2 avg. 34.5% ± 4.2. Both were lower than the recovery of SARS-CoV-2, with the detection of the reference material being less in plant 1 wastewater (p = 0.0047). This may indicate there are some differences between the recovery of the alphavirus reference material from the wastewater matrices, but overall, the same trend was observed regardless of concentration and detection method. As seen for the SARS-CoV-2 virus, detection by RT-qPCR, revealed that the Luna® Universal master mix resulted in significantly (p < 0.05) higher detection rates as compared to TaqPath™ for plant 1 wastewater.
Fig. 4.
The mean recovery rates of the AccuPlex™ SARS-CoV-2 reference material (SeraCare, USA) from wastewater. The virus reference material was concentrated from wastewater samples using (A) PEG6000 precipitation and (B) the adsorption-extraction method. Viral RNA extraction was carried out using the NucleoSpin™ RNA Virus kit and detection by either the Luna® Universal Probe One-Step RT-qPCR Kit or the TaqPath™ 1-Step Multiplex Master Mix as indicated. Values are means of three triplicates and error bars indicate standard deviation. P values for the paired t-test comparing the RT-qPCR master mixes following virus precipitation/concentration and RNA extraction are also presented.
3.5. Presence of RT-qPCR inhibitors
To evaluate the presence of inhibitors co-purified by RNA extraction, wastewater from plant 1 and 2, verified to be free of the SARS-CoV-2, was used in PEG6000 precipitation and in the adsorption-extraction method. Following extraction, carried out by both kits trialed in this study, a known concentration of the control plasmid was added to each sample, and the concentration of the N1 and N2 targets detected by RT-qPCR using TaqPath™ and the Luna® Universal master mixes. Results generated by PEG6000 precipitation (Table 3 ) showed comparable Ct scores across all samples, regardless of extraction kit or master mix used, when compared to the positive control. In contrast, neither N1 or N2 gene targets could be amplified from plant 1 RNA extracts, following the adsorption-extraction method, extraction with both kits and RT-qPCR using the Luna® Universal master mix (Table 3). This result was consistent with the copurification of PCR inhibitors during filtration. Although possibly containing co-purified inhibitors, the resulting RNA extracts did not appear to affect the performance of TaqPath™, as comparable Ct scores were obtained for all samples. The possible presence of co-purified inhibitors obtained using the adsorption-extraction method may help to explain the difficulties detecting the SARS-CoV-2 and the AccuPlex™ reference material from wastewater sourced from plant 1.
Table 3.
Inhibitor assessment of plant 1 and plant 2 wastewater samples following PEG6000 precipitation, concentration by adsorption-extraction and viral RNA extraction. Ct scores of the N plasmid control in RNAse-free water were used as a measure of no inhibition.
| Ct scores |
|||||
|---|---|---|---|---|---|
| N target/ polymerase | Plant 1 | Plant 2 | Plasmid | ||
| PEG precipitation | MN | N1 TaqPath™ | 23.0 | 23.0 | 22.9 |
| N2 TaqPath™ | 24.6 | 24.5 | 24.6 | ||
| N1 Luna® Universal | 23.0 | 22.8 | 23.0 | ||
| N2 Luna® Universal | 25.2 | 25.4 | 25.7 | ||
| Adsorption-extraction | QK | N1 TaqPath™ | 21.1 | 21.0 | 21.3 |
| N2 TaqPath™ | 21.1 | 21.1 | 20.8 | ||
| N1 Luna® Universal | – | 24.2 | 25.7 | ||
| N2 Luna® Universal | – | 28.2 | 26.2 | ||
| MN | N1 TaqPath™ | 25.3 | 25.5 | 25.0 | |
| N2 TaqPath™ | 25.4 | 25.7 | 25.6 | ||
| N1 Luna® Universal | – | 25.1 | 25.7 | ||
| N2 Luna® Universal | – | 26.0 | 26.1 | ||
QK: RNeasy PowerWater Kit (Qiagen).
MN: NucleoSpin™ RNA Virus (Macherey-Nagel™).
3.6. Comparison of master mix efficiencies
Amplification efficiencies for the TaqPath™ and Luna® Universal master mixes, were based on SARS-CoV-2 standard curves generated for the both the N1 and N2 target sequences, using the amplification efficiency formula (Wong and Medrano, 2005):
The calculated efficiencies were shown to be significantly lower for the TaqPath™ master mix (N1 = 71.5%, N2 = 64.4%) in comparison to the Luna® Universal (N1 = 97.7%, N2 = 97.3%) (Table 4 ).
Table 4.
Comparison of TaqMan™ and Luna® Universal master mix efficiencies.
| Target | Standard curve | R () | Efficiency (%) | |
|---|---|---|---|---|
| TaqPath™ | N1 | y = −4.2691x + 38.113 | 0.998 | 71.5 |
| N2 | y = −4.6344x + 39.56 | 0.991 | 64.4 | |
| Luna® Universal | N1 | y = −3.3791x + 35.243 | 0.996 | 97.7 |
| N2 | y = −3.3885x + 36.321 | 0.996 | 97.3 |
3.7. Application of methods
Since PEG6000 precipitation was found to be the most robust method to reproducibly concentrate SARS-CoV-2 regardless of the extraction kit or PCR master mix used, this method was finally applied by measuring the yield of the virus from wastewater sourced from a third wastewater plant with different physiochemical properties (Table 1). This wastewater was consequently seeded with the gamma-irradiated SARS-CoV-2 virus which was then detected with high recovery rates (N1 and N2 avg.: 45.0% + 2.2) using the PEG6000 precipitation and extraction method described in this study. In addition, the PEG6000 precipitation and Trizol/chloroform adapted RNA extraction method was also tested on wastewater sourced from a site in Queensland, Australia, confirmed to contain the SARS-CoV-2. Following precipitation of 200 ml of this wastewater, the virus was detected at Ct 37.19 for the N1 target, and 35.25 for the N2 target, corresponding with an average of 3 copies of viral RNA per reaction.
As a final quality control, the PEG6000 precipitation and Trizol/chloroform adapted RNA extraction method was used to detect the norovirus in wastewater sourced from plant 1 and 2 using two established GI and GII probe/primer sets (da Silva AK et al., 2007; Miura et al., 2013). RT-qPCR analysis revealed the presence of this virus (both genogroups) in both samples tested (GI: Ct – 33.2 and 33.7, GII: Ct – 29.2 and 32.7 for plant 1 and 2, respectively) indicating this methodology was sufficiently sensitive to detect a virus that is naturally present at low concentrations in wastewater.
4. Discussion
WBE contains many strengths, such as ease of acquisition of wastewater and the ability to carry out population-level assessment of viral load and spread. One of the biggest limitations of WBE lies with the diverse methodologies used to concentrate and subsequently detect the virus from wastewater samples (Rusiñol et al., 2020). Ultimately, there are two key components that need consideration when developing a standard method of detection; the type of virus being detected and the wastewater matrix itself. Variability of wastewater including the presence of PCR inhibitors such as inorganic and organic compounds, total suspended solids, residual pharmaceutical drugs and microbial load will ultimately impact viral survival and its ability to be detected. Rates and factors which affect viral RNA decay are also in need of consideration. Studies investigating these factors have revealed that temperature influences SARS-CoV-2 RNA decay rates (Ahmed et al., 2020b; Hokajarvi et al., 2021), however, pH, which can impact RNA degradation (Wozniak et al., 2020) and hence its subsequent detection by RT-qPCR, has not been addressed. In this study, wastewater from plant 1 presented with a high pH range of 8.8 to 9.3, which when compared to other sites (Yadav et al., 2019), appears to be higher than the norm. However, without further studies, the effect of high wastewater pH on SARS-CoV-2 detection is unknown.
Virus particles are relatively unstable in wastewater and enveloped viruses such as SARS-CoV-2 even more so compared to non-enveloped enteric viruses (Wang et al., 2005), hence concentration and extraction steps are important to obtain and subsequently purify viral particles to a level suitable for detection by molecular techniques. Methods described in current literature generally rely on filtration or precipitation. Since high throughput and reliable recovery rates are crucial for the routine detection of SARS-CoV-2, methods for the concentration of actual virus particles were investigated and optimized as a priority.
Our early investigations also examined other published viral concentration methods such as ultrafiltration (Ahmed et al., 2020a; Medema et al., 2020; Nemudryi et al., 2020) and ultracentrifugation (Wurtzer et al., 2020). Although both being shown to be suitable for SARS-CoV-2 detection, early in the pandemic these methods presented several logistical problems. In the case of ultrafiltration this included the inability to obtain filtration units and the number of units needed to process highly turbid samples. This was further complicated by the propensity of certain types of wastewater to block the membrane of the filtration units very quickly resulting in difficulties in concentrating viruses from larger volumes of wastewater. Ultracentrifugation also requires access to expensive equipment which may be prohibitive to some laboratories and can take a long time to process many samples. Although their value is recognized, these methods were not pursued any further.
Initial assay development was performed on two concentration and extraction methods, namely PEG and adsorption-extraction. Most significantly, of these was the addition of Trizol/chloroform prior to RNA extraction as low yields were obtained for both methods when this step was omitted. Trizol/chloroform is effective in lysing tissue samples and microorganisms and inhibiting the degradation of RNA (Gautam et al., 2016; Jureka et al., 2020; Li-Korotky et al., 2007). Adsorption-extraction has successfully been used for the concentration and extraction of murine hepatitis viral particles by other research groups (Ahmed et al., 2020c) yielding a mean virus recovery of 65.7% ± 23. In our present study, lower overall SARS-CoV-2 recovery rates were achieved using the adsorption-extraction method and the recovery rates varied significantly with the wastewater source (Fig. 3) using our optimized adsorption-extraction method. This was attributed to the likely co-concentration of inhibitors in this sample.
The PEG precipitation and extraction method was also evaluated for viral recovery from whole wastewater (liquid fraction) and from the pre-spin pellets (solid fraction). The inclusion of both fractions led to lower overall results, in particular for wastewater sourced from plant 1 (Table 2). However, the low recovery rates may be due to coprecipitated inhibitors which interfered with RT-qPCR. Removal of the solid fraction appeared to significantly improve the recovery rate of SARS-CoV-2 from plant 2 wastewater, as detected by Luna® Universal master mix. The low recovery rates observed from the pre-spun pellets of plant 2 wastewater may indicate that the virus was not present in the pellets. However, the more likely explanation was due to carry-over of a high level of inhibitors into the qPCR reaction, which interfered in the detection of the virus.
Although the Luna® Universal master mix had a higher detection efficiency of SARS-CoV-2 compared to TaqPath™, this master mix seemed to be more sensitive to inhibitors co-purified using the adsorption-extraction method as no SARS-CoV-2 was detected in wastewater from plant 1 (Fig. 3 and Table 3). Introducing a pre-spin step to remove solids and associated PCR inhibitors led to even lower recovery rates for wastewaters from both plant 1 and plant 2 with either of the RNA extraction kits or PCR master mixes used (Table 2). The difference in the observed recovery efficiencies may be due to the flow rate during filtration, which was very high after the solid phase was removed by centrifugation, leading to decreased time for the virus to bind to the negatively charged membrane. Adjusting the wastewater pH to 3.5 to promote viral adsorption to the membrane did not result with increased recovery yields. However, it has been postulated that SARS-CoV-2 may be sensitive to low pH, with more research needing to be carried out to determine the effect of acidification on SARS-CoV-2 recovery from wastewater (Ahmed et al., 2020c).
To further elucidate the efficiency of these master mixes in combination with the N1/N2 primer/probe sets used in this study, their amplification efficiencies were calculated. These revealed that under optimal conditions the Luna® Universal master mix functioned at above 95% whilst TaqPath™ ranged from approximately 65 to 72% (Table 4). These results may explain why detection by TaqPath™ resulted in lower recovery rates for samples. However, its ability to function in the presence of coprecipitated inhibitors may explain the significantly higher (p < 0.05) recovery rates of samples processed by the adsorption-extraction method, though overall, the recovery rates remained low (avg. < 20%). This demonstrated that enzyme/master mix efficiency should be determined prior to use, as efficiency is influenced by numerous variables. Final enzyme and master mix efficiency and virus detection will depend on the extraction kit and its effectiveness at removing inhibitors, as well as the characteristics of the wastewater itself, as some sources carry more inhibitors making their removal more challenging.
Of the two SARS-CoV-2 concentration and extraction methods tested, precipitation with PEG6000 and detection with the Luna® Universal master mix delivered significantly higher (p < 0.05) recovery yields irrespective of the wastewater source (Fig. 2). PEG precipitation also minimized the carryover of wastewater inhibitors which interfered in RT-qPCR. These results were consistent with the trend observed in the recovery of the AccuPlex™ SARS-CoV-2 reference material extracted from plant 1 and 2 wastewater, indicating that this reference material is indeed a suitable surrogate for SARS-CoV-2 to standardize and optimize virus extraction protocols (Fig. 4). The suitability of the optimized PEG precipitation method was further confirmed by demonstrated reliable recovery rates (N1 and N2 avg.: 45. 0% + 2.2) of SARS-CoV-2 from a third, and distinct, wastewater sample (plant 3).
This study is the first to evaluate the success of PEG precipitation and the adsorption-extraction method for their use in SARS-CoV-2 detection in wastewaters which have varying physiochemical properties. This study exemplifies the importance of the wastewater matrix itself and shows that it can greatly impact virus recovery rates. As determined here, the presence of inhibitors and the concentration of both the total and dissolved suspended solids can play a significant role in virus detection. As such it is vital that a methodology used to concentrate viruses and extract viral RNA be robust enough to mitigate these parameters. PEG precipitation, coupled with a Trizol and chloroform adapted extraction method, has been shown here to consistently recover the SARS-CoV-2 from three distinctly different types of wastewater with high efficiency. It was also efficient at recovering the AccuPlex™ SARS-CoV-2 reference material as well as the norovirus.
5. Conclusions
Despite the immense potential of WBE for the surveillance of the spread of disease its effectiveness and usability rely on having methods that are efficient and robust enough to be suitable for use with wastewater with varying physiochemical properties, a variety of extraction kits and PCR master mixes and hence would be easily adaptable for use in other laboratories. We have optimized and described a technique that fits all those parameters. Our optimized PEG precipitation method, coupled with a Trizol and chloroform extraction step, was shown to be most consistent in its ability to recover the SARS-CoV-2 from wastewater which presented with different physiochemical properties. Using this method, we were able to detect as few as 3 copies of SARS-CoV-2 viral material from a wastewater sample using different RNA extraction kits and PCR master mixes. In addition, we did not rely on a commercial kit for the primer and probes used. This method was also shown to be successful in the concentration and detection of the AccuPlex™ SARS-CoV-2 reference material and the norovirus from differently sourced wastewater samples. The flexibility of our method would be paramount for maintaining a surveillance program during a pandemic when access to specific molecular reagents could be severely restricted.
The following is the supplementary data related to this article.
Alternative virus concentration methods trailed in this study.
CRediT authorship contribution statement
Sylvia A. Sapula: Validation, Investigation, Writing – original draft, Writing – review & editing, Visualization. Jonathan J. Whittall: Validation, Investigation, Writing – original draft, Writing – review & editing, Visualization. Aaron J. Pandopulos: Investigation, Writing – review & editing. Cobus Gerber: Conceptualization, Resources, Writing – review & editing, Supervision, Funding acquisition. Henrietta Venter: Conceptualization, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare not conflict of interest.
Acknowledgments
Funding: This work was supported by SAHealth, Adelaide, Australia.
The authors would also like to thank the operators of the various wastewater treatment plants, Queensland Alliance for Environmental Health Science for supplying wastewater samples and Melbourne Health for providing gamma-irradiated SARS-CoV-2.
Finally, we would like to thank Dr. Steven Polyak for helpful discussions and the University of South Australia for providing support during the COVID-19 pandemic.
Editor: Damia Barcelo
References
- Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020:9. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar-Oliveira M.D.L., Campos A.R., Matos A., Rigotto C., Sotero-Martins A., PFP Teixeira, et al. Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: a valuable tool for COVID-19 surveillance-a brief review. Int. J. Environ. Res. Public Health. 2020;17:9251. doi: 10.3390/ijerph17249251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. (138764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. (110092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. (139960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761:144216. doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsalu A., Sapula S.A., De Barros Lopes M., Hart B.J., Nguyen A.H., Drigo B., et al. Efflux pump-driven antibiotic and biocide cross-resistance in Pseudomonas aeruginosa isolated from different ecological niches: a case study in the development of multidrug resistance in environmental hotspots. Microorganisms. 2020;8:1647. doi: 10.3390/microorganisms8111647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987; 162: 156–9. [DOI] [PubMed]
- da Silva AK, J-C Le Saux, Parnaudeau S., Pommepuy M., Elimelech M., Le Guyader F.S. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 2007;73:7891. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020;726 doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- Farkas K., Hillary L.S., Malham S.K., McDonald J.E., Jones D.L. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr. Opin. Environ. Sci. Health. 2020;17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouladvand F., Bemani P., Mohammadi M., Amini R., Azizi Jalilian F. A review of the methods for concentrating M13 phage. J. Appl. Biotechnol. Rep. 2020;7:7–15. [Google Scholar]
- Gautam A., Kumar R., Dimitrov G., Hoke A., Hammamieh R., Jett M. Identification of extracellular miRNA in archived serum samples by next-generation sequencing from RNA extracted using multiple methods. Mol. Biol. Rep. 2016;43:1165–1178. doi: 10.1007/s11033-016-4043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller C., Varbanov M., Duval R.E. Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses. 2012;4:3044–3068. doi: 10.3390/v4113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Schwab KJ, Spencer SK, Borchardt MA. Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 2012; 46: 4281–91. [DOI] [PubMed]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020:737. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejkal TW, Wellings FM, Lewis AL, LaRock PA. Distribution of viruses associated with particles in waste water. Appl. Environ. Microbiol. 1981; 41: 628–34. [DOI] [PMC free article] [PubMed]
- Hokajarvi A.M., Rytkonen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.M., et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4:41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Jureka A.S., Silvas J.A., Basler C.F. Propagation, inactivation, and safety testing of SARS-CoV-2. Viruses. 2020:12. doi: 10.3390/v12060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. (115899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. (139652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Korotky HS, Kelly LA, Piltcher O, Hebda PA, Doyle WJ. Evaluation of microbial RNA extractions from Streptococcus pneumoniae. J. Microbiol. Methods 2007; 68: 342–8. [DOI] [PMC free article] [PubMed]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Miura T, Parnaudeau S, Grodzki M, Okabe S, Atmar RL, Le Guyader FS. Environmental detection of genogroup I, II, and IV noroviruses by using a generic real-time reverse transcription-PCR assay. Appl. Environ. Microbiol. 2013; 79: 6585–92. [DOI] [PMC free article] [PubMed]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel Coronovirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes. U.S. Centers for Disease Control and Prevention; 2020. (2020) [Google Scholar]
- Perez-Cataluna A., Cuevas-Ferrando E., Randazzo W., Falco I., Allende A., Sanchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrich A., Mahony J., Chong S., Broukhanski G., Gharabaghi F., Johnson G., et al. Multicenter comparison of nucleic acid extraction methods for detection of severe acute respiratory syndrome coronavirus RNA in stool specimens. J. Clin. Microbiol. 2006;44(8):2681. doi: 10.1128/JCM.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Kitajima M., Campillo M.E., Gerba C.P., Pepper I.L. Virus reduction during advanced Bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 2016;50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S., Sherchan S.P., Haramoto E. Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep. 2020;10:3616. doi: 10.1038/s41598-020-60547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton M.R., Andrews R.C., Hofmann R. Particle-associated viruses in water: impacts on disinfection processes. Crit. Rev. Environ. Sci. Technol. 2008;38:137–164. [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda B.P. Concentration and purification of viruses and bacteriophages with polyethylene glycol. Folia Microbiol. (Praha) 1978;23:88–96. doi: 10.1007/BF02876605. [DOI] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B.Y., Guo B.Z., Liu C., Ou G.R., Wang M.N., Fang T.Y., Chao F.H., Li J.W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.L., Medrano J.F. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- Wozniak A., Cerda A., Ibarra-Henriquez C., Sebastian V., Armijo G., Lamig L., et al. A simple RNA preparation method for SARS-CoV-2 detection by RT-qPCR. Sci. Rep. 2020;10:16608. doi: 10.1038/s41598-020-73616-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020:5. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., et al. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M.K., Short M.D., Gerber C., Awad J., van den Akker B., Saint C.P. Removal of emerging drugs of addiction by wastewater treatment and water recycling processes and impacts on effluent-associated environmental risk. Sci. Total Environ. 2019;680:13–22. doi: 10.1016/j.scitotenv.2019.05.068. [DOI] [PubMed] [Google Scholar]
- Ye Y, Ellenberg RM, Graham KE, Wigginton KR. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016; 50: 5077–85. [DOI] [PubMed]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alternative virus concentration methods trailed in this study.