Abstract
The complement component 4 (C4) gene is linked to schizophrenia and synaptic refinement. In humans, greater expression of C4A in the brain is associated with an increased risk of schizophrenia. To investigate this genetic finding and address how C4A shapes brain circuits in vivo, here, we generated a mouse model with primate-lineage-specific isoforms of C4, human C4A and/ or C4B. Human C4A bound synapses more efficiently than C4B. C4A (but not C4B) rescued the visual system synaptic refinement deficits of C4 knockout mice. Intriguingly, mice without C4 had normal numbers of cortical synapses, which suggests that complement is not required for normal developmental synaptic pruning. However, overexpressing C4A in mice reduced cortical synapse density, increased microglial engulfment of synapses and altered mouse behavior. These results suggest that increased C4A-mediated synaptic elimination results in abnormal brain circuits and behavior. Understanding pathological over-pruning mechanisms has important therapeutic implications in disease conditions such as schizophrenia.
Schizophrenia is a heritable neuropsychiatric disorder of unknown etiology and affects 1% of the adult population. Schizophrenia is associated with impaired cognition, perception and motivation, which become clinically apparent in late adolescence and early adulthood. The concurrent excessive loss of gray matter and dendritic spines during this period suggests that the reduction in synaptic connections might contribute to the behavioral and cognitive deficits1–5. However, it is unclear when synaptic defects in the cerebral cortex arise and how reduced synaptic connections relate to genes and alleles and the proteins they encode.
The human genome’s largest population-level influence on the risk of schizophrenia arises in substantial part from the C4 genes6. The C4A and C4B genes, which reside in the major histocompatibility complex (MHC) locus, encode two highly conserved C4 isoforms, C4A and C4B. Although the peptide sequences of C4A and C4B differ by only four amino acids7, the isoforms exhibit differential binding to unique chemical targets8,9. Thus, their molecular structures could dictate the corresponding binding sites in tissues. In addition, both genes exhibit common variations in copy number, with one to four total C4 genes per haplotype, and vary in length due to insertion of an endogenous retroviral element in an intron, most commonly present in the C4A isoform10. Alleles that increase C4A expression correlate with increased schizophrenia risk, but ones that increase C4B expression do not6. In this study, we sought to explain these findings by distinguishing the roles of C4A and C4B in the brain and exploring whether C4A overexpression affects brain development and synaptic pruning.
Synaptic pruning is a process by which supernumerary neuronal connections are eliminated, which allows appropriate connections to strengthen and mature11–13. Previous studies have identified some key players of the pruning processes, including microglia through CD47 (ref. 14) and trogocytosis15, astrocytes by MERTK and MEGF10 (ref. 16) pathways, and many immune system molecules such as MHC I, pentraxins, paired immunoglobulin-like receptor B and complement17–21. The classical complement cascade, including C1q, C3 and C4, is critical to pruning, as mice deficient in these components show defects in synaptic refinement in the retinogeniculate system6,19. C4 can localize to subsets of synapses; however, functional and behavioral consequences of increased C4 expression and its downstream pathways are unknown and challenging to characterize6. Moreover, although synaptic refinement in the visual system is well characterized, the time line of synaptic pruning, the critical periods of plasticity and the role of complement in regions relevant to schizophrenia remain to be identified.
Here, we constructed humanized mouse models to examine the functional differences between human C4A and C4B in the brain and to determine whether C4A overexpression can pathologically affect brain development. In this model, variable copy numbers of C4A and C4B genes were introduced into the mouse genome to investigate their physiological effects in vivo. We found distinct roles for C4A and C4B in the developing brain, and further discovered that C4A overexpression affected microglia engulfment, synaptic pruning and mouse behavior. These findings highlight the importance of C4A in synaptic pruning and brain development, and draw attention to the parallels observed between C4A overexpression and schizophrenia pathology.
Results
C4A is more efficient than C4B in synaptic pruning.
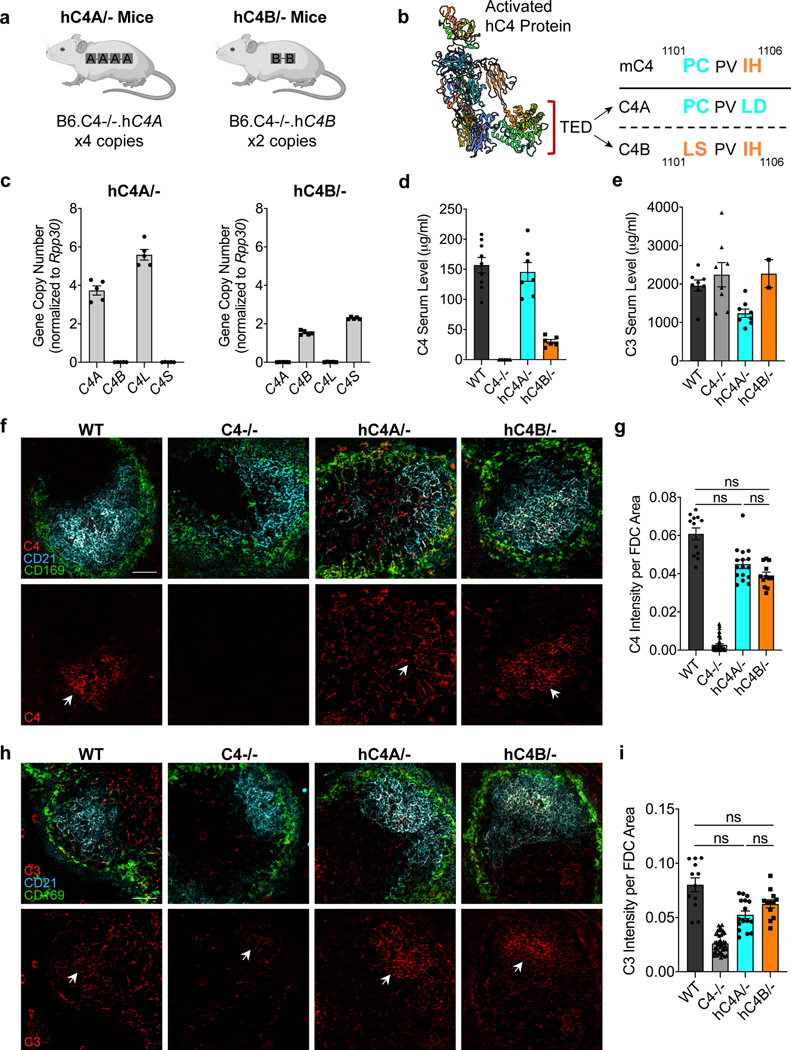
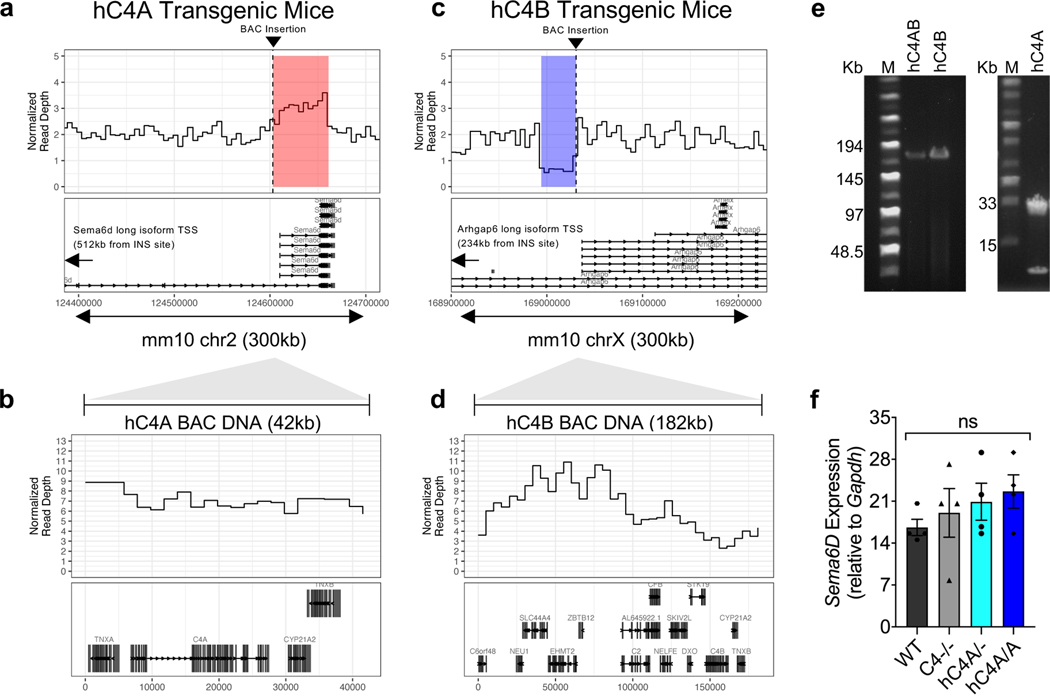
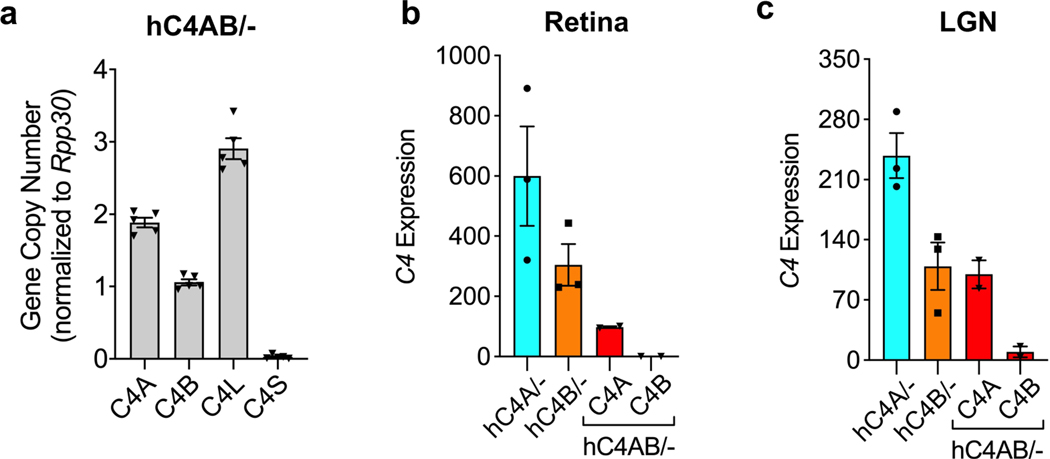
Genetic analyses of patients with schizophrenia made the surprising prediction that C4A and C4B differ in their effects on brain development. Although within primates there exists two isoforms of C4, which differ in four amino acid residues in the TED domain of activated C4 protein, mice have only one corresponding C4 gene (mC4), from which primate C4A and C4B evolved (Fig. 1b). mC4 is made up of a chimeric C4A/C4B sequence in the key isotypic residues: positions 1101–1102 are identical to C4A residues (proline and cysteine), whereas positions 1105–1106 resemble residues in C4B (isoleucine and histidine). The mouse Slp gene, which is a mC4-like gene, is a sequence outlier and not functional in the classical complement pathway as its C1s cleavage site is mutated and cannot be activated6,22. Given the limitations of the mC4 lacking isotypes, we introduced human C4A (hC4A) or C4B (hC4B) genes into the mouse genome using bacterial artificial chromosome (BAC) DNA transgenesis. The use of BACs enabled the delivery of human genes with their introns and cis-regulatory elements. The two resulting mouse strains were backcrossed onto a mC4-deficient background, thus creating hC4A/− and hC4B/− mice that express C4 only from the human transgene (Fig. 1a). The number of gene copies for each mouse strain was determined using droplet digital PCR (ddPCR), and this showed that hC4A/− mice had four copies of C4A long (C4A-L) and hC4B/− mice had two copies of C4B short (C4B-S) (Fig. 1c). Whole-genome sequencing analysis to locate the integration site of the BACs showed tandem insertion in chromosome 2 and chromosome X in hC4A/− and hC4B/− mice, respectively (Extended Data Fig. 1a–d). Of note, the C4A BAC DNA insertion resulted in the duplication of the endogenous gene Sema6d; however, its expression was unchanged between hC4A/− and wild-type (WT) mice (Extended Data Fig. 1f).
Fig. 1 |. Human C4A and C4B are functionally active in a mouse model.
a, Schematic of hC4A/− and hC4B/− transgenic mice. b, A three-dimensional (3D) structure of activated hC4 protein showing the TED domain and the amino-acid sequence differences between C4A and C4B. The mC4 protein is a hybrid of C4A and C4B, gaining positions 1,101–1,102 from C4A and 1,105–1,106 from C4B. c, Gene copy numbers for C4A, C4B, C4A-L and C4B-S were determined by ddPCR using Rpp30 as a reference gene (n = 5 representatives per group). d, C4 serum levels were measured by ELISA in adult (P60) WT (n = 9), C4−/− (n = 10), hC4A/− (n = 7) and hC4B/− (n = 6) mice. e, C3 serum levels were measured by ELISA in adult (P60) WT (n = 8), C4−/− (n = 8), hC4A/− (n = 8) and hC4B/− (n = 2) mice. f, Representative confocal images of spleen showing C4 deposition on CD21+ FDCs in follicles delineated by CD169+ macrophages in WT, C4−/−, hC4A/−and hC4B/− mice (red, C4; cyan, CD21; green, CD169). White arrows indicate C4 puncta deposited onto FDCs. Scale bar, 50 μm. g, C4 deposition was quantified by fluorescence intensity per FDC area (in each FOV) for WT (n = 3), C4−/− (n = 11), hC4A/− (n = 4) and hC4B/− (n = 3) mice. Kruskal–Wallis test with Dunn’s multiple comparison test. h, Representative images showing C3 deposition on FDCs (white arrows) in WT, C4−/−, hC4A/− and hC4B/− mice (red, C3; cyan, CD21; green, CD169). Scale bar, 50 μm. i, C3 deposition was quantified by fluorescence intensity per FDC area (in each FOV) for WT (n = 3), C4−/− (n = 11), hC4A/− (n = 4) and hC4B/− (n = 3) mice. Kruskal–Wallis test with Dunn’s multiple comparison test. Bar graphs show the mean ± s.e.m. NS, not significant (P > 0.05).
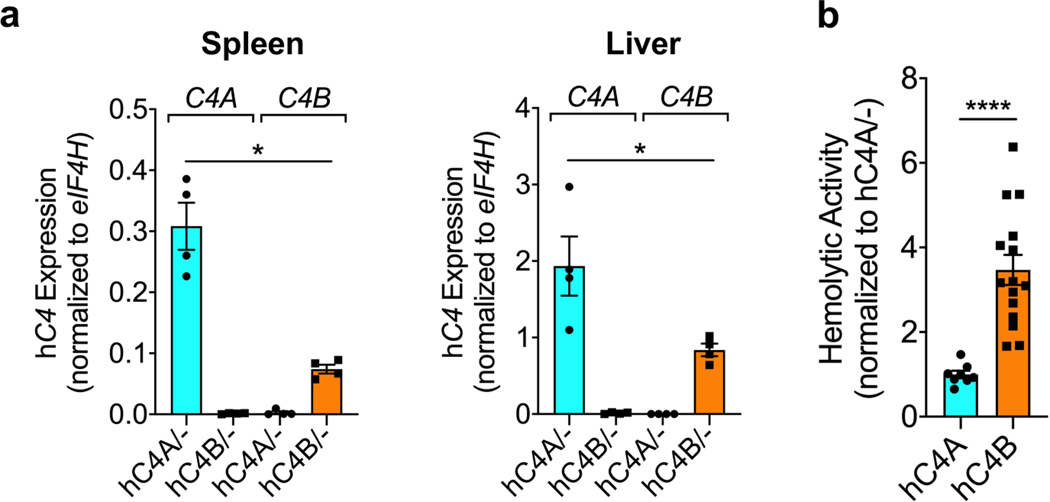
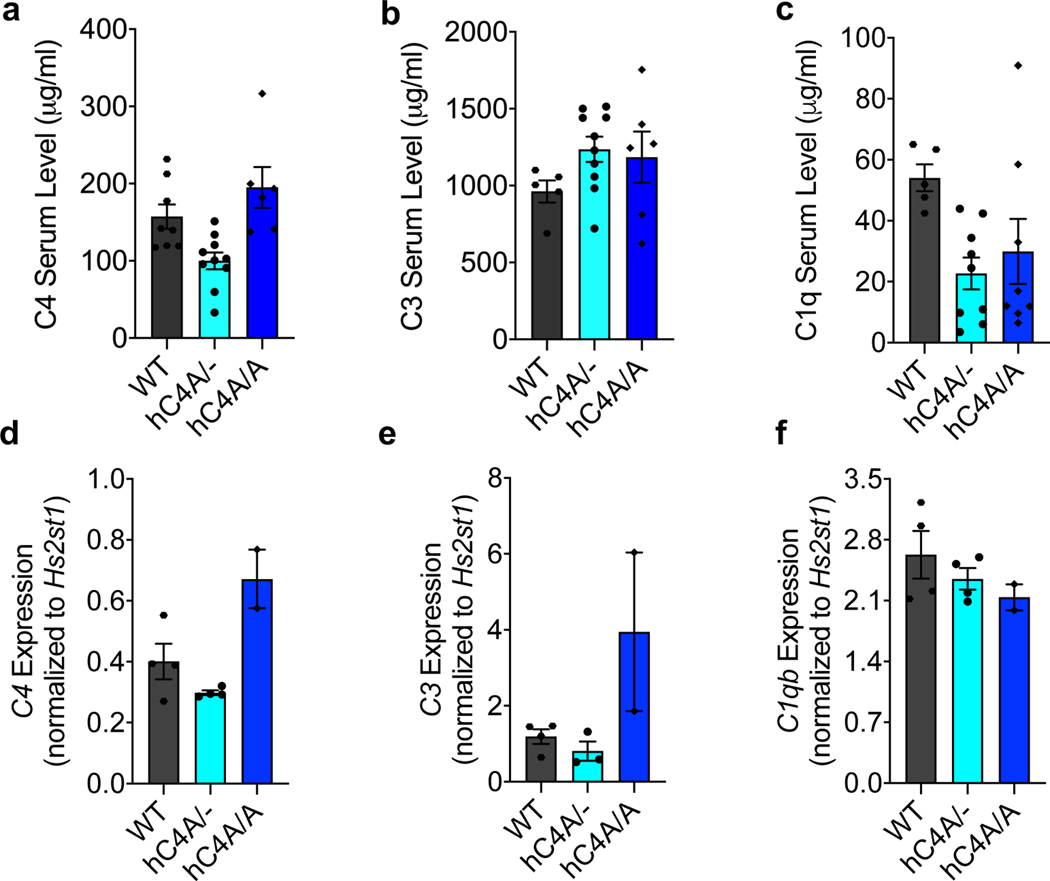
Both hC4A/− and hC4B/− strains had peripheral C4 mRNA expression and serum protein levels consistent with their relative gene copy number23 (Fig. 1d and Extended Data Fig. 2a). In the periphery, C4 functions to opsonize pathogens and cellular debris as a part of the classical complement cascade. C4, together with C2a, forms the C3 convertase complex to cleave and activate the downstream protein C3. To investigate whether the known function of C4 was maintained in this model, C3 serum levels were measured. Similar levels of C3 were observed in WT, hC4A/− and hC4B/− mice (Fig. 1d). Furthermore, the deposition of C4 and C3 on splenic stroma was quantified in normal resting mice of all four genotypes (WT, C4−/−, hC4A/− and hC4B/−)24. Immunostaining revealed equivalent deposits of both C4 and C3 on the splenic stroma (Fig. 1f,h), which showed similar C3 activation in hC4A/−, hC4B/− and WT mice (Fig. 1g,i). Only residual C3 activation was observed in C4−/− mice (Fig. 1i). In addition, complement system functionality was measured by hemolysis assay. Both C4A and C4B bound to sheep red blood cells and induced hemolysis by activating downstream complement proteins (Extended Data Fig. 2b). As previously described9, C4B was more potent than C4A in binding to red blood cells due to its molecular structure determined by the isotypic region. Validating the peripheral expression and known functions of C4A and C4B in the mouse models allowed us to study the isotype differences in the brain.
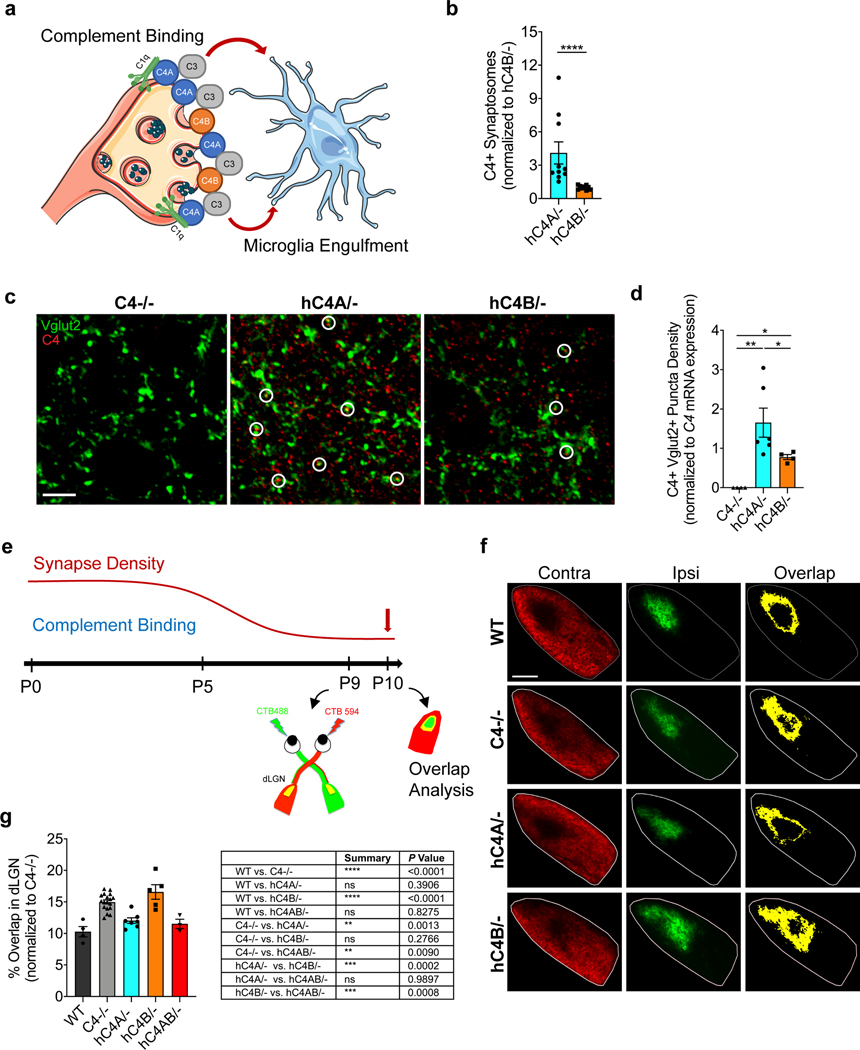
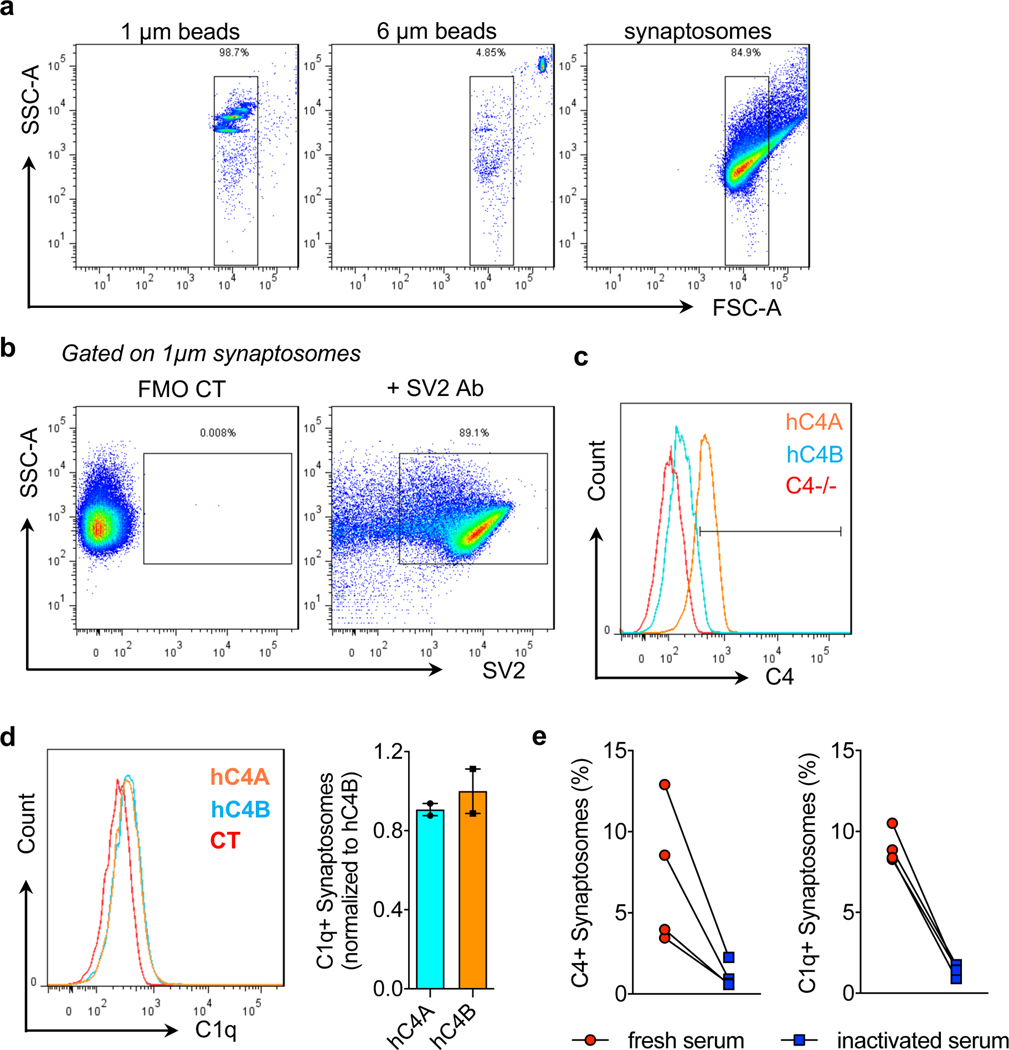
Although hC4 has been shown to deposit onto synapses, its isoform effects on synaptic deposition and pruning have not6. To test whether hC4A/− and hC4B/− mice exhibit differences in activation after synaptic deposition (Fig. 2a), we established an in vitro assay and used flow cytometry to measure C4 binding efficiency onto synaptosomes (Extended Data Fig. 3a,b). Purified synaptosomes from C4−/− mice were incubated with serum from hC4A/− and hC4B/− mice, with equal C4 levels, and stained for C4 together with C1q as a control. Staining of the synaptosome membranes showed higher levels of C4A binding than C4B (Fig. 2b and Extended Data Fig. 3c). Conversely, bound C1q levels were similar in both groups (Extended Data Fig. 3d). Neither C1q nor C4 binding was observed when heat-inactivated serum was used, which highlights the specificity of complement binding (Extended Data Fig. 3e). Together, these results show that C4A binds more efficiently to synaptosomes than C4B in vitro.
Fig. 2 |. Human C4A is more efficient than C4B in synaptic pruning.
a, At the synapse, complement-dependent pruning is carried out by the classical complement cascade. After C1q tagging, C4 binds the synapse and C3 is then activated for microglia recognition by the receptor CR3. Microglia engulf the complement-bound synapses for refinement. b, Synaptosomes from C4−/− mice were isolated and incubated with serum containing the same amount of C4 from hC4A/− (n = 10) or hC4B/− (n = 9) mice. C4 deposition on synaptosomes was detected and quantified by flow cytometry (serum from three independent experiments; Mann–Whitney test, two-tailed, ****P < 0.0001). c,d, C4 deposition onto Vglut2+ synaptic terminals in the dLGN was identified by immunofluorescence staining at P5 in C4−/− (n = 4), hC4A/− (n = 6) and hC4B/− (n = 4) mice. c, Representative images of C4 (red) and Vglut2 (green) staining in the dLGN showing C4+Vglut2+ synapses (white circles). Scale bar, 5 μm. d, Quantification of C4+Vglut2+ synapse density in P5 hC4 transgenic mice (three independent experiments; results were normalized to hC4 mRNA expression in the LGN (Mann–Whitney test, two tailed: **PC4−/− vs hC4A/− = 0.0095, *PC4−/− vs hC4B/− = 0.0286, *PhC4A/− vs hC4B/− = 0.0190). e, To investigate complement-dependent synaptic pruning in the retinogeniculate system, the eye-specific segregation assay was performed. Mice were injected with fluorescently tagged CTB (CTB-488 and CTB-594) in each eye at P9 and brains were collected at P10. f, Representative images of WT, C4−/−, hC4A/− and hC4B/− mice at P10 are shown. Inputs from contralateral (Contra; red) and ipsilateral (Ipsi; green) eyes and overlapping area (yellow) are shown. Scale bar, 100 μm. g, The percentage overlap of the contralateral and ipsilateral areas were calculated for P10 WT (n = 4), C4−/− (n = 18), hC4A/− (n = 7), hC4B/− (n = 6) and hC4AB/− (n = 3) mice (2 independent experiments; all results were normalized to the C4−/− group to retain littermate controls; one-way ANOVA with Tukey’s multiple comparisons test). Bar graphs show the mean ± s.e.m.
To visualize hC4 deposition and investigate synaptic refinement processes in hC4A/− and hC4B/− mice in vivo, the retinogeniculate system was utilized. The visual system in the developing mouse brain has been extensively used as a model to not only delineate synaptic elimination pathways25 but to also examine the involvement of the complement system in synaptic pruning6,19,26. In early postnatal mice, retinal ganglion cells (RGCs) from both eyes form synapses onto the same relay neurons within the dorsolateral geniculate nucleus (dLGN)27. During a key subsequent critical period (postnatal day (P) 3–9)28–30, many of these synaptic connections are refined due to spontaneous activity, which results in RGCs from either eye to synapse onto different relay neurons and create nonoverlapping, eye-specific territories in the dLGN. P5 was determined as the most robust period of pruning, when most C3 deposition has been observed26. At this time point, differential deposition of C4A and C4B onto the RGC synaptic terminals was investigated. dLGN sections were stained for hC4 and vesicular glutamate transporter 2 (Vglut2; Fig. 2c), and a colocalization analysis was performed. We observed greater numbers of C4+ puncta colocalizing with Vglut2+ synapses in hC4A/− mice compared with hC4B/− mice (Fig. 2d) after normalizing for C4 mRNA expression in the LGN (Extended Data Fig. 4c). As expected, C4−/− mice showed only background levels of staining (Fig. 2c,d). These results demonstrate that C4A and C4B have differing binding capacities in the brain, with C4A binding to synapses at a higher level than C4B.
Furthermore, eye-specific territories of the dLGN were used to determine the functionality of C4A and C4B in synaptic pruning. The segregation of RGC projections was traced by fluorescently tagged cholera toxin subunit B (CTB) molecules, which were intravitreally injected (Fig. 2e). Previous studies using global knockout mice showed that C1q-, C4- and C3-deficient mice fail to fully segregate the axonal projections of RGCs from each eye into nonoverlapping territories, thereby indicating defects in synaptic refinement6,19. Strikingly, hC4A/− mice showed a significantly lower percentage of overlapping territories compared with C4−/− littermates and similar levels to WT mice (Fig. 2f,g). Conversely, hC4B/− mice had a similar percentage of overlap as C4−/− littermates, but significantly higher levels than WT mice (Fig. 2f,g). The difference is unlikely explained by the C4 gene copy number differences between hC4A/− and hC4B/− mice, since a single copy of the C4A transgene in the hC4AB/− mouse model (Extended Data Fig. 4a) was sufficient to rescue the defect observed in C4−/− mice (Fig. 2g). These results suggest that C4A can rescue the synaptic refinement deficits of C4−/− mice and that C4B, unlike C4A, is functionally less active in synaptic pruning. Overall, these in vitro binding assays and early development experiments highlight the unique part that C4A plays in synaptic pruning compared with C4B. These findings, taken together with the established connection of C4A to schizophrenia, made C4A and its overexpression the focus of the remainder of our study and led us to question whether increased C4A expression alters synaptic refinement.
C4A overexpression increases synaptic refinement via microglia in the dLGN.
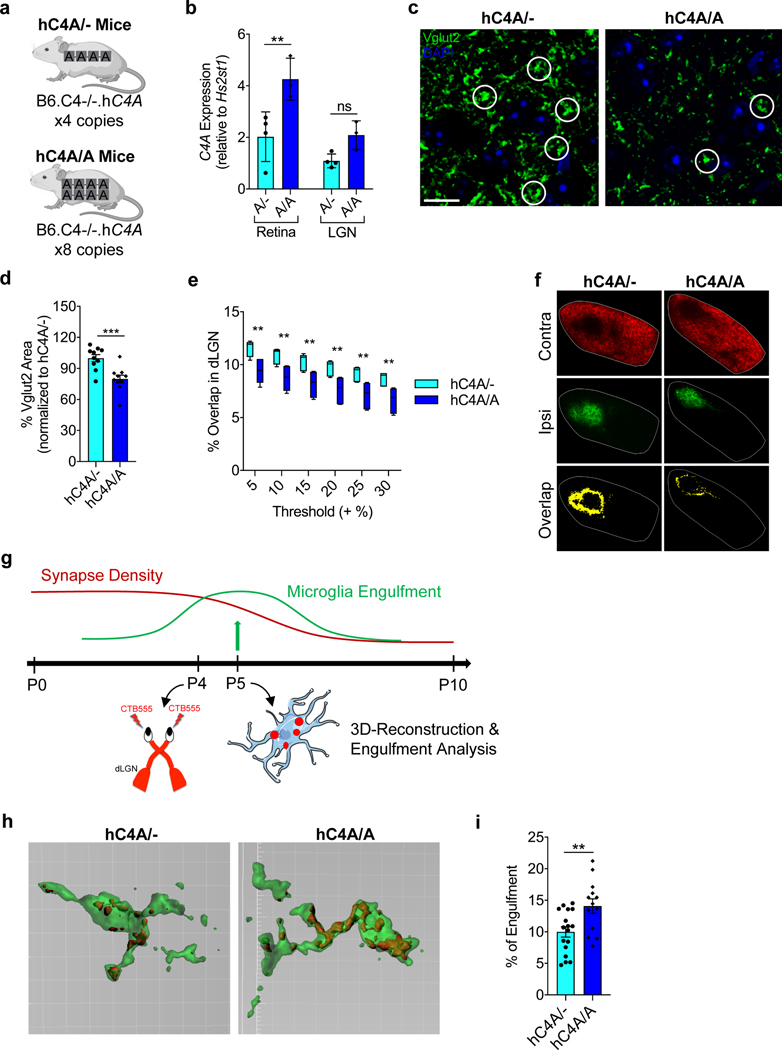
An increased C4A gene copy number is associated with schizophrenia6, which is characterized by an excessive reduction in cortical gray matter and dendritic spine density on cortical neurons1,3. To understand the underlying biology of this genetic association and to study the effects of C4A copy number on synaptic pruning, the hC4A mouse model was used to generate hC4A-overexpressing mice. This overexpression model (hC4A/A) was obtained by crossing hC4A/− mice together, which resulted in eight C4A copies per strain (Fig. 3a). It was confirmed that hC4A/A mice had approximately twice the level of serum C4 protein than hC4A/− mice (Extended Data Fig. 5a). In the retinogeniculate system, hC4A/A mice expressed more C4A mRNA in the retina, and, although not significant, in the LGN (Fig. 3b).
Fig. 3 |. increased C4A copy number induces excessive synaptic pruning via microglia engulfment.
a, Schematic of hC4A/− and hC4A/A transgenic mice. b, C4A mRNA expression was measured at P5 in the retina and the LGN from hC4A/− (A/−; n = 4) and hC4A/A (A/A; n = 3) mice by ddPCR (littermates from two independent cohorts; two-way ANOVA with Sidak’s multiple comparisons test, **P = 0.0039). c,d, The presynaptic marker Vglut2 was stained by immunofluorescence at P10 for hC4A/− (n = 10) and hC4A/A (n = 10) mice. c, Representative images of Vglut2 immunostaining are shown. White circles indicate examples of Vglut2 synapses. Scale bar, 10 μm. d, Percentage area of Vglut2 staining was measured (littermates from three independent cohorts; unpaired, two-tailed t-test, ***P = 0.0009). e,f, The eye-specific segregation assay was performed for hC4A/− (n = 6) and hC4A/A (n = 6) mice. e, The percentage overlap over multiple thresholds was used for analysis in P10 hC4A/− and hC4A/A mice (n = 6 in each group, littermates from 3 independent experiments; two-way ANOVA with Sidak’s multiple comparisons test, 0.0026 ≤ **P ≤ 0.0054). Box-and-whisker plot displays the median (center line), the 25th to 75th percentile (box), and the minimum and maximum values (whiskers). f, Representative images of hC4A/− and hC4A/A littermates at P10 from three independent experiments showing inputs from contralateral (red) and ipsilateral (green) eyes and the overlapping area (yellow). Scale bar, 100 μm. g, Schematic of the experiment. Mice received an intravitreal injection of fluorescently conjugated CTB (CTB-555) at P4 and brains were collected at P5, when microglia engulfment is at its peak. Microglia were stained with Iba1 for confocal imaging and 3D reconstruction. h, Representative surface-rendered microglia (green) and engulfed retinogeniculate inputs (red) from the dLGN. Grid line increments are 5 μm. i, Quantification of the percentage of CTB engulfment within individual microglia volume for hC4A/− (n = 3) and hC4A/A (n = 4) mice (three independent experiments; unpaired, two-tailed t-test, **P = 0.0060). Bar graphs show the mean ± s.e.m.
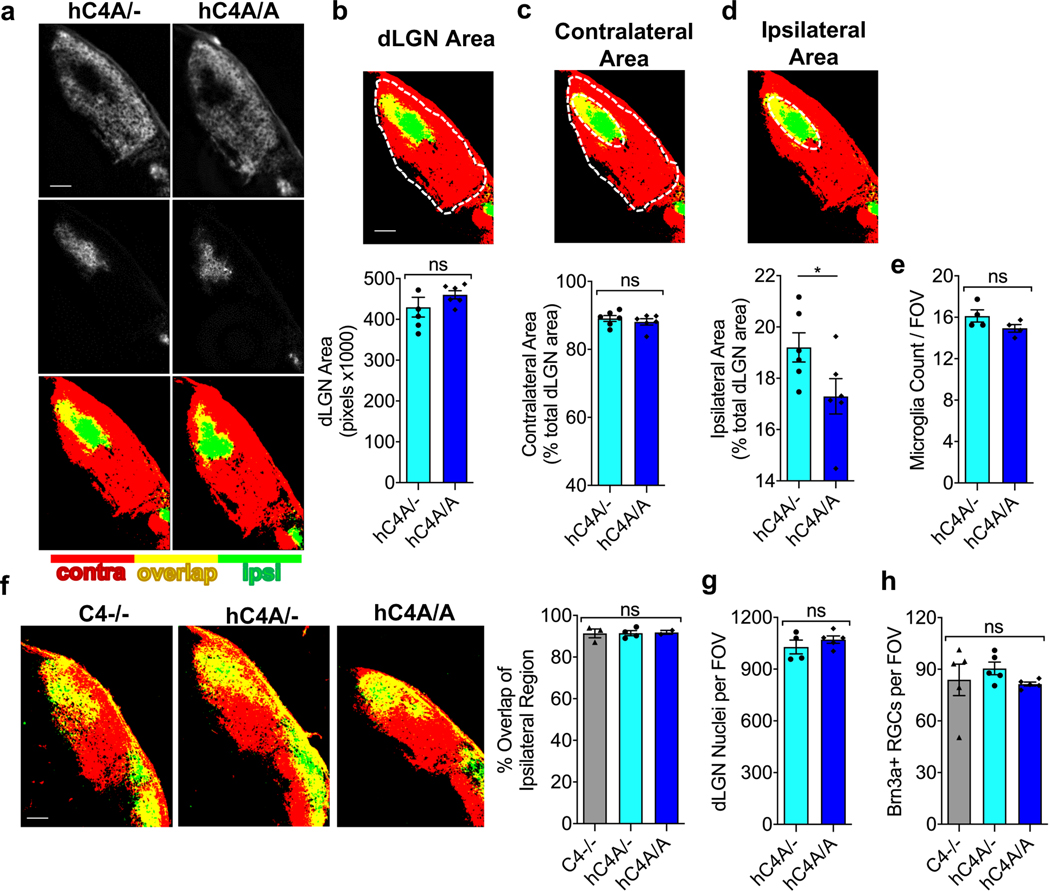
Considering the increase in copy number and mRNA expression in hC4A/A mice, higher levels of synaptic pruning were expected. In the dLGN, the level of synaptic pruning was measured by quantifying the RGC inputs31. RGC synapses (Vglut2+) are morphologically distinct from other terminals in the dLGN as they form clusters and appear larger32. By measuring the area of Vglut2+ inputs, we found that the RGC axon terminal surface in hC4A/A mice was significantly reduced compared with hC4A/− mice at P10 (Fig. 3c,d), which indicated higher levels of synaptic pruning. These results were supported by the reduced overlapping territories in the dLGN in hC4A/A mice compared with hC4A/− littermates (Fig. 3e,f). To control for morphological changes in retinal arborization, projections from each eye were examined (Extended Data Fig. 6a). Notably, the ipsilateral area was smaller in hC4A/A mice than in hC4A/− mice, thus confirming that ipsilateral projections were excessively pruned in hC4A-overexpressing mice (Extended Data Fig. 6b–d). To test whether C4A overexpression leads to abnormal developmental processes before the critical period, the eye-specific segregation assay was performed at P5. At this time point, we found that projections from each eye almost completely overlapped in C4−/−, hC4A/− and hC4A/A mice (Extended Data Fig. 6f), which suggests that C4A overexpression does not affect synaptic inputs before the critical pruning period. This result could not be attributed to differing numbers of cells, as cells stained positive for 4,6-diamidino-2-phenylindole (DAPI+) in the dLGN and Brn3a+ RGCs in the retina were equivalent in number (Extended Data Fig. 6g,h). Observing the morphological and functional effects of C4A expression on synaptic inputs led us to investigate the role of microglia in the complement-dependent synaptic pruning process.
Microglia are crucial for development of the brain, and in the retinogeniculate system, they participate in synapse elimination by phagocytizing complement-tagged synapses via complement receptor 3 (CR3; also known as CD11b)25. In the dLGN, P5 is the time point at which microglia are most phagocytic26. To determine whether excessive synaptic loss is accompanied by increased microglial engulfment, mice were injected with fluorescent CTB, and microglial phagocytosis of synaptic material in the dLGN of P5 mice was measured (Fig. 3g). Importantly, microglia from hC4A/A mice engulfed more inputs than those in hC4A/− mice (Fig. 3h,i), while having similar densities in each group (Extended Data Fig. 6e). This suggests that increased C4A expression induces a greater degree of synaptic engulfment per microglia and excessive synaptic refinement in the dLGN. Overall, utilizing the robust retinogeniculate system gave insight into the isoform differences of hC4, the role of C4A in the mechanism of complement-mediated synaptic pruning and its copy number effects on synapse elimination via microglia. However, to better examine the schizophrenia-related effects of C4A, the focus was shifted to the medial prefrontal cortex (mPFC), a region highly associated with schizophrenia pathology.
C4A overexpression affects microglia-mediated synaptic pruning in the mPFC.
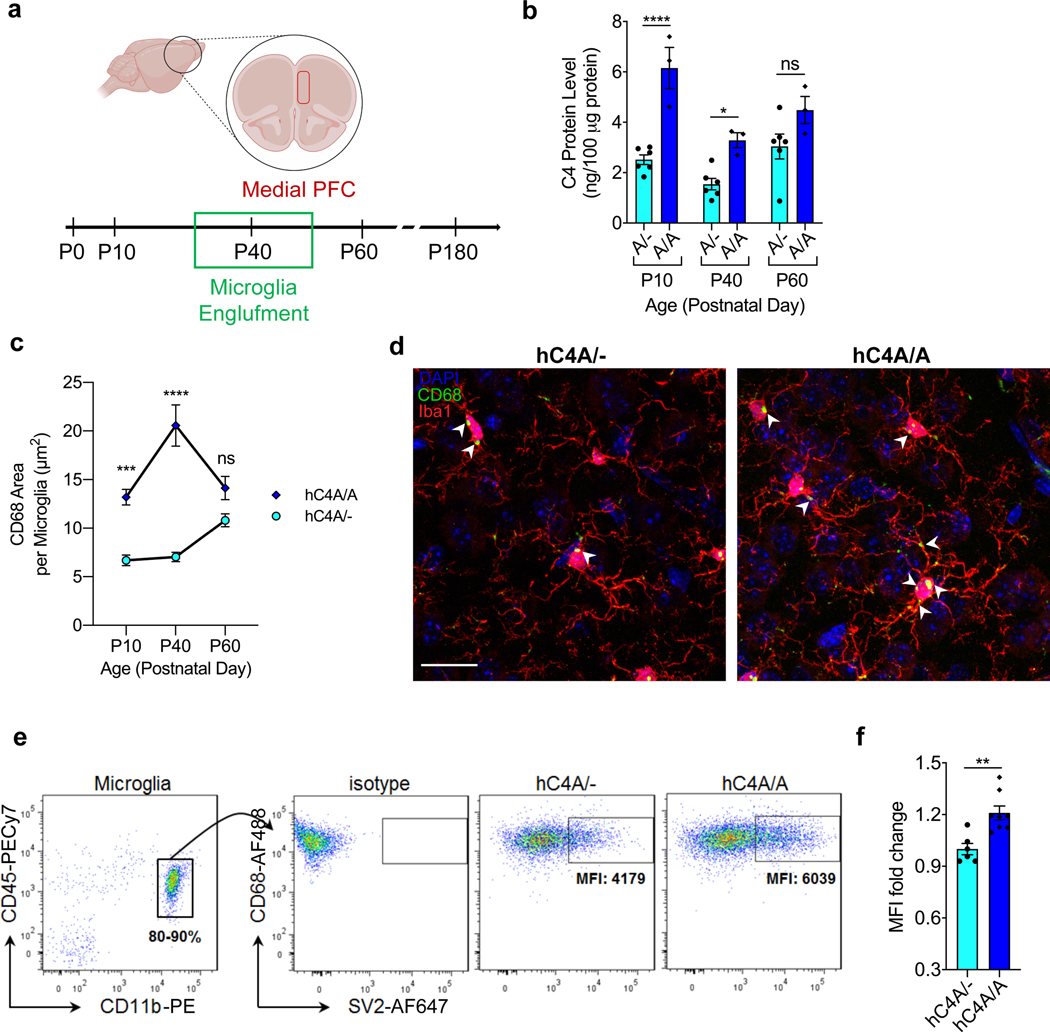
The complex organization and circuitry of the frontal cortex (FC) make it a difficult region to study synaptic pruning processes. Due to the limited knowledge of the role of C4 in the FC and how it is regulated, C4 protein levels were measured at different time points from development to adulthood. Forebrains from hC4A/A mice had significantly higher levels of C4 compared with hC4A/− mice at P10 (development) and P40 (adolescence; Fig. 4b). Although not significant, a trend of an increased C4 level was observed in hC4A/A mice at P60 (adulthood).
Fig. 4 |. hC4A-overexpressing mice have increased synaptic material uptake by microglia in the mPFC.
a, Schematic of the experiment. The role of C4A was examined in the mPFC across different time points for protein expression and microglia engulfment. Further microglia analyses were conducted at the adolescence stage for synaptic material engulfment. b, C4 protein expression in the forebrain was measured by ELISA for hC4A/− (n = 6) and hC4A/A (n = 3) mice at different time points: development (P10), adolescence (P40) and adulthood (P60; two-way ANOVA with Sidak’s multiple comparisons test, ****P < 0.0001, *P = 0.0322). c,d, Microglia engulfment in the mPFC was analyzed using Iba1+ and CD68+ markers across different time points. The peak difference between hC4A/− and hC4A/A mice was observed at the adolescence stage (P40). c, Quantification of CD68+ puncta area per microglia using Cell Profiler at P10 (hC4A/− (301 microglia), hC4A/A (368 microglia)), P40 (hC4A/− (265 microglia), hC4A/A (320 microglia)) and P60 time points (hC4A/− (104 microglia), hC4A/A (121 microglia); two-way ANOVA with Sidak’s multiple comparisons test; x–y plot shows the mean ± s.e.m. ***P = 0.001, ****P < 0.0001). d, Representative images of microglia with the CD68 signature. White arrowheads indicate CD68+ puncta in microglia. Scale bar, 20 μm. e,f, Comparison of microglial engulfment in the FC of P40 hC4A/− (n = 6) and hC4A/A (n = 8) mice. e, Purified microglia were intracellularly stained for the detection of SV2 synaptic protein. The gating strategy for microglia is shown (CD45low and CD11bhigh). f, The SV2 mean fluorescence intensity (MFI) was measured in hC4A/− (n = 6) and hC4A/A (n = 8) mice (three independent experiments; Mann–Whitney test, two-tailed, **P = 0.0047). Bar graphs show the mean ± s.e.m.
In humans, schizophrenia symptoms arise during adolescence, the period in which the FC matures33. Given this, the complement profile both in the periphery and the FC was investigated at the analogous time point in mice, P40. In serum, levels of C4, C3 and C1q were similar between WT and hC4A/− mice (Extended Data Fig. 5a–c). In the FC, RNA expression levels of mC4, hC4A, C3 and C1qb were also similar between the two groups (Extended Data Fig. 5d–f). Thus, the hC4A/− model presents a context whereby the effect of C4A overexpression can be assessed in the brain.
As microglia play a crucial role in complement-dependent synaptic pruning26,34, their engulfment levels were investigated in the mPFC of hC4A/− and hC4A/A mice (Fig. 4a). Microglia that are highly phagocytic show increased levels of CD68, a monocytic cytochemical biomarker found on the lysosomal membrane35,36. Microglial engulfment levels were measured in the mPFC by immunostaining for the markers Iba1 and CD68 (Fig. 4d). Cell Profiler was used to perform morphology analyses and to quantify the CD68-stained area in each microglia (Extended Data Fig. 7a). Similar to C4 protein expression, the CD68+ area was significantly higher in the hC4A/A mice at P10 and P40, when the peak difference was observed, but not at P60 (Fig. 4c). Microglia density and morphology were unchanged (Extended Data Fig. 7b–h). Although these results suggest that microglia are more phagocytic under C4A overexpression during adolescence, they do not indicate engulfment of synaptic material. To overcome this issue, we developed a fluorescence-activated cell sorting (FACS)-based assay to detect intracellular synaptic proteins in microglia. Purified microglia from the FC of P40 mice were permeabilized, stained for SV2 and quantified by FACS (Fig. 4e). Microglia isolated from hC4A/A mice contained more synaptic material than microglia from hC4A/− mice (Fig. 4f), thereby strengthening our previous findings that C4A overexpression induces increased microglial engulfment of synaptic material in the mPFC.
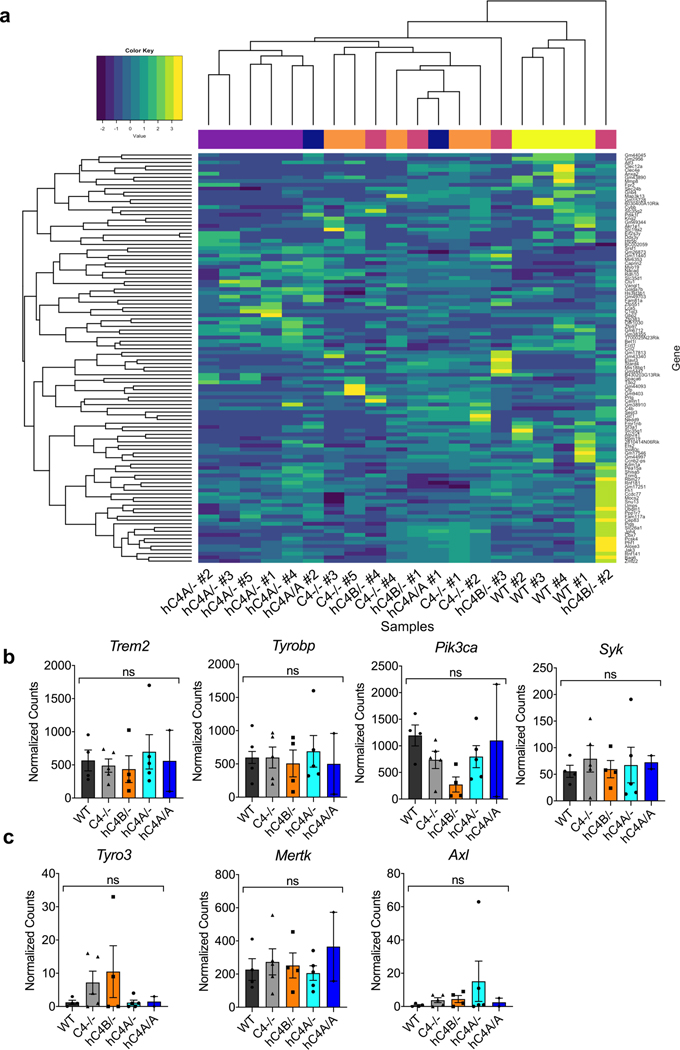
The pronounced phagocytic state of microglia in hC4A/A mice raised the question of whether these microglia engage in transcriptional programs for regulating synapse plasticity under C4A overexpression. To investigate whether C4A overexpression affected gene expression during adolescence, FC microglia from mice were sequenced. Bulk-sequencing analysis showed an unchanged RNA profile in C4−/−, hC4B/−, hC4A/− and hC4A/A mice compared with WT mice (Extended Data Fig. 8a). Moreover, the normalized counts of phagocytic pathway genes showed similar levels among all groups (Extended Data Fig. 8b,c). These results suggest that the increased synapse engulfment observed in the hC4A/A microglia is not due to a change in the microglial transcriptional profile, but instead may be guided by complement tagging of synapses.
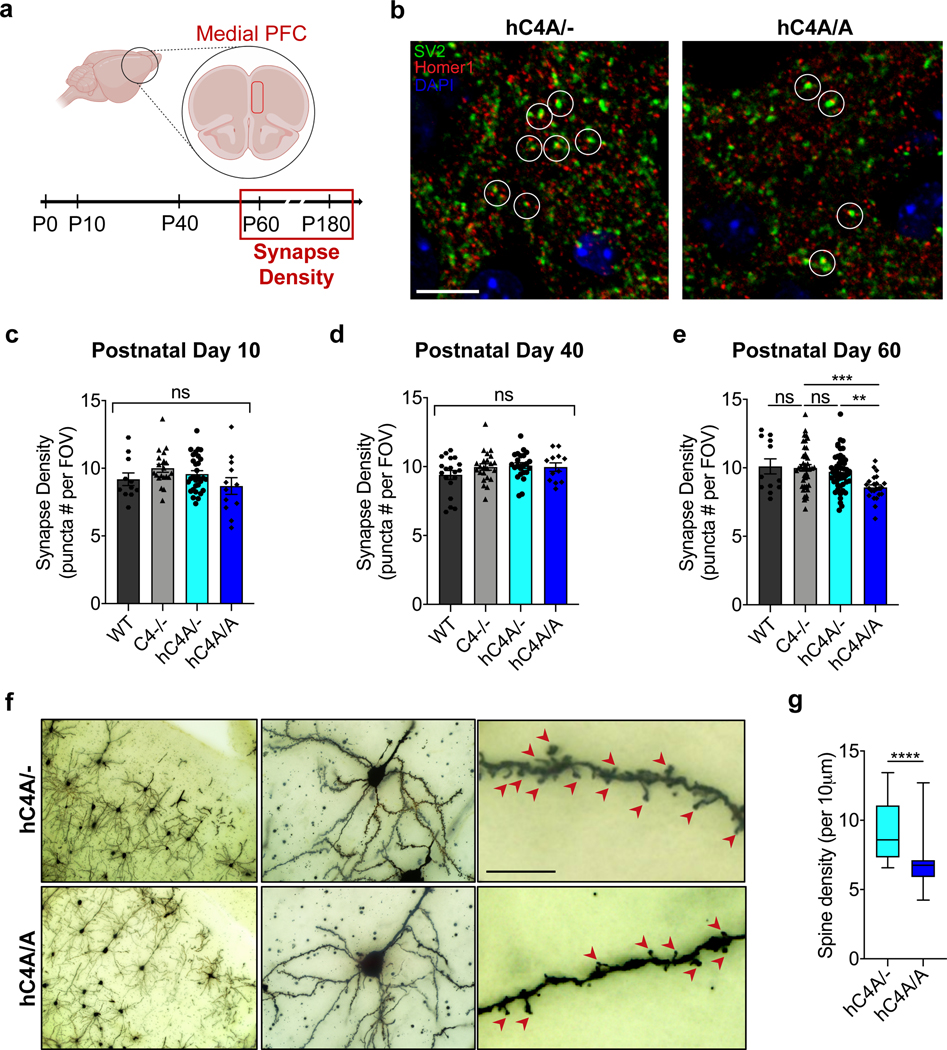
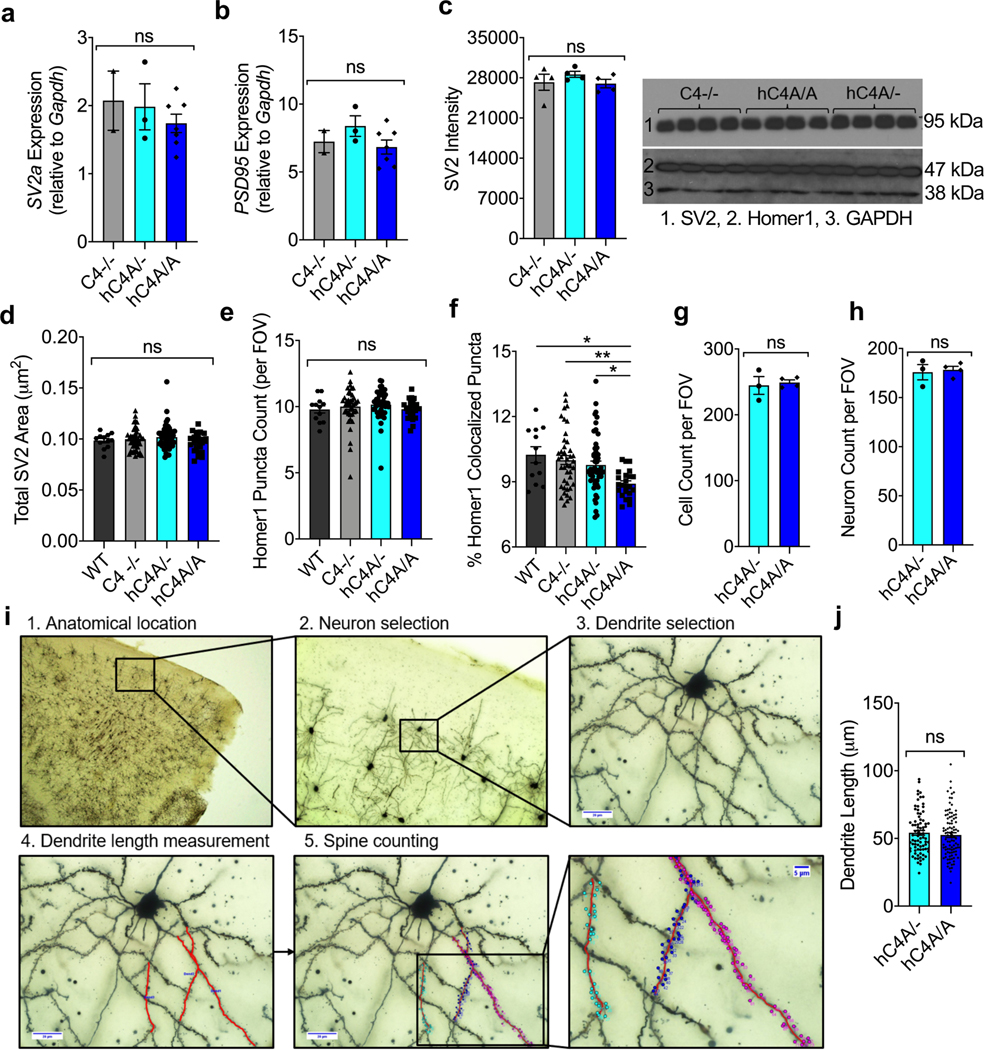
To test whether C4A overexpression and increased microglial engulfment resulted in synapse loss in the mPFC, the synapse density was quantified in WT, C4−/−, hC4A/− and hC4A/A mice by immunostaining for pre- and post-synaptic markers (SV2 and Homer-1, respectively) at P10, P40 and P60 (Fig. 5a). hC4A/A mice did not exhibit a decrease in synapse density at P10 and P40 (Fig. 5c,d); however, a striking reduction in synapse density in hC4A/A mice was observed at P60 (Fig. 5b,e). Additional analyses revealed that expression of the synaptic proteins was not altered in P60 hC4A/A mice (Extended Data Fig. 9a–e), but rather their colocalization was reduced (Extended Data Fig. 9f). Surprisingly, synapse density did not differ between WT and C4−/− mice in this region across all time points, which suggests that complement might not be the only mechanism involved in synaptic pruning in the mPFC.
Fig. 5 |. Adult hC4A-overexpressing mice have decreased synapse density in the mPFC.
a, Schematic of the experiment. The extent of synaptic pruning in the mPFC was evaluated by synapse-density quantification at different time points (P10, P40, P60 and P180). b, Synapses are defined as SV2+ and Homer-1+ colocalized puncta (indicated by white circles) in P60 hC4A/− and hC4A/A mice. Scale bar, 10 μm. c–e, Synapse-density quantification by immunofluorescence in the mPFC at P10 (c), P40 (d) and P60 (e) mice showed a reduction in hC4A/A mice only at the P60 time point. All results were normalized to the C4−/− group to retain littermate controls. c, Synapse density in the mPFC of P10 WT (n = 3), C4−/− (n = 5), hC4A/− (n = 8) and hC4A/A (n = 3) mice was quantified (3–4 FOVs per mouse; 4 independent cohorts; Kruskal–Wallis test with Dunn’s multiple comparisons test). d, Synapse density in the mPFC of P40 WT (n = 3), C4−/− (n = 6), hC4A/− (n = 6) and hC4A/A (n = 3) mice was quantified (3–4 FOVs per mouse; 4 independent cohorts; ordinary one-way ANOVA with Tukey’s multiple comparisons test). e, Synapse density in the mPFC of P60 WT (n = 3), C4−/− (n = 11), hC4A/− (n = 13) and hC4A/A (n = 6) mice was quantified (3–4 FOVs per mouse; 5 independent cohorts; Kruskal–Wallis test with Dunn’s multiple comparisons test, ***P = 0.0007, **P = 0.0039). f, Golgi staining was used to identify pyramidal neurons in the mPFC (left and middle), and high-magnification imaging revealed dendritic spines (right; red arrowheads) in P180 hC4A/− and hC4A/A mice. Scale bar, 10 μm. g, The spine density for each neuron is represented (5–6 neurons per mouse from 5 hC4A/− and 5 hC4A/A littermates from 2 independent cohorts; Mann–Whitney test, two-tailed, ****P < 0.0001). Bar graphs show the mean ± s.e.m. Box-and-whisker plots display the median (center line), the 25th to 75th percentile (box), and minimum to maximum values (whiskers).
To investigate whether the reduced synapse density observed at P60 in C4A-overexpressing mice was sustained through later stages of adulthood, a dendritic spine density analysis was performed using the Golgi–Cox staining method in the mPFC. Pyramidal neurons from layer 2/3 were analyzed from 6-month-old (P180) mice (Extended Data Fig. 9i). Dendritic spine density was significantly reduced in hC4A/A mice compared with hC4A/− mice (Fig. 5f,g), whereas the total cell number, the neuron number and the neuronal dendrite size analyzed were unchanged (Extended Data Fig. 9g,h,j). Altogether, these results show that increased C4A expression leads to excessive synaptic loss in the adult mPFC via increased, cumulative microglia engulfment.
C4A overexpression alters mouse behaviors.
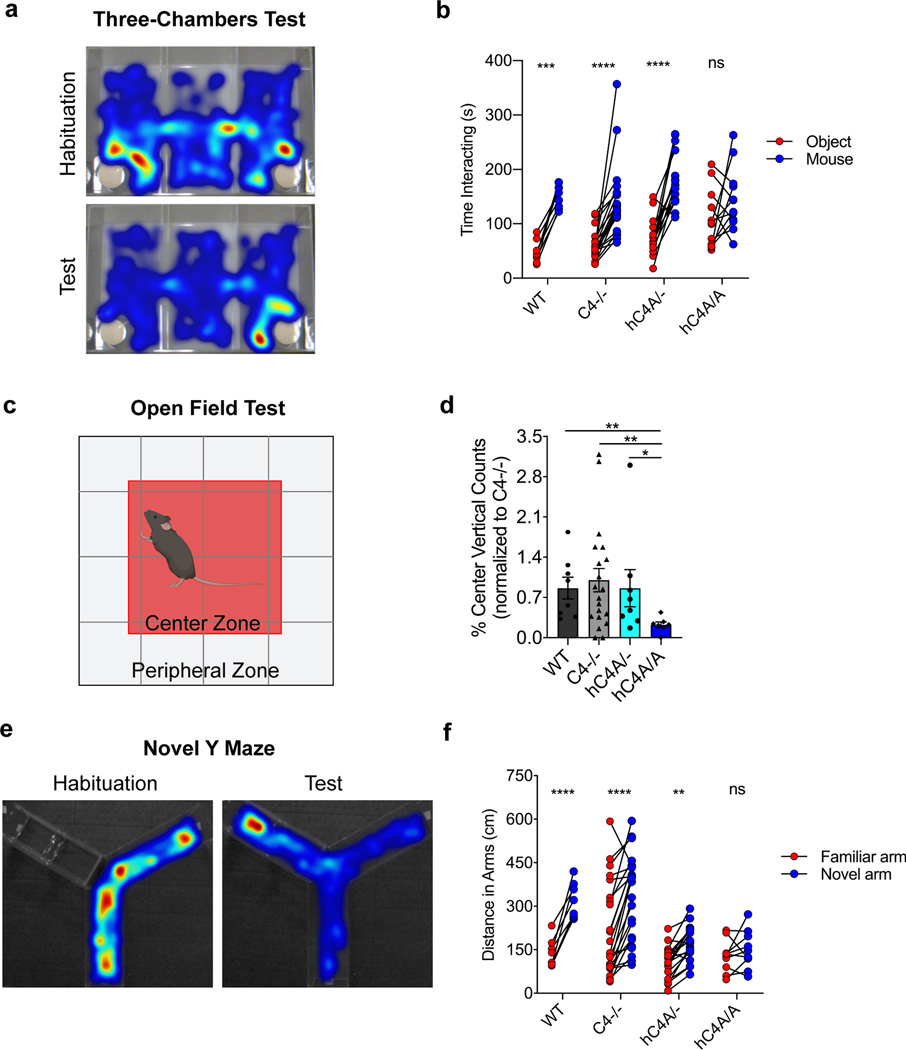
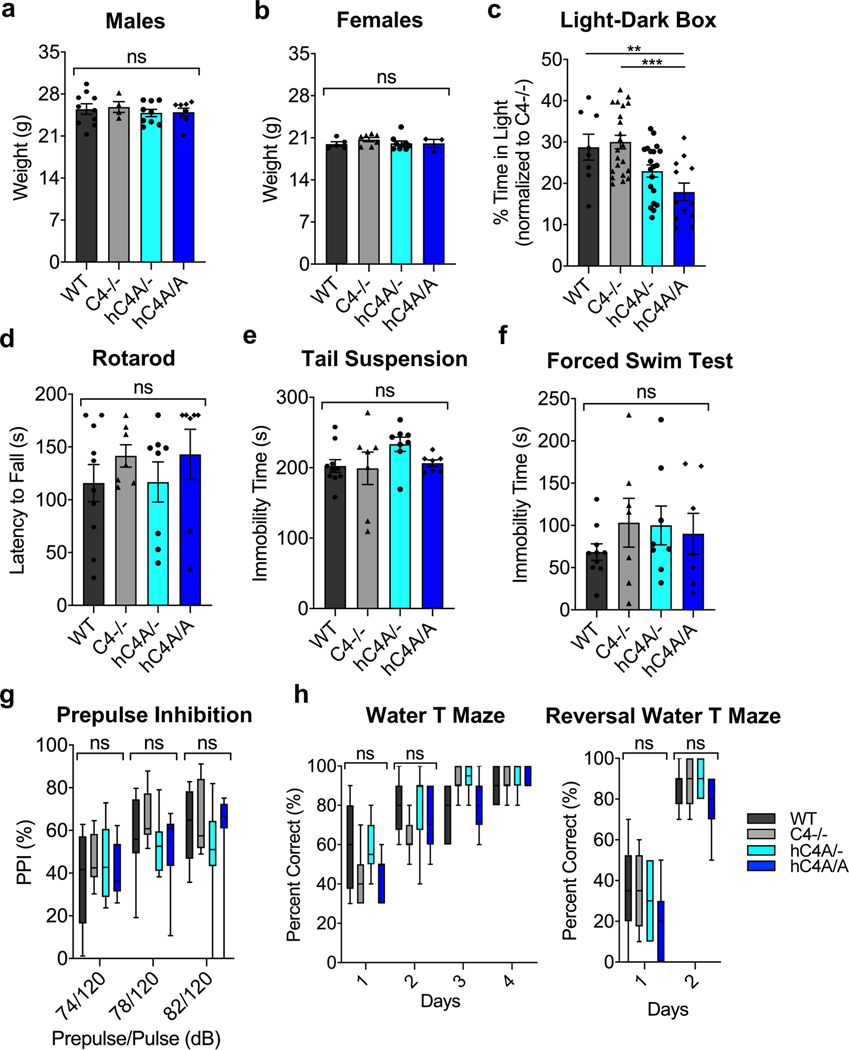
Given the microglial engulfment and synapse density phenotypes, we focused on investigating whether the molecular and cellular effects of C4A overexpression on the murine brain were accompanied by behavioral phenotypes analogous to schizophrenia symptoms. Patients with schizophrenia suffer from cognitive dysfunction, social withdrawal and decreased emotional expression manifesting as amotivation, anxiety and depression37,38. To test for behavior alterations in the hC4A-overexpressing mice, 10- to 12-week-old mice were examined using a battery of common behavior tests. After undergoing the three-chamber social-interaction test (Fig. 6a), C4−/− and hC4A/− mice showed similar social tendencies as WT mice (Fig. 6b). hC4A/A mice, however, had altered social behavior by not showing a preference toward interacting with the mouse as opposed to the object. To measure anxiety levels, mice were placed in the open-field test (Fig. 6c). The hC4A/A mice displayed anxiety-like behavior compared with WT, C4−/− and hC4A/− mice, as measured by the number of rearing events in the center zone (Fig. 6d). Similarly, hC4A/A mice exhibited an anxiety phenotype in the light–dark box test compared with WT and C4−/− mice, although the difference between hC4A/− and hC4A/A mice did not reach significance (Extended Data Fig. 10c). Furthermore, when exposed to the novel Y-maze (Fig. 6e), hC4A/A mice displayed spatial working memory deficits compared with WT, C4−/− and hC4A/− mice by showing no preference toward the novel arm of the maze (Fig. 6f). By contrast, prepulse inhibition (PPI), motor defects, coordination defects or depression-like phenotypes were not detected in hC4A/A mice (Extended Data Fig. 10d–h). These findings suggest that C4A overexpression and its downstream effects can be associated with abnormal mouse behaviors in social, emotional and cognitive contexts, which closely resemble the negative symptoms experienced by patients with schizophrenia.
Fig. 6 |. Human C4A overexpression alters mouse behavior.
a,b, Mice were tested for social-interaction behavior with the three-chambers test. a, Representative heatmaps of the three-chambers test during the habituation period, when mice travel the same amount of time in each chamber, and during the test period, when WT mice spend more time interacting with stranger mouse (right chamber) than with an object (left chamber). b, Representation of the amount of time spent interacting with a novel object (red) or a stranger mouse (blue) for WT ( n = 8), C4−/− (n = 22), hC4A/− (n = 16) and hC4A/A (n = 12) mice (two-way ANOVA with Sidak’s multiple comparisons test, ***PWT = 0.0002, ****P < 0.0001). c,d, The anxiety-like phenotype was tested with the open-field test. c, Representative map of the arena, divided into two zones: center and periphery. d, The percentage of vertical counts in the center for WT (n = 8), C4−/− (n = 20), hC4A/− (n = 8) and hC4A/A (n = 7) mice (results were normalized to the C4−/− group to retain littermate controls; two-tailed Mann–Whitney test, **PWT versus hC4A/A = 0.0022, **PC4−/−vs hC4A/A = 0.0093, *PhC4A/−vs hC4A/A = 0.0140). e,f, Short-term memory was tested using the novel Y-maze. e, Representative heatmaps of the novel Y-maze test during the habituation period, when mice are only allowed to travel in an open arm (right arm), and the during test period, when WT mice spend more time in the novel arm (left arm). f, Distance traveled in the familiar (red) and the novel arm (blue) for WT (n = 8), C4−/− (n = 24), hC4A/− (n = 19) and hC4A/A (n = 10) mice (two-way ANOVA with Sidak’s multiple comparisons test, ****P < 0.0001, **PhC4A/− = 0.0034). For all behavior analyses, WT and C4−/−, and C4−/− and hC4A transgenic mice littermates (two cohorts) were used. Bar graph shows the mean ± s.e.m.
Discussion
To investigate the genetic findings that link the human C4 gene locus to schizophrenia risk, we constructed two mouse models that express human C4A or C4B, and one that overexpresses C4A. We found that C4A and C4B result in different outcomes in synaptic pruning and that the C4A overexpression not only induces increased microglia engulfment, resulting in a reduced synapse density, but also alters mouse behavior. Thus, our findings establish functional differences between C4A and C4B and identifies the impacts of C4A overexpression on the brain.
C4A and C4B have different covalent binding properties, which are driven by the amino acid residue differences in the isotypic region. C4A preferentially forms amine bonds with proteins, whereas hC4B binds hydroxyl groups of carbohydrates7,8,39, as observed in the hemolytic assays. Although there have been various studies showing C4A and C4B differences in vitro, the number of investigations in vivo is limited40. The hC4A and hC4B mouse models serve as important tools as they allow the isotype differences to be tested in the context of synaptic pruning within the brain.
Synaptic pruning and complement components have previously been challenging to study in the mouse cortex. However, the narrow synaptic refinement period of the retinogeniculate circuits in dLGN make it a desirable brain region to investigate complement-dependent synaptic pruning. For this, our initial investigations focused on the dLGN to characterize the functional differences of C4A and C4B and the effects of C4A overexpression. By utilizing these circuits, we found that C4A is functionally more efficient in synaptic pruning than C4B, as it binds to synapses at a higher level. This increased hC4A deposition could be due to its binding properties arising from the hydrophilic amino acid sequence at positions 1101–1102, which bears the exact sequence as mC4, as well as the molecular properties of its target at the synapse. However, the lack of knowledge regarding the binding partner of C4, in addition to the unknown binding properties of C4A and C4B at the synapse, is a caveat when addressing the preferential deposition of C4A. Therefore, in future studies, it will be important to investigate targets of C4A to define the signals that make a synapse vulnerable to complement-mediated elimination. Moreover, experiments using the C4A-overexpressing model demonstrate that an increased copy number of C4A leads to excessive synaptic pruning by microglia engulfment in the dLGN, which is enhanced most likely by increased opsonization of synapses by C3. Importantly, the discovery of an excessive pruning phenotype due to C4A overexpression provides an impetus to explore C4A in regions with higher brain functions.
Previous findings highly associate schizophrenia phenotypes to processes in the mPFC, which makes it an appealing region for investigating synaptic pruning in the context of C4A overexpression. Unlike the retinogeniculate system, the critical refinement period in the mouse FC is likely to span over several weeks, as it occurs over several years during adolescence and early adulthood in humans33. The limited knowledge in synaptic pruning time lines and complement expression in the mPFC necessitates testing multiple time points across different stages of the life of the mouse. We found that the order of events (C4 deposition, microglia engulfment and synapse elimination) in the dLGN is similar in the FC, although the pace and range of the developmental time line are markedly different. The highest levels of C4 protein expression in the forebrain of C4A-overexpressing mice was observed during development (P10). Microglia engulfment in response to hC4 protein expression and deposition in the mPFC peaked during adolescence (P40) in the hC4A-overexpressing mice. Reduced synapse density was observed in hC4A-overexpressing mice only after adolescence (at the P60 time point). Taking into consideration the trends of hC4 protein expression and microglia engulfment, it is likely that the effects of C4A overexpression on synaptic pruning are not instantaneous, but rather cumulative in the mPFC. In the C4A-overexpressing mice, there was an increased C4 protein level, and likely deposition onto synapses, at a level that exceeds a threshold for pathology during development and adolescence. Consequently, there is a continuous increased engulfment of synapses over a period from P10 to P40, thereby resulting in the reduced synapse density pathology observed after adolescence and retained during later stages of adulthood. This study not only demonstrates an ‘excessive pruning’ phenotype in the mPFC, a novel concept in the field, but also establishes the time line of onset and relative order of events regarding synaptic pruning in C4A-overexpressing mice.
The hC4A transgenic mice also helped us identify mouse behavior associated with C4A overexpression. Abnormal social behavior, spatial working memory defects and anxiety phenotypes were observed in mice that overexpress C4A; strikingly, these behaviors resemble negative symptoms of schizophrenia in humans. The observed behaviors are associated with circuits in the mPFC; however, they can also be linked to other various brain regions such as the parietal cortex, the basal ganglia, the hippocampus and the amygdala41–43. Our results showing reduced synapse density in the mPFC can only start to explain the behavior phenotypes observed in C4A-overexpressing mice, and future electrophysiological and behavioral studies are needed to explore specific mPFC circuitry and other brain regions in relation to the aforementioned behaviors.
In contrast to overexpression, the loss of C4 did not affect cortical synapse density, which indicates that C4 is not required for circuit maturation in the mPFC and that other mechanisms may be involved. Moreover, C4−/− mice did not exhibit behavioral defects. Thus, our data suggest that the excessive elimination of synapses after C4A overexpression has pathological consequences on brain development, whereas C4 is not necessary, or has compensatory pathways, for normal brain development in the mPFC. Therefore, the use of therapeutic strategies targeting C4 downregulation can prevent and address the abnormal physiology resulting from C4A overexpression, while not distorting the endogenous synaptic refinement processes in the brain.
The effects of C4A overexpression manifest in molecular, cellular and behavioral phenotypes from development into adolescence and early adulthood. The strongest effects of human C4A became apparent only at substantial numbers of C4A gene copies, which suggests that there is a threshold for pathology. An intriguing possibility proposed by these results is that C4A-related pathology might arise primarily in the context of unknown biological events that provoke greater-than-usual C4 expression and result in C4 levels above the threshold. Individuals with more C4A gene copies could exhibit particularly strong increases in C4 expression in response to such events, and therefore increased vulnerability in particular contexts, but without manifesting brain phenotypes under normal circumstances.
The pathophysiology of schizophrenia is poorly understood, and animal models that recapitulate key molecular events have long been needed. Our findings from creating C4-humanized mice show that C4A is more effective in synapse refinement compared with C4B. We also demonstrated that C4A overexpression is linked to increased microglial engulfment, reduced cortical synapses and altered mouse behaviors that resemble the negative symptoms of schizophrenia. Our findings on the time line of onset, impaired synaptic pruning processes and anomalous behavior in hC4A-overexpressing mice draws important parallels between our overexpression mouse model and schizophrenia pathology. Using mouse models such as described here will be important to further understand the impact of C4A overexpression on other brain regions and ways in which excessive synapse loss affects brain circuits in the context of disease.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41593–020-00763–8.
Methods
Mice.
All animal experiments were carried out in agreement with the institutional guidelines provided by the Institutional Animal Care and Use Committee at Harvard Medical School and Boston Children’s Hospital (protocol numbers IS00000748, IS00000111, IS00004762 and IS00002660), per applicable laws and regulations and in accordance with the guidelines of the Laboratory Animal Center at the NIH for the humane treatment of animals. C57Bl/6J mice (WT; The Jackson Laboratory, 000664), C4−/− mice (The Jackson Laboratory, 003643) and four new transgenic mice strains, hC4A/−, hC4A/A, hC4B/− and hC4AB/−, were bred and maintained under specific pathogen-free conditions. Mice were kept at an ambient temperature of 21 °C under a 12-h light–dark cycle with a standard chow diet at the AAALAC-accredited animal facility at Harvard Medical School. C4−/− mice were generated by our laboratory and previously described44. C4+/− heterozygous parent mice were used to generate litters where WT and C4−/− littermates could be compared. Mice from both sexes were analyzed in all of the experiments. For experiments involving the dLGN, mice at the ages of P5 and P10 were used. For experiments involving the mPFC, mice at the ages of P10, P40, P60, P90 and P180 were used. Further details of the ages can be found in the figure legends. Mice that showed a state of hydrocephalus were excluded from the study. No data points were excluded from any analyses.
Generation of hC4 transgenic mice.
Standard microinjection procedures were used to produce the transgenic mice45. Fertilized mouse eggs were flushed from the oviducts of superovulated 3-week-old C57BL/6J DBA/2 F1 mice (The Jackson Laboratory), and male pronuclei were injected with BAC DNA (2 ng μl−1) that had been purified using a cesium chloride gradient and linearized with the PI-SceI enzyme (New England Biolabs) at its cognate restriction site located in the pBACe3.6 vector. BAC DNA containing hC4A, hC4B or both sequences from a common MHC haplotype were selected (Supplementary Table 1) and purchased from CHORI (https://bacpacresources.org). BAC DNA size and linearization were validated by pulse-field gel electrophoresis (Extended Data Fig. 1e). The injected eggs that developed into the two-cell stage were implanted in the oviducts of pseudopregnant recipient females. At 3 weeks of age, the animals were tested for the presence of the hC4A and hC4B genes by PCR analysis of their genomic DNA (see primer sequences in Supplementary Table 2). hC4A/−, hC4B/− and hC4AB/− strains were generated from one founder for each transgene. Both strains were backcrossed into the C4−/− strain for at least ten generations before use in experiments to ensure a C57BL/6J genetic background. All animals used for experiments were tested for the presence of the transgene by ddPCR (see below).
Determining gene copy number variation by ddPCR.
hC4A, hC4B, C4A-L and C4B-S gene copy numbers in the transgenic mice were determined by ddPCR as previously described for human samples6. Tail specimens were incubated with DirectPCR lysis buffer (Viagen Biotech) containing 0.5 μg ml−1 Proteinase K (Fisher Bioreagents) overnight at 55 °C for tissue lysis. DNA was purified with a DNA Clean & Concentrator kit (Zymo Research) following the manufacturer’s instructions, and the DNA concentration was measured using a Nanodrop-1000 spectrophotometer (Thermo Fisher Scientific). Purified genomic DNA was diluted to 25 ng μl−1 and digested with AluI (New England Biolabs) for 1 h at 37 °C. We mixed 30 ng of digested DNA with 2× ddPCR Supermix for Probes (no dUTP, Bio-Rad) and 20× primer–probe mix for hC4A or hC4B and the mouse gene Rpp30 as a reference (18 μM of forward and reverse primers each; 5 μM of fluorescent probe). The sequences for primers and probes are listed in Supplementary Table 2. For each sample, this reaction mixture was then emulsified into approximately 20,000 droplets in an oil/aqueous emulsion using a microfluidic droplet generator (Bio-Rad). The droplets containing this reaction mixture were subjected to PCR using the following cycling conditions: 95 °C for 10 min, 40 cycles of 94 °C for 30 s and 60 °C (for C4A and C4A-L) or 59 °C (for C4B and C4B-S) for 1 min, followed by 98 °C for 10 min. After PCR, the fluorescence (both colors) in each droplet was read using a QX100 droplet reader (Bio-Rad). Data were analyzed using QuantaSoft software (Bio-Rad, v.1.7.4.0917), which estimates the absolute concentration of DNA templates by Poisson-correcting the fraction of droplets that are positive for each amplicon (C4 or Rpp30). Since there are two copies of Rpp30 (the control locus) in each diploid genome, the ratio of the concentration of the C4 amplicon to that of the reference (Rpp30) amplicon is multiplied by two to yield the measurement of copy number of the C4 sequence per diploid genome.
Whole-genome sequencing.
For each transgenic mouse strain, we created a custom reference genome consisting of the mm10 mouse reference and the sequences of the inserted BAC and cloning vector used. Reads were aligned to the custom genome for each mouse using BWA MEM (v.0.7.12)46 and duplicates were marked using Picard Mark Duplicates (http://broadinstitute.github.io/picard/). We identified candidate insertion locations for the BACs by running DELLY (v.0.7.6, default parameters)47 and Manta (v.1.0.3, default parameters)48 and searching for candidate translocation calls linking the BAC contig or cloning vector contig to canonical contigs in the mm10 genome. All candidate translocation calls were manually reviewed and discarded if they appeared to be due to misalignments of reads due to sequence homology between the mm10 reference and the BAC sequence. To assess the read depth, we collected coverage metadata for each whole-genome sequencing sample using Genome STRiP (v.r1609)49 and generated read depth profiles (command: ComputeDepthProfiles) using a custom uniqueness mask generated for each transgenic genome reference (command: ComputeGenomeMask) that masked non-uniquely alignable 100-mers in the reference and then plotted read depth profiles near the candidate insertion sites and across the inserted BAC sequence.
RNA expression analysis by ddPCR and PCR with reverse transcription.
hC4A, hC4B, C3 and C1qb RNA levels were determined by reverse transcription followed by ddPCR as previously described in human samples6. Briefly, RNA was isolated using the TRIzol protocol, and for each sample, 1 μg of RNA was reverse transcribed using an iScript kit (Bio-Rad). A similar ddPCR mix and PCR program as for gene copy number was used. Primers and probes for expression are listed in Supplementary Table 2. Mouse Hs2st1 for brain samples and Eif4h for spleen and liver samples was used as a normalization control to calculate the ratio of hC4A (or hC4B) to Hs2st1 (or Eif4h) expression. PCR with reverse transcription reactions were assembled for genes of interest (Sv2a, Sema6d and Psd95) using the iTaq universal SYBR Green supermix system (Bio-Rad) and run on a CFX96 machine (Bio-Rad). Gene expression was normalized to Gapdh and compared using the ΔΔCt method. All primer sequences are listed in Supplementary Table 2.
Serum ELISA.
C4 protein levels were detected in serum from WT (C57BL/6J) and hC4 transgenic mice. Details of all the antibodies used are provided in Supplementary Table 3. High binding microplates were coated overnight with rat anti-mouse C4 (16D2, in-house) for mC4 or goat antisera to hC4 antibody (Quidel) for hC4 at 4 °C. All washes were done with 0.05% Tween 20 in PBS, repeated three times. Plates were then blocked with 1% BSA in PBS for 1 h at 37 °C. Samples were applied in a twofold dilution series, starting at 1:2,000 dilution in PBS for hC4 transgenic mouse serum, 1:100 for C57BL/6J mouse serum, and incubated for 2 h at 37 °C. Purified hC4 (Complement Technology) or mC4 (provided by G. R. Andersen, Aarhus, Denmark) was used as standards starting at 100 ng ml−1 and 500 ng ml−1, respectively, followed by twofold dilutions. C4 in samples was detected with rabbit anti-C4c antibody (Dako), followed by goat anti-rabbit horseradish peroxidase, both incubated at 1 h at 37 °C. The signal was developed by the addition of TMB substrate, and the reaction was stopped with 2 M H2SO4. Absorbance at 450 nm was measured, and absolute quantification was measured based on a standard curve.
To detect C3 levels, plates were coated with goat anti-C3 and detected by rabbit anti-C3 (Abcam). As standard, purified mouse C3 (Complement Technology, M113) was used, starting at 50 ng ml−1 followed by twofold dilutions. C1q levels were measured by coating with rat anti-C1q (Abcam) and detecting by rabbit anti-C1q (Abcam). As standard, purified mouse C1q (Complement Technology, M099) was used, starting at 50 ng ml−1 followed by twofold dilutions. For both proteins, ELISA was performed with the same protocol described above.
Tissue ELISA.
To measure C4 protein levels in the CNS, hC4 transgenic mice were transcardially perfused with PBS and brains were collected as fresh-frozen. The forebrain was cryosectioned as 100-μm sections and collected. Total protein from tissue was extracted using N-PER neuronal extraction buffer (Thermo Fisher) containing 1× protease inhibitor cocktail (Pierce, A32965) according to the manufacturer’s protocol. Total protein (200 μg; determined using a BCA kit) was loaded as the starting volume and followed by twofold dilutions. C4 amount was detected as described above. Results are plotted as C4 amount (ng) in 100 μg total protein.
Immunohistochemistry.
Brains were collected from mice after transcardial perfusion with PBS followed by 4% paraformaldehyde (PFA). Tissue was then immersed in 4% PFA overnight following perfusion, cryoprotected in 30% sucrose, embedded in OCT and frozen. Tissue was cryosectioned (14 μm), sections were dried, washed three times in PBS and blocked with 5% BSA and 0.2% Triton X-100 in PBS for 1 h. Primary antibodies were diluted in the antibody buffer (containing 0.2% Triton X-100 and 5% BSA). Details of all the primary antibodies used are provided in Supplementary Table 3. The antibodies were incubated overnight at 4 °C, and secondary Alexa-conjugated antibodies (Invitrogen) were added at 1:200 in the antibody buffer for 2 h at room temperature. Slides were mounted in Vectashield (containing DAPI).
Spleen sections (14 μm) were fixed with acetone, blocked with 10% goat serum and 0.1% Triton X-100 in PBS for 1 h. Details of all the antibodies used are provided in Supplementary Table 3. Primary antibodies were diluted in the antibody buffer (containing 10% goat serum and 0.1% Triton X-100). The antibodies were incubated for 2 h at room temperature, and secondary Alexa-conjugated antibodies (Invitrogen) were added at 1:200 in the antibody buffer for 2 h at room temperature. Slides were mounted in Vectashield (containing DAPI).
All sections were imaged with an Olympus IX 81 microscope with a FluoView FV1000 confocal system at Boston Children’s Hospital (Program in Cellular and Molecular Medicine Microscopy Core). All images from confocal microscopy were processed using Fiji (Image J, NIH) unless otherwise specified.
Eye-specific segregation analysis.
Visualization and analysis of RGC synaptic inputs in the mouse dLGN was performed as previously described26. CTB conjugated to Alexa 488 (green label) and CTB conjugated to Alexa 594 (red label; Supplementary Table 4) were intravitreally injected into the left and right eyes, respectively, of P9 mice, which were killed the following day. Images were acquired using a Zeiss Axiocam and quantified blinded to experimental conditions and compared to age-matched littermate controls. The percentage of overlap was measured as previously described19,29.
Synapse density analysis.
FC sections were stained with pre- and post-synaptic markers (SV2 and Homer-1, respectively). Details of all the antibodies used are provided in Supplementary Table 3. Sections were imaged using an Olympus FV1000 confocal microscope at ×60 magnification and ×2 optical zoom. Z-stacks were taken 0.5-μm above and below the midplane with 0.2-μm increments. Four fields of view (FOVs) were imaged per animal from two separate brain sections. Images were analyzed using Fiji software. Projection images were subjected to background subtraction with a rolling ball radius of 10 pixels. To identify SV2+ and Homer-1+ puncta, each image was thresholded according to the software’s auto function, followed by the Analyze particles function. Colocalized puncta were determined by the image calculator - “AND” function for the SV2 and Homer-1 images, with a lower limit of 15 pixels for size.
To compare WT and transgenic mice groups, the common genotype from each litter (C4−/− group) was used as the baseline. C4-deficient animals from both WT and hC4A litters were used to normalize data points for their respective litters, allowing us to represent our data as relative changes in synapse density in WT, hC4A/− and hC4A/A groups with respect to their C4−/− littermates.
Synaptic C4 deposition quantification.
dLGN sections from P5 mice were stained for the pre-synaptic marker Vglut2 and for hC4. Details for all the antibodies used are provided in Supplementary Table 3. The sections were imaged, quantified and analyzed using the same method as for synapse density analysis. Vglut2-stained areas were recorded for further analysis. The groups were normalized to the number of hC4 gene copies: four copies for hC4A/− and two copies for hC4B/−.
Splenic C4 and C3 deposition quantification.
Spleen sections were stained for CD169, CD21 and C4 or C3. Sections were imaged using an Olympus FV1000 confocal microscope at ×20 magnification. Z-stacks were taken 5-μm above and below the midplane with 1-μm increments. Four FOVs were imaged per animal from two separate spleen sections. Z-projection images were analyzed using Cell Profiler (Broad Institute of Harvard and MIT, v.4.0.2)50. The follicular dendritic cell (FDC) networks in each FOV were identified and used as a mask to identify C4 or C3 puncta. The fluorescence intensity was measured using the MeasureImageIntensity tool in Cell Profiler. All fluorescence intensity measurements were normalized by the FDC area.
Golgi–Cox staining and dendritic spine density analysis.
Brains were collected from P180 transgenic mice and processed according to the protocol provided with the FD Rapid Golgi Stain kit (FD Neurotechnologies) for the Golgi–Cox staining method. The brains were then sectioned (100 μm per slice) using a cryostat. Sections were collected on gelatin-coated microscope slides, dehydrated in absolute alcohol, cleared in xylene and cover slipped. The stained pyramidal neurons from layer 2/3 of the mPFC were examined, and Z-stack images were captured using bright-field imaging on an Olympus Bx63 microscope at ×4, ×20 and ×100 magnification (Harvard Medical School Neurobiology Imaging Facility) and captured using cellSens software (Olympus). Images at ×4 magnification were used for mapping the region of interest and ×20 images were used for selecting neurons (Extended Data Fig. 9i). To be selected for analysis, Golgi-impregnated neurons had to meet the following characteristics: (1) dark and consistent impregnation throughout the extent of all of the dendrites; (2) relative isolation from neighboring impregnated cells that could interfere with analysis; and (3) a cell body in the middle third of the tissue section to avoid analysis of impregnated neurons that extend largely into other sections. Images at ×100 magnification were used for dendritic spine analysis. Secondary and tertiary dendrites were measured for length. Dendritic spines were manually counted by an experimenter blinded to the mouse genotype, regardless of shape. The density of dendritic spines was calculated as the number of spines per 10 μm of dendrite. All image processing was carried out using Fiji (Image J, NIH).
Microglia isolation.
To prepare single-cell suspensions, the FC was dissected, and the tissue was digested in 330 U papain in DPBS at 35 °C for 30 min. Papain was neutralized in an ovomucoid solution and cells were dissociated by gentle trituration and filtered through a 70-μm nylon mesh to remove clumps. Microglia were then purified by magnetic purification using anti-CD45 microbeads (Miltenyi).
Flow cytometry.
Cells were treated with the antibody clone 2.4G2 to block Fc receptors before staining, and staining was performed in a PBS solution containing 5% fetal calf serum and 2 mM EDTA. For intracellular staining, microglia were fixed and permeabilized (fixation and permeabilization buffers from BioLegend) before antibody staining. Anti-SV2 (DSHB/UIowa) was prepared and conjugated to fluorochromes in-house. Monoclonal antibodies against CD45, CD11b and CD68 were purchased from BioLegend, and viability dye (eBioscience, 65–0865-14) was used to exclude dead cells. Stained cells were acquired using a FACS Canto II and FACSDiva software (BD Biosciences), and data were analyzed using FlowJo (Tree Star, v.10.4) software.
Synaptosome isolation.
All procedures were done at 4 °C using precooled reagents. Brains were removed from the cranium into HEPES-buffered sucrose (0.32 M sucrose, 5 mM HEPES pH 7.4) containing protease inhibitor cocktail (Roche mini complete tablets) and homogenized in a glass dounce. The homogenate was centrifuged at 1,200 × g at 4 °C for 10 min and the resulting supernatant was spun at 15,000 × g for 15 min to yield the crude synaptosomal pellet. The pellet was resuspended in 1 ml of HEPES-buffered sucrose and layered on top of a discontinuous gradient containing 5 mM HEPES and 0.8 to 1.0 to 1.2 M sucrose (top to bottom, which equals 27%/34%/41%) in an ultracentrifuge tube then centrifuged at ~150,000 × g for 2 h in a swinging bucket rotor. Synaptosomes were recovered in the layer between 1.0 and 1.2 M sucrose.
Complement binding assay.
Synaptosomes were incubated with 10% fresh serum from C4−/−, hC4B/− or hC4A/− mice (diluted in half with C4−/− serum) for 30 min at 37 °C in gelatin veronal buffer (Complement Technology) then washed three times in PBS. Synaptosomes were fixed and permeabilized (fixation and permeabilization buffers from BioLegend) before antibody staining with anti-SV2 (DSHB/UIowa). For C4 detection, synaptosomes were stained with polyclonal rabbit anti-C4 (Dako) and acquired using a FACS Canto (BD Biosciences), and data were analyzed using FlowJo (Tree Star, v.10.4) software.
Hemolytic assay.
EA sheep red blood cells (Complement Technology) were washed with gelatin veronal buffer (Complement Technology) and counted with a spectrophotometer at 541 nm. To obtain 109 cells per ml, the following formula was used: final volume = initial volume × OD541/0.62. Cells were then diluted to 108 cells per ml, incubated with 2% C4-deficient guinea pig serum mixed with serial dilutions of fresh serum from hC4A/− and hC4B/− mice for 30 min at 37 °C. Absorbance was read at 415 nm. The percentage of hemolysis was measured based on 100% lysis control. In parallel, C4 concentration was measured by ELISA for each sample, and hemolytic activity per g of C4 was calculated and normalized to hC4A samples.
Western blotting.
Equal amounts of proteins (20 μg as determined using a BCA protein assay) were diluted 1:1 with 2× Laemmli buffer with 1 mM dithiothreitol. Samples were heated at 95 °C for 5 min, except for the samples used for SV2 detection. Protein electrophoresis was performed using a Mini-Trans-blot cell (Bio-Rad). Proteins were then transferred onto polyvinylidene difluoride membranes (Immun-BlotPVDF, Bio-Rad) for western blot analysis. Membranes were blocked in a 5% milk solution in TBS-T (0.1% Tween 20) for 1 h at room temperature and then incubated with anti-SV2 (DSHB/UIowa), anti-Homer-1 (Synaptic Systems) and anti-GAPDH (BioLegend) primary antibody overnight at 4 °C. Following washes in TBS-T, secondary antibody conjugated with horseradish peroxidase (Southern Biotech) was hybridized for 1 h at room temperature. Membranes were washed in TBS-T again and then reactivity was revealed via a chemiluminescence reaction performed with ECL detection reagents (Pierce) and film exposure.
Microglia engulfment analysis in the mPFC.
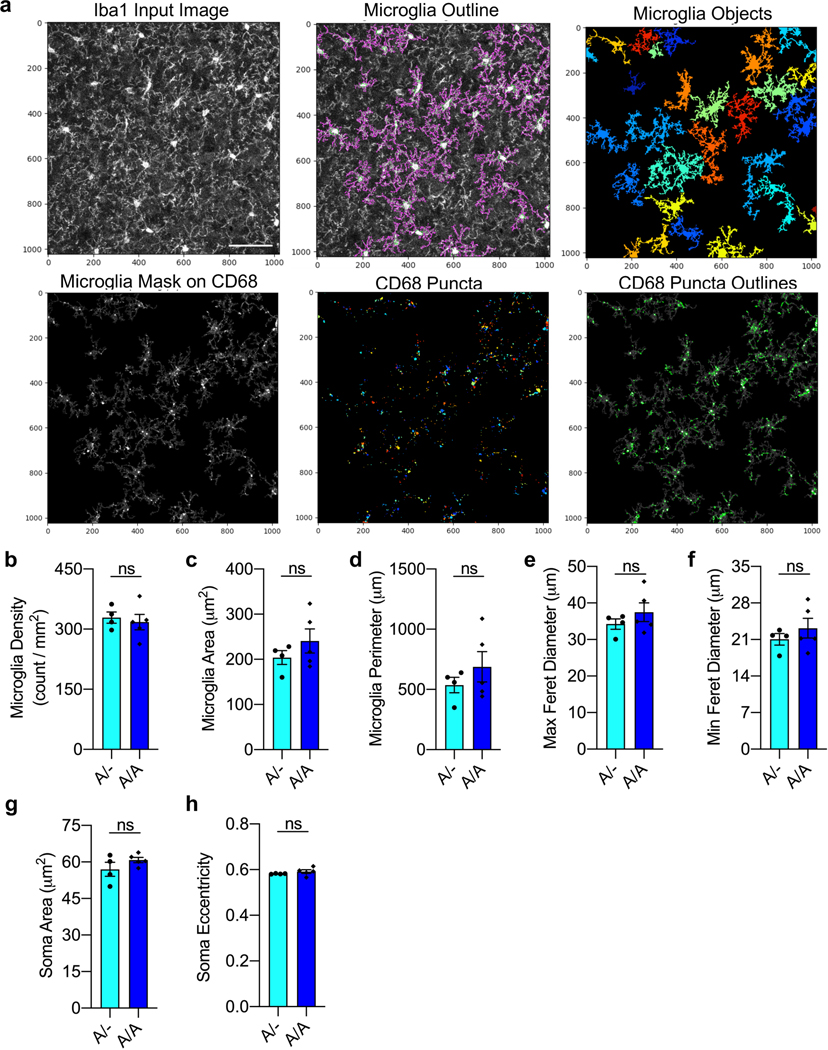
We immunostained 40-μm sagittal sections containing the regions of interest of the brain for Iba1 (1:400, Wako) and CD68 (1:300, BioLegend). A confocal microscope was used to collect four ×40 FOVs per mouse with 1-μm interval Z-steps. The analysis was performed on maximum-intensity projections of the 40-μm section using the software Cell Profiler (Broad Institute of Harvard and MIT, v.4.0.2)50. The microglial soma and processes were mapped out based on Iba1 signature, and the microglia identified were used as a mask for identifying intracellular CD68 puncta. The engulfment levels were evaluated by the number and area of CD68+ puncta (Extended Data Fig. 7a). Additional morphological parameters were measured using the Cell Profiler module MeasureObjectSizeShape. All analyses were performed blinded to the genotype and the treatment group.
Microglia engulfment analysis in the dLGN.
The in vivo microglia phagocytosis assay and analyses were performed as previously described in detail26,51. Briefly, mice received an intraocular injection of fluorescent-conjugated anterograde tracer CTB (Supplementary Table 4) at P4. Animals were killed at P5 and brains were fixed overnight in 4% PFA at 4 °C, followed by cryoprotection with 30% sucrose. We prepared 40-μm sections from only those brains with sufficient dye fill. For each animal, three to four adjacent dLGN sections were imaged for reconstruction of RGC inputs, Iba1 staining and subsequent engulfment analysis. Images were acquired using a confocal microscope at ×60 magnification with 0.2-μm interval Z-steps. Three to five FOVs were collected from each section (10–15 FOVs per animal). Images were processed using ImageJ (NIH) and analyzed using Imaris software (Bitplane).
Bulk RNA-sequencing analysis.
Microglia were first sorted from FC homogenates using a FACSAria. For each mouse, 1,000 microglia were sorted based on CD11bhigh and CD45low expression into 20 μl guanidine thiocyanate buffer (Qiagen) supplemented with 1% β-mercaptoethanol. RNA was isolated using RNAClean XP beads (Agencourt, A63987). Template switching, complementary DNA synthesis, cDNA amplification and libraries were prepared using a Nextera kit (Illumina). Libraries were sequenced with a MiSeq (Illumina), giving 76-bp paired-end reads. STAR Aligner was used to align the paired-end reads to the mouse reference genome (Ensembl)52. EdgeR (Bioconductor) was used for differential gene analysis53. R and RStudio were used in the process of differential gene expression analysis and in creating heatmaps54.
Behavior analyses.
All behavioral analyses were performed in the Mouse Behavior Core of Harvard Medical School and Brigham and Women’s Hospital. Before each test, mice were allowed to acclimate in their housing cages in a separate room for 30 min.
Three-chamber social-interaction test.
The three-chamber sociability test included two consecutive 10-min sessions and was performed in a rectangular arena constructed of clear Plexiglas that was 63-cm long × 40-cm wide × 22-cm high. The test arena contained three equally sized chambers (40-cm long × 20-cm wide × 22-cm high), and the outer left and right chambers each contained an inverted wire cup. During the habituation session, test mice were initially placed into the center chamber and allowed to freely explore the three-chambered arena. For the social-interaction session, test mice were returned to the center chamber, an unfamiliar target mouse was placed under one of the two wire cups, an object was put under the second cup, and test mice were again allowed to move freely among the three chambers. The position of the first target mouse was alternated and equally distributed between the left and right cups for both transgenic and control mice. The test operator was blinded to the test mouse genotype and remained outside the test room during all sessions. Test subject performance was automatically recorded using TopScan software (CleverSys) or EthoVision XT 14 software (Noldus), and the following behaviors were scored: amount of time spent in close proximity (3-cm radius) of the wire cups, amount of time spent in each chamber of the three-chamber arena and latency to approach each wire cup.
Light–dark box test.
The experimental setup included an ENV-510 open-field arena with dimensions 53.34 × 34.93 × 1.27 cm (Med Associates) that was equally portioned into two chambers with an opening in between. One chamber was brightly illuminated (950 lux) and the other was covered with an opaque, black, plastic insert to prevent light from passing through. The mice were placed in the middle of the light chamber and were allowed to freely explore both chambers for 10 min. The movements of the mice were tracked by infrared beams, and all the data were recorded and analyzed using Activity Monitor software (Med Associates). The percentage of time spent in the light box is reported.
Novel Y-maze.
The experimental setup included a clear, three-arm, Y-shaped maze with a 120° angle between the arms. One of the arms was denoted as the start arm throughout all trials, where all mice were placed in the beginning of each trial. Each mouse trial was run in two stages, habituation (forced choice) and test (free choice), with an inter-trial interval of 2 min. During habituation, one of the arms besides the starting arm was blocked, and the blocked arm was randomized for each trial. The mice were allowed to explore the maze for 3 min. After habituation, during the inter-trial interval when the mice were taken out of the maze, the maze was cleaned, and the blocked arm was re-exposed. For the test run, mice were placed back in the maze at the start arm and were allowed to explore both the novel (previously blocked) and familiar (previously explored) arms. All trials were video recorded, and the data were acquired using EthoVision XT 14 software (Noldus). The time spent in the familiar and the novel arm during the test run is reported.
Open-field test.
The mouse open-field chambers (Med Associates) were made of Plexiglas and consisted of a square base (27 cm × 27 cm) with walls that were 20-cm high. All walls were clear, but opaque barriers were added between arenas so that subjects did not see one another during testing. For each testing session, the mouse was allowed free exploration in the environment for 1 h. A computer-assisted infrared tracking system and software (Activity Monitor, Med Associates) were used to record the number of beam breaks, time and entries into the center and peripheral zones. The total distance traveled (cm) was used as a measurement of general locomotor activity.
Water T-maze.
This behavioral paradigm, encompassing similar principles to the Morris water maze, was used to assess spatial learning and memory. The mouse uses spatial cues to navigate and find an escape platform submerged just below the surface of the water. In addition, the test measures cognitive flexibility through a reversal trial requiring the mice to learn a new platform location. The testing apparatus was a plus maze (each arm 14.1-cm long and 4.6-cm wide) made of clear Plexiglas with each arm designated as north (N), south (S), east (E) or west (W). For the mice to choose only the E or W arm for escape, dividers were placed to block off either the N or S arm. Mice were placed in the N or S arms, in a semi-random order, at the start of each trial. The maze is filled with 25–26 °C water and made opaque by adding nontoxic white paint so the mouse cannot see the submerged escape platform. Initially for the acquisition trial, the hidden escape platform was placed in the E arm. Mice were carried to the starting point and the experimenter scored a correct or incorrect response for each trial based on whether the arm with the escape platform was chosen. Regardless of the response, mice were allowed to remain on the platform for 10 s at the end. Mice were given ten trials per day and the percentage of correct responses was calculated by averaging the correct responses across the ten trials for each day. The acquisition criterion was achieved when a mouse scored 80% or more correct responses over two consecutive days. After all mice reached the acquisition criterion, the reversal trial began, whereby the platform was moved to the opposite side (W) and the same procedure was repeated until all mice learn the new location of the platform.
PPI test.
PPI was monitored using Med Associates chambers. Mice were acclimated to the PPI apparatus for 5 min in the presence of a ~63-dB white-noise background. After this acclimation period, the test session was automatically started. The session began with a habituation block of six presentations of the startle stimulus alone, followed by ten PPI blocks of six different types of trials. The following trial types were used: null (no stimuli), startle (120 dB), startle plus prepulse (variable dB over a background noise of 65, 75 or 85 dB) and prepulse alone. Each trial began with a 50-ms null period during which baseline movements were recorded. There was a subsequent 20-ms period during which prepulse stimuli were presented and responses to the prepulse measured. After a further 100 ms, the startle stimuli were presented for 40 ms and responses recorded for 100 ms from startle onset. Responses were sampled every ms. The inter-trial interval varied, with an average of 15 s (range from 10 to 20 s). The percentage PPI = (1 − (prepulse trials/startle-only trials)) × 100.
Rotarod test.
The purpose of the rotarod test is to assess the sensorimotor coordination and/or fatigue resistance of the mouse. The test is sensitive to damage of the basal ganglia and cerebellum, and to drugs that affect motor functions. The rotarod apparatus consisted of a gritted or textured plastic roller (Ugo Basile) flanked by large round plates on each end to prevent the animal from escaping. The plastic rod sat at a height of approximately 20-cm above individual electronic sensing platforms. The sensing platforms were not cushioned, but mice do not appear to experience any discomfort from falling from that height. Before actual testing, mice were habituated to the rotarod apparatus by placing them onto the rod as it revolved at a very low rotation speed (4 r.p.m. for 5 min) (the number of times a mouse falls was recorded and the mouse was placed back on the rotarod if it falls). Following the habituation session, mice were tested in the accelerated rotarod (4–40 r.p.m. in 3 min). The latency until the mouse falls from the rod onto the sensing platform below was recorded automatically. This test was repeated after a 15-min break.
Tail-suspension test.
This procedure is an alternative to the forced-swim test and is used to induce a despair-like state and to test the effects of antidepressants in mice. Antidepressants will reduce the amount of time the mouse spends immobile in this test. At the beginning of a trial, mice were suspended by the tail (taped onto a suspension hook so that the animal hangs with its tail in a straight line) above a flat surface covered with soft padding material. During testing (10-min trial), the duration of immobility was automatically scored (Med Associates) or by an experimenter.
Forced-swim test.
This procedure is used to induce a despair-like state and to test the effects of antidepressants in mice. Antidepressants will reduce the amount time the mouse spends immobile in this test. Mice were placed for 6 min in a glass cylinder (height, 35 cm; diameter, 17 cm) filled with water (25 ± 2 °C) to a depth of 25 cm. The water depth was adjusted so that the animals must swim or float without their hind limbs or tail touching the bottom. During testing, the duration of immobility (the time during which the subject makes only the small movements necessary to keep their heads above water) was scored. The experimenter monitored each mouse continuously during the entire session. The mouse was immediately taken out of the cylinder and excluded from the study if it failed to swim or keep its head above water. After completion of the trial, the subject was dried using a paper towel and placed back in its home cage. After every trial, the water was changed, and the cylinder rinsed with clean water.
Statistical analysis and study design.
All data in dot plots are presented as the mean ± s.e.m. All box-and-whisker plots display the median (center line), the 25th to 75th percentile (box), and the minimum to maximum values (whiskers). To determine the distribution of datasets, a D’Agostino–Pearson normality test was performed in all analyses. For data that passed the normality test, a two-tailed parametric unpaired t-test was used; for the datasets that were not normally distributed, a two-tailed Mann–Whitney test was used. One-way (or Kruskal– Wallis test for non-normally spread data followed by Dunn’s multiple comparisons tests) and two-way analysis of variance (ANOVA) tests were followed by Tukey’s multiple comparisons and Sidak’s multiple comparisons tests, respectively. Details of statistical analyses are provided in Supplementary Table 5. All statistical analyses were carried out using GraphPad Prism 8 and Microsoft Excel.
Experiments were repeated with two to three independent cohorts, and the obtained results were replicable. Details of the number of independent experiments are provided in the figure legends. Investigators involved in conducting experiments, collecting data and performing analyses were blinded to the mice genotypes. No statistical method was used to predetermine sample sizes. However, our sample sizes were similar to those reported in previous publications6,19,26,55. All mice in the study were randomly allocated in experimental groups. Randomization protocols in behavior experiments are further explained in their respective subsections. Additional specifics regarding study design and methods are provided in the Nature Research Reporting Summary.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Characteristics of C4 BAC DNA integration.
a-d, Insertion sites and associated rearrangements are shown for each mouse strain. a, c, The BAC DNA insertion site is shown for each mouse strain. Normalized read depth in 2-kb genomic windows at the insertion sites of the BACs. Dashed vertical lines indicate the insertion site. The hC4A insertion was associated with a duplication, and the hC4B insertion was associated with a small deletion. b, d, The BAC DNA-associated rearrangements are shown for each mouse strain. Normalized read depth in 1-kb genomic windows shows the copy number of the inserted constructs. Gene models are shown beneath the read depth plots. Human C4A was likely inserted into hC4A transgenic mice at a normalized read depth between 5 and 9. Human C4B was likely inserted into hC4B transgenic mice at a copy number between 2 and 5. The inserted hC4B BAC constructs contain additional internal copy number variants. e, Pulse-field gel showing linearized hC4B and hC4A BAC DNA used for microinjection into zygotes (representative of 3 independent experiments). Unprocessed versions of the gels can be found in Supplementary Figure 1a. f, Sema6D mRNA expression level in cortex was not changed by the region duplication caused by hC4A BAC DNA insertion. Gapdh was used as a control housekeeping gene (n = 4 mice per group; ns = P > 0.05, Kruskal-Wallis test with Dunn’s multiple comparisons test). Bar graph shows mean ± s.e.m.
Extended Data Fig. 2 |. Peripheral expression and function of C4 in hC4 transgenic mice.
a, C4A- and C4B-specific mRNA level was measured by ddPCR in the spleen (left) and liver (right) of hC4A/- and hC4B/- mice. eiF4H was used as the control housekeeping gene (n = 4 mice per group; * PSpleen = 0.0286, * PLiver = 0.0286; two-tailed Mann-Whitney test). b, C4 hemolytic activity was measured with equal amounts of hC4A or hC4B protein using sensitized sheep red blood cells. Data were normalized to hC4A samples (n = 8 hC4A and n = 15 hC4B combined from 3 independent experiments; **** P < 0.0001, Unpaired, two-tailed t test). Bar graphs show mean ± s.e.m.
Extended Data Fig. 3 |. Complement activation and binding to synaptosomes.
a, Representative dot plots showing the FSC-A / SSC-A of 1-μm beads, 6-μm beads, and synaptosomes. Further analysis will be gated on 1-μm synaptosomes. b, Synaptosomes were permeabilized and stained with anti-SV2 antibody (+ SV2 Ab) or no antibody (FMO CT). More than 85% of the particles analyzed contain SV2 protein. c, Representative histogram plot of C4 staining on synaptosomes isolated from C4−/− mice and incubated with serum from C4−/− (red), hC4A (orange), and hC4B (blue) mice. d, C1q deposition is shown and quantified using serum from hC4A mice (orange; n = 2), hC4B mice (blue; n = 2), or no serum CT (red). e, C4 (left) and C1q (right) deposition was detected on synaptosomes using fresh (red) or heat-inactivated (blue) serum. Bar graph shows mean ± s.e.m.
Extended Data Fig. 4 |. Human C4 in the retinogeniculate system.
a, hC4AB/- mice: gene copy numbers for C4A, C4B, C4A-L (C4L), and C4B-S (C4S) were determined by ddPCR using Rpp30 as a reference gene. It showed an insertion of two C4A-L genes and one C4B-L gene (n = 5 representative mice). Whole-genome sequencing revealed the hC4AB BAC DNA was inserted in chromosome 3 and that one C4A copy was truncated resulting in only one C4A coding copy (data not shown). b-c, Absolute quantification by ddPCR was used to measure C4A- and C4B-specific mRNA level in the retina and LGN from P5 hC4A/- (n = 3), hC4B/- (n = 3), and hC4AB/- mice (n = 2). Bar graphs show mean ± s.e.m.
Extended Data Fig. 5 |. Complement profile in hC4 transgenic mice during adolescence (P40).
a-c, Level of classical complement cascade proteins were measured in serum by ELISA in adolescent (P40) WT, hC4A/-, and hC4A/A mice. a, C4 serum level was measured in WT (n = 8), hC4A/- (n = 10), and hC4A/A mice (n = 6). b, C3 serum level was measured in WT (n = 5), hC4A/- (n = 10), and hC4A/A mice (n = 6). c, C1q serum level was measured in WT (n = 5), hC4A/- (n = 9), and hC4A/A mice (n = 8). d-f, RNA expression of classical complement cascade components in the FC were measured by ddPCR in adolescent (P40) WT, hC4A/-, and hC4A/A mice. All RNA measurements were normalized to Hs2st1 expression level. d, C4 mRNA expression level in FC was measured in WT (n = 4), hC4A/- (n = 4), and hC4A/A mice (n = 2). e, C3 mRNA expression level in FC was measured in WT (n = 4), hC4A/- (n = 3), and hC4A/A mice (n = 2). f, C1qb mRNA expression level in FC was measured in WT (n = 4), hC4A/- (n = 4), and hC4A/A mice (n = 2). Bar graphs show mean ± s.e.m.
Extended Data Fig. 6 |. C4A overexpression doesn’t change LGN cellularity or morphology.
a, Representative images of contralateral (top), ipsilateral (middle), and overlapping regions (bottom; scale = 20 μm). b-d, dLGN size (b), contralateral area (c), and ipsilateral area (d) were measured in P10 hC4A/- and hC4A/A mice (n = 6 hC4A/- and n = 6 hC4A/A littermates from 3 independent cohorts; two-tailed Mann-Whitney test; ns = P > 0.05, * P = 0.0411; scale = 20 μm). e, Iba1 staining was used to count the total number of microglia in the dLGN FOV (n = 4 mice per group; two-tailed Mann-Whitney test; ns = P > 0.05). f, Mice were injected with fluorescent-conjugated cholera toxin subunit-B (CTB) in each eye at P4 and brains were harvested at P5. Representative pseudo-colored images of P5 dLGN from C4−/−, hC4A/-, and hC4A/A littermates of contralateral (red), ipsilateral territories (green), and overlap (yellow) between the two territories (scale = 20 μm). Percent of overlap in ipsilateral region was compared in C4−/− (n = 3), hC4A/- (n = 4), and hC4A/A mice (n = 2; littermates). g, DAPI was used to count the total number of cells in the dLGN field of view (FOV) (n = 6 hC4A/- and n = 5 hC4A/A littermates from 3 independent cohorts; two-tailed Mann-Whitney test; ns = P > 0.05). h, The number of Brn3a+ RGCs was compared between C4−/−, hC4A/-, and hC4A/A mice (n = 5 mice per group; Kruskal-Wallis test with Dunn’s multiple comparisons test; ns = P > 0.05; scale = 20 μm). Bar graphs show mean ± s.e.m.
Extended Data Fig. 7 |. Cell Profiler microglia morphological analysis.
a, Cell Profiler software was used to analyze microglia morphology and lysosomal activity in the frontal cortex of P40 hC4A/- and hC4A/A mice. Microglial soma and processes are identified by Iba1 signature by using the image-based watershed method (top row). Microglia are used as a mask to select and quantify intracellular CD68 puncta (bottom row; scale = 50 μm). b-h, Morphological parameters for mPFC microglia (b-f) and their soma (g-h) from hC4A/- (n = 4) and hC4A/A (n = 5) mice were calculated by Cell Profiler software at P40 timepoint (ns = P > 0.05, two-tailed Mann-Whitney test). Bar graphs show mean ± s.e.m.
Extended Data Fig. 8 |. Microglial RNA sequencing analysis reveals no transcriptomic alterations in hC4 transgenic mice.
a, Bulk RNA sequencing analysis of microglia isolated from the frontal cortex of adolescent (P40) mice from WT (n = 4), C4−/− (n = 5), and hC4A/- (n = 5), hC4B/- (n = 4), and hC4A/A (n = 2) groups. Heatmap representation of differentially expressed genes between all experimental groups shows no significant transcriptional profile difference between any two groups. b-c, Normalized gene counts for the TREM2/DAP12 signaling pathway (b) and TAM receptor genes (c) were calculated for WT (n = 4), C4−/− (n = 5), hC4A/- (n = 5), hC4B/- (n = 4), and hC4A/A (n = 2; ns = P > 0.05, Kruskal-Wallis test with Dunn’s test). Bar graphs show mean ± s.e.m.
Extended Data Fig. 9 |. Synaptic protein expression is not affected by C4A overexpression in mice.
a-b, rt-PCR was used to measure mRNA expression of Sv2a (a) and Psd95 (b) in the FC in adult mice (C4−/− n = 2, hC4A/- n = 3, and hC4A/A littermates n = 7). RNA expression was normalized to Gapdh expression. c, SV2 and PSD95 protein level were analyzed by western blot. GAPDH was used as a loading control protein (C4−/− n = 4, hC4A/- n = 4, and hC4A/A n = 4 littermates from one experiment; Kruskal-Wallis test with Dunn’s multiple comparisons test; ns = P > 0.05). Unprocessed versions of the western blots can be found in Supplementary Figure 1b. d, Total SV2 area per FOV in the mPFC was calculated from immunofluorescence staining between WT (n = 3), C4−/− (n = 11), hC4A/- (n = 13), and hC4A/A (n = 6) groups (Kruskal-Wallis test with Dunn’s multiple comparisons test). e-f, Total Homer1 puncta and percentage of colocalized Homer1 puncta from the mPFC in adult mice were calculated for WT (n = 3), C4−/− (n = 11), hC4A/- (n = 13), and hC4A/A (n = 6) groups (Kruskal-Wallis test with Dunn’s multiple comparisons test; ns = P > 0.05, * PWT vs A/A = 0.0139, ** PWT vs A/A = 0.0139). g-h, Brain sections were stained with DAPI and NeuN, and cellularity was measured in frontal cortex at P180. Total cells (g) and neurons (h) per FOV in frontal cortex is represented for hC4A/- (n = 3) and hC4A/A mice (n = 4; two-tailed Mann-Whitney test; ns = P > 0.05). i, Method used for the manual counting of dendritic spines in mPFC of hC4 mice. j, Length of dendrites that have been used for spine density analysis for hC4A/- (80 dendrites) and hC4A/A (96 dendrites; two-tailed Mann-Whitney test; ns = P > 0.05). Bar graphs show mean ± s.e.m.
Extended Data Fig. 10 |. Human-C4A overexpression alters mouse behavior.
a-h, WT, C4−/−, hC4A/-, and hC4A/A mice were subjected to a battery of behavioral tests. a-b, Weight of male and female mice were compared between WT (n = 10), C4−/− (n = 4), hC4A/- (n = 9), and hC4A/A mice (n = 8; Kruskal-Wallis test with Dunn’s multiple comparisons test; ns = P > 0.05). c, Anxiety levels were measured in the light-dark box test by time spent in the light-zone between WT (n = 8), C4−/− (n = 22), hC4A/- (n = 20), and hC4A/A (n = 12) groups (two-tailed, unpaired t test; ** P = 0.0085; two-tailed Mann-Whitney test, *** P = 0.0009). Results were normalized to the C4−/− group to retain littermate controls. d, In the rotarod test, latency to fall was measured in seconds for WT (n = 10), C4−/− (n = 7), hC4A/- (n = 8), and hC4A/A (n = 7) groups (Kruskal-Wallis test with Dunn’s multiple comparisons test, ns = P > 0.05). e-f, Immobility time in seconds was compared in the tail suspension test (e) and the forced swim test (f) for WT (n = 10), C4−/− (n = 7), hC4A/- (n = 8), and hC4A/A (n = 7) groups (Kruskal-Wallis test with Dunn’s multiple comparisons test, ns = P > 0.05). g, Prepulse inhibition was measured between WT (n = 10), C4−/− (n = 7), hC4A/- (n = 8), and hC4A/A (n = 7) groups (two-way ANOVA with Tukey’s multiple comparisons test; ns = P > 0.05). h, Percent of correct decisions were recorded in the water t maze and the reversal water t maze for WT (n = 10), C4−/− (n = 7), hC4A/- (n = 8), and hC4A/A (n = 7) groups (two-way ANOVA with Tukey’s multiple comparisons test; ns = P > 0.05). Bar graphs show mean ± s.e.m. Box-and-whisker plots display the median (center line), 25th to 75th percentile (box), and minimum to maximum values (whiskers).
Supplementary Material
Acknowledgements
We thank members of the Carroll, Stevens and McCarroll laboratories for helpful comments on experimental design and feedback on the manuscript; E. M. Carroll for invaluable assistance; G. R. Andersen for providing the purified mouse C4 protein; X. Chen, D. Ghitza and L. Prince-Wright for technical support; B. Caldarone at the Mouse Behavior Core of Harvard Medical School and Brigham and Women’s Hospital; H. Leung at the Boston Children’s Hospital/PCMM microscopy core; the Boston Children’s Hospital/PCMM flow cytometry core; K. Holton at the HMS research computing core; and C. Usher and D. E. Potts for help with editing the manuscript. Parts of Figs. 2a and Fig. 3 were drawn by using pictures from Servier Medical Art (http://smart.servier.com/), licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/). Parts of Figs. 1a,b, 3a, 4a, 5a and 6c were created with BioRender (https://biorender.com/). This research was supported by funding from NIH P50MH112491–01 (to B.S., S.A.M. and M.C.C.) and AR072965 (to M.C.C.).
Footnotes
Data availability
The bulk RNA-sequencing data have been uploaded to the NCBI GEO under accession code GSE161557. Source data for Extended Data Figs. 1a and 9c are provided in Supplementary Fig. 1. The datasets generated and analyzed during the current study, along with the detailed protocols and codes for the experiments, are available upon request from the corresponding author.
Code availability
The study utilized simple codes commonly used for bulk RNA sequencing. All scripts and codes are available upon request from the corresponding author.
Competing interests
B.S. serves on the scientific advisory board of Annexon LLC and is a minor shareholder of Annexon LLC; however, this is unrelated to the submitted work. All other authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41593–020-00763–8.
Supplementary information is available for this paper at https://doi.org/10.1038/s41593–020-00763–8.
Peer review information Nature Neuroscience thanks Marie-Eve Tremblay, Michisuke Yuzaki, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Cannon TD et al. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc. Natl Acad. Sci. USA 99, 3228–3233 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon TD et al. Progressive reduction in cortical thickness as psychosis develops: a multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol. Psychiatry 77, 147–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glantz LA & Lewis DA Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry 57, 65–73 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Glausier JR & Lewis DA Dendritic spine pathology in schizophrenia. Neuroscience 251, 90–107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feinberg I. Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J. Psychiatr. Res 17, 319–334 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Sekar A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belt KT, Carroll MC & Porter RR The structural basis of the multiple forms of human complement component C4. Cell 36, 907–914 (1984). [DOI] [PubMed] [Google Scholar]
- 8.Law SK, Dodds AW & Porter RR A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 3, 1819–1823 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isenman DE & Young JR The molecular basis for the difference in immune hemolysis activity of the Chido and Rodgers isotypes of human complement component C4. J. Immunol 132, 3019–3027 (1984). [PubMed] [Google Scholar]
- 10.Presumey J, Bialas AR & Carroll MC Complement system in neural synapse elimination in development and disease. Adv. Immunol 135, 53–79 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Katz LC & Shatz CJ Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Sanes JR & Lichtman JW Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci 22, 389–442 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Hua JY & Smith SJ Neural activity and the dynamics of central nervous system development. Nat. Neurosci 7, 327–332 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Lehrman EK et al. CD47 protects synapses from excess microglia-mediated pruning during development. Neuron 100, 120–134.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinhard L. et al. Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat. Commun 9, 1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung WS et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datwani A. et al. Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron 64, 463–470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjartmar L. et al. Neuronal pentraxins mediate synaptic refinement in the developing visual system. J. Neurosci 26, 6269–6281 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens B. et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Corriveau RA, Huh GS & Shatz CJ Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron 21, 505–520 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Bochner DN et al. Blocking PirB up-regulates spines and functional synapses to unlock visual cortical plasticity and facilitate recovery from amblyopia. Sci. Transl. Med 6, 258ra140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nonaka M. et al. Identification of the 5-flanking regulatory region responsible for the difference in transcriptional control between mouse complement C4 and Slp genes. Proc. Natl Acad. Sci. USA 83, 7883–7887 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y. et al. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J. Immunol 171, 2734–2745 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Gadjeva M. et al. Macrophage-derived complement component C4 can restore humoral immunity in C4-deficient mice. J. Immunol 169, 5489–5495 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Huh GS et al. Functional requirement for class I MHC in CNS development and plasticity. Science 290, 2155–2159 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer DP et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guido W. Refinement of the retinogeniculate pathway. J. Physiol 586, 4357–4362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godement P, Salaün J. & Imbert M. Prenatal and postnatal development of retinogeniculate and retinocollicular projections in the mouse. J. Comp. Neurol 230, 552–575 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Jaubert-Miazza L. et al. Structural and functional composition of the developing retinogeniculate pathway in the mouse. Vis. Neurosci 22, 661–676 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Hooks BM & Chen C. Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron 52, 281–291 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Werneburg S. et al. Targeted complement inhibition at synapses prevents microglial synaptic engulfment and synapse loss in demyelinating disease. Immunity 52, 167–182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh R. et al. Fibroblast growth factor 22 contributes to the development of retinal nerve terminals in the dorsal lateral geniculate nucleus. Front. Mol. Neurosci 4, 61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petanjek Z. et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl Acad. Sci. USA 108, 13281–13286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zotova E. et al. Inflammatory components in human Alzheimer’s disease and after active amyloid-β42 immunization. Brain 136, 2677–2696 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Rabinowitz SS & Gordon S. Macrosialin, a macrophage-restricted membrane sialoprotein differentially glycosylated in response to inflammatory stimuli. J. Exp. Med 174, 827–836 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Correll CU & Schooler NR Negative symptoms in schizophrenia: a review and clinical guide for recognition, assessment, and treatment. Neuropsychiatr. Dis. Treat 16, 519–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mccutcheon RA, Marques TR & Howes OD Schizophrenia—an overview. JAMA Psychiatry 77, 201–210 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Isenman DE & Young JR Covalent binding properties of the C4A and C4B isotypes of the fourth component of human complement on several C1-bearing cell surfaces. J. Immunol 136, 2542–2550 (1986). [PubMed] [Google Scholar]
- 40.Finco O, Li S, Cuccia M, Rosen FS & Carroll MC Structural differences between the two human complement C4 isotypes affect the humoral immune response. J. Exp. Med 175, 537–543 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko J. Neuroanatomical substrates of rodent social behavior: the medial prefrontal cortex and its projection patterns. Front. Neural Circuits 11, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eriksson J, Vogel EK, Lansner A, Bergström F. & Nyberg L. Neurocognitive architecture of working memory. Neuron 88, 33–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourin M. & Hascoët M. The mouse light/dark box test. Eur. J. Pharmacol 463, 55–65 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Fischer MB et al. Regulation of the B cell response to T-dependent antigens by classical pathway complement. J. Immunol 157, 549–556 (1996). [PubMed] [Google Scholar]
- 45.Van Keuren ML, Gavrilina GB, Filipiak WE, Zeidler MG & Saunders TL Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 18, 769–785 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv https://arxiv.org/abs/1303.3997 (2013). [Google Scholar]
- 47.Rausch T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Handsaker RE et al. Large multiallelic copy number variations in humans. Nat. Genet 47, 296–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpenter AE et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schafer DP, Lehrman EK, Heller CT & Stevens B. An engulfment assay: a protocol to assess interactions between CNS phagocytes and neurons. J. Vis. Exp 10.3791/51482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunningham F. et al. Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy DJ, Chen Y. & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koyama Y, Hattori T, Nishida T, Hori O. & Tohyama M. Alterations in dendrite and spine morphology of cortical pyramidal neurons in DISC1-binding zinc finger protein (DBZ) knockout mice. Front Neuroanat. 9, 52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.