Abstract
Accurate assessment of fetal gestational age (GA) is critical to the clinical management of pregnancy. Industrialized countries rely upon obstetric ultrasound (US) to make this estimate. In low- and middle- income countries, automatic measurement of fetal structures using a low-cost obstetric US may assist in establishing GA without the need for skilled sonographers. In this report, we leverage a large database of obstetric US images acquired, stored and annotated by expert sonographers to train algorithms to classify, segment, and measure several fetal structures: biparietal diameter (BPD), head circumference (HC), crown rump length (CRL), abdominal circumference (AC), and femur length (FL). We present a technique for generating raw images suitable for model training by removing caliper and text annotation and describe a fully automated pipeline for image classification, segmentation, and structure measurement to estimate the GA. The resulting framework achieves an average accuracy of 93% in classification tasks, a mean Intersection over Union accuracy of 0.91 during segmentation tasks, and a mean measurement error of 1.89 centimeters, finally leading to a 1.4 day mean average error in the predicted GA compared to expert sonographer GA estimate using the Hadlock equation.
Keywords: Fetal ultrasound, GA estimation, Machine learning
1. INTRODUCTION
The lack of access to care during pregnancy and childbirth in low-resource settings represents a significant challenge to improving maternal and perinatal health outcomes. A clear disparity exists between industrialized and developing regions in the world, where the least developed countries have greater maternal mortality and complications related to pregnancy and childbirth are more than 300 times greater in low- and middle- income countries (LMICs)1.2 Obstetric ultrasound (US) is the primary diagnostic modality for several conditions impacting maternal and fetal outcomes, including multiple gestation, congenital anomalies, fetal growth restriction, abnormalities of placental implantation, and amniotic fluid disturbances. Additionally, US is critical for determining fetal gestational age (GA), a key piece of information upon which much obstetric decision-making is based.3 Improved access to obstetric US is hoped to improve accurate targeting of risk-reducing health care interventions and decrease the burden of specific pregnancy-related complications in LMICs. Conventional obstetric US requires skilled operators to obtain specific images of uterine, placental, and fetal anatomy. With the advent of artificial intelligence (AI) and deep learning algorithms, many image recognition tasks can be automated, thus reducing the skill required by the device operator. However, large, annotated image databases are required for training these algorithms effectively.
In this paper, we propose an end-to-end framework to automate image recognition and measurement of major anatomical structures in fetal US images. We start by training a UNET4 to remove calipers and text annotations from clinically obtained images. Most commercial ultrasound machines do not collect these annotations in a separate layer; rather, they are “burned in” to the image. This has historically limited the utility of routinely-collected ultrasound image data in training machine learning classifiers, since it is imperative to ensure that models are trained solely on raw features, rather than their interpretation by experts. The burned-in annotations are removed to let the classifiers solely rely on US image features, and to facilitate use of the vast routine-care retrospective data.
Once the annotations are removed, the UNET output is used to train a RESNET5 classifier to identify images of the fetal head, abdomen, femur, and in early pregnancy, fetal body length (a.k.a. crown-rump length; CRL). Next, we train a modified residual UNET (RUNET)6 for image segmentation of the anatomical structures. Finally, we measure the segmented structure of interest which can be subsequently used for gestational age estimation by applying any of a number of established formulas7,8,9 to the estimated biometry measurements.
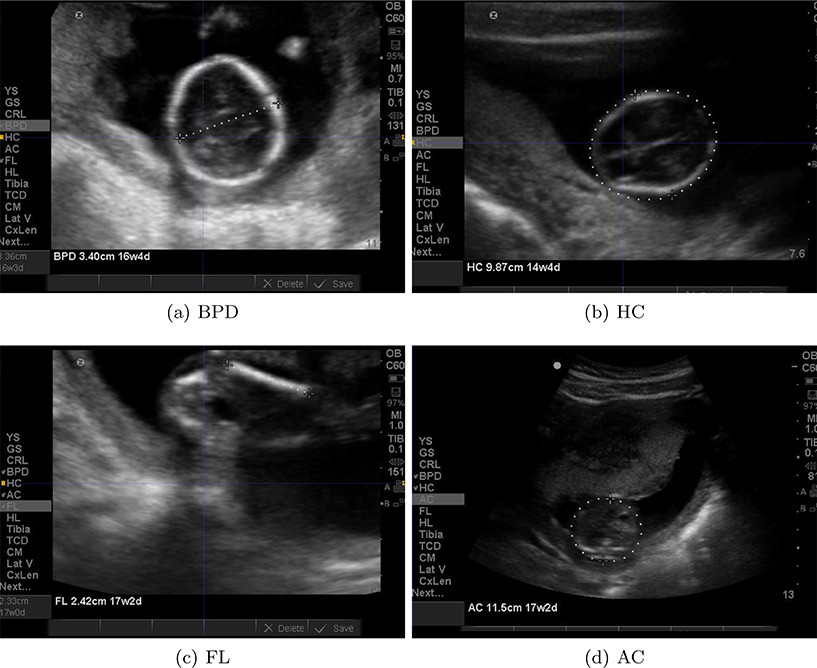
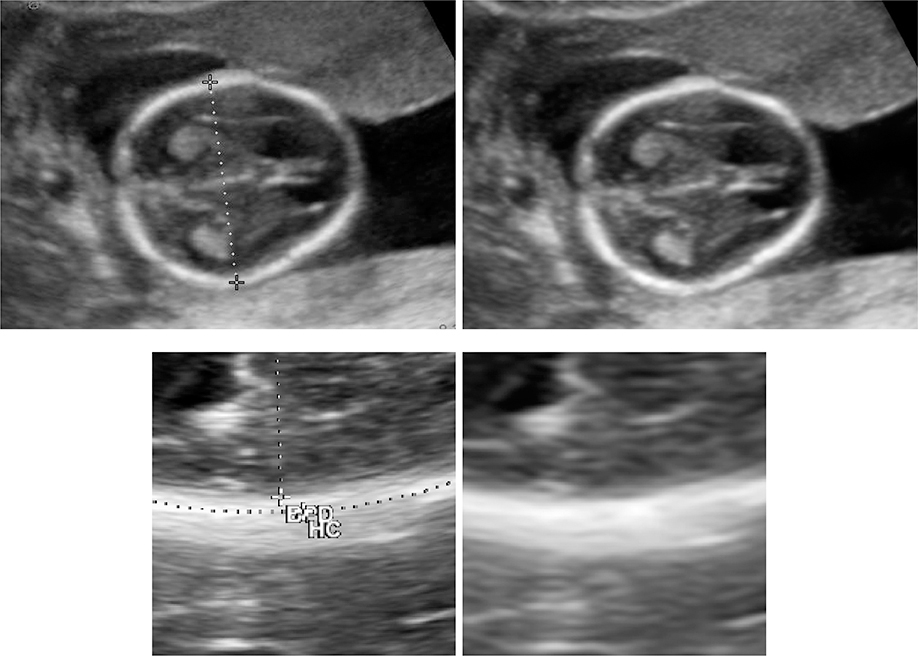
Figure 1 shows examples of US images with different calipers and annotations by experts.
Figure 1.
Examples of historical images that could be used for training machine learning algorithms. The image annotations/calipers are removed to preserve image features for future analysis.
To the best of our knowledge this is a first attempt to create an end-to-end pipeline that includes classification, segmentation, and measurement of multiple fetal structures for GA prediction. Previous work has evaluated state-of-the-art convolutional neural networks(CNNs)for the task of fetal structure classification using 2D still frame images (BPD=99.5%, FL=89.1%, AC=91.3%).10 Others have explored the classification and measurement of the fetal head from images selected from free-hand US sweeps11, 12 with high accuracy. We report comparable high classification accuracy, and a low measurement error compared to expert measurements.
2. MATERIALS
We used three data sources for this work. The Zambian Preterm Birth Prevention Study (ZAPPS) is a prospective pregnancy cohort established at the Women and Newborn Hospital of the University Teaching Hospitals (UTH) in Lusaka, Zambia.13 Enrolled participants receive routine antenatal and postnatal care, lab testing, mid-trimester cervical length measurement, serial fetal growth monitoring, and careful assessment of birth outcomes. Between August 2015 and September 2017 (study phase 1), 1450 women were enrolled at a median gestational age of 16 weeks (IQR 13–18). 23,209 fetal biometry images from 3,369 studies were collected in this phase of the ZAPPS Study. We call this retrospective data set ZAPPS.
The second data set used for training was a historical archive of studies performed for clinical care at the University of North Carolina Hospitals during the years 2012 through 2018. This database includes 124,646 2D images from 2,983 ultrasound sessions. Ultrasound studies of women who received obstetric ultrasound by the University of North Carolina Maternal-Fetal Medicine group are included in this data set. We call this retrospective data set UNC.
We used optical character recognition (OCR)* on the ZAPPS and UNC datasets to identify burned-in labels and measurement calipers placed on images by sonographers at the time of collection, including 7274 head images labeled with “BPD” and/or “HC”, 3152 embryo/fetus images labeled with “CRL”, 7216 femur images labeled with “FL” and 6717 abdominal images labeled with “AC”. The BPD, HC, FL, AC and CRL labels refer to biparietal diameter, head circumference, femur length, abdominal circumference and crown-rump length, respectively. These are the standardized ultrasound images with specific anatomic landmarks used universally in clinical practice to estimate fetal size and gestational age. These images were used to train the generative networks.
Finally, we evaluated our classification and segmentation methods on a third, prospectively collected dataset obtained as part of the ongoing Fetal Age Machine Learning Initiative (FAMLI), a prospective study funded by the Bill and Melinda Gates Foundation (OPP1191684). FAMLI enrolled adult pregnant women in both Lusaka and Chapel Hill, North Carolina, and collected a variety of fetal ultrasound data, including raw images without burned-in annotation. Our evaluation set included images collected between September 2018 and October 2019, and included 1646 BPD/HC, 2622 AC, 2466 FL, and 499 CRL images without labels or measurement calipers. We call this data set FAMLI.
3. METHODS
3.1. Caliper removal via inpaint/UNET
As a preprocessing step to prepare the ZAPPS and UNC datasets for model training, calipers and extra text were removed from each image. As mentioned above, this ensured the classification and segmentation algorithms rely only on native image features during the training process.
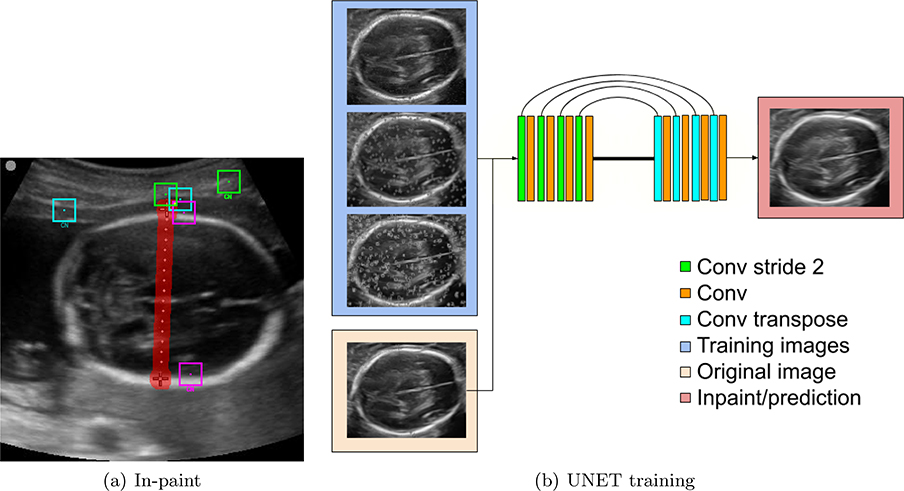
Caliper removal is performed in two stages. First, an inpainting technique for object removal is employed to erase calipers from a set of images.14, 15 This algorithm takes as input an image and the target region. Starting from the outer edges, pixels in the labeled region are replaced by averaging pixels from similar patches. Figure 2(a) shows example patches and their corresponding closest neighbor(CN) in the image. Next, a UNET4 is trained to automatically remove the calipers. Ideally, the caliper removal should not rely on identifying the text or calipers, therefore, we train a neural network to automate this procedure. Figure 2 summarizes the caliper and text removal steps and the UNET architecture.
Figure 2.
Caliper removal: The in-paint technique replaces the labeled region (red) using redundant information in the image. The UNET is trained using the in-paint images as target and the source images (blue) are modified by randomly adding small calipers, big calipers and text (top to bottom).
The images produced in this step are used as input for the rest of the methods described in subsequent sections.
3.2. Image classification via RESNET
The first step towards a fully automated system for GA prediction is to classify images into their respective category. We trained a RESNET5 to classify images into four categories: head (BPD and HC), abdomen (AC), femur (FL), and fetus (crown rump length: CRL). During training, the dropout rate is set to 0.4; the learning rate is set to 1e−4 and to decay exponentially for 10000 steps, at a decay rate of 0.96. The batch size is set to 16, and the network is trained for 10 epochs. The loss function measures the probability of classification error in the mutually exclusive classes.
3.3. Image segmentation via RUNET
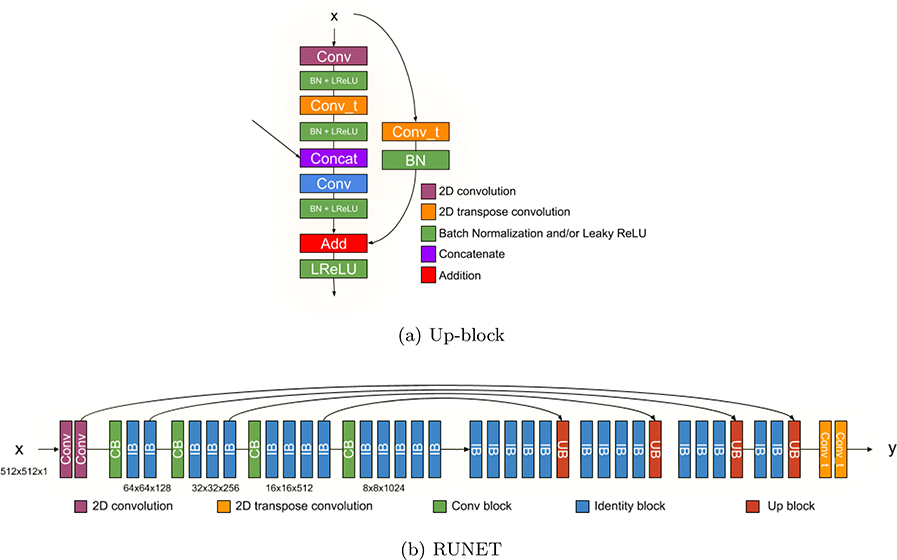
A residual UNET or RUNET was proposed by,6 however, for segmenting the fetal structures, the implementation of the up-sampling block is modified as shown in Figure 4(a), mirroring the original down-sampling block of the RESNET architecture.5 The full architecture is shown in Figure 4(b). We train 3 different RUNETs to segment the regions of interest (i.e. head, abdomen and femur). The learning rate is set to 1e−3, with an exponential decay of 0.96 for 10000 steps, and the dropout rate is 0.2. The network is trained for 150 epochs. Crown rump length images are not included because of lack of ground-truth data.
Figure 4.
Architecture of the RUNET for image segmentation.
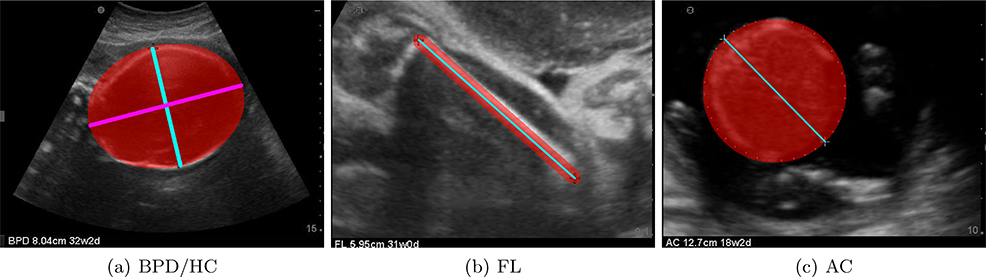
Ground truth label maps were generated using the original position of the calipers as shown in Figure 3. Head images used the position of the calipers to calculate the minor radius minr (cyan) and orientation of the ellipse; the major radius of the ellipse was empirically set to 1.3 ∗ minr (magenta). Femur image label maps are generated by interpolating between the calipers. Abdominal images used the calipers to compute the radius and center of a circle. Since such automatic label map generating steps cannot be perfomed on full fetal images, crown rump length images were not included in the segmentation training process.
Figure 3.
Ground truth label maps generated for all classes. BPD/HC, FL and AC use the original position of the calipers to generate an ellipse/circle or a line.
We use this RUNET implementation to segment the structures of interest. The loss function used for all segmentation tasks is the Intersection over Union (IoU) metric, which is defined as
| (1) |
where Pr is the predicted segmentation and Gt is the ground truth label map. The optimization minimizes 1 − IoU.
3.4. Measurement via Ellipse/circle/line fitting
After the segmentation is complete, an ellipse, circle, or line was fit to the segmented region based on the image type.
Scaling information in the input US images was used to measure the corresponding (a) biparietal diameter and head circumference from ellipses fit on head images, (b) abdominal circumference from a circle fit on abdomen images, and (c) femur length from the line fit in the femur images. The results biometry measurements were then used to estimate GA using the established Hadlock polynomial equation that is commonly used in clinical practice.7
4. RESULTS
4.1. Caliper removal
Figure 5 shows examples from the caliper removal step. The algorithm preserves structures from the original US images while removing calipers and text.
Figure 5.
Examples for the UNET caliper removal step.
4.2. Classification via RESNET
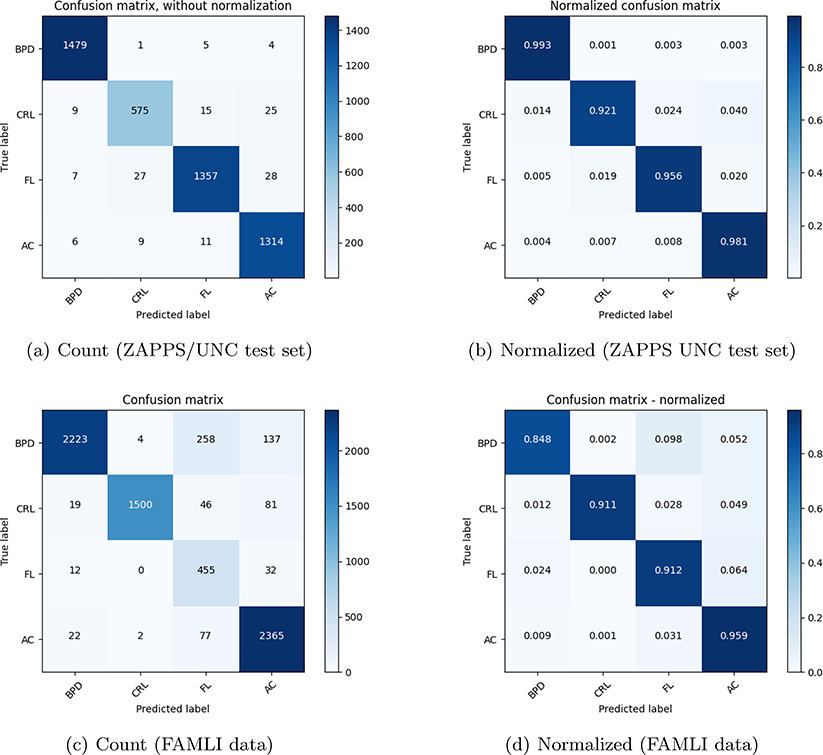
Figure 6 shows the classification results on the evaluation data set for the combined ZAPPS/UNC data, and FAMLI dataset. The combined ZAPPS/UNC data had a total of 24,359 images out of which 20% (for each class) was randomly selected and used for evaluating the performance of image classification. Evaluation of the RESNET model on the never-annotated and without calipers, prospectively collected FAMLI ultrasound image data illustrates the performance of the model when trained on retrospectively collected data.
Figure 6.
Confusion matrices showing the classification results for the ZAPPS/UNC test set and the FAMLI validation set.
The classification performed well on both evaluation sets with a mean accuracy of 93%.
4.3. Image segmentation and ellipse/circle/line fitting
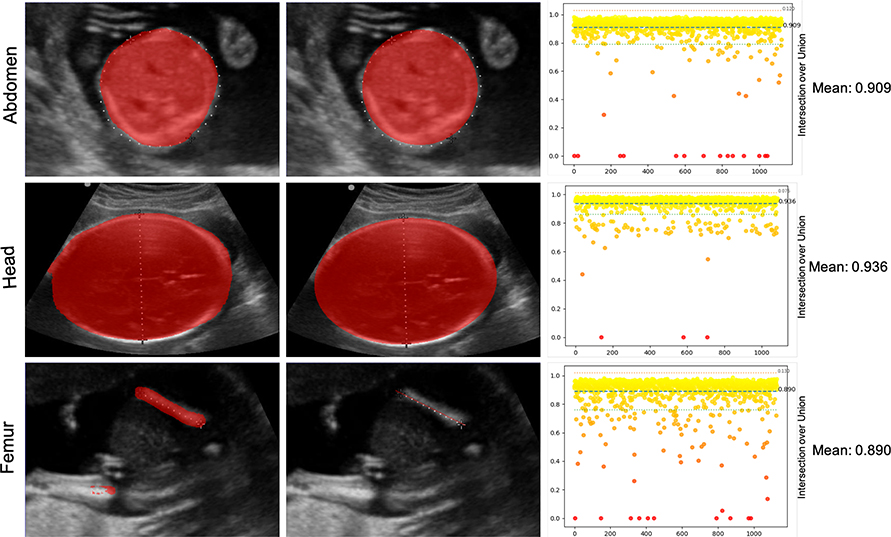
Figure 7 shows the performance of the segmentation task on the evaluation set from the ZAPPS/UNC data. As before, 20% (4,871 images) of the ZAPPS/UNC data was used for evaluation. The mean IoU was 0.91. Since the ground truth labelmaps were not available for the FAMLI dataset, IoU analysis was not performed.
Figure 7.
Segmentation accuracy for head, femur and abdomen images using the evaluation data set. From left to right: the segmentation output from the RUNET; the fitted element; and the IoU error distribution
4.4. Measurement and GA estimation
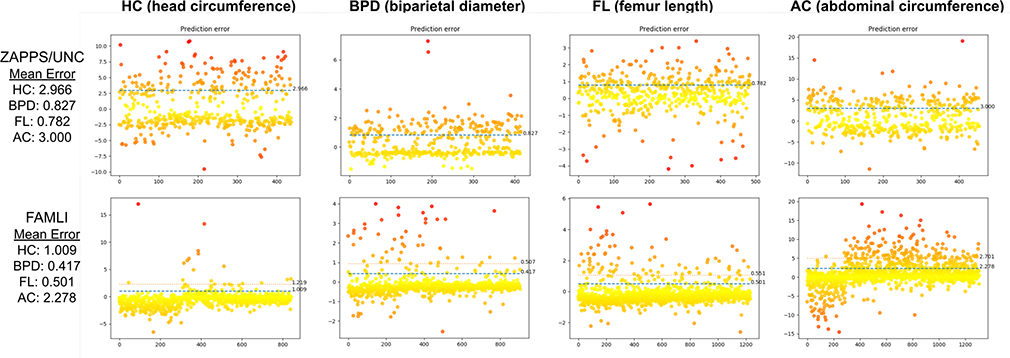
For the ZAPPS/UNC set, the ground truth measurements (circumference, diameter, and length) were extracted using OCR as physical size for pixels was not available in the ZAPPS images. The ground truth measurements were compared with scaled predicted measurements. Real world pixel size was available for the FAMLI Dataset, so the predicted measurements were directly compared to the ground truth (obtained from concurrent clinical data). Measurement errors for both evaluation sets are depicted in Figure 8.
Figure 8.
Measurement error for the structures in Zambia/UNC and FANLI dataset. Since FAMLI images have real world pixel size, the measurement accuracy is better.
For the FAMLI dataset the mean absolute error (in cm) was 0.9 for HC, 0.41 for BPD, 2.21 for AC, and 0.49 for FL with standard deviations of 1.14, 0.5, 2.61, and 0.55 respectively. The prediction error for all categories lies between two standard deviations of the mean, which is an acceptable accuracy for clinical GA prediction7,16.17
Finally, using these predicted measurements, GA was estimated with the Hadlock formula7 for studies that had all BPD, HC, AC and FL measures available (716 ultrasound sessions). This was compared to the clinician-determined GA at the time of the study. The mean error in GA prediction was 1.4 days.
5. CONCLUSION
In this paper we describe a fully automated machine learning framework that accurately recognizes and measures important fetal structures (HC, AC, BPD, FL) for GA prediction, and can recognize fetal images (CRL) accurately. The classification task recognizes structures with an average accuracy of 93%, while the average IoU for segmentation was 0.91. The mean absolute errors for the predicted measurements in cm were BPD=0.82, HC=2.96, FL=0.78, and AC=3.0, which are acceptable for GA prediction7,16.17 Predicting GA using these measures gives a mean error of 1.4 days. The fully automated framework presented here has diverse applications. Among our most important findings there are three key findings. First, we determined that burned-in annotations – formerly thought to render routine images of little use for model training – can be successfully removed with a 2-step inpainting and UNET approach. The resulting raw images can then be used for model training, unlocking vast databases of expertly collected and annotated images. Secondly, image classification of specific fetal US images can be performed accurately with AI, not only in stored 2D images but also in prospectively acquired ultrasound images. Thirdly, accurate automated segmentation and measurement of specific major fetal structures of interest are possible in order to estimate GA.
Future work will aim to continue and increase the inclusion of images of all the available data sets (historical and prospective) for the image classification task to further validate this model in other populations. Additionally, we will seek to improve our methods to generate ground truth label maps from expert annotations to improve segmentation and measurement accuracy. Finally, we aim to create semi-autonomous real-time feedback system for training purposes, driven by the location of the probe, which will permit identification and measurement of structures of interest during blind sweep acquisition.
We expect that a machine learning-based sonography approach allowing au- tomated GA determination will serve as an opportunity to improve health out- comes of women and children in low-resource settings who otherwise lack access to routine and often life-saving sonography. Ultimately, our hope is to develop a system that accurately determines GA and other high-risk prenatal diagnoses, allowing providers to make appropriate clinical decisions that direct limited resources toward the highest risk pregnancies.
Supplementary Material
Table 1.
Description of data sets used in this work
| Name | no. of studies | no. of images | Calipers/annotations present | Training/evaluation |
|---|---|---|---|---|
| ZAPPS | 3,369 | 23,209 | present | training |
| UNC | 2,983 | 124,646 | present | training |
| FAMLI | 2,491 | 7,233 | absent | evaluation |
ACKNOWLEDGMENTS
This work was supported by grants from the Bill and Melinda Gates Foundation (OPP1191684, INV003266) with additional support from the US NIH (UL1TR002489).
Footnotes
REFERENCES
- [1].UNICEF., [The state of the world’s children 2009: maternal and newborn health], vol. 9, Unicef; (2008). [Google Scholar]
- [2].Unicef et al. , “The state of the world’s children 2012: children in an urban world,” tech. rep., eSocialSciences (2012). [Google Scholar]
- [3].Alkema L, Chou D, Hogan D, Zhang S, Moller A-B, Gemmill A, Fat DM, Boerma T, Temmerman M, Mathers C, et al. , “Global, regional, and national levels and trends in maternal mortality between 1990 and 2015, with scenario-based projections to 2030: a systematic analysis by the un maternal mortality estimation inter-agency group,” The Lancet 387(10017), 462–474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in [International Conference on Medical image computing and computer-assisted intervention], 234–241, Springer (2015). [Google Scholar]
- [5].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” CoRR abs/1512.03385 (2015). [Google Scholar]
- [6].Zhang Z, Liu Q, and Wang Y, “Road extraction by deep residual u-net,” IEEE Geoscience and Remote Sensing Letters 15(5), 749–753 (2018). [Google Scholar]
- [7].Hadlock FP, Deter RL, Harrist RB, and Park S, “Estimating fetal age: computer-assisted analysis of multiple fetal growth parameters.,” Radiology 152(2), 497–501 (1984). [DOI] [PubMed] [Google Scholar]
- [8].A.T., P. B, K. W, S. EO, O. SH, K. M, P. LJ, S. DG, A. JA, N. E,B, M.G. G, R. P,L, I. FC, B. A,L, and Victoria CG, J. Y., Z.A. B, and J. V, “Ultrasound-based gestational-age estimation in late pregnancy,” Ultrasound in Obstetrics and Gynecology 48(6), 719–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].D.W., S. J, O. S, K. K, F. P, A. K,G, and E., K, “Estimating gestational age from ultrasound fetal biometries,” Obstetrics and Gynecology 133(2) (2019). [DOI] [PubMed] [Google Scholar]
- [10].Burgos-Artizzu XP, Coronado-Gutiérrez D, Valenzuela-Alcaraz B, Bonet-Carne E, Eixarch E, Crispi F, and Gratacós E, “evaluation of deep convolutional neural networks for automatic classification of common maternal fetal ultrasound planes,” Scientific Reports 10(1), 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van den Heuvel TL, Petros H, Santini S, de Korte CL, and van Ginneken B, “Automated fetal head detection and circumference estimation from free-hand ultrasound sweeps using deep learning in resource-limited countries,” Ultrasound in medicine & biology 45(3), 773–785 (2019). [DOI] [PubMed] [Google Scholar]
- [12].Sridar P, Kumar A, Quinton A, Nanan R, Kim J, and Krishnakumar R, “Decision fusion-based fetal ultrasound image plane classification using convolutional neural networks,” Ultrasound in medicine & biology 45(5), 1259–1273 (2019). [DOI] [PubMed] [Google Scholar]
- [13].Castillo MC, Fuseini NM, Rittenhouse K, Price JT, Freeman BL, Mwape H, Winston J, Sindano N, Baruch-Gravett C, Chi BH, et al. , “The zambian preterm birth prevention study (zapps): Cohort characteristics at enrollment,” Gates Open Research 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Criminisi A, Pérez P, and Toyama K, “Region filling and object removal by exemplar-based image inpainting,” IEEE Transactions on image processing 13(9), 1200–1212 (2004). [DOI] [PubMed] [Google Scholar]
- [15].Prieto J-C, Revol-Muller C, Peyrin F, Camelliti P, and Odet C, “3d texture synthesis for modeling realistic organic tissues.,” VISAPP (2), 60–65 (2012). [Google Scholar]
- [16].Gao Y, N. J., “Learning and understanding deep spatio-temporal representations from free-hand fetal ultrasound sweeps,” in [Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019.], 11768, Lecture Notes in Computer Science, Springer; (2019). [Google Scholar]
- [17].Baumgartner CF, Kamnitsas K, Matthew J, Fletcher TP, Smith S, Koch LM, Kainz B, and Rueckert D, “Sononet: Real-time detection and localisation of fetal standard scan planes in freehand ultrasound,” IEEE Transactions on Medical Imaging 36(11), 2204–2215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.