Since its recognition, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has caused 31.2 million cases of coronavirus disease (COVID-19) worldwide (1). The surge in infections has overwhelmed the healthcare systems, and identifying patients who have a high risk for poor outcomes is critically important (2). Host factors have been associated with mortality (3–7), whereas the data on the association of viral factors with COVID-19 outcomes remain conflicting (8–11). We aimed to study the association of SARS-CoV-2 genomic load in nasopharyngeal samples with clinical outcomes. We used the cycle threshold (Ct) value, the number of amplification cycles needed to yield a positive fluorescent signal in a real-time reverse transcription–polymerase chain reaction (RT-PCR), as a surrogate for viral load.
Methods
We conducted a retrospective cohort study at the New York University Langone Medical Center, a tertiary academic medical center in New York City. We evaluated all patients who presented to the emergency department between March 31, 2020, and April 10, 2020, with clinical and radiographic findings of viral pneumonia and positive screening for SARS-CoV-2 who required hospitalization. We excluded patients who were tested more than 24 hours into the admission, as our goal was to study the association of the genomic load at the time of admission to the hospital with patient outcomes.
The qualitative Cepheid Xpert Xpress SARS-CoV-2 assay was used for in-house diagnosis of COVID-19 (12). This assay detects two nucleic acid targets, namely, N2 and E, and reports the Ct values. The Ct values provide a semiquantitative measure of genomic load, with an inverse relationship between the genomic load and Ct value (13). The N2 target is specific for SARS-CoV-2, whereas the E nucleic acid can also be found in SARS-CoV-1. A positive assay result implies that either N2 and E or N2 target alone were detected, whereas detection of the E nucleic acid alone is considered a presumptive positive result (the latter were excluded from this study).
The primary study outcome was the association of the genomic load in patients admitted to the hospital with COVID-19–related pneumonia with disease outcomes. We used a composite outcome of death or discharge to hospice care and use of mechanical ventilation or extracorporeal membrane oxygenation. All patients were followed up until April 30, 2020. The Charlson Comorbidity Index (CCI) and the pneumonia severity index (PSI) were calculated. Patients were divided into five classes based on the PSI, with a higher class at the time of admission being associated with worse outcomes (14). The duration of symptoms before presentation was also extrapolated.
We categorized Ct values into the following three SARS-CoV-2 genomic load groups based on tertiles: low (≥34.2), intermediate (27.7–34.2), and high (≤27.7). We compared the patients in the three genomic load groups on the basis of demographic characteristics, body mass index (BMI), smoking history, CCI, comorbidities, immunosuppressive diseases, duration of symptoms, and the PSI using the χ2 test. A multivariate logistic regression analysis was performed to examine the association of the SARS-CoV-2 genomic load with the primary composite outcome adjusted for patient demographics, BMI, smoking history, comorbidities, transplant status, PSI, and duration of symptoms. The marginal method was used to estimate the probability of the composite outcome among patients with low, intermediate, and high genomic loads when all the other variables were fixed at their means (15). All calculations were performed using the Stata version 14.2 software package (Stata Corporation). A P value of less than 0.05 was considered statistically significant. This study was approved with a waiver of informed consent by the New York University Institutional Review Board.
Results
Of the 457 patients who presented to our emergency department with positive for SARS-CoV-2 between March 31, 2020, and April 10, 2020, 314 met the inclusion criteria and were included in the final analysis. Among the included patients, the median age was 64 years (interquartile range [IQR], 54–72 yr), 205 (65.3%) were male, 140 (44.6%) were white, and the median BMI was 28.3 (IQR, 25.1–32.3). In terms of comorbidities, the median CCI was 3 (IQR, 1–5), and 117 patients (37.3%) were obese (i.e., BMI ≥30 kg/m2). In addition, 50 patients (15.9%) had at least one pulmonary comorbidity, 72 (23.5%) were active or former smokers, 21 (6.7%) were transplant recipients (20 solid organ transplantations and one hematopoietic stem cell transplantation) and four had human immunodeficiency virus (three of them virologically suppressed). The median duration of symptoms before presentation was 7 days (IQR, 5–10 d). Nine patients were classified in class I based on their PSI (2.9%), 78 were classified in class II (24.8%), 84 were classified in class III (26.8%), 102 were classified in class IV (32.5%) and 41 were classified in class V (13%).
Of the 314 included patients, 107 (34.1%) were categorized into the low, 103 (32.8.%) into the intermediate, and 104 (33.1%) into the high SARS-CoV-2 genomic load category (Table 1). Patients with high genomic loads had higher CCI scores (P = 0.006), were more likely to be transplant recipients (P < 0.001), and had a significantly shorter duration of symptoms (P = 0.004). The PSI was significantly higher in patients with high genomic loads (P = 0.03). Transplant history (odds ratio [OR], 5.37; 95% confidence interval [CI], 1.15–25.0) and duration of symptoms (OR, 0.93; 95% CI, 0.88–0.97) remained significantly associated with high genomic load in multivariate analysis.
Table 1.
Association of SARS-CoV-2 genomic load with patient characteristics
| Patient characteristics | Low Genomic Load (n = 107) | Intermediate Genomic Load (n = 103) | High Genomic Load (n = 104) |
|---|---|---|---|
| Age, yr, n (%) | |||
| 18–44 | 14 (13.1) | 10 (9.7) | 13 (9.6) |
| 45–64 | 52 (48.6) | 35 (34.0) | 38 (36.5) |
| ≥65 | 41 (38.3) | 58 (56.3) | 53 (50.9) |
| Race, n (%) | |||
| White | 44 (41.1) | 54 (52.4) | 42 (40.4) |
| Black | 11 (10.3) | 12 (11.7) | 18 (17.3) |
| Hispanic | 11 (10.3) | 10 (9.7) | 14 (13.5) |
| Other/Unknown | 41 (38.3) | 25 (35.2) | 30 (28.8) |
| Sex, M, n (%) | 68 (63.6) | 65 (63.1) | 72 (69.2) |
| Obesity (BMI ≥ 30), n (%) | 44 (41.1) | 39 (37.9) | 34 (32.7) |
| Smoking (current/former), n (%) | 21 (20.2) | 30 (29.1) | 21 (20.2) |
| Any pulmonary comorbidity, n (%) | 15 (14.0) | 23 (22.3) | 12 (11.5) |
| Transplant,†n (%) | 2 (1.9) | 4 (3.9) | 15 (14.4) |
| CCI,*†n (%) | |||
| Low | 58 (54.2) | 41 (39.8) | 35 (33.7) |
| Medium | 33 (30.8) | 28 (27.2) | 36 (34.6) |
| High | 16 (15.0) | 34 (33.0) | 33 (31.7) |
| Symptoms for ≤7 d,†n (%) | 47 (43.9) | 63 (61.2) | 68 (65.4) |
Definition of abbreviations: BMI = body mass index; CCI = Charlson Comorbidity Index; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Low CCI 1–2, Medium CCI 3–4, High CCI ≥5.
Statistically significant.
The follow-up period was a median of 25 days (IQR, 21–28 d). At the end of follow-up, the composite outcome was reached by 74 patients (23.6%). Median time to primary outcome was 3.5 days (IQR, 1–6 d). On the day of censoring, 309 patients (98.4%) had either reached the primary outcome or were discharged. Compared with patients with low genomic load, patients with high genomic load had a significantly higher unadjusted risk to die (P < 0.001) and reach the composite outcome of death, intubation, or extracorporeal membrane oxygenation (P = 0.004) (Table 2).
Table 2.
Association of SARS-CoV-2 genomic load with patient outcomes
| Patient characteristics | Low (n = 107) | Intermediate (n = 103) | High (n = 104) |
|---|---|---|---|
| Fever, n (%) | 78 (72.9) | 79 (76.7) | 77 (74.0) |
| CRP, n (%) | |||
| ≤80 | 35 (32.7) | 35 (34.0) | 37 (35.6) |
| 80–160 | 38 (35.5) | 34 (33.0) | 36 (34.6) |
| ≥160 | 34 (31.8) | 34 (33.0) | 31 (29.8) |
| Pulmonary severity index,*n (%) | |||
| I–II | 41 (38.3) | 25 (24.3) | 21 (20.2) |
| III | 28 (26.2) | 29 (28.2) | 27 (26.0) |
| IV–V | 38 (35.5) | 49 (47.5) | 56 (53.8) |
| Death,*n (%) | 8 (7.5) | 9 (8.7) | 21 (20.2) |
| Composite outcome,*†n (%) | 17 (15.9) | 21 (20.4) | 36 (34.6) |
Definition of abbreviations: CRP = C-reactive protein; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Statistically significant.
Composite outcome = death or discharge to hospice or intubation or extracorporeal membrane oxygenation.
In the multivariate model, controlling for patient age, sex, BMI, CCI, smoking and transplant history, duration of symptoms, and PSI, high genomic load remained an independent risk factor for the composite outcome (OR, 1.59; P = 0.02). In addition, the duration of symptoms (OR, 0.93; P = 0.05) and PSI at the time of admission (OR, 3.7; P < 0.01) were also significantly associated with the composite outcome in multivariate analysis. The margins analysis indicated that the average probability of the composite outcome would be 24% (95% CI, 0.14–0.33) if everyone had a high genomic load compared with 11% (95% CI, 0.05–0.18) if everyone had a low genomic load (Figures 1A and 1B). Among patients with a high PSI, the expected probability of the composite outcome was 49% (95% CI, 0.36–0.62) for those with a high genomic load as opposed to 31% (95% CI, 0.17–0.44) for those with a low genomic load (Figure 1C).
Figure 1.
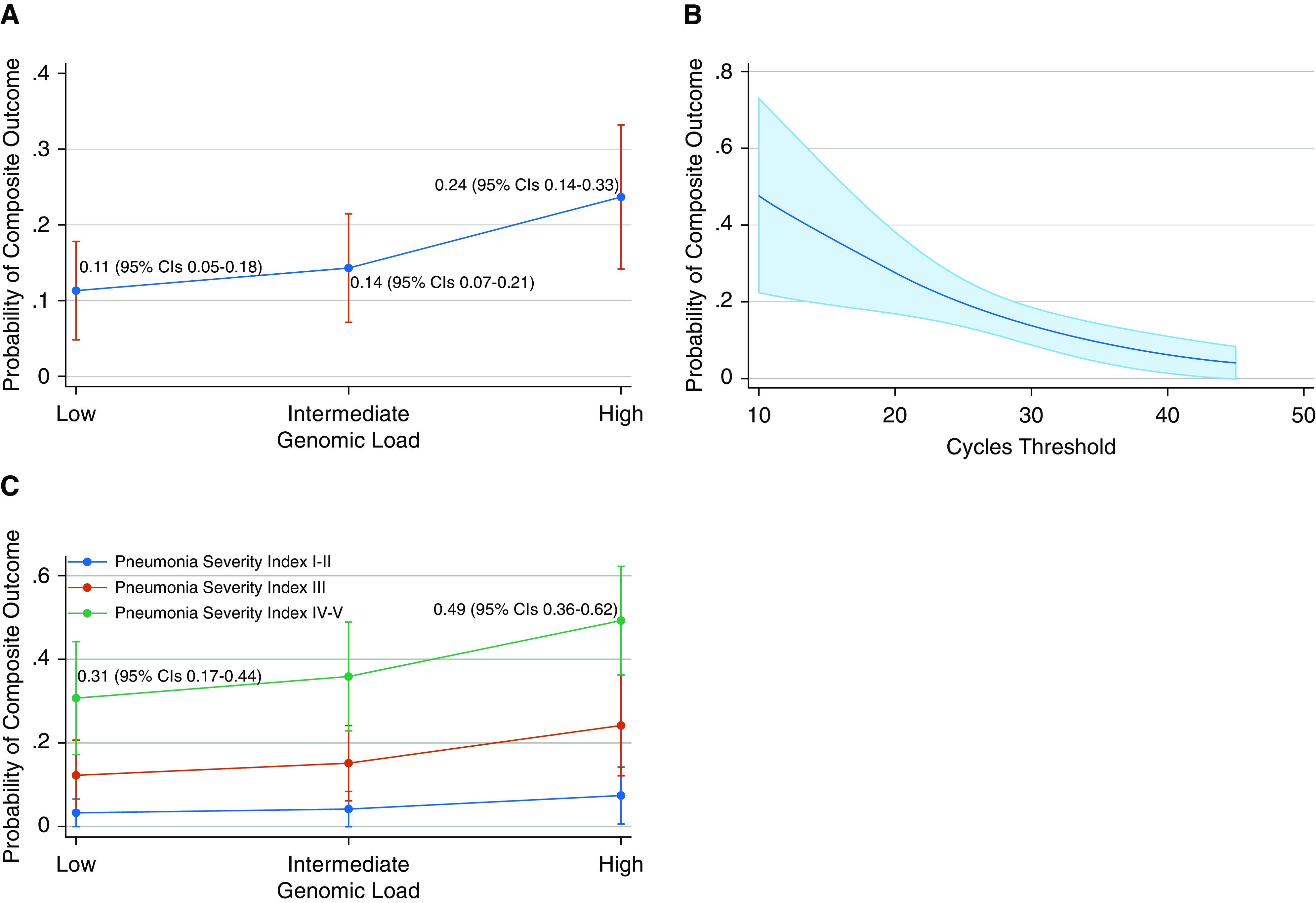
(A) Prediction of outcomes based on genomic load category. (B) Prediction of outcomes based on cycles threshold. (C) Prediction of outcomes based on genomic load and pneumonia severity index. CI = confidence interval.
Discussion
We found that patients with a short duration of symptoms and high comorbidity index, as well as transplant recipients, were more likely to have a high SARS-CoV-2 genomic load at the time of hospital admission. The patients with high genomic load had a more severe clinical presentation and two times higher odds of dying or being intubated, independent of age, comorbidities, and severity of illness on presentation. Among patients with a severe clinical presentation at the time of hospital admission, patients with high genomic load were almost twice as likely to die or get intubated.
The contribution of viral factors in disease severity is less understood, with conflicting evidence in the literature (9, 11, 16, 17). In our study, we examined the utility of genomic load from the upper respiratory tract in making inferences for the disease outcomes. Although still unclear, it is plausible that lower respiratory samples may be more closely associated with clinical outcomes than nasopharyngeal samples (18). However, the difficulty in obtaining such samples makes it unlikely that this will be of significant value in daily clinical practice. Current evidence suggests that there is active replication of SARS-CoV-2 in the upper respiratory tissues during the first 5 days after the onset of symptoms (19), a finding that correlates with our observation of higher genomic load in patients presenting within 7 days of symptom onset.
Limitations of this study should be acknowledged and arise primarily from its retrospective design. However, both the primary outcome and the genomic load are objective measures that would not be influenced by incomplete reporting. Second, this study relies on Ct values obtained through a single assay, and the generalizability of the outcomes across different RT-PCR methods should be examined. Next, variation in the technique of obtaining the nasopharyngeal swab or collection of the specimen at different phases of the respiratory cycle could potentially cause fluctuation in the genomic load detected by the assay.
In summary, we showed that SARS-CoV-2 genomic load is an independent predictor of adverse outcomes in patients admitted to the hospital with COVID-19–related pneumonia and that above and beyond age, comorbidities, and severity of illness on presentation, genomic load may be used to risk-stratify patients in an era in which appropriate triaging is of utmost importance.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank all New York University Langone Health healthcare providers and laboratory technologists for their selfless devotion to patient care during the coronavirus disease (COVID-19) pandemic.
Footnotes
Author Contributions: I.M.Z. and M.E.A.-R. conceptualized the study. I.M.Z., P.J.P., A.B., K.I., and S.A.W. collected the data. I.M.Z. and F.N.Z. conducted the analysis. All authors participated in interpretation of data, drafting of the work; final version of the manuscript was approved by all authors.
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Centers for Systems Science and Engineering Coronavirus COVID-19 global cases Baltimore, MD: Johns Hopkins University; 2019[accessed 2020 Sep 20]. Available from: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html [Google Scholar]
- 2.Phua J, Weng L, Ling L, Egi M, Lim CM, Divatia JV, et al. Asian Critical Care Clinical Trials Group Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations Lancet Respir Med 20208506–517.[Published erratum appears in Lancet Respir Med 8:e42.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Wang X, Jia X, Li J, Hu K, Chen G, et al. Risk factors for disease severity, unimprovement, and mortality in COVID-19 patients in Wuhan, China. Clin Microbiol Infect. 2020;26:767–772. doi: 10.1016/j.cmi.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji D, Zhang D, Xu J, Chen Z, Yang T, Zhao P, et al. Prediction for progression risk in patients with COVID-19 pneumonia: the CALL score. Clin Infect Dis. 2020;71:1393–1399. doi: 10.1093/cid/ciaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97:829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Argyropoulos KV, Serrano A, Hu J, Black M, Feng X, Shen G, et al. Association of initial viral load in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) patients with outcome and symptoms. Am J Pathol. 2020;190:1881–1887. doi: 10.1016/j.ajpath.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shlomai A, Ben-Zvi H, Glusman Bendersky A, Shafran N, Goldberg E, Sklan EH. Nasopharyngeal viral load predicts hypoxemia and disease outcome in admitted COVID-19 patients. Crit Care. 2020;24:539. doi: 10.1186/s13054-020-03244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng MP, Papenburg J, Desjardins M, Kanjilal S, Quach C, Libman M, et al. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann Intern Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa K, Jartti T, Mansbach JM, Laham FR, Jewell AM, Espinola JA, et al. Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the United States and Finland. J Infect Dis. 2015;211:1550–1559. doi: 10.1093/infdis/jiu658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fine MJ, Auble TE, Yealy DM, Hanusa BH, Weissfeld LA, Singer DE, et al. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336:243–250. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 15.Muller CJ, MacLehose RF. Estimating predicted probabilities from logistic regression: different methods correspond to different target populations. Int J Epidemiol. 2014;43:962–970. doi: 10.1093/ije/dyu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, et al. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am J Respir Crit Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


