Abstract
Background
The shRNA lentiviral vector was constructed to silence c-Ski expression in cardiac muscle cells, with the aim of exploring the role of c-Ski in transforming growth factor β1 (TGF-β1)-induced epithelial-mesenchymal transitions (EMT) in H9C2 cells.
Methods
Real-time polymerase chain reaction (RT-PCR) and western blot were used to detect c-Ski expression at protein and messenger ribonucleic acid (mRNA) levels in 5 different cell lines. Then, lentiviral vector was constructed to silence or overexpress c-Ski in H9C2 cells. MTT and/or soft agar assay and transwell assay were used to detect cell proliferation and migration, respectively. The expression levels of c-Ski under different concentrations of TGF-β1 stimulation were detected by RT-qPCR and immunocytochemical analysis. In the presence or absence of TGF-β1 stimulation, the proteins’ expression levels of α-SMA, FN and E-cadherin, which are closely correlated with the process of EMT, were measured by western blot after c-Ski silencing or overexpression. Meanwhile, the effect of c-Ski on Samd3 phosphorylation with TGF-β1 stimulation was investigated.
Results
There is a high expression of c-Ski at protein and mRNA levels in H9C2 cell line, which first demonstrated the presence of c-Ski expression in H9C2 cells. Overexpression of c-Ski significantly increased H9C2 cell proliferation. The ability of c-Ski gene silencing to suppress cell proliferation was gradually enhanced, and inhibition efficiency was the highest after 6 to 7 d of transfection. Moreover, H9C2 cells with c-Ski knockdown gained significantly aggressive invasive potential when compared with the control group. TGF-β1 stimulation could dose-independently reduce c-Ski expression in H9C2 cells and lead to obvious down-regulated expression of E-cadherin. Interestingly, c-Ski could restore E-cadherin expression while suppressing α-SMA and/or FN expression stimulated by TGF-β1. However, shRNA-induced c-Ski knockdown aggravated only the TGF-β1-induced EMT. Moreover, c-Ski-shRNA also promoted the phosphorylation of Samd3 induced by TGF-β1.
Conclusions
c-Ski expression in cardiac muscle cells could be down-regulated by TGF-β1. Silencing of c-Ski gene was accompanied by down-regulation of E-cadherin, up-regulation of α-SMA and/or FN and Smad3 phosphorylation induced by TGF-β1, promoting EMT process. Therefore, c-Ski may be closely associated with TGF-β1-induced EMT and play an important role in cardiac fibrosis development and progression.
Keywords: cardiac muscle cells, c-Ski, proliferation, migration, TGF-β1-induced epithelial-mesenchymal transition
Introduction
Epithelial-mesenchymal transition (EMT), a pathological process that is highly regulated, has already been identified as a critical mechanism for controlling either cardiac development or cardiac fibrosis [1]. Although the traditional view that the resident fibroblasts and their derived myofibroblasts play a crucial role in fibrosis still holds, increasing amounts of data collected in the past decade suggest that EMT is a direct contributor to the fibroblast/myofibroblast pool during fibrogenesis [2]. In adults, abnormal activation of EMT and differentiation of EMT-derived fibroblast-like cells to collagen-producing myofibroblasts play a significant role in the development and progression of fibrosis in organs such as heart and lung [3, 4]. Therefore, in addition to targeting fibroblast activation and proliferation, prevention of EMT may be another effective strategy to mediate cardiac fibrosis.
The c-Ski gene was first found in chicken embryonic cells infected with the avian leukaemia transformation defect virus, Bratislava77, by Li et al. [5] in 1986. c-Ski expression was found in all tissues of mice and was most abundant in nerve and muscle tissues. The expression level of c-Ski in other tissues was low, suggesting that c-Ski may play an important role in neurological and muscular tissue function [6]. Previous studies have shown that in the post-traumatic repair process, c-Ski can promote fibroblast proliferation [7].
There is increasing evidence that cytokines play a critical role in EMT, especially transforming growth factor β1 (TGF-β1). TGF-β plays a key role in EMT progression [8]. TGF-β1-induced EMT can promote myofibroblast transformation, which is prominently involved in fibrotic processes [9]. To further explore the role of c-Ski in TGF-β1-induced EMT and its related function in cardiac fibrosis, the present study used the lentivirus system to silence expression of c-Ski and applied TGF-β1 to stimulate EMT in H9C2 cells to evaluate the biological function of c-Ski in EMT.
Methods
Reagents
Reagents included: RPMI1640/DMEM medium and foetal bovine serum (FBS) (HyClone, USA); transfection reagent X-tremeGENE™ HP, (Roche, Germany); TRIzol (Life Technologies, USA); polymerase chain reaction (PCR) instrument (Bio-Rad, USA); cDNA synthesis kit (Fermentas, USA); SYBR qPCR Mix/HiScript II (Vazyme Biotech Co., Ltd, Nanjing, China); TGF-β1 (R&D, USA); Thiazolyl blue (MTT) (Sigma, USA); anti-c-Ski (CST, 1: 1000), anti-α-SMA (Abcam, 1: 1000), anti-fibronectin (FN) (Abcam, 1: 1000), anti-p-Smad3 (Abcam, 1: 1000), anti-Smad3 (Abcam, 1: 1000), anti-E-cadherin (Abcam, 1: 1000), anti-β-actin (Abcam, 1: 3000) and HRP labelled secondary antibody (CST, 1: 3000).
The design of shRNA
The synthetic oligonucleotides of shRNA (3 pairs of c-Ski shRNA synthesized by Sangon Biotech, Co.) were designed by using shRNA online design software (http://www.ambion.com). As listed in Table 1, the specific sequences were verified by BLAST analysis, and no homology with other human coding sequences was found. shRNA without homology to c-Ski mRNA and other genes were selected as the control (shScramble).
Table 1.
The gene sequences of shRNA oligo for vector construction and the primers for polymerase chain reaction.
| Primers | Gene sequences |
|---|---|
| c-Ski-shRNA1 | F1: 5′-CCG GAC GTC TTA CCG TGC CTA TTA CCT CGA GGT AAT AGG CAC GGT AAG ACG TTT TTG-3′ |
| R1: 5′-AAT TCA AAA AAC GTC TTA CCG TGC CTA TTA CCT CGA GGT AAT AGG CAC GGT AAG ACGT-3′ | |
| c-Ski-shRNA2 | F2: 5′-CCG GGA ATC TGC CAC TCT CAG AAT ACT CGA GTA TTC TGA GAG TGG CAG ATT CTT TTT G-3′ |
| R2: 5′-AAT TCA AAA AGA ATC TGC CAC TCT CAG AAT ACT CGA GTA TTC TGA GAG TGG CAG ATT C-3′ | |
| c-Ski-shRNA3 | F3: 5′-CCG GCA GTG TCA GCG AGT GAG AAA GCT CGA GCT TTC TCA CTC GCT GAC ACT GTT TTT G-3′ |
| R3: 5′-AAT TCA AAA ACA GTG TCA GCG AGT GAG AAA GCT CGA GCT TTC TCA CTC GCT GAC ACT G-3′ | |
| shScramble | 5′-CCG GCC TAA GGT TAA GTC GCC CTC GCT CGA GCG AGG GCG ACT TAA CCT TAG GTT TTT G-3′ |
| c-Ski | F 5′-CGA GAC AGC TTC TAC TCC TAC A-3′ |
| R 5′-TAG TAA GGG GTT CTG GTG CC-3′ | |
| GAPDH | F 5′-GGA GAT TGT TGC CAT CAA CG-3′ |
| R 5′-TTG GTG GTG CAG GAT GCA TT-3′ |
Cell culture
Cells were cultured in RPMI1640 and DMEM medium, containing 10% FBS and ampicillin-streptomycin (100 U/mL), at 37°C in a 5% CO2 saturated humidity environment.
Construction of pLKO.1/c-Ski or sh-c-Ski vector
Oligo sh-c-Ski or c-Ski was diluted with ultra-pure water to the appropriate concentration. The annealing conditions were 94°C 10 min, 70°C 10 min, and 50°C water at room temperature. The vector plko-GFP-TRC was digested with Age I, EcoR I restriction endonuclease for 1 h at 37°C. The treated sh-c-Ski or c-Ski was mixed with vector, and T4 ligase was added and then reacted at 22°C for 3 h. The ligated products were added into competent cells, and 1 mL LB medium was added. After activation at 37°C for 40 min at 300 r/min, the cells were centrifuged at 4000 r/min for 3 min, and then the supernatant was discarded. The next day, a single clone was selected and inoculated in LB medium, shaking at 37°C 300 r/min overnight. Plasmids were extracted from the bacterial solution, and the purity was determined. The empty vector (Control), shScramble vector and c-Ski-shRNA vector were digested with restriction endonuclease Xho I at 37°C for 1 h. Then, 4 μL digested products or markers was added to 10 × loading buffer and then loaded on 0.8% agarose gel for gel electrophoresis.
The immunofluorescence assay for c-Ski expression after transfection
293T cells were cultured in DMEM medium containing 10% FBS and ampicillin-streptomycin (100 U/mL) at 37°C in a saturated humidity environment with 5% CO2 and digested with 0.25% trypsin. The culture medium was replaced by MEM medium 2 h before the experiment. Next, 2.5 μg of c-Ski-shRNA vector, 2.5 μg of Δ8.91 and 0.25 μg of vesicular stomatitis virus G glycoprotein (VsV-g) were homogeneously mixed. At the same time, the shScramble group and GFP control group were set up. A total of Δ8.91 and VsV-g helper plasmids were used, then 10 μL of X-tremeGENE HP DNA Transfection Reagent was added and let stand for 20 min at room temperature. The transfection complex was added to the culture medium for 24 h and 48 h, and the supernatant was added to the H9C2 cell culture medium. After adding puromycin (1 μg/mL), cells were screened for fluorescence. c-Ski expression in the c-Ski-shRNA group, shScramble group and GFP control group was detected by immunofluorescence.
Cell proliferation was detected by MTT assay
Four groups of H9C2 cells in logarithmic growth phase were inoculated in 96-well plates (5 × 103 cells per well) at 24, 48, 72 and 96 h, and three replicates were set at each time point. After incubating at 37°C for 4 h with 20 μL MTT (5 mg/mL), 200 μL DMSO was added. After 10 min vibration, the absorbance (A) value of the mixture was measured at 560 nm using a microplate reader. The proliferation rate of these four groups of cells was calculated.
Soft agar colony formation assay
Four groups of cells were digested with trypsin when the fusion degree reached 70% to 80% and were then washed 3 times with PBS. The cells were re-suspended in a medium containing low-melting agar and then plated at a density of 1 × 103 cells per 6 cm dish, and 3 duplicate dishes per set. After 4 d, 5 d, 6 d and 7 d, cells were fixed with 4% paraformaldehyde and stained with crystal violet; the cell clones in each dish were counted for the colony-formation rate calculation, and the experiment was repeated 3 times. The colony-formation rate = (cell clones/inoculum density) × 100%.
Invasion assay
Three groups of H9C2 cells were incubated with 0.25% trypsin/EDTA and re-suspended in serum-free RPMI1640 to 5 × 105 cells per mL after 72 h culture. Then, 100 μL of cell suspension was added to the transwell chamber, and the lower layer was added with 600 μL RPMI 1640 containing 10% FBS; each set of 3 duplicates. The transwell chamber was removed after 6 h incubation, and cells were wiped that did not pass through the membrane with a cotton swab. Then, cells were immobilized with ethanol and air dried and stained with 0.1% crystal violet. The cell numbers in the upper, lower, left, right and middle visual fields were all counted under light microscopy.
RT-PCR
The expression of c-Ski mRNA in different cell lines and different concentrations of TGF-β1-treated groups was detected. The cells were collected, and the total RNA was extracted with TRIzol (according to the manufacturer instructions). The total RNA was reverse transcribed to cDNA using MMLV reverse transcriptase. PCR was carried out by adding PCR buffer, primers (Table 1) and the cDNA (as a template). The PCR reaction conditions were as follows: a pre-denaturation at 95°C for 2 min followed by 35 cycles of denaturing at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. Three replicate wells were set up for each sample.
Western blot assay
After discarding the culture medium, cells were rinsed twice with cold PBS, and then lysed in RIPA lysate on ice. After incubation at 95°C for 5 to 10 min, 3 μL was taken for quantification by the BCA method. Boiled proteins were stored at −20°C until used. A 10% SDS-PAGE gel was prepared, the sample loaded, and the gel was run. After electrophoresis, the proteins were transferred to PVDF membrane at 200 mA constant current for 1.5 h. Then, the PVDF membrane was blocked with 5% non-fat milk for 1 h and incubated overnight at 4°C after adding the primary antibody. The next day, after adding the secondary antibody for 1 h co-incubation at room temperature, the membrane was washed 3 times with TBST. The membrane was removed, ECL substrate was added, and then the luminescence signal was recorded in a dark chamber.
Immunocytochemical analysis
The logarithmic phase H9C2 cells were divided into four groups, and the cell climbing slides were prepared for immunocytochemical analysis. Cells were treated with different concentrations of TGF-β1 (0, 2, 4 and 8 ng/mL) for 48 h. After washing 3 times with PBS, the cells were immediately fixed with 4% paraformaldehyde for 15 min. After removing paraformaldehyde by washing, the cells were permeabilized with 0.1% Triton-X 100, blocked with 3% boovine serum albumin, and incubated with c-Ski antibody overnight at 4°C. All slides were analyzed using an Olympus BX51 microscope to evaluate c-Ski protein expression after 1 h of co-incubation with HRP-labelled secondary antibodies.
Statistical analysis
The measurement data were normally distributed with homogeneity of variance and were expressed as the mean ± standard deviation. Independent data between the two groups were compared by paired t-test design. Comparison among groups was made by one-way ANOVA. If the variance was not uniform, Dunnett’s T3 method was used. Count data were compared by χ2 test. SPSS13.0 software was used for statistical analysis. p < 0.05 was considered significant.
Results
c-Ski expression in cardiomyocytes
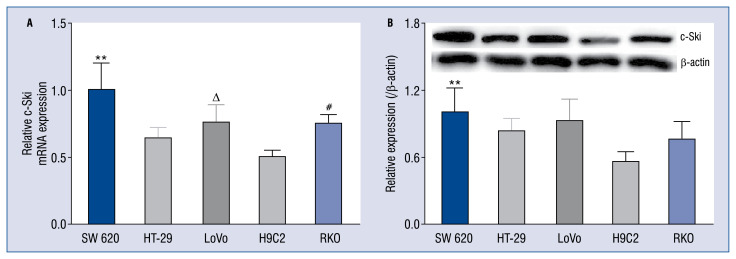
The present study investigated the expression of c-Ski at protein and mRNA levels in different cell lines, among which, the colorectal cancer cell line, SW620, showed significantly higher c-Ski expression than that in H9C2 cell line; wherein, both the expression of mRNA (t = 3.880, p < 0.05) and protein (t = 14.29, p < 0.001) was significantly different. Notably, in the H9C2 cell line, a high expression of c-Ski at protein and mRNA level was also detected, which first demonstrated the presence of c-Ski expression in H9C2 cells (Fig. 1). The results of the expression of c-Ski mRNA and protein were the average of 5 duplicates, each sample N = 5. The data were consistent with homogeneity of variance by Levene test, and K-S normal test results suggest that the data were Gaussian distribution.
Figure 1.
The expression of c-Ski mRNA and protein in different cell lines. A. mRNA expression level. B. Protein expression level; using software to calculate the gray value of the bands; **vs. SW620, p < 0.01; #vs. RKO, p < 0.05; Δvs. LoVo, p < 0.05.
Down-regulation of c-Ski in H9C2 cells
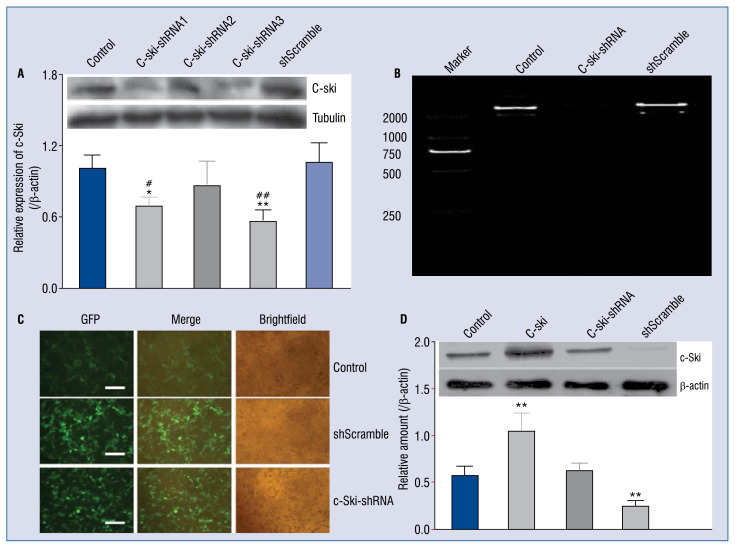
c-Ski was silenced or overexpressed with lentivirus technology to observe the effect of c-Ski on the biological function of cardiomyocyte. Three pairs of c-Ski-shRNA sequences were designed, linked to pLKO vector and then transformed to the competent cells. The plasmids were extracted and identified by enzyme digestion. The expression of c-Ski protein was detected by western blot after 48 h of transfection. As Figure 2A shows, these three pairs of sequences had a function of interference. In addition, the shRNA-3 gene silencing of the c-Ski gene in H9C2 cells was the most efficient (p < 0.01, compared with control) and was used in follow-up experiments. The results of gel electrophoresis showed that c-Ski expression in the c-Ski-shRNA group was significantly lower than that of the control group (Fig. 2B, p < 0.001, F = 126.8). From the fluorescence results, the cell infection rate reached 98% (Fig. 2C). Meanwhile, to confirm whether the decreased expression of c-Ski could still present in cells after 7 d, the cells were collected after 7 d of transfection with empty vector, c-Ski vector or sh-c-Ski vector to measure the protein expression of c-Ski. As shown in Figure 2D, the transfection of c-Ski induced an overexpression of c-Ski (p < 0.001 vs. control group), while in the c-SkishRNA group, c-Ski could be significantly suppressed (p < 0.001 vs. control group). Therefore, the above results indicated successful overexpression and down-regulation of c-Ski in H9C2 cells.
Figure 2.
The construction of c-Ski or sh-c-Ski lentiviral vector and the transfection of H9C2 cells. A. Western blot assay of the c-Ski protein expression in H9C2 cells in different groups. B. The c-Ski-shRNA vector was confirmed by gel electrophoresis. C. The fluorescent intensity of three groups of H9C2 cells 48 h post transfection (scale bar indicated 100 μm). D. The knock down efficiency of c-Ski was detected by western blot in H9C2 cells after 7 d transfection. Upper panel: the original pictures; lower panel: the quantification of the gray scale of the bands in different treated groups by software ImageJ; *p < 0.05, **p < 0.01, compared with the GFP control; ##p < 0.01, compared with the shScramble group. #p < 0.05, compared with the shScramble group.
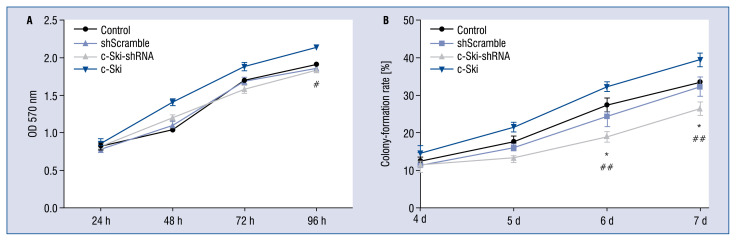
Effect of c-Ski and sh-c-Ski on cell proliferation
The changes of cell proliferation after c-Ski silencing or overexpression were analyzed by MTT assay and soft agar colony formation assay. Results showed that a significant increase of cell proliferation could be observed in c-Ski overexpression group. However, after stably silencing c-Ski, the proliferation rates of H9C2 cells were not significantly different within 96 h transfection compared with the GFP control and shScramble groups at different time points, but the difference was significant compared with the overexpression group at 96 h after the transfection (p < 0.05), as shown in Figure 3A. Meanwhile, the ability of c-Ski gene silencing-suppressed cell proliferation was gradually enhanced, and the inhibition efficiency was the highest after 6 to 7 d of transfection, as shown in the results of the soft agar colony formation assay (Fig. 3B; p < 0.05, compared with GFP control, p < 0.01, compared with the c-Ski overexpression group).
Figure 3.
Effect of c-Ski or c-Ski-shRNA on cell proliferation in H9C2 cells. A. MTT assay for detecting the cell viability after transfection. B. The long-term efficiency of c-Ski on cell proliferation by soft agar colony formation assay after stable silencing; *p < 0.05, compared with the GFP control; #p < 0.05 and ##p < 0.01, compared with the c-Ski overexpression group.
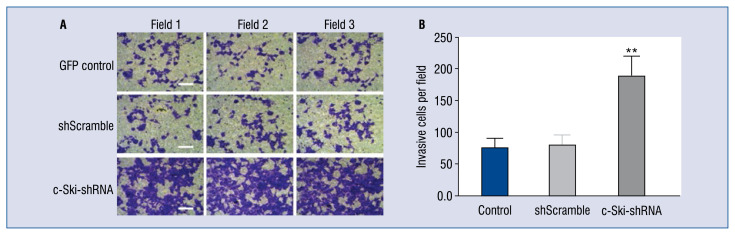
Effect of c-Ski-shRNA on migration ability of cardiomyocytes
As shown in Figure 4A, the transwell assay showed that the number of invasive cells in the c-Ski-shRNA group was (278 ± 36/field) significantly higher than in the control group (52 ± 15/ field) and the shScramble group (45 ± 13/field) (F = 155.9, p < 0.001). This result indicated that down-regulation of c-Ski could enhance the invasion of the H9C2 cells at almost 6 times greater than the control group (Fig. 4B).
Figure 4.
Effect of c-Ski-shRNA on cell proliferation in H9C2 cells. A. Results of the transwell migration test (scale bar indicated 50 μm). B. Invasive cells per field were counted; **p < 0.001, compared with the GFP control and shScramble groups.
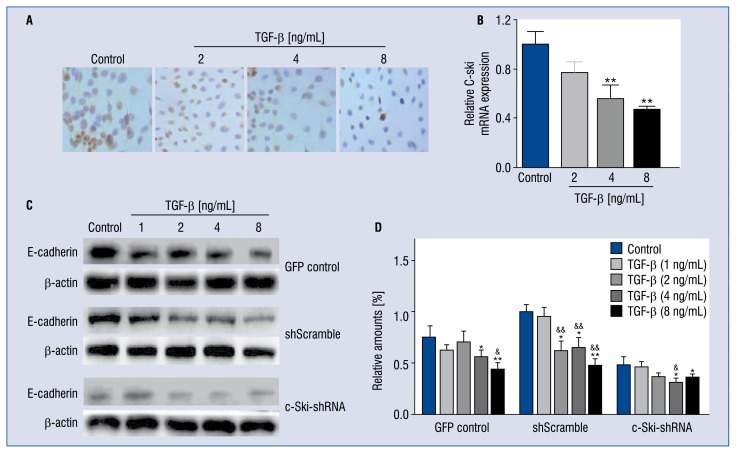
Effect of c-Ski-shRNA on E-cadherin expression in H9C2 cells
As shown in Figure 5A, after stimulation with the increased concentration of TGF-β1, the expression of c-Ski protein was significantly suppressed, as shown by the gradual decrease of brown-yellow area. The qPCR results further confirmed that TGF-β1 could down-regulate the c-Ski at mRNA level in a dose-dependent manner (Fig. 5B). E-cadherin was down-regulated during TGF-β1-induced EMT; herein, the expression of E-cadherin in the c-Ski-shRNA group (Fig. 5C); after stimulation with TGF-β1 was also evaluated; in the control and shScramble groups, E-cadherin expression was normal in the absence of TGF-β stimulation, but it could be down-regulated when stimulated with TGF-β in a dose-dependent manner. However, the expression of E-cadherin was significantly abolished after down-regulation of c-Ski, and the down-regulation tendency of E-cadherin was aggregated after TGF-β stimulation. This result suggests that TGF-β1 could also down-regulate the expression of E-cadherin dose-dependently, and this inhibitory effect could be enhanced by c-Ski-shRNA.
Figure 5.
Effect of TGF-β1 on c-Ski expression and c-Ski-shRNA on E-cadherin expression in H9C2 cells. Immunocytochemical analysis (A) and RT-qPCR assay (B) for the protein and mRNA expression of c-Ski with TGF-β1 stimulation. C. Effect of c-Ski-shRNA on E-cadherin expression under different concentrations of TGF-β1 stimulation; D. The quantification of the gray scale of the bands in different treated groups by software ImageJ; *p < 0.05, **p < 0.01, compared with the GFP control; &p < 0.05, &&p < 0.01, compared with the non-TGF-β1-treated group.
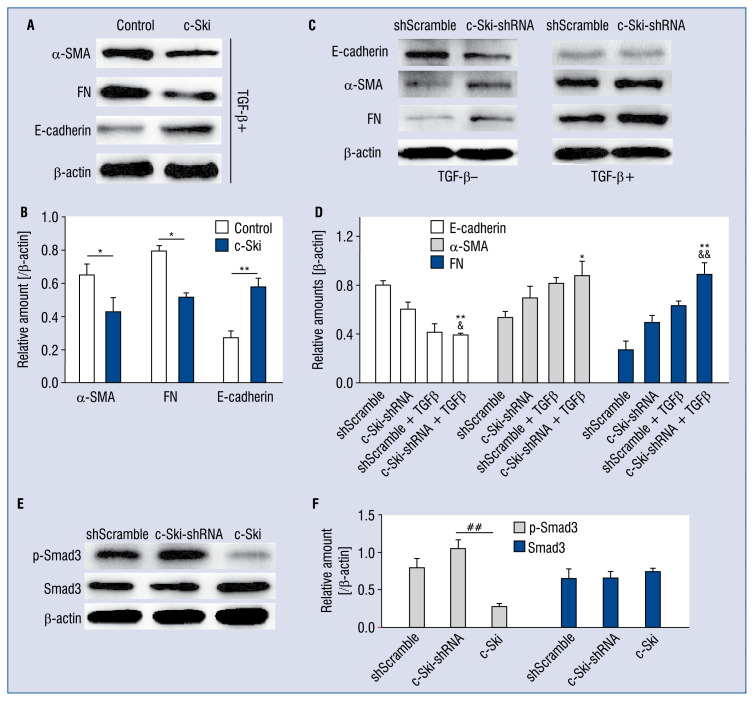
Effect of c-Ski-shRNA on EMT-related proteins
TGF-β1, an important regulatory factor in the fibrosis process, can lead to EMT and finally induce fibroblasts to transform into myofibroblasts and produce extracellular matrix (ECM). Here, post 48 h transfection of c-Ski, c-Ski-shRNA or shScramble, the H9C2 cells were stimulated with TGF-β1 (8 ng/mL) for 24 h. After being washed with PBS 3 times, cells were collected for western blot assay to detect the expression of EMT-related proteins (α-SMA, FN and E-cadherin). As shown in Figure 6A, overexpression of c-Ski could up-regulate the expression of E-cadherin down-regulated by TGF-β1 stimulation while suppressing the expression of α-SMA and FN increased by TGF-β1 stimulation. However, the knockdown of c-Ski could achieve a similar efficiency, as TGF-β1 induced on EMT, showing as the down-regulated E-cadherin and up-regulated α-SMA and FN in H9C2 cells without TGF-β1 stimulation. Moreover, for TGF-β1-stimulated E-cadherin suppression or α-SMA and/or FN up-regulation, c-Ski-shRNA could enhance the effects of TGF-β1. Additionally, c-Ski-shRNA promoted the phosphorylation of Smad3, but overexpressed c-Ski exerted the opposite effect in H9C2 cells stimulated with TGF-β1, suggesting that shc-Ski could aggravate the TGF-β1-induced EMT.
Figure 6.
Effect of c-Ski on the expression of TGF-β1-induced EMT-related proteins and the phosphorylation of Smad3. A. The effect of overexpressed c-Ski on α-SMA, FN and E-cadherin expression in the absence of TGF-β1 stimulation. B. Histogram show the quantitative results of Figure 6A. C. The effect of c-Ski-shRNA on expression of α-SMA, FN and E-cadherin in the presence or absence of 8 ng/mL TGF-β1 stimulation. D. Histogram shows the quantitative results of Figure 6C. E. The influence of c-Ski and c-Ski-shRNA on Smad3 phosphorylation with TGF-β1 stimulation; *p < 0.05, **p < 0.01, compared with GFP control; &p < 0.05, &&p < 0.01, compared with the c-Ski-shRNA-alone-treated group; ##p < 0.01, compared with overexpressed c-Ski-treated group.
Discussion
Studies have shown that c-Ski protein plays an important role in nerve, muscle development and organ fibrosis and promotes the malignant transformation process [6, 7, 10, 11]. High levels of expression of c-Ski were reported in several tumor cell lines [12, 13]. In the present study, a high expression of c-Ski was also observed in different colon cancer cell lines, including highly metastatic SW620 and HT-29 cells, lowly metastatic RKO cells and undifferentiated LoVo cells. Interestingly, in the H9C2 cell line, a rat cardiac muscle cell, a high expression of c-Ski was also detected, which first demonstrated the presence of c-Ski expression in H9C2 cells, was distributed in the nucleus and cytoplasm and it could play a role in cytological biological function. c-Ski was reported to function as a negative regulator of TGF-β signalling [14, 15], which is closely related to the occurrence and development of myocardial fibrosis, yet the direct and precise effect of c-Ski on managing myofibroblast function and cardiac fibrosis remains unclear.
In this study, 3 pairs of c-Ski-shRNA sequences were designed and synthesized in vitro using online software, according to the design principle of shRNA. c-Ski in H9C2 cells were silenced or overexpressed through constructing a lentiviral vector of c-Ski or sh-c-Ski to detect the expression of c-Ski mRNA and protein by RT-PCR and western blot after transfection into 293T cells by liposome transfection. Data collected confirmed that these 3 pairs of sequences had a function of interference. Therein, the shRNA-3 gene silencing of the c-Ski in H9C2 cells could obtain a stable and desirable knockout efficiency and the decreased expression of c-Ski could still be present in cells after 7 d of transfection. Therefore, this method was used in the follow-up experiments. Overexpression of c-Ski abolished TGF-β — induced growth inhibition [16, 17]. Inhibition of TGF-β signalling is thus regarded as part of the mechanism of oncogenesis by Ski proteins. Similarly, it also proved that c-Ski had a positive effect on promoting H9C2 cell growth, but compared with the control and shScramble groups after stably silencing c-Ski, the cell proliferation rates were not significantly different within 96 h transfection. However, the ability of c-Ski gene silencing to suppress cell proliferation was gradually enhanced, and inhibition efficiency was the highest after 6 to 7 d of transfection. This outcome suggests that c-Ski-shRNA could induce the proliferation inhibition of cardiac muscle cells in the long term. Its possible mechanism was regulating the transcription of nuclear factor, and suppressing the transcription of TGF-β downstream Smad2 and Smad3. These genes regulated many cell signals related to proliferation and differentiation; for example, Smad3 promoted apoptosis by promoting the process of P21 and P15 inhibiting cell proliferation [18–20]. Therefore, c-Ski may participate in cell proliferation through the abovementioned pathways, affecting cell viability. However, the potential molecular mechanism of this process remains to be investigated. This study found that the ability of cell migration was increased within a short time (≤ 24 h) after c-Ski knockdown, suggesting that H9C2 cells may express some skeletal proteins or turn to fibroblasts after c-Ski silencing. Previous studies have demonstrated that TGF-β, as the best characterized fibrogenic growth factor [21], is markedly and consistently activated in experimental models of cardiac fibrosis [22] and in fibrotic human hearts [23]
For nearly a decade, much research showed that EMT is involved in a variety of tissue fibrosis and is a direct contributor to the fibroblast/myofibroblast population in the development of tissue fibrosis, such as myocardial, renal and liver fibrosis [24]. TGF-β1-induced EMT can promote myofibroblast transformation, which is prominently involved in the fibrotic processes [9]. In this study, activation and phenotype transformation of H9C2 cells from resident cardiac muscle cells into myofibroblasts could have resulted from the knockdown of c-Ski, and showed as the up-regulated expression of α-SMA, a specific marker of myofibroblasts. The proportional reduction and fibrosis of cardiac muscle cells may cause heart failure, which is characterized as pathological ventricular remodeling and is the main cause of death in patients with ischemic heart disease. The present results suggest that c-Ski-shRNA may induce cardiac disease by reducing the proportion of H9C2 cells, increasing the cell migration capacity and inducing phenotype transformation of H9C2 cells.
TGF-β1 affects the entire epithelial cell EMT process. TGF-β1-induced EMT plays an important role in cardiac fibrosis. Thus, this study further investigated the detailed function and mechanisms of c-Ski in TGF-β-induced EMT by stimulating H9C2 cells with TGF-β1. In the study of human proximal tubule epithelial cells [25], the down-regulation of c-Ski reduced the response threshold of TGF-β, creating an ideal “fibrogenic” environment for TGF-β, which amplified the TGF-β effect. In contrast, by overexpression of c-Ski, the amount of TGF-β increased to 10-fold, and could not completely inhibit the expression of E-cadherin and induce the expression of α-SMA [26, 27]. In the present study, findings showed that c-Ski expression was down-regulated by TGF-β in a concentration-dependent manner. EMT is characterized by the loss of epithelial characteristics and the acquisition of mesenchymal characteristics [28]; loss of epithelial markers, such as E-cadherin and the induction of mesenchymal markers including α-SMA and vimentin, which are hallmarks of early- and late-stage events of EMT, respectively [29]. In this study, the influence of c-Ski on the expression levels of EMT-related proteins were measured (α-SMA, FN and E-cadherin) to evaluate c-Ski-shRNA auxo-action on TGF-β1-induced EMT. The results indicated that TGF-β1 stimulation could lead to obvious down-regulated expression of E-cadherin, an important functional adhesive molecule, which affects the adhesion among cells [30]. In addition, the expression of E-cadherin could also be significantly down-regulated by c-Ski-shRNA. TGF-β activation in the cardiac interstitium which promotes α-SMA transcription in fibroblasts through activation of the Smad3 signalling cascade [31]. Additionally, further investigation of the effect of c-Ski on Samd3 phosphorylation found that c-Ski-shRNA also promoted the phosphorylation of Samd3 induced by TGF-β1. Interestingly, c-Ski could restore E-cadherin expression while suppressing α-SMA and/or FN expression stimulated by TGF-β1. However, shRNA-induced c-Ski knockdown aggravated only the TGF-β1-induced EMT. These results suggest that the knockdown of c-Ski in H9C2 may result in an increased TGF-β biologic effect on the occurrence and development of cardiac fibrosis by inhibiting cardiac muscle cell proliferation, promoting cell migration, down-regulating E-cadherin expression and increasing Smad3 phosphorylation.
Conclusions
The above results indicated that c-Ski-shRNA silenced the c-Ski gene and regulated the proliferation and phenotype transformation of H9C2 cells through TGF-β1-induced EMT. Therefore, c-Ski may be closely associated with TGF-β1-induced EMT and play an important role in cardiac fibrosis development and progression.
Acknowledgements
This work was supported by grants from the Excellent Youth Medical Talent Training Plan of Health System in Shanghai Pudong New Area (NO. PWRq2014-03).
Footnotes
Conflict of interest: None declared
References
- 1.Yu W, Liu Z, An S, et al. The endothelial-mesenchymal transition (EndMT) and tissue regeneration. Curr Stem Cell Res Ther. 2014;9(3):196–204. doi: 10.2174/1574888x09666140213154144. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arciniegas E, Frid MG, Douglas IS, et al. Perspectives on endothelial-to-mesenchymal transition: potential contribution to vascular remodeling in chronic pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2007;293(1):L1–L8. doi: 10.1152/ajplung.00378.2006. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh AK, Bradham WS, Gleaves LA, et al. Genetic deficiency of plasminogen activator inhibitor-1 promotes cardiac fibrosis in aged mice: involvement of constitutive transforming growth factor-beta signaling and endothelial-to-mesenchymal transition. Circulation. 2010;122(12):1200–1209. doi: 10.1161/CIRCULATIONAHA.110.955245. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Turck CM, Teumer JK, et al. Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J Virol. 1986;57(3):1065–1072. doi: 10.1128/jvi.57.3.1065-1072.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong CC, Poon WH, Tsim TY, et al. Gene expressions during the development and sexual differentiation of the olfactory bulb in rats. Brain Res Dev Brain Res. 2000;119(2):187–194. doi: 10.1016/s0165-3806(99)00173-x. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Hou Y, Sun Y, et al. c-Ski activates cancer-associated fibroblasts to regulate breast cancer cell invasion. Mol Oncol. 2013;7(6):1116–1128. doi: 10.1016/j.molonc.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooley BC, Nevado J, Mellad J, et al. TGF-β signaling mediates endothelial-to-mesenchymal transition (EndMT) during vein graft remodeling. Sci Transl Med. 2014;6(227):227ra34. doi: 10.1126/scitranslmed.3006927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cell Mol Life Sci. 2014;71(4):549–574. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Zhao Li, Yang T, et al. c-Ski inhibits autophagy of vascular smooth muscle cells induced by oxLDL and PDGF. PLoS One. 2014;9(6):e98902. doi: 10.1371/journal.pone.0098902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasui T, Kadono Y, Nakamura M, et al. Regulation of RANKL-induced osteoclastogenesis by TGF-β through molecular interaction between Smad3 and Traf6. J Bone Miner Res. 2011;26(7):1447–1456. doi: 10.1002/jbmr.357. [DOI] [PubMed] [Google Scholar]
- 12.Nomura N, Sasamoto S, Ishii S, et al. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res. 1989;17(14):5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fumagalli S, Doneda L, Nomura N, et al. Expression of the c-ski proto-oncogene in human melanoma cell lines. Melanoma Res. 1993;3(1):23–27. doi: 10.1097/00008390-199304000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, Yagi K, Kondo M, et al. c-Ski inhibits the TGF-beta signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene. 2004;23(29):5068–5076. doi: 10.1038/sj.onc.1207690. [DOI] [PubMed] [Google Scholar]
- 15.Cunnington RH, Nazari M, Dixon IMC. c-Ski, Smurf2, and Arkadia as regulators of TGF-beta signaling: new targets for managing myofibroblast function and cardiac fibrosis. Can J Physiol Pharmacol. 2009;87(10):764–772. doi: 10.1139/Y09-076. [DOI] [PubMed] [Google Scholar]
- 16.Luo K, Stroschein SL, Wang W, et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13(17):2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Angelis K, Danielpour D, et al. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor. Proc Natl Acad Sci U S A. 2000;97(11):5924–5929. doi: 10.1073/pnas.090097797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M, Shi J, Zhang W, et al. MiR-23b controls TGF-β 1 induced airway smooth muscle cell proliferation via direct targeting of Smad3. Pulm Pharmacol Ther. 2017;42:33–42. doi: 10.1016/j.pupt.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Gui Q, Hui X, et al. TGF-β 1/Smad3 Signaling Pathway Suppresses Cell Apoptosis in Cerebral Ischemic Stroke Rats. Med Sci Monit. 2017;23:366–376. doi: 10.12659/MSM.899195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Gu H, Gan Lu, et al. Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget. 2017;8(6):9267–9279. doi: 10.18632/oncotarget.14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biernacka A, Cavalera M, Wang J, et al. TGF-β signaling in fibrosis. Growth Factors. 2011;29(5):196–202. doi: 10.3109/08977194.2011.595714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J Mol Cell Cardiol. 2011;51(4):600–606. doi: 10.1016/j.yjmcc.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li RK, Li G, Mickle DA, et al. Overexpression of transforming growth factor-beta1 and insulin-like growth factor-I in patients with idiopathic hypertrophic cardiomyopathy. Circulation. 1997;96(3):874–881. doi: 10.1161/01.cir.96.3.874. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Luan F, Zhao Y, et al. Epithelial-mesenchymal transition: An emerging target in tissue fibrosis. Exp Biol Med (Maywood) 2016;241(1):1–13. doi: 10.1177/1535370215597194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Wang Y, Xiao Y, et al. SnoN as a key regulator of the high glucose-induced epithelial-mesenchymal transition in cells of the proximal tubule. Kidney Blood Press Res. 2012;35(6):517–528. doi: 10.1159/000339172. [DOI] [PubMed] [Google Scholar]
- 26.Xiao C, Ogle SA, Schumacher MA, et al. Hedgehog signaling regulates E-cadherin expression for the maintenance of the actin cytoskeleton and tight junctions. Am J Physiol Gastroin-test Liver Physiol. 2010;299(6):G1252–G1265. doi: 10.1152/ajpgi.00512.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Zhan L, Yang T, et al. Ski prevents TGF-β-induced EMT and cell invasion by repressing SMAD-dependent signaling in non-small cell lung cancer. Oncol Rep. 2015;34(1):87–94. doi: 10.3892/or.2015.3961. [DOI] [PubMed] [Google Scholar]
- 28.Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50(6):2007–2013. doi: 10.1002/hep.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izaguirre MF, Casco VH. E-cadherin roles in animal biology: A perspective on thyroid hormone-influence. Cell Commun Signal. 2016;14(1):27. doi: 10.1186/s12964-016-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashrafian H, McKenna WJ, Watkins H. Disease pathways and novel therapeutic targets in hypertrophic cardiomyopathy. Circ Res. 2011;109(1):86–96. doi: 10.1161/CIRCRESAHA.111.242974. [DOI] [PubMed] [Google Scholar]