Abstract
Emerging evidence indicates that microRNAs (miRNAs) play a critical role in breast cancer development. We recently reported that a higher expression of miR-374b in tumor tissues was associated with a better disease-free survival of triple-negative breast cancer (TNBC). However, the functional significance and molecular mechanisms underlying the role of miR-374b in breast cancer are largely unknown. In this current study, we evaluated the biological functions and potential mechanisms of miR-374b in both TNBC and non-TNBC. We found that miR-374b was significantly downregulated in breast cancer tissues, compared to adjacent tissues. MiR-374b levels were also lower in breast cancer cell lines, as compared to breast epithelial cells. In vitro and in vivo studies demonstrated that miR-374b modulates the malignant behavior of breast cancer cells, such as cell proliferation in 2D and 3D, cell invasion ability, colony-forming ability and tumor growth in mice. By using bioinformatics tools, we predicted that miR-374b plays a role in breast cancer cells through negatively regulating cyclin D1 (CCND1) and transforming growth factor alpha (TGFA). We further confirmed that CCND1 and TGFA contribute to the malignant behavior of breast cancer cells in vitro and in vivo. Our rescue experiments showed that overexpressing CCND1 or TGFA reverses the phenotypes caused by miR-374b overexpression. Taken together, our studies suggest that miR-374b modulates malignant behavior of breast cancer cells by negatively regulating CCND1 and TGFA genes. The newly identified miR-374b-mediated CCND1 and TGFA gene silencing may facilitate a better understanding of the molecular mechanisms of breast cancer progression.
We explored the potential mechanisms of miR-374b in breast cancer development. Our in vitro and in vivo studies demonstrate that miR-374b modulates malignant behavior of breast cancer cells by negatively regulating CCND1 and TGFA.
Introduction
MicroRNAs (miRNAs) regulate gene expression to affect various biological processes such as development, differentiation and apoptosis (1–3). These small non-coding RNAs bind to the 3′ untranslated regions (3′ UTR) of target messenger RNAs (mRNAs) and disrupt target gene expression by preventing its translation or promoting mRNA degradation (4). Emerging evidence indicates that miRNAs play a critical role in a variety of pathological processes, including cancer. MiRNAs can function as oncogenes or tumor suppressors by targeting different genes that regulate the cell cycle, migration and apoptosis. Dysregulation of miRNA expression is correlated with cancer initiation and progression (5,6).
MiRNAs have been shown to be associated with breast cancer metastasis and prognosis (7–10). Tumor suppressor miRNAs, such as miR-126, miR-31, miR-34 and the miR-200 family, suppress breast cancer metastasis (8,11–13). Oncogene miRNAs, such as miR-9 and miR-10b, are elevated in metastatic breast cancer cells and promote cell migration/invasion, leading to metastasis (9,10). Therefore, miRNAs may serve as therapeutic targets or prognostic biomarkers.
MiRNAs may play different roles in different types of cancers, or different subsets of a particular type of cancer, depending on their targeting genes. For example, a previous study showed that miR-374a promotes breast cancer metastasis by activating the Wnt/β-catenin pathway (14). However, in our previous study of 455 triple-negative breast cancer (TNBC) cases, we found that higher expressions of both miR-374a and miR-374b were associated with better breast cancer disease-free survival (15). These results were consistent with findings of an independent study on TNBC (16). In this current study, we further explored the biological functions and potential molecular mechanisms of miR-374b in breast cancer.
Materials and methods
Cell lines, cell culture and breast cancer tissues
Human breast epithelial cells (MCF-10A and MCF-10F) and human breast cancer cell lines (MDA-MB-231, MDA-MB-468, MCF-7, T47D, BT549 and MDA-MB-453) were obtained from the American Type Culture Collection (ATCC, Manassas, VA) within 6 months of purchase. MCF-10A and MCF-10F (ATCC® CRL10318™) cells were cultured in a complete medium suggested by ATCC. For MCF-10A, the kit MEGM (CC-3150) was purchased from Lonza/Clonetics, and GA-1000 was replaced by 100 ng/ml cholera toxin. For MCF-10F, base medium of MCF-10F (1 L) included 14.8 g/L Dulbecco’s modified Eagle’s medium and Ham’s F12 base (Sigma, D-9785), 1.2 g NaHCO3, 0.365 g l-glutamine, 0.059 g l-leucine, 0.0912 g l-lysine, 0.017 g l-methionine, 0.0612 g MgCl2.6H2O, 0.0488 g MgSO4.7H2O, 0.006 g CaCl2.2H2O and 0.0086 g Phenol Red (Sigma, P-3532). Complete growth medium included base medium with 20 ng/ml epidermal growth factor (Sigma, E-9644), 100 ng/ml cholera toxin (Sigma, C-8052), 0.01 mg/ml human insulin (Sigma, I-2643), 500 ng/ml hydrocortisone (Sigma, H-0888) and 5% horse serum (Sigma, H0146). Other breast cancer cell lines were cultured in RPMI 1640 medium (Gibco/Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.
A total of 21 paired breast cancer tissues, adjacent cancer tissues (normal tissue adjacent to the tumor) (17) and adjacent normal tissues (Human Paraffin-Embedded Tissue Arrays, Breast BC08032a) were purchased from US Biomax, Inc.
Transfection
Cells were transiently transfected with miR-374b or miR-control (Ambion/Life Technologies) using Lipofectamine RNAiMAX Reagent (Life Technologies), according to the manufacturer’s protocol. For migration, colony formation, 3D culture, quantitative real-time PCR and western blotting, two transfections (0 and 24 h) were performed in 48 h, and cells were collected 48 h after the first transfection. Cells were then plated for functional assays. Co-transfection of miRNA and plasmid DNA was conducted using Lipofectamine 2000 Reagent (Life Technologies) with the same design as the miRNA transfection.
siRNAs against CCND1 (5′-ACUUGAAGUAAGAUACGGAGGGCGC-3′) or TGFA (5′-UCGCCCUGUUCGCUCUGGGUAUUGU-3′ and 5′-CAGAAGAAGCAGGCCAUCACCGCU-3′) were introduced into cells using RNAiMAX (Life Technologies), according to the manufacturer’s protocol.
We evaluated transfection efficiency at the beginning of the study. Cell lines with high transfection efficiency were chosen. For miRNA, we used Cy3™ Dye-Labeled Pre-miR Negative Control (ThermoFisher). Both MDA-MB-468 and T47D have a transfection efficiency around 50–60% after the first transfection and 70–80% after the second transfection. For plasmid transfection, we used GFP construct to check transfection efficiency. Both MDA-MB-468 and T47D have a transfection efficiency around 30–40% after the first transfection and 50–60% after the second transfection.
Cell proliferation assay
Breast cancer MDA-MB-468 and T47D cells were cultured in 96-well plates with a density of 2 × 103 cells per well and tested with 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assays at different time points, as described previously (18).
Trans-well migration assay
Cell migration assays were performed in a 24-well plate with 8-µm pore size chamber inserts (Corning, New York, NY), as described previously (15,18). MDA-MB-468 cells were cultured in serum-free medium, with a density of 5 × 104 per well, and placed in the upper compartments of trans-well chambers. The lower chambers were filled with culture medium containing 10% fetal bovine serum. After 12 h of incubation at 37°C, cells on the top surface of the inserts were removed by cotton swab. Cells that migrated to the bottom surface of the insert were fixed in 100% pre-cooling methanol for 30 min, stained with crystal violet and then subjected to microscopic inspection. Five visual fields of each insert were randomly chosen and counted under a light microscope. Three independent experiments were performed, and the data were presented as mean ± SD.
Clonogenic assay
MDA-MB-468 and T47D cells were seeded with a density of 500 cells per plate and cultured for 14 days. Cells were then washed twice with phosphate-buffered saline (PBS), fixed in ice cold methanol and stained with crystal violet. Three independent experiments were performed, and the results were quantified as percentage to the control group (mean ± SD).
3D cell culture and multicellular tumor spheroid formation
Six-well plates were coated with 3% sterile agarose in PBS buffer. When the agarose floor dried, cell culture medium containing 5 × 104 cells was seeded. The cells were maintained at 37°C in a 5% CO2 incubator for 14 days. Spheroid volume was calculated according to the formula v = ½ * length * width * width. Results were captured by digital camera. To compare the number of spheroids under different conditions, we performed the same experiments using 10-cm plates. After 14 days, the number of spheroids was counted under a microscope. Three independent experiments were performed, and the data were presented as mean ± SD.
Bioinformatic analysis of miR-374b targets
The miR-374b targets were predicted using TargetScan and miRDB (19–22).
RNA isolation and quantitative real-time PCR
Tissue cores from Human Paraffin-Embedded Tissues arrays were removed from tissue array blocks for total RNA extraction. Total nucleic acid, including miRNA, from Human Paraffin-Embedded Tissues was isolated using the RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (Thermo Fisher Scientific, Waltham, MA), following the manufacturer’s introduction. Total RNA was isolated from cells using the miRNeasy Mini Kit (Qiagen, Valencia, CA), and 500 ng of the total RNA from each sample were subjected to reverse transcription using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocol. Levels of cyclin D1 (CCND1), fibroblast growth factor receptor 2 (FGFR2) and transforming growth factor alpha (TGFA) mRNA were detected using RT2 qPCR Primer Assays (CCDN1: F: 5′-AGGCGGATGAGAACAAGCAGA-3′ and R: 5′ CAGGCTTGACTCCAGAAGGG-3′; FGFR2: F: 5′- GATAAATACTTCCAATGCAGAAGTGCT-3′ and R: 5′-TGCCCTATATAATTGGAGACCTTACA-3′; TGFA: F: 5′-AGCATGTGTCTGCCACTCT-3′ and R: 5′-GGATCAGCACACAGGTGAT -3′; GAPDH: F: 5′-GCACCGTCAAGGCTGAGAAC-3′ and R: 5′-ATGGTGGTG AAGACGCCAGT-3′) and RT2 SYBR Green Mastermixes (Qiagen) in an ABI 7900HT Sequence Detection System (Applied Biosystems). Relative mRNA expression was normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated using the 2(−ΔΔCT) method. MiR-374b primers (F: 5′-ATATAATACAACCTGCTAAGTG-3′; R: 5′-GTGCAGGGTCCGAGGTATTC-3′) and control primers (F: 5′-AGAGAAGATTAGCATGGCCCCTG-3′; R: 5′-ATCCAGTGCGGGTCCGAGG-3′) were obtained from Millipore Sigma (St. Louis, MO).
Luciferase reporter assay
The 3′ UTR sequences, or mutated sequences, for CCND1 and TGFA genes interacting with miR-374b were synthesized and inserted into the PmeI and XbaI sites of the pmirGLO Vectors (Promega, Madison, WI), as described previously (18). The sequences for wild-type (WT) and mutated TGFA gene 3′ UTR were: 5′-AAACTAGCGGCCGCTAGTTATCTTTATTATAATAAACCT-3′, and 5′-AAACTAGCGGCCGCTAGTTATCTTGCTGCTCCT AAACCT-3′. The sequences for WT and mutated CCDN1 gene 3′ UTR were 5′- TGCATAATTA TTATTATTAT TATTATAACA AGTGTGTCTT-3′ and 5′-TGCATAATTA TTATTATTAT GAGGAGCCCA AGTGTGTCTT-3′. WT or mutant luciferase reporter constructs, together with the pRL-TK Vector (Promega) and miR-374b or miR-control, were co-transfected into MDA-MB-468 cells by Lipofectamine 2000 (Life Technologies). Firefly and Renilla luciferase activity were measured by the dual-luciferase reporter assay system (Promega) 48 h post-transfection. Firefly luciferase activity was normalized to Renilla luciferase activity. Three independent experiments were performed, and the data were presented as mean ± SD.
Western blotting
Cells were lysed with RIPA Buffer (Life Technologies), supplemented with protease inhibitors (Roche, Indianapolis, IN). Total protein was quantified with Pierce BCA Protein Assay (Pierce Biotechnology, Rockford, IL). Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The membrane was blocked in a blocking buffer (0.1% Tween-20 and 5% non-fat milk in PBS) at room temperature for 1 h and then incubated at 4°C overnight using human Cyclin D1 (Cell Signaling, Danvers, MA) or TGFA antibody (R&D Systems, Minneapolis, MN) primary antibodies. Western blots were then incubated with a secondary antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Dallas, TX). GAPDH was used as a loading control (Cell Signaling Technology). Protein was visualized by LumiGLO chemiluminescent substrate (Cell Signaling Technology).
DNA constructs of CCND1 and TGFA
CCND1 or TGFA cDNA was cloned into pENTR plasmid. The cDNA was then introduced into pLenti6 using Gateway Technology, according to the manufacturer’s introduction (Life Technologies). Both pLenti6-CCND1 and pLenti6-TGFA were used for rescue experiments.
Generation of stably transfected cell lines
A lentivirus overexpressing human miR-374b was obtained from Genomedtech (Shanghai, China) and used to infect MDA-MB-468 cells. Another lentivirus overexpressing siRNA targeting human CCND1 or TGFA was generated using the BLOCK-iT™ Lentiviral RNAi Expression System (Thermo Fisher Scientific) and used to infect MDA-MB-468 cells. All transfected cells were selected by adding 2 µg/ml puromycin to a culture medium 48 h after addition of the virus.
Animal xenograft study
MDA-MB-468 cells overexpressing miR-374b, si-CCND1, si-TGFA, or control miRNA/siRNA were injected subcutaneously into nude mice with a density of 5 × 106. Six mice were included in each group. After 5 weeks, the tumor weight was recorded. Animal use and care was approved by the Institutional Animal Care and Use Committee, Tongji Medical College, Huazhong University of Science and Technology.
Statistical analysis
Data were shown as mean ± SD. Differences between experimental groups and controls were assessed by Student’s or Welch’s t-test. Two-tailed tests were used, and a P value of ≤0.05 was considered statistically significant.
Results
MiR-374b was significantly downregulated in breast cancer tissues and cell lines
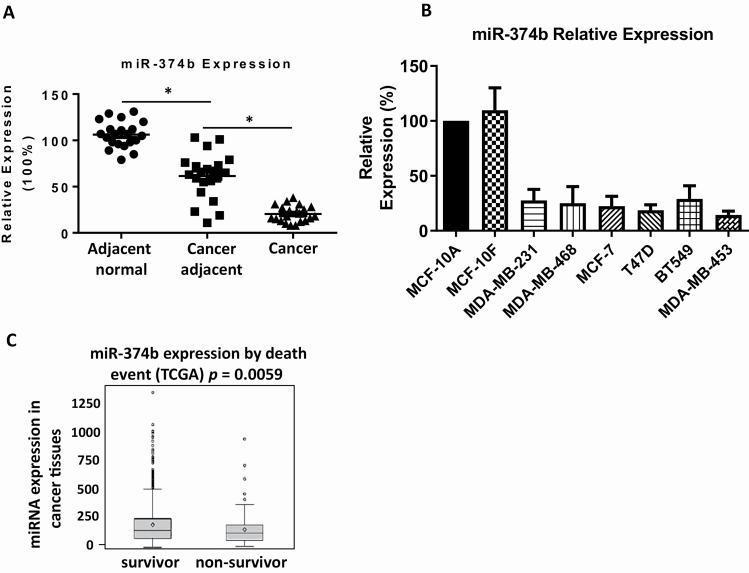
We have identified miR-374b as a prognostic biomarker in TNBC. To investigate whether miR-374b is dysregulated in breast cancer, we analyzed the level of miR-374b in 21 paired samples of breast cancer tissues, adjacent cancer tissues and adjacent normal tissues. Breast cancer tissues had the lowest levels of miR-374b (Figure 1A). We also compared miR-374b expression in multiple human breast epithelial cells and cancer cells and found breast cancer cells had lower expression of miR-374b compared with human epithelial cells, which is consistent with the results observed in tissue samples (Figure 1B). We further performed analysis on miR-374b expression in 1058 breast cancer patients included in TCGA data. MiR-374b expression was associated with better survival (P = 0.0059; Figure 1C), suggesting that miR-374b may play similar roles in all types of breast cancer.
Figure 1.
MiR-374b is dysregulated in breast cancer tissues and breast cancer cell lines. (A) The levels of miR-374b in breast cancer tissues, paired adjacent cancer tissues and paired adjacent normal tissues were measured by quantitative real-time PCR. (B) The levels of miR-374b in human breast epithelial cells (MCF-10A and MCF-10F) and breast cancer cells (MDA-MB-231, MDA-MB-468, MCF-7, T47D, BT549 and MDA-MB-453) were examined by real-time PCR. (C) MiR-374b expression stratified by death event in TCGA (1058 patients). *P < 0.05.
Overexpressing miR-374b disrupts the malignant behavior of breast cancer cells
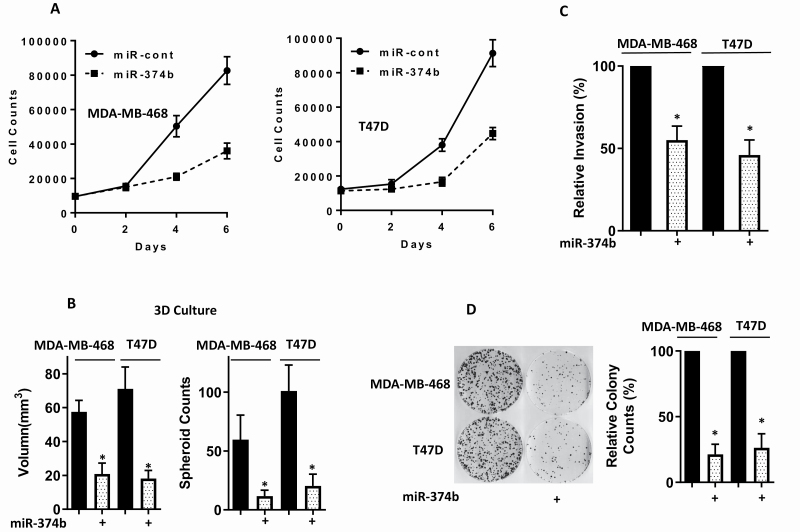
To explore whether miR-374b would affect the biological behavior of breast cancer cells, we overexpressed miR-374b in MDA-MB-468 and T47D breast cancer cells and evaluated its effects on cell proliferation under 2D and 3D conditions, migration and colony-forming ability. Overexpression of miR-374b hampered tumor cell proliferation in breast cancer cells under 2D and 3D conditions (Figure 2A and B), cell migration (Figure 2C) and colony formation (Figure 2D).
Figure 2.
Overexpressing miR-374b disrupts the malignant behavior of breast cancer cells. MiR-374b was transfected into MDA-MB-468 and T47D cells. Forty-eight hours post-transfection, functional assays were performed. (A) Cell proliferation was measured by MTT assay at different time points. (B) Multicellular tumor spheroids formation using 3D cell culture was examined. (C) Cell migration ability was measured by trans-well migration assay. (D) Colony-forming ability was examined by clonogenic assay. Three independent experiments were performed, and the data were presented as mean ± SD. *P < 0.05.
MiR-374b modulates the malignant behavior of breast cancer cells through regulating CCND1 and TGFA
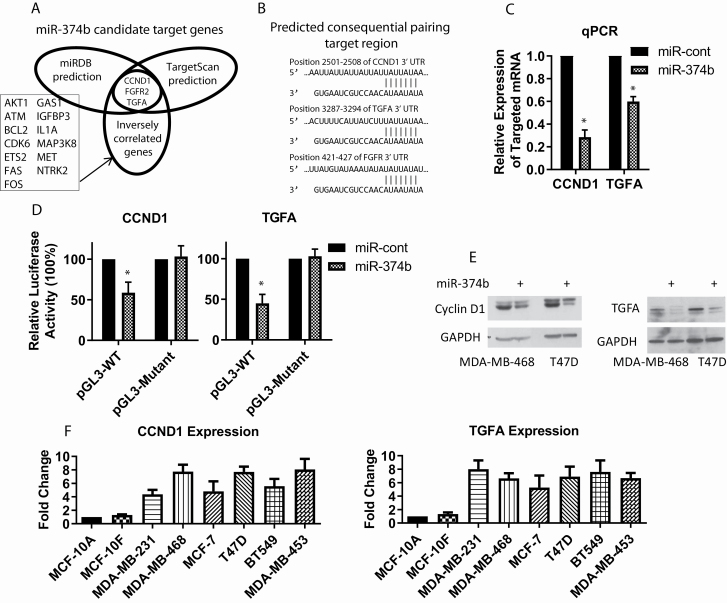
Cascione et al. (16) reported that miR-374b expression was inversely associated with expression of 16 genes, including CCND1, FGFR2, TGFA, AKT serine/threonine kinase 1 (AKT1), ATM serine/threonine kinase (ATM), BCL2 apoptosis regulator (BCL2), cyclin-dependent kinase 6 (CDK6), interleukin 1 alpha (IL1A), neurotrophic receptor tyrosine kinase 2 (NTRK2), ETS proto-oncogene 2, transcription factor (ETS2), fas cell surface death receptor (FAS), fos proto-oncogene, AP-1 transcription factor subunit (FOS), growth arrest specific 1 (GAS1), mitogen-activated protein 3-kinase 8 (MAP3K8), insulin-like growth factor-binding protein 3 (IGFBP3) and MET proto-oncogene, receptor tyrosine kinase (MET). Utilizing two prediction algorithms (TargetScan and miRDB) and gene lists from Cascione et al. (16), we predicted that CCND1, FGFR2 and TGFA were candidate targets for miR-374b (Figure 3A and B). We conducted real-time qRT-PCR analysis and found that mRNA expression levels of CCND1 and TGFA were reduced in response to an overexpression of miR-374b in MDA-MB-468 cells (Figure 3C), but expression of FGFR2 was not detected in this cell line (data not shown).
Figure 3.
MiR-374b negatively regulates the expression of CCND1 and TGFA in breast cancer cells. (A) CCND1 and TGFA were predicted as direct targets of miR-374b (TargetScan and miRDB). (B) The potential miR-374b targeting sites at the 3′ UTR of CCND1, TGFA and FGFR2 genes. (C) The expression of CCND1 and TGFA mRNA in miR-374b-transfected MDA-MB-468 cells was measured by real-time PCR. (D) Relative luciferase level was examined after luciferase reporter plasmids with CCND1 or TGFA 3′ UTR (WT or mutant), or control reporter plasmid miR-374b or miR-control, were co-transfected in MDA-MB-468 cells. (E) The levels of CCND1 and TGFA in miR-374b-transfected cells were examined by western blotting. (F) The levels of CCND1 and TGFA mRNA in human breast epithelial cells and cancer cells were analyzed by real-time PCR. *P < 0.05.
To further determine whether CCND1 and TGFA would be regulated by miR-374b via direct binding to their 3′ UTRs, we employed a dual-luciferase reporter system with luciferase reporter vectors, containing either WT or mutant 3′ UTRs of CCND1 or TGFA genes. Relative luciferase activity was reduced in cells co-transfected with luciferase reporter pmirGLO-3′ UTR-WT and miR-374b mimic compared to control cells. However, the suppressive effects were abolished by the miR-374b target sequence mutation (Figure 3D). Western blotting analysis consistently showed that the expression of CCNDI and TGFA was reduced by the overexpression of miR-374b (Figure 3E). We demonstrate in Figure 1B that breast cancer cell lines have lower levels of miR-374b compared to human breast epithelial cells. To investigate whether these breast cancer cell lines would have higher levels of CCND1 and TGFA, due to the loss of miR-374b, we analyzed their expression using real-time PCR. Our results demonstrate that both CCND1 and TGFA have higher expression in breast cancer cell lines compared to human breast epithelial cells (Figure 3F).
CCND1 and TGFA regulate the malignant behavior of breast cancer cells
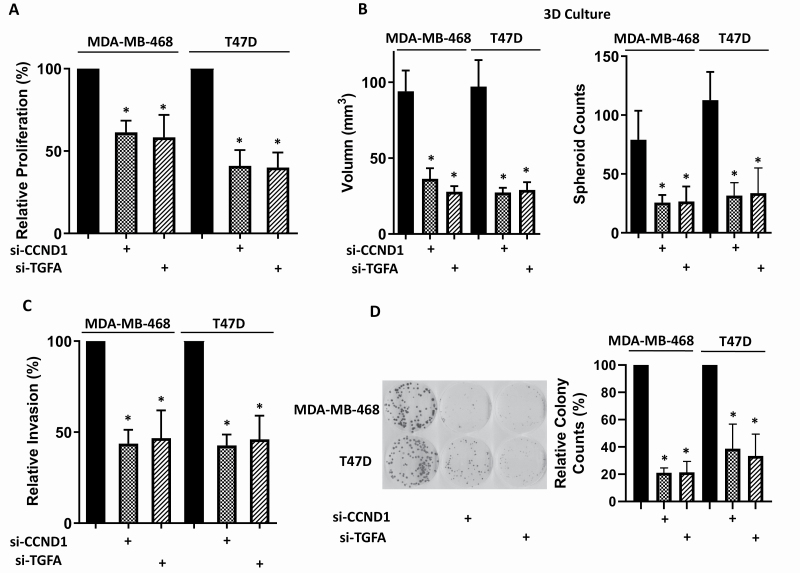
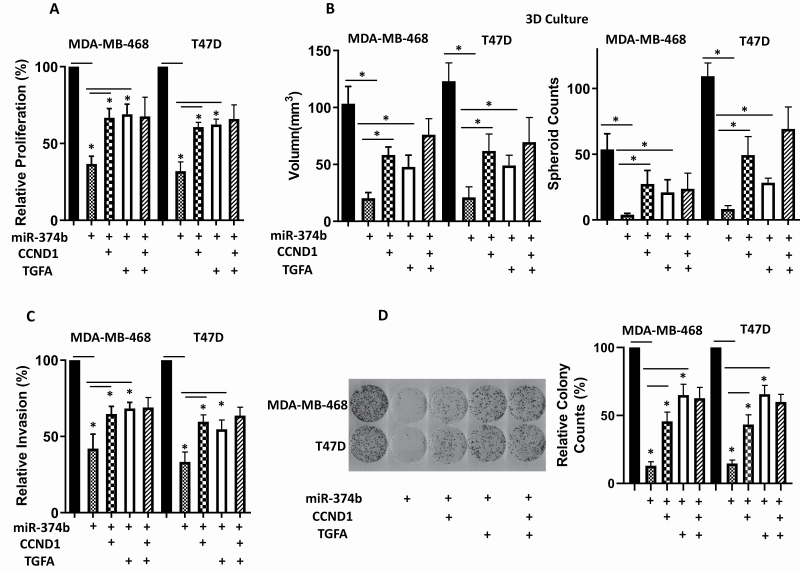
We have demonstrated that miR-374b directly targets CCND1 and TGFA. To further explore whether CCND1 and TGFA influence tumor malignance, we knocked down CCND1 or TGFA. Supplementary Figure S1 (available at Carcinogenesis Online) shows that CCND1 and TGFA siRNAs reduced mRNA expression of CCND1 and TGFA, respectively. We found that knocked down CCND1 or TGFA expression reduced cell proliferation in 2D (Figure 4A) and 3D (Figure 4B), cell migration (Figure 4C) and colony formation (Figure 4D) in both MDA-MB-468 and T47D cells. To investigate whether overexpressing CCND1 or TGFA could partially rescue malignant behaviors of breast cancer cells caused by the overexpression of miR-374b, we co-transfected CCND1 or TGFA with miR-374b and found partially rescued cell proliferation in 2D and 3D (Figure 5A and B, respectively), cell migration (Figure 5C) and colony formation (Figure 5D) for the overexpression of both CCND1 and TGFA in both MDA-MB-468 and T47D cells.
Figure 4.
Knocking down CCND1 or TGFA disrupts the malignant behavior of breast cancer cells. MDA-MB-468 and T47D cells were transfected with si-CCND1, si-TGFA or siRNA negative control. Forty-eight hours post-transfection, functional assays were performed. (A) Cell proliferation was measured by MTT assay at 96 h post-transfection. (B) Multicellular tumor spheroids formation using 3D cell culture was examined. (C) Cell migration ability was analyzed by trans-well migration assay. (D) Colony-forming ability was investigated by clonogenic assay. Three independent experiments were performed, and the data were presented as mean ± SD. *P < 0.05.
Figure 5.
Overexpressing CCND1 or TGFA partially disrupts the effects of overexpressing miR-374b on the malignant behavior of breast cancer cells. MiR-374b was co-transfected into breast cancer cells with expression construct of CCND1 or TGFA, or both. (A) Cell proliferation was measured by MTT assay at 96 h post-transfection. (B) Multicellular tumor spheroids formation using 3D cell culture was examined. (C) Cell migration ability was analyzed by trans-well migration assay. (D) Colony-forming ability was investigated by clonogenic assay. Three independent experiments were performed, and the data were presented as mean ± SD. *P < 0.05.
Overexpressing miR-374b represses tumor growth in vivo through regulating CCND1 and TGFA
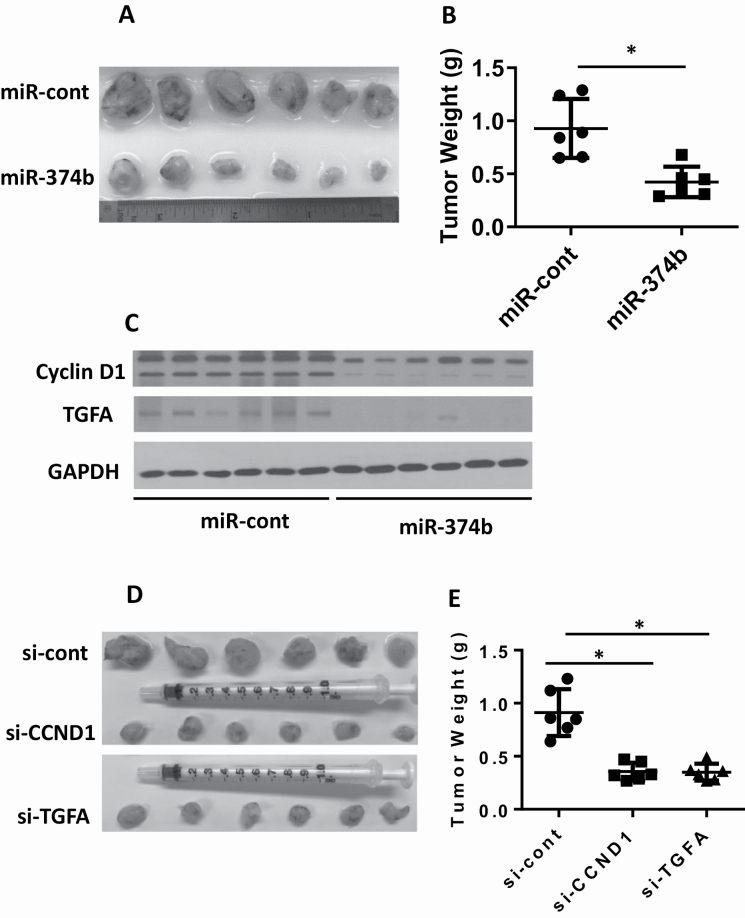
To confirm our in vitro results, we also performed in vivo studies and found that overexpressing miR-374b reduced tumor weights by around 50% (Figure 6A and B). Analyzing gene expression in the tumor tissues, we also found that overexpression of miR-374b reduced the levels of both CCNDI and TGFA (Figure 6C). In addition, decreasing either CCND1 or TGFA significantly reduced tumor weights (Figure 6D and E).
Figure 6.
Overexpressing miR-374b represses tumor growth in vivo through regulating CCND1 and TGFA. MDA-MB-468 stable cells were generated to constitutively express miR-374b or miR-control. (A and B) MDA-MB-468 stable cells expressing miR-374b or miR-control were injected subcutaneously into nude mice. After 5 weeks, tumor weight was evaluated. (C) Total protein lysates were extracted from tumor xenografts and the levels of cyclin D1 and TGFA were evaluated by western blotting. MDA-MB-468 stable cells were generated to constitutively express si-CCND1, si-TGFA or si-control. (D and E) 5 × 106 cells were injected subcutaneously into nude mice. After 5 weeks, tumor weight was evaluated.
Discussion
MiR-374a was previously described as an oncogene to promote breast cancer metastasis through upregulating Wnt/β-catenin signaling (14). This study found that miR-374a was overexpressed in breast cancer tissues from patients with metastases compared to those without metastases, and a high miR-374a expression level was significantly associated with short metastasis-free survival (14). However, that study was based on results comparing miRNA expression in parental MDA-MB-435 cells with their lung metastatic derivatives (14). This is problematic because, while the MDA-MB-435 cell line was previously considered a breast cancer cell line, recently it was confirmed to be a melanoma cell line (23) (http://www.atcc.org/products/all/HTB-129.aspx). MiR-374a (12 – uuauaauacaaccugauaagug – 33) and miR-374b (11 – auauaauacaaccugcuaagug – 32) have very similar RNA sequences, according to miRBase: the microRNA database (http://www.mirbase.org/index.shtml). Unlike the previous study, we found that a high expression of either miR-374a or miR-374b in tumor tissues was associated with better survival in TNBC cases from the Shanghai Breast Cancer Survival Study (SBCSS), although miR-374b was more statistically significant. Our results were supported by a previous study, which included 173 TNBC cases (16). Our analysis from TCGA data, which included 1058 breast cancer patients, showed that miR-374b expression is associated with better survival, suggesting that it may play similar roles in all types of breast cancer.
The expression of miR-374b was significantly downregulated in breast cancer tissues compared with adjacent normal tissues. Functional studies revealed that the overexpression of miR-374b inhibited cell proliferation, migration and colony formation in breast cancer cells, as well as suppressed tumor growth in vivo. The data suggest that miR-374b functions as a tumor suppressor in breast cancer.
Using two bioinformatics tools, we found that CCND1, FGFR2 and TGFA genes could be potential targets of miR-374b. Our in vitro experiments confirmed that CCND1 and TGFA were direct targets of miR-374b, suggesting that miR-374b-mediated CCND1 and TGFA gene silencing resulted in significant inhibitory effects on the malignant behavior of breast cells and affected patient prognosis. A previous study also reported that miR-374a and miR-374b are both inversely associated with expressions of AKT1, ATM, BCL2, CCND1, CDK6, ETS2, FAS, FGFR2, FOS, GAS1, IGFBP3, IL1A, MAP3K8, MET, NTRK2 and TGFA (16). We previously reported that miR-374a overexpression inhibited lung adenocarcinoma cell proliferation, migration and invasion via targeting of TGFA gene expression (18).
It has been well established that CCND1 and TGFA play important roles in breast cancer, including tumor growth, cell cycle regulation and migration (24–29). In addition, both CCND1 and TGFA serve as prognostic biomarkers for breast cancer patients (30–35). The CCND1 protein is overexpressed and has many functions in breast cancer, including carcinogenesis (27–29). In addition, both mRNA and protein expression of CCND1 correlate with breast cancer patient prognosis (33,36,37). Using publicly available microarray gene expression and breast cancer outcome data, we found that higher CCND1 mRNA levels were associated with poor metastasis- or recurrence-free survival. TGFA is a growth factor and member of the epidermal growth factor (EGF) family that regulates autocrine breast cancer growth (24,38). Animal models have demonstrated that TGFA promotes epithelium growth in mammary glands by binding to the EGF receptor, activating its kinase cell signaling processes (25). TGFA has been detected in 50–70% of primary breast cancer tissues, and its activity is 2-to 3-fold higher in breast cancer patients (24). High TGFA expression is associated with axillary lymph node metastasis and poor prognosis in breast cancer patients (30). These data further support the notion that miR-374b functions as a tumor suppressor in breast cancer by targeting CCND1 and TGFA genes.
In summary, our in vitro and in vivo studies demonstrate that miR-374b modulates malignant behavior of breast cancer cells by negatively regulating CCND1 and TGFA genes. The newly identified miR-374b-mediated CCND1 and TGFA gene silencing may facilitate a better understanding of the molecular mechanisms of breast cancer progression.
Funding
This work was supported by the Department of Defense Breast Cancer Research Program (DAMD 17-02-1-0607); the National Institutes of Health (R01CA118229, P50CA098131); the Vanderbilt Molecular and Genetic Epidemiology of Cancer training program (R25CA160056) and General Program from the National Natural Science Foundation of China (81472706 and 81874187). Part of the lab assays were performed at the Survey and Biospecimen Shared Resource, which is supported in part by the Vanderbilt-Ingram Cancer Center (P30CA068485).
Supplementary Material
Acknowledgements
We thank Regina Courtney and Dr Jie Wu for their laboratory support and Dr Mary Shannon Byers and Mr Marshal Younger for assistance with editing and manuscript preparation. The authors would like to thank the investigators of the Cancer Cell Line Encyclopedia who have made their invaluable data available to the scientific community.
Conflict of Interest Statement: Authors declare no conflict of interest.
Glossary
Abbreviations
- CCND1
cyclin D1
- EGF
epidermal growth factor
- FGFR
fibroblast growth factor receptor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- miRNA
microRNA
- mRNA
messenger RNA
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- PBS
phosphate-buffered saline
- SBCSS
the Shanghai Breast Cancer Survival Study
- TCGA
The Cancer Genome Atlas
- TGFA
transforming growth factor alpha
- TNBC
triple-negative breast cancer
- UTR
untranslated region
- WT
wild type
References
- 1. Brennecke, J., et al. (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell, 113, 25–36. [DOI] [PubMed] [Google Scholar]
- 2. Chen, C.Z., et al. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science, 303, 83–86. [DOI] [PubMed] [Google Scholar]
- 3. Schratt, G.M., et al. (2006) A brain-specific microRNA regulates dendritic spine development. Nature, 439, 283–289. [DOI] [PubMed] [Google Scholar]
- 4. Bartel, D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Croce, C.M. (2009) Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet., 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garzon, R., et al. (2009) MicroRNAs in cancer. Annu. Rev. Med., 60, 167–179. [DOI] [PubMed] [Google Scholar]
- 7. Png, K.J., et al. (2011) A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature, 481, 190–194. [DOI] [PubMed] [Google Scholar]
- 8. Tavazoie, S.F., et al. (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature, 451, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ma, L., et al. (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature, 449, 682–688. [DOI] [PubMed] [Google Scholar]
- 10. Ma, L., et al. (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol., 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valastyan, S., et al. (2009) A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell, 137, 1032–1046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Yang, S., et al. (2013) MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene, 32, 4294–4303. [DOI] [PubMed] [Google Scholar]
- 13. Gregory, P.A., et al. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol., 10, 593–601. [DOI] [PubMed] [Google Scholar]
- 14. Cai, J., et al. (2013) MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J. Clin. Invest., 123, 566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu, Y., et al. (2015) Tumor tissue microRNA expression in association with triple-negative breast cancer outcomes. Breast Cancer Res. Treat., 152, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cascione, L., et al. (2013) Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One, 8, e55910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aran, D., et al. (2017) Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun., 8, 1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu, H., et al. (2016) MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis, 37, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewis, B.P., et al. (2003) Prediction of mammalian microRNA targets. Cell, 115, 787–798. [DOI] [PubMed] [Google Scholar]
- 20. Wong, N., et al. (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res., 43, D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krek, A., et al. (2005) Combinatorial microRNA target predictions. Nat. Genet., 37, 495–500. [DOI] [PubMed] [Google Scholar]
- 22. Kertesz, M., et al. (2007) The role of site accessibility in microRNA target recognition. Nat. Genet., 39, 1278–1284. [DOI] [PubMed] [Google Scholar]
- 23. Holliday, D.L., et al. (2011) Choosing the right cell line for breast cancer research. Breast Cancer Res., 13, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ciardiello, F., et al. (1991) Expression of transforming growth factor alpha (TGF alpha) in breast cancer. Ann. Oncol., 2, 169–182. [DOI] [PubMed] [Google Scholar]
- 25. Humphreys, R.C., et al. (2000) Transforming growth factor alpha and mouse models of human breast cancer. Oncogene, 19, 1085–1091. [DOI] [PubMed] [Google Scholar]
- 26. Ciardiello, F., et al. (1989) mRNA expression of transforming growth factor alpha in human breast carcinomas and its activity in effusions of breast cancer patients. J. Natl Cancer Inst., 81, 1165–1171. [DOI] [PubMed] [Google Scholar]
- 27. Bartkova, J., et al. (1994) Cyclin D1 protein expression and function in human breast cancer. Int. J. Cancer, 57, 353–361. [DOI] [PubMed] [Google Scholar]
- 28. Velasco-Velázquez, M.A., et al. (2011) Examining the role of cyclin D1 in breast cancer. Future Oncol., 7, 753–765. [DOI] [PubMed] [Google Scholar]
- 29. Roy, P.G., et al. (2006) Cyclin D1 and breast cancer. Breast, 15, 718–727. [DOI] [PubMed] [Google Scholar]
- 30. Auvinen, P.K., et al. (1996) Prognostic significance of TGF-alpha expression in breast cancer. Acta Oncol., 35, 995–998. [DOI] [PubMed] [Google Scholar]
- 31. Bebök, Z., et al. (1994) Prognostic relevance of transforming growth factor alpha (TGF-alpha) and tumor necrosis factor alpha (TNF-alpha) detected in breast cancer tissues by immunohistochemistry. Breast Cancer Res. Treat., 29, 229–235. [DOI] [PubMed] [Google Scholar]
- 32. Murray, P.A., et al. (1993) The prognostic significance of transforming growth factors in human breast cancer. Br. J. Cancer, 67, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gillett, C., et al. (1996) Cyclin D1 and prognosis in human breast cancer. Int. J. Cancer, 69, 92–99. [DOI] [PubMed] [Google Scholar]
- 34. Kenny, F.S., et al. (1999) Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin. Cancer Res., 5, 2069–2076. [PubMed] [Google Scholar]
- 35. Turner, B.C., et al. (2000) Cyclin D1 expression and early breast cancer recurrence following lumpectomy and radiation. Int. J. Radiat. Oncol. Biol. Phys., 47, 1169–1176. [DOI] [PubMed] [Google Scholar]
- 36. van Diest, P.J., et al. (1997) Cyclin D1 expression in invasive breast cancer. Correlations and prognostic value. Am. J. Pathol., 150, 705–711. [PMC free article] [PubMed] [Google Scholar]
- 37. Tobin, N.P., et al. (2011) Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer, 11, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sandgren, E.P., et al. (1990) Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell, 61, 1121–1135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.