Abstract
Background:
Due to the ubiquitous use of chemicals in modern society, humans are increasingly exposed to thousands of chemicals that contribute to a major portion of the human exposome. Should a comprehensive and risk-based human exposome database be created, it would be conducive to the rapid progress of human exposomics research. In addition, once a xenobiotic is biotransformed with distinct half-lives upon exposure, monitoring the parent compounds alone may not reflect the actual human exposure. To address these questions, a comprehensive and risk-prioritized human exposome database is needed.
Objectives:
Our objective was to set up a comprehensive risk-prioritized human exposome database including physicochemical properties as well as risk prediction and develop a graphical user interface (GUI) that has the ability to conduct searches for content associated with chemicals in our database.
Methods:
We built a comprehensive risk-prioritized human exposome database by text mining and database fusion. Subsequently, chemicals were prioritized by integrating exposure level obtained from the Systematic Empirical Evaluation of Models with toxicity data predicted by the Toxicity Estimation Software Tool and the Toxicological Priority Index calculated from the ToxCast database. The biotransformation half-lives () of all the chemicals were assessed using the Iterative Fragment Selection approach and biotransformation products were predicted using the previously developed BioTransformer machine-learning method.
Results:
We compiled a human exposome database of chemicals, prioritized 13,441 chemicals based on probabilistic hazard quotient and 7,770 chemicals based on risk index, and provided a predicted biotransformation metabolite database of metabolites. In addition, a user-interactive Java software (Oracle)-based search GUI was generated to enable open access to this new resource.
Discussion:
Our database can be used to guide chemical management and enhance scientific understanding to rapidly and effectively prioritize chemicals for comprehensive biomonitoring in epidemiological investigations. https://doi.org/10.1289/EHP7722
Introduction
The human exposome, defined as the totality of exposures throughout the human lifespan (Wild 2005), has raised concerns in environmental health studies in recent years. Due to the ubiquitous use of manmade chemicals in modern society, people are potentially exposed to thousands of chemicals via multiple pathways, such as air, water, food, and soil (Wambaugh et al. 2014). As an important part of the overall human exposome, chemical exposure is challenging to characterize because it requires exposure data to be obtained under multiple scenarios (Dai et al. 2017). We are still at the early stage of figuring out what we are exposed to and it is also difficult to determine which chemicals pose high exposure risk to human health. These data gaps necessitate the human exposome database as well as chemical risk prioritization.
To date, some yet insufficient efforts have been directed to collect and organize data on hazard and exposure for thousands of chemicals. For known exposure biomarkers, the Exposome-Explorer Database (http://exposome-explorer.iarc.fr/) with 691 chemicals has been established to provide comprehensive data on exposure to dietary factors, pollutants, and contaminants measured in population studies (Neveu et al. 2017). More recently, Barupal and Fiehn (2019) conducted a more in-depth literature mining and database fusion for the blood exposome, yielding 49,940 unique chemicals pooled from 676,643 papers. Our earlier work (Dong et al. 2019) established a dust exposome database with 511 chemicals with measured concentrations by an extensive text mining approach. However, most of these studies are either based on studies with previous measurements for only limited numbers of chemicals or databases without associated toxicity information. It is still possible that many compounds remain uncovered by existing databases, such as the Exposome-Explorer Database and the Blood Exposome Database (http://bloodexposome.org/). Hence, a comprehensive human exposome database, as well as a potential biomarker database, is needed. In addition, the enormous number of chemicals make biomonitoring using traditional methods, such as targeted or nontargeted mass spectrometry methods, almost impossible. Therefore, it is of great scientific value to sort and prioritize chemicals based on their exposure and risk in the context of exposome research for activities such as chemical management and environmental epidemiological investigation.
To date, a few studies have tried to prioritize limited numbers of chemicals using either exposure level or in vitro toxicity screening methods. For example, the U.S. Environmental Protection Agency (EPA) has established the ToxCast Program combining in vitro high-throughput assays to facilitate rapid hazard assessments based on chemical bioactivities (Williams et al. 2017). In one study, the prioritization of 309 environmental chemicals was characterized by the Toxicological Priority Index (ToxPi) model using the data from 467 in vitro high-throughput screening (HTS) assays (Reif et al. 2010). In another example, the U.S. EPA initiated the Exposure Forecasting (ExpoCast) program, which contained information on exposure to ensure the need for rapid characterization of exposure potential, and a total of 1,936 chemicals were ranked by the predicted exposure level (Wambaugh et al. 2013). Subsequently, the U.S. EPA developed an updated consensus, meta-model using the Systematic Empirical Evaluation of Models (SEEM) approach to calibrate various exposure predictors from ExpoCast with the output of relevant exposure pathway(s), median intake rate, and credible interval for 479,926 chemicals (Ring et al. 2019). In general, the prioritization approaches were conducted based on either only exposure or toxicity alone. One previous study presented a risk-based method for chemical prioritization; however, only 180 chemicals were prioritized (Shin et al. 2015). Some efforts were made to develop quantitative approaches to translate in vitro toxicity potencies to in vivo equivalent doses using in vitro–in vivo extrapolation (IVIVE) models, which can be used to conduct in vitro toxicity screening for chemical prioritization (Ring et al. 2017; Sipes et al. 2017; Wambaugh et al. 2015; Wetmore et al. 2012, 2014; Wetmore 2015). However, the prioritization was only based on the National Institutes of Health (NIH) Toxicology in the 21st Century (Tox21) and ToxCast HTS data, which are far from sufficient given the overwhelmingly and ever-increasingly large number of environmental chemicals. Thus, a more widely applicable and high-throughput methodological approach incorporating exposure data with toxicity data is still needed to evaluate and prioritize chemicals for potential risk to human health.
Besides risk prioritization, another knowledge gap for the current human exposome database is that most of the focus is on the prioritization for parent chemicals (Reif et al. 2010; Wambaugh et al. 2013, 2014). Chemicals can be often biotransformed in several organs, such as the gut and liver, and excreted through the feces or urine (Djoumbou-Feunang et al. 2019). For example, the biotransformation half-lives () of the common plasticizers bisphenol A (BPA) and phthalates are only within several hours (Koch et al. 2004; Thayer et al. 2015). For these chemicals with a short , human biomonitoring (HBM) of the parent compounds becomes challenging and even partially meaningless without knowing their xenobiotic biotransformation products. Biotransformation, which could either detoxify or activate the toxic potential, greatly impacts on toxicity (Bland 2007). Thus, this knowledge gap necessitates the human exposome database containing possible biotransformation metabolites, which will be very beneficial for the human exposure biomarkers biomonitoring of epidemiological investigation and untargeted exposomics surveys.
Consequently, in the present study, we aimed to establish an upgraded human exposome database that includes both parent compounds and predicted biotransformation metabolites and prioritizes the parent compounds based on risk. The results will guide the chemical management and enhance the understanding to rapidly and effectively prioritize chemicals for HBM in epidemiological investigations on knowledge-based risk ranking. The specific aims were a) the buildup of a human exposome database using comprehensive text mining and a database fusion approach; b) prioritization of chemicals in our human exposome database using models of exposure and toxicity prediction; c) evaluation of the of human exposome chemicals and prediction of their biotransformation metabolites; and d) development of an interactive interface for allowing access to the newly established Human Exposome and Metabolite Database (HExpMetDB).
Methods
Compiled List of Human Exposome Database
To set up the prioritized human exposome database, we screened as many chemicals as possible in different previously published databases and sources relevant to exposome research including environmental pollutants, toxicants, gut microbiome-derived metabolites, disinfection and combustion by-products, carcinogens, and food nutrients and additives (Table 1). Endogenous human metabolites and inorganic chemicals were excluded in this study. Because publicly available databases, such as the U.S. EPA High Production Volume (HPV) List, mostly include registered industrial chemicals, we also searched the literature for additional chemicals in various environmental media such as drinking water, indoor dust, and air that are closely associated with routes of exposure. Literature mining was conducted by manually searching research articles or reviews on the Web of Science and PubMed using the combined keywords such as human exposome, drinking water, air, and disinfection or combustion by-products to collect studies focusing on cataloging environmental contaminants as shown in Excel Table S1. Due to the large numbers of search hits, we preferentially selected review articles with an extensive summary of chemicals from environmental matrices (e.g., indoor air exposome, dust exposome, or waterborne chemicals) and used their summary as the input for the chemical database fusion if available. We compiled the database by gathering publicly available databases and literature tabulated in Table 1. After removing the inorganic compounds and organic mixtures, as well as replicates by Chemical Abstracts Service Registry Number (CASRN) and canonical Simplified Molecular Input Line Entry Specification (SMILES), a total of 20,756 unique chemicals with CASRN were included in our database. Additional information was also obtained from the U.S. EPA Chemistry Dashboard, including chemical identifiers [Distributed Structure-Searchable Toxicity substance identifier (DTXSID), chemical name, CASRN, condensed version of the International Chemical Identifier (InChIKey), and International Union of Pure and Applied Chemistry (IUPAC) name], structures (SMILES and InChI string), and intrinsic properties (molecular formula, average mass, and monoisotopic mass).
Table 1.
Coverage of chemicals in different databases and resources relevant to exposome research.
| Source category | Source name and description | a | Website | Reference |
|---|---|---|---|---|
| Government databases | U.S. EPA: High Production Volume List | 3,146 | https://comptox.epa.gov/dashboard/chemical_lists/EPAHPV | U.S. EPA 2020c |
| European inventory of existing commercial chemical substances | 7,301 | https://echa.europa.eu/information-on-chemicals/ec-inventory | ECHA 2008 | |
| Candidate List of substances of very high concern for authorization | 233 | https://echa.europa.eu/candidate-list-table | ECHA 2020 | |
| USDA: FoodData Central data | 154 | https://fdc.nal.usda.gov/index.html | USDA 2019 | |
| European Commission, Food Additives Database and Food Flavorings Database | 2,543 | https://webgate.ec.europa.eu/foods_system/main/?sector=FAD&auth=SANCAS | EC 2017 | |
| https://webgate.ec.europa.eu/foods_system/main/?event=display | EC 2012 | |||
| Toxicological databases | U.S. EPA: Chemical Inventory for ToxCast | 6,350 | https://comptox.epa.gov/dashboard/chemical_lists/CHEMINV | U.S. EPA 2007 |
| European Commission: priority list of endocrine disruptors | 385 | https://ec.europa.eu/environment/chemicals/endocrine/strategy/substances_en.htm | EC 2020 | |
| NIH: Toxicology in the 21st Century (Tox21) | 7,632 | https://ncats.nih.gov/tox21 | — | |
| IARC: Agents Classified by the IARC Monographs | 845 | https://monographs.iarc.fr/agents-classified-by-the-iarc/ | — | |
| NIH: 14th Report on Carcinogens | https://ntp.niehs.nih.gov/whatwestudy/assessments/cancer/roc/index.html | — | ||
| U.S. EPA: Pesticides | 3,265 | https://www.epa.gov/pesticides | U.S. EPA 2017b | |
| Exposure biomarker databases | Exposome-Explorer: database on biomarkers of exposure to environmental risk factors for diseases | 233 | http://exposome-explorer.iarc.fr/ | — |
| CDC: The NHANES National Report on Human Exposure to Environmental Chemicals | 425 | https://www.cdc.gov/exposurereport/ | — | |
| Literature data | Environmental pollutants detected in water | 96 | — | Andrianou et al. 2019; Remucal and Manley 2016; Sjerps et al. 2016; U.S. EPA 2016a; Vikesland and Raskin 2016 |
| Environmental pollutants detected in dust | 470 | — | Dong et al. 2019 | |
| Environmental pollutants detected in the air | 205 | — | WHO 2016; U.S. EPA 2018a, 2018b, 2020a, 2020b | |
| Environmental by-products (e.g., disinfection and combustion) | 43 | — | Castaño-Vinyals et al. 2011; Hebert et al. 2010; Nieuwenhuijsen et al. 2009 | |
| Mycotoxins | 40 | — | Shephard 2008; Warth et al. 2012 | |
| Gut microbiome-related metabolites | 26 | — | Donia and Fischbach 2015; Wang et al. 2011; Wilmanski et al. 2019; Zhang and Davies 2016 |
Note: —, not applicable; CDC, Centers for Disease Control and Prevention; ECHA, European Chemical Agency; EPA, Environmental Protection Agency; IARC, International Agency for Research on Cancer; NHANES, National Health and Nutrition Examination Survey; NIH, National Institutes of Health; USDA, U.S. Department of Agriculture.
Number of chemicals.
Exposure Estimates
Given that most chemicals lack measured data of human exposure, we obtained the human exposure predictions from the SEEM consensus exposure model predictions, which yielded a coefficient of determination value of (i.e., 80% of the data fit the regression model) for high-throughput exposure assessment as reported by Ring et al. (2019) and used by Wambaugh et al. (2019). The prediction of median exposure level [in milligrams per kilogram of body weight (BW) per day] with uncertainty [95% confidence interval (CI)] for each chemical, if available, was retrieved from the subset of the Bayesian inferences reported by Ring et al. (2019) for a consensus model of 12 exposure predictors that were calibrated based on their ability to predict intake rates inferred from the National Health and Nutrition Examination Survey (NHANES).
Chemical Biotransformation Half-Life () Prediction
The prediction of chemical was based on the quantitative structure−activity relationship (QSAR) approach called Iterative Fragment Selection (IFS) (Arnot et al. 2014). According to a previous study, the , , and root-mean-square errors for the QSAR were 0.89, 0.73, and 0.75, respectively; 96% and 76%, and 82% and 52% of the predicted values were within a factor of 10 and 3 of the expected values for the training and testing sets, respectively (Arnot et al. 2014). Because the IFS algorithm cannot be applied to ionogenic chemicals and siloxanes, they were not included in the training set of the IFS model (Brown et al. 2012; Papa et al. 2014). The and , which were used to estimate , were calculated using the Helmholtz Centre for Environmental Research Linear Solvation Energy Relationship (UFZ-LSER) database (version 3.2.1; Ulrich et al. 2017).
Chemical Toxicity Prediction
Chemical toxicities were estimated using the Toxicity Estimation Software Tool (TEST, version 4.2.1) (U.S. EPA 2016b). The prediction was applicable to compounds containing only the following element symbols: carbon (C), hydrogen (H), oxygen (O), nitrogen (N), fluorine (F), chorine (Cl), bromine (Br), iodine (I), sulfur (S), phosphorus (P), silico (Si), or arsenic (As). The QSAR-ready SMILES code for each chemical was submitted to the models. The rat oral toxicological properties were selected to evaluate the chemical toxicities in this study.
Due to the possible uncertainties from in silico toxicity predictions, we included the highly ranked toxicants from ToxCast bioassay and U.S. EPA chemicals of concerns, such as flame retardants and chemicals of interest to the U.S. EPA Endocrine Disruption Screening Program (EDSP) for the 21st Century (EDSP21) (Richard et al. 2016; U.S. EPA 2007). These chemicals were arbitrarily included in the final exposome database as high-risk groups.
To further expand our toxicity data, we applied the Toxicological Priority Index (ToxPi) model in HTS data to prioritize 8,845 environmental chemicals with potential toxicological activities, providing a transparent visualization of the relative contribution of all information sources to an overall priority ranking (Filer et al. 2014; Marvel et al. 2018; Reif et al. 2010). For the ToxPi modeling, we used 97 in vitro HTS assays, including the targets of the estrogen, androgen, and thyroid pathways and the glucocorticoid receptor, peroxisome proliferator-activated receptors (PPARs), and monoamine signaling, as well as two physicochemical properties, the octanol-water partitioning coefficient (log P) and bioconcentration factor (BCF) (Filer et al. 2014). We used the potency [concentration for half-maximal activity ()] and efficacy (Emax) estimates provided by the ToxCast program (Kavlock et al. 2012), as well as estimates for the log P and BCF retrieved from the U.S. EPA Chemistry Dashboard to calculate the ToxPi scores of 8,845 chemicals by ToxPi graphical user interface (GUI) (version 2.0; Marvel et al. 2018).
Chemical Risk Prioritization and Uncertainty Analysis
Noncancer risk was expressed in terms of a probabilistic hazard quotient (PrHQ) for each substance to prioritize its risk in this study. PrHQ is the ratio of estimated human exposure (EHE; milligrams per kilogram of BW per day) and probabilistic reference dose (RfD), a target human dose (); milligrams per kilogram of BW per day) was calculated according to Equation (1) (Chiu et al. 2018; WHO/IPCS 2017). The is the probabilistic estimate of the human dose associated with an effect magnitude M and population incidence I. Using this definition, we derived estimates for the for a population incidence of , that is, the (median) is calculated by dividing the benchmark dose for a magnitude of effect M () by the product of uncertainty factors (UFs: , a probabilistic factor of 4.5 for interspecies BW scaling; , a probabilistic factor of 1 for interspecies toxicokinetic (TK) and toxicodynamic (TD) differences (after BW scaling); , a probabilistic factor of 9.7 for human variability in sensitivity for a population incidence I) (Equation 2) (Chiu et al. 2018; WHO/IPCS 2017). A (a benchmark response of 10% extra risk) was considered to estimate PrHQ, which was modeled according to calculated rat oral by the product of UFs (: a factor of 10 for interspecies; : a factor of 10 for instead of and ) (Equation 3) (WHO/IPCS 2017).
| (1) |
| (2) |
| (3) |
In addition, to further assess the distribution of uncertainty when estimating the PrHQ, Monte Carlo (MC) simulation was conducted to simulate the impact of exposure and uncertainty on calculating the PrHQ 10,000 times, using a similar model as in our previous studies (Jia et al. 2019; Zhang et al. 2020). Three separate MC simulations were performed referring to another previous study (Wambaugh et al. 2019): exposure prediction uncertainty only, prediction uncertainty only, and both exposure and prediction uncertainty.
We also defined a risk index (RI) as the product of normalized exposure and ToxPi score (Equation 4) to estimate the potential hazard of a chemical. The log-transformed exposure values predicted by SEEM were normalized to the interval [0, 1]. Thus, the RIs ranged from zero to one, with values near one indicating high potential risks. The uncertainties of risk index calculated from the ToxPi Score and Exposure followed the MC simulation mentioned above.
| (4) |
Pearson correlation coefficients were also calculated to assess the bivariate relationship between the overlapping substances rank-based PrHQ and RI by IBM SPSS (version 22.0; IBM).
Biotransformation Metabolite Prediction
The open-access biotransformation prediction tool BioTransformer was used to predict the biotransformation metabolites of the chemicals in the prioritized human exposome database (Djoumbou-Feunang et al. 2019). This method offers a knowledge-based approach to predict small-molecule biotransformation in human tissues, the human gut, and the environment based on chemical structure and/or physicochemical properties. BioTransformer has been reported to achieve a better prediction (49%) and recall (88%) than Meteor Nexus, which is considered to be the gold standard for predicting biotransformations of xenobiotics, at the equivocal level of confidence (35% precision, and 71% recall) (Djoumbou-Feunang et al. 2019). Only compounds with a molecular weight Da and containing a limited set of 64 different structural motifs are included in the training set of the model, whereas a number of chemical classes, including ether lipids, glycerolipids, and glycerophospholipids, sphingolipids, and acyl coenzyme A conjugates are excluded from the training set (Djoumbou-Feunang et al. 2019). We retrieved biotransformation predictions and compound identification data using the modules of a) the Enzyme Commission-based (EC-based) transformation; b) the CYP450 (phase I) transformation; c) the phase II transformation; and d) human gut microbial transformation.
Development of a GUI of the HExpMetDB
We used Java (version 8; Oracle) to construct the HExpMetDB GUI client program linked to a series of data that provides the ability to search for content associated with chemicals in our database. The GUI allows users to search compounds by CASRN, formula, mass-charge-ratio (m/z), adduct search, and accuracy (in parts per million), and retrieve the corresponding metadata including chemical identifiers, structures, and predicted data of , exposure and rat oral . Users can further search the candidate metabolites of the searched parent compound. HExpMetDB (version 1.0) of either Windows or Mac OS is available as an open-access Java library in the Supplemental Material (file, HExpMetDB_for_Win.zip or HExpMetDB_for_Mac.zip) and at https://github.com/FangLabNTU/HExpMetDB. A user tutorial was also provided in the Supplemental Material (Text S1, “Graphical User Interface (GUI) installation” and Text S2, “Database functionality”).
Results
Chemical List Screening and Merging
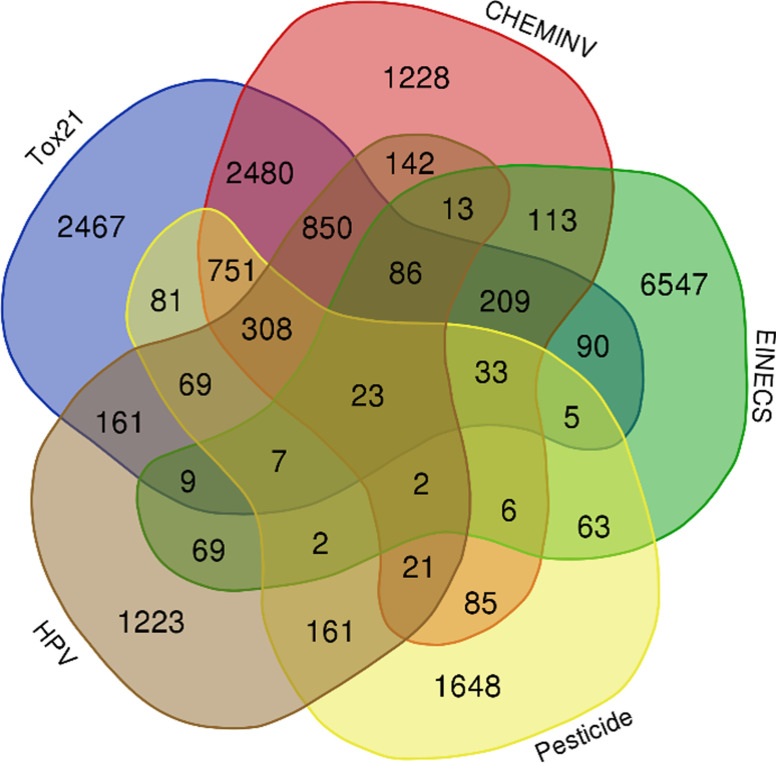
The workflow used to compile the set of chemicals included in the new prioritized human exposome database is illustrated in Figure 1. To enrich the database, we attempted to collect and merge relevant chemicals from databases and literature for a broad exposome-scale resource (Table 1). In total, a consolidated list of 20,756 compounds was successfully mapped to the prioritized human exposome database in the present study. The database is available in the Supplemental Material (Excel Table S2) and at https://github.com/FangLabNTU/HExpMetDB. To date, U.S. EPA HPV chemicals (U.S. EPA 2020c), European Inventory of Existing Commercial Chemical Substances (EINECS) (ECHA 2008), U.S. EPA Chemical Inventory for ToxCast (CHEMINV) (U.S. EPA 2007), NIH Tox21 (https://tripod.nih.gov/tox21/samples) chemicals and U.S. EPA pesticides (U.S. EPA 2017b) are the dominant contributors of the exposome database, which covered 93% (18,909/20,756) of the whole database (Figure 2). For each chemical compound with CASRN, we obtained metadata—such as the InChIKey, DTXSID, IUPAC name, molecular formula, QSAR-ready SMILES, average mass, and monoisotopic mass—from the U.S. EPA Chemistry Dashboard. The remaining prioritized chemicals without CASRN were not included for downstream exposure and toxicity evaluation.
Figure 1.
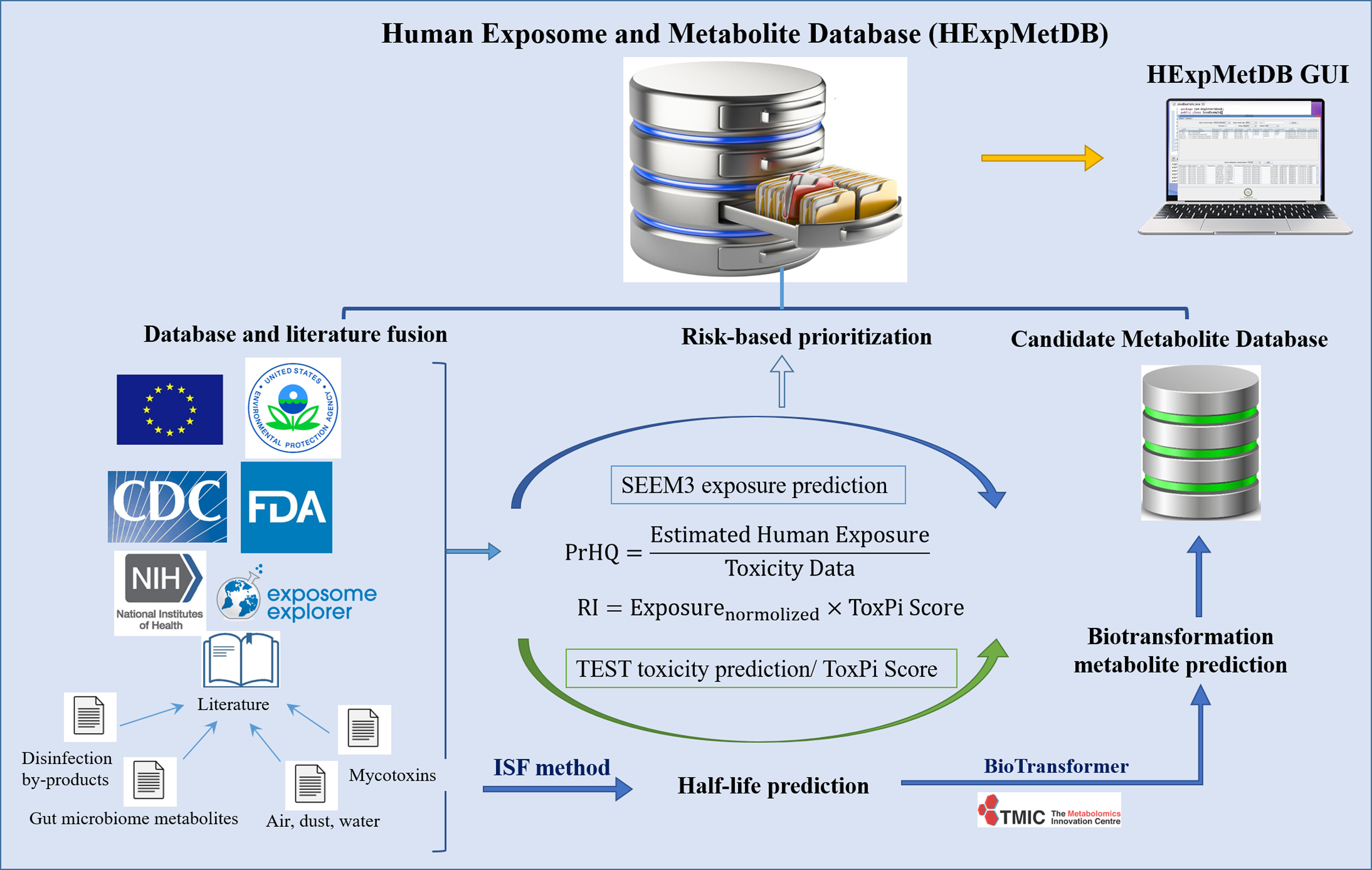
Schematic workflow for Human Exposome and Metabolite Database (HExpMetDB) establishment. Note: CDC, Centers for Disease Control and Prevention; FDA, U.S. Food and Drug Administration; GUI, graphical user interface; ISF, Iterative Fragment Selection; NIH, National Institutes of Health; PrHQ, probabilistic hazard quotient; RI, risk index; SEEM3, Systematic Empirical Evaluation of Models; TEST, Toxicity Estimation Software Tool; ToxPi, Toxicological Priority Index.
Figure 2.
Overlap analysis of five major database mapping in HExpMetDB. High production volume (HPV) chemicals, European Inventory of Existing Commercial Chemical Substances (EINECS), U.S. EPA Chemical Inventory for ToxCast (CHEMINV), NIH toxicology in the 21st Century (Tox21) chemicals and U.S. EPA pesticides contain a total of 18,909 chemicals, covering 91% of the whole database (20,756). Note: EPA, Environmental Protection Agency; HExpMetDB, Human Exposome and Metabolite Database; NIH, National Institutes of Health.
Evaluation
For the 20,756 chemicals with both CASRN and SMILES in the database, we used the QSAR approach to predict their . The of 19,406 chemicals listed in Excel Table S2 were successfully predicted; the calculation was not feasible for the rest of the 1,350 chemicals because they contained atoms outside of the training set or have molecular weights Da. Of those 19,406 chemicals, the median and were estimated to be 2.67 (range: to 28.2) and 8.96 ( to 62.8), respectively (Excel Table S2). The median was predicted to be 4.96 h. Chemicals such as matairesinol and l-cichoric acid were predicted to have the shortest of 0.05 h. On the other hand, some perfluorinated compounds, such as perfluorotributylamine and perfluorotetradecanoic acid, as well as some polychlorinated biphenyls (PCBs), such as decachlorobiphenyl, were predicted to have the longest of h, with a wide range of eight orders of magnitude. The of the typical environmental pollutants di(2-ethylhexyl) phthalate (DEHP) and BPA were predicted to be 4.5 h and 7.1 h, respectively, which were close to the empirical 2.0 h and 6.4 h, respectively (Koch et al. 2004; Thayer et al. 2015). The of persistent bioaccumulative toxic chemicals, such as perfluorooctanoic acid (PFOA) and 2,3,3′,4,4′-pentachlorobiphenyl (PCB-105), were predicted to be 7,786 h (0.89 y) and 21,809 h (2.5 y), respectively, with an order of magnitude lower than the experimentally determined total eliminations of 4.37 y and 13.7 y, but within the same order of magnitude (Kudo and Kawashima 2003; Seegal et al. 2011). Overall, (13,733/19,406) of the substances were estimated to have half-lives of and (15,624/19,406) with , which suggested that most of the substances are effectively biotransformed with short .
Toxicity Estimation
We estimated oral for rat values of all substances using U.S. EPA’s TEST software (U.S. EPA 2016b). Predicted oral of rats were available for 14,786/20,756 substances, whereas the values for the other 5,970 substances were unable to be obtained through TEST (Excel Table S2). The -transformed predicted values for most of the chemicals—such as BPA ( BW), DEHP ( BW), and triphenyl phosphate (TPHP, BW)—were similar to their experimental values as retrieved by TEST, which were BW, BW, and BW, respectively (U.S. EPA 2016b). After replacing the predicted values with the experimental values, the median rat oral was (range: –) mol/kg BW, among which 2,3-dibromo-7,8-dichlorodibenzo-p-dioxin (50,585-40-5) was predicted to have the highest toxicity [ (90% prediction interval: , ) mol/kg BW] (Figure 3A). For PFOA, DEHP, and BPA, our values were of BW, BW, and BW, respectively, which were consistent with the experimental data of BW, BW, and BW, respectively (Kamel et al. 2018; Kennedy et al. 2004; U.S. CPSC 2010). Our database also provides the number of assays in which the chemical was tested as well as the percent of assays for which the chemical was active in ToxCast (U.S. EPA 2017a). Finally, 8,201 of the 20,756 originally included substances were available in the database, with the percentage of active assays ranging from 0% to 73.8% (175/237). This information allowed us to expand toxicity data on relevant end points, such as endocrine disruption, carcinogenicity, and receptor binding, among others.
Figure 3.
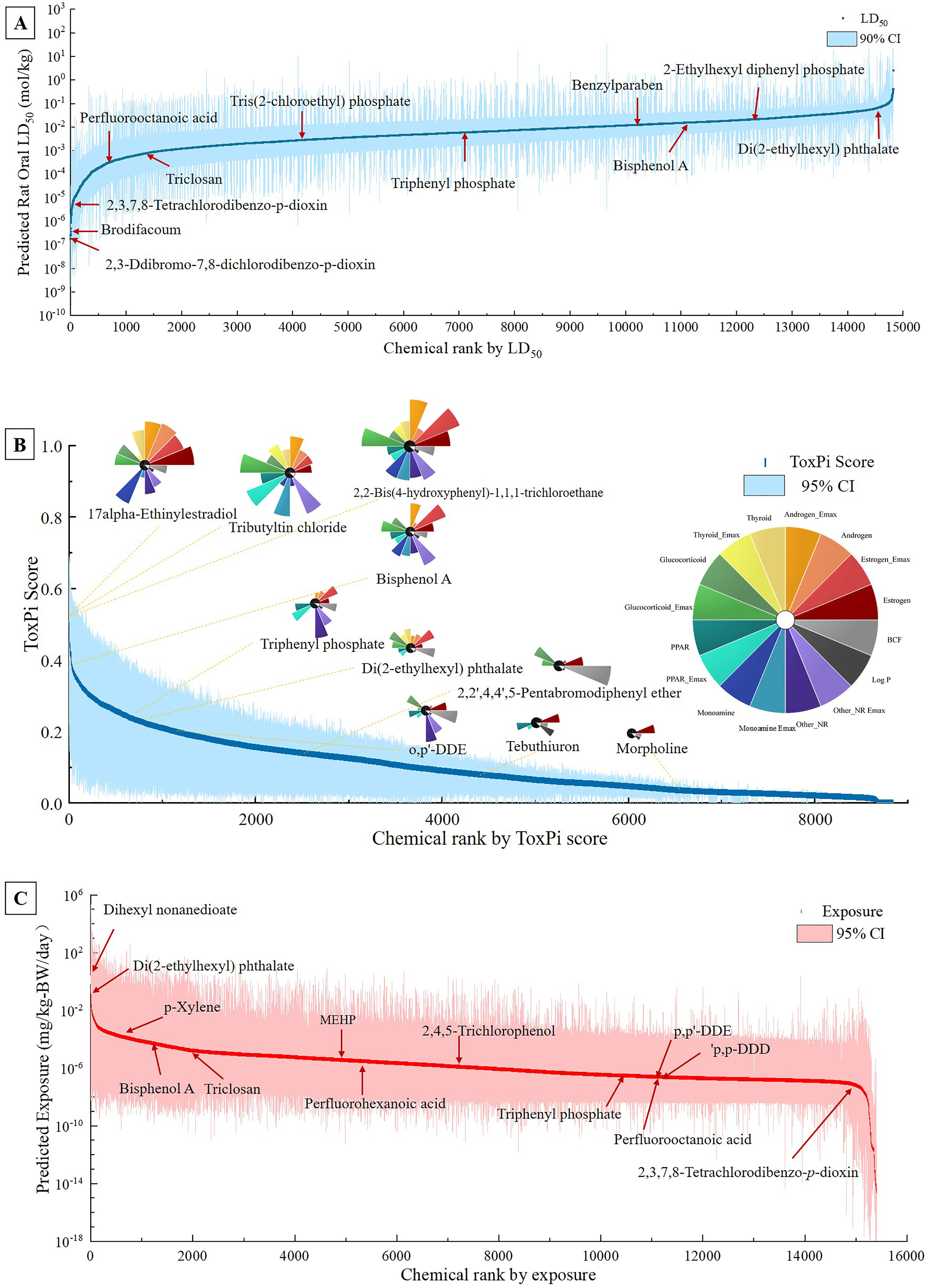
The cumulative distribution of (A) predicted rat oral (); (B) Toxicological Priority Index (ToxPi) score (); (C) Systematic Empirical Evaluation of Model (SEEM) predicted exposure values (). Some typical environmental pollutants are labeled. The summary data are listed in Table S2 and Excel Table S2. Note: BCF, bioconcentration factor; BW, body weight; CI, confidence interval; Emax, efficacy; , median lethal dose; NR, nuclear receptor; PPAR, peroxisome proliferator-activated receptor; ToxPi, Toxicological Priority Index.
Compared with the result of , only 8,845 ToxPi scores of chemicals were successfully calculated, ranging from 0.00 to 0.50 (Excel Table S3). Figure 3B shows the individual ToxPi profiles for these reference chemicals and their ranking along with the ToxPi score distribution for 8,845 chemicals; individual slice scores for all 8,845 chemicals generated by ToxPi GUI (version 2.0; Marvel et al. 2018) were available in Excel Table S3. (EE2) and tributyltin chloride are among the chemicals with the highest ToxPi values of 0.50 (95% CI: 0.22, 0.62) and 0.50 (95% CI: 0.26, 0.67), respectively. 2,2-Bis-(p-hydroxyphenyl)-1,1,1-trichloroethane (HPTE) ranked third in the present study.
Human Exposure Evaluation
A total of 15,408/20,756 substances had predicted exposure values obtained from SEEM (Excel Table S2). The estimated human daily exposure to chemicals ranged from (95% CI: , ) to (95% CI: , ) mg/kg BW per day, spanning across 15 orders of magnitude (Figure 3C). Dihexyl nonanedioate (DHND; CASRN 109-31-9), a plasticizer for food packaging material, was shown to have the highest predicted exposure value. For BPA, DEHP, and PFOA, the exposures were estimated to be (95% CI: , ), (95% CI: , ), and (95% CI: , ) mg/kg BW per day, which were similar to the inferred exposures of , , , and BW per day, respectively, in the previous studies (Lakind and Naiman 2008; Müller et al. 2003; Zhang et al. 2019).
Chemical Risk-Based Prioritization
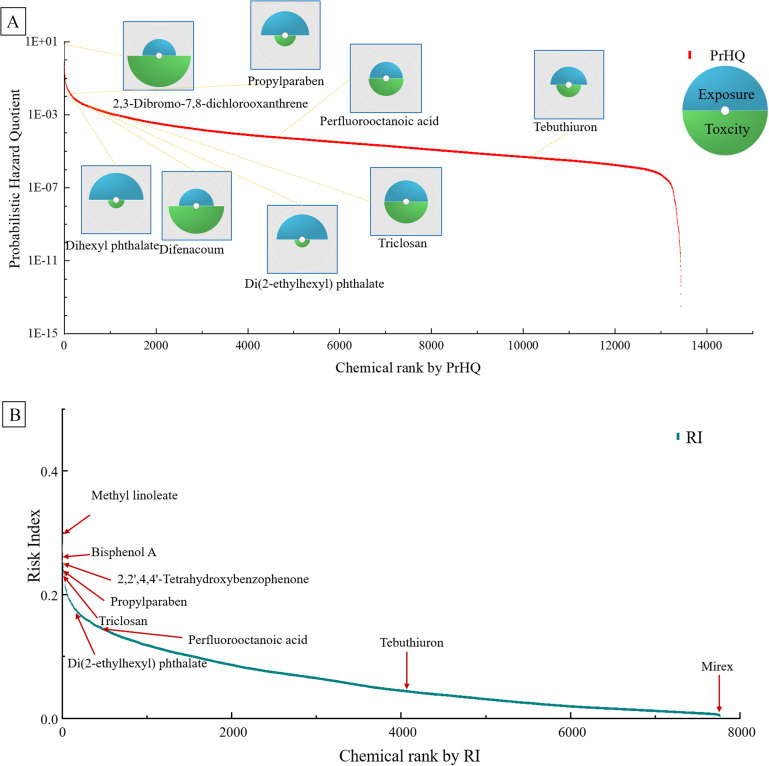
To characterize the chemicals in the HExpMetDB according to their potential risk, we used PrHQs that were based on both exposure and toxicity estimates to prioritize the gathered chemicals for their potential risk. Because some chemicals lack exposure or prediction data, a total of 13,441 chemicals were prioritized by ranking risk (Excel Table S2), among which the PrHQs ranged from to , covering 14 orders of magnitude (Figure 4A). Our approach predicted that DHND would have the highest risk ranking, and N,N′-di-2-naphthyl-p-phenylenediamine (CASRN 93-46-9) was predicted to have the least risk potential. Consistent with the previous study using bioactivity quotients (BQs) for risk-based prioritization, chemicals with BQs also showed a higher risk ranking in our study, such as naphthalene (85/13,441) and phenoxyethanol (118/13,441) (Shin et al. 2015).
Figure 4.
(A) The cumulative distribution of chemical probabilistic hazard quotients (PrHQs) (). The inset shows the PrHQs for typical environmental pollutants represented as exposure (blue) and toxicity (green) component slices. For each slice, the distance from the origin is proportional to the normalized value. (B) The cumulative distribution of chemical risk indexes (RIs) (). The summary data are listed in Excel Table S2.
In addition, we used 95th percentile PrHQ to characterize the uncertainty of the prediction. Three MC simulations (SEEM prediction uncertainty alone, prediction uncertainty alone, and both) were performed to determine the PrHQ upper 95th percentile. The ratio of the PrHQ for the 95th percentile to the median indicates the relative contribution uncertainty with larger ratios indicating greater uncertainty. We observed that the ratio values of prediction uncertainty were relatively small (median value of 5.75; Figure S1). Although the ratio of exposure prediction uncertainty and both uncertainty were roughly close, which indicated that exposure prediction predominated the main uncertainty for the PrHQ estimate.
RIs of 7,770 chemicals calculated from the ToxPi Score were observed ranging from 0 to 0.30 (Excel Table S2). Methyl linoleate (112-63-0, ) was estimated, with the highest potential risk contributed by both high prediction exposure value ( BW per day) and ToxPi score (0.35). BPA (8/7,770), 2,2′,4,4′-tetrahydroxybenzophenone (21/7,770), propylparaben (33/7,770) and triclosan (TCS, 34/7,770) showed higher priority ranking based on RI (Figure 4B). Interestingly, the ranks based on RI were significantly correlated with the PrHQ ranking order but with a small Pearson correlation coefficient (, ).
Biotransformation Metabolite Prediction
We used the BioTransformer software to predict the biotransformation metabolites of 20,756 chemicals in the our updated human exposome database. Of which, 4,225 solicited chemicals could not be applied to the BioTransformer algorithm because of either their large molecular weights ( Da) or the nonmatched chemical categories in the training set of the model. Thus, a total of 95,976 predicted metabolites were obtained from the prediction of 20,061 chemicals, of which, 19,212 were for EC-based metabolism, 72,193 for CYP 450 metabolism, 15,762 for phase II metabolism, and 6,337 for human gut microbial metabolism. To provide the further possibility for exposure biomarker development and identification, we developed a biotransformation predicted metabolite database of a total of 95,976 metabolites derived from the originally employed 20,756 xenobiotics.
HExpMetDB GUI
We used Java plugins to construct a GUI linking the series of data above, aiming to provide the ability for fast searches from our database. The initial interface is a search box allowing for a chemical search using a CASRN, molecular formula, or m/z (Figure 5). The corresponding metadata including chemical identifiers, structures, predicted , exposure, and rat oral can be retrieved for the searched chemical. The user can further search the candidate metabolites of the searched parent compound. Our GUI also allows a search for candidate metabolites by m/z or formula. A successful search result displays the metabolites of the chemical entered with molecular formula or m/z. According to the predicted metabolites, we can figure out the possible metabolism pathways of the chemical. Figure 5 shows the predicted metabolites using DEHP (CASRN 117-81-7) as an example. The predicted results were consistent with the DEHP metabolites observed in the previous study (Koch et al. 2006).
Figure 5.
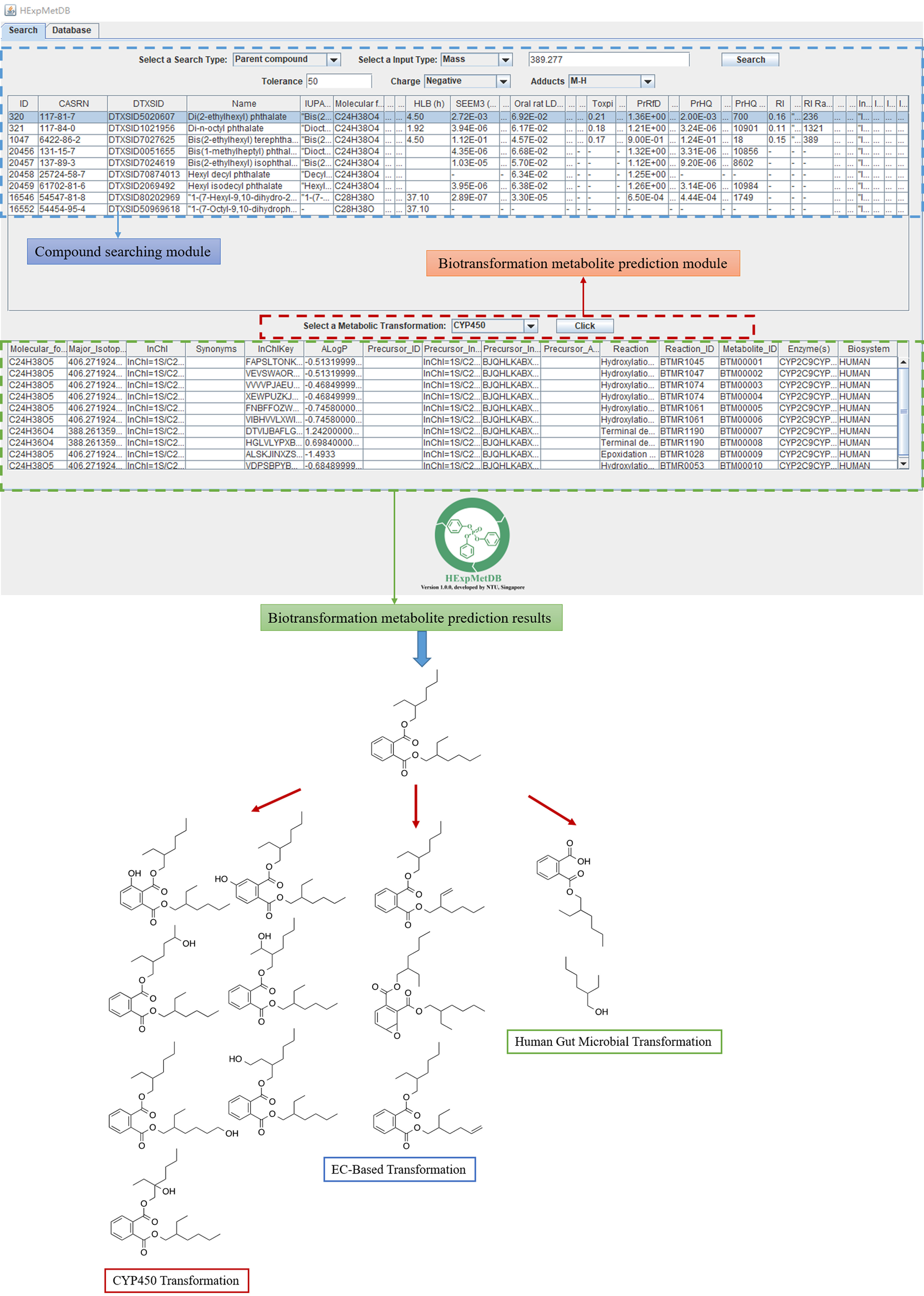
The graphical user interface (GUI) of our developed HExpMetDB. The compound search module can perform searches based on CASRN, formula, mass-charge-ratio (m/z), adduct search with mass accuracy (in ppm), and retrieve the corresponding metadata including chemical identifiers, structures, and predicted data of , exposure and rat oral . The biotransformation metabolite prediction module can further search the candidate metabolites of the searched compound. Di(2-ethylhexyl) phthalate (CASRN 117-81-7) biotransformation metabolite prediction was used as an example. Note: ALogP, predicted values of the logarithm transformed 1-octanol/water partition coefficient; CASRN, Chemical Abstracts Service Registry Number; DTXSID, Distributed Structure-Searchable Toxicity substance identifier; EC-based, enzyme commission based; HExpMetDB, Human Exposome and Metabolite Database; , biotransformation half-life; ID, identifier; InChI, International Chemical Identifier; InChIKey, condensed version of the InChI; IUPAC, International Union of Pure and Applied Chemistry; , median lethal dose; PrRD, probabilistic reference dose; PrHQ, probabilistic hazard quotient; RI, risk index; SEEM3, Systematic Empirical Evaluation of Models.
Discussion
The present study aimed to establish a database of human exposome for the screening of xenobiotic compounds, as well as their possible metabolites in humans, to provide a resource for chemical annotation of the exposome and to prioritize chemicals based on their risk. In the present study, we established a comprehensive database and literature fusion to generate the HExpMetDB with 20,756 chemicals. Besides the intrinsic physicochemical properties, predicted , toxicity data, and exposure values were based on the IFS approach, U.S. EPA’s TEST software/ToxPI GUI, and SEEM, respectively. We further prioritized 13,441 chemicals in our database by both PrHQs and RIs. In addition, we also established the predicted metabolite database with a total number of 121,767 small molecules as the preparatory database to extend the present exposome database. Our HExpMetDB supports ongoing efforts in the field of exposomics research with a wide range of applications, such as compound identification in untargeted exposomics, mass spectral library development, a meta-analysis of chemicals, prioritizing chemicals for pollution mitigation control, and opening a new opportunity for providing candidate exposure biomarkers for HBM in epidemiological cohort studies (Barupal and Fiehn 2019).
Our database was established by aggregating and curating existing chemical databases and the literature. The database includes not only the parent compounds, but also their corresponding metabolites that are likely to be present in humans. However, given the fast-increasing number of new chemicals and the dynamic human exposure levels with time, the need for a systematic compilation of emerging chemicals to gain more insight into the human exposome is highlighted. Therefore, our database should be periodically updated to incorporate new chemicals in the future. In addition, we also provide users with an email inquiry service, which can be used to calculate the predicted values in our server based on the chemical information provided by the user, and further updating our database. Similar chemical text mining and database fusion research were carried out in the recently published blood and dust exposome databases (Barupal and Fiehn 2019; Dong et al. 2019). However, our database focused on the external chemical exposure and excluded endogenous compounds (which can be a response to xenobiotic exposure and thus also part of the exposome), trying to avoid the overlap with the human metabolome database. As a result, our database contains 20,756 parent compounds, which is less than that of the Blood Exposome Database with 41,474 compounds (Barupal and Fiehn 2019). Hence, more detailed and comprehensive data mining still need to be carried out in subsequent GUI updates.
The prediction of chemical was based on an automated IFS approach to develop and evaluate various QSARs, which was well used to predict human (Arnot et al. 2014). The of 19,406 chemicals were successfully predicted in the present study. Interestingly, the median predicted was relatively short (5.0 h), and 70.8% (13,733/19,406) of the substances had half-lives of , which suggests that most of the chemicals in our database have the potential to be easily metabolized in the human body. Chemicals with short show the potential to be easily eliminated via urine or feces as their metabolite forms. The fact that most compounds have a short suggests that the HBM of the parent compounds could be very challenging and even meaningless without knowing their xenobiotic biotransformation products. The measured levels of parent compounds in the blood or urine might be only a snapshot of the entire exposure. This highlights the need for more refined information on urinary exposure biomarkers in holistic exposomics studies. However, only limited metabolites were known for the chemicals in the current exposome database. Thus, in the present study, we included predicted biotransformation metabolites in our exposome database for the further application in the compound identification in untargeted exposomics and developing new exposome biomarkers for epidemiological investigation.
To estimate the toxicity of chemicals, the predicted oral values for rats by TEST were used in the present study. The results indicated that our predicted oral data can well support the prioritization of chemicals. For example, the (median) calculated by our predicted values of BPA, DEHP, tributyl phosphate, and tris(1,3-dichloro-2-propyl) phosphate (TDCPP) were , , , and , respectively, which are within 10 times of the (median) retrieved from the APROBAweb probabilistic RfD database (Chiu 2018) of , , , and , calculated by different species and toxicity end points. Such a simple and convenient approach could be used as a first step to facilitate the assessment of chemical priority in terms of overall toxicity. However, the disadvantage of this method is that it may underestimate the risk of the chemicals for their chronic and subchronic toxicity. Some studies have made great progress in the prioritization method based on the Tox21/ToxCast HTS assay using IVIVE (Ring et al. 2017; Sipes et al. 2017; Wambaugh et al. 2015; Wetmore et al. 2012, 2014; Wetmore 2015). Therefore, we further calculated the ToxPi score in order to more comprehensively estimate the toxicity ranking of chemicals.
A total of 97 in vitro HTS assays, as well as BCF and Log P, were used in our ToxPi score calculation, which could be used to indicate the overall endocrine-disrupting toxicity of a chemical. As seen in the ranking distribution for 8,845 chemicals in Figure 3B, similar ranking results were also observed in the previous study, which prioritized 309 ToxCast Phase I chemicals by their ToxPi score (Reif et al. 2010). For example, the reference chemical HPTE and BPA were shown to be high-ranked chemicals, whereas tebuthiuron was low-ranked, which was similar to the results from our study, with a much larger number of tested chemicals.
Figure 4 shows the individual profiles for some well-known typical environmental chemicals and their positions along the exposome and toxicity distribution for the 13,441 prioritized chemicals. DHND had the highest PrHQ among the chemicals. DHND’s high PrHQ is due to both the high exposure and strong toxicity predicted in our study. From the distributional dot plot, it can be seen that the PrHQ of a compound was not determined solely by toxicity or exposure, but by both (Figure 4). When only the exposure was considered, 2,3-dibromo-7,8-dichlorooxanthrene (2,3-B-7,8-CDD; CASRN: 50585-40-5) was found to rank only 11,972nd among the 13,441 compounds using the exposure-based chemical prioritization; however, this compound ranked 122nd using the PrHQ risk-based chemical prioritization due to its strong toxicity. A similar pattern was also observed for chemicals such as TCS (3380-34-5) and difenacoum (56073-07-5). Although their exposure levels are relatively low, their strong toxicity increases priority (798/13,441 for TCS, and 314/13,441 for difenacoum). Although for chemicals such as phthalates (PAEs), organophosphate flame retardants (OPFRs), and parabens, their higher priority was mainly contributed by their higher exposure. Thus, 2-ethylhexyl diphenyl phosphate, propylparaben, dihexyl phthalate, diethyl phthalate (DEP) and DEHP showed relatively higher priority. All three MC simulation results indicated that exposure prediction uncertainties were mainly contributed by exposure prediction, rather than toxicity prediction, for PrHQ estimate when prioritizing chemicals. This is also expected because of the high complexity of human exposure scenarios.
RI was calculated based on the combination of ToxPi score (i.e., the results of various assays and parameters) and the assessment of human exposure. Compared with PrHQ, RI could better reflect the overall risk evaluation for multiple specific toxicity end points. However, it should be noted that we only used the predicted external exposure to calculate RI, and external exposure to internal exposure estimation conversion is much needed in future studies although we considered BCF and log P when calculating ToxPi. The results further implied that there was some consistency between PrHQ and ToxPi RI-based ranking, but the ranking of some substances can be very different due to the different priority principles. Both prioritization methods should be considered in the risk assessment. Users can select either PrHQ or RI or both provided in “my database” to filter out the substances that meet their requirements.
In the present study, we also constructed a GUI that can conduct searches for chemical-specific results documented in our database. Due to the integrated content, the dashboard can be further used to search for the following information: a) a parent compound and its intrinsic properties by m/z, CASRN or formula; b) predicted data of , toxicity and exposure through m/z, CASRN or formula; c) predicted metabolites of a chemical in our database; and d) candidate metabolites by m/z or formula of metabolites. Our database opens a new opportunity to develop exposure biomarkers for environmental epidemiology and to propose probable identifications for untargeted exposomics surveys. The usage examples for our software can be found in the Supplemental Material (Text S2, “Database functionality”).
This study has several limitations. First, the SEEM database was unable to cover all the compounds summarized in the present study, and SEEM merely represents the exposure of the general Americans for their historical exposure. This highlights the need for more studies on exposure estimates. Second, as our HTS method is based on the U.S. EPA Chemistry Dashboard, only chemicals with CASRN were successful in retrieving diverse types of relevant domain data from the U.S. EPA Chemistry Dashboard. Third, we cannot predict the of chemicals with a metal atom or molecular weight of using the IFS approach and the portion of chemicals that failed to meet the applicability domain of prediction. Fourth, the risk score was calculated using the external exposure, and the IVIVE model should be considered in future studies. Fifth, it should be also noted that the risk assessment of chemicals in the present framework is only based on each individual one and we cannot exclude its toxicity due to chemical interaction (e.g., synergistic effect) in the mixture effects, which has been shown in several recent studies (Hsieh et al. 2021; Liu et al. 2020; Zhang et al. 2020). Last, BioTransformer cannot predict all metabolites, and the prediction results may also be inaccurate for some chemicals. In addition, there is still a knowledge gap on the identification of the exposure biomarker among all possible biotransformation products.
In conclusion, we have established a human prioritized exposome database, which included both parent compounds and predicted potential metabolites, and developed a systematic approach that integrates exposure and toxicity information in a holistic framework for chemical risk-based prioritization. Our study would assist in a broad array of human exposure research by facilitating chemical and metabolite identification. Nonbiased targeted and untargeted chemical screening is still needed in the future to fully prioritize the chemicals.
Supplementary Material
Acknowledgments
This work was funded by the Singapore Ministry of Education Academic Research Fund Tier 1 (04MNP000567C120), the Nanyang Technological University Harvard Sus Nano (M4082370), and the Singapore Ministry of Health’s National Medical Research Council under its Clinician-Scientist Individual Research Grant (CS-IRG) (MOH-000141) and Open Fund-Individual Research Grant (OFIRG/0076/2018).
References
- Andrianou XD, van der Lek C, Charisiadis P, Ioannou S, Fotopoulou KN, Papapanagiotou Z, et al. 2019. Application of the urban exposome framework using drinking water and quality of life indicators: a proof-of-concept study in Limassol, Cyprus. PeerJ 7:e6851, PMID: 31179170, 10.7717/peerj.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnot JA, Brown TN, Wania F. 2014. Estimating screening-level organic chemical half-lives in humans. Environ Sci Technol 48(1):723–730, PMID: 24298879, 10.1021/es4029414. [DOI] [PubMed] [Google Scholar]
- Barupal DK, Fiehn O. 2019. Generating the Blood Exposome Database using a comprehensive text mining and database fusion approach. Environ Health Perspect 127(9):97008, PMID: 31557052, 10.1289/EHP4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland J. 2007. Managing biotransformation: introduction and overview. Altern Ther Health Med 13(2):S85–S87, PMID: 17405682. [PubMed] [Google Scholar]
- Brown TN, Arnot JA, Wania F. 2012. Iterative fragment selection: a group contribution approach to predicting fish biotransformation half-lives. Environ Sci Technol 46(15):8253–8260, PMID: 22779755, 10.1021/es301182a. [DOI] [PubMed] [Google Scholar]
- Castaño-Vinyals G, Cantor KP, Villanueva CM, Tardon A, Garcia-Closas R, Serra C, et al. 2011. Socioeconomic status and exposure to disinfection by-products in drinking water in Spain. Environ Health 10(1):18, PMID: 21410938, 10.1186/1476-069X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WA. 2018. APROBAweb: an interactive web application for probabilistic hazard characterization/dose-response assessment. https://wchiu.shinyapps.io/APROBAweb/ [accessed 12 February 2020].
- Chiu WA, Axelrad DA, Dalaijamts C, Dockins C, Shao K, Shapiro AJ, et al. 2018. Beyond the RfD: broad application of a probabilistic approach to improve chemical dose–response assessments for noncancer effects. Environ Health Perspect 126(6):067009, PMID: 29968566, 10.1289/EHP3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Prussin AJ II, Marr LC, Vikesland PJ, Edwards MA, Pruden A. 2017. Factors shaping the human exposome in the built environment: opportunities for engineering control. Environ Sci Technol 51(14):7759–7774, PMID: 28677960, 10.1021/acs.est.7b01097. [DOI] [PubMed] [Google Scholar]
- Djoumbou-Feunang Y, Fiamoncini J, Gil-de-la-Fuente A, Greiner R, Manach C, Wishart DS. 2019. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Cheminform 11(1):2, PMID: 30612223, 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Zhang Y, Jia S, Shang H, Fang W, Chen D, et al. 2019. Human indoor exposome of chemicals in dust and risk prioritization using EPA’s ToxCast database. Environ Sci Technol 53(12):7045–7054, PMID: 31081622, 10.1021/acs.est.9b00280. [DOI] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. 2015. Human microbiota. Small molecules from the human microbiota. Science 349(6246):1254766, PMID: 26206939, 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EC (European Commission). 2012. Food flavorings database: part I of Annex I of Regulation (EC) NO 1334/2008. https://webgate.ec.europa.eu/foods_system/main/?event=display [accessed 20 July 2019].
- EC. 2017. Food additives database: Annex II of Regulation (EC) no 1333/2008. https://webgate.ec.europa.eu/foods_system/main/?sector=FAD&auth=SANCAS [accessed 20 July 2019].
- EC. 2020. Priority list of endocrine disruptors. https://ec.europa.eu/environment/chemicals/endocrine/strategy/substances_en.htm [accessed 12 February 2020].
- ECHA (European Chemical Agency). 2008. EC Inventory. https://echa.europa.eu/information-on-chemicals/ec-inventory [accessed 12 February 2020].
- ECHA. 2020. Candidate list of substances of very high concern for authorisation. https://echa.europa.eu/candidate-list-table [accessed 12 February 2020].
- Filer D, Patisaul HB, Schug T, Reif D, Thayer K. 2014. Test driving ToxCast: endocrine profiling for 1858 chemicals included in phase II. Curr Opin Pharmacol 19:145–152, PMID: 25460227, 10.1016/j.coph.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert A, Forestier D, Lenes D, Benanou D, Jacob S, Arfi C, et al. 2010. Innovative method for prioritizing emerging disinfection by-products (DBPs) in drinking water on the basis of their potential impact on public health. Water Res 44(10):3147–3165, PMID: 20409572, 10.1016/j.watres.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Hsieh NH, Chen Z, Rusyn I, Chiu WA. 2021. Risk characterization and probabilistic concentration–response modeling of complex environmental mixtures using new approach methodologies (NAMs) data from organotypic in vitro human stem cell assays. Environ Health Perspect 129(1):17004, PMID: 33395322, 10.1289/EHP7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Sankaran G, Wang B, Shang H, Tan ST, Yap HM, et al. 2019. Exposure and risk assessment of volatile organic compounds and airborne phthalates in Singapore’s child care centers. Chemosphere 224:85–92, PMID: 30818198, 10.1016/j.chemosphere.2019.02.120. [DOI] [PubMed] [Google Scholar]
- Kamel AH, Foaud MA, Moussa HM. 2018. The adverse effects of bisphenol A on male albino rats. J Basic Appl Zool 79:6, 10.1186/s41936-018-0015-9. [DOI] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, et al. 2012. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25(7):1287–1302, PMID: 22519603, 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Kennedy GL, Butenhoff JL, Olsen GW, O’Connor JC, Seacat AM, Perkins RG, et al. 2004. The toxicology of perfluorooctanoate. Crit Rev Toxicol 34(4):351–384, PMID: 15328768, 10.1080/10408440490464705. [DOI] [PubMed] [Google Scholar]
- Koch HM, Bolt HM, Angerer J. 2004. Di(2-ethylhexyl)phthalate (DEHP) metabolites in human urine and serum after a single oral dose of deuterium-labelled DEHP. Arch Toxicol 78(3):123–130, PMID: 14576974, 10.1007/s00204-003-0522-3. [DOI] [PubMed] [Google Scholar]
- Koch HM, Preuss R, Angerer J. 2006. Di(2-ethylhexyl)phthalate (DEHP): human metabolism and internal exposure—an update and latest results. Int J Androl 29(1):155–165, PMID: 16466535, 10.1111/j.1365-2605.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- Kudo N, Kawashima Y. 2003. Toxicity and toxicokinetics of perfluorooctanoic acid in humans and animals. J Toxicol Sci 28(2):49–57, PMID: 12820537, 10.2131/jts.28.49. [DOI] [PubMed] [Google Scholar]
- Lakind JS, Naiman DQ. 2008. Bisphenol A (BPA) daily intakes in the United States: estimates from the 2003–2004 NHANES urinary BPA data. J Expo Sci Environ Epidemiol 18(6):608–615, PMID: 18414515, 10.1038/jes.2008.20. [DOI] [PubMed] [Google Scholar]
- Liu M, Jia S, Dong T, Zhao F, Xu T, Yang Q, et al. 2020. Metabolomic and transcriptomic analysis of MCF-7 cells exposed to 23 chemicals at human-relevant levels: estimation of individual chemical contribution to effects. Environ Health Perspect 128(12):127008, PMID: 33325755, 10.1289/EHP6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvel SW, To K, Grimm FA, Wright FA, Rusyn I, Reif DM. 2018. ToxPi Graphical User Interface 2.0: dynamic exploration, visualization, and sharing of integrated data models. BMC Bioinformatics 19(1):80, PMID: 29506467, 10.1186/s12859-018-2089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AK, Nielsen E, Ladefoged O. 2003. Human Exposure to Selected Phthalates in Denmark. Glostrup, Denmark: Danish Veterinary and Food Administration. [Google Scholar]
- Neveu V, Moussy A, Rouaix H, Wedekind R, Pon A, Knox C, et al. 2017. Exposome-explorer: a manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res 45(D1):D979–D984, PMID: 27924041, 10.1093/nar/gkw980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuijsen MJ, Martinez D, Grellier J, Bennett J, Best N, Iszatt N, et al. 2009. Chlorination disinfection by-products in drinking water and congenital anomalies: review and meta-analyses. Environ Health Perspect 117(10):1486–1493, PMID: 20019896, 10.1289/ehp.0900677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa E, van der Wal L, Arnot JA, Gramatica P. 2014. Metabolic biotransformation half-lives in fish: QSAR modeling and consensus analysis. Sci Total Environ 470–471:1040–1046, PMID: 24239825, 10.1016/j.scitotenv.2013.10.068. [DOI] [PubMed] [Google Scholar]
- Reif DM, Martin MT, Tan SW, Houck KA, Judson RS, Richard AM, et al. 2010. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ Health Perspect 118(12):1714–1720, PMID: 20826373, 10.1289/ehp.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remucal CK, Manley D. 2016. Emerging investigators series: the efficacy of chlorine photolysis as an advanced oxidation process for drinking water treatment. Environ Sci Water Res Technol 2(4):565–579, 10.1039/C6EW00029K. [DOI] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. 2016. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 29(8):1225–1251, PMID: 27367298, 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Ring CL, Arnot JA, Bennett DH, Egeghy PP, Fantke P, Huang L, et al. 2019. Consensus modeling of median chemical intake for the U.S. population based on predictions of exposure pathways. Environ Sci Technol 53(2):719–732, PMID: 30516957, 10.1021/acs.est.8b04056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring CL, Pearce RG, Setzer RW, Wetmore BA, Wambaugh JF. 2017. Identifying populations sensitive to environmental chemicals by simulating toxicokinetic variability. Environ Int 106:105–118, PMID: 28628784, 10.1016/j.envint.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegal RF, Fitzgerald EF, Hills EA, Wolff MS, Haase RF, Todd AC, et al. 2011. Estimating the half-lives of PCB congeners in former capacitor workers measured over a 28-year interval. J Expo Sci Environ Epidemiol 21(3):234–246, PMID: 20216575, 10.1038/jes.2010.3. [DOI] [PubMed] [Google Scholar]
- Shephard GS. 2008. Determination of mycotoxins in human foods. Chem Soc Rev 37(11):2468–2477, PMID: 18949120, 10.1039/b713084h. [DOI] [PubMed] [Google Scholar]
- Shin HM, Ernstoff A, Arnot JA, Wetmore BA, Csiszar SA, Fantke P, et al. 2015. Risk-based high-throughput chemical screening and prioritization using exposure models and in vitro bioactivity assays. Environ Sci Technol 49(11):6760–6771, PMID: 25932772, 10.1021/acs.est.5b00498. [DOI] [PubMed] [Google Scholar]
- Sipes NS, Wambaugh JF, Pearce R, Auerbach SS, Wetmore BA, Hsieh JH, et al. 2017. An intuitive approach for predicting potential human health risk with the Tox21 10k library. Environ Sci Technol 51(18):10786–10796, PMID: 28809115, 10.1021/acs.est.7b00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjerps RMA, Vughs D, van Leerdam JA, ter Laak TL, van Wezel A. 2016. Data-driven prioritization of chemicals for various water types using suspect screening LC-HRMS. Water Res 93:254–264, PMID: 26921851, 10.1016/j.watres.2016.02.034. [DOI] [PubMed] [Google Scholar]
- Thayer KA, Doerge DR, Hunt D, Schurman SH, Twaddle NC, Churchwell MI, et al. 2015. Pharmacokinetics of bisphenol A in humans following a single oral administration. Environ Int 83:107–115, PMID: 26115537, 10.1016/j.envint.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. CPSC (Consumer Product Safety Commission). 2010. Toxicity review of di(2-ethylhexyl) phthalate (DEPH). https://www.cpsc.gov/s3fs-public/ToxicityReviewOfDBP.pdf [accessed 21 April 2020].
- U.S. EPA. 2007. Chemical inventory for ToxCast. https://comptox.epa.gov/dashboard/chemical_lists/CHEMINV [accessed 12 February 2020].
- U.S. EPA. 2016a. The Data Management and Quality Assurance/Quality Control Process for the Third Six-Year Review Information Collection Rule Dataset. https://nepis.epa.gov/Exe/ZyPURL.cgi?Dockey=P100QO9Q.TXT [accessed 10 October 2020].
- U.S. EPA. 2016b. User’s Guide for T.E.S.T. (version 4.2) (Toxicity Estimation Software Tool): A Program to Estimate Toxicity from Molecular Structure. https://www.epa.gov/chemical-research/users-guide-test-version-42-toxicity-estimation-software-tool-program-estimate [accessed on 5th Feb. 2020].
- U.S. EPA. 2017a. Chemistry dashboard. https://comptox.epa.gov/dashboard/dsstoxdb/batch_search [accessed 12 February 2020].
- U.S. EPA. 2017b. EPA: Pesticide Chemical Search Database. https://comptox.epa.gov/dashboard/chemical_lists/EPAPCS [accessed 13 April 2022].
- U.S. EPA. 2018a. ICIS-Air. https://ofmpub.epa.gov/sor_internet/registry/substreg/searchandretrieve/searchbylist/search.do?search=&searchCriteria.substanceList=79&searchCriteria.substanceType=-1 [accessed 10 October 2020].
- U.S. EPA. 2018b. Organic Hazardous Air Pollutants National Emission Standards. https://ofmpub.epa.gov/sor_internet/registry/substreg/searchandretrieve/searchbylist/search.do?search=&searchCriteria.substanceList=180&searchCriteria.substanceType=-1 [accessed 10 October 2020].
- U.S. EPA. 2020a. CAA112(b) HAP—Hazardous Air Pollutants. https://ofmpub.epa.gov/sor_internet/registry/substreg/searchandretrieve/searchbylist/search.do?search=&searchCriteria.substanceList=165&searchCriteria.substanceType=-1 [accessed 10 October 2020].
- U.S. EPA. 2020b. List of Lists: Consolidated List of Chemicals Subject to the Emergency Planning and Community Right-to-Know Act (EPCRA), Comprehensive Environmental Response, Compensation and Liability Act (CERCLA) and Section 112(R) of the Clean Air Act. https://www.epa.gov/sites/production/files/2015-03/documents/list_of_lists.pdf [accessed 10 October 2020].
- U.S. EPA. 2020c. EPA: High Production Volume List. https://comptox.epa.gov/dashboard/chemical_lists/EPAHPV [accessed 12 February 2020].
- Ulrich N, Endo S, Brown TN, Watanabe N, Bronner G, Abraham MH, et al. 2017. UFZ-LSER database v 3.2.1 [internet]. http://www.ufz.de/lserd [accessed on 4th Feb. 2020].
- USDA (U.S. Department of Agriculture). 2019. FoodData Central. https://fdc.nal.usda.gov/index.html [accessed 12 February 2020].
- Vikesland P, Raskin L. 2016. The drinking water exposome. Environ Sci Water Res Technol 2(4):561–564, 10.1039/C6EW90016J. [DOI] [Google Scholar]
- Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, et al. 2013. High-throughput models for exposure-based chemical prioritization in the ExpoCast project. Environ Sci Technol 47(15):8479–8488, PMID: 23758710, 10.1021/es400482g. [DOI] [PubMed] [Google Scholar]
- Wambaugh JF, Wang A, Dionisio KL, Frame A, Egeghy P, Judson R, et al. 2014. High throughput heuristics for prioritizing human exposure to environmental chemicals. Environ Sci Technol 48(21):12760–12767, PMID: 25343693, 10.1021/es503583j. [DOI] [PubMed] [Google Scholar]
- Wambaugh JF, Wetmore BA, Pearce R, Strope C, Goldsmith R, Sluka JP, et al. 2015. Toxicokinetic triage for environmental chemicals. Toxicol Sci 147(1):55–67, PMID: 26085347, 10.1093/toxsci/kfv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh JF, Wetmore BA, Ring CL, Nicolas CI, Pearce RG, Honda GS, et al. 2019. Assessing toxicokinetic uncertainty and variability in risk prioritization. Toxicol Sci 172(2):235–251, PMID: 31532498, 10.1093/toxsci/kfz205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472(7341):57–63, PMID: 21475195, 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B, Sulyok M, Fruhmann P, Mikula H, Berthiller F, Schuhmacher R, et al. 2012. Development and validation of a rapid multi‐biomarker liquid chromatography/tandem mass spectrometry method to assess human exposure to mycotoxins. Rapid Commun Mass Spectrom 26(13):1533–1540, PMID: 22638970, 10.1002/rcm.6255. [DOI] [PubMed] [Google Scholar]
- Wetmore BA. 2015. Quantitative in vitro-to-in vivo extrapolation in a high-throughput environment. Toxicology 332:94–101, PMID: 24907440, 10.1016/j.tox.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Allen B, Clewell HJ III, Parker T, Wambaugh JF, Almond LM, et al. 2014. Incorporating population variability and susceptible subpopulations into dosimetry for high-throughput toxicity testing. Toxicol Sci 142(1):210–224, PMID: 25145659, 10.1093/toxsci/kfu169. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, et al. 2012. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol Sci 125(1):157–174, PMID: 21948869, 10.1093/toxsci/kfr254. [DOI] [PubMed] [Google Scholar]
- WHO (World Health Organization). 2016. Ambient Air Pollution: A Global Assessment of Exposure and Burden Of Disease. Geneva, Switzerland: WHO. [Google Scholar]
- WHO/IPCS (International Programme on Chemical Safety). 2017. Guidance Document on Evaluating and Expressing Uncertainty in Hazard Characterization. 2nd ed. Geneva, Switzerland: WHO. https://apps.who.int/iris/bitstream/handle/10665/259858/9789241513548-eng.pdf;jsessionid=751F81EC4EAD03E3C58D59BC1EE5ECB1?sequence=1 [accessed 17 November 2020]. [Google Scholar]
- Wild CP. 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 14(8):1847–1850, PMID: 16103423, 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, et al. 2017. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform 9(1):61, PMID: 29185060, 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, et al. 2019. Blood metabolome predicts gut microbiome α-diversity in humans. Nat Biotechnol 37(10):1217–1228, PMID: 31477923, 10.1038/s41587-019-0233-9. [DOI] [PubMed] [Google Scholar]
- Zhang LS, Davies SS. 2016. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med 8(1):46, PMID: 27102537, 10.1186/s13073-016-0296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Kang Q, Peng H, Ding M, Zhao F, Zhou Y, et al. 2019. Relationship between perfluorooctanoate and perfluorooctane sulfonate blood concentrations in the general population and routine drinking water exposure. Environ Int 126:54–60, PMID: 30776750, 10.1016/j.envint.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu M, Peng B, Jia S, Koh D, Wang Y, et al. 2020. Impact of mixture effects between emerging organic contaminants on cytotoxicity: a systems biological understanding of synergism between tris(1,3-dichloro-2-propyl)phosphate and triphenyl phosphate. Environ Sci Technol 54(17):10722–10734, PMID: 32786581, 10.1021/acs.est.0c02188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.