Abstract
Background:
Humans are exposed to tens of thousands of chemical substances that need to be assessed for their potential toxicity. Acute systemic toxicity testing serves as the basis for regulatory hazard classification, labeling, and risk management. However, it is cost- and time-prohibitive to evaluate all new and existing chemicals using traditional rodent acute toxicity tests. In silico models built using existing data facilitate rapid acute toxicity predictions without using animals.
Objectives:
The U.S. Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) Acute Toxicity Workgroup organized an international collaboration to develop in silico models for predicting acute oral toxicity based on five different end points: Lethal Dose 50 ( value, U.S. Environmental Protection Agency hazard (four) categories, Globally Harmonized System for Classification and Labeling hazard (five) categories, very toxic chemicals [ ()], and nontoxic chemicals ().
Methods:
An acute oral toxicity data inventory for 11,992 chemicals was compiled, split into training and evaluation sets, and made available to 35 participating international research groups that submitted a total of 139 predictive models. Predictions that fell within the applicability domains of the submitted models were evaluated using external validation sets. These were then combined into consensus models to leverage strengths of individual approaches.
Results:
The resulting consensus predictions, which leverage the collective strengths of each individual model, form the Collaborative Acute Toxicity Modeling Suite (CATMoS). CATMoS demonstrated high performance in terms of accuracy and robustness when compared with in vivo results.
Discussion:
CATMoS is being evaluated by regulatory agencies for its utility and applicability as a potential replacement for in vivo rat acute oral toxicity studies. CATMoS predictions for more than 800,000 chemicals have been made available via the National Toxicology Program’s Integrated Chemical Environment tools and data sets (ice.ntp.niehs.nih.gov). The models are also implemented in a free, standalone, open-source tool, OPERA, which allows predictions of new and untested chemicals to be made. https://doi.org/10.1289/EHP8495
Introduction
Acute systemic toxicity studies are required by regulators around the world to inform chemical hazard classification, labeling, and risk management. The testing to assess acute systemic toxicity is conducted in vivo through a predefined route of exposure (oral, dermal, or via inhalation) during a fixed observation period as described in test guidelines issued by the Organization for Economic Cooperation and Development (OECD) (OECD 2002a, 2002b, 2002c, 2008). Five U.S. agencies [Consumer Product Safety Commission (CPSC), Department of Defense (DoD), Department of Transportation (DoT), Environmental Protection Agency (U.S. EPA), Occupational Safety and Health Administration (OSHA)], as well as Registration, Evaluation, Authorization, and Restriction of Chemicals (REACH) in Europe use the median Lethal Dose 50 (; the dose of a substance that would be expected to kill half the animals in a test group) from acute oral toxicity data for the classification and labeling of chemical substances (ECHA 2008; Kleinstreuer et al. 2018; Strickland et al. 2018). However, in vivo acute oral toxicity testing is cost- and time-prohibitive and raises ethical concerns related to the use of many animals. Given the large number of new and existing substances requiring assessment, there is a pressing need for cost-effective and rapid nonanimal alternatives.
Recent technological advances in computational resources and artificial intelligence have increased the accuracy and speed of machine learning algorithms. As a result, in silico approaches such as quantitative structure–activity relationships (QSARs) are being increasingly recognized as alternatives to bridge the lack of knowledge about chemical properties and their biological activities. QSARs are being promoted for their ability to accurately predict toxicological end points at low cost but also for being reliable, reproducible, and broadly applicable to the diversity of chemicals requiring testing (Dearden et al. 2009; Worth et al. 2005). Consequently, the integration of nonanimal methods for assessing chemical toxicity is gaining momentum. In Europe, REACH regulations call for the use of nonanimal methods to assess chemical toxicity (Benfenati et al. 2011; European Commission, Environment Directorate General 2007; Lahl and Hawxwell 2006). Similarly, in 2020, U.S. EPA created a New Approach Methods (NAMs) Work Plan to prioritize agency efforts and resources toward activities that will reduce the use of animal testing while continuing to protect human health and the environment (U.S. EPA 2020). Furthermore, the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM), consisting of representatives from 16 U.S. federal agencies, has several workgroups focused on the development or validation of NAMs. These workgroups contribute to the goals of the ICCVAM Strategic Roadmap for Establishing New Approaches to Evaluate the Safety of Chemicals and Medical Products in the United States (Interagency Coordinating Committee on the Validation of Alternative Methods 2018). One of the ICCVAM ad hoc workgroups established was the Acute Toxicity Workgroup (ATWG), which sought to develop an implementation plan for identifying, evaluating, and applying alternative methods for acute systemic toxicity (Kleinstreuer et al. 2018; Lowit et al. 2017). An initial ATWG study was conducted to assess the acute toxicity data regulatory requirements, needs, and decision contexts of member agencies as well as to understand the current acceptance of alternative methods (Strickland et al. 2018). Subsequent charges of the ATWG were to identify, acquire, and curate high-quality data from reference test methods that could be used to evaluate existing models for acute toxicity as well as investigate the feasibility of developing new models. Focusing initially on the oral route of exposure to evaluate existing in silico models, the ATWG organized an international collaborative project to develop new in silico models for predicting acute oral systemic toxicity (Kleinstreuer et al. 2018; Strickland et al. 2018).
International consortia have successfully developed collaborative computational solutions for challenging toxicological problems. Examples in the area of endocrine disruption screening include the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP) (Mansouri et al. 2016a) and the Collaborative Modeling Project for Androgen Receptor (CoMPARA) (Mansouri et al. 2020). The predictive consensus models from these projects have been integrated to assess the endocrine activity potential of organic chemicals within the EPA’s Endocrine Disruptor Screening Program (EDSP) (U.S. EPA-NCCT 2014b). The global network of experts represented by these successful consortia was leveraged for the current acute oral systemic toxicity modeling project, and the legacy workflows from CERAPP and CoMPARA were adapted and applied for the data analysis and modeling conducted herein.
For the current project, the U.S. National Toxicology Program (NTP) Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) and the U.S. EPA’s Center for Computational Toxicology and Exposure (CCTE) collected and curated rat oral data for more than 15,000 substances from public sources to produce data sets that were used during the project as training and evaluation sets (Karmaus et al. 2019; Kleinstreuer et al. 2018). Thirty-five international collaborators representing various sectors, including government, industry, and academia, participated in this effort, which produced a total of 139 different models. All submitted models were both quantitatively and qualitatively evaluated. A workshop was convened (https://ntp.niehs.nih.gov/go/atwksp-2018) to bring contributing computational modelers and regulatory decision makers together to discuss the feasibility of using in silico predictions for regulatory use in lieu of in vivo acute oral systemic toxicity testing (Kleinstreuer et al. 2018). Ultimately, predictions within the applicability domains of the developed models were combined into consensus predictions based on a weight-of-evidence (WoE) approach, forming the Collaborative Acute Toxicity Modeling Suite (CATMoS). CATMoS was then implemented into the open-source, open-data OPERA [OPEn (q)saR App] tool to enable further screening of new chemicals (Mansouri et al. 2016b, 2018). This paper provides a description of the data on which the CATMoS models are based, the evaluation process, and development of consensus models. We close with a discussion of the limitations of CATMoS and a description of implementation and additional evaluation of the model.
Materials and Methods
U.S. Regulatory Uses for Acute Oral Toxicity Data
Prior to identifying any existing alternative methods or investing in the development, optimization, and validation of new ones, it is important to understand the current regulatory needs and decision contexts, including the use and acceptance of nonanimal data for the toxicological end point of concern. Strickland et al. (2018) described the use of acute oral toxicity data by ICCVAM regulatory agencies to provide a basis for identifying opportunities for flexibility with regard to replacing or reducing the need for in vivo acute oral toxicity studies (Strickland et al. 2018). The regulatory needs of these agencies require three different types of acute toxicity outcomes, as detailed in Table 1: a) an value estimate; b) a binary outcome based on a single threshold; and c) a multiclass scheme based on different thresholds. Two binary models were relevant to U.S. agencies: a) the identification of whether a chemical was “very toxic” (i.e., ); and b) identification of whether a chemical was “nontoxic” (i.e., ). Multiclass schemes in use by several agencies included hazard categories defined by the U.S. EPA and the U.N. Globally Harmonized System of Classification and Labeling of Chemicals (GHS), which consist of four or five categories, respectively (Table 2) (Strickland et al. 2018).
Table 1.
Acute oral toxicity classification strategies used by U.S. regulatory agencies.
| Requirement | Description | Agencies |
|---|---|---|
| Binary | values above or below specific threshold | CPSC, DoD |
| Multi-class | Multiple ranges of values | EPA, OSHA, DOT |
| value | Discrete values | EPA, CPSC, DoD |
Note: See (Strickland et al. 2018). CPSC, Consumer Product Safety Commission; DoD, U.S. Department of Defense; DOT, U.S. Department of Transportation; EPA, U.S. Environmental Protection Agency; OSHA, U.S. Occupational Safety and Health Administration.
Table 2.
U.S. EPA and GHS hazard labeling categories.
| Categories | EPA thresholds | GHS thresholds |
|---|---|---|
| 1 | ||
| II | ||
| III | ||
| IV | ||
| V | NA |
Note: NA, No EPA Cat V; See (Strickland et al. 2018). EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; ,dose of a substance that would be expected to kill half the animals in a test group.
Based on this information, for this project we asked participants to develop models to predict one or more of the following end points:
Very toxic (VT; vs. all others)
Nontoxic (NT; vs. all others)
U.S. EPA hazard categories (U.S. EPA 2016)
GHS hazard categories (United Nations 2015)
Point estimate values.
Data Sets
Data collection and preprocessing.
The data set underlying the modeling effort for this project was initially compiled by NICEATM and U.S. EPA’s CCTE. Briefly, data were collected from rat acute oral systemic toxicity tests, including limit tests, ranges/confidence intervals, and discrete values. The full data set included 21,200 entries for 15,688 substances. These data came from a variety of publicly available databases, including OECD’s eChemPortal, the National Library of Medicine’s Hazardous Substances Data Bank (NLM HSDB), ChemIDplus databases, and the European Commission Joint Research Center’s (JRC) AcutoxBase (Karmaus et al. 2019; Kleinstreuer et al. 2018; NTP 2018). Data were reviewed to ensure that values with obvious errors in the extracted data such as unit conversion errors (e.g., comma and decimal separator misplacements) were either fixed or removed. After this review, 16,209 values remained. Many of the chemicals represented had multiple entries, requiring that a single representative value per Chemical Abstracts Service Registry Number (CASRN) identifier be defined to facilitate modeling efforts. Based on ATWG feedback and to define the representative as a protective value while accounting for the distribution across multiple s, the median of the lowest quartile was computed using only discrete values (omitting limit test data and range and confidence interval data). A detailed summary of the data compilation is available online on the NICEATM webpage dedicated to the collaborative modeling project (NTP 2020).
To obtain chemical structure information, CASRNs served as identifiers to search the U.S. EPA’s DSSTox database hosted in the CompTox Chemicals Dashboard (Grulke et al. 2019; Richard and Williams 2002; U.S. EPA-NCCT 2014a; Williams et al. 2017) as well as other cross-checked online databases: ChemIDPlus (NIH 2016), PubChem (Bolton et al. 2008) and ChemSpider (Royal Society of Chemistry 2015). The collected structures were then processed using a standardization workflow developed for the purpose of generating QSAR-ready structures compatible with most modeling approaches (Mansouri et al. 2016a; McEachran et al. 2018). In fact, this workflow was first developed in KNIME (Berthold et al. 2008) for the CERAPP project and was also employed for CoMPARA (Mansouri et al. 2016a, 2018, 2020). The workflow is a multistep process that includes:
A filter to remove inaccurate chemical representations, inorganics/metallo-organics, mixtures, and general representations that are not specific (Markush structures, repeating monomers, connection points)
A standardization step for ring representations, isomers/mesomers, and other tautomeric forms
A step to identify salts/solvents, counterions, and duplicate structures.
The workflow can be downloaded from GitHub or KNIME hub as KNIME workflow or used in command line within a Docker container (https://github.com/NIEHS/QSAR-ready, https://kni.me/w/_iyTwvXi6U3XTFW1, https://hub.docker.com/r/kamelmansouri). After the standardization process, the final data set included 11,992 chemical structures amenable for the modeling project with encoded SMILES and SDF formats.
This data set, complete with chemical structures and representative values, was further processed to ensure that each chemical had only one representative value/call for all remaining modeling end points (i.e., the different binary and multiclass categories). For categorical designations, limit test and range/confidence interval data were integrated wherever possible. As such, the number of chemicals with end point values/calls varies across the modeling end points (Table 3) based on whether chemicals had representative values and/or other data. If a chemical had both, the representative value was used to determine categorical calls rather than limit test data. Some data were useable only for certain end points and categories depending on the thresholds. For example, ranges that spanned multiple hazard categories were considered only for the binary (very toxic/nontoxic) end points and omitted from rendering a determination for hazard category assignment.
Table 3.
Number of chemicals modeled for each end point, used for the training and evaluation of models.
| End point | Number of chemicals |
|---|---|
| VT | 11,886 |
| NT | 11,871 |
| EPA categories | 11,755 |
| GHS categories | 11,845 |
| Discrete values | 8,908 |
Note: See Supplemental Material 1 (training set) and Supplemental Material 2 (evaluation set). EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , dose of a substance that would be expected to kill half the animals in a test group; NT, non-toxic/toxic; VT, very toxic/not very toxic.
Training and evaluation sets: source, compilation, splitting.
The final data set comprising 11,992 chemicals was split into training and evaluation sets consisting of 75% (8,994 chemicals) and 25% (2,895 chemicals), respectively. This process was performed semirandomly by ordering chemicals based on the five end points (categorical and continuous values) and partitioning every fourth record into the evaluation set accordingly. This approach was taken to ensure an equivalent distribution of the values and the different hazard categories between the training and the evaluation sets without supervised sampling of the chemicals based on structures (Figure 1A). The sources of the chemical structures were also kept equivalent between the two sets, with chemical structures obtained from DSSTox, being the highest quality, representing over 75% of each set (Figure 1B).
Figure 1.
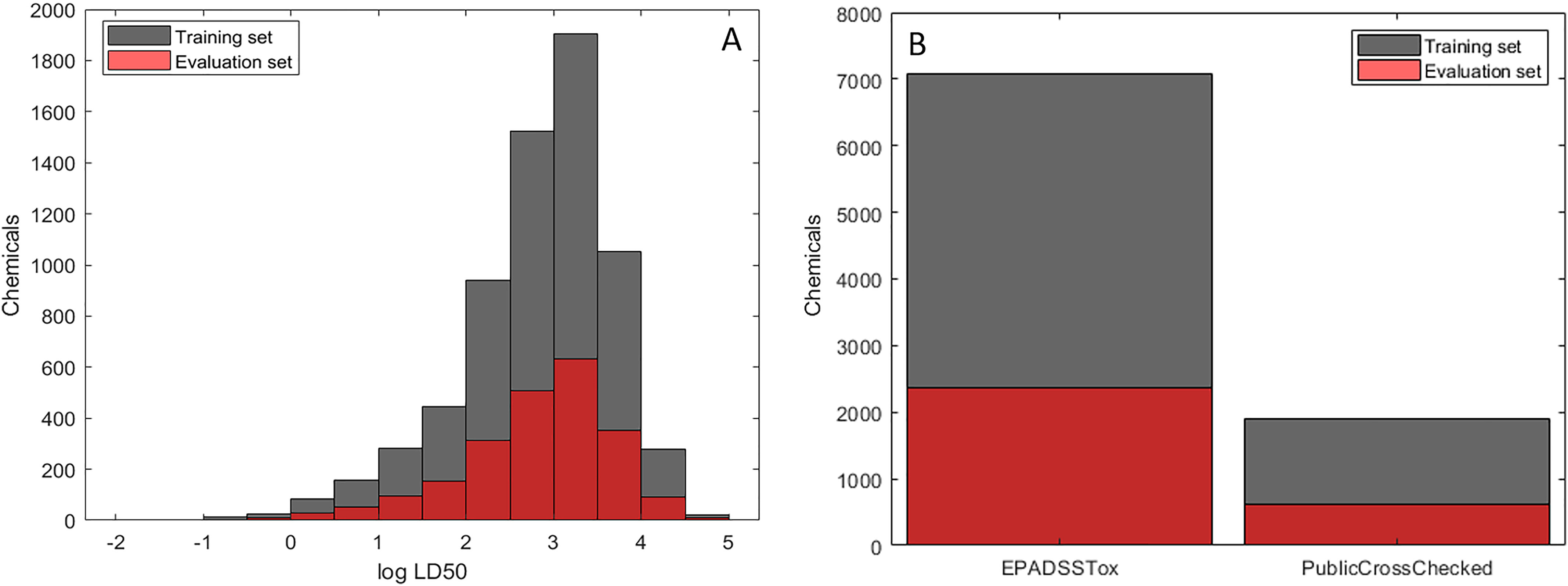
Characteristics of training (Supplemental Material 1) and evaluation sets (Supplemental Material 2). (A) Distribution of values. (B) Sources of the chemical structures.
The training set (Supplemental Material 1: TrainingSet.sdf, TrainingSet.xlsx, TrainingSet_Original.sdf) was made available for collaborators on the project webpage along with an explanation of the proper use of the data for the modeling steps (https://ntp.niehs.nih.gov/iccvam/methods/acutetox/model/qna.pdf). Modelers were encouraged to use the provided training set but were given the flexibility to make any modifications and apply post processing to suit their own modeling approaches. For example, modelers might choose to augment the data set with additional toxicity data or use undersampling approaches to reduce the number of low-potency chemicals to achieve a more balanced data set.
The empirical data (Supplemental Material 2: EvaluationSet.sdf, EvaluationSet.xlsx, EvaluationSet_Original.sdf) of the evaluation set were initially withheld from the project website so that NICEATM could perform an independent assessment of the validity of the models submitted by the collaborators. The chemical structures of this evaluation set were, however, provided to the participants to generate model predictions that would serve as an external validation set. These structures were provided as part of a much larger prediction set of chemical structures, as described below, ensuring that participants were not privy to the identities of the evaluation set chemicals during model development.
Prediction set: structure collection and curation.
The list of evaluation set chemicals was contained within a comprehensive chemical list for which participants were asked to generate predictions using their optimized models. This prediction set encompassed lists of interest to the ICCVAM ATWG regulatory agencies who use acute oral data, as well as to stakeholders and other chemical screening programs, including ToxCast™/Tox21, EDSP, the Toxic Substances Control Act (TSCA), and a general list of substances on the market from the U.S. EPA CompTox chemicals dashboard (Dix et al. 2007; Grulke et al. 2019 p. 21; Kavlock et al. 2012; U.S. EPA-NCCT 2014b, 2019).
The QSAR-ready KNIME workflow was applied to standardize the chemical structures and remove duplicates. After integration of the evaluation set, the final prediction set (see Supplemental Material 3) included 48,137 chemical structures (including the hidden evaluation set) and was made available for download on the project webpage (NTP 2020). Participating groups were encouraged to generate predictions for as many chemicals as possible.
Participants and Modeling Methods
Multiple modeling approaches were applied by the 35 international participating groups to predict the above mentioned acute oral toxicity end points. The list of participating groups with the abbreviations that are used in this manuscript to identify the different models is provided in Table 4 . The list of participants is provided in Supplemental Material 4. For transparency, the modelers were encouraged to use the provided training set and apply free and open-source tools to develop new models. However, the use of existing and proprietary commercial tools and/or other data was also permitted. The various molecular descriptors/tools and modeling approaches employed are summarized in Tables 4 and 5. For further information about the methods and detailed descriptions of modeling processes as provided by the participating groups, see https://doi.org/10.22427/NTP-DATA-002-00090-0001-0000-2 and the respective modeling references in Table 4.
Table 4.
Methods and tools applied by the different participating groups.
| Group ID | Institution | Country | Methods* | Descriptors/tool | References |
|---|---|---|---|---|---|
| ATSDR | Agency for Toxic Substances and Disease Registry, CDC | USA | ADMET | ||
| COLPHA | Collaborations Pharmaceuticals, Inc. | USA | NB | ECFP6 fingerprints | Clark et al. 2015; Minerali et al. 2020 |
| DOW | The Dow Chemical Company | USA | Mechanistic profiler | MACCS, KNIME | Anderson 1984; Berthold et al. 2008 |
| DOW_AGRO | Dow Agrosciences | USA | XGBoost | MOE, DataWarrior | CCG 2016; Chen and Guestrin 2016; Sander et al. 2015 |
| DUT | Dalian University of Technology | China | RF | DRAGON, Pipeline Pilot | Mauri et al. 2006 |
| ECUST | East China University of Science and Technology, China | China | CDK | ||
| UFG | Universidade Federal de Goiás | Brazil | RF | RDKit, MACCS | Anderson 1984 |
| IRFMN (5 groups) | Istituto di Ricerche Farmacologiche Mario Negri, IRCCS | Italy | GLM, RF, kNN, SARpy | KNIME, H2O, CDK, VEGA, DRAGON 7, Caret | Ferrari et al. 2013; Floris et al. 2014; Gadaleta et al. 2019; Hussami et al. 2015; Kuhn 2008; Manganaro et al. 2016; Vukovic et al. 2019 |
| VCCLAB | Virtual Computational Chemistry Laboratory, Helmholtz Zentrum München (GmbH) | Germany | ASNN | OCHEM, DRAGON, ISIDA | Mauri et al. 2006; Salmina et al. 2015; Sushko et al. 2012; Tetko and Tanchuk 2002; Varnek et al. 2008 |
| KU | Kyoto University Graduate School of Medicine | Japan | AL | MACCS fingerprint | Anderson 1984; Reker et al. 2017 |
| LOREAL | L’Oréal R&I | France | Ctox- | ACD, EPISUITE | ACDLabs 2019; U.S. EPA 2015 |
| LSINC | LeadScope Inc. | USA | Leadscope fingerprints | Cross et al. 2003; Roberts et al. 2000; Yang et al. 2004 | |
| MSU | Michigan State University | USA | ANN | TensorFlow | Google, Inc. 2019 |
| NCATS | National Center for Advancing Translational Sciences, National Institutes of Health | USA | ANN | Keras, TensorFlow, ADAM, RDKit | Google, Inc. 2019; Kingma and Ba 2017; Ramsundar et al. 2015 |
| NCCT | National Center for Computational Toxicology (Currently Center for Computational Toxicology and Exposure), U.S. EPA | USA | ANN | Keras, SciKit-learn, CORINA, Toxprints, MOE, RDKit | |
| NCSTATE | North Carolina State University | USA | RF | RDKit, Caret | Berthold et al. 2008; Kuhn 2008 |
| NRMRL | National Risk Management Research Laboratory (Currently Center for Computational Toxicology and Exposure), USEPA | USA | TEST | Martin et al. 2008; Zhu et al. 2009 | |
| PNNL | Pacific Northwest National Laboratory | USA | ANN | RDKit, ToxNet | Goh et al. 2017a, 2017c, 2017b, 2018 |
| ROSETTAC | Rosettastein Consulting UG | Germany | ASNN, kNN, MLR, PLS, RF, SVM, DT | OCHEM, CDK, DRAGON, ISIDA, Adriana, ChemAxon | Berthold et al. 2008; ChemAxon; Mauri et al. 2006; Sushko et al. 2011; Varnek et al. 2008 |
| RUT (2 groups) | Rutgers University | USA | kNN, RF, SVM, ANN | DRAGON, RDKit, scikit | Mauri et al. 2006 |
| SIMPLUS | Simulations Plus, Inc. | USA | ANNE | ADMET | |
| UCOL | University of Colorado | USA | RF | Semantic knowledgebase | Tripodi et al. 2017 |
| UL | Underwriters Laboratories | USA | RF | PubChem fingerprints, ULCT | REACHAcross software |
| UNC | UNC Eshelman School of Pharmacy | USA | RF | RDKit, ISIDA, DRAGON 7, KNIME | Berthold et al. 2008; Varnek et al. 2008 |
| USAFSAM | U.S. Air Force School of Aerospace Medicine | USA | OCHEM, ISIDA, CDK, Dragon 6, WEKA | Hall et al. 2009; Mauri et al. 2006; Sushko et al. 2011; Varnek et al. 2008; Willighagen et al. 2017 | |
| UNIBARI | Università degli Studi di Bari | Italy | Similarity search | RDKit, Pybel, CDK | Alberga et al. 2019; O’Boyle et al. 2008 |
| UNICAMB | University of Cambridge | UK | NB, RF | KNIME | Berthold et al. 2008; Wedlake et al. 2020 |
| UNIMIB | University of Milano-Bicocca | Italy | DRAGON 7, ECFP, MATLAB | Ballabio et al. 2019; Rogers and Hahn 2010 | |
| UNISTRA | Université de Strasbourg | France | ISIDA, MOE, WEKA, KNIME | Bonachéra and Horvath 2008; Varnek et al. 2005, 2008 |
See Table 5 for definitions of abbreviations.
Table 5.
Modeling approaches applied by the participating groups.
| Abbreviation | Approach | References |
|---|---|---|
| ASNN | Associative artificial neural networks | Tetko 2002; Tetko and Tanchuk 2002 |
| ANN | Artificial neural networks | Dreyfus 1990; Schmidhuber 2015 |
| NN | Nearest neighbor | Martin et al. 2008 |
| kNN | k-nearest neighbors | Cover and Hart 1967; Kowalski and Bender 1972; Todeschini et al. 2015 |
| SVM | Support-vector machines | Cortes and Vapnik 1995 |
| PLS | Partial least squares | Wold et al. 2001 |
| MLR | Multilinear regression | |
| DT/RT | Decision trees/Regression trees | Breiman et al. 1984; Loh 2011 |
| PLSDA | Partial least squares discriminative approach | Frank and Friedman 1993; Nouwen et al. 1997 |
| HC | Hierarchical clustering | Martin et al. 2008 |
| DF | Decision forest | Hong et al. 2004, 2005; Tong et al. 2003; Xie et al. 2005 |
| RF | Random forest | Breiman 2001 |
| SCR | Self-consistent regression | Lagunin et al. 2011 |
| RBF | Radial basis function | Zakharov et al. 2014 |
| NB | Naïve Bayes | Murphy 2006 |
| BNN | Binned-Nearest Neighbors | Mauri et al. 2016; Todeschini et al. 2015 |
| GBM | Gradient boosting method | Berk 2008 |
| GLM | Generalized linear model | Generalized Linear Model (GLM) |
| AL | Active learning | Rakers et al. 2018; Reker et al. 2017 |
| XGB | Extreme gradient boosting | Chen et al. 2019; Chen and Guestrin 2016; XGBoost 2019 |
Evaluation Procedure
The project timeline and guidelines for submission were published in an online document posted to the project webpage on the NICEATM website (NTP 2020). The guidelines included recommendations about the modeling process as well as detailed instructions about information to be included with each submission. Qualitative and quantitative evaluation procedures for the submitted models and predictions were based on the five OECD principles for QSAR modeling (OECD 2005, 2007; OECD n.d.) Models and predictions were evaluated by an organizing committee of scientists from NICEATM, CCTE, and industry.
Qualitative evaluation.
The qualitative evaluation process assessed the transparency of the submitted models. The criteria used for this evaluation (Table 6) satisfied four of the five OECD principles and added a category for general documentation, as shown in Table 6. Participants who did not provide sufficient information for analysts to understand and interpret their results were asked either to provide additional clarification until all requirements were met or to withdraw/resubmit the model.
Table 6.
Criteria for the qualitative evaluation.
| Criteria | Description |
|---|---|
| Documentation |
Documentation submitted to facilitate review of the following criteria:
|
| Defined end point(s) |
|
| Unambiguous algorithm |
|
| Domain of applicability |
|
| Mechanistic interpretation, if possible |
|
Quantitative evaluation.
This step of evaluation satisfied the OECD principle of QSAR validation addressing appropriate measures of goodness-of-fit, robustness, and predictivity. To be fully inclusive for high- and low-throughput modeling approaches, the participating groups were not required to predict the entire prediction set but were encouraged to provide predictions for as many chemicals as possible. This approach was designed to ensure sufficient predictions for the 2,895 evaluation set chemicals that were hidden within the 48,137 structures of the prediction set. Although this flexibility could lead to models being evaluated for varying portions of the evaluation set, the results of the evaluation set were used for comparison purposes and to check for mistakes and mismatches so any corrections could be made prior to consensus modeling.
The quantitative evaluation considered only predictions within the applicability domain (AD) of the models. Models predicting the binary and multiclass end points were evaluated separately from those used to evaluate models predicting discrete values using appropriate statistical parameters. The parameters of the scoring functions included the three criteria from the OECD principles:
Goodness of fit: statistics on the training set (Tr)
Predictivity: statistics on the evaluation set (Eval)
Robustness: balance between goodness of fit and predictivity.
Based on these parameters, each model produced a score (S) ranging from 0 to 1 for predictions of chemicals within its AD. This score was used in the consensus modeling step as a weighting scheme. The parameter multipliers (for the global and subparameter functions) were assigned based on importance to the evaluation procedure as established in the CoMPARA project (Mansouri et al. 2020):
| (1) |
Quantitative evaluation of binary and multi-class models.
The performance of models for binary and multiclass end points was evaluated using statistical indices proposed in the literature (Consonni et al. 2009; Dearden et al. 2009; Todeschini et al. 2016). The indices used were calculated from a confusion matrix, which summarizes the number of observed and predicted classes in the rows and columns, respectively. For the current evaluation, classifications based on experimental data were used as truth. The classification parameters were defined using the number of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). The performance measures calculated for consideration during the evaluation step included balanced accuracy (BA), specificity (Sp), and sensitivity (Sn).
BA is given by:
| (2) |
Sn, or True Positive Rate (TPR), is given by:
| (3) |
and the Sp, or True Negative Rate (TNR), is given by:
| (4) |
For multicategory end points, these parameters were calculated for each category and then averaged. The balance between Sn and Sp was also included in the calculation of goodness of fit and predictivity. The three parameters of the scoring function S were calculated as follows:
| (5) |
| (6) |
| (7) |
Quantitative evaluation of discrete prediction models:
The performance of the discrete value predictions was evaluated using the experimental values from the embedded evaluation set. The commonly used parameter root mean square error (RMSE) and the coefficient of determination () were calculated for all predictions (Consonni et al. 2009; Todeschini et al. 2016).
| (8) |
where and are the estimated and observed responses of the element, respectively; is the mean; and is the number of training compounds.
The three parameters of the scoring function S were calculated as follows:
| (9) |
| (10) |
| (11) |
Consensus Modeling
After being evaluated according to the defined strategy, each model was assigned a score (S) for the predictions within its AD. This score was used in the consensus modeling step as a weighting scheme to combine the predictions from all the submitted models to produce a single consensus prediction for each end point. The majority rule was applied for binary and multiclass end points, whereas the weighted average value in the regression was applied to generate the consensus predictions for the discrete end point, as detailed below. This approach resulted in each chemical in the prediction set being assigned a consensus prediction for each of the five end points.
Consensus for binary and multiclass end points.
For each chemical in the prediction set, the consensus category/call was decided by the weighted majority rule: the class with the highest average score of the models predicting it. This average score was calculated excluding the models that did not provide a prediction within AD for the specific chemical.
Consensus for discrete predictions.
For each chemical in the prediction set, the consensus predicted value was calculated as the average of the predictions within the AD from the different models weighted by their S scores.
The predicted consensus value (C) of the chemical i was calculated as:
| (12) |
where N is the number of models that provided predictions within AD for the chemical i, and is the predicted from each model. The weight (w), summing to 1, for each model j is calculated as:
| (13) |
The consensus model predictions for each end point were first evaluated using the same evaluation set used to evaluate the individual models. The defined ADs of the different models were taken into consideration to investigate the accuracy of the final predictions generated by the consensus model. Analysis of the coverage trends and concordance among the individual models’ predictions were also conducted as part of the evaluation of the consensus models.
WoE approach to combine all models.
The consensus modeling combined the submitted single models for each of the five end points, resulting in five consensus predictions for each chemical in the prediction set. Because the respective models for the five end points, were trained separately on the data set prepared for each end point (Table 3), the consensus predictions of each of the five end points could disagree for an individual chemical. Examples of discordant predictions were especially likely between the two multiclass end points (U.S. EPA and GHS categorizations), which are based on multiple thresholds with overlapping ranges. Discrete value predictions could also be slightly inconsistent with the predicted categories. To produce final consensus predictions that were consistent across all five end points for each chemical, it was important to apply correcting rules for the outlier predictions that over or underestimated a specific end point. Thus, a WoE approach was developed to optimize the consensus based on the majority rule and obtain more robust predictions. The fact that there was an odd number of end points and consequent predictions (five) also helped when applying the majority rule to determine the predictions in agreement and minimize the needed degree of correction. This WoE approach also served to combine all five consensus model results into a single prediction per chemical for acute oral toxicity.
In an effort to quantify inherent variability in the animal data used in this work and to determine a confidence interval (CI) representing the uncertainty that should accompany experimentally derived values, we have leveraged our large compendium of rat acute oral values to compute a 95% CI across standard deviations (SDs) for chemicals having multiple point estimate values. SDs for 1,120 chemicals with at least three independent values (excluding limit test data) were bootstrapped 1 million times, and the result provided a representative SD that takes into account the range of SDs in the data set.
Additional Evaluation of the Consensus Models
As a further evaluation step for the consensus predictions, highly curated experimental in vivo acute oral toxicity data were used to assess concordance between predictions and experimental outcomes. Briefly, a data set was tandemly generated for the study of variability in acute oral toxicity animal data (Nelms et al. 2020; Ly Pham et al. 2020). This data set was limited to chemicals associated with multiple values (including discrete values, limit tests, and ranges/CIs) as well as additional entries pulled from the European Chemicals Agency (ECHA) database for risk assessment (ECHA 2020). The chemicals in this data set were not used in the original CATMoS modeling project. After an initial consistency analysis between the two data sets, a thorough manual curation resulted in a total of 916 chemicals with at least two discrete values to be used for the evaluation of the predicted values, and a data set of 1,323 chemicals with at least two entries including discrete values, limit tests, ranges, and CIs to be used for the evaluation of the predicted binary and multiclass categories.
To adapt the curated in vivo experimental data to the five end points studied in this project, the raw formatting was processed using a KNIME workflow to convert the entries into a computer-readable, format. For each chemical (unique CASRN), the replicate data were processed to assign consistent hazard categories, which were unanimous across replicates. Then, to produce high confidence data for model evaluation, the entries were grouped by CASRN to determine the final category based on the majority rule where there was an agreement between the different entries above a certain threshold. For each of the multiclass end points, the agreement was calculated as a concordance percentage, and the threshold for assigning a call was set to 75%. For example, if a chemical A was associated with four entries, three of them in U.S. EPA Category II and one in U.S. EPA Category III, the concordance would be 75%, and the overall assignment would be U.S. EPA Category II. For discrete values, the median of only the discrete values was taken (within 1.5 SD threshold to account for outliers). The total number of chemicals having in vivo consensus data, per end point, is summarized in Table 7.
Table 7.
Chemicals with in vivo experimental acute oral toxicity data used for the additional predictivity analysis.
| Modeled end point | Total number of chemicals in curated data set |
|---|---|
| VT | 1,296 |
| NT | 1,153 |
| EPA categories | 1,089 |
| GHS categories | 1,083 |
| value | 916 |
Note: The number of chemicals represents the total for that data set, not the number that fell beneath the threshold (e.g. for Very Toxic). See Supplemental Material 8. EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , dose of a substance that would be expected to kill half the animals in a test group; NT, nontoxic/toxic; VT, very toxic/not very toxic.
Generalization of the Consensus and Implementation in OPERA
To apply the consensus models beyond the initial prediction set, the combined predictions were used to train generalized models capable of replicating the original consensus. This procedure was achieved by applying a weighted k-nearest neighbor (kNN) approach to fit the classification models based on the majority vote of the nearest neighbors. This approach has the advantage of resembling read-across, a broadly accepted data gap filling tool within regulatory agencies (Cover and Hart 1967; Kowalski and Bender 1972; Todeschini et al. 2015). kNN also fulfills the OECD principles for QSAR modeling, given its nonambiguous algorithm, high accuracy, and interpretability.
To increase the sensitivity of the models for more conservative predictions, all toxic chemicals from the prediction set (s less than or equal to , i.e., U.S. EPA Categories I and II) were included, whereas less toxic chemicals were included with an 85% concordance threshold among the predicting models for the binary models (VT and NT) and 75% for the remaining modeled end points. Each one of the data sets was divided semirandomly into training and test sets representing 75% and 25%, respectively.
PaDEL (version 2.2) and CDK2 (CDK version 2.0) were first used to calculate two-dimensional molecular descriptors. Because PaDEL uses a previous version of CDK (1.5), duplicate descriptors were excluded. The union of the PaDEL descriptors (1,444) and CDK2 (287) resulted in a total of 1,616 variables that were later filtered for low variance. Subsequently, kNN was coupled with genetic algorithms (GAs) to select a minimized optimal subset of molecular descriptors (form the combined PaDEL-CDK list) for calculating the similarity in the kNN model based on the Euclidean distance. GAs start with an initial random population of binary vectors representing the presence or absence of molecular descriptors. Then an evolutionary process is simulated to optimize a defined fitness function in 5-fold cross-validation, in which new vectors are created by coupling the binary vectors of the initial population with genetic operations such as crossover and mutation (Ballabio et al. 2011; Leardi and Lupiáñez González 1998).
This procedure was applied separately for each of the modeled end points. The best models were selected and implemented in combination using the WoE approach in the free, standalone, open-source QSAR modeling suite OPERA (Mansouri et al. 2016b, 2018). Both OPERA’s global and local AD approaches, as well as the accuracy estimation procedure, were applied to all predictions. The global AD is a Boolean index based on the leverage approach for the whole training set, whereas the local AD is a continuous index in the 0–1 range based on the most similar chemical structures from the training set (Mansouri et al. 2018; Sahigara et al. 2012).
The extended, similarity-based predictive approach as well as the WoE consensus are implemented as the CATMoS consensus model in the OPERA application (version 2.5) (Mansouri et al. 2018). OPERA can be downloaded from the National Institute of Environmental Health Sciences GitHub repository (https://github.com/NIEHS/OPERA) and used locally via a command-line interface or user-friendly graphical interface. The use of this standalone application facilitates the generation of CATMoS predictions by providing different user input options. The simplest way for the user to input chemicals would be via a text file with chemical identifiers such as CASRN, DTXSID (U.S. EPA’s DSSTox database public identifier) or InChIKey. In fact, OPERA contains an internal database with the complete list of DSSTox chemical structures (periodically updated) stored in QSAR-ready format and ready to use for prediction with any of its models. In addition, users can provide their own chemical structures in SMILES or SDF formats as described in Mansouri et al. (2018). Since version 2.6, OPERA has been equipped with an embedded version of the above-mentioned structure standardization workflow and can generate QSAR-ready structures prior to prediction. The local nearest neighbors–based and the global leverage-based AD approaches implemented in OPERA help the user determine whether their chemicals are within the model’s interpolation space, where it is safe to generate predictions. In fact, a local AD index of 1 means that the chemical being predicted was one of the 48,137 chemicals of the prediction set and that the initial combined predictions of the single end point models were used to make the final consensus call.
Results and Discussion
Submitted Consortium Models: Prediction Review
The 35 participating groups submitted a total of 139 models across the five end points, as summarized in Table 8. Each group submitted predictions for the full or partial prediction set for at least one end point. The two binary end points were predicted by the highest number of models; fewer submitted models predicted the multiclass end points and the value end point. The number of submitted models was likely an indication of the level of difficulty for each end point, a finding consistent with the previous collaborative projects, CERAPP (Mansouri et al. 2016a) and CoMPARA (Mansouri et al. 2020). The main difficulties that participants faced while modeling these end points were generally related to the skewed nature of the training set, a challenge encountered in many toxicity modeling projects. As shown in Figure 1, most of the data represented nontoxic and low toxicity chemicals (i.e., high values), which was also reflected in the binary and multi-categorical end points. However, 11/35 participating groups still submitted models for all five requested end points. The full list of submitted predictions files and model details is available at https://doi.org/10.22427/NTP-DATA-002-00090-0001-0000-2. A KNIME workflow (https://doi.org/10.22427/NTP-DATA-002-00090-0001-0000-2) was used to process the predictions from all models and combine them in a single file per end point for further evaluation and analysis.
Table 8.
Models received for the five end points from the different participating groups.
| End point | Number of models |
|---|---|
| VT | 32 models |
| NT | 33 models |
| EPA categories | 26 models |
| GHS categories | 23 models |
| value | 25 models |
Note: For additional information see https://doi.org/10.22427/NTP-DATA-002-00090-0001-0000-2; EPA, Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , lethal dose, 50%, or dose of a substance that would be expected to kill half the animals in a test group; NT, nontoxic/toxic VT, very toxic/not very toxic.
Results of Qualitative and Quantitative Evaluation
All submitted models were reviewed and evaluated by the organizing committee using the criteria described above. The qualitative evaluation ensured clarity of the submitted information, confirmed that all models fulfilled the OECD requirements for computational models as well as the goals of this project, and provided a basis to facilitate the use of the predictions in the following consensus modeling (i.e., compute S score for weighing schema). The subsequent quantitative evaluation step assessed the quality of the predictions prior to the consensus modeling, which was the main goal of the project.
This evaluation was not intended to provide comparisons between the models, especially with the uneven coverages of the prediction set (and consequently the hidden evaluation set) due to AD differences. In fact, the total number of provided predictions per model and the predictions within the AD varied substantially depending on the type of model and the employed AD approach. Thus, the quantitative evaluation served mainly as a first checkpoint to reveal data mishandling issues that might have occured during the initial modeling steps. These issues included mismatches between structures, identifiers, and associated data as well as misinterpretation and inversion of the different data fields/end points, which could potentially lead to a severe decrease in prediction accuracy. Models with such issues were returned to participants to withdraw or replace their submission with updated models. The final evaluated submissions were used to generate summary statistics (Figure 2A; Supplemental Material 5 with detailed parameters).
Figure 2.
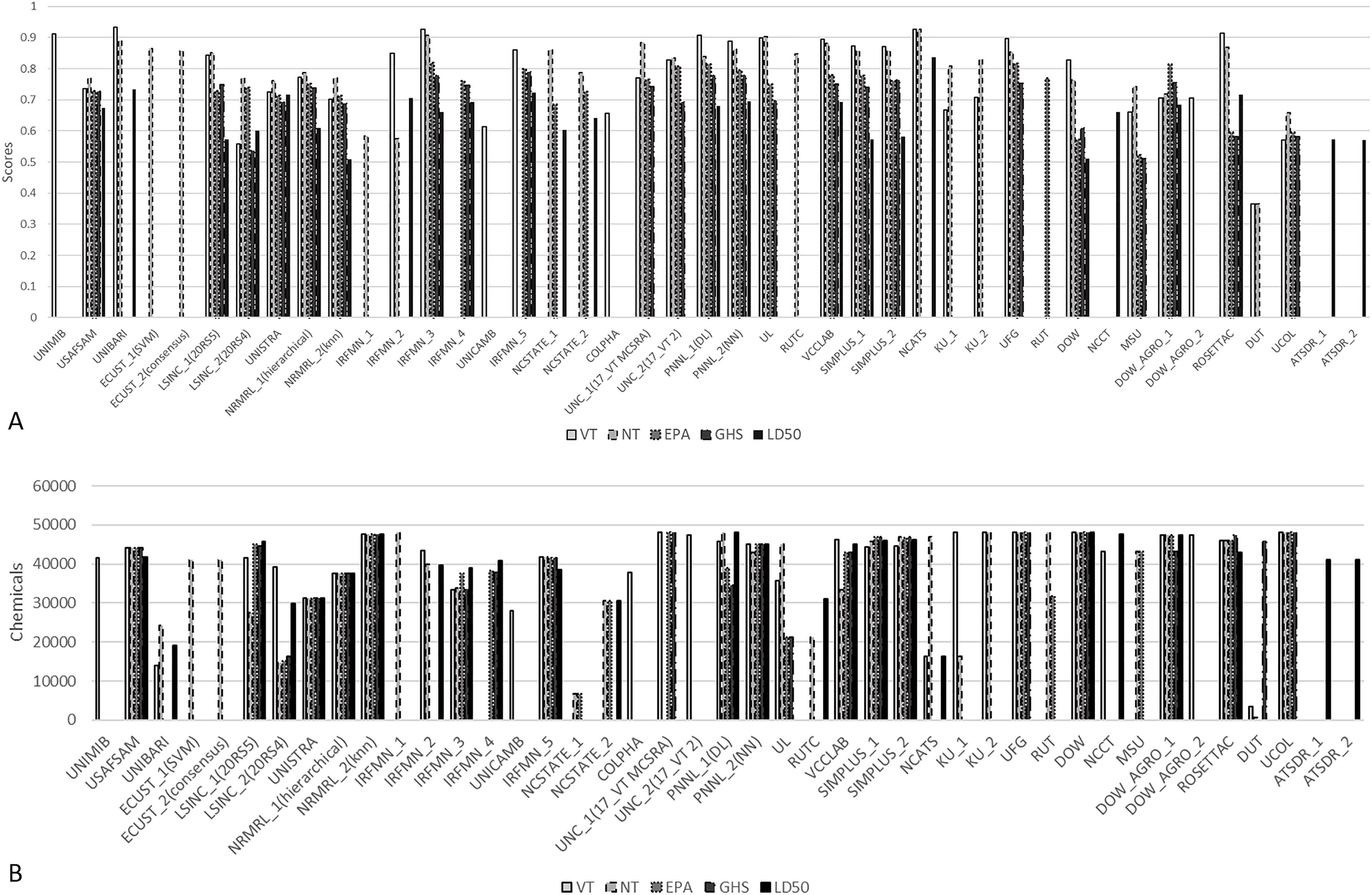
Evaluation scores (A) and coverage of the prediction set (B) by the submitted models. See Supplemental Material 5 and 6 for Figure 2A and B, respectively. Modeling groups along the x-axis are defined in Table 4.
Most of the models achieved high predictivity scores on the evaluation set (Figure 2A). The relatively low score for the Dalian University of Technology (DUT) binary models was related to the fact that the submitted predictions covered only small portions of the evaluation set representing only one of two classes for both modeled end points (VT and NT), which led to a BA of 0.5 that was not directly informative as to the real predictivity of this model. Although coverage was not included as a qualifying parameter during evaluation and did not affect quantitative scores, to be inclusive for low-throughput approaches, models with limited coverage had only a marginal influence on the consensus calls and statistics of a prediction set with over 48,000 structures. The remaining models covered most of the prediction set with median coverage ranging from to chemicals (out of 48,137) per end point. This showed that most of the employed AD approaches were rather permissive. The coverage of the different models on the prediction set for the five end points is summarized in Figure 2B and in more details in Supplemental Material 6.
In general, the binary and multiclass models achieved higher scores (median S scores ranging from 0.74 to 0.82) than the discrete prediction models (median 0.66). This finding was expected for such a challenging end point with high variability, a low number of toxic compounds, and different data sources leading to a decrease in precision. As noted with the total number of submitted models per end point, the relatively higher statistics of the binary models confirmed that multiclass end points are more difficult to model. The median S scores for VT and NT reached 0.80 and 0.82, respectively, whereas the median scores for the EPA and GHS category end points were 0.75 and 0.74, respectively.
Consensus Modeling
Prior to combining the predictions into consensus calls for evaluation, it was important to check the coverage and concordance among the models. Figure 3A shows that all chemicals in the prediction set were predicted by at least 10 models. Moreover, most chemical structures were predicted by about 20 models for the multiclass and value end points and at least 25 models for the binary VT and NT models. This high coverage for all five end points provided a solid basis for the consensus modeling step and strengthened the statistical relevance of the combined predictions.
Figure 3.
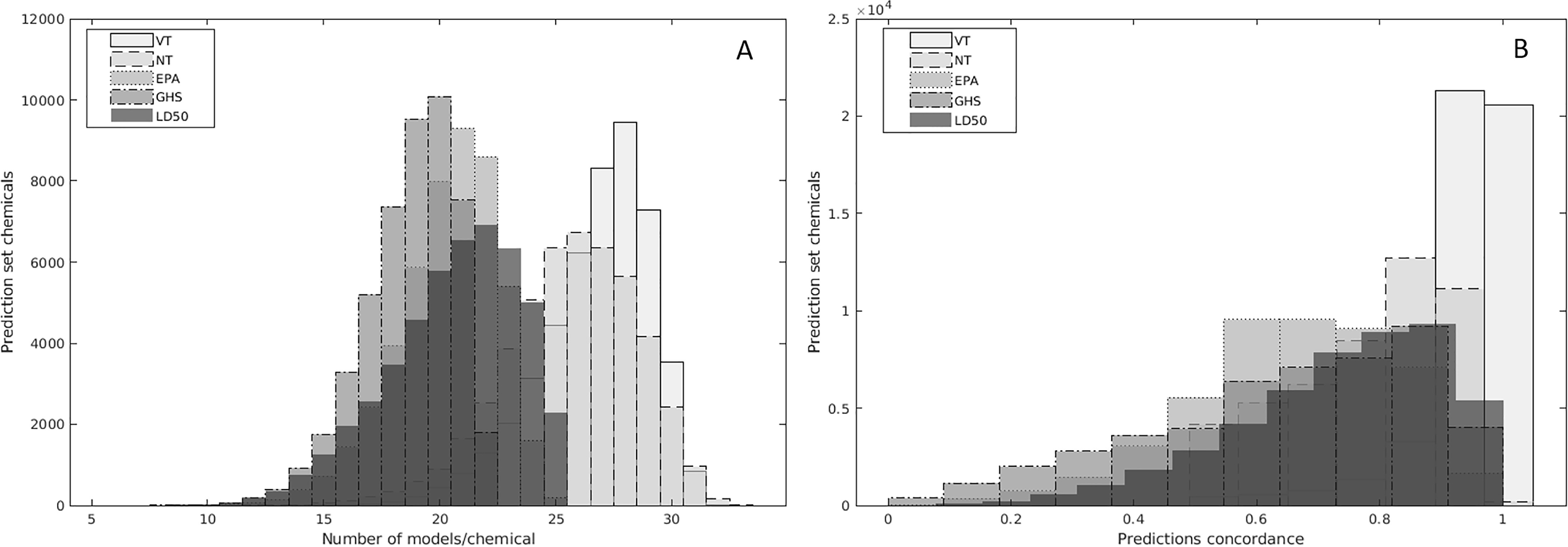
Distributions of the coverage of the prediction set chemicals (A) and concordance among the single models (B) across the five end points. See Supplemental Material 7.
The concordance among the models was an equally important criterion for combining the predictions into consensus calls. In fact, it was demonstrated during the previous collaborative modeling projects, CERAPP and CoMPARA, that higher concordance among numerous models built using different modeling approaches corresponded to higher accuracy (Mansouri et al. 2016a, 2020). Figure 3B showed that the concordance among the binary, multiclass, and value models was about 0.8 for most of the prediction set chemicals. This high concordance simplified the process of generating consensus calls for the prediction set, especially for the binary and multiclass models for which the consensus classification was largely driven by the majority rule. The exceptions to this were chemicals with cross-model concordance near 0.5, for which only a subset of all models would be driving the classification. In sum, based on the analyses of coverage and concordance between models, it can be concluded that the data were amenable to combining the different model predictions into consensus predictions.
Consensus modeling step 1: combining predictions per end point.
The predictions from the five modeled end points were first combined independently based on the defined rules. As described, the models had different contributions in terms of chemical coverage, and each prediction from each model was associated with different weights across the prediction set. After consensus calls were generated for each chemical in the prediction set, the same evaluation procedure was applied for each one of the five end points. The resulting statistical details are reported in Tables 9–12 under the main heading “Step 1: End point.”
Table 9.
Evaluation parameters for the consensus predictions after the WoE approach.
| Step 1: End point | Step 2: WoE | |||
|---|---|---|---|---|
| Training | Evaluation | Training | Evaluation | |
| 0.83 | 0.67 | 0.85 | 0.65 | |
| RMSE | 0.31 | 0.47 | 0.30 | 0.49 |
Note: , coefficient of determination; RMSE, root mean square error; WoE, weight of evidence.
Table 12.
Evaluation parameters for the GHS category consensus predictions after the WoE approach.
| Hazard category | Step 1: End point | Step 2: WoE | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GHS training | GHS evaluation | GHS training | GHS evaluation | |||||||||||||||||||||
| I | II | III | IV | V | Overall | I | II | III | IV | V | Overall | I | II | III | IV | V | Overall | I | II | III | IV | V | Overall | |
| BA | 0.68 | 0.74 | 0.79 | 0.81 | 0.84 | 0.77 | 0.52 | 0.73 | 0.68 | 0.69 | 0.73 | 0.67 | 0.83 | 0.87 | 0.89 | 0.85 | 0.92 | 0.88 | 0.74 | 0.75 | 0.72 | 0.7 | 0.78 | 0.74 |
| Sn | 0.37 | 0.49 | 0.63 | 0.91 | 0.71 | 0.62 | 0.04 | 0.50 | 0.45 | 0.77 | 0.55 | 0.46 | 0.67 | 0.76 | 0.85 | 0.80 | 0.88 | 0.79 | 0.50 | 0.53 | 0.56 | 0.66 | 0.67 | 0.58 |
| Sp | 1.00 | 0.99 | 0.96 | 0.72 | 0.98 | 0.93 | 1.00 | 0.97 | 0.91 | 0.62 | 0.92 | 0.88 | 0.99 | 0.99 | 0.94 | 0.90 | 0.97 | 0.96 | 0.99 | 0.97 | 0.89 | 0.74 | 0.90 | 0.90 |
Note: Corresponding confusion matrices available in Supplemental Material 9. BA, balanced accuracy; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; WoE, weight of evidence.
Table 10.
Evaluation parameters for the VT and NT consensus predictions after the WoE approach.
| Step 1: End point | Step 2: WoE | |||||||
|---|---|---|---|---|---|---|---|---|
| VT | NT | VT | NT | |||||
| Training | Evaluation | Training | Evaluation | Training | Evaluation | Training | Evaluation | |
| BA | 0.96 | 0.87 | 0.94 | 0.80 | 0.93 | 0.84 | 0.92 | 0.78 |
| Sn | 0.94 | 0.77 | 0.92 | 0.70 | 0.87 | 0.70 | 0.88 | 0.67 |
| Sp | 0.99 | 0.96 | 0.97 | 0.89 | 0.99 | 0.97 | 0.97 | 0.90 |
Note: Corresponding confusion matrices available in Supplemental Material 9. BA, balanced accuracy; NT, nontoxic/toxic; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; VT, very toxic/not very toxic; WoE, weight of evidence.
The statistics on the consensus predictions for each of the five end points followed the same general trend as for the single models. The two binary models, VT and NT, showed the highest accuracy for both training and evaluation sets. As the end points increased in precision, going from binary to four or five categories and ultimately to discrete value prediction, the performance of the consensus predictions decreased accordingly.
Consensus modeling step 2: WoE approach.
The second step of consensus modeling was to generate a single consistent acute oral toxicity prediction per chemical by reconciling the five independent consensus end point predictions. The WoE approach combined predictions from the five end points based on a majority rule. To use binary and multiclass end points with different thresholds, the overlapping ranges of categories were used as bins, resulting in a total of seven bins (Figure 4). To extend the range of the discrete values, the inherent animal data variability was considered. The resulting statistical details are reported in Tables 9–12 under the main heading “Step 2: WoE.” These statistics represent the evaluation of the consensus model for that end point following the weight of evidence integration of all consensus models across the five end points.
Figure 4.
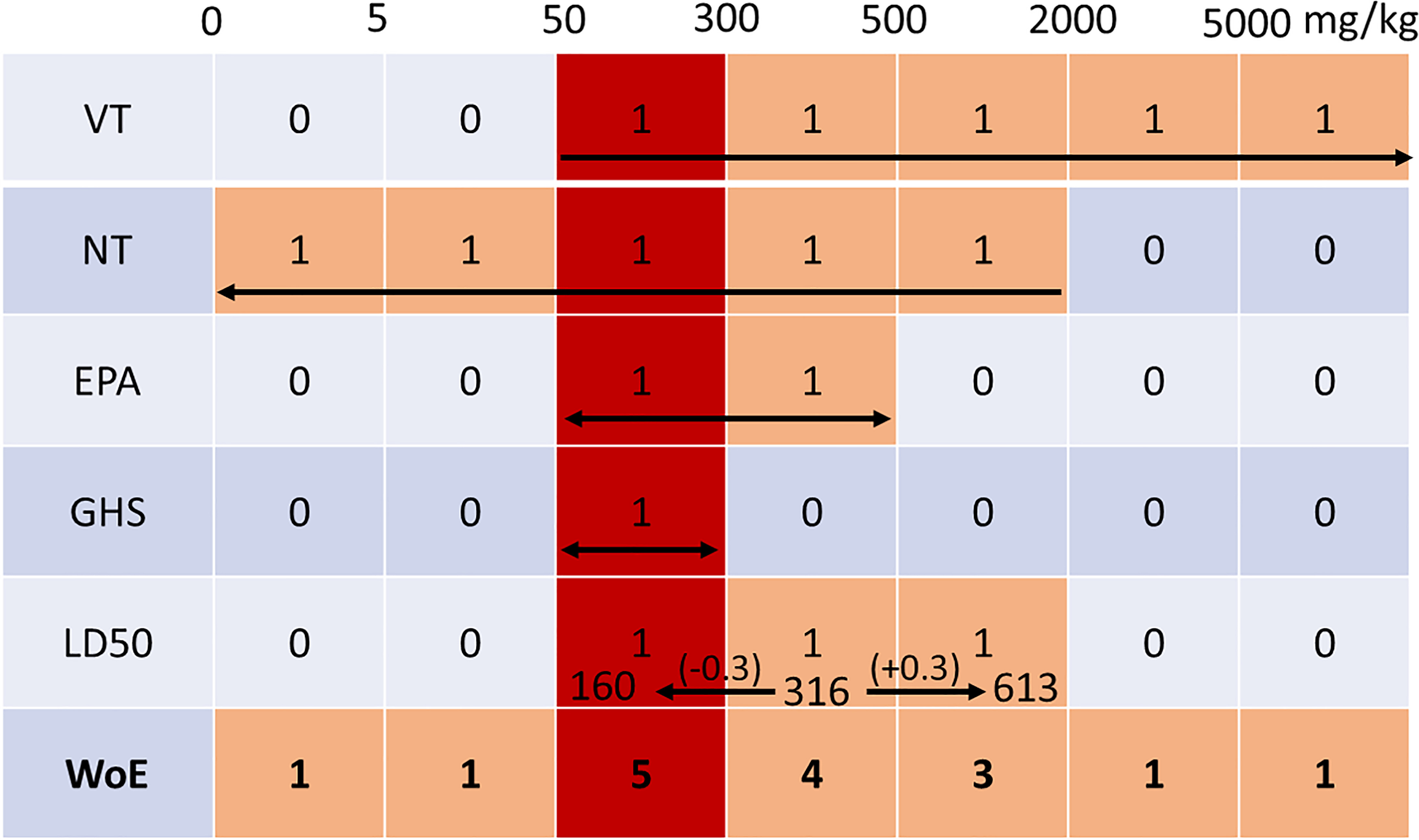
Example of identifying the winning bin and reconciling the consensus predictions across five end points using the WoE (weight of evidence) approach. The columns represent the different bins from the EPA and GHS categories combined. The rows represent the five different end points and the WoE prediction. The arrows in each row represent the range of the prediction for each end point which were attributed a value of 1 and outside of it a value of 0. For the end point, the range limits are calculated by adding and subtracting the confidence interval of 0.3 in log value to the original prediction of resulting in a range of 160 to . The winning bin is determined by the maximum of the sum of each column in the WoE row. Note: EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , dose of a substance that would be expected to kill half the animals in a test group; NT, nontoxic/toxic; VT, very toxic/not very toxic; WoE, weight of evidence.
After quantifying the inherent variability based on the bootstrap analysis, the resulting margin of was considered the 95% CI for acute oral values. This approach not only quantified and defined a confidence margin for the experimental values, but also informed on an acceptable range to apply around predictions.
This CI was applied to computing a range for every predicted value. Once the winning bin was determined based on the maximum overlap among the five end points, corrections were made on the outlier prediction(s) by adjusting the corresponding category for the multiclass predictions. For the discrete value predictions, if the prediction did not fall within the range of the winning bin, a new was calculated depending on the reach of the extended CI range. To explain this further, an example is illustrated in Table 13, with corresponding prediction ranges represented by arrows in Figure 4.
Table 13.
Example of consensus predictions, corresponding to bins highlighted in Figure 4.
| End point | Prediction | |
|---|---|---|
| VT | Not very toxic | |
| NT | Not nontoxic | |
| EPA | Category II | (50–500) |
| GHS | Category III | (50–300) |
| (160–613) |
Note: EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , dose of a substance that would be expected to kill half the animals in a test group; NT, Nontoxic/toxic; VT, very toxic/not very toxic.
To computationally automate this process, the concept was translated by an algorithm that converted the bins within each end point prediction ranges to “ones” and the remaining bins to “zeros.” The winning bin of the WoE approach was determined by summing the bins and selecting the maximum. In this example, the bin having a total of five overlapping bins is the winner. This means that only the value requires adjustment to ensure that the predicted value falls within the WoE-identified “correct” bin range. In this case, the new is calculated by taking the average of the lower CI () and the upper threshold of the winning bin (), resulting in an adjusted of . In general, the rule for adjusting the point estimate if it does not fall within the winning bin would be the average of the covered threshold and the corresponding CI boundary. For example, if the winning bin here was , the adjusted would be . If the CI should span the entire winning bin or be completely nonoverlapping, then the adjusted would be its center. In cases where there was more than one winning bin, the most conservative bin was selected.
The statistics of the WoE-adjusted predictions were recalculated and are summarized in Table 9 (Step 2: WoE). In many cases, the calculated parameters did not show a significant difference. However, the performance for the lower categories (highly toxic) of the U.S. EPA and GHS multicategory end points increased significantly, with the WoE approach demonstrating higher sensitivity (Tables 11–12). This improvement is in part because the available data were skewed toward the upper categories (less toxic). Thus, the difference was more noticeable in categories with a lower number of data points. All results of the consensus analysis are available in Supplemental Material 7.
Table 11.
Evaluation parameters for the U.S. EPA category consensus predictions after the WoE approach.
| Hazard category | Step 1: End point | Step 2: WoE | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EPA training | EPA evaluation | EPA training | EPA evaluation | |||||||||||||||||
| I | II | III | IV | Overall | I | II | III | IV | Overall | I | II | III | IV | Overall | I | II | III | IV | Overall | |
| BA | 0.80 | 0.89 | 0.83 | 0.80 | 0.83 | 0.77 | 0.72 | 0.71 | 0.68 | 0.72 | 0.93 | 0.89 | 0.83 | 0.80 | 0.87 | 0.83 | 0.72 | 0.71 | 0.68 | 0.74 |
| Sn | 0.61 | 0.84 | 0.92 | 0.63 | 0.75 | 0.57 | 0.59 | 0.81 | 0.39 | 0.59 | 0.87 | 0.83 | 0.91 | 0.63 | 0.81 | 0.70 | 0.56 | 0.81 | 0.40 | 0.62 |
| Sp | 1.00 | 0.94 | 0.75 | 0.98 | 0.92 | 0.98 | 0.86 | 0.61 | 0.97 | 0.85 | 0.99 | 0.95 | 0.75 | 0.98 | 0.92 | 0.97 | 0.88 | 0.62 | 0.97 | 0.86 |
Note: Corresponding confusion matrices available in Supplemental Material 9. BA, balanced accuracy; EPA, U.S. Environmental Protection Agency; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; WoE, weight of evidence.
Generalization of the Consensus and Implementation in OPERA
The two-step approach for combining predictions from the 139 submitted models resulted in a robust consensus model that covered the entire prediction set of 48,137 chemical structures. To make the model applicable for further screening of new chemicals, an additional step was required. A weighted-kNN modeling approach was implemented in OPERA (version 2.5) to mimic the initial consensus predictions and generate new ones. This was achieved by training extended models based on the existing experimental data and predictions with high concordance. To facilitate the training process, the five end points were processed separately. Then, the WoE approach was similarly applied to the generated predictions to make a final consistent consensus call. The prefiltered PaDEL and CDK descriptors were used in a GA-kNN procedure to select the most informative variables in a supervised, end point–dependent approach. The resulting minimized numbers of descriptors selected during the training process and performance of the best kNN models are summarized in Table 14.
Table 14.
Parameters of the selected generalized weighted-kNN models as implemented in OPERA.
| End point | Number of chemicals from the prediction set | Descriptors | Training (5-fold cross-validation) * | Test set * |
|---|---|---|---|---|
| VT | 23,767 | 21 | 0.79 | 0.77 |
| NT | 30,971 | 11 | 0.90 | 0.89 |
| EPA categories | 25,487 | 15 | 0.79 | 0.81 |
| GHS categories | 25,720 | 15 | 0.78 | 0.79 |
| values | 28,954 | 23 | 0.79 | 0.81 |
Balanced accuracy (BA) values are reported for performance metrics, except for the value end point for which the coefficient of determination and its equivalent for cross-validation are reported for the training and test sets, respectively. Note: EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , dose of a substance that would be expected to kill half the animals in a test group; NT, nontoxic/toxic; VT, very toxic/not very toxic. The number of prediction set chemicals represents the total for that data set, not the number that fell beneath the threshold (e.g., for VT).
The descriptors were selected based on the importance ranking performed by the GA during multiple independent runs of generation optimization. The selection of the best models was simultaneously based on maximizing the performance in 5-fold cross-validation as well as minimizing both the number of meaningful descriptors and the model complexity.
The performance statistics of the models summarized in Table 14 showed a high level of accuracy in training set cross-validation in terms of BA and . This high performance was equally complemented, and confirmed, by performances on the test set as further validation of the models. The balance, stability, and robustness of the five end point models were sufficient to simulate the original combined predictions without overfitting the initial set. Thus, the resulting models could be combined via the WoE approach and applied to generate predictions for new chemicals that have sufficient similarity to the original prediction set.
Additional Evaluation Using a Highly Curated Data Set
Preparation of the curation data set and consistency analysis.
Prior to combining the experimental data for chemicals with multiple entries from the data collected for the CATMoS project and the ECHA data set to produce a curated data set, a review of the different values revealed a number of inconsistencies between the two data sets. This finding can be partly attributed to the variability of animal data, but in some cases, were also due to errors that may have been introduced and propagated during reporting, publishing, or data retrieval. To estimate the disagreement between replicate studies per chemical, the different values and binary/multiclass calls were compared using a representative from each of the two data sets (as described above). As shown in Table 15, the discordance was highest for the very toxic compounds, as represented by the Sn of the VT end point. This is due to the fact that 25 out of the 38 chemicals that are considered very toxic in the CATMoS data set are associated with an in the ECHA data set. Discordance between the Sn and Sp parameters was less apparent for the NT end point. However, a closer look at the confusion matrix generated during the calculation of these parameters revealed a disagreement on a total of 126 chemicals, with 109/126 classified as toxic in the CATMoS data but not in the ECHA data set. A similar level of disagreement was observed for U.S. EPA and GHS categorizations; the confusion matrices in Tables 13 and 14 showed that most of the discordant classifications differed by one category. Even for the agreeing categories, there was disagreement between the corresponding values that increased with the wider range categories. These inconsistencies could be due to a number of factors, including the sources and the data interpretation and processing. For example, in the ECHA data set, CASRN 14,489-75-9 was associated with a range of , which placed it outside the “Very Toxic” class. However, in the CATMoS data, the same CASRN is associated with a unique point estimate of and was consequently classified as very toxic.
Table 15.
Concordance between ECHA and CATMoS data sets for the categorical end points.
| VT | NT | EPA | GHS | |
|---|---|---|---|---|
| Sn* | 0.34 | 0.98 | 0.70 | 0.56 |
| Sp* | 1.00 | 0.86 | 0.93 | 0.96 |
| BA | 0.67 | 0.92 | 0.82 | 0.76 |
*Sn and Sp of ECHA calculated based on CATMoS data.
Note: BA, balanced accuracy; CATMoS, Collaborative Acute Toxicity Modeling Suite; ECHA, European Chemicals Agency; EPA, U.S. Environmental Protection Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; , lethal dose, 50% or dose of a substance that would be expected to kill half the animals in a test group; NT, nontoxic/toxic; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; VT, very toxic/not very toxic.
To help with the detection of the outlier entries and the assessment of the data, CATMoS consensus predictions were also considered during the analysis. It was expected that the predictions would diverge even further from the ECHA data set due to the differences with CATMoS experimental data (i.e., data used to train the CATMoS models), as noted above. However, an examination of the values revealed several chemicals that were associated with low values in ECHA but were predicted as less toxic by CATMoS with a high concordance among the submitted models. A closer look at 28 of these chemicals with available source toxicity reports on the ECHA website revealed a number of reporting errors in the source database, such as unit conversion (grams vs. milligrams), typos, decimal misplacement (“,” vs. “.”) and estimated doses using read-across, among others. Such findings highlighted how robust and highly concordant CATMoS predictions are because they helped to identify where the compiled in vivo data had typographical errors. These types of errors most likely affect other chemicals in the data set and therefore affect the statistics in Tables 15–17 even further. For this reason, a CI was applied to all CATMoS predictions that covered the observed range of the animal data variability (i.e., estimated at log values). When this range was applied to the predictions, 96.6% of the ECHA in vivo values fell within the confidence interval of the CATMoS predictions. This could also be considered an indication that empirical values should consistently be accompanied by a CI.
Table 17.
Concordance between ECHA and CATMoS experimental data sets for GHS categories.
| ECHA\CATMoS | I | II | III | IV | V | Category concordance |
|---|---|---|---|---|---|---|
| I | 0 | 0 | 0 | 0 | 0 | — |
| II | 0 | 12 | 13 | 3 | 4 | 35.29 |
| III | 0 | 0 | 84 | 40 | 5 | 65.11 |
| IV | 0 | 0 | 15 | 467 | 100 | 80.24 |
| V | 0 | 0 | 0 | 32 | 1,323 | 97.78 |
Note: —, no data; CATMoS, Collaborative Acute Toxicity Modeling Suite; ECHA, European Chemicals Agency; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals.
Table 16.
Concordance between ECHA and CATMoS experimental data sets for U.S. EPA categories.
| ECHA\CATMoS | I | II | III | IV | Category concordance |
|---|---|---|---|---|---|
| I | 13 | 16 | 4 | 2 | 37.14 |
| II | 0 | 175 | 92 | 2 | 57.56 |
| III | 0 | 2 | 1,126 | 46 | 95.91 |
| IV | 0 | 0 | 71 | 401 | 84.96 |
Note: CATMoS, Collaborative Acute Toxicity Modeling Suite; ECHA, European Chemicals Agency; EPA, U.S. Environmental Protection Agency.
This assessment step also revealed certain CATMoS predictions that were in high disagreement with the original CATMoS empirical data but turned out to agree with the ECHA in vivo data. For example, the of CASRN 108-91-8 was predicted by CATMoS to have an of and reported on the ECHA website as (https://echa.europa.eu/brief-profile/-/briefprofile/100.003.300). However, in the experimental data underlying the CATMoS training set, the of this chemical was only , which was also the value reported for the chemical on the U.S. EPA CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/dsstoxdb/results?search=DTXSID1023996#toxicity-values). This chemical, although present in the training set and therefore seen by the fitting algorithms during the learning process, was consistently predicted differently by most models based on its structural features and was initially counted as an inaccurate prediction in the evaluation process. The fact that the other chemicals in the data set and the modeling algorithms predicted a value closer to the true value identified from the curated in vivo data set rather than the possibly erroneous value that made its way into the training set revealed that the size of the data set and approaches used could overcome outlier values and that predictions are robust.
The noted differences and inconsistencies in the collected experimental values required a deeper manual curation effort to remove the outlier entries for each chemical that could be erroneous. The resulting highly curated multientry data with an acceptable degree of concordance were used as an additional evaluation set (Supplemental Material 8).
Assessment of the consensus and WoE predictions vs. curated in vivo data.
The curated in vivo data set was a subset of chemicals with multiple (at least two unique) experimental data entries that was used as an external set to assess the accuracy of the binary, multiclass, and value CATMoS predictions. The resulting classification and regression statistical parameters are summarized in Tables 18–21. As for Tables 9–12 above, the entries under the main heading “Step 1: End point” represent the evaluation of the consensus model developed for the specific end point, whereas entries under the main heading “Step 2: WoE” represent the evaluation of the consensus model for that end point following the WoE integration of all consensus models across the five end points.
Table 18.
Evaluation parameters for the point estimate consensus predictions of the curated in vivo data.
| Step 1: End point | Step 2: WoE | |
|---|---|---|
| 0.76 | 0.73 | |
| 0.89 | 0.86 | |
| RMSE | 0.36 | 0.37 |
Note: , lethal dose, 50% or dose of a substance that would be expected to kill half the animals in a test group; , coefficient of determination; , Pearson’s correlation coefficient; RMSE, root mean square error; WoE, weight of evidence.
Table 21.
Evaluation parameters for (D): assessment of the GHS category predictions for the curated in vivo data set.
| Step 1: End point | Step 2: WoE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | Overall | I | II | III | IV | V | Overall | |
| BA | 0.71 | 0.84 | 0.77 | 0.75 | 0.78 | 0.77 | 0.87 | 0.85 | 0.80 | 0.78 | 0.84 | 0.83 |
| Sn | 0.42 | 0.69 | 0.62 | 0.85 | 0.60 | 0.64 | 0.75 | 0.72 | 0.72 | 0.76 | 0.75 | 0.74 |
| Sp | 1.00 | 0.99 | 0.93 | 0.66 | 0.96 | 0.91 | 0.99 | 0.98 | 0.89 | 0.81 | 0.94 | 0.92 |
Note: Corresponding confusion matrices available in Supplemental Material 9. BA, balanced accuracy; GHS, U.N. Globally Harmonized System of Classification and Labeling of Chemicals; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; WoE, weight of evidence.
The statistical parameters in all four tables show high accuracy performances for all five end points, and these metrics are in fact higher than the results on the evaluation set. A similar observation was also reported during the evaluation of the previously mentioned collaborative projects, CERAPP and CoMPARA (Mansouri et al. 2016a, 2020), which showed an increased agreement between the predictions of the consensus models and the evaluation data with the increase of concordant sources. This finding can be also interpreted as an indication of the lower quality and noise in the single-source data points, which cause a decrease in performance especially between training and evaluation sets as noted earlier (Tables 9–12).
There was a slight decrease in the statistical parameters for predictions after the application of the WoE approach (Table 18). This could have been because some of the newly assigned values were based on semi-arbitrary calculations within the winning bin. The adjustment was intended to place the value within the correct category, but there is always the possibility that the value could become skewed further from the experimental value in comparison to the initially predicted on the other side of the category threshold.
Table 19 showed similar statistics before and after the application of the WoE approach, implying that the binary end point consensus predictions did not require substantive adjustment. However, higher predictive performances for the multiclass models were noted after the application of the WoE adjustments (Tables 20–21). This was particularly clear for categories representing the most toxic compounds and indicated that the WoE consensus predictions were more conservative than the initial multiclass predictions. The GHS WoE (Table 21) seemed to have more balanced predictivity/sensitivity in comparison with the U.S. EPA WoE (Table 20), which showed a drop in sensitivity for U.S. EPA Category IV similar to that observed in initial evaluation (Table 11). This was mostly due to the lower number of chemicals tested in U.S. EPA Category IV () in comparison with GHS Category 5 ().
Table 19.
Evaluation parameters for the VT and NT binary end point predictions for the curated in vivo data set.
| Step 1: End point | Step 2: WoE | |||
|---|---|---|---|---|
| VT | NT | VT | NT | |
| BA | 0.95 | 0.85 | 0.93 | 0.85 |
| Sn | 0.92 | 0.76 | 0.88 | 0.75 |
| Sp | 0.98 | 0.94 | 0.99 | 0.94 |
Note: Corresponding confusion matrices available in Supplemental Material 9. BA, balanced accuracy; NT, nontoxic/toxic; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; VT, very toxic/not very toxic; WoE, weight of evidence.
Table 20.
Evaluation parameters for the U.S. EPA category predictions for the curated in vivo data set.
| Step 1: End point | Step 2: WoE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | Overall | I | II | III | IV | Overall | |
| BA | 0.84 | 0.81 | 0.79 | 0.67 | 0.78 | 0.93 | 0.79 | 0.79 | 0.67 | 0.80 |
| Sn | 0.70 | 0.76 | 0.84 | 0.36 | 0.66 | 0.88 | 0.72 | 0.84 | 0.36 | 0.70 |
| Sp | 0.99 | 0.86 | 0.75 | 0.98 | 0.89 | 0.99 | 0.87 | 0.75 | 0.98 | 0.90 |
Note: Corresponding confusion matrices available in Supplemental Material 9. BA, balanced accuracy; EPA, U.S. Environmental Protection Agency; Sn, sensitivity or true negative rate; Sp, specificity or true positive rate; WoE, weight of evidence.
In addition to the demonstrated high performances, this evaluation showed the utility of CATMoS both for providing accurate predictions for new chemicals and for revisiting and filtering existing data for additional curation. Regulatory agencies could benefit from both application of the model to consider new predictions and associated confidence intervals as well as checking previous decisions for additional assessment of the data and the predictions. Both of these applications are currently being considered by members of ICCVAM and industry stakeholders.
Limitations
The predictive ability of CATMoS is limited to the quality of the data used to train and evaluate the contributing models. Certainly, the lack of metadata of the collected training and evaluation set is a limiting factor to delineate any study differences or sources of variability in the in vivo assays. Although the predictions are able to identify specific erroneous data points, it still leaves uncertainty regarding the overall reliability of in vivo data, which is still not well-characterized. As discussed, the skewness of the in vivo data caused limitations in the predictivity of the single models and the initial consensus for the highly toxic chemicals. However, the data still contained more than 400 chemicals in the very toxic class (), and as demonstrated in Tables 11–12, these limitations at the lower end of the multiclass prediction models were largely remediated by the final WoE approach.
Similar to most QSAR/QSPR models, the CATMoS can be applied only to single organic compounds. Mixtures of organic chemicals should be studied separately. However, to accommodate mixtures of multiple compounds, the GHS system provides an additivity rule to help classify chemicals that can use CATMoS predictions as input (http://www.unece.org/fileadmin/DAM/trans/danger/publi/ghs/GHS_presentations/English/health_env_e.pdf).
Additionally, as most molecular descriptors are developed for small- and medium-size molecules, CATMoS and other QSAR models cannot process large biomolecules, long polymeric chains, and nanomaterials. To help identify and use the most adequate chemical structures for predictions and to avoid unpredictable substances, OPERA users wishing to generate CATMoS predictions can use either of two input options:
Provide a text file with a list of chemical identifiers (CASRN, DTXSID, InChI) and let OPERA pull the correct QSAR-ready structure from its database of 830,000 highly curated DSSTox chemicals; or,
Provide their own structures but apply the embedded standardization workflow to generate QSAR-ready structures prior to curation.
A possible limiting aspect of the OPERA standalone application is that it must be installed locally, requiring access to the user’s computer operating system and hardware to perform calculations. Users can, however, avoid this process and access predictions by visiting the Integrated Chemical Environment (ICE) dashboard of the NTP (https://ice.ntp.niehs.nih.gov/) and querying its internal database of predictions for the DSSTox chemicals. These predictions will also be made available on the U.S. EPA CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/).
Conclusion
This project was organized by the ICCVAM ATWG as an implementation of the strategic roadmap for the development and validation of new alternative methods for acute oral toxicity testing. The resulting CATMoS models provide consensus predictions for 48,137 chemicals of interest to regulatory agencies and stakeholders. CATMoS combined contributions from 35 internationally renowned groups in the field of in silico modeling. This is the third such collaborative project for most of the consortium members, following endocrine disruption modeling efforts CERAPP and CoMPARA. However, the uniqueness of CATMoS comes from the fit-for-purpose end points resulting from the upfront participation of regulatory agencies. Thus the needs of the ICCVAM ATWG member agencies and international partners were considered when defining the end points for predictive modeling, ensuring that there would be regulatory interest in potentially using the predicted acute oral toxicity models, and supporting agencies’ interest in alternative methods.
This early stage involvement of regulators not only identified the five modeled end points representing the different uses of the data but also facilitated open stakeholder dialog during the project’s workshop held at the National Institutes of Health in Bethesda, Maryland (Kleinstreuer et al. 2018). At the workshop, participating groups presented their approaches to overcoming the different challenges of the project, such as the skewed data distribution or tackling particular chemistries. The consensus modeling and the implementation of the final CATMoS model and its uses were also discussed among the modelers and the stakeholders. Currently, consensus predictions on specific chemicals of interest are being assessed by different regulatory agencies for potential use as alternative sources of data. For example, a list of more than 100 chemicals and corresponding values derived from existing regulatory studies were identified by the U.S. EPA and are being checked, curated, and compared with CATMoS consensus predictions. The preliminary analysis shows that in 96% of the cases, the CATMoS predictions are either overlapping or more conservative than the existing . In the few cases where the disagreement is highest, a closer look showed potential issues with the considered sources of the in vivo studies and in some cases disagreement with the in vivo values used to train CATMoS models.
The details of the CATMoS model and predictions are available in a QSAR Model Report Format (QMRF) that was submitted to the European Commission’s JRC for review and publication on their QMRF Inventory for easy access by the international community (European Commission 2013; JRC 2017).
In addition to the initial prediction set, CATMoS was implemented in OPERA and used to screen the list of 837,000 chemical substances in the U.S. EPA’s DSSTox database (underpinning the Dashboard application). These predictions are made available via the Integrated Chemical Environment dashboard (https://ice.ntp.niehs.nih.gov/) of the NTP and in the future via the U.S. EPA’s CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/).
CATMoS is an example of how toxicological problems can be solved collaboratively using computational approaches. In fact, the resulting consensus models leverage the strengths and overcome the limitations of any single approach, proving to be as good as or better than animal data. Such successful collaborative projects support international collaboration, a legacy of free and open-source code and workflows, and increasing consideration and adoption from regulators interested in implementing NAMs. Finally, it is worth noting that the international aspect of these collaborations can also help with harmonizing global regulatory processes toward a universal system. We now have a solid foundation for future collaborations to establish globally accepted alternative methods for assessing acute toxicity end points.
Supplementary Material
Acknowledgments
The authors thank C. Sprankle for editorial assistance. This work was supported by the intramural research program of the NIEHS, NIH and NCATS, NIH. Technical support was provided by Integrated Laboratory Systems under NIEHS contract HHSN273201500010C.
The views expressed in this article are those of the authors and do not necessarily reflect the views of policies of the U.S. EPA, NIH, Agency for Toxic Substances and Disease Registry, Centers for Disease Control and Prevention, or any other agency and should not be construed to represent any agency determination or policy. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Collaborations Pharmaceuticals Inc. acknowledges NIH funding: R44GM122196-02A1 from the National Institute of General Medical Sciences and 1R43ES031038-01 NIEHS. Research reported in this publication was supported by the NIEHS of the NIH under award R43ES031038.
H.C., D.P.R., and H.Z. at Rutgers University at Camden were partially supported by the NIEHS (grants R01ES031080 and R15ES023148).
References
- ACDLabs. 2019. Chemistry Software for Analytical and Chemical Knowledge Management. https://www.acdlabs.com/ [accessed 21 May 2019].
- Alberga D, Trisciuzzi D, Mansouri K, Mangiatordi GF, Nicolotti O. 2019. Prediction of acute oral systemic toxicity using a multifingerprint similarity approach. Toxicol Sci 167(2):484–495, PMID: 30371864, 10.1093/toxsci/kfy255. [DOI] [PubMed] [Google Scholar]
- Anderson S. 1984. Graphical representation of molecules and substructure-search queries in MACCStm. J Mol Graph 2(3):83–90, 10.1016/0263-7855(84)80060-0. [DOI] [Google Scholar]
- Ballabio D, Grisoni F, Consonni V, Todeschini R. 2019. Integrated QSAR models to predict acute oral systemic toxicity. Mol Inf 38(8–9):1800124, PMID: 30549437, 10.1002/minf.201800124. [DOI] [PubMed] [Google Scholar]
- Ballabio D, Vasighi M, Consonni V, Kompany-Zareh M. 2011. Genetic algorithms for architecture optimisation of counter-propagation artificial neural networks. Chemometr Intell Lab Syst 105(1):56–64, 10.1016/j.chemolab.2010.10.010. [DOI] [Google Scholar]
- Benfenati E, Diaza RG, Cassano A, Pardoe S, Gini G, Mays C, et al. . 2011. The acceptance of in silico models for REACH: requirements, barriers, and perspectives. Chem Cent J 5:58, PMID: 21982269, 10.1186/1752-153X-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk RA. 2008. Statistical Learning from a Regression Perspective. New York, NY: Springer-Verlag. [Google Scholar]
- Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, et al. 2008. KNIME: The Konstanz Information Miner. In: Studies in Data Analysis, Machine Learning and Applications. Preisach C, Burkhardt, H, Schmidt-Thieme, L, and Decker, R, eds., Berlin, Germany: Springer, 319–326. [Google Scholar]
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. 2008. Chapter 12-PubChem: Integrated Platform of Small Molecules and Biological Activities. Wheeler RA, Spellmeyer, DC, eds. Annu Rep Comput Chem 4:217–241, 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- Bonachéra F, Horvath D. 2008. Fuzzy tricentric pharmacophore fingerprints. 2. Application of topological fuzzy pharmacophore triplets in quantitative structure-activity relationships. J Chem Inf Model 48(2):409–425, PMID: 18254617, 10.1021/ci7003237. [DOI] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Machine Learning 45(1):5–32, 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Breiman L, Friedman J, Stone CJ.Olshen RA. 1984. Classification and Regression Trees. 1st ed. Boca Raton, FL: Chapman and Hall/CRC. [Google Scholar]
- CCG (Chemical Computing Group). 2016. Molecular Operating Environment (MOE). Montreal, QC, Canada: Chemical Computing Group Inc. [Google Scholar]
- ChemAxon. Calculator Plugins–Documentation. https://docs.chemaxon.com/display/docs/Calculator_Plugins.html [accessed 16 July 2020].
- Chen T, Guestrin C. 2016. XGBoost: a Scalable Tree Boosting System. KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, August 2016, 785–794, 10.1145/2939672.2939785. [DOI]
- Chen T, He T, Benesty M, Khotilovich V, Tang Y, Cho H, et al. . 2019. XGBoost: Extreme Gradient Boosting. https://CRAN.R-project.org/package=xgboost [accessed 21 May 2019].
- Clark AM, Dole K, Coulon-Spektor A, McNutt A, Grass G, Freundlich JS, et al. . 2015. Open source Bayesian models. 1. Application to ADME/tox and drug discovery datasets. J Chem Inf Model 55(6):1231–1245, PMID: 25994950, 10.1021/acs.jcim.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni V, Ballabio D, Todeschini R. 2009. Comments on the definition of the Q2 parameter for QSAR validation. J Chem Inf Model 49(7):1669–1678, PMID: 19527034, 10.1021/ci900115y. [DOI] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. Mach Learn 20(3):273–297, 10.1007/BF00994018. [DOI] [Google Scholar]
- Cover T, Hart P. 1967. Nearest neighbor pattern classification. IEEE Trans Inform Theory 13(1):21–27, 10.1109/TIT.1967.1053964. [DOI] [Google Scholar]
- Cross KP, Myatt G, Yang C, Fligner MA, Verducci JS, Blower PE. 2003. Finding discriminating structural features by reassembling common building blocks. J Med Chem 46(22):4770–4775, PMID: 14561096, 10.1021/jm0302703. [DOI] [PubMed] [Google Scholar]
- Dearden JC, Cronin MTD, Kaiser KLE. 2009. How not to develop a quantitative structure–activity or structure–property relationship (QSAR/QSPR). SAR QSAR Environ Res 20(3–4):241–266, PMID: 19544191, 10.1080/10629360902949567. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. 2007. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci 95(1):5–12, PMID: 16963515, 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- Dreyfus SE. 1990. Artificial neural networks, back propagation, and the Kelley-Bryson gradient procedure. J Guid Control Dyn 13(5):926–928, 10.2514/3.25422. [DOI] [Google Scholar]
- ECHA (European Chemicals Agency). 2008. Chapter R6: QSARs and grouping of chemicals In: Guidance on Information Requirements and Chemical Safety Assessment. Helsinki, Finland: European Chemicals Agency. [Google Scholar]
- ECHA. 2020. ECHA. https://echa.europa.eu/en [accessed 7 July 2020].
- European Commission. 2007. REACH in brief. https://www.fecc.org/wp-content/uploads/2020/01/reach_in_brief_dec06.pdf [accessed 15 July 2020].
- European Commission. 2013. QSAR Model Reporting Format (QMRF). EU Science Hub. https://ec.europa.eu/jrc/en/scientific-tool/qsar-model-reporting-format-qmrf [accessed 18 August 2017].
- Ferrari T, Cattaneo D, Gini G, Golbamaki Bakhtyari N, Manganaro A, Benfenati E. 2013. Automatic knowledge extraction from chemical structures: the case of mutagenicity prediction. SAR QSAR Environ Res 24(5):365–383, PMID: 23710765, 10.1080/1062936X.2013.773376. [DOI] [PubMed] [Google Scholar]
- Floris M, Manganaro A, Nicolotti O, Medda R, Mangiatordi GF, Benfenati E. 2014. A generalizable definition of chemical similarity for read-across. J Cheminform 6(1):39, PMID: 25383097, 10.1186/s13321-014-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank IE, Friedman JH. 1993. A statistical view of some chemometrics regression tools. Technometrics 35(2):109–135, 10.1080/00401706.1993.10485033. [DOI] [Google Scholar]
- Gadaleta D, Vuković K, Toma C, Lavado GJ, Karmaus AL, Mansouri K, et al. . 2019. SAR and QSAR modeling of a large collection of LD50 rat acute oral toxicity data. J Cheminform 11(1):58, PMID: 33430989, 10.1186/s13321-019-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generalized Linear Model (GLM). H2O 3.30.0.6 documentation. http://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/glm.html [accessed 15 July 2020].
- Goh GB, Hodas NO, Siegel C, Vishnu A. 2017a. SMILES2Vec: an Interpretable General-Purpose Deep Neural Network for Predicting Chemical Properties. http://arxiv.org/abs/1712.02034 [accessed 7 July 2020]. [Google Scholar]
- Goh GB, Siegel C, Vishnu A, Hodas NO. 2017b. ChemNet: A Transferable and Generalizable Deep Neural Network for Small-Molecule Property Prediction. https://www.arxiv-vanity.com/papers/1712.02734/ [accessed 15 July 2020].
- Goh GB, Siegel C, Vishnu A, Hodas NO, Baker N. 2017c. Chemception: a deep neural network with minimal chemistry knowledge matches the performance of expert-developed QSAR/QSPR models. http://arxiv.org/abs/1706.06689 [accessed 15 July 2020]. [Google Scholar]
- Goh GB, Siegel C, Vishnu A, Hodas NO, Baker N. 2018. How much chemistry does a deep neural network need to know to make accurate predictions? http://arxiv.org/abs/1710.02238 [accessed 15 July 2020]. [Google Scholar]
- Google, Inc. 2019. TensorFlow. https://www.tensorflow.org/ [accessed 21 May 2019].
- Grulke CM, Williams AJ, Thillanadarajah I, Richard AM. 2019. EPA’s DSSTox database: history of development of a curated chemistry resource supporting computational toxicology research. Comput Toxicol 12:100096, PMID: 33426407, 10.1016/j.comtox.2019.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. 2009. The WEKA data mining software: an update. SIGKDD Explor Newsl 11(1):10–18, 10.1145/1656274.1656278. [DOI] [Google Scholar]
- Hong H, Tong W, Perkins R, Fang H, Xie Q, Shi L. 2004. Multiclass Decision Forest–a novel pattern recognition method for multiclass classification in microarray data analysis. DNA Cell Biol 23(10):685–694, PMID: 15585126, 10.1089/dna.2004.23.685. [DOI] [PubMed] [Google Scholar]
- Hong Y, Hsu K-L, Sorooshian S, Gao X. 2005. Self-organizing nonlinear output (SONO): a neural network suitable for cloud patch-based rainfall estimation at small scales. Water Resour Res 41(3):1–15, 10.1029/2004WR003142. [DOI] [Google Scholar]
- Hussami N, Kraljevic T, Nykodym T, Rao A, Lanford J, Wang A. 2015. Generalized Linear Modeling with H2O. http://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/glm.html [accessed 15 July 2020].
- JRC. 2017. (Q)SAR Model Reporting Format (QMRF) Inventory. http://qsardb.jrc.ec.europa.eu/qmrf/ [accessed 18 August 2017].
- Interagency Coordinating Committee on the Validation of Alternative Methods. 2018. A Strategic Roadmap for Establishing New Approaches to Evaluate the Safety of Chemicals and Medical Products in the United States. 10.22427/NTP-ICCVAM-ROADMAP2018 [accessed 15 July 2020]. [DOI]
- Karmaus AL, Patlewicz G, Mansouri K, Fitzpatrick J, Allen DG, Kleinstreuer NC. 2019. Developing predictive models for acute oral systemic toxicity: lessons learned from a global collaboration. CICSJ Bull 37(1):23, 10.11546/cicsj.37.23. [DOI] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, et al. . 2012. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25(7):1287–1302, PMID: 22519603, 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Kingma DP, Ba J. 2017. Adam: a method for stochastic optimization. arXiv:14126980. [Google Scholar]
- Kleinstreuer NC, Karmaus AL, Mansouri K, Allen DG, Fitzpatrick JM, Patlewicz G. 2018. Predictive models for acute oral systemic toxicity: a workshop to bridge the gap from research to regulation. Comput Toxicol 8(11):21–24, PMID: 30320239, 10.1016/j.comtox.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski BR, Bender CF. 1972. The K-nearest neighbor classification rule (pattern recognition) applied to nuclear magnetic resonance spectral interpretation. Anal Chem 44(8):1405–1411, 10.1021/ac60316a008. [DOI] [Google Scholar]
- Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Soft 28(5):1–26, 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- Lagunin A, Zakharov A, Filimonov D, Poroikov V. 2011. QSAR modelling of rat acute toxicity on the basis of PASS prediction. Mol Inform 30(2–3):241–250, PMID: 27466777, 10.1002/minf.201000151. [DOI] [PubMed] [Google Scholar]
- Lahl U, Hawxwell KA. 2006. REACH-The new European chemicals law. Environ Sci Technol 40(23):7115–7121, PMID: 17180957, 10.1021/es062984j. [DOI] [PubMed] [Google Scholar]
- Leardi R, Lupiáñez González A. 1998. Genetic algorithms applied to feature selection in PLS regression: how and when to use them. Chemometr Intell Lab Syst 41(2):195–207, 10.1016/S0169-7439(98)00051-3. [DOI] [Google Scholar]
- Loh W. 2011. Classification and regression trees. WIREs Data Mining Knowl Discov 1(1):14–23, 10.1002/widm.8. [DOI] [Google Scholar]
- Lowit A, Schlosser C, Myska A, Patlewicz G, Paris M, Karmaus A, et al. . 2017. Replacing Animals for Acute Systemic Toxicity Testing: A U.S. Strategy and Roadmap. Baltimore, MD: Society of Toxicology. [Google Scholar]
- Manganaro A, Pizzo F, Lombardo A, Pogliaghi A, Benfenati E. 2016. Predicting persistence in the sediment compartment with a new automatic software based on the k-Nearest neighbor (k-NN) algorithm. Chemosphere 144:1624–1630, PMID: 26517391, 10.1016/j.chemosphere.2015.10.054. [DOI] [PubMed] [Google Scholar]
- Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, et al. . 2016a. CERAPP: collaborative estrogen receptor activity prediction project. Environ Health Perspect 124(7):1023–1033, PMID: 26908244, 10.1289/ehp.1510267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Grulke CM, Richard AM, Judson RS, Williams AJ. 2016b. An automated curation procedure for addressing chemical errors and inconsistencies in public datasets used in QSAR modelling. SAR QSAR Environ Res 27(11):911–937, PMID: 27885862, 10.1080/1062936X.2016.1253611. [DOI] [PubMed] [Google Scholar]
- Mansouri K, Grulke CM, Judson RS, Williams AJ. 2018. OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminform 10(1):10, PMID: 29520515, 10.1186/s13321-018-0263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Kleinstreuer N, Abdelaziz AM, Alberga D, Alves VM, Andersson PL, et al. . 2020. CoMPARA: collaborative modeling project for androgen receptor activity. Environ Health Perspect 128(2):27002, PMID: 32074470, 10.1289/EHP5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TM, Harten P, Venkatapathy R, Das S, Young DM. 2008. A hierarchical clustering methodology for the estimation of toxicity. Toxicol Mech Methods 18(2–3):251–266, PMID: 20020919, 10.1080/15376510701857353. [DOI] [PubMed] [Google Scholar]
- Mauri A, Ballabio D, Todeschini R, Consonni V. 2016. Mixtures, metabolites, ionic liquids: a new measure to evaluate similarity between complex chemical systems. J Cheminform 8:49, PMID: 28316647, 10.1186/s13321-016-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri A, Consonni V, Pavan M, Todeschini R. 2006. DRAGON software: an easy approach to molecular descriptor calculations. Match 56:237–248. [Google Scholar]
- McEachran AD, Mansouri K, Grulke C, Schymanski EL, Ruttkies C, Williams AJ. 2018. “MS-Ready” structures for non-targeted high-resolution mass spectrometry screening studies. J Cheminform 10(1):45, PMID: 30167882, 10.1186/s13321-018-0299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minerali E, Foil DH, Zorn KM, Ekins S. 2020. Evaluation of assay central machine learning models for rat acute oral toxicity prediction. ACS Sustainable Chem Eng 8(42):16020–16027, 10.1021/acssuschemeng.0c06348. [DOI] [Google Scholar]
- Murphy KP. 2006. Naive Bayes Classifiers. Vancouver, Canada: University of British Columbia, 8. https://www.ic.unicamp.br/∼rocha/teaching/2011s2/mc906/aulas/naive-bayes.pdf. [Google Scholar]
- Nelms MD, Karmaus AL, Patlewicz G. 2020. An evaluation of the performance of selected (Q)SARs/expert systems for predicting acute oral toxicity. Comput Toxicol 16:100135, PMID: 33163737, 10.1016/j.comtox.2020.100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH (National Institutes of Health). 2016. ChemIDPlus. http://chem.sis.nlm.nih.gov/chemidplus [accessed 26 January 2015].
- Nouwen J, Lindgren F, Hansen B, Karcher W, Verhaar HJM, Hermens JLM. 1997. Classification of environmentally occurring chemicals using structural fragments and PLS discriminant analysis. Environ Sci Technol 31(8):2313–2318, 10.1021/es9609213. [DOI] [Google Scholar]
- NTP (National Toxicology Program). 2018. Workshop: Predictive Models for Acute Oral Systemic Toxicity. https://ntp.niehs.nih.gov/whatwestudy/niceatm/3rs-meetings/past-meetings/tox-models-2018/index.html?utm_source=direct&utm_medium=prod&utm_campaign=ntpgolinks&utm_term=atwksp-2018 [accessed 7 July 2020].
- NTP. 2020. Predictive Models for Acute Oral Systemic Toxicity. https://ntp.niehs.nih.gov/whatwestudy/niceatm/test-method-evaluations/acute-systemic-tox/models/index.html?utm_source=direct&utm_medium=prod&utm_campaign=ntpgolinks&utm_term=tox-models [accessed 15 October 2020].
- O’Boyle NM, Morley C, Hutchison GR. 2008. Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit. Chem Cent J 2:5, PMID: 18328109, 10.1186/1752-153X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co-operation and Development). 2002a. Guidance Document on Acute Oral Toxicity Testing. Paris, France: OECD Publishing. [Google Scholar]
- OECD. 2002b. Test No. 420: Acute Oral Toxicity–Fixed Dose Procedure. OECD Guidelines for the Testing of Chemicals, Section 4. Paris, France: OECD Publishing. 10.1787/9789264070943-en [accessed 7 July 2020]. [DOI] [Google Scholar]
- OECD. 2002c. Test No. 423: Acute Oral toxicity–Acute Toxic Class Method. OECD Guidelines for the Testing of Chemicals, Section 4. Paris, France: OECD Publishing. 10.1787/9789264071001-en [accessed 7 July 2020]. [DOI] [Google Scholar]
- OECD. 2005. Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment. ENV/JM/MONO(2005)14. http://www.oecd.org/ [accessed 15 July 2020].
- OECD. 2007. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. ENV/JM/MONO(2007)2. http://search.oecd.org/officialdocuments/displaydocumentpdf/?cote=env/jm/mono(2007)2&doclanguage=en [accessed 15 July 2020].
- OECD. N. D. Validation of (Q)SAR Models. https://www.oecd.org/env/ehs/risk-assessment/validationofqsarmodels.htm [accessed 7 July 2020].
- OECD. 2008. Test No. 425: Acute Oral Toxicity: Up-and-Down Procedure. https://www.oecd-ilibrary.org/content/publication/9789264071049-en [accessed 15 July 2020].
- Ly Pham L, Watford S, Pradeep P, Martin MT, Thomas R, Judson R, et al. . 2020. Variability in in vivo studies: defining the upper limit of performance for predictions of systemic effect levels. Comput Toxicol 15(August 2020):1–100126, PMID: 33426408, 10.1016/j.comtox.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakers C, Najnin RA, Polash AH, Takeda S, Brown JB. 2018. Chemogenomic active learning’s domain of applicability on small, sparse qHTS matrices: a study using cytochrome P450 and nuclear hormone receptor families. Chem Med Chem 13(6):511–521, PMID: 29211346, 10.1002/cmdc.201700677. [DOI] [PubMed] [Google Scholar]
- Ramsundar B, Kearnes S, Riley P, Webster D, Konerding D, Pande V. 2015. Massively Multitask Networks for Drug Discovery. https://arxiv.org/abs/1502.02072v1 [accessed 15 July 2020].
- Reker D, Schneider P, Schneider G, Brown J. 2017. Active learning for computational chemogenomics. Future Med Chem 9(4):381–402, PMID: 28263088, 10.4155/fmc-2016-0197. [DOI] [PubMed] [Google Scholar]
- Richard AM, Williams CR. 2002. Distributed structure-searchable toxicity (DSSTox) public database network: a proposal. Mutat Res 499(1):27–52, PMID: 11804603, 10.1016/s0027-5107(01)00289-5. [DOI] [PubMed] [Google Scholar]
- Roberts G, Myatt GJ, Johnson WP, Cross KP, Blower PE. 2000. LeadScope: software for exploring large sets of screening data. J Chem Inf Comput Sci 40(6):1302–1314, PMID: 11128088, 10.1021/ci0000631. [DOI] [PubMed] [Google Scholar]
- Rogers D, Hahn M. 2010. Extended-connectivity fingerprints. J Chem Inf Model 50(5):742–754, PMID: 20426451, 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Royal Society of Chemistry. 2015. ChemSpider. http://www.chemspider.com/ [accessed 29 January 2015].
- Sahigara F, Mansouri K, Ballabio D, Mauri A, Consonni V, Todeschini R. 2012. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 17(5):4791–4810, PMID: 22534664, 10.3390/molecules17054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmina ES, Haider N, Tetko IV. 2015. Extended functional groups (EFG): an efficient set for chemical characterization and structure-activity relationship studies of chemical compounds. Molecules 21(1):1, PMID: 26703557, 10.3390/molecules21010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander T, Freyss J, von Korff M, Rufener C. 2015. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55(2):460–473, PMID: 25558886, 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- Schmidhuber J. 2015. Deep learning in neural networks: an overview. Neural Netw 61:85–117, PMID: 25462637, 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Strickland J, Clippinger AJ, Brown J, Allen D, Jacobs A, Matheson J, et al. . 2018. Status of acute systemic toxicity testing requirements and data uses by U.S. regulatory agencies. Regul Toxicol Pharmacol 94:183–196, PMID: 29408321, 10.1016/j.yrtph.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sushko I, Novotarskyi S, Körner R, Pandey AK, Rupp M, Teetz W, et al. . 2011. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J Comput Aided Mol Des 25(6):533–554, PMID: 21660515, 10.1007/s10822-011-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sushko I, Salmina E, Potemkin VA, Poda G, Tetko IV. 2012. ToxAlerts: a web server of structural alerts for toxic chemicals and compounds with potential adverse reactions. J Chem Inf Model 52(8):2310–2316, PMID: 22876798, 10.1021/ci300245q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetko IV. 2002. Neural network studies. 4. Introduction to associative neural networks. J Chem Inf Comput Sci 42(3):717–728, PMID: 12086534, 10.1021/ci010379o. [DOI] [PubMed] [Google Scholar]
- Tetko IV, Tanchuk VY. 2002. Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J Chem Inf Comput Sci 42(5):1136–1145, PMID: 12377001, 10.1021/ci025515j. [DOI] [PubMed] [Google Scholar]
- Todeschini R, Ballabio D, Cassotti M, Consonni V. 2015. N3 and BNN: two new similarity based classification methods in comparison with other classifiers. J Chem Inf Model 55(11):2365–2374, PMID: 26479827, 10.1021/acs.jcim.5b00326. [DOI] [PubMed] [Google Scholar]
- Todeschini R, Ballabio D, Grisoni F. 2016. Beware of unreliable Q2! a comparative study of regression metrics for predictivity assessment of QSAR models. J Chem Inf Model 56(10):1905–1913, PMID: 27633067, 10.1021/acs.jcim.6b00277. [DOI] [PubMed] [Google Scholar]
- Tong W, Hong H, Fang H, Xie Q, Perkins R. 2003. Decision Forest: combining the predictions of multiple independent decision tree models. J Chem Inf Comput Sci 43(2):525–531, PMID: 12653517, 10.1021/ci020058s. [DOI] [PubMed] [Google Scholar]
- Tripodi I, Cohen KB, Hunter LE. 2017. A semantic knowledge-base approach to drug-drug interaction discovery. 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) Kansas City, MO. 1123–1126.
- United Nations. 2015. Globally Harmonized System of Classification and Labelling of Chemicals: GHS. 6th rev. ed. New York, NY: United Nations. [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2015. EPI Suite. https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface [accessed 15 July 2020].
- U.S. EPA. 2016. Process for Establishing & Implementing Alternative Approaches to Traditional in Vivo Acute Toxicity Studies. https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/process-establishing-implementing-alternative [accessed 1 March 2021].
- U.S. EPA. 2020. New Approach Methods Work Plan. https://www.epa.gov/chemical-research/new-approach-methods-work-plan [accessed 14 October 2020].
- U.S. EPA-NCCT (U.S. Environmental Protection Agency–National Center for Computational Toxicology). 2014a. DSSTox. http://www.epa.gov/ncct/dsstox/ [accessed 25 November 2014].
- U.S. EPA-NCCT. 2014b. U.S. EPA’s Endocrine Disruptor Screening Program (EDSP) home page. http://www.epa.gov/endo/#universe [accessed 12 January 2015].
- U.S. EPA-NCCT. 2019. EPA TSCA: TSCA inventory, active non-confidential portion. https://comptox.epa.gov/dashboard/chemical_lists/tscaactivenonconf [accessed 21 May 2019].
- Varnek A, Fourches D, Hoonakker F, Solov’Ev VP. 2005. Substructural fragments: an universal language to encode reactions, molecular and supramolecular structures. J Comput Aided Mol Des 19(9–10):693–703, PMID: 16292611, 10.1007/s10822-005-9008-0. [DOI] [PubMed] [Google Scholar]
- Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, et al. . 2008. ISIDA–platform for virtual screening based on fragment and pharmacophoric descriptors. CAD 4(3):191–198, 10.2174/157340908785747465. [DOI] [Google Scholar]
- Vukovic K, Gadaleta D, Benfenati E. 2019. Methodology of aiQSAR: a group-specific approach to QSAR modelling. J Cheminform 11(1):27, PMID: 30945010, 10.1186/s13321-019-0350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlake AJ, Folia M, Piechota S, Allen TEH, Goodman JM, Gutsell S, et al. . 2020. Structural alerts and random Forest models in a consensus approach for receptor binding molecular initiating events. Chem Res Toxicol 33(2):388–401, PMID: 31850746, 10.1021/acs.chemrestox.9b00325. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, et al. . 2017. The CompTox chemistry dashboard: a community data resource for environmental chemistry. J Cheminform 9(1):61, PMID: 29185060, 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willighagen EL, Mayfield JW, Alvarsson J, Berg A, Carlsson L, Jeliazkova N, et al. . 2017. The chemistry development kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching. J Cheminform 9(1):33, PMID: 29086040, 10.1186/s13321-017-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Sjöström M, Eriksson L. 2001. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 58(2):109–130, 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- Worth AP, Bassan A, Gallegos A, Netzeva TI, Patlewicz G, Pavan M, et al. . 2005. The Characterisation of (Quantitative) Structure-Activity Relationships: Preliminary Guidance. Ispra, Italy: European Commission Institute for Health and Consumer Protection, Toxicology and Chemical Substances Unit. [Google Scholar]
- XGBoost. 2019. XGBoost Documentation. https://xgboost.readthedocs.io/en/latest/ [accessed 21 May 2019].
- Xie L, Thrippleton K, Irwin MA, Siemering GS, Mekebri A, Crane D, et al. . 2005. Evaluation of estrogenic activities of aquatic herbicides and surfactants using an rainbow trout vitellogenin assay. Toxicol Sci 87(2):391–398, PMID: 16049272, 10.1093/toxsci/kfi249. [DOI] [PubMed] [Google Scholar]
- Yang C, Cross K, Myatt GJ, Blower PE, Rathman JF. 2004. Building predictive models for protein tyrosine phosphatase 1B inhibitors based on discriminating structural features by reassembling medicinal chemistry building blocks. J Med Chem 47(24):5984–5994, PMID: 15537353, 10.1021/jm0497242. [DOI] [PubMed] [Google Scholar]
- Zakharov AV, Peach ML, Sitzmann M, Nicklaus MC. 2014. A new approach to radial basis function approximation and its application to QSAR. J Chem Inf Model 54(3):713–719, PMID: 24451033, 10.1021/ci400704f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Martin TM, Ye L, Sedykh A, Young DM, Tropsha A. 2009. Quantitative structure-activity relationship modeling of rat acute toxicity by oral exposure. Chem Res Toxicol 22(12):1913–1921, PMID: 19845371, 10.1021/tx900189p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


