Abstract
Naturally derived antimicrobial peptides have been an area of great interest because of high selectivity against bacterial targets over host cells and the limited development of bacterial resistance to these molecules throughout evolution. There are also a significant number of venom-derived peptides that exhibit antimicrobial activity in addition to activity against mammals or other organisms. Many venom peptides share the same net cationic, amphiphilic nature as host-defense peptides, making them an attractive target for development as potential antibacterial agents. The peptide ponericin L1 derived from Neoponera goeldii was used as a model to investigate the role of cationic residues and net charge on peptide activity. Using a combination of spectroscopic and microbiological approaches, the role of cationic residues and net charge on antibacterial activity, lipid bilayer interactions, and bilayer and membrane permeabilization were investigated. The L1 peptide and derivatives all showed enhanced binding to lipid vesicles containing anionic lipids, but still bound to zwitterionic vesicles. None of the derivatives were especially effective at permeabilizing lipid bilayers in model vesicles, in-tact Escherichia coli, or human red blood cells. Taken together the results indicate that the lack of facial amphiphilicity regarding charge segregation may impact the ability of the L1 peptides to effectively permeabilize bilayers despite effective binding. Additionally, increasing the net charge of the peptide by replacing the lone anionic residue with either Gln or Lys dramatically improved efficacy against several bacterial strains without increasing hemolytic activity.
Keywords: antimicrobial peptides, fluorescence, lipid binding, membrane permeabilization, venom peptides
1 |. INTRODUCTION
The widespread development of antimicrobial resistance (AMR) in bacteria has been identified and widely publicized as a major threat to global health in the coming decades.[1] The ability of numerous types of bacteria to rapidly develop resistance mechanisms to commercially available antibiotics has proven a major challenge in both clinical settings and in the research and development of new antibacterial agents. Thus, there is significant interest in the development of novel antibacterial molecules that are both therapeutically viable and exhibit reduced potential for resistance development.
Venoms are a rich source of biologically active peptides with a myriad of activities and targets including neurotoxins, membrane permeabilizing agents, and GPCR agonists and antagonists.[2] Often venoms contain a mixture of peptides or other biomolecules that act in concert to impair or kill the target.[3–5] Nonetheless, when isolated, the individual components can have significant potential as therapeutic agents.[6–9] Notably, a number of commercially available drugs such as captopril, eptifibatide, tirofiban, ziconotide, and exenatide all have origins in venoms from a variety of organisms.[2]
Pore forming venom peptides have been an area of significant study from both biological and biophysical perspectives. Some of the most well studied examples include melittin, mastoparan, and pardaxin.[10–14] The relative ease of solid-phase peptide synthesis coupled with the high aqueous solubility of these peptides have made them a model system for investigating the biophysical processes involved in peptide-bilayer binding interactions, pore-formation, and helix-formation at the bilayer water interface.[15–17] These synthetically accessible systems are also amenable to incorporation of fluorescent or other spectroscopic probes as well as nonnatural amino acids, further expanding the utility as a biophysical model system.[14,18–21] Among these, by far, melittin from Apis mellifera is the most widely studied and well understood with several high resolution structures solved in both solution and bound forms[22,23] as well as numerous molecular dynamics simulations of the peptide interaction with bilayers.[15,24–26]
Ponericins are a class of venom peptides originally derived from the predatory South American ponerine ant Neoponera goeldii (previously misclassified as Pachycondyla goeldii).[27] The ponericins fall into three classes based on amino acid sequence patterns and similarities: G, W, and L. All three classes are cationic, amphiphilic sequences and nearly all (12/15) contain a Trp residue in the N-terminal region of the peptide. The ponericin G and W families both contain a conserved Pro residue that lies near the middle of the sequence in 11 of 13 sequences (the other two sequences, G6 and G7, the Pro is near the C-terminus due to the shorter overall length). A number of groups have investigated the peptides from the G- and W-families of ponericins,[28–33] and other groups have investigated novel peptides that show similarity to the G- and W-families.[34–37] A comparison of ponericin sequences are shown in Supplemental Table S1.
The most poorly understood ponericins are those that belong to the L-family, ponericins L1 and L2. The only report of the activity and characteristics of these peptides are found in the original report by Orivel et al., and even in this study, only the L2 peptide was examined. Ponericin L2 exhibited broad spectrum antimicrobial activity, no detectable hemolytic activity and was shown to adopt a random coil conformation (possibly polyproline-II helix) in water. Ponericins L1 (SwissProt P82421) differs from L2 by only a single amino acid at position 9, with L1 containing a Met at this position while L2 contains an Ile. The L1 sequence is shown in Figure 1. Nonetheless, there is a general lack of understanding and investigation of the L-subfamily of ponericins and their mechanism
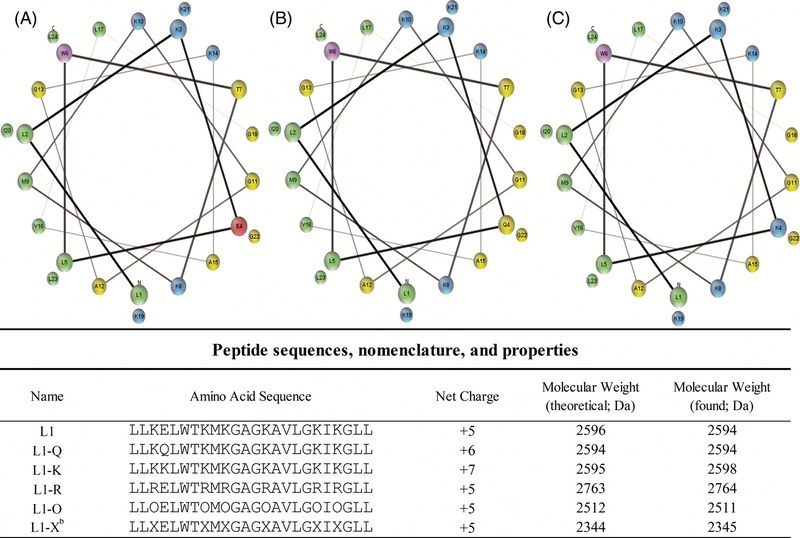
FIGURE 1.
Peptide sequences and helical wheel diagrams. Helical wheel diagram of L1, A, L1-Q, B, and L1-K, C. Cationic residues are shown in blue, anionic in red, hydrophobic in green, polar uncharged in yellow, and aromatic are in purple. Peptides sequences and relevant properties are shown below the helical wheels. Helical wheel representations were made using MPEx[50]
This work presents the first in-depth investigation into the characteristics and behavior of ponericin L1 and extends to an investigation of the role of cationic residues and overall charge on L1 activity. A series of peptides was synthesized in which either the net charge of the peptide or the identity of the cationic amino acids were varied. Using a series of well-characterized biophysical/spectroscopic measurements as well as microbiological and cellular assays, the activity of these L1 sequences was investigated.[14,17,38–44] Due to the inherent environmental sensitivity of Trp fluorescence emission, the native Trp in the L1 sequence (W6) was exploited as a spectroscopic probe for the oligomerization in solution, bilayer binding, and bilayer orientation of peptides. Antibacterial, bacterial membrane permeabilization, and hemolysis assays were also conducted to evaluate the activity of these sequences against natural biological membranes.
2 |. METHODS
2.1 |. Materials
Lipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; PC), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG; PG), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE; PE) were purchased from Avanti Polar Lipids (Alabaster, Alabama), and stored as stocks in chloroform at −20 °C. Lipids were prepared as 100% DOPC, 3:1 DOPC:DOPG (PC:PG), or 3:1 POPE:DOPG (PE:PG). Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Chem-Impex Int’l INC.), ortho-nitrophenyl-β-galactoside (ONPG) (Research Products International Co.), nitrocefin (Biovision, Milpitas, California). All other chemicals and reagents were from Thermo Fisher Scientific (Waltham, Massachusetts), VWR (Radnor, Pennsylvania), or Sigma-Aldrich (St. Louis, Missouri).
Peptides L1-Q, L1-K, and L1-O (ornithine) were synthesized using standard FMOC-chemistry in-house. Peptide L1-R was purchased from Genscript (Piscataway, New Jersey). The peptide L1-X (L1 with lysine replaced with di-amino-propionic acid, Dap) presented significant synthetic challenges and was thus purchased from Synthetic Proteomics (Carlsbad, California) along with the L1 parent sequence. All peptides were subsequently purified via RP-HPLC using a Zorbax C3 column and eluted by a linear gradient of water to acetonitrile (both supplemented with 0.1% TFA). Peptide identity was confirmed using ESI-MS. HPLC fractions containing peptide were pooled, lyophilized, and stored at −20 °C. Prior to use, peptides were dissolved in 3:1 H2O:ethanol to create a stock solution of 150 to 200 μM and stored at 4 °C.
Small unilamellar vesicles (SUVs) were formed by sonication or by ethanol dilution methods. In all cases, appropriate volumes of lipids in chloroform were mixed in a glass test tube, dried under a gentle stream of N2, and further dried under vacuum for at least 1 hour to create a lipid film in the tube. For the ethanol dilution vesicles, 10 μM of pure ethanol was added to the film, vortexed until the film was dissolved at which point the appropriate amount of PBS was added while vortexing. For sonicated vesicles, the PBS was added directly to the lipid film and vortexed to create multilamellar vesicles (MLVs). This MLV suspension was then subjected to sonication in a high-powered bath sonicator (Avanti Lipids) for 20 minutes to create SUVs.
2.2 |. Fluorescence spectroscopy
All fluorescence spectroscopy was performed on a JY-Horiba Fluoromax4 instrument with excitation and emission slit widths at 2.5 nm. Measurements were taken in semi-micro quartz cuvettes. Spectral barycenters were calculated using the following formula:
| 1 |
where B is the barycenter, λ is the wavelength, and i is the intensity at that wavelength. The summation is carried out over the entire emission spectrum collected, in this case 300 to 400 nm.
Lipid binding assays were carried out as performed previously.[44] Briefly, the samples contained 2 μM peptide in PBS buffer (150 mM NaCl, 50 mM Na2HPO4; pH 7.0) and were titrated with a lipid vesicle stock (0.5 or 1.0 mM concentration). All binding assays used excitation at 280 nm and an emission range of 300 to 400 nm. The spectra were corrected from the background and the dilution after each addition.
Red edge excitation shift (REES) experiments used excitation at 280, 290, 295, 300, 305, and 307 nm with an emission range of 310 to 410 nm. Again, samples contained 2 μM peptide in PBS buffer. The spectra were corrected from the background and the dilution after each addition.
Trichloroethanol (TCE) quenching assays were performed by titrating 10 μL aliquots of 10 M TCE (Alfa Aesar, Haverhill, Massachusetts) into samples containing 2 μM peptide in PBS (150 mM NaCl, 50 mM Na2HPO4; pH 7.0). The Trp fluorescence emission was recorded after each addition with excitation at 280 nm and emission at 340 nm. Data were corrected from background and dilution. The Stern-Volmer quenching constant (Ksv) was determined from the slope of a linear best fit equation to the data.
Acrylamide quenching was performed by titrating 10 μL aliquots of 4 M acrylamide and measuring Trp emission after each addition. Samples contained 2 μM peptide and 250 μM total lipid in PBS. The Trp fluorescence emission was recorded after each addition with excitation at 295 nm and emission at 340 nm. Data were corrected for background, dilution, and inner filter effects in the excitation path as described previously.[39] The Ksv was calculated as above.
Dual quencher analysis (DQA) experiments were performed as previously described.[39] Briefly, samples were prepared as above, with the exception that in one set of samples 10% of the lipid (molar basis) was replaced with the quencher 10-doxyl nonadecane (10-DN). Samples contained either no 10-DN (F0) or 20 μM 10-DN (Fdox). Similarly, peptide was added to a final concentration of 2 μM and allowed to equilibrate for 30 minutes. Fluorescence measurements were taken, and then 50 μL of acrylamide was added to each F0 sample, allowed to equilibrate, and remeasured. After inner filter correction (only for acrylamide containing samples) and background subtraction, the Q-ratio was calculated as previously described.[39]
2.3 |. Circular dichroism spectroscopy
Circular dichroism (CD) spectra were collected using Jasco J-810 spectropolarimeter. Samples contained 5 μM peptide in 0.1X PBS buffer or 0.1X PBS buffer with 50% trifluoroethanol (Alfa Aesar). Lipid containing samples consisted of 200 μM lipid vesicles with 3 μM peptide in 0.1X PBS buffer. Each sample had 64 scans performed with correction from background without peptide.
2.4 |. Minimal inhibitory concentration/minimal bactericidal concentration
Bacteria were streaked on LB-Miller agar plates from a frozen glycerol stock made from the original samples shipped from CGSC (Coli Genetic Stock Center, Yale University) or ATCC (American Type Culture Collection) (Escherichia coli D31 CGSC 5165,[45] Staphylococcus aureus ATCC 35556, Pseudomonas aeruginosa PA-01 ATCC 47085,[46] Acinetobacter baumannii ATCC 19606). An overnight was prepared using a single colony of each bacterial strain into fresh LB broth and placed into a shaking incubator 37 °C at 225 rpm for ~18 hours. After incubating overnight, a fresh 1:200 dilution was made in LB broth and used for the experiment. Minimum inhibitory concentration (MIC) was performed using mid-log phase bacteria diluted to 5 × 105 CFU/mL. Then, 90 μL of the diluted culture was added to the wells of a sterile 96-well plate containing serially diluted aliquots of each peptide for a total volume of 100 μL. The plate was incubated at 37 °C for 18 hours. After incubation, bacterial growth was determined by measuring OD600 using a Spectramax M5 multimode plate reader. Minimum bactericidal concentration (MBC) was performed by removing 1 μL of culture from each well of the MIC plate and plating on a fresh LB agar plate which was subsequently incubated overnight at 37 °C. MBC was determined by the growth or absence of growth from each well on the agar plate.
2.5 |. Lipid vesicle leakage
Lipid films were prepared as described above. A 75 mM stock of calcein was prepared in HEPES buffer, pH 7. The lipid film was resuspended in 700 μL of calcein solution for a final lipid concentration of 20 mM. The resuspended lipid vesicle sample was subjected to seven freeze-thaw cycles by alternating between a liquid N2 bath and a 42 °C water bath, culminating with a final thaw of the sample. The vesicles were then sonicated using a VCX 130 Probe sonicator (Sonics & Materials, Inc.) for 1 minute, using 1 second pulses at 45% intensity.
Vesicles were separated from unincorporated calcein by size exclusion chromatography using G25 Sephadex. The column was equilibrated in HBS (50 mM HEPES, 150 mM NaCl, pH 7) under gravity flow for approximately 1 hour before the vesicle solution was loaded onto the column. Upon loading, ~40 1 mL Fractions were then collected from the column. Fractions containing vesicles with entrapped calcein exhibited a cloudy, orange color and were separated from the unincorporated calcein, which has a clear yellow color. Vesicles were used on the same day as prepared. Vesicle concentrations/dilution factor were determined using a ratiometric assay with vesicles containing a known concentration of fluorescent lipid.[47]
The two most concentrated fractions were diluted 1:1 with HBS buffer. Solutions were dispensed into a 96 well plate in the subsequent order: 10 μL of peptide with serial dilutions starting at a final concentration from 15 μM (excluding the last row which had 10 μL of 0.01% Acetic Acid as a negative control), 20 μL of the diluted vesicle fractions, and 70 μL of PBS buffer (50 mM sodium phosphate, 150 mM sodium chloride pH 7) for a final lipid concentration of ~200 μM. The samples were excited at 495 nm and fluorescence emission was measured at 520 nm using a 510 nm cutoff filter in a Spectramax M5 (Molecular Devices) multimode plate reader.
Following the initial reading, 20 μL of Triton X-100 was added the last row as a positive control. The plate was placed on a plate shaker for 1 hour at 400 rpm, shielded from light. Calcein fluorescence emission was then remeasured and used as the 100% leakage normalization value. Data reported are the average of three replicates.
2.6 |. Inner membrane permeabilization
An overnight culture of E. coli D31 was prepared by inoculating a single colony into fresh LB broth and placed in a 37 °C shaking incubator (225 rpm) for ~18 hours. Following the overnight incubation, a 1:250 dilution of the culture to fresh LB broth was made. This was supplemented with 100 μL of 100 mM IPTG to induce the expression of β-galactosidase. The dilution was incubated with shaking until an OD600 of 0.2 to 0.4 was reached.
Solutions were dispensed into a 96 well plate in the subsequent order: 10 μL of peptide with serial dilutions starting a final concentration from 15 μM (excluding the last row which had 10 μL of 0.01% acetic acid as a negative control), 56 μL Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7), 19 μL of E. coli culture, and 15 μL of 4 mg/mL ONPG dissolved in Z-buffer. Immediately following the addition of ONPG, absorbance was recorded at 420 nm every 5 minutes for 90 minutes. The detergent cetyltrimethylammonium bromide was added in place of the peptide as a positive control. Data reported are the average of three replicates.
2.7 |. Outer membrane permeabilization
An overnight culture of E. coli D31 was prepared by inoculation a single colony into fresh LB broth with 100 μg/mL ampicillin (LB-Amp) and placed in a 37 °C shaking incubator (225 rpm) for ~18 hours. Following the overnight incubation, a 1:250 dilution of the culture to fresh LB-Amp was made. The dilution was incubated with shaking until an OD600 of 0.2 to 0.4 was reached. The bacterial culture was then centrifuged at 2500 rpm for 15 minutes in a tabletop clinical centrifuge. The supernatant was discarded and the pellet was resuspended in an equal amount of PBS (100 mm NaH2PO4, 200 mM NaCl, pH 7).
The experimental samples were prepared in a 96 well plate in the subsequent order: 10 μL of peptide with serial dilutions starting at a final concentration from 15 μM (excluding the last row which had 10 μL of 0.01% acetic acid as a negative control), 80 μL of the resuspended E. coli culture, and 10 μL of 50 μg/mL Nitrocefin in PBS. Immediately following the addition of nitrocefin, absorbance was recorded at 486 nm every 5 minutes for 90 minutes. The antimicrobial polymyxin B was used as a positive control. Data reported are the average of three replicates.
2.8 |. Hemolysis
Hemolysis of human red blood cells (RBCs) was used to quantify membrane destabilization by leakage of hemoglobin. Blood was collected in vacutainer tubes (10 mL) with EDTA coating as an anticoagulant (provided by the Rowan Department of Health and Exercise Science from a fresh draw ~1 hour prior to use, IBC protocol 2019–11). A 7 mL aliquot of whole human blood was mixed with 7 mL of sterile PBS. The solution was then sedimented via centrifugation for 7 minutes at 2500 rpm. The supernatant was removed and the pellet containing RBCs was resuspended to the original volume of 14 mL with PBS. This process was repeated three times. The final cell pellet was resuspended to a final volume of 14 mL and 90 μL of this cell suspension was added to all wells of a conical-bottom 96-well plate. Prior to addition of RBCs, 10 μL of serially diluted peptide or the detergent Triton-X 100 (as a positive control) were added to the wells. The covered plate was placed into a shaking incubator for 60 minutes at 37 °C while shaking (150 rpm). The conical plate was centrifuged for 10 minutes at 200 rpm and 4 °C. Subsequently, 6 μL of the supernatant was added to 94 μL of fresh PBS in wells of a flatbottom plate. Analysis of hemolysis was performed by measuring absorbance at 415 nm using a Molecular Devices Spectramax M5 plate reader. Percent hemolysis was calculated based on the absorbance of each well compared to those wells with no additive and those with Triton-X 100. All data reported are the average of three replicates.
2.9 |. Data analysis
Data analysis and visualization was aided by Daniel’s XL Toolbox addin for Excel, version 7.3.4, by Daniel Kraus, Würzburg, Germany (www.xltoolbox.net).
3 |. RESULTS
3.1 |. Peptides
The sequences of the original L1 sequence and the variants examined are listed in Figure 1, along with the peptide net charge, molecular mass. Regarding nomenclature, standard amino acid abbreviations are used except in the case of L1-O and L1-X which represent cationic substitutions of lysine with ornithine or di-amino-propionic acid, respectively. All peptides were synthesized using solid-phase FMOC chemistry methods and purified by HP LC. Helical wheel diagrams depicting the peptides and how the amino acids would orient along the helical axis are shown in Figure 1. In general, the L1 sequence does not adopt a canonical amphiphilic helix with a hydrophobic face and a cationic face. Instead, in a helical conformation, the L1 peptide has a hydrophilic face, but the cationics are flank that face with a number of polar, uncharged, and the lone anionic group interspaced on this face. Like L1, numerous venom peptides do not have a strictly cationic face, but instead a polar face with cationic and polaruncharged residues.[40,48,49]
3.2 |. Antimicrobial activity
The antimicrobial activity of the L1 and derivative peptides was examined using a broth microdilution assay to determine the MIC (Table 1). All variants of the peptide exhibited similar (fourfold range) of activity against E. coli, and all but L1-X showed similar activity against A. baumannii. Interestingly, the pattern of activity was more complex for S. aureus and P. aeruginosa. Compared to E. coli and A. baumannii, the parent L1 sequence showed weak activity against these species but L1R, L1-K, and L1-Q were more effective against S. aureus and L1-K and L1-Q also being active against P. aeruginosa. The peptide MBC, the concentration required to kill the bacteria in solution, was also measured by a plating assay after the MIC assay. In most cases where MBC could be determined, the MBC value was at or at most 2× greater than the MIC. These results indicate that the L1 peptides are likely acting by a bactericidal mechanism and not a bacteriostatic mechanism.
TABLE 1.
MIC and MBC (μM)
|
E. coli |
S. aureus |
P. aeruginosa |
A. baumannii |
|||||
|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| L1 | 0.94 | 0.94 | 15.00 | 15.00 | 15.00 | 15.00 | 1.88 | 1.88 |
| L1-Q | 0.94 | 0.94 | 1.88 | 1.88 | 7.50 | 7.50 | 0.94 | 1.88 |
| L1-K | 1.88 | 1.88 | 3.75 | 3.75 | 3.75 | 3.75 | 0.94 | 0.94 |
| L1-R | 0.94 | 0.94 | 1.88 | 1.88 | 15.00 | 15.00 | 0.94 | 1.88 |
| L1-O | 1.88 | 1.88 | >15 | >15 | 15.00 | 15.00 | 1.88 | 1.88 |
| L1-X | 3.75 | 7.50 | >15 | >15 | 15.00 | >15 | >15 | >15 |
Abbreviations: MIC, minimal inhibitory concentration; MPC, minimal bactericidal concentration.
3.3 |. Solution behavior
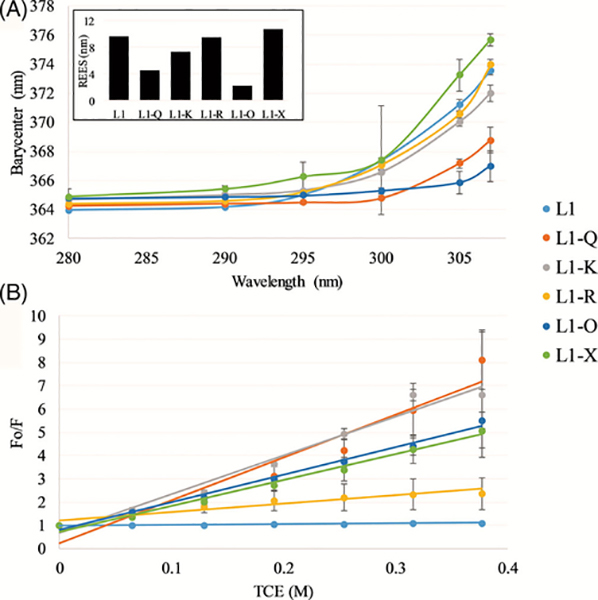
As with the majority of antimicrobial peptide (AMPs), the antimicrobial activity of the L1 peptide begins with the peptide in solution before interacting with the bacterial membrane. The behavior of the peptide in solution, and how sequence variation can affect these properties, can inherently affect the equilibrium between the free and bound states of the peptide. REES has been commonly used to indicate the mobility of the Trp fluorophore, and by extension yield insights into relative changes in peptide aggregation and/or oligomerization.[17,18] The REES results are shown in Figure 2A. All peptides display some extent of increase in the spectral barycenter as a function of excitation wavelength, indicating some level of restricted motion around the fluorophore (Trp). Of the sequences tested, L1-X, L1-R, L1-K, and L1 all showed significant shifts in the emission, between 7 and 11 nm at the highest excitation wavelength (inset Figure 2A). These red edge effects of this magnitude are strongly indicative of restricted motion around the Trp, likely resulting from peptide oligomerization or aggregation.
FIGURE 2.
Peptide solution assays—red edge excitation shift (REES), A, and trichloroethanol (TCE) quenching, B. The inset in panel A depicts the total shift for each peptide. In both panels, L1 is shown in light blue, L1-Q in orange, L1-K in gray, L1-R in yellow, L1-O in dark blue, and L1-X in green. All data are averages of three samples with correction from the background and error bars represent the SD
Quenching of Trp fluorescence by TCE was also used to investigate aggregation of the L1 peptides. TCE has been shown to partition to the interior of protein aggregates and quench Trp more efficiently at the interior of these aggregates. The Ksv, the Stern-Volmer quenching constant calculated from the slope of a linear fit to the quenching data, was used to analyze the quenching with higher Ksv values indicating increased quenching. The pattern of TCE quenching as determined by the Ksv (Figure 2B, Supplemental Table S2) is somewhat different from the REES results. L1-X and L1-K which showed significant REES were also quenched strongly by TCE. However, L1 and L1-R which showed significant REES were minimally quenched by TCE. This indicates potential differences in packing or organization of the L1 solution aggregation state as a function of sequence.
3.4 |. Interactions with lipid bilayers
The peptide interaction with lipid bilayers is a critical early step in the widely accepted membrane-permeabilization mechanism for AMPs. The affinity and orientation of membrane interacting peptides has been shown to be impacted by bilayer thickness, headgroup composition, pH, and other factors. As compared to traditional host defense peptides, L1 is a venom-derived peptide and thus there is no inherent evolutionary need for the peptide to preferentially interact with anionic bilayers, despite the net positive charge.
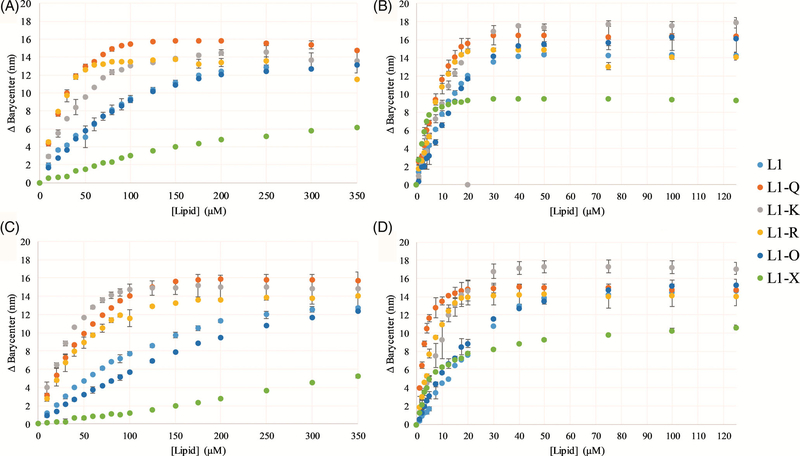
The affinity for lipid bilayers was investigated utilizing the environmental sensitivity of the naturally occurring Trp residue in the L1 sequence. The Trp fluorescence emission spectrum exhibits a shift from “red” to “blue” when the polarity of the local environment around the Trp shifts from more polar (i.e., in aqueous solution) to less polar (i.e., at the surface of the bilayer or imbedded in the bilayer core). The spectral shift is determined by the shift in the spectral barycenter, or center of mass, as a function of titrated lipid vesicles.[51] The data from these binding experiments are shown in Figure 3, with representative spectra of the free and bound peptides shown in Supplemental Figure S1. All of the peptides exhibited the ability to bind all lipid compositions tested (100% PC; 3:1 PC:PG; 3:1 PE:PG; and 7:3 PC:Cholesterol), but clearly bound with higher affinity to those bilayers containing anionic lipids (note the difference in X-axis scaling in Panels A and C vs B and D in Figure 3). The L1-X peptide, with the shortest side chain, exhibited notably lower total shift in barycenter when completely bound in all lipids, indicating this substitution impacts the depth of insertion in the bilayer. However, this did not seem to affect the general affinity for vesicles with anionic lipids.
FIGURE 3.
Binding to lipid vesicles—Peptide binding was assayed using Trp fluorescence emission by titrating SUVs into a sample containing 2 μM peptide. The spectral barycenter was calculated after each addition of lipid vesicle and the Δbarycenter is the difference between the initial barycenter and that after each addition. Binding was assayed with, A, PC, B, 3:1 PC:PG, C, 3:1 PC:Cholesterol, and, D, 3:1 PE:PG. In all panels, L1 is shown in light blue, L1-Q in orange, L1-K in gray, L1-R in yellow, L1-O in dark blue, and L1-X in green. Note the difference in X-axis scaling for panels A and C vs B and D. All data are averages of three replicates and the error bars represent the SD
To further investigate how the peptides interact with bilayers, a series of fluorescence quenching experiments were performed. First, acrylamide quenching was carried out on the peptides in solution and when bound to bilayers with various composition. Acrylamide is effective at quenching Trp residues that are exposed to the aqueous environment, but not those which are buried in the nonpolar core of the bilayer. Thus, the quenchability of the Trp is directly related to its exposure to the aqueous environment. The Stern-Volmer quenching constants for the L1 peptides quenched by acrylamide under different conditions are shown in Table 2, with the full quenching profiles shown in Supplemental Figure S2. In all cases, the peptides show a marked decrease in the Ksv between aqueous and any bilayer-interacting condition, supporting the binding results showing the peptides interact with all bilayers.
TABLE 2.
Fluorescence quenchinga
| Acrylamide quenching (Ksv [M −1]) |
Q-ratio |
|||||||
|---|---|---|---|---|---|---|---|---|
| PBS | DOPC | PC:PG | PC:Chol | PE:PG | DOPC | PC:PG | PE:PG | |
| L1 | 10.14 ± 1.15 | 2.49 ± 0.32 | 2.08 ± 0.27 | 3.18 ± 1.27 | 1.51 ± 0.08 | 3.46 | 1.29 | 0.15 |
| L1-Q | 15.43 ± 0.17 | 1.67 ± 0.19 | 1.23 ± 0.01 | 1.72 ± 0.16 | 1.63 ± 0.08 | 1.68 | 2.12 | 0.80 |
| L1-K | 16.48 ± 1.97 | 1.44 ± 0.13 | 1.07 ± 0.07 | 1.77 ± 0.37 | 1.86 ± 0.01 | 4.09 | 2.51 | 1.78 |
| L1-R | 18.90 ± 1.49 | 4.64 ± 0.65 | 2.20 ± 0.21 | 4.82 ± 0.81 | 1.46 ± 0.23 | 3.13 | 0.13 | 1.00 |
| L1-O | 22.68 ± 0.84 | 3.86 ± 0.09 | 2.96 ± 1.40 | 4.54 ± 1.14 | 2.47 ± 0.39 | 4.01 | 1.86 | 1.41 |
| L1-X | 19.69 ± 4.29 | 7.09 ± 2.38 | 4.28 ± 1.17 | 7.56 ± 0.20 | 4.17 ± 0.72 | 4.08 | 1.86 | 7.16 |
Abbreviations: DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPG, 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol).
Data are averages of three to six replicates with SD.
An extension of the acrylamide quenching assay is the DQA assay, which utilizes acrylamide and a second, membrane-imbedded quencher, 10-DN. The ratio of quenching between the two quenchers has been shown to be directly dependent on Trp depth in the bilayer, which can inform on the orientation in the bilayer or more qualitatively to identify orientation differences between different bilayer compositions.[39] In the DQA assay, the more exposed the Trp residue is to the aqueous environment, the higher the Q-ratio. The Q-ratios for the L1 peptides are shown in Table 2. Interestingly, the Q-ratio for all peptides in DOPC were similar, regardless of sequence or affinity, but lower values were observed for peptides in bilayers with anionic lipids except L1-X in PE:PG bilayers.
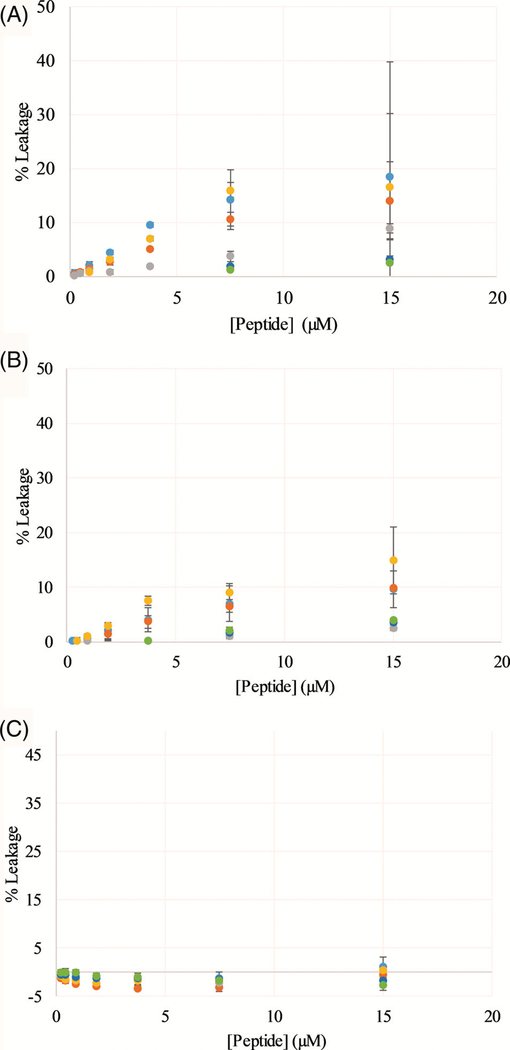
The ability of the peptides to permeabilize model lipid vesicles was also investigated using a fluorescent dye-release assay. The fluorophore calcein undergoes self-quenching when entrapped in the lumen of the vesicle and thus exhibits low fluorescence emission which increases dramatically upon bilayer permeabilization and release of the fluorophore into the aqueous milieu. Again, peptides were exposed to lipid vesicles composed of either PC, PC:PG, or PE: PG lipid mixtures. Percent leakage was calculated based on a subsequent treatment of the vesicles with the bilayer disrupting detergent Triton X-100. The leakage data are shown in Figure 4. In all lipid compositions, the L1 peptides did not induce leakage to a large extent, however at the highest concentrations tested several of the L1 sequences (L1, L1-R, and L1-Q) induced up to ~15% leakage in vesicles containing PC. Interestingly, none of the peptides induced any discernable leakage in vesicles composed of PE:PG lipids which are commonly found in bacterial membranes.
FIGURE 4.
Vesicle permeabilization assays—Peptide permeabilization of model lipid vesicles determined by dye leakage assays using calcein. Vesicles were composed of, A, PC, B, 3:1 PC:PG, C, 3:1 PE:PG. In all panels, data were normalized based on untreated vesicles (0% leakage) and vesicles treated with the detergent Triton X-100 (100% leakage). L1 is shown in light blue, L1-Q in orange, L1-K in gray, L1-R in yellow, L1-O in dark blue, and L1-X in green. All data are averages of at least three replicates and the error bars represent the SD
3.5 |. Interactions with cellular membranes
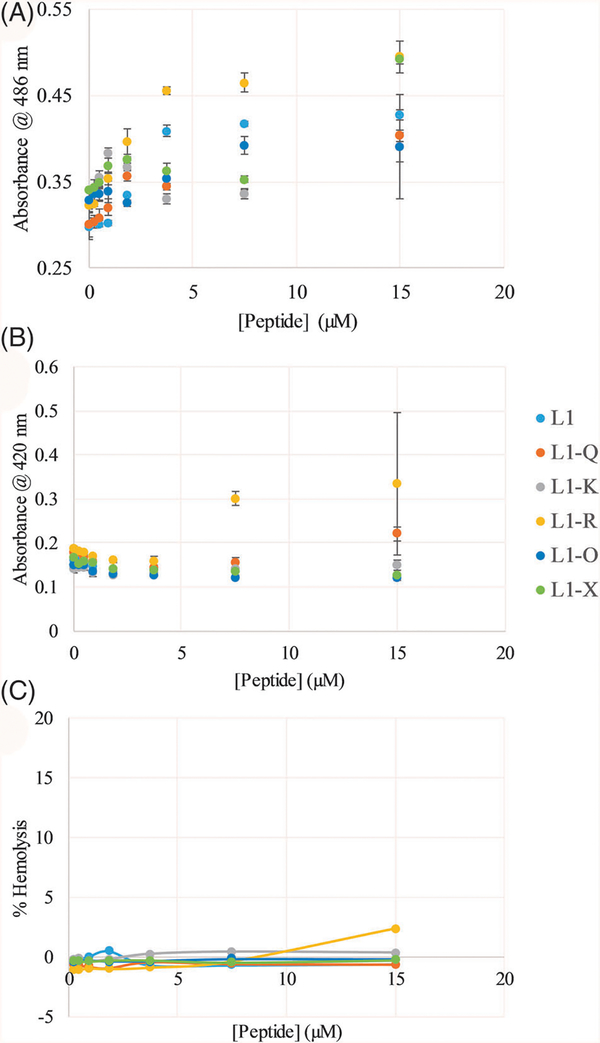
Peptide interactions with model lipid vesicles is informative; however, model vesicles only replicate a small fraction of the complexity of natural lipid membranes which inherently contain greater lipid diversity, lipid asymmetry, and integral membrane proteins. Thus, the L1 peptides were investigated for the ability to permeabilize natural bacterial and mammalian membranes. Using chromogenic substrates of enzymes localized in the E. coli periplasmic space (nitrocefin and β-lactamase) or cytoplasm (ONPG and β-galactosidase), the peptide effects on the permeability of the bacterial outer or inner membrane can be investigated. Outer membrane permeability is shown in Figure 5A while inner membrane permeability is shown in Figure 5B. Notably, these data represent snapshots of leakage after 30 minutes of exposure to the peptides, but full 90-minute time course data is shown in Supplemental Figures S3 and S4. All peptides exhibited the ability to disrupt the outer membrane in a concentration dependent manner, although L1-X only did so at the highest concentration tested (Figure 5A). Conversely, none of the peptides induced any significant permeabilization of the E. coli inner membrane other than L1-R at the highest concentrations tested (Figure 5B).
FIGURE 5.
Membrane permeabilization of E. coli and hemolysis of human red blood cells (RBCs)—E. coli, A, outer membrane and, B, inner membrane permeabilization, and, C, hemolysis of RBCs. Data in, C, were normalized based on untreated RBCs (0% hemolysis) and RBCs treated with the detergent Triton X-100 (100% hemolysis). L1 is shown in light blue, L1-Q in orange, L1-K in gray, L1-R in yellow, L1-O in dark blue, and L1-X in green. All data are averages of at least three replicates and error bars represent the SD. In some cases, error bars are smaller than the size of the symbols
Bacterial membrane permeabilization may be linked to antimicrobial activity; the L1 peptides are venom components, many of which have been shown to also permeabilize mammalian membranes. The ability to disrupt human RBC membranes was investigated using a hemolysis assay monitoring the release of hemoglobin after exposure to the peptides for 1 hour. Percent leakage was calculated based on a subsequent treatment of the RBCs with the bilayer disrupting detergent Triton X-100. The results are shown in Figure 5C, with none of the L1 peptides inducing any significant disruption of RBCs, consistent with the previously published results on the related L2 peptide.[27]
3.6 |. Peptide secondary structure
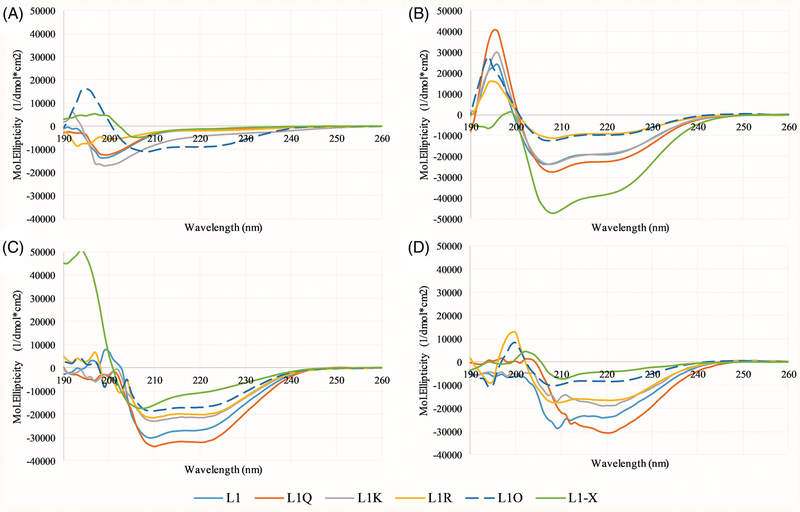
The changes in amino acid sequence can inherently impact the structure that the peptides adopt in solution or when bound to membranes. Due to their small size, the L1 peptides are unlikely to adopt any tertiary structures, so secondary structure analysis by CD spectroscopy provides a good picture of the overall peptide structure. CD spectra of the L1 peptides in phosphate buffer alone or in buffer with 3:1 PC:PG vesicles are shown in Figure 6. In aqueous solution, all peptides exhibit CD spectra consistent with random / disordered structures except L1-O which exhibits a helical signature. When bound to lipid vesicles, all variants exhibited spectra indicative of α-helix formation.
FIGURE 6.
Circular dichroism (CD) spectroscopy—Spectra of all peptides were collected in, A, 10X diluted PBS, B, 50:50 PBS:TFE using the 10X diluted PBS, C, in the presence of 250 μM 3:1 PC:PG vesicles, or, D, in the presence of 250 μM 3:1 PC:Cholesterol vesicles. In 10X diluted PBS and 50:50 PBS:TFE, the peptide concentration was 5 μM, while in the presence of lipid vesicles, the peptide concentration was 3 μM. L1 is shown in light blue, L1-Q in orange, L1-K in gray, L1-R in yellow, L1-O in dark blue, and L1-X in green. All data represent the average of 64 scans with correction of background spectra lacking peptide
4 |. DISCUSSION
When analyzing the context of the data, it is important to remember that the L1 peptide is derived from venom of an omnivorous species of ant which forages for both plants and insects as food.[52] Thus, the primary function of the venom peptides is not evolved for antimicrobial activity, but instead for offensive or defensive purposes.[27,52] Additionally, venoms often contain multiple components which can act synergistically or act on different and unrelated targets.[3–5] This can be further complicated by variations in the diet and habitat of the insect as well as the season.[5,53]
4.1 |. Net charge
One of the goals of the current study was to investigate the role of net charge on AMP properties. Although the majority of AMPs that have been studied have a net positive charge, many, including L1, have anionic amino acids in the sequence. If net positive charge is a driving force for antimicrobial activity, evolutionarily there would be no selective pressure to retain these anionic amino acids in the sequence. However, this evolutionary pressure is not as clear in venom peptides, which can also act as antimicrobials.
In solution, the change of net charge from +5 to +6 or +7 did not have a significant impact on the REES results but were dramatically different for TCE quenching profiles. As the REES and TCE assays report on different aspects of Trp behavior (mobility and accessibility, respectively), the differences are potentially rooted in structural or aggregation state changes in the peptide in solution.[17,18,54] The CD spectra in Figure 6 indicate these three peptides (L1, L1-Q, L1-K) are not helical when in solution alone. However, the amino acid substitutions to change the net charge of the peptide are all at position 4, while the native Trp is at position 6 (Figure 1). These substitutions inherently likely do not impact the rotational freedom of the Trp, but may impact the local environment regarding polarity and/or the local packing of any aggregated form. Previous studies have shown that local interaction between anionic residues and Trp can impact spectral position and shifts,[55,56] as well as peptide-peptide interactions.[57–59]
The effect of increasing L1 net charge had marked effects on the antimicrobial efficacy against S. aureus and P. aeruginosa (Table 1). Increasing the charge from +5 to +6 resulted in an eightfold enhancement of activity against S. aureus and a twofold enhancement against P. aeruginosa while further increase of net charge to +7 further twofold increased the efficacy against P. aeruginosa. Notably, these changes also enhanced activity against E. coli and A. baumannii, but the parent L1 was already quite effective against those strains. This is consistent with previous reports that demonstrate cationic charge and net charge are important determinants of antimicrobial activity in AMPs.[41,54,60,61] These changes, however, do not appear to be driven by increased ability to permeabilize vesicles or native membranes. While increasing the net charge did result in moderate enhancements to lipid binding, this enhancement was not dependent on bilayer charge. These results may point to a nonpermeabilizing role of L1 peptides in antimicrobial activity.
Additionally, altering the net charge on the peptide can alter the hydrophobic/hydrophilic balance of the sequence. Numerous studies on AMPs and peptidomimetic polymers have shown that this balance plays a key role in antimicrobial activity, cytotoxicity, and interactions with lipid bilayers.[41,43,49,62] In the peptides presented here, the overall hydrophobic balance is not significantly impacted as E→Q→K changes at position 4 are all polar. Notably, Gln is an uncharged polar residue which is naturally less polar than Lys, but does not result in major differences in lipid bilayer binding or antimicrobial activity. As such, any small changes in the hydrophobic/hydrophilic balance, if at all, are swamped by the change in net charge.
4.2 |. Cationic side chain identity
While the net charge of the peptide does appear to play some role in activity and may have strain-specific influences, previous studies on peptides and peptidomimetic polymers have shown that not all cationic groups behave identically in AMPs. Side chain length and cationic moiety identity have both been shown to dramatically impact characteristics of antimicrobials.[51,63–66] Additionally the well-studied TAT sequence, originally derived from HIV, has shown a necessity for Arg residues to function in membrane translocation.[67,68] In the L1 background, switching Lys to Arg had minimal effects on MIC and MBC for all strains tested, except P. aeruginosa which had a marked decrease in efficacy. Interestingly, the Lys to Arg substitution resulted in a twofold decrease in MIC (increased efficacy) against E. coli, consistent with several Trp-rich sequences described by Arias et al.[63] A number of studies have shown that Arg substitutions for Lys would enhance antimicrobial activity,[69,70] likely from an increased H-bonding network and ability to interact with more lipid molecules for each cationic moiety.[71]
Side chain length also clearly plays a role in activity and characteristics of the L1 peptides. Reducing the side chain length by one methyl group (Lys to Orn) had little effect on efficacy or binding, but reduction by three methyl groups (Lys to Dap) dramatically increased the MIC values. This is consistent with previous results on AMPs and peptidomimetic polymers.[49,51,64,65] However, a recent report by Hodges and coworkers has shown the inclusion of Dap in AMPs can cause a small reduction of antibacterial activity but can have a significant reduction in hemolytic activity, improving the overall therapeutic potential.[72] The loss of activity from shortening the length of the cationic side chain has been attributed to peptide penetration depth in the bilayer, the Dap side chain forcing a shallower, less disruptive conformation.[51,64] The DQA results indicate that the L1-X does appear to adopt a shallower orientation in PE:PG membranes (model of bacterial membranes) but shows less of a difference compared to the L1-K in PC or PC:PG membranes. This is consistent with the best performing sequences against E. coli also exhibiting the greatest OM disruption and lowest Q-ratio in PE:PG membranes.
5 |. CONCLUSIONS
The data show that the L1 peptide is a membrane-binding peptide that can adopt an α-helical conformation when bound to lipid bilayers. Both the net charge on the peptide and the identity of the cationic groups play a significant role in activity, with increasing net charge from +5 to +7 improving antimicrobial activity and shortening cationic side chains decreasing antimicrobial activity. The peptides clearly adopt different conformations in zwitterionic and anionic vesicles which somewhat correlates with antimicrobial activity; however, E. coli OM disruption correlated the best with MIC. Taken together, the venom peptide L1 has good potential as a selective antimicrobial platform due to tunable activity and low hemolytic activity. Future studies on the hydrophobic/hydrophilic balance and the role of the polar face of the peptide should provide important fundamental understanding and allow the design of more effective and selective antimicrobial molecules.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this work was provided in part by NIH R15 GM094330 to G. A. C.. A. S. S. was supported by a Rowan University Graduate Research Fellowship. M. R. N. was supported in part by the Summer Undergraduate Research Program at Rowan University.
Funding information
National Institute of General Medical Sciences, Grant/Award Number: R15 GM094330; Summer Undergraduate Research Program; Rowan University Graduate Research Fellowship
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interests.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- [1].World Health Organization, Food and Agriculture Organization of the United Nations and World Organisation for Animal Health, Monitoring Global Progress on Addressing Antimicrobial Resistance (AMR) 2018, p. 66. https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;jsessionid=77040347A0CAD29A244CEB9453A7BC75?sequence=1
- [2].Pennington MW, Czerwinski A, Norton RS, Bioorg. Med. Chem 2018, 26, 2738. [DOI] [PubMed] [Google Scholar]
- [3].Ceolin Mariano DO, de Oliveira UC, Zaharenko AJ, Pimenta DC, Radis-Baptista G, Prieto-da-Silva ARB, Toxins 2019, 11, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Amorim FG, Longhim HT, Cologna CT, Degueldre M, Pauw E, Quinton L, Arantes EC, J. Venom. Anim. Toxins Incl. Trop. Dis 2019, 25, e148218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Santos PP, Games PD, Azevedo DO, Barros E, de Oliveira LL, de Oliveira Ramos HJ, Baracat-Pereira MC, Serrao JE, Arch. Insect Biochem. Physiol 2017, 96, e21424. [DOI] [PubMed] [Google Scholar]
- [6].Chaves-Moreira D, Matsubara FH, Schemczssen-Graeff Z, De Bona E, Heidemann VR, Guerra-Duarte C, Gremski LH, Chavez-Olortegui C, Senff-Ribeiro A, Chaim OM, Arni RK, Veiga SS, Toxins 2019, 11, pii: E355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Undheim EA, Jenner RA, King GF, Expert Opin. Drug Discov 2016, 11, 1139. [DOI] [PubMed] [Google Scholar]
- [8].Chatterjee B, Curr. Top. Med. Chem 2018, 18, 2555. [DOI] [PubMed] [Google Scholar]
- [9].Rivera-de-Torre E, Palacios-Ortega J, Gavilanes JG, Martinez-Del-Pozo A, Garcia-Linares S, Toxins 2019, 11, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shai Y, Fox J, Caratsch C, Shih YL, Edwards C, Lazarovici P, FEBS Lett 1988, 242, 161. [DOI] [PubMed] [Google Scholar]
- [11].Tang J, Signarvic RS, DeGrado WF, Gai F, Biochemistry 2007, 46, 13856. [DOI] [PubMed] [Google Scholar]
- [12].DeGrado WF, Musso GF, Lieber M, Kaiser ET, Kezdy FJ, Biophys. J 1982, 37, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shai Y, Toxicology 1994, 87, 109. [DOI] [PubMed] [Google Scholar]
- [14].Ridgway Z, Picciano AL, Gosavi PM, Moroz YS, Angevine CE, Chavis AE, Reiner JE, Korendovych IV, Caputo GA, Biopolymers 2015, 104, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen CH, Wiedman G, Khan A, Ulmschneider MB, Biochim. Biophys. Acta 2014, 1838, 2243. [DOI] [PubMed] [Google Scholar]
- [16].Hristova K, Dempsey CE, White SH, Biophys. J 2001, 80, 801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Raghuraman H, Chattopadhyay A, Biopolymers 2006, 83, 111. [DOI] [PubMed] [Google Scholar]
- [18].Haldar S, Raghuraman H, Chattopadhyay A, J. Phys. Chem. B 2008, 112, 14075. [DOI] [PubMed] [Google Scholar]
- [19].Runnels LW, Scarlata SF, Biophys. J 1995, 69, 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jamasbi E, Ciccotosto GD, Tailhades J, Robins-Browne RM, Ugalde CL, Sharples RA, Patil N, Wade JD, Hossain MA, Separovic F, Biochim. Biophys. Acta 2015, 1848, 2031. [DOI] [PubMed] [Google Scholar]
- [21].Moroz YS, Binder W, Nygren P, Caputo GA, Korendovych IV, Chem. Commun 2013, 49, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hewish DR, Barnham KJ, Werkmeister JA, Kirkpatrick A, Bartone N, Liu ST, Norton RS, Curtain C, Rivetta DE, Protein Chem J. 2002, 21, 243. [DOI] [PubMed] [Google Scholar]
- [23].Ramirez LS, Pande J, Shekhtman A, J. Phys. Chem. B 2019, 123, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Leveritt III JM., Pino-Angeles A, Lazaridis T, Biophys. J 2015, 108, 2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun D, Forsman J, Woodward CE, J. Phys. Chem. B 2017, 121, 10209. [DOI] [PubMed] [Google Scholar]
- [26].Woo SY, Lee H, Phys. Chem. Chem. Phys 2017, 19, 7195. [DOI] [PubMed] [Google Scholar]
- [27].Orivel J, Redeker V, Le Caer JP, Krier F, Revol-Junelles AM, Longeon A, Chaffotte A, Dejean A, Rossier J, J. Biol. Chem 2001, 276, 17823. [DOI] [PubMed] [Google Scholar]
- [28].Garcia F, Villegas E, Espino-Solis GP, Rodriguez A, Paniagua-Solis JF, Sandoval-Lopez G, Possani LD, Corzo G, J. Antibiot 2012, 66, 3. [DOI] [PubMed] [Google Scholar]
- [29].Nolasco M, Biondi I, Pimenta DC, Branco A, Toxicon 2018, 150, 74. [DOI] [PubMed] [Google Scholar]
- [30].Rodriguez A, Villegas E, Montoya-Rosales A, Rivas-Santiago B, Corzo G, PLoS One 2014, 9, e101742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shukla A, Fleming KE, Chuang HF, Chau TM, Loose CR, Stephanopoulos GN, Hammond PT, Biomaterials 2010, 31, 2348. [DOI] [PubMed] [Google Scholar]
- [32].Zhang H, Jiao J, Jin H, Artif. Cells Nanomed. Biotechnol. 2019, 47, 2391. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Y, Zhang L, Li B, Han Y, ACS Appl. Mater. Interfaces 2017, 9, 9449. [DOI] [PubMed] [Google Scholar]
- [34].Graham C, Richter SC, McClean S, O’Kane E, Flatt PR, Shaw C, Peptides 2006, 27, 1313. [DOI] [PubMed] [Google Scholar]
- [35].Johnson SR, Copello JA, Evans MS, Suarez AV, Toxicon 2010, 55, 702. [DOI] [PubMed] [Google Scholar]
- [36].Kozic M, Fox SJ, Thomas JM, Verma CS, Rigden DJ, Proteins 2018, 86, 548. [DOI] [PubMed] [Google Scholar]
- [37].Pluzhnikov KA, Kozlov SA, Vassilevski AA, Vorontsova OV, Feofanov AV, Grishin EV, Biochimie 2014, 107(Pt B), 211. [DOI] [PubMed] [Google Scholar]
- [38].Caputo GA, Methods Mol. Biol 2013, 1063, 95. [DOI] [PubMed] [Google Scholar]
- [39].Caputo GA, London E, Methods Mol. Biol 2019, 2003, 351. [DOI] [PubMed] [Google Scholar]
- [40].Chrom CL, Renn LM, Caputo GA, Antibiotics 2019, 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kuroda K, Caputo GA, DeGrado WF, Chemistry 2009, 15, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mensa B, Kim YH, Choi S, Scott R, Caputo GA, DeGrado WF, Antimicrob. Agents Chemother 2011, 55, 5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Saint Jean KD, Henderson KD, Chrom CL, Abiuso LE, Renn LM, Caputo GA, Probiotics Antimicro. Proteins 2018, 10, 408. [DOI] [PubMed] [Google Scholar]
- [44].Shirley DJ, Chrom CL, Richards EA, Carone BR, Caputo GA, Pept. Sci 2018, 110, pii: e24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Burman LG, Nordstrom K, Boman HG, Bacteriol J. 1968, 96, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Capilato JN, Philippi SV, Reardon T, McConnell A, Oliver DC, Warren A, Adams JS, Wu C, Perez LJ, Bioorg. Med. Chem 2017, 25, 153. [DOI] [PubMed] [Google Scholar]
- [47].Caputo GA, London E, Biochemistry 2003, 42, 3265. [DOI] [PubMed] [Google Scholar]
- [48].Wu Y, Han M-F, Liu C, Liu T-Y, Feng Y-F, Zou Y, Li B, Liao H-L, RSC Adv 2017, 7, 17514. [Google Scholar]
- [49].Zelezetsky I, Tossi A, Biochim. Biophys. Acta 2006, 1758, 1436. [DOI] [PubMed] [Google Scholar]
- [50].Snider C, Jayasinghe S, Hristova K, White SH, Protein Sci 2009, 18, 2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kohn EM, Shirley DJ, Arotsky L, Picciano AM, Ridgway Z, Urban MW, Carone BR, Caputo GA, Molecules 2018, 23, pii: E329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schmidt CA, Shattuck SO, Zootaxa 2014, 3817, 1. [DOI] [PubMed] [Google Scholar]
- [53].Cologna CT, Rodrigues RS, Santos J, de Pauw E, Arantes EC, Quinton L, J. Venom. Anim. Toxins Incl. Trop. Dis 2018, 24, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hitchner MA, Santiago-Ortiz LE, Necelis MR, Shirley DJ, Palmer TJ, Tarnawsky KE, Vaden TD, Caputo GA, Biochim. Biophys. Acta Biomembr 2019, 1861, 182984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Caputo GA, London E, Biochemistry 2004, 43, 8794. [DOI] [PubMed] [Google Scholar]
- [56].Jones JD, Gierasch LM, Biophys. J 1994, 67, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lew S, Caputo GA, London E, Biochemistry 2003, 42, 10833. [DOI] [PubMed] [Google Scholar]
- [58].Rajagopal K, Lamm MS, Haines-Butterick LA, Pochan DJ, Schneider JP, Biomacromolecules 2009, 10, 2619. [DOI] [PubMed] [Google Scholar]
- [59].Wang Y, Truex NL, Vo NDP, Nowick JS, Bioorg. Med. Chem 2018, 26, 1151. [DOI] [PubMed] [Google Scholar]
- [60].Jiang Z, Vasil AI, Hale JD, Hancock RE, Vasil ML, Hodges RS, Biopolymers 2008, 90, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rosenfeld Y, Lev N, Shai Y, Biochemistry 2010, 49, 853. [DOI] [PubMed] [Google Scholar]
- [62].Avery CW, Palermo EF, McLaughlin A, Kuroda K, Chen Z, Anal. Chem. 2011, 83, 1342. [DOI] [PubMed] [Google Scholar]
- [63].Arias M, Piga KB, Hyndman ME, Vogel HJ, Biomolecules 2018, 8, pii: E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Palermo EF, Kuroda K, Biomacromolecules 2009, 10, 1416. [DOI] [PubMed] [Google Scholar]
- [65].Palermo EF, Kuroda K, Appl. Microbiol. Biotechnol 2010, 87, 1605. [DOI] [PubMed] [Google Scholar]
- [66].Choi S, Isaacs A, Clements D, Liu D, Kim H, Scott RW, Winkler JD, DeGrado WF, Proc. Natl. Acad. Sci. U. S. A 2009, 106, 6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B, J. Biol. Chem 2003, 278, 585. [DOI] [PubMed] [Google Scholar]
- [68].Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y, J. Biol. Chem 2001, 276, 5836. [DOI] [PubMed] [Google Scholar]
- [69].Nguyen LT, de Boer L, Zaat SA, Vogel HJ, Biochim. Biophys. Acta 2011, 1808, 2297. [DOI] [PubMed] [Google Scholar]
- [70].Schmidt NW, Wong GC, Curr. Opin. Solid State Mater. Sci 2013, 17, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Li L, Vorobyov I, Allen TW, J. Phys. Chem. B 2013, 117, 11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mant CT, Jiang Z, Gera L, Davis T, Nelson KL, Bevers S, Hodges RS, J. Med. Chem 2019, 62, 3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.