Abstract
The EphA4 receptor tyrosine kinase is well-known for its pivotal role in development, cancer progression, and neurological disorders. However, how EphA4 kinase activity is regulated in time and space still remains unclear. To visualize EphA4 activity in different membrane microdomains, we developed a sensitive EphA4 biosensor based on Förster resonance energy transfer (FRET), and targeted it in or outside raft-like microdomains in the plasma membrane. We showed that our biosensor can produce a robust and specific FRET response upon EphA4 activation, both in vitro and in live cells. Interestingly, we observed stronger FRET responses for the non-raft targeting biosensor than for the raft targeting biosensor, suggesting that stronger EphA4 activation may occur in non-raft regions. Further investigations revealed the importance of the actin cytoskeleton in suppressing EphA4 activity in raft-like microdomains. Therefore, our FRET-based EphA4 biosensor could serve as a powerful tool to visualize and investigate EphA4 activation and signaling in specific subcellular compartments of single live cells.
Keywords: EphA4, FRET biosensor, membrane microdomain, cytoskeleton, live cell imaging
Graphical Abstract
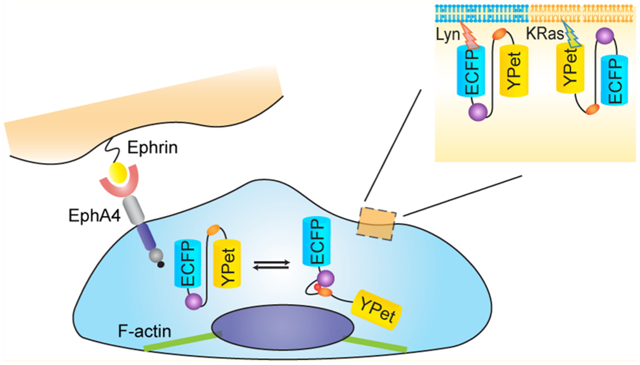
EphA4 belongs to the Eph receptor family, which is the largest family of tyrosine kinase receptors. The Eph receptors are subdivided into two classes, EphAs and EphBs, according to sequence similarities and ligand binding preferences.1 Eph receptors bind to another family of membrane tethered proteins, the ephrin ligands, which include the GPI-linked ephrinAs and the transmembrane ephrinBs. As a general rule, EphAs bind to ephrinA ligands, while EphBs bind preferentially to ephrinBs. EphA4 is an exception, since it binds to both ephrinA and ephrinB ligands.2 Structurally, all Eph receptors share a conserved domain composition. The extracellular region contains the N-terminal ephrin-binding domain, a cysteine-rich portion, and two fibronectin type III repeats. The intracellular region includes a membrane-spanning juxtamembrane (JM) segment, a tyrosine kinase domain, a sterile-α motif (SAM) domain, and a C-terminal Psd-95, Dlg, and ZO1 (PDZ)-binding motif.3
Eph receptors were initially found to guide growing neuronal processes during development through repulsive effects.4 They have also been implicated in other cell communication processes, including angiogenesis, cell morphogenesis, cell migration, tissue patterning, and neuronal plasticity.2 For example, in the nervous system EphA4 is well-known as a classic guidance receptor that promotes axon growth cone collapse5 and also regulates synaptic plasticity.2 Recent studies have also revealed important roles of EphA4 in many types of human cancers,6,7 such as breast8 and lung,9 and glioblastoma,10 by regulating cell migration and invasion.9–11 To mediate such a variety of functions, the activation and signaling of the EphA4 kinase domain require a highly coordinated regulation in space and time. Upon ligand dependent activation, Eph receptors form higher order clusters,12,13 which are critical for its signaling function.5 However, how EphA4 activation is regulated in different microdomains of the plasma membrane in live cells remains unclear.
The cell plasma membrane has long been proposed to be more mosaic than fluid.14 It is composed of discrete microdomains with unique physical and biological properties. Raft-like domains are small nanosize compartments enriched with cholesterol, sphingomyelin, and saturated fatty acids.15 These microcompartments are involved in intracellular trafficking and signaling.16 Because of their detergent-resistant property, raft-like structures were mainly studied by the detergent extraction method.17 EphA4 was reported to be present in both raft-like and non-raft regions by this method.18 However, the detergent extraction method is disruptive to membrane structure, and protein contamination among different fractions during the extraction process is likely to happen.19 Furthermore, how EphA4 activity is dynamically regulated at the membrane compartments also remains unclear. Therefore, the development of more advanced methods is needed to study EphA4 regulation at different subcompartments of the plasma membrane in live cells.
One of the first events in Eph receptor activation is the autophosphorylation of two conserved JM tyrosine residues, including Tyr596 in EphA4. In this study, we have developed a novel EphA4 biosensor to specifically detect phosphorylation of Tyr596 and its subsequent binding to the Src SH2 domain. An improved FRET pair, enhanced CFP (ECFP) and YPet, was utilized to enhance the sensitivity of the biosensor.20 To monitor EphA4 activity in different microdomains of the plasma membrane, we targeted the EphA4 biosensor either in or outside raft-like regions through lipid modifications, including acylation and prenylation, respectively.21 Using this strategy, this biosensor can be directed to tether in different compartments of the plasma membrane, where local EphA4 activity can be monitored in real time. Our results revealed that EphA4 activation induced by ephrin ligands is differentially regulated in different subdomains of the plasma membrane. Further results indicate that the cytoskeleton plays an important role in regulating differential EphA4 activation at the plasma membrane.
RESULTS
Design of an EphA4 Biosensor and Its Characterization in Vitro.
We developed a FRET-based EphA4 biosensor, with a central segment containing the SH2 domain of c-Src, a flexible linker peptide, and a specific substrate sequence, concatenated between ECFP and YPet (Figure 1A,B). We tested 3 candidate substrate sequences,22 including two substrates derived from EphA4 juxtamembrane (JM) domain and one substrate from EphB2 JM region (see Materials and Methods). Due to its overall efficiency and specificity among the substrate sequences tested, the EphA4 JM region encompassing Tyr596 was selected as our biosensor substrate (Figure S1). In our design, the active EphA4 kinase can phosphorylate Tyr596 in the substrate peptide, which subsequently binds intramolecularly to the SH2 domain, causing a conformational change. This conformational change alters the distance or relative orientation between cyan and yellow fluorescent proteins (ECFP and YPet, respectively), inducing a decrease in FRET strength (increase in ECFP/FRET emission ratio) (Figure 1B). The purified EphA4 biosensor protein showed a robust FRET response upon stimulation: upon incubation with EphA4 kinase, the ECFP emission peak of the biosensor increased, while the YPet emission peak decreased (Figure 1C). A lambda protein phosphatase was subsequently added to dephosphorylate the substrate peptide. We observed a decrease in ECFP/FRET emission ratio back to its basal level before EphA4 kinase phosphorylation (Figure S2). These results suggest that signals from the EphA4 FRET biosensor are reversible and dependent on the substrate phosphorylation. Active EphA4 kinase induced FRET response was eliminated by mutation of Tyr596 to Phe in the substrate peptide, or mutation of Arg175 to Val in the binding site of the SH2 peptide (Figure 1D). In addition, we monitored the emission spectra of the EphA4 biosensor before and after adding trypsin. After trypsin cleavage of the biosensor, the ECFP emission peak of the biosensor increased dramatically, while the YPet emission peak decreased (Figure S3). These results suggest that the biosensor FRET response is indeed resulted from the intramolecular interactions between the phosphorylatable substrate and the SH2 domain within the biosensor, as we designed. Importantly, no changes in FRET signal was observed upon incubation with the platelet-derived growth factor (PDGF) receptor. Only moderate and slow biosensor responses occurred upon incubation with Src kinase (Figure 1D). Collectively, these results suggest that the EphA4 biosensor reports EphA4 activity with relatively high efficiency and specificity in vitro.
Figure 1.
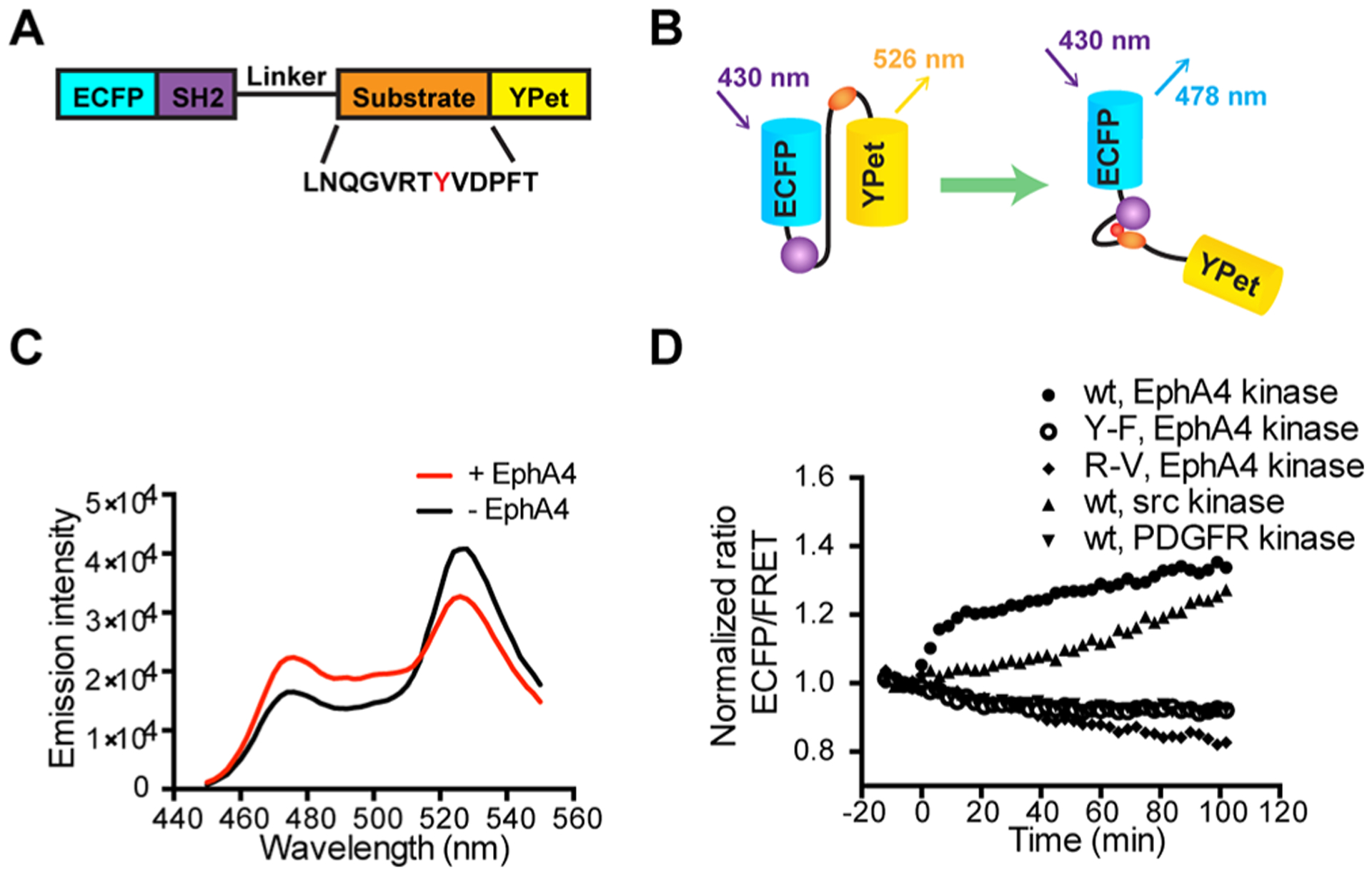
Design and in vitro characterization of FRET-based EphA4 biosensor. (A) The EphA4 biosensor is composed of ECFP, the c-Src SH2 domain, a flexible linker (see Materials and Methods), the EphA4 substrate peptide containing Tyr596 (indicated as a red Y) and YPet. (B) Design principle of the EphA4 biosensor. Active EphA4 phosphorylates the substrate peptide of the EphA4 biosensor, which then intramolecularly binds to the SH2 domain, causing a decrease in the FRET signal. (C) Emission spectra of the purified EphA4 biosensor before (black) and after (red) phosphorylation by EphA4 for 2 h. (D) Time courses of the ECFP/FRET emission ratio of the EphA4 biosensor (1 μM) before and after incubation with EphA4 (1 μg/mL), Src kinase (1 μg/mL), or PDGFR (1 μg/mL) (N = 3). Time courses of EphA4 Y596F (Y–F) mutant biosensor (1 μM) and SH2 R175 V (R–V) mutant biosensor (1 μM) before and after EphA4 stimulation is also shown (N = 3). The ECFP/FRET ratios were normalized to the average values before enzyme incubation. “N” means the number of independent experiments.
Characterization of the EphA4 Biosensor in Mammalian Cells.
Next we examined the performance of the EphA4 biosensor in mammalian cells. The ROS generator pervanadate (PVD, 20 μM) inhibits tyrosine phosphatases and hence activates Eph receptors.23 Therefore, PVD was used to stimulate the biosensor in live HeLa cells, which are known to express endogenous EphA4.24 Upon PVD stimulation, a 100% FRET change was observed (Figure 2A,B). This suggests that the EphA4 biosensor is capable of undergoing a FRET change upon phosphorylation in live cells.
Figure 2.
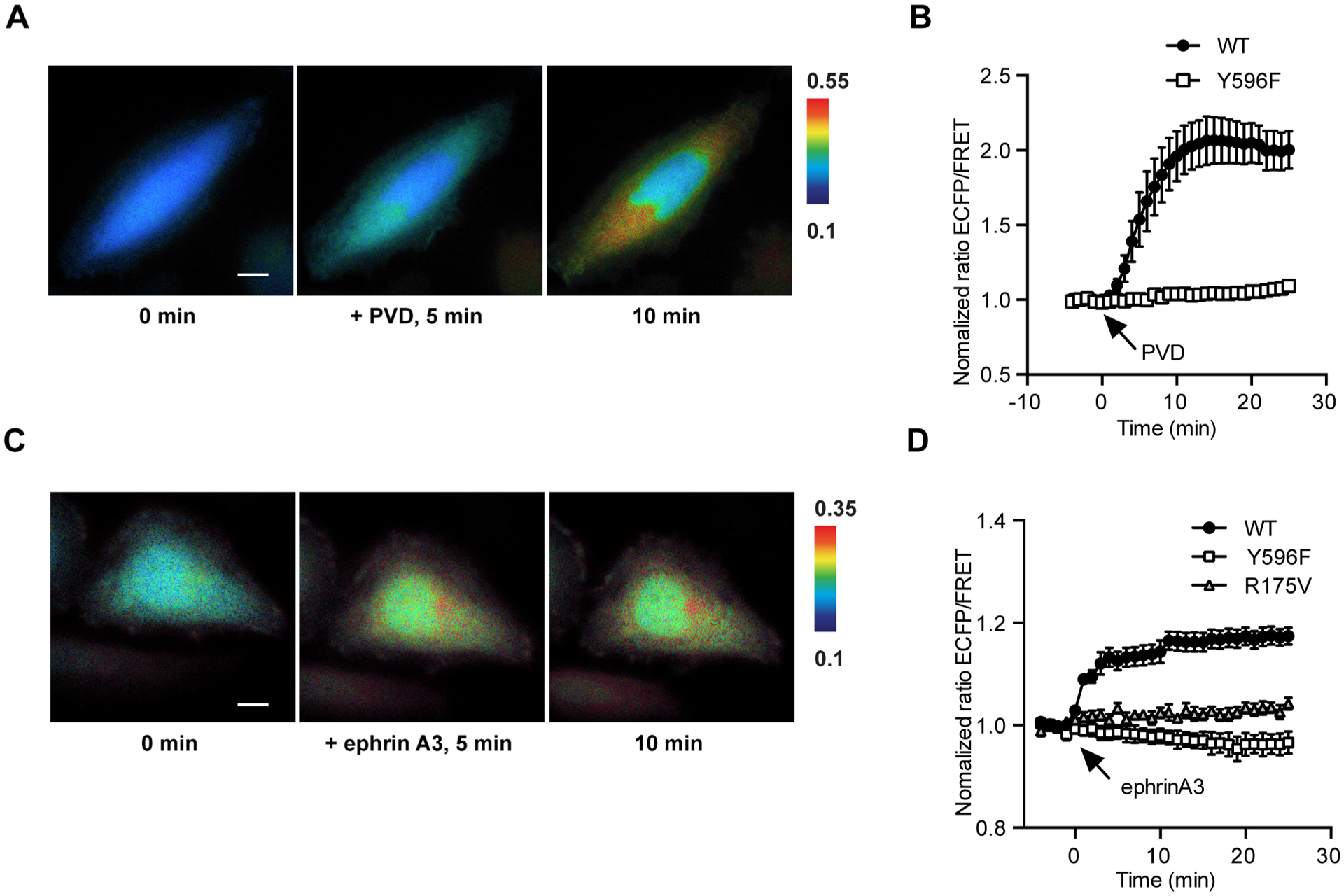
FRET responses of the EphA4 biosensor in mammalian cells. (A) Representative images of ECFP/FRET ratios for the EphA4 biosensor in HeLa cells before and after PVD stimulation (100× objective lens, scale bar, 10 μm). Color scale bars in the figures are to show the ECFP/FRET ratios, with cold and hot colors representing low and high ratios, respectively. (B) Time courses of normalized ECFP/FRET ratios (mean ± SEM) for EphA4 wild-type EphA4 (n = 12, N = 3) or Y596F (n = 7, N = 3) mutant biosensor before and after PVD stimulation in HeLa cells. (C) Representative images of ECFP/FRET ratios for the EphA4 biosensor in HeLa cells before and after ephrinA3 stimulation (100× objective lens, scale bar, 10 μm). (D) Time courses of normalized ECFP/FRET ratios (mean ± SEM) for EphA4 wild -type (black squares, n = 21, N = 4), Y596F mutant (white squares, n = 6, N = 3) or R175 V mutant (triangle, n = 9, N = 3) biosensors before and after PVD stimulation in HeLa cells. Normalized ECFP/FRET ratio is normalized by the basal level of the averaged ratio of the biosensor in the same cell before stimulation. “n” means the total cell number. “N” means the number of individual experiment repeats.
Following this initial test, different ephrin ligands were used to activate EphA4 and examine biosensor responsiveness. Upon EphA4 activation by stimulation with ephrinA3 or ephrinA1, two ligands known to activate EphA4, a 20% FRET change was observed (Figure 2C–D, Figure S4). We also tested a lower affinity ligand ephrin B2 to EphA4 receptors.25 Upon ephrinB2 stimulation with the same dosage, we observed a significantly lower FRET change of the EphA4 biosensor (Figure S4B). These results suggest that the EphA4 biosensor can detect the ligand-induced EphA4 activation in mammalian cells, and report the different levels of EphA4 activations. The mutation of either Tyr596 to Phe (Y596F) in the substrate, or Arg175 to Val (R175 V) in the SH2 domain of the biosensor, eliminated the FRET response of the EphA4 biosensor upon ephrinA3 stimulation (Figure 2D). These results corroborate our strategy based on the idea that the FRET response of the biosensor following EphA4 activation is due to the intramolecular interaction between the phosphorylated Tyr596 substrate and the Src SH2 domain.
To further examine the specificity of the EphA4 biosensor, we applied the peptide antagonist APY-d3 for EphA4 in HeLa cells.26 Cells were pretreated with 2 μM APY-d3 for 20 min and the EphA4 biosensor responses were observed upon ligand ephrinA1 stimulation. We observed that the response of EphA4 biosensor was completely suppressed for the first 5 min before a gradual and moderate response afterward (Figure S4). We reasoned that the delayed response could be due to the endocytosis of the inhibited EphA4 receptors, and the newly recycled and fresh EphA4 receptors deposited at the plasma membrane. We also examined the effect of Src family kinase by using a Src family kinase inhibitor PP1. Cells were pretreated with 500 nM PP1 for 1 h.27 Then we monitored the EphA4 biosensor response upon ligand ephrinA1 stimulation. While the response magnitude is slightly lower than the control group without PP1, we observed a rapid and significant FRET response of EphA4 biosensor in cells pretreated by PP1 (Figure S4), suggesting that the Src inhibition cannot block the EphA4 biosensor response. In addition, 293AD cells engineered to express the exogenous EphA4 receptor were also tested together with the parental 293AD cells which do not express endogenous EphA4. Upon ligand stimulation, a significant FRET response was observed in the EphA4 expressing cells while no obvious signal was detected in the EphA4 null cells (Figure S5). Collectively, these results suggest that the EphA4 biosensor can detect the EphA4 activity with high sensitivity in mammalian cells.
EphA4 Activity in Different Compartments of the Plasma Membrane.
EphA4 is a transmembrane receptor and it is activated at the plasma membrane, which contains different microdomains. To study EphA4 regulation in these subcompartments, we further developed EphA4 biosensors targeting to different microdomains of the plasma membranes by modifying the biosensor with the KRas or Lyn membrane-targeting motifs. The KRas biosensor, which localizes in membrane regions outside raft-like regions, was constructed by fusing a prenylation motif derived from KRAS to the C-terminus of the cytosolic EphA4 (cyto-EphA4) biosensor (Figure 3A). In parallel, a raft-like microdomain targeting motif containing myristoylation and palmitoylation sites (glycine and cysteine) derived from Lyn kinase, was fused to the N-terminus of the cytosolic EphA4 biosensor to generate the EphA4 biosensor that can specifically be located in raft-like regions (Figure 3A).
Figure 3.
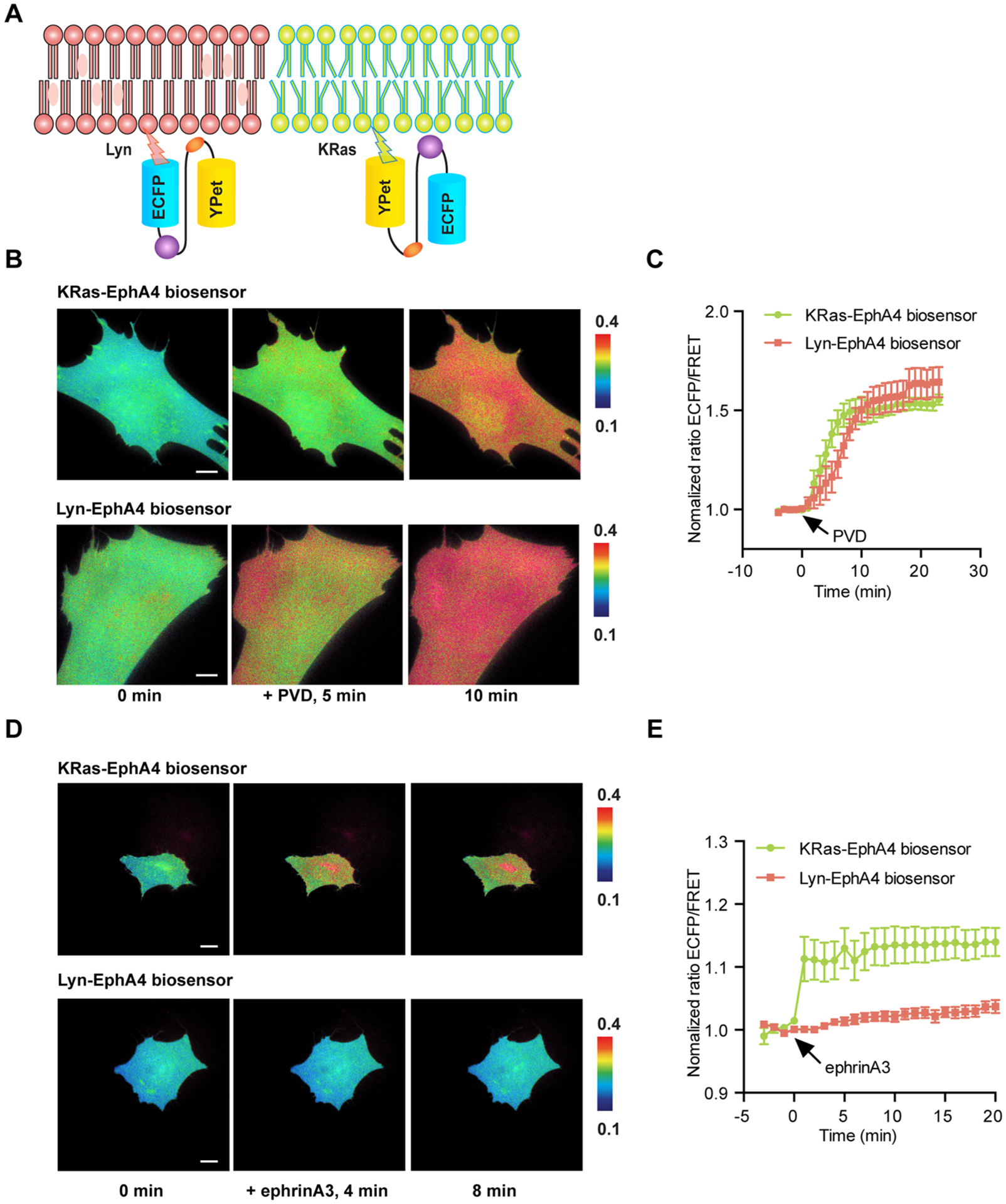
EphA4 activation in raft-like microdomains and non-raft regions. (A) The design principle of non-raft targeted (KRas-EphA4) and raft-targeted (Lyn-EphA4) biosensors. (B) Representative images KRas-EphA4 of ECFP/FRET ratios for the KRas-EphA4 and Lyn-EphA4 biosensors in MEF cells before and after PVD stimulation (100× objective lens, scale bar, 10 μm). (C) Time courses of normalized ECFP/FRET ratios (mean ± SEM) for the KRas-EphA4 biosensor (green line, n = 10, N = 3) and the Lyn-EphA4 biosensors (red line, n = 6, N = 3) before and after PVD stimulation in MEF cells. (D) Representative images of ECFP/FRET ratios for KRas-EphA4 and Lyn-EphA4 biosensors in MEF cells before and after ephrinA3 stimulation (40× objective lens, scale bar, 20 μm). (E) Time courses of normalized ECFP/FRET ratios (mean ± SEM) for the KRas-EphA4 biosensors (green line, n = 16, N = 3) and the Lyn-EphA4 biosensors (red line, n = 14, N = 3) before and after ephrinA3 stimulation in MEF cells. “n” means the total cell number. “N” means the number of individual experiment repeats.
For this experiment, we examined EphA4 activation in membrane microdomains of MEF cells. At first, the KRas- and Lyn-EphA4 biosensors were examined by PVD stimulation in MEF. Similar FRET responses were observed (Figure 3B,C), suggesting that the different localization did not affect activation capability of EphA4 in different microdomains. However, in response to ephrinA3 stimulation, the KRas-EphA4 biosensor in nonlipid raft regions showed a rapid 20–25% increase in EphA4 activation, whereas the Lyn-EphA4 biosensor showed only a minor FRET change even 10 min after stimulation (Figure 3D,E). KRas- or Lyn-EphA4 biosensor with the Y596F or R175 V inactivating mutations did not show a FRET response in MEF cells upon ligand stimulation (Figure S6). There was also no significant difference in the non-normalized ratios between Cyto-, KRas-, or Lyn-EphA4 biosensors (Figure S6C). Consistent results were obtained when the biosensor was tested with ephrinA1 stimulation of the HEK293AD-EphA4 system (Figure S7). Altogether, the results suggest that the differential FRET responses of KRas-EphA4 and Lyn-EphA4 biosensors are not due to their different membrane targeting tags, but rather reflect the differential localization of activated EphA4 in response to ephrin ligands in specific membrane microdomains. In particular, the results indicate a faster and stronger EphA4 activation upon ligand stimulation in non-raft region than in raft-like microdomains.
Regulation of the Actin Cytoskeleton Is Involved in Differential EphA4 Activation in Different Compartments of the Plasma Membrane.
The plasma membrane is directly connected with and supported by cytoskeletal structures, including polymerized actin and microtubule filaments.28 Actin filaments interact with raft-like microdomains through proteins such as annexins and polybasic proteins.17 Therefore, in order to investigate the downstream mechanisms responsible for the differential EphA4 activation patterns in different plasma membrane microdomains, we examined the role of the actin cytoskeleton.
MEF cells were pretreated with cytochalasin D (cytoD) for 1 h to block actin polymerization.29 Interestingly, upon ephrinA1 stimulation of cells treated with cytoD, the KRas-EphA4 biosensor in nonlipid raft regions still showed 20–25% increase in the ECFP/FRET ratio, whereas the Lyn-EphA4 biosensor had a much increased response with a 35–40% change in ECFP/FRET ratio (Figure 4), since the cell morphology also changed significantly upon treatment. In order to eliminate the possibility that the FRET response change was due to cell morphology, we also reduced the CytoD concentration to 200 nM and treatment time to 30 min. This lower concentration of CytoD and shorter treatment time nevertheless caused a clear disruption of actin cytoskeleton, as shown in the GFP images of the MEF cells transfected with actin-GFP before and after treatment (Figure S7B). We showed that there were no more drastic changes in cell morphology at this condition (Figure S7C). Lyn-EphA4 biosensor still showed stronger responses than those of KRas EphA4 biosensor (Figure S7D), consistent with the results we presented in the original condition with higher CytoD concentrations. These results suggest that actin filaments, which connect to raft-like microdomains, may inhibit EphA4 receptor activation in the raft microdomains by ephrin ligands.
Figure 4.
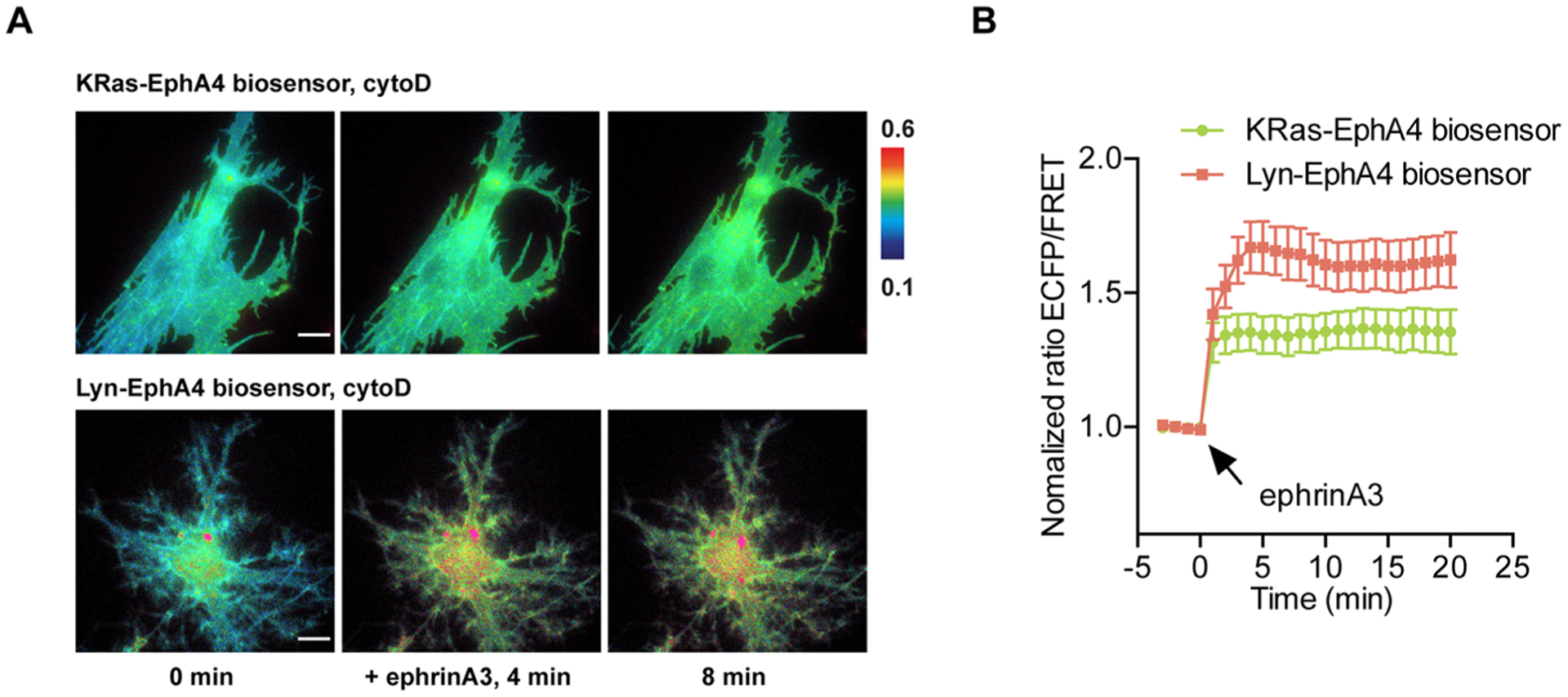
Actin filaments differentially regulate EphA4 activation at the plasma membrane. (A) Representative images of ECFP/FRET emission ratios for the KRas-EphA4 biosensor and the Lyn-EphA4 biosensor in cytochalasin D (cytoD) pretreated MEF cells before and after ephrinA3 stimulation (100× objective lens, scale bar, 10 μm). (B) Time courses of normalized ECFP/FRET emission ratios (mean ± SEM) for the KRas-EphA4 biosensor (green line, n = 10, N = 3) and the Lyn-EphA4 biosensor (red line, n = 6, N = 3) before and after ephrinA3 stimulation in CytoD pretreated MEF cells. “n” means the total cell number. “N” means the number of individual experiment repeats.
DISCUSSIONS
EphA4 functions are closely dependent on the subcellular localization of the receptor. However, it remains unclear how EphA4 activation is regulated in different membrane microdomains. In this study, we developed a FRET based EphA4 biosensor capable of monitoring EphA4 phosphorylation of the Tyr596 motif (found in the EphA4 JM segment) and its subsequent intramolecular binding to the Src SH2 domain. Our EphA4 biosensor has several advantages for studying signaling hierarchy in single live cells. First, the substrate peptide in the EphA4 biosensor encompasses Tyr596, which can be specifically phosphorylated by EphA4 but not as much by other tyrosine kinases abundant at the plasma membrane such as Src or the PDGFR. In addition, the biosensor mimics the molecular events of EphA4 activation to detect receptor activity, but does not contain the EphA4 kinase domain, which may perturb the endogenous signaling and physiology in live cells. Therefore, the EphA4 FRET biosensor should enable us to monitor EphA4 activity with minimal perturbation of host cells. Finally, EphA4 biosensors with Lyn- and KRas tags are ideal for visualizing local EphA4 activity at dynamic membrane microdomains in live cells with high accuracy.
We observed stronger FRET responses of the non-raft targeting EphA4 biosensor in response to ligand stimulation, while the raft-targeting EphA4 biosensor showed minor activation. However, when we blocked the polymerization of actin filaments, which interact with raft-like domains, EphA4 activation at the raft-like regions significantly increased. This activation pattern may be explained by the fact that raft-like microdomains are known to be more rigid and less fluid than non-raft region.30 Since EphA4 activation requires receptor molecules to cluster upon ligand binding, a more fluid membrane structure in the non-raft region may allow and in fact facilitate the clustering event. In fact, actin filaments have been proposed to anchor raft-like microdomains to regulate lateral diffusion of proteins on the membrane.31 It is hence likely that the interaction between actin filaments and raft-like microdomains could inhibit clustering of EphA4 molecules by raft-associated proteins.
In summary, we have developed a novel FRET-based EphA4 biosensor that allows the detection of EphA4 activity with high spatial and temporal resolution in live cells. Targeting of the EphA4 biosensor to specific membrane microdomains through acylation and prenylation further provides powerful tools to monitor the dynamic molecular activities of EphA4 in specific subcellular compartments. Our results suggest that EphA4 can be activated in different membrane microdomains with distinct kinetics in response to physiological stimulations.
MATERIALS AND METHODS
DNA Construction and Plasmid.
The construct for the cytosolic EphA4 biosensor was generated by polymerase chain reaction (PCR) of the complementary DNA encoding the c-Src SH2 domain using a forward primer containing a SphI site and a reverse primer containing the gene sequence for a flexible linker (GSTSGSGKPGSGEGS), a substrate peptide containing the Y596 motif derived from the EphA4 juxtamembrane (JM) region and a SacI site. Similar biosensors including candidate substrates corresponding to Y602 from the EphA4 juxtamembrane region and Y610 from the EphB2 JM region were also generated. PCR products were fused with N-terminal enhanced CFP (ECFP) and C-terminal YPet. Y596F, R175V mutations were conducted using the Quickchange site-directed mutagenesis kit (Stratagene). Constructs were cloned using BamHI and EcoRI into the pRSETB plasmid (Invitrogen) for bacterial expression and into pcDNA3 for mammalian cell expression (Invitrogen).
The membrane-targeted Lyn-EphA4 biosensor was constructed by fusing 16 amino acids from the Lyn kinase (MGCIKSKRKDNLNDDE) to the N-terminus of the cytosolic EphA4 biosensor by PCR. The K-Ras-EphA4 biosensor was constructed by fusing the 14 amino acids of K-Ras-prenylation site (KKKKKKSKTKCVIM) to the C-termini of the cytosolic EphA4 biosensor by PCR. The PCR products for the Lyn- and K-Ras-EphA4 biosensors were subcloned into pcDNA3′ for mammalian cell expression.
In Vitro Protein Characterization.
The EphA4 biosensors were expressed with an N-terminal 6 × His tag in Escherichia coli and purified by nickel chelation chromatography. The fluorescence emission ratio of ECFP/FRET (478 nm/526 nm) of the biosensors was measured at an excitation wavelength of 430 nm using a fluorescence plate reader (TECAN, Sapphire II) before and after the addition of 1 mM ATP and EphA4 (1 μg mL−1, Sigma), c-Src (1 μg mL−1, Upstate), or PDGFR (1 μg mL−1, Sigma) kinases in kinase buffer (50 mM Tris·HCl, 100 mM NaCl, 10 mM MgCl2, 2 mM DTT, pH 8), respectively. Lambda protein phosphatase (New England Biolabs) was added at a final concentration of 12 units/μL. Trypsin (Gibco) was used at a final concentration of 0.25%.
Cell Culture and Reagents.
HeLa cells and mouse embryonic fibroblasts (MEF) were purchased from ATCC. The HEK293AD cell line stably transfected with a vector encoding human EphA4 or a control empty vector have been previously described.32 Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 unit/mL penicillin, 100 μg/mL streptomycin, and 1 mM sodium pyruvate. Cell culture reagents were obtained from Invitrogen. Cells were cultured in a humidified 95% air, 5% CO2 incubator at 37 °C. Lipofectamine 2000 (Invitrogen) was used for transfection of DNA plasmids.
Pervanadate Preparation.
1 mM pervanadate solution was prepared as previously described.33 10 mL of 100 mM Na3VO4 and 50 mL 0.3% H2O2 in 20 mM HEPES (pH 7.3) were mixed in 940 mL H2O. After 5 min a small scoop of 100 μg catalase (CalBiochem, 260 U/mL) was added to release excess H2O2.
Recombinant human ephrinA3 Fc chimera (R&D systems, Minneapolis, MN) was preclustered using anti-human IgG (Jackson Immuno Research, West Grove, PA) for 1 h at room temperature in a 5:1 molar ratio. Recombinant mouse ephrinA1 Fc chimera, ephrin B2 Fc chimera (R&D systems, Minneapolis, MN) was used as a stimulant for the EphA4 biosensors without preclustering (since similar activation was observed with and without preclustering). Platelet-derived growth factor (PDGF) was purchased from Sigma. PP1 src inhibitor was purchased from Sigma.
Image Acquisition.
Cells were starved in 0.5% fetal bovine serum for 36–48 h before imaging. For imaging, the cells were cultured in cover-glass-bottom dishes and maintained in 0.5% FBS DMEM medium with 5% CO2 supplement at 37 °C. Images were collected with a Nikon microscope with 40× or 100× objective lens and a cooled charge-coupled device (CCD) camera using MetaFluor 6.2 and MetaMorph software (Universal Imaging) with a 420DF20 excitation filter, a 455DCXRU dichroic mirror, and two emission filters controlled by a filter changer (480DF40 for CFP and 535DF25 for YFP). A neutral density filter was used to control the intensity of the excitation light. The fluorescence intensity of nontransfected cells was quantified as the background signals and subtracted from the CFP and YFP signals of transfected cells. The pixel-by-pixel CFP/FRET ratios for the images were calculated based on the background-subtracted fluorescence intensity images of CFP and YFP using the Metafluor program to allow quantification and statistical analysis of FRET responses. The emission ratio images were shown in the intensity modified display (IMD) mode.34
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by NIH CA204704, NSF/NIH Math/Bio Initiative DMS-1361421 (S.L. and Y.W.), NIH HL121365, GM125379, CA209629, NSF CBET1360341 (Y. Wang) and NIH NS087070 (E.B.P.). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.8b00465.
Characterization of EphA4 biosensor candidates; FRET responses; EphA4 activation in raft-like microdomains and non-raft regions (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Gale NW; Holland SJ; Valenzuela DM; Flenniken A; Pan L; Ryan TE; Henkemeyer M; Strebhardt K; Hirai H; Wilkinson DG; Pawson T; Davis S; Yancopoulos GD Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 1996, 17 (1), 9–19. [DOI] [PubMed] [Google Scholar]
- (2).Pasquale EB Eph-ephrin bidirectional signaling in physiology and disease. Cell 2008, 133 (1), 38–52. [DOI] [PubMed] [Google Scholar]
- (3).Klein R Eph/ephrin signalling during development. Development 2012, 139 (22), 4105–4109. [DOI] [PubMed] [Google Scholar]
- (4).Flanagan JG; Vanderhaeghen P The ephrins and Eph receptors in neural development. Annu. Rev. Neurosci 1998, 21, 309–345. [DOI] [PubMed] [Google Scholar]
- (5).Egea J; Nissen UV; Dufour A; Sahin M; Greer P; Kullander K; Mrsic-Flogel TD; Greenberg ME; Kiehn O; Vanderhaeghen P; Klein R Regulation of EphA4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron 2005, 47 (4), 515–528. [DOI] [PubMed] [Google Scholar]
- (6).Surawska H; Ma PC; Salgia R The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev 2004, 15 (6), 419–433. [DOI] [PubMed] [Google Scholar]
- (7).Pasquale EB Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10 (3), 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fox BP; Kandpal RP Invasiveness of breast carcinoma cells and transcript profile: Eph receptors and ephrin ligands as molecular markers of potential diagnostic and prognostic application. Biochem. Biophys. Res. Commun 2004, 318 (4), 882–892. [DOI] [PubMed] [Google Scholar]
- (9).Saintigny P; Peng SH; Zhang L; Sen B; Wistuba II; Lippman SM; Girard L; Minna JD; Heymach JV; Johnson FM Global Evaluation of Eph Receptors and Ephrins in Lung Adenocarcinomas Identifies EphA4 as an Inhibitor of Cell Migration and Invasion. Mol. Cancer Ther 2012, 11 (9), 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Fukai J; Yokote H; Yamanaka R; Arao T; Nishio K; Itakura T EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mol. Cancer Ther 2008, 7 (9), 2768–2778. [DOI] [PubMed] [Google Scholar]
- (11).Hachim IY; Villatoro M; Canaff L; Hachim MY; Boudreault J; Haiub H; Ali S; Lebrun JJ Transforming Growth Factor-beta Regulation of Ephrin Type-A Receptor 4 Signaling in Breast Cancer Cellular Migration. Sci. Rep 2017, 7, 1 DOI: 10.1038/s41598-017-14549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Davis S; Gale NW; Aldrich TH; Maisonpierre PC; Lhotak V; Pawson T; Goldfarb M; Yancopoulos GD Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 1994, 266 (5186), 816–819. [DOI] [PubMed] [Google Scholar]
- (13).Kania A; Klein R Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat. Rev. Mol. Cell Biol 2016, 17 (4), 240–256. [DOI] [PubMed] [Google Scholar]
- (14).Klausner RD; Kleinfeld AM; Hoover RL; Karnovsky MJ Lipid domains in membranes. Evidence derived from structural perturbations induced by free fatty acids and lifetime heterogeneity analysis. J. Biol. Chem 1980, 255 (4), 1286–1295. [PubMed] [Google Scholar]
- (15).Pike LJ Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res 2006, 47 (7), 1597–1598. [DOI] [PubMed] [Google Scholar]
- (16).Jacobson K; Mouritsen OG; Anderson RGW Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol 2007, 9 (1), 7–14. [DOI] [PubMed] [Google Scholar]
- (17).Edidin M The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct 2003, 32, 257–283. [DOI] [PubMed] [Google Scholar]
- (18).Yumoto N; Wakatsuki S; Kurisaki T; Hara Y; Osumi N; Frisen J; Sehara-Fujisawa A Meltrin beta/ADAM19 interacting with EphA4 in developing neural cells participates in formation of the neuromuscular junction. PLoS One 2008, 3 (10), No. e3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chichili GR; Rodgers W Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci 2009, 66 (14), 2319–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ouyang M; Sun J; Chien S; Wang Y Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. U. S. A 2008, 105 (38), 14353–14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zacharias DA; Violin JD; Newton AC; Tsien RY Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 2002, 296 (5569), 913–916. [DOI] [PubMed] [Google Scholar]
- (22).Warner N; Wybenga-Groot LE; Pawson T Analysis of EphA4 receptor tyrosine kinase substrate specificity using peptide-based arrays. FEBS J 2008, 275 (10), 2561–2573. [DOI] [PubMed] [Google Scholar]
- (23).Wimmer-Kleikamp SH; Janes PW; Squire A; Bastiaens PI; Lackmann M Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J. Cell Biol 2004, 164 (5), 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Winter J; Roepcke S; Krause S; Muller EC; Otto A; Vingron M; Schweiger S Comparative 3′UTR analysis allows identification of regulatory clusters that drive Eph/ephrin expression in cancer cell lines. PLoS One 2008, 3 (7), No. e2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pasquale EB Eph-ephrin promiscuity is now crystal clear. Nat. Neurosci 2004, 7 (5), 417–418. [DOI] [PubMed] [Google Scholar]
- (26).Olson EJ; Lechtenberg BC; Zhao C; Rubio de la Torre E; Lamberto I; Riedl SJ; Dawson PE; Pasquale EB Modifications of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor To Achieve High Plasma Stability. ACS Med. Chem. Lett 2016, 7 (9), 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shah K; Vincent F Divergent roles of c-Src in controlling platelet-derived growth factor-dependent signaling in fibroblasts. Mol. Biol. Cell 2005, 16 (11), 5418–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Fletcher DA; Mullins RD Cell mechanics and the cytoskeleton. Nature 2010, 463 (7280), 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lu S; Wang Y Fluorescence resonance energy transfer biosensors for cancer detection and evaluation of drug efficacy. Clin. Cancer Res 2010, 16 (15), 3822–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Gallegos AM; McIntosh AL; Atshaves BP; Schroeder F Structure and cholesterol domain dynamics of an enriched caveolae/raft isolate. Biochem. J 2004, 382, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Head BP; Patel HH; Insel PA Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta, Biomembr 2014, 1838 (2), 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lamberto I; Lechtenberg BC; Olson EJ; Mace PD; Dawson PE; Riedl SJ; Pasquale EB Development and structural analysis of a nanomolar cyclic peptide antagonist for the EphA4 receptor. ACS Chem. Biol 2014, 9 (12), 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Huyer G; Liu S; Kelly J; Moffat J; Payette P; Kennedy B; Tsaprailis G; Gresser MJ; Ramachandran C Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J. Biol. Chem 1997, 272 (2), 843–851. [DOI] [PubMed] [Google Scholar]
- (34).Wang Y; Botvinick EL; Zhao Y; Berns MW; Usami S; Tsien RY; Chien S Visualizing the mechanical activation of Src. Nature 2005, 434 (7036), 1040–1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


