Abstract
Worldwide decline in biodiversity during the Holocene has impeded a comprehensive understanding of pre-human biodiversity and biogeography. This is especially true on islands, because many recently extinct island taxa were morphologically unique, complicating assessment of their evolutionary relationships using morphology alone. The Caribbean remains an avian hotspot but was more diverse before human arrival in the Holocene. Among the recently extinct lineages is the enigmatic genus Nesotrochis, comprising three flightless species. Based on morphology, Nesotrochis has been considered an aberrant rail (Rallidae) or related to flufftails (Sarothruridae). We recovered a nearly complete mitochondrial genome of Nesotrochis steganinos from fossils, discovering that it is not a rallid but instead is sister to Sarothruridae, volant birds now restricted to Africa and New Guinea, and the recently extinct, flightless Aptornithidae of New Zealand. This result suggests a widespread or highly dispersive most recent common ancestor of the group. Prior to human settlement, the Caribbean avifauna had a far more cosmopolitan origin than is evident from extant species.
Keywords: Holocene extinction, island diversity, Anthropocene, avian phylogeny, Gruiformes
1. Introduction
Islands have been recognized as places to study evolution, ecology and biogeography [1,2]. The natural biota of all islands, no matter how remote, has been altered by humans, leaving a residual modern diversity that is different from its pre-human condition [3]. Most island diversity has been lost relatively recently because the mass extinction events that began on continents during the late Pleistocene did not affect islands until human colonization in the Holocene (e.g. [4]). Because of this delay, islands acted as refugia of diversity in the recent past [4,5]. Still, we have a limited understanding of the phylogenetic affinities of species lost after human contact, thereby limiting what we know about the evolutionary and biogeographic mechanisms underpinning these communities.
The unique morphology of many island birds has been driven by niche availability and the lack of mammalian predators, making them susceptible to predation by humans and other invasive predators [6,7]. Doves, rails, passerines and certain seabirds had the highest overall numbers of insular avian extinctions [8,9], although losses occurred across all groups of island birds, involving entire subfamilies (Raphinae—dodo and solitaire), families (Aptornithidae—adzebills) and even orders (Aepyornithiformes—elephant birds; Dinornithiformes—moas). The extreme morphological diversity that was lost, exemplified by large herbivores (elephant birds) or predators (adzebills), complicates the recovery of their evolutionary relationships with living taxa. Recently, ancient DNA (aDNA) has illuminated the phylogenetic placement of previously enigmatic extinct island birds. For example, aDNA has revealed that elephant birds are sister to Apterygiformes [10], and adzebills (Aptornithidae) are sister to flufftails (Sarothruridae; [11]) from sub-Saharan Africa and New Guinea. These results provide context for lost biogeographic connections, long-distance dispersal events, and the evolution of gigantism and flightlessness [12,13].
Nesotrochis is an extinct genus with three flightless species once found in the Greater Antilles. Nesotrochis debooyi was described from bones in archaeological middens in the Virgin Islands, suggesting it survived until European arrival [14]; later it was discovered in Puerto Rico [15,16]. Nesotrochis was believed to be related to the rallid genus Aramides, with a South American origin [15]. Later, Wetmore considered Nesotrochis related to gallinules (Gallinula, Rallidae; [17]), which Olson [16] supported. Nesotrochis picipicensis on Cuba was described first as a coot, Fulica picipicensis (Rallidae; [18]), but then placed in Nesotrochis from osteological characters by Olson [16], who also described Nesotrochis steganinos from Hispaniola. However, morphological and ecological convergence is commonplace across rails (Rallidae) and the closely related flufftails (Sarothruridae) [19,20]. Recent work suggests that Nesotrochis shares diagnostic hypotarsus characters with flufftails (Sarothruridae) and related taxa (Heliornithidae) that differ from Rallidae, indicating that Nesotrochis may not be a member of the Rallidae [21].
The affinities of Nesotrochis based on skeletal morphology have been conjectural since its description. Here, we use aDNA from N. steganinos to evaluate the osteology-based hypothesis that Nesotrochis is closely related to Gallinula (Rallidae) or to other closely related families, and discuss the systematic and biogeographic implications of our findings.
2. Methods
A Nesotrochis steganinos pedal phalanx (Florida Museum, University of Florida (UF) 431763; Haiti: Trouing Marassa; 20 July 1983, 10X.31Y.26Z subunit A; GenBank accession no. MW145005.1) was used for DNA extraction and sequencing. DNA extraction, library preparation, target capture enrichment and post-sequencing data cleaning followed the methods in Oswald et al. ([22,23]; full details are also provided in the electronic supplementary material). An associated phalanx of UF 431763 (same catalogue number) was radiocarbon dated (Beta Analytic Testing Laboratory ID: Beta-502522) at 6430 ± 30 BP (conventional radiocarbon age) and calendrically calibrated (using INTCAL13) to 7424 to 7289 cal BP (early Holocene). Radiocarbon pretreatment was a collagen extraction with alkali and ultrafiltration. For target capture enrichment, we used a mitochondrial DNA bait set designed by Arbor Biosciences (Ann Arbor, MI) based on the rallid Porphyrio melanotus (NC025508.1; a hypothetical close relative of Nesotrochis based on osteology; see electronic supplementary material, table S1 for sample and taxonomic information).
In Geneious (v. 11.1.4; https://www.geneious.com), the Map to Reference feature set to default settings (Medium-Low Sensitivity; five iterations) was used to map the cleaned, unpaired reads to P. melanotus. An initial BLAST [24] search of the recovered cytochrome b gene indicated that Nesotrochis was not closely related to P. melanotus so we repeated the Map to Reference approach using other Gruiformes as references including Aptornis otidiformis (MK434262.1; Aptornithidae), Sarothrura ayresi (NC034316.1; Sarothruridae) and Canirallus oculeus (MK434261.1; Rallidae). mapDamage [25] was used to determine if the base patterns in our reads were consistent with those found in a DNA, using paired reads mapped to the P. melanotus mitochondrial genome as a reference.
The majority consensus sequences for each of the four references were aligned to discern possible discrepancies between the reference used and the resultant base calls. Sites where more than two of the consensus sequences had an ambiguous base call (e.g. M or Y, not including the D loop) were reviewed by eye in the read pile-ups to evaluate the number of reads that supported the ambiguity and whether it was a possible site of deamination or degradation, i.e. at the end of the read, where degradation is more prevalent in ancient data. The greatest area of disagreement across different references was located in the D loop, which we subsequently removed from the N. steganinos data before further analyses.
Following the eBird/Clements checklist [26], we downloaded all available Gruiformes mitochondrial genome data from GenBank. The Gruiformes comprise 192 currently recognized extant or very recently extinct species. Whole or nearly complete mitochondrial genome sequences were available for 21% of gruiform species. Rallids represented 89% of the missing species. All recognized extant families were represented by at least one species in our dataset. The mitochondrial genome of Rallicula forbesi (Rallina per [26]) was obtained by mapping the raw read data from Garcia-R et al. [27] to the mitochondrial genome of C. oculeus in Geneious (v. 11.1.4) using the Medium-Low Sensitivity default settings. Along with extant species, we included partial mitochondrial genomes of the extinct New Zealand adzebills: A. otidiformis and Aptornis defossor (Aptornithidae; [11]). To increase our sampling of Sarothruridae, we also included three mitochondrial gene regions (ATP6, tRNA-Gly ND3 and CytB) of Mentocrex kioloides and Mentocrex beankaensis. Tringa semipalmata and Tringa ochropus (Scolopacidae) were used as the outgroup. GenBank accession numbers are given in electronic supplementary material, table S1.
Mitochondrial sequences were aligned using the Geneious multiple sequence alignment algorithms. Because of the phylogenetic diversity in our dataset, the D loop region remaining in the other taxa aligned poorly and was removed (dataset 1; 15 954 bp). Gblocks [28] was used to remove poorly aligning regions (dataset 2; 14 514 bp). With each dataset, we performed a maximum-likelihood analysis in RAxML (8.2.11; [29]) with three partitions: (i) protein-coding sequences (CDS), (ii) rRNA and tRNA sequences, and (iii) non-coding sequences. We assessed topological support with 1000 bootstrap replicates. Dataset 1 recovered low support for Nesotrochis and putative sister taxa, so we visualized alternative topologies by calculating the Robinson–Foulds distance matrix [30] using the package phytool [31] in R v. 3.6.1 [32]. We used classical multidimensional scaling to reduce this matrix to a single axis representing tree similarity. We coloured each tree according to topological similarity based on this metric (similarity index).
3. Results
Using Aptornis otidiformis, Sarothrura ayresi, Canirallus oculeus (MK434261.1) and Porphyrio melanotus as references, we recovered 329 330, 330 367, 325 644 and 336 218 on-target reads, respectively, of Nesotrochis steganinos. Average read length ranged from 93.5 to 93.9 bp with a coverage mean of 1892–1903 reads across the four references. mapDamage fragment misincorporation plots indicate the expected C→T and A→G substitutions characteristic of a DNA (electronic supplementary material, figure S1). The only missing data, regardless of the reference used, were within the D loop as expected based on high divergence within and across taxa.
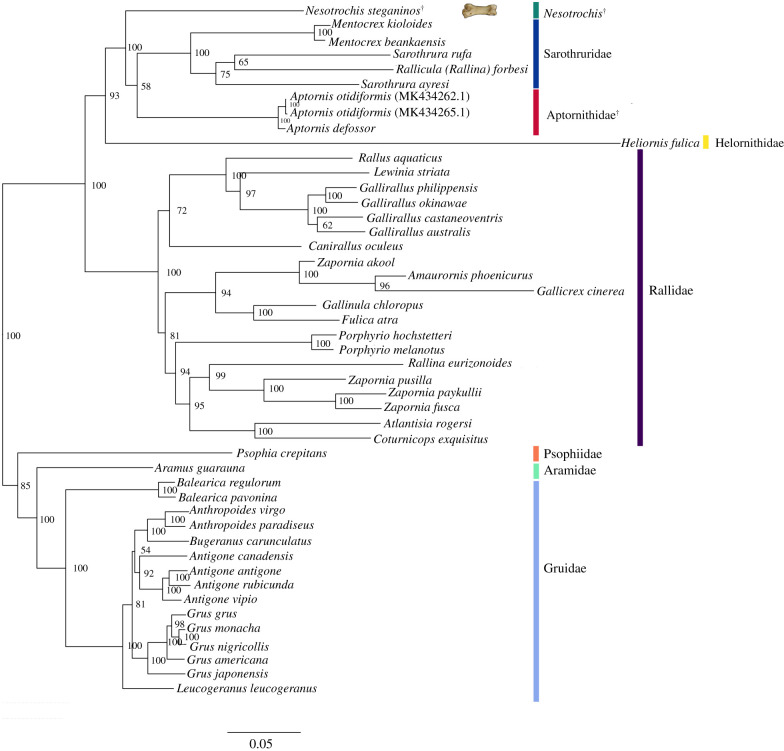
Our mitochondrial phylogeny, based on both datasets, recovered the same topology of extant gruiform families as Prum et al. [33] using targeted genome sequencing. Relationships within Gruidae differ between datasets 1 and 2. The dataset 1 RAxML phylogeny is the same as that of Krajewski et al. [34]. The Rallidae topology does not differ between datasets. Both RAxML phylogenies indicate that N. steganinos does not fall within the Rallidae and instead is in the clade composed of Sarothruridae and the extinct Aptornithidae (represented by A. otidiformis and Aptornis defossor). The relationships among Nesotrochis, Sarothruridae and Aptornithidae are not resolved. Dataset 1 suggests Nesotrochis is sister to Sarothruridae, albeit with only marginal support (52%; electronic supplementary material, figure S2), and this clade is sister to Aptornithidae. The alternative topology (48% of bootstrap replicates) of dataset 1 recovered Nesotrochis as sister to Aptornithidae + Sarothruridae (electronic supplementary material, figure S3). Dataset 2 resulted in 100% topological support of a clade comprising Nesotrochis, Aptornithidae and Sarothruridae and 58% topological support for Nesotrochis as sister to Aptornithidae and Sarothruridae (figure 1).
Figure 1.
RAxML phylogeny of the Gruiformes based on the Gblocks dataset (dataset 2). The bootstrap support for the sister relationship of Nesotrochis steganinos to Sarothruridae + Aptornithidae is 100% yet the phylogenetic relationships within this clade are not resolved. See electronic supplementary material, figures S1 and S2 for the alternative topology, where N. steganinos is sister to Sarothruridae (based on dataset 1). The N. steganinos pedal phalanx from which aDNA was extracted is shown next to the tip name.
4. Discussion
The Cenozoic fossil record of Gruiformes is relatively species-rich, but the repeated convergent evolution of rail-like body plans, along with convergence due to flightlessness, hinders the phylogenetic placement of extinct species. Our aDNA results support Nesotrochis as sister to the clade Sarothruridae + the extinct Aptornithidae, or just to the Sarothruridae, both of which inhabit(ed) the Old World (figure 2). They are part of a larger clade that also includes the Heliornithidae. Nesotrochis is the only DNA-based example of a Caribbean avian genus that is sister to families restricted to the Old World. As such, it provides a dramatic example of human-caused extinction erasing complex biogeographic history.
Figure 2.
The distribution of Nesotrochis (green) within the Caribbean, Sarothruridae (blue) in Africa and New Guinea, and Aptornithidae (red) in New Zealand. Both Nesotrochis and Aptornithidae are extinct and are the most closely related lineages to Sarorthruridae. Image credits: ‘Nesotrochis': T. Michael Keesey and Hutty Mcphoo, https://creativecommons.org/licenses/by-sa/3.0/; ‘Sarothrura': Ryan S. Terrill; and Aptornis otidiformis: N. Tamura.
(a). Biogeography of families convergent on Rallidae
The finfoots/sungrebe (Heliornithidae), adzebills (Aptornithidae), flufftails (Sarothruridae) and Nesotrochis clade contains many species that are morphologically convergent with rallids, with which they shared a common ancestor likely during the early Cenozoic [11,27]. Holocene extinctions have greatly diminished this clade; combined data from morphology and aDNA indicate 50% family level extinction during the Holocene (herein; [11]). Here, we briefly discuss the clade's diversity and biogeography.
Heliornithidae consists of three monotypic genera, each in a single tropical region (Neotropics—Heliornis fulica; Afrotropics—Podica senegalensis; and SE Asia—Heliopais personatus). The relationships among these species have not been evaluated with large-scale genomic data [35], but the family's pan-tropical distribution suggests either extinction of a more widespread temperate taxa or a highly dispersive most recent common ancestor. Aptornithidae is an extinct family from New Zealand; its fossil record extends to the Miocene [36], with divergence from the Sarothruridae conjectured during the late Eocene [11]. The two Holocene species of aptornithids were 0.8 m tall, approximately 17 kg, flightless predators until their extinction following human arrival 600 years ago [37–39].
The greatest extant diversity within the larger clade occurs in the Sarothruridae, which comprises 15 species in the genera Sarothrura, Mentocrex and Rallicula (Rallina in [26]). (Rallina ‘rails' are likely all sarothrurids; Rallina ‘crakes' are rallids; see [26,27]). Sarothrurids have been considered to be rallids based on morphology [19], although hypotarsal characters support the close relationship of Sarothruridae and Heliornithidae [21]. Further study may disclose that some modern and fossil taxa currently considered to be rallids are actually sarothrurids (e.g., [20]). Learning just how deeply divergent Nesotrochis is from the Sarothruridae will require further genetic sampling of sarothrurids and the two other species of Nesotrochis. If Nesotrochis truly is sister to the Sarothruridae, the divergence of the two lineages likely took place during the Eocene or Oligocene based on recent calibrations [11,27].
(b). Caribbean biogeography
Modern Caribbean bird diversity largely consists of New World families such as Trochilidae (hummingbirds), Mimidae (mockingbirds, tremblers), Thraupidae (tanagers) and Parulidae (wood-warblers). It also includes four endemic families: the Todidae (todies), Dulidae (palmchat), Calyptophilidae (chat-tanagers) and Phaenicophilidae (palm-tanagers). The phylogenetic relationship of Nesotrochis to Sarothruridae and Aptornithidae indicates a novel historical biogeographic connection between the Caribbean and the Old World. A similar hypothesis has been proposed, based on morphology, for the endemic Cuban dove Starnoenas [40], an idea waiting to be tested with DNA. Nesotrochis could be a relictual taxon that survived in the Caribbean after extinction on the adjacent mainland, or an example of long-distance dispersal from the Old World to the Caribbean.
This is the first avian Caribbean–Old World connection supported by DNA-based evidence, yet other non-avian taxa have been proposed to have dispersed from the Old World. For example, New World monkeys and caviomorph rodents arrived from Africa between or during the mid Eocene to mid Oligocene [41,42]. The enigmatic hoatzin (Opisthocomus hoazin) of the Amazon Basin is the last living representative of an avian order found in Europe and Africa from the late Eocene to Miocene [43–45]. The African Ptilopachus is the sole genus of New World quail (Odontophoridae) outside of the Americas, with a divergence that also dates to the mid Eocene to mid Oligocene [46]. Another American bird with Old World affinities is the wrentit (Paradoxornithidae), which colonized the New World in the late Miocene [47]. Others include the bushtit (Aegithalidae), verdin (Remizidae), and South American painted-snipe (Rostratulidae). For taxa with calculated divergence times, all divergences occurred well before the significant global cooling that began during the Pliocene, which suggests that warmer, less seasonal climatic conditions may have been favourable for long-distance dispersal and more widespread distributions.
5. Conclusion
Island taxa have been disproportionately affected by human-caused extinction compared with their continental relatives, which has limited our understanding of processes shaping insular biota. Ancient DNA approaches are critical for the phylogenetic placement of extinct island taxa, which in turn supports a broader understanding of biogeography and evolution of often-enigmatic species. New insights from such research suggest that while islands can act as refugia for previously widely distributed clades, they may have also harboured unique, deeply divergent island-endemic groups such as Nesotrochis. As the fossil record of island vertebrates grows, we see rich opportunities for combining both morphological and molecular data within a geographically wide-open framework.
Acknowledgements
We thank Oona Takano for cataloguing fossils from Hispaniola. We also thank Alvaro Hernandez and Chris Wright at the Roy J. Carver Biotechnology Center (University of Illinois Urbana-Champaign) for sequencing advice.
Ethics
The authors had permission from the Vertebrate Paleontology curatorial staff at the Florida Museum of Natural History to sample UF 431763 Nesotrochis steganinos.
Data accessibility
The UF 431763 Nesotrochis steganinos mitochondrial genome is available on GenBank (accession no. MW145005.1). Raw reads are available on NCBI SRA; BioProject accession number: PRJNA680380. Individual sequencing efforts are accessioned as: SAMN16913466 (HiSeq, shotgun sequences), SAMN16913467 (MiSeq, shotgun sequences), SAMN16913468 (MiSeq, mitochondrial genome enriched sequences).
Authors' contributions
J.A.O., B.J.S., M.J.L., D.W.S., R.P.G. and J.M.A conceived and designed the study. J.A.O. and R.S.T. performed analyses. J.A.O. and B.J.S. performed laboratory work. All authors contributed towards data interpretation and writing of the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed herein.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by a Florida Museum Natural History Research Grant to M.J.L., J.A.O., B.J.S., J.M.A., D.W.S. and R.P.G.; NSF grants BCS-1118369 and GSS-1461496 to D.W.S.; NSF grant DEB-2033905 to R.P.G., M.J.L. and B.J.S.; and NSF grant DEB-2034316 to J.M.A. and J.A.O.
References
- 1.Wallace AR. 1880. Island life; or, the phenomena and causes of insular faunas and floras. London, UK: Macmillan. [Google Scholar]
- 2.Darwin C. 1859. On the origin of species by means of natural selection, or, the preservation of favoured races in the struggle for life. London, UK: J. Murray. [PMC free article] [PubMed] [Google Scholar]
- 3.Steadman DW. 2006. Extinction and biogeography of tropical Pacific birds. Chicago, IL: University of Chicago Press. [Google Scholar]
- 4.Steadman DW, Martin PS, MacPhee RDE, Jull AJT, McDonald HG, Woods CA, Iturralde-Vinent M, Hodgins GWL. 2005. Asynchronous extinction of late Quaternary sloths on continents and islands. Proc. Natl Acad. Sci. USA 102, 11 763-11 768. ( 10.1073/pnas.0502777102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCulloch GA, Waters JM. 2019. Phylogenetic divergence of island biotas: molecular dates, extinction, and ‘relict’ lineages. Mol. Ecol. 28, 4354-4362. ( 10.1111/mec.15229) [DOI] [PubMed] [Google Scholar]
- 6.Kirchman JJ. 2012. Speciation of flightless rails on islands: a DNA-based phylogeny of the typical rails of the Pacific. Auk 129, 56-69. ( 10.1525/auk.2012.11259) [DOI] [Google Scholar]
- 7.Wright NA, Steadman DW, Witt CC. 2016. Predictable evolution toward flightlessness in volant island birds. Proc. Natl Acad. Sci. USA 113, 4765-4770. ( 10.1073/pnas.1522931113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyrberg T, Milberg P. 1993. Naïve birds and noble savages - a review of man-caused prehistoric extinctions of island birds. Ecography 16, 229-250. ( 10.1111/j.1600-0587.1993.tb00213.x) [DOI] [Google Scholar]
- 9.Steadman DW. 1995. Prehistoric extinctions of Pacific island birds: biodiversity meets zooarchaeology. Science 267, 1123-1131. ( 10.1126/science.267.5201.1123) [DOI] [PubMed] [Google Scholar]
- 10.Mitchell KJ, Llamas B, Soubrier J, Rawlence NJ, Worthy TH, Wood J, Lee MSY, Cooper A. 2014. Ancient DNA reveals elephant birds and kiwi are sister taxa and clarifies ratite bird evolution. Science 344, 898-900. ( 10.1126/science.1251981) [DOI] [PubMed] [Google Scholar]
- 11.Boast AP, et al. 2019. Mitochondrial genomes from New Zealand's extinct adzebills (Aves: Aptornithidae: Aptornis) support a sister-taxon relationship with the Afro-Madagascan Sarothruridae. Diversity 11, 24. ( 10.3390/d11020024) [DOI] [Google Scholar]
- 12.Bunce M, Szulkin M, Lerner HRL, Barnes I, Shapiro B, Cooper A, Holdaway RN. 2005. Ancient DNA provides new insights into the evolutionary history of New Zealand's extinct giant eagle. PLoS Biol. 3, e9. ( 10.1371/journal.pbio.0030009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knapp M, et al. 2019. Mitogenomic evidence of close relationships between New Zealand's extinct giant raptors and small-sized Australian sister-taxa. Mol. Phylogenet. Evol. 134, 122-128. ( 10.1016/j.ympev.2019.01.026) [DOI] [PubMed] [Google Scholar]
- 14.Wetmore A. 1918. Bones of birds collected by Theodoor de Booy from kitchen midden deposits in the islands of St. Thomas and St. Croix. Proc. US Natl Mus. 54, 513-523. ( 10.5479/si.00963801.54-2245.513) [DOI] [Google Scholar]
- 15.Wetmore A. 1922. Bird remains from the caves of Porto Rico. Bull. Am. Mus. Nat. Hist. 46, 297-333. ( 10.2307/4073610) [DOI] [Google Scholar]
- 16.Olson SL. 1974. A new species of Nesotrochis from Hispaniola, with notes on other fossil rails from the West Indies (Aves: Rallidae). Proc. Biol. Soc. Wash. 87, 439-450. [Google Scholar]
- 17.Wetmore A. 1937. Ancient records of birds from the island of St. Croix with observations on extinct and living birds of Puerto Rico. J Agr. Univ. Puerto Rico. 21, 5-16. ( 10.46429/jaupr.v21i1.14340) [DOI] [Google Scholar]
- 18.Fischer K, Stephan B. 1971. Weitere Vogelreste aus dem Pleistozän der Pio-Domingo-Höhle in Kuba [More bird remains from the Pleistocene of the Pio Domingo Cave in Cuba]. Wiss. Z. Humboldt Univ. Berlin. Math. Naturwiss. Rei. 20, 593-607. [In German.] [Google Scholar]
- 19.Livezey BC. 1998. A phylogenetic analysis of the Gruiformes (Aves) based on morphological characters, with an emphasis on the rails (Rallidae). Phil. Trans. R. Soc. Lond. B 353, 2077-2151. ( 10.1098/rstb.1998.0353) [DOI] [Google Scholar]
- 20.De Pietri VL, Mayr G. 2014. Reappraisal of early Miocene rails (Aves, Rallidae) from central France: diversity and character evolution. J. Zool. Syst. Evol. Res. 52, 312-322. ( 10.1111/jzs.12074) [DOI] [Google Scholar]
- 21.Mayr G. 2019. Hypotarsus morphology of the Ralloidea supports a clade comprising Sarothrura and Mentocrex to the exclusion of Canirallus. Acta Ornithol. 54, 51-58. ( 10.3161/00016454AO2019.54.1.005) [DOI] [Google Scholar]
- 22.Oswald JA, Allen JM, Witt KE, Folk RA, Albury NA, Steadman DW, Guralnick RP. 2019. 2,500-year-old aDNA from Caribbean fossil places an extinct bird (Caracara creightoni) in a phylogenetic context. Mol. Phylogenet. Evol. 140, 106576. ( 10.1016/j.ympev.2019.106576) [DOI] [PubMed] [Google Scholar]
- 23.Oswald JA, Allen JM, LeFebvre MJ, Stucky BJ, Folk RA, Albury NA, Morgan GS, Guralnick RP, Steadman DW. 2020. Ancient DNA and high-resolution chronometry reveal a long-term human role in the historical diversity and biogeography of the Bahamian hutia. Scient. Rep. 10, 1373. ( 10.1038/s41598-020-58224-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 25.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682-1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clements JF, Schulenberg TS, Iliff MJ, Billerman SM, Fredericks TA, Sullivan BL, Wood CL. 2019. The eBird/Clements Checklist of Birds of the World: v2019. See https://www.birds.cornell.edu/clementschecklist/download/.
- 27.Garcia-R JC, Lemmon EM, Lemmon AR, French N. 2020. Phylogenomic reconstruction sheds light on new relationships and timescale of rails (Aves: Rallidae) evolution. Diversity 12, 70. ( 10.3390/d12020070) [DOI] [Google Scholar]
- 28.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540-552. ( 10.1093/oxfordjournals.molbev.a026334) [DOI] [PubMed] [Google Scholar]
- 29.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math. Biosci. 53, 131-147. ( 10.1016/0025-5564(81)90043-2) [DOI] [Google Scholar]
- 31.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217-223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 32.R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/. [Google Scholar]
- 33.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526, 569-577. ( 10.1038/nature15697) [DOI] [PubMed] [Google Scholar]
- 34.Krajewski C, Sipiorski JT, Anderson FE. 2009. Complete mitochondrial genome sequences and the phylogeny of cranes (Gruiformes: Gruidae). Auk 127, 440-452. ( 10.1525/auk.2009.09045) [DOI] [Google Scholar]
- 35.Fain MG, Krajewski C, Houde P. 2007. Phylogeny of ‘‘core Gruiformes’’ (Aves: Grues) and resolution of the Limpkin–Sungrebe problem. Mol. Phylogenet. Evol. 43, 515-529. ( 10.1016/j.ympev.2007.02.015) [DOI] [PubMed] [Google Scholar]
- 36.Worthy TH, Tennyson AJD, Scofield RP. 2011. Fossils reveal an early Miocene presence of the aberrant gruiform Aves: Aptornithidae in New Zealand. J. Ornithol. 152, 669-680. ( 10.1007/s10336-011-0649-6) [DOI] [Google Scholar]
- 37.Worthy TH, Holdaway RN. 2002. The lost world of the moa: prehistoric life of New Zealand. Bloomington, IN: Indiana University Press. [Google Scholar]
- 38.Perry GL, Wheeler A, Wood JR, Wilmshurst JM. 2014. A high-precision chronology for the rapid extinction of New Zealand moa (Aves, Dinornithiformes). Q. Sci. Rev. 105, 126-135. ( 10.1016/j.quascirev.2014.09.025) [DOI] [Google Scholar]
- 39.Wood JR, Scofield RP, Hamel J, Lalas C, Wilmshurst JM. 2017. Bone stable isotopes indicate a high trophic position for New Zealand's extinct South Island adzebill (Aptornis defossor) (Gruiformes: Aptornithidae). NZ J. Ecol. 41, 240-244. ( 10.20417/nzjecol.41.24) [DOI] [Google Scholar]
- 40.Olson SL, Wiley JW. 2016. The blue-headed quail-dove (Starnoenas cyanocephala): an Australasian dove marooned in Cuba. Wilson. J. Ornithol. 128, 1-21. ( 10.1676/1559-4491-128.1.1) [DOI] [Google Scholar]
- 41.Poux C, Chevret P, Huchon D, de Jong WW, Douzery EJP. 2006. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst. Biol. 55, 228-244. ( 10.1080/10635150500481390) [DOI] [PubMed] [Google Scholar]
- 42.Seiffert ER, Tejedor MF, Fleagle JG, Novo NM, Cornejo FM, Bond M, de Vries D, Campbell KE Jr. 2020. A parapithecid stem anthropoid of African origin in the Paleogene of South America. Science 368, 194-197. ( 10.1126/science.aba1135) [DOI] [PubMed] [Google Scholar]
- 43.Mayr G, Alvarenga H, Mourer-Chauviré C. 2011. Out of Africa: fossils shed light on the origin of the hoatzin, an iconic Neotropic bird. Sci. Nat. 98, 961-966. ( 10.1007/s00114-011-0849-1) [DOI] [PubMed] [Google Scholar]
- 44.Mayr G. 2014. A hoatzin fossil from the middle Miocene of Kenya documents the past occurrence of modern-type Opisthocomiformes in Africa. Auk 131, 55-60. ( 10.1642/AUK-13-134.1) [DOI] [Google Scholar]
- 45.Mayr G, De Pietri VL. 2014. Earliest and first Northern Hemispheric hoatzin fossils substantiate Old World origin of a ‘Neotropic endemic’. Sci. Nat. 101, 143-148. ( 10.1007/s00114-014-1144-8) [DOI] [PubMed] [Google Scholar]
- 46.Hosner PA, Braun EL, Kimball RT. 2015. Land connectivity changes and global cooling shaped the colonization history and diversification of New World quail (Aves: Galliformes: Odontophoridae). J. Biogeogr. 42, 1883-1895. ( 10.1111/jbi.12555) [DOI] [Google Scholar]
- 47.Cai T, Shao S, Kennedy JD, Alström P, Moyle RG, Qu Y, Lei F, Fjeldså J. 2020. The role of evolutionary time, diversification rates and dispersal in determining the global diversity of a large radiation of passerine birds. J. Biogeogr. 47, 1612-1625. ( 10.1111/jbi.13823) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The UF 431763 Nesotrochis steganinos mitochondrial genome is available on GenBank (accession no. MW145005.1). Raw reads are available on NCBI SRA; BioProject accession number: PRJNA680380. Individual sequencing efforts are accessioned as: SAMN16913466 (HiSeq, shotgun sequences), SAMN16913467 (MiSeq, shotgun sequences), SAMN16913468 (MiSeq, mitochondrial genome enriched sequences).