Abstract
Glutathione is the major thiol-containing species in both prokaryotes and eukaryotes and plays a wide variety of roles, including detoxification of metals by sequestration, reduction, and efflux. ABC transporters such as MRP1 and MRP2 detoxify the cell from certain metals by exporting the cations as a metal–glutathione complex. The ability of the bacterial Atm1 protein to efflux metal–glutathione complexes appears to have evolved over time to become the ABCB7 transporter in mammals, located in the inner mitochondrial membrane. No longer needed for the role of cellular detoxification, ABCB7 appears to be used to transport glutathione-coordinated iron–sulfur clusters from mitochondria to the cytosol.
Graphical Abstract
Graphical Abstract.
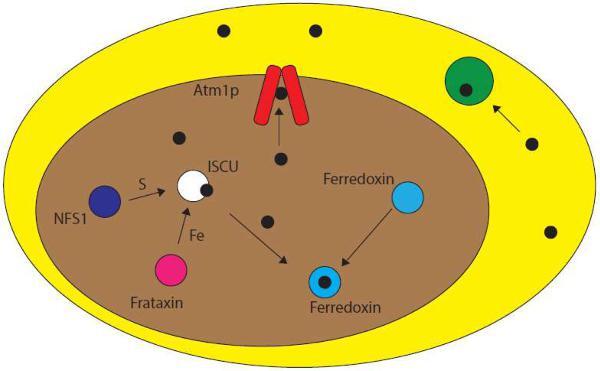
Glutathione has long been implicated in the trafficking and export of cellular metal species. Recent evidence suggests a connection to Fe-S cluster assembly and trafficking. In eukaryotes, Fe–S cluster biosynthesis is mediated through the mitochondrial scaffold protein, ISCU. Clusters are then transferred to apo targets within the mitochondria or exported to the cytosol via an ABCB7-type protein, such as Atm1p (yeast). This transport protein is thought to have evolved from heavy metal transporters via endosymbiotic evolution from the alphaproteobacteria bacterial family.
Significance to Metallomics
Glutathione adducts of metal ions and complex metallocofactors serve as substrates for cellular detoxification or mobilization by membrane-spanning transporters. Eukaryotic mitochondrial ABC-type exporters appear to be based on bacterial heavy metal detoxification transporters, consistent with the endosymbiotic hypothesis. In particular, bacterial heavy metal transporters display striking homologies and glutathione-binding chemistry to human ABCB7, localized to the inner mitochondrial membrane, which is suggested to export glutathione-coordinated iron–sulfur clusters from mitochondria for use in the cytosol.
Background
Glutathione (GSH; (2S)-2-amino-4-{[(1R)-1-[(carboxymethyl)carbamoyl]-2-sulfanyl ethyl]carbamoyl}butanoic acid) is the major thiol-containing species in both prokaryotic and eukaryotic cells, with levels reaching ∼10 mM.1,2 It is biosynthesized from three amino acids, glutamate, glycine, and cysteine, in reactions that are catalyzed by two enzymes, namely γ-glutamylcysteine synthetase and GSH synthetase. The former catalyzes the dehydration reaction between the γ-carboxyl group of glutamate and the amino group of cysteine. After the dipeptide is formed through the γ-linkage, glutathione synthetase adds glycine to the carboxyl group of cysteine, forming glutathione. Reduced glutathione exists in equilibrium with its oxidized dimer (GSSG), with the GSH:GSSG ratio varying depending on the organism (∼100-fold greater GSH in mammalian cells and 50 000 times greater in Saccharomyces cerevisiae) and cellular compartment.3,4
Glutathione performs many cellular roles, including reduction of proteins, neutralization of reactive oxygen species and free radicals, regulation of the nitric cycle, DNA replication and repair, protein production and activation, amino acid transport, iron metabolism, and cell differentiation and death.5–11 In animals, glutathione has been found to play a role in preventing and treating acetaminophen overdose, and detoxification of hydrophobic compounds in the liver and methylglyoxal in the glyoxalase system.12–16 In plants, glutathione takes part in the glutathione–ascorbate cycle, is a phytochelatin precursor, helps with defense against pathogens, and plays a role in flower development.10,17–20
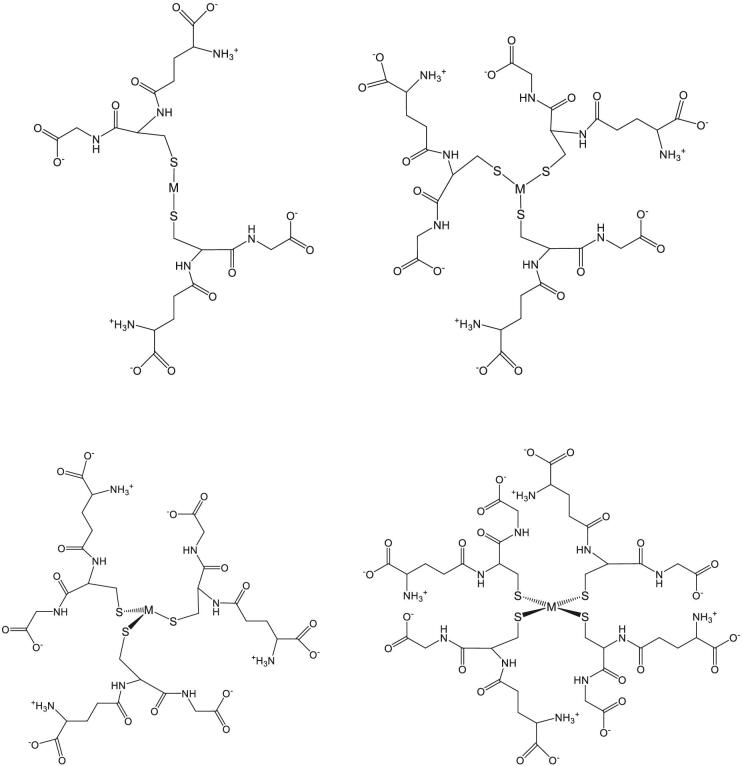
A principal reason for the ability of glutathione to serve the cell in such a large variety of roles stems from the redox activity of the sulfhydryl group and its ability to both bind to metals and react with other thiol groups. The structure of various metal–glutathione complexes is shown in Fig. 1. The sulfur of reduced glutathione has a high affinity for softer metal cations and complexes, such as mercury, lead, arsenic, and cisplatin.21 Glutathione has also been shown to coordinate iron–sulfur (Fe–S) clusters (Fig. 2).22 The high affinity for metals and metal complexes allows for glutathione to be an efficient molecule for heavy metal detoxification by one of three methods, namely sequestration, reduction, or efflux.23 A summary of metals able to form such complexes is shown in Fig. 3 and transporter proteins identified for a variety of metal–glutathione complexes are detailed in> Table 1.
Fig. 1.
Metal–glutathione complex structures. Structures of different metal–glutathione complexes described in the text. The metal in each structure is indicated by “M”. Top left: linear; top right: trigonal planar; bottom left: trigonal pyramidal; bottom right: tetrahedral.
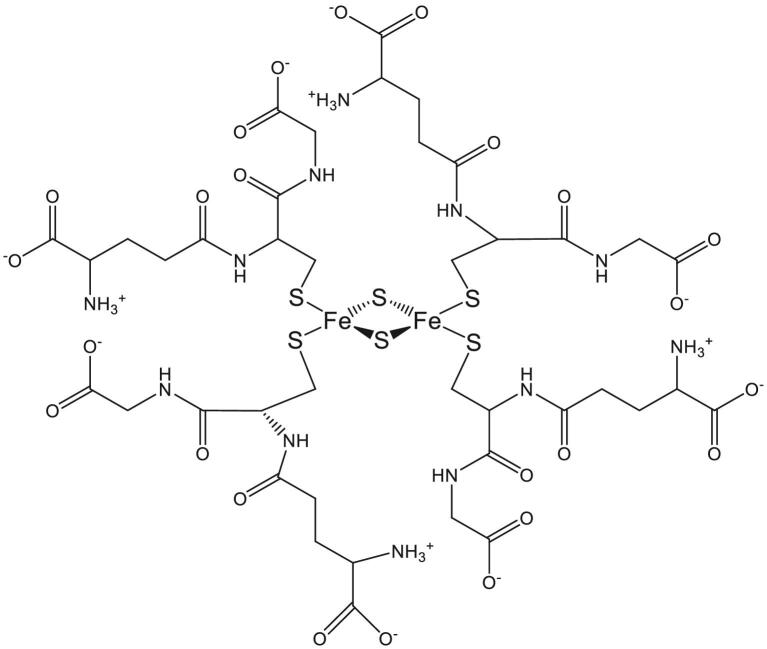
Fig. 2.
Structure of glutathione-coordinated iron–sulfur cluster. A 2D representation of the “macrocyclic-like” tetrameric glutathione network in [2Fe-2S](GS)4, stabilized by intramolecular salt bridges between the amine and carboxylate groups of neighboring glutathiones.109
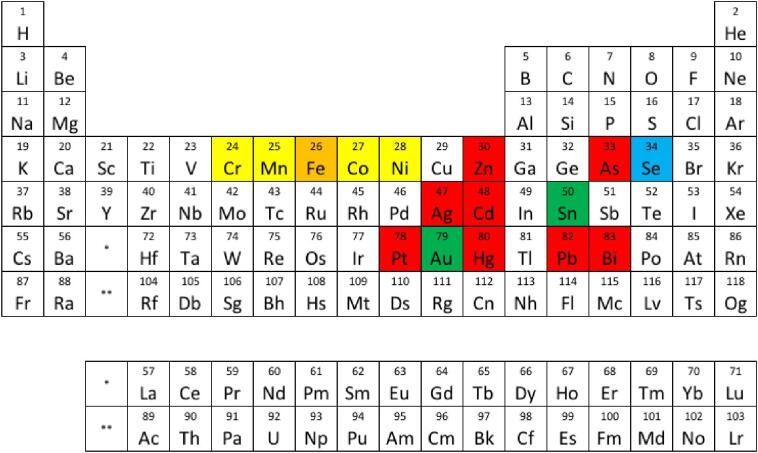
Fig. 3.
Metal–glutathione complexes and ABC transporters. Metals in red are known to be transported and/or stimulate ATPase activity as a metal–glutathione complex.21,25–27,32,50,58,69,72,74,77,110–112 Metals highlighted in yellow have been shown to not be transported as a metal–glutathione complex in known heavy metal transporters.23,31,81,83,113 Metals highlighted in green most likely bind to phytochelatin in plants.68 GSH plays a role in tolerance to the metals highlighted in blue, but the mechanism of tolerance is unlikely to be through export via ABC transporters.83 Metals highlighted in orange have been shown to be transported by ABC transporters as a metal–sulfur cluster complexed by glutathione.114
Table 1.
Summary of metals transported by ABC transporters as a GSH complex
| Protein | Metal transported | Organism of tested protein |
|---|---|---|
| MRP1/2 | Hg | S. purpuratus,25Rattus,28,29Drosphilia30 |
| As | Rattus,35C. elegans49 | |
| Zn | Rattus 69 | |
| Bi | H. sapiens 77 | |
| Pt | H. sapiens 80 | |
| Ycf1 | Hg | S. cerevisiae 33 |
| As | S. cerevisiae 37 | |
| Pb | S. cerevisiae 53 | |
| Cd | S. cerevisiae 62 | |
| HMT-1/ABCB6 | As | C. elegans 49 |
| Cd | H. sapiens 65 | |
| PGPa (MRPA) | As | L. tarentolae 47 |
| Yor1 | Cd | S. cerevisiae 62 |
Metal–glutathione complex transport
Mercury
Methyl mercury rapidly combines with GSH to form a stable tripeptide complex that circumvents heavy metal poisoning.24 The first evidence that glutathione may play a role in the secretion of metals into the bile was obtained from animal studies with rats, where mercury was found to be complexed by glutathione following administration of methyl mercury. Glutathione levels have also been correlated to the amount of both methyl mercury and inorganic mercury transported into rat bile. Organic anions such as sulfobromophthalein (BSP), indocyanine green (ICG), and phenol-3,6-dibromphthalein disulfonate (DBSP) were observed to inhibit both GSH and methyl mercury biliary secretion without affecting the normal flow of bile or levels of Hg and GSH in the liver. Moreover, the rate of biliary excretion of methyl mercury is independent of methyl mercury concentration but is dependent on GSH, consistent with the importance of GSH in protection against mercury poisoning.24
The mechanism by which mercury is exported from the cell is likely to involve an ABC-type transport protein and mercury as a GSH complex.21,25–27 Hg-GSH conjugates were found to stimulate the ATPase activity of MRP1 and MRP2, with GSH alone having no additional effect on ATPase activity. Mercuric chloride resulted in upregulation of GSH levels, and normal levels of GSH efflux were diminished in the presence of mercury, while HgCl2 inhibited transport of the substrate calcein acetoxymethylester by both human MRP1 and MRP2 in MDCKII cells, suggesting that the Hg–GSH complex is exported through MRP1 and MRP2.21 Mercury export by MRP has also been demonstrated in sea urchins25 and mammals.28,29
Drosophila MRP is highly homologous to human MRP1 (50% identity and 67% similarity), making it a good model to study the effect of methyl mercury on organism development. MRP mutant flies exposed to mercury were less likely to develop into an adult. Knockdown studies show that methyl mercury accumulated in the Malpighian tubules, gut, and nervous system, suggesting that MRP1 may help prevent neurotoxicity of methyl mercury in humans.30
In Escherichia coli, it has long been known that GSH plays a role in resistance to mercury,31 but it was not until recently that a bacterial ABC transporter with the potential to transport a Hg–GSH complex was identified.32 The protein identified in Novosphingobium aromaticivorans, Atm1, is a homolog to other heavy metal transporters in yeast and humans and conveys resistance to mercury in bacteria. The form of mercury that appears to be the substrate for this protein is Hg(GS)2, which is the same form that has been identified as a substrate in eukaryotes.32 Additionally, Ycf1 has been shown to detoxify S. cerevisiae of mercury by transport of Hg(GS)2 via an ATP-dependent mechanism. Ycf1 also plays a role in detoxification of other heavy metals, as discussed later.33
Arsenic
As–GSH complexes have been shown to form at physiological and clinically relevant levels between 0.2 and 0.5 µM but appear to be unstable.21,34 In canine cells transfected with genes for human MRP1 and MRP2, administration of As2O3 resulted in upregulation of MRP2 and to a lesser extent MRP1. These cells lines also had elevated levels of GSH when exposed to arsenic, and As2O3 inhibited calcein transport.21 These data suggest a role for GSH in mediating the export of arsenic from the cell by MRP1 and MRP2, potentially as As(GS)3; however, the identity of the transported species is yet to be confirmed.21,35 In rats, GSH also mediates the biliary secretion of As(III), while GSH is also involved in the metabolic processing of arsenate to arsenite and formation of monomethylated arsenic.35 The reduction of arsenate to arsenic has also been reported in human erythrocytes and is of physiological relevance as As(V) is the most common form of environmental arsenic.35,36
Yeast cells may have multiple ways of dealing with arsenic poisoning, two of which involve export proteins (Acr3 and Ycfl).37 Acr3 yields higher tolerance as Ycf1 cells were less sensitive to arsenic. However, this is likely concentration dependent, as As efflux through Acr3 saturates quickly. Acute arsenic stress results in arsenic coordination to proteins. However, long-term exposure upregulates GSH and leads to the formation of cellular As(GS)3. In Acr3 knockout cells, Ycf1 becomes upregulated when exposed to arsenic due to the need for Ycf1 to transport more As(III) in the cell. Similar to other metals, As(III) is transported into vacuoles by Ycf1 as a glutathione-coordinated metal complex, and most of the metal is found in vacuoles. Even with the As(III) exporter knocked out, yeast cells can rid themselves (albeit slowly) of arsenic, suggesting a mechanism for arsenic export from the vacuole.37
Arsenic export is especially of interest in humans as As2O3 is used to induce apoptosis to treat a range of cancers, yet resistance to As2O3 has been observed.38–43 Resistance has been linked to higher expression levels of GSTP1-1, a glutathione transferase, in leukemia, lymphoma, and prostate cancers.38,39,41,42,44 GSH reduces arsenate to arsenite, while GSTP1-1 is needed to form As(GS)3, which has been established as a MRP1 substrate in cancer cells.45 As(GS)3 has a very high affinity for MRP1, with a KM of 0.32 µM, and MRP2 can transport As(GS)346 in addition to the mercury–glutathione complexes discussed earlier.
Glutathione also plays a key role in arsenic detoxification in lower eukaryotes. The identity of the ABC transporter for the As(GS)3 complex has been a topic of debate since it was observed that PGPa (phosphatidylglycerophosphatase A) located in the plasma membrane of Leishmania did not convey arsenic resistance, but the protein does have the ability to transport the complex.47,48 In Leishmania, the mechanism of arsenic detoxification is similar to that of Ycf1 in yeast, sequestering the As(GS)3 complex in vesicles within the organism.48 In C. elegans, arsenic tolerance has been linked to the expression of MRPs (as seen with mercury) and HMT-1.49 Based on studies of homologous proteins in other eukaryotes, it is likely that arsenic is transported by these proteins as an As(GS)3 complex.
Lead
When S. cerevisiae is exposed to lead, free GSH levels decrease, most likely due to Pb(II) forming a cytosolic complex with glutathione as observed with other heavy metals [Hg(II), As(III), etc.]. The lack of glutathione did not raise the susceptibility of S. cerevisiae to lead, indicating that GSH itself does not detoxify the cell.50 Glutathione transferase deletion in yeast results in increased sensitivity to 0.5–1 mM Pb(II) compared with wild type, suggesting that glutathione transferases are needed to form Pb–GSH complexes. However, as a result of the kinetic lability of the complex, it is likely that the dependence arises through an indirect pathway.51,52 The mechanism for lead detoxification in S. cerevisiae most likely involves compartmentalization of Pb–GSH complexes inside vacuoles, as knockouts of ABC transporters Ycf1, VMR1, YBT1, and BPT1 resulted in increased sensitivity to lead, increasing the number of exporters that can potentially transport glutathione-coordinated metals.51 Other studies conducted with S. cerevisiae show that Pb(II) levels in the cell reach the same levels as the environment and confirmed that Pb(II) tolerance is Ycf1 dependent.53
Relative to cadmium and mercury, little is known concerning transport of glutathione-coordinated lead complexes via ABC-type transporters. In rat bile, the form of lead that is transported appears to be a lead–glutathione complex, although the protein through which this occurs has not been identified but could potentially be the same protein involved in the biliary secretion of mercury–glutathione conjugates.54
Antimony
Antimony resistance is of interest to the scientific community due to its use in drugs. Leishmaniasis is caused by Leishmania, which is prevalent in Nepal and Northern India. Antimonial drugs were used to treat Leishmaniasis until 2005; however, Leishmania has developed resistance to antimony-based drugs, as indicated in 60% of Indian patients.55 Overexpression of two ABC transporters, MRPA (PGPa) and ABCI4, has been linked to antimony resistance in Leishmania via sequestration in vesicles and export from the cell, respectively.55,56 These two transporters, in addition to ABCG2, have been shown to transport metal–thiol complexes.55 ABCG2 has also been shown to transport glutathione, increasing the likelihood that the thiol species is a component of the transported metal complex.57
Cadmium
Cadmium is a toxin that causes mutations and, like many other metals, GSH plays a key role in the natural detoxification pathway. When cells are exposed to cadmium, the primary form that is prevalent in the cell is Cd(GS)2.58 Similar to lead, it is thought that glutathione transferases play a role in formation of the Cd(GS)2 complex via an indirect pathway.59–61 ABC transporters Yor1p and Ycf1 play a role in cadmium detoxification in S. cerevisiae, although by different mechanisms. Yor1 exports Cd(GS)2 directly from the cell, while Ycf1 transports it into vacuoles. At normal levels, both proteins are required to convey cadmium tolerance in S. cerevisiae when exposed to 40 µM Cd(II). However, overexpression of Yor1 was able to restore ΔYcf1p cells to wild-type growth rates.62
Multiple Cd(GS)2 efflux proteins have also been found in animal cells. In the case of the cystic fibrosis transmembrane conductance regulator protein (CFTR), a chloride channel, exposure to Cd(II) of CFTR knockout mouse cells prompted necrosis, while normal cells undergo apoptosis. When exposed to 5 µM Cd(II), ABCC7 (CFTR) conductance was stimulated, suggesting cadmium to be exported by CFTR, potentially as a Cd(GS)2 complex.63,64 ABCC1 (MRP1) is another candidate for Cd(GS)2 efflux.64 Similar to Ycf1 in yeast, and as observed with arsenic, it is thought that HMT1 (ABCB6) can transport Cd(GS)2 into human vacuoles to sequester the toxic metal.65
Similar to mercury, detoxification in plants and bacteria appears to be conveyed by a different mechanism than in yeast and animals.66 Based on transport studies with proteins proposed to convey cadmium resistance in E. coli, CzcP, and CadA, transport of the metal was inhibited at glutathione concentrations of 3 mM, demonstrating that the proteins do not transport a glutathione coordinated metal, and likely transport the metal as the ion itself.67 In plants, the mode of detoxification is by sequestration through phytochelatins, as seen with other metals.66,68
Zinc
MRP2 mutant rats are unable to export endogenous zinc efficiently, while bile in the same mutants contained no zinc even after IV injection of the metal ion.69 Other studies have suggested that biliary zinc excretion is GSH dependent.70,71 Mutant rats injected with cadmium ions behaved similarly, suggesting that the biliary excrement of Zn(II) and Cd(II) share a similar mechanism of export through MRP2.69 It is likely that Zn(II) is excreted as a Zn–GSH complex, in comparison with other divalent metals that are biliary excreted as glutathione complexes through the ABC transporter MRP2.72
In drosophila, an ABC transporter with 26% identity to scYCF1 is upregulated in the presence of zinc ions, as well as copper and cadmium.73 YCF1 transports Cd–GSH complexes into vesicles, sequestering cadmium inside.62 It is possible that zinc tolerance is conveyed in a similar manner as glutathione S-transferase genes are upregulated in the presence of both cadmium and zinc.73
Copper
Cu(I) is exported through another ABC transporter, cCOP; however, this transporter does not transport a Cu–GSH complex. CopB was shown to transport copper without GSH, but it was DTT dependent.69Saccharomyces cerevisiae proteins of the multidrug resistance associated family, which have been shown to transport glutathione conjugated metals, do not appear to play a role in copper tolerance.74 However, while copper transporters in humans do not appear to transport copper via a glutathione complex, transport still appears to be GSH dependent.75,76
In the bacteria Pneumococci, GSH conveys Cu resistance as it was observed that copper tolerance decreased in gshT and gor mutants. These two genes, encoding an ABC transporter that transports GSH into the cell and a glutathione reductase, are required for normal cellular levels of GSH to be achieved. GSH can convey copper tolerance by complex formation and likely export via copA.31
Bismuth
Bismuth has a high affinity for glutathione, readily forming complexes with glutathione in cells where it is transported into vesicles via MRP, as observed in human HK-2 cells.77 Bismuth upregulates GSH levels, potentially forming a positive feedback of bismuth uptake, as the more the GSH is present in the cells, the more bismuth is absorbed. Similar to other post-transition metals, formation of the bismuth–glutathione complex, Bi(GS)3, is thermodynamically favorable, but kinetically labile, as the exchange rate between GSH molecules is 1500 s–1.77
Platinum
Resistance to cisplatin, a common anti-cancer drug, is glutathione dependent. From cisplatin, Pt(GS)2 can be formed as first observed in leukemia cells. The export of this compound is ATP dependent.78,79 MRP2 (ABCC2) exports Pt(GS)2 as HEK-293 cells expressing MRP2 were found to be resistant to cisplatin, and this resistance is GSH dependent. Both γ-GCSh and MRP2 are expressed at higher levels when exposed to cisplatin. Increased expression of γ-GCSh, the catalytic domain of γ-GCS, and one of the enzymes required for GSH biosynthesis, results in higher levels of GSH in the cell. However, it is unlikely that higher levels of cellular GSH would lead to increased resistance to cisplatin, as GSH is at millimolar levels in the cell, while platinum concentration would be significantly lower (micromolar levels at most).80 As stated earlier, MRP2 can transport a variety of metal–glutathione complexes, including glutathione complexes containing mercury and arsenic.21
Silver
The ATPase activity of N. aromaticivorans was stimulated in the presence of several compounds, including metal–glutathione complexes. Although transport of a silver–glutathione complex was not directly tested, resistance to Ag and Hg was conveyed in E. coli through expression of NaAtm1 and was consistent with transport of a metal–glutathione complex. The Km was 10-fold lower for Ag(GS)2 than for Hg(GS)2, suggesting that the protein can detoxify the cell of silver more effectively than mercury.32 Other proteins with the ability to transfer glutathione-coordinated silver have not been identified, although phytochelatins have been implicated in silver sequestration.68
Other metals
Glutathione is important for the detoxification of several other metals, but the proteins identified to transport glutathione-conjugated metals have not been shown to play a role in tolerance to cobalt, manganese, nickel, or chromium. In Arabidopsis, elevated levels of GSH confer nickel tolerance, most likely by protecting against the oxidative stress induced by nickel.81 Glutathione does not have an effect on chromium resistance in S. cerevisiae, but it does in E. coli.23,31 In addition, cancer cells expressing MRP1, which were resistant to arsenic and mercury, showed no resistance to chromate.82 GSH has also been shown to play a role in selenium tolerance, but this is likely a result of limiting uptake.83 In plants, phytochelatin synthase is upregulated when exposed to tin and gold, suggesting the ability of phytochelatins to protect plants against the toxicity of these metals.68
Iron–sulfur clusters
Biogenesis
There are three classes of iron–sulfur cluster assembly systems used in prokaryotes, namely ISC, NIF, and SUF.84 The ISC and SUF apparatus are found in both prokaryotes and eukaryotes, further suggesting that the mitochondria of eukaryotes retained the Fe–S cluster biosynthesis pathway following endosymbiosis from prokaryotic precursors.84,85 In all systems, the same general mechanism of synthesis is at play. An iron donor, sulfur donor, and electron donor all interact with a scaffold protein on which the cluster is built, and then subsequently transferred to a wide variety of apo Fe–S cluster acceptor proteins (Fig. 4).84
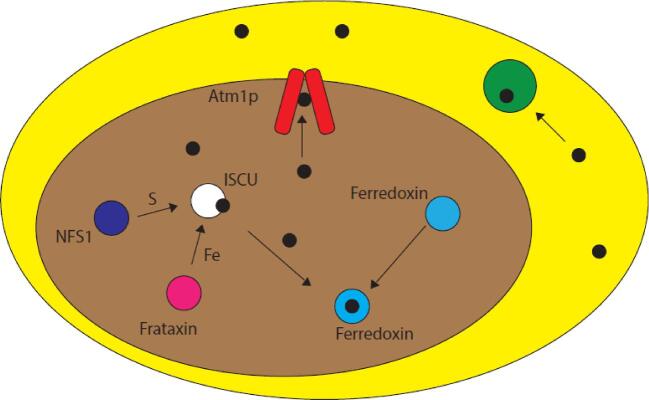
Fig. 4.
Eukaryotic Fe–S cluster biogenesis. In eukaryotes, iron–sulfur cluster biogenesis takes place in the mitochondria (brown) on the scaffold protein ISCU (white). Iron is donated to ISCU from frataxin (pink) and sulfur is donated by the cysteine desulfurase NFS1 (purple). Fe–S clusters (black) built on ISCU can then be transferred to apo targets within the mitochondria (e.g. ferredoxin, blue) or exported out of the mitochondria to the cytosol (yellow) via an ABCB7-type protein (red) such as Atm1p (yeast) for use in cytosolic Fe–S cluster proteins (green). Cluster transfer may be facilitated by other proteins.
Atm1p homology
The yeast and human homologs of the same protein that transport glutathione conjugates of mercury and silver have shown the ability to transfer glutathione-coordinated iron–sulfur clusters.86–88 Although these results have yet to be confirmed in vivo (a technically challenging problem), it has been shown that [2Fe–2S](GS)4 can reconstitute a number of apo iron–sulfur cluster proteins,89–91 and it is known that ABCB7-type proteins are involved in cytosolic Fe–S cluster maturation.92 Although other chemical species have been shown to bind or be transported by Atm1/ATM3, such as GSSG and glutathione trisulfide (GS-S-SG),92 these molecules are either already present in the cytosol and/or are not known to be directly involved in Fe–S cluster maturation. Binding by glutathione adducts is consistent with binding sites characterized for oxidized glutathione in NaAtm1p, where metal–glutathione adducts were observed to bind, and were likely conserved.32 The [2Fe-2S](GS)4 complex appears to be a more promising candidate to bridge the mitochondrial and cytosolic cluster maturation pathways as it has been shown to be biosynthetically accessible by glutathione extraction from the scaffold protein ISCU1, and demonstrated to be a molecular substrate that is transported by Atm1/ABCB7.83,84
In eukaryotes, the biosynthesis of Fe–S clusters is compartmentalized within the mitochondria, and in S. cerevisiae, Atm1p is located in the inner mitochondrial membrane (Fig. 4). As the mitochondria are hypothesized to have originated from a bacterial cell, the iron–sulfur cluster assembly machinery in the mitochondria (ICA) has been highly conserved from bacteria to man.93 The ability for NaAtm1p to transport glutathione-coordinated metals, having two binding sites for oxidized glutathione and mimicking a glutathione-coordinated iron–sulfur cluster, combined with the high homology with ScAtm1p, all support a structural model where the genes for NaAtm1p and ScAtm1p diverge from a common ancestral gene.32
The common ancestral gene is most likely the one found in the bacteria that originally became mitochondria in eukaryotes. Saccharomyces cerevisiae Atm1p shares a high % similarity with N. aromaticivorans (Na) Atm1p (Fig. 5),32 a bacterial strain from the alphaproteobacteria family that is thought to have developed into mitochondria through the endosymbiotic theory.94–102 BLAST analysis shows that the NaAtm1p gene is very similar to other Atm1p genes in the alphaproteobacteria family, including sphingomondae.88,103 The NaAtm1p gene was most closely related to this genus of bacteria, which is commonly found in environments containing toxic compounds.104 Since glutathione is involved in detoxification of several different types of compounds, including metals, this is the likely need for such proteins in these bacteria. In bacteria, there is no need to transport iron–sulfur clusters out of the cell, as they would be retained for use with cytoplasmic proteins.
Fig. 5.
Alignment of ScAtm1p, NaAtm1p, and HsABCB7. ScAtm1p, NaAtm1p, and HsABCB7 have high sequence identity, with NaAtm1p showing 42.74% and 45.39% sequence identity with ScAtm1p and HsABCB7, respectively.
The top 100 matches for the NaAtm1p sequence were all found in alphaproteobacteria. When expanded to 500 matched results, the protein sequence also matched to several betaproteobacteria, mainly Bordetella and Achromobacter.88,103 The bacteria of both of these genera are pathogenic in humans, with Bordetella being more aggressive and Achromobacter affecting immunocompromised individuals.105,106 These bacteria are in the same phylum as alphaproteobacteria, proteobacteria, which also have Atm1p genes. Interestingly, of the 416 organisms that appeared in the BLAST results when omitting proteobacteria, 383 are eukaryotes, including some of the top matches (Streptomyces cavourensis and Nephila clavipes).88 This supports the idea of the Atm1p genes having a common bacterial ancestor that evolved after mitochondria originated.
The BLAST results in the animal kingdom that most closely matched the protein sequence for NaAtm1p were those of the arthropods Ixodes scapularis (deer tick) and N. clavipes (spider). Other arthropods that matched the protein sequence include multiple species of drosophila (fly), Nilaparvata lugens (Brown planthopper), and non-chordates Hypsibius dujardini (tardigrade), Acanthamoeba castellanii, (amoeba), and Acanthaster planci (starfish) also matched to NaAtm1. Higher animals in the chordate phylum also matched to the bacterial gene, including Python bivittatus (snake) and multiple species of turtles (Chrysemys picta bellii and Terrapene mexicana triunguis) and birds (Cyanistes caeruleus and Amazona aestiva). The highest organism to show up in the results was a koala (Phascolarctos cinereus).88
Not surprisingly, several different fungi also appeared in the BLAST results. This was expected, as ScAtm1p is a protein known to reside in the inner mitochondrial membrane and has high homology with NaAtm1p. Several species of Aspergillus contained proteins with similar sequences to NaAtm1p. Other fungi that appeared in the BLAST results include Basidiobolus ranarum, Rhizophagus irregularis, Cercospora beticola, Emergomyces pasteuriana, and Paecilomyces variotii.88
Organisms from the plant kingdom were the least represented when conducting a BLAST analysis that omitted all proteobacteria, as only red and green algae and mosses (Physcomitrella patens and Selaginella moellendorffii) appeared in the results. Selaginella moellendorffii, which is considered a primitive moss, is the plant with the highest homology. The lower representation of plants in the eukaryotic group most likely reflects the higher homology that several fungi and animals share with NaAtm1p, as fungi and animals are more closely related to each other than to plants. Additional evidence suggests that the Fe–S cluster assembly machinery in plants evolved from cyanobacteria, again based on the endosymbiotic theory.107,108 The BLAST analysis suggests that plant genes related to Atm1p have diverged from bacteria more so than fungi and animal genes, as the moss Selaginella moellendorffii is considered to be a primitive plant.88,103 This also explains the different roles for glutathione in mediating metal transport in bacteria, animals, plants, and fungi, as summarized earlier. In bacteria, animals, and fungi, glutathione is used by some proteins to transport glutathione-conjugated metals, but in plants, phytochelatins, a glutathione derivative, is the main mechanism by which metals are chelated and trafficked.
Conclusions
In conclusion, there appears to be clear evolutionary development of eukaryotic mitochondrial ABC-type exporters from bacterial heavy metal detoxification transporters, as expected from the endosymbiotic theory. Metal–glutathione complexation arose in bacterial cells as a means for heavy metal detoxification, by sequestration and efflux. In plants, phytochelatins, for which glutathione is a precursor, are used for heavy metal detoxification. The mode of heavy metal detoxification is more conserved in yeast and animals, as complex formation, followed by efflux through an ABC transporter, is similar to that of bacteria. Glutathione plays multiple roles in this process. In addition to binding to metals to reduce toxicity, it also facilitates recognition of the metal–glutathione complex through contacts between glutathione and the transporter. It would be evolutionarily unfavorable for heavy metal exporters to export metal ions themselves, as this would likely require multiple proteins, whereas similar metal–glutathione complexes can be recognized by the same protein. Proteins such as NaAtm1 contain multiple sites that can bind glutathione, providing flexibility to recognize several different types of compounds, including multiple metal–glutathione complexes. Following evolution from prokaryotes to eukaryotes, Atm1 was no longer required for the export of heavy metals, as it is located in the mitochondrial membrane. Instead, the binding pocket appears to have evolved to recognize a related molecular complex (glutathione-complexed iron–sulfur clusters) for export of these essential cofactors to the cytosol.
Contributor Information
Stephen A Pearson, The Ohio State University Biophysics Program, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210, USA.
J A Cowan, The Ohio State University Biophysics Program, The Ohio State University, 484 West 12th Avenue, Columbus, OH 43210, USA; Department of Chemistry and Biochemistry, The Ohio State University, 100 West 18th Avenue, Columbus, OH 43210, USA.
Funding
This work was supported by a grant from the National Institutes of Health (AI072443).
Conflicts of interest
The authors declare no conflicts of interest.
Data availability
No new data were generated or analyzed in support of this research.
References
- 1. Griffith O. W., Biologic and pharmacologic regulation of mammalian glutathione synthesis, Free Radic. Biol. Med., 1999, 27 (9–10), 922–935. [DOI] [PubMed] [Google Scholar]
- 2. Jones D. P., Redox potential of GSH/GSSG couple: assay and biological significance, Methods Enzymol., 2002, 348, 93–112. [DOI] [PubMed] [Google Scholar]
- 3. Giustarini D., Colombo G., Garavaglia M. L., Astori E., Portinaro N. M., Reggiani F., Badalamenti S., Aloisi A. M., Santucci A., Rossi R., Milzani A., Dalle-Donne I., Assessment of glutathione/glutathione disulphide ratio and S-glutathionylated proteins in human blood, solid tissues, and cultured cells, Free Radic. Biol. Med., 2017, 112, 360–375. [DOI] [PubMed] [Google Scholar]
- 4. Horn T., Bettray W., Slusarenko A. J., Gruhlke M. C. H., S-allylmercaptoglutathione is a substrate for glutathione reductase (E.C. 1.8.1.7) from yeast (Saccharomyces cerevisiae), Antioxidants, 2018, 7 (7), 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu S. C., Glutathione synthesis, Biochim. Biophys. Acta Gen. Subj., 2013, 1830 (5), 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grant C. M., Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions, Mol. Microbiol., 2001, 39 (3), 533–541. [DOI] [PubMed] [Google Scholar]
- 7. Dringen R., Metabolism and functions of glutathione in brain, Prog. Neurobiol., 2000, 62 (6), 649–671. [DOI] [PubMed] [Google Scholar]
- 8. Scholz R. W., Graham K. S., Gumpricht E., Reddy C. C., Mechanism of interaction of vitamin-E and glutathione in the protection against membrane lipid-peroxidation, Ann. N. Y. Acad. Sci., 1989, 570 (1), 514–517. [Google Scholar]
- 9. Hughes R. E., Reduction of dehydroascorbic acid by animal tissues, Nature, 1964, 203 (4949), 1068–1069. [DOI] [PubMed] [Google Scholar]
- 10. Ha S. B., Smith A. P., Howden R., Dietrich W. M., Bugg S., O'Connell M. J., Goldsbrough P. B., Cobbett C. S., Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe, Plant Cell, 1999, 11 (6), 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar C., Igbaria A., D'Autreaux B., Planson A. G., Junot C., Godat E., Bachhawat A. K., Delaunay-Moisan A., Toledano M. B., Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control, EMBO J., 2011, 30 (10), 2044–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steullet P., Neijt H. C., Cuenod M., Do K. Q., Synaptic plasticity impairment and hypofunction of NMDA receptors induced by glutathione deficit: relevance to schizophrenia, Neuroscience, 2006, 137 (3), 807–819. [DOI] [PubMed] [Google Scholar]
- 13. Varga V., Jenei Z., Janaky R., Saransaari P., Oja S. S., Glutathione is an endogenous ligand of rat brain N-methyl-d-aspartate (NMDA) and 2-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptors, Neurochem. Res., 1997, 22 (9), 1165–1171. [DOI] [PubMed] [Google Scholar]
- 14. Janaky R., Ogita K., Pasqualotto B. A., Bains J. S., Oja S. S., Yoneda Y., Shaw C. A., Glutathione and signal transduction in the mammalian CNS, J. Neurochem., 1999, 73 (3), 889–902. [DOI] [PubMed] [Google Scholar]
- 15. Freitas H. R., Ferraz G., Ferreira G. C., Ribeiro-Resende V. T., Chiarini L. B., do Nascimento J. L. M., Oliveira K., Pereira T. D., Ferreira L. G. B., Kubrusly R. C., Faria R. X., Herculano A. M., Reis R. A. D., Glutathione-induced calcium shifts in chick retinal glial cells, PLoS One, 2016, 11 (4), e0153677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oja S. S., Janaky R., Varga V., Saransaari P., Modulation of glutamate receptor functions by glutathione, Neurochem. Int., 2000, 37 (2–3), 299–306. [DOI] [PubMed] [Google Scholar]
- 17. Noctor G., Foyer C. H., Ascorbate and glutathione: keeping active oxygen under control, Annu. Rev. Plant Biol., 1998, 49 (1), 249–279. [DOI] [PubMed] [Google Scholar]
- 18. Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F., Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis, Plant J., 2007, 49 (1), 159–172. [DOI] [PubMed] [Google Scholar]
- 19. Rouhier N., Lemaire S. D., Jacquot J. P., The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation, Annu. Rev. Plant Biol., 2008, 59 (1), 143–166. [DOI] [PubMed] [Google Scholar]
- 20. Panigrahi G. K., Verma N., Singh N., Asthana S., Gupta S. K., Tripath A., Das M., Interaction of anthraquinones of Cassia occidentalis seeds with DNA and glutathione, Toxicol. Rep., 2018, 5, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wortelboer H. M., Balvers M. G. J., Usta M., van Bladeren P. J., Cnubben N. H. P.., Glutathione-dependent interaction of heavy metal compounds with multidrug resistance proteins MRP1 and MRP2, Environ. Toxicol. Pharmacol., 2008, 26 (1), 102–108. [DOI] [PubMed] [Google Scholar]
- 22. Qi W. B., Li J. W., Chain C. Y., Pasquevich G. A., Pasquevich A. F., Cowan J. A., Glutathione complexed Fe-S centers, J. Am. Chem. Soc., 2012, 134 (26), 10745–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Helbig K., Bleuel C., Krauss G. J., Nies D. H., Glutathione and transition-metal homeostasis in Escherichia coli, J. Bacteriol., 2008, 190 (15), 5431–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ballatori N., Clarkson T. W., Biliary-secretion of glutathione and of glutathione–metal complexes, Fundam. Appl. Toxicol., 1985, 5 (5), 816–831. [DOI] [PubMed] [Google Scholar]
- 25. Bosnjak I., Uhlinger K. R., Heim W., Smital T., Franekic-Colic J., Coale K., Epel D., Hamdoun A., Multidrug efflux transporters limit accumulation of inorganic, but not organic, mercury in sea urchin embryos, Environ. Sci. Technol., 2009, 43 (21), 8374–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Long Y., Li Q., Wang Y. H., Cui Z. B., MRP proteins as potential mediators of heavy metal resistance in zebrafish cells, Comp. Biochem. Physiol. C Toxicol. Pharmacol., 2011, 153 (3), 310–317. [DOI] [PubMed] [Google Scholar]
- 27. Leslie E. M., Deeley R. G., Cole S. P. C., Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense, Toxicol. Appl. Pharmacol., 2005, 204 (3), 216–237. [DOI] [PubMed] [Google Scholar]
- 28. Bridges C. C., Joshee L., Zalups R. K., MRP2 and the handling of mercuric ions in rats exposed acutely to inorganic and organic species of mercury, Toxicol. Appl. Pharmacol., 2011, 25 (1), 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bridges C. C., Joshee L., Zalups R. K., Multidrug resistance proteins and the renal elimination of inorganic mercury mediated by 2,3-dimercaptopropane-1-sulfonic acid and meso-2,3-dimercaptosuccinic acid, J. Pharmacol. Exp. Ther., 2008, 324 (1), 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prince L., Korbas M., Davidson P., Broberg K., Rand M. D., Target organ specific activity of drosophila MRP (ABCC1) moderates developmental toxicity of methylmercury, Toxicol. Sci., 2014, 140 (2), 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Potter A. J., Trappetti C., Paton J. C., Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity, J. Bacteriol., 2012, 194 (22), 6248–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J. Y., Yang J. G., Zhitnitsky D., Lewinson O., Rees D. C., Structural basis for heavy metal detoxification by an Atm1-type ABC exporter, Science, 2014, 343 (6175), 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gueldry O., Lazard M., Delort F., Dauplais M., Grigoras I., Blanquet S., Plateau P., Ycf1p-dependent Hg(II) detoxification in Saccharomyces cerevisiae, Eur. J. Biochem., 2003, 270 (11), 2486–2496. [DOI] [PubMed] [Google Scholar]
- 34. Rey N. A., Howarth O. W., Pereira-Maia E. C., Equilibrium characterization of the As(III)-cysteine and the As(III)-glutathione systems in aqueous solution, J. Inorg. Biochem., 2004, 98 (6), 1151–1159. [DOI] [PubMed] [Google Scholar]
- 35. Csanaky I., Gregus Z., Role of glutathione in reduction of arsenate and of gamma-glutamyltranspeptidase in disposition of arsenite in rats, Toxicology, 2005, 207 (1), 91–104. [DOI] [PubMed] [Google Scholar]
- 36. Nemeti B., Gregus Z., Glutathione-dependent reduction of arsenate in human erythrocytes: a process independent of purine nucleoside phosphorylase, Toxicol. Sci., 2004, 82 (2), 419–428. [DOI] [PubMed] [Google Scholar]
- 37. Talemi S. R., Jacobson T., Garla V., Navarrete C., Wagner A., Tamas M. J., Schaber J., Mathematical modelling of arsenic transport, distribution and detoxification processes in yeast, Mol. Microbiol., 2014, 92 (6), 1343–1356. [DOI] [PubMed] [Google Scholar]
- 38. Gaffey M. J., Iezzoni J. C., Meredith S. D., Boyd J. C., Stoler M. H., Weiss L. M., Zukerberg L. R., Levine P. A., Arnold A., Williams M. E., Cyclin D1 (PRAD1, CCND1) and glutathione-S-transferase-Pi gene-expression in head and neck squamous-cell carcinoma, Hum. Pathol., 1995, 26 (11), 1221–1226. [DOI] [PubMed] [Google Scholar]
- 39. Gilbert L., Elwood L. J., Merino M., Masood S., Barnes R., Steinberg S. M., Lazarous D. F., Pierce L., Dangelo T., Moscow J. A., Townsend A. J., Cowan K. H., A pilot study of Pi-class glutathione S-transferase expression in breast cancer: correlation with estrogen-receptor expression and prognosis in node-negative breast cancer, J. Clin. Oncol., 1993, 11 (1), 49–58. [DOI] [PubMed] [Google Scholar]
- 40. Haga N., Fujita N., Tsuruo T., Involvement of mitochondrial aggregation in arsenic trioxide (As2O3)-induced apoptosis in human glioblastoma cells, Cancer Sci., 2005, 96 (11), 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harries L. W., Stubbins M. J., Forman D., Howard G. C. W., Wolf C. R., Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer, Carcinogenesis, 1997, 18 (4), 641–644. [DOI] [PubMed] [Google Scholar]
- 42. Helzlsouer K. J., Selmin O., Huang H. Y., Strickland P. T., Hoffman S., Alberg A. J., Watson M., Comstock G. W., Bell D., Association between glutathione S-transferase M1, P1, and T1 genetic polymorphisms and development of breast cancer, J. Natl. Cancer Inst., 1998, 90 (7), 512–518. [DOI] [PubMed] [Google Scholar]
- 43. Jia P. M., Chen G. Q., Huang X. J., Cai X., Yang J., Wang L., Zhou Y. H., Shen Y. L., Zhou L., Yu Y., Arsenic trioxide induces multiple myeloma cell apoptosis via disruption of mitochondrial transmembrane potentials and activation of caspase-3, Chin. Med. J., 2001, 114 (1), 19–24. [PubMed] [Google Scholar]
- 44. Russo D., Marie J. P., Zhou D. C., Faussat A. M., Damiani D., Michelutti A., Michieli M., Fanin R., Baccarani M., Zittoun R., Melli C., Evaluation of the clinical relevance of the anionic glutathione-S-transferase (GST-PI) and multidrug-resistance (MDR-1) gene coexpression in leukemias and lymphomas, Leuk. Lymphoma, 1994, 15 (5–6), 453–468. [DOI] [PubMed] [Google Scholar]
- 45. Shukalek C. B., Swanlund D. P., Rousseau R. K., Weigl K. E., Marensi V., Cole S. P. C., Leslie E. M., Arsenic triglutathione As(GS)(3) transport by multidrug resistance protein 1 (MRP1/ABCC1) is selectively modified by phosphorylation of Tyr920/Ser921 and glycosylation of Asn19/Asn23, Mol. Pharmacol., 2016, 90 (2), 127–139. [DOI] [PubMed] [Google Scholar]
- 46. Leslie E. M., Haimeur A., Waalkes M. P., Arsenic transport by the human multidrug resistance protein 1 (MRP1/ABCC1): evidence that a tri-glutathione conjugate is required, J. Biol. Chem., 2004, 279 (31), 32700–32708. [DOI] [PubMed] [Google Scholar]
- 47. Dey S., Ouellette M., Lightbody J., Papadopoulou B., Rosen B. P., An ATP-dependent As(III)-glutathione transport system in membrane vesicles of Leishmania tarentolae, Proc. Natl. Acad. Sci. USA, 1996, 93 (5), 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Legare D., Richard D., Mukhopadhyay R., Stierhof Y. D., Rosen B. P., Haimeur A., Papadopoulou B., Ouellette M., The Leishmania ATP-binding cassette protein PGPA is an intracellular metal–thiol transporter ATPase, J. Biol. Chem., 2001, 276 (28), 26301–26307. [DOI] [PubMed] [Google Scholar]
- 49. Martinez-Finley E. J., Aschner M., Revelations from the nematode Caenorhabditis elegans on the complex interplay of metal toxicological mechanisms, J. Toxicol., 2011, 2011, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perez R. R., Sousa C. A., Vankeersbilck T., Machado M. D., Soares E. V., Evaluation of the role of glutathione in the lead-induced toxicity in Saccharomyces cerevisiae, Curr. Microbiol., 2013, 67 (3), 300–305. [DOI] [PubMed] [Google Scholar]
- 51. Sousa C. A., Hanselaer S., Soares E. V., ABCC subfamily vacuolar transporters are involved in Pb (lead) detoxification in Saccharomyces cerevisiae, Appl. Biochem. Biotechnol., 2015, 175 (1), 65–74. [DOI] [PubMed] [Google Scholar]
- 52. Mah V., Jalilehvand F., Lead(II) complex formation with glutathione, Inorg. Chem., 2012, 51 (11), 6285–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wysocki R., Tamas M. J., How Saccharomyces cerevisiae copes with toxic metals and metalloids, FEMS Microbiol. Rev., 2010, 34 (6), 925–951. [DOI] [PubMed] [Google Scholar]
- 54. Alexander J., Aaseth J., Mikalsen A., Excretion of lead in rat bile: the role of glutathione, Acta Pharmacol. Toxicol., 1986, 59 (S7), 486–489. [DOI] [PubMed] [Google Scholar]
- 55. Perea A., Manzano J. I., Castanys S., Gamarro F., The LABCG2 Transporter from the protozoan parasite Leishmania is involved in antimony resistance, Antimicrob. Agents Chemother., 2016, 60 (6), 3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Manzano J. I., Garcia-Hernandez R., Castanys S., Gamarro F., A new ABC half-transporter in Leishmania major is involved in resistance to antimony, Antimicrob. Agents Chemother., 2013, 57 (8), 3719–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Perea A., Manzano J. I., Kimura Y., Ueda K., Castanys S., Gamarro F., Leishmania LABCG2 transporter is involved in ATP-dependent transport of thiols, Biochem. J., 2018, 475 (1), 87–97. [DOI] [PubMed] [Google Scholar]
- 58. Leverrier P., Montigny C., Garrigos M., Champeil P., Metal binding to ligands: cadmium complexes with glutathione revisited, Anal. Biochem., 2007, 371 (2), 215–228. [DOI] [PubMed] [Google Scholar]
- 59. Rai R., Tate J. J., Cooper T. G., Ure2, a prion precursor with homology to glutathione S-transferase, protects Saccharomyces cerevisiae cells from heavy metal ion and oxidant toxicity, J. Biol. Chem., 2003, 278 (15), 12826–12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lewinska A., Bartosz G., Protection of yeast lacking the Ure2 protein against the toxicity of heavy metals and hydroperoxides by antioxidants, Free Radic. Res., 2007, 41 (5), 580–590. [DOI] [PubMed] [Google Scholar]
- 61. Adamis P. D. B., Gomes D. S., Pinto M., Panek A. D., Eleutherio E. C. A., The role of glutathione transferases in cadmium stress, Toxicol. Lett., 2004, 154 (1–2), 81–88. [DOI] [PubMed] [Google Scholar]
- 62. Nagy Z., Montigny C., Leverrier P., Yeh S., Goffeau A., Garrigos M., Falson P., Role of the yeast ABC transporter Yor1p in cadmium detoxification, Biochimie, 2006, 88 (11), 1665–1671 [DOI] [PubMed] [Google Scholar]
- 63. L'Hoste S., Chargui A., Belfodil R., Duranton C., Rubera I., Mograbi B., Poujeol C., Tauc M., Poujeol P., CFTR mediates cadmium-induced apoptosis through modulation of ROS level in mouse proximal tubule cells, Free Radic. Biol. Med., 2009, 46 (8), 1017–1031. [DOI] [PubMed] [Google Scholar]
- 64. Sigel A., Sigel H., Sigel R. K. O. (eds)., Cadmium: From Toxicity to Essentiality, Vol. 11, Springer International Publishing, Cham, 2013, pp. 1–560. [Google Scholar]
- 65. Rakvacs Z., Kucsma N., Gera M., Igriczi B., Kiss K., Barna J., Kovacs D., Vellai T., Bencs L., Reisecker J. M., Szoboszlai N., Szakacs G., The human ABCB6 protein is the functional homologue of HMT-1 proteins mediating cadmium detoxification, Cell. Mol. Life Sci., 2019, 76 (20), 4131–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Preveral S., Gayet L., Moldes C., Hoffmann J., Mounicou S., Gruet A., Reynaud F., Lobinski R., Verbavatz J. M., Vavasseur A., Forestier C., A common highly conserved cadmium detoxification mechanism from bacteria to humans: heavy metal tolerance conferred by the ATP-binding cassette (ABC) transporter SpHMT1 requires glutathione but not metal-chelating phytochelatin peptides, J. Biol. Chem., 2009, 284 (8), 4936–4943. [DOI] [PubMed] [Google Scholar]
- 67. Scherer J., Nies D. H., CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34, Mol. Microbiol., 2009, 73 (4), 601–621. [DOI] [PubMed] [Google Scholar]
- 68. Schat H., Llugany M., Vooijs R., Hartley-Whitaker J., Bleeker P. M., The role of phytochelatins in constitutive and adaptive heavy metal tolerances in hyperaccumulator and non-hyperaccumulator metallophytes, J. Exp. Bot., 2002, 53 (379), 2381–2392. [DOI] [PubMed] [Google Scholar]
- 69. Dijkstra M., Havinga R., Vonk R. J., Kuipers F., Bile secretion of cadmium, silver, zinc and copper in the rat. Involvement of various transport systems, Life Sci., 1996, 59 (15), 1237–1246. [DOI] [PubMed] [Google Scholar]
- 70. Alexander J., Aaseth J., Refsyik T., Excretion of zinc in rat bile: a role of glutathione, Acta Pharmacol. Toxicol., 1981, 49 (3), 190–194. [DOI] [PubMed] [Google Scholar]
- 71. Gregus Z., Varga F., Role of glutathione and hepatic glutathione S-transferase in the biliary-excretion of methyl mercury, cadmium and zinc: a study with enzyme inducers and glutathione depletors, Acta Pharmacol. Toxicol., 1985, 56(5), 398–403. [DOI] [PubMed] [Google Scholar]
- 72. Fardel O., Kolasa E., Le Vee M., Environmental chemicals as substrates, inhibitors or inducers of drug transporters: implication for toxicokinetics, toxicity and pharmacokinetics, Expert Opin. Drug Metab. Toxicol., 2012, 8 (1), 29–46. [DOI] [PubMed] [Google Scholar]
- 73. Yepiskoposyan H., Egli D., Fergestad T., Selvaraj A., Treiber C., Multhaup G., Georgiev O., Schaffner W., Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc, Nucleic Acids Res., 2006, 34 (17), 4866–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cui Z. F., Hirata D., Tsuchiya E., Osada H., Miyakawa T., The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions, J. Biol. Chem., 1996, 271 (25), 14712–14716. [DOI] [PubMed] [Google Scholar]
- 75. Gupta A., Lutsenko S., Human copper transporters: mechanism, role in human diseases and therapeutic potential, Future Med. Chem., 2009, 1 (6), 1125–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maryon E. B., Molloy S. A., Yu K. I. H., Kaplan J. H., Rate and regulation of copper transport by human copper transporter 1 (hCTR1), J. Biol. Chem., 2013, 288 (25), 18035–18046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hong Y. F., Lai Y. T., Chan G. C. F., Sun H. Z., Glutathione and multidrug resistance protein transporter mediate a self-propelled disposal of bismuth in human cells, Proc. Natl. Acad. Sci. USA, 2015, 112 (11), 3211–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ishikawa T., Aliosman F., Glutathione-associated cis-diamminedichloroplatinum(II) metabolism and ATP-dependent efflux from leukemia cells. Molecular characterization of glutathione–platinum complex and its biological significance, J. Biol. Chem., 1993, 268 (27), 20116–20125. [PubMed] [Google Scholar]
- 79. Ishikawa T., Wright C. D., Ishizuka H., GS-X pump is functionally overexpressed in cis-diamminedichloroplatinum(II)-resistant human leukemia HL-60 cells and down-regulated by cell differentiation, J. Biol. Chem., 1994, 269 (46), 29085–29093. [PubMed] [Google Scholar]
- 80. Chen H. H. W., Kuo M. T.., Role of glutathione in the regulation of cisplatin resistance in cancer chemotherapy, Met. Based Drugs, 2010, 2010, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Freeman J. L., Persans M. W., Nieman K., Albrecht C., Peer W., Pickering I. J., Salt D. E., Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators, Plant Cell, 2004, 16 (8), 2176–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vernhet L., Allain N., Bardiau C., Anger J. P., Fardel O., Differential sensitivities of MRP1-overexpressing lung tumor cells to cytotoxic metals, Toxicology, 1999, 142 (2), 127–134. [DOI] [PubMed] [Google Scholar]
- 83. Gharieb M. M., Gadd G. M., Role of glutathione in detoxification of metal(loid)s by Saccharomyces cerevisiae, Biometals, 2004, 17 (2), 183–188. [DOI] [PubMed] [Google Scholar]
- 84. Blanc B., Gerez C., de Choudens S. A., Assembly of Fe/S proteins in bacterial systems: biochemistry of the bacterial ISC system, Biochim. Biophys. Acta, 2015, 1853 (6), 1436–1447. [DOI] [PubMed] [Google Scholar]
- 85. Lill R., Dutkiewicz R., Elsasser H. P., Hausmann A., Netz D. J. A., Pierik A. J., Stehling O., Urzica E., Muhlenhoff U., Mechanisms of iron–sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes, Biochim. Biophys. Acta Mol. Cell Res., 2006, 1763 (7), 652–667. [DOI] [PubMed] [Google Scholar]
- 86. Li J. W., Cowan J. A., Glutathione-coordinated 2Fe-2S cluster: a viable physiological substrate for mitochondrial ABCB7 transport, Chem. Commun., 2015, 51 (12), 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pearson S. A., Wachnowsky C., Cowan J. A., Defining the mechanism of the mitochondrial Atm1p 2Fe-2S cluster exporter, Metallomics, 2020, 12 (6), 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pearson S. A., Cowan J. A., Evolution of the human mitochondrial ABCB7 2Fe-2S (GS)4 cluster exporter and the molecular mechanism of an E433K disease-causing mutation, Arch. Biochem. Biophys., 2021, 697, 108661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fidai I., Wachnowsky C., Cowan J. A., Mapping cellular Fe-S cluster uptake and exchange reactions: divergent pathways for iron–sulfur cluster delivery to human ferredoxins, Metallomics, 2016, 8 (12), 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fidai I., Wachnowsky C., Cowan J. A., Glutathione-complexed 2Fe-2S clusters function in Fe-S cluster storage and trafficking, J. Biol. Inorg. Chem., 2016, 21 (7), 887–901. [DOI] [PubMed] [Google Scholar]
- 91. Qi W., Li J., Chain C. Y., Pasquevich G. A., Pasquevich A. F., Cowan J. A., Glutathione complexed Fe-S centers, J. Am. Chem. Soc., 2012, 134 (26), 10745–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Schaedler T. A., Thornton J. D., Kruse I., Schwarzlander M., Meyer A. J., van Veen H. W., Balk J., A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly, J. Biol. Chem., 2014, 289 (34), 23264–23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Muhlenhoff U., Lill R., Biogenesis of iron–sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria, Biochim. Biophys. Acta Bioenerg., 2000, 1459 (2–3), 370–382. [DOI] [PubMed] [Google Scholar]
- 94. Gray M. W., Burger G., Lang B. F., Mitochondrial evolution, Science, 1999, 283 (5407), 1476–1481. [DOI] [PubMed] [Google Scholar]
- 95. Wang Z., Wu M., An integrated phylogenomic approach toward pinpointing the origin of mitochondria, Sci. Rep., 2015, 5, 7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Williams K. P., Sobral B. W., Dickerman A. W., A robust species tree for the Alphaproteobacteria, J. Bacteriol., 2007, 189 (13), 4578–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Fitzpatrick D. A., Creevey C. J., McInerney J. O., Genome phylogenies indicate a meaningful alpha-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales, Mol. Biol. Evol., 2006, 23 (1), 74–85. [DOI] [PubMed] [Google Scholar]
- 98. Sassera D., Lo N., Epis S., D'Auria G., Montagna M., Comandatore F., Horner D., Pereto J., Luciano A. M., Franciosi F., Ferri E., Crotti E., Bazzocchi C., Daffonchio D., Sacchi L., Moya A., Latorre A., Bandi C., Phylogenomic evidence for the presence of a flagellum and cbb3 oxidase in the free-living mitochondrial ancestor, Mol. Biol. Evol., 2011, 28 (12), 3285–3296. [DOI] [PubMed] [Google Scholar]
- 99. Andersson S. G. E., Zomorodipour A., Andersson J. O., Sicheritz-Ponten T., Alsmark U. C. M., Podowski R. M., Naslund A. K., Eriksson A. S., Winkler H. H., Kurland C. G.., The genome sequence of Rickettsia prowazekii and the origin of mitochondria, Nature, 1998, 396 (6707), 133–140. [DOI] [PubMed] [Google Scholar]
- 100. Ferla M. P., Thrash J. C., Giovannoni S. J., Patrick W. M., New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability, PLoS One, 2013, 8 (12), e83383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Darby A. C., Cho N. H., Fuxelius H. H., Westberg J., Andersson S. G. E., Intracellular pathogens go extreme: genome evolution in the Rickettsiales, Trends Genet., 2007, 23 (10), 511–520. [DOI] [PubMed] [Google Scholar]
- 102. Renvoise A., Merhej V., Georgiades K., Raoult D., Intracellular Rickettsiales: insights into manipulators of eukaryotic cells, Trends Mol. Med., 2011, 17 (10), 573–583. [DOI] [PubMed] [Google Scholar]
- 103. Boratyn G. M., Schaffer A. A., Agarwala R., Altschul S. F., Lipman D. J., Madden T. L., Domain enhanced lookup time accelerated BLAST, Biol. Direct, 2012, 7 (1), 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Aires T., Serrao E. A., Engelen A. H., Host and environmental specificity in bacterial communities associated to two highly invasive marine species (genus Sparagopsis), Front. Microbiol., 2016, 7, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ray C. G., Sherris J. C., Ryan K. J., Sherris Medical Microbiology: An Introduction to Infectious Diseases, McGraw-Hill, New York, 2003. [Google Scholar]
- 106. Swenson C. E., Sadikot R. T., Achromobacter respiratory infections, Ann. Am. Thorac. Soc., 2015, 12 (2), 252–258. [DOI] [PubMed] [Google Scholar]
- 107. Archibald J. M., Endosymbiosis and eukaryotic cell evolution, Curr. Biol., 2015, 25 (19), R911–R921. [DOI] [PubMed] [Google Scholar]
- 108. Balk J., Lobreaux S., Biogenesis of iron–sulfur proteins in plants, Trends Plant Sci., 2005, 10 (7), 324–331. [DOI] [PubMed] [Google Scholar]
- 109. Li J. W., Pearson S. A., Fenk K. D., Cowan J. A., Glutathione-coordinated 2Fe-2S cluster is stabilized by intramolecular salt bridges, J. Biol. Inorg. Chem., 2015, 20 (8), 1221–1227. [DOI] [PubMed] [Google Scholar]
- 110. Odermatt A., Suter H., Krapf R., Solioz M., Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae, J. Biol. Chem., 1993, 268 (17), 12775–12779. [PubMed] [Google Scholar]
- 111. Salerno M., Petroutsa M., Garnier-Suillerot A., The MRP1-mediated effluxes of arsenic and antimony do not require arsenic-glutathione and antimony-glutathione complex formation, J. Bioenerg. Biomembr., 2002, 34 (2), 135–145. [DOI] [PubMed] [Google Scholar]
- 112. Parker L. J., Bocedi A., Ascher D. B., Aitken J. B., Harris H. H., Lo Bello M., Ricci G., Morton C. J., Parker M. W., Glutathione transferase P1-1 as an arsenic drug-sequestering enzyme, Protein Sci., 2017, 26 (2), 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jozefczak M., Remans T., Vangronsveld J., Cuypers A., Glutathione is a key player in metal-induced oxidative stress defenses, Int. J. Mol. Sci., 2012, 13 (3), 3145–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Qi W. B., Li J. W., Cowan J. A., A structural model for glutathione-complexed iron–sulfur cluster as a substrate for ABCB7-type transporters, Chem. Commun., 2014, 50 (29), 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research.