Abstract
Pseudomonas aeruginosa is a ubiquitous opportunistic pathogen that causes considerable human morbidity and mortality, particularly in nosocomial infections and individuals with cystic fibrosis. P. aeruginosa can adapt to surface growth by undergoing swarming motility, a rapid multicellular movement that occurs on viscous soft surfaces with amino acids as a nitrogen source. Here we tested the small synthetic host defense peptide, innate defense regulator 1018, and found that it inhibited swarming motility at concentrations as low as 0.75 μg/ml, well below the MIC for strain PA14 planktonic cells (64 μg/ml). A screen of the PA14 transposon insertion mutant library revealed 29 mutants that were more tolerant to peptide 1018 during swarming, five of which demonstrated significantly greater swarming than the WT in the presence of peptide. Transcriptional analysis (RNA-Seq) of cells that were inoculated on swarming plates containing 1.0 μg/ml peptide revealed differential expression of 1,190 genes compared to cells swarming on plates without peptide. Furthermore, 1018 treatment distinctly altered the gene expression profile of cells when compared to that untreated cells in the centre of the swarm colonies. Peptide-treated cells exhibited changes in the expression of genes implicated in the stringent stress response including those regulated by anr, which is involved in anaerobic adaptation, indicative of a mechanism by which 1018 might inhibit swarming motility. Overall, this study illustrates potential mechanisms by which peptide 1018 inhibits swarming surface motility, an important bacterial adaptation associated with antibiotic resistance, virulence, and dissemination of P. aeruginosa.
Introduction
The accelerated emergence of antibiotic resistance mechanisms in bacterial pathogens threatens our ability to treat major, and even minor, infections. Intrinsic and adaptive properties of some pathogenic species of bacteria contribute to their resistance in the clinic. For example, the opportunistic pathogen Pseudomonas aeruginosa has an outer membrane permeability 12–100 times less than that of Escherichia coli and an impressive suite of multidrug efflux pumps [1–3]. Adaptability is associated with transient growth states, including biofilm formation and swarming motility, in which many genes change expression, including those leading to multidrug resistance [3–6]. P. aeruginosa adaptability is reflected by its pathogenicity in diverse areas of the human body; it can cause acute and/or chronic infections in the lungs, burn wounds, urinary tract and bloodstream [7–9]. Swarming motility manifests as a form of rapid surface translocation in which groups of cells move coordinately using both flagella and pili to propel themselves, assisted by production of the surface-wetting agent rhamnolipids [10]. Additionally, swarming motility is involved in bacterial disemmination in a murine abscess model [11] and P. aeruginosa exhibits increased resistance to most conventional antibiotics during swarming motility [3, 12]. The mechanism underlying swarm-associated multi-drug resistance is not fully understood but is independent of efflux pump activity and synthesis of periplasmic glucans, which contribute to resistance in planktonic and biofilm growth states respectively [13].
Biofilm and swarming growth states rely on the stringent stress response signaling pathway that is triggered by the alarmone (p)ppGpp, levels of which increase under the nutrient-limiting or anoxic conditions that are characteristic of chronic infection sites [14, 15]. In turn, bacteria downregulate the expression of genes involved in central metabolism and divert cells toward a growth arrest phenotype [14, 15]. The stringent stress response, however, is also functional under optimal growth conditions [14]. In fact, swarming motility and the production of rhamnolipids, which can occur in oxygen- and nutrient-replete media, are dependent on the stringent-stress response [10, 14, 16].
Innate defense regulator 1018 is a host-defense peptide derived from bovine bactenecin [17]. Host-defense peptides with multifaceted mechanisms of action have received increasing attention for their potential as adjuvants for conventional antibiotic therapies [18, 19]. Numerous studies have investigated the anti-biofilm properties of a subset of these peptides and the clinical utility of peptide derivatives (reviewed in ref. 18). Peptide 1018 has a broad range of beneficial immunomodulatory effects, including suppression of lipopolysaccharide-induced macrophage-mediated inflammatory responses, and antimicrobial effects in various infection models [11, 19, 20], as well as broad-spectrum activity against biofilms [21] through a mechanism dependent on the stringent stress response. Interestingly, it has more potent antibiofilm than antibacterial activity, with a minimal biofilm inhibitory concentration (MBIC) of 2–10 μg/ml, cf. an MIC of 64 μg/ml for strain PA14 [21]. We tested here its activity against another adaptive growth state, swarming motility.
To identify how peptide 1018 inhibited swarming at a remarkably low concentration (0.75 μg/ml), we compared the transcriptome of cells inoculated onto swarming plates containing 1018 to that of cells swarming in the absence of peptide. We identified modulation of metabolic adaptations implicated in the stringent stress response. This modulation was at least partially mediated by the metabolic regulator Anr, since 114 of 253 genes in the Anr regulon were differentially expressed following peptide treatment. Furthermore, the transcriptional profile of cells inoculated on swarming plates with peptide was different from that of quiescent cells in the centre of swarming colonies inoculated on plates without peptide. Swarming in the presence of peptide was restored by the addition of serine hydroxamate (SHX), which promotes (p)ppGpp synthesis, providing evidence that 1018 works at least in part through the stringent stress response.
Materials and methods
Bacterial strains and plasmids
The PA14NR library, consisting of 5,850 transposon insertion mutants corresponding to 4,596 predicted PA14 genes [22], was used to screen for mutants that swarmed in the presence of peptide 1018. Phenotypes are described relative to that of the PA14 wild type (WT). All bacterial strains used in this study are listed in S1 Table.
Growth conditions
Bacterial strains were grown overnight in Luria-Bertani (LB) broth at 37°C with aeration. Unless specified, overnight cultures were sub-cultured into either LB broth or basal medium 2 (BM2) [62 mM potassium phosphate buffer (pH 7), 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose] at a normalized optical density at 600 nm (OD600) of 0.1 and grown to mid-log phase (OD600 of 0.4–0.6) at 37°C with aeration. 15 μg/ml gentamicin was included when required for transposon selection and maintenance.
Peptide
Peptide 1018 was dissolved in distilled water, aliquoted and stored at -20°C until use. All experiments were performed using batch CL-03-00140 of peptide 1018 (>95% purity), synthesized by CPC Scientific using solid-phase 9-fluorenylmethoxy carbonyl (Fmoc) chemistry and purified using reverse-phase high-performance liquid chromatography (HPLC).
Broad library swarming screen
A high throughput swarming screen was modified from Overhage et al. [23] to examine the inhibition of swarming motility by peptide 1018 in the PA14NR transposon insertion mutant library. Bacteria were grown in LB overnight in 96-well plates. A custom-made 96-pin stamp was used to transfer approximately 1 μl of overnight culture onto agar plates containing BM2 glucose swarming medium [62 mM potassium phosphate buffer (pH 7), 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose, 0.1% (wt/vol) Casamino acids (CAA), and 0.5% (wt/vol) Difco agar] [23]. Peptide 1018 was incorporated directly into the agar at 1 μg/ml. All swarming plates were incubated at 37°C for 18–20 h. Colonies were visually assessed for their ability to swarm in the presence of the peptide. Two to three replicates were conducted for each plate of mutants in the PA14NR library.
Standard swarming assay
The standard swarming assay for P. aeruginosa involved inoculating 1 μl of mid-log phase sub-culture into the centre of a polystyrene plate containing BM2 glucose swarming agar supplemented with 0.1% CAA. This assay was used to verify the results of the broad library screen and subsequent experiments. When necessary, peptide 1018 (0–2 μg/ml) or serine hydroxamate (SHX) (0–1000 μM) were incorporated directly into the agar at concentrations indicated in the Figure legends. Plates were incubated at 37°C for 18–20 h. At the endpoint, an image of each plate was captured using a BioRad ChemiDoc, and the surface area of each swarming colony was quantified using ImageJ software (v1.8.0; Redwood, CA, USA). Each swarming assay was carried out three to five times.
Microscopy
Transmission electron microscopy (TEM) was used to investigate differences in cellular morphology between untreated cells from the leading edge of the swarm front and swarm centre, when compared to peptide-treated cells. The TEM protocol was modified from Köhler et al. [15]. Briefly, cells were picked with a sterile pipette tip and gently resuspended in 10 μl water. Formvar and carbon-coated copper TEM grids (200-mesh) were placed on top of the suspension for 30 s to allow cell adherence. Excess liquid was removed using filter paper. The grids were stained with 5 μl of a 2% aqueous uranyl acetate solution for 30 s and then washed for 5 s in 10 μl water. Excess liquid was removed from the grids with filter paper, and they were allowed to air dry. Images from multiple grid sections were taken with a Hitachi H7600 TEM at the UBC Bioimaging facility.
RNA isolation
One μl of PA14 WT mid-log phase sub-culture was inoculated onto the centre of each BM2 swarming agar plate, with or without 1 μg/ml peptide 1018. Plates were incubated for 20 h at 37°C, after which cells were harvested for RNA isolation. Cells were collected in RNAprotect Bacteria Reagent (Qiagen) with sterile swabs from the leading 2–3 mm of the swarming edge, and centres of swarming colonies grown on standard BM2 swarming plates. The entire colony was collected from plates treated with peptide 1018. Cells from three independent biological replicates in each condition were harvested. Total RNA extracted from these cells by resuspending in 100 μl of 3 mg/ml lysozyme in Tris-EDTA (TE) buffer (pH 8.0; Thermo Fisher), followed by extraction using the RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. Purified RNA underwent deoxyribonuclease (DNase) treatment with the TURBO DNA-free kit (Ambion) to remove chromosomal DNA. RNA purity was assessed using the Agilent 2100 Bioanalyzer.
RNA-Seq and identification of DE genes
To enrich for mRNA, we performed rRNA depletion on the purified total RNA with the RiboZero Bacteria kit (Illumina), followed by the Kapa stranded Total RNA kit (Kapa Biosystems) and the indexing kit (Bio Scientific, USA) to construct multiplexed cDNA libraries for sequencing. The libraries were sequenced on an Illumina HiSeq 2500 platform in one lane of a high-output flowcell to generate 100-bp single-end reads at the UBC Sequencing and Bioinformatics Consortium (SBC). Subsequently, FastQC v0.11.5 and MulitQC v0.8 were used to obtain the quality score for FASTQ files [24]. STAR aligner was used to align reads to the UCBPP-PA14 genome (GCF_000014625.1) [25, 26]. Read count tables were then generated with HTseq-count v0.6.1p1 [27]. DESeq2 v1.18 was used to estimate the fold-change (FC) in gene expression between untreated cells from the edge or centre and peptide-treated cells under swarming conditions [28] with thresholds of adjusted P ≤ 0.05 and absolute FC ≥ 1.5. The list of differentially expressed (DE) genes is available in S2 Table. RNA-Seq data were deposited in NCBI GEO under the accession number GSE151264.
Functional enrichment of DE genes
Enrichment of gene ontology (GO) terms was performed using the R package GofuncR [29], testing the DE genes against a custom set of GO annotations downloaded from the Pseudomonas genome database [30]. Results were filtered using a significance-threshold of P ≤ 0.05 and a multi-test corrected family-wise error rate (FWER) ≤ 0.1. The full list of functionally enriched GO terms is available in S3 Table. DE genes belonging to the anr regulon [31] in the 1018 vs. edge comparison were tested for enrichment using Fisher’s Exact test. All analyses described were performed in R (v4.0.3; Boston, MA, United States).
Statistical analysis
Unless otherwise stated, statistics were performed using GraphPad Prism 8.0 (La Jolla, CA, United States). P-values were calculated using one-way ANOVA followed by Dunnett’s correction, Kruskal–Wallis nonparametric test followed by Dunn’s post-hoc analysis, or Student’s two-tailed t-test as indicated in the Figure legends. Data were considered statistically significant when P ≤ 0.05.
Results
1018 inhibited swarming motility in P. aeruginosa
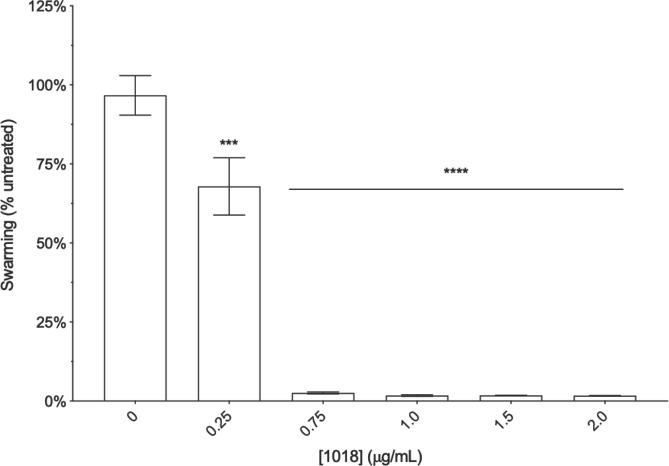
Titration of 1018 (0–2 μg/ml) into swarming agar revealed that this peptide inhibited swarming motility of P. aeruginosa strain PA14 at low concentrations (Fig 1). Swarming motility was significantly reduced by 28.8% when cells were inoculated on plates containing as little as 0.25 μg/ml 1018 when compared to cells inoculated on plates without peptide (0 μg/ml). In subsequent experiments, 0.75 or 1.0 μg/ml 1018 was used since it reduced swarming by 94.1 and 95.0% respectively.
Fig 1. Peptide 1018 inhibited swarming motility in PA14 at low concentrations.
BM2 swarm plates containing 1018 or water only were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Mean and standard error of the mean are shown. One-way ANOVA followed by Dunn’s correction was used to determine statistical significance. *** P < 0.001, **** P < 0.0001. Each result was obtained from at least three independent biological replicates.
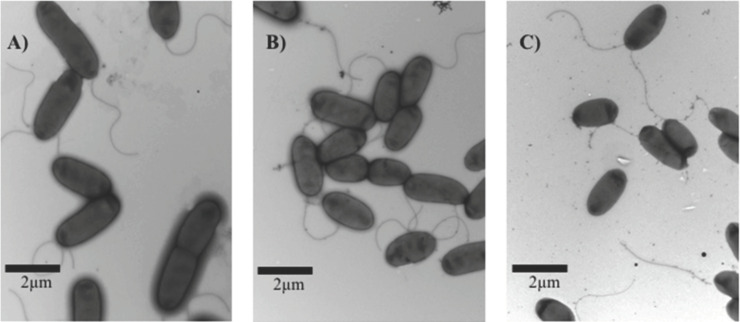
Strain PA14 was harvested from peptide-containing swarm plates and examined by TEM (Fig 2). TEM revealed that bacteria from swarming plates with peptide 1018 were flagellated and resembled cells taken from the centre of untreated swarming colonies (Fig 2). More specifically, cells from the leading edge of the untreated swarm front were elongated and septated, indicative of cellular division, whereas cells from the centre of the untreated swarm colony and peptide-treated cells were neither elongated or septated.
Fig 2. Representative images of cells captured using Transmission Electron Microscopy (TEM).
Briefly, BM2 swarm plates (±0.75 μg/ml 1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were transferred to a mesh grid for TEM. (A) Actively swarming WT cells from the edge of a swarming colony. (B) Non-swarming cells taken from the centre of a swarming colony. (C) Cells from a swarming plate treated with 0.75 μg/ml of 1018.
Twenty-nine transposon mutants swarmed in the presence of peptide 1018
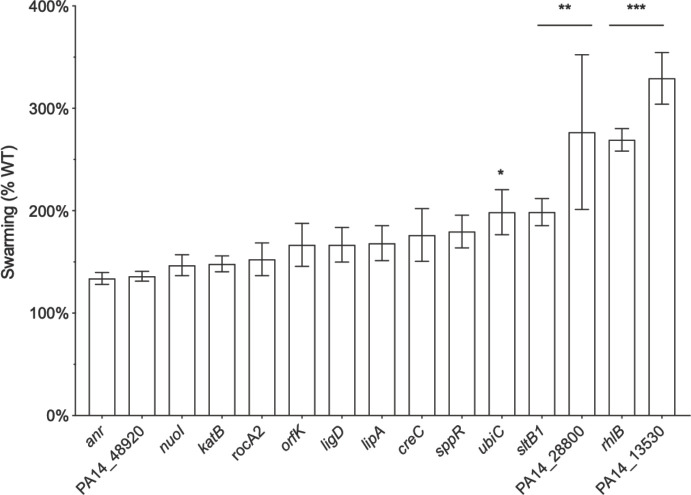
The entire PA14 transposon insertion mutant library [22] was screened for swarming in the presence of an inhibitory concentration of 1018 (1 μg/ml). This screen revealed 29 transposon mutants that upon retesting were tolerant to peptide 1018 under swarming conditions. Five of these exhibited significantly increased (2.0- to 3.3-fold) swarming when compared to WT (Fig 3). Nonetheless, these peptide tolerant mutants swarmed 75% less than that of the untreated WT. In particular, mutations in genes implicated in the stringent stress response (creC, lipA) [32, 33], metabolism (anr) [31, 34] and rhamnolipid biosynthesis (rhlB) also conferred moderate peptide tolerance under swarming conditions that could be complemented with the respective gene in the transposon mutants (S1 Fig). Furthermore, transposon interruption of genes directly involved in coping with stress, including the hydrogen peroxide-inducible katB, creC, lipA, and sppR, conferred tolerance to peptide under swarming conditions. Other peptide-tolerant mutants had transposon insertions in genes that belonged to diverse functional categories including regulatory genes (orfK and PA3045), and genes involved in iron acquisition and virulence factor production (Fig 3).
Fig 3. Selected PA14 transposon mutants that swarmed in the presence of 0.75 μg/ml peptide 1018.
Briefly, BM2 swarm plates (with 0.75 μg/ml 1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. At least three biological replicates for each transposon mutant were examined. Mean and standard error of the mean are shown. * P < 0.05, ** P < 0.01, *** P < 0.001 according to Kruskal–Wallis test followed by Dunnett’s post hoc analysis.
The transcriptome of peptide-treated cells was distinct from that of swarming cells
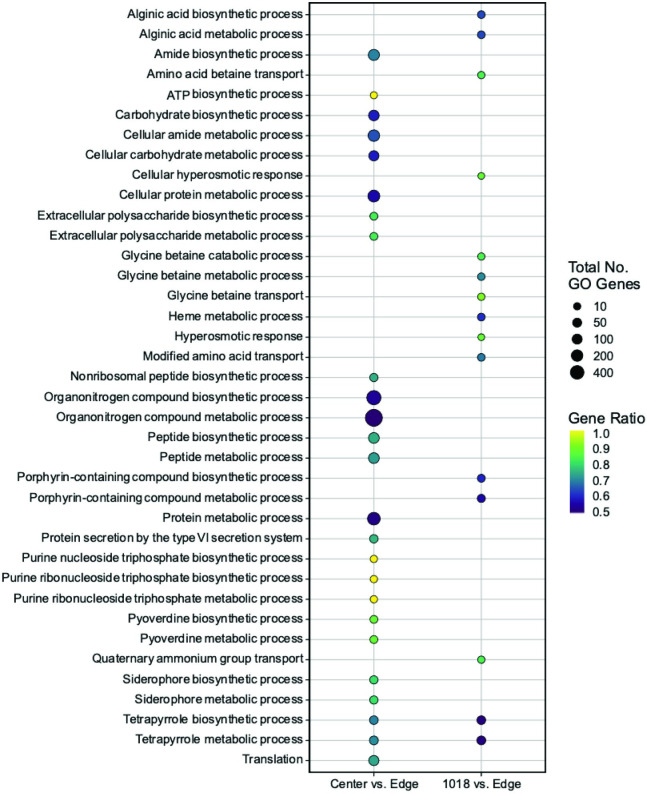
To characterize the cellular or molecular mechanisms by which 1018 influenced swarming motility, RNA-Seq was performed. The transcriptome of cells inoculated on swarm plates containing peptide (1.0 μg/ml) was compared to that of cells at the swarm front of colonies inoculated on plates without peptide (1018 vs. edge). The transcriptome of cells taken from the centre of colonies inoculated on peptide-free swarm plates was also compared to that of cells at the swarm front of the same plates (centre vs. edge). Peptide treatment resulted in the differential expression of 1,190 genes (S2 Table). DE genes in both treatment groups were functionally mapped according to gene ontology (Fig 4). Relative to swarming cells from the swarm leading edge, there was enrichment for DE genes encoding proteins involved in the transport of small molecules, heme metabolism and biosynthesis and metabolism of specific amino acids. Non-swarming cells from the swarm centre differentially expressed 2,369 genes when compared to cells from the swarm edge (S2 Table). In both peptide-treated and non-swarming cells, ~68% of all DE genes overlapped (S2 Table). However, a statistical test showed significant dysregulation of the anr regulon (P = 3.86E-18). As well, regulators of swarming and virulence were expressed differently in the two groups (Table 1) [31, 35]. Of note, rhlA and rhlB were 3-fold less upregulated in peptide-treated cells than untreated swarming cells when compared to the control.
Fig 4. Functional classification of Differentially Expressed (DE) genes using Gene Ontology (GO) term enrichment.
GO term enrichment was performed using GofuncR on the list of DE genes in cells treated with 1018 and cells at the centre of swarming colonies relative to cells at the edge of swarming colonies. GO terms were considered significant with a P ≤ 0.05 and a multi-test corrected family-wise error rate (FWER) ≤ 0.1. Cells treated with 1018, but not cells at the centre of swarming colonies, cf. swarming edge cells, exhibited functional enrichment in transport of small molecules, heme metabolism and amino acid biosynthesis/metabolism.
Table 1. Selected categories of DE genes, mapped to PAO1 orthologs, as determined by RNA-Seq in peptide-treated and swarming colony centre cells under swarming conditions.
| PAO1 locus (name) | Product | FC in gene expression | |
|---|---|---|---|
| 1018 treated vs. edge | Centre vs. edge | ||
| Subset of anr regulon genes [31] (of the 141 DE genes in S4 Table) | |||
| PA0141 | hypothetical protein | 3.5 | -2.6 |
| PA0200 | hypothetical protein | 3.3 | -2.2 |
| PA0201 | hypothetical protein | NC | -12 |
| PA0408 (pilG) | twitching motility protein | NC | -1.7 |
| PA0459 (clpC) | ClpA/B protease ATP binding subunit | 5.3 | 1.9 |
| PA0482 (glcB) | malate synthase G | 1.6 | NC |
| PA0519 (nirS)D | nitrite reductase | NC | -18 |
| PA0524 (norB)D | nitric-oxide reductase subunit B | NC | -9.4 |
| PA0797 | transcriptional regulator | -1.9 | NC |
| PA0836 (ackA) | acetate kinase | 1.9 | NC |
| PA0962 (dps) | DNA-binding stress protein | 2.3 | NC |
| PA1300 | RNA polymerase ECF-subfamily sigma-70 factor | NC | -28 |
| PA1414 | hypothetical protein | 3.8 | 1.6 |
| PA1429 | cation-transporting P-type ATPase | 3.9 | -3 |
| PA1546 (hemN) | coproporphyrinogen III oxidase | 1.9 | -4.8 |
| PA1557 (ccoN2) | cbb3-type cytochrome c oxidase subunit I | 3.5 | -5.5 |
| PA1561 (aer) | aerotaxis receptor | 2.4 | -1.5 |
| PA1673 | hypothetical protein | 3.4 | -5.5 |
| PA1745 | hypothetical protein | 2.3 | 4.2 |
| PA1746 | hypothetical protein | 3.5 | -3.8 |
| PA1920 (nrdD) | anaerobic ribonucleoside triphosphate reductase | 2.4 | -9.9 |
| PA2126 | conserved hypothetical protein | 2.4 | -1.9 |
| PA2127 (cgrA) | hypothetical protein | 1.9 | -3.4 |
| PA2575 | hypothetical protein | 2 | NC |
| PA2753 | hypothetical protein | 4.4 | -3.3 |
| PA2754 | conserved hypothetical protein | 4.8 | NC |
| PA3309 (hepA) | hypothetical protein | 3.1 | -1.7 |
| PA3337 (rfaD) | ADP-L-glycero-D-manno-heptose-6-epimerase | 2 | -5.4 |
| PA3391 (nosR)D | regulatory protein | NC | -4.4 |
| PA3878 (narX)D | two-component sensor | NC | 2.3 |
| PA3879 (narL) | two-component response regulator NarL | 1.7 | 1.6 |
| PA3930 (cioA) | cyanide insensitive terminal oxidase | 2 | 2.5 |
| PA4205 (mexG) | hypothetical protein | 1.9 | NC |
| PA4328 | hypothetical protein—response to stress | 3.1 | NC |
| PA4348 | conserved hypothetical protein | NC | -5.5 |
| PA4352 | hypothetical protein—response to stress | 4.8 | -3.3 |
| PA4577 | hypothetical protein | 4.2 | NC |
| PA4587 (ccpR) | cytochrome c551 peroxidase | 3.5 | -17 |
| PA4675 (chtA) | TonB-dependent receptor | -1.8 | -5.1 |
| PA5027 | hypothetical protein | 2.6 | -2.8 |
| PA5170 (arcD) | arginine/ornithine antiporter | 2.7 | NC |
| PA5171 (arcA) | arginine deiminase | 6.8 | NC |
| PA5172 (arcB) | ornithine carbamoyltransferase, catabolic | 5.6 | NC |
| PA5173 (arcC) | carbamate kinase | 4.6 | NC |
| PA5426 (purE) | phosphoribosylaminoimidazole carboxylase catalytic subunit | -2.1 | -1.9 |
| PA5427 (adhA) | alcohol dehydrogenase | 5.8 | -1.8 |
| PA5475 | hypothetical protein | 3.8 | -2.5 |
| Select genes encoding regulatory proteins implicated in virulence [35] | |||
| PA0652 (vfr) | cAMP-regulatory protein, virulence factor regulator | -1.7 | -1.6 |
| PA5550 (glmR)D | transcriptional regulator of polysaccharide biosynthesis | -2 | -2.3 |
| PA2426 (pvdS) | extracytoplasmic-function σ-70 factor | -2 | -52 |
| PA3879 (narL) | transcriptional regulator | 1.7 | 1.6 |
| PA5483 (algB) | two-component response regulator | 3.6 | NC |
| PA0610 (prtN) | transcriptional regulator | 2.6 | NC |
| PA3630 (gfnR) | glutathione-dependent formaldehyde neutralization regulator (sarcosine metabolism) | 2.3 | NC |
| PA5274 (rnk) | nucleoside diphosphate kinase regulator | -1.6 | NC |
| PA0795 prpR | propionate catabolism operon regulator | 1.8 | NC |
| PA4080 rcsB | two-component response regulator, CupD activation | 1.7 | -1.6 |
| Regulators of swarming (of 20 total in S5 Table) | |||
| PA4853 (fis)1D | DNA-binding protein | -1.8 | -2.1 |
| PA0905 (rsmA)1D | RNA binding protein translational regulator | 1.6 | NC |
| PA4725 (cbrA)1D | two-component sensor | -1.5 | NC |
| PA5261 (algR)1D | alginate biosynthesis regulatory protein | 2.1 | 2.3 |
| PA0762 (algU) | RNA polymerase σ factor | 2.1 | NC |
| PA2895 (sbrR)D | anti-σ factor | 2.6 | NC |
| PA2896 (sbrI) | RNA polymerase σ factor (inhibits swarming) | 2.5 | NC |
| PA3391 (nosR)D | regulatory protein | NC | -4.4 |
| PA1713 (exsA)D | transcriptional regulator | -1.7 | NC |
| PA4546 (pilS)D | two-component sensor (twitching, biofilm, swarming) | -1.7 | NC |
| PA04791D | LysR family transcriptional regulator | 2.1 | 1.9 |
| PA20721D | sensory box protein | 2 | 4.6 |
| PA25711D | signal transduction histidine kinase | 1.7 | 4.2 |
| PA19761D | two-component sensor | 2.5 | NC |
| PA11961D | transcriptional regulator | 2.1 | NC |
| PA14581D | two-component sensor | 1.5 | NC |
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (± 0.75 μg/ml 1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from the leading swarming edge, the centres of swarming colonies, and 1018-treated colonies. NC = no significant change in gene expression.
1: Transcriptional regulators controlling swarming motility identified by Yeung et al. [36]
Compared to actively swarming cells from the leading edge of a swarming colony, the expression of Anr-regulated genes was different in peptide-treated and colony centre cells. A total of 141 of 253 known genes from the Anr regulon were dysregulated in either peptide-treated cells or cells from the centre of swarming colonies (S4 Table; see Table 1 for representative genes). Of these, 30 were dysregulated only by peptide treatment, 66 were dysregulated only at the swarm centre (44 of which were downregulated), 23 showed an opposite pattern of regulation between both conditions, and 22 showed similar regulation. Overall, treatment with peptide significantly triggered expression of the Anr regulon, as a total of 114 of these genes were more highly expressed in peptide-treated cells relative to untreated swarming cells. Gene expression of several other regulatory proteins that are involved in the stringent stress response, virulence and adaptive lifestyles (such as biofilm formation or swarming motility, both of which are inhibited by 1018 treatment) was influenced by peptide treatment but not by lack of motility at the centre of swarming colonies (Table 1, S5 Table). These virulence-associated or adaptation genes included algB, algR, cbrA, prtN, gfnR, rnk and prpR. Some of these genes encode regulators that are essential for swarming motility, such as CbrA [36]. Other swarming regulators for which transcription was altered by peptide treatment included the RNA-binding translational regulatory protein RsmA, as well as sigma and anti-sigma factors AlgU/MucA and SbrI [11, 36, 37].
Serine Hydroxamate (SHX) enhanced the tolerance of certain mutants to 1018
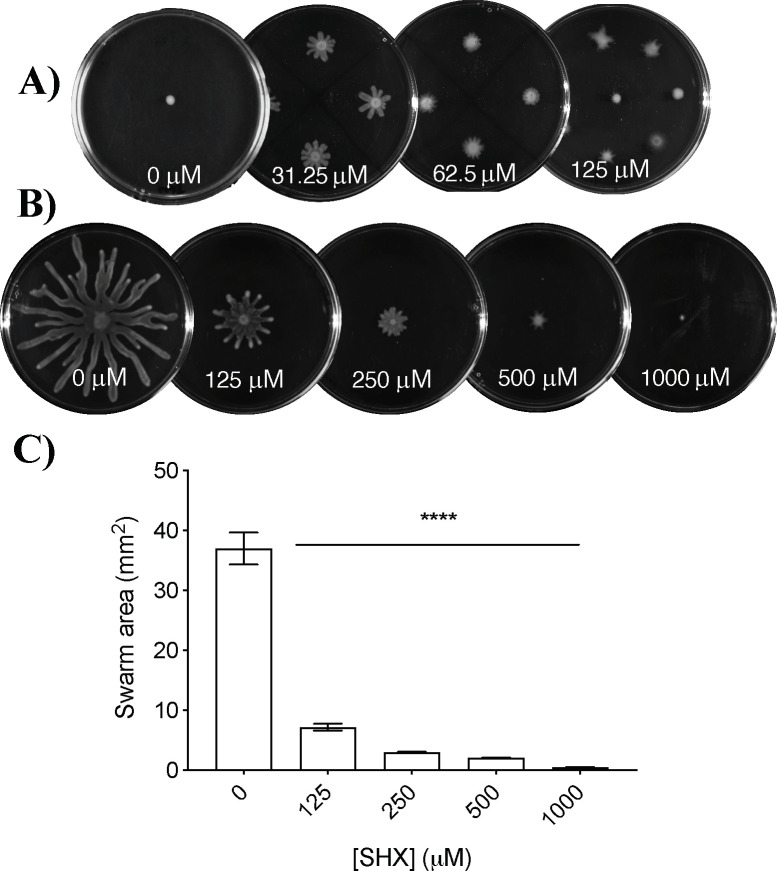
It was previously shown that peptide 1018 inhibited biofilm formation by modulating the stringent stress response through direct interaction with (p)ppGpp promoting its degradation [21]. Thus, we examined the effect of peptide treatment on swarming in the presence of SHX, a structural analogue of L-serine that induces the stringent stress response by inhibiting seryl-tRNA synthetase from incorporating amino acids into proteins [38], leading to induction of (p)ppGpp synthesis (via RelA) in excess of 1018. Prior studies showed that SHX induced (p)ppGpp accumulation over a concentration range of ~50–500 μM [21, 38]. Here, we treated swarming cells with SHX in addition to 1018 (0.75 μg/ml) and observed that 31.25 μM SHX partially restored the swarming ability of WT cells in the presence of peptide (Fig 5A) without inhibiting swarming of cells in the absence of peptide. Although we observed that 62.5–125 μM SHX did not fully restore swarming in peptide treated cells, we also observed that SHX inhibited swarming motility of untreated cells in a dose-dependent manner (Fig 5B and 5C). These results indicate that high concentrations of SHX inhibited swarming motility independent of peptide treatment, which supports the importance of amino acid metabolism in swarming.
Fig 5. SHX partially restored swarming of peptide-inhibited cells in an inverse dose-dependent fashion and inhibited swarming of PA14 WT at higher doses.
Briefly, BM2 swarm plates (±0.75 μg/ml 1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. The stringent stress response ((p)ppGpp production) was induced by supplementing swarm medium with SHX. (A) Peptide 1018 was antagonized by treatment with SHX at a low concentration (31.25 μM). (B) In the absence of peptide, SHX partially inhibited swarming at 125 μM. (C) Swarm colony area of SHX-treated cells (in the absence of peptide 1018). Mean and standard error of the mean are shown. One-way ANOVA followed by Dunn’s correction was used to determine statistical significance. **** P < 0.0001. Each experiment had at least three biological replicates.
Interestingly, the peptide tolerance of some transposon mutants (including sltB1::MAR2xT7, creC::MAR2xT7, nuoI::MAR2xT7 and PA4400::MAR2xT7) was even more pronounced in the presence of SHX (125 μM) and peptide (0.75 μg/ml) than it was in the presence of peptide alone, when compared to WT grown in respective conditions (Fig 6). The genes for these mutants encode a soluble lytic transglycosylase (SltB1) involved in cell wall biosynthesis [39]; the two-component sensor CreC, that together with CreB regulates β-lactam resistance, biofilm formation, fitness and anaerobic respiration [33, 40]; the NADH dehydrogenase NuoI; and a probable pyrophosphohydrolase PA4400 [30]. These data were consistent with the conclusion that these genes either had a role in the stringent stress response or worked in synergy with the stringent response.
Fig 6. SHX enhanced the swarming phenotype of peptide 1018-tolerant transposon insertion mutants.
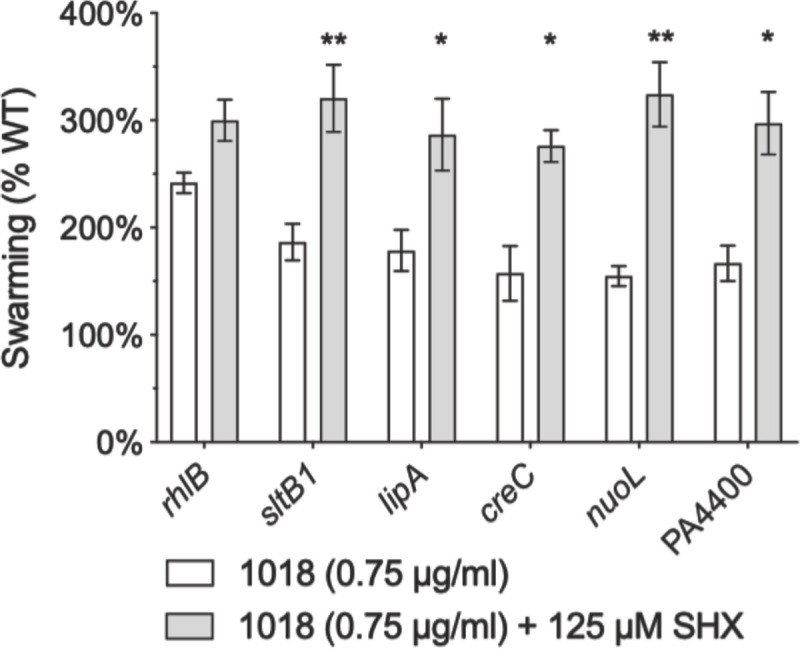
BM2 swarm plates (+ 0.75 μg/ml 1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C (A) Peptide-tolerant mutants showed enhanced swarming in the presence of SHX (125 μM). Mean and standard error of the mean are shown. Two-tailed student t-tests were used to determine statistical significance between treatments with or without SHX. * P < 0.05, ** P < 0.01. (B) Representative image of peptide-tolerant mutants swarming in the presence of SHX and peptide. Each experiment had at least three biological replicates.
Discussion
Peptide 1018 is a known inhibitor of biofilm formation and can induce biofilm dispersion in a diverse range of species, including P. aeruginosa, at concentrations well below its MIC [21]. Here it was shown that 1018 completely inhibited swarming motility at a concentration of 0.75 μg/ml. This data aligned with observations from a prior study [11] that extended the screening to other P. aeruginosa motilities including swimming and twitching, and showed that neither were impacted by 1018 at concentrations up to 20 μg/ml. Building on our knowledge of 1018 as a swarming-specific inhibitor of motility, we identified a subset of genes that conferred partial peptide tolerance on P. aeruginosa under swarming conditions by screening a comprehensive transposon insertion mutant library. Similar effects have been observed when identifying genes that contribute to tobramycin resistance, where it was shown that 41 mutants contributed partially to adaptive resistance to this antibiotic [3]. We propose here that susceptibility to peptide 1018 is multigenic, whereby multiple genes collectively contribute to susceptibility. A number of the genes identified in the peptide tolerance screen have been implicated in SpoT-mediated activation of the stringent response. For example, LipA is a lipase involved in both fatty acid metabolism and the regulation of the iron-starvation sigma factor, PvdS [32]; the latter was two-fold downregulated in peptide-treated cells but strongly downregulated in swarming centre cells (S2 Table). Indeed, lipA expression is essential for full pyoverdine production under iron-limiting conditions and is lower in stringent response mutants [32]. Likewise, a sensor of carbon catabolites (CreC) activates PhoB, which regulates, through SpoT, adaptation to phosphate starvation in response to low levels of inorganic phosphate [33]. Inorganic phosphate is required for swarming motility in P. aeruginosa, and reduced levels are associated with motile-sessile switching and decreased expression of the T3SS [41]. KatB is a catalase induced by exogenous hydrogen peroxide and an important enzyme for stationary phase survival and antibiotic tolerance mediated by the stringent response [34]. Thus, peptide treatment might be inhibiting adaptations necessary for swarming motility since we showed peptide susceptibility could be reversed by inducing the synthesis of (p)ppGpp.
The data also provided evidence for a role of anr and rhlB in adaptation and tolerance to 1018 (S1 Fig). These genes were of particular interest because they modulate physiological processes associated with adaptive lifestyles such as biofilm formation, swarming motility, anaerobic growth, quorum sensing and the stringent stress response [11]. More specifically, rhlB is required for rhamnolipid biosynthesis, which is reduced in a non-swarming RelA/SpoT mutant [14]. Corroborating evidence was provided here, since rhlA and rhlB were 3-fold less upregulated in peptide treated swarming cells than untreated swarming cells when compared to the control (S2 Table).
The global regulator Anr influences metabolic processes that transform carbohydrates into energy across a range of oxygen levels [42, 43]. Our data revealed that treatment with peptide 1018 significantly triggered expression of the Anr regulon, since a total of 114 of these genes were more highly expressed in peptide-treated cells relative to untreated swarming centre cells. Intriguingly a connection has been made between the stringent stress response and anr, whereby the universal stress proteins are upregulated in anr mutants in a stringent stress response-dependent manner [44]. Furthermore, an anr mutant demonstrated a modest but complementable decrease in susceptibility to peptide 1018 under swarming conditions (Fig 3). This indicates that peptide 1018 works in part by modulating the Anr regulon to interfere with the stringent stress response. Consistent with this, some Anr-regulated genes that are required for swarming were dysregulated by peptide treatment, namely PA1196, clpS, cspD, amrZ, nosR, and PA4958 (S4 and S5 Tables), and anr deletion led to reduced swarming under anaerobic conditions [31].
There were significant differences between cells at the centre of swarming colonies and peptide treated cells that appeared in the same position of swarming plates. Indeed, the metabolically quiescent characteristics of centre cells did not appear to occur in the presence of peptide. Thus, transcripts for nitrogen metabolism genes including nor (nitric oxide), nir (nitrite reductases), and nosR (regulatory protein) were significantly downregulated (by 4.4- to 18-fold) in cells from the centre of swarming colonies, but not in peptide-treated cells (S2 Table). A similar trend was observed for transcripts encoding cytochrome oxidases (Ccp and Cco) that serve as terminal electron acceptors and are important for energy production [45]. Corroborating evidence was observed in genes implicated in amino acid (argS, arcD) and carbon (rfaD, glcB, gltP, ackA) metabolism as well as detoxification of reactive oxygen species (dnr, katA). No differences in expression for genes previously implicated in peptide resistance (phoPQ, pmrAB, cprRS, parRS, arn operon) or those important for adaptive resistance [3] were detected between peptide-treated cells and cells from the swarm front, suggesting that 1018 did not trigger canonical resistance mechanisms to peptides. However, the multidrug efflux pump mexGHI-opmD was upregulated by 1.8- to 2-fold in peptide-treated cells (S4 Table).
Examination of genes that were uniquely dysregulated in the presence of peptide revealed 50 other genes that are known to be required for swarming motility [37], including those for 19 regulators (Fig 3, Table 1, S4 Table). These included the regulatory genes exsA, cbrA, pilS, glmR, fis and the negative regulator sbrI (S5 Table). ExsA is a master regulator of the T3SS, and through interactions with the DNA-binding protein Fis has a variety of roles in optimizing bacterial adaptation and virulence [46]. T3SS is an important virulence factor in acute infections, and its expression is positively associated with swarming motility [10, 47]. The global regulator CbrA is involved in regulating swarming motility, biofilm formation, cytotoxicity against human bronchial epithelial cells, antibiotic resistance, and the adaptability of P. aeruginosa to nutritionally depleted environments [48]. The observed alterations in the expression of these genes and downstream effectors could collectively contribute to the inhibition of swarming motility by peptide 1018.
Evidence of the (at least partial) involvement of the stringent stress response was corroborated by observations that (p)ppGpp induction via SHX restored swarming in the presence of peptide (Fig 5) as well as enhanced swarming in the presence of peptide in some 1018-tolerant transposon insertion mutants (Fig 6). The alarmone (p)ppGpp is required for optimal swarming, and ΔrelAΔspoT double mutants, which lack the ability to synthesize (p)ppGpp, are deficient in swarming [11]. During biofilm formation, 1018 has been proposed to bind (p)ppGpp and mark it for degradation [21]. High levels of SHX inhibited swarming, likely because SHX inhibits the use of amino acids [21] that were supplied as the nitrogen source during swarming assays.
Preventing bacteria from adapting to their environment can make them vulnerable to antibiotic treatment and clearance by the immune system [48, 49]. As a complex and tightly regulated lifestyle adaptation, swarming motility confers fitness in situ by promoting mucosal surface colonization, dissemination to distal tissues [11], and antibiotic resistance [37], which can be blocked by 1018 treatment. Furthermore, in vivo models revealed that 1018 has protective immunomodulatory effects, including attenuated inflammation, immune cell recruitment, and enhanced wound healing and reduced bacterial spread [18, 20]. Therefore, peptide 1018 sensitizes bacteria to innate immune processes, bolsters these processes, and limits potentially harmful inflammatory responses. The data presented herein suggest 1018 as a promising anti-swarming compound that could be used in combination with anti-infective therapies to improve treatment of P. aeruginosa infections by inhibiting bacterial spread or adaptation.
Supporting information
(DOCX)
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (±1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from tips of swarm tendrils, centres of swarming colonies, and 1018-treated colonies. 0 = no significant change in gene expression.
(XLSX)
Number of genes that were dysregulated for a particular GO term in each condition, as well as total number of genes annotated to that GO term, are shown.
(XLSX)
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (±1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from tips of swarm tendrils, centres of swarming colonies, and 1018-treated colonies. 0 = no significant change in gene expression. References: [31, 35].
(XLSX)
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (±1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from tips of swarm tendrils, centres of swarming colonies, and 1018-treated colonies. 0 = no significant change in gene expression.
(XLSX)
(A, B) In the presence of peptide 1018 (0.75 μg/ml), rhlB-Tn and anr-Tn mutants swarmed more than the PA14 wild-type (WT). Non-swarming was restored by complementation of the respective gentic loci in trans since no differences were detected between complemented mutants (shown as mutant+) and WT transformed with empty vector (ev) (C, D) Representative images showing swarming phenotypes of mutants and complemented mutants in the presence or absence of peptide 1018, as indicated. Mean and standard error of the mean are shown. One-way ANOVA followed by Dunn’s correction was used to determine statistical significance. *** P < 0.001. Each experiment had at least three biological replicates.
(DOCX)
Data Availability
All RNA-Seq data files are available from the NCBI GEO database (accession number GSE151264). All differentially expressed genes are listed in S2 Table, and all other relevant data are within the paper and its Supporting Information files.
Funding Statement
REWH received a grant from the Canadian Institutes for Health Research (CIHR) FDN-154287 and the Cystic Fibrosis (CF) Canada Award Number 3177. REWH also holds a Canada Research Chair in Health and Genomics and a UBC Killam Professorship. LVW was the recipient of a Sports Canada AAP Tuition Support award. SRC was the recipient of CIHR Frederick Banting and Charles Best Canada Graduate Scholarship Master’s (CGS-M) and Doctoral Awards (CGS-D), and a Four-Year Fellowship for PhD students from UBC. MAA was the recipient of a CIHR Vanier Graduate Scholarship, UBC Killam Doctoral Scholarship and Cystic Fibrosis Canada Studentship (#617081). BCW is supported by the John Richard Turner Fellowship in Microbiology and Immunology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Canadian Institutes for Health Research.
References
- 1.Fernández L, Hancock REW. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. 10.1128/CMR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. 10.1016/j.tim.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 3.Coleman SR, Blimkie T, Falsafi R, Hancock REW. 2020a. Multidrug adaptive resistance of Pseudomonas aeruginosa swarming cells. Antimicrob Agents Chemother 64: e01999–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alford MA, Baghela A, Yeung ATY, Pletzer D, Hancock REW. 2020. NtrBC regulates invasiveness and virulence of Pseudomonas aeruginosa during high-density infection. Front Microbiol 11:773. 10.3389/fmicb.2020.00773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173. 10.1016/j.femsle.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 6.Sun E, Gill EE, Falsafi R, Yeung A, Liu S, Hancock REW. 2018. Broad-spectrum adaptive antibiotic resistance associated with Pseudomonas aeruginosa mucin-dependent surfing motility. Antimicrob Agents Chemother 62:1–12. 10.1128/AAC.00848-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. 2009. Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J Infect Public Health 2:101–111. 10.1016/j.jiph.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 8.Davies JC. 2002. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev 3:128–134. 10.1016/s1526-0550(02)00003-3 [DOI] [PubMed] [Google Scholar]
- 9.Murray TS, Egan M, Kazmierczak BI. 2007. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr Opin Pediatr 19:83–88. 10.1097/MOP.0b013e3280123a5d [DOI] [PubMed] [Google Scholar]
- 10.Yeung ATY, Torfs ECW, Jamshidi F, Bains M, Wiegand I, Hancock REW, et al. 2009. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol 191:5592–5602. 10.1128/JB.00157-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman SR, Pletzer D, Hancock REW. 2020b. Contribution of swarming motility to dissemination in a Pseudomonas aeruginosa murine skin abscess infection model. J Infect Dis. jiaa778 [online ahead of print 10.1093/infdis/jiaa778 ]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overhage J, Bains M, Brazas MD, Hancock REW. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679. 10.1128/JB.01659-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai A, Tremblay J, Déziel E. 2009. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ Microbiol 11(1):126–36. 10.1111/j.1462-2920.2008.01747.x [DOI] [PubMed] [Google Scholar]
- 14.Pletzer D, Blimkie TM, Wolfmeier H, Li Y, Baghela A, Lee AHY, et al. 2020. The stringent stress response controls proteases and global regulators under optimal growth conditions in Pseudomonas aeruginosa. mSystems 5(4):e00495–20 10.1128/mSystems.00495-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pletnev P, Osterman I, Sergiev P, Bogdanov A, Dontsova O. 2015. Survival guide: Escherichia coli in the stationary phase. Acta Naturae 7(4):22–33. [PMC free article] [PubMed] [Google Scholar]
- 16.Köhler T, Curty LK, Barja F, van Delden C, Pechère J-C. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. 10.1128/jb.182.21.5990-5996.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour SC, de la Fuente-Núñez C, Hancock REW. 2015. Peptide IDR-1018: modulating the immune system and targeting bacterial biofilms to treat antibiotic-resistant bacterial infections. J Pept Sci 21:323–9. 10.1002/psc.2708 [DOI] [PubMed] [Google Scholar]
- 18.Pletzer D, Coleman SR, Hancock REW. 2016. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr Opin Microbiol 33:35–40. 10.1016/j.mib.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pletzer D, Mansour SC, Hancock REW. 2018. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog 14: e100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas-Santiago B, Castañeda-Delgado JE, Rivas Santiago CE, Waldbrook M, González-Curiel I, León-Contreras JC, et al. 2013. Ability of innate defence regulator peptides IDR-1002, IDR-HH2 and IDR-1018 to protect against Mycobacterium tuberculosis infections in animal models. PLoS One 8: e59119. 10.1371/journal.pone.0059119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Fuente-Núñez C, Reffuveille F, Haney EF, Straus SK, Hancock REW. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10: e1004152. 10.1371/journal.ppat.1004152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–8. 10.1073/pnas.0511100103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overhage J, Lewenza S, Marr AK, Hancock REW. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 Mini-Tn5-lux mutant library. J Bacteriol 189:2164–2169. 10.1128/JB.01623-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. 2012. GenBank. Nucleic Acids Res 41: D36–D42. 10.1093/nar/gks1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. 2015. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grote S. 2018. GofuncR: Gene ontology enrichment using FUNC. R package, version 1(0). [Google Scholar]
- 30.Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, Yu NY, et al. 2011. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:596–600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trunk K, Benkert B, Quäck N, Münch R, Scheer M, Garbe J, et al. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: Definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. 10.1111/j.1462-2920.2010.02252.x [DOI] [PubMed] [Google Scholar]
- 32.Funken H, Knapp A, Vasil ML, Wilhelm S, Jaeger KE, Rosenau F. 2011. The lipase lipA (PA2862) but not lipC (PA4813) from Pseudomonas aeruginosa influences regulation of pyoverdine production and expression of the sigma factor PvdS. J Bacteriol 193:5858–5860. 10.1128/JB.05765-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao NN, Liu S, Kornberg A. 1998. Inorganic polyphosphate in Escherichia coli: The phosphate regulon and the stringent response. J Bacteriol 180:2186–2193. 10.1128/JB.180.8.2186-2193.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. 10.1128/JB.02061-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, et al. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol 5:1294–1308. 10.1046/j.1462-2920.2003.00542.x [DOI] [PubMed] [Google Scholar]
- 36.Yeung ATY, Janot L, Pena OM, Neidig A, Kukavica-Ibrulj I, Hilchie A, et al. 2014. Requirement of the Pseudomonas aeruginosa CbrA sensor kinase for full virulence in a murine acute lung infection model. Infect Immun 82:1256–1267. 10.1128/IAI.01527-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tribelli PM, Lujan AM, Pardo A, Ibarra JG, Fernández Do Porto D, Smania A, et al. 2019. Core regulon of the global anaerobic regulator Anr targets central metabolism functions in Pseudomonas species. Sci Rep 9:1–13. 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 190(3): 1084–1096. 10.1128/JB.01092-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolaidis I, Izoré T, Job V, Thielens N, Breukink E, Dessen A. 2012. Calcium-dependent complex formation between PBP2 and lytic transglycosylase SltB1 of Pseudomonas aeruginosa. Microb Drug Resist 18:298–305. 10.1089/mdr.2012.0006 [DOI] [PubMed] [Google Scholar]
- 40.Zamorano L, Moyà B, Juan C, Mulet X, Blázquez J, Oliver A. 2014. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother 58:5084–5095. 10.1128/AAC.02556-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. 10.1073/pnas.060030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. 2010. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol 12:1399–1412. 10.1111/j.1462-2920.2009.02109.x [DOI] [PubMed] [Google Scholar]
- 43.Ray A, Williams HD. 1997. The effects of mutation of the anr gene on the aerobic respiratory chain of Pseudomonas aeruginosa. FEMS Microbiol Lett 156:227–232. 10.1111/j.1574-6968.1997.tb12732.x [DOI] [PubMed] [Google Scholar]
- 44.Boes N, Schreiber K, Schobert M. 2008. SpoT-triggered stringent response controls usp gene expression in Pseudomonas aeruginosa. J Bacteriol 190:7189–7199. 10.1128/JB.00600-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vázquez-Torres A, Bäumler A. 2016. Nitrate, nitrite and nitric oxide reductases: from the last universal common ancestor to modern bacterial pathogens. Curr Opin Microbiol 29:1–8. 10.1016/j.mib.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hauser AR. 2010. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung ATY, Bains M, Hancock REW. 2011. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol 193:918–931. 10.1128/JB.00911-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de la Fuente-Núñez C, Cardoso MH, de Souza Cândido E, Franco OL, Hancock REW. 2016. Synthetic antibiofilm peptides. Biochim Biophys Acta—Biomembr 1858:1061–1069. 10.1016/j.bbamem.2015.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alford MA, Pletzer D, Hancock REW. 2019. Dismantling the bacterial virulence program. Microb Biotechnol 12:409–413. 10.1111/1751-7915.13388 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (±1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from tips of swarm tendrils, centres of swarming colonies, and 1018-treated colonies. 0 = no significant change in gene expression.
(XLSX)
Number of genes that were dysregulated for a particular GO term in each condition, as well as total number of genes annotated to that GO term, are shown.
(XLSX)
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (±1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from tips of swarm tendrils, centres of swarming colonies, and 1018-treated colonies. 0 = no significant change in gene expression. References: [31, 35].
(XLSX)
Gene expression is reported as fold-change (FC) relative to swarm front cells. Briefly, BM2 glucose swarm plates (±1018) were inoculated with 1 μl of planktonic cells suspended at an OD600 of 0.4–0.6 in BM2 and then incubated for 20 h at 37°C. Cells were harvested from tips of swarm tendrils, centres of swarming colonies, and 1018-treated colonies. 0 = no significant change in gene expression.
(XLSX)
(A, B) In the presence of peptide 1018 (0.75 μg/ml), rhlB-Tn and anr-Tn mutants swarmed more than the PA14 wild-type (WT). Non-swarming was restored by complementation of the respective gentic loci in trans since no differences were detected between complemented mutants (shown as mutant+) and WT transformed with empty vector (ev) (C, D) Representative images showing swarming phenotypes of mutants and complemented mutants in the presence or absence of peptide 1018, as indicated. Mean and standard error of the mean are shown. One-way ANOVA followed by Dunn’s correction was used to determine statistical significance. *** P < 0.001. Each experiment had at least three biological replicates.
(DOCX)
Data Availability Statement
All RNA-Seq data files are available from the NCBI GEO database (accession number GSE151264). All differentially expressed genes are listed in S2 Table, and all other relevant data are within the paper and its Supporting Information files.